Highlights
-
•
This finding may provide a novelty insight to help elucidate the mechanisms of the relapses.
-
•
Relapsed tumours from stage IA LUAD patients exhibited a weakened immune phenotype.
-
•
A targeted 395-gene expression NGS assay helps assess tumour immunity.
Keywords: Stage IA, Lung adenocarcinoma, Relapse, Tumour immunity, Molecular alteration
Abbreviations: NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; FFPE, formalin-fixed paraffin-embedded; DEGs, differentially expressed genes; DFS, disease-free survival; TMB, tumour mutation burden; CNI, copy number instability; MATH, mutant-allele tumour heterogeneity; WES, whole-exome sequencing; IO, immune oncology; SNV, single nucleotide variation; ssGSEA, single sample gene set enrichment analysis; CTA, cancer-testis antigen; CTL, cytotoxic T lymphocyte
Abstract
Patients with early-stage non-small cell lung cancer (NSCLC), even stage IA, are at substantial risk of relapse and death. We explored the distinct features of molecular alterations and immune-related gene expression in Formalin-fixed paraffin-embedded (FFPE) samples from 25 relapsed patients compared with 25 non-relapsed patients through using whole-exome sequencing and an immune oncology panel RNA sequencing platform. Results showed that the chemokine, cytolytic activity and tumour-associated antigen gene signatures exhibited significantly higher expression in non-relapsed tumours from stage IA lung adenocarcinoma (LUAD) than that in relapsed tumours. Besides, Kaplan–Meier survival analysis revealed that the gene signatures of chemokines and tumour-associated antigens were significantly associated with the patients’ disease-free survival (DFS), indicating their prognostic value in early-stage LUAD. Cytolytic activity displayed a similar trend but failed to reach statistical significance. These findings revealed a weakened immune phenotype in relapsed tumours and provide valuable information for improving the treatment management of these high-risk patients. Due to the overall small patient number in this study, these differences should be further validated in a larger cohort.
Introduction
Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer cases and is the leading cause of death worldwide [1]. The proportion of patients with early-stage NSCLC has dramatically increased with the prevalence of low-dose computed tomography (CT) for screening [2]. Unfortunately, the rates of locoregional recurrence (LRR) after surgery for stage I NSCLC ranges from 5% to 19% [3]. Reports show that nearly 27% of stage I NSCLC patients eventually die of recurrence or metastasis within five years [4]. Thus, understanding the comprehensive genomic and immunological characterisation of early-stage patients with high risk and further providing valuable information for the precise treatment of these patients are of great importance.
Previous studies have shown that certain driver mutations are associated with tumour progression, with evidence that KRAS mutations are associated with disease recurrence in patients with stage I lung adenocarcinoma (LUAD) [5]. However, the association of genomic alterations beyond gene mutations, such as the tumour mutation burden (TMB), copy number instability (CNI) and mutant-allele tumour heterogeneity (MATH), with recurrence in stage IA LUAD is poorly understood. At the same time, with the enrichment of our knowledge and insights into the hallmarks of cancer, we further realise that the collaborative interaction between malignant cancer cells and their supporting stroma drives tumour development and progression [6]. Hence, quite a lot of prognostic immune markers in the tumour microenvironment and peripheral blood, as well as tumour expression of immune genes have been identified in NSCLC [7]. Kei Suzuki and colleagues reported that the tumoural immune markers IL-12Rβ2 and IL-7R and the stromal FoxP3 to CD3 ratio were independently associated with recurrence in patients with stage I LUAD [4]. In addition, several groups have developed gene expression-based signatures, including immune-related gene signatures, for identifying patients at high risk for recurrence after the resection of early-stage disease [8], [9], [10]. However, these studies examined very few markers [4] or used OS as the main study endpoint [8], [9], [10], rendering their application difficult to characterise the relapse of stage IA patients when recurrence is more clinically relevant to patients with stage I disease.
Thus, in our study, we performed whole-exome sequencing (WES) and immune oncology (IO)-panel RNA sequencing to comprehensively investigate the difference in genomic profile and immune microenvironment between relapsed and non-relapsed tumours from patients with stage IA LUAD. By analysing sequencing data, a detailed molecular characterisation of relapse tumours may help to gain novel insight into the mechanisms by which cancer cells evade immune surveillance and to improve the individual management of relapsed patients.
Material and methods
Patient enrolment and sample collection
Up to April 9th, 2018, we collected both tumour and normal formalin-fixed paraffin-embedded (FFPE) samples from 25 pairs of stage IA LUAD patients with recurrence and non-recurrence. The corresponding flow chart was shown in the (Supplementary Table S1). All patients underwent radical surgery with systematic lymph node dissection from October 2013 to April 2018 at the National Cancer Centre/National Clinical Research Centre for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China). Informed consent was obtained from each patient and their family for experimentation. In addition, the recurrence and non-recurrence pairs were selected by a 1-to-1 match. All eligible patients were pathologically diagnosed with stage IA LUAD using the American Joint Committee on Cancer (AJCC) 8th TNM system and did not receive neo or adjuvant chemotherapy or other systemic therapies.
Disease-free survival (DFS) was defined as the length of time from lung cancer radical surgery to any kind of recurrence, including regional and distant recurrences. Regional recurrence was recorded as recurrence on resection margins such as bronchial stumps or stapler lines or within the ipsilateral of subcarinal lymph nodes. Distant recurrence was defined as any recurrence occurring in the contralateral lung, brain, liver, adrenal gland, bone, and other locations. Disease recurrence was assessed by CT scan, magnetic resonance imaging (MRI), and bone scanning. Positron emission tomography (PET)-CT was also permitted for the screening or confirmation of recurrences. Follow-up information was obtained through hospital visits or telephone contact with patients or relatives. The follow-up schedule consisted of a clinic visit and chest CT examination every 3 months during the first 2 years, every 3–6 months from the third to fifth year and 1-year intervals thereafter. Moreover, patients would be recommended to screen brain MRI, bone scanning or PET-CT every year in the first 5 years. The longest interval from the time of radical surgery to the last follow-up was 5 years. The median follow-up time was 38.8 months (95%CI 32.6–45.1).
WES
WES was performed for tumour and matched normal tissue from 25 pairs of patients and then analysed with a mean depth of 146. DNA from FFPE specimens from tumour and matched normal tissue were extracted and quantified. And extracted DNA was sheared into fragments and built into DNA libraries. Then DNA libraries were captured, and the captured samples were subjected to paired-end sequencing on an Illumina NovaSeq 6000 platform. High-quality paired-end reads were aligned to the hg19 reference genome using the Burrows-Wheeler Aligner (BWA). The VarDict and FreeBayes programs were used for single nucleotide variation (SNV) and indel calling, while the ANNOVAR assay was used for the functional annotation of genetic variants. The somatic SNVs and indels were filtered as previously reported [11].
Calculation of TMB
For the determination of TMB, the number of somatic nonsynonymous SNVs in the whole exome (with depth> 40X and allele frequency ≥0.05) detected by next generation sequencing (NGS) was quantified. Alterations known to be oncogenic drivers were excluded. TMB was measured in mutations per Mb.
CNI score calculation
The read depth was transformed into log2 ratios after correcting the GC content and length of the target region and then converted into Z-scores based on Gaussian transformations versus a normal baseline (n = 494). The CNI score was summed by the Z-score of the target regions (whose Z-score is greater than the 95th percentile) plus two times the absolute standard deviation of the normal baseline.
MATH score calculation
The MATH score was calculated by including all somatic variants with a variant allele frequency (VAF) between 0.05 and 1, with the formula 100 × median absolute deviation (MAD)/median of the VAF.
Calculation of neoantigen burden
Somatic mutation immunogenicity was predicted according to previous reports [12]. All mutations with binding scores below 500 nM are defined as neoantigens. The strong predicted binders were those peptides that had a predicted binding affinity < 50 nM and a ratio of wild-type sequences to mutated sequences>1.5. Neoantigen burden was summed by the number of strong predicted binders.
RNA IO platform for gene expression analysis
Total RNA was isolated from the FFPE tumour tissues of 50 patients using the RNeasy FFPE Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Finally, 43 samples met the requirements of quality control (QC). Ten ng of RNA was reverse transcribed into cDNA using the SuperScript VILO cDNA Synthesis Kit (Life Technologies, Carlsbad, CA), and targets were amplified with the primer pool targeting 395 IO-related genes, including 10 housekeeping (HK) genes. Sequencing was performed with the Ion Chef Instrument and the Ion GeneStudio S5 Prime System using the Ion Torrent ™ Ion 540 ™ Kit (Life Technologies, Carlsbad, CA). RNA-seq absolute reads were generated with Torrent Suite's plugin immuneResponseRNA version 5.8.0.1 (Thermo Fisher Scientific). The absolute digital gene expression counts of all samples in the same run were automatically generated in the in-house bioinformatics pipeline. Only sequencing data meeting the QC criteria for mapped reads, on-target reads and mean reads were included in the study.
Differentially expressed genes (DEGs) and gene signature analysis
The R package “limma” (version 3.29.0) was used for the analysis of DEGs between patients with relapsed and non-relapsed LUAD. Genes with a fold change greater than 2 and an adjusted P<0.05 were reported. The functional and pathway enrichment analysis of DEGs was conducted by Metascape.
The gene signature scores were calculated based on the average expression of all the component genes in each signature. CTAG1B, GAGE2C, MAGEA3, MAGEA12, CTAG2 and MAGEC2 were selected as the tumour-associated antigen gene signature and NKG7, KLRF1 and NCR3 were selected as the natural killer (NK) cell signalling-associated gene signature. Three previously reported gene signatures, including the cytolytic activity gene signature (GZMA and PRF1) [13], the T-effector and interferon-γ (Teff) gene signature (CD8A, GZMA, GZMB, IFNγ, EOMES, CXCL9, CXCL10, and TBX21) [14] and the chemokine signature (CXCL9, CXCL10, CCL5 and CX3CL1) [15] were also selected here to examine the distinct immune response in relapsed tumours (Supplementary Table S2).
Activated CD8+ T cell analysis
Marker genes for activated CD8+ T cells were selected on the basis of a previous report [16] (Supplementary Table S2). The enrichment score of activated CD8+ T cells was calculated with the RNA IO sequencing data using the single sample gene set enrichment analysis (ssGSEA) method as previously described [17].
Statistical analysis
Statistical analyses were conducted using GraphPad Prism (version 7.01) and SPSS version 22.0 (SPSS, Inc., Chicago, IL, USA). The frequencies of mutations identified by WES were compared between tumours from patients with relapse and those without relapse by Fisher's exact test. The TMB, CNI and MATH were analysed by the Mann–Whitney U test. The survival curves for DFS from the Kaplan–Meier analysis were compared using the log-rank test. The cut-off value was obtained by X-tile 3.6.1 software (Yale University, New Haven, CT, USA) [18]. All reported P values are two-tailed, and for all analyses, P<0.05 was considered statistically significant unless otherwise specified.
Results
Genomic differences between LUAD patients with and without relapsed tumours at baseline
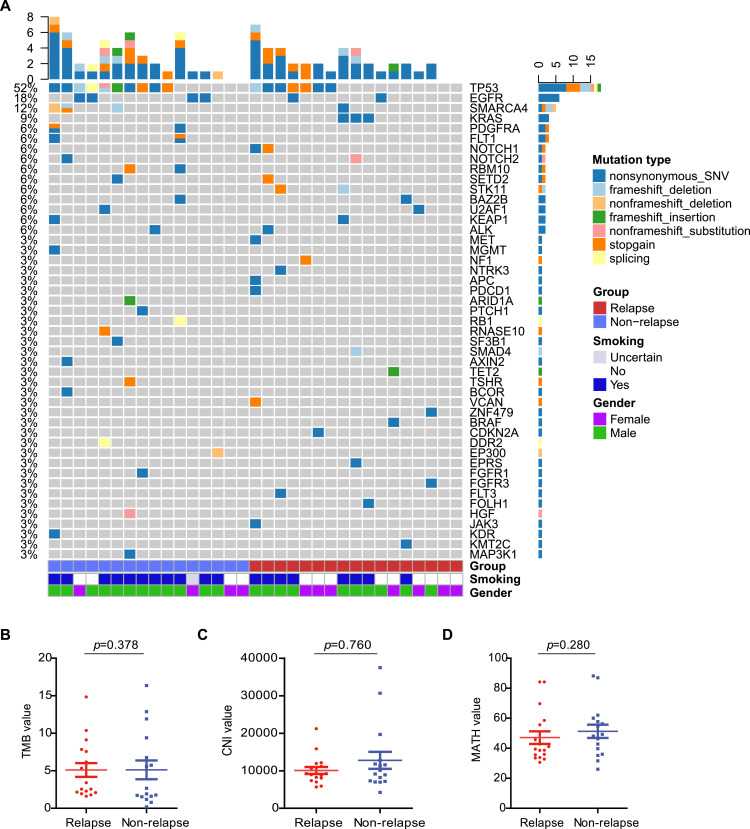
WES was successfully performed on 33 stage IA LUAD tumour samples and paired normal lung tissues. The number of relapsed and non-relapsed patients was 17 and 16, respectively. Somatic mutation analysis of these 33 tumour/normal pairs showed that TP53 (48%), EGFR (18%), and KRAS (9%) were among the top mutated genes (Fig. 1A), which was consistent with the findings of previous studies [19, 20]. TP53 and EGFR alterations showed no significant differences between non-relapse patients and relapse patients (TP53, 56% vs. 41%, p = 0.494; EGFR, 25% vs. 12%, p = 0.398), while KRAS mutations only occurred in the relapsed tumours (0% vs. 18%, p = 0.227) (Supplementary Fig. S1). The co-mutation of TP53 and KRAS was not seen in this cohort, and only one patient in each group was observed to harbour a co-mutation of TP53 and EGFR (Fig. 1A). Mutations in KRAS were mutually exclusive with those in EGFR, in line with the findings of previous investigations [20] (Fig. 1A). The TMB, as assessed by nonsynonymous somatic mutations per Mb, was similar between patients with relapse and those without relapse (p = 0.378, Fig. 1B). The CNI and MATH were further analysed to explore whether there was any difference between relapsed and non-relapsed tumours. No statistically significant difference was observed between these two data sets in terms of the CNI and the MATH value (p = 0.760, p = 0.280, Fig. 1C and D). These results suggest that somatically acquired DNA alterations, including gene somatic mutations, TMB, CNI and MATH, were not associated with stage IA LUAD patient relapse.
Fig. 1.
Characterisation of the genomic profile in lung adenocarcinoma. (A) 43 significantly mutated genes in ADC and the genes are ranked by their mutation frequencies. Tumour samples were classified into two groups: blue for non-relapse and red for relapse. Smoking status and gender are annotated in the bottom panel. (B)-(D) Comparison of TMB (B), CNI (C), and MATH (D) between the relapse and non-relapse subgroups (Mann–Whitney U test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Association of relapse with pre-existing tumour immunity
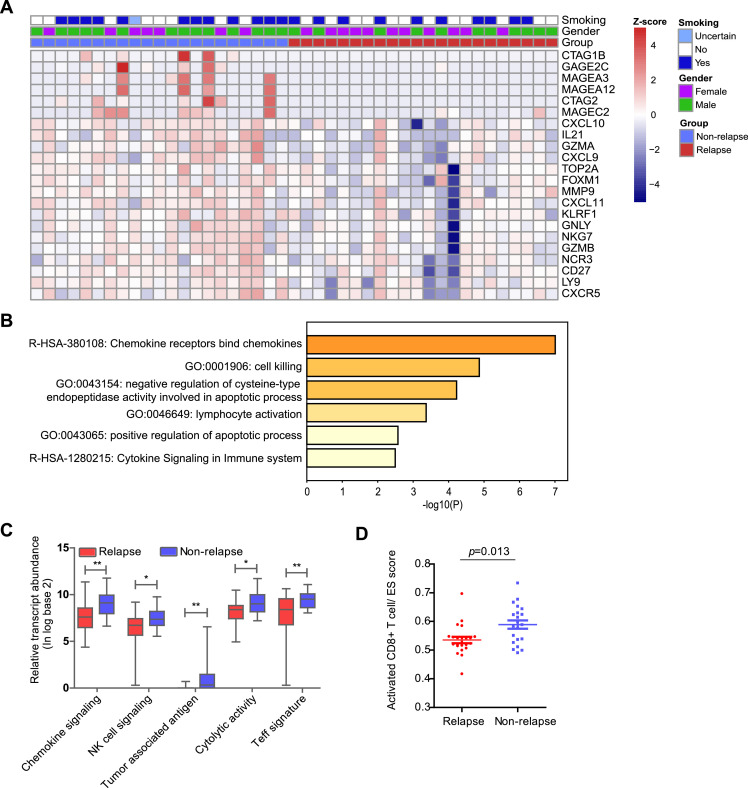
Since genomic alterations, which represent the intrinsic factors critical for tumour progression, could not fully account for the relapse of early-stage LUAD, we further investigated the antitumour immunity variation in the TME underpinning relapsed and non-relapsed tumours. Tumours from 22 patients with relapse and 21 without relapse were successfully evaluated using the RNA IO profiling assay. In total, 22 DEGs were identified between the two groups, and all these 22 genes exhibited significantly higher expression in the non-relapse group than in the relapse group (Fig. 2A). Pathway enrichment analysis conducted by Metascape (http://metascape.org/) [21] indicated that 22 upregulated genes in non-relapsed tumours were related to immune response processes, including chemokine receptors that bind chemokines, cell killing and lymphocyte activation (Fig. 2B). Based on our enrichment results and previous reports [15], [16], [17], five immune-related gene signatures were further analysed between the tumours with relapse or non-relapse, namely, the tumour-associated antigen, chemokine, NK cell signalling, cytolytic activity and Teff gene signatures. The results showed that patients with relapsed tumours had significantly lower expression levels among all five gene signatures than those with non-relapsed tumours (Fig. 2C). Consistent with the observation that the immune-related gene signatures are associated with relapse, ssGSEA revealed that a lower fraction of activated CD8+ T cells was seen in relapsed tumours than in non-relapsed tumours (Fig. 2D). The expression differences of genes in each gene signature are shown in Fig. S2.
Fig. 2.
Immune oncology RNA profiling of lung adenocarcinoma. (A) Heatmap of differentially expressed genes (DEGs) between relapse and non-relapse subgroups. (B) Histogram of the enrichment pathways of the DEGs. Comparison of immune gene signatures (C) and activated CD8+ T cells (D) between the relapse and non-relapse subgroups (Mann–Whitney U test). ES score, ssGSEA enrichment score; “*”, P<0.05, “**”, P<0.01.
Positive correlation of pre-existing tumour immunity with patients’ DFS
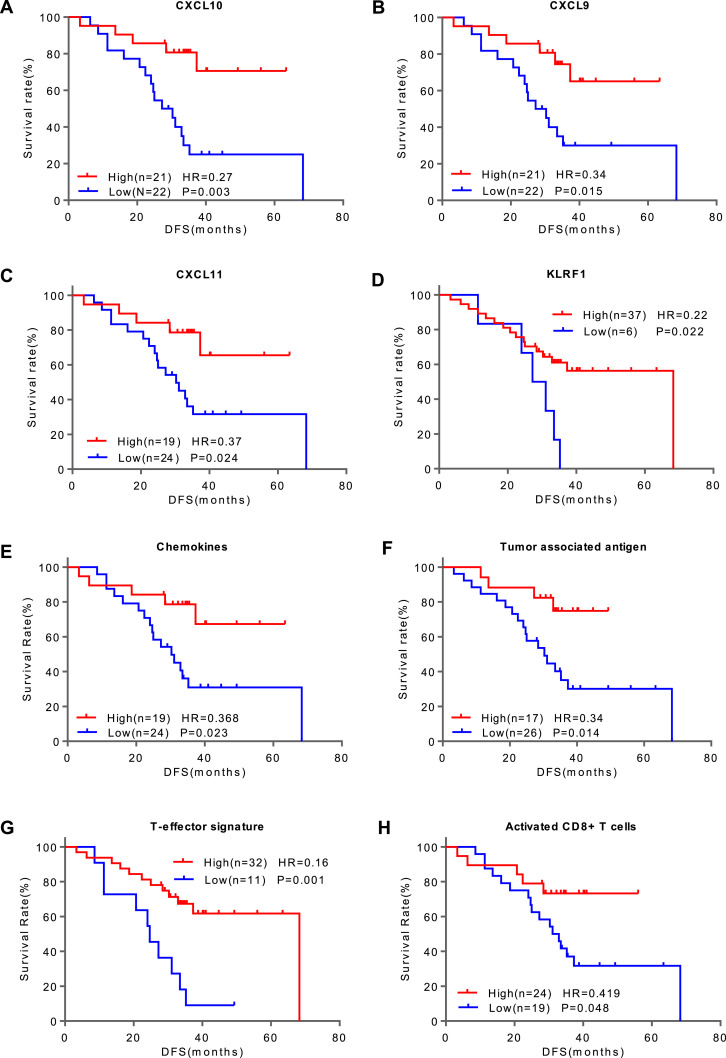
Since early relapses usually lead to poor survival, we further demonstrated the prognostic value of these DEGs and gene signatures in LUAD patients using Kaplan–Meier analysis on the 43 patients (relapse, N = 22; non-relapse, N = 21). X-tile 3.6.1 software (Yale University, New Haven, CT, USA) was applied to determine the best gene expression cut-off value to divide the patients into two groups. The Kaplan–Meier analysis showed that CXCL9, CXCL10, CXCL11 and KLRF1 were significantly associated with patients’ DFS, and the group with a higher expression of these genes had a longer DFS than the group with lower expression in early LUAD patients (Fig. 3A–D). The same trend was seen in the three gene signatures (the chemokine, T-effector and tumour-associated antigen signatures) as well as in the immune cell subpopulation of activated CD8+ T cells (Fig. 3E–H). The cytolytic activity showed a similar trend but failed to reach statistical significance (p = 0.098) (Supplementary Fig. S3). Altogether, these results indicate that the poor prognosis of relapsed patients is probably due to the immunosuppressive microenvironment.
Fig. 3.
Kaplan–Meier curves of disease-free survival (DFS) in patients with stage IA LUAD. Patients were stratified by the expression levels (High vs. Low) of the DEGs, including CXCL10 (A), CXCL9 (B), CXCL11 (C), and KLRF1 (D); gene signatures, including the chemokine signalling (E), tumour-associated antigen (F) and T-effector (G) signatures; and the immune cell subpopulation of activated CD8+ T cells (H). Hazard ratios (HRs) are for high vs. low expression levels. P values were calculated with the log-rank test. In (F), patients with at least one tumour associated antigen gene positive in the tumour-associated antigen signature were classified as the “High” group, otherwise as the “Low” group.
Association of driver gene mutations with tumour immunity
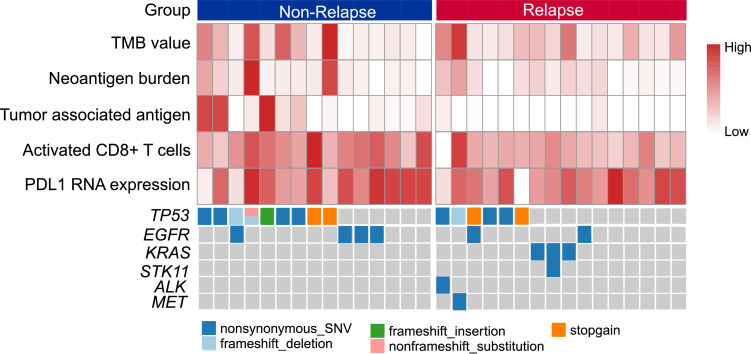
Previous studies have indicated that TP53, KRAS and EGFR mutations may influence the immune microenvironment of lung tumours. Therefore, we investigated whether the host immune response differed in the wild-type group and their mutant group. Tumours from 16 patients with relapse and 15 patients without relapse who had both WES sequencing data and RNA expression data were enroled in the analyses (Fig. 4).
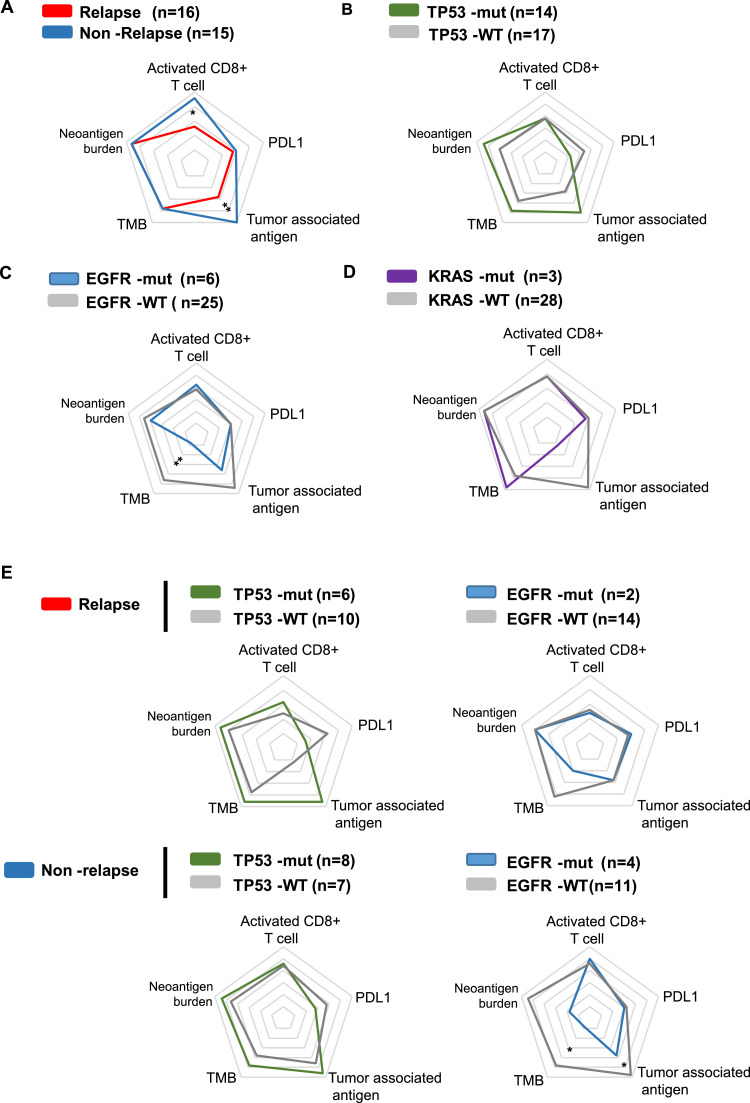
Fig. 4.
Landscape of gene alterations and immune profiles in patients with or without relapse (N = 31).
The tumour immunity levels, including TMB, neoantigen burden, PD-L1 mRNA expression, activated CD8+ T cells and tumour-associated antigen, are shown on a radar map. In line with the above results, activated CD8+ T cells and tumour-associated antigen showed significantly lower levels in relapsed tumours than in non-relapsed tumours, while TMB, neoantigen burden and PD-L1 expression were similar between the two groups (Fig. 5A). In the TP53-mutated tumours, there was a trend of higher TMB, neoantigen burden and tumour-associated antigen and lower PD-L1 expression, though these differences were not statistically significant (Fig. 5B). In contrast, EGFR-mutated tumours were characterised by a significantly lower TMB than wild-type counterparts, and neoantigen burden and tumour-associated antigen showed similar trends but failed to reach statistical significance (Fig. 5C). Elevated TMB was also observed in KRAS-mutated tumours, but the difference was not significant (Fig. 5D). Because the KRAS mutation was identified in only the relapsed tumours, we further explored the different impacts of TP53 and EGFR mutations on the tumour immunity of the relapse and non-relapse tumours. We found that in both relapsed and non-relapsed tumours, TP53-mutated tumours showed a similar trend of higher tumour immunity and lower PD-L1 expression, while EGFR-mutated tumours showed the opposite trend (Fig. 5E). Accordingly, TP53-mutated tumours were associated with an intense immune phenotype, while EGFR-mutated tumours conferred a weakened immune response in both relapsed and non-relapsed patients.
Fig. 5.
Differential impact of TP53, EGFR, and KRAS mutations on the tumour immune profiles. Radar plots show the immune profile of TMB, neoantigen burden, PD-L1 expression, activated CD8+ T cells and tumour-associated antigen between patients with relapse or without relapse (A), in TP53-mutated tumours (B), EGFR-mutated tumours (C), and KRAS-mutated tumours (D) compared to their wild-type counterparts, and in relapsed tumours and non-relapsed tumours respectively (E).
Discussion
Our study was designed to identify the distinct molecular bases with respect to the relapses of early-stage lung cancer patients. The results showed that pre-existing tumour immunity features was the significant determinant of relapse. This finding was consistent with the growing research showing that the host immune response plays a more important role in tumour initiation and progression in lung carcinoma and that mutationally corrupted cancer cells cannot drive tumour development without the contribution of their supporting stroma in the TME [7].
One of the most important tumour immunity features of the relapsed tumours in this cohort was the significantly decreased expression level of chemokines and impaired cytolytic activity. Chemokines play a crucial role in the trafficking of T cells to tumours and determine the density of intratumoural immune cells [15, 22]. According to another study, chemotaxis was enriched among genes in the immune signature of 25 gene pairs consisting of 40 unique genes that can significantly stratify patients into high- vs. low-risk groups with stage I, IA, IB, or II NSCLC [8]. In addition, numerous studies have reported that higher expression levels of these chemokine genes or gene signatures were associated with prolonged DFS in patients with various cancers, including lung cancer, colonrectal cancer, breast cancer, melanoma and so on [23], [24], [25], [26]. In accordance with these data, a higher expression of the chemokine genes and gene signature was associated with a longer DFS time in this study.
According to a recent study, CTL function seems to be more important than CTL density in the immune response [27], so we further examined the immune cell cytolytic activity represented by two critical genes, GZMA and PRF1, as previously described [13]. The results showed that non-relapsed tumours showed significantly higher cytolytic activity than relapsed tumours (Fig. 2C). Overall, CTL density and CTL function were both elevated in non-relapsed tumours compared to relapsed tumours. In addition, a higher expression of the NK cell-mediated cytotoxicity signature observed in this study was also expected to protect the patients from suffering relapse. In summary, an activated immune response of both innate and adaptive immunity in the TME may together contribute to non-relapses.
Another tumour immunity feature of relapsed tumours was the significantly lower expression level of the cancer-testis antigen (CTA) genes. According to previous reports, many CTA genes showed significant immunogenicity in various human cancers, including NSCLC [28, 29]. CTAG1B was one of the differentially expressed CTA genes that we identified in this study (Figs. 2A and S2). Cancer/testis antigen 1 (CTAG1B), better known as NY-ESO-1, is a protein encoded by the CTAG1B gene, and NY-ESO-1 was reported to be the most immunogenic CT antigen in terms of eliciting humoural and cellular immune responses in cancer patients [30]. Though most investigations suggested a poor outcome in lung cancer patients with CTA expression, including patients with stage I LUAD [31], the prognostic impact of CTA expression in NSCLC remains controversial [32]. Our results suggesting a better prognosis in patients with CTA expression need to be verified in a larger cohort. Nonetheless, for the first time, the CT antigen gene signature consisting of six CTA genes was identified to be associated with the recurrence of patients with stage IA LUAD in this study, and the patients with higher expression level of the CT antigen gene signature achieved prolonged DFS time. Since the CT antigens are immunogenic and highly restricted to tumours [28], these CTA genes are attractive targets for vaccine immunotherapy, and several clinical trials with MAGE-A and NY-ESO-1 are ongoing in NSCLC (NCT03132922, NCT03709706, and NCT03029273).
Though this study showed no association of gene alterations such as KRAS, EGFR and TP53, as well as TMB, CNI and MATH, with DFS in patients with stage IA LUAD, we found that oncogenic driver mutations in KRAS, TP53 and EGFR do have a major impact on the immune microenvironment in LUAD, which was in line with previous reports [33, 34]. Considering the limited sample size in our study, the influence of gene mutation must be interpreted cautiously. We comprehensively examined TMB, neoantigen burden and tumour-associated antigen, which represent the immunogenicity of tumours, and activated CD8+ T cells that can recognise a specific antigen and kill cancer cells, and PD-L1 expression, which is a negative regulator and can prevent the immune system from killing cancer cells [35] to characterise antitumour immunity. To the best of our knowledge, no investigations have examined these antitumour immunity factors at the same time in stage IA LUAD.
In conclusion, the distinct tumour immune profile of relapsed tumours from patients with stage IA LUAD was characterised, which helps elucidate the mechanisms of the relapses, thus offering potential therapeutic strategies suitable for these patients. Certainly, these findings must be validated prospectively.
Authors’ contributions
Lu Yang, Jing Zhang, Henghui Zhang, Jianming Ying and Yan Wang designed this study. Guangjian Yang, Haiyan Xu, Junling Li, Lei Guo and Xin Li collected the FFPE samples and clinical data. Yane Song, Xinying Shi, Ying Yang, Lijia Wu and Jiyu Wei performed the statistical analysis. Lu Yang, Jing Zhang, Yane Song and Beibei Mao drafted the manuscript. Beibei Mao, Henghui Zhang, Jianming Ying and Yan Wang provided critical comments, suggestions and revised the manuscript.
CRediT authorship contribution statement
Lu Yang: Methodology, Supervision, Resources, Writing - original draft. Jing Zhang: Methodology, Resources, Writing - original draft. Yane Song: Methodology, Writing - original draft, Data curation. Guangjian Yang: Resources. Haiyan Xu: Resources. Junling Li: Resources. Lei Guo: Resources. Xin Li: Resources. Xinying Shi: Data curation. Beibei Mao: Writing - review & editing. Ying Yang: Data curation. Lijia Wu: Data curation. Jiyu Wei: Data curation. Henghui Zhang: Conceptualization, Writing - review & editing. Jianming Ying: Conceptualization, Writing - review & editing. Yan Wang: Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgements
The present study was funded by the CAMS Innovation Fund for Medical Sciences (CIFMS) (2017-12M-2-003), the National Key Sci-Tech Special Project of China (No. 2018ZX10302207) and Peking Union Medical College Graduate Innovation Fund (grant number 2019-1002-55).
Ethics approval and consent
This study has been approved by the ethics committee of Cancer Hospital Chinese Academy of Medical Sciences. Informed consent was obtained from all individual participants included in the study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100942.
Contributor Information
Henghui Zhang, Email: zhhbao@ccmu.edu.cn.
Jianming Ying, Email: jmying@hotmail.com.
Yan Wang, Email: wangyanyifu@163.com.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Miller K.D., Nogueira L., Mariotto A.B., Rowland J.H., Yabroff K.R., Alfano C.M., Jemal A., Kramer J.L., Siegel R.L. Cancer treatment and survivorship statistics. CA Cancer J. Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 3.Saynak M., Veeramachaneni N.K., Hubbs J.L., Nam J., Qaqish B.F., Bailey J.E., Chung W., Marks L.B. Local failure after complete resection of N0-1 non-small cell lung cancer. Lung Cancer. 2011;71:156–165. doi: 10.1016/j.lungcan.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki K., Kadota K., Sima C.S., Nitadori J., Rusch V.W., Travis W.D., Sadelain M., Adusumilli P.S. Clinical impact of immune microenvironment in stage I lung adenocarcinoma: tumor interleukin-12 receptor beta2 (IL-12Rbeta2), IL-7R, and stromal foxp3/CD3 ratio are independent predictors of recurrence. J. Clin. Oncol. 2013;31:490–498. doi: 10.1200/JCO.2012.45.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo T., Okudela K., Yazawa T., Wada N., Ogawa N., Ishiwa N., Tajiri M., Rino Y., Kitamura H., Masuda M. Prognostic value of KRAS mutations and Ki-67 expression in stage I lung adenocarcinomas. Lung Cancer. 2009;65:355–362. doi: 10.1016/j.lungcan.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D., Coussens L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki K., Kachala S.S., Kadota K., Shen R., Mo Q., Beer D.G., Rusch V.W., Travis W.D., Adusumilli P.S. Prognostic immune markers in non-small cell lung cancer. Clin. Cancer Res. 2011;17:5247–5256. doi: 10.1158/1078-0432.CCR-10-2805. [DOI] [PubMed] [Google Scholar]
- 8.Li B., Cui Y., Diehn M., Li R. Development and validation of an individualized immune prognostic signature in early-stage nonsquamous non-small cell lung cancer. JAMA Oncol. 2017;3:1529–1537. doi: 10.1001/jamaoncol.2017.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kratz J.R., He J., Van Den Eeden S.K., Zhu Z.H., Gao W., Pham P.T., Mulvihill M.S., Ziaei F., Zhang H., Su B., Zhi X., Quesenberry C.P., Habel L.A., Deng Q., Wang Z., Zhou J., Li H., Huang M.C., Yeh C.C., Segal M.R., Ray M.R., Jones K.D., Raz D.J., Xu Z., Jahan T.M., Berryman D., He B., Mann M.J., Jablons D.M. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379:823–832. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beer D.G., Kardia S.L., Huang C.C., Giordano T.J., Levin A.M., Misek D.E., Lin L., Chen G., Gharib T.G., Thomas D.G., Lizyness M.L., Kuick R., Hayasaka S., Taylor J.M., Iannettoni M.D., Orringer M.B., Hanash S. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 11.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R., Schrock A., Campbell B., Shlien A., Chmielecki J., Huang F., He Y., Sun J., Tabori U., Kennedy M., Lieber D.S., Roels S., White J., Otto G.A., Ross J.S., Garraway L., Miller V.A., Stephens P.J., Frampton G.M. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szolek A., Schubert B., Mohr C., Sturm M., Feldhahn M., Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehrenbacher L., Spira A., Ballinger M., Kowanetz M., Vansteenkiste J., Mazieres J., Park K., Smith D., Artal-Cortes A., Lewanski C., Braiteh F., Waterkamp D., He P., Zou W., Chen D.S., Yi J., Sandler A., Rittmeyer A., Group P.S. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387:1837–1846. doi: 10.1016/S0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- 15.Chen D.S., Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X.C., Wang J., Shao G.G., Wang Q., Qu X., Wang B., Moy C., Fan Y., Albertyn Z., Huang X., Zhang J., Qiu Y., Platero S., Lorenzi M.V., Zudaire E., Yang J., Cheng Y., Xu L., Wu Y.L. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat. Commun. 2019;10:1772. doi: 10.1038/s41467-019-09762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.N. Cancer Genome Atlas Research Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A., Moulton H.M., DeJesus P., Che J., Mulder L.C., Yanguez E., Andenmatten D., Pache L., Manicassamy B., Albrecht R.A., Gonzalez M.G., Nguyen Q., Brass A., Elledge S., White M., Shapira S., Hacohen N., Karlas A., Meyer T.F., Shales M., Gatorano A., Johnson J.R., Jang G., Johnson T., Verschueren E., Sanders D., Krogan N., Shaw M., Konig R., Stertz S., Garcia-Sastre A., Chanda S.K. Meta- and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galon J., Angell H.K., Bedognetti D., Marincola F.M. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39:11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Moran C.J., Arenberg D.A., Huang C.C., Giordano T.J., Thomas D.G., Misek D.E., Chen G., Iannettoni M.D., Orringer M.B., Hanash S., Beer D.G. RANTES expression is a predictor of survival in stage I lung adenocarcinoma. Clin. Cancer Res. 2002;8:3803–3812. [PubMed] [Google Scholar]
- 24.Mlecnik B., Tosolini M., Charoentong P., Kirilovsky A., Bindea G., Berger A., Camus M., Gillard M., Bruneval P., Fridman W.H., Pages F., Trajanoski Z., Galon J. Biomolecular network reconstruction identifies T-cell homing factors associated with survival in colorectal cancer. Gastroenterology. 2010;138:1429–1440. doi: 10.1053/j.gastro.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 25.Ascierto M.L., Kmieciak M., Idowu M.O., Manjili R., Zhao Y., Grimes M., Dumur C., Wang E., Ramakrishnan V., Wang X.Y., Bear H.D., Marincola F.M., Manjili M.H. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res. Treat. 2012;131:871–880. doi: 10.1007/s10549-011-1470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messina J.L., Fenstermacher D.A., Eschrich S., Qu X., Berglund A.E., Lloyd M.C., Schell M.J., Sondak V.K., Weber J.S., Mule J.J. 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci. Rep. 2012;2:765. doi: 10.1038/srep00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong Z., Jia Q., Chen J., Diao X., Gao J., Wang X., Zhu B. Impaired cytolytic activity and loss of clonal neoantigens in elderly patients with lung adenocarcinoma. J. Thorac Oncol. 2019;14:857–866. doi: 10.1016/j.jtho.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 28.Scanlan M.J., Gure A.O., Jungbluth A.A., Old L.J., Chen Y.T. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol. Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura Y., Noguchi Y., Satoh E., Uenaka A., Sato S., Kitazaki T., Kanda T., Soda H., Nakayama E., Kohno S. Spontaneous remission of a non-small cell lung cancer possibly caused by anti-NY-ESO-1 immunity. Lung Cancer. 2009;65:119–122. doi: 10.1016/j.lungcan.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Gnjatic S., Atanackovic D., Jager E., Matsuo M., Selvakumar A., Altorki N.K., Maki R.G., Dupont B., Ritter G., Chen Y.T., Knuth A., Old L.J. Survey of naturally occurring CD4+ T cell responses against NY-ESO-1 in cancer patients: correlation with antibody responses. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8862–8867. doi: 10.1073/pnas.1133324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gure A.O., Chua R., Williamson B., Gonen M., Ferrera C.A., Gnjatic S., Ritter G., Simpson A.J., Chen Y.T., Old L.J., Altorki N.K. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin. Cancer Res. 2005;11:8055–8062. doi: 10.1158/1078-0432.CCR-05-1203. [DOI] [PubMed] [Google Scholar]
- 32.Djureinovic D., Hallstrom B.M., Horie M., Mattsson J.S.M., La Fleur L., Fagerberg L., Brunnstrom H., Lindskog C., Madjar K., Rahnenfuhrer J., Ekman S., Stahle E., Koyi H., Branden E., Edlund K., Hengstler J.G., Lambe M., Saito A., Botling J., Ponten F., Uhlen M., Micke P. Profiling cancer testis antigens in non-small-cell lung cancer. JCI Insight. 2016;1:e86837. doi: 10.1172/jci.insight.86837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biton J., Mansuet-Lupo A., Pecuchet N., Alifano M., Ouakrim H., Arrondeau J., Boudou-Rouquette P., Goldwasser F., Leroy K., Goc J., Wislez M., Germain C., Laurent-Puig P., Dieu-Nosjean M.C., Cremer I., Herbst R., Blons H., Damotte D. TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin. Cancer Res. 2018;24:5710–5723. doi: 10.1158/1078-0432.CCR-18-0163. [DOI] [PubMed] [Google Scholar]
- 34.Dong Z.Y., Zhong W.Z., Zhang X.C., Su J., Xie Z., Liu S.Y., Tu H.Y., Chen H.J., Sun Y.L., Zhou Q., Yang J.J., Yang X.N., Lin J.X., Yan H.H., Zhai H.R., Yan L.X., Liao R.Q., Wu S.P., Wu Y.L. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 35.Syn N.L., Teng M.W.L., Mok T.S.K., Soo R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017;18:e731–e741. doi: 10.1016/S1470-2045(17)30607-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.