Abstract
Temporal resolution of cellular features associated with a severe COVID-19 disease trajectory is needed for understanding skewed immune responses and defining predictors of outcome. Here, we performed a longitudinal multi-omics study using a two-center cohort of 14 patients. We analyzed the bulk transcriptome, bulk DNA methylome, and single-cell transcriptome (>358,000 cells, including BCR profiles) of peripheral blood samples harvested from up to 5 time points. Validation was performed in two independent cohorts of COVID-19 patients. Severe COVID-19 was characterized by an increase of proliferating, metabolically hyperactive plasmablasts. Coinciding with critical illness, we also identified an expansion of interferon-activated circulating megakaryocytes and increased erythropoiesis with features of hypoxic signaling. Megakaryocyte- and erythroid-cell-derived co-expression modules were predictive of fatal disease outcome. The study demonstrates broad cellular effects of SARS-CoV-2 infection beyond adaptive immune cells and provides an entry point toward developing biomarkers and targeted treatments of patients with COVID-19.
Keywords: COVID-19, virus, blood, immune response, disease trajectory, acute respiratory distress, methylation, RNA-seq, scRNA-seq, infectious disease
Graphical Abstract

Highlights
-
•
SARS-CoV2 infection elicits dynamic changes of circulating cells in the blood
-
•
Severe COVID-19 is characterized by increased metabolically active plasmablasts
-
•
Elevation of IFN-activated megakaryocytes and erythroid cells in severe COVID-19
-
•
Cell-type-specific expression signatures are associated with a fatal COVID-19 outcome
Bernardes et al. explore COVID-19 disease trajectories by performing longitudinal multi-omics analyses in peripheral blood samples from hospitalized patients. The analyses identify increased numbers of plasmablasts, interferon-activated megakaryocytes, and erythroid cells as hallmarks of severe disease and define molecular signatures linked to a fatal COVID-19 disease outcome.
Introduction
COVID-19 shows a heterogeneous clinical course, ranging from asymptomatic cases to overwhelming inflammatory responses leading to organ failure and death. Severe, and ultimately fatal, COVID-19 is associated with a dysfunctional immune response. Critically ill COVID-19 patients show higher blood plasma amounts of numerous cytokines and chemokines (Chen et al., 2020; Giamarellos-Bourboulis et al., 2020; Huang et al., 2020; Ong et al., 2020) than do less severe cases. Patients with severe COVID-19 exhibit an impaired type I interferon (IFN) response with low IFN production and heterogeneous regulation of IFN-stimulated genes (Blanco-Melo et al., 2020; Hadjadj et al., 2020). Peripheral blood of patients with severe COVID-19 exhibits high frequencies of interleukin-6 (IL-6)-secreting CD14hiCD16hi monocytes as well as a decrease of non-classical (CD14loCD16hi) monocytes (Hadjadj et al., 2020; Schulte-Schrepping et al., 2020). Hyperinflammatory COVID-19 is associated with the appearance of proliferating, type-I-IFN-activated CD14+HLAlo suppressive monocytes and emergency granulopoiesis with elevated pre-neutrophil counts. T cell lymphopenia and exhaustion are suggested as hallmarks of severe COVID-19 as well (Diao et al., 2020; Giamarellos-Bourboulis et al., 2020; Guan et al., 2020; Huang et al., 2020; Zheng et al., 2020). SARS-CoV-2 infection can elicit specific T cell and B cell responses (Braun et al., 2020; Grifoni et al., 2020; Long et al., 2020; Ni et al., 2020). How these changes in immune cell populations and functions are related to disease outcome lasting immunity are areas of active investigation.
A significant disease burden is mounted by thrombotic complications associated with COVID-19. Pulmonary embolism and thrombosis are frequent clinical features of critically ill COVID-19 cases (Deshpande, 2020), sometimes despite sufficient anticoagulation. Alveolar capillary microthrombi are nine times more prevalent in COVID-19 than in influenza autopsies (Lax et al., 2020). Patients exhibit elevated D-dimer levels and widespread thrombotic microvascular injury (De Voeght et al., 2020; Rapkiewicz et al., 2020). Several studies suggest altered platelet immune cell interactions (Leppkes et al., 2020, Manne et al., 2020) and the presence of megakaryocytes (MKs) in affected lungs (Meyerholz and McCray, 2020).
Longitudinal analyses on the dynamics of circulating immune cells in COVID-19 so far have mostly investigated one information layer (e.g., fluorescence-activated cell sorting [FACS]) at a time. Here, we aimed to provide a comprehensive, longitudinal view of cellular features by using an integrated multi-omics approach. We analyzed up to five peripheral blood samples from hospitalized COVID-19 patients throughout their disease course by single-cell RNA sequencing (scRNA-seq), transcriptome and DNA methylome profiling, multi-color flow cytometry, as well as multiplex cytokine ELISA. In addition to complex changes of immune cells, our study linked circulating MKs and responses of erythroid cells to COVID-19 clinical outcome.
Results
Study Design
We applied a multi-omics approach using up to 5 longitudinal peripheral blood samples of 13 hospitalized COVID-19 patients and 1 additional recovery control from 2 University hospitals in Germany (Cologne and Kiel). We employed parallel scRNA-seq (10x Genomics), single-cell B cell receptor (BCR) profiling, bulk mRNA sequencing (RNA-seq), BCR amplicon sequencing, and multicolor flow cytometry. Array-based DNA methylation profiling and multiplex cytokine ELISA analysis was performed in a subset of seven patients (Figure 1 A). All patients were recruited at admission and samples were taken at days 0, 2, 7, 10, 13 and/or at discharge. Three patients were diagnosed with Acute Respiratory Distress Syndrome (ARDS), two of which had a fatal disease course. Five patients received remdesivir after inclusion into this study. Patient demographics and clinical characteristics are described in Table S1. Fourteen age- and gender-matched healthy controls were processed in parallel. To describe the heterogenous disease trajectories over time, we used a modified WHO ordinal scale (WHO, 2020), which also considers several inflammatory markers (serum c-reactive protein [CRP], serum IL-6, and ferritin) (Table S2). The score was used to classify patients along their disease course (Figures 1B and 1C).
Figure 1.
Clinical Definition of Disease Phases for Disease Trajectory Analyses
(A) Graphical overview of the study cohorts.
(B) Concept of disease phase pseudotimes. Clinical disease phases (pseudotimes) were defined based on inflammatory markers and ventilation need (according to WHO ordinal scale). They reflect temporal disease severity and distinguish between incremental and recovering disease stages. Abbreviation is a follows: Hosp, Hospitalization. For detailed explanation of the scoring and pseudotimes, see Table S2.
(C) Overview of the cohort 1. All patients are temporally aligned to the day of initial admission. Squares mark the day of COVID-19-related symptom onset. Frames mark days of in-patient care, and the color represents the disease pseudotime. Sampling days were marked with a white circle. Intubations and extubations are depicted with triangle symbols, if applicable.
scRNA-seq Analysis Identifies Cellular Changes along the COVID-19 Disease Trajectory
We first analyzed scRNA-seq data from 358,930 cells with 10,900 cells on average per sample (Figure 2 A). Up to four longitudinal samples were analyzed per patient. Uniform Manifold Approximation And Projection (UMAP) was used to visualize the structure of cellular populations (Figures 2B–2E). Graph-based clustering identified 37 distinct cell clusters in the dataset (Figure S1A). Cell type classification was performed on the basis of signature genes of each cluster and confirmed by using reference transcriptomes (Aran et al., 2019; Wilk et al., 2020; Zheng et al., 2017) (Figures 2B, 2C, S1B, and S1C). Cells from individual patients were dispersed evenly in the UMAP representation, with the exception of a higher degree of interindividual heterogeneity in the monocytic subpopulations (Figure 2D, 2E, and S2). Enumeration across time points revealed changes of several cell types including monocytes, proliferative lymphocytes, and natural killer (NK) cells (Figures 2E, 2F, and S1D). We observed an increase of classical CD14+ monocytes especially in convalescence stages, whereas nonclassical CD16+ monocytes amounts were increased in pseudotimes 2 and 3 (Figure S2). Moreover, we confirmed suppression of human leukocyte antigen-DR (HLA-DR) expression in the critical phase (pseudotime 2) (Schulte-Schrepping et al., 2020). Lymphopenia of CD4+ cells was present, but less pronounced than in other datasets. We also noted increased plasmablast (PB) proportions from the incremental phase to early convalescence, whereas B cells were depleted during incremental and complicated stages (Wilk et al., 2020). Of note, we recognized the increased presence of several bone-marrow-derived precursor cell types in the peripheral blood of COVID-19 patients. Prominently, MK proportions were elevated throughout the course of the disease (Figure 2F). To define potential cell populations, which might be directly infected by SARS-CoV-2, we specifically interrogated ACE2 mRNA amounts in the dataset, which were below detection limit (<7 reads) in all cell populations (data not shown).
Figure 2.
Cellular Changes along COVID-19 Disease Trajectories
(A) Schematic workflow.
(B) Cell type UMAP representation of all merged samples. Twelve cell types were identified by cluster gene signatures. In total, 358,930 cells are depicted.
(C) Dot plot for cell-type-specific signature genes. Genes were selected on the basis of the expression amounts of the ten most characteristic genes. Color discriminates genes with increased (red) or decreased (blue) expression, and point size represents the number of cells per group expressing the corresponding gene.
(D) Sample of origin UMAP representation of all merged samples. Cells were colored by the sample. Samples nomenclature is based on patient ID (001–014) and time points of sample collection day 1 (after admission TA), day 3 (TA2), day 8 (TB), day 11 (TB2), day 14 (TC), and day 20 (TE).
(E) Pseudotime UMAP representation of all merged samples, colored by pseudotime.
(F) Cell proportions grouped by pseudotime. Cell proportions depicted as points referring to percentages based on the total cell numbers of individual samples and horizontal bars depicting the mean. Pseudotimes are represented by colors. p values are based on longitudinal linear mixed model for comparison among COVID-19 pseudotimes and p values are based on Mann-Whitney test for comparisons between healthy and COVID-19 samples.
(G) Correlation heatmap between cell-specific proportions and clinical parameters included in routine tests and multiplex ELISA. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 in Spearman’s correlation. Color intensity corresponds to correlation coefficient.
Abbreviations are as follows: DC, dendritic cells; PB, PBs; MK, megakaryocytes; HSC, hematopoietic stem cell; MEP, megakaryocyte-erythroid progenitor cell; CMP, common myeloid progenitor cell; GMP, granulocyte-monocyte progenitor.
See also Figure S1.
A correlation analysis of relative cell proportions with clinical activity parameters and multiplex serum ELISA revealed that PB proportions were correlated with serum amounts of e.g., tumor necrosis factor (TNF), IL-10, and IL-21, a factor critically involved in B cell differentiation to PBs via STAT3 and BLIMP-1 (also known as PRDM1) (Ozaki et al., 2004). Increased bone marrow precursor cells were associated with increased amounts of IL-33, a Th2 cytokine involved in regulating hematopoietic stem cell regeneration (Kim et al., 2014), and elevated MKs were linked to heightened inflammatory parameters, e.g., serum CRP, IL-6, and IFN-α (Figure 2G).
Whole-Blood Transcriptome Signatures Vary Dynamically along COVID-19 Disease Trajectory
To delineate the transcriptional response to SARS-CoV-2 infection at higher temporal resolution and to instruct further cell type selection from the single-cell dataset, we next analyzed whole-blood transcriptomic data of the 13 hospitalized patients from up to 5 time points along with the recovery control and compared the signatures against 14 healthy controls (Figure 3 A). Principal component analysis showed a separation between healthy controls and COVID-19 patients on the first principal component (PC1) (Figure 3B). In total, 5,915 genes were differentially expressed between healthy controls and COVID-19 patients in a pairwise comparison between controls and each of the disease trajectory pseudotimes (Figures 3C and S3A). Most of the differentially expressed genes (DEGs) were expressed at higher levels in COVID-19 patients than in healthy controls (Figures 3C and S3A). Notably, early changes comprised increase of immunoglobulin transcripts (IGHA1 and IGHG1) and lactoferrin (LTF), whereas RORC mRNA levels, encoding for a Th17-specific transcription factor (TF) and class II HLA transcripts were decreased throughout the disease course.
Figure 3.
Dynamics of Whole-Blood Gene Expression in COVID-19
(A) Schematic workflow.
(B) PCA plot of all control and COVID-19 samples on the basis of the expression of all genes. Samples are color-coded by their pseudotime and labeled by the patient or individual ID.
(C) Log2-fold change (y axis) of DEGs between controls and each of the COVID-19 pseudotimes (x axis). Color discriminates genes with increased (red) or decreased (blue) expression, and point size represents statistical significance (adjusted p value). The transparency of the points denotes the number of comparisons in which the genes is significantly differentially expressed. The numbers of genes with increased and decreased expression are written at the top and bottom of the plot, respectively.
(D) Heatmap of top 500 longitudinal DEGs across COVID-19 pseudotimes (pseudotimes 1, 3, 4, 5, and 6). Gene expression in controls and pseudotime 2 are shown for comparison on the left and right ends of the heatmap, respectively. The row-wise z-scores of the normalized counts are plotted in the heatmap. Genes are hierarchically clustered by using their adjacency scores as distance.
See also Figure S2.
Next, we employed ImpulseDE2 to construct a model of intra-individual variation of DEGs over time (Sander et al., 2017). Given that two of three patients reaching pseudotime 2 were deceased soon after reaching this time point and did not experience a convalescent disease course, this category was excluded from this analysis and was only used to contrast identified transcripts post hoc. We identified 935 DEGs following impulse-like progression patterns across the disease trajectory (Figures 3D and S3A). Genes involved in IL-1β and vasodilatory signaling were highly expressed during the incremental trajectory (pseudotime 1). A broad decrease in transcripts encoding ribosomal structural proteins was present from the incremental to the late convalescent state (pseudotimes 1, 3, and 4), which might reflect a general suppression of the protein synthetic apparatus by type I IFN (Jiang et al., 1997). Opposingly, there was a strong increase in IFN-related transcripts during the peak of disease activity, which was, however, suppressed in the critical disease category (Figures 3D and S3B), confirming earlier reports (Blanco-Melo et al., 2020; Hadjadj et al., 2020). Transcripts involved in myeloid-cell-mediated immunity and neutrophil degranulation were modestly increased during pseudotimes 1, 3, and 4, but were strongly increased in the critical pseudotime 2. Notably, during late convalescence (pseudotimes 5 and 6) a strong signal of erythroid cell differentiation (e.g., HBD, OPTN, and FIS1) was detectable indicating a response to hypoxia (Ashrafi and Schwarz, 2013).
We used the gene set resource DoRothEA to infer transcription factor (TF) activity by enriched regulon analysis. TFs in active disease were related to inflammation and IFN signaling (e.g., IRF1 and STAT3) as well as hypoxic signaling (HIF1A) (Figure S3C). Supporting the IL-21-PB axis identified in the first part of the single-cell analysis, PRDM1 (encoded by BLIMP-1) was predicted as significantly active both at the critical as well as at convalescent time points. A hypergeometric test of all active TFs against the REACTOME database showed an enrichment of “cell differentiation pathway” (q = 0.03) and “MK development and platelet production” (q = 0.05). Using upstream network topology of gene expression changes by using signed and directed protein-protein interactions (Liu et al., 2019), we could show that MAPK3 (also known as ERK1) and MAPK1 (also known as ERK2) had the highest centralities across all pseudotimes (Figure S3D). We further constructed metabolic models of each pseudotime by using the respective DEGs (Gebauer et al., 2016) and found that the inflammatory disease states were associated with higher numbers of predicted metabolically active pathways than in convalescence and healthy individuals (Figure S3E). We also investigated the bulk RNA dataset for presence of viral reads and did not detect relevant amounts (>10) of viral reads in any sample. This finding was also confirmed by negative routine real-time PCR (E gene and S gene amplicon) (data not shown).
Longitudinal Co-expression Modules Identify Impaired IFN Response in Critical Disease and Signatures of Increased Thrombo- and Erythropoiesis
We next used all DEGs identified from the combination of pairwise and longitudinal analysis (6,317 genes) for weighted gene co-expression network analysis (Langfelder and Horvath, 2008; Lee et al., 2004). The analysis identified a total of 10 modules, which we refer to as M1–M10, of co-expressed genes following distinct expression patterns throughout the COVID-19 disease phases (Figures 4 A and S3F; Table S3). We calculated the eigengene values, which represent a single expression profile for all genes within a module, to assess the individual correlation between the modules and scRNA-seq-derived cellular composition (Figure 4B) and clinical parameters (Figure 4C). Projecting the expression of the hub genes of each module on the scRNA-seq data revealed cell-type- and pseudotime-specific expression patterns (Figure S4). Gene set enrichment analysis revealed biological processes and pathways enriched in each of the modules. Transcription factor binding site (TFBS) inference (Breuer et al., 2013) was used to depict putatively involved TFs. M2 represented a signature of a type I IFN response and proliferative activity (G2M transition) (Figure 4D) and was enriched in binding sites for IRF1, STAT1, and STAT2 (Figure S3G). M4 was associated with the presence of MKs in the peripheral blood as well as D-dimers and had two expression peaks, in the critical state (pseudotime 2) and in the early convalescent state (pseudotime 4), respectively. M7 contained transcripts related to hemoglobin biosynthesis and coincided with TFBS motifs for GATA-1 and GATA-2, which are TFs related to erythroid cell differentiation (Figure S3G). This biphasic pattern is likely related to hypoxia during highly acute inflammation (first peak) and weaning off supplemental oxygen during convalescence (second peak). Altogether, analysis of temporal gene expression patterns in whole blood of patients clearly depicted pathophysiological consequences of a SARS-CoV-2 virus infection along an idealized disease trajectory.
Figure 4.
Co-expression Analysis of Differentially Expressed Genes and Integration with Changes in Methylation
(A) Group eigengene heatmap of co-expression modules constructed by using all pairwise and longitudinal DEGs. The average eigengene values of all samples within each pseudotime are plotted. The number of genes is plotted as a bar plot (right).
(B) Correlation heatmap showing Spearman’s rank correlation coefficients between gene co-expression modules (rows) and cell-specific proportion from scRNA-seq data (columns). ∗p < 0.05, ∗∗ p < 0.01, and ∗∗∗ p < 0.001 in Spearman’s correlation. Color intensity corresponds to correlation coefficient.
(C) Correlation heatmap showing Spearman’s rank correlation coefficients between gene co-expression modules (rows) and clinical parameters (columns). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 in Spearman’s correlation. Color intensity corresponds to correlation coefficient.
(D) Dot plot showing the gene set enrichment analysis (GSEA) of gene co-expression modules against GO (Biological Processes), Hallmark, and Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets. Size of the dots is proportional to the normalized enrichment score (NES), and the color corresponds to the false discovery rate (FDR). Selected top terms are visualized.
(E) Schematic workflow of the analysis performed on the whole-blood EPIC array data and its integration with bulk RNA-seq data.
(F) Number of significantly DMPs between controls and each of the COVID-19 pseudotimes. Colors discriminate hypermethylated (red) and hypomethylated (blue) positions in COVID-19 samples compared with those from controls.
(G) DMP comparisons between top 30,000 DMPs at each pseudotime. Vertical bar plots indicate the number of specific DMPs (left) shared between time points (right) indicated as connected dots (bottom). Only selected overlaps are visualized.
(H) Heatmap showing the significant enrichment, quantified by odds ratio, of TFBS in the DMPs identified at different COVID-19 pseudotimes. Selected top TFs are visualized.
(I) Dot plot showing GO terms enriched in DMP-DEG pairs. Size of the dots is proportional to the gene ratio and the color corresponds to the p value of the enrichment. Selected top terms are visualized.
See also Figures S2, S3, and S4 and Table S3.
Whole-Blood DNA Methylation Profiling Reveals Genome-Wide Hypomethylation in COVID-19 Associated with Gene Expression
Epigenetic changes have been shown to contribute to the pathophysiology of systemic inflammatory states (Lorente-Sorolla et al., 2019). We thus investigated DNA methylation (DNAm) patterns along the COVID-19 disease course following the workflow depicted in Figure 4E in a subset of the same patients (n = 6) (Table S1) and compared them with a cohort of six healthy age and gender-matched controls. A pairwise comparison with healthy controls and between pseudotimes identified between 46,071 and 69,733 differentially methylated CpG sites (Figures 4F, 4G, and S5B). A preponderance of hypomethylated sites was present at each time point compared with in healthy controls. Cellular deconvolution analysis (Müller et al., 2019) identified that major parts of the COVID-19-associated DNAm signatures originated from granulocytes, B cells, NK cells, and monocytes (Figure S5A). Using Locus Overlap Analysis for inferring differentially methylated TF binding sites (Sheffield and Bock, 2016), we observed a significant overrepresentation of binding sites of the CCAAT Enhancer Binding Protein Beta (CEBPB) in hypomethylated regions (Figure 4H), which has a critical role for emergency granulopoiesis (Hirai et al., 2006) and B-lymphocyte-to-granulocyte trans-differentiation, a process that has been proposed in severe COVID-19 (Wilk et al., 2020).
We employed a hierarchical testing approach (Pan et al., 2018) to identify interactions between transcriptome and DNAm signatures in cis (Figure 4E). To that end, we screened all DEGs (combined set) for differentially methylated positions (DMPs) within a 5 kb window up- and downstream of their respective transcription start site. Of all 3,280 DMP-DEG pairs, 68.3% showed increased expression with reduced methylation or decreased expression with increased methylation, which is in line with previous studies (Häsler et al., 2012). We next investigated the representation of DMP-DEG pairs in the co-expression modules M1–M10. Rank-based correlation analysis identified DMP-DEG pairs in all modules, and there was a significant overrepresentation in M3 and M8 (Figures S5C and S5D). Gene ontology (GO) term enrichment analysis of DMP-DEG pairs showed an enrichment of innate-immunity-related terms in the DEGs with increased expression (e.g., TNF and IL-6 signaling and Toll-like receptor [TLR] pathway), and also identified gene sets related to platelet function and metabolic processes (ATP metabolism and autophagy) (Figure 4I). DMP-DEG pair genes, which were either increased in the inflammatory phase (pseudotimes 1–4) or decreased in late convalescence (pseudotimes 5 and 6) were mapped to scRNA-seq data, identifying larger cell-type-specific clusters of DMP-DEG pairs for PBs, monocytes, and MKs (Figures S5E and S5F).
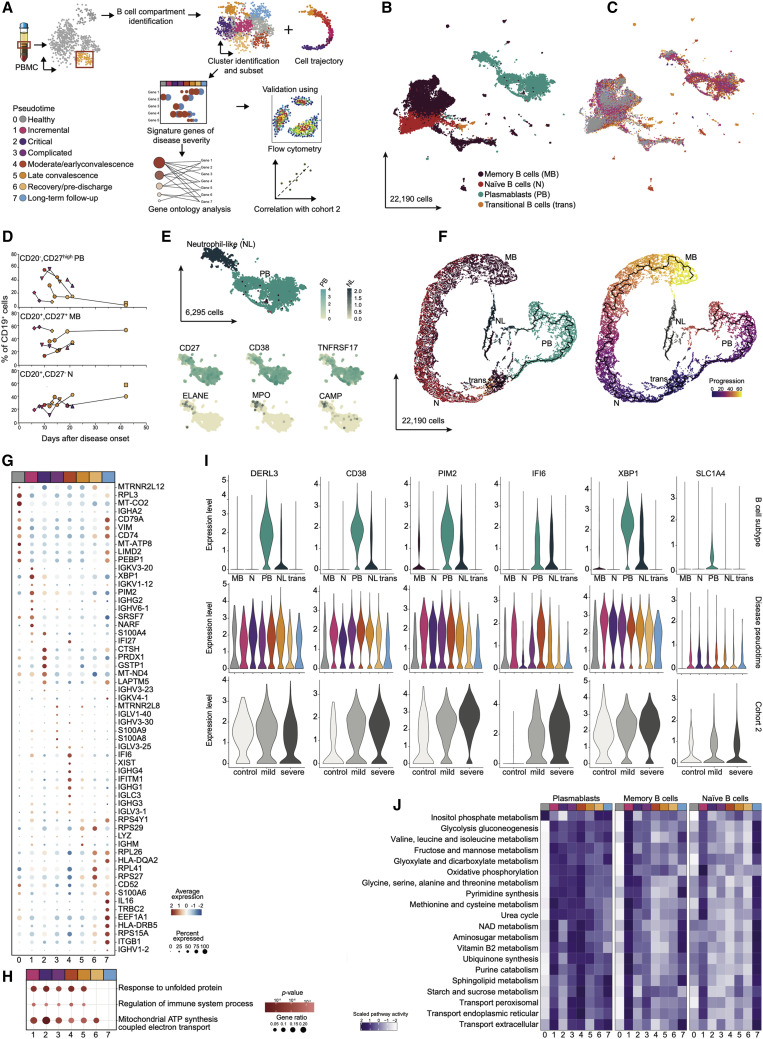
Plasmablast and B Cell Changes across the COVID-19 Disease Trajectory
Given that our findings pointed to changes of the B cell compartment along the disease trajectory of COVID-19, we next investigated the B cell lineage in greater detail (Figure 5 A). We first extracted 22,190 cells identified as part of the B cell lineage from scRNA-seq data in cohort 1. UMAP embedding identified two distinct large clusters reflecting bona fide B cells, which included 11,509 memory B cells, 3,993 naive B cells, 383 transitional B cells, and 6,295 PBs (Figure 5B). PBs were largely expanded during COVID-19 with highest levels in the hyperinflammatory phase (Figure 5C). Multicolor flow cytometry confirmed high amounts of circulating CD27hiCD20− cells in the fraction of CD19+ cells, which dropped along the disease convalescence. Likewise, the early relative decrease of naive (CD20+CD27−) and memory B cell (CD20+CD27+) amounts normalized at later time points (Figure 5D). We also identified a large fraction of HLA-DR+CD138+ double-positive PBs in the inflammatory phase (pseudotimes 1–4). Whereas CD138+ cells were only abundant in the active disease and not in the convalescence phase, 82%–98% of all CD27hiCD20− cells remained HLA-DR+ (Figure S6F). Of note, because PBs are notoriously sensitive to manipulation, we recognized that the handling procedures for scRNA-seq, also for cohort 2, diminished the amounts of intact PBs compared to flow-cytometry data, whereas the longitudinal dynamics of increased PB numbers was retained between the two methods.
Figure 5.
B Cell Compartment Analysis Identifies Plasmablast Changes across the COVID-19 Disease Trajectory
(A) Schematic workflow.
(B) B cell compartment subtypes represented as a UMAP. In total, 22,190 cells are depicted. Memory B cells (MB) (dark red), naive B cells (N) (red), transitional B cells (trans) (orange), and plasmablasts (PB) (blue).
(C) B cell compartment pseudotimes represented as a UMAP.
(D) Flow-cytometry analysis of B cell subtypes. CD19+ B cells were stained for CD20 and CD27. CD20−CD27high B cells classified as PB, CD20+CD27+ cells as MB, and CD20+CD27− cells as N. Proportions of each cell type among CD19+ B cells is relative to the disease onset, colored by corresponding pseudotime, and connected by patient. (n = 7 individuals).
(E) PB-specific UMAP highlighted neutrophil-like cells (NL). Smaller UMAPs corresponding to expression of PB markers (CD27, CD38, and TNFRSF17) and neutrophil-like markers (ELANE, MPO, and CAMP).
(F) Cell trajectory analysis of B cell compartment. Cell trajectory was calculated by using Monocle3. The analysis rooted on transitional B cells (purple) and differentiated into 2 branches: B cells naive and memory branch (gray line, culminating in yellow) and an over imposed PB branch (black line, culminating in orange).
(G) Dot plot for pseudotime signature genes in PBs. Genes selected on the basis of the increased expression of the ten most characteristic genes. Color discriminates genes with increased (red) or decreased (blue) expression, and point size represents the number of cells per group expressing the corresponding gene.
(H) GO enrichment analysis for genes with increased expression during disease trajectory. Dot size is proportional to the gene ratio and the color corresponds to the p value of the enrichment. Selected top terms are visualized.
(I) Gene expression of genes of interest in B cell subtypes. Genes of interest selected on the basis of their high expression in PB or NL. For each gene, top violin plot depicting B cell subtypes expression, center violin plot based on pseudotime, and bottom violin plot based on the expression of cohort 2 (healthy control [white], mild disease [light gray], and severe disease [dark gray]).
(J) Metabolic pathways enriched in B cell compartment subtypes. Top 20 active metabolic pathways for context-specific metabolic networks reconstructed in PBs are shown. For each B cell subtype, significant differences in metabolic activity were determined by using a Kruskal-Wallis test. Number of reaction counts found per pathway is displayed as color intensity.
See also Figure S6L.
We could distinguish a smaller cluster of PBs, which expressed genes associated with neutrophils (e.g., ELANE, MPO, and CAMP) (Figure 5E). Unlike in the previous study (Wilk et al., 2020), we also found such cells in healthy subjects, albeit at lower frequency (Figure 5C). Using monocle3 (Qiu et al., 2017), we highlighted the cellular trajectory of transitional B cells into naive and memory B cells, with a separate memory B cell cluster being most likely CD45RB− cells (Glass et al., 2020) (Figure 5F). We could retrace the progression of PBs, which did not cluster on the basis of immunoglobulin classes but rather on the basis of disease state, with cells from hyperinflammatory phases being more distant from the root of the trajectory (Figures S6A and S6B). Neutrophil-like PB cells were not continuously linked to the PB trajectory (Figure 5F).
We next analyzed the BCR repertoire in the different B cell compartments by using heavy-chain bulk- and scBCR-seq (Figures S6D, S6E, and S6G–S6P). In the bulk dataset, we identified all together 596,882 unique BCR CDR3 heavy-chain sequences, whereas for scBCR-seq we had information referring to 14,785 cells. Diversity analysis showed a heightened clonality, which sharply increased at early time points and then gradually decreased in the convalescent pseudotimes until normalization in follow-up (Figures S6D and S6E). Bulk BCR identified an expansion of IgA+ and IgG+ cells (Figures S5G and S5I) in both memory B cells and PBs (Figures S5H and S5J). Increased IGHA1 and IGHG1 expression was reached earlier in PBs than in memory B cells. Expanded IgA+ and IgG+ PBs were confirmed in the scBCR-seq data (Figures S5K and S5N). Analysis of immunoglobulin heavy-chain variable region (IGHV) family subunits (Figures S5L, S5M, S5O, and S5P) showed a preponderance of specific V regions in COVID-19 patients compared with in controls, e.g., IGHV3-30 and IGHV3-23 were overrepresented in PBs and neutrophil-like cells during disease. In summary, we observed an increase in B cell clonality in COVID-19, and there was an increase of memory B cells and PBs, dominated by the IgA and IgG isotypes and a skewed use of the IGHV gene early during the disease course.
We next analyzed the longitudinal gene expression patterns of the 6,295 PBs (Figure 5G). The incremental inflammatory phase of COVID-19 patients was characterized by transcripts related to endoplasmic reticulum (ER) stress and protein folding (e.g., XBP1) and cell proliferation (e.g., PIM2 and S100A4). Type I IFN response genes (IFI27, IFI6, and IFITM1) were present in the PBs until late in the disease (up to pseudotime 5), yet such genes were absent from PBs in critically ill patients (pseudotime 2). Throughout disease, we also identified an increase in SLC1A4 expression, a potential upstream regulator of metabolic changes (Figure 5I). High IL-16 expression, which supports migration of CD4+ T cells and circulating blood dendritic cells (DCs) into lymphoid organs during the initiation of a humoral immune response (Kaser et al., 2000), was a feature of long-term recovery PBs (Figure 5G). GO enrichment analysis identified increased unfolded protein response and mitochondrial ATP synthesis during active disease (Figure 5H). These findings were also corroborated in an independent cohort (cohort 2) of patients with mild and severe COVID-19 by using another scRNA-seq technology (Schulte-Schrepping et al., 2020). The 2,263 PBs extracted from this dataset confirmed a strong increase of PBs in severe versus mild COVID-19 (30% versus 8% of entire B cell lineage) as well as increased expression of CD38, PIM2, IFI6, XBP1, and SLC1A4 and similarly enriched GO terms (Fisher test, p = 1.80 × 10−5 for unfolded protein response and p = 2.50 × 10−8 for mitochondrial ATP synthesis).
PBs can modulate immune responses by serving as a nutrient sink (Vijay et al., 2020). Thus, we used constraint-based modelling to reconstruct the metabolic state of individual cells from scRNA-seq data (Joshi et al., 2020; Pacheco and Sauter, 2018). PBs from inflammatory states displayed a high metabolic activity, which was reduced only upon recovery, whereas memory and naive B cells displayed no significant differences in overall metabolic activity between disease states (Figure S7L). Increased metabolic processes in PBs were oxidative phosphorylation, glyoxylate and dicarboxylate metabolism, NAD synthesis, and an increase of various amino acid metabolic pathways (including glycine, serine, alanine, threonine, valine, leucine, and isoleucine). Glycolysis was predicted to have a low activity state in inflammatory disease phases, whereas it was highly active at clinical recovery (pseudotime 6) (Figure 5J). Altogether, the analysis identified the broad activation of PBs and suggested a strong immunometabolic shift of the cells toward amino acid metabolism, which might contribute to the immunopathology seen in severe COVID-19.
Elevated Megakaryocyte Amounts as a Feature of the Systemic Inflammatory Response to COVID-19
Systemic inflammatory responses are known to consume platelets, which exert broad immune and inflammatory functions in addition to their well-established hemostatic role (Semple et al., 2011; Yeaman, 2014). Pulmonary and cerebral embolism is an important contributor to morbidity and mortality in COVID-19 (Liao et al., 2020). Given that we had observed a transient increase of circulating MKs, the cellular source of platelets (Figure 2B), in the single-cell data and identified co-expression modules (M3 and M4) related to platelet counts and D-dimer levels, we hypothesized that altered presence and function of MKs might be a distinct feature of COVID-19. We thus performed sub-clustering of 6,512 cells identified as MKs and their respective hematopoietic stem cell precursors (HSCs) and MK-Erythroid precursors (MEPs) by using the k-nearest neighbor method (Figures 6 A and S7A). The cells clustered into 2 distinct subgroups, 5,870 identified as bona fide MKs and another smaller cluster containing all HSCs and MEPs (Figures 6B, S7B, and S7C). The relatively low number of cell precursors complicated the comparison between individual pseudotimes (Figures S7D–S7G). However, we could discern a significant increase of HSCs and MEPs at the convalescence state in comparison with healthy controls (Figure S7J).
Figure 6.
Elevated Megakaryocyte Levels as a Feature of COVID-19
(A) Schematic workflow.
(B) MKs and their precursors as a UMAP. In total, 6,512 cells are depicted. MKs (green), HSCs (pink), and MEPs (yellow) are shown.
(C) MKs pseudotimes represented as a UMAP. In total, 5,870 cells are depicted.
(D) MKs across COVID-19 disease trajectory. For each UMAP, pseudotime-specific cells were highlighted by color.
(E) Dot plot for disease trajectory signature genes in MKs. Genes selected on the basis of the expression amount of the ten most characteristic genes. Color discriminates genes with increased (red) or decreased (blue) expression, and point size represents the number of cells per group expressing the corresponding gene.
(F) Expression of genes of interest in MKs. Top violin plot based on pseudotime and bottom violin plot based on cohort 2 by disease classification; healthy control (white), mild disease (light gray), and severe disease (dark gray).
(G) GO enrichment analysis for genes with increased expression during disease trajectory. Dot size is proportional to gene ratio and the color corresponds to the p value of the enrichment. Selected top terms are visualized.
(H) Cohort 2 MK proportions grouped by disease severity. Healthy control (white), mild disease (light gray) and severe disease (dark gray) were depicted. AIC and p value are based on linear mixed model.
(I) Metabolic pathways enriched in MKs. Top differentially active metabolic pathways for context-specific metabolic networks reconstructed are shown. Significant differences in metabolic activity were determined by using a Kruskal-Wallis test. Number of reaction counts per pathway are displayed as color intensity.
(J) Pyruvate metabolism in MKs. Number of pyruvate metabolic pathway active reactions by pseudotime were depicted. p value based on Kruskal-Wallis test. Number of models per pseudotime were denoted as n above each column.
See also Figure S6L.
By focusing exclusively on MKs, we found there was a clear separation between samples belonging to healthy controls and patients with active COVID-19 (Figure 6C), particularly cells from complicated disease phases form distinct subclusters from cells of healthy and long-term recovery disease phases (Figures 6D and 6E). Strong increase in PKM transcript amounts (PKM2), encoding a pyruvate kinase that is involved in ATP formation, interacts with HIF1A and promotes its activity, was observed in critical patients (pseudotime 2). In this group, high expression of FCER1G, encoding the common FcR γ-chain adaptor responsible for integrin (ITGA2-GP6)-mediated platelet adhesion was also present (Figure 6E). GO enrichment analysis identified broad terms related to immune responses, type I IFN response, and platelet aggregation to be altered along the disease trajectory (Figure 6G).
Transiently decreased transcripts comprised of ODC1 (ornithine decarboxylase), the rate-limiting enzyme for polyamine synthesis (Kanerva et al., 2008), and TGFB1 (Figure 6F). IFITM3, IFI27, and IFITM2 had increased expression in the inflammatory pseudotimes 1, 2, 3, and 4, indicating a lasting IFN response throughout the disease trajectory in MKs. Furthermore, we validated our findings in an independent cohort of mild and severe COVID-19 (Schulte-Schrepping et al., 2020), which confirmed not only higher numbers of MKs in severe patients (Figure 6H), but also confirmed increased expression of IFITM3, PKM, ITGA2B (also known as CD41), and IFITM2, and decreased expression of ODC1 and TGFB1 in severely ill patients (Figure 6F, bottom).
Metabolic modeling identified an increased metabolic activity of MKs along the disease trajectory compared with that in healthy controls, albeit at a lower level than in PBs (Figure 6I). Notable predicted processes were related to energy metabolism (pyruvate metabolism, glycolysis, and reactive oxygen species [ROS] detoxification) (Figures 6J and 6I). The metabolic model inferred a pronounced increase of glycolytic flux toward lactate, an induction of the methylglyoxal pathway (Kalapos, 2008) and a decrease of spermidine-polyamine products known to inhibit platelet aggregation (de la Peña et al., 2000).
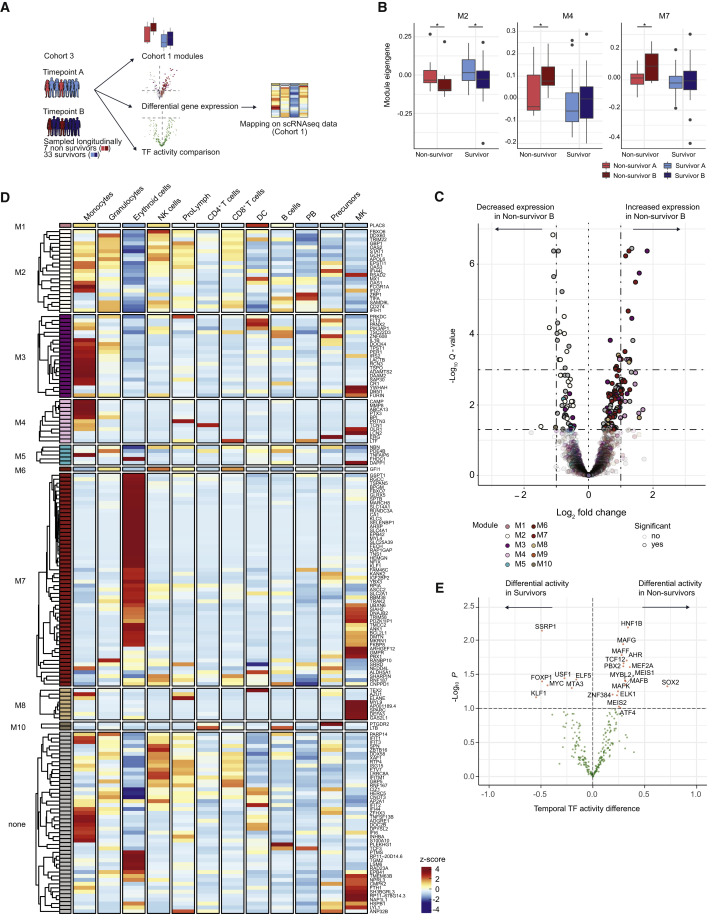
Association of Co-expression Modules with Clinical Outcome in a Cohort of Severely Ill COVID-19 Patients
Lastly, we focused on the potential importance of the obtained signatures in a clinical context in a longitudinal cohort of 40 mechanically ventilated, critically ill COVID-19 patients from Radboud University Medical Center (UMC) in Nijmegen (cohort 3) (Table S4).
Bulk RNA sequencing data in this cohort were obtained at two time points early upon admission to the ICU. We used matched sample pairs with similar increasing inflammatory activity changes from survivors (n = 33) and non-survivors (n = 7) and interrogated the change of expression amounts of module genes (M1–M10, as defined in the German longitudinal cohort [cohort 1] initially shown in Figure 4A) between the two time points (Figure 7 A). The first sample was obtained in median 3 days after ICU admission, the median period between the time points was 2 days without systematic differences between surviving and non-surviving patients. The second time point of non-survivors varied between 4–35 days before death. Three modules were significantly regulated in this longitudinal comparison. M2 transcripts related to failing type I IFN response were significantly decreased in both survivors and non-survivors, corroborating the association of IFN dysregulation and severe disease (Blanco-Melo et al., 2020; Hadjadj et al., 2020). M4, indicative of MKs in the peripheral blood as well as elevated D-dimer levels and M7 transcripts, associated with erythroid differentiation and MKs, were significantly increased only in COVID-19 non-survivors when comparing the change between sampling at ICU admission and the follow-up time point 2 days later (Figure 7B).
Figure 7.
Clinical Significance of Co-expression Modules in a Longitudinal Cohort of Severe COVID-19 Patients
(A) Schematic workflow.
(B) Module eigengene comparisons between the two sampled time points in survivors and non-survivors for M2, M4 and M7. Error bars depict 1.5 of interquartile distance and *p < 0.05 (Mann Whitney tests).
(C) Volcano plot depicting log2-fold changes and FDR-adjusted p values between the two sampled time points in non-survivors. Genes are color-coded by the corresponding co-expression modules. Darker colors represent significantly DEGs.
(D) Heatmap showing the average expression of DEGs identified in non-survivors in different cell-types of cohort 1 (from scRNA-seq data). The average expression in severe stages of the disease (pseudotimes 1, 2, and 3) is shown. Row-wise z-scores of the average gene counts are plotted in the heatmap and hierarchically clustered for each module separately.
(E) Volcano plot depicting the differential TF activity over time in non-survivors (right) versus survivors (left) versus the −log10 transformed p value. Significant TFs (p < 0.1) are marked in red.
See also Table S4.
In a reverse approach, we asked which transcripts displayed longitudinally different expression patterns between the two time points in survivors versus non-survivors. We found that whereas in survivors only 3 transcripts were regulated, 182 transcripts were significantly different in non-survivors (Figure 7C). Of these transcripts, 130 were contained in the previously defined temporal co-expression modules from cohort 1 with a significant enrichment of transcripts related to M2, M4, and M7. Cell-type-specific expression patterns of these fatality-associated transcripts (average expression value per cell type, derived from scRNaseq cohort 1) showed that the signature for M2 genes marked a broad array of cell types, such as monocytes, granulocytes, NK cells, proliferative lymphocytes, and CD8+ T cells, M3 and M4 genes were mostly specific for monocytes, with few highly expressed transcripts attributed to MKs. A large proportion of M7 genes painted the erythroid lineage, a separate cluster was expressed specifically in MKs, e.g., PBX1, TRIM58, and PDZK1IP1 (Figure 7D).
Lastly, we quantified the differential regulation of TFs over time in survivors versus non-survivors via a moderated t test using limma (Ritchie et al., 2015). The analysis identified 16 TFs that were differentially regulated in non-survivors only and 7 TFs in survivors (Figure 7E). Pathway analysis (REACTOME) revealed significant enrichment of the terms “MK development and platelet production” (p = 0.0001) and “TRAF6-mediated induction of pro-inflammatory cytokines” (p = 0.001) in the non-survivor TFs.
Discussion
The clinically heterogeneous disease presentation renders individual molecular dynamics of the hematopoietic and immune cell compartments system in response to COVID-19 an important topic with regard to understanding the pivot points of the disease. Our longitudinal analysis provided a chronological rank order to changes observed in prior cross-sectional studies. We observed an early and lasting depletion of NK cells and lymphopenia of the CD4+ T cell compartment. Our data corroborated a sustained increase and shift of the monocytic compartment (Schulte-Schrepping et al., 2020; Wilk et al., 2020). Interestingly, we found in our DNAm data that hypomethylated positions were highly enriched in cis of transcripts with increased expression related to positive regulation of TNF secretion, IL-1β release, and innate immune signaling. Vice versa, transcripts with decreased expression comprised T cell receptor signaling and negative regulation of ATP metabolism, indicating a potential long-term regulation of the immunological misfiring (Lee et al., 2020; Lucas et al., 2020) by epigenetic processes. We observed a transient increase of PBs and a relative decrease of memory and naive B cells, which coincides with inflammatory severity and normalizes with convalescence (De Biasi et al., 2020; Kuri-Cervantes et al., 2020; Stephens and McElrath, 2020; Mathew et al., 2020). Similar to other studies, the changes were not correlated with levels, but preceded the appearance of SARS-CoV-2-recognizing IgG antibodies (Kuri-Cervantes et al., 2020; Woodruff et al., 2020). The expression of CD138 on a large fraction of PBs during the disease supports the theory that these cells are non-specifically mobilized from the bone marrow or other tissues. Of note, we found that the COVID-19 PBs were predicted to be highly metabolically active in a systems biology modeling approach (Joshi et al., 2020). The observed changes suggest a role of PBs as a nutrient sink, which was already observed in extrafollicular PBs as a hallmark of a systemic inflammatory response in severe malaria (Vijay et al., 2020). The predicted lower energy availability at the peak of COVID-19 might indicate excessive shuttling of glucose into antibody glycosylation, which might contribute to metabolic exhaustion of the cells (Corcoran and Nutt, 2016; Lam et al., 2016) and/or altered glycosylation patterns of antibodies, which were linked to severe COVID-19 (Hoepel et al., 2020; Larsen et al., 2020).
Our longitudinal approach identified two other cellular features induced by COVID-19, which were unrecognized in previous studies. First, we found a significant increase of MKs, which carried a strong type I IFN signature. The change was associated with two longitudinal co-expression modules from the bulk RNA sequencing data (M3 and M4). Both modules were positively correlated with serum levels of D-dimers, suggesting a link to the inflammation-induced pro-coagulative state as a potentially fatal complication in COVID-19 patients. One of the top transcripts with increased expression in MKs was IFITM3, which confers antiviral activity in MKs and platelets (Campbell et al., 2019). Metabolic modeling suggested that in the hyperinflammatory states, a shift toward higher rates of pyruvate metabolism and glycolysis occurs, which sensitizes platelets toward activation and aggregation (Nayak et al., 2019).
Although thrombopenia has been observed as a clinical correlate of critical COVID-19 (metanalysis in Lippi et al., 2020), it is unlikely that direct infection of MKs as seen in Dengue fever is responsible for this alteration. We found no evidence of either ACE2 expression in MKs or any virus-associated reads in the cell population (data not shown). The observed increase of circulating precursors including MEPs argues for an influence of COVID-19, which could not only represent a bystander response to increased platelet consumption but might rather reflect inflammation-induced emergency megakaryopoiesis (Haas et al., 2015). We did not observe a direct correlation of MK levels with neither platelet counts nor D-Dimers, which might be limited by the small number of scRNaseq datasets. A continuous presence of type I IFN signals, as seen in our dataset, might additionally increase of the aggregation potential of platelets (Leppkes et al., 2020; Middleton et al., 2020). In line with this, the MK-associated module M4 is strongly correlated with D-Dimer levels in the larger bulk dataset from cohort 1.
The second cellular feature was primarily observed in the bulk RNA-seq data and was related to the co-expression module M7. We saw a biphasic upregulation of a transcript group that comprises canonical components of erythropoiesis, which are most likely related to the presence of reticulocytes. GATA-1 TF binding motifs linked to hypoxia-induced stress erythropoiesis (Zhang et al., 2012) were significantly enriched in the module. We thus reasoned that this feature reflects a canonical response to hypoxia because it is present in critically ill patients and at a later stage when patients are weaned of supplemental oxygen. Post-hypoxia polycythaemia and the presence of different erythroid progenitors in the circulation has been studied as a response mechanism of the bone marrow to acute hypoxic insults and critical illness for decades (Loeffler et al., 1984; Peschle et al., 1977). Mobilization of erythroid progenitor cells and their presence in the circulation has been linked to augmented immune responses (Serebrovskaya et al., 2011). Although we did not directly profile erythrocytes in the scRNA-seq data because of red blood cell lysis and size and feature filtering steps in the data processing, we found evidence for the increased presence of committed erythroid-progenitor-like cells in the scRNA-seq data in two of our patients at the stage of oxygen-weaning. From the gene content and the number of cells identified, it is unlikely that these cells represented reticulocytes. Together, the results indicate a profound reaction of the erythroid lineage to COVID-19 at different phases of the disease. These features of the computed disease trajectory were linked to clinical outcome in a larger retrospective cohort of 40 mechanically ventilated COVID-19 patients. We could show that two modules from cohort 1, M4 (related to MK numbers in cohort 1) and M7 (indicative of erythropoiesis in cohort 1), were significantly correlated with a fatal outcome in the independent cohort. Decrease in expression of the M2 module (associated with hypomorphic type I IFN in cohort 1) in a broad array of cell types including monocytes, NK cells, CD8+ T cells, and PBs was present in both survivors and non-survivors, corroborating the observation of failing type I IFN in severe COVID-19, but questioning its clear relation to fatal outcome. Our results clearly suggest that regulatory events in megakaryocytic and erythroid cells might act as pivotal components of an unfavorable course of COVID-19, which mandates further prospective exploration.
Limitations of Study
Limitations of our study are given by the relatively low sample size of the initial two-center cohort (cohort 1), which we aimed to compensate by two independent validation cohorts. The initial findings, which point to functional alterations of cellular features, mandate prospective validation and currently can only be interpreted in light of mechanistic findings in other systemic inflammatory disorders. We saw no evidence for a correlation of PB numbers with delayed development of SARS-CoV-2-recognizing antibodies, which is in line with another study (Kuri-Cervantes et al., 2020). However, an interference of the broad PB activation with other specific B cell reactions, such as affinity maturation of neutralizing antibodies or memory formation, as in severe malaria (Vijay et al., 2020) should not be excluded. Metabolic hyperactivation of PBs will have to be confirmed by other methods, yet from the comparison of the models between subpopulations of the B cell lineage, the observed pattern in PBs does not appear as a general immunometabolic consequence of COVID-19. Likewise, the presence, altered function, and inferred metabolic skewing of MKs and other hematopoietic precursors (e.g., granulocytes-macrophage progenitors [GMPs]) has to be confirmed in larger cohorts and by orthogonal methods (e.g., FACS and proliferation assays). Such studies should definitely also take long-term consequences of microvascular complications (e.g., neurological deficits) into account. Our study furthermore suggests several cytokines (e.g., IL-10 and GDF-15 [Myhre et al., 2020]) as markers of severe COVID-19 trajectories. Large prospective, longitudinal biomarker trials are mandated to come up with clinically actionable predictors of severe COVID-19 trajectories.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human-CD3 Pacific Blue (clone OKT3) | Biolegend | Cat#317314; RRID: AB_571909 |
| Anti-human-CD14 Pacific Blue (clone M5E2) | Biolegend | Cat#301828; RRID: AB_2275670 |
| Anti-human-CD19 Pe-Vio770 (clone REA657) | Miltenyi Biotec | Cat#130-113-647; RRID: AB_2726200 |
| Anti-human-CD20 REA PerCP-Vio700 (clone REA780) | Miltenyi Biotec | Cat#130-111-342; RRID: AB_2656078 |
| Anti-human-CD27 APC (clone M-T271) | Biolegend | Cat#356410; RRID: AB_2561957 |
| Anti-human-CD62L Brilliant Violet 650 (clone DREG-56) | Biolegend | Cat#304832; RRID: AB_2563821 |
| Anti-human-CD95 Alexa Fluor 700 (clone DX2) | Biolegend | Cat#305648; RRID: AB_2750145 |
| Anti-human-CD138 Brilliant Violet 605 (clone MI15) | Biolegend | Cat#356519; RRID: AB_2562861 |
| Anti-human-CCR10 PE (clone REA326) | Biolegend | Cat#130-120-407; RRID: AB_2752080 |
| Anti-human-HLA-DR VioGreen (clone REA805) | Miltenyi Biotec | Cat#130-111-795; RRID: AB_2652164 |
| Anti-human-IgA Heavy Chains FITC (clone M24A) | Millipore | Cat#CBL114F; RRID: AB_92852 |
| Anti-TREML1 polyclonal antibody | Thermo Fischer | Cat#PA5-53647; RRID: AB_2648897 |
| Anti-IFITM3 XP® (clone D8E8G) | Cell Signaling Technologies | Cat#59212S; RRID: AB_2799561 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| PAXgene™ | PreAnalytiX | Cat#762165 |
| BD Vacutainer® CPT™ Mononuclear Cell Preparation Tube – Sodium Citrate | BD | Cat#362761 |
| K3-EDTA Monovette® 7.5 ml | Sarstedt | Cat#01.1605.001 |
| Serum Monovette® 7.5 ml | Sarstedt | Cat#01.1601.001 |
| DPBS | Thermo Fischer/GIBCO | Cat#15326239 |
| DMSO | Merck | Cat#472301-100ML |
| Fetal bovine serumBS | Merck | Cat#F7524; Lot# BCBV0083 |
| RPMI 1640 Medium | Thermo Fischer/GIBCO | Cat#11875093 |
| Sodium azide extra pure | Merck | Cat#1.0668 |
| Titriplex III (EDTA) | Merck | Cat#1.08421.1000 |
| Albumin Bovine Fraction V, pH 7.0 | Serva | Cat#11930.03 |
| Critical Commercial Assays | ||
| 22-plex Luminex assay | Bio-techne | Cat#LXSAHM-22 Config code: JVRbY86E |
| Human Thrombopoietin Quantikine ELISA Kit | R&D Systems | Cat#DPT00B |
| Anti-SARS-CoV-2-specific IgA ELISA | EUROIMMUN | Cat#EI 2606-9601 A |
| Anti-SARS-CoV-2-specific IgG ELISA | EUROIMMUN | Cat#EI 2606-9601 G |
| PAXgene Blood miRNA Kit | PreAnalytiX | Cat#763134 |
| TruSeq® Stranded mRNA Library Prep Kit | Illumina | Cat#20020595 |
| NovaSeq 6000 S1 Reagent Kit v1 (200 cycles) | Illumina | Cat#20012864 |
| NovaSeq 6000 S2 Reagent Kit v1 (200 cycles) | Illumina | Cat#20012861 |
| NovaSeq 6000 S4 Reagent Kit v1 (200 cycles) | Illumina | Cat#20027466 |
| QIAamp DNA Blood Mini Kit | QIAGEN | Cat#51106 |
| Infinium© MethylationEPIC BeadChip Kit | Illumina | Cat#WG-317-1003 |
| KAPA Real-Time PCR library amplification kit | Peqlab | Cat#KK2702 |
| RealStar® SARS-CoV-2 RT-PCR Kit RUO | Altona Diagnostics | Cat#821005 |
| Chromium Next GEM Single Cell 5′ Library & Gel Bead Kit v1.1, 16 rxns | 10x genomics | Cat#1000165 |
| Chromium Single Cell V(D)J Enrichment Kit, Human B Cell, 96 rxns | 10x genomics | Cat#1000016 |
| Chromium Single Cell 5′ Library Construction Kit | 10x genomics | Cat#1000020 |
| BOND Polymer Refine Detection Kit | Leica Biosystems | Cat#DS9800 |
| BOND Epitope Retrieval Solution 1 | Leica Biosystems | Cat#AR9961 |
| BOND Epitope Retrieval Solution 2 | Leica Biosystems | Cat#AR9640 |
| Deposited Data | ||
| Cohort 1 Bulk RNA-seq and Bulk BCR-seq raw and processed data | This paper | GEO: GSE161777 |
| Cohort 1 Illumina EPIC Array raw and processed data | This paper | GEO: GSE161678 |
| Cohort 1 scRNA-seq and scBCR-seq raw data | This paper | N/A |
| Cohort 1 scRNA-seq and scBCR-seq processed data | This paper | FastGenomics https://github.com/Systems-Immunology-IKMB/COVIDOMICs/tree/main/scRNAseq/Prop_Celltypes_github.csv |
| Cohort 2 data | Schulte-Schrepping et al., 2020 | https://dx.doi.org/10.1016/j.cell.2020.08.001 |
| Supplemental Tables | This paper | https://doi.org/10.17632/7686ww5z33.2 |
| Software and Algorithms | ||
| XPonent Software | Luminex | v3.1 |
| FlowJo | https://www.flowjo.com | v10 |
| Prism | https://www.graphpad.com | v8 |
| RNA-seq pipeline | https://github.com/nf-core/rnaseq | N/A |
| Trim Galore | https://github.com/FelixKrueger/TrimGalore | v0.4.4 |
| STAR | Dobin et al., 2013 | v2.5.2b |
| featureCounts | Liao et al., 2014 | v1.5.2 |
| DESeq2 (Bioconductor package) |
Love et al., 2014 http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
v1.20.0 |
| ImpulseDE2 (Bioconductor package) |
Fischer et al., 2018 http://bioconductor.org/packages/release/bioc/html/ImpulseDE2.html |
v1.4.0 |
| R | https://cran.r-project.org | v3.6.3 |
| WGCNA (R package) | Langfelder and Horvath, 2008 | v1.69 |
| MiXCR | Bolotin et al., 2015 | v.3.0.12 |
| vegan (R package) | https://cran.r-project.org/web/packages/vegan/index.html | v1.5-6 |
| tcR (R package) | https://cran.r-project.org/web/packages/tcR/vignettes/tcrvignette.html | v2.3.2 |
| RnBeads (Bioconductor package) |
Assenov et al., 2014 https://rnbeads.org/ |
v1.12.1 |
| GSEA (desktop application) | https://www.gsea-msigdb.org/gsea/ | v4.0.3 |
| topGO (Bioconductor package) | https://bioconductor.org/packages/release/bioc/html/topGO.html | v2.32.0, v2.38.2 |
| LOLA (Bioconductor package) | Sheffield and Bock, 2016 | v1.14.0 |
| CellRanger | 10x genomics | v3.1.0 |
| CellRanger VDJ function | 10x genomics | v3.1.1 |
| Seurat (R package) | Butler et al., 2018 | v3.1.5 |
| SingleR (R package) | Aran et al., 2019 | v1.0.6 |
| monocle3 | Qiu et al., 2017 | v3 |
| MAST | Finak et al., 2015 | v.1.16.0 |
| Sybil (R package) | Gelius-Dietrich et al., 2013 | v2.1.5 |
| sybilSBML (R package) | Gelius-Dietrich et al., 2013 | v3.0.5 |
| Recon 2.2 | https://github.com/varnerlab/Recon-2.2-Model | v2.2 |
| FASTCORE algorithm/COBRA Toolbox |
Vlassis et al., 2014 https://opencobra.github.io/cobratoolbox/stable/modules/dataIntegration/transcriptomics/FASTCORE/index.html |
v.3.0 |
| coin (R package) | https://cran.r-project.org/web/packages/coin/index.html | v1.3-1 |
| viper (R package) | Alvarez et al., 2016 | v1.22.0 |
| OmnipathR (R package) | https://github.com/saezlab/OmnipathR | v1.2.1 |
| CARNIVAL | https://bioconductor.org/packages/release/bioc/html/CARNIVAL.html | v1.0.1 |
| Limma | Ritchie et al., 2015 | v3.44.3 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Philip Rosenstiel, p.rosenstiel@mucosa.de
Material Availability
This study did not generate unique reagents.
Data and Code Availability
The bulk RNA-seq and BCR-seq data generated during this study is available in GEO database (GSE161777).
The Illumina EPIC Array data generated during this study is available in GEO database (GSE161678).
The raw scRNA-seq data and scBCR-seq data generated during this study is available in EGA (Accession number to be confirmed).
The processed scRNA-seq and scBCR-seq data generated during this study is available in FastGenomics (https://beta.fastgenomics.org/p/565003).
Additional processed data is available at GitHub (https://github.com/Systems-Immunology-IKMB/COVIDOMICS).
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/7686ww5z33.2.
The custom codes used in this study are available in https://github.com/Systems-Immunology-IKMB/COVIDOMICs. Please note that there is a diverse set of codes depending on the OMICs layer in question: bulk RNA-seq, Methylation, scRNaseq, BCR, Metabolic modeling, TF enrichment and Data integration.
Experimental Model and Subject Details
Patients and specimen collection (cohort 1)
14 patients from two independent University hospitals (Cologne, Kiel) were recruited for the longitudinal multi-Omics study. Eligibility criteria included age ≥18 years and admission to the respective hospitals (either normal ward or ICU) with a positive SARS-CoV-2 nasopharyngeal swab by RT-PCR. Sex, age and additional demographic information, as well as patient comorbidities can be found in Table S1. This study did not particularly focused on impact of sex on patient outcome due to the limit number of individuals; larger cohorts have been used to study the influence of sex upon SARS-CoV-2 infection. Five patients were co-enrolled in an ongoing clinical trial (remdesivir) at UKSH. Enrolment for longitudinal molecular phenotyping occurred between 1st April 2020 and 6th May 2020 (seven patients at the University Medical Center Schleswig-Holstein, Campus Kiel (UKSH) and seven patients at the University Hospital Cologne (UKK), with the last sample taken on 20th May 2020). One patient with a mild disease course was recruited after his recovery to serve as an additional recovery control. The patients consented to the sampling of biomaterials, analytic processing of the biomaterials and genetic analysis, the study was approved by the independent ethical review board of Kiel University (ref no°: D 466/20) and Cologne (identifier: 20-1295). Eight healthy donors were included as controls at a single time point in the framework of the DZHK study (ethics vote ref no°: D 441/16). While the patients were in inpatient health care, the sampling scheme was day 0 (at admission), day 2, day 7, day 10, and day 13. At each sampling day, blood was collected in PAXgene tubes, CPT tubes, EDTA and serum tubes (except from day 10 1: no CPT tubes were taken). Clinical parameters were retrieved from the electronic patients record systems or from written discharge letters from transferred patients by the COVID-19 clinical consultants (T.B., J.R.) and clinical research fellows (F.T. and P.K.).
To describe the heterogenous disease trajectories over time, a modified WHO ordinal scale (WHO, 2020), which also considers the behavior of several inflammatory markers (serum CRP, serum IL-6 and ferritin) was used (Table S2) to classify patients along their disease course (Figures 1B and 1C) enabling the interrogation of molecular states associated with transition between phases (e.g., complicated to early convalescent). Phases were defined as pseudotimes in accordance with WHO and the LEOSS register (https://leoss.net) to depict the longitudinal course of the disease: incremental (pseudotime 1, where clinical symptoms and inflammatory markers were increasing, ICU or non-ICU), critical (pseudotime 2, ICU, mechanically ventilated with signs of ARDS), complicated (pseudotime 3, state with severe signs of a systemic inflammatory response, ICU, high-flow oxygen, intubation readiness), moderate or early convalescent (pseudotime 4, supplemental oxygen, significant signs of systemic inflammation), late convalescent (pseudotime 5, intermittent supplemental oxygen, minor signs of inflammation), recovery/pre-discharge (pseudotime 6, no supplemental oxygen, absent inflammation markers) and long-term follow-up (pseudotime 7, at least two weeks after hospital discharge) (Table S2). Note that pseudotimes 2 and 3 do not directly reflect the chronological order, but can also represent peak levels, i.e., a given patient might have gone from 1 to 3 without intubation/mechanical ventilation.
Validation cohorts
For validation of cellular findings from our prospective cohorts, data from two independent cohorts were analyzed:
The scRNA-seq data from 18 patients admitted to the University Hospital Bonn (Schulte-Schrepping et al., 2020) (cohort 2), and RNA-seq data from 40 SARS-CoV-2-positive patients admitted to the Intensive Care Unit of the Radboud university medical center in Nijmegen (cohort 3). Age, sex, and patient outcome information included in Table S4. COVID-19 was diagnosed by a positive SARS-CoV-2 RT-PCR test in nasopharyngeal and throat swabs and/or by typical chest CT scan findings. Within cohort 3, seven patients deceased. Blood was collected in PAXgene tubes. The frozen tubes were shipped to Bonn University for mRNA sequencing. Sampling in cohort 3 was carried out in accordance with the applicable rules concerning the review of research ethics committees and informed consent in the Netherlands. All patients or legal representatives were informed about the study details and could decline to participate.
Method Details
ELISA
Serum cytokines were analyzed using a “Human Magnetic Luminex assay” (Bio-techne, Minneapolis, Minnesota, US) with 22 analytes: APRIL, BAFF, CCL2, CCL3, CD40L, CD138, CXCL9, CXCL10, IL-1β, IL-2, IL-4 IL-6, IL-10, IL-12, IL-13, IL-17, IL-18, IL-21, IL-33, IFN-α, IFN-γ, and TNF. Frozen patient serum samples were thawed and diluted before the experiment with an equal amount of dilution buffer and the experiment was performed according to the manufacturer’s instructions. All samples were measured on a Lifematch Fluoroanalyzer (Tepnel Life Science PLC, Wythenshawe, UK) equipped with XPonent 3.1 Software (Luminex Corporation, Austin, Texas, US). Serum TPO was quantified by ELISA using Human Thrombopoietin Quantikine ELISA Kit (R&D Systems, Minneapolis, Minnesota, US) according to manufacturer’s protocol.
Anti-SARS-CoV-2 specific antibodies
Anti-SARS-CoV-2-specific IgA and IgG was quantified using commercial ELISA kits (EUROIMMUN, Lübeck, Germany).
PAXgene Blood RNA Isolation and TruSeq® messenger RNA (mRNA) sequencing
Blood (2.5 mL) was taken from each patient into a PAXgene Blood RNA Tube, containing a patented RNA stabilizer reagent composition. RNA was automated isolated in QIAGEN’s QIAcube using the PAXgene Blood miRNA Kit from QIAGEN PreAnalytiX. RNA sequencing libraries were prepared according to the Illumina TruSeq® messenger (mRNA) sequencing protocol (TruSeq® RNA Seq Library Prep Kit v2). The resulting libraries were sequenced on the NovaSeq 6000 (2 × 50 bp, S2 chemistry).
DNA isolation and methylation profiling
Blood (2.7 mL) was taken from each patient into an Ethylenediaminetetraacetic acid (EDTA) monovette (Sarstedt) to decelerate blood coagulation. The monovette was centrifuged for 15 min at 3000 rpm and the buffy coat was frozen in micronic tubes at −80°C. DNA was extracted using the QIAamp DNA Blood Mini Kit (QIAGEN) according to manufacturer’s protocol with a QIACube (QIAGEN). The Infinium© MethylationEPIC BeadChip was used to measure the DNA methylation levels. The Infinium© MethylationEPIC BeadChip targets the following regions: CpG islands, CpG sites, open chromatin, transcription factor binding sites, enhancer and mRNA promoter regions. The EPIC arrays were processed according to Illumina recommendations.
Bulk BCR sequencing
Bulk BCR libraries were prepared starting from 100 ng of total RNA isolated from PAXGene tubes. Library construction protocol was performed as previously described (Bashford-Rogers et al., 2019). In brief, primers for the constant (C) regions of the BCR were used during cDNA synthesis. Product was then amplified via PCR using a multiplex primer set for the variable (V) regions using the Real-Time PCR library amplification kit from KAPA Biosystems. Libraries were sequenced on Illumina MiSeq machine 2 × 300 bp.
Experimental virus mRNA detection
SARS-CoV-2-specific viral RNA from the RNA extracts from the PAXGene tubes was quantified by RT-PCR using the RealStar® SARS-CoV-2 RT-PCR Kit RUO (altona diagnostics, Hamburg, Germany). The two amplicons were in the E gene and in the S glycoprotein in Spike protein 2 gene.
Isolation of peripheral blood mononuclear cell (PBMC)
Blood (2 × 8 mL) was collected using venipuncture technique and processed within maximum 30 min. PBMCs were isolated using the BD vacutainer® cell preparation tube (CPT) with sodium citrate according to the manufacturer’s protocol. Briefly, CPT tubes were centrifuged at 1,650 × g for 20 min at room temperature. PBMCs were collected and washed two times with phosphate-buffered saline (PBS) and then resuspended in PBS. Half of the suspension was washed and resuspended in flow cytometry washing buffer (containing fetal bovine serum, EDTA and sodium azide in PBS) and prepared for flow cytometry within 3-5 h. The rest of the suspension was washed once in PBS and the pellet was resuspended in resuspension medium (Roswell Park Memorial Institute (RPMI) + 40% fetal bovine serum (FBS)), followed by freezing medium (30% DMSO in medium containing 40% FCS) according the 10 × Chromium Demonstrated Protocol (Fresh Frozen Human Peripheral Blood Mononuclear Cells for Single-Cell ribonucleic acid (RNA) Sequencing, Document CG00039 Rev D). PBMCs were stored at −80°C and thawed when needed also according to the 10 × Chromium Demonstrated Protocol (Document CG00039 Rev D). To prevent batch effects, all samples from different time points from one patient were thawed and sequenced together.
Flow cytometry
After preparation of freshly isolated PBMCs in flow cytometry buffer, PBMCs were stained with fluorescent labeled antibodies and measured on a MACSQuant 16 flow cytometer (Miltenyi Biotec, Bergisch Gladbach, Germany). B cell subsets were stained using antibodies against CD19 (clone REA675, Miltenyi), CD20 (clone REA780, Miltenyi), CD27 (clone M-T271, Biolegend, San Diego, California, US), CD138 (clone BB4/MI15, Biolegend), HLA-DR (clone REA332, Miltenyi), IgD (clone REA740, Miltenyi), IgM (clone MHM-88, Biolegend), IgA (clone M24A, Merck Millipore), CD95 (clone DX2, Biolegend) as well as CD3 (clone OKT3, Biolegend) and CD14 (clone M5E2, Biolegend) for the dump channel. Definition of B cell populations: naive B cells: CD19+CD20+CD27-; memory B cells: CD19+CD20+CD27+; Plasmablasts and plasma cells: CD19+CD20-CD27hi).
Immunohistochemistry staining
Tissue material was obtained in the autopsy procedure of the deceased patient 002 and from an age- and gender-matched patient (control), who had suffered from pulmonary adenocarcinoma and died from bronchopneumonia. SARS-CoV-2-testing was negative repeatedly in the control patient. Immunostaining was done on formalin-fixed and paraffin-embedded tissue sections with the Bond Max Leica immunostainer using the Bond Polymer Refine Detection Kit. Antigen retrieval was carried out with the Leica ER1-Bond Epitope Retrieval Solution 1 (IFITM3) or the Leica ER2-Bond Epitope Retrieval Solution 2 (TREML1)(all Leica Biosystems, Wetzlar, Germany). Histological slides were then immunostained anti-IFITM3 and anti-TREML1 antibodies and counterstained with hematoxylin.
Single-cell RNA sequencing (scRNA-seq)
Single-cell libraries were generated using the Chromium Next GEM Single Cell 5′ Library & Gel bead Kit v1.1 according to the manufacturer’s user guide targeting 20,000 cells per sample. The libraries were sequenced on an Illumina NovaSeq 6000 (2 × 100 bp, S4 chemistry) to generate > 500 million reads per library. Additionally, the Chromium Single Cell V(D)J Enrichment Kit for human B cells were applied together with the Chromium Single Cell 5′ Library Construction Kit. Those resulting libraries were sequenced on an Illumina NovaSeq 6000 (2 × 150 bp, S4 chemistry) to generate > 50 million reads per library.
Quantification and Statistical Analysis
Bulk RNA-seq data analysis
An in-house RNA-seq pipeline was used to map and align the sequenced data (https://github.com/nf-core/rnaseq). Adapters and low-quality bases from the RNA-seq reads were removed using Trim Galore (version 0.4.4), which is a wrapper tool for Cutadapt and FastQC. Reads that were shorter than 35 bp after trimming were discarded. The filtered reads were mapped to the human genome (GRCh38, gencode version 25) using STAR aligner (version 2.5.2b) (Dobin et al., 2013). featureCounts (version 1.5.2) was used to estimate the expression counts of the genes. The expression counts were normalized across samples using the DESeq normalization method.
Differential expression analysis
Differentially expressed genes between healthy controls and each of the COVID pseudotime samples were identified using the Bioconductor package DESeq2 (version 1.20.0). Genes with FDR adjusted p value of less than 0.05, log fold change greater than 0.5 or less than −0.5 and average expression counts of more than 100 were regarded as differentially expressed (DEGs). Longitudinal differential expression analysis of the COVID samples was performed by applying the case-only analysis from the Bioconductor package ImpulseDE2 (version 1.4.0). Pseudotimes 1, 3, 4, 5, and 6 were used as single time points of a time-course experiment and the patient IDs were regarded as batch effects in order to perform a paired analysis. To identify the transcripts regulated longitudinally in survivors and non-survivors from the Nijmegen cohort, differentially expressed genes between in the two selected time points were identified using DESeq2 for survivors and non-survivors separately. To perform a paired analysis, patient ID was used as batch effect.
Co-expression analysis
Modules of co-expressed genes were identified using the WGCNA package for R (version 1.69). All differentially expressed genes identified from the pairwise and longitudinal analysis (6,318 genes in total) were used to generate the gene co-expression modules. First, pairwise gene correlations were calculated based on the log transformed normalized expression counts across all samples. A signed adjacency matrix was constructed by applying a soft threshold function with a power of 14. The Topology Overlap Matrix (TOM) constructed using the adjacency matrix was used to construct a gene tree by hierarchical clustering. Genes were then split into modules based on the gene tree by using the function cutreeDynamic with the minimum module size set to 15. Modules that were closely related were then merged using the function mergeCloseModules with parameter cutHeight set to 0.45.
To associate gene co-expression modules with clinical parameters and cell type fractions and to visualize the expression profile of the genes in a module, the module eigengene values for the samples were calculated. Spearman’s rank correlation coefficients were calculated between the module eigengenes and different clinical parameters and cell type fractions.
DNA methylation data analysis
DNA methylation data were analyzed using the Bioconductor package RnBeads (version 1.12.1). Sites that overlapped with SNPs and had unreliable measurements were filtered resulting in the removal of 17,371 sites and 19,745 probes. 2,977 Context-specific probes, 18,976 probes on the sex chromosomes, and 4 probes with missing values were also removed. In total 41,702 out of 866,895 probes were filtered. The signal intensity values were normalized using the dasen method. Differentially methylated positions (DMPs) between healthy controls and each of the COVID-19 pseudotime samples as well as between sequential COVID-19 pseudotime samples were identified using the automatically selected rank cutoff of RnBeads.
Functional enrichment analysis
Gene set enrichment analysis (GSEA) was conducted for the co-expression modules using GSEA desktop application (version 4.0.3). Pre-ranked analyses against Hallmark, KEGG and GO (Biological Processes) genes sets were conducted for each of the modules by ranking all genes by the module membership score. FDR of 0.05 was used as the significance threshold.
All gene ontology enrichment analyses were conducted using the Bioconductor package topGO (version 2.32.0), with all expressed genes as the universe set. In the topGO analysis, the Fisher.elim p value, calculated using the weight algorithm, of 0.05 was used as the significance threshold.
Transcription factor binding sites (TFBS) enriched in the promoter regions of the co-expression module genes were identified by conducting enrichment analysis using the Bioconductor package LOLA (version 1.14.0). Promoter regions were defined as the region between 1,500 bp upstream to 500 bp downstream of the transcription start site.
Predicted transcription factor binding sites (TFBS) enriched in DMPs were identified by conducting enrichment analysis using the Bioconductor package LOLA (version 1.14.0).
DNA methylation-transcriptome integrated analysis
For the integrated analysis of gene expression with DNA methylation, we first identified DMPs located 5,000 bp upstream and downstream of the transcription start sites of DEGs. Spearman’s rank correlation coefficient between the normalized expression count of each DEG and the methylation intensity (β values) of its corresponding DMPs were calculated. To test the statistical significance of the correlations, we calculated the false discovery rate (FDR) using a permutation approach.
Bulk BCR analysis
Sequencing reads were aligned to BCR gene reference and clonotypes were identified and grouped using the software MiXCR (Bolotin et al., 2015). Relative proportions of IGH classes were calculated. Alpha diversity measures were calculated using the R packages vegan and tcR (versions 1.5-6 and 2.3.2).
Computational virus mRNA detection
To quantify the amount of virus present in the blood of COVID-19 patients, reads from whole blood RNA-seq data were first aligned to the human reference genome (GRCh38) using STAR with default parameters. The reads that did not map to the human genome were then aligned to the SARS-CoV-2 reference genome (NC_045512.2) using STAR with slightly relaxed parameters (–outFilterScoreMinOverLread 0.2–outFilterMatchNminOverLread 0.2–outFilterMatchNmin 0–outFilterMismatchNmax 4). The reads that aligned to the SARS-CoV-2 genome with at least 40 consecutive matches were then aligned locally to the human genome using the Smith-Waterman algorithm (Smith and Waterman, 1981) in order to filter any reads of human origin. The reads that aligned locally to the human genome or were composed largely of homopolymers were filtered and the remaining reads were considered as viral reads.
Data analysis for ELISA
Standard curves and cytokine concentrations were calculated using linear regression in Microsoft Excel GraphPad Prism (Graphpad Software Inc, San Diego, US).
Data analysis for Flow cytometry data
Analyses were performed using FlowJo v10 (FlowJo LLC, Beckton Dickinson, Ashland, Oregon, US) and Graphpad Prism 8 (GraphPad Software, San Diego, California USA).
scRNA-seq data quality control and data analysis
The sequences were processed using cell ranger v3.1.0 (10 × Genomics). Each sample was mapped to GRCh38 Homo sapiens reference genome, in order to produce their respective count matrices. Raw feature-barcode matrixes were filtered using Seurat package (version 3.1.5) in R environment (Butler et al., 2018; Stuart et al., 2019); low quality cells that were potentially disrupted or doublets cells were removed from the analysis using number of features (number of reads mapping to gene between [200;5000]) or percentage of mitochondria (lower than 25%) (Figure S1E). We used a broad threshold in order to encompass cells from a variety of cell-types.
Each filtered sample matrix was then merged into a single object containing 358,930 cells with overall reads mapping to 22,519 human genes. The merge object was normalized and scaled using LogNormalized() and ScaleData() functions respectively. Principal component analysis was performed utilizing the top 2,000 variable genes. We identified the clusters using the standard k-nearest neighbor method based on 80 dimensions with a 0.2 resolution. In total 37 clusters were displayed as a Uniform Manifold Approximation and Projection (UMAP).
scRNA-seq signature genes
Cluster cell types were identified by their corresponding gene signatures using the Wilcoxon rank sum, with a cut-off based on genes expressed in more than 25% of the cluster cells and exhibiting a 0.25-fold difference between clusters. Clusters of interest were identified based on marker genes (Figure S1C); In general, monocytes (CD14, ITGAM, S100A8 and A100A9), granulocytes (FCGR3B), erythroid cells (HBA1, HBA2 and HBB), NK cells (GNLY, NCAM1), proliferative lymphocytes (MKI67 and TUBA1B), CD4+ T cells (CD3G, CD4 and CCR7), CD8+ T cells (CD3G and CD8A), dendritic cells (PLD4, IL3RA and LILRA4), B cells (CD19), plasmablasts (CD27, CD38 and MZB1), megakaryocytes (ITGA2B, TUBB1 and GP9) and cell precursors (CD34, ITGA4 and TUBA1A), such as, hematopoietic stem cells (HSCs), megakaryocyte-erythroid progenitor cells (MEPs), common myeloid progenitors (CMPs) and granulocytes-macrophage progenitors (GMPs). To confirm our findings, we used SingleR (version 1.0.6) Bioconductor package that assigns each individual cell to a known cell type based on transcriptome reference datasets (NovershternHematopoieticData and BlueprintEncodeData reference datasets) (Aran et al., 2019). Cell type proportions were quantified per sample and grouped based on pseudotimes. We compared cell type proportions of healthy controls against patients using a Mann-Whitney non-parametric test and measured cell proportion changes between pseudotimes by comparing a linearlinear mixed model with pseudotime (proportion of cell type∼pseudotime +[1|patientID]) and compared it against a reduce model without pseudotime (proportion of cell type +[1|patientID]) by the means of an ANOVA (Figure 2F). We found most cell types to be impacted by disease trajectory, with only proliferative lymphocytes and cell precursors not having a significant difference between pseudotimes. Furthermore, we correlated cell type proportion with the clinical parameters available for each sample using spearman correlation (Figure 2G).
Cell type specific analysis
The clusters identified as cell types of interest—B cell compartment, megakaryocytes, and cell precursors were pulled from the merged object. Each cell type of interest was re-clustered and analyzed separately. B cell compartment and cell precursors were re-clustered using 80 PCs, while megakaryocytes and their respective precursors (HSCs and MEPs) were re-cluster using 60 PCs. B cell compartment clusters were assigned based on expression of marker genes, with memory B cell expressing CD73/NT5E, naive B cells expressing IGHD and CD185/CXCR5, transitional B cells expressing CD9, plasmablasts expressing CD27 and CD38) and neutrophil-like cell expressing ELANE, MPO and CAMP (Glass et al., 2020). We presented the B cell compartment as a cell trajectory analysis using monocle3 (Qiu et al., 2017). Trajectories were calculated and the cells displayed based on monocle3 pseudotime approach rooted on the previously identified transitional B cells. Plasmablasts (with neutrophil-like cells included), megakaryocytes and cell precursors signature genes for the individual disease groups were selected based on genes expressed in more than 25% and 0.25-fold difference between pseudotimes. Differentially expressed genes between healthy controls and each of the COVID pseudotimes was identified using MAST (Finak et al., 2015) and GO enrichment analysis was performed with TopGO package for R (version 2.38.1) (Alexa et al., 2006) and GO terms of interest selected based on fisher classic test statistic (p value < 0.05).
To validate our findings, we performed a parallel analysis from an independent cohort (cohort 2) of mild and severe COVID-19 patients using another scRNA-seq technology (Rhapsody BD) (Schulte-Schrepping et al., 2020). Similarly, we identified plasmablasts and megakaryocytes, identified differences in cell proportions and tested if genes of interest were expressed differently based on their disease classification (control, mild COVID-19 or severe COVID-19 patients).
single cell BCRseq (scBCR-seq) analysis
BCR sequences were processed using cellranger v3.0.1 vdj function. Relative proportions of IGH classes were calculated. Alpha diversity measures were calculated using the R packages vegan and tcR (versions 1.5-6 and 2.3.2). scBCR-seq information was merged with B cell compartment containing both B cells and plasmablasts using barcode information. BCR information was merged with scRNA-seq expression, thus, we were able to discern IG class information per cell type and displayed it in the form of a UMAP or monocle3 cell trajectory.
Metabolic modeling
Blood metabolites originating from the Human Metabolome Database (HMDB) (Wishart et al., 2018); specifically, an advanced search of blood metabolites from healthy adults was initially conducted in March 2019. In the same time period, a list of all human metabolites (Brunk et al., 2018) was downloaded from Virtual Metabolic Human (VMH) database (Noronha et al., 2019). The latter contains the HMDB indices of the compounds, which enabled the merging of the two databases into one (Spring 2020). Compounds were removed from the database (e.g., drug-related ones or without FooDB IDs [https://foodb.ca/]) and the metabolites nomenclature was altered to be compatible with an adapted version of the model Recon 2.2 (Swainston et al., 2016). All values were transformed to mM to correspond with mean values of the HMDB database (cut-off of values > 10−6). Water was set to 55,000 mM, and pH was set to 7.4. All calculations were conducted with the R packages sybil (version 2.1.5) (Gelius-Dietrich et al., 2013) and sybilSBML (version 3.0.5) (Gelius-Dietrich et al., 2013) along with their dependencies.
Reconstruction of tissue-specific metabolic models from bulk sequencing data
For the reconstruction of metabolic models from bulk sequencing data we used a previously described two-step approach that first discretizes gene expression based on differential gene expression analysis and subsequently reconstructs metabolic models based on gene expression states (Gebauer et al., 2016; Yang et al., 2019). Differentially expressed genes between each pairwise set of COVID pseudotimes were determined via the DESeq2 algorithm with pseudotime as the main independent variable. Wherever possible, i.e., when models reached full rank, we included the donor identifier as a covariable to control for paired samples with similar genetic background in the data. The significance cut-off used for optimizing independent filtering was adjusted to α = 0.05 before differentially expressed genes were extracted from the tests. For each gene, based on the directionality of changes in gene expression activity between conditions and the significance of changes, we determined a binary activity (on or off) if a gene had at least one case of significant change in activity (adjusted p value < 0.05) between any pair of conditions.
Subsequently, we used the binary gene activity as an input into the iMAT approach (Zur et al., 2010) on a generic metabolic model of humans (Recon 2.2) (Swainston et al., 2016) constrained with the serum metabolic environment. In order to test method-inherent uncertainties in the reconstructed context-specific metabolic network, the model reconstruction procedure was repeated fifty times while leaving out gene activity data for 5% of the genes each time.
Reconstruction of cell-specific metabolic models from single-cell sequencing data
Reconstructions of cell-specific metabolic models were created by integrating single-cell transcriptomics with a human genome-scale metabolic network (Swainston et al., 2016) conditioned with the serum metabolic environment. We employed a two-step approach, in which StanDep (Joshi et al., 2020) first identifies a core reaction list across cell types. Second, the FASTCORE algorithm (Pacheco and Sauter, 2018) in the COBRA Toolbox v.3.0 (Heirendt et al., 2019) then builds context-specific models defined by sets of active core reactions in the extracted model. scRNA counts from patient, megakaryocytes, plasmablasts, memory B cells and naive B cells were used as input for StanDep. Normalized counts were converted into TPM-values, and ENSEMBL gene names were mapped to Recon 2.2 (Cunningham et al., 2019; Swainston et al., 2016). For StanDep, expression data from identified core genes across cell types were used to calculate enzyme type and expression within the model. Enzyme expressions were log10 transformed and counted as a binary matrix (rows representing enzymes and columns as bins) to identify the minimum and maximum enzyme expression values. A complete linkage metric for hierarchical clustering with Euclidean distance was used to cluster (number of clusters = 40) genes based on gene expression. Assembled core reaction matrices were defined and input into FASTCORE to reconstruct context-specific metabolic models. Based on an updated version of Recon 2.2 simulated in the serum metabolic environment, a consistent model was generated using FASTCORE’s FASTcc algorithm in MATLAB. With this consistent model, the assembled core reaction matrices, and additional optional core reactions, such as the biomass objective function from Recon 2.2, an input file was generated for every cell-specific model comprising at least 30 core reactions. FASTCORE processed these input files, together with the consistent Recon 2.2 model, and the default value 10−4 for ε to generate a list of all required reactions for each cell-specific metabolic model. With these lists, new metabolic models were curated and optimized for the biomass objective function and/or the viral biomass objective function.
Identification of disease-specific metabolic pathways
Tissue- and cell-specific models were stratified by cell type (megakaryocytes, plasmablasts, memory B cells, and naive B cells) and annotated with clinical metadata (COVID-19 pseudotime) according to donor and sampling time points. Reactions per pathway and cell were counted for a total of 82 metabolic pathways that were identified in the models. For each of the four model types, pathways were filtered out if reaction counts were zero across all models of that type. Differential pathway activity was determined for each pathway by comparing reaction counts across all eight pseudotimes via Kruskal-Wallis test as implemented in the R-package coin (parameters: two-sided test, unpaired, average-scores for ties, and without continuity correction). Resulting probability values were corrected for multiple testing via the Benjamini and Hochberg method. Significantly differential active pathways across disease states (pseudotimes) were determined via this method for all four model types separately with an FDR cut-off of <0.05. For the heatmap representation of B cell subtypes, the top 20 pathways that were identified as significantly differential active in all three model types were selected and clustered by the mean reaction counts of plasmablast models.
Transcription factor activity analysis
Putative transcription factor activity from RNA-seq data were assessed per pseudotime against healthy controls using the human gene set resource DoRothEA v1, which provides a curated collection of transcription factor and target genes interactions (the regulon) from different sources (Garcia-Alonso et al., 2019). Only interactions with high, likely, and medium confidence (levels A, B, and C) were considered. Regulons were statistically evaluated using the R package viper (v1.22.0; row-wise t tests) and regulons having at least 15 expressed gene targets were considered (Alvarez et al., 2016). Identification of upstream regulatory signaling pathways from downstream gene expression was performed on t-statistic values from viper against the Omnipath interaction database (R package OmnipathR version 1.2.1) applying CARNIVAL (version 1.0.1 with IBM Cplex solver as network optimizer) (Liu et al., 2019). For the resulting network, only edges with an inferred weight > 50 (on a scale from 1 to 100) were considered. The similarity between the transcription factor activity of cohort 1 and cohort 3 was accessed using Pearson correlation. The significance of the correlation was confirmed by a bootstrapping approach. We permuted gene names row wise 1,000 times and correlated each time the inferred TF activity between the original and permuted gene expression. The resulting distribution of correlation values was fitted to a skewed Gaussian normal distribution, finding a standard deviation of 0.08.The significance in the differential regulation of transcription factors over time in the non-survivor versus the survivor groups of cohort 3 was quantified via a moderated t test using limma (Ritchie et al., 2015), while accounting for patient correlation using a block design and limma’s duplicateCorrelation function.
Consortia
The members of the HCA lung biological network are: Nicholas E. Banovich, Tushar Desai, Oliver Eickelberg, Muzlifa Haniffa, Peter Horvath, Jonathan A. Kropski, Robert Lafyatis, Joakim Lundeberg, Kerstin Meyer, Martijn C. Nawijn, Marko Nikolic, Jose Ordovas Montanes, Dana Pe’er, Purushothama Rao Tata, Emma Rawlins, Aviv Regev, Paul Reyfman, Christos Samakovlis, Joachim L. Schultze, Alex Shalek, Douglas Shepherd, Jason Spence, Sarah Teichmann, Fabian Theis, Alexander Tsankov, Maarten van den Berge, Michael von Papen, Jeffrey Whitsett, and Laure Emmanuelle Zaragosi
The members of the Deutsche COVID-19 Omics Initiative (DeCOI) are: Angel Angelov, Robert Bals, Alexander Bartholomäus, Anke Becker, Daniela Bezdan, Ezio Bonifacio, Peer Bork, Thomas Clavel, Maria Colme-Tatche, Andreas Diefenbach, Alexander Dilthey, Nicole Fischer, Konrad Förstner, Julia-Stefanie Frick, Julien Gagneur, Alexander Goesmann, Torsten Hain, Michael Hummel, Stefan Janssen, Jörn Kalinowski, René Kallies, Birte Kehr, Andreas Keller, Sarah Kim-Hellmuth, Christoph Klein, Oliver Kohlbacher, Jan O. Korbel, Ingo Kurth, Markus Landthaler, Yang Li, Kerstin Ludwig, Oliwia Makarewicz, Manja Marz, Alice McHardy, Christian Mertes, Markus Nöthen, Peter Nürnberg, Uwe Ohler, Stephan Ossowski, Jörg Overmann, Silke Peter, Klaus Pfeffer, Anna R. Poetsch, Alfred Pühler, Niklaus Rajewsky, Markus Ralser, Olaf Rieß, Stephan Ripke, Ulisses Nunes da Rocha, Philip Rosenstiel, Antoine-Emmanuel Saliba, Leif Erik Sander, Birgit Sawitzki, Philipp Schiffer, Eva-Christina Schulte, Joachim L. Schultze, Alexander Sczyrba, Oliver Stegle, Jens Stoye, Fabian Theis, Janne Vehreschild, Jörg Vogel, Max von Kleist, Andreas Walker, Jörn Walter, Dagmar Wieczorek, and John Ziebuhr
Acknowledgments
We thank K. Greve, M. Rohm, M. Hansen, S. Kock, D. Oelsner, S. Baumgarten, M. Reffelmann, M. Schlapkohl, N. Braun, T. Wesse, M. Basso, Y. Dolschanskaya, X. Yi, C. Lancken, and M. Vollstedt for perfect technical assistance. This work was supported by the German Research Foundation (DFG) CCGA Nr. 07495230, a COVID-response grant of the state SH, ExC 2167 Precision Medicine in Chronic Inflammation (RTF-VI), the research group miTARGET and the CRC1182 C2 project to P.R.; the IMI2 Project 3TR to P.R. and S.S.; INST 37/1049-1, INST 216/981-1, INST 257/605-1, INST 269/768-1, INST 217/988-1, INST 217/577-1, and EXC2151/1 (390873048) to J.L.S.; SFB TR57 and SPP1937 to J.N.; Helmholtz-Gemeinschaft Deutscher Forschungszentren, Germany (sparse2big) to J.L.S.; EU projects SYSCID (733100) to P.R. and J.L.S.; the DZIF, Germany (TTU 04.816 and 04.817) to J.N.; and the Hector Foundation) (M89) to J.N. A.D. acknowledges support by the DFG (EXC 2124-390838134 Controlling Microbes to Fight Infections) and the German Center for Infection Research (DZIF grant N° 8020708703). C.K. acknowledges support by the DFG through EXC 2167, RTF-VIII, and the CRC 1182. This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications). We are indebted to the patients, their families, and the hospital staff for support, without whom this study would not have been possible.
Author Contributions
S.S., J.L.S., and P.R. conceived study concept and design. J.P.B., N.M., F.T., T. Bahmer, U.G., P.K., P.B., G.E., A.F., N.F., R.J., K.F.R., J. Rupp, A.S., H.B., B.F.H., C.K., J. Heyckendorf, M.K., and J. Rybniker contributed to study concept. J.P.B., N.M., F.T., L.B., U.G., A.K, E.R., J.J.S, H.B., B.F.H., C.K., J.L.S., and P.R. contributed to literature search, data interpretation, and writing the initial manuscript. All authors contributed to reviewing and editing of the manuscript. F.T., T. Bahmer, J.I.B., J.F., U.G., P.K., E.R., J.D., G.E., J. Franzenburg, S.F., N.F., J. Fuß, A.G., J. Hamm, F.H., S.I., S.K., C.L., G.L., M.L., R.M., J.N., P.P., C.R., J. Rupp, A.S., D. Skowasch, J. Heyckendorf, M.K., and P.R. conducted sample collection and processing. J.P.B., N.M., F.T., L.B., U.G., J.J.S., A.K., E.R., A.C.A, N.B., T. Boysen, B.B., A.D., D.E., M.F., M.P.H., Y.H.K., R.K., C.L., G.M., A.R., J.S.S, T.U., K.P.W., M.W., J.Z., H.B., and C.K. conducted data analysis. J.P.B., N.M., F.T., L.B., D.B., U.G., J.J.S., A.K., C.R., E.R., H.B., and P.R. made figures and tables.
Declaration of Interests
The authors declare no conflicting interests.
Published: December 15, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.11.017.
Contributor Information
HCA Lung Biological Network:
Nicholas E. Banovich, Tushar Desai, Oliver Eickelberg, Muzlifa Haniffa, Peter Horvath, Jonathan A. Kropski, Robert Lafyatis, Joakim Lundeberg, Kerstin Meyer, Martijn C. Nawijn, Marko Nikolic, Jose Ordovas Montanes, Dana Pe’er, Purushothama Rao Tata, Emma Rawlins, Aviv Regev, Paul Reyfman, Christos Samakovlis, Joachim Schultze, Alex Shalek, Douglas Shepherd, Jason Spence, Sarah Teichmann, Fabian Theis, Alexander Tsankov, Maarten van den Berge, Michael von Papen, Jeffrey Whitsett, and Laure Emmanuelle Zaragosi
the Deutsche COVID-19 Omics Initiative (DeCOI):
Angel Angelov, Robert Bals, Alexander Bartholomäus, Anke Becker, Daniela Bezdan, Ezio Bonifacio, Peer Bork, Thomas Clavel, Maria Colme-Tatche, Andreas Diefenbach, Alexander Dilthey, Nicole Fischer, Konrad Förstner, Julia-Stefanie Frick, Julien Gagneur, Alexander Goesmann, Torsten Hain, Michael Hummel, Stefan Janssen, Jörn Kalinowski, René Kallies, Birte Kehr, Andreas Keller, Sarah Kim-Hellmuth, Christoph Klein, Oliver Kohlbacher, Jan O. Korbel, Ingo Kurth, Markus Landthaler, Yang Li, Kerstin Ludwig, Oliwia Makarewicz, Manja Marz, Alice McHardy, Christian Mertes, Markus Nöthen, Peter Nürnberg, Uwe Ohler, Stephan Ossowski, Jörg Overmann, Silke Peter, Klaus Pfeffer, Anna R. Poetsch, Alfred Pühler, Niklaus Rajewsky, Markus Ralser, Olaf Rieß, Stephan Ripke, Ulisses Nunes da Rocha, Philip Rosenstiel, Antoine-Emmanuel Saliba, Leif Erik Sander, Birgit Sawitzki, Philipp Schiffer, Eva-Christina Schulte, Joachim L. Schultze, Alexander Sczyrba, Oliver Stegle, Jens Stoye, Fabian Theis, Janne Vehreschild, Jörg Vogel, Max von Kleist, Andreas Walker, Jörn Walter, Dagmar Wieczorek, and John Ziebuhr
Supplemental Information
References
- Alexa A., Rahnenführer J., Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Alvarez M.J., Shen Y., Giorgi F.M., Lachmann A., Ding B.B., Ye B.H., Califano A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 2016;48:838–847. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenov Y., Müller F., Lutsik P., Walter J., Lengauer T., Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford-Rogers R.J.M., Bergamaschi L., McKinney E.F., Pombal D.C., Mescia F., Lee J.C., Thomas D.C., Flint S.M., Kellam P., Jayne D.R.W. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature. 2019;574:122–126. doi: 10.1038/s41586-019-1595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F. Presence of SARS-CoV-2 reactive T cells in COVID-19 patients and healthy donors. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Breuer K., Foroushani A.K., Laird M.R., Chen C., Sribnaia A., Lo R., Winsor G.L., Hancock R.E., Brinkman F.S., Lynn D.J. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk E., Sahoo S., Zielinski D.C., Altunkaya A., Dräger A., Mih N., Gatto F., Nilsson A., Preciat Gonzalez G.A., Aurich M.K. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 2018;36:272–281. doi: 10.1038/nbt.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.A., Schwertz H., Hottz E.D., Rowley J.W., Manne B.K., Washington A.V., Hunter-Mellado R., Tolley N.D., Christensen M., Eustes A.S. Human megakaryocytes possess intrinsic antiviral immunity through regulated induction of IFITM3. Blood. 2019;133:2013–2026. doi: 10.1182/blood-2018-09-873984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran L.M., Nutt S.L. Long-Lived Plasma Cells Have a Sweet Tooth. Immunity. 2016;45:3–5. doi: 10.1016/j.immuni.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S. Ensembl 2019. Nucleic Acids Res. 2019;47(D1):D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasi S., Tartaro D.L., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Mattioli M., Paolini A., Gozzi L. Expansion of plasmablasts and loss of memory B cells in peripheral blood from COVID-19 patients with pneumonia. Eur. J. Immunol. 2020;50:1283–1294. doi: 10.1002/eji.202048838. [DOI] [PubMed] [Google Scholar]
- de la Peña N.C., Sosa-Melgarejo J.A., Ramos R.R., Méndez J.D. Inhibition of platelet aggregation by putrescine, spermidine, and spermine in hypercholesterolemic rabbits. Arch. Med. Res. 2000;31:546–550. doi: 10.1016/s0188-4409(00)00238-1. [DOI] [PubMed] [Google Scholar]
- De Voeght A., Calmes D., Beck F., Sylvestre J.-B., Delvenne P., Peters P., Vertenoeil G., Baron F., Layios N., Canivet J.-L. Thrombotic microvascular injury is not mediated by thrombotic microangiopathy despite systemic complement activation in Covid-19 patients. medRxiv. 2020 doi: 10.1101/2020.10.07.20208603. [DOI] [Google Scholar]
- Deshpande C. Thromboembolic Findings in COVID-19 Autopsies: Pulmonary Thrombosis or Embolism? Ann Intern Med. 2020 doi: 10.7326/M20-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D.S., Theis F.J., Yosef N. Impulse model-based differential expression analysis of time course sequencing data. Nucleic Acids Res. 2018;46:e119. doi: 10.1093/nar/gky675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alonso L., Holland C.H., Ibrahim M.M., Turei D., Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019;29:1363–1375. doi: 10.1101/gr.240663.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer J., Gentsch C., Mansfeld J., Schmeißer K., Waschina S., Brandes S., Klimmasch L., Zamboni N., Zarse K., Schuster S. A Genome-Scale Database and Reconstruction of Caenorhabditis elegans Metabolism. Cell Syst. 2016;2:312–322. doi: 10.1016/j.cels.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Gelius-Dietrich G., Desouki A.A., Fritzemeier C.J., Lercher M.J. Sybil--efficient constraint-based modelling in R. BMC Syst. Biol. 2013;7:125. doi: 10.1186/1752-0509-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass D.R., Tsai A.G., Oliveria J.P., Hartmann F.J., Kimmey S.C., Calderon A.A., Borges L., Glass M.C., Wagar L.E., Davis M.M., Bendall S.C. An Integrated Multi-omic Single-Cell Atlas of Human B Cell Identity. Immunity. 2020;53:217–232. doi: 10.1016/j.immuni.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas S., Hansson J., Klimmeck D., Loeffler D., Velten L., Uckelmann H., Wurzer S., Prendergast A.M., Schnell A., Hexel K. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 2015;17:422–434. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häsler R., Feng Z., Bäckdahl L., Spehlmann M.E., Franke A., Teschendorff A., Rakyan V.K., Down T.A., Wilson G.A., Feber A. A functional methylome map of ulcerative colitis. Genome Res. 2012;22:2130–2137. doi: 10.1101/gr.138347.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heirendt L., Arreckx S., Pfau T., Mendoza S.N., Richelle A., Heinken A., Haraldsdóttir H.S., Wachowiak J., Keating S.M., Vlasov V. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 2019;14:639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H., Zhang P., Dayaram T. C/EBPβ is required for 'emergency' granulopoiesis. Nat. Immunol. 2006;7:732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- Hoepel W., Chen H.-J., Allahverdiyeva S., Manz X., Aman J., Bonta P., Brouwer P., de Taeye S., Caniels T., van der Straten K. Anti-SARS-CoV-2 IgG from severely ill COVID-19 patients promotes macrophage hyper-inflammatory responses. bioRxiv. 2020 doi: 10.1101/2020.07.13.190140. [DOI] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Lin J.J., Tao J., Fisher P.B. Suppression of human ribosomal protein L23A expression during cell growth inhibition by interferon-beta. Oncogene. 1997;14:473–480. doi: 10.1038/sj.onc.1200858. [DOI] [PubMed] [Google Scholar]
- Joshi C.J., Schinn S.M., Richelle A., Shamie I., O’Rourke E.J., Lewis N.E. StanDep: Capturing transcriptomic variability improves context-specific metabolic models. PLoS Comput. Biol. 2020;16:e1007764. doi: 10.1371/journal.pcbi.1007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalapos M.P. Methylglyoxal and glucose metabolism: a historical perspective and future avenues for research. Drug Metabol. Drug Interact. 2008;23:69–91. doi: 10.1515/dmdi.2008.23.1-2.69. [DOI] [PubMed] [Google Scholar]
- Kanerva K., Mäkitie L.T., Pelander A., Heiskala M., Andersson L.C. Human ornithine decarboxylase paralogue (ODCp) is an antizyme inhibitor but not an arginine decarboxylase. Biochem. J. 2008;409:187–192. doi: 10.1042/BJ20071004. [DOI] [PubMed] [Google Scholar]
- Kaser A., Dunzendorfer S., Offner F.A., Ludwiczek O., Enrich B., Koch R.O., Cruikshank W.W., Wiedermann C.J., Tilg H. B lymphocyte-derived IL-16 attracts dendritic cells and Th cells. J. Immunol. 2000;165:2474–2480. doi: 10.4049/jimmunol.165.5.2474. [DOI] [PubMed] [Google Scholar]
- Kim J., Kim W., Le H.T., Moon U.J., Tran V.G., Kim H.J., Jung S., Nguyen Q.-T., Kim B.-S., Jun J.-B. IL-33-induced hematopoietic stem and progenitor cell mobilization depends upon CCR2. J. Immunol. 2014;193:3792–3802. doi: 10.4049/jimmunol.1400176. [DOI] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., Kuthuru O., Apostolidis S.A., Bershaw L., Dougherty J., Greenplate A.R., Pattekar A., Kim J., Han N., Gouma S., Weirick M.E., Arevalo C.P., Bolton M.J., Goodwin E.C., Anderson E.M., Hensley S.E., Jones T.K., Mangalmurti N.S., Luning Prak E.T., Wherry E.J., Meyer N.J., Betts M.R. Comprehensive mapping of immune perturbations associated with severe COVID-19. Science Immunology. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W.Y., Becker A.M., Kennerly K.M., Wong R., Curtis J.D., Llufrio E.M., McCommis K.S., Fahrmann J., Pizzato H.A., Nunley R.M. Mitochondrial Pyruvate Import Promotes Long-Term Survival of Antibody-Secreting Plasma Cells. Immunity. 2016;45:60–73. doi: 10.1016/j.immuni.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.D., de Graaf E.L., Sonneveld M.E., Plomp H.R., Linty F., Visser R., Brinkhaus M., Sustic T., deTaeye S.W., Bentlage A.E. Afucosylated immunoglobulin G responses are a hallmark of enveloped virus infections and show an exacerbated phenotype in COVID-19. bioRxiv. 2020 doi: 10.1101/2020.05.18.099507. [DOI] [Google Scholar]
- Lax S.F., Skok K., Zechner P., Kessler H.H., Kaufmann N., Koelblinger C., Vander K., Bargfrieder U., Trauner M. Pulmonary Arterial Thrombosis in COVID-19 With Fatal Outcome: Results From a Prospective, Single-Center, Clinicopathologic Case Series. Ann Intern Med. 2020 doi: 10.7326/M20-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Hsu A.K., Sajdak J., Qin J., Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.-S. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppkes M., Knopf J., Naschberger E., Lindemann A., Singh J., Herrmann I., Stürzl M., Staats L., Mahajan A., Schauer C. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58:102925. doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liao S.-C., Shao S.-C., Chen Y.-T., Chen Y.-C., Hung M.-J. Incidence and mortality of pulmonary embolism in COVID-19: a systematic review and meta-analysis. Crit. Care. 2020;24:464. doi: 10.1186/s13054-020-03175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G., Plebani M., Henry B.M. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clinica Chimica Acta. 2020 doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Trairatphisan P., Gjerga E., Didangelos A., Barratt J., Saez-Rodriguez J. From expression footprints to causal pathways: contextualizing large signaling networks with CARNIVAL. NPJ Syst. Biol. Appl. 2019;5:40. doi: 10.1038/s41540-019-0118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler M., Herkenrath P., Wichmann H.E., Lord B.I., Murphy M.J., Jr. The kinetics of hematopoietic stem cells during and after hypoxia. A model analysis. Blut. 1984;49:427–439. doi: 10.1007/BF00320485. [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Lorente-Sorolla C., Garcia-Gomez A., Català-Moll F., Toledano V., Ciudad L., Avendaño-Ortiz J., Maroun-Eid C., Martín-Quirós A., Martínez-Gallo M., Ruiz-Sanmartín A. Inflammatory cytokines and organ dysfunction associate with the aberrant DNA methylome of monocytes in sepsis. Genome Med. 2019;11:66. doi: 10.1186/s13073-019-0674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne B.K., Denorme F., Middleton E.A., Portier I., Rowley J.W., Stubben C.J., Petrey A.C., Tolley N.D., Guo L., Cody M.J. Platelet Gene Expression and Function in COVID-19 Patients. Blood. 2020 doi: 10.1182/blood.2020007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz D.K., McCray P.B., Jr. Illuminating COVID-19 lung disease through autopsy studies. EBioMedicine. 2020;57:102865. doi: 10.1016/j.ebiom.2020.102865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton E.A., He X.-Y., Denorme F., Campbell R.A., Ng D., Salvatore S.P., Mostyka M., Baxter-Stoltzfus A., Borczuk A.C., Loda M. Neutrophil Extracellular Traps (NETs) Contribute to Immunothrombosis in COVID-19 Acute Respiratory Distress Syndrome. Blood. 2020 doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Scherer M., Assenov Y., Lutsik P., Walter J., Lengauer T., Bock C. RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol. 2019;20:55. doi: 10.1186/s13059-019-1664-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhre P.L., Prebensen C., Strand H., Røysland R., Jonassen C.M., Rangberg A., Sørensen V., Søvik S., Røsjø H., Svensson M., Berdal J.E., Omland T. Growth Differentiation Factor-15 Provides Prognostic Information Superior to Established Cardiovascular and Inflammatory Biomarkers in Unselected Patients Hospitalized with COVID-19. Circulation. 2020 doi: 10.1161/circulationaha.120.050360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak M.K., Ghatge M., Dhanesha N., Flora G.D., Jain M., Rodriguez O., Markan K., Potthoff M., Lentz S.R., Chauhan A.K. American Society of Hematology Washington; DC: 2019. Targeting Metabolic Enzyme Pyruvate Kinase M2: A Novel Strategy to Inhibit Platelet Function and Arterial Thrombosis. [Google Scholar]
- Ni L., Ye F., Cheng M.-L., Feng Y., Deng Y.-Q., Zhao H., Wei P., Ge J., Gou M., Li X. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noronha A., Modamio J., Jarosz Y., Guerard E., Sompairac N., Preciat G., Daníelsdóttir A.D., Krecke M., Merten D., Haraldsdóttir H.S. The Virtual Metabolic Human database: integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 2019;47(D1):D614–D624. doi: 10.1093/nar/gky992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E.Z., Chan Y.F.Z., Leong W.Y., Lee N.M.Y., Kalimuddin S., Haja Mohideen S.M., Chan K.S., Tan A.T., Bertoletti A., Ooi E.E., Low J.G.H. A Dynamic Immune Response Shapes COVID-19 Progression. Cell Host Microbe. 2020;27:879–882. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K., Spolski R., Ettinger R., Kim H.-P., Wang G., Qi C.-F., Hwu P., Shaffer D.J., Akilesh S., Roopenian D.C. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 2004;173:5361–5371. doi: 10.4049/jimmunol.173.9.5361. [DOI] [PubMed] [Google Scholar]
- Pacheco M.P., Sauter T. The FASTCORE Family: For the Fast Reconstruction of Compact Context-Specific Metabolic Networks Models. Methods Mol. Biol. 2018;1716:101–110. doi: 10.1007/978-1-4939-7528-0_4. [DOI] [PubMed] [Google Scholar]
- Pan W.-H., Sommer F., Falk-Paulsen M., Ulas T., Best P., Fazio A., Kachroo P., Luzius A., Jentzsch M., Rehman A. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med. 2018;10:27. doi: 10.1186/s13073-018-0534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschle C., Magli M.C., Cillo C., Lettieri F., Genovese A., Pizzella F., Soricelli A. Kinetics of erythroid and myeloid stem cells in post-hypoxia polycythaemia. Br. J. Haematol. 1977;37:345–352. doi: 10.1111/j.1365-2141.1977.tb01005.x. [DOI] [PubMed] [Google Scholar]
- Qiu X., Hill A., Packer J., Lin D., Ma Y.A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapkiewicz A.V., Mai X., Carsons S.E., Pittaluga S., Kleiner D.E., Berger J.S., Thomas S., Adler N.M., Charytan D.M., Gasmi B. Megakaryocytes and platelet-fibrin thrombi characterize multi-organ thrombosis at autopsy in COVID-19: A case series. EClinicalMedicine. 2020:100434. doi: 10.1016/j.eclinm.2020.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J., Schultze J.L., Yosef N. ImpulseDE: detection of differentially expressed genes in time series data using impulse models. Bioinformatics. 2017;33:757–759. doi: 10.1093/bioinformatics/btw665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., Deutsche COVID-19 OMICS Initiative (DeCOI) Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple J.W., Italiano J.E., Jr., Freedman J. Platelets and the immune continuum. Nat. Rev. Immunol. 2011;11:264–274. doi: 10.1038/nri2956. [DOI] [PubMed] [Google Scholar]
- Serebrovskaya T.V., Nikolsky I.S., Nikolska V.V., Mallet R.T., Ishchuk V.A. Intermittent hypoxia mobilizes hematopoietic progenitors and augments cellular and humoral elements of innate immunity in adult men. High Alt. Med. Biol. 2011;12:243–252. doi: 10.1089/ham.2010.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield N.C., Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Stephens D.S., McElrath M.J. COVID-19 and the Path to Immunity. JAMA. 2020;324:1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainston N., Smallbone K., Hefzi H., Dobson P.D., Brewer J., Hanscho M., Zielinski D.C., Ang K.S., Gardiner N.J., Gutierrez J.M. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay R., Guthmiller J.J., Sturtz A.J., Surette F.A., Rogers K.J., Sompallae R.R., Li F., Pope R.L., Chan J.-A., de Labastida Rivera F. Infection-induced plasmablasts are a nutrient sink that impairs humoral immunity to malaria. Nat. Immunol. 2020;21:790–801. doi: 10.1038/s41590-020-0678-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassis N., Pacheco M.P., Sauter T. Fast reconstruction of compact context-specific metabolic network models. PLoS Comput. Biol. 2014;10:e1003424. doi: 10.1371/journal.pcbi.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2020. https://www.who.int/blueprint/priority-diseases/key-action/COVID-19_Treatment_Trial_Design_Master_Protocol_synopsis_Final_18022020.pdf
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020 doi: 10.1101/2020.04.17.20069930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vázquez-Fresno R., Sajed T., Johnson D., Li C., Karu N. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Petersen C., Pees B., Zimmermann J., Waschina S., Dirksen P., Rosenstiel P., Tholey A., Leippe M., Dierking K. The Inducible Response of the Nematode Caenorhabditis elegans to Members of Its Natural Microbiota Across Development and Adult Life. Front. Microbiol. 2019;10:1793. doi: 10.3389/fmicb.2019.01793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman M.R. Platelets: at the nexus of antimicrobial defence. Nat. Rev. Microbiol. 2014;12:426–437. doi: 10.1038/nrmicro3269. [DOI] [PubMed] [Google Scholar]
- Zhang F.L., Shen G.M., Liu X.L., Wang F., Zhao Y.Z., Zhang J.W. Hypoxia-inducible factor 1-mediated human GATA1 induction promotes erythroid differentiation under hypoxic conditions. J. Cell. Mol. Med. 2012;16:1889–1899. doi: 10.1111/j.1582-4934.2011.01484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur H., Ruppin E., Shlomi T. iMAT: an integrative metabolic analysis tool. Bioinformatics. 2010;26:3140–3142. doi: 10.1093/bioinformatics/btq602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNA-seq and BCR-seq data generated during this study is available in GEO database (GSE161777).
The Illumina EPIC Array data generated during this study is available in GEO database (GSE161678).
The raw scRNA-seq data and scBCR-seq data generated during this study is available in EGA (Accession number to be confirmed).
The processed scRNA-seq and scBCR-seq data generated during this study is available in FastGenomics (https://beta.fastgenomics.org/p/565003).
Additional processed data is available at GitHub (https://github.com/Systems-Immunology-IKMB/COVIDOMICS).
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/7686ww5z33.2.
The custom codes used in this study are available in https://github.com/Systems-Immunology-IKMB/COVIDOMICs. Please note that there is a diverse set of codes depending on the OMICs layer in question: bulk RNA-seq, Methylation, scRNaseq, BCR, Metabolic modeling, TF enrichment and Data integration.