Graphical abstract
Keywords: Recursive partitioning analysis, Cancer staging, Clinical predictive ability, Performance comparison, Web services
Abstract
Cancer staging provides a common language that is used to describe the severity of an individual's cancer, which plays a critical role in optimizing cancer treatment. Recursive partitioning analysis (RPA) is the most widely accepted method for cancer staging. Despite its widespread use, to date, only limited tools have been developed to implement the RPA algorithm for cancer staging. Moreover, most of the available tools can be accessed only from command lines and also lack visualization, making them difficult for clinical investigators without programing skills to use. Therefore, we developed a web server called autoRPA that is dedicated to supporting the construction of prognostic staging models and performance comparisons among different staging models. Based on the RPA algorithm and log-rank test statistics, autoRPA can establish a decision-making tree from survival data and provide clinicians an intuitive method to further prune the decision tree. Moreover, autoRPA can evaluate the contribution of each submitted covariate that is involved in the grouping process and help identify factors that significantly contribute to cancer staging. Four indicators, including hazard consistency, hazard discrimination, percentage of variation explained, and sample size balance, are introduced to validate the performance of the designed staging models. In addition, autoRPA can also be used to compare the performance of different prognostic staging models using a standard bootstrap evaluation method. The web server of autoRPA is freely available at http://rpa.renlab.org.
1. Introduction
Cancer staging is an important task in cancer treatment. Staging describes the severity of an individual’s cancer based on the magnitude of the primary tumor as well as whether the cancer has spread to nearby lymph nodes or other different parts of the body. Understanding the stage of a patient’s cancer is essential for many aspects. First, doctors will design treatment plans for individual patients based on their cancer stages and may also assess the prognosis using the cancer stage. For the research component, the cancer stage is usually used to assess treatment results among patient groups, to compare treatment outcomes among different centers and to plan future research studies.
Currently, the TNM staging system maintained by the American Joint Committee on Cancer (AJCC)/Union for International Cancer Control (UICC) is the most authoritative solution for cancer staging in the world. The TNM staging system categorizes cancer based on the extent of the tumor (T), the extent of spread to the lymph nodes (N), and the presence of metastasis (M). In many cancer types, TNM scores are combined to create overall staging groups from I to IV that stratify patients according to survival outcomes. However, due to the complexity of cancer, researchers have shown that the anatomical TNM stage does not always adequately predict survival for some cancer types [1], [2]. Therefore, including nonanatomical factors to further differentiate prognosis while maintaining anatomical stage grouping is necessary for improving the staging performance of cancers.
Currently, recursive partitioning analysis (RPA) is the most widely used approach to achieve this goal. RPA was first proposed as a tree-based regression model by Morgan and Sonquist in 1963 [3]. Later, in 1985, Gordon and Olshen extended the algorithm to adapt it to censored data by using log-rank statistics as splitting criteria [4]. RPA can easily combine prognostic factors, anatomical factors and even genetic characteristics in a decision tree, making it a most suitable technique for creating cancer staging models. Recently, the RPA method has been applied in multiple clinical studies and has assisted the refinement of many staging systems, such as breast cancer [5], [6], non-small cell lung cancer [6], [7], [8], oropharyngeal cancer [1], [9], [10], nasopharyngeal cancer [11], [12], and so on [13], [14], [15]. In contrast, with its widespread use, only limited tools have been developed to implement the RPA algorithm for cancer staging to date. Most clinical investigators use the R package rpart or the STREE or RECPAM program to build a staging model for cancer patients. However, these tools can only be accessed from command lines, and they lack the ability to visualize the analytical results, making them difficult to use by clinicians without programming skills. Moreover, the original tree constructed by the RPA algorithm often needs further refinements as it is applied in clinical treatment. Clinical investigators will usually prune the tree structure according to their clinical experiences and regroup tree leaves with relatively homogeneous survival performance metrics. As such, the above tools that do not interact will also greatly hinder the work efficiency of clinical investigators. Some researchers also apply the SPSS Modeler for cancer staging in their studies. Although a basic graphical user interface is available in SPSS Modeler, it still lacks certain interactivity for users to refine their model in real time. More importantly, all the above-mentioned tools do not provide a model comparison functionality, which further limits their application in the area of clinical research. In view of this, the development of an interactive tool that not only implements the RPA algorithm but also provides model comparison is still necessary in current clinical studies.
To address these challenges, we have developed a web server called autoRPA in this paper. Based on the RPA algorithm and log-rank test statistics, autoRPA will establish a decision-making tree from survival data and provide clinicians an intuitive way to further refine the tree. Using a permutation test, autoRPA can evaluate the contribution of each submitted clinical factor that is involved in the grouping process and help identify clinical factors that significantly contribute to cancer staging. Four indicators, including hazard consistency, hazard discrimination, percentage of variation explained, and balance, are introduced in autoRPA to validate the staging performance. In addition, autoRPA can also compare the performances of different prognostic staging models using a standard bootstrap evaluation method. To facilitate the use of autoRPA, we implemented a web server using Java and PHP and made it freely available at http://rpa.renlab.org.
2. Material and methods
2.1. Implementation of recursive partitioning analysis
The RPA algorithm consists of two major stages. First, it scans through all the available covariates and uses all the possible values to split the observations into two parts. The impurity of each part is then evaluated by the log-rank test statistic. The covariate that has the largest log-rank test statistic is preserved as the final splitting criterion. Second, the RPA algorithm fits a regression model in each node of the resulting partition and predicts the survival rate. This process is applied recursively until the regression tree has grown with sufficient depth. We provide an interactive GUI to further prune the regression tree. According to the survival rate of each node, users can easily regroup the nodes with relatively homogeneous survival performance and assign the final cancer stage for the inputted patients.
2.2. Evaluation of the RPA model
Based on the defined stages, the Kaplan-Meier curve for each stage is drawn, and the statistical significance is calculated using a log-rank test strategy. A receiver-operator characteristic (ROC) curve is also implemented in autoRPA using methods proposed by Patrick et al. [16]. To further evaluate the constructed staging model, hazard consistency, hazard discrimination, percentage of variation explained (PVE) and sample size balance are computed based on the method of Groome et al. [17].
2.3. Computation of the variable contribution for the RPA model
To investigate which covariate contributes most to the staging of cancer patients, we develop an algorithm to quantitatively compute such value. When considering an RPA tree, it is intuitively clear that for each patient, the tree will predict its survival through a path from the root of the tree to the leaf. This path consists of a series of decisions guarded by a particular covariate, each of which will contribute to the final predictions. When summing the survival rate change in each node and assigning it to the corresponding covariate, we can therefore compute the contribution of each covariate (see also the supplementary method for more details).
To determine whether the contribution of a given covariate is significant for survival prediction, we further introduced a permutation test in our algorithm [18]. To preserve the relationships between covariates, we randomly permute the last follow-up date and status of each patient. For each permutation, the contribution of each covariate is assessed with the above approach. After repeating the permutation n times, we construct a null distribution of covariate contributions. Given this distribution, the probability of observing a contribution value higher than x under the null hypothesis can be computed. This p-value is then used as an indicator to determine statistical significance.
2.4. Construction of the model comparison module in autoRPA
In most studies, comparison to a previous staging model is a necessary step for proving the validity of the RPA-adjusted model. To provide such functionality, we have developed a model comparison module in autoRPA. The bootstrap validation is applied in the comparison, and four criteria, such as hazard consistency, hazard discrimination, percentage of variation explained (PVE) and sample size balance, are calculated as the performance indicators. The hazard consistency measures the similarity of survival rate for each subgroup within each stage group. Lower scores indicate better consistency between subgroups that make up the groups. Hazard discrimination evaluates the differences in survival rate between different stage groups. Hazard discrimination is ranging from 0 to 1 with a higher score indicating better discrimination between survival curves. The PVE calculates the percentage of survival rate variation explained by the stage grouping. A higher score indicates better prediction power of the grouping scheme. Sample size balance assesses the differences in sample size across stage groups. A lower score of sample size balance indicates a better grouping scheme. To compare between models, the normalized score and normalized rank are also computed using the method of Groome et al. [17]. In addition, in autoRPA, the ROC curves of different staging models together with their pairwise p-values are also provided.
3. Results
3.1. A user-friendly web server for constructing the RPA staging model
We created a webserver called autoRPA to assist in the building of a cancer staging tree using the RPA method. A set of analytical tools and visualization tools were implemented. Detailed information about the autoRPA system, download instructions, additional videos, and a detailed user guide are available at http://rpa.renlab.org.
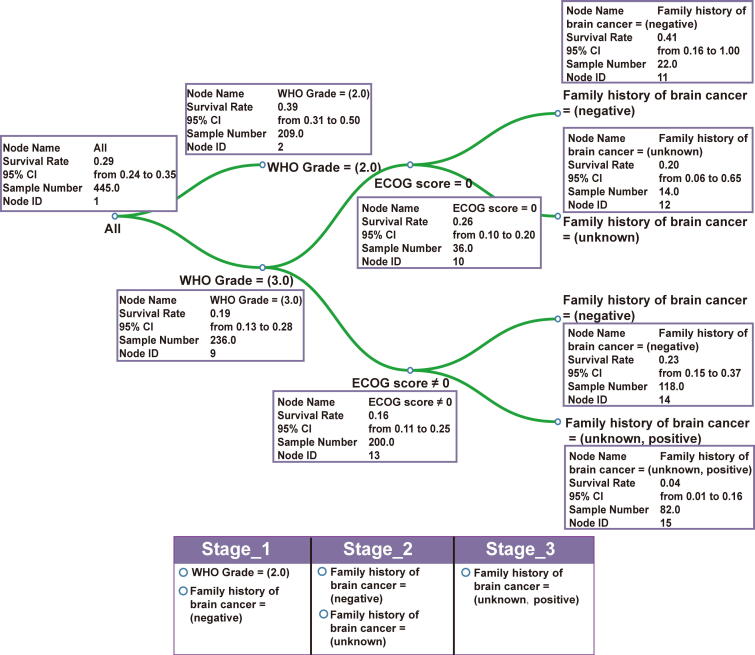
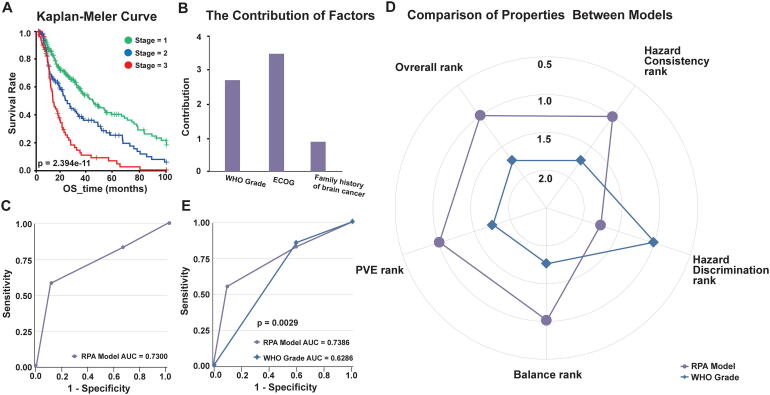
To further illustrate the main features of autoRPA, we used a follow-up data set of human low-grade gliomas (LGGs) from The Cancer Genome Atlas (TCGA) as a research case. From the clinical data set, we first extracted the World Health Organization (WHO) grading score of each patient as an anatomical factor. Additionally, the performance status score proposed by the Eastern Cooperative Oncology Group (ECOG score) and the family history of cancer from the TCGA database were included as clinical characteristics to further improve the prediction capability of the RPA staging model. To accurately represent outcomes in patients with low ECOG scores, we used the progress free survival other than the overall survival to construct RPA staging models. Fig. 1 and Fig. S1 shows the staging result of LGG. Using the d3.js library, we implement a visualization panel for the survival tree constructed by the RPA algorithm. In the tree model, we present the splitting criteria of each node on the tree branches. The survival rate together with its 95% confidence interval are also shown in an interactive box for each node. To allow manual refinement of the original tree structure, an interactive pruning method is provided in this panel. Below the visualization panel, a staging form is developed. By dragging tree leaves into the corresponding column, one can easily regroup the patients with a homogeneous survival rate and establish a staging strategy for the cancer under investigation. In this example, we regrouped the LGG patients into three stages based on the WHO grade and two clinical factors. Next, the Kaplan-Meier curve was plotted to evaluate the association between the derived stages and survival (Fig. 2A). Then, the contribution of each covariate was calculated, and the statistical significance was be evaluated (Fig. 2B). Furthermore, based on the user-defined RPA stage, a ROC curve is drawn in autoRPA to evaluate the prediction performance of survival rate (Fig. 2C).
Fig. 1.
The interface developed for RPA tree visualization. The RPA tree model of LGG patients was displayed. The tree structure is visualized using the d3.js library, and tree leaves can be manually regrouped in the interactive table.
Fig. 2.
The main features provided in autoRPA for assisting the building of a cancer staging model. (A) The K-M curves of our proposed LGG stages are plotted in autoRPA. (B) The covariate contributions of the RPA staging model constructed for LGG patients. (C) The ROC curve of our proposed LGG staging model. (D) The radar map showing the normalized rank of four performance indicators for the RPA model and the original WHO grade. (E) Comparison of the ROC curves between the proposed RPA model and the WHO grade. RPA, Recursive partitioning analysis. LGG, low-grade gliomas. K-M curves, Kaplan–Meier curves. ROC, receiver operating characteristic curve. WHO, World Health Organization.
3.2. An interactive module for comparing performance between different staging models
After building the RPA staging model, a major task was to compare the prediction performance with other existing models. In autoRPA, we have developed a web-based module for further model comparisons. This model will first calculate the hazard consistency, hazard discrimination, PVE and sample size balance for different staging models. Specifically, the hazard consistency evaluates the similarity of survival rates for subgroups defined by grouping factors within each staging group. A larger value of hazard consistency indicates a higher prediction consistency. Hazard discrimination is defined as the differences in survival rates across the staging groups and is used to assess the discriminatory power for different cancer stages. PVE is the percentage of overall survival variation explained by the stage groupings. A larger PVE indicates higher predictive power. Sample size balance measures the difference in sample sizes across stage groups. To facilitate the comparison between different models, the normalized score and rank are calculated, and the detailed values are listed in an interactive table in autoRPA (Table S1). Using the normalized rank, autoRPA plots a radar map for the four performance indicators. An overall rank is also calculated to report the overall performance. Compared with the WHO grading model for LGG, our RPA model performed superiorly in hazard consistency, PVE and sample size balance (Fig. 2D). Finally, the ROC curves of all the inputted models are plotted, and the p-values measuring the significant differences between different ROC curves are computed (Fig. 2E).
3.3. A case study on melanoma
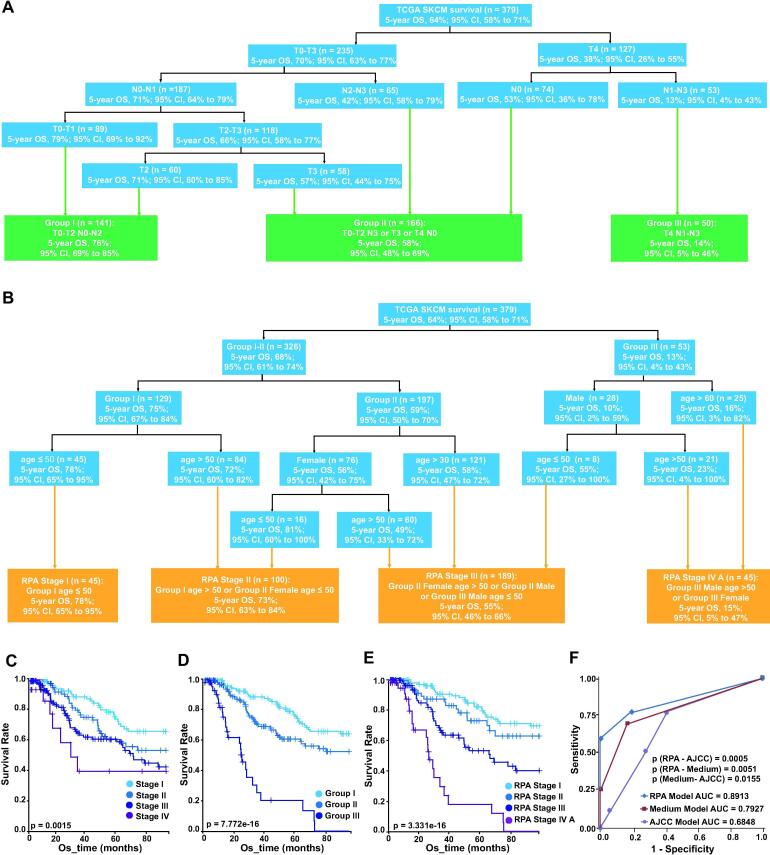
Next, we applied autoRPA to stage patients with melanoma. We collected a total of 379 patients from the Cancer Genome Atlas (TCGA) project. Using autoRPA, we first refined the stage strategy for melanoma using the clinical T/N categories (Fig. 3A). The number of patients in the derived group and the 5-year overall survival rate (5-year OS) and its 95% confidence interval are listed in each tree node. According to the 5-year OS, we classified patients into the following three RPA stage groups: Group I (T0-2 N0-1), Group II (T3N0-1 or T0-3 N2-3) and Group III (T4N1-3), with corresponding 5-year OS rates of 75%, 59%, 13%, respectively. In this case, patients with metastatic tumours (M1) are grouped into a separate stage and are not classified in this model.
Fig. 3.
A case study on melanoma. (A) The refined RPA stage of melanoma. The T/N scores are used to build the staging model. Patients are regrouped into three stages according to their survival. (B) The final proposed stage of melanoma. This stage model is built upon the refined RPA stage. The refined RPA stage together with age and gender are combined by autoRPA. (C) The K-M curve of the 8th edition AJCC/UICC staging for melanoma. (D) The K-M curve of the refined RPA staging model for melanoma. (E) The K-M curve of the final proposed stage for melanoma which combining age and gender in the staging model. (E) Comparison of the ROC curves among the final proposed stage, the refined RPA stage and the AJCC/UICC staging strategy. RPA, Recursive partitioning analysis. K-M curves, Kaplan–Meier curves. AJCC, the American Joint Committee on Cancer. UICC, Union for International Cancer Control. ROC, receiver operating characteristic curve.
In addition to the RPA stage groups, age and gender are also known as independent survival predictor for melanoma [19]. Therefore, we further combined them with the RPA stage groups to derive a more prognostic model. In another RPA analysis, we used the variables included RPA stage (I, II or III), age and gender (female or male) to construct the following four prognostic groups (Fig. 3B): Stage I (Group I, age ≤ 50), Stage II (Group I, age > 50 or Group II, female age ≤ 50), Stage III (Group II, female, age > 50 or Group II, male or Group III, male, age ≤ 50) and Stage IVA (Group III, male, age > 50 or Group III, female), with 5-year OS rate of 78%, 73%, 55%, 15%, respectively. Metastatic (M1) disease would be classified as prognostic group IVB. In this analysis, we observed that the prognosis of melanoma decreased with advancing age. An age of 50 will be considered as a distinct cutoff in our final staging model. What’s more, this decrease of prognosis seemed more pronounced among females than males.
To further demonstrate the superiority of our constructed RPA model, we compared it against the eight edition AJCC/UICC stage groups using the comparison module in autoRPA. The K-M curves of both the AJCC/UICC stages (Fig. 3C), the refined RPA stage (Fig. 3D) and the final combined stage (Fig. 3E) were plotted. Although both staging models are significantly associated with overall survival, the refined RPA stage and the final combined stage provides a more refined classification. The bootstrap validation also confirms this point. Both the two RPA-derived stages outperform the AJCC/UICC stage (Table S2). Furthermore, the AUC of the two RPA-derived stages are significantly larger than that of the AJCC/UICC stage, suggesting a better predictive capability of the RPA model in our case study (Fig. 3F).
4. Discussion
autoRPA is a user-friendly web service that enables the user to construct and visualize a cancer staging model from follow-up data. With the interactive characteristics of web-based applications, autoRPA allows the user to participate in building the staging model. By manually pruning the decision-making tree and regrouping patients with homogeneous survival performance, users may construct a more refined staging model according to their clinical experiences. This interactive feature is a unique characteristic provided in autoRPA compared to other state-of-art tools that may provide curative functionality for clinical investigators. We have also illustrated other important characteristics of autoRPA using example data of lower-grade glioma and skin cutaneous melanoma from the TCGA projects. In these examples, we show that the standard performance indicators implemented in autoRPA can greatly assist the user in evaluating the proposed staging model and comparing it with other existing strategies. Beyond that, the rich set of visualization tools available in autoRPA can also help the user interpret staging results in a more intuitive way.
It is worth mentioning that the RPA algorithm is essentially a variant of decision tree model. In the decision tree algorithm, a key parameter for controlling the prediction performance is the tree depth. Tree depth is a measure of how many splits a tree can make before coming to a prediction. A larger tree depth can split the tree as pure as possible, however, it may also leads to overfitting on the training dataset by constructing a very complicated tree with many nodes. Therefore, to reduce overfitting, an optimal tree depth should be found to balance the self-consistency and generalization. Considering that the analysis of clinical data by RPA method is still largely empirical, a practical way to determine optimal tree depth is to use independent dataset. By varying the tree depth, we can evaluate the performance indicator under different tree depth threshold, and select the best performance to determine this parameter. In general, the tree depth should not larger than the number of inputted factors. Alternatively, setting the tree depth equal to the number of inputted factors may also be acceptable.
In the future, we will implement more types of recursive partitioning algorithms in autoRPA web servers, including conditional inference trees (CTrees) and model-based recursive partitioning (MOB). More splitting criteria, such as those employing a Kaplan-Meier survival function or likelihood ratio statistic, will also be introduced in the future version. As a major vulnerability in RPA method, the overfitting of training dataset is an important issue that need to be addressed. Therefore, a cross-validation module will be developed in the future version. Applying cross-validation, we expected that users may be able to evaluate the generalization ability of RPA staging model. Besides, by constructing an ensemble of RPA models, the Random Forest (RF) algorithm is believed to be more stable than a single RPA tree, thus can help to efficiently reduce the overfitting of training data. In near future, a RF algorithm will be implemented in autoRPA for constructing of more powerful staging model in multiple cancers. We also plan to develop a module that implements the latest AJCC/UICC staging strategy to help users assign the TNM score or overall stage of collected patients.
In conclusion, autoRPA is a useful tool for clinical investigators. In autoRPA, we provide complete functionalities for cancer staging studies, covering model building, model evaluation, model comparison and result visualization. Using autoRPA, one can easily build a staging model for specific cancer patients and perform manual refinement of the proposed model. The visualization module of autoRPA can further help users generate publication-quality figures. We expect that autoRPA can serve as a gateway to the building of cancer stages and support the decisions of personalized therapeutic strategies for multiple cancers.
5. Availability
autoRPA is an online websever available in http://rpa.renlab.org.
Funding
This work was supported by the National Natural Science Foundation of China [91753137, 31771462, 81772614, U1611261, 31801105]; Program for Guangdong Introducing Innovative and Entrepreneurial Teams [2017ZT07S096]; Guangdong Basic and Applied Basic Research Foundation [2018A030313323, 2020A1515011219]; Fundamental Research Funds for the Central Universities [SYSU: 19ykpy184].
CRediT authorship contribution statement
Yubin Xie: Conceptualization, Methodology, Funding acquisition, Writing - original draft. Xiaotong Luo: Formal analysis, Investigation, Data curation, Software, Visualization. Huiqin Li: Software, Visualization. Qingxian Xu: Software, Visualization. Zhihao He: Software. Qi Zhao: Conceptualization. Zhixiang Zuo: Conceptualization, Funding acquisition. Jian Ren: Conceptualization, Supervision, Funding acquisition, Methodology, Resources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.10.038.
Contributor Information
Qi Zhao, Email: zhaoqi@sysucc.org.cn.
Zhixiang Zuo, Email: zuozhx@sysucc.org.cn.
Jian Ren, Email: renjian.sysu@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huang S.H., Xu W., Waldron J., Siu L., Shen X., Tong L.i., Ringash J., Bayley A., Kim J., Hope A. Refining American Joint Committee on Cancer/Union for International Cancer Control TNM stage and prognostic groups for human papillomavirus–related oropharyngeal carcinomas. JCO. 2015;33(8):836–845. doi: 10.1200/JCO.2014.58.6412. [DOI] [PubMed] [Google Scholar]
- 2.Ward M.J., Mellows T., Harris S., Webb A., Patel N.N., Cox H.J., Piper K., Ottensmeier C.H., Thomas G.J., King E.V. Staging and treatment of oropharyngeal cancer in the human papillomavirus era: Staging and Treatment of HPV-related oropharyngeal cancer. Head Neck. 2015;37(7):1002–1013. doi: 10.1002/hed.23697. [DOI] [PubMed] [Google Scholar]
- 3.Morgan J.N., Sonquist J.A. Problems in the analysis of survey data, and a proposal. J Am Stat Assoc. 1963;58(302):415–434. doi: 10.1080/01621459.1963.10500855. [DOI] [Google Scholar]
- 4.Gordon L., Olshen R.A. Tree-structured survival analysis. Cancer Treat Rep. 1985;69:1065–1069. [PubMed] [Google Scholar]
- 5.Subbiah I.M., Lei X., Weinberg J.S., Sulman E.P., Chavez-MacGregor M., Tripathy D. Validation and development of a modified breast graded prognostic assessment as a tool for survival in patients with breast cancer and brain metastases. JCO. 2015;33(20):2239–2245. doi: 10.1200/JCO.2014.58.8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chansky K., Sculier J.-P., Crowley J.J., Giroux D., Van Meerbeeck J., Goldstraw P. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thoracic Oncol. 2009;4(7):792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 7.Ashworth A.B., Senan S., Palma D.A., Riquet M., Ahn Y.C., Ricardi U. An individual patient data metaanalysis of outcomes and prognostic factors after treatment of oligometastatic non–small-cell lung cancer. Clin Lung Cancer. 2014;15(5):346–355. doi: 10.1016/j.cllc.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Komaki R., Scott C.B., Byhardt R., Emami B., Asbell S.O., Russell A.H., Roach M., Parliament M.B., Gaspar L.E. Failure patterns by prognostic group determined by recursive partitioning analysis (RPA) of 1547 patients on four radiation therapy oncology group (RTOG) studies in inoperable nonsmall-cell lung cancer (NSCLC) Int J Radiat Oncol Biol Phys. 1998;42(2):263–267. doi: 10.1016/S0360-3016(98)00213-2. [DOI] [PubMed] [Google Scholar]
- 9.O'Sullivan B., Huang S.H., Su J., Garden A.S., Sturgis E.M., Dahlstrom K. Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): a multicentre cohort study. Lancet Oncol. 2016;17:440–451. doi: 10.1016/S1470-2045(15)00560-4. [DOI] [PubMed] [Google Scholar]
- 10.Keane F.K., Chen Y.-H., Tishler R.B., Schoenfeld J.D., Haddad R.I., Goguen L.A. Population-based validation of the recursive partitioning analysis-based staging system for oropharyngeal cancer: SEER validation of the recursive partitioning analysis-based staging system. Head Neck. 2016;38(10):1530–1538. doi: 10.1002/hed.24470. [DOI] [PubMed] [Google Scholar]
- 11.Guo R., Tang L.-L., Mao Y.-P., Du X.-J., Chen L., Zhang Z.-C. Proposed modifications and incorporation of plasma Epstein-Barr virus DNA improve the TNM staging system for Epstein-Barr virus-related nasopharyngeal carcinoma: Refined TNM Staging for EBV-Related NPC. Cancer. 2019;125(1):79–89. doi: 10.1002/cncr.31741. [DOI] [PubMed] [Google Scholar]
- 12.Lee V.-F., Kwong D.-W., Leung T.-W., Choi C.-W., O'Sullivan B., Lam K.-O. The addition of pretreatment plasma Epstein–Barr virus DNA into the eighth edition of nasopharyngeal cancer TNM stage classification. Int. J. Cancer. 2019;144(7):1713–1722. doi: 10.1002/ijc.31856. [DOI] [PubMed] [Google Scholar]
- 13.Yuan S.Q., Chen Y.T., Huang Z.P. Equipping the 8th Edition American Joint Committee on Cancer Staging for Gastric Cancer with the 15-Node Minimum: a Population-Based Study Using Recursive Partitioning Analysis. J Gastrointest Surgery. 2017;21:1591–1598. doi: 10.1007/s11605-017-3504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y.-T., Huang Z.-P., Zhou Z.-W., He M.-M. Equipping the American Joint Committee on Cancer staging for resectable pancreatic ductal adenocarcinoma with tumor grade: a recursive partitioning analysis. Med Oncol. 2016;33(11) doi: 10.1007/s12032-016-0839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park S.Y., Cho Y.Y., Kim H.I., Choe J.-H., Kim J.-H., Kim J.S. Clinical validation of the prognostic stage groups of the eighth-edition TNM staging for medullary thyroid carcinoma. J Clin Endocrinol Metab. 2018;103:4609–4616. doi: 10.1210/jc.2018-01386. [DOI] [PubMed] [Google Scholar]
- 16.Heagerty P.J., Lumley T., Pepe M.S. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 17.Groome P.A., Schulze K., Boysen M., Hall S.F., Mackillop W.J. A comparison of published head and neck stage groupings in carcinomas of the oral cavity. Head Neck. 2001;23(8):613–624. doi: 10.1002/hed.1087. [DOI] [PubMed] [Google Scholar]
- 18.Altmann A., Tolosi L., Sander O., Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics (Oxford, England) 2010;26:1340–1347. doi: 10.1093/bioinformatics/btq134. [DOI] [PubMed] [Google Scholar]
- 19.Enninga E.A.L., Moser J.C., Weaver A.L., Markovic S.N., Brewer J.D., Leontovich A.A., Hieken T.J., Shuster L., Kottschade L.A., Olariu A., Mansfield A.S., Dronca R.S. Survival of cutaneous melanoma based on sex, age, and stage in the United States, 1992-2011. Cancer Med. 2017;6(10):2203–2212. doi: 10.1002/cam4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.