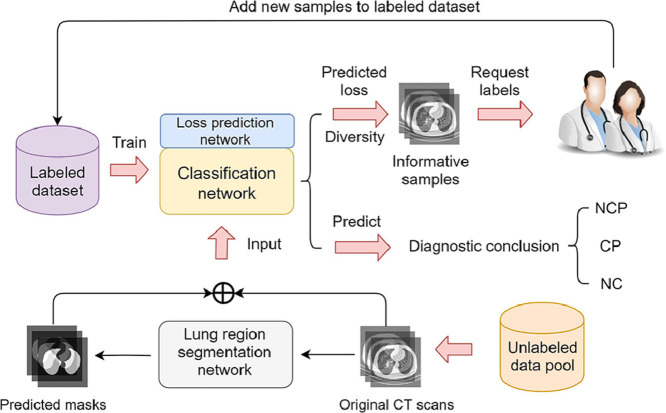
Graphical abstract
Keywords: COVID-19, Deep active learning, Computer-aided diagnosis, Sample diversity, Predicted loss
Abstract
The efficient diagnosis of COVID-19 plays a key role in preventing the spread of this disease. The computer-aided diagnosis with deep learning methods can perform automatic detection of COVID-19 using CT scans. However, large scale annotation of CT scans is impossible because of limited time and heavy burden on the healthcare system. To meet the challenge, we propose a weakly-supervised deep active learning framework called COVID-AL to diagnose COVID-19 with CT scans and patient-level labels. The COVID-AL consists of the lung region segmentation with a 2D U-Net and the diagnosis of COVID-19 with a novel hybrid active learning strategy, which simultaneously considers sample diversity and predicted loss. With a tailor-designed 3D residual network, the proposed COVID-AL can diagnose COVID-19 efficiently and it is validated on a large CT scan dataset collected from the CC-CCII. The experimental results demonstrate that the proposed COVID-AL outperforms the state-of-the-art active learning approaches in the diagnosis of COVID-19. With only 30% of the labeled data, the COVID-AL achieves over 95% accuracy of the deep learning method using the whole dataset. The qualitative and quantitative analysis proves the effectiveness and efficiency of the proposed COVID-AL framework.
1. Introduction
The outbreak of the novel coronavirus epidemic in the world has gotten everyone’s attention. This new coronavirus called SARS-CoV-2 can cause serious infectious diseases which are also termed as Coronavirus Disease 2019 (COVID-19). Many patients develop novel coronavirus pneumonia and progress into serious acute respiratory illness Guan et al. (2020). The pandemic has propagated fast due to person-to-person transmission Phan et al. (2020) and has resulted in around 17,023,229 cases all over the world with over 667,106 death till July 30, 2020, which is a great threat to the global human health. Therefore, the early recognition of COVID-19 risk factors is critical for not only the cure of individual infected patients but also public disease containment.
Currently, a reverse transcriptase polymerase chain reaction (RT-PCR) is a gold standard for SARS-CoV-2 confirmation. However, the laboratory test may take up to days because of the limited supply and strict condition requirements, which may delay the diagnosis of suspected patients. While chest computed tomography (CT) is not so accurate as RT-PCR test in virus detection, it may be more efficient and sensitive for clinical evaluation of SARS-CoV-2 pneumonia and is better for quantitative measurement of the severity of lung involvement (Ai et al., 2020). The significance of chest CT manifestations in the diagnostic procedure has been shown in several recent reports (Chung, Bernheim, Mei, Zhang, Huang, Zeng, Cui, Xu, Yang, Fayad, et al., 2020, Lei et al., 2020, Song, Shi, Shan, Zhang, Shen, Lu, Ling, Jiang, Shi, 2020). The combination of CT scans and clinical findings can make a quick and effective diagnosis possible.
Recent advances in the applications of artificial intelligence (AI) Shi et al. (2020) have inspired the development of AI-based computer-aided diagnosis (CAD) system (Esteva et al., 2019, Wang et al., 2020, Zhang et al., 2020, Xie, Ma, Robson, Chung, Bernheim, Mani, Calcagno, Li, Li, Shan, et al.) in many healthcare areas. As one of the important techniques of AI, deep learning has shown significant diagnostic accuracy in disease detection of medical imaging (Ardila et al., 2019, Suzuki, 2017). Previous works have reviewed the effectiveness of quantitative models of chest CT in lung disease diagnosis (Chen et al., 2020).
Although deep learning can make an accurate diagnosis, the performance of deep neural networks depends on large numbers of labeled CT scans. However, the labeling work of chest CT scans requires not only lots of time and effort but also the domain expertise of medical professionals. Due to the fast spread of the COVID-19, large scale annotation of CT scans is impossible because of limited time and heavy burden on the healthcare system. To meet this challenge, the active learning method is introduced to effectively cut the cost of labeling by actively selecting the most informative samples according to certain strategies. The effectiveness of deep active learning methods is shown in early studies (Zhou, Shin, Gurudu, Gotway, Liang, Yoo, Kweon, 2019, Yang, Zhang, Chen, Zhang, Chen, 2017, Smailagic, Costa, Gaudio, Khandelwal, Mirshekari, Fagert, Walawalkar, Xu, Galdran, Zhang, et al.,). What’s more, large scale lesion annotation of COVID-19 is also impossible because of the fast spread of the disease. Thus, a better choice is performing weakly-supervised COVID-19 detection. To train a deep neural network with fewer labeled data and patient-level labels, we propose a weakly-supervised deep active learning framework called COVID-AL to interactively diagnose COVID-19. The proposed COVID-AL is validated on the CT scans constructed from cohorts from the China Consortium of Chest CT Image Investigation (CC-CCII). The experimental results demonstrate that COVID-AL can greatly cut the annotation cost for training a deep neural network and consistently outperforms the state-of-the-art active learning methods in the diagnosis of COVID-19.
The main contributions of this paper can be summarized as follows:
-
•
We propose the COVID-AL framework that can simultaneously consider the data diversity and data uncertainty to improve the efficiency of active learning methods. Furthermore, a discretization method is also proposed to map the values of different scales to scale-free ranks. Thus the problem of sampling efficiency in active learning methods is addressed from the basic principle of data infomativeness.
-
•
We tailor-design a 2D U-Net for the lung region segmentation and a 3D residual network for the diagnosis of COVID-19.
-
•
We perform weakly supervised active learning with patient-level labels in the proposed COVID-AL framework, which outperforms the state-of-the-art active learning approaches in cutting the labeling cost and achieves effective diagnosis of COVID-19.
The remaining parts of this paper are arranged as follows. The related studies are discussed in Section 2. The detailed description of the experimental materials, proposed framework, and algorithms will be shown in Section 3. Section 4 demonstrates the experimental results and corresponding empirical evaluation. Section 5 presents a discussion about the motivation and originality of this work. This paper is concluded with future work in Section 6.
2. Related work
2.1. Medical AI application
There are many effective medical AI applications in previous studies. Wu et al. (2020) integrated deformable convolutional neural networks into attentive encoder-decoder, which can be employed in the diagnosis of gastroenteric diseases. Ardila et al. (2019) trained a deep learning model on 42,290 CT scans from 14,851 candidates and achieved 0.944 ROC-AUC. Chilamkurthy et al. (2018) proposed a deep learning method on the head CT task and obtained 0.920 ROC-AUC. For COVID-19 detection, Zhang et al. (2020) utilized a DeepLabv3 model to segment the lung region and applied a 3D ResNet model to perform 3-way classification. The overall performance obtained an ROC-AUC of 0.981. Xie et al. (2020) trained a convolutional neural network to learn the features of CT scans and combined the features with clinical information data using a multi-layer perceptron classifier. Wang et al. (2020) proposed a weakly-supervised deep neural network that could be trained using patient-level labels. They used only 499 scans to obtain 0.959 ROC-AUC.
Although there have been some deep learning-based segmentation or detection methods for COVID-19, there are few deep active learning methods for the diagnosis of it.
2.2. Deep active learning
In the field of medical image analysis, it is hard to build large labeled datasets since the labeling of medical images is time-consuming as well as labor-intensive. For this reason, the research of active learning in medical imaging keeps attracting intensive interests. Yang et al. (2017) proposed a suggestive deep active learning framework for medical image segmentation. The framework is based on a fully convolutional network (FCN) architecture and selects the informative samples based on uncertainty. Zhou et al. (2017) combined deep active learning with transfer learning and proposed an active fine-tuning framework called AIFT. However, this framework is only suitable for binary classification tasks. Zhou et al. (2018) improved the AIFT and proposed a superior approach AFT* for adapting multi-class cases. AFT* is also used in a carotid intima-media thickness video interpretation task (Zhou et al., 2019). Yoo and Kweon (2019) proposed a method to learn the loss of classification. They attached a loss prediction module to the target classifier model and jointly trained them in each round of active learning. The above-mentioned approaches only considered a single sampling strategy in the active learning process. Yuan et al. (2019) proposed a multiple criteria deep active learning framework. However, due to excessive computational consumption, they could only use a relatively shallow neural network. OMelAL Smailagic et al. (2019) was a state-of-the-art active learning framework for medical image analysis, which selected the most distant data points from the centroid in embedding space. But this framework may not perform well when only weak labels are available.
In a word, there has not yet been an effective weakly-supervised active learning framework for COVID-19 diagnosis.
3. Materials and methods
3.1. The datasets
In real world applications, it is natural to apply the active learning framework on unlabeled datasets. However, in the experiment of this research, it will not be able to evaluate the performance of the proposed active learning framework without ground-truth labels. Thus, we introduce a labeled dataset to evaluate the performance of proposed COVID-AL framework. When the framework queries labels from medical experts, it is provided with the ground-truth labels. This experimental setup helps us to conduct the experiments without the costly interaction of medical experts and enables quantitative evaluation of the framework performance.
3.1.1. The dataset for COVID-19 diagnosis
The dataset for COVID-19 diagnosis was constructed from cohorts from the China Consortium of Chest CT Image Investigation (CC-CCII) Zhang et al. (2020). All CT scans are classified into 3 classes including novel coronavirus pneumonia (NCP) due to SARS-CoV-2 virus infection, common pneumonia (CP), and normal controls (NC). The human subjects were considered clinically appropriate for chest CT scans. The association of age and gender were not taken into consideration. The CC-CCII dataset contains 617,775 axial slices of CT scans from 4154 human subjects. Since the active learning iterations on the entire dataset would be too expensive in terms of computational resources and time, we use a subset of the CT scans from 962 patients (304 for NCP, 316 for CP, and 342 for NC respectively). The selection of the subset of CT scans is not in contradiction to the following data augmentation. The selection is to reducing computing cost of experiments, whereas the data augmentation is to imitate real world active learning applications.
To support the diversity-based sampling strategy as well as to avoid overfitting, the CT scans are preprocessed with data augmentation. Random affine transformation and color jittering are applied before the training process. On the one hand, the affine transformation is composed of rotation, scaling, and shearing. On the other hand, the color jittering randomly adjusts brightness and contrast of CT scans.
3.1.2. The dataset for lung region segmentation
To improve the accuracy of COVID-19 diagnosis, the lung region segmentation is indispensable, which is an essential preprocessing step to minimize the analytical region and system computation. However, there is a lack of the lung region annotation for the CC-CCII dataset. Thus the NSCLC dataset Kiser et al. (2020) is introduced to train a tailor-designed 2D U-Net Ronneberger et al. (2015) for lung region segmentation.
The NSCLC dataset has 402 CT scans with corresponding lung region annotations. In each volume of the dataset, the label of the backgrounds is 0; the label of the left lung is 1; the label of the right lung is 2. In the training of the segmentation network, the label of both left and right lung regions is set to 1. The axial slices in the volumes is cropped to the size of 352*320 for model training.
3.2. The lung region segmentation
3.2.1. The framework of lung region segmentation
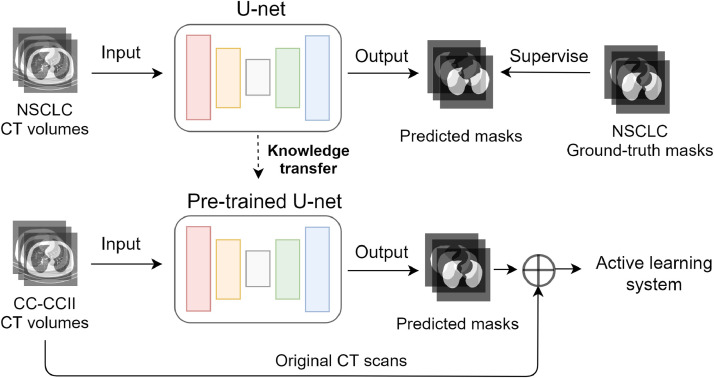
Lung region segmentation is a critical preprocessing step in the COVID-19 diagnosis. Since lung region annotation is not available for CC-CCII dataset, the key idea of the proposed framework of lung region segmentation is knowledge transfer. As shown in Fig. 1 , a 2D U-Net is pre-trained on the NSCLC dataset as the segmentation network and the knowledge is transferred to the CC-CCII dataset for lung region segmentation.
Fig. 1.
The knowledge transfer for lung region segmentation.
On the one hand, the CT scans in the NSCLC dataset are split into 2D slices to train the segmentation network. On the other hand, the masks for the CT scan volumes are obtained by the pre-trained segmentation network slice by slice without using the temporal information on the CC-CCII dataset. In this step, each CT volume in the CC-CCII dataset is provided with its corresponding masks before the active learning procedure. Because the original data volumes in the CC-CCII dataset are preprocessed with lung region segmentation, the background information is removed for CT scans and leads to better detection and diagnosis of COVID-19.
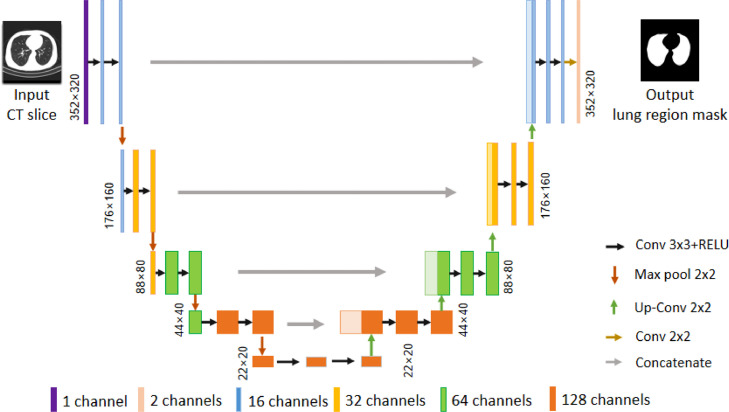
3.2.2. The network architecture
We tailor-design a 2D U-Net architecture as shown in Fig. 2 to segment the lung regions for each CT slice. The slices are first cropped to and input into the network. The encoder of the segmentation network extracts image features by double convolutional layers and pooling layers in 4 steps of down sampling. The down sampling reduces each slice to 1/16 of its original size. Then, in 4 steps of up sampling, the decoder of the segmentation network uses skip connection to concatenate the feature map in the same stage. Finally, the output layer gives out a lung region mask whose size is the same as the size of the input CT slice. Thus, the segmentation network is capable of utilizing multi-scale image features to learn a lung region mask for each input CT slice.
Fig. 2.
The network architecture for lung region segmentation.
3.3. The diagnosis of COVID-19 with the COVID-AL
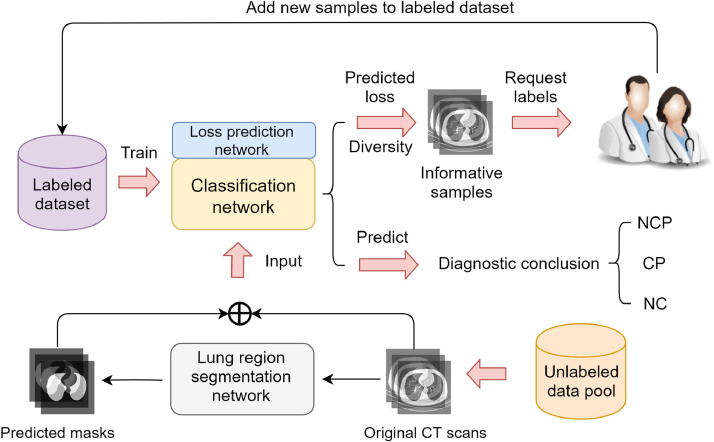
In practice, with the fast spread of COVID-19, there will be large numbers of CT scans waiting for labeling, which can be considered as a large unlabeled data pool. It is unacceptable for medical experts to carefully annotate all these CT scans, which is extremely time-consuming and labor-intensive. As demonstrated in Fig. 3 , the proposed COVID-AL reads CT scans and picks the most informative data samples with a hybrid sampling strategy. The COVID-AL then requests medical experts for labels of selected data samples. In each active learning cycle, the new samples selected by the framework will be added into the labeled dataset and removed from the unlabeled data pool.
Fig. 3.
The COVID-AL framework for weakly-supervised active learning.
The details of the COVID-AL algorithm is demonstrated in Algorithm 1 . The central idea of the proposed COVID-AL is to consider both the diversity and the predicted loss for sample selection simultaneously. The classification network draws the diagnostic conclusion according to the original CT volumes. The prediction confidence can be used to compute the diversity of CT volumes. The loss prediction network is responsible for predicting the classification loss for each CT volume. The informativeness of samples can be precisely represented by fusing the diversity and the predicted loss.
Algorithm 1.
COVID-AL: Active learning framework for COVID-19 diagnosis.
3.3.1. Diversity-based sampling strategy
The diversity-based sampling strategy is supported by data augmentation techniques discussed in Section 3.1.1. With data augmentation, the CT volume of one patient can generate multiple CT volumes with the same ground truth labels. That is to say, only the patient-level labels are available for the COVID-AL framework. However, the classification network may make different decisions in the classification of these augmented CT volumes, which results in inconsistent prediction. A higher degree of inconsistency indicates that more information is contained in the data sample.
The diversity of one candidate can be represented by the divergence of the generated volumes:
| (1) |
In Eq. (1), is the category number of the classification task. is the augmented volume number of each patient.
Data augmentation can effectively improve model performance when the training data is insufficient, but it will also inevitably produce uncertain noisy data in CT volumes. The above-mentioned diversity-based method utilizes the probability information of all outputted categories from the classification network in which the result will be affected by noises. Thus, it is indispensable to remove noisy data.
A processing approach is adopted to select only a portion of the CT volumes generated by each data sample for calculating the diversity. Due to the inconsistency of classification prediction mentioned above, the classification network will produce different classification predictions for multiple augmented CT volumes of a single patient. The main category of a certain candidate is defined as the category where the classification network has the largest average prediction confidence.
| (2) |
The classification network outputs the probability distribution of each category. If the probability for an input CT volume being the main category is very low, then this input CT volume is more likely to be noisy data. The augmented volumes are arranged in descending order of confidence on the main category and the top of the volumes are selected for calculating the divergence . This method can effectively eliminate the influence of noisy data and accelerate the calculation of diversity.
3.3.2. Loss-prediction-based sampling strategy
Intuitively, the classification network will have higher classification loss when deciding on hard samples. The classification loss directly depicts the informativeness of data samples. For the real world application of COVID-AL, it is infeasible to compute the classification loss without ground-truth labels. Thus, our loss-prediction-based method is to train a regression network to dynamically fit the loss of the classification network. In this way it’s possible to predict the classification loss on unlabeled data.
A loss prediction network is used to select candidate samples based on the predicted loss. The loss prediction network is designed for a regression task to predict the current classification loss of the input unlabeled data. The loss prediction network is attached to the classification network to obtain the 3D feature maps as input. The 3D residual blocks of the classification network can extract volume features. The classification loss is computed by comparing the predicted probability and ground truth labels with certain criteria. The loss prediction network uses the feature maps as input and it takes the classification loss as ground truth labels for training.
The loss prediction task is different from traditional regression problems in many aspects. The distribution and the scale of the classification loss vary significantly during the learning of the classification network. In the early stages of active learning, the classification loss is relatively large; while in the later stages, the classification loss becomes very small. Under this circumstance, using mean square error (MSE) as the loss function may not lead to good results. What’s more, the ultimate goal of loss prediction is to correctly sort the samples according to the predicted loss, not to precisely predict loss values. Thus it is intuitive to enlarge the margin between a small loss and a large loss.
A data pair-based loss function is designed to discard the impact of scale changes of regression. The COVID-AL considers a pair of input data samples at the same time and compares their loss. In deep learning, training data is input into the network in the form of mini-batches in most cases. Supposing the batch size is an even number the samples of one batch can be treated as sample pairs. The loss prediction network can be trained by comparing a pair of predicted losses. In this way, the influence of scale change is eliminated. The loss function of the loss prediction network is designed as follows:
| (3) |
In Eq. (3), represents the loss pair and represents the predicted loss pair . is a positive constant which represents the margin between large loss and small loss. The meaning of Eq. (3) is as follows: when the expected loss prediction network should give the prediction and the difference between them exceeds otherwise a penalty is imposed to the loss prediction network to update its weights.
3.3.3. Hybrid sampling strategy
The key idea of the hybrid sampling strategy is to integrate the diversity-based sampling strategy and the loss-prediction-based sampling strategy mentioned above. The problem is that different types of informativeness metrics have different data scales and cannot be combined directly. In the hybrid sampling strategy, a discretization method is proposed to map the values of predicted loss and diversity to scale-free ranks. The candidate samples are sorted in ascending order according to the predicted loss and computed diversity, respectively. The sorted indices are and . The candidate sample with larger predicted loss value has larger . Similarly, the candidate sample with larger diversity value has larger . Since the ranks are scale-free, we can get hybrid rank by summing up and in an element-wise manner as illustrated in Eq. (4).
| (4) |
The weight of the diversity-based sampling strategy and loss-prediction-based sampling strategy is controlled by the parameters and . It is easy to adjust the weight in the hybrid strategy to adapt to the tasks.
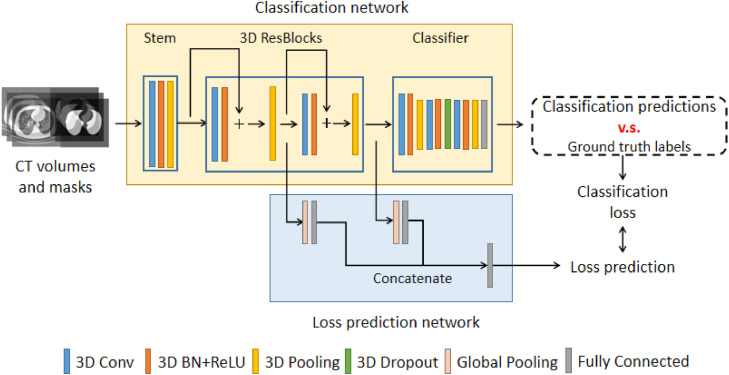
3.3.4. The network architecture
The detailed information of the network architecture is illustrated in Fig. 4 , which consists of a classification network and a loss prediction network. The classification network is a 3D deep residual network Hara et al. (2017) trained with 3D CT volumes. As shown in the upper part of Fig. 4, the classification network consists of a network stem, two 3D residual blocks, and a classifier. The network stem is constructed with a 3D convolutional layer, a batch normalization layer, and a pooling layer. In each residual block He et al. (2016), a 3D feature map is passed into a 3D convolutional layer with a batch normalization layer and a shortcut connection. The output feature maps of each residual block are added in an element-wise manner. The high-level feature maps are input into the classifier, which contained three 3D convolutional layers with pooling and a fully-connect (FC) layer activated by softmax function. The adaptive 3D max-pooling layer makes the network capable of learning in a weakly-supervised manner. The classifier finally outputs the predicted probability of being NCP (novel coronavirus pneumonia), CP (common pneumonia), and NC (normal control).
Fig. 4.
The network architecture of COVID-AL.
In the training process, the loss prediction network extracts the 3D feature maps from the residual blocks of the classification network. As shown in the lower part of Fig. 4. The feature maps are passed into global pooling layers and fully connected layers and converted to feature vectors. The output feature vectors are concatenated into a complete feature vector that contained feature representation of different levels. The feature vector is input into another fully connected layer to make loss prediction accordingly.
4. Experimental evaluation
4.1. Experimental setup
The active learning process is conducted in 30 cycles. In each active learning cycle, 10 patient-level labels are requested from the medical experts. The number of data augmentation for each patient is 8. The training batch size is set to 2. In each cycle, the classification network is trained until its convergence. The network is considered converged when the accuracy on validation set does not increase within 20 epochs. The learning rate of the classification network and the loss prediction network is set to 0.001. The momentum is set to 0.9 and the weight decay is set to 0.0005. The loss prediction margin threshold is set to 0.8. In the diversity-based sampling strategy, the majority selection ratio is set to 0.5. Randomization is injected into sampling by selecting a random subset of the unlabeled data pool before applying the hybrid active learning strategy. The random subset size is fixed to 200. The weight parameters and are set to 0.5.
We run 3 trials for the COVID-AL and the state-of-the-art active learning methods. We report the average performance on 3 trials for each method. In each trial, the dataset is randomly shuffled. 70% of the data is used to form an unlabeled data pool. Another 10% is for validation and the last 20% is for performance evaluation.
4.2. Comparisons against the state-of-the-art methods
The proposed COVID-AL is compared against the following active learning sampling strategies: (1) Random sampling strategy (RAND) randomly selects samples to be manually labeled in each cycle. (2) Entropy-based sampling strategy (ENT) Wang et al. (2016) selects samples with highest softmax output entropy in each cycle. (3) Diversity-based sampling strategy proposed in AFT* (AFT*-DIV) Zhou et al. (2018) selects samples with highest softmax output diversity computed with augmented data in each cycle. (4) Loss-prediction-based sampling strategy (LP) Yoo and Kweon (2019) selects samples with highest predicted loss in each cycle. (5) Core-set sampling strategy (CSET) Sener and Savarese (2017) selects samples with largest distance from current set in each cycle. (6) OMedAL framework proposed a embedding-distance-based sampling strategy (OMEDAL) Smailagic et al. (2019) to select samples which is the farthest from the centroid in the embedding space in eacy cycle. For all methods, we use the masks predicted by the same segmentation network. We also use the same random seed for a fair comparison during the active learning procedure.
For the evaluation metrics, the diagnostic accuracy, precision-recall curve, and ROC curve are used to represent the performance of different methods. The accuracy is reported for three way classification, while the precision-recall curve and ROC curve show the capability of the COVID-AL framework to differentiate NCP from the other two classes (CP and NC). We also report PR-AUC and ROC-AUC in Table 1 .
Table 1.
The performance of the active learning methods.
| Method | Accuracy | PR-AUC | ROC-AUC |
|---|---|---|---|
| RAND | 0.797 | 0.839 | 0.909 |
| ENT | 0.829 | 0.902 | 0.935 |
| AFT*-DIV | 0.850 | 0.929 | 0.955 |
| LP | 0.856 | 0.939 | 0.955 |
| CSET | 0.840 | 0.928 | 0.940 |
| OMEDAL | 0.840 | 0.939 | 0.951 |
| COVID-AL | 0.866 | 0.962 | 0.968 |
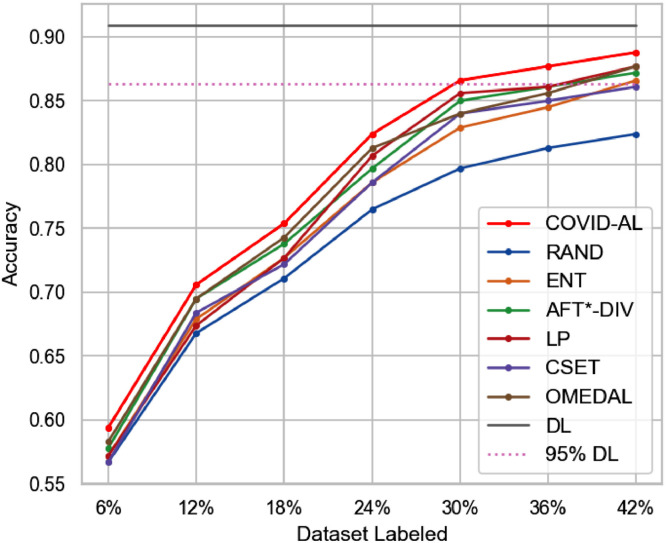
The Fig. 5 shows the diagnostic accuracy of different active learning methods. All the active learning methods outperform random sampling strategy (RAND). It means that deep active learning improves the labeling efficiency in the diagnosis of COVID-19. Our proposed COVID-AL with hybrid sampling strategy achieves best performance among all the active learning methods for the same percentage of labeled data. In Fig. 5, the horizontal solid line at the top represents the classification accuracy achieved by training the classification network on the whole dataset. The dashed line represents 95% accuracy of that performance. On the CC-CCII dataset, with only about 30% of labeled data, the COVID-AL is able to obtain an diagnostic accuracy of 0.867, which is over 95% of the performance achieved when the classification network is trained over the whole dataset (accuracy of 0.909).
Fig. 5.
Comparison of the proposed COVID-AL with the state-of-the-art active learning methods using the accuracy.
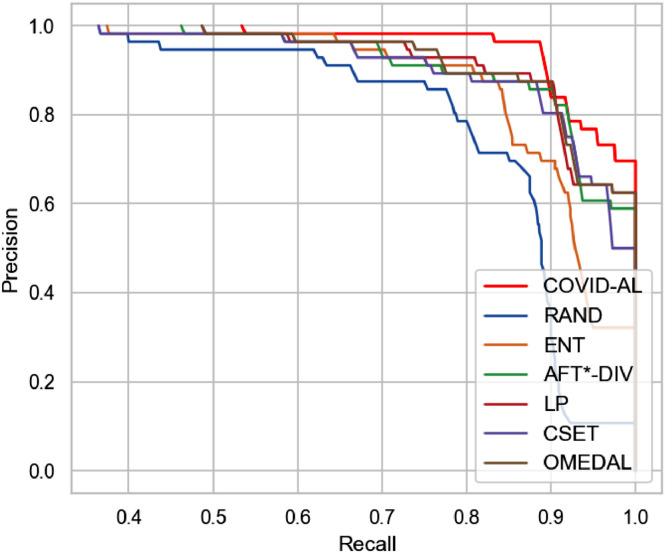
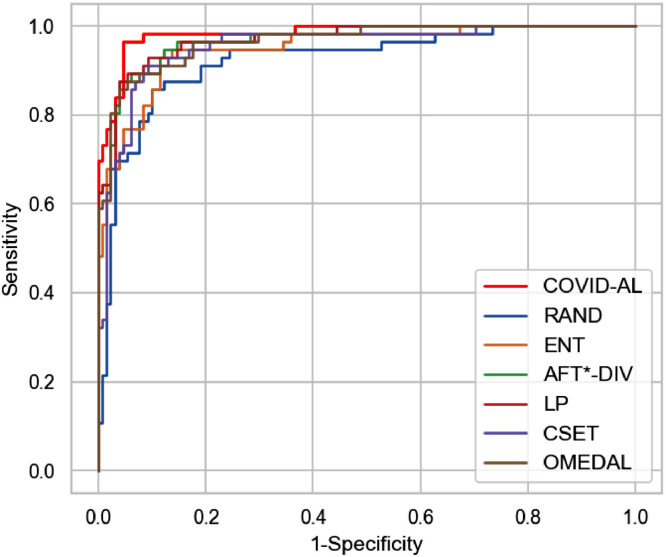
We select the classification networks trained with different active learning methods when 30% of the dataset labels are queried. For every testing CT scan, the classification networks predict its probability of being NCP. By comparing the predicted probability with the ground-truth labels, we plot precision-recall curve and ROC curve in Figs. 6 and 7 respectively.
Fig. 6.
Comparison of the proposed COVID-AL with the state-of-the-art active learning methods using the precision-recall curve.
Fig. 7.
Comparison of the proposed COVID-AL with the state-of-the-art active learning methods using the ROC curve.
The detailed information of the performance of each active learning method is illustrated in Table 1 when 30% of the labels are available. The proposed COVID-AL achieves 0.866 in accuracy, 0.962 in PR-AUC and 0.968 in ROC-AUC, which is the best performance among all the active learning methods.
5. Discussion
The motivation of this study is to reduce the cost of labeling when learning a deep neural network for COVID-19 diagnosis. There are many effective applications of COVID-19 diagnosis in previous studies. However, these AI applications heavily relied on large amounts of labeled data and lacked interaction with human experts. The main challenge is how to reduce the labeling cost of different AI applications when time and human resources are limited. The first solution is to train deep learning models with weak labels. In this way, it is not necessary to annotate the lesions of CT scans, which may save lots of time and labors. The second solution is to find out the most informative data samples to decrease unnecessary labeling efforts. Thus we merge these two solutions to design and build the COVID-AL framework.
We compute the informativeness of data samples from two different aspects: uncertainty and diversity. In the COVID-AL, uncertainty is represented by the predicted classification loss while diversity is represented by the divergence of input CT volumes. Data samples with high uncertainty are near the decision boundary of the classifier. The classifier tends to make wrong decisions on these data samples. It means that these data samples can effectively boost the training of the classifier. The diversity indicates the degree of difference between multiple CT volumes augmented by a single candidate. Samples with high diversity can cover the sample space and fully represent the features of samples. COVID-AL utilizes a hybrid sampling strategy to select samples with both high uncertainty and diversity in each round of active learning.
6. Conclusion and future work
We propose a weakly-supervised deep active learning framework called COVID-AL for the diagnosis of COVID-19, which can greatly reduce the cost of manual labeling for training models in chest CT scans and can further relieve the burden of the healthcare system when the pandemic is spreading fast. To the best of our knowledge, this is the first framework to actively learn a deep neural network model using patient-level labels to diagnose COVID-19. The proposed framework is capable of boosting the performance of deep neural networks by simultaneously considering diversity and predicted loss of unlabeled candidates. To verify the effectiveness of COVID-AL, extensive experiments have been conducted on CT scans collected from the CC-CCII. The experimental results demonstrate that the COVID-AL outperforms the existing state-of-the-art active learning methods.
In the future, we plan to improve COVID-AL by combining clinical information with CT scans to achieve more reliable diagnostic conclusions.
CRediT authorship contribution statement
Xing Wu: Conceptualization, Methodology, Funding acquisition, Writing - review & editing. Cheng Chen: Conceptualization, Methodology, Software, Writing - original draft. Mingyu Zhong: Software, Writing - review & editing. Jianjia Wang: Validation, Investigation, Writing - review & editing. Jun Shi: Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key R&D Program of China (Grant no. 2019YFE0190500), the Natural Science Foundation of Shanghai, China (Grant no. 20ZR1420400), the State Key Program of National Natural Science Foundation of China (Grant no. 61936001 ), the Shanghai Science and Technology Foundation (Grant no.18010500600) and the 111 Project (Grant no. D20031).
References
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):32–40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila D., Kiraly A.P., Bharadwaj S., Choi B., Reicher J.J., Peng L., Tse D., Etemadi M., Ye W., Corrado G., et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nature medicine. 2019;25(6):954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- Chen A., Karwoski R.A., Gierada D.S., Bartholmai B.J., Koo C.W. Quantitative CT analysis of diffuse lung disease. Radiographics. 2020;40(1):28–43. doi: 10.1148/rg.2020190099. [DOI] [PubMed] [Google Scholar]
- Chilamkurthy S., Ghosh R., Tanamala S., Biviji M., Campeau N.G., Venugopal V.K., Mahajan V., Rao P., Warier P. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet. 2018;392(10162):2388–2396. doi: 10.1016/S0140-6736(18)31645-3. [DOI] [PubMed] [Google Scholar]
- Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., et al. Ct imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A., Robicquet A., Ramsundar B., Kuleshov V., DePristo M., Chou K., Cui C., Corrado G., Thrun S., Dean J. A guide to deep learning in healthcare. Nature medicine. 2019;25(1):24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S., et al. Clinical characteristics of coronavirus disease 2019 in China. New England journal of medicine. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Kataoka H., Satoh Y. Proceedings of the IEEE International Conference on Computer Vision Workshops. 2017. Learning spatio-temporal features with 3d residual networks for action recognition; pp. 3154–3160. [Google Scholar]
- He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- Kiser K., Ahmed S., Stieb S., et al. Data from the thoracic volume and pleural effusion segmentations in diseased lungs for benchmarking chest CT processing pipelines [dataset] The Cancer Imaging Archive. 2020 doi: 10.1002/mp.14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Li J., Li X., Qi X. Ct imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295(1) doi: 10.1148/radiol.2020200236. 18-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q., Nguyen T.T., Cao T.M., Pham Q.D. Importation and human-to-human transmission of a novel coronavirus in vietnam. New England journal of medicine. 2020;382(9):872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O., Fischer P., Brox T. International Conference on Medical image Computing and Computer-Assisted Intervention. Springer; 2015. U-Net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Sener O., Savarese S. Active learning for convolutional neural networks: acore-set approach. arXiv:1708.00489
- Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE reviews in biomedical engineering. 2020 doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- Smailagic A., Costa P., Gaudio A., Khandelwal K., Mirshekari M., Fagert J., Walawalkar D., Xu S., Galdran A., Zhang P., et al. O-medal: online active deep learning for medical image analysis. arXiv:1908.10508
- Song F., Shi N., Shan F., Zhang Z., Shen J., Lu H., Ling Y., Jiang Y., Shi Y. Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295(1):210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. Overview of deep learning in medical imaging. Radiological physics and technology. 2017;10(3):257–273. doi: 10.1007/s12194-017-0406-5. [DOI] [PubMed] [Google Scholar]
- Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19classification and lesion localization from chest ct. IEEE Transactions on Medical Imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Ma, Y., Robson, P. M., Chung, M., Bernheim, A., Mani, V., Calcagno, C., Li, K., Li, S., Shan, H., et al., 2020. Artificial intelligence for rapid identification of the coronavirus disease 2019 (COVID-19). medRxiv.
- Wang K., Zhang D., Li Y., Zhang R., Lin L. Cost-effective active learning for deep image classification. IEEE Transactions on Circuits and Systems for Video Technology. 2016;27(12):2591–2600. [Google Scholar]
- Wu X., Zhong M., Guo Y., Fujita H. The assessment of small bowel motility with attentive deformable neural network. Information Sciences. 2020;508:22–32. [Google Scholar]
- Yang L., Zhang Y., Chen J., Zhang S., Chen D.Z. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2017. Suggestive annotation: a deep active learning framework for biomedical image segmentation; pp. 399–407. [Google Scholar]
- Yoo D., Kweon I.S. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2019. Learning loss for active learning; pp. 93–102. [Google Scholar]
- Yuan J., Hou X., Xiao Y., Cao D., Guan W., Nie L. Multi-criteria active deep learning for image classification. Knowledge-Based Systems. 2019;172:86–94. [Google Scholar]
- Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., et al. Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Shin J., Feng R., Hurst R.T., Kendall C.B., Liang J. Integrating active learning and transfer learning for carotid intima-media thickness video interpretation. Journal of digital imaging. 2019;32(2):290–299. doi: 10.1007/s10278-018-0143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Shin J., Zhang L., Gurudu S., Gotway M., Liang J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Fine-tuning convolutional neural networks for biomedical image analysis: actively and incrementally; pp. 7340–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Shin, J. Y., Gurudu, S. R., Gotway, M. B., Liang, J., Aft*: integrating active learning and transfer learning to reduce annotation efforts. arXiv:1802.00912