Abstract
Plants regularly experience multiple abiotic and biotic pressures affecting their normal development. The 90-kDa heat shock protein (HSP90) plays a dynamic role in countering abiotic and biotic stresses via a plethora of functional mechanisms. The HSP90 has been investigated in many plant species. However, there is little information available about this gene family in the cultivated Mediterranean olive tree, Olea europaea subsp. europaea var. europaea. In the current study, we systematically performed genome-wide identification and characterization of the HSP90 gene family in O. europaea var. europaea (OeHSP90s). Twelve regular OeHSP90s were identified, which were phylogenetically grouped into two major clusters and four sub-clusters, showing five paralogous gene pairs evolving under purifying selection. All of the 12 proteins contained a Histidine kinase-like ATPase (HATPase_c) domain, justifying the role played by HSP90 proteins in ATP binding and hydrolysis. The predicted 3D structure of OeHSP90 proteins provided information to understand their functions at the biochemical level. Consistent with their phylogenetic relationships, OeHSP90 members were predicted to be localized in different cellular compartments, suggesting their involvement in various subcellular processes. In consonance with their spatial organization, olive HSP90 family members were found to share similar motif arrangements and similar number of exons. We found that OeHSP90 promoters contained various cis-acting elements associated with light responsiveness, hormone signaling pathways and reaction to various stress conditions. In addition, expression sequence tags (ESTs) analysis offered a view of OeHSP90 tissue- and developmental stage specific pattern of expression. Proteins interacting with OeHSP90s were predicted and their potential roles were discussed. Overall, our results offer premises for further investigation of the implication of HSP90 genes in the physiological processes of the olive and its adaptation to stresses.
Electronic supplementary material
The online version of this article (10.1007/s12298-020-00888-x) contains supplementary material, which is available to authorized users.
Keywords: Gene family, Genome-wide analysis, HSP90, Phylogenetic analysis, Olea europaea
Introduction
The Mediterranean olive tree (Olea europaea subsp. europaea) is one of the oldest tree crops in human history and is recently of major agricultural importance in the Mediterranean region. It is the source of olive oil, which forms the basis of the Mediterranean diet, attracting special attention because of its proven health benefits (Gavahiana et al. 2019). Olive is a perennial, mainly clonally propagated, diploid (2n = 2x = 46) tree (Mousavi et al. 2017). The complete genome assembly of the cultivated Mediterranean olive tree, O. europaea subsp. europaea cv. Farga, has been recently released, giving a total length of 1.31 Gb, with 56,349 unique protein-coding genes (Cruz et al. 2016).
Plants encounter several abiotic (drought, chilling, light intensity, chemical pollutants, salinity, heavy metal stresses, heat shock, etc.) and biotic stresses (attacks by pathogens and pests), which have a serious impact on their normal growth and development (Prasad et al. 2011; Ramegowda and Senthil-Kumar 2015). During their evolution, plants have acquired a wide range of stress response factors playing central roles in biotic and abiotic stress sensing and signalling (Swindell et al. 2007). Among the main factors influencing the stress response in plants, against harmful environmental stimuli are the highly conserved proteins called heat shock proteins (HSPs) (Queitsch et al. 2002; Sangster and Queitsch 2005). These proteins act primarily through assisting in refolding of unfolded/misfolded proteins, which accumulate in cells as a consequence of cell homeostasis perturbation following stress (Park and Seo 2015). Heat shock proteins have been categorized into five families, HSP100/ClpB, HSP90, HSP70/DnaK, chaperonin (HSP60/GroEL), and small heat-shock proteins (sHSP), based on their approximate molecular weights (Scharf et al. 2012). Heat shock proteins are widely distributed among fungi, animals and plants (Zhang et al. 2017). They are mainly located in the cytosol under normal physiological conditions, but are rapidly conveyed to the nucleus under stress conditions (Xu et al. 2012). For example, there are seven HSP90s in Arabidopsis, of which AtHSP90–1, AtHSP90–2, AtHSP90–3, and AtHSP90–4 are located in the cytosol, and AtHSP90–5, AtHSP90–6, and AtHSP90–7 are located in the chloroplast, mitochondria and endoplasmic reticulum, respectively (Krishna and Gloor 2001).
HSP90 (heat shock protein 90) is a highly conserved and essential molecular chaperone with an approximate molecular weight of 90-kDa, which helps other signaling proteins to fold properly in eukaryotes, stabilizes client proteins against stress, and aids in protein degradation (Theodoraki and Caplan 2012; Hoter et al. 2018). HSP90 comprises mainly three highly conserved domains: the C-terminal domain that binds to the substrate, the intermediate domain, and the N-terminal domain of the ATP-binding (NTD) (Pearl and Prodromou 2006). In plants, the transcriptional patterns of HSP90 expression have been extensively analyzed and observed as diverse in different plant organs and tissues (Xu et al. 2013; Chen et al. 2018; Song et al. 2019). It is, therefore, reasonable to assume that different isoforms can play specific roles in cell growth and development as well as in reacting to several types of stresses. HSP90 frequently functions together with co-chaperones that regulate the conformational cycle and/or load the “client” protein onto HSP90 (Kadota and Shirasu 2012). HSP90 clients including, among others, kinases, transcription factors, steroid hormone receptors and E3 ubiquitin ligases (Kadota and Shirasu 2012). Nucleotide-binding domain and leucine-rich repeat-containing proteins, which recognize pathogen effectors in plant specific immunity, are also among HSP90 client proteins (Kadota and Shirasu 2012).
HSP90 genes were identified and analyzed from several dicotyledonous plant species. For example in Rosidae, seven HSP90 genes were identified in A. thaliana (Krishna and Gloor 2001), whereas in Asteridae, seven and twenty-one HSP90 genes have been identified in tomato (Liu et al. 2014) and tobacco (Song et al. 2019), respectively. In spite of the recent availability of the whole genome assembly of the cultivated Mediterranean olive tree, genome-scale identification and analysis of the O. europaea HSP90 gene family has not yet been completed. As a stress-related gene family, a deep and multifaceted understanding of HSP90s will be helpful to increase the possibilities of using them to improve the stress resistance and adaptations in olive. In the present study, we identified the complete set (12) of HSP90 candidate proteins and their encoding genes from the whole Mediterranean olive genome data. We submitted the gene set to a comprehensive computational study covering various structural (annotation of functional protein domains and conserved motifs, exon/intron gene structures, physicochemical features and 3D structure), evolutionary (phylogenetic relationships, gene duplications and selection pressure) and functional (subcellular localization, promoter regulatory elements and interaction networks) analyses. We have also performed digital expression analysis using an expressed-sequence tags (ESTs) database. From our results together, HSP90 genes appear as a genome-wide enriched family in olive, with various biophysical features and are suggested to play a wide array of stress-related functions shaped by the subcellular and organelle-specific context, contributing to the environmental robustness of O. europaea. Our findings already provide an improved understanding of HSP90 genes in olive, while building a strong foundation for future analyses of candidate genes.
Materials and methods
Database search and sequence retrieval
InterProScan annotations for 79.910 O. europaea var. europaea protein products (Cruz et al. 2016) were downloaded from GigaDB data repository (http://gigadb.org/dataset/100201) as of Dec. 2017. This tabular data resource (56.64 Mega Bytes) contained domain annotations for the Mediterranean olive whole predicted protein set. Proteins with HSP90 domain were filtered out with the Pfam ID PF00183. The HSP90 amino acid sequences were analyzed using the Pfam database version 32.0 (https://pfam.xfam.org/), to verify domain size and sequence conservation in comparison with the Pfam family PF00183 HMM model (version 19) of 518 amino acids (https://pfam.xfam.org/family/PF00183#tabview=tab6). The protein sequences lacking or presenting an abnormally short HSP90 domain or a partial coding sequence (CDS) were manually excluded. Then, each of the final set of regular O. europaea HSP90 proteins was systematically named OeHSP90n. In addition to protein sequences, protein coding sequences (CDS) were downloaded from GigaDB (http://gigadb.org/dataset/100201). Besides, gene sequences of all OeHSP90 proteins were extracted from their corresponding scaffolds according to gene coordinates available in GFF3 format gene annotation file. These scaffolds (11038) are registered in NCBI GenBank under accession codes LT560395.1 to LT571432.1 (Cruz et al. 2016). These genes are derived from the reference genome assembly and annotation of the O. europaea millenial tree named “Santander” (cv. Farga) located in Santander, Spain (Cruz et al. 2016) (NCBI assembly OE6; https://www.ncbi.nlm.nih.gov/assembly/GCA_900603015.1). For comparison purposes, protein sequences of the seven A. thaliana HSP90 genes were directly taken from Zhang et al. (2013).
Phylogenetic analysis and duplication dates
Multiple sequence alignment of the full-length protein sequences was performed using ClustalW implemented in MEGA 5.05 software (Tamura et al. 2011), with default settings. A maximum likelihood (ML) phylogenetic tree was constructed using MEGA 5.05 with the JTT amino acid substitution model with 1000 bootstrap replicates. A discrete Gamma distribution was used to model evolutionary rate differences between sites (4 categories).
The nonsynonymous/synonymous ratio is an important indicator of the mode and magnitude of natural selection expected to act on nonsynonymous mutations in protein-coding genes. In general, Ka/Ks = 1, > 1 and < 1 indicate neutral, positive, and purifying selection, respectively. The nucleotide coding sequences (CDSs), in duplicate gene pairs, were aligned by ClustalW and the values of Ka, Ks as well as Ka/Ks were computed by MEGA 5.05, using the method described by Nei and Gojobori (1986). The Ks value of each pair was then used to estimate the timing of gene duplication (million years ago, Mya), by the following formula: duplication time = Ks/2λ, based on 1.8 × 10−9 synonymous substitutions per year (λ) for O. europaea, as reported by Barghini et al. (2015).
Protein physico-chemical features and subcellular localization
Chemical and physical characteristics of OeHSP90 amino acid sequences [molecular weight (MW), number of amino acids, grand average of hydropathicity (GRAVY), isoelectric point (pI) and percentage of positively/negatively charged residues] were analysed by the ProtParam tool of the Expert Protein Analysis System program (ExPASy) (http://web.expasy.org/protparam/) (Wilkins et al. 1999) and by EMBOSS Pepstats (https://www.ebi.ac.uk/Tools/seqstats/emboss_pepstats/). WoLF PSORT (https://wolfpsort.hgc.jp/) was used to predict subcellular localization of the proteins.
Three dimensional (3D) protein structure
The three dimensional (3D) structures of predicted proteins were deduced using homology modelling. For this purpose, we used SWISS-MODEL (https://swissmodel.expasy.org/), a fully automated protein structure homology-modelling server, which can be accessed through the ExPASy website. Once the amino acid sequence of the target protein is submitted, the SWISS-MODEL Template Library (SMTL) derived from Protein Data Bank (Berman et al. 2000) was searched in parallel both with BLAST (Camacho et al. 2009) and HHblits (Remmert et al. 2012) to identify templates and to obtain target–template alignments. The coupled use of these two approaches ensures good alignments at high and low sequence identity levels. The best ranked template for each gene was recruited by SWISS-MODEL server, based on the predicted consistency of the resulting models, as calculated by Global Model Quality Estimation (GMQE), which is a model quality estimator combining properties of the target–template alignment and the template search method. The GMQE score is displayed as a number between 0 and 1, expressing the expected validity of a model, where higher numbers imply higher reliability. These best templates were used to generate the 3D structures of the predicted proteins, based on the target-template alignment using ProMod3 Version 1.3.0 embedded in SWISS-MODEL. Finally, the function of OeHSP90 proteins at the cellular and biochemical level was predicted from the 3D structures of proteins using ProFunc server (https://www.ebi.ac.uk/thornton-srv/databases/profunc/) (Laskowski et al. 2005).
Functional domains, conserved motifs and gene structure
Sequences from O. europaea that were confirmed to contain HSP90 domain were also checked for the presence of additional domains using Pfam database version 32.0 (https://pfam.xfam.org/), and the NCBI’s Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al. 2017).
MEME online version 5.0.5 (http://meme-suite.org/) was used to predict motifs in the O. europaea HSP90 protein sequences. The optimized parameters of MEME (number of repetitions: any; maximum number of motifs: 20; and optimum motif width: 30–70 residues) were used. Then, the predicted motifs of OeHSP90s were annotated by searching against InterProScan database (http://www.ebi.ac.uk/Tools/pfa/iprscan/). In addition, the exon–intron structures of the OeHSP90 genes were determined using Gene Structure Display Server (GSDS 2.0; http://gsds.cbi.pku.edu.cn/) by aligning cDNA to their corresponding genomic DNA sequences.
Cis-acting regulatory element prediction in promoter regions and digital expression analysis of OeHSP90 genes
To explore probable stress-related cis-elements in promoter sequences of OeHSP90 genes, the upstream regions (1.5 kb) of OeHSP90 genes were submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Rombauts et al. 1999; Lescot et al. 2002) for prediction. To gain insight into the expression profiles of HSP90 genes in O. europaea in different tissues, developmental stages and stress levels, the genomic sequences of the OeHSP90s were searched against the EST database provided at the NCBI web site (https://www.ncbi.nlm.nih.gov/genbank/) by BLASTN with an e-value cut-off of 1e−10. The final expression pattern was obtained by removing EST hits shorter than 200 base pairs, lesser than 90% EST sequence coverage as well as lesser than 90% identity with their similar OeHSP gene (Arya et al. 2014). EST data were classified into the different expression profiles according to the tissue or developmental stage specified in the EST library biosample description.
Protein–Protein interaction networks
To investigate the relationship between OeHSP90s and related proteins, the protein interaction networks were constructed by STRING (https://string-db.org/) version 11.0 (Szklarczyk et al. 2019), using medium confidence level (0.400), experiments and co-expression as active interaction sources.
Results
Identification of the HSP90 gene family in O. europaea and gene copy number analysis
To identify HSP90 family in the Mediterranean olive, O. europaea subsp. europaea var. europaea, protein sequences were scanned with the Pfam ID PF00183, resulting in 27 prospective HSP90 proteins (Supplementary file S1). Among this preliminary set, two sequences were characterized by an incomplete coding sequence (CDS) while 13 others were found to possess an incomplete HSP90 domain. We, therefore, excluded these sequences, retaining a final unique set of 12 regular O. europaea HSP90 proteins (OeHSP90s) that had a conserved HSP90 domain and an intact coding sequence.
Comparison of the number of HSP90 genes in the olive with two other sequenced dicot and monocot genomes has shown that olive possesses a number of genes which is higher than A. thaliana, Solanum lypersicum and Oryza sativa (Table 1). Among four compared genomes, the number of HSP90 genes was proportional to the total protein coding genes, suggesting that the higher number of HSP90 members in olive was a consequence of the expansion of gene families during the genome evolution. Accordingly, the density of HSP90 genes decreased from 0.051/Mb to 0.077/Mb from Arabidopsis to tomato, 0.051/Mb to 0.0087/Mb from Arabidopsis to olive, and 0.051/Mb to 0.018/Mb from Arabidopsis to rice, consistent with the shrunken genome of A. thaliana.
Table 1.
The total number of HSP90 genes and their genomic proportions in different genomes
| Olea europaea (Mediterranean olive) | Solanum lycopersicum (Tomato) | Arabidopsis thaliana (Thale cress) | Oryza sativa (Rice) | |
|---|---|---|---|---|
| Total HSP90 family proteins | 12a | 7c | 7e | 9f |
| Total protein-coding genes in genome | 56,349b | 34,727d | 27,416d | 37,960g |
| HSP90 genes (%) | 2.12 10−4 | 2.01 10−4 | 2.55 10−4 | 2.37 10−4 |
| Genome size (Mb) | 1 380b | ~900d | 135d | 500g |
| Average number of HSP90 genes per Mb | 0. 0087 | 0.0077 | 0.051 | 0.018 |
Phylogenetic relationships and duplication events of HSP90 genes
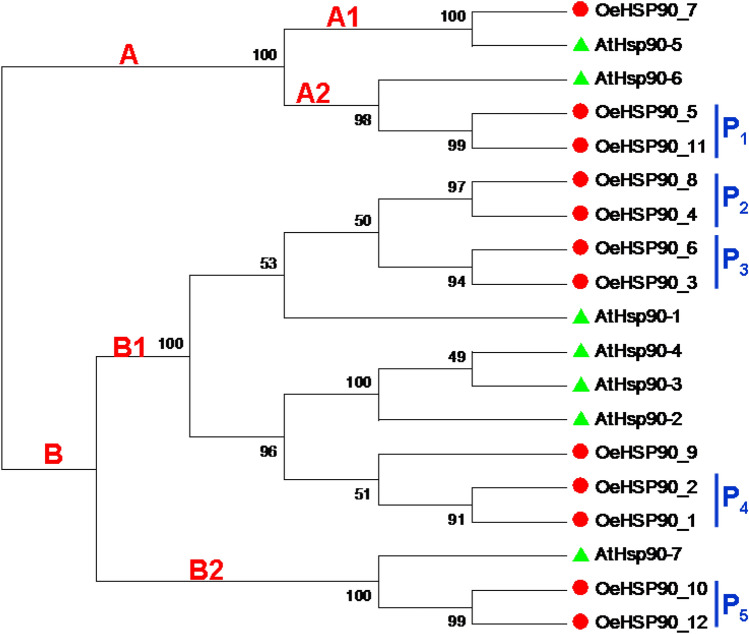
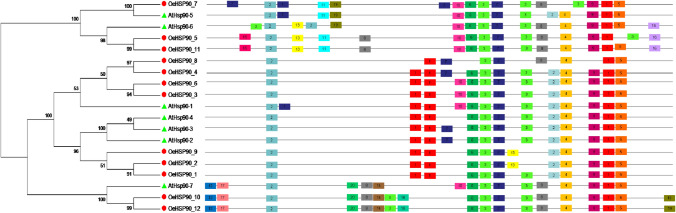
The full amino acid sequences of 12 and seven HSP90 genes of olive and Arabidopsis, respectively, were aligned using the ClustalW program. An unrooted phylogenetic tree of the HSP90 protein sequences from O. europaea and A. thaliana was constructed using the maximum likelihood (ML) method (Fig. 1). The resulting tree topology revealed two (i.e. a minor and a major) clades designated as A and B, which comprised five and 14 HSP90 proteins, respectively. The minor cluster A was further divided in two sub clusters A1 and A2 with 2 and 3 proteins respectively. The major cluster B could also be divided into two sub-clusters, namely B1 and B2, with 11 and 3 HSP90 proteins, respectively. Among these latter ones, the largest sub-cluster, B1, could be further subdivided into at least four groups, giving an idea on the genetic diversity and evolutionary dynamics of subcluster B1.
Fig. 1.
Phylogenetic relationships of HSP90 genes in O. europaea (Red circle) and A. thaliana (Green triangle). P1–P5 refer to close paralogous gene pairs in O. europaea. Bootstrap support is indicated at each node
To identify the selection mode exerted on duplicates of HSP90 genes divergence, the ratios of non-synonymous substitution (Ka) to synonymous substitution (Ks) for pairs of close paralogs were estimated (Table 2). Five OeHSP90 closely-related paralogous gene pairs were identified (Fig. 1) and their ratios of Ka/Ks ranged from 0.067 to 0.222, suggesting that these genes were under strong purifying selection pressure (Ka/Ks < 1). Four gene pairs were characterized by Ka/Ks < 0.01, and a unique gene pair (i.e. OeHSP3/-6) was clearly under more relaxed selection. The estimated time of duplication for paralogous genes indicated that all paralogs were ancient (from ~ 80 to 136 million years ago, Mya), except OeHSP3/-6, which is a recently duplicated gene pair (2.5 Mya).
Table 2.
Duplicated HSP90 genes, selection pressure estimates, and the dating of gene duplication in olive tree
| Gene 1 | Gene 2 | aKs | Ka | Ka/Ks | bDate (Mya) |
|---|---|---|---|---|---|
| OeHSP90-5 | OeHSP90-11 | 0.291 | 0.027 | 0.093 | 80.833 |
| OeHSP90-8 | OeHSP90-4 | 0.490 | 0.039 | 0.080 | 136.111 |
| OeHSP90-6 | OeHSP90-3 | 0.009 | 0.002 | 0.222 | 2.5 |
| OeHSP90-2 | OeHSP90-1 | 0.362 | 0.011 | 0.030 | 100.556 |
| OeHSP90-10 | OeHSP90-12 | 0.330 | 0.022 | 0.067 | 91.667 |
aKa and Ks were calculated using the program MEGA 5.05
bThe divergence time was estimated according to formula: T = Ks/2λ. The clock like rate (λ) was 1.8 × 10−9 substitutions per site per year. Mya, million years ago
Physicochemical features and 3D structure of the olive HSP90 proteins
The lengths of predicted proteins for 12 OeHSP90 genes were more variable (593–824 aa) than the seven AtHSP90 proteins (699–823 aa) (Table 3; Supplementary file S1), suggesting lower level of conservation of OeHSP90s. The isoelectric point (pI) values of all OeHSP90 proteins were within the acidic range. However, the pI ranged from 4.86 to 5.48, with an average pI for all proteins was 5.04, which is more variable compared to AtHSP90s all ranging from 4.93 to 4.96 except AtHSP90-6 (5.26), once again suggesting higher level of conservation of AtHSP90s compared to OeHSP90s. In all OeHSP90s, the grand average of hydropathicity was less than zero, indicating that OeHSP90 proteins were hydrophilic. The percentage of aliphatic amino acids was almost threefold higher than that of aromatic amino acids in OeHSP90 proteins. The negatively charged amino acids in OeHSP90s were more represented than the positively charged amino acids in each OeHSP90 protein.
Table 3.
Basic physicochemical features and information of putative 12 HSP90 family genes identified in Olea europaea
| Protein ID | Alternative protein ID | Protein length | HSP90 domain amino acid span | Theoretical isoelectic point (pI) | Negativey charged residues (Asp + Glu) (%) | Positively charged residues (Arg + Lys) (%) | Aliphatic (%) | Aromatic (%) | Grand average of hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|---|---|
| OE6A019954P1 | OeHSP90-1 | 699 | [184,696] | 4.96 | 19.74 | 14.88 | 26.896 | 9.442 | − 0.585 |
| OE6A043783P2 | OeHSP90-2 | 700 | [184,695] | 4.99 | 22.40 | 17.05 | 26.714 | 9.571 | − 0.591 |
| OE6A047856P1 | OeHSP90-3 | 703 | [189,701] | 5.01 | 20.06 | 15.65 | 26.743 | 9.246 | − 0.634 |
| OE6A049986P2 | OeHSP90-4 | 703 | [189,701] | 5.03 | 23.61 | 18.72 | 26.031 | 9.388 | − 0.62 |
| OE6A060752P4 | OeHSP90-5 | 791 | [276,786] | 5.48 | 16.94 | 14.63 | 27.307 | 10.114 | − 0.539 |
| OE6A071045P1 | OeHSP90-6 | 703 | [189,701] | 5.03 | 19.91 | 15.65 | 27.027 | 9.246 | − 0.617 |
| OE6A076054P7 | OeHSP90-7 | 794 | [263,777] | 4.92 | 17.65 | 13.24 | 26.826 | 9.446 | − 0.553 |
| OE6A078529P1 | OeHSP90-8 | 593 | [141,591] | 4.98 | 20.40 | 15.68 | 27.319 | 8.769 | − 0.568 |
| OE6A104208P3 | OeHSP90-9 | 705 | [184,702] | 4.93 | 22.87 | 17.07 | 26.809 | 9.362 | − 0.612 |
| OE6A113947P1 | OeHSP90-10 | 824 | [259,802] | 4.87 | 20.39 | 15.05 | 25.728 | 9.345 | − 0.746 |
| OE6A117167P1 | OeHSP90-11 | 791 | [276,786] | 5.42 | 15.93 | 13.91 | 26.802 | 9.861 | − 0.535 |
| OE6A119484P2 | OeHSP90-12 | 824 | [259,802] | 4.86 | 25.11 | 18.50 | 25.85 | 9.709 | − 0.735 |
| Mean | – | 735,83 | – | 5.04 | 20.42 | 15.83 | 26.67 | 9.46 |
The detailed information and physicochemical features are available in Supplementary file S1
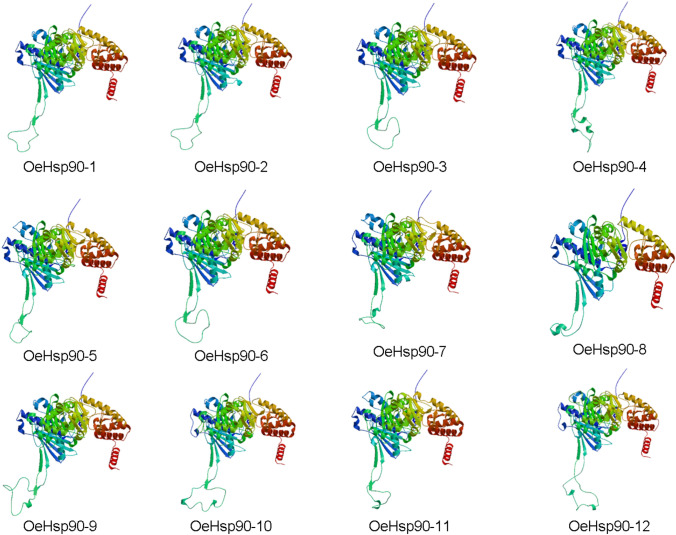
In silico 3D structures were determined using Swiss-Model algorithm for 12 OeHSP90 proteins (Fig. 2), which shared 45–72% identity to the corresponding proteins used as template. This degree of structural similarity was appropriate for the analysis of 3D protein structures, as a minimum of 30% sequence identity is widely accepted as a threshold for successful homology modeling (Xiang 2006). Global Model Quality Estimation (GMQE) ranged from 0.63 to 0.77 and QMEAN Z-scores were comprised between − 0.83 and − 2.32 (i.e. within the [0−4] interval), suggesting satisfactory geometry of the predicted 3D structures (Table 4). These 3D structures obtained from OeHSP90 proteins were archived in Protein Model Database (Castrignanò et al. 2006), which can be freely accessed (https://bioinformatics.cineca.it/PMDB/) through accession numbers shown in Table 4.
Fig. 2.
Three-dimensional (3D) structures of OeHSP90 proteins. In all figures, spirals are helices, broad strips with arrows are β-sheets and thin loops are coils
Table 4.
Homology modelling and model quality estimation of OeHSP90 proteins using SWISS-MODEL along with models PMDB accessions
| Homology modeling | Model quality estimation | PMDB Id | |||||
|---|---|---|---|---|---|---|---|
| Protein (target) | Template SMTLa Id | Template description | Target/Template identity (%) | Model oligo-state | GMQEb | QMEANc
 /
/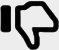
|
|
| OeHSP90-1 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 70.79 | Monomer | 0.77 | − 1.00
|
PM0082241 |
| OeHSP90-2 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 70.88 | Monomer | 0.76 | − 1.25
|
PM0082244 |
| OeHSP90-3 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 72.47 | Monomer | 0.77 | − 0.83
|
PM0082245 |
| OeHSP90-4 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 71.45 | Monomer | 0.76 | − 1.19
|
PM0082246 |
| OeHSP90-5 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 45.20 | Monomer | 0.64 | − 1.87
|
PM0082247 |
| OeHSP90-6 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 72.47 | Monomer | 0.77 | − 0.89
|
PM0082248 |
| OeHSP90-7 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 46.67 | Monomer | 0.65 | − 1.53
|
PM0082255 |
| OeHSP90-8 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 68.27 | Monomer | 0.77 | − 1.43
|
PM0082250 |
| OeHSP90-9 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 71.20 | Monomer | 0.76 | − 1.22
|
PM0082251 |
| OeHSP90-10 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 51.76 | Monomer | 0.64 | − 2.43
|
PM0082256 |
| OeHSP90-11 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 45.65 | Monomer | 0.65 | − 2.00
|
PM0082253 |
| OeHSP90-12 | 5fwl.1.A | HEAT SHOCK PROTEIN HSP 90 BETA | 51.69 | Monomer | 0.63 | − 2.32
|
PM0082254 |
aSMTL: SWISS-MODEL template library
bGMQE: “Global Model Quality Estimation” is a quality prediction, combining properties from the target–template alignment and the template search method. It is expressed as a score (GMQE score) comprised between 0 and 1, reflecting model accuracy, where higher numbers indicate higher reliability
cQMEAN: QMEAN (Benkert et al. 2011) provides both global (i.e. for the entire structure) and local (i.e. per residue) absolute consistency estimates on the basis of one single model. QMEAN Z-scores around zero indicate a strong agreement between the model structure and experimental structures of similar size. Scores of − 4.0 or below are a sign of models with low quality, which is also emphasized by the shift of the “thumb-up” symbol to a “thumb-down” symbol next to the score
Functional domains associated with the HSP90
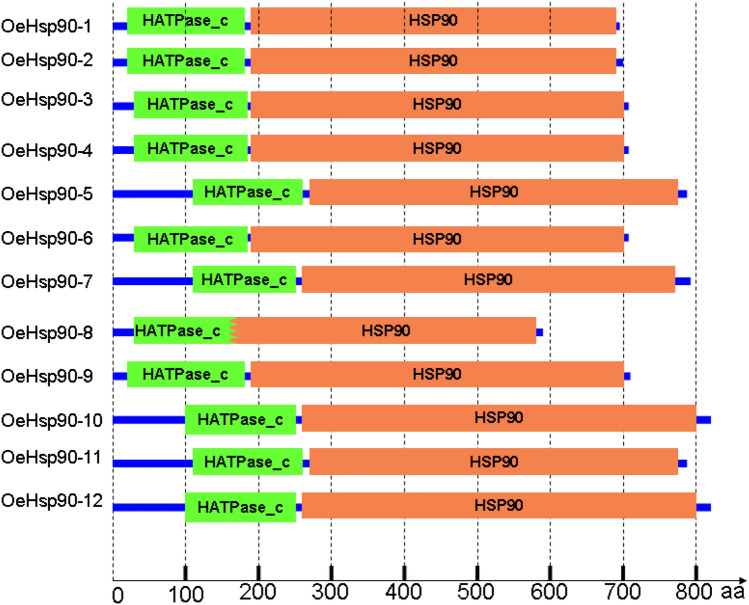
We predicted the major domains of these proteins in O. europaea using Pfam 32.0. All of the 12 proteins contained a Histidine kinase-like ATPase (HATPase_c) domain (Pfam: PF02518; SMART: SM00387) and an HSP90 family domain (Supplementary file 2; Fig. 3).
Fig. 3.
HATPase-c domains predicted in 12 olive HSP90 proteins reported in this study and their relative position to the HSP90 one. In OeHSP90-8, HATPase_c [32-184 aa] and HSP90 [141-591 aa] are overlapping. Relative proteins lengths are respected. All protein sizes and domain spans are presented in Table 3 and Supplementary file S2, respectively
Subcellular localization, gene structure and conserved motifs of the HSP90 gene family
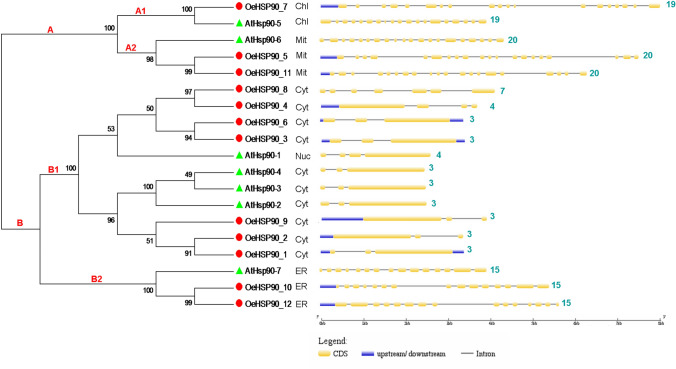
Protein subcellular localization is crucial information to elucidate protein functions. In silico analysis of the cellular location of HSP90 proteins from O. europaea indicated that seven out of 12 proteins were predicted to be localized in cytosol, 2/12 in the endoplasmic reticulum, while the others were localized in mitochondria (2/12) and chloroplast (1/12) (Supplementary file S1, Fig. 4). As shown in Fig. 4, the minor phylogenetic clade A consisted of organellar, while the major clade B consisted of cytoplasmic HSP90 proteins. Within clade A, subclades A1 and A2 contained chloroplastic and mitochondrial proteins, respectively; and within clade B, subclades B1 and B2 contained cytosolic and endoplasmic reticulum proteins, respectively.
Fig. 4.
Intron-exon structure of HSP90 genes in O. europaea var. europaea (OeHSP90s) and A. thaliana (AtHSP90s). The yellow blocks indicate exons, the black lines indicate introns and the blue blocks indicated upstream or downstream. The number of exons is indicated on the right hand side. Cyt, Cytosol; Chl, Chloroplast; ER, Endoplasmic reticulum; Mit, Mitochondria; Nuc, Nucleus
To further investigate the structural characters of the HSP90 gene family in O. europaea, the exon–intron structure was investigated using the online GSDS. Our results revealed that in the same group, the gene members had similar exon–intron structures and exon numbers (Fig. 4). The average number of exons in subclade A1 (chloroplastic members) was exactly 19. Compared with subclade A1, mitochondrial proteins (subclade A2) contained one additional exon (20). In contrast, endoplasmic reticulum proteins (subclade B2) contained clearly fewer exons (15), while the average number of exons in cytosolic proteins (subclade B1) was 3.55 and most members of this group contained 3 exons. Interestingly, all paralogous gene pairs identified above (Table 2), showed conserved exon/intron structures, except OeHSP90-8/OeHSP-4 that differed in terms of numbers of exons (7 and 4, respectively) and introns (6 and 3, respectively).
In order to determine the motif patterns of the HSP90 proteins in O. europaea, and to explore whether the cytosolic, endoplasmic reticulum, mitochondrial and chloroplastic groups shared motifs, the proteins were examined using the program MEME. A total of 20 distinct motifs were discovered using MEME (Table 5 and Fig. 5). As shown in Fig. 4, motifs 1, 2, 3 and 7 were uniformly distributed with a standard one occurrence in all proteins; yet, in cytosolic proteins, motif 1 was present in 2–3 sites. Motifs 4, 5, 6, and 9 were also widely present across the OeHSP90 and AtHSP90 proteins (in 18 proteins out of 19). Motifs 8, 10, 15, 18 and 19 did not show a specific distribution pattern in relation to the subcellular localization and phylogenetic position. In contrast, members within the same sub cluster and sharing the same subcellular location were found to share a number of motifs suggesting possible functional similarity among these HSP90 proteins (Fig. 4). For example, motifs 13 and 16 are unique and uniformly found in mitochondrial proteins (sub cluster A2), whereas motif 11 is specific to organellar proteins (cluster A) except AtHSP90-6. Finally, sub cluster B2 (endoplasmic reticulum proteins) was characterized by the generalized presence of four motifs: 12, 14, 17 and 20. Overall, the similar motif arrangements among HSP90 proteins within sub clusters indicated that the protein architecture is conserved within a specific sub family, suggesting that these similar motif features may be related to their functional attributes in genome. Yet, the functions of most of these conserved motifs remain to be elucidated.
Table 5.
Multilevel consensus sequence identified by MEME among OeHSP90 in O. europaea var. europaea
| Motifs | Number of amino acids | Number of occurrences | E-value | Consensus sequence |
|---|---|---|---|---|
| 1 | 65 | 39 | 6.1e−1147 | ESLGDKVEKVVVSERFVDSPCCLVTGEYGWSANMERIMKAQALGDGSKIAYMSSKKTMEINPENA |
| 2 | 42 | 30 | 3.0e−660 | FAFQAEINQLLSLIINTFYSNKEJFJRELISNASDALDKJRF |
| 3 | 42 | 41 | 1.8e−594 | YLSFYKGLVBSDELPLAVSHFTLZGQLEFKAILKVPKRAPFD |
| 4 | 70 | 17 | 5.3e−687 | HSTKSGDELTSLKDYVTRMKEGQKDIYYITGESKKAVENSPFLEKLKKKGYEVLYMVDAIDEYAVGQLKE |
| 5 | 59 | 18 | 3.1e−509 | IMEELRKRAEADKNDKSVKDLVLLLYETALLTSGFSLDDPNTFGSRIHRMLKLGLSIDE |
| 6 | 42 | 18 | 9.7e−338 | DEEKEEKKKKKKKIKEVSHEWELVNKQKPIWLRKPEEITKEE |
| 7 | 30 | 33 | 4.6e−314 | LFDTNKKPNNIKLYVRRVFIMDNCEEEIPE |
| 8 | 42 | 14 | 4.5e−136 | DISISENKEDYIKFWENFGKSLKLGIIEDTQNSKRJAPLLRF |
| 9 | 30 | 18 | 7.6e−141 | YEGKKLVSATKEGLKLGEEEEEKKKKEELK |
| 10 | 30 | 8 | 5.4e−085 | IKDLVKNYSZFVSFPIYTWQEKGYTKEVEV |
| 11 | 42 | 4 | 5.7e−055 | LGTIAQSGTAKFLKALKENKDLGADNNLIGQFGVGFYSAFLV |
| 12 | 42 | 3 | 2.5e−026 | MRKWTIPSVLFLLCLLFLLPDQGRKIQAHAESDSDAPVDPPK |
| 13 | 42 | 5 | 2.6e−026 | LSVTEPDLDKDALDLDIRIZTDKDNGIITITDSGIGMTRQEL |
| 14 | 30 | 3 | 1.5e−024 | GVGFYSVYLVADYVEVISKHNDDKQHVWES |
| 15 | 69 | 2 | 2.6e−017 | MHRVVSRRAVSAILRNGGLRHRNSAVPLSSSNPFDQSADEGKKEKKWLSFLAAGSCNATIKPFNVRNVP |
| 16 | 30 | 3 | 8.1e−009 | WGSAQQVVPPSHVPETFEAEVVEPAEAGSP |
| 17 | 30 | 3 | 1.4e−007 | GAVPSGLSTDSDVTKREAESISRRSLRANA |
| 18 | 30 | 2 | 2.3e−001 | NFPIHLWASKEVDEEVPADEDESNDEDETS |
| 19 | 30 | 4 | 2.5e−001 | WENEAESSSKTEEEPTDPADTEPRGVKDEL |
| 20 | 30 | 3 | 3.2e−008 | TDKEVLGEGDNAKLEIQIKLDKEKKILSIR |
Fig. 5.
Distribution of twenty distinct conserved motifs across HSP90 proteins from O. europaea (OeHSP90s) and A. thaliana (AtHSP90s)
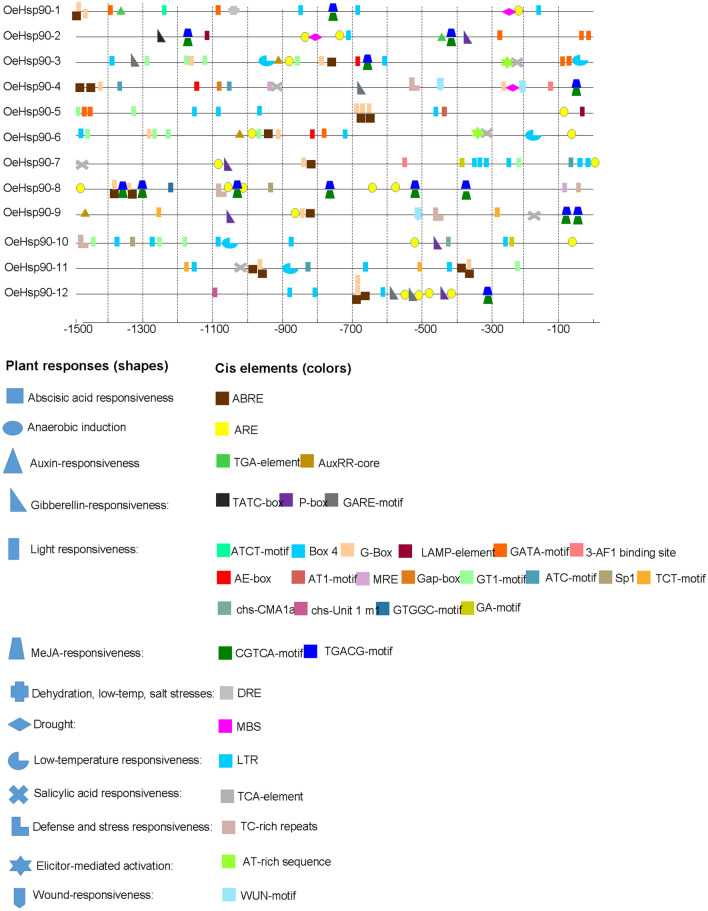
Stress-related cis-elements in the OeHSP90 gene promoters
In order to further explore the regulation of OeHSP90 gene expression in reaction to stress conditions, the cis-elements in the promoter region (the 1.5 kb upstream sequences from translation starting sites) of the OeHSP90 genes were analyzed. As illustrated in Fig. 6, the promoters of all OeHSP90 genes contained light responsiveness elements, such as ATCT-motif, Box 4, G-Box, GATA-motif, GT1-motif, LAMP-element, etc. In addition, one to two LTRs existed in 4 OeHSP90s (OeHSP90-3, OeHSP90-6, OeHSP90-10 and OeHSP90-11), and 1 DRE was found in OeHSP90-1, which indicated that the expressions of these HSP90s were associated with dehydration, low-temperature and salt stresses responsiveness. In total, 10 out of 12 OeHSP90s promoters contained anaerobic responsive elements (ARE), suggesting a potential role of OeHSP90 in reponse to oxygen limitation. We also identified a large number of other cis-acting elements related to a variety of plant responses to abiotic and biotic stress; namely, drought-inducibility element (MBS), defense and stress responsiveness elements (TC-rich repeats), wounding responsive element (WUN-motif), and elicitor-mediated activation (AT-rich sequence) were all found in 25%, 33.3%, 41.6%, and 16.6% of OeHSP90 promoters, respectively (Fig. 6). Some of the OeHSP90 promoters also contained phytohormone-induced elements, such as AuxRR-core or TGA-element (auxin responsive), TATC-box, GARE-motif, or P-box element (gibberellin-responsive), ABRE element (abscisic acid responsive), TCA-element (salicylic acid responsive), and TGACG-motif or CGTCA-motif element (MeJA-responsive), suggesting that OeHSP90s are part of the vast phytohormone signaling network leading to stress responsiveness. Our analysis clearly showed that OeHSP90 genes, in olive, are inducible under different stress conditions.
Fig. 6.
Predicted stress-related cis-elements in OeHSP90 promoters. Promoter sequences (1500 pb upstream region) of 12 OeHSP90 genes were analyzed online at PlantCARE web server
Expression evidence of OeHSP90 genes
The spatio-temporal expression of members of the HSP90 gene family in olive was investigated using publicly available O. europaea expressed sequence tags (ESTs). Of 12 regular olive HSP90 genes, seven were supported by at least one EST from O. europaea tissue-specific and developmental stage-related libraries (Table 6). The maximum number of OeHSP90s was expressed in “leaves and fruits” (6/7), while only two genes were supported by ESTs expressed in “seedlings”, and a unique gene matched an EST expressed in “immature fruit”. Evidence of expression in more than two tissues was inferred for a single gene (OeHSP90-2) that matched ESTs expressed in the leaves, fruits and seedlings.
Table 6.
Spatial and temporal expression profiles of O. europaea HSP90 genes, based on BLAST to publicly available Expressed Sequence Tags (ESTs)
| OeHSP90 gene (Query) | Corresponding target in Olea europaea (taxid: 4146) ESTs | Spatial and temporal expression features | ||
|---|---|---|---|---|
| EST ID | Definition | Tissue | Developmental stage | |
| OeHSP90-1 | JK748014.1 | se015_D7 One-month old Olive (Olea europaea) cv. Koroneiki seedlings library Olea europaea cDNA, mRNA sequence | Seedlings | One-month old |
| OeHSP90-2 | JK748014.1 | se015_D7 One-month old Olive (Olea europaea) cv. Koroneiki seedlings library Olea europaea cDNA, mRNA sequence | Seedlings | One-month old |
| GO245607.1 | OEAA-070810_Plate7m15.b1 cDNA library from Olive leaves and fruits Olea europaea cDNA, mRNA sequence | Leaves and fruits | – | |
| OeHSP90-3 | GO244160.1 | OEAA-070810_Plate3o08.b1 cDNA library from Olive leaves and fruits Olea europaea cDNA, mRNA sequence | Leaves and fruits | – |
| GO244048.1 | OEAA-070810_Plate3j16.b1 cDNA library from Olive leaves and fruits Olea europaea cDNA, mRNA sequence | Leaves and fruits | – | |
| FL684093.1 | C_P23_H12_0414F_p7 Olea europaea cv. Leccino fruitlet Olea europaea cDNA, mRNA sequence | Fruit | Immature | |
| OeHSP90-6 | GO244160.1 | OEAA-070810_Plate3o08.b1 cDNA library from Olive leaves and fruits Olea europaea cDNA, mRNA sequence | Leaves and fruits | – |
| GO244048.1 | OEAA-070810_Plate3j16.b1 cDNA library from Olive leaves and fruits Olea europaea cDNA, mRNA sequence | Leaves and fruits | – | |
| GO246288.1 | OEAA-070810_Plate9j13.b1 cDNA library from Olive leaves and fruits Olea europaea cDNA, mRNA sequence | Leaves and fruits | – | |
| FL684093.1 | C_P23_H12_0414F_p7 Olea europaea cv. Leccino fruitlet Olea europaea cDNA, mRNA sequence | Fruit | Immature | |
| OeHSP90-7 | GO244489.1 | OEAA-070810_Plate4m08.b1 cDNA library from Olive leaves and fruits Olea europaea cDNA, mRNA sequence | Leaves and fruits | – |
| OeHSP90-10 | GO245204.1 | OEAA-070810_Plate6l07.b1 cDNA library from Olive leaves and fruits Olea europaea cDNA, mRNA sequence | Leaves and fruits | – |
| OeHSP90-12 | GO245204.1 | OEAA-070810_Plate6l07.b1 cDNA library from Olive leaves and fruits Olea europaea cDNA, mRNA sequence | Leaves and fruits | – |
Functional annotation of 3D structures and interaction networks of OeHSP90 family
Gene Ontology (GO) terms suggested the involvement of OeHSP90s in multiple biological processes including protein folding, as well as cellular, metabolic and macromolecule processes (Supplementary file S3). In order to further investigate the relationship between OeHSP90s and related proteins in the olive interactome, the protein interaction networks were analyzed by STRING software using directly the A. thaliana model. All the 12 HSP90 proteins matched their counterparts in A. thaliana with identities ranging between 77 and 100% (Supplementary file S4). As shown in Fig. 7, five proteins were predicted as functional partners of OeHSP90s, indicating these proteins may co-regulate some biological processes. In particular, results indicate that Calnexin homolog 1 (CNX1), SGT1 homolog A and heat shock proteins (HSP70s) are pivotal proteins that interact directly with all OeHSP90 proteins. We have found that OeHSP90-1, OeHSP90-2 and OeHSP90-9 interact directly with Heat Shock Transcription Factor A2 (HSFA2). In addition to interaction with functional partners, there are interactions between OeHSP90 proteins (Fig. 7). Overall, our analysis showed the involvement of HSP90 chaperones in complex biological signalling networks in olive.
Fig. 7.
Functional interaction network of 12 O. europaea var. europaea HSP90 proteins, based on their A. thaliana orthologs. Nodes are connected by lines that indicate experimentally determined interactions. Line thickness indicates the strength of data support. Framed proteins are functional partners interacting with OeHSP90s. Unconnected OeHSP90s have no known relationship. The original figure was constructed in STRING 11.0 (Szklarczyk et al. 2019) using medium confidence level (0.400), and experiments and co-expression as active sources of interaction
Discussion
The HSP90 has been studied at genome-scale in various of monocot and dicot species, such as Brachypodium distachyon (Zhang et al. 2017), Nicotiana tabacum (Song et al. 2019), Populus (Zhang et al. 2013) and different legumes (Agarwal et al. 2016). However, our study is the first to identify and characterize HSP90 proteins from the Mediterranean olive. This extensive survey of the HSP90 gene family has been facilitated by the recent completion of O. europaea genome sequencing and its annotation (Cruz et al. 2016), providing a wealth of information on the functional genomics of this economically important tree. Based on a wide range of computational methods, we suggest that the olive genome is enriched in heat shock genes, with 12 HSP90 genes, compared to seven HSP90 genes in each Arabidopsis (Zhang et al. 2013) and tomato (Liu et al. 2014; Zai et al. 2015). Differences in gene family sizes often correspond to differences in the functional array of roles of the members of the gene families. Therefore, we suggest that the comparatively high number of olive HSP90 proteins are linked to their numerous roles in O. europaea. This is also supported by the wide diversity of the olive HSP90 members, as denoted by their diverse biophysical features detected in our study.
Gene duplication events are thought to take place frequently in genome evolution and range in size from small-scale (SSD) to whole-genome events (WGD) (Carretero-Paulet and Fares 2012; Fares et al. 2013), all of them contributing to the expansion of gene families in the genome. Paralogs are types of homologous genes that are related by duplication and tend to diverge in function after duplication (Gabaldón and Koonin 2013). There were five pairs of duplicated HSP90 genes identified in O. europaea in the present study, indicating that this gene family evolution has been strongly marked by species-specific gene duplication. The estimated time of duplication for the paralogous OeHSP90 genes indicated that all paralogs occurred before the oldest WGD that is at the base of the family Oleaceae (~ 33–72 Mya), except for OeHSP3/-6, which is a recently duplicated gene pair (2.5 Mya) after the last WGD event specific to the O. europaea lineage, about 10 Mya (Julca et al. 2018), suggesting that the latter was a small scale event. Phylogenetic analysis is typically used to gain insight into the evolutionary relationships of species and to infer orthologs and paralogs, between and within species, respectively. Such evolutionary relationships may be of great importance to understand and describe current functional features of a set of studied genes. We discovered that the members within the same phylogenetic sub-cluster shared the same subcellular location, consistent with HSP90 genes in other species as Populus trichocarpa (Zhang et al. 2013), A. thaliana (Krishna and Gloor 2001) and Brachypodium distachyon (Zhang et al. 2017). Such conserved subcellular or organelle localization of HSP90s in the same phylogenetic subgroup suggests that they present roles in stress response or organelle-specific development, providing clues for their specific cellular functions. The largest number of OeHSP90 proteins were predicted to be localized in cytosol; this feature is shared with Arabidopsis (Sarkar et al. 2009) and Brachypodium distachyon (Zhang et al. 2017), and indicates that the cytoplasm, where protein assembly occurs, may be the primary site of action for HSP90 proteins (Vabulas et al. 2010).
The Histidine kinase-like ATPase (HATPase_c) domain (Pfam: PF02518; SMART: SM00387) is an evolutionary conserved protein domain, which is found in several ATP-binding proteins including histidine kinase, DNA gyrase B, topoisomerases (Bellon et al. 2004), molecular chaperones HSP90 (Immormino et al. 2004), phytochrome-like ATPases and DNA mismatch repair proteins. The HATPase_c domain has already been reported in HSP90 proteins from algae (Chlamydomonas reinhardtii), mosses (Physcomitrella patens), monocots (Brachypodium distachyon, Oryza sativa, Triticum aestivum, and Zea mays) and dicots (Arabidopsis thaliana, Glycine max, Medicago truncatula and Gossypium raimondii) (Zhang et al. 2017); and its association with the HSP90 justifies the role played by HSP90 proteins in ATP binding and hydrolysis, as previously demonstrated (Panaretou et al. 1998). This domain functions as an ATP/ADP binding site with ATPase activity (Young et al. 2001).
The gene structure difference between HSP90 groups may be associated with their functions in different biological processes in subcellular compartments. Of the OeHSP90 genes containing introns, 10 were found to contain only two introns, and their lengths were highly variable. The connection between the occurrence of introns and the extent of gene expression is questionable (Parra et al. 2011). In some studies, the lack of introns, or a short intron length, has been observed to increase the level of gene expression in plants (Ren et al. 2006; Jeffares et al. 2008). In our study, the variation of the number of introns suggested that some intron loss, along with intron gain events, may have occurred throughout the structural evolution of OeHSP90 encoding genes.
Structural plasticity of OeHSP90 biomolecules could be inferred from their physicochemical features. On the other hand, produced 3D structures and their gene ontology annotations supported a wide functional array, and may, in the long term, give rise to docking studies aimed at fine-tuning the molecular properties to promote binding interactions (Baloji et al. 2019).
Cis-regulatory elements (CREs) are implicated in the control of gene expression and provide an indication of the physiological process in which members of a gene family may be involved. Promoter regions of OeHSP90 genes included a variety of CREs indicating that OeHSP90 genes are involved in O. europaea growth, development and responses to abiotic and biotic stresses via the regulation of hormone signaling pathways. In addition, we observed that anaerobic-stress response elements (ARE), previously studied in the promoters of maize (Walker et al. 1987), were present extensively in OeHSP90 promoters. Likewise, ARE have been observed to be the most widespread cis-regulatory elements in Arabidopsis (Sadeghnezhad et al. 2014) and cotton (Wu et al. 2019) genomes. Anaerobic stress usually corresponds to hypoxic or anoxic conditions created around the roots usually as a consequence of flooding (Dennis et al. 2000). Finally, we revealed the occurrence of a putative AT region, involved in elicitor-mediated activation, in heat-shock 90 promoters of O. europaea. AT-rich elements have been outlined as being actually involved in the expression control of GmHSP17.6-L gene in reaction to nematode disease in soybean (Fuganti et al. 2010). It will now be interesting to uncover whether these AT rich regions (in particular the number of AT repetitions) can influence the resistance and/or susceptibility responses to plant-parasitic nematodes associated with olive tree.
Although CRE sequences have been generally conserved by mutation-eliminating selection, their activity is likely to be erased during evolution due to variability in number, order or position. Therefore, it is important to study expression in order to infer spatial and temporal profiles of olive HSP90 genes and their eventual responsiveness to any stimuli. It is known that individual members of broad gene families are variable in sequence and function exhibiting different patterns of expression across different tissues. Gene tissue-specific expression analysis may therefore offer valuable information on gene functions and gene regulations (Wen et al. 2017). Typically, our results have shown that more than half of the OeHSP90 genes were specifically expressed in one or more tissues, suggesting that these genes play multiple roles in the growth of olive and may be actively implicated in the development of various tissues and organs. In particular, HSP90 genes were supported by ESTs expressed in the leaf, fruit and seedlings of the olive plant, similar to recent reports in Arabidopsis, tomato and soybean (Xu et al. 2013; Liu et al. 2014; Wang et al. 2016).
It is helpful to predict interactome networks in order to get an integrative view of the cellular functions in which olive HSP90s may be involved. Homologs of A. thaliana CNX1, SGT1, HSP70s and HSFA2 were predicted to be of high importance as functional partners of OeHSP90s, as they occupied central positions in the protein–protein interaction network. A. thaliana Calnexin homolog 1 (CNX1) is a Calcium-binding protein that interacts with newly synthesized glycoproteins in the endoplasmic reticulum (ER). It may contribute to the folding, assembly and/or retention within the ER of unassembled protein subunits, thus playing a major role in the quality control machinery of the ER (Bergeron et al. 1994; Liu et al. 2017). SGT1 homologs (A and B) are functionally redundant proteins playing a role in resistance protein accumulation during plant resistance against pathogens (Azevedo et al. 2006). Besides, SGT1 is probably required for SCF-mediated ubiquitination, by coupling HSP90 to SCF complex for ubiquitination of HSP90 client proteins (Zhang et al. 2010). In A. thaliana, it has been observed that SGT1 associates with the N-terminal ATPase domain of HSP90 and another co-factor, RAR1 (Required for Mla12 resistance) (Botër et al. 2007; Zhang et al. 2008). SGT1 and RAR1 assist HSP90 in stabilizing plant disease resistance (R) proteins (Austin et al. 2002; Azevedo et al. 2006). The 70-kDa heat shock proteins (HSP70s) could cooperate with other chaperones and key regulators of many signal transduction pathways, to assist in wide range of folding processes, including refolding of misfolded stress-denatured proteins (Mayer and Bukau 2005), assisting translocation of precursor proteins into organelles, and preventing protein aggregation, thus controlling cell homeostasis, proliferation, differentiation and cell death. Finally, HSFA2 is one of 21 HSF genes identified in A. thaliana (Scharf et al. 2012). It is strongly heat inducible and seems to be involved in environmental stress response including response to salt (Ogawa et al. 2007) and oxidative stress (Nishizawa-Yokoi et al. 2010).
Conclusion
The present study provides an in silico integrative analysis of the Mediterranean olive HSP90 gene family covering physicochemical features, conserved domains and motifs, protein 3D, gene and promoter structures, evolutionary relationships, expression patterns and protein–protein interaction networks. We provided evidence of the relationship between the phylogeny, structural and cellular localization of this gene family. Olive HSP90 showed different levels of expression among different tissues with a rich array of regulation controls, all suggesting that their functional diversity has been a backbone of the olive physiological and adaptive processes. Our study sets groundwork for further experimental gene expression and gene manipulation studies of OeHSP90 gene candidates, which will be relevant to improving stress-related and developmental responses, in olive.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Physico-chemical features and basic information of 12 regular HSP90 family genes identified in Olea europaea var. europaea. The list comprises seven Arabidopsis HSP90 sequences (Zhang et al. 2013) used in this study. (XLS) (XLS 108 kb)
Conserved domains of HSP90 proteins in Olea europaea var. europaea. The major domains were identified using Pfam (https://pfam.xfam.org/). (XLS) (XLS 19 kb)
Gene ontology terms of OeHsp90 proteins, based on their 3D structures, using ProFunc server. (XLS 51 kb)
Proteins in Arabidopsis thaliana that match the OeHSP90 proteins in olive tree by STRING 11.0. (XLS) (XLS 21 kb)
Acknowledgments
The authors thank all lab members for assistance.
Abbreviations
- 3D
Three dimensional
- ABRE element
ACGT-containing abscisic acid response element
- At
Arabidopsis thaliana
- CDD
Conserved Domain Database
- CDS
Coding sequences
- CNX1
Calnexin homolog 1
- CS domain
CHORD-containing proteins
- DRE
Dehydration responsive element
- ER
Endoplasmic reticulum
- EST
Expressed sequence tags
- ExPASy
Expert Protein Analysis System program
- GO
Gene Ontology
- GMQE
Global Model Quality Estimation
- GRAVY
Grand average of hydropathicity
- GSDS
Gene Structure Display Server
- HATPase_c
Histidine kinase-like ATPase
- HSF
Heat shock transcription factors
- HSP
Heat shock proteins
- Ka
Non synonymous substitution rate
- Ks
Synonymous substitution rate
- LTRs
Long terminal repeats
- MBS
MYB binding site
- ML
Maximum likelihood
- MW
Molecular weight
- Mya
Million years ago
- NLRs
NOD-like receptors
- NTD
N-terminal domain
- Oe
Olea europaea
- OeHSP90
Olea europaea HSP90
- P-box
Gibberellin-responsive element
- pI
Isoelectric point
- SMTL
SWISS-MODEL Template Library
- SSD
Small-scale duplications
- WGD
Whole-genome duplication
- WUN-motif
Wound-responsive element
Authors’ contributions
IB contributed by conceptualization, data curation, writing original draft. JH helped in performing bioinformatics analyses and contributed to discussions. DB contributed by the conceptualization, methodology, software, data curation, supervision, study administration, manuscript editing. All authors have read and approved the final manuscript.
Funding
This study was financially supported by the Ministry of Higher Education and Scientific Research (Tunisia). The funder had no role in the design of the study, the collection, analysis, and interpretation of data and in writing the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interests.
Data archiving statement
All data analysed during this study are included in this article and its Additional files.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Agarwal G, Garg V, Kudapa H, Doddamani D, Pazhamala LT, Khan AW, Thudi M, Lee S-H, Varshney RK. Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea. Plant Biotechnol J. 2016;14:1563–1577. doi: 10.1111/pbi.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya P, Kumar G, Acharya V, Singh AK. Genome-wide identification and expression analysis of NBS-encoding genes in Malus x domestica and expansion of NBS genes family in Rosaceae. PLoS ONE. 2014;9:e107987. doi: 10.1371/journal.pone.0107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE. Regulatory role of SGT1 in early R gene-mediated plan defenses. Science. 2002;295:2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noël L, Sadanandom A, Casais C, Parker J, Shirasu K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006;25(9):2007–2016. doi: 10.1038/sj.emboj.7601084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloji G, Pasham S, Mahankali V, Garladinne M, Ankanagari S. Insights from the molecular docking analysis of phytohormone reveal brassinolide interaction with HSC70 from Pennisetum glaucum. Bioinformation. 2019;15(2):131–138. doi: 10.6026/97320630015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barghini E, Natali L, Giordani T, Cossu RM, Scalabrin S, Cattonaro F, Šimková H, Vrána J, Doležel J, Morgante M, Cavallini A. LTR retrotransposon dynamics in the evolution of the olive (Olea europaea) genome. DNA Res. 2015;22(1):91–100. doi: 10.1093/dnares/dsu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon S, Parsons JD, Wei Y, Hayakawa K, Swenson LL, Charifson PS, Lippke JA, Aldape R, Gross CH. Crystal structures of Escherichia coli topoisomerase IV ParE Subunit (24 and 43 Kilodaltons): a single residue dictates differences in novobiocin potency against Topoisomerase IV and DNA Gyrase. Antimicrob Agents Chemother. 2004;48(5):1856–1864. doi: 10.1128/aac.48.5.1856-1864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron JJM, Brenner MB, Thomas DY, Williams DB. Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19(3):124–128. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig I, Shindyalov I, Bourne PE. The proten data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botër M, Amigues B, Peart J, Breuer C, Kadota Y, Casais C, Moore G, Kleanthous C, Ochsenbein F, Shirasu K, Guerois R. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell. 2007;19:3791–3804. doi: 10.1105/tpc.107.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421–430. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Fares MA. Evolutionary dynamics and functional specialization of plant paralogs formed by whole and small-scale genome duplications. Mol Biol Evol. 2012;29(11):3541–3551. doi: 10.1093/molbev/mss162. [DOI] [PubMed] [Google Scholar]
- Castrignanò T, De Meo PD, Cozzetto D, Talamo IG, Tramontano A. The PMDB protein model database. Nucleic Acids Res. 2006;34:D306–D309. doi: 10.1093/nar/gkj105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Gao T, Wan S, Zhang Y, Yang J, Yu Y, Wang W. Genome-wide identification, classification and expression analysis of the HSP gene superfamily in tea plant (Camellia sinensis) Int J Mol Sci. 2018;19:2633. doi: 10.3390/ijms19092633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F, Julca I, Gómez-Garrido J, Loska D, Marcet-Houben M, Cano E, Galán B, Frias L, Ribeca P, Derdak S, Gut M, Sánchez-Fernández M, García JL, Gut IG, Vargas P, Alioto TS, Gabaldón T. Genome sequence of the olive tree, Olea europaea. GigaScience. 2016;5:29. doi: 10.1186/s13742-016-0134-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Dolferus R, Ellis M, Rahman M, Wu Y, Hoeren FU, Grover A, Ismond KP, Good AG, Peacock WJ. Molecular strategies for improving waterlogging tolerance in plants. J Exp Bot. 2000;51(342):89–97. doi: 10.1093/jexbot/51.342.89. [DOI] [PubMed] [Google Scholar]
- Fares MA, Keane OM, Toft C, Carretero-Paulet L, Jones GW. The roles of whole-genome and small-scale duplications in the functional specialization of Saccharomyces cerevisiae genes. PLoS Genet. 2013;9(1):e1003176. doi: 10.1371/journal.pgen.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuganti R, Machado MFPS, Lopes VS, Silva JFV, Arias CAA, Rockenbach-Marin SR, Binneck E, Abdelnoor RV, Marcelino FC, Nepomuceno AL. Size of AT(n) insertions in promoter region modulates GmHsp17.6-L mRNA transcript levels. J Biomed Biotechnol. 2010;10:1–9. doi: 10.1155/2010/847673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Koonin EV. Functional and evolutionary implications of gene orthology. Nat Rev Genet. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavahiana M, Khaneghah AM, Lorenzo JM, Munekata PES, Garcia-Mantrana I, Collado MC, Meléndez-Martínez AJ, Barbaf FJ. Health benefits of olive oil and its components: impacts on gut microbiota antioxidant activities, and prevention of noncommunicable diseases. Trends Food Sci Technol. 2019;88:220–227. doi: 10.1016/j.tifs.2019.03.008. [DOI] [Google Scholar]
- Hoter A, El-Sabban ME, Naim HY. The HSP90 family: structure, regulation, function, and implications in health and disease. Int J Mol Sci. 2018;19(9):2560. doi: 10.3390/ijms19092560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immormino RM, Dollins DE, Shaffer PL, Soldano KL, Walker MA, Gewirth DT. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J Biol Chem. 2004;279(44):46162–46171. doi: 10.1074/jbc.m405253200. [DOI] [PubMed] [Google Scholar]
- Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24(8):375–378. doi: 10.1016/j.tig.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Julca I, Marcet-Houben M, Vargas P, Gabaldón T. Phylogenomics of the olive tree (Olea europaea) disentangles ancient allo- and autopolyploidizations in Lamiales. BMC Biol. 2018;16:15. doi: 10.1186/s12915-018-0482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K. The HSP90 complex of plants. Biochim Biophys Acta. 2012;1823(3):689–697. doi: 10.1016/j.bbamcr.2011.09.016. [DOI] [PubMed] [Google Scholar]
- Krishna P, Gloor G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6(3):238–246. doi: 10.1379/1466-1268(2001)006<0238:thfopi>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wan H, Yang Y, Wei Y, Li Z, Ye Q, Wang R, Ruan M, Yao Z, Zhou G. Genome wide identification and analysis of heat shock protein 90 in tomato. Yi Chuan = Hered. 2014;36(10):1043–1052. doi: 10.3724/sp.j.1005.2014.1043. [DOI] [PubMed] [Google Scholar]
- Liu DYT, Smith PMC, Barton DA, Day DA, Overall RL. Characterisation of Arabidopsis calnexin 1 and calnexin 2 in the endoplasmic reticulum and at plasmodesmata. Protoplasma. 2017;254(1):125–136. doi: 10.1007/s00709-015-0921-3. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi S, Mariotti R, Bagnoli F, Costantini L, Cultrera NGM, Arzani K, Pandolfi S, Vendramin GG, Torkzaban B, Hosseini-Mazinani M, Baldoni L. The eastern part of the Fertile Crescent concealed an unexpected route of olive (Olea europaea L.) differentiation. Ann Bot. 2017;119(8):1305–1318. doi: 10.1093/aob/mcx027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Tainaka H, Yoshida E, Tamoi M, Yabuta Y, Shigeoka S. The 26S proteasome function and Hsp90 activity involved in the regulation of HsfA2 expression in response to oxidative stress. Plant Cell Physiol. 2010;51:486–496. doi: 10.1093/pcp/pcq015. [DOI] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot. 2007;58:3373–3383. doi: 10.1093/jxb/erm184. [DOI] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17(16):4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Seo YS. Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol J. 2015;31(4):323–333. doi: 10.5423/ppj.rw.08.2015.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Rose AB, Korf I. Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res. 2011;39:5328–5337. doi: 10.1093/nar/gkr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75(75):271. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Prasad PVV, Pisipati SR, Momčilović I, Ristic Z. Independent and combined effects of high temperature and drought stress during grain filling on plant yield and chloroplast EF-Tu expression in spring wheat. Agron Crop Sci. 2011;197:6. doi: 10.1111/j.1439-037x.2011.00477.x. [DOI] [Google Scholar]
- Queitsch C, Sangstert TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417(6889):618. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Ramegowda V, Senthil-Kumar M. The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic under standing from drought and pathogen combination. J Plant Physiol. 2015 doi: 10.1016/j.jplph.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM–HMM alignment. Nat Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- Ren XY, Vorst O, Fiers MWEJ, Stiekema WJ, Nap JP. In plants, highly expressed genes are the least compact. Trends Genet. 2006;22:528–532. doi: 10.1016/j.tig.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Rombauts S, Déhais P, Van Montagu M, Rouzé P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27:295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghnezhad E, Askari H, Soltani S, Honarvar F. Identification and distribution of anaerobic responsive elements (AREs) in genes functional categorization of Arabidopsis thaliana. J Appl Biotechnol Rep. 2014;1(4):135–141. [Google Scholar]
- Sangster TA, Queitsch C. The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr Opin Plant Biol. 2005;8(1):86–92. doi: 10.1016/j.pbi.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Sarkar NK, Kim YK, Grover A. Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genom. 2009;393:1471–2164. doi: 10.1186/1471-2164-10-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat Stress Transcription Factor (HSF) family: structure, function and evolution. Biochim Biophys Acta Gene Regul Mech. 2012;1819:104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Song Z, Pan F, Yang C, Jia H, Jiang H, He F, Li N, Lu X, Zhang H. Genome-wide identification and expression analysis of HSP90 gene family in Nicotiana tabacum. BMC Genet. 2019;20:35. doi: 10.1186/s12863-019-0738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genom. 2007;8:125. doi: 10.1186/1471-2164-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, von Mering C. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Steecher G, Nei M, Kumar S. MEGA: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoraki MA, Caplan AJ. Quality control and fate determination of Hsp90 client proteins. Biochim Biophys Acta. 2012;1823(3):683–688. doi: 10.1016/j.bbamcr.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol. 2010;2(12):a004390. doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Howard EA, Dennis ES, Peacock WJ. DNA sequences required for anaerobic expression of the maize Adh1 gene. Proc Nat Acad Sci USA. 1987;84:6624–6629. doi: 10.1073/pnas.84.19.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat Commun. 2016;7:10269. doi: 10.1038/ncomms10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, Wu X, Li T, Jia M, Liu X, Li P, et al. Genome-wide survey of heat shock factors and heat shock protein 70s and their regulatory network under abiotic stresses in Brachypodium distachyon. PLoS ONE. 2017;12(7):e0180352. doi: 10.1371/journal.pone.0180352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Wu A, Hao P, Wei H, Sun H, Cheng S, Chen P, Ma Q, Gu L, Zhang M, Wang H, Yu S. Genome-wide identification and characterization of glycosyltransferase family 47 in cotton. Front Genet. 2019;10:824. doi: 10.3389/fgene.2019.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z. Advances in homology protein structure modeling. Curr Protein Pept Sci. 2006;7(3):217–227. doi: 10.2174/138920306777452312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Li ZY, Chen Y, Chen M, Li LC, Ma YZ. Heat shock protein 90 in plants: molecular mechanisms and roles in stress responses. Mol Sci. 2012;13:15706–15723. doi: 10.4238/2015.july.14.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xue C, Xue D, Zhao J, Gai J, Guo N, Xing H. Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS ONE. 2013;8(7):e69810. doi: 10.1371/journal.pone.0069810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Moarefi I, Ulrich Hartl F. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154(2):267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zai WS, Miao LX, Xiong ZL, Zhang HL, Ma YR, Li YL, Chen YB, Ye SG. Comprehensive identification and expression analysis of Hsp90s gene family in Solanum lycopersicum. Genet Mol Res. 2015;14(3):7811–7820. doi: 10.4238/2015.july.14.7. [DOI] [PubMed] [Google Scholar]
- Zhang M, Botër M, Li K, Kadota Y, Panaretou B, Prodromou C, Shirasu K, Pearl LH. Structural and functional coupling of Hsp90- and Sgt1-centred multi-protein complexes. EMBO J. 2008;27:2789–2798. doi: 10.1038/emboj.2008.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Kadota Y, Prodromou C, Shirasu K, Pearl LH. Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: implications for chaperoning of NLR innate immunity receptors. Mol Cell. 2010;39:269–281. doi: 10.1016/j.molcel.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li J, Liu B, Zhang L, Chen J, Lu M. Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genom. 2013;14:532. doi: 10.1186/1471-2164-14-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li L, Ye T, Chen R, Gao X, Xu Z. Molecular characterization, expression pattern and function analysis of the OsHSP90 family in rice. Biotechnol Biotechnol Equip. 2016;30(4):669–676. doi: 10.1080/13102818.2016.1184588. [DOI] [Google Scholar]
- Zhang M, Shen Z, Meng G, Lu Y, Wang Y. Genome-wide analysis of the Brachypodium distachyon (L.) P. Beauv. Hsp90 gene family reveals molecular evolution and expression profiling under drought and salt stresses. PLoS ONE. 2017;12(12):e0189187. doi: 10.1371/journal.pone.0189187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Physico-chemical features and basic information of 12 regular HSP90 family genes identified in Olea europaea var. europaea. The list comprises seven Arabidopsis HSP90 sequences (Zhang et al. 2013) used in this study. (XLS) (XLS 108 kb)
Conserved domains of HSP90 proteins in Olea europaea var. europaea. The major domains were identified using Pfam (https://pfam.xfam.org/). (XLS) (XLS 19 kb)
Gene ontology terms of OeHsp90 proteins, based on their 3D structures, using ProFunc server. (XLS 51 kb)
Proteins in Arabidopsis thaliana that match the OeHSP90 proteins in olive tree by STRING 11.0. (XLS) (XLS 21 kb)