Abstract
Accurate mechanical characterization of adherent cells and their substrates is important for understanding the influence of mechanical properties on cells themselves. Recent mechanobiology studies outline the importance of mechanical parameters, such as stress relaxation and strain stiffening on the behavior of cells. Numerous techniques exist for probing mechanical properties and it is vital to understand the benefits of each technique and how they relate to each other. This mini review aims to guide the reader through the toolbox of mechanical characterization techniques by presenting well-established and emerging methods currently used to assess mechanical properties of substrates and cells.
Keywords: biointerfaces, mechanical properties, mechanotransduction, cell-substrate interactions, atomic force microscopy, traction force microscopy, Brillouin microscopy, magnetic tweezers
Introduction
When engineering mechanical cues to study cell mechanotransduction, it is important to take into account how the cells sense the mechanical properties. Material mechanical properties ought to be measured at timescales and frequencies relevant to the properties sensed by adhering cells (Cameron et al., 2014; Charrier et al., 2018). When cells interact with a substrate, they can apply forces parallel and perpendicular to the surface (Janmey et al., 2020). This implies that the method used for assessing cellular forces may affect the results in mechanotransduction studies. One more factor to consider is the depth into a material that cells can sense the mechanical properties. Modeling, and studies on thin gels, suggest that cells sense mechanical properties in the range of their radius (Sen et al., 2009; Buxboim et al., 2010; He et al., 2014). The range within which cells can sense mechanical properties may be material-dependent though, and it has been shown that strain stiffening fibrous networks enable long-range force sensing of up to hundreds of microns (Leiphart et al., 2019). It is therefore relevant to consider the sensing depth of surface and bulk mechanical characterization techniques, respectively. How cells sense their environment is also cell-type dependent and, while many excellent model studies have been performed in 2D, cell sensing is likely to be different in three-dimensional (3D) culture environments (Doyle and Yamada, 2016).
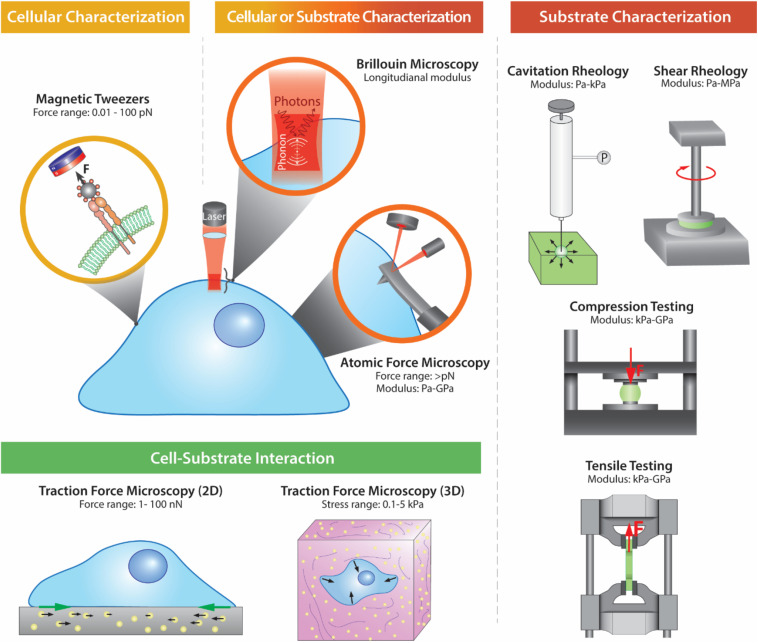
Many model substrates have been adopted to study cellular mechanotransduction. In 1980, silicone rubber was first used as a deformable elastic substrate for cell culture, which revealed that cells constantly exert forces on substrates (Harris et al., 1980). Building on this work, gels gained interest as model substrates due to their tunable viscoelastic properties and close resemblance, mechanically, to the extracellular matrix (ECM) (Tibbitt and Anseth, 2009; Chaudhuri, 2017). However, thin films and liquids have also been used as substrates (Yang et al., 2014; Kong et al., 2018). To precisely determine the mechanical cues that substrates display to cells is crucial for biological studies. It is also important to assess the changes in the mechanical properties of cells in response to various biophysical and biochemical cues. Therefore, this review describes and contrasts the most commonly used techniques to determine the mechanical properties of materials and cells (Figure 1 and Table 1).
FIGURE 1.
Schematic illustrations of the different methods to characterize the mechanical properties of substrates and/or cells.
TABLE 1.
Considerations and relevant examples for the various characterization techniques to assess mechanical properties at different length scales, ranging from cm to nm.
| Method | Considerations | Examples |
| Global scale (cm-mm) Shear rheology |
|
|
| Compression testing |
|
|
| Tensile testing |
|
|
| Cavitation rheology |
|
|
| Brillouin microscopy |
|
|
| Traction force microscopy |
|
|
| Probe-based techniques |
|
|
| Magnetic tweezers Local scale (nm) |
|
|
Assessment of the Global Mechanical Properties
The mechanical properties of substrates such as gels used in mechanotransduction studies are usually characterized globally by rheology, tensile, and compression testing.
Shear rheology can be used to monitor the gelation process, or to measure the elastic and viscous properties of gels (denoted by storage and loss modulus) by applying small amplitude oscillatory shear forces (Zuidema et al., 2014). Time-dependent aspects of viscoelasticity are quantified by relaxation time and relaxation modulus (Fitzgerald et al., 2015; Chaudhuri et al., 2016). Strain stiffening materials are characterized by monitoring the elastic modulus as a function of increasing stress. It is important to note the significance of the geometries used for probing the sample. For instance, a recent study showed that a crosshatched geometry revealed the strain stiffening property of agarose, while a parallel plate geometry obscured it due to the wall slippage effect (Bertula et al., 2019). The main limitation of rheology to characterize gels is that the spatial variation in moduli arising from sample heterogeneities cannot be probed.
Compression and tensile testing are also techniques commonly used to probe the global mechanical properties, especially for biomaterials (Xiao et al., 2013; Vedadghavami et al., 2017). The elastic, or Young’s modulus, of the sample is generally obtained from the slope of the stress-strain curve, but the viscoelastic properties can also be accessed through this kind of characterization (Mirahmadi et al., 2013; Bosnjak et al., 2020). Variations in the testing protocols can, however, influence the results (Patel et al., 2019) and difficulties can also arise in tensile testing from the breakage or slipping of the sample at the clamps (Oyen, 2014).
Several of the aforementioned mechanical parameters can be probed using different techniques, and it is important to understand how the parameters are measured. For example, tensile testing measures the elastic properties (Young’s modulus) by applying tensile forces, while in rheology shear forces are applied (storage modulus).
Assessment of the Local Mechanical Properties
The local mechanical properties are relevant for measuring what properties cells sense at the surface of a material. The resolution of the techniques described below enable the mechanical properties of cells to be probed in real time due to their specificity and sensitivity.
Probe-Based Techniques
The atomic force microscope (AFM) belongs to a family of instruments known as scanning probe microscopes. These instruments are commonly used for obtaining high resolution, 3D images of a sample surface using a tip which scans across an area of interest whilst measuring the interactions with the surface. Force-indentation is one major application of the AFM, measuring the interaction forces between the probe tip (sharp or colloidal) and a sample surface (Yablon, 2013). A force-indentation curve is obtained by measuring the displacement of the tip at the end of a cantilever, monitored by a laser and photodiode while controlling the force of the indentation. AFM has been extended for studying biological samples and their processes (Alsteens et al., 2017; Krieg et al., 2019).
The main advantage AFM force-indentation has over traditional nanoindentation is the ability to apply a lower range of well-defined forces (pico-Newtons to nano-Newtons) by changing the cantilever spring constant or tip, which is particularly useful for hydrogel or soft biological samples. Sub-pico-Newton forces have also been achievable by improving tip stability (King et al., 2009; Churnside and Perkins, 2014). Some challenges associated with AFM force-indentation are that the indentation parameters and cantilever stiffness must be appropriately selected and accurately calibrated to model the data to mechanical information. A variety of methods exist for accurate cantilever calibration, the most common being the thermal noise calibration method (Hutter and Bechhoefer, 1993). Laser Doppler vibrometry has also been used to achieve more accurate calibrations (Ohler, 2007; Gates and Pratt, 2012). Radmacher and colleagues have developed a standardized nanomechanical procedure for AFM on polyacrylamide hydrogels and MDCK-C11 cells showing improvement in consistency and reproducibility across eleven research groups in Europe (Schillers et al., 2017).
The tip and cantilever used for indentation are important for modeling the force-indentation curves obtained from AFM. Tips are usually made from stiff materials (silicon, silicon nitride or, in the case of colloids, glass or polystyrene) to be suitable for modeling, and to avoid deforming or damaging the tip when performing indentations. AFM tips can generally be classified as blunt or sharp tips. Blunt or rounded tips have a larger contact area and can achieve higher forces for the same indentation depth compared to a sharp tip (Qian and Zhao, 2018). Single cells have also been used as probes to achieve pico-Newton forces (Helenius et al., 2008; Li et al., 2019). Several different models are available to model force-indentation curves, each associated with its own assumptions and limitations (Lin and Horkay, 2008). Force-indentation data is preferably fitted in the first few hundreds of nanometers of the approach curve. The elastic modulus is usually obtained by fitting Hertz model with a rounded tip; although, sharp tips are commonly used for their availability and simplicity (Sirghi et al., 2008; MacKay and Kumar, 2012). It has also been demonstrated that the shear modulus can be obtained from Hertz modeling of force-indentation curves (Berry et al., 2020).
AFM viscoelastic measurements such as stress relaxation and creep (Yango et al., 2016; Efremov et al., 2017), strain stiffening (Van Helvert and Friedl, 2016) and nano-rheology (Li et al., 2014) are also possible. Oscillatory AFM has been used to determine the complex shear modulus of cells (Alcaraz et al., 2003). The storage stiffness, loss stiffness, and loss tangent of polyacrylamide hydrogels have similarly been determined using a magnetically driven cantilever (Nalam et al., 2015). Nano-rheology is still a relatively new technique, but future improvements of nano-rheological modeling in AFM software are expected to enhance the field significantly.
Viscoelastic modeling can be performed using a variety of models such as the Maxwell and the Voigt-Kelvin models (Cheng et al., 2005; Fischer-Cripps, 2011) or Ting’s viscoelastic model (Ting, 1964), which has been only recently applied to AFM force-indentation curves (Brückner et al., 2017; Efremov et al., 2017, 2019b). For a recent review that highlights the various models and properties obtainable from soft biological samples with AFM, the reader is directed to Efremov et al. (2019a).
Nanomechanical Mapping
AFM modes to map mechanical properties on the nanoscale, while imaging, have also been developed. Generally, nanomechanical mapping modes are capable of mapping the elastic and loss moduli in a range between 1 kPa and 100 GPa across an entire imaging area (100 nm2 to 100 μm2) (Morales-Rivas et al., 2015; Viji Babu et al., 2019). This technique offers high spatial resolution and can be operated in air, liquid and vacuum. The main benefit of this method compared to force-indentation is the imaging feature and the ability to overlay mechanical data and topography. Different AFM manufacturers provide integrated modes for nanomechanical mapping.
Generally, in nanomechanical mapping, individual force curves are recorded as the tip is oscillating in tapping mode and scanning the surface. The maximum force exerted on the sample is kept constant by the feedback loop, which ensures the protection of the tip and the sample. The resulting force curves are converted into force indentation curves, which can subsequently be analyzed with regards to mechanical properties such as modulus, adhesion, dissipation or deformation. The spring constant of the cantilever and the tip radius have to be calibrated prior to quantitative measurements. The operating range of this technique ranges from very soft samples (low kPa) to hard materials (100 GPa) and has been demonstrated on amyloid nanofibrils (Sweers et al., 2011), proteoglycan mimetic nanoparticles (Hedayati and Kipper, 2018), and microvilli on living cells (Schillers et al., 2016). A protocol to analyze the mechanical properties of very soft materials (cells) can be found in reference (Hu et al., 2020).
We believe the use of fast, spatially resolved, mechanical, and topographical analysis will be increasingly valuable for mechanotransduction studies in the future.
Traction Force Microscopy
While the mechanical properties of substrates are important to characterize, an understanding of the forces that cells can exert on their surroundings is central for our understanding of mechanotransduction. Traction force microscopy (TFM) is a technique used to assess forces that cells apply to the ECM or substrate they grow on/in, by monitoring the displacement of fluorescent beads embedded in the substrate (Hur et al., 2020). Some studies also measure these forces through the deflection of elastic micropillar arrays onto which cells are adhering (Schoen et al., 2010), or use the cell-induced alignment of collagen fibers to deduce the applied traction forces (Shakiba et al., 2020).
Different kinds of TFM exist according to the number of dimensions considered, and the accuracy of the method actually increases with the number of dimensions involved in the calculations. So far, most of the materials used as substrates are hydrogels that exhibit linearly elastic and isotropic properties such as silicon or polyacrylamide hydrogels (Polacheck and Chen, 2016). However, these materials do not always reflect the behavior of native tissues. Some studies develop TFM methods that take into account the non-linear behaviors of matrices, such as fibrillar networks like collagen, or strain-stiffening materials (Steinwachs et al., 2016; Song et al., 2020). Integrating these parameters often complicates the modeling, but significantly improves the applicability of the results. TFM-based methods are also developed to focus on cell-cell and intracellular traction forces (Maruthamuthu et al., 2011), which is particularly relevant for cells of tissues with cell junctions, such as the endothelium. Recently, traction force microscopy was combined with microarray-based techniques to enable high throughput screening of cell generated forces (Kaylan et al., 2017).
Emerging Techniques
Recent research has propagated the development of modern characterization techniques, all with unique advantages and limitations. Some of the most promising emerging techniques for mechanical characterization are summarized in this section.
Brillouin microscopy was originally demonstrated on cells over a decade ago (Scarcelli and Yun, 2008) and has since garnered attention within the biological and medical research fields for its novel approach to non-destructive viscoelastic measurements of biological samples. The method involves interacting the intrinsic acoustic waves of a material with probe laser pulses and interpreting how the light is scattered – which is heavily linked to the material’s viscoelastic properties (Gusev and Ruello, 2018). Recent works have demonstrated a submicrometric spatial resolution (Caponi et al., 2020). Brillouin microscopy is also largely non-destructive while still possessing resolution capabilities for cellular characterization (μm scale) (Prevedel et al., 2019). Since its pioneering application in 2005 (Koski and Yarger, 2005), Brillouin microscopy has been utilized in tissue-level sensing (Mathieu et al., 2011; Margueritat et al., 2019) and subcellular-level characterization (Steelman et al., 2015; Bevilacqua et al., 2019). However, interpretation of results requires specific knowledge of the refractive index and material density (Prevedel et al., 2019) which is difficult to experimentally attain (Liu et al., 2016), and the weakness of the measured signal causes extended scanning and data acquisition times (Ballmann et al., 2019). Scrutinous evaluation of Brillouin output is also required, especially in biological matter, due to the spatiotemporal inhomogeneity causing differences in the material’s intrinsic acoustic wave behavior (Wu et al., 2018; Ballmann et al., 2019).
Microfluidic devices constitute an efficient and relatively high-throughput method to characterize the mechanical properties of cells. For instance, micropipette aspiration on microfluidic chips using giant vesicles as cell models have been developed to access cell membrane mechanics (Elias et al., 2020). Other techniques use flow field analysis to study the deformations applied by fluids to cells to deduce their viscoelastic properties (Guillou et al., 2016). The influence of fluid flows on cells has also been combined with computational models to determine the elastic modulus of cells (Song et al., 2012) and such flows have also been demonstrated to impact cell behavior and maturation (Song et al., 2013; Shemesh et al., 2015).
Optical tweezers (OTs) is a single-molecule technique initially designed in 1986 (Ashkin et al., 1986). OTs involve the precise manipulation of beads (0.1–100 pN) with molecules tethered to them by trapping them with strongly focused light. In the context of mechanobiology, OTs have been used to assess ligand-receptor bond properties using ligand-functionalized trapped particles (Jiang et al., 2003), to explore forces on the structures at the surface of the cell involved in mechanotransduction (Choquet et al., 1997), to assess forces on structures within the cell (Welte et al., 1998), and to investigate the mechanical properties of the cell (Tan et al., 2012). OTs are, however, accompanied by drawbacks such as photo-entrapment of impurity particles, local heating and photodamage (Peterman et al., 2003). Some of these challenges are solved by magnetic tweezers (MTs). This single-molecule technique is relevant for biological applications such as single-molecule force measurements and cellular micromanipulation (Basoli et al., 2018). It operates by utilizing superparamagnetic beads (0.1–100 μm) as a force transducer for a tethered single large molecule by applying a magnetic field (Xin et al., 2017). Advantages of MT include its cost-effectiveness and ease of implementation, superior to other single-molecule techniques (Kemmerich et al., 2016). Furthermore, MTs can generate forces inside closed opaque bodies, such as living cells, which is not the case with other non-invasive methods (Timonen and Grzybowski, 2017). MTs have been used extensively to generate tension on cell adhesion molecules, and then observing the cellular response to reveal mechanotransduction pathways(Guilluy et al., 2014; Marjoram et al., 2016). The main challenge in MT development has been their subpar resolution that is limited by relying on camera-based measurements of magnetic beads (De Vlaminck and Dekker, 2012). However, high spatiotemporal resolution magnetic tweezers (Å scale) are being implemented with help from more sophisticated cameras and computer hardware (Huhle et al., 2015; Ostrofet et al., 2020). There are also difficulties working with particles susceptible to magnetic fields. Furthermore, the requirement that bulky magnets must be situated close to the sample complicates the combination of MTs with other applications (Sarkar and Rybenkov, 2016).
Concerning substrates, a relatively new and inexpensive technique to probe local mechanical properties is cavitation rheology (Zimberlin et al., 2007). Cavitation rheology quantifies the elastic properties by growing a bubble in a gel or fluid and monitoring the pressure dynamics (Hashemnejad and Kundu, 2017). The resolution of cavitation rheology can go as low as 1 μm and requires only small sample volumes (Barney et al., 2020). One drawback of this method is that it is destructive to the sample. Questions also remain around relating gel fracture to cavitation and the influence of local network structures on cavitation bubble growth (Barney et al., 2020).
Conclusion and Outlooks
One of the fundamental challenging aspects of characterizing substrates is the relevance of a particular technique in capturing the properties of substrates experienced by the cell. Each technique probes the mechanical properties in a different way, and it is necessary to compare the parameters obtained using different techniques. Rheology or compression tests, for instance, can provide the elastic and loss modulus of a material, but only as a mean value for the whole material, whereas probe-based techniques such as AFM assess the local mechanical properties at the microscopic scale, which can differ from the macroscopic one. A recent study showed that the bulk elastic modulus obtained for stiffer gels using rheology and AFM correlate well but showed marked differences for softer gels (Megone et al., 2018). There has also been a recent trend in using simple and inexpensive techniques for probing mechanical properties. For instance, using fluorescent bead density as a readout for stiffness gradients in gels instead of using expensive techniques like AFM (Barber-Pérez et al., 2020). Recent technology also allows assessment of mechanical properties of materials with extremely precise force (resolution down to 10 nN claimed by the manufacturer) through micro-scale compression testing, particularly for soft biomaterials (Kerscher et al., 2017).
Emerging techniques are now focusing on probing these properties even more precisely and in-depth. Cell-substrate interactions are also highly relevant to assess mechanotransduction and techniques such as traction force microscopy and magnetic tweezers can be applied to survey such interactions. Forces in the order of pico-Newtons, applied by integrins on ligands, have also been assessed by DNA mechanotechnology (Brockman et al., 2018; Glazier et al., 2019; Ma et al., 2019). However, significant challenges regarding the integrity of the DNA probes in cell culture conditions and the type of receptor-ligand interactions that can be probed remain (Ma and Salaita, 2019; Yasunaga et al., 2019).
Many of the existing tools for measuring the mechanical properties of cells are limited in terms of throughput. To mitigate this issue, high throughput based methods have been explored to separate large populations of cells based on mechanical properties using microfluidics and shear stress (Otto et al., 2015). A combination of smart design of microfluidic geometries and powerful computational approaches such as microparticle image velocimetry and machine learning promise to further progress the field of mechanobiology (Song et al., 2010; Guillou et al., 2016).
The influence of substrate mechanical properties on processes such as nascent protein deposition has been explored in recent studies by using metabolic labeling techniques (Loebel et al., 2019, 2020). It would be interesting to combine such labeling with 3D traction force microscopy in order to directly visualize the effects of cell traction forces on nascent protein deposition.
Going forward, standard mechanical characterization techniques combined with newer, more accurate and more local assessments of the mechanical properties, will enable the development of more complex engineered matrices. Such materials with spatiotemporally-controlled mechanical properties will further the understanding of how cells interact with their environment in vitro, and indirectly in vivo. This is important to fully use the potential of stem cells in tissue engineering and stem cell therapy.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The authors gratefully acknowledge financial support from the Marsden Fund Council and the Rutherford Discovery Fellowship, from Government funding, managed by Royal Society Te Apārangi, and support from the MacDiarmid Institute for Advanced Materials and Nanotechnology, and from The University of Auckland.
References
- Alcaraz J., Buscemi L., Grabulosa M., Trepat X., Fabry B., Farré R., et al. (2003). Microrheology of human lung epithelial cells measured by atomic force microscopy. Biophys. J. 84 2071–2079. 10.1016/S0006-3495(03)75014-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsteens D., Gaub H. E., Newton R., Pfreundschuh M., Gerber C., Müller D. J. (2017). Atomic force microscopy-based characterization and design of biointerfaces. Nat. Rev. Mater. 2:17008 10.1038/natrevmats.2017.8 [DOI] [Google Scholar]
- Ashkin A., Dziedzic J. M., Bjorkholm J. E., Chu S. (1986). Observation of a single-beam gradient force optical trap for dielectric particles. Opt. Lett. 11:288. 10.1364/ol.11.000288 [DOI] [PubMed] [Google Scholar]
- Balaban N. Q., Schwarz U. S., Riveline D., Goichberg P., Tzur G., Sabanay I., et al. (2001). Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3 466–472. 10.1038/35074532 [DOI] [PubMed] [Google Scholar]
- Ballmann C. W., Meng Z., Yakovlev V. V. (2019). Nonlinear Brillouin spectroscopy: what makes it a better tool for biological viscoelastic measurements. Biomed. Opt. Express 10:1750. 10.1364/boe.10.001750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber-Pérez N., Georgiadou M., Guzmán C., Isomursu A., Hamidi H., Ivaska J. (2020). Mechano-responsiveness of fibrillar adhesions on stiffness-gradient gels. J. Cell Sci. 133:jcs242909. 10.1242/jcs.242909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barney C. W., Dougan C. E., McLeod K. R., Kazemi-Moridani A., Zheng Y., Ye Z., et al. (2020). Cavitation in soft matter. Proc. Natl. Acad. Sci. U.S.A. 117 9157–9165. 10.1073/pnas.1920168117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basoli F., Giannitelli S. M., Gori M., Mozetic P., Bonfanti A., Trombetta M., et al. (2018). Biomechanical characterization at the cell scale: present and prospects. Front. Physiol. 9:1449. 10.3389/fphys.2018.01449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz K. C., Sultan N., Savin D. A. (2018). Quantitative relationship between cavitation and shear rheology. Soft Matter 14 8395–8400. 10.1039/c8sm01560k [DOI] [PubMed] [Google Scholar]
- Bergert M., Lendenmann T., Zündel M., Ehret A. E., Panozzo D., Richner P., et al. (2016). Confocal reference free traction force microscopy. Nat. Commun. 7:12814. 10.1038/ncomms12814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J. D., Biviano M., Dagastine R. R. (2020). Poroelastic properties of hydrogel microparticles. Soft Matter 16 5314–5324. 10.1039/d0sm00191k [DOI] [PubMed] [Google Scholar]
- Bertula K., Martikainen L., Munne P., Hietala S., Klefström J., Ikkala O., et al. (2019). Strain-Stiffening of Agarose Gels. ACS Macro Lett. 8 670–675. 10.1021/acsmacrolett.9b00258 [DOI] [PubMed] [Google Scholar]
- Bevilacqua C., Sánchez-Iranzo H., Richter D., Diz-Muñoz A., Prevedel R. (2019). Imaging mechanical properties of sub-micron ECM in live zebrafish using Brillouin microscopy. Biomed. Opt. Express 10:1420. 10.1364/boe.10.001420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnjak N., Nadimpalli S., Okumura D., Chester S. A. (2020). Experiments and modeling of the viscoelastic behavior of polymeric gels. J. Mech. Phys. Solids 137:103829 10.1016/j.jmps.2019.103829 [DOI] [Google Scholar]
- Brockman J. M., Blanchard A. T., Pui-Yan V., Derricotte W. D., Zhang Y., Fay M. E., et al. (2018). Mapping the 3D orientation of piconewton integrin traction forces. Nat. Methods 15 115–118. 10.1038/nmeth.4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner B. R., Nöding H., Janshoff A. (2017). Viscoelastic properties of confluent MDCK II cells obtained from force cycle experiments. Biophys. J. 112 724–735. 10.1016/j.bpj.2016.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby G. A., Grant M. H., MacKay S. P., Riches P. E. (2013). Confined compression of collagen hydrogels. J. Biomech. 46 837–840. 10.1016/j.jbiomech.2012.11.048 [DOI] [PubMed] [Google Scholar]
- Buxboim A., Rajagopal K., Brown A. E. X., Discher D. E. (2010). How deeply cells feel: methods for thin gels. J. Phys. Condens. Matter. 22:194116 10.1088/0953-8984/22/19/194116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron A. R., Frith J. E., Gomez G. A., Yap A. S., Cooper-White J. J. (2014). The effect of time-dependent deformation of viscoelastic hydrogels on myogenic induction and Rac1 activity in mesenchymal stem cells. Biomaterials 35 1857–1868. 10.1016/j.biomaterials.2013.11.023 [DOI] [PubMed] [Google Scholar]
- Caponi S., Fioretto D., Mattarelli M. (2020). On the actual spatial resolution of Brillouin Imaging. Opt. Lett. 45:1063. 10.1364/ol.385072 [DOI] [PubMed] [Google Scholar]
- Charrier E. E., Pogoda K., Wells R. G., Janmey P. A. (2018). Control of cell morphology and differentiation by substrates with independently tunable elasticity and viscous dissipation. Nat. Commun. 9:449. 10.1038/s41467-018-02906-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O. (2017). Viscoelastic hydrogels for 3D cell culture. Biomater. Sci. 5 1480–1490. 10.1039/c7bm00261k [DOI] [PubMed] [Google Scholar]
- Chaudhuri O., Gu L., Klumpers D., Darnell M., Bencherif S. A., Weaver J. C., et al. (2016). Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 15 326–334. 10.1038/nmat4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Xia X., Scriven L. E., Gerberich W. W. (2005). Spherical-tip indentation of viscoelastic material. Mech. Mater. 37 213–226. 10.1016/j.mechmat.2004.03.002 [DOI] [Google Scholar]
- Choquet D., Felsenfeld D. P., Sheetz M. P. (1997). Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell 88 39–48. 10.1016/S0092-8674(00)81856-5 [DOI] [PubMed] [Google Scholar]
- Churnside A. B., Perkins T. T. (2014). Ultrastable atomic force microscopy: improved force and positional stability. FEBS Lett. 588 3621–3630. 10.1016/j.febslet.2014.04.033 [DOI] [PubMed] [Google Scholar]
- De Vlaminck I., Dekker C. (2012). Recent advances in magnetic tweezers. Annu. Rev. Biophys. 41 453–472. 10.1146/annurev-biophys-122311-100544 [DOI] [PubMed] [Google Scholar]
- Doyle A. D., Yamada K. M. (2016). Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp. Cell Res. 343 60–66. 10.1016/j.yexcr.2015.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov Y. M., Okajima T., Raman A. (2019a). Measuring viscoelasticity of soft biological samples using atomic force microscopy. Soft Matter 16 64–81. 10.1039/c9sm01020c [DOI] [PubMed] [Google Scholar]
- Efremov Y. M., Shpichka A. I., Kotova S. L., Timashev P. S. (2019b). Viscoelastic mapping of cells based on fast force volume and PeakForce Tapping. Soft Matter 15 5455–5463. 10.1039/c9sm00711c [DOI] [PubMed] [Google Scholar]
- Efremov Y. M., Wang W. H., Hardy S. D., Geahlen R. L., Raman A. (2017). Measuring nanoscale viscoelastic parameters of cells directly from AFM force-displacement curves. Sci. Rep. 7:1541. 10.1038/s41598-017-01784-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M., Dutoya A., Laborde A., Lecestre A., Montis C., Caselli L., et al. (2020). Microfluidic characterization of biomimetic membrane mechanics with an on-chip micropipette. Micro Nano Eng. 8:100064 10.1016/j.mne.2020.100064 [DOI] [Google Scholar]
- Fischer-Cripps A. C. (2011). Nanoindentation. New York, NY: Springer; 10.1007/978-1-4419-9872-9 [DOI] [Google Scholar]
- Fitzgerald M. M., Bootsma K., Berberich J. A., Sparks J. L. (2015). Tunable stress relaxation behavior of an alginate-polyacrylamide hydrogel: comparison with muscle tissue. Biomacromolecules 16 1497–1505. 10.1021/bm501845j [DOI] [PubMed] [Google Scholar]
- Garcia R. (2020). Nanomechanical mapping of soft materials with the atomic force microscope: methods, theory and applications. Chem. Soc. Rev. 49 5850–5884. 10.1039/D0CS00318B [DOI] [PubMed] [Google Scholar]
- Gates R. S., Pratt J. R. (2012). Accurate and precise calibration of AFM cantilever spring constants using laser Doppler vibrometry. Nanotechnology 23:375702 10.1088/0957-4484/23/37/375702 [DOI] [PubMed] [Google Scholar]
- Glazier R., Brockman J. M., Bartle E., Mattheyses A. L., Destaing O., Salaita K. (2019). DNA mechanotechnology reveals that integrin receptors apply pN forces in podosomes on fluid substrates. Nat. Commun. 10:4507. 10.1038/s41467-019-12304-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L., Dahl J. B., Lin J. M. G., Barakat A. I. I., Husson J., Muller S. J., et al. (2016). Measuring cell viscoelastic properties using a microfluidic extensional flow device. Biophys. J. 111 2039–2050. 10.1016/j.bpj.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C., Osborne L. D., Van Landeghem L., Sharek L., Superfine R., Garcia-Mata R., et al. (2014). Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 16 376–381. 10.1038/ncb2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev V. E., Ruello P. (2018). Advances in applications of time-domain Brillouin scattering for nanoscale imaging. Appl. Phys. Rev. 5 1–55. 10.1063/1.5017241 [DOI] [Google Scholar]
- Harris A., Wild P., Stopak D. (1980). Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208 177–179. 10.1126/science.6987736 [DOI] [PubMed] [Google Scholar]
- Hashemnejad S. M., Kundu S. (2017). Probing Gelation and Rheological Behavior of a Self-Assembled Molecular Gel. Langmuir 33 7769–7779. 10.1021/acs.langmuir.7b01531 [DOI] [PubMed] [Google Scholar]
- He S., Su Y., Ji B., Gao H. (2014). Some basic questions on mechanosensing in cell-substrate interaction. J. Mech. Phys. Solids 70 116–135. 10.1016/j.jmps.2014.05.016 [DOI] [Google Scholar]
- Hedayati M., Kipper M. J. (2018). Atomic force microscopy of adsorbed proteoglycan mimetic nanoparticles: toward new glycocalyx-mimetic model surfaces. Carbohydr. Polym. 190 346–355. 10.1016/j.carbpol.2018.02.023 [DOI] [PubMed] [Google Scholar]
- Helenius J., Heisenberg C. P., Gaub H. E., Muller D. J. (2008). Single-cell force spectroscopy. J. Cell Sci. 121 1785–1791. 10.1242/jcs.030999 [DOI] [PubMed] [Google Scholar]
- Hu J., Chen S., Huang D., Zhang Y., Lü S., Long M. (2020). Global mapping of live cell mechanical features using PeakForce QNM AFM. Biophys. Rep. 6 9–18. 10.1007/s41048-019-00103-9 [DOI] [Google Scholar]
- Huhle A., Klaue D., Brutzer H., Daldrop P., Joo S., Otto O., et al. (2015). Camera-based three-dimensional real-time particle tracking at kHz rates and Ångström accuracy. Nat. Commun. 6:5885. 10.1038/ncomms6885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S. S., Jeong J. H., Ban M. J., Park J. H., Yoon J. K., Hwang Y. (2020). Traction force microscopy for understanding cellular mechanotransduction. BMB Rep. 53 74–081. 10.5483/BMBRep.2020.53.2.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter J. L., Bechhoefer J. (1993). Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64 1868–1873. 10.1063/1.1143970 [DOI] [Google Scholar]
- Janmey P. A., Fletcher D. A., Reinhart-King C. A. (2020). Stiffness sensing by cells. Physiol. Rev. 100 695–724. 10.1152/physrev.00013.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Giannone G., Critchley D. R., Fukumoto E., Sheet M. P. (2003). Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424 334–337. 10.1038/nature01805 [DOI] [PubMed] [Google Scholar]
- Kaylan K. B., Kourouklis A. P., Underhill G. H. (2017). A high-throughput cell microarray platform for correlative analysis of cell differentiation and traction forces. J. Vis. Exp. 121:55362. 10.3791/55362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerich F. E., Kasaciunaite K., Seidel R. (2016). Modular magnetic tweezers for single-molecule characterizations of helicases. Methods 108 4–13. 10.1016/j.ymeth.2016.07.004 [DOI] [PubMed] [Google Scholar]
- Kerscher P., Kaczmarek J. A., Head S. E., Ellis M. E., Seeto W. J., Kim J., et al. (2017). Direct production of human cardiac tissues by pluripotent stem cell encapsulation in gelatin Methacryloyl. ACS Biomater. Sci. Eng. 3 1499–1509. 10.1021/acsbiomaterials.6b00226 [DOI] [PubMed] [Google Scholar]
- King G. M., Carter A. R., Churnside A. B., Eberle L. S., Perkins T. T. (2009). Ultrastable atomic force microscopy: atomic-scale stability and registration in ambient conditions. Nano Lett. 9 1451–1456. 10.1021/nl803298q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., Peng L., Di Cio S., Novak P., Gautrot J. E. (2018). Stem cell expansion and fate decision on liquid substrates are regulated by self-assembled nanosheets. ACS Nano 12 9206–9213. 10.1021/acsnano.8b03865 [DOI] [PubMed] [Google Scholar]
- Koski K. J., Yarger J. L. (2005). Brillouin imaging. Appl. Phys. Lett. 87:061903 10.1063/1.1999857 [DOI] [Google Scholar]
- Krieg M., Fläschner G., Alsteens D., Gaub B. M., Roos W. H., Wuite G. J. L., et al. (2019). Atomic force microscopy-based mechanobiology. Nat. Rev. Phys. 1 41–57. 10.1038/s42254-018-0001-7 [DOI] [Google Scholar]
- Leiphart R. J., Chen D., Peredo A. P., Loneker A. E., Janmey P. A. (2019). Mechanosensing at cellular interfaces. Langmuir 35 7509–7519. 10.1021/acs.langmuir.8b02841 [DOI] [PubMed] [Google Scholar]
- Li M., Xi N., Wang Y., Liu L. (2019). Advances in atomic force microscopy for single-cell analysis. Nano Res. 12 703–718. 10.1007/s12274-018-2260-0 [DOI] [Google Scholar]
- Li T. D., Chiu H. C., Ortiz-Young D., Riedo E. (2014). Nanorheology by atomic force microscopy. Rev. Sci. Instrum. 85:123707 10.1063/1.4903353 [DOI] [PubMed] [Google Scholar]
- Lin D. C., Horkay F. (2008). Nanomechanics of polymer gels and biological tissues: a critical review of analytical approaches in the Hertzian regime and beyond. Soft Matter 4 669–682. 10.1039/b714637j [DOI] [PubMed] [Google Scholar]
- Liu P. Y., Chin L. K., Ser W., Chen H. F., Hsieh C. M., Lee C. H., et al. (2016). Cell refractive index for cell biology and disease diagnosis: past, present and future. Lab Chip 16 634–644. 10.1039/c5lc01445j [DOI] [PubMed] [Google Scholar]
- Loebel C., Kwon M. Y., Wang C., Han L., Mauck R. L., Burdick J. A. (2020). Metabolic labeling to probe the spatiotemporal accumulation of matrix at the chondrocyte–hydrogel interface. Adv. Funct. Mater. 2020:1909802 10.1002/adfm.201909802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel C., Mauck R. L., Burdick J. A. (2019). Local nascent protein deposition and remodelling guide mesenchymal stromal cell mechanosensing and fate in three-dimensional hydrogels. Nat. Mater. 18 883–891. 10.1038/s41563-019-0307-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R., Kellner A. V., Ma V. P. Y., Su H., Deal B. R., Brockman J. M., et al. (2019). DNA probes that store mechanical information reveal transient piconewton forces applied by T cells. Proc. Natl. Acad. Sci. U.S.A. 116 16949–16954. 10.1073/pnas.1904034116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma V. P. Y., Salaita K. (2019). DNA nanotechnology as an emerging tool to study mechanotransduction in living systems. Small 15:e1900961. 10.1002/smll.201900961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay J. L., Kumar S. (2012). “Measuring the elastic properties of living cells with atomic force microscopy indentation,” in Methods in Molecular Biology Methods in Molecular Biology, eds Taatjes D. J., Roth J. (Totowa, NJ: Humana Press; ), 313–329. 10.1007/978-1-62703-056-4_15 [DOI] [PubMed] [Google Scholar]
- Margueritat J., Virgone-Carlotta A., Monnier S., Delanoë-Ayari H., Mertani H. C., Berthelot A., et al. (2019). High-frequency mechanical properties of tumors measured by brillouin light scattering. Phys. Rev. Lett. 122:018101. 10.1103/PhysRevLett.122.018101 [DOI] [PubMed] [Google Scholar]
- Marjoram R. J., Guilluy C., Burridge K. (2016). Using magnets and magnetic beads to dissect signaling pathways activated by mechanical tension applied to cells. Methods 94 19–26. 10.1016/j.ymeth.2015.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthamuthu V., Sabass B., Schwarz U. S., Gardel M. L. (2011). Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc. Natl. Acad. Sci. U.S.A. 108 4708–4713. 10.1073/pnas.1011123108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu V., Fukui K., Matsukawa M., Kawabe M., Vayron R., Soffer E., et al. (2011). Micro-brillouin scattering measurements in mature and newly formed bone tissue surrounding an implant. J. Biomech. Eng. 133:021006 10.1115/1.4003131 [DOI] [PubMed] [Google Scholar]
- Megone W., Roohpour N., Gautrot J. E. (2018). Impact of surface adhesion and sample heterogeneity on the multiscale mechanical characterisation of soft biomaterials. Sci. Rep. 8:6780. 10.1038/s41598-018-24671-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirahmadi F., Tafazzoli-Shadpour M., Shokrgozar M. A., Bonakdar S. (2013). Enhanced mechanical properties of thermosensitive chitosan hydrogel by silk fibers for cartilage tissue engineering. Mater. Sci. Eng. C 33 4786–4794. 10.1016/j.msec.2013.07.043 [DOI] [PubMed] [Google Scholar]
- Morales-Rivas L., González-Orive A., Garcia-Mateo C., Hernández-Creus A., Caballero F. G., Vázquez L. (2015). Nanomechanical characterization of nanostructured bainitic steel: peak Force Microscopy and Nanoindentation with AFM. Sci. Rep. 5:17164. 10.1038/srep17164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam P. C., Gosvami N. N., Caporizzo M. A., Composto R. J., Carpick R. W. (2015). Nano-rheology of hydrogels using direct drive force modulation atomic force microscopy. Soft Matter 11 8165–8178. 10.1039/c5sm01143d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler B. (2007). Cantilever spring constant calibration using laser Doppler vibrometry. Rev. Sci. Instrum. 78:063701 10.1063/1.2743272 [DOI] [PubMed] [Google Scholar]
- Ostrofet E., Papini F. S., Dulin D. (2020). High spatiotemporal resolution data from a custom magnetic tweezers instrument. Data Br. 30:105397. 10.1016/j.dib.2020.105397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto O., Rosendahl P., Mietke A., Golfier S., Herold C., Klaue D., et al. (2015). Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nat. Methods 12 199–202. 10.1038/nmeth.3281 [DOI] [PubMed] [Google Scholar]
- Oyen M. L. (2014). Mechanical characterisation of hydrogel materials. Int. Mater. Rev. 59 44–59. 10.1179/1743280413Y.0000000022 [DOI] [Google Scholar]
- Patel J. M., Wise B. C., Bonnevie E. D., Mauck R. L. (2019). A systematic review and guide to mechanical testing for Articular cartilage tissue engineering. Tissue Eng. Part C Methods 25 593–608. 10.1089/ten.tec.2019.0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman E. J. G., Gittes F., Schmidt C. F. (2003). Laser-induced heating in optical traps. Biophys. J. 84(2 Pt 1), 1308–1316. 10.1016/S0006-3495(03)74946-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck W. J., Chen C. S. (2016). Measuring cell-generated forces: a guide to the available tools. Nat. Methods 13 415–423. 10.1038/nmeth.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevedel R., Diz-Muñoz A., Ruocco G., Antonacci G. (2019). Brillouin microscopy: an emerging tool for mechanobiology. Nat. Methods 16 969–977. 10.1038/s41592-019-0543-3 [DOI] [PubMed] [Google Scholar]
- Qian L., Zhao H. (2018). Nanoindentation of soft biological materials. Micromachines 9:654. 10.3390/mi9120654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randriantsilefisoa R., Hou Y., Pan Y., Camacho J. L. C., Kulka M. W., Zhang J., et al. (2020). Interaction of human mesenchymal stem cells with soft nanocomposite hydrogels based on polyethylene glycol and dendritic polyglycerol. Adv. Funct. Mater. 30:1905200 10.1002/adfm.201905200 [DOI] [Google Scholar]
- Sarkar R., Rybenkov V. V. (2016). A guide to magnetic tweezers and their applications. Front. Phys. 4:48 10.3389/fphy.2016.00048 [DOI] [Google Scholar]
- Scarcelli G., Yun S. H. (2008). Confocal Brillouin microscopy for three-dimensional mechanical imaging. Nat. Photonics 2 39–43. 10.1038/nphoton.2007.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli G., Yun S. H. (2018). Reply to ‘Water content, not stiffness, dominates Brillouin spectroscopy measurements in hydrated materials.’. Nat. Methods 15 562–563. 10.1038/s41592-018-0075-2 [DOI] [PubMed] [Google Scholar]
- Schillers H., Medalsy I., Hu S., Slade A. L., Shaw J. E. (2016). PeakForce Tapping resolves individual microvilli on living cells. J. Mol. Recognit. 29 95–101. 10.1002/jmr.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillers H., Rianna C., Schäpe J., Luque T., Doschke H., Wälte M., et al. (2017). Standardized nanomechanical atomic force microscopy procedure (snap) for measuring soft and biological samples. Sci. Rep. 7:5117. 10.1038/s41598-017-05383-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen I., Hu W., Klotzsch E., Vogel V. (2010). Probing cellular traction forces by micropillar arrays: contribution of substrate warping to pillar deflection. Nano Lett. 10 1823–1830. 10.1021/nl100533c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S., Engler A. J., Discher D. E. (2009). Matrix strains induced by cells: computing how far cells can feel. Cell. Mol. Bioeng. 2 39–48. 10.1007/s12195-009-0052-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakiba D., Alisafaei F., Savadipour A., Rowe R. A., Liu Z., Pryse K. M., et al. (2020). The balance between actomyosin contractility and microtubule polymerization regulates hierarchical protrusions that govern efficient fibroblast–collagen interactions. ACS Nano 14 7868–7879. 10.1021/acsnano.9b09941 [DOI] [PubMed] [Google Scholar]
- Shemesh J., Jalilian I., Shi A., Heng Yeoh G., Knothe Tate M. L., Ebrahimi Warkiani M. (2015). Flow-induced stress on adherent cells in microfluidic devices. Lab Chip 15 4114–4127. 10.1039/c5lc00633c [DOI] [PubMed] [Google Scholar]
- Sirghi L., Ponti J., Broggi F., Rossi F. (2008). Probing elasticity and adhesion of live cells by atomic force microscopy indentation. Eur. Biophys. J. 37 935–945. 10.1007/s00249-008-0311-2 [DOI] [PubMed] [Google Scholar]
- Song D., Dong L., Gupta M., Li L., Klaas O., Loghin A., et al. (2020). Recovery of tractions exerted by single cells in three-dimensional nonlinear matrices. J. Biomech. Eng. 142:081012 10.1115/1.4046974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. J., Brady-Kalnay S. M., McBride S. H., Phillips-Mason P., Dean D., Tate M. L. K. (2012). Mapping the mechanome of live stem cells using a novel method to measure local strain fields in situ at the fluid-cell interface. PLoS One 7:e43601. 10.1371/journal.pone.0043601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. J., Dean D., Knothe Tate M. L. (2010). In situ spatiotemporal mapping of flow fields around seeded stem cells at the subcellular length scale. PLoS One 5:e12796. 10.1371/journal.pone.0012796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. J., Dean D., Knothe Tate M. L. (2013). Mechanical modulation of nascent stem cell lineage commitment in tissue engineering scaffolds. Biomaterials 34 5766–5775. 10.1016/j.biomaterials.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steelman Z., Meng Z., Traverso A. J., Yakovlev V. V. (2015). Brillouin spectroscopy as a new method of screening for increased CSF total protein during bacterial meningitis. J. Biophotonics 8 408–414. 10.1002/jbio.201400047 [DOI] [PubMed] [Google Scholar]
- Steinwachs J., Metzner C., Skodzek K., Lang N., Thievessen I., Mark C., et al. (2016). Three-dimensional force microscopy of cells in biopolymer networks. Nat. Methods 13 171–176. 10.1038/nmeth.3685 [DOI] [PubMed] [Google Scholar]
- Subhash G., Liu Q., Moore D. F., Ifju P. G., Haile M. A. (2011). Concentration dependence of tensile behavior in agarose gel using digital image correlation. Exp. Mech. 51 255–262. 10.1007/s11340-010-9354-2 [DOI] [Google Scholar]
- Sweers K., van der Werf K., Bennink M., Subramaniam V. (2011). Nanomechanical properties of α-synuclein amyloid fibrils: a comparative study by nanoindentation, harmonic force microscopy, and Peakforce QNM. Nanoscale Res. Lett. 6:270. 10.1186/1556-276X-6-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Kong C. W., Chen S., Cheng S. H., Li R. A., Sun D. (2012). Probing the mechanobiological properties of human embryonic stem cells in cardiac differentiation by optical tweezers. J. Biomech. 45 123–128. 10.1016/j.jbiomech.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Thomson L., Schweins R., Draper E. R., Adams D. J. (2020). Creating transient gradients in supramolecular hydrogels. Macromol. Rapid Commun. 41:2000093 10.1002/marc.202000093 [DOI] [PubMed] [Google Scholar]
- Tibbitt M. W., Anseth K. S. (2009). Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 103 655–663. 10.1002/bit.22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timonen J. V. I., Grzybowski B. A. (2017). Tweezing of magnetic and non-magnetic objects with magnetic fields. Adv. Mater. 29:1603516. 10.1002/adma.201603516 [DOI] [PubMed] [Google Scholar]
- Ting T. C. T. (1964). The contact stresses between a rigid indenter and a viscoelastic half-space. J. Appl. Mech. 33 845–854. 10.1115/1.3625192 [DOI] [Google Scholar]
- Van Helvert S., Friedl P. (2016). Strain stiffening of fibrillar collagen during individual and collective cell migration identified by AFM Nanoindentation. ACS Appl. Mater. Interfaces 8 21946–21955. 10.1021/acsami.6b01755 [DOI] [PubMed] [Google Scholar]
- van Loenhout M. T. J., De Vlaminck I., Flebus B., den Blanken J. F., Zweifel L. P., Hooning K. M., et al. (2013). Scanning a DNA molecule for bound proteins using hybrid magnetic and optical tweezers. PLoS One 8:e65329. 10.1371/journal.pone.0065329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedadghavami A., Minooei F., Mohammadi M. H., Khetani S., Rezaei Kolahchi A., Mashayekhan S., et al. (2017). Manufacturing of hydrogel biomaterials with controlled mechanical properties for tissue engineering applications. Acta Biomater. 62 42–63. 10.1016/j.actbio.2017.07.028 [DOI] [PubMed] [Google Scholar]
- Viji Babu P. K., Rianna C., Mirastschijski U., Radmacher M. (2019). Nano-mechanical mapping of interdependent cell and ECM mechanics by AFM force spectroscopy. Sci. Rep. 9:12317. 10.1038/s41598-019-48566-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M. A., Gross S. P., Postner M., Block S. M., Wieschaus E. F. (1998). Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell 92 547–557. 10.1016/S0092-8674(00)80947-2 [DOI] [PubMed] [Google Scholar]
- Wu P. J., Kabakova I. V., Ruberti J. W., Sherwood J. M., Dunlop I. E., Paterson C., et al. (2018). Water content, not stiffness, dominates Brillouin spectroscopy measurements in hydrated materials. Nat. Methods 15 561–562. 10.1038/s41592-018-0076-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Friis E. A., Gehrke S. H., Detamore M. S. (2013). Mechanical testing of hydrogels in cartilage tissue engineering: beyond the compressive modulus. Tissue Eng. Part B Rev. 19 403–412. 10.1089/ten.teb.2012.0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Q., Li P., He Y., Shi C., Qiao Y., Bian X., et al. (2017). Magnetic tweezers for the mechanical research of DNA at the single molecule level. Anal. Methods 9 5720–5730. 10.1039/c7ay01495c [DOI] [Google Scholar]
- Yablon D. G. (ed.) (2013). Scanning Probe Microscopy for Industrial Applications: Nanomechanical Characterization. (Hoboken, NJ: John Wiley & Sons, Inc; ). 10.1002/9781118723111 [DOI] [Google Scholar]
- Yang K., Jung H., Lee H. R., Lee J. S., Kim S. R., Song K. Y., et al. (2014). Multiscale, hierarchically patterned topography for directing human neural stem cells into functional neurons. ACS Nano 8 7809–7822. 10.1021/nn501182f [DOI] [PubMed] [Google Scholar]
- Yango A., Schäpe J., Rianna C., Doschke H., Radmacher M. (2016). Measuring the viscoelastic creep of soft samples by step response AFM. Soft Matter 12 8297–8306. 10.1039/c6sm00801a [DOI] [PubMed] [Google Scholar]
- Yasunaga A., Murad Y., Li I. T. S. (2019). Quantifying molecular tension—classifications, interpretations and limitations of force sensors. Phys. Biol. 17:011001. 10.1088/1478-3975/ab38ff [DOI] [PubMed] [Google Scholar]
- Zimberlin J. A., Sanabria-DeLong N., Tew G. N., Crosby A. J. (2007). Cavitation rheology for soft materials. Soft Matter 3 763–767. 10.1039/b617050a [DOI] [PubMed] [Google Scholar]
- Ziv K., Nuhn H., Ben-Haim Y., Sasportas L. S., Kempen P. J., Niedringhaus T. P., et al. (2014). A tunable silk-alginate hydrogel scaffold for stem cell culture and transplantation. Biomaterials 35 3736–3743. 10.1016/j.biomaterials.2014.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuidema J. M., Rivet C. J., Gilbert R. J., Morrison F. A. (2014). A protocol for rheological characterization of hydrogels for tissue engineering strategies. J. Biomed. Mater. Res. Part B Appl. Biomater. 102 1063–1073. 10.1002/jbm.b.33088 [DOI] [PubMed] [Google Scholar]