Abstract
In wheat, 25 Rht genes for dwarfness are known, which include both, GA-insensitive and GA-responsive genes. The GA-insensitive Rht genes have been widely used, although, their suitability under abiotic stress conditions has been questioned. This necessitated a search for alternative GA-responsive, spontaneous and induced dwarfing genes. We earlier reported an induced dwarf mutant (‘dwarf mutant-3′; 44 cm), and the mutant allele was named Rht4c allele (2BL). This dwarf mutant was not suitable for cultivation due to its extra dwarf nature. Therefore, we searched for naturally occurring QTLs, which would modify the phenotype of ‘dwarf-mutant-3′ using ‘mutant-assisted gene identification and characterization’ (MAGIC) approach. For this purpose, the ‘dwarf mutant-3′ was crossed with a tall wheat cv. NP114 and homozygous mutant F2 plants (~ 25% of the progeny) were selected, which were phenotyped for plant height and genotyped using SSR markers. The data were utilized for QTL analysis and plant height. Six modifier QTLs were identified on chromosomes 2A, 2B and 4A. Two QTLs each on 2A and 2B were responsible for increase in plant height (described as ‘enhancer modifiers’), whereas the remaining two QTLs located on 4A were responsible for reducing the plant height (described as ‘suppressor modifiers’). It was hypothesized that the enhancer QTLs could be exploited for the development of semi-dwarf high yielding genotypes containing Rht4c allele. This is the first study of its kind in wheat demontsrating that the MAGIC approach could be used for identification of modifiers of the mutant phenotypes of other traits for wheat improvement.
Keywords: Rht genes, Dwarf mutant, Plant height, MAGIC (mutant-assisted gene identification and characterization), Modifier QTLs, Triticum aestivum L
Introduction
In wheat, 25 major dwarfing genes called Rht genes (Rht1-Rht25) are known (McIntosh et al. 2017; Mo et al. 2018). Majority of these Rht genes occur in natural populations; some were also obtained using physical/chemical mutagens. The Rht genes are known to be responsible for reduced plant height, lodging resistance, increased tillers, higher harvest index and higher yield (Hedden 2003; Ellis et al. 2005; see Jobson et al. 2019 for references). The most important of these Rht genes, are the two GA-insensitive dwarfing genes, namely Rht1 (RhtB1b) located on chromosome arm 4BS and Rht2 (RhtD1b), located on 4DS. These two genes have similar and additive effect on plant height (see Jobson et al. 2019 for references), and are described as ‘green revolution genes’ due to their wide exploitation in breeding bread wheat cultivars around the world. Using molecular markers, the distribution of these genes has been examined in popular wheat cultivars in India and elsewhere (Elllis et al. 2002; Sheoran et al. 2013). Some of the popular semi-dwarf wheat cultivars containing the above two genes include the following: Jaz, Inia66, Cranbrook, Avocet, Ciano67, Westonia, HD1941, HD1981, HD2428, MACS2496, UP368 etc.
At present, almost entire land area under wheat cultivation is occupied by semi-dwarf varieties (https://croplife.org/news/semi-dwarf-wheat-the-game-changer/). A majority of these semi-dwarf durum and bread wheat cultivars contain either of the above two ‘green revolution genes’, although following other Rht genes have also been exploited: Rht8, Rht9, Rht14, Rht15, Rht16, Rht18 and Rht19 (Konzak 1988; Maluszynski et al. 2001). Due to lack of response to endogenous GA (gibberellins), the plants containing GA-insensitive genes have shorter coleoptile length, seedling roots and smaller seedling leaf area, making them unsuitable for deep sowing under condition of water stress (Rebetzke et al. 2001; Mathews et al. 2006; Bai et al. 2013; Amram et al. 2015; for references see Grover et al. 2018). There are also GA-sensitive Rht genes, which include the following: Rht4, Rht5, Rht8, Rht12, Rht13, Rht14, Rht18, Rht24 and Rht25 (Ellis et al. 2004; Wurschum et al. 2017; Mo et al. 2018; Grover et al. 2018). Among these GA-sensitive genes, Rht8 is known to carry no adverse effect on coleoptile length, and has therefore, been exploited in the development of wheat varieties in Europe and China (Rebetzke and Richards 2000; Borojevic and Borojevic 2005; Casperini et al. 2012). Other GA-sensitive genes, that have been recommended for deployment include the following: Rht4, Rht5, Rht7, Rht9, Rht12, Rht13 and Rht18. These genes do not reduce the coleoptile length and have a positive effect on grain number and harvest index (for references see Dan et al. 2020). In order to exploit the known additive and epistatic interactions between known Rht loci, following combinations have also been recommended for use in wheat breeding: Rht8 + Rht13, Rht4 + Rht8, Rht13 + Rht-D1b (Rebetzke et al. 2011; Wang et al. 2014; Du et al. 2018; Sannemann et al. 2018).
Novel GA-sensitive dwarfing genes/alleles could also be produced through mutagenesis to increase the number of available dwarfing genes (Parry et al. 2009). In order to achieve this objective, we earlier conducted a study involving induced mutagenesis in wheat cv. PBW550 using chemical mutagen ethyl nitrosourea (ENU), and identified a variety of mutations including some dwarf mutants (Agarwal et al. 2013). One of these mutants called ‘dwarf mutant-3′ was later mapped on chromosome arm 2BL, close to the SSR markers Xwmc361 and Xmc317, the latter associated with Rht4, a GA-responsive dwarfing gene (Agarwal et al. 2015). We assumed that the phenotype of ‘dwarf mutant-3′ may be due to an induced novel allele of the gene Rht4, and therefore named it Rht4c.
The ‘dwarf mutant-3′, was further characterized using the approach called “mutant-assisted gene identification and characterization” (MAGIC), which was developed for the identification of modifiers of the mutant phenotypes in maize (Johal et al. 2008; Chintamanani et al. 2010). This approach makes use of readily scorable phenotype of a mutant gene affecting the trait of interest as a ‘reporter’. The strategy involves crossing of the recessive mutant with one or more wild type genotypes to obtain F2 population, which is screened for transgressive segregants (both reduced and increased) (Johal et al. 2008; Chintamanani et al. 2010; Chaikam et al 2011). However, only 25% homozygous mutant type F2 plants are used for the identification of QTLs, which have a modifying effect on the mutant phenotype. MAGIC approach has been successfully utilized in maize, where the following three maize mutants proved useful: shrunken2 (sh2), opaque2 (o2) and les23; sh2 was used for the development of commercially important ‘super-sweet’ corn, o2 was used for ‘high-quality protein’ maize (Tracy 1997; Prasanna et al. 2001) and les23 allowed identification of four minor QTLs, suggesting the utility of MAGIC approach for the identification of the modifier QTLs. Several QTLs which modify the hypersensitive response (HR) of the partially dominant and auto-active maize disease resistance gene Rp1-D21 were also identified using MAGIC approach (Chaikam et al. 2011).
The ‘dwarf mutant-3′ has a mean plant height of only 44 cm, thus making it unsuitable for commercial cultivation, since there is a positive correlation between plant height and grain yield (Qaseem et al. 2018; Baye et al. 2020). This prompted us to identify modifier QTLs, which may then be used for alteration of the phenotype of ‘dwarf mutant -3′ and make it suitable for use in breeding programmes. The present study helped us to identify QTLs, which we would like to describe as ‘enhancer’ and ‘suppressor’ modifiers of the induced mutant allele Rht4c. The results of this study are summarized in this communication.
Material and methods
Plant material and recording of data
The ‘dwarf mutant-3′ carrying allele Rht-4c (44 cm plant height) was crossed with a tall wheat cv. NP-114 (131 cm plant height) in the crop season 2010–11. The F1 derived from the above cross was raised and selfed during the off-season summer nursery in 2011 and F2 seed was harvested. During the next crop season (2011–12), a population of 162 F2 plants was raised and data recorded on plant height. The F3 seed harvested from individual F2 plants was used to raise the F3 progenies along with the parental genotypes in 1.5 m plots during the crop season 2012–13; 37 F3 progenies (shorter than the tall parent cv. NP-114) that did not segregate for plant height were identified, so that their parent F2 plants were selected as homozygous for Rht4c allele. The data on plant height of these 37 F2 plants and their genotyping data were used for mapping of modifiers following QTL analysis (for details see below).
Marker analysis of the selected F2 plants
For QTL analysis, we used the selected 37 F2 plants (producing non-segregating F3 progenies) and the parents. Genomic DNA was extracted from the leaves of 1 month-old plants using a modified CTAB method (Saghai-Maroof et al. 1984). The quality and quantity of DNA were checked on agarose gel through a comparison with known quantities of lambda DNA marker. A polymorphism survey was conducted for 537 simple sequence repeats (SSRs), distributed on all 42 arms of 21 wheat chromosomes (Somers et al. 2004). For this polymorphism survey, two parental genotypes, namely ‘dwarf mutant-3′ and the tall cv. ‘NP-114′ were used; as many as 164 SSRs were found to be polymorphic.
For genotyping, DNA amplification was carried out in 25 µl reaction mixtures, each carrying 50 ng template DNA, 0.2 µM SSR primers, 200 µM dNTPs, 1.5 mM MgCl2, 1 × PCR buffer and 0.5 U Taq DNA polymerase (iNtRON Biotechnology Inc., Korea); double distilled water was used to make 25 µl volume in an Eppendorf Mastercycler (Hamburg, Germany). PCR profile included initial denaturation at 95 °C for 5 min followed by 40 cycles of 94 °C for 1 min (denaturation), 50/55/58/60 °C for 1 min (annealing) and 72 °C for 2 min (extension) with a final extension for 10 min. The PCR products were resolved on 10% polyacrylamide denaturing gels (PAGE) in Mega Gel Dual High-Throughput Vertical System, CDASG-400–50 (CBS Scientific Company, CA, USA) followed by silver staining (Tegelstrom 1992). Segregation data for the marker alleles for all SSRs were recorded manually.
Construction of molecular map
Using the genotypic data from the above 37 F2 plants for the above 164 polymorphic SSRs, a framework linkage map containing SSR loci was constructed following Lincoln et al. (1993) using ‘MAPMAKER/EXP version 3.0′ (Lander et al.1987).
Composite interval mapping
QTLs for plant height along with their positions and effects were determined using composite interval mapping (CIM) using the software QTL Cartographer V2.5 (Wang et al. 2012). For this purpose, the CIM Zmapqtl module model 6 with a window size of 10 cM and the 5 markers for the background control were used (Wang et al. 2012). QTLs were identified using LOD score of ≥ 3 as the threshold (probability of 1000–1) (Kocherina et al. 2011). The proportion of observed phenotypic variation explained (PVE) due to a particular QTL was estimated by the coefficient of determination (R2) using maximum likelihood approach (Basten et al. 1994).
Results and discussion
The present study is an extension of our earlier study on ENU-induced ‘dwarf mutant-3′ (Agarwal et al. 2013, 2015), which was subjected to further analysis for identification of modifiers using MAGIC approach that was earlier proposed and successfully utilized in maize for identification of enhancer modifiers of two maize mutants (Johal et al. 2008). In the present study, this approach has largely been successful in identification of enhancer and suppressor types of modifier QTLs for plant height using dwarf phenotype due to Rht4c as a ‘reporter’. The present study, thus also reaffirms that a dwarf mutant allele (Rht4c in the present study), when transferred into the unrelated genetic backgrounds, may produce a range of phenotypes due to interactions with differences in genetic background, as also earlier discussed in a review article by Chandler et al. (2013).
It is also known that the cryptic modifying QTLs responsible for modification of the phenotype due to mutant allele may be distributed throughout the genome and that the alleles at these loci are dispersed in the natural germplasm and their effect is not easy to analyse. MAGIC approach has been developed to detect and characterise such QTLs that occur in the germplasm and modify the mutant phenotype caused due to mutant allele. For detection of these QTLs, ideally the mutants should be crossed with a large number of genotypes, followed by the analysis of an equal number of segregating populations for the identification of modifier QTLs. However, in MAGIC approach, a solitary F2 population or a few segregating F2 populations may also allow identification of modifier QTLs. One of the advantages of MAGIC approach is that in F2 population, only 25% homozygous plants, which visually resemble the mutant type plants are phenotyped and genotyped, saving time and resources (Johal et al 2008). Another characteristic useful feature of MAGIC is that it begins with a clearly defined mutation followed by identification of QTLs that contribute to the variation in the trait controlled by the mutant allele. Because of the distinguishable known effects of the initial mutation, the relationship between the starting mutant allele and the interacting QTLs can be more clearly defined. In a typical QTL interval mapping, a priori information of genes involved in a trait is missing, and an understanding of the QTLs and their interactions emerge subsequent to initial analysis.
In the present study, the plant height of the ‘dwarf mutant-3′ was 44 cm, suggesting 60% reduction in plant height compared to the parent cv. PBW550 (110 cm). On the basis of our results, we expected the presence of enhancer and suppressor modifiers that modify the expression of Rht4c. In order to identify these modifiers, we generated F2/F3 populations from the cross ‘dwarf mutant-3′x NP-114 (wild type tall wheat). Thirty seven (37) homozygous dwarf F2 individuals (validated as homozygous dwarf type on the basis of lack of segregation for plant height in F3) were available and exhibited considerable variability (range 30–110 cm; mean = 62.8 cm; CV = 30%) accompanied with normal distribution for plant height (Fig. 1). These were therefore used for QTL mapping. As obvious, the plant height of the F2 plants transgressed the mean plant height of the ‘dwarf mutant-3′, suggesting the occurrence of suppressor and enhancer modifier QTL alleles of plant height, which were carried by the tall parent cv. NP114.
Fig. 1.
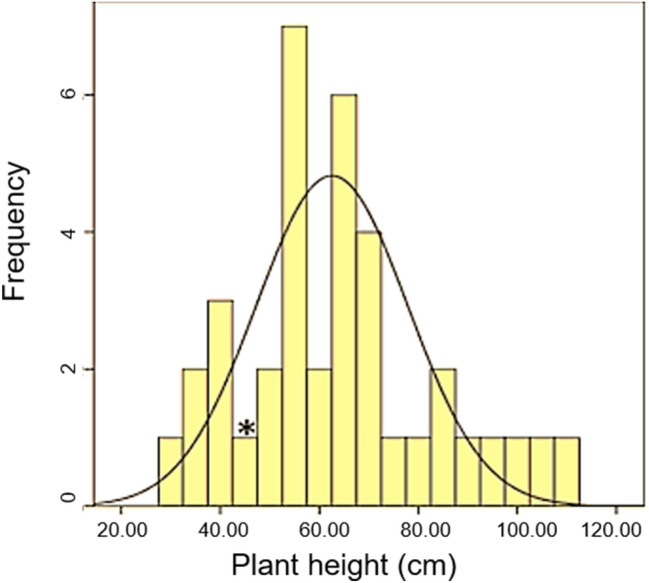
Frequency distribution of plant height of 37 F2 dwarf plants containing Rht4c allele used for QTL analyses through CIM. The asterisk indicates mean plant height of ‘dwarf mutant-3′ parent
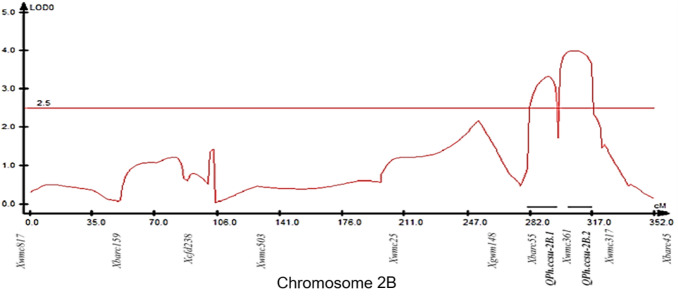
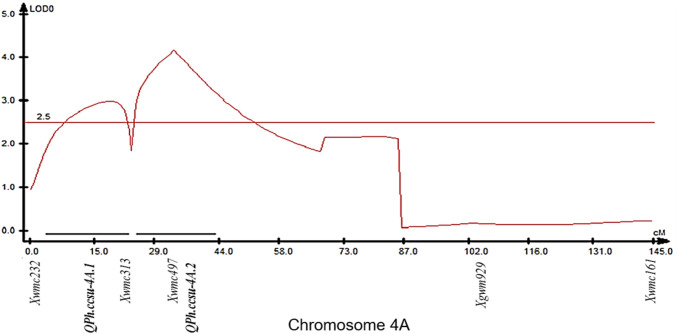
Six QTLs for plant height were detected, with the LOD (Logarithm of the odds) score ranging from 3.00 to 4.48. These QTLs were located on chromosomes 2A (two QTLs), 2B (two QTLs) and 4A (two QTLs) (Table 1 and Figs. 2 and 3). A flanking marker (Xwmc361) associated with two QTLs on 2B (QPh.ccsu-2B.1 and QPh.ccsu-2B.2) was also linked with Rht4c mutant locus, that controls plant height of ‘dwarf mutant-3′. QTLs/genes for plant height on chromosome 2A, 2B and 4A of wheat were also previously reported in several studies (Goud and Sridevi 1988; Borner et al. 2002; Huang et al. 2003; Marza et al. 2006; Jia et al. 2013; Liu et al. 2014, 2019; Lozada et al. 2017). The precise positions of the QTLs reported during the present study and those reported in the previous studies, however, could not be compared due to lack of shared markers between the linkage maps used in these different studies. Meta-QTLs for plant height have also been reported on 2A and 2B (Griffiths et al. 2012). The closest marker locus Xgwm311 for each of the two QTLs on 2A (QPh.ccsu-2A.1 and QPh.ccsu-2A.2) was only 3 cM away from the marker locus Xgwm382 (Sommers et al. 2004) associated with a meta-QTL. This suggested that the genomic region of chromosome 2A defined by the markers reported during our study and that of Griffiths et al. (2012) is an important region controlling plant height in wheat. Two of the above six QTLs (Ph.ccsu-4A.1 and Ph.ccsu-4A.2) each also explained ~ 57% phenotypic variance (PVE) in plant height and were thus considered to be major QTLs, the remaining four QTLs being minor QTL (PVE ranging from 2.91 to 9.8%). An analysis of the additive effect of each of the six QTLs suggested that the alleles of these four QTLs (QPh.ccsu-2A.1, QPh.ccsu-2A.2, QPh.ccsu-2B.1 and QPh.ccsu-2B.2) carried by NP-114 had an additive effect on the dwarf plant height due to Rht4c, whereas the alleles at the remaining two QTLs on chromosome 4A (QPh.ccsu-4A.1 and QPh.ccsu-4A.2) had an inhibitory effect on the plant height of the ‘dwarf mutant-3′ due to Rht4c.
Table 1.
Results of QTL analysis by CIM for plant height in bread wheat conducted using homozygous dwarf F2 plants for Rht4c allele derived from the cross of ‘dwarf mutant-3′ × cv. NP114
| Modifier alleles contributed by wild type parent cv. NP114 | ||||||||
|---|---|---|---|---|---|---|---|---|
| QTL | Chromosome | Flanking markers | Position (cM) |
LOD Score | A | R2(%) | Enhancer allele | Suppresser allele |
| QPh.ccsu-2A.1 | 2A | Xwmc407-Xgwm311 | 12 | 4.30 | − 29.04 | 9.80 | + | – |
| QPh.ccsu-2A.2 | 2A | Xwmc407-Xgwm311 | 18 | 4.48 | − 28.86 | 9.50 | + | – |
| QPh.ccsu-2B.1 | 2B | Xbarc55-Xwmc361 | 291.7 | 3.08 | − 34.21 | 2.91 | + | – |
| QPh.ccsu-2B.2 | 2B | Xwmc361-Xwmc317 | 305 | 3.93 | − 34.37 | 5.57 | + | – |
| QPh.ccsu-4A.1 | 4A | Xwmc232-Xwmc313 | 18 | 3.00 | 12.63 | 57.23 | – | + |
| QPh.ccsu-4A.2 | 4A | Xwmc313-Xgwm929 | 33.4 | 4.18 | 12.36 | 57.93 | – | + |
Fig. 2.
Representative QTL Cartographer plot for the two modifier QTLs for plant height identified on chromosome 2B of bread wheat
Fig. 3.
Representative QTL Cartographer plot for the two modifier QTL for plant height identified on chromosome 4A of bread wheat
The QTLs having a modifying effect on plant height, identified during the present study, also explain a wide range of variation (30–110 cm) observed in plant height of the Rht4c homozygous dwarf F2 plants. These results suggested a random distribution of alleles in the germplasm (like cv. NP114 used during the present study), which may be used to modify the phenotypic expression of the induced dwarf mutants in the breeding programmes in a directed manner. We suggest that the ‘dwarf mutant-3′ and similar other mutants may be crossed with other wild type tall genotypes to discover more such modifiers, which together or individually may be used in developing genotypes with suitable plant height containing the dwarfing alleles like Rht4c. The improved genotypes thus developed may be tested for their suitability even under abiotic stress environments such as water/drought stress.
Acknowledgements
HSB was awarded the Senior Scientist position by INSA, New Delhi. Thanks are due to Head of the Department for providing facilities.
Author contributions
PKG and HSB conceived the study and wrote the manuscript. PA conducted the field and laboratory experiment and analysed the data.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Agarwal P, Kumar S, Mir RR, Balyan HS, Gupta PK. Some ENU induced mutations: a resource for functional genomics in bread wheat. Plant Mutat Rep. 2013;3:9–17. [Google Scholar]
- Agarwal P, Jaiswal V, Kumar S, Balyan HS, Gupta PK. Chromosome mapping of four novel mutants in bread wheat (Triticum aestivum L.) Acta Physiol Plant. 2015;37:66. [Google Scholar]
- Amram A, Faidida-Myers A, Golan G, Nashef K, Ben-David PZ. Effect of GA-sensitivity on wheat early vigor and yield components under deep sowing. Front Plant Sci. 2015;6:487. doi: 10.3389/fpls.2015.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Liang Y, Hawkesford MJ. Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. J Exp Bot. 2013;64:1745–1753. doi: 10.1093/jxb/ert041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye A, Berihun B, Bantayehu M, Derebe B. Genotypic and phenotypic correlation and path coefficient analysis for yield and yield related traits in advanced bread wheat (Triticum aestivum L.) lines. Cogent Food Agric. 2020;6:1752603. [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B. Zmap-a QTL cartographer. In: Smith C, Gavora JS, Benkel B, Chesnais J, Fairfull W, Gibson JP, Kennedy BW, Burnside EB, editors. Proc 5th World congress of genetics applied to livestock production: computing strategies and software. Guelph, Ontario: 5th World Congress on Genetics Applied to Livestock Production; 1994. pp. 65–66. [Google Scholar]
- Borner A, Schumann E, Fürste A, Cöster H, Leithold B, Röder MS, Weber WE. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.) Theor Appl Genet. 2002;105:921–936. doi: 10.1007/s00122-002-0994-1. [DOI] [PubMed] [Google Scholar]
- Borojevic K, Borojevic K. The transfer and history of “reduced height genes” (Rht) in wheat from Japan to Europe. J Hered. 2005;96:455–459. doi: 10.1093/jhered/esi060. [DOI] [PubMed] [Google Scholar]
- Casperini D, Greenland A, Hedden P, Dreos R, Harwood W, Griffiths S. Genetic and physiological analysis of Rht8 in bread wheat: an alternative source of semi-dwarfism with a reduced sensitivity to brassinosteroids. J Exp Bot. 2012;63:4419–4436. doi: 10.1093/jxb/ers138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler CH, Chari C, Dworkin I. Does your gene need a background check? how genetic background impacts the analysis of mutations, genes, and evolution. Trends Genet. 2013;29:358–366. doi: 10.1016/j.tig.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikam V, Negeri A, Dhawan R, Puchaka B, Ji J, Chintamanani S, Gachomo EW, Zillmer A, Doran T, Weil C, Balint-Kurti P, Johal G. Use of mutant-assisted gene identification and characterization (MAGIC) to identify novel genetic loci that modify the maize hypersensitive response. Theor Appl Genet. 2011;123:985–997. doi: 10.1007/s00122-011-1641-5. [DOI] [PubMed] [Google Scholar]
- Chintamanani S, Hulbert SH, Johal GS, Balint-Kurti PJ. Identification of a maize locus that modulates the hypersensitive defense response, using mutant-assisted gene identification and characterization. Genetics. 2010;184:813–825. doi: 10.1534/genetics.109.111880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan S, Zhao Z, Qiao Y, Cui C, Morgunov A, Condon AG, Chen L, Hu Y-G. GAR dwarf gene Rht14 reduced plant height and affected agronomic traits in durum wheat (Triticum durum) Field Crops Res. 2020;248:107721. doi: 10.1016/j.fcr.2020.107721. [DOI] [Google Scholar]
- Du YY, Chen L, Wang YS, Yang ZY, Saeed I, Daoura BG, Hu YG. The combination of dwarf genes Rht4 and Rht8 reduced plant height, improved yield traits of rainfed bread wheat (Triticum aestivum L.) Field Crops Res. 2018;215:149–155. [Google Scholar]
- Ellis MH, Spielmeyer W, Gale KR, Rebetzke GJ, Richards RA. “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet. 2002;105:1038–1042. doi: 10.1007/s00122-002-1048-4. [DOI] [PubMed] [Google Scholar]
- Ellis MH, Rebetzke GJ, Chandler P, Bonnett D, Spielmeyer W, Richards RA. The effect of different height reducing genes on the early growth of wheat. Funct Plant Biol. 2004;31:583–589. doi: 10.1071/FP03207. [DOI] [PubMed] [Google Scholar]
- Ellis MH, Rebetzke GJ, Azanza F, Richards RA, Spielmeyer W. Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theor Appl Genet. 2005;111:423–430. doi: 10.1007/s00122-005-2008-6. [DOI] [PubMed] [Google Scholar]
- Goud JV, Sridevi O. Cytogenetics investigation of some quantitative characters in hexaploid wheat (Triticum aestivum L) using F2 monosomic analysis. In: Miller TE, Koenbner RMD, editors. Proc VIIth int wheat genetics symposium. Cambridge: Institute of Pl Sci Res, Cambridge Lab, Trumpington; 1988. pp. 521–525. [Google Scholar]
- Griffiths S, Simmonds J, Leverington M, Wang YK, Fish L, Sayers L, Alibert L, Orford S, Wingen L, Snape J. Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Mol Breed. 2012;29:159–171. doi: 10.1007/s00122-009-1046-x. [DOI] [PubMed] [Google Scholar]
- Grover G, Sharma A, Gill HS, Srivastava P, Bains NS. Rht8 gene as an alternate dwarfing gene in elite Indian spring wheat cultivars. PLoS ONE. 2018;13:e0199330. doi: 10.1371/journal.pone.0199330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. The genes of the green revolution. Trends Genet. 2003;19:5–9. doi: 10.1016/s0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Cöster H, Ganal MW, Röder MS. Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.) Theor Appl Genet. 2003;106:1379–1389. doi: 10.1007/s00122-002-1179-7. [DOI] [PubMed] [Google Scholar]
- Jia H, Wan H, Yang S, et al. Genetic dissection of yield-related traits in a recombinant inbred line population created using a key breeding parent in China's wheat breeding. Theor Appl Genet. 2013;126:2123–2139. doi: 10.1007/s00122-013-2123-8. [DOI] [PubMed] [Google Scholar]
- Jobson EM, Johnston RE, Oiestad AJ, Martin JA, Gioux MJ. The impact of the wheat Rht-B1b semi-dwarfing allele on photosynthesis and seed development under field conditions. Front Plant Sci. 2019;10:51. doi: 10.3389/fpls.2019.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal GS, Balint-Kurti P, Weil CF. Mining and harnessing natural variation: a little MAGIC. Crop Sci. 2008;48:2066–2073. [Google Scholar]
- Kocherina NV, Artem’eva AM, Chesnokov YuV. Use of LOD score technology in mapping quantitative trait loci in plants. Russ Agric Sci. 2011;37:201–204. [Google Scholar]
- Konzak CF (1988) Genetic analysis, genetic improvement and evaluation of induced semi-dwarf mutants in wheat. In: Semidwarf cereal mutants and their use in cross-breeding III, research coordination meeting, 16–20 December 1985. International Atomic Energy Agency, Vienna, p. 77–94
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lincoln SE, Daly MJ, Lander ES. Constructing genetic linkage maps with MAPMAKER/EXP. Cambridge, MA: Whitehead Institute for Biomedical Research; 1993. [Google Scholar]
- Liu G, Jia L, Lu L, et al. Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor Appl Genet. 2014;127:2415–2432. doi: 10.1007/s00122-014-2387-7. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu B, Singh RP, Velu G. QTL mapping for micronutrients concentration and yield component traits in a hexaploid wheat mapping population. J Cereal Sci. 2019;88:57–64. doi: 10.1016/j.jcs.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada DN, Mason RE, Babar MA, et al. Association mapping reveals loci associated with multiple traits that affect grain yield and adaptation in soft winter wheat. Euphytica. 2017;213:222. [Google Scholar]
- Maluszynski M, Szarejko I, Maluszynska J. Induced mutations in wheat. In: Bonjean AP, Angus WJ, editors. The world wheat book. London: Lavoisier Publishing; 2001. pp. 939–977. [Google Scholar]
- Marza F, Bai G-H, Carver BF, Zhou W-C. Quantitative trait loci for yield and related traits in the wheat population Ning7840 × Clark. Theor Appl Genet. 2006;112:688–698. doi: 10.1007/s00122-005-0172-3. [DOI] [PubMed] [Google Scholar]
- Mathews KL, Chapman SC, Trethowan R, Singh RP, Crossa J, Pfieffer W, van Ginkel M, DeLacy I. Global adaptation of spring bread wheat and durum wheat lines near isogenic for major reduced height genes. Crop Sci. 2006;46:603–613. [Google Scholar]
- McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Xia XC (2017) Catalogue of gene symbols for wheat: 2017 supplement. https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2017.pdf
- Mo YJ, Vanzetti LS, Hale I, Spagnolo EJ, Guidobaldi F, Al-Oboudi J, Odle N, Pearce S, Helguera M, Dubcovsky J. Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theor Appl Genet. 2018;131:2021–2035. doi: 10.1007/s00122-018-3130-6. [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Madgwick PJ, Bayon C, et al. Mutation discovery for crop improvement. J Exp Bot. 2009;60:2817–2825. doi: 10.1093/jxb/erp189. [DOI] [PubMed] [Google Scholar]
- Penning BW, Johal GS, McMullen MD. A major suppressor of cell death, slm1, modifies the expression of the maize (Zea mays L.) lesion mimic mutation les23. Genome. 2004;47:961–969. doi: 10.1139/g04-046. [DOI] [PubMed] [Google Scholar]
- Prasanna BM, Vasal SK, Kassahun B, Singh NN. Quality protein maize. Curr Sci. 2001;81:1308–1319. [Google Scholar]
- Qaseem MF, Qureshi R, Muqaddasi QH, Shaheen H, Kousar R, Roder MS. Genome wide association in bread wheat subjected to independent and combined heat and drought stress. PLoS ONE. 2018;13:e0199121. doi: 10.1371/journal.pone.0199121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebetzke GJ, Richards RA. Gibberellic acid-sensitive dwarfing genes reduce plant height to increase kernel number and grain yield of wheat. Aust J Agric Res. 2000;51:235–245. [Google Scholar]
- Rebetzke GJ, Appels R, Morrison AD, Richards RA, McDonald G, Ellis MH, Spielmeyer W, Bonnett DG. Quantitative trait loci on chromosome 4B for coleoptile length and early vigour in wheat (Triticum aestivum L.) Aust J Agric Res. 2001;52:1221–1234. [Google Scholar]
- Rebetzke GJ, Ellis MH, Bonnett DG, Condon AG, Falk D, Richards RA. The Rht13 dwarf gene reduces peduncle length and plant height to increase grain number and yield of wheat. Field Crops Res. 2011;124:323–331. [Google Scholar]
- Saghai-Maroof MA, Biyashev RM, Yang GP, Zhang Q, Allard W. Extraordinarily polymorphic microsatellite DNA in barley: species diversity, chromosomal locations, and population dynamics. Proc Natl Acad Sci. 1984;91:5466–5470. doi: 10.1073/pnas.91.12.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannemann W, Lisker A, Maurer A, Léon J, Kazman E, Cöster H, Holzapfel J, Kempf H, Korzun V, Ebmeyer E, Pillen K. Adaptive selection of founder segments and epistatic control of plant height in the MAGIC winter wheat population WM-800. BMC Genomics. 2018;19:559. doi: 10.1186/s12864-018-4915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheoran S, Singh V, Malik R, Kundu S, Tiwari R, Kumar R, Shoran J. Distribution of dwarfing genes Rht-B1b and Rht-D1b in Indian wheat (Triticum aestivum) cultivars detected by functional markers. Indian J Agric Sci. 2013;83:820–825. [Google Scholar]
- Somers DJ, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) Theor Appl Genet. 2004;109:1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- Tracy WF. History, genetics and breeding of super sweet (shrunken2) sweet corn. Plant Breed Rev. 1997;14:189–236. [Google Scholar]
- Tegelstrom H. Detection of mitochondrial DNA fragments. In: HoeIzel AR, editor. Molecular genetic analysis of populations: a practical approach. Oxford: IRL Press; 1992. pp. 89–114. [Google Scholar]
- Wang S, Basten CJ, Zeng ZB (2012) Windows QTL cartographer 2.5. Depaertment of statistics, North Carolina State University, Raleigh, NC (https://statgen.ncsu.edu/qtlcart/WQTLCart.htm)
- Wang Y, Chen L, Du Y, Yang Z, Condon AG, Hu YG. Genetic effect of dwarf gene Rht13 compared with Rht-D1b on plant height and some agronomic traits in common wheat (Triticum aestivum L.) Field Crops Res. 2014;162:39–47. [Google Scholar]
- Wurschum T, Langer SM, Longin CFH, Tucker MR, Leiser WL. A modern green revolution gene for reduced height in wheat. Plant J. 2017;92:892–903. doi: 10.1111/tpj.13726. [DOI] [PubMed] [Google Scholar]