Abstract
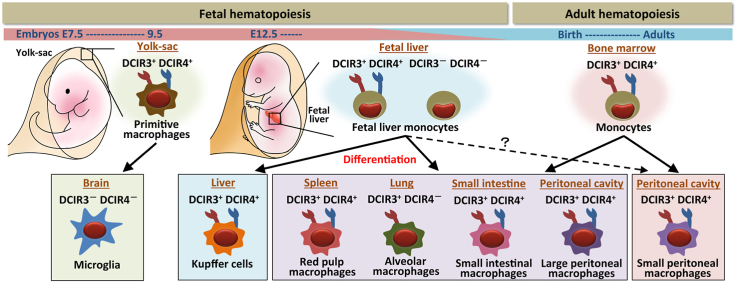
Dendritic cell inhibitory receptor 3 (DCIR3, Clec4a3) and dendritic cell inhibitory receptor 4 (DCIR4, Clec4a1) are C-type lectin receptors that belong to mouse dendritic cell immunoreceptor (DCIR) family. We recently showed that DCIR3 and DCIR4 are co-expressed on inflammatory and patrolling monocytes. In this study, we investigated the expression of DCIR3 and DCIR4 on tissue-resident macrophages. We found that spleen red pulp macrophages, liver Kupffer cells, large and small peritoneal macrophages and small intestinal macrophages expressed both DCIR3 and DCIR4. By contrast, lung alveolar macrophages expressed DCIR3 but not DCIR4 and brain microglia expressed neither DCIR3 nor DCIR4. Considerable part of tissue-resident macrophages are derived from embryonic precursors. We, therefore, examined the expression of DCIR3 and DCIR4 on the embryonic precursors. Yolk-sac macrophages from embryonic day (E) 8.5 embryos expressed both DCIR3 and DCIR4, while DCIR3 and DCIR4 were expressed on subpopulations of fetal liver monocytes from E14.5 embryos. Our results, together with previous data, indicate that the expression of DCIR3 and DCIR4 is widely shared by mononuclear phagocytes, including monocytes and macrophages, and that the expression of DCIR3 and DCIR4 on the embryonic precursors are not always retained by their progenies, suggesting that expression of DCIR3 and DCIR4 on tissue-resident macrophages might be regulated by environment of the tissues where the embryonic precursors differentiate into macrophages.
Keywords: Monocytes, Macrophages, C-type lectin receptors, Mononuclear phagocytes, Embryonic precursors
Graphical abstract
Highlights
-
•
The majority of tissue-resident macrophages express DCIR3 and DCIR4.
-
•
Brain microglia lack the expression of DCIR3 and DCIR4.
-
•
Lung alveolar macrophages express DCIR3 but not DCIR4.
-
•
Macrophages do not always retain DCIR3/DCIR4 expression on their precursors.
1. Introduction
Dendritic cell inhibitory receptor 3 (DCIR31, Clec4a3) and dendritic cell inhibitory receptor 4 (DCIR4, Clec4a1) are C-type lectin receptors that belong to mouse dendritic cell immunoreceptor (DCIR) family [1]. Mouse DCIR family consists of four inhibitory receptors, DCIR1 ~ 4, and two activating receptors, DCAR1 and DCAR2 [1]. We recently found that DCIR3 and DCIR4 are co-expressed on Ly-6C+ inflammatory monocytes and Ly-6C- patrolling monocytes [2,3]. Monocytes have a capacity to differentiate into dendritic cells (DCs) and macrophages in vitro and in vivo. In vitro culture of mouse Ly-6C+ monocytes in the presence of GM-CSF and IL-4 induces differentiation into DCs and macrophages, while that in the presence of M-CSF induces differentiation into macrophages [4]. When Ly-6C+ DCIR4+ monocytes differentiate into DCs, DCIR4 expression is diminished, whereas when they differentiate into macrophages, DCIR4 expression is retained [2]. These observations raise the possibility that DCIR3 and DCIR4 might be co-expressed on macrophages in vivo.
The majority of tissues in the body contain tissue-resident macrophages, which exert tissue-specific functions [5]. Most tissue-resident macrophages develop from embryonic precursors in the fetal stage and considerable part of them are maintained by self-renewal in adults [[6], [7], [8]]. Recently, Liu et al. showed that lung alveolar macrophages (AMs), spleen red pulp macrophages, large peritoneal macrophages and small intestinal macrophages are replaced by the macrophages derived from adult bone marrow monocytes to some extent after birth, resulting in a mixture of embryo- and adult-derived macrophages in adult mice, while brain microglia and liver Kupffer cells maintain their population by self-renewal after differentiation from embryonic precursors and are hardly replaced by the macrophages derived from adult bone marrow monocytes [9].
In this study, we investigated the expression of DCIR3 and DCIR4 on various tissue-resident macrophages and their embryonic precursors. We demonstrate that DCIR3 and DCIR4 are co-expressed by majority of tissue-resident macrophages except brain microglia and lung AMs. We also show that the expression of DCIR3 and DCIR4 on the embryonic precursors is not always retained by their progenies. Our data suggest that the expression of DCIR3 and DCIR4 on tissue-resident macrophages is independent of DCIR3 and DCIR4 expression on their precursors, but might be influenced by the environments of the tissues where precursors differentiate into macrophages.
2. Materials and methods
2.1. Animals
C57BL/6J mice were obtained from Nippon SLC (Shizuoka, Japan) and housed under specific pathogen-free conditions. B6. SJL-Ptprca/BoyYuoRbrc mice obtained from Bioresource Research Center of RIKEN (Ibaraki, Japan) were bred and housed under specific pathogen-free conditions. All the animal experiments were conducted in accordance with the University of Tokyo's rules regarding animal experimentation and the University's animal-experimentation manual.
2.2. Cells
Mouse fibroblast cell line L929 was obtained from Cell Resource Center for Biomedical Research, Tohoku University (Miyagi, Japan). L929 cells were maintained in RPMI1640 medium (Sigma-Aldrich, St Louis, MO) containing 10% heat-inactivated fetal calf serum (FCS, Sigma-Aldrich), 50 μM 2-mercaptoethanol (Sigma-Aldrich), 100 U/mL of penicillin G (Sigma-Aldrich) and 100 μg/mL of streptomycin sulfate (Sigma-Aldrich). The culture supernatants from L929 cells were prepared by culturing 80–90% confluent L929 cells in the medium described above for 2 or 3 days. The collected culture supernatants were filtered through a 0.22 μm filter (Merck Millipore Ltd., Co. Cork, IRL) and used for preparation of bone marrow-derived macrophages (BMDMs).
2.3. Preparation of BMDMs
BMDMs were prepared as described by Zhang et al. [10]. In brief, bone marrow (BM) cells from C57BL/6J mice were cultured in the RPMI1640-based medium as described above in the presence of 20% culture supernatant from L929 fibroblasts. The medium was changed every 2 or 3 days and the adherent cells obtained after 7 days of culture were used for flow cytometry analysis.
2.4. Preparation of single cell suspensions of immune cells from various organs and mouse embryos
8–10 weeks-old female C57BL6/J mice were used. To obtain immune cells from small intestine, small intestines removed of Peyer's patches were longitudinally opened, cut into 10 mm pieces and incubated in PBS containing 1.3 mM EDTA at 37 °C to remove epithelial cells, then digested with collagenase in the collagenase solution (RPMI1640 containing 100 U/mL of type IV collagenase (Sigma-Aldrich), 40 μg/mL of DNaseI (Sigma-Aldrich), 10% heat-inactivated FCS, 50 μM 2-mercaptoethanol, 100 U/mL of penicillin G and 100 μg/mL of streptomycin sulfate) for 1 h at 37 °C. Single cell suspension from the small intestines were prepared by passing through cell strainers. The cells were layered on a 30/50% Percoll (GE Healthcare Japan Corporation, Tokyo, Japan) density-gradient and centrifuged for 20 min at 600×g. The cells enriched in the 30/50% interface were collected.
Lungs, brains and livers from mice were minced and digested with collagenase as described above. Single cell suspensions from lungs, brains and livers were prepared by passing through cell strainers, followed by treatment with red blood cell lysis solution. The red blood cells-depleted lung cells were used for flow cytometry analysis.
The brain cells were layered on a 30/37/70% Percoll density-gradient and centrifuged for 40 min at 300×g. The cells enriched in the 37/70% interface, containing microglia, were collected and analyzed.
The red blood cells-depleted liver cells were suspended in 33% Percoll and centrifuged for 30 min at 800×g. The precipitated cells containing Kupffer cells were collected and analyzed.
Peritoneal exudate cells were collected by injecting PBS into the peritoneal cavity of mice and removing the fluid from the peritoneal cavity.
To obtain mouse embryos, C57BL/6J female mice (CD45.2) were crossed with B6. SJL-Ptprca/BoyYuoRbrc male mice (CD45.1). Yolk-sac was obtained from fetuses at 8.5 days after fertilization (E8.5) and fetal livers were obtained from fetuses at 14.5 days after fertilization (E14.5). Single cell suspensions from yolk-sac and fetal livers were prepared by digesting with collagenase as described above and passing through cell strainers followed by treatment with red blood cell lysis solution.
All the cells were suspended in PBS containing 1 mM EDTA, 0.1% BSA (Wako, Osaka, Japan) and 0.1% NaN3.
2.5. Antibodies and flow cytometry
FITC-conjugated mAbs against mouse F4/80 (BM8), CD11c (N418) and Ly6C (HK1.4), and PE-conjugated mAbs against mouse F4/80 (BM8), CD11b (M1/70) and CD45.1 (A20), and APC-conjugated mAbs against mouse F4/80 (BM8) and CD64 (X54-5/7.1) were obtained from BioLegend (San Diego, CA). PE-conjugated mAb against Siglec-F (ES22-10D8) and MerTK (REA477) were from Milteny-Biotec (Bergisch Gladbach, Germany). mAbs against mouse Fcγ receptors (2.4G2) and CD11b (M1/70) were purified from supernatants of hybridomas obtained from ATCC (Manassas, VA) as described [2]. The purified anti-CD11b mAbs were then labeled with FITC (Sigma-Aldrich) or DyLight 650 NHS-ester (Thermo-Fischer Scientific K·K., Tokyo, Japan). Biotinylated anti-DCIR3 (7E7F2) and anti-DCIR4 mAbs (MH7E7) were described previously [2,3].
For staining of mouse immune cells, PBS containing 1 mM EDTA, 0.1% BSA and 0.1% NaN3 was used. The single cell suspensions prepared from various organs were incubated with anti-mouse Fcγ receptors mAb (2.4G2) before the addition of mAbs. The binding of biotinylated anti-DCIR3 (7E7F2) and anti-DCIR4 (MH7E7) mAbs was detected with APC-conjugated streptavidin (BioLegend) or PE-conjugated streptavidin (BioLegend). Propidium iodide (Sigma-Aldrich) was added at the final concentration of 1 μg/mL immediately before flow cytometry acquisition to exclude dead cells from analysis. The data were acquired with a FACSCalibur system (BD Biosciences, San Jose, CA) and were analyzed with FlowJo software (TreeStar, San Carlos, CA). Background staining was estimated using fluorochrome-conjugated or biotinylated isotype control mAbs: rat IgG2a (anti-human AICL, 3G72) or rat IgG1 control mAbs (anti-human KLRF1, H206), both of which were prepared in our laboratory [11], rat IgG1 (RTK2071), mouse IgG2a (eBM2a) or rat IgG2c (RTK4174) control mAbs obtained from BioLegend or REA control (REA293) obtained from Milteny-Biotec.
2.6. Microarray data analysis
To examine mRNA expression of DCIR3 and DCIR4 in embryonic precursors, BM monocytes and alveolar macrophages, a microarray data set normalized by quantile normalization (accession number: GSE66970) and a log2 converted microarray data set normalized by quantile normalization (accession number: GSE76999) were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/gds). The data sets of GSE66970 were converted to log10 values. Heatmaps of the indicated mRNA expression were prepared by Multi Experiment Viewer (MeV) version 4.8.1 (TM4 Software Suite).
3. Results
3.1. Expression of DCIR3 and DCIR4 on bone marrow-derived macrophages (BMDMs)
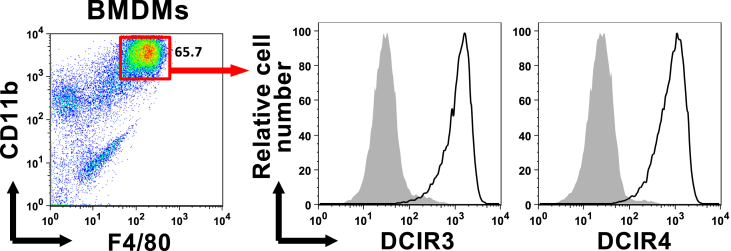
We recently showed that macrophages induced from bone marrow (BM) Ly-6C+ monocytes express DCIR4 [2]. To further extend this observation, we examined the expression of DCIR3 and DCIR4 on BMDMs. The BMDMs expressed both DCIR3 and DCIR4 (Fig. 1), suggesting the possibility that DCIR3 and DCIR4 might be expressed widely among macrophage populations, including tissue-resident macrophages.
Fig. 1.
Expression of DCIR3 and DCIR4 on bone marrow-derived macrophages (BMDM).
BMDMs were stained with the indicated mAbs and analyzed by flow cytometry. Expression of DCIR3 or DCIR4 (solid line histograms) on CD11b+F4/80+ BMDMs are shown. Shaded histograms are staining with isotype control mAbs. Data shown are one of three independent experiments with similar results.
3.2. Expression of DCIR3 and DCIR4 on various tissue-resident macrophages
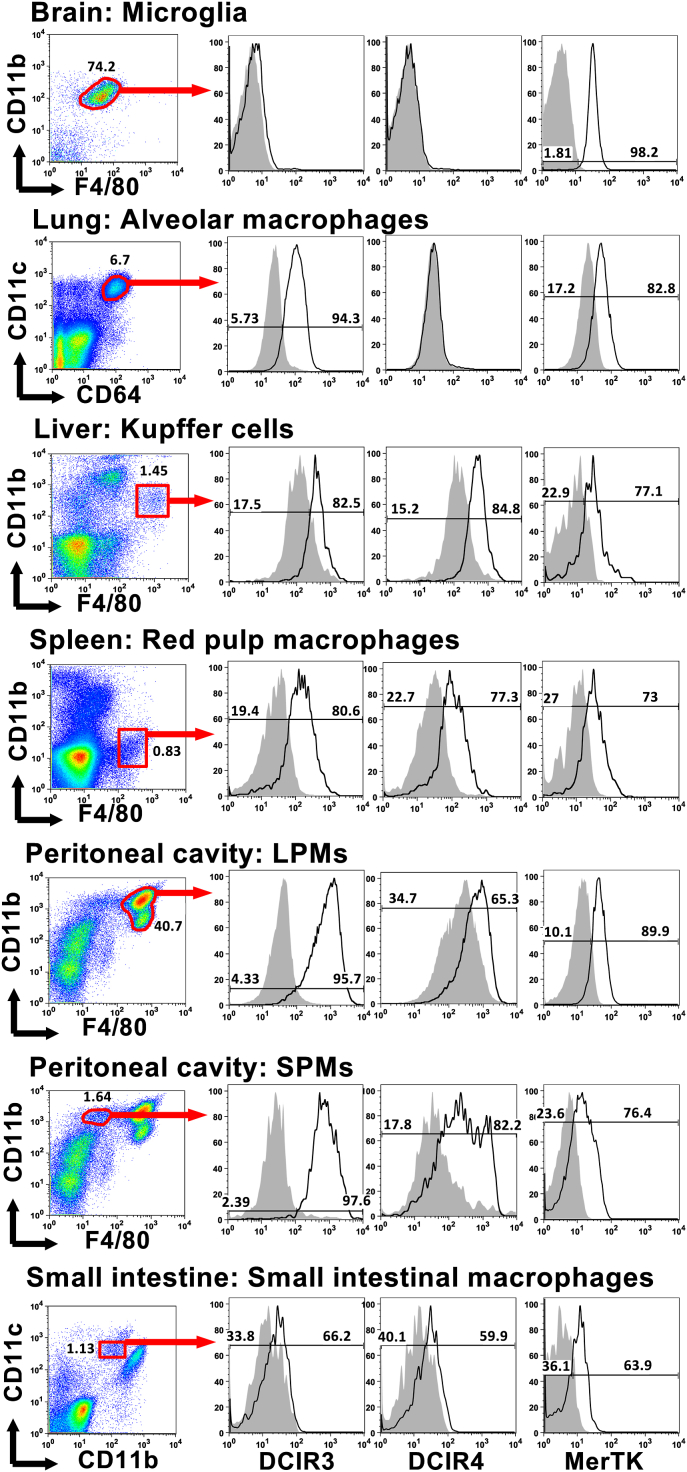
To investigate the expression of DCIR3 and DCIR4 on tissue-resident macrophages, we prepared immune cells from various organs, including brain, lung, liver, spleen, peritoneal cavity and small intestine. According to the literatures, brain microglia are defined as CD11b+F4/80Lo cells [12], lung alveolar macrophages (AMs) are defined as CD11cHiCD64+ cells [13,14] and spleen red pulp macrophages, liver Kupffer cells are defined as F4/80+CD11b+ cells [15]. Macrophages in peritoneal cavity are divided into two subsets, large peritoneal macrophages (LPMs) and small peritoneal macrophages (SPMs), which are defined as F4/80HiCD11b+ and F4/80LoCD11b+ cells, respectively [16]. Small intestinal lamina propria macrophages are defined as F4/80HiCD11bInt cells [17]. We stained the immune cells obtained from the organs with mAbs against the indicated markers and anti-MerTK, anti-DCIR3 or anti-DCIR4 mAbs (Fig. 2). All the tissue-resident macrophages defined as above expressed MerTK, a marker of macrophages [14], indicating that they were bona fide macrophages. Red pulp macrophages, Kupffer cells, LPMs and SPMs expressed both DCIR3 and DCIR4. Small intestinal macrophages also expressed both DCIR3 and DCIR4, although their expression was low. By contrast, AMs expressed DCIR3 but not DCIR4 and microglia expressed neither DCIR3 nor DCIR4. These data indicate that DCIR3 and DCIR4 are widely expressed by tissue-resident macrophages, except brain microglia and lung AMs, in both of which expression of either one or both of DCIR3 and DCIR4 were absent.
Fig. 2.
Expression of DCIR3 and DCIR4 on various populations of tissue-resident macrophages. Immune cells prepared from various organs were stained with the indicated mAbs and analyzed by flow cytometry. Expression of DCIR3, DCIR4 or MerTK (solid line histograms) on each population of the tissue-resident macrophages are shown. Shaded histograms are staining with isotype control mAbs. Data shown are one of three independent experiments with similar results.
3.3. Expression of DCIR3 and DCIR4 on embryonic precursors of tissue-resident macrophages
Since microglia lacked the expression of DCIR3 and DCIR4 and AMs lacked that of DCIR4 (Fig. 2), we investigated the expression of DCIR3 and DCIR4 on their precursors.
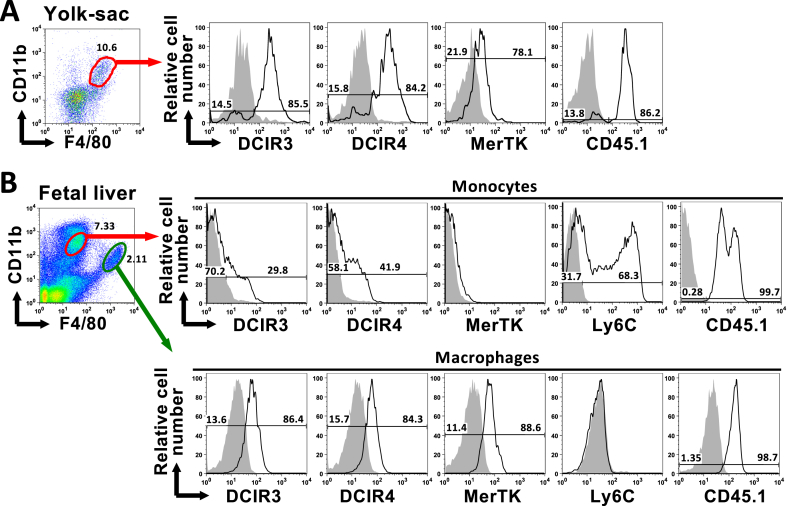
In mice, microglia are derived from E7.5 ~ E9.5 yolk-sac macrophages (YS-MFs) [8,18,19]. To investigate the expression of DCIR3 and DCIR4 on YS-MFs, yolk-sac cells from E8.5 embryos were analyzed by flow cytometry (Fig. 3A). Despite the result that microglia expressed neither DCIR3 nor DCIR4 (Fig. 2), their precursor, CD11b+F4/80+ YS-MFs, expressed both DCIR3 and DCIR4.
Fig. 3.
Expression of DCIR3 and DCIR4 on embryonic precursors of tissue-resident macrophages. C57BL6/J female mice (CD45.2) were crossed with B6. SJL-Ptprca/BoyYuoRbrc male mice (CD45.1) to obtain fetuses. Single cell suspensions prepared from yolk-sac from E8.5 embryos (A) or fetal livers from E14.5 embryos (B) were stained with the indicated mAbs and analyzed by flow cytometry. Expressions of DCIR3, DCIR4, MerTK or CD45.1 on CD11b+F4/80+ YS-MFs, CD11bHiF4/80Lo FL-MOs and CD11bLoF4/80Hi fetal liver macrophages are shown. Expressions of Ly-6C on FL-MOs and fetal liver macrophages are also shown. Shaded histograms are staining with isotype control mAbs. Data shown are one of three independent experiments with similar results.
The fetal liver begins to serve as the major hematopoietic organ around E11.5 and generates all hematopoietic cells [8,20]. CD11bHiF4/80Lo cells generally called fetal liver monocytes (FL-MOs) and CD11bLoF4/80HiLy6C- cells defined as early tissue macrophages appear in the fetal liver after E12.5 [20]. FL-MOs are the major embryonic precursors of most tissue-resident macrophages except microglia [20]. To investigate the expression of DCIR3 and DCIR4 on FL-MOs, fetal liver cells from E14.5 embryos were analyzed by flow cytometry (Fig. 3B). Consistent with the previous report, the heterogeneous expression of Ly6C and the lack of the MerTK expression on CD11bHiF4/80Lo cells and the MerTK expression on CD11bLoF4/80Hi cells enabled us to distinguish FL-MOs from fetal liver macrophages. The CD45.1 expression on CD11bHiF4/80Lo and CD11bLoF4/80Hi cells indicate the fetal origin of these populations. The CD11bLoF4/80Hi fetal liver macrophages expressed both DCIR3 and DCIR4, while CD11bHiF4/80Lo FL-MOs were the mixture of DCIR3+ and DCIR3- populations and, similarly, the mixture of DCIR4+ and DCIR4- populations.
These results indicate that DCIR3 and DCIR4 are expressed on YS-MFs, which are the precursors of microglia, and on subpopulations of FL-MOs, which are the precursors of most tissue-resident macrophages.
3.4. DCIR3 and DCIR4 mRNA were expressed in FL-MOs and AMs derived from YS-MFs, FL-MOs and adult BM monocytes
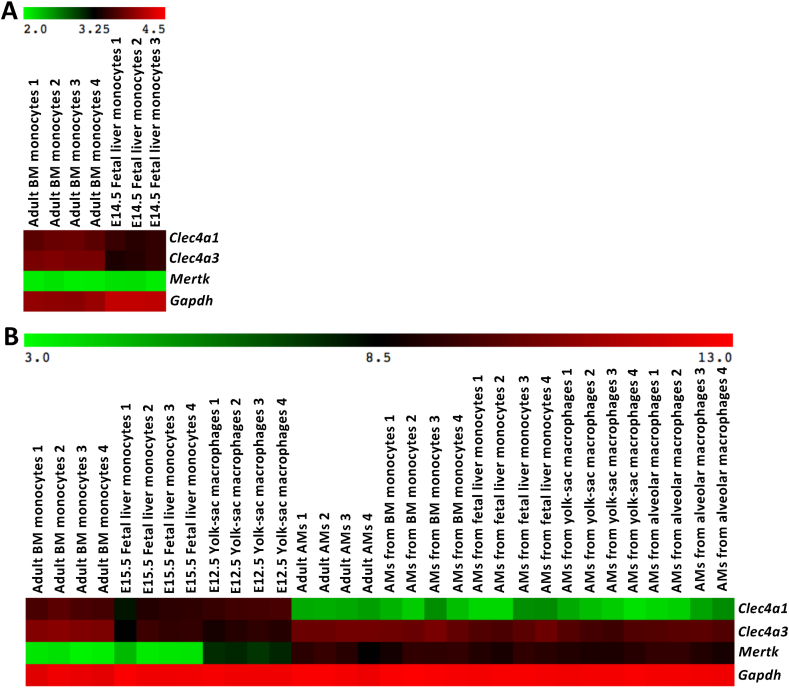
Given that DCIR3 and DCIR4 were expressed on subpopulations of FL-MOs, we examined the expression of DCIR3 and DCIR4 at mRNA level in FL-MOs. G. Hoeffel et al. performed the microarray analysis of E14.5 Ly6C+ FL-MOs and adult Ly6C+ BM monocytes (BM-MOs) [20]. We obtained these data from the Gene Expression Omnibus database and analyzed the mRNA expression of DCIR3 (Clec4a3) and DCIR4 (Clec4a1). While Mertk, a macrophage marker, was minimally expressed in either BM-MOs or FL-MOs, Clec4a3 and Clec4a1 were expressed in adult BM-MOs and also in FL-MOs at levels lower than those in BM-MOs (Clec4a3: p < 0.005, Clec4a1: p < 0.001, Welch's t-test) (Fig. 4A), consistent with the result that DCIR3 and DCIR4 proteins were expressed on subpopulations of FL-MOs (Fig. 3B).
Fig. 4.
mRNA expression of DCIR3 and DCIR4 in adult monocytes, FL-MOs and alveolar macrophages (AMs). A set of microarray data (accession number: GSE66970) of E14.5 Ly6CHi FL-MOs and adult Ly6CHi BM monocytes (BM-MOs), which were normalized by quantile normalization (A) and a set of log2 converted microarray data (accession number: GSE76999) of E15.5 Ly6CHi FL-MOs, E12.5 YS-MFs, adult Ly6CHi BM-MOs, adult alveolar macrophages (AMs) and AMs differentiated from FL-MOs, YS-MFs, adult BM-MOs or adult lung AMs (B) were obtained from the GEO database and analyzed. The data of GSE66970 were converted to log10 values.
L. van de Laar et al. showed that not only FL-MOs, which are the embryonic precursors for AMs in wild type mice, but also YS-MFs and adult BM-MOs have capacity to differentiate into AMs when they are introduced into the empty AM niche of neonatal Csf2rb−/- mice [21]. To get insight into the regulation of the DCIR3/DCIR4 expression during the differentiation of the precursors into AMs, we examined the expression of the Clec4a3 and Clec4a1 genes in AMs developed from YS-MFs, FL-MOs and adult BM-MOs by analyzing a microarray data set provided by L. van de Laar et al. (Fig. 4B). Importantly, Clec4a3 was expressed in the AMs developed from any of the three precursors, while Clec4a1 was negligibly expressed in the AMs. The expression profile of Clec4a3 and Clec4a1 genes in the AMs differentiated from YS-MFs, FL-MOs and adult BM-MOs were quite similar to that of DCIR3 and DCIR4 proteins in AMs from wild type mice (Fig. 2). These results suggest that the expression of DCIR3 and DCIR4 on AMs is independent of their precursors and is regulated by the environments of the niche where the precursors differentiate into AMs.
4 Discussion
We recently showed that DCIR3 and DCIR4 are expressed on two major populations of monocytes, inflammatory and patrolling monocytes [2,3]. Our current study extends the observation in monocytes to that in macrophages to clearly demonstrate that DCIR3 and DCIR4 are widely expressed by macrophages including tissue-resident macrophages, except brain microglia and lung AMs (Fig. 2). Thus, our data together with the previous findings in monocytes indicate that DCIR3 and DCIR4 are widely distributed among phagocytic mononuclear cells. How DCIR3 and DCIR4 regulate the function of phagocytic mononuclear cells and what are the ligands recognized by DCIR3 and DCIR4 are important questions remain to be examined in future studies.
Our current study also showed that microglia lack the expression of DCIR3 and DCIR4, while AMs express DCIR3 but not DCIR4 (Fig. 2). The dichotomic expression of DCIR3 and DCIR4 in monocytes raised the possibility of coordinated regulation of DCIR3 and DCIR4 expressions [3], however, the current observation in AMs indicates that the expressions of DCIR3 and DCIR4 are independently regulated.
We also investigated the expression of DCIR3 and DCIR4 on fetal precursors of tissue-resident macrophages (Fig. 3). Microglia are derived from E7.5~E9.5 YS-MFs [8,18,19] and are hardly replaced by microglia derived from blood monocytes [9]. Interestingly, DCIR3 and DCIR4 were expressed on YS-MFs, but not on microglia (Fig. 2, Fig. 3A). These data suggest that the expression of DCIR3 and DCIR4 is lost during differentiation of YS-MFs into microglia.
Z. Liu et al. showed that tissue-resident macrophages including AMs, spleen red pulp macrophages, LPMs and small intestinal macrophages in adult mice are the mixture of macrophages derived from embryonic precursors and those derived from adult blood monocytes by the fate-mapping technique [9]. Notably, in 8–10 weeks-old mice, 10–20% of AMs were derived from adult blood monocytes [9]. This observation together with our current result that AMs from 8–10 weeks-old mice expressed DCIR3 but not DCIR4 (Fig. 2) suggests that, upon the differentiation of blood monocytes into AMs, the expression of DCIR3 is sustained, while the expression of DCIR4 is lost.
The data AMs derived from YS-MFs, FL-MOs and adult BM-MOs upon reconstitution in neonatal Csf2rb−/- mice showed similar patterns regarding the expression of Clec4a3 and Clec4a1 genes (Fig. 4B) suggest that the expression of DCIR3 and DCIR4 might be influenced by the environment where precursors differentiate into macrophages irrespective of the types of the precursors.
The two populations of peritoneal macrophages, LPMs and SPMs, both expressed DCIR3 and DCIR4 (Fig. 2). LPMs are derived from embryonic precursors [6] and up to 40% of them are replaced with the macrophages derived from blood monocytes after birth [9], while SPMs are short-lived and the majority of them are originated from blood inflammatory monocytes [22,23].
Regarding the expression of DCIR3 and DCIR4, FL-MOs included positive and negative populations (Fig. 3B). This result precluded us to determine whether the precursors of the FL-MOs-derived tissue-resident macrophages express DCIR3 and DCIR4 or not. Fate mapping experiments by making DCIR3-cre or DCIR4-cre mice and crossing them with fluorescent reporters such as Rosa26-LSL-YFP, Rosa26-LSL-RFP, Rosa26-LSL-GFP and Rosa26-LSL-Tdtomato lines to generate reporter mice would be useful to answer the question in the future, as used in the fate mapping studies of fetal monocytes/macrophages [24].
In contrast with the adult monocytes, which express both DCIR3 and DCIR4 [2,3], only subpopulations of FL-MOs expressed DCIR3 and DCIR4 (Fig. 3B), suggesting that FL-MOs might have gene expression profile distinct from adult monocytes. Indeed, mRNA expression levels of Clec4a3 (DCIR3) and Clec4a1 (DCIR4) were significantly lower in the FL-MOs than in the adult BM monocytes (Fig. 4). Moreover, Hoeffel et al. reported that FL-MOs selectively express genes related to cell cycle and differentiation, while adult monocytes selectively express genes related to immune responses and pathogen recognition [20].
Collectively, our results demonstrate that the majority of tissue-resident macrophages except microglia and AMs express DCIR3 and DCIR4. Our results also indicate that when the embryonic precursors differentiate into tissue-resident macrophages, they sometimes retain the expression of DCIR3 and DCIR4 and sometimes lose the expression of DCIR3 and/or DCIR4, depending on the organs where the embryonic precursors differentiate into tissue-resident macrophages. Our observations suggest that the expression of DCIR3 and DCIR4 on tissue-resident macrophages does not depend on the presence or absence of DCIR3 and DCIR4 expression on their precursors and that the expression of DCIR3 and DCIR4 might be influenced by environmental factors of the tissues where the precursors differentiate into macrophages.
Author contributions
Naoki Matsumoto conceived and designed the study. Ryo Okada acquired the data. Ryo Okada, Naoki Matsumoto and Kazuo Yamamoto analyzed and interpreted the data. Ryo Okada and Naoki Matsumoto drafted and revised the manuscript. Ryo Okada, Naoki Matsumoto and Kazuo Yamamoto approved the submission of the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by a research grant from Kobayashi foundation to NM.
Footnotes
Abbreviations: AM, alveolar macrophage; BM, bone marrow; BMDMs, bone marrow-derived macrophages; DCIR, dendritic cell immunoreceptor; DCIR3, dendritic cell inhibitory receptor 3; DCIR4, dendritic cell inhibitory receptor 4; FL-MOs, fetal liver monocytes; YS-MFs, yolk-sac macrophages.
References
- 1.Flornes L.M., Bryceson Y.T., Spurkland A., Lorentzen J.C., Dissen E., Fossum S. Identification of lectin-like receptors expressed by antigen presenting cells and neutrophils and their mapping to a novel gene complex. Immunogenetics. 2004;56:506–517. doi: 10.1007/s00251-004-0714-x. [DOI] [PubMed] [Google Scholar]
- 2.Kameda Y., Hanayama M., Kishimoto A., Kume M., Yamamoto K., Matsumoto N. Dendritic cell inhibitory receptor 4 (DCIR4) is preferentially expressed on inflammatory and patrolling monocytes. Biochem. Biophys. Res. Commun. 2016;480:215–221. doi: 10.1016/j.bbrc.2016.10.030. [DOI] [PubMed] [Google Scholar]
- 3.Hsu Y., Okada R., Nishimura T., Kawasaki N., Yamamoto K., Matsumoto N. DCIR3 and DCIR4 are co-expressed on inflammatory and patrolling monocytes. Biochem. Biophys. Res. Commun. 2017;494:440–445. doi: 10.1016/j.bbrc.2017.10.067. [DOI] [PubMed] [Google Scholar]
- 4.León B., Martínez del Hoyo G., Parrillas V., Vargas H.H., Sánchez-Mateos P., Longo N., López-Bravo M., Ardavín C. Dendritic cell differentiation potential of mouse monocytes: monocytes represent immediate precursors of CD8- and CD8+ splenic dendritic cells. Blood. 2004;103:2668–2676. doi: 10.1182/blood-2003-01-0286. [DOI] [PubMed] [Google Scholar]
- 5.Davies L.C., Jenkins S.J., Allen J.E., Taylor P.R. Tissue-resident macrophages. Nat. Immunol. 2013;14:986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., Hume D.A., Perlman H., Malissen B., Zelzer E., Jung S. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D., Greter M., Mortha A., Boyer S.W., Forsberg E.C., Tanaka M., van Rooijen N., García-Sastre A., Stanley E.R., Ginhoux F., Frenette P.S., Merad M. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., Samokhvalov I.M., Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z., Gu Y., Chakarov S., Bleriot C., Kwok I., Chen X., Shin A., Huang W., Dress R.J., Dutertre C.A., Schlitzer A., Chen J., Ng L.G., Wang H., Su B., Ginhoux F. Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell. 2019;178:1509–1525. doi: 10.1016/j.cell.2019.08.009. e1519. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Im. 2008;14 doi: 10.1002/0471142735.im1401s83. Unit 14.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akatsuka A., Ito M., Yamauchi C., Ochiai A., Yamamoto K., Matsumoto N. Tumor cells of non-hematopoietic and hematopoietic origins express activation-induced C-type lectin, the ligand for killer cell lectin-like receptor F1. Int. Immunol. 2010;22:783e790. doi: 10.1093/intimm/dxq430. [DOI] [PubMed] [Google Scholar]
- 12.Lund H., Pieber M., Parsa R., Han J., Grommisch D., Ewing E., Kular L., Needhamsen M., Espinosa A., Nilsson E., Överby A.K., Butovsky O., Jagodic M., Zhang X.M., Harris R.A. Competitive repopulation of an empty microglial niche yields functionally distinct subsets of microglia-like cells. Nat. Commun. 2018;9:4845. doi: 10.1038/s41467-018-07295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y.R., O'Koren E.G., Hotten D.F., Kan M.J., Kopin D., Nelson E.R., Que L., Gunn M.D. A protocol for the comprehensive flow cytometric analysis of immune cells in normal and inflamed murine non-lymphoid tissues. PLoS One. 2016;11 doi: 10.1371/journal.pone.0150606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., Mazloom A.R., Ma'ayan A., Chua W.J., Hansen T.H., Turley S.J., Merad M., Randolph G.J., Consortium I.G. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Movita D., Kreefft K., Biesta P., van Oudenaren A., Leenen P.J., Janssen H.L., Boonstra A. Kupffer cells express a unique combination of phenotypic and functional characteristics compared with splenic and peritoneal macrophages. J. Leukoc. Biol. 2012;92:723–733. doi: 10.1189/jlb.1111566. [DOI] [PubMed] [Google Scholar]
- 16.Ghosn E.E., Cassado A.A., Govoni G.R., Fukuhara T., Yang Y., Monack D.M., Bortoluci K.R., Almeida S.R., Herzenberg L.A. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uematsu S., Fujimoto K., Jang M.H., Yang B.G., Jung Y.J., Nishiyama M., Sato S., Tsujimura T., Yamamoto M., Yokota Y., Kiyono H., Miyasaka M., Ishii K.J., Akira S. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 2008;9:769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- 18.Kierdorf K., Erny D., Goldmann T., Sander V., Schulz C., Perdiguero E.G., Wieghofer P., Heinrich A., Riemke P., Hölscher C., Müller D.N., Luckow B., Brocker T., Debowski K., Fritz G., Opdenakker G., Diefenbach A., Biber K., Heikenwalder M., Geissmann F., Rosenbauer F., Prinz M. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 19.Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., Frampton J., Liu K.J., Geissmann F. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 20.Hoeffel G., Chen J., Lavin Y., Low D., Almeida F.F., See P., Beaudin A.E., Lum J., Low I., Forsberg E.C., Poidinger M., Zolezzi F., Larbi A., Ng L.G., Chan J.K., Greter M., Becher B., Samokhvalov I.M., Merad M., Ginhoux F. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van de Laar L., Saelens W., De Prijck S., Martens L., Scott C.L., Van Isterdael G., Hoffmann E., Beyaert R., Saeys Y., Lambrecht B.N., Guilliams M. YS-MFs, FL, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity. 2016;44:755–768. doi: 10.1016/j.immuni.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Cain D.W., O'Koren E.G., Kan M.J., Womble M., Sempowski G.D., Hopper K., Gunn M.D., Kelsoe G. Identification of a tissue-specific, C/EBPβ-dependent pathway of differentiation for murine peritoneal macrophages. J. Immunol. 2013;191:4665–4675. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okabe Y., Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi J., Hua L., Harmer D., Li P., Ren G. Cre driver mice targeting macrophages. Methods Mol. Biol. 2018;1784:263–275. doi: 10.1007/978-1-4939-7837-3_24. [DOI] [PMC free article] [PubMed] [Google Scholar]