Abstract
NFU1, a late-acting iron–sulfur (Fe–S) cluster carrier protein, has a key role in the pathogenesis of the disease, multiple mitochondrial dysfunctions syndrome. In this work, using genetic and biochemical approaches, we identified the initial scaffold protein, mitochondrial ISCU (ISCU2) and the secondary carrier, ISCA1, as the direct donors of Fe–S clusters to mitochondrial NFU1, which appears to dimerize and reductively mediate the formation of a bridging [4Fe–4S] cluster, aided by ferredoxin 2. By monitoring the abundance of target proteins that acquire their Fe–S clusters from NFU1, we characterized the effects of several novel pathogenic NFU1 mutations. We observed that NFU1 directly interacts with each of the Fe–S cluster scaffold proteins known to ligate [2Fe–2S] clusters, ISCU2 and ISCA1, and we mapped the site of interaction to a conserved hydrophobic patch of residues situated at the end of the C-terminal alpha-helix of NFU1. Furthermore, we showed that NFU1 lost its ability to acquire its Fe–S cluster when mutagenized at the identified site of interaction with ISCU2 and ISCA1, which thereby adversely affected biochemical functions of proteins that are thought to acquire their Fe–S clusters directly from NFU1, such as lipoic acid synthase, which supports the Fe–S-dependent process of lipoylation of components of multiple key enzyme complexes, including pyruvate dehydrogenase, alpha-ketoglutarate dehydrogenase and the glycine cleavage complex.
Introduction
Iron–sulfur (Fe–S) clusters (ISCs) are ancient protein metallo-cofactors found in virtually all living organisms that are required for many fundamental biochemical processes, including respiration, DNA replication and heme biosynthesis. ISCs generally exist in two main forms: [2Fe–2S] and [4Fe–4S] clusters. These cofactors are assembled by a highly conserved and complex biochemical pathway that operates independently in the cytosol and mitochondria of mammalian cells (1,2). The current understanding of the molecular mechanisms of mitochondrial Fe–S protein biogenesis has been worked out in bacterial, yeast and human cells as well as in animal models. De novo generation of a basic rhombic [2Fe–2S] cluster occurs through assembly on a scaffold protein, ISCU, which has a mitochondrial (ISCU2) and a cytosolic/nuclear isoform (ISCU1) (3). The reaction requires sulfane sulfur, which is provided by the cysteine desulfurase (NFS1) aided by two ancillary proteins, ISD11 and ACP1 (4–7). It remains unclear how iron is provided to the nascent Fe–S cluster. Two other proteins that are important for the de novo assembly of the cluster on ISCU2 include frataxin (FXN), which is thought to regulate NFS1 activity (8), and ferredoxin2 (FDX2), which is proposed to donate electrons for sulfur reduction (9–11). The newly assembled cluster is then transferred from ISCU2 to recipient proteins via multimolecular complexes composed of a chaperone/co-chaperone system and of intermediary transfer and secondary carrier proteins (1).
Multiple pathways for transfer of nascent clusters from ISCU2 to recipient proteins have been proposed. As elucidated in bacteria, yeast and human model systems, cluster release from mitochondrial ISCU to a subset of late-acting carrier proteins (12–14) is mediated by the ATPase activity of a chaperone (HSPA9 in humans) aided by a co-chaperone (HSC20 or HSCB in humans). HSC20 is thought to facilitate the transfer of the nascent Fe–S cluster from the scaffold protein to downstream carriers such as the monothiol glutaredoxin, GLRX5, found in the mitochondrial matrix (15,16) or directly to the target proteins such as SDHB (B subunit of succinate dehydrogenase) (12,17–19). GLRX5 is then proposed to heterodimerize with BOLA3 through shared bridging of a [2Fe–2S] cluster. Tetramerization of the BOLA3-GLRX5 dimers was recently shown in vitro to enable two [2Fe–2S] clusters to coalesce into a [4Fe–4S] cluster in the presence of reducing agents such as DTT and GSH, and the [4Fe–4S] cluster was subsequently transferred to downstream acceptors such as NFU1 (20). An alternative mechanism for [4Fe–4S] cluster formation proposed that the A-type ISC proteins, ISCA1 and ISCA2, and IBA57 received two [2Fe–2S] clusters from GLRX5 and assembled them into a [4Fe–4S] cluster that was transferred to NFU1 or BOLA3 or NUBPL (16,21). Recently, the assembly of a [4Fe–4S] cluster on ISCU in vitro and its direct transfer from the scaffold protein to purified NFU1 was also proposed, introducing yet another mechanism of [4Fe–4S] cluster assembly (22). Pathogenic mutations in NFU1, ISCA2, IBA57, BOLA3 and ISCA1 cause fatal infantile Multiple Mitochondrial Dysfunctions Syndrome (MMDS) types 1–5 (1,12,23). However, a clear pathway for Fe–S cluster transfer, likely mediated by these late-acting Fe–S cluster carriers (including ISCA proteins, NFU1, BOLA3 and GLRX5), from the primary scaffold to the target proteins in vivo has thus far remained elusive.
Studies in yeast and bacteria have characterized NFU1 as a non-essential, late-acting [4Fe–4S] cluster carrier protein (24,25). In these unicellular organisms, NFU1 appeared to play an essential role only under oxidative stress conditions. In humans, functional isoforms of NFU1 have been reported in both cytosol and mitochondria, and the purified NFU1 dimer has been shown to ligate a bridging [4Fe–4S] cluster (26–28). NFU1 is a bimodular protein, which contains two domains: a degenerate N-terminal domain (NTD) and a highly conserved C-terminal domain (CTD), which likely ligates the Fe–S cluster through its CXXC motif (26). The structures of both NTD-NFU1 and CTD-NFU1 were resolved by nuclear magnetic resonance (NMR) studies. Pathogenic mutations resulting in the depletion of functional NFU1 cause a rare infantile disease, Multiple Mitochondrial Dysfunctions Syndrome 1 (MMDS1) (29–32). The clinical symptoms usually include lactic acidosis, hyperglycinemia and reduced activities of mitochondrial respiratory complexes (29,30). Pulmonary hypertension and cardiomyopathy have also been reported in some of the patients with NFU1 mutations (29–31,33,34). Biochemical experiments on patient cells with NFU1 mutations have revealed decreased activities of several Fe–S cluster containing enzymes including lipoic acid synthase (LIAS), Complex-I (CI) and Complex-II (CII) (30,31). However, it remains unclear whether NFU1 directly delivers [4Fe–4S] clusters to a subset of recipient proteins or if it indirectly mediates Fe–S cluster transfer to downstream secondary carriers, which then transfer Fe–S clusters to recipient apo-proteins. The mechanism by which NFU1 acquires its Fe–S cluster in vivo has also thus far remained unknown. In this study, we report that NFU1 interacts directly with the main scaffold protein ISCU2 and with ISCA1 through a conserved hydrophobic motif to dimerize and build its [4Fe–4S] cluster. Dimerization enables NFU1 to assemble a [4Fe–4S] cluster when the two [2Fe–2S] clusters from each Fe–S donor, namely ISCU2 and ISCA1, coalesce to generate a [4Fe–4S] cluster that bridges the dimer, aided by the likely contribution of reducing equivalents from FDX2. Additionally, NFU1 appears to be a major carrier of [4Fe–4S] clusters, which is able to engage, through direct physical interaction, with a partially defined subset of target apo-proteins to deliver [4Fe–4S] clusters to recipient proteins, including LIAS and SDHB.
Results
A [4Fe–4S] cluster ligating human NFU1 interacted with the main mitochondrial Fe–S cluster scaffold protein ISCU2
A dimer of NFU1 has been reported to ligate a [4Fe–4S] cluster in vitro likely via the two conserved cysteine residues (C210 and C213) present at the C-terminus of each monomer (26–28). To validate this observation in vivo, we overexpressed and purified human NFU1 and the NFU1C210A_C213A mutant in which the two cysteines were replaced by alanines from Escherichia coli cells that co-expressed the ISC operon from Azotobacter vinelandii (Fig. 1A). As shown in Figure 1B, the as-purified NFU1-WT protein showed a broad shoulder at 425 nm, a distinctive feature of [4Fe–4S] cluster, which was clearly missing from the mutant NFU1C210A_C213A (Fig. 1B). Furthermore, the NFU1C210A_C213A mutant was unable to ligate the cluster even upon in vitro chemical reconstitution (Rc_NFU1C210A_C213A) (Fig. 1B). These results confirmed that NFU1 ligated a [4Fe–4S] cluster in vivo as a dimer via its conserved cysteine residues (C210 and C213) on each monomer.
Figure 1.
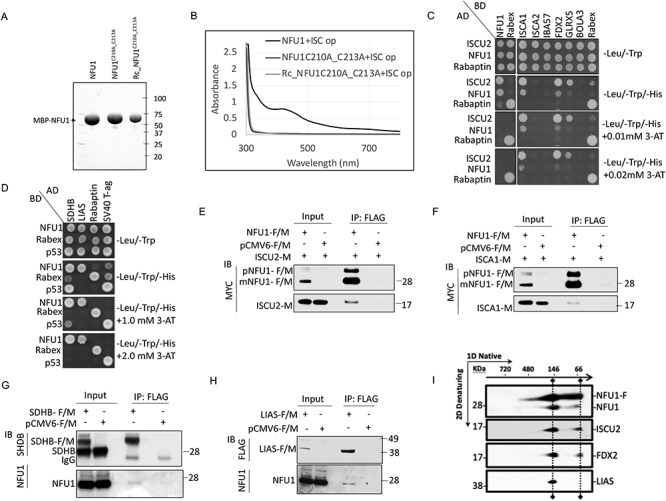
Mammalian NFU1 ligates a [4Fe–4S] cluster and directly interacts with ISCU2. (A) SDS-PAGE separation of the purified proteins on a stain-free denaturing gel. (B) UV–visible spectra of as-purified MBP-NFU1, MBP-NFU1C210_C213A and chemically reconstituted-Rc_MBP-NFU1C210_C213A proteins from E. coli co-expressing the ISC operon from A. vinelandii. The MBP-tagged proteins were purified using amylose resin affinity purification followed by the HiTrap Q column ion-exchange purification. ISC, Iron-Sulfur Cluster; Op, operon; Rc, chemically reconstituted. (C and D) Y2H assays. (C) Prey vectors expressing GAL4-AD fused with ISCU2, NFU1 or Rabaptin were co-expressed with the bait vectors expressing GAL4-BD fused with NFU1, ISCA1, ISCA2, IBA57, FDX2, GLRX5, BOLA3 or Rabex, as indicated in AH109 yeast cells. Direct interactions between preys and baits were tested by Y2H assay in -Leu/-Trp/-His media. 3-AT, a competitive inhibitor of HIS3, was used to increase the stringency of the assay. (D) SDHB, LIAS, Rabaptin or SV40-T-Ag, each cloned in the prey vector, were co-transformed with NFU1, RABEX5 or p53 in the bait vector in AH109 yeast cells and tested for direct interactions via Y2H assay. (E and F) Total cell lysates from HeLa cells co-transfected with (E) pCMV6-NFU1-F/M + ISCU2-M or pCMV6-F/M + ISCU2-M or (F) pCMV6-NFU1-F/M + ISCA1-M or pCMV6-F/M + ISCA1-M were subjected to IP with anti-FLAG beads (mouse) followed by immunoblot (IB) with anti-MYC (rabbit). (G and H) Total cell lysates from HeLa cells transfected with (G) pCMV6-SDHB-F/M or (H) pCMV6-LIAS-F/M were subjected to IP with anti-FLAG beads (mouse) followed by IB with anti-NFU1 (rabbit), anti SDHB (mouse) or anti-FLAG (rabbit). Total cell lysates expressing the empty vector (pCMV6-F/M) were used as controls. The input loaded on the gel is a small fraction (5–10%) of the total sample loaded for IP, and therefore, an enrichment of the FLAG-tagged proteins is observed in the IP samples. (I) 2D native/SDS-PAGE immunoblots identified two NFU1-containing complexes isolated from mitochondrial extracts of HEK293 cells transfected with FLAG-tagged NFU1. Abbreviations: pNFU1-F/M: FLAG and MYC tagged precursor-NFU1 (1–254); mNFU1-F/M: FLAG and MYC tagged mature mitochondrial isoform of NFU1; NFU1-F: FLAG-tagged NFU1.
To establish the position of NFU1 in the sequence of events that lead to Fe–S cluster biogenesis/transfer and to determine whether it functioned as a late-acting carrier protein that acquired its cluster from intermediary chaperones or directly acquires its Fe–S cluster from the initial mitochondrial Fe–S cluster scaffold protein (ISCU2), we examined its potential direct interacting partners among the candidate ISC biogenesis proteins (ISCU2, NFU1, ISCA1, ISCA2, IBA57, FDX2, GLRX5, BOLA3) by yeast two-hybrid (Y2H) assay. Our results revealed that NFU1 interacted directly with NFU1, ISCU2, ISCA1 and FDX2 (Fig. 1C). While the interaction between NFU1-bait and NFU1-prey was consistent with the already known ability of NFU1 to dimerize, the interaction of NFU1 with the electron donor, FDX2, and the two scaffold proteins ISCU2 and ISCA1 (Fig. 1C) suggested the possibility that assembly of the [4Fe–4S] cluster on NFU1 required acquisition of two [2Fe–2S] clusters, each donated by ISCU2 and ISCA1, which coalesced through reductive coupling into a [4Fe–4S] cluster, utilizing electrons provided by FDX2 (9,15,35).
We also detected direct interaction of ISCU2 with NFU1, ISCA1, FDX2 and GLRX5 (Fig. 1C). These results suggest that NFU1, ISCA1 and GLRX5 may independently acquire their clusters directly from ISCU2 before diverging to serve their target proteins. Additionally, we tested the interaction of NFU1 with its two proposed target proteins, SDHB and LIAS (30,31), and observed direct interactions between NFU1 and SDHB as well as LIAS via the Y2H assay (Fig. 1D). Remarkably, the interaction of NFU1 with its targets was observed under highly stringent conditions, with 3-AT concentrations as high as 2 mm, whereas its interactions with ISCU2 and ISCA1 were detected only on plates with negligible amounts of 3-AT, suggesting that the affinity of the interaction between NFU1 and either ISCU2 or ISCA1 was low, as expected for a transient interaction (36). Although it is possible that the interactions of apo-NFU1 (likely a less stable form) with ISCU2 and ISCA1 are weak and transient as opposed to its interaction with the targets when NFU1 has been stabilized by the [4Fe–4S] cluster, it is also plausible that the interaction of NFU1 with an individual donor (ISCU2 or ISCA1) that appears weak in the binary setting of the Y2H assay could be strengthened if the donor proteins were present as a complex with the right ancillary/partner protein, and therefore, the strength of the interaction of NFU1 with its donors or targets may be different in vivo. The binding affinities between the interacting proteins are a subject for further investigation.
Co-immunoprecipitation (co-IP) assays in mammalian cells confirmed these interactions between Fe–S donors and NFU1 and the NFU1 targets in vivo (Fig. 1E–H and Supplementary Material, Fig. S1A–C). The interactions between NFU1 and ISCU2 (Fig. 1E and Supplementary Material, Fig. S1A) and between NFU1 and ISCA1 (Fig. 1F and Supplementary Material, Fig. S1B) were confirmed by reciprocal co-IP experiments. Similarly, reciprocal co-IP experiments validated the interaction of NFU1 with two target proteins, SDHB (Fig. 1G and Supplementary Material, Fig. S1C) and LIAS (Fig. 1H and Supplementary Material, Fig. S1D), in the mammalian cells. Moreover, by analyzing the protein complexes that co-immunoprecipitated with FLAG-tagged NFU1 (NFU1-F) by 2D native/SDS-PAGE, we were able to characterize the composition of two main complexes containing mitochondrial NFU1 in vivo (Fig. 1I). The lower molecular weight (MW) complex that had an apparent MW of 66 kDa on the native gel (first dimension) contained endogenous NFU1, which confirmed that NFU1 dimerized in vivo, and also bound ISCU2 and FDX2. Although we were unable to detect ISCA1 in the complex, due to the low sensitivity of the anti-ISCA1 antibody, the direct interaction observed between NFU1 and ISCA1 in Y2H and mammalian co-IP experiments (Fig. 1C and F and Supplementary Material, Fig. S1B) suggests that ISCA1 co-exists along with an NFU1 dimer, ISCU2 and FDX2 in a quaternary complex. A second complex of higher MW (~146 kDa) contained LIAS in addition to the components detected in the complex at ~66 kDa. The co-existence of LIAS with NFU1 and the Fe–S cluster scaffold, ISCU2, in a single complex suggests that the assembly and transfer of the [4Fe–4S] cluster from NFU1 occur almost contemporaneously, thereby likely protecting the Fe–S cluster from degradation by cellular oxidants during the assembly and transfer process. Interestingly, FDX2 was found to associate with the complex at ~146 kDa (Fig. 1I). The presence of FDX2 in this complex may be required to enable the reductive coupling of the two [2Fe–2S] clusters donated by ISCU2 and ISCA1 into the [4Fe–4S] ligated by NFU1. Another interesting possibility for the presence of FDX2 in the higher MW complex comes from studies in bacteria that reported the association of bacterial ferredoxins with the S-adenosyl-methionine (SAM) [4Fe-4S] cluster in the prokaryotic radical SAM enzyme MiaB, in order to maintain the cluster in the catalytically active +1 oxidation state (37–40). FDX2 may perhaps donate one electron to the SAM-ligating [4Fe–4S]2+ of LIAS to reduce it to [4Fe–4S]+1. Taken together, these results demonstrate that NFU1, which ligates a [4Fe–4S] cluster, interacted with two potential Fe–S cluster donors, ISCU2 and ISCA1, with the electron donor, FDX2, and with two Fe–S recipient proteins, SDHB and LIAS.
NFU1 interacted with ISCU2 via a hydrophobic domain identified by site-directed mutagenesis
To map the site of interaction with its potential Fe–S cluster donor proteins on NFU1, we used mutational/deletional analysis approaches in conjunction with Y2H screens. We first investigated the site of interaction between NFU1 and ISCU2. To identify the domain of NFU1 involved in the interaction with ISCU2, the constructs encoding NTD-NFU1 (25–150) and CTD-NFU1 (150–254) along with the full-length NFU1 (25-254) were tested in Y2H assays. Consistent with the previous observations (22), we detected a strong interaction between ISCU2 and NFU1 mediated by its CTD but not its NTD (Fig. 2A). Similarly, only CTD-NFU1 and not NTD-NFU1 appeared to dimerize with NFU1 (Supplementary Material, Fig. S2A), which argued against the possibility of formation of a holo-NFU1 oligomer mediated by its NTD residues (26). Deletion of the NTD from NFU1 appeared to strengthen the interaction of the CTD of NFU1 with ISCU2, likely because the NTD caused steric hindrance. Thus, our data suggest that the NTD may contribute to the transient nature of the interaction between NFU1 and ISCU2. In contrast, the target protein, SDHB, interacted strongly with both the NTD and CTD of NFU1 (Fig. 2A). Interestingly, LIAS interacted only with specific conserved residues of CTD-NFU1 (Supplementary Material, Fig. S2B), unlike the bacterial homologue NfuA, which has a unique NTD that differs significantly from human NFU1 (41). To summarize, these results suggest that the Fe–S cluster ligating CTD of NFU1 physically interacted with both the Fe–S cluster donor and recipient proteins.
Figure 2.
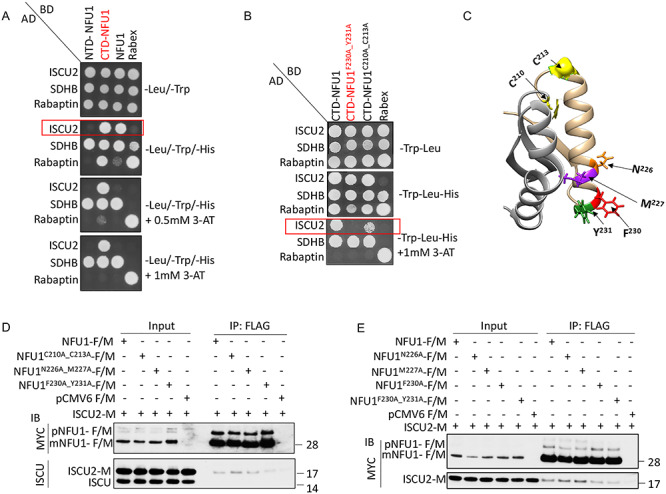
NFU1 interacts with ISCU2 via a hydrophobic FY motif in its CTD. (A) NTD-NFU1 (25–150), CTD-NFU1 (150–254; highlighted in red) and NFU1 (25–254) were cloned into the bait vector and assayed for a direct interaction with ISCU2 or SDHB cloned in the prey vector via Y2H assay. (B) Y2H assay to test the interaction between baits (CTD-NFU1, NFU1F230A_Y231A (highlighted in red), NFU1C210A_C213A or Rabex) and preys (ISCU2, SDHB or Rabaptin). The red boxes highlight the distinct interaction patterns of the baits with ISCU2-prey. Rabex and Rabaptin were used as controls. 3-AT, a competitive inhibitor of HIS3, was used to increase the stringency of the assay. (C) Ribbon representation of the NMR structure of CTD-NFU1 (PDB: 2M5O) highlighting the residues (F230, Y231, N226, M227) involved in its interaction with ISCU2. Cysteines C210 and C213 that ligate the [4Fe–4S] cluster are shown in yellow. CTD-NFU1 (150–211) and CTD-NFU1 (212–254) are represented in grey and tan, respectively. (D and E) HeLa cell extracts co-expressing ISCU2-M and NFU1-F/M or alanine mutants of NFU1-F/M or the empty vector (pCMV6) were immunoprecipitated with anti-FLAG beads (mouse) and immunoblotted with anti-MYC (rabbit) or anti-ISCU (rabbit) antibodies. The input loaded on the gel is a small fraction (5–10%) of the total sample loaded for IP, and therefore, an enrichment of the FLAG-tagged precursor (pNFU1) and mitochondrial NFU1 (mNFU1) bands is observed in the IP samples. These figures are a representative of a result that was obtained at least three times.
We then performed extensive mutational analysis of several conserved residues and peptide sequences in the CTD of NFU1. A comprehensive Y2H screening of the mutants revealed that a surface-exposed, hydrophobic motif, consisting of F230 and Y231, mediated an interaction between CTD-NFU1 and ISCU2 (Supplementary Material, Figs S2C–E and S3). Additionally, the polar residue N226 also appeared to be important for this interaction (Supplementary Material, Fig. S2D). The double mutation of F230A_Y231A on CTD-NFU1 completely abrogated the interaction between NFU1 and ISCU2. In contrast, the interaction with the recipient protein, SDHB, was not affected (Fig. 2B). Additionally, the double mutations, C210A_C213A, did not affect the interaction of NFU1 with either ISCU2 or SDHB, indicating that the site for cluster ligation (CXXC) is distinct from the site that mediates initial cluster acquisition (FY) by NFU1 (Fig. 2B and C).
When the effect of these mutations on the interaction of NFU1 with ISCU2 was measured en bloc in HeLa cells, similar results were observed. While NFU1C210A_C213A and NFU1N226A_M227A interacted with ISCU2 similar to the WT-NFU1; NFU1F230A_Y231A exhibited a significant loss of interaction with ISCU2 (Fig. 2D). We tested the effects of N226A and M227A mutations separately and found that the substitution of either of these two residues individually resulted in a stronger interaction between NFU1 and ISCU2 (Fig. 2E), suggesting that N226 and M227 may contribute to the transient nature of this interaction, perhaps because their polarity diminished affinity. Taken together, these results suggest that the residues N226M227 appeared to play an important indirect role in maintaining the transient nature of the interaction between NFU1 and ISCU2, whereas the F230Y231 motif of NFU1 was involved in a direct interaction with ISCU2 (Fig. 2C).
Reciprocally, the major molecular determinants of the CTD-NFU1/ISCU2 interaction were also identified on the primary scaffold ISCU2 using deletional/mutational analyses of conserved residues on ISCU2 in conjunction with Y2H assays. Six residues (V72, M73, V79, G83, F90 and F94) on three different peptides of ISCU2 were found to play a significant role in its interaction with CTD-NFU1 (Fig. 3A and B). Interestingly, four of these residues (namely, V72, M73, F90 and F94) have been previously identified as being involved in the mutually exclusive interaction of ISCU2 with either the cysteine desulfurase NFS1 or with the co-chaperone Jac1, the yeast ortholog of HSC20 (13). The residues on Peptide #2 of ISCU2 (comprising amino acid residues V79-G83) appeared to be far from the Fe–S cluster binding site on the main scaffold and thus might contribute to anchoring the NFU1 dimer to ISCU2 (Fig. 3B). Nevertheless, the site of interaction of ISCU2 with NFU1 appeared to be positionally advantageous for the transfer of the Fe–S cluster to the interacting NFU1 protein (Fig. 3C).
Figure 3.
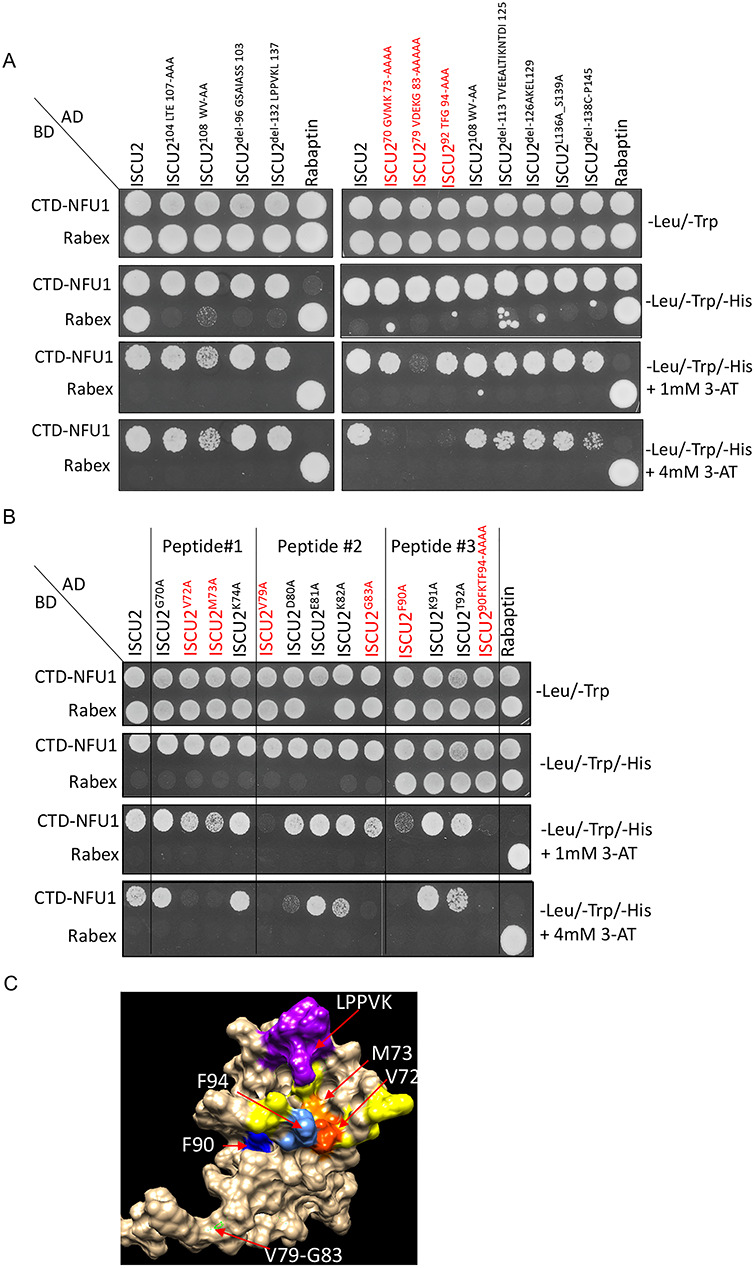
Identification of the amino acid residues on ISCU2 involved in its interaction with NFU1. (A) Mutational and deletional analyses of several stretches on ISCU2 protein sequence yielded three conserved peptides that showed interaction with CTD-NFU1 by Y2H assay. (B) Mutagenesis of individual amino acids present on the three peptides of the protein that were identified to interact with CTD-NFU1 using Y2H assay. ISCU2 mutants shown in red indicate the amino acids involved in its interaction with CTD-NFU1. CTD-NFU1 and Rabex were cloned into the bait vector and ISCU2 and Rabaptin were cloned into the prey vector. Rabaptin-Prey and Rabex-Bait were used as controls. (C) Surface representation of the NMR structure of ISCU2 (PDB: 2L4X) highlighting the residues involved in its interaction with NFU1 (red: V72, orange: M73, green: V79-G83, deep blue: F90, corn blue: F94) , the Fe–S cluster ligating residues (yellow) and the LPPVK motif (purple), which is the binding site for HSPA9.
The ISCA1 binding site on NFU1 overlapped with the binding site for ISCU2
We then investigated the potential involvement of the FY motif of NFU1 in its interactions with the other binding partners identified in our study, ISCA1 and FDX2 (Fig. 1 and Supplementary Material, Fig. S1). The Y2H screening on the NFU1 mutants (NFU1F230A_Y231A and NFU1C210A_C213A) revealed that while these mutations had no effect on the interaction of NFU1 with FDX2, they impaired the interaction between NFU1 and ISCA1 (Fig. 4A). Similar observations in mammalian cells by co-IP experiments confirmed that the loss of C210_C213 as well as F230_Y231 motifs of NFU1 significantly inhibited its interaction with ISCA1 in vivo (Fig. 4B). Interestingly, the substitution of N226M227 residues (which influenced binding to ISCU2) into alanines did not affect the interaction of NFU1 with ISCA1 (Fig. 4B). Overall, these results indicated that ISCA1 interacted with NFU1 at the same hydrophobic site (F230_Y231) as ISCU2, suggesting that this motif of NFU1 functioned in engaging its two potential donors, namely ISCU2 and ISCA1, each able to interact with an NFU1 monomer of the dimeric functional [4Fe–4S]-NFU1 through binding to the FY motif. Additionally, the loss of interaction between ISCA1 and NFU1C210A_C213A mutant suggests the possibility that ligation of the Fe–S cluster between the ISCA1 and NFU1 heterodimer may stabilize the interaction between these two proteins, as was previously reported for the bacterial heterodimer formed by the A-type carrier ErpA and NfuA (42). Alternatively, it is possible that ISCA1 is recruited by NFU1 once the C210 and C213 of NFU1 are engaged in ligating a Fe–S cluster.
Figure 4.
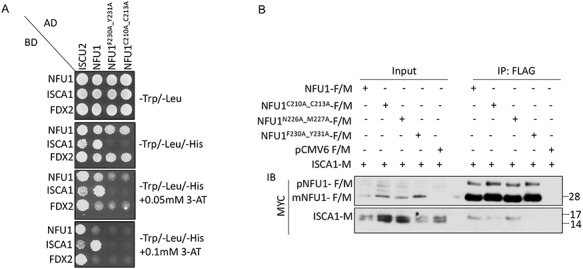
ISCA1 interacts with NFU1 via the same hydrophobic motif (F230_Y231) that mediated the interaction between NFU1 and ISCU2 and also requires C210_C213. (A) Alanine mutants of NFU1 (NFU1F230A_Y231A, NFU1C210A_C213A) along with full-length NFU1 and ISCU2 cloned in the prey vector were assessed for an interaction with full-length NFU1, ISCA1 or FDX2 cloned in the bait vector via Y2H assay. (B) HeLa cell extracts co-expressed with ISCA1-M and NFU1-F/M or alanine mutants of NFU1-F/M or the empty vector (pCMV6-F/M) were immunoprecipitated with anti-FLAG beads (mouse) and immunoblotted with anti-MYC antibody (rabbit). The input loaded on the gel is a small fraction (5–10%) of the total sample loaded for IP, and therefore, the FLAG-tagged precursor (pNFU1) and mitochondrial NFU1 (mNFU1) bands are enriched in the IP samples. The co-IP data presented here are a representative of results that were obtained at least three times.
Human [2Fe–2S] cluster-ISCA1 is required for cluster formation on NFU1
To address whether the interaction between [2Fe–2S]-ISCA1 and NFU1 was physiologically relevant for cluster acquisition by NFU1, we first investigated the stoichiometry of the Fe–S cluster ligated by human ISCA1. We co-expressed full-length human ISCA1-MBP with the ISC operon and purified the protein anaerobically from E. coli. The as-purified ISCA1-MBP protein eluted as a single band in a brown colored solution (Fig. 5A and B), which showed the UV–visible spectrum (a peak at 320 as well as 420 nm) characteristic of [2Fe–2S] clusters (Fig. 5C), as was observed previously for the murine ISCA1 and (43). These results support that human ISCA1 is a [2Fe–2S] cluster ligating protein.
Figure 5.
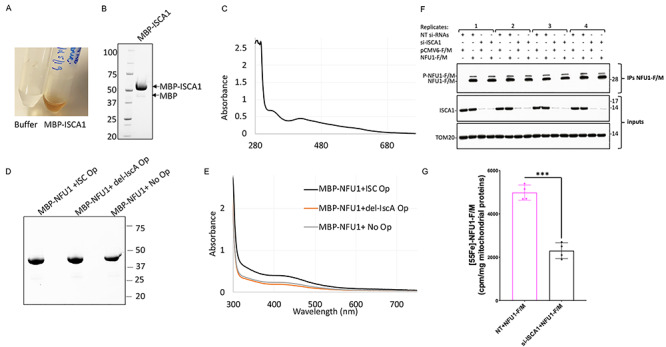
Human ISCA1 ligates a [2Fe–2S] cluster and is required for cluster formation on NFU1. (A) The as-purified MBP-ISCA1 protein from E. coli co-expressing the ISC operon from A. vinelandii eluted as a brown colored solution from the amylose resin. (B) SDS-PAGE separation of purified MBP-ISCA1 protein on a stain-free denaturing gel. (C) UV–visible spectrum of as-purified MBP-ISCA1 shows the characteristic features of a [2Fe–2S] cluster (43). (D) SDS-PAGE separation and (E) UV–visible spectrum of MBP-NFU1 purified from E. coli co-expressing either WT ISC operon (ISC Op), ISC operon with deleted IscA (del-IscA Op) or no operon (No Op) on a stain-free denaturing gel. The MBP-tagged proteins were purified using amylose resin affinity purification followed by the HiTrap Q column ion-exchange purification. (F) IBs on samples as those analyzed in (G) for the 55Fe incorporation into NFU1-F/M showed effective knockdown of ISCA1 in mammalian cells transfected with siRNAs targeting the ISCA1 mRNA and efficient immunoprecipitation of recombinant NFU1-F/M. TOM20 was used as a loading control (G) 55Fe incorporation into NFU1-F/M assessed by liquid scintillation counting showed significantly decreased levels of radioactive iron incorporated into NFU1-F/M upon KD of ISCA1. The background, corresponding to 55Fe measurements of eluates after anti-FLAG immunoprecipitations on mitochondrial extracts from cells transfected with the empty vector, was subtracted from each reading. Unpaired t-test analyses of 55Fe labeling experiments were performed with GraphPad Prism 7. ***P < 0.001.
Subsequently, to determine if the [2Fe–2S] cluster bound ISCA1 was a physiological Fe–S cluster donor to NFU1, we monitored the effect of loss of IscA (the bacterial homologue of ISCA1) on cluster acquisition by overexpressing the recombinant human NFU1 in E. coli cells that overexpressed the ISC operon. Accordingly, we modified the ISC operon from A. vinelandii by making an in-frame deletion of the IscA gene from the operon (del-IscA Op) and then co-expressing it with human NFU1 (MBP-NFU1) in E. coli. Cultures of MBP-NFU1 co-expressed with (+ISC Op) or without (No Op) the ISC operon were used as positive and negative controls, respectively. Highly pure MBP-NFU1 proteins were anaerobically purified from all three bacterial cell lysates (Fig. 5D). Interestingly, the UV–visible spectrum of MBP-NFU1 purified from cells expressing the ISC operon lacking IscA (IscA-del Op) showed similar cluster occupancy as the MBP-NFU1 purified from cells without the operon, which was ~50% lower than the levels of cluster occupancy of NFU1 purified from cells expressing the WT-ISC operon (Fig. 5E). The residual cluster occupancy observed in the spectrum of NFU1 purified in the absence of the operon or in the IscA-del Op cells was likely to have been contributed by the endogenous ISC proteins of E. coli. These results suggest that IscA is required for cluster acquisition by NFU1 in vivo in E. coli. We did not perform the deletion of the main scaffold IscU from the ISC operon as it is essential for Fe–S cluster assembly (44–46).
To confirm the role of ISCA1 as the Fe–S cluster donor to NFU1 in mammalian cells, we examined 55Fe-incorporation by NFU1 in cells in which ISCA1 expression had been silenced. HEK293 cells cultured in the presence of 55Fe-Tf were transfected with small interfering RNAs (siRNAs) targeting ISCA1 mRNA to knockdown its expression and co-transfected at the time of the second transfection with siRNAs against ISCA1 with either NFU1-F/M or pCMV6-Entry plasmids. The incorporation of 55Fe on immunoprecipitated NFU1-F/M from control or ISCA1 knock down cell lines was then measured by liquid scintillation counting. We observed a 54% decrease in the 55Fe signal associated with NFU1-F/M immunoprecipitated from ISCA1 knockdown cells as opposed to control, supporting that ISCA1 was involved in Fe–S cluster donation to NFU1 (Fig. 5F and G).
NFU1FY-AA mutant was unable to acquire the [4Fe–4S] cluster from the donor proteins
To investigate if the FY motif (F230_Y231) of NFU1 that mediated its interaction with ISCU2 and ISCA1 was in fact necessary for the Fe–S cluster acquisition by NFU1, we purified MBP-NFU1 and MBP-NFU1FY-AA mutant anaerobically from E. coli co-expressing the ISC operon (Fig. 6A). As opposed to the WT-NFU1, the NFU1FY-AA mutant lacked the brown color (Fig. 6B) as well as the shoulder around 425 nm as observed on the UV–visible spectrum of WT-NFU1, indicating the absence of a [4Fe–4S] cluster on the NFU1FY-AA mutant (Fig. 6C). To determine if the absence of cluster ligation on NFU1FY-AA mutant was in fact due to its inability to acquire Fe–S clusters from its physiological donors, we performed in vitro chemical reconstitution of the cluster on apo-NFU1 and apo-NFU1FY-AA. Both WT and NFU1FY-AA, after chemical reconstitution, showed the shoulder at 425 nm, which is a characteristic feature of a [4Fe–4S] cluster, on their UV–visible spectrum (Fig. 6D and E). These results suggest that while NFU1FY-AA was able to ligate a [4Fe–4S] cluster in vitro, the FY motif on NFU1 was important for the physiological acquisition of the Fe–S cluster from the biogenesis machinery in vivo. Additionally, we purified the NFU1NM-AA mutant and observed a slightly lower cluster occupancy than the WT on the as-purified mutant protein (Fig. 6F and G). Taken together, these data suggest that the NM motif (N226_M227), which indirectly participated in the interaction of NFU1 with ISCU2, also contributed to the cluster acquisition by NFU1. Furthermore, the FY motif of NFU1, which mediated direct interactions with both ISCU2 and ISCA1, was essential for the in vivo Fe–S cluster acquisition from these donors.
Figure 6.
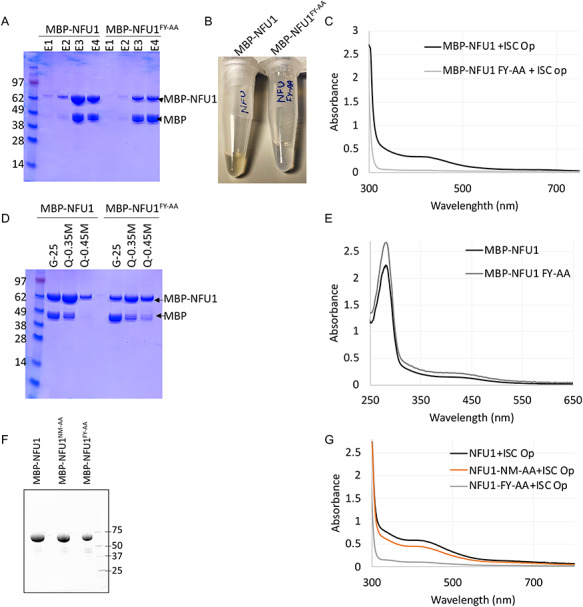
NFU1FY-AA mutant cannot acquire an Fe–S cluster in vivo, but the NFU1FY-AA mutant can acquire the Fe–S cluster upon in vitro chemical reconstitution. (A) Coomassie-stained SDS gel of the eluates (E1, E2, E3, E4) obtained upon affinity purification by amylose resin of MBP-NFU1 (WT) and MBP-NFU1FY-AA proteins. (B) E3-MBP-NFU1 solution was brown in color, whereas E3- MBP-NFU1FY-AA was not. (C) UV–visible spectra of anaerobically purified (E3) MBP-NFU1 (WT) and (E3) MBP-NFU1FY-AA from E. coli co-expressing the ISC operon from A. vinelandii. (D) Coomassie-stained SDS gel of the chemically reconstituted purified proteins obtained from the desalting column (G-25) and the HiTrap Q column at 0.35 and 0.45 M NaCl concentration. (E) UV–visible spectra of anaerobically reconstituted and HiTrap Q column purified MBP-NFU1 (WT) and MBP-NFU1FY-AA at 0.35 M NaCl concentration. (F) Stain-free SDS PAGE separation and (G) UV–visible spectra of as-purified MBP-NFU1 (WT) and MBP-NFU1FY-AA and MBP-NFU1NM-AA proteins.
Complementation with WT-NFU1 restored the biochemical defects of NFU1-deficient patient cells whereas complementation with either NFU1FY-AA or NFU1NM-AA did not
NFU1 is believed to serve as a [4Fe–4S] cluster carrier for several mitochondrial proteins including SDHB and LIAS (30), and the defects in NFU1 have been shown to result in biochemical defects such as reduced activities of CI, CII and LIAS. Based on the direct interactions observed between NFU1 and SDHB/LIAS and the defects displayed by NFU1FY-AA and NFU1NM-AA mutants in the cluster acquisition, we speculated that these mutants would mirror the NFU1-deficiency phenotype resulting in loss of SDHB and LIAS activities. To investigate the in vivo effects of NM-AA and FY-AA substitutions on the function of NFU1, we used a previously characterized NFU1 patient-derived fibroblast cell line (NFU1 Pt: c.545G>A (p.Arg182Gln)) (30) and assessed the levels of various Fe–S cluster containing proteins upon complementation with either the WT (+NFU1-F/M) or NFU1 mutants (+NFU1NM-AA-F/M and + NFU1FY-AA-F/M). Lysates from MCH46 and the NFU1 Pt cells transfected with the empty vector (+pCMV6-F/M) were used as positive and negative controls, respectively. As observed in Figure 7A, a decrease in the LIAS activity reflected by diminished lipoylated pyruvate dehydrogenase complex (LA-PDH) and lipoylated α-ketoglutarate dehydrogenase (LA-KGDH) in the NFU1 Pt cells was partially restored by the complementation with the WT-NFU1 but not upon complementation with NFU1FY-AA or NFU1NM-AA (Fig. 7A) and a subtle but similar trend was observed in the protein levels of SDHB. Protein levels of another [4Fe–4S] cluster containing enzyme, ACO2, did not change, and SDHA (which functions in close association with SDHB) protein levels were stable, whereas levels of the extra-mitochondrial [4Fe–4S] cluster protein ABCE1 increased in the NFU1 Pt cells as compared to the WT, for unknown reasons (Fig. 7A). Interestingly, the protein levels of both ISCU and ISCA1 were consistently upregulated in the NFU1 Pt cells, suggesting that an uncharacterized mechanism of regulating in the ISC biogenesis pathway was induced to increase expression of donor Fe–S proteins to compensate for the loss of functional NFU1 (Fig. 7A). We also assessed the activities of respiratory complexes (CI and CII) by the in-gel activity assay. While CI seemed to be more adversely affected than CII in the NFU1 Pt cells, the activities of both complexes were only marginally restored by complementation with WT-NFU1 (Fig. 7B). The fact that the complementation of the Pt cells with WT-NFU1 restored lipoylation more efficiently than the activities of CI and CII at the time of analysis post-complementation suggests that NFU1 may be directly involved in cluster transfer to LIAS, but perhaps indirectly involved in cluster transfer to the CI or CII subunits. The differences in the degree of complementation can be partially attributed to direct versus indirect effects due to loss of functional NFU1 on CI and CII activities, as well as to the fact that the complementation assays were performed by transiently transfecting WT-NFU1 in the patient cells. Taken together, these results indicate that the association and dissociation of NFU1 with its Fe-S cluster donor proteins mediated by FY and NM residues, respectively, were critical for the physiological role of NFU1, i.e., to deliver the Fe-S clusters to its target proteins.
Figure 7.
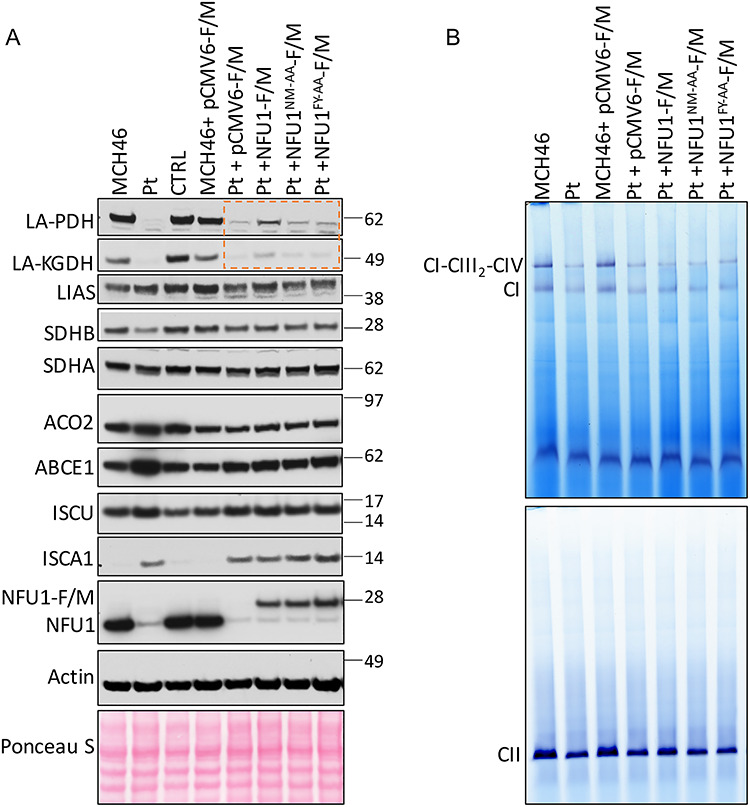
Biochemical deficiencies in NFU1R182Q patient-derived fibroblasts can be partially rescued by WT-NFU1 but not by NFU1N226A_M227A or NFU1F230A_Y231A. (A) Immunoblots on total cell lysate were performed with antibodies to the proteins labeled on the vertical axis, and (B) in-gel assay for CI and CII on the mitochondrial extracts of MCH46, CTRL and NFU1R182Q patient (Pt) fibroblast mitochondrial lysates complemented with constructs expressing WT-NFU1-F/M, NFU1N226A_M227A-F/M, NFU1F230A_Y231A-F/M, NFU1C210A_C213A-F/M or the empty vector pCMV6-F/M. Rescue of lipoylation by transfection with NFU1-F/M, but not by NFU1FY-AA-F/M or NFU1NM-AA-F/M mutants, is highlighted by the orange box in A.
ISCA1 functioned upstream of NFU1 and responded to the loss of function of NFU1 in vivo
To address if ISCA1 functioned upstream of NFU1 as an Fe–S cluster donor, we studied the biochemical effects elicited by silencing ISCA1 in WT and NFU1 Pt cells. An effective knockdown of ISCA1 was achieved by performing two rounds of siRNA transfections over 6 days. As previously reported (21), the levels of both NFU1 and ISCU did not seem to be significantly affected by the loss of ISCA1 (Fig. 8A). Conversely, the loss of NFU1 in Pt cells correlated with increased levels of ISCU and ISCA1 (Fig. 8A). Furthermore, we observed that loss of either NFU1 or ISCA1 caused a decrease in protein levels of LA-PDH and LA-KGDH, which was much more pronounced when both ISCA1 and NFU1 were simultaneously decreased through silencing of ISCA1 in NFU1 Pt cells (Fig. 8A). The profound loss in the activity of LIAS caused by simultaneous loss of functional NFU1 and ISCA1 suggested that ISCA1 was involved in the process of lipoylation, independent of NFU1. Furthermore, a profound loss of SDHB protein levels upon the knockdown of ISCA1 but a marginal decrease in the NFU1 Pt cells suggested that the two proteins are engaged in different Fe–S cluster delivery pathways. Thus, based on these results, LIAS appeared to be a clear primary target of NFU1. Taken together, these results indicate that ISCA1 functioned as an upstream Fe–S cluster donor that donated its cluster to other mitochondrial targets in addition to
Figure 8.
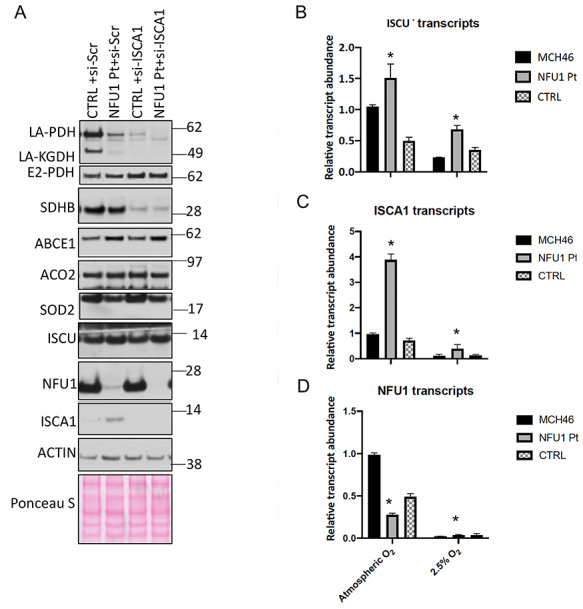
ISCA1 is required for NFU1-mediated lipoylation and SDHB stabilization in human cells. (A) NFU1 and ISCA1 independently affected Fe–S cluster proteins in human cells. Control (CTRL) and NFU1R182Q patient (Pt) fibroblasts were transfected twice for 6 days with siRNAs targeting ISCA1 (si-ISCA1) or non-specific scramble si-RNAs (si-Scr). Immunoblots of total cell lysate demonstrate the effects on various proteins involved in Fe–S cluster pathway. (B–D) Relative expression levels of (B) ISCU, (C) ISCA1 and (D) NFU1 transcripts were measured by quantitative real-time polymerase chain reaction on the control (MCH46 and CTRL) and NFU1R182Q patient (NFU1-Pt) fibroblasts grown in 21% (atmospheric oxygen) or at 2.5% oxygen (hypoxia) conditions. Transcript levels were normalized to β-actin. Asterisks (*) represent a significant change in the transcript levels in NFU1 Pt cells relative to the control cells (P < 0.05).
While NFU1 in humans is an indispensable protein, in other organisms such as yeast and bacteria, its orthologs are dispensable but are induced under oxidative stress conditions (24,25,42,47). To determine if NFU1 expression was regulated by oxygen concentration in human cells, we grew the NFU1-Pt and WT fibroblasts under atmospheric oxygen as well as hypoxia conditions (2.5% O2) and measured the transcript levels of NFU1, ISCU and ISCA1 in these cells. The transcript levels of NFU1 were reduced in the patient cells as expected (30), but irrespective of the oxygen levels, the transcripts of both ISCU and ISCA1 were significantly upregulated compared to WT in the NFU1 Pt cells, suggesting that cells have a mechanism to transcriptionally respond to loss of functional NFU1 at all times (Fig. 8B–D). Nevertheless, the presence of small but detectable transcripts in the WT and upregulated ISCU and ISCA1 transcripts in NFU1 Pt cells under hypoxic conditions points towards the indispensable nature of NFU1 in humans.
Identification and characterization of new pathogenic mutations in NFU1
Upon the discovery of the significance of the NM and FY residues of NFU1 for Fe–S cluster acquisition and delivery, we inspected the SNP databases to identify human variants with mutations in the region of interest. In our search, we found three variants available (N226D, M227I and Y231C) on the NCBI SNP database, one of which (N226D) was annotated as likely pathogenic. To functionally characterize these variants, we generated NFU1-F/M mutants corresponding to NFU1N226D-F/M, NFU1M227I-F/M and NFU1Y231C-F/M. We analyzed the interaction of these mutants with ISCU2 and ISCA1 as well as their ability to complement the loss of NFU1 in the NFU1 Pt cells. While all three variants showed marginal reduction in the interaction with ISCA1 (Fig. 9A), NFU1N226D-F/M showed increased binding with ISCU2 (Fig. 9B). Phenotypically, NFU1N226D-F/M showed impaired ability to complement the loss of NFU1 as demonstrated by its inability to restore lipoylation of PDH and KGDH or to restore SDHB protein levels (Fig. 9C). Complementation by NFU1M227I-F/M and NFU1Y231C-F/M was also compromised. Thus, these results revealed new mutations in human NFU1, which exhibited pathogenicity due to their inability to effectively acquire and subsequently deliver the Fe–S cluster to the target proteins (LIAS and SDHB).
Figure 9.
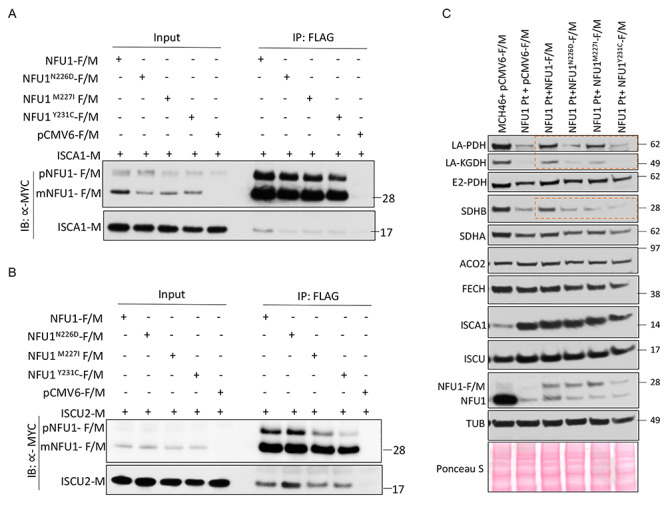
Characterization of natural pathogenic NFU1 mutations. HeLa cell extracts co-transfected with NFU1-F/M or SNP mutants of NFU1-F/M (NFU1N226D, NFU1M227I, NFU1Y231C) or the empty vector (pCMV6-F/M) along with (A) ISCA1-M or (B) ISCU2-M were immunoprecipitated with anti-FLAG beads (mouse) and immunoblotted with anti-MYC (rabbit) antibody. pNFU1-F/M:FLAG and MYC tagged precursor NFU1, m-NFU1-F/M:FLAG and MYC tagged mature mitochondrial NFU1 isoform. (C) Biochemical deficiencies of NFU1R182Q Pt fibroblast were partially rescued by WT-NFU1 (NFU1-F/M) but not by NFU1 SNP mutants as shown by the western blots probing for lipoylated PDH and KGDH as well as SDHB (orange boxes).
Discussion
The significance of Fe–S clusters in cellular metabolism and their association with the etiologies of several rare diseases has made it essential to have a complete understanding of the mechanisms that govern Fe–S cluster biogenesis and trafficking of the clusters to recipient proteins. De novo biosynthesis of Fe–S clusters begins with the assembly of a nascent [2Fe–2S] cluster upon the main scaffold protein ISCU2, which functions as a building block to generate [3Fe–4S] and [4Fe–4S] clusters that are subsequently distributed to recipient Fe–S apo-proteins by additional proteins and complex pathways. The recognition that a tripeptide motif (Leu-Tyr-Arg) in the sequence of SDHB binds the co-chaperone, HSC20, and enables the HSC20/HSPA9 chaperone-co-chaperone complex to transfer the [2Fe–2S] cluster from ISCU to a target protein demonstrated that an energy consuming pathway is employed by cells to specifically transfer Fe–S clusters from the initial biogenesis machinery to recipient proteins (17). Here, we report another pathway that illustrates how [2Fe–2S] cluster building blocks are used in mammalian cells to assemble a [4Fe–4S] cluster, which is eventually transferred to specific recipient proteins. We employed genetic and biochemical techniques to show that the assembly of a [4Fe–4S] cluster on NFU1 requires coordinated interactions with two [2Fe–2S] cluster-containing proteins, the main scaffold ISCU2 and the intermediate carrier ISCA1 in vivo. The cluster thus assembled on NFU1 is then directly transferred to its target proteins, such as LIAS. By studying direct protein–protein interactions and performing co-IPs, our study reveals that direct protein interactions are required for the physiologically relevant acquisition and delivery of a [4Fe–4S] cluster from the initial biogenesis proteins to the recipient proteins in cells.
An in vitro study recently reported that NFU1 acquires its [4Fe–4S] cluster directly from ISCU (22). Notably, that study demonstrated that only ISCU that bound a [4Fe–4S] cluster and not ISCU with a [2Fe–2S] cluster was able to transfer the cluster to NFU1. While the physiological relevance of ISCU ligating a [4Fe–4S] cluster remains controversial (48–50), our study agrees with the observation that ISCU ligating a [2Fe–2S] cluster alone cannot deliver [4Fe–4S] cluster to NFU1. In our experiments, we found that NFU1 engaged directly with two [2Fe–2S] cluster containing proteins, ISCU2 and ISCA1, via a common and conserved hydrophobic motif (F230_Y231; FY) (Figs 1, 2, 4). The initial screening that identified the role of NFU1-FY residues as the binding site for ISCU2 was primarily based on the Y2H assay. Mutagenesis of several residues buried in the core (including I224A, L228A and I231A, V234A) of the CTD-NFU1 protein structure resulted in inconclusive, non-specific growth patterns relative to the negative control (Supplementary Material, Fig. S2D and E), perhaps due to the misfolding of the mutated proteins in yeast (36). Thus, while more residues on NFU1 might play a supporting role in its interactions with ISCU2 and ISCA1 (22), the mutagenesis of FY residues of NFU1 specifically reduced its interaction with either ISCU2 or ISCA1 and suggests that the FY motif is directly involved in the interaction with each of the two scaffold proteins (Figs 2 and 4). The FY motif of NFU1 was also shown to be essential for the biological acquisition of the Fe–S cluster, thereby confirming the roles of the interacting proteins as the physiological Fe–S cluster donors (Fig. 6). Furthermore, the reduced cluster occupancy on NFU1 upon the knockdown of mammalian ISCA1 or bacterial IscA in the ISC operon, as well as the loss of the residues of NFU1 (N226M227) that specifically engaged in an interaction with ISCU2, provided direct evidence that supported the role of both ISCA1 and ISCU2 as the Fe–S cluster donors to NFU1 (Figs 5 and 6). Finally, the inability of NFU1FY-AA and NFU1NM-AA mutants to restore the biochemical defects of the NFU1 Pt cells in the complementation assays provided strong evidence that both ISCU and ISCA1 were essential to assemble a functional cluster on NFU1 (Fig. 7). These data collectively suggest that ISCU2 and ISCA1 each donate a [2Fe–2S] cluster to the NFU1 dimer, which is then potentially assembled into a [4Fe–4S] cluster by the reductive coupling mediated by FDX2, another direct interacting partner of NFU1 detected in this study.
In our study, we did not detect interactions of NFU1 with other presumed Fe–S scaffolds/donors including ISCA2, IBA57, GLRX5 or BOLA3. A complex composed of ISCA1–ISCA2–IBA57 has been suggested to function as the intermediate carrier that assembles the [4Fe–4S] cluster for the downstream late-acting carrier proteins including NFU1 and BOLA3 (21,25,51). However while ISCA1 and ISCA2 can interact with each other, the two proteins appear to have distinct binding partners in vivo, including IBA57, which was shown to interact only with ISCA2 and to be regulated by the levels of ISCA2 (43,52). Furthermore, only ISCA1, and not ISCA2, was shown to be essential for [4Fe–4S] cluster assembly in skeletal and neuronal tissue under physiological conditions (43). Our work also supports that only ISCA1 and not ISCA2 or IBA57 is essential for the assembly of a [4Fe–4S] cluster on NFU1 (Figs 4 and 5). In vitro studies recently showed that a heterotetrameric complex between dimers of GLRX5 and BOLA3 transferred a [4Fe–4S] cluster to NFU1 (20). This pathway may serve as an alternative pathway, given that one patient with GLRX5 mutation was reported to have a NFU1-like phenotype (23). However, patients with NFU1 mutations develop MMDS1, whereas the clinical presentation of two GLRX5-deficient patients was mainly limited to sideroblastic anemia, suggesting that although Fe–S cluster trafficking pathways that involve GLRX5 and NFU1 may partially overlap, the pathway outlined in this article may represent the predominant pathway in most cells (53–55).
While the existence of other pathways to deliver the Fe–S cluster to NFU1 is plausible, the cluster transfer from ISCU2 and ISCA1 mediated by FY motif is physiologically relevant and appears to be absolutely essential for the function of targets such as LIAS (Fig. 7). In this study, we showed that NFU1 interacts directly with two known [4Fe–4S] cluster recipient proteins, SDHB and LIAS in vivo, a novel previously unreported observation (Fig. 1). Despite interacting directly with NFU1, SDHB protein levels do not appear to be significantly modulated by the levels of NFU1 (Fig. 7). Further investigations will be required to establish the role of NFU1 in Fe–S cluster delivery from NFU1 to SDHB. In bacteria, the mechanism for the cluster delivery to LIAS was recently elucidated in E. coli (47). LipA, the LIAS counterpart in bacteria, ligates two [4Fe–4S] clusters (56). NfuA, the bacterial homologue of NFU1 that is larger (24) and multifunctional, was shown to replenish the rapidly turned over auxiliary cluster in LipA that donates sulfur for lipoic acid formation; however, the source for the stable SAM [4Fe–4S] cluster was not discovered (47). Although this mechanism remains to be investigated in humans, NFU1 may supply the auxiliary Fe–S cluster in LIAS and thereby enable LIAS to maintain its activity. ISCA1, which appears to independently contribute to LIAS activity (Fig. 8), could be involved in the delivery of the SAM cluster in LIAS. According to our data, ISCA1 appears to function as an upstream protein that is involved in donating its cluster to other mitochondrial proteins in addition to NFU1 (Fig. 8). Similarly, SDHB appears to depend on ISCA1 for acquisition of one or more of its cluster(s), which in turn affects its stability in the mammalian cells in which ISCA1 is silenced (Fig. 8). Further studies are required to establish if an ISCA1 dimer directly donates a [4Fe–4S] cluster to these targets or if it mediates transfer through another terminal donor. Identification of shared sequence features in the binding sites for Fe–S cluster carrier proteins in multiple recipients may help to define a motif that can be used informatically to identify other potential unrecognized [4Fe–4S] cluster containing proteins.
In conclusion, we showed that NFU1 directly interacts with two Fe–S cluster biogenesis proteins (ISCU2 and ISCA1) through its conserved FY motif, and these interactions are required for the physiological acquisition of the cluster by NFU1. The analysis of the multimeric protein complexes containing NFU1 by native/2D SDS-PAGE provided further insights into the mechanism of the assembly of the [4Fe–4S] cluster on NFU1 and its delivery to its target proteins (Fig. 1I). Although the transfer of Fe–S clusters from holo-ISCU (22) and holo-ISCA1 (57) to apo-NFU1 has been previously reported in vitro, further investigations are required to establish the sequence of events required for the Fe–S cluster transfer which may reveal alternative pathways under specific conditions. Based on our observations, we postulate that both ISCU2 and ISCA1 interact with the FY motif of NFU1 and donate their respective [2Fe–2S] clusters to NFU1, which coalesce into a [4Fe–4S] cluster upon the donation of electrons by FDX2 (Fig. 10). The fact that both the CXXC and FY motifs are essential for the interaction between NFU1 and ISCA1 (Fig. 4) suggests that NFU1 may acquire its first [2Fe–2S] building block from ISCU2, which may be ligated transiently between ISCU2 and one monomer of NFU1 or between the two monomers of NFU1 via the C210_C213 motif of each monomer. This proposed partial loading may then permit ISCA1 containing the second [2Fe–2S] cluster to be to recruited and bind to NFU1 at its FY motif (Fig. 4). The interaction between ISCA1 and ISCU2 (Figs 1 and 4) may facilitate the recruitment of ISCA1 by NFU1. We envision that the delivery of the second cluster from ISCA1 allows the two [2Fe–2S] clusters to coalesce into a [4Fe–4S] cluster when FDX2 provides a reducing equivalent and enhances the formation of a stable cubane [4Fe–4S] cluster. This assembly complex containing holo-NFU1 may then recruit the apo-target proteins such as LIAS and thus act as a terminal Fe–S cluster donor to recipient proteins (Figs 1I and 10), as was observed in the case of bacterial LipA (58). Finally, the identification and characterization of new NFU1 pathogenic mutations in the human population provide genetic information that may facilitate early diagnosis of MMDS1 (Fig. 9). Thus, our study elucidates a complete pathway employed by mammalian cells that illustrates how basic [2Fe–2S] clusters are converted into [4Fe–4S] clusters on an intermediate scaffold protein, which then conveys its Fe–S cluster to recipient proteins through direct protein-binding interactions.
Figure 10.
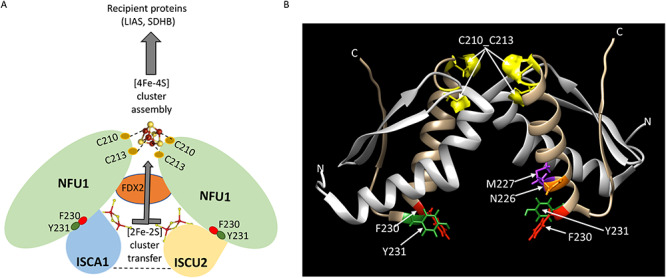
Proposed model for the assembly of a [4Fe–4S] cluster on NFU1. (A) Two [2Fe–2S] clusters, each ligated by holo-ISCA1 and holo-ISCU2, are transferred to dimeric NFU1 and assembled into a [4Fe–4S] cluster by a reductive coupling mediated by FDX2. The [4Fe–4S] cluster assembled on NFU1 is subsequently transferred to recipient proteins such as LIAS and SDHB. We postulate that NFU1 initially binds ISCU2 via its F230_Y231 residues and transiently ligates the [2Fe–2S] cluster via C210_C213. This partial loading of the cluster on NFU1 via C210_C213 is necessary to recruit ISCA1 (mediated by the FY and the CXXC motifs of NFU1) and thereby incorporate the second [2Fe–2S] cluster from ISCA1. This process may be facilitated by the affinity of ISCA1 for binding to ISCU2 (represented by the dashed line between ISCA1 and ISCU2; Figs 1 and 4). The delivery of the second cluster from ISCA1 allows two [2Fe–2S] clusters to coalesce into a single [4Fe–4S] cluster that forms a bridge between the two NFU1 monomers, aided by donation of reducing equivalents from FDX2, which permits the formation of a stable bridging [4Fe–4S] cluster. It is possible that the [2Fe–2S] cluster is transiently ligated between the two NFU1 monomers, which is then converted into a more stable [4Fe–4S] cluster by FDX2 upon the recruitment of ISCA1 and acquisition of its cluster. The assembly complex containing holo-NFU1 subsequently recruits the apo-target proteins such as LIAS (as shown in Fig. 1I) and thus acts as a terminal Fe–S cluster donor to recipient proteins (B) The ribbon model representation of a dimer of CTD-NFU1 (PDB:2M5O) displaying the residues involved in the binding of ISCA1 (F230, Y231; left) and ISCU (N226, M227, F230, Y231; right) on each monomer. The [4Fe–4S] is ligated via C210 and C213 between the two NFU1 monomers. Color coding of the residues: NFU1—red, F230; green, Y231; purple, M227; orange, N226; yellow, C210_C213.
Materials and Methods
Primary cultures
Two immortalized WT fibroblast cell lines (MCH46 and CTRL), which were used as controls, and NFU1R182Q fibroblasts derived from patients (NFU1 Pt) were previously generated (30). Fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 5 mm glucose and 1 mm sodium pyruvate (Thermo Fisher Scientific) and supplemented with 10% fetal bovine serum (FBS) (CellGro) and 2 mm glutamine. HeLa or HEK293 cells purchased from ATCC were cultured in DMEM supplemented with 10% FBS, 4.5 g/l glucose along with 2 mm glutamine. Cells were grown at 37°C and 5% CO2 in a humidified incubator at atmospheric oxygen concentration unless mentioned otherwise.
Cloning and plasmid constructs
The plasmids used to transfect human cells were all pCMV6-Entry-based vectors (Origene) to allow the expression of the full-length gene of interest under the CMV promoter (Supplementary Material, Table S1). The constructs were C-terminally tagged with FLAG/MYC. Plasmid transfections into human cells were routinely performed using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Point mutations/deletions were introduced using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent).
The plasmids, pGADT7 and pGBKT7 (Takara), were used as prey and bait vectors, respectively, for the Y2H screens. The cDNAs encoding ISCU2 and NFU1 were subcloned in the prey vector (pGADT7) and the cDNAs encoding ISCA1, ISCA2, IBA57, FDX2, GLRX5, BOLA3 and NFU1 were subcloned in the bait vector (pGBKT7) from their respective pCMV6 constructs or the HeLa cell cDNA (Supplementary Material, Table S1) using HiFi cloning kit [New England Biolabs (NEB)] according to the manufacturer’s instructions. These cDNAs were cloned at the 3′ end of GAL4 activation domain (AD) and the GAL4 DNA binding domain (BD) in the prey and bait vectors, respectively, such that the proteins encoded by these inserts were fused to the C-terminus of either GAL4 AD or BD, which rendered their mitochondrial targeting sequences non-functional. SDHB-prey and LIAS-prey constructs were obtained from the Mate & Plate™ Library (Takara). Rabaptin/RABEP1-prey, SV40 T-antigen-prey, RABEX5-bait and p53-bait were used as controls as previously reported (17). The AD as well as DNA BD of GAL4 contained the nuclear localization signal, which ensured the translocation of each expressed construct into the nucleus upon translation, thereby facilitating possible interactions in the same cellular compartment.
The plasmids used for bacterial overexpression and protein purification encoding NFU1 and ISCA1 were subcloned into pMAL-c5x vector (NEB) expressing maltose binding protein (MBP) at the N-terminus of the fusion construct generating MBP-NFU1 or MBP-ISCA1 using HiFi cloning kit (NEB) following the manufacturer’s instructions. pBDR1421 (a kind gift from Dr Caroline Philpott who obtained the operon from Dr Dennis Dean) was used to overexpress the ISC operon (ISC operon encodes: IscR, IscS, IscU, IscA, hscB, hscA, fdx, iscX).
The ISC operon deleted for IscA (IscA-del-Op) was generated by site-directed mutagenesis (Agilent). To avoid the polar effects on the expression of downstream genes in the operon, an in-frame deletion of IscA in the ISC operon was performed by looping out a majority of IscA CDS, leaving behind at least 18–21 nucleotides, corresponding to 6–7 amino acids intact on each end of the gene in the ISC operon. In-frame deletions or substitutions generally do not adversely affect the expression of downstream genes (59,60).
SiRNA transfection of human cells
On-TARGETPlus siRNA pools against human ISCA1 (L-014678-02-0005) and the control non-targeting pool (D-001810-10-05) were purchased from Dharmacon. Knockdown of ISCA1 was achieved by transfecting cells twice with siRNAs at a 48-h interval using Dharmafect 1 according to manufacturer’s instructions.
Y2H assay
150 ng each of the prey and the bait constructs were co-transformed into the AH109 competent cells using the EZ transformation kit following the manufacturer’s instructions (MP Biomedical) and plated on leucine and tryptophan drop-out media (-Leu/-Trp) for 4 days. This media provided the selection pressure to select the cells harboring both the bait and prey plasmids, which encoded Leu and Trp biosynthesis genes, respectively, that are otherwise absent from the cell. The interaction of the bait and prey constructs was engineered to activate the transcription of HIS3 gene, thus allowing the cells to grow in the media devoid of histidine (His). 3-Aminotriazole (3-AT), a competitive inhibitor of HIS3, was added to the media to neutralize leaky expression of HIS3, thus increasing the stringency of the media for the interaction. To test the interactions between preys and baits, four transformed colonies from each Leu/-Trp plate were pooled and resuspended in ultrapure water. The concentration of the cells in the solution was subsequently normalized to OD = 0.05. 5 μl of the cells were spotted on -Leu/-Trp, -Leu/-Trp/-His, -Leu/-Trp/-His/+ 3-AT with increasing 3-AT concentrations as indicated along the individual figures. The growth of the cells was monitored over 5 days. Two pairs of known interacting proteins, Rabaptin-Rabex and SV40 Tag-p53, were used as controls.
IP in transfected mammalian cells
12 μg of NFU1-FLAG/MYC (NFU1-F/M) along with 10 μg ISCU2-MYC (ISCU2-M) or 10 μg ISCA1-MYC (ISCA1-M) were co-expressed in HeLa cells (ATCC). Twenty-four hours post transfection, cells were harvested in a lysis buffer (25 mm Tris, 0.15 mm NaCl, 1 mm EDTA, 1% NP-40, 5% glycerol, protease inhibitor cocktail, pH 7.4) and 750 μg of the total proteins were incubated overnight at 4°C with either EZ View Red Anti-Flag Beads (Sigma) or beads chemically crosslinked with anti-ISCU (Origene), anti-ISCA1 (Sigma) or anti-NFU1 antibodies. The beads were subsequently harvested by centrifugation, washed in the lysis buffer followed by Dulbecco’s phosphate-buffered saline (DPBS) washes and finally boiled to elute the proteins bound to the beads. Five percent of the post-transfection lysates were loaded as inputs for the respective samples and ~17% of the eluted samples were loaded in the IP lanes per sample. Immunoblots (IBs) were performed with antibodies as indicated on the figure panels and in Supplementary Material, Table S2.
Protein purification and Fe–S cluster reconstitution
Recombinant pMAL-c5X vector was co-expressed with ISC operon-pBDR1421 in BL(DE)21 bacteria. The pre-inoculum was used to grow a 250 ml culture expressing MBP-NFU1 or MBP-ISCA1 with the ISC operon vector. The ISC operon was induced with 0.2% D-arabinose at the OD = 0.3 and then with 0.3 mm IPTG at OD = 0.6. The culture was supplemented with 0.1 mm l-cysteine and 25 μm ferric ammonium citrate (58) and grown with shaking at 140 rpm at 15°C overnight. Cells were pelleted by centrifugation at 4°C at 5000g. The cell pellet was lysed with 1× BugBuster™ (ENDMillipore) in PBS for 30 min at 4°C and then centrifuged at 16 000g for 25 min to extract the soluble protein lysate. The soluble lysate was incubated with 1 ml amylose beads (NEB) anaerobically (batch purification) overnight and subsequently washed and eluted with 10 mm maltose. For further purification, the buffer of the eluted protein was exchanged to 50 mm HEPES, pH 6.2 using PD-10 column (Sigma), and the eluted protein was then further purified using a HiTrap Q column (Sigma) from which the purified protein was subsequently eluted by stepwise increasing NaCl concentrations. NFU1 was eluted at 0.3–0.35 M NaCl, 50 mm HEPES. ISCA1 was eluted at 0.15–0.2 M NaCl. 100 μm purified protein was anaerobically reconstituted with Fe–S cluster as described (20,58). All the buffers contained 5 mm DTT and 10% glycerol (58). The purified protein was then visualized on the stain-free SDS gel or Coomassie-stained SDS gel; the UV–visible spectrum was recorded using Nanodrop OneC (Thermo Fisher Scientific).
Iron incorporation assay
The 55Fe incorporation assay was performed essentially as previously described (17,19), with minor modifications. HEK293 cells were grown in the presence of 1 μm 55Fe-Tf for the duration of the experiment and transfected twice at 48-h interval with siRNAs targeting ISCA1 or with non-targeting siRNAs, as indicated. At the time of the second transfection with siRNAs, cells were co-transfected with FLAG/MYC-tagged NFU1 or with the empty vector (pCMV6-Entry; Origene CAT#: PS100001), which was used to normalize the results. Mitochondrial extracts were prepared as previously described (17) and subjected to immunoprecipitation with anti-FLAG M2 agarose beads to immunocapture recombinant NFU1-F/M.
55Fe incorporation into recombinant FLAG-MYC-tagged NFU1 was measured by scintillation counting of FLAG-beads after immunoabsorption of FLAG-tagged recombinant NFU1-F/M, followed by extensive washing with buffer I (25 mm Tris, 0.15 M NaCl, 1 mm EDTA, 1% NP-40, 5% glycerol (pH 7.4), protease and phosphatase inhibitor cocktail with no EDTA (Roche)). The background, corresponding to 55Fe measurements of eluates after anti-FLAG immunoprecipitations on mitochondrial extracts from cells transfected with the empty vector, was subtracted from each reading. Unpaired t-test analyses of 55Fe labeling experiments were performed with GraphPad Prism 7.
ISCA1 knockdown and complementation analysis of NFU1R182Q patient (NFU1 Pt) fibroblasts
ISCA1 knockdown was performed by transfecting control (CTRL) or NFU1 Pt cells with si-ISCA1 (Dharmacon) according to the manufacturer’s instructions. Si-SCR (non-targeting pool of siRNAs; Dharmacon) were used as controls. Cells were transfected twice over 6 days to achieve an efficient knockdown. For complementation studies, MCH46 and NFU1 Pt fibroblasts (30) were transfected with either pCMV6-F/M or pCMV6-NFU1-F/M (WT or mutants, as indicated) for 60 h in serum-free media using Lipofectamine 2000 according to the manufacturer’s instruction. Total lysates were prepared in lysis buffer (25 mm Tris, 0.15 mm NaCl, 1 mm EDTA, 1% NP-40, 5% glycerol, protease inhibitor cocktail, pH 7.4) and 30–40 μg total proteins were loaded per lane on SDS-PAGE (Thermo Fisher Scientific) and probed with the different antibodies, as indicated (Supplementary Material, Table S2). The CI and CII in-gel activity assays were performed on the mitochondrial extracts of the aforementioned fibroblasts as previously reported (17).
Quantitative real-time polymerase chain reaction
Fibroblasts cultured at 21% O2 or in a hypoxic chamber (2.5%) for 1 week were used to isolate RNA using Qiagen RNeasy kit according to the manufacturer’s instructions (Qiagen). 2 μg of RNA was used to prepare cDNA using Applied Biosystems High Capacity cDNA Reverse transcription kit (Fisher Scientific) and the real-time transcript levels of NFU1, ISCU2 and ISCA1 relative to β-actin were measured and analyzed using Fast SYBR Green Master mix according to the manufacturer’s instructions (Applied Biosystems).
IP of NFU1-F for native/2D SDS PAGE
Mitochondrial fractions were prepared from HEK293 cells transfected with FLAG-tagged NFU1 (NFU1-F) as previously described (17). IP of NFU1-F was performed using M2-FLAG beads (Sigma). Washed FLAG M2 beads were added to mitochondrial lysates and incubated for 2 h at room temperature. Beads were recovered after extensive washing, and proteins were competitively eluted with 100 μg/ml 3xFLAG peptide (Sigma).
Two-dimensional native/SDS-PAGE
Two-dimensional native/SDS-PAGE was performed by resolving the eluates after competitive elution of NFU1-F from M2 agarose beads in the first dimension, by BN-PAGE. The NativePAGE Novex Bis-Tris gel system (Thermo Fisher Scientific) was used, with the following modifications: only the light blue cathode buffer was used; the electrophoresis was performed at 150 V for 1 h and 250 V for 2 h. Each lane of the native gel was excised, equilibrated in SDS buffer supplemented with reducing agent and then immersed in the alkylating solution for 15 min, before quenching for an additional 15 min. For the second dimension, the gel strip was fixed horizontally onto the NuPAGE 4–12% Bis-Tris Zoom Gel (Thermo Fisher Scientific), and classical SDS immunoblots were performed.
Supplementary Material
Acknowledgements
We thank Dr Caroline Philpott for pBDR1421 construct and colleagues in the Rouault laboratory for helpful discussions.
Funding
This work was supported by the intramural funding ZIAHD008814 for Mammalian iron-sulfur cluster biogenesis from the National Institutes of Child Health and Human Development and National Institutes of Health.
Conflict of Interest statement. The authors declare no conflict of interest.
Contributor Information
Anshika Jain, Molecular Medicine Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA.
Anamika Singh, Molecular Medicine Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA.
Nunziata Maio, Molecular Medicine Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA.
Tracey A Rouault, Molecular Medicine Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA.
Authors’ Contributions
A.J. and T.A.R. designed the study and wrote the manuscript. A.J., A.S. and N.M. performed the experiments and analyzed the data. A.J. generated the clones and performed most of the experiments. A.S. performed the co-IP experiments in HeLa cells. N.M. performed the co-IP experiments in HEK293 cells, the 2D-native/SDS-PAGE, 55Fe-incorporation assay and the activity assays of the respiratory complexes in mammalian cells; intellectually contributed to the study and edited the manuscript. T.A.R. analyzed the data and supervised the study. All the authors reviewed the final manuscript.
References
- 1. Maio N., Jain A. and Rouault T.A. (2020) Mammalian iron–sulfur cluster biogenesis: recent insights into the roles of frataxin, acyl carrier protein and ATPase-mediated transfer to recipient proteins. Curr. Opin. Chem. Biol., 55, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim K.S., Maio N., Singh A. and Rouault T.A. (2018) Cytosolic HSC20 integrates de novo iron–sulfur cluster biogenesis with the CIAO1-mediated transfer to recipients. Hum. Mol. Genet., 27, 837–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tong W.H. and Rouault T.A. (2006) Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron–sulfur cluster biogenesis and iron homeostasis. Cell Metab., 3, 199–210. [DOI] [PubMed] [Google Scholar]
- 4. Adam A.C., Bornhövd C., Prokisch H., Neupert W. and Hell K. (2006) The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J., 25, 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cory S.A., Van Vranken J.G., Brignole E.J., Patra S., Winge D.R., Drennan C.L., Rutter J. and Barondeau D.P. (2017) Structure of human Fe–S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc. Natl. Acad. Sci. USA., 114, e5325–e5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Van Vranken J.G., Jeong M.Y., Wei P., Chen Y.C., Gygi S.P., Winge D.R. and Rutter J. (2016) The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. Elife, 5:e17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wiedemann N., Urzica E., Guiard B., Müller H., Lohaus C., Meyer H.E., Ryan M.T., Meisinger C., Mühlenhoff U., Lill R. and Pfanner N. (2006) Essential role of Isd11 in mitochondrial iron–sulfur cluster synthesis on Isu scaffold proteins. EMBO J., 25, 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fox N.G., Yu X., Feng X., Bailey H.J., Martelli A., Nabhan J.F., Strain-Damerell C., Bulawa C., Yue W.W. and Han S. (2019) Structure of the human frataxin-bound iron–sulfur cluster assembly complex provides insight into its activation mechanism. Nat. Commun., 10, 2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai K., Tonelli M., Frederick R.O. and Markley J.L. (2017) Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron–sulfur cluster biosynthesis. Biochemistry, 56, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gervason S., Larkem D., Mansour A.B., Botzanowski T., Muller C.S., Pecqueur L., Le Pavec G., Delaunay-Moisan A., Brun O., Agramunt J. et al. (2019) Physiologically relevant reconstitution of iron–sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat. Commun., 10, 3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi Y., Ghosh M., Kovtunovych G., Crooks D.R. and Rouault T.A. (2012) Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron–sulfur cluster biogenesis. Biochim. Biophys. Acta, 1823, 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maio N. and Rouault T.A. (2015) Iron–sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery. BBA–Mol. Cell Res., 1853, 1493–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Majewska J., Ciesielski S.J., Schilke B., Kominek J., Blenska A., Delewski W., Song J.Y., Marszalek J., Craig E.A. and Dutkiewicz R. (2013) Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron–sulfur cluster scaffold Isu protein is mutually exclusive. J. Biol. Chem., 288, 29134–29142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vickery L.E. and Cupp-Vickery J.R. (2007) Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron–sulfur protein maturation. Crit. Rev. Biochem. Mol. Biol., 42, 95–111. [DOI] [PubMed] [Google Scholar]
- 15. Rouault T.A. (2012) Biogenesis of iron–sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis. Model. Mech., 5, 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Braymer J.J. and Lill R. (2017) Iron–sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem., 292, 12754–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maio N., Singh A., Uhrigshardt H., Saxena N., Tong W.-H. and Rouault T.A. (2014) Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab., 19, 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maio N., Ghezzi D., Verrigni D., Rizza T., Bertini E., Martinelli D., Zeviani M., Singh A., Carrozzo R. and Rouault T.A. (2016) Disease-causing SDHAF1 mutations impair transfer of Fe–S clusters to SDHB. Cell Metab., 23, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maio N., Kim K.S., Singh A. and Rouault T.A. (2017) A single adaptable cochaperone-scaffold complex delivers nascent iron–sulfur clusters to mammalian respiratory chain complexes I–III. Cell Metab., 25, 945–953.e946. [DOI] [PubMed] [Google Scholar]
- 20. Nasta V., Suraci D., Gourdoupis S., Ciofi-Baffoni S. and Banci L. (2020) A pathway for assembling [4Fe–4S]2+ clusters in mitochondrial iron–sulfur protein biogenesis. FEBS J., 287, 2312–2327. [DOI] [PubMed] [Google Scholar]
- 21. Sheftel A.D., Wilbrecht C., Stehling O., Niggemeyer B., Elsasser H.P., Muhlenhoff U. and Lill R. (2012) The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe–4S] protein maturation. Mol. Biol. Cell, 23, 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cai K., Frederick R.O. and Markley J.L. (2020) ISCU interacts with NFU1, and ISCU[4Fe–4S] transfers its Fe–S cluster to NFU1 leading to the production of holo-NFU1. J. Struct. Biol., 210, 107491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baker P.R. 2nd, Friederich M.W., Swanson M.A., Shaikh T., Bhattacharya K., Scharer G.H., Aicher J., Creadon-Swindell G., Geiger E., MacLean K.N. et al. (2014) Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5. Brain J. Neurol., 137, 366–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bandyopadhyay S., Naik S.G., O'Carroll I.P., Huynh B.H., Dean D.R., Johnson M.K. and Dos Santos P.C. (2008) A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron–sulfur cluster carrier. J. Biol. Chem., 283, 14092–14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Melber A., Na U., Vashisht A., Weiler B.D., Lill R., Wohlschlegel J.A. and Winge D.R. (2016) Role of Nfu1 and Bol3 in iron–sulfur cluster transfer to mitochondrial clients. Elife, 5, e15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cai K., Liu G., Frederick R.O., Xiao R., Montelione G.T. and Markley J.L. (2016) Structural/functional properties of human NFU1, an intermediate [4Fe–4S] carrier in human mitochondrial iron–sulfur cluster biogenesis. Structure, 24, 2080–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nasta V., Suraci D., Gourdoupis S., Ciofi-Baffoni S. and Banci L. (2019) A pathway for assembling [4Fe–4S](2+) clusters in mitochondrial iron–sulfur protein biogenesis. FEBS J. 287, 2312–2327. [DOI] [PubMed] [Google Scholar]
- 28. Tong W.-H., Jameson G.N.L., Huynh B.H. and Rouault T.A. (2003) Subcellular compartmentalization of human Nfu, an iron–sulfur cluster scaffold protein, and its ability to assemble a [4Fe–4S] cluster. Proc. Natl. Acad. Sci., 100, 9762–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ahting U., Mayr J.A., Vanlander A.V., Hardy S.A., Santra S., Makowski C., Alston C.L., Zimmermann F.A., Abela L., Plecko B. et al. (2015) Clinical, biochemical, and genetic spectrum of seven patients with NFU1 deficiency. Front. Genet., 6, 123–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cameron J.M., Janer A., Levandovskiy V., Mackay N., Rouault T.A., Tong W.H., Ogilvie I., Shoubridge E.A. and Robinson B.H. (2011) Mutations in iron–sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am. J. Hum. Genet., 89, 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Navarro-Sastre A., Tort F., Stehling O., Uzarska M.A., Arranz J.A., Del Toro M., Labayru M.T., Landa J., Font A., Garcia-Villoria J. et al. (2011) A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe–S proteins. Am. J. Hum. Genet., 89, 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wachnowsky C., Wesley N.A., Fidai I. and Cowan J.A. (2017) Understanding the molecular basis of multiple mitochondrial dysfunctions syndrome 1 (MMDS1)-impact of a disease-causing Gly208Cys substitution on structure and activity of NFU1 in the Fe/S cluster biosynthetic pathway. J. Mol. Biol., 429, 790–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Invernizzi F., Ardissone A., Lamantea E., Garavaglia B., Zeviani M., Farina L., Ghezzi D. and Moroni I. (2014) Cavitating leukoencephalopathy with multiple mitochondrial dysfunction syndrome and NFU1 mutations. Front. Genet., 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niihori M., Eccles C.A., Kurdyukov S., Zemskova M., Varghese M.V., Stepanova A.A., Galkin A., Rafikov R. and Rafikova O. (2019) Rats with human mutation of NFU1 develop pulmonary hypertension. Am. J. Respir. Cell Mol. Biol.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rouault T.A. (2014) Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nat. Rev. Mol. Cell Biol., 16, 45–55. [DOI] [PubMed] [Google Scholar]
- 36. Brückner A., Polge C., Lentze N., Auerbach D. and Schlattner U. (2009) Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci., 10, 2763–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arcinas A.J., Maiocco S.J., Elliott S.J., Silakov A. and Booker S.J. (2019) Ferredoxins as interchangeable redox components in support of MiaB, a radical S-adenosylmethionine methylthiotransferase. Protein Sci., 28, 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dong G., Cao L. and Ryde U. (2018) Insight into the reaction mechanism of lipoyl synthase: a QM/MM study. JBIC Journal of Biological Inorganic Chemistry, 23, 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harmer J.E., Hiscox M.J., Dinis P.C., Fox S.J., Iliopoulos A., Hussey J.E., Sandy J., Van Beek F.T., Essex J.W. and Roach P.L. (2014) Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem. J., 464, 123–133. [DOI] [PubMed] [Google Scholar]
- 40. McLaughlin M.I., Lanz N.D., Goldman P.J., Lee K.-H., Booker S.J. and Drennan C.L. (2016) Crystallographic snapshots of sulfur insertion by lipoyl synthase. Proc. Natl. Acad. Sci., 113, 9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCarthy E.L., Rankin A.N., Dill Z.R. and Booker S.J. (2019) The A-type domain in Escherichia coli NfuA is required for regenerating the auxiliary [4Fe–4S] cluster in Escherichia coli lipoyl synthase. J. Biol. Chem., 294, 1609–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Py B., Gerez C., Huguenot A., Vidaud C., Fontecave M., Ollagnier de Choudens S. and Barras F. (2018) The ErpA/NfuA complex builds an oxidation-resistant Fe–S cluster delivery pathway. J. Biol. Chem., 293, 7689–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beilschmidt L.K., Ollagnier de Choudens S., Fournier M., Sanakis I., Hograindleur M.-A., Clémancey M., Blondin G., Schmucker S., Eisenmann A., Weiss A. et al. (2017) ISCA1 is essential for mitochondrial Fe4S4 biogenesis in vivo. Nat. Commun., 8, 15124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ayala-Castro C., Saini A. and Outten F.W. (2008) Fe–S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev., 72, 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nordin A., Larsson E., Thornell L.-E. and Holmberg M. (2011) Tissue-specific splicing of ISCU results in a skeletal muscle phenotype in myopathy with lactic acidosis, while complete loss of ISCU results in early embryonic death in mice. Hum. Genet., 129, 371–378. [DOI] [PubMed] [Google Scholar]
- 46. Vanlander A.V. and Van Coster R. (2018) Clinical and genetic aspects of defects in the mitochondrial iron–sulfur cluster synthesis pathway. JBIC Journal of Biological Inorganic Chemistry, 23, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McCarthy E.L. and Booker S.J. (2017) Destruction and reformation of an iron–sulfur cluster during catalysis by lipoyl synthase. Science, 358, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fox N.G., Chakrabarti M., McCormick S.P., Lindahl P.A. and Barondeau D.P. (2015) The human iron–sulfur assembly complex catalyzes the synthesis of [2Fe–2S] clusters on ISCU2 that can be transferred to acceptor molecules. Biochemistry, 54, 3871–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Webert H., Freibert S.A., Gallo A., Heidenreich T., Linne U., Amlacher S., Hurt E., Muhlenhoff U., Banci L. and Lill R. (2014) Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin. Nat. Commun., 5, 5013. [DOI] [PubMed] [Google Scholar]
- 50. Chandramouli K., Unciuleac M.C., Naik S., Dean D.R., Huynh B.H. and Johnson M.K. (2007) Formation and properties of [4Fe–4S] clusters on the IscU scaffold protein. Biochemistry, 46, 6804–6811. [DOI] [PubMed] [Google Scholar]
- 51. Muhlenhoff U., Richter N., Pines O., Pierik A.J. and Lill R. (2011) Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe–4S] proteins. J. Biol. Chem., 286, 41205–41216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nasta V., Da Vela S., Gourdoupis S., Ciofi-Baffoni S., Svergun D.I. and Banci L. (2019) Structural properties of [2Fe–2S] ISCA2-IBA57: a complex of the mitochondrial iron–sulfur cluster assembly machinery. Sci. Rep., 9, 18986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Camaschella C., Campanella A., De Falco L., Boschetto L., Merlini R., Silvestri L., Levi S. and Iolascon A. (2007) The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood, 110, 1353–1358. [DOI] [PubMed] [Google Scholar]
- 54. Ye H., Jeong S.Y., Ghosh M.C., Kovtunovych G., Silvestri L., Ortillo D., Uchida N., Tisdale J., Camaschella C. and Rouault T.A. (2010) Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J. Clin. Invest., 120, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu G., Guo S., Anderson G.J., Camaschella C., Han B. and Nie G. (2014) Heterozygous missense mutations in the GLRX5 gene cause sideroblastic anemia in a Chinese patient. Blood, 124, 2750–2751. [DOI] [PubMed] [Google Scholar]
- 56. Lanz N.D. and Booker S.J. (2015) Auxiliary iron–sulfur cofactors in radical SAM enzymes. BBA-Mol. Cell Res., 1853, 1316–1334. [DOI] [PubMed] [Google Scholar]
- 57. Wesley N.A., Wachnowsky C., Fidai I. and Cowan J.A. (2017) Analysis of NFU-1 metallocofactor binding-site substitutions-impacts on iron–sulfur cluster coordination and protein structure and function. FEBS J., 284, 3817–3837. [DOI] [PubMed] [Google Scholar]
- 58. McCarthy E.L. and Booker S.J. (2018) Chapter eight—biochemical approaches for understanding iron–sulfur cluster regeneration in Escherichia coli lipoyl synthase during catalysis In Bandarian V. (ed), Methods in Enzymology. Academic Press, 606: pp. 217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chaperon D.-N. (2006) Construction and complementation of in-frame deletions of the essential Escherichia coli thymidylate kinase gene. Appl. Environ. Microbiol., 72, 1288–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nelson C.E. and Gardner J.G. (2015) In-frame deletions allow functional characterization of complex cellulose degradation phenotypes in Cellvibrio japonicus. Appl. Environ. Microbiol., 81, 5968–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


