Highlights
-
•
Mutations in the Wnt, Notch, and Ras-ERK signaling pathways in C. elegans lead to infertility, sterility, and multivulva formation.
-
•
Phenotypic assays using C. elegans mutant strains can be used as in vivo models for drug repurposing.
-
•
Itraconazole, disulfiram, etodolac, and ouabain have anticancer potential that can specifically target the Wnt, Notch, and RAS-ERK signaling pathways.
Keywords: Drug repurposing, Cancer, Caenorhabditis elegans
Abstract
Drug repurposing is used as a strategy for finding new drugs for cancer. The process is a faster and a more cost-effective way of providing new indications for drugs that can address emerging drug resistance and numerous side effects of chemotherapeutic drugs. In this study, the in vivo anticancer potential of itraconazole, disulfiram, etodolac, and ouabain were assessed using five different C. elegans mutant strains. Each strain contains mutations in genes involved in different signaling pathways such as Wnt (JK3476), Notch (JK1107 and BS3164), and Ras-ERK (SD939 and MT2124) that result to phenotypes of sterility, infertility, and multivulva formation. These same signaling pathways have been shown to be defective in several human cancer types. The four candidate drugs were tested on the C. elegans mutant strains to determine if they rescue the mutant phenotypes. Both ouabain and etodolac significantly reduced the sterile and infertile phenotypes of JK3476, JK1107, BS3164, and SD939 strains (p=0.0010). Finally, itraconazole and etodolac significantly reduced multivulva formation (p=0.0021). The degrees of significant phenotypic rescues of each mutant were significantly higher than vehicle only (1% DMSO). Therefore, this study demonstrated that the four candidate drugs have anticancer potential in vivo, and etodolac had the highest anticancer potential.
Introduction
Drug repurposing is being used as a strategy for finding new drugs for cancer [1]. This approach is a faster and a more cost-effective way of finding new drugs for certain diseases [2]. Compared to the conventional way of drug discovery, concerns on toxicity are circumvented in drug repurposing because the selected drugs already have well-documented safety profiles. Thus, there would be a shorter amount of time needed from compound identification up to pre-clinical studies in drug development [3]. The main principle behind this approach is polypharmacy [4], which assumes that all drugs can influence different pathways. The numerous off-target effects become an opportunity to find new indications for the existing drugs.
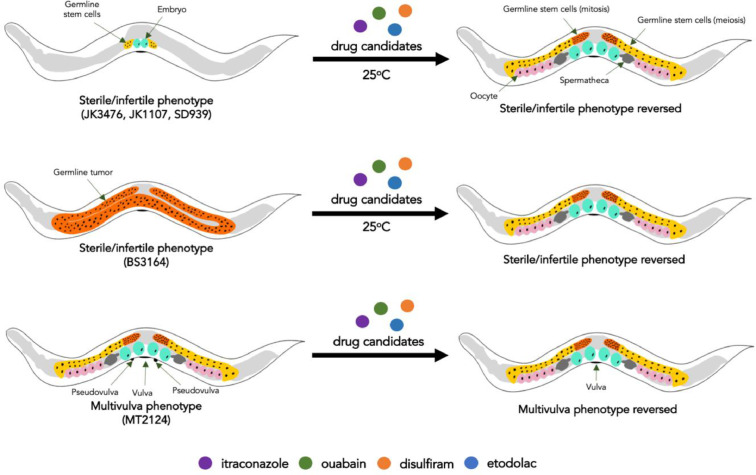
Despite the developments in understanding the agents that target pathways of tumorigenesis, many types of cancer lack efficacious treatment. Thus, alternative approaches are being sought to find new cancer treatments that may be effective, available, and affordable. One of these approaches is the use of simple in vivo models, such as the free-living transparent roundworm Caenorhabditis elegans, in elucidating the mechanisms of repurposed drugs. Mutations in the signaling pathways, such as in Wnt, Notch, and Ras-ERK signaling cascades, may lead to cancer in humans. These mutations that lead to defective signaling pathways can also be observed in C. elegans, however, instead of forming cancer, the C. elegans mutant strains become sterile, infertile, and form multivulva (Muv) [5]. Thus, drugs that can reverse these mutant phenotypes in C. elegans may also potentially reverse cancer in humans (Fig. 1).
Fig. 1.
Screening potential anticancer drugs using C. elegans
A phenotypic screen using different C. elegans mutant strains can be developed for discovering drugs for cancer. Each strain corresponds to a signaling pathway mutation, which lead to the development of a mutant phenotype that is a surrogate outcome for cancer. The anticancer potential of drug candidates can be determined by the reversal of the mutant phenotypes.
The mutations in the Wnt, Notch, and Ras-ERK signaling pathways exemplified by C. elegans strains are already established to promote carcinogenesis in humans since these pathways control different aspects of cell proliferation, cell differentiation, cell cycle progression, cell fate and cell death [6], [7], [8]. The development of tissue stem cells necessitates activation of the Wnt pathway, which acts as a signaling cascade that mediates the proliferation of cells through growth factors [9]. Loss of function mutations of β-catenin, an essential component of this pathway, is associated with several types of cancer such as colorectal cancer, hepatocellular carcinoma, melanoma, pancreatic cancer, and adrenocortical carcinoma [10]. Similarly, the Ras-ERK pathway also acts as a mediator of growth factors. Point mutations of the Ras receptor and several downstream kinases (e.g. RAF, MEK, and ERK) lead to constitutive activation of this pathway. The aberrant cell proliferation from the prolonged activation of this pathway is observed in non-small cell lung cancer, hairy cell leukemia, melanoma, and papillary thyroid cancer [11]. Lastly, different cues from the cellular microenvironment are processed by the Notch pathway through the direct contact of cells. Patterns of mutations in the different domain of the Notch receptor are associated with different types of cancers such as T cell acute lymphocytic leukemia, adenoid cystic carcinoma, triple negative breast cancer, chronic lymphocytic leukemia, mantle cell leukemia, B cell lymphoma, and squamous cell carcinoma of the skin, lung, head and neck [12].
Previously, itraconazole (anti-fungal drug), ouabain (cardiac glycoside), disulfiram (aldehyde dehydrogenase inhibitor), and etodolac (non-steroidal anti-inflammatory drug) were shown to have anticancer activities in vitro. Itraconazole is originally used as an anti-fungal drug by inhibiting fungal cytochrome P450, specifically 14-α demethylase, which is an enzyme involved in ergosterol synthesis [13]. The anticancer potential of itraconazole has been demonstrated in basal cell carcinoma [14,15], leukemia [16], and breast cancer cell lines. Ouabain is a member of a class of drugs called cardiac glycosides. These are drugs used for the treatment of heart failure by inhibiting the Na-K-ATPase pump found in cardiomyocytes [17]. Specific anticancer effects of ouabain were shown in in vitro studies using lung tumor cells and adrenocortical cells wherein these cancer cell types were suppressed by inhibiting focal adhesion kinase and apoptosis, respectively [18,19]. Disulfiram is used to treat chronic alcoholism by irreversibly inhibiting aldehyde dehydrogenase [20]. Combination of this drug with copper is an effective anticancer regimen because this complex induces aldehyde dehydrogenase activity, suppresses NF-kB signaling and creates an imbalance of ROS levels, which contribute to cytotoxicity in different cancer cell types [20]. In vitro data have shown the effectivity of the drug against temozolomide-resistant glioblastoma [21], inflammatory breast cancer [22], prostate cancer [23], and non-small cell lung cancer [24]. Lastly, etodolac is classified as a non-steroidal anti-inflammatory drug. This drug inhibits cyclooxygenase, which synthesized prostaglandins from arachidonic acid. This drug affects tumor formation by decreasing the expression of cyclin D and inhibits thymidylate synthase, which prevents progression of cell cycle and increases the sensitivity of the tumor to other chemotherapeutic drugs such as 5-fluorouracil [25]. These activities were shown in experiments involving breast cancer [26] and head and neck squamous cells [27].
In spite of the prevalence of in vitro data, in vivo evidence on the anticancer potential of these four candidate drugs is lacking. This study determined the in vivo anticancer potential of itraconazole, disulfiram, etodolac, and ouabain by their ability to rescue the mutant phenotypes of the C. elegans mutant strains.
Materials and methods
Cultivation of C. elegans strains
A wild type (N2) and five mutant C. elegans strains were obtained from the Caenorhabditis Genetics Center (CGC) at the University of Minnesota. The mutant strains were JK3476 (ceh-22(q632) sma-1(e30) V; nT1(qIs51) (IV;V)), JK1107 (glp-1(q224) III), BS3164 (unc-31(e189) glp-1(ar202) III), SD939 (mpk-1(ga111) unc-79(e1068) III) and MT2124 (let-60(n1046) IV). These strains are described in Table 1. The E. coli OP50 strain, which is used to feed the worms, was also obtained from the Caenorhabditis Genetics Center (CGC) at the University of Minnesota.
Table 1.
Characteristics of different C. elegans mutant strains and their associated human cancer types.
| Strain | Characteristics | Examples of Associated Human Cancer |
|---|---|---|
| JK3476* | Signaling pathway: Wnt/β-catenin [29] Type of mutation: Loss-of-function Mutant phenotype: Sterile and infertile at 25 oC |
Papillary thyroid carcinoma [30] Pediatric acute lymphoblastic leukemia [31] Skin squamous cell carcinoma [32] |
| JK1107* | Signaling pathway: Notch [33,34] Type of mutation: Loss-of-function Mutant phenotype: Sterile and infertile at 25 oC |
Cutaneous squamous cell carcinoma [35] Small cell lung carcinoma [36] Urothelial carcinoma [37] |
| BS3164* | Signaling pathway: Notch [38] Type of mutation: Gain-of-function Mutant phenotype: Sterile and infertile at 25 oC |
Diffuse large B-cell lymphoma [39] Acute lymphoblastic leukemia [40] Breast carcinoma [41] |
| SD939* | Signaling pathway: Ras-ERK [42,43] Type of mutation: Loss-of-function Mutant phenotype: Sterile and infertile at 25 oC |
Melanoma [44] Non-small cell lung cancer [45] Colorectal cancer [46] |
| MT2124 | Signaling pathway: Ras-ERK [47] Type of mutation: Gain-of-function Phenotype: Multivulva formation |
Cutaneous melanoma [48] Acute myeloid leukemia [49] Juvenile myelomonocytic leukemia [50] |
Wild type phenotype at 15 °C.
All of the C. elegans strains were maintained using a modified protocol by T. Stiernagle from Wormbook [28]. Briefly, 6-10 nematodes were transferred into a single culture plate containing nematode growth medium (NGM) (NaCl, peptone, bacteriological agar, 1M CaCl2, 5mg/mL ethanol, 1M MgSO4, 1M KPO4) layered with a condensed E. coli OP50 solution. The culture plates were kept at 15 °C.
Preparation of candidate drugs, positive controls, and negative control
The four candidate drugs and the anticancer drug controls were purchased from Abcam and AdooQ Bioscience. For each bioassay, an anticancer drug that targets the relevant signaling pathway was used as the positive control: PRI-724 (Wnt control), DAPT-GSI (Notch control) and U0126 (Ras-ERK control). All the drugs were dissolved in 1% dimethyl sulfoxide (DMSO) to produce the desired concentrations for each bioassay (Table 2). The negative control used was 1% DMSO for all assays.
Table 2.
Concentrations used for each candidate drug (µg/mL) and their corresponding conversion in molarity (µM).
| Itraconazole |
Disulfiram |
Etodolac |
Ouabain |
PRI-724 |
DAPT-GSI |
U0126 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| µg/mL | µM | µg/mL | µM | µg/mL | µM | µg/mL | µM | µg/mL | µM | µg/mL | µM | µg/mL | µM |
| 0 | 0.1 | 0 | 0.3 | 0 | 0.3 | 0 | 0.2 | 0 | 0.2 | 0 | 0.2 | 0 | 0.3 |
| 1 | 1.4 | 1 | 3.4 | 1 | 3.5 | 1 | 1.7 | 1 | 1.5 | 1 | 2.3 | 1 | 2.6 |
| 10 | 14 | 10 | 34 | 10 | 35 | 10 | 17 | 10 | 15 | 10 | 23 | 10 | 26 |
| 100 | 142 | 100 | 337 | 100 | 348 | 100 | 171 | 100 | 152 | 100 | 231 | 100 | 263 |
| 200 | 283 | 200 | 674 | 200 | 696 | 200 | 342 | 125 | 190 | 125 | 289 | 145 | 381 |
| 400 | 567 | 370 | 1248 | 390 | 1357 | 400 | 684 | 200 | 304 | 200 | 462 | 290 | 762 |
| 600 | 850 | 400 | 1349 | 400 | 1392 | 600 | 1026 | 250 | 380 | 250 | 578 | 580 | 1524 |
| 800 | 1134 | 600 | 2023 | 600 | 2088 | 800 | 1368 | 400 | 607 | 400 | 925 | 1000 | 2628 |
| 1000 | 1417 | 800 | 2698 | 800 | 2784 | 1000 | 1710 | 470 | 714 | 500 | 1156 | ||
| 1000 | 3372 | 1000 | 3480 | 500 | 759 | 600 | 1387 | ||||||
| 600 | 911 | 800 | 1850 | ||||||||||
| 800 | 1215 | 1000 | 2312 | ||||||||||
| 1000 | 1518 | ||||||||||||
Sublethal bioassay
The sublethal concentration is the highest possible concentration that would produce a percent survival of more than 90% (or less than 10% death) at 72 h of incubation with the drug candidate. The bioassay is from a modified protocol by Jiang et al. [51]. Briefly, 15–20 age-synchronous nematodes in the L4 stage were added to culture plates containing NGM and OP50 E. coli. Different concentrations of each candidate drug were prepared in 1% DMSO (Table 2). A volume of 100 µL of the test solution containing the drug were added to the culture plates. The plates were incubated at 15 °C for 24, 48, or 72 h. Then, the number of live, missing, and dead worms were recorded. Experimental set-ups for each time point and drug concentration were done in triplicates. The percentage survival for each set-up was calculated using the following formula:
For each candidate drug, the percent survival was interpolated based on the line of best fit calculated using the values for each concentration. Additional concentrations were added as needed to give further resolution. Then, the percent survival for the interpolated concentration was experimentally determined.
Determination of anticancer potential of the drug candidates
Egg-laying bioassay
Sterility is the inability of the worm to produce and lay eggs. This bioassay is used to measure the decrease in sterility of the sterile strains JK3476, JK1107, BS3164, and SD939 when treated with the candidate drugs. The bioassay was based on a protocol by A. Hart in Wormbook [52] with some modifications. Twenty late L4 larvae were transferred to culture plates containing NGM and 100 µl of condensed OP50 E. coli solution. The plates were incubated at 15 °C (permissive temperature) or 25 °C (restrictive temperature) with or without the corresponding 100 µl of candidate drug using the sublethal concentration. The restrictive temperature is the temperature at which the sterile phenotype can be produced for each of the mutant strain, while the permissive temperature is the optimal temperature for egg production. Then, the number of eggs were counted for 120 h in 24-h intervals. Each experimental set-up for each temperature and each treatment group was done in triplicates.
Fertility bioassay
Infertility is the inability to produce eggs that hatch. This bioassay is used to measure the increase in fertility of the infertile strains JK3476, JK1107, BS3164, and SD939 when treated with the candidate drugs. The bioassay was based on a protocol by A. Killeen and C. Marin de Evsikova [53] with some modifications. Twenty L1 to L2 larvae were transferred to culture plates containing NGM and 100 µl of condensed OP50 E. coli solution. The larvae were allowed to grow until the L4 stage with shifts in incubation temperature from 15 °C (permissive temperature) to 25 °C (restrictive temperature) with or without the corresponding 100 µl of candidate drug using the sublethal concentration. The restrictive temperature is the temperature at which the infertile phenotype can be produced for each of the mutant strain, while the permissive temperature is the optimal temperature for egg hatching. Fertility was measured by counting the number of F1 progeny. The number of hatched and unhatched eggs were also counted for 120 h in 18-h intervals. Each experimental set-up for each temperature shift and each treatment group was done in triplicates. The percentage of hatched eggs in each of the experimental set-up was calculated using the following formula:
Multivulva reduction bioassay
The multivulva (Muv) phenotype is the presence of at least one pseudovulva, in addition to the normal vulva, along the ventral side of the worms when viewed with a stereoscope at 40X magnification. This bioassay is used to measure the reduction of the Muv phenotype of the MT2124 mutant strain when treated with the candidate drugs. The assay was based on a protocol by Hara and Han [54]. Twenty larvae of the MT2124 strain were transferred to culture plates containing NGM and 100 µl of condensed OP50 E. coli solution. The nematodes were incubated for 120 h at 15 °C with or without the corresponding 100 µl of candidate drug using five varying concentrations that differ between drugs. Vulva induction was observed during the development of the worms from the L3 stage to adulthood. Each experimental set-up for each treatment group was done in triplicates. The percentage of worms with Muv phenotype was calculated using the following formula:
Statistical analyses
Data were processed using Graph pad (Prism 8) software. The data from treatment groups of each candidate drug were compared to the positive and negative control groups using one-way ANOVA with post-hoc Tukey HSD (Honestly Significance Difference) and Bonferroni and Holm test for multiple comparison.
Results
Sublethal concentrations of candidate drugs and positive controls
The sublethal concentrations for each candidate drug and the positive control anticancer drugs were initially determined in order to use as basis for dosing in the different bioassays.
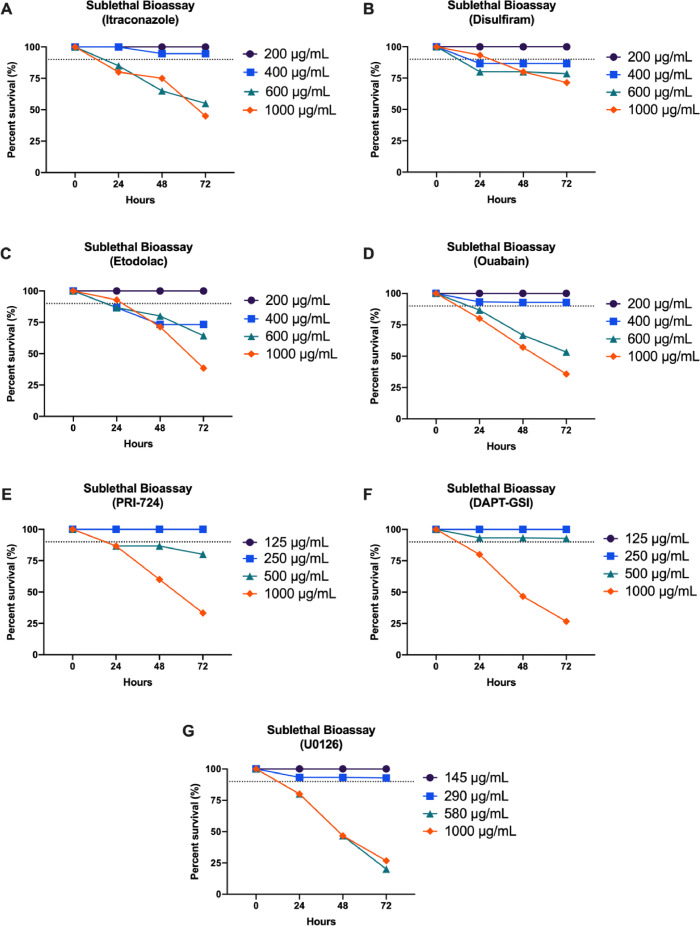
The C. elegans N2 strain demonstrated a decrease in the survival when exposed to increasing concentrations of itraconazole, disulfiram, etodolac, and ouabain in a dose-dependent manner (Fig. 2). This same trend of reduced survival was also observed in the five C. elegans mutant strains when treated with the candidate drugs. The sublethal concentrations of each of the candidate drugs for every C. elegans strain were determined by interpolation (Table 3). These concentrations had consistent results in all of the strains.
Fig. 2.
Percent Survival of C. elegans N2 strain exposed to the different drug candidates (A)–(D) and positive controls for each of the signaling pathway (E)–(G) for 72 h (n = 15–20 per concentration). The broken line represents 90% survival where concentrations above this value are considered as sublethal.
Table 3.
Sublethal concentrations of candidate drugs and positive controls with the corresponding percent survival for each C. elegans strain at 72 h of incubation.
| Drug | Sublethal concentration* | C. elegans strain | Percent survival (%) |
|---|---|---|---|
| Itraconazole | 400 µg/mL | N2 | 94.74 |
| JK3476 | 93.33 | ||
| JK1107 | 92.86 | ||
| BS3164 | 92.86 | ||
| SD939 | 90.00 | ||
| MT2124 | 92.31 | ||
| Disulfiram | 370 µg/mL | N2 | 92.31 |
| JK3476 | 92.31 | ||
| JK1107 | 92.31 | ||
| BS3164 | 93.33 | ||
| SD939 | 92.86 | ||
| MT2124 | 92.86 | ||
| Etodolac | 390 µg/mL | N2 | 92.31 |
| JK3476 | 92.86 | ||
| JK1107 | 92.86 | ||
| BS3164 | 92.86 | ||
| SD939 | 92.86 | ||
| MT2124 | 92.86 | ||
| Ouabain | 400 µg/mL | N2 | 92.86 |
| JK3476 | 92.86 | ||
| JK1107 | 92.86 | ||
| BS3164 | 92.86 | ||
| SD939 | 92.86 | ||
| MT2124 | 92.86 | ||
| PRI-724 | 470 µg/mL | N2 | 93.33 |
| JK3476 | 92.86 | ||
| DAPT-GSI | 500 µg/mL | N2 | 92.86 |
| JK1107 | 92.86 | ||
| BS3164 | 92.86 | ||
| U0126 | 290 µg/mL | N2 | 92.86 |
| SD939 | 92.86 | ||
| MT2124 | 92.86 |
See Table 2 for the molarity equivalents of each concentration.
Candidate drugs reduces the sterility of C. elegans mutant strains
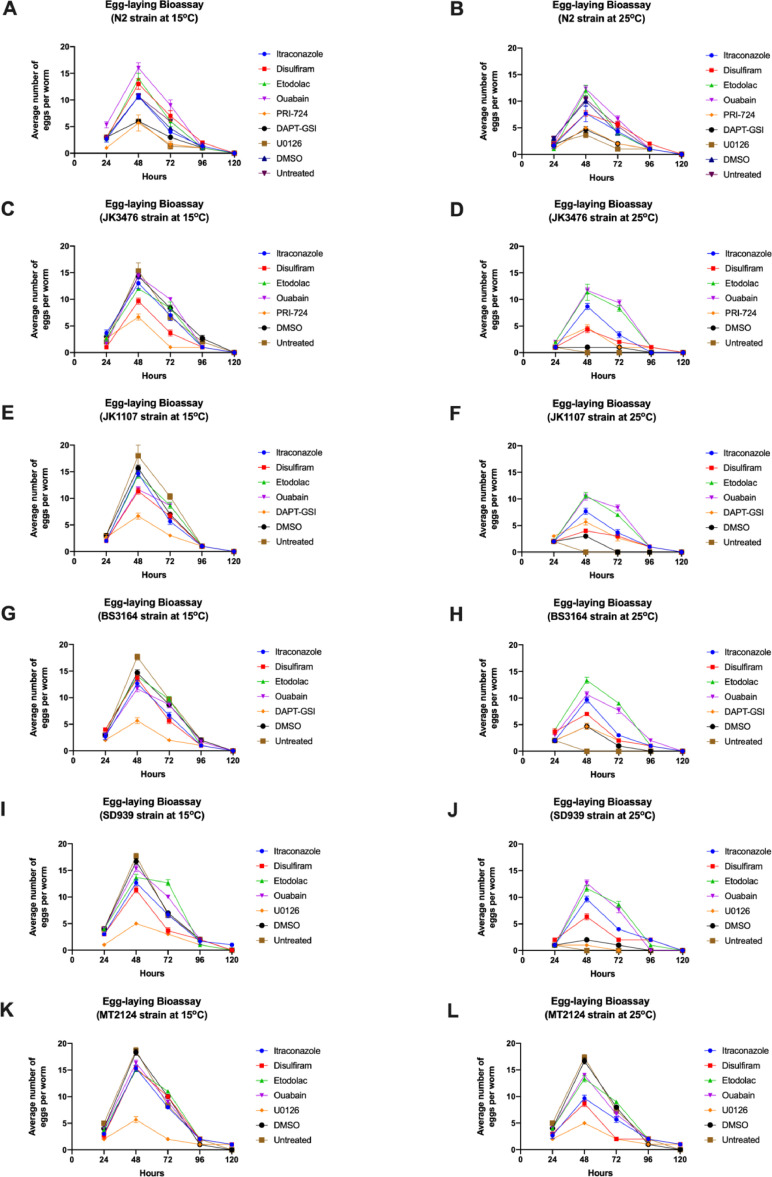
To determine the effect of itraconazole, disulfiram, etodolac, and ouabain on the Wnt, Notch, and Ras-ERK signaling pathways, the sterility of the JK3476, JK1107, BS3146, and SD939 mutant strains was observed by counting the number of eggs produced by each strain when exposed to each drug candidate for 120 h at 15 °C (permissive temperature) or 25 °C (restrictive temperature). The highest average number of eggs laid by all C. elegans strains was at 48 h of exposure with each drug candidate and anticancer drug control at 15 °C and 25 °C (Figure 3). Thus, the specific reference period used to compare the effects of the drugs across the wild type and mutant strains was selected at 48 h.
Fig. 3.
Average number of eggs produced by the wild type (A) and (B) and mutant C. elegans strains (C)–(L) incubated with the corresponding candidate drugs and controls for 120 h at 15 °C and 25 °C. (n=20 per strain)
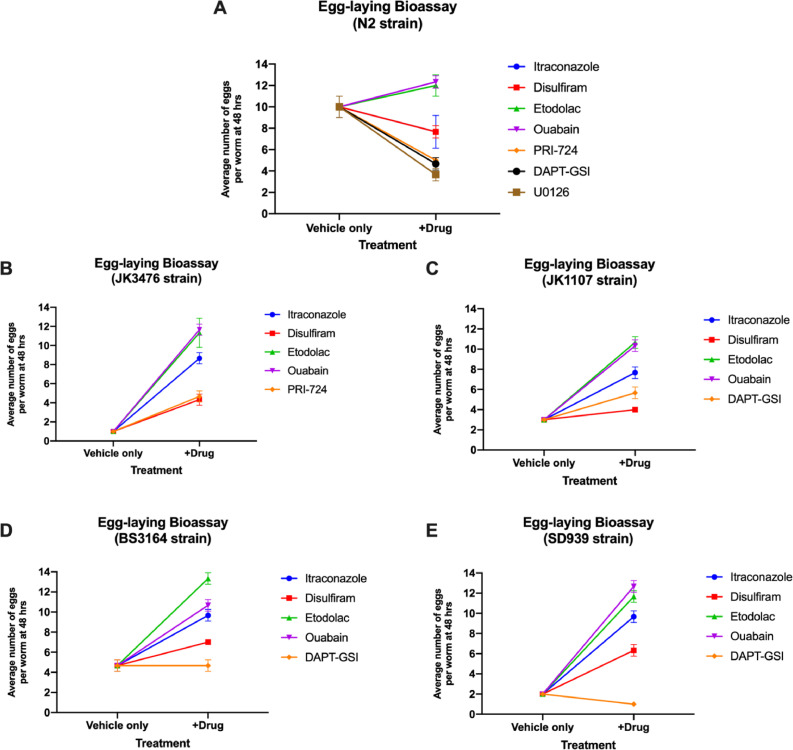
The effects of treatment with the different candidate drugs for 48 h on the average number of eggs laid was compared using the C. elegans strains at 25 °C (Fig. 4, Supplementary Table 1). None of the drugs including the positive control significantly affected the egg laying capacity of the N2 strain. Relative to the vehicle only, all of the candidate drugs significantly increased egg production of all the mutant strains. Etodolac treatment produced the significantly highest egg production in JK1107 and BS3164 (p=0.0010), while ouabain treatment produced the significantly highest egg production in JK3476 and SD939 (p=0.0010).
Fig. 4.
Average number of eggs produced by the wild type (A) and mutant (B)–(E) C. elegans strains incubated with the corresponding candidate drugs and controls for 120 h at 15 °C and 25 °C. (n=20 per strain).
Candidate drugs increases fertility of C. elegans mutant strains
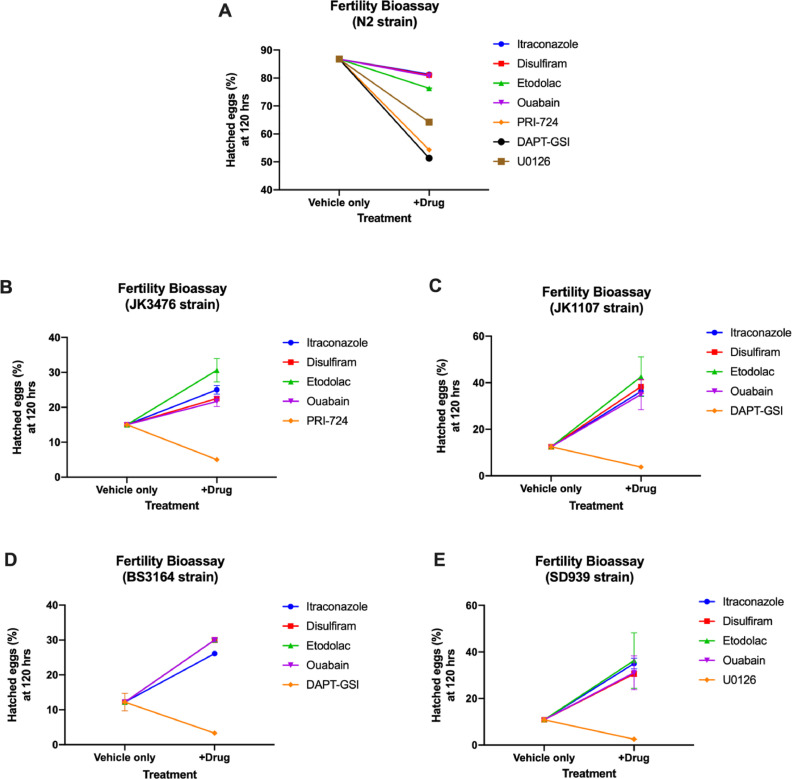
To further investigate the effect of the candidate drugs on the Wnt, Notch, and Ras-ERK signaling pathways, the fertility of the JK3476, JK1107, BS3146, and SD939 mutant strains was observed by counting the number of hatched and unhatched eggs produced by each strain when exposed to each drug candidate for 120 h at 15 °C (permissive temperature) or 25 °C (restrictive temperature). Then, the percent of hatched eggs were calculated.
The candidate drugs and anticancer drug controls significantly decreased the percentage of hatched eggs of the N2 strain (Fig. 5, Supplementary Table 2). Relative to the vehicle only, all of the candidate drugs significantly increased the percentage of hatched eggs of all the mutant strains. Etodolac treatment produced the significantly highest percentage of hatched eggs in JK3476, JK1107, and SD939 (p=0.0010), while ouabain treatment produced the significantly highest egg production in BS3164 (p=0.0010).
Fig. 5.
Percent hatched eggs based on the number of hatched and unhatched eggs of wild type (A) and mutant (B-E) C. elegans strains incubated with the corresponding candidate drugs and controls for 120 h at 15 °C and 25 °C. (n=20 per strain).
Candidate drugs prevent the formation of the Muv phenotype in the MT2124 mutant strain
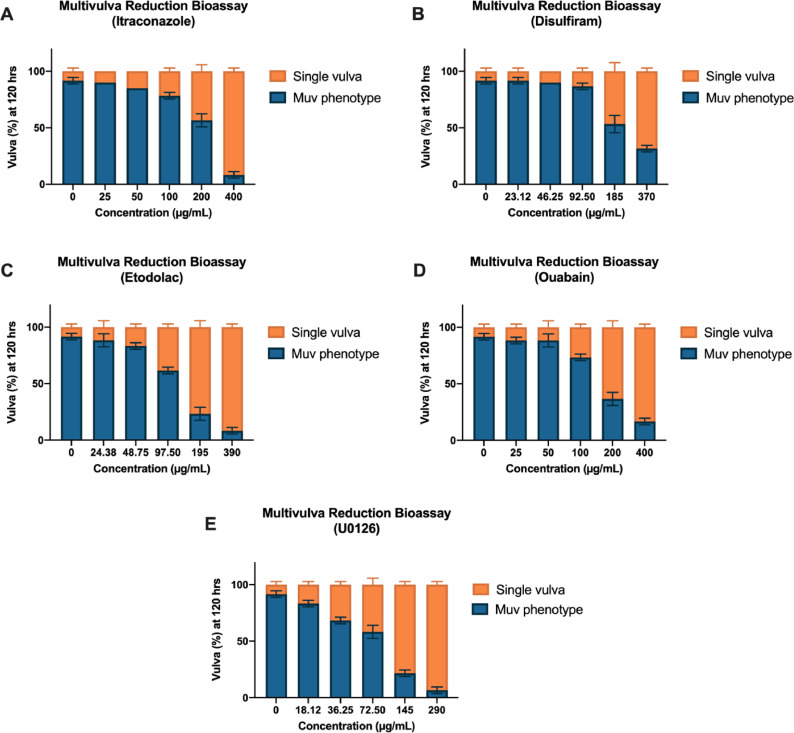
To augment the sterile and infertile phenotypes, another phenotypic endpoint, multivulva formation, was used to assess the effect of the candidate drugs on the Ras-ERK pathway. The effect of the candidate drugs on the development of the Muv phenotype was observed by counting the number of worms that produced the multivulva phenotype for 120 h at 15 °C (Figure 6, Supplementary Table 3). All of the candidate drugs produced significantly decreased percentage of Muv formation in the MT2124 strain (p=0.0021) compared to vehicle only in a dose-dependent manner. Etodolac and itraconazole treatment of the MT2124 strain produced the significantly lowest percentage of Muv formation (p=0.0021).
Fig. 6.
Percent Muv formation of MT2124 mutant strain incubated with different concentrations of the corresponding candidate drugs (A)–(D) and positive control (E) for 120 h at 15 °C. (n=20 per strain). The Muv phenotype is the presence of at least one pseudovulva, in addition to the normal vulva, along the ventral side of the worms when viewed with a stereoscope at 40X magnification. The difference between single vulva % and Muv % per concentration is the value of strain that reverted to wild-type vulva.
Ranking of each candidate drug based on rescuing of the mutant phenotype of each C. elegans mutant strain
Etodolac treatment consistently ranked the highest among all the candidate drugs in rescuing the mutant phenotypes of the different mutant strains (Table 4). Additionally, ouabain consistently ranked second in rescuing 3 out of 5 mutant strains.
Table 4.
Ranking of drug candidates based on the degree of phenotypic rescue observed in each mutant strain using the different bioassays. (1 – highest, 4 – lowest).
|
C. elegans mutant strains |
|||||
|---|---|---|---|---|---|
| Rank | JK3476 | JK1107 | BS3164 | SD939 | MT2124 |
| 1 | Etodolac | Etodolac | Etodolac | Etodolac | Etodolac |
| 2 | Ouabain | Disulfiram | Ouabain | Ouabain | Itraconazole |
| 3 | Itraconazole | Itraconazole | Disulfiram | Itraconazole | Ouabain |
| 4 | Disulfiram | Ouabain | Itraconazole | Disulfiram | Disulfiram |
Discussion
This study aimed to evaluate the anticancer potential of itraconazole, disulfiram, etodolac and ouabain using the mutant strains of C. elegans. The results of the assays show that all of the candidate drugs were able to significantly rescue the mutant phenotypes in each strain. Among these, etodolac was able to revert all of the mutant phenotypes with the consistently highest level. This drug may affect tumor formation by decreasing the expression of cyclin D and inhibits thymidylate synthase, which prevents progression of cell cycle and increases the sensitivity of the tumor to other chemotherapeutic drugs such as 5-fluorouracil [55]. Several mechanisms have been hypothesized to explain the antitumor effects of etodolac in terms of the Wnt, Notch, and Ras-ERK pathways. An analogue of etodolac introduced to hepatoma [56] and multiple myeloma cells [57] showed inhibition of the Wnt pathway by preventing the nuclear translocation of β-catenin. Meanwhile, similar COX-2 inhibitors have been shown to inhibit the Notch [58] and Ras-ERK [59] signaling by inhibition of NICD and MAPK phosphorylation activity, respectively. Additionally, the effectivity of ouabain may be due to its inhibitory effect on the Na+/K+ ATPase pump. This is exemplified by the hypothesized interaction of the pump to activators of the Ras-ERK signaling pathway such as Src [60].
There are still certain aspects of the anticancer properties of candidate drugs that are difficult to harness in the C. elegans model such as safety, dose range, and delivery [61,62]. Thus, further evaluation is warranted using other in vivo models such as rodents. Given the significant effects of etodolac and ouabain using the C. elegans model, these can be recommended for further evaluation using other in vivo models.
Aside from determining anticancer potential of the candidate drugs, this study was also able to demonstrate the role of C. elegans as a model for drug discovery. The transparent body and short life cycle of C. elegans enable faster generation of an observable response, simplicity in detecting outcomes (e.g. fluorescence imaging), and ease in generating large amounts of worms [6–8;[63], [64]–65]. As observed in the phenotypic assays, results can be produced within 168 h. (7 days) wherein worm cultivation can be done in 48 hours and the subsequent observation period for 120 h.. With these known features, the nematode can be used as a model that allows large scale screening of drugs, which may reduce the number of drugs for further testing in more complicated in vivo models. This may significantly to less consumption of time and resources. In terms of drug repurposing, this may also further reduce the cost of re-evaluating the new indication for the drug and the time spent for evaluation. Furthermore, several studies have suggested improvements on the C. elegans model to make it more efficient for drug screening. More fluorescent markers can be added not only as a food additive, but also as a transgene, which may make counting of progeny more effective especially when used in an automated setting [63;[66], [67], [68]].
Conclusions
The anticancer potential of itraconazole, disulfiram, etodolac, and, ouabain was demonstrated in this study using phenotypic assays of C. elegans mutant strains. Specifically, the specific ability of these drugs to affect the Wnt, Notch and Ras-ERK signaling pathways were shown since the drugs were able to significantly rescue sterility, infertility, and Muv formation compared to vehicle only. Several in vitro studies have already confirmed much of the anticancer effects of the four candidate drugs by promoting apoptosis, activating autophagy [69], or producing reactive oxygen species [70]. Some evidences also demonstrated that these drugs can target specific signaling pathways such as Wnt and NF-κB [71]. The findings of this study augments these previous studies by showing the robustness of these drugs in targeting the pathways, which lead to carcinogenesis in humans.
The conventional process of drug discovery entails the use of many resources, which would take years before application to human studies [3]. The in vivo system using C. elegans can effectively bridge in vitro and mammalian in vivo studies because the model can complement the simplicity of the system in cell cultures, while providing a faster and more perceptible development of phenotypic endpoints, which are more complex and harder to attain in rodents and non-human primates [72]. These aspects are clearly depicted in the phenotypic assays used in this study. Differences in egg-laying, fertility, and Muv formation were already observable as early as 48 hours post exposure to the drug at a certain restrictive temperature with the use of a stereoscope. Furthermore, the advantages of using C. elegans is elaborated by the role of the model as part of the drug screening process. Assessment of potential drugs may reduce the pool of candidate anticancer drugs to be tested, so that the drugs chosen to be tested in more expensive and longer validation studies in mammals may be more likely to provide a beneficial effect.
CRediT authorship contribution statement
Paul Mark Medina: Conceptualization, Methodology, Formal analysis, Writing - original draft, Writing - review & editing, Supervision. Jozelle Marie Ponce: Conceptualization, Methodology, Formal analysis, Investigation, Writing - original draft. Christian Alfredo Cruz: Formal analysis, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Department of Science and Technology (DOST) under the Accelerated Science and Technology Human Resource Development Program (ASTHRDP). Additionally, all the C. elegans strains were obtained and provided by the Caenorhabditis Genetics Centre (CGC) of the University of Minnesota, which is funded by the NIH Office of Research Infrastructure Programs [P40OD010440].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100940.
Appendix. Supplementary materials
References
- 1.Nowak-Sliwinska P., Scapozza L., Ruiz i Altaba A. Drug repurposing in oncology: compounds, pathways, phenotypes and computational approaches for colorectal cancer. Biochim. Biophys. Acta (BBA) – Rev. Cancer. 2019;1871(2):434–454. doi: 10.1016/j.bbcan.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown A.S., Patel C.J. A standard database for drug repositioning. Sci Data. 2017;4 doi: 10.1038/sdata.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 4.Car B.D. John Wiley & Sons, Inc; 2012. Polypharmacology in Drug Discovery. [Google Scholar]
- 5.Kobet R.A., Pan X., Zhang B., Pak S.C., Asch A.S., Lee M.-H. Caenorhabditis elegans: a model system for anti-cancer drug discovery and therapeutic target identification. Biomol Ther. 2014;22(5):371–383. doi: 10.4062/biomolther.2014.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M.-H., Yoon D.S. A phenotype-based RNAi Screening for Ras-ERK/MAPK signaling-associated stem cell regulators in C. elegans. Methods Mol. Biol. 2017;1622:207–221. doi: 10.1007/978-1-4939-7108-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorrepati L., Krause M.W., Chen W., Brodigan T.M., Correa-Mendez M., Eisenmann D.M. Identification of wnt pathway target genes regulating the division and differentiation of larval seam cells and vulval precursor cells in Caenorhabditis elegans. G3: Genes Genom. Genet. 2015;5(8):1551–1566. doi: 10.1534/g3.115.017715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng X., Michaelson D., Tchieu J., Cheng J., Rothenstein D., Feldman R. Targeting homologous recombination in notch-driven C. elegans stem cell and human tumors. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0127862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duchartre Y., Kim Y.-M., Kahn M. The Wnt signaling pathway in cancer. Crit. Rev. Oncol. Hematol. 2016;99:141–149. doi: 10.1016/j.critrevonc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nusse R., Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Samatar A.A., Poulikakos P.I. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 2014;13(12):928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 12.Aster J.C., Pear W.S., Blacklow S.C. The varied roles of notch in cancer. Annu. Rev. Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang G., Liu M., Wang Q., Shen Y., Mei H., Li D. Itraconazole exerts its anti-melanoma effect by suppressing Hedgehog, Wnt, and PI3K/mTOR signaling pathways. Oncotarget. 2017;8(17):28510–28525. doi: 10.18632/oncotarget.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei X., Liu W., Wang J.Q., Tang Z. “Hedgehog pathway”: a potential target of itraconazole in the treatment of cancer. J. Cancer Res. Clin. Oncol. 2020;146(2):297–304. doi: 10.1007/s00432-019-03117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsubamoto H., Ueda T., Inoue K., Sakata K., Shibahara H., Sonoda T. Repurposing itraconazole as an anticancer agent. Oncol. Lett. 2017;14(2):1240–1246. doi: 10.3892/ol.2017.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pantziarka P., Sukhatme V., Bouche G., Meheus L., Sukhatme V.P. Repurposing drugs in oncology (ReDO)-itraconazole as an anti-cancer agent. Ecancermedicalscience. 2015;9:521. doi: 10.3332/ecancer.2015.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botelho A.F.M., Pierezan F., Soto-Blanco B., Melo M.M. A review of cardiac glycosides: Structure, toxicokinetics, clinical signs, diagnosis and antineoplastic potential. Toxicon. 2019;158:63–68. doi: 10.1016/j.toxicon.2018.11.429. [DOI] [PubMed] [Google Scholar]
- 18.Schneider N.F.Z., Cerella C., Simões C.M.O., Diederich M. Anticancer and immunogenic properties of cardiac glycosides. Molecules. 2017;22(11) doi: 10.3390/molecules22111932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilubol N., Zhang L., Shen M., Zhang Y.-Q., He M., Austin C.P. Four clinically utilized drugs were identified and validated for treatment of adrenocortical cancer using quantitative high-throughput screening. J. Transl. Med. 2012;10:198. doi: 10.1186/1479-5876-10-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Chen F., Chen J., Chan S., He Y., Liu W. Disulfiram/copper induces antitumor activity against both nasopharyngeal cancer cells and cancer-associated fibroblasts through ROS/MAPK and ferroptosis pathways. Cancers. 2020;12(1) doi: 10.3390/cancers12010138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lun X., Wells J.C., Grinshtein N., King J.C., Hao X., Dang N.-H. Disulfiram when combined with copper enhances the therapeutic effects of temozolomide for the treatment of glioblastoma. Clin. Cancer Res. 2016;22(15):3860–3875. doi: 10.1158/1078-0432.CCR-15-1798. [DOI] [PubMed] [Google Scholar]
- 22.Allensworth J.L., Evans M.K., Bertucci F., Aldrich A.J., Festa R.A., Finetti P. Disulfiram (DSF) acts as a copper ionophore to induce copper-dependent oxidative stress and mediate anti-tumor efficacy in inflammatory breast cancer. Mol. Oncol. 2015;9(6):1155–1168. doi: 10.1016/j.molonc.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majera D., Skrott Z., Bouchal J., Bartkova J., Simkova D., Gachechiladze M. Targeting genotoxic and proteotoxic stress-response pathways in human prostate cancer by clinically available PARP inhibitors, vorinostat and disulfiram. Prostate. 2019;79(4):352–362. doi: 10.1002/pros.23741. [DOI] [PubMed] [Google Scholar]
- 24.Duan L., Shen H., Zhao G., Yang R., Cai X., Zhang L. Inhibitory effect of Disulfiram/copper complex on non-small cell lung cancer cells. Biochem. Biophys. Res. Commun. 2014;446(4):1010–1016. doi: 10.1016/j.bbrc.2014.03.047. [DOI] [PubMed] [Google Scholar]
- 25.Schwab R.B., Kato S., Crain B., Pu M., Messer K., Weidner N. A window-of-opportunity biomarker study of etodolac in resectable breast cancer. Cancer Med. 2015;4(10):1583–1588. doi: 10.1002/cam4.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy D., Arason G.A., Chowdhury B., Mitra A., Calaf G.M. Profiling of cell cycle genes of breast cells exposed to etodolac. Oncol. Rep. 2010;23(5):1383–1391. doi: 10.3892/or_00000775. [DOI] [PubMed] [Google Scholar]
- 27.Murata S., Adachi M., Kioi M., Torigoe S., Ijichi K., Hasegawa Y. Etodolac improves 5-FU sensitivity of head and neck cancer cells through inhibition of thymidylate synthase. Anticancer Res. 2011;31(9):2893–2898. [PubMed] [Google Scholar]
- 28.Stiernagle T. WormBook. 2006. Maintenance of C. elegans; pp. 1–11. 2007/12/01 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam N., Chesney M.A., Kimble J. Wnt signaling and CEH-22/tinman/Nkx2. 5 specify a stem cell niche in C. elegans. Curr. Biol. 2006;16:3. doi: 10.1016/j.cub.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penha, R. C. C., Buexm, L. A., Rodrigues, F. R., de Castro, T. P., Santos, M. C. S., Fortunato, R. S., & Ferreira, A. C. NKX2. 5 is expressed in papillary thyroid carcinomas and regulates differentiation in thyroid cells. BMC Cancer 208;18:1 [DOI] [PMC free article] [PubMed]
- 31.Nagel S., Kaufmann M., Drexler H.G., MacLeod R.A. The cardiac homeobox gene NKX2-5 is deregulated by juxtaposition with BCL11B in pediatric T-ALL cell lines via a novel t (5; 14)(q35. 1; q32. 2) Cancer Res. 2003;63:17. [PubMed] [Google Scholar]
- 32.Hwang C., Jang S., Choi D.K., Kim S., Lee J.H., Lee Y., Lee J.H. The role of nkx2. 5 in keratinocyte differentiation. Ann. Dermatol. 2009;21:4. doi: 10.5021/ad.2009.21.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cinquin A., Zheng L., Taylor P.H., Paz A., Zhang L., Chiang M., Cinquin O. Semi-permeable diffusion barriers enhance patterning robustness in the C. elegans germline. Dev. Cell. 2015;35:4. doi: 10.1016/j.devcel.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C., Sorensen E.B., Lynch T.R., Kimble J.C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. Elife. 2016;5:e18370. doi: 10.7554/eLife.18370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang N.J., Sanborn Z., Arnett K.L., Bayston L.J., Liao W., Proby C.M., Pennypacker S. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc. Natl. Acad. Sci. 2011;108:43. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.George J., Lim J.S., Jang S.J., Cun Y., Ozretić L., Kong G., Müller C. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:7563. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rampias T., Vgenopoulou P., Avgeris M., Polyzos A., Stravodimos K., Valavanis C., Klinakis A. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat. Med. 2014;20:10. doi: 10.1038/nm.3678. [DOI] [PubMed] [Google Scholar]
- 38.Pepper A.S.R., Killian D.J., Hubbard E.J.A. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics. 2003;163:1. doi: 10.1093/genetics/163.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arcaini L., Rossi D., Lucioni M., Nicola M., Bruscaggin A., Fiaccadori V., Casaluci G.M. The NOTCH pathway is recurrently mutated in diffuse large B-cell lymphoma associated with hepatitis C virus infection. Haematologica. 2015;100:2. doi: 10.3324/haematol.2014.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Vlierberghe P., Ambesi-Impiombato A., Perez-Garcia A., Haydu J.E., Rigo I., Hadler M., Wiernik P.H. ETV6 mutations in early immature human T cell leukemias. J. Exp. Med. 2011;208:13. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson D.R., Kalyana-Sundaram S., Wu Y.M., Shankar S., Cao X., Ateeq B., Lonigro R.J. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 2011;17:12. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee M.H., Yoon D.S. A phenotype-based RNAi screening for Ras-ERK/MAPK signaling-associated stem cell regulators in C. elegans. RNAi Small Regul. RNAs in Stem Cells. 2017 doi: 10.1007/978-1-4939-7108-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lackner M.R., Kim S.K. Genetic analysis of the Caenorhabditis elegans MAP kinase gene mpk-1. Genetics. 1998;150:1. doi: 10.1093/genetics/150.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colombino M., Capone M., Lissia A., Cossu A., Rubino C., De Giorgi V., Pagani E. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol. 2012;30:20. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 45.Seo J.S., Ju Y.S., Lee W.C., Shin J.Y., Lee J.K., Bleazard T., Yu S.B. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:11. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:7407. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beitel G.J., Clark S.G., Horvitz H.R. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990;348:6301. doi: 10.1038/348503a0. [DOI] [PubMed] [Google Scholar]
- 48.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 2014;13:11. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brendel C., Teichler S., Millahn A., Stiewe T., Krause M., Stabla K., Barckhausen C. Oncogenic NRAS primes primary acute myeloid leukemia cells for differentiation. PloS One. 2015;10:4. doi: 10.1371/journal.pone.0123181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cirstea I.C., Gremer L., Dvorsky R., Zhang S.C., Piekorz R.P., Zenker M., Ahmadian M.R. Diverging gain-of-function mechanisms of two novel KRAS mutations associated with Noonan and cardio-facio-cutaneous syndromes. Hum. Mol. Genet. 2013;22:2. doi: 10.1093/hmg/dds426. [DOI] [PubMed] [Google Scholar]
- 51.Jiang Y., Chen J., Wu Y., Wang Q., Li H. Sublethal toxicity endpoints of heavy metals to the nematode Caenorhabditis elegans. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0148014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hart A. WormBook. 2006. Behavior. [DOI] [Google Scholar]
- 53.Killeen A., Marin de Evsikova C. Effects of sub-lethal teratogen exposure during larval development on egg laying and egg quality in adult Caenorhabditis elegans. F1000Res. 2016;5:2925. doi: 10.12688/f1000research.8934.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara M., Han M. Ras farnesyltransferase inhibitors suppress the phenotype resulting from an activated ras mutation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1995;92(8):3333–3337. doi: 10.1073/pnas.92.8.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwab RB., Kato S., Crain B., Pu M., Messer K., Weidner N. A window-of-opportunity biomarker study of etodolac in resectable breast cancer. Cancer Med. 2015;4(10):1583–1588. doi: 10.1002/cam4.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Behari J., Zeng G., Otruba W., Thompson M.D., Muller P., Micsenyi A., Monga S.P. (2007). R-Etodolac decreases β-catenin levels along with survival and proliferation of hepatoma cells. J. Hepatol. 2007;46(5):849–857. doi: 10.1016/j.jhep.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasui H., Hideshima T., Ikeda H., Ocio E.M., Kiziltepe T., Vallet S., Chauhan D. Novel etodolac analog SDX-308 (CEP-18082) induces cytotoxicity in multiple myeloma cells associated with inhibition of β-catenin/TCF pathway. Leukemia. 2007;21(3):535–540. doi: 10.1038/sj.leu.2404561. [DOI] [PubMed] [Google Scholar]
- 58.Gallicchio M., Rosa A.C., Dianzani C., Brucato L., Benetti E., Collino M., Fantozzi R. Celecoxib decreases expression of the adhesion molecules ICAM‐1 and VCAM‐1 in a colon cancer cell line (HT29) Br. J. Pharmacol. 2008;153(5):870–878. doi: 10.1038/sj.bjp.0707634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu S., Chen Q., Lin T., Hong W., Wu W., Wu M., Jin R. (2018). The function of Notch1 intracellular domain in the differentiation of gastric cancer. Oncol. Lett. 2018;15(5):6171–6178. doi: 10.3892/ol.2018.8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J., Akkuratov E.E., Bai Y., Gaskill C.M., Askari A., Liu L. Cell signaling associated with Na+/K+-ATPase: activation of phosphatidylinositide 3-kinase IA/Akt by ouabain is independent of Src. Biochemistry. 2013;52:9059–9067. doi: 10.1021/bi4011804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu T., Xu H., Liang X., Tang M. Caenorhabditis elegans as a complete model organism for biosafety assessments of nanoparticles. Chemosphere. 2019;221:708–726. doi: 10.1016/j.chemosphere.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 62.Hunt PR. The C. elegans model in toxicity testing. J. Appl. Toxicol. 2017;37(1):50–59. doi: 10.1002/jat.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frank B.Y., Hamza A., Singh T., Flibotte S., Hieter P., O'Neil N.J. A multimodal genotoxic anticancer drug characterized by pharmacogenetic analysis in Caenorhabditis elegans. Genetics. 2020;215:3. doi: 10.1534/genetics.120.303169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwendeman A.R., Shaham S. A high-throughput small molecule screen for C. elegans linker cell death inhibitors. PloS One. 2016;11:10. doi: 10.1371/journal.pone.0164595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gosai S.J., Kwak J.H., Luke C.J., Long O.S., King D.E., Kovatch K.J., Silverman G.A. Automated high-content live animal drug screening using C. elegans expressing the aggregation prone serpin α1-antitrypsin Z. PloS One. 2010;5:11. doi: 10.1371/journal.pone.0015460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heppert J.K., Dickinson D.J., Pani A.M., Higgins C.D., Steward A., Ahringer J. Comparative assessment of fluorescent proteins for in vivo imaging in an animal model system. Mol. Biol. Cell. 2016;27(22):3385–3394. doi: 10.1091/mbc.E16-01-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gosai S.J., Kwak J.H., Luke C.J., Long O.S., King D.E., Kovatch K.J. Automated high-content live animal drug screening using C. elegans expressing the aggregation prone serpin α1-antitrypsin Z. PLoS One. 2010;5(11):e15460. doi: 10.1371/journal.pone.0015460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benson J.A., Cummings E.E., O'Reilly L.P., Lee M.-H., Pak S.C. A high-content assay for identifying small molecules that reprogram C. elegans germ cell fate. Methods. 2014;68(3):529–535. doi: 10.1016/j.ymeth.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 69.Wang X., Wei S., Zhao Y., Shi C., Liu P., Zhang C. Anti-proliferation of breast cancer cells with itraconazole: hedgehog pathway inhibition induces apoptosis and autophagic cell death. Cancer Lett. 2017;385:128–136. doi: 10.1016/j.canlet.2016.10.034. [DOI] [PubMed] [Google Scholar]
- 70.Pongrakhananon V., Chunhacha P., Chanvorachote P. Ouabain suppresses the migratory behavior of lung cancer cells. PLoS One. 2013;8(7):e68623. doi: 10.1371/journal.pone.0068623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee C., Sorensen E.B., Lynch T.R., Kimble J.C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. Elife. 2016;5:e18370. doi: 10.7554/eLife.18370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carretero M., Solis G.M., Petrascheck M. C. elegans as model for drug discovery. Curr. Top. Med. Chem. 2017;17(18):2067–2076. doi: 10.2174/1568026617666170131114401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.