Abstract
Long non-coding RNAs (lncRNAs) are characterized as key layers of the genome in various cancers. TSPEAR-AS2 was highlighted to be a candidate lncRNA potentially involved in gastric cancer (GC) progression. However, the clinical significance and mechanism of TSPEAR-AS2 in GC required clarification. The clinical significance of TSPEAR-AS2 was elucidated through Kaplan-Meier Plotter. The mechanism of TSPEAR-AS2 in GC was clarified in vitro and in vivo using luciferase reporter, chromatin immunoprecipitation, RNA immunoprecipitation assays, and animal models. TSPEAR-AS2 elevation was closely correlated with overall survival of GC patients. A basic transcription element-binding protein 2 (BTEB2)-activated TSPEAR-AS2 model was first explored in this study. TSPEAR-AS2 silencing substantially reduced tumorigenic capacities of GC cells, while TSPEAR-AS2 elevation had the opposite effect. Mechanistically, TSPEAR-AS2 bound with both polycomb repressive complex 2 (PRC2) and argonaute 2 (Ago2). TSPEAR-AS2 knockdown significantly decreased H3K27me3 levels at promoter regions of gap junction protein alpha 1 (GJA1). Ago2 was recruited by TSPEAR-AS2, which was defined to sponge miR-1207-5p, contributing to the repression of claudin 4 (CLDN4) translation. The axis of EZH2/GJA1 and miR-1207-5p/CLDN4 mediated by BTEB2-activated-TSPEAR-AS2 plays an important role in GC progression, suggesting a new therapeutic direction in GC treatment.
Keywords: long non-coding RNA, TSPEAR-AS2, BTEB2, gastric cancer
Graphical Abstract
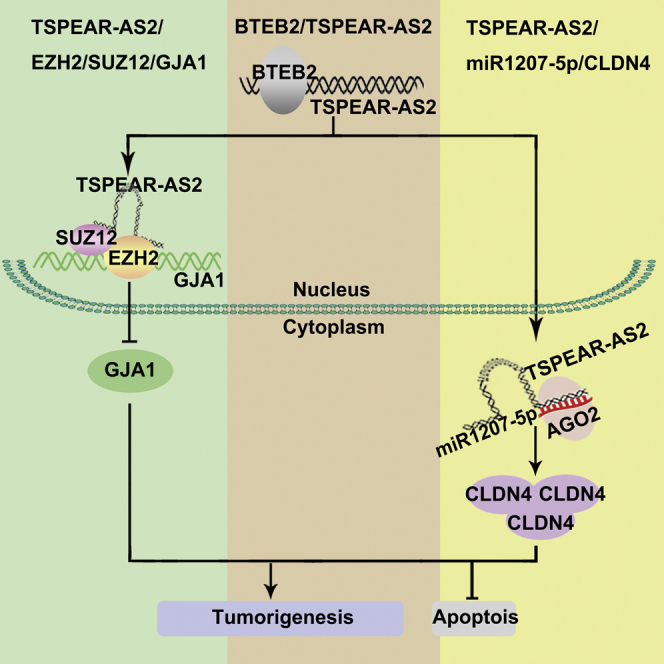
Ma and colleagues elucidated the clinical significance and mechanism of lncRNA TSPEAR-AS2 in gastric cancer progression. The regulatory axis of EZH2/GJA1 and miR-1207-5p/CLDN4 mediated by BTEB2-activated-TSPEAR-AS2 suggested a new therapeutic direction for patients with gastric cancer.
Introduction
Gastric cancer (GC) is an important issue strongly linked to public health, ranking as the third leading cause of cancer death globally.1 Despite tremendous progress in the clinical detection and treatment of GC in recent decades, the prognosis remains unsatisfactory, with a 5-year survival rate of less than 30% in most countries.2,3 The most important reasons for poor prognosis are largely due to late diagnosis, a high postoperative recurrence rate, and metastasis.4,5 Thus, a great challenge lies ahead in understanding the molecular mechanism of GC in identifying novel prognostic molecular biomarkers that can facilitate the development of appropriate therapeutic strategies earlier in GC.
Integrative genomic studies have shown that only 2% of DNA sequences can encode proteins, with more than 90% of these transcripts being actively transcribed; most of these transcripts are referred to as non-coding RNAs (ncRNAs).6,7 Based on size, ncRNAs are divided into two groups, long non-coding RNAs (lncRNAs) over 200 nucleotides and small ncRNAs such as microRNAs (miRNAs).8 Epigenetic modifiers, including lncRNAs and miRNAs, act to impact on human malignancies.9, 10, 11 Currently, increasing studies show that lncRNAs usually interact with RNA-binding protein (RBP) to participate in a variety of biological processes, such as chromatin remodeling, transcriptional regulation, and RNA degradation.12,13 Identification of lncRNA-dependent mechanisms of carcinogenesis is essential for understanding additional complexities of various tumors. Importantly, combined targeting of lncRNA-modulated key axes may provide a prospective rationale for cancer therapy.11,14
lncRNA TSPEAR-AS2 was previously reported to be involved in the regulation of hypoxia-induced pulmonary artery hypertension in vitro.15 In this study, TSPEAR-AS2 is defined as a GC-associated lncRNA that we identified by analyzing The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets. Coding potential assessment tool (CPAT)16 and coding potential calculator (CPC) analyses17 predicted the low coding potential of lncRNA TSPEAR-AS2 (Figure 1B). It was shown that high TSPEAR-AS2 level closely associated with the overall survival (OS) of patients with GC, suggesting the predictive value of TSPEAR-AS2 in the prognosis of GC patients. Then, we demonstrated a mechanism by which overexpression of TSPEAR-AS2 in GC cells was activated by basic transcription element binding protein 2 (BTEB2). This mechanistic model of BTEB2/TSPEAR-AS2 was first elucidated in this research. In vitro and in vivo assays found that silencing of TSPEAR-AS2 markedly inhibited cell growth, migration, and invasion. By contrast, ectopic expression of TSPEAR-AS2 played an oncogenic role in GC progression. Based on the analysis of high-throughput RNA sequencing (RNA-seq), we identified the contribution of TSPEAR-AS2 and its key target gene gap junction protein alpha 1 (GJA1) in GC progression. Specifically, TSPEAR-AS2 epigenetically inhibited GJA1 expression through the interaction with polycomb repressive complex 2. Moreover, argonaute 2 (Ago2) was recruited by TSPEAR-AS2, which was defined to sponge miR-1207-5p, thereby contributing to the repression of claudin 4 (CLDN4) translation. In conclusion, our study aimed to characterize novel lncRNAs closely correlated with GC. The axis of enhancer of zeste 2 (EZH2)/GJA1 and miR-1207-5p/CLDN4 mediated by BTEB2-activated-TSPEAR-AS2 may provide new clues facilitating the identification of therapeutic targets as well as effective biomarkers for patients with GC.
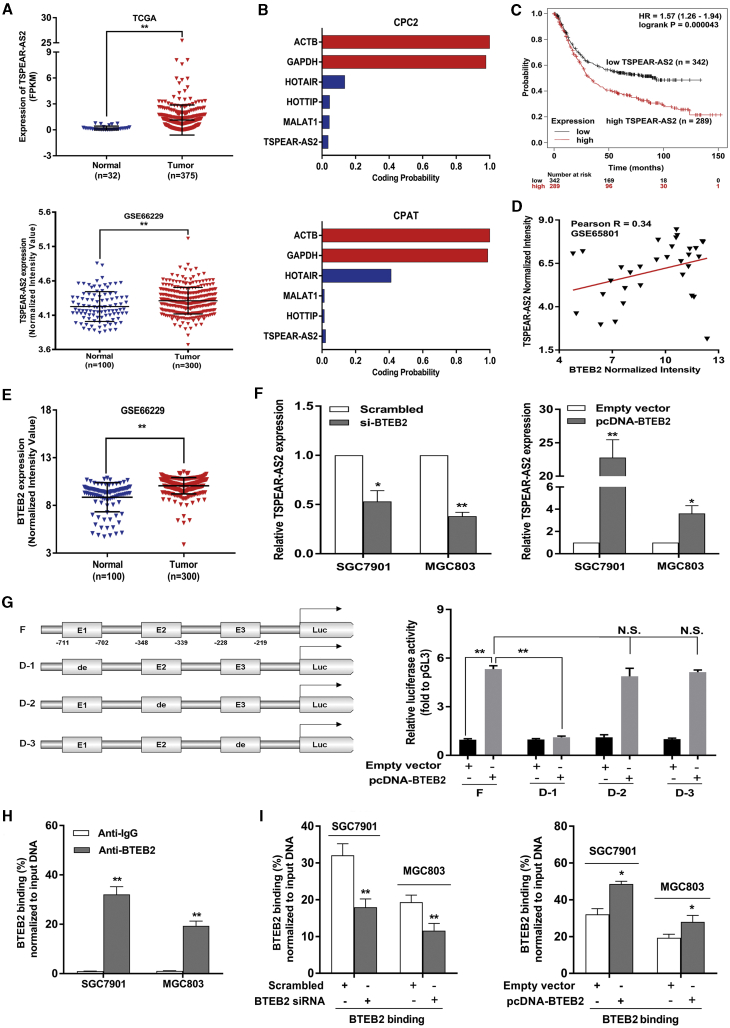
Figure 1.
High TSPEAR-AS2 Level Closely Associated with Poor Outcome of Patients with GC and the Transcription Factor BTEB2 Critically Activated the Transcription of TSPEAR-AS2
(A) Relative expression of TSPEAR-AS2 in GC tissues and normal tissues was measured in the data provided from GEO (GEO: GSE66229) and TCGA. (B) Evaluating the protein coding capacity of TSPEAR-AS2 through the Coding Potential Calculator 2 (CPC2) and Coding Potential Assessment Tool (CPAT). (C) The analysis of the correlation between TSPEAR-AS2 level and overall survival of GC patients (n = 631) using Kaplan-Meier Plotter. (D) The relationship between BTEB2 and TSPEAR-AS2 in GC tissue specimens was analyzed based on GSE65801 database. (E) BTEB2 levels in GC tissues and normal tissues were detected by analyzing data from GEO (GEO: GSE66229) database. (F) The effects of BTEB2 alteration on regulating the expression level of TSPEAR-AS2 in GC cells. (G) Dual-luciferase reporter assay was performed by cotransfecting the full TSPEAR-AS2 promoter fragment (pGL3-TSPEAR-AS2-F) or deleted TSPEAR-AS2 promoter fragment (pGL3-TSPEAR-AS2-D1, pGL3-TSPEAR-AS2-D2, pGL3-TSPEAR-AS2-D3) with pcDNA-BTEB2 or empty vector in HEK293T cells. (The predicted binding regions by JASPAR are represented as E1, E2, and E3.) (H) ChIP-qPCR assay showed direct binding of BTEB2 to endogenous TSPEAR-AS2 promoter regions in GC cells. (I) ChIP-qPCR assay showed BTEB2 enrichment on TSPEAR-AS2 promoter in GC cells transfected with BTEB2 siRNA or overexpression vector. ∗p < 0.05, ∗∗p < 0.01.
Results
lncRNA TSPEAR-AS2 Was Upregulated in GC and Tightly Associated with Survival Rates of Patients with GC
As an effort to identify lncRNAs closely correlated with GC, the publicly available data were downloaded from TCGA and GEO datasets. It was revealed that lncRNA TSPEAR-AS2 exhibited obvious upregulation in GC tissues (n = 375), as compared with normal tissues (n = 32) (Figure 1A). Moreover, the analysis of GSE66229 datasets demonstrated that TSPEAR-AS2 was obviously increased in GC tissues (n = 300) compared with normal tissues (n = 100) (Figure 1A). Additionally, TSPEAR-AS2 was markedly upregulated in GC cells compared with GES-1 cell line (Figure S1A). SGC7901 and MGC803 cells were selected for further research owing to their remarkable elevation of TSPEAR-AS2. Next, CPAT and CPC analyses were performed to reveal the low coding potential of lncRNA TSPEAR-AS2 (Figure 1B). To explore the correlation between TSPEAR-AS2 level and OS of patients, we also analyzed the publicly available data from 631 GC patients using Kaplan-Meier Plotter (http://kmplot.com/analysis/). As shown in Figure 1C, higher TSPEAR-AS2 expression closely associated with worse OS, highlighting the prognostic value of TSPEAR-AS2 in GC (Figure 1C).
TSPEAR-AS2 Was Transcriptionally Activated by Transcription Factor BTEB2 in GC
Currently, transcription factors (TFs) have been found to be capable of driving the expression of lncRNAs with tumor-promoting functions.18,19 Therefore, we made assumptions that certain transcription factors were responsible for ectopic expression of TSPEAR-AS2. BTEB2, a zinc-finger transcription factor, has been identified to modulate activities correlated with various functions such as cell growth, proliferation, differentiation, and tumorigenesis.20, 21, 22 It was shown that BTEB2 was significantly enriched in GC tissue samples (Figure 1E). Further analysis discovered the positive correlation between TSPEAR-AS2 and BTEB2 in GC tissue samples (Figure 1D). Then, we investigated the transcriptional regulation of TSPEAR-AS2 using JASPAR (http://jaspardev.genereg.net/) databases and found that BTEB2 can possibly bind to the promoter region of the TSPEAR-AS2 gene. The predicted binding regions are represented as E1, E2, and E3, shown in Figure 1G. In this regard, we deduced that BTEB2 might activate the transcription of TSPEAR-AS2. Therefore, the protein level of BTEB2 was first examined in GC cells with BTEB2 knockdown or overexpression. Compared with the control group, the protein level of BTEB2 was dramatically increased in GC cells with transfection of pcDNA-BTEB2 (Figure S1B). By contrast, small interfering RNAs (siRNAs) against BTEB2 obviously decreased the protein level of BTEB2 in GC cells (Figure S1B). Then, qRT-PCR assays were used to test the alteration of TSPEAR-AS2 in BTEB2-depleted or BTEB2-overexpressed GC cells. As expected, BTEB2 knockdown obviously decreased TSPEAR-AS2 expression, and BTEB2 overexpression markedly increased TSPEAR-AS2 expression, identifying BTEB2 as potential upstream regulator of TSPEAR-AS2 (Figure 1F).
To assess the transcription activation of BTEB2 on the promoter of TSPEAR-AS2, we cloned the promoter region of TSPEAR-AS2 into a luciferase reporter plasmid and made deletions at the promoter of TSPEAR-AS2 (Figure 1G). Then, cotransfection was performed in HEK293T cells with pcDNA-BTEB2/empty vector and luciferase reporter vectors TSPEAR-AS2 promoter full length (F), TSPEAR-AS2 promoter deletion 1# (D-1), TSPEAR-AS2 promoter deletion 2# (D-2), or TSPEAR-AS2 promoter deletion 3# (D-3) (Figure 1G). Dual-luciferase reporter analysis showed that D-1 caused a significant downregulation in promoter activity compared with F, D-2, and D-3 (Figure 1G). These findings elucidated the binding of BTEB2 to this region and its efficacy of luciferase activation. Meanwhile, chromatin immunoprecipitation (ChIP) experiments were implemented. Our data demonstrated that BTEB2 directly bound to their binding sites on TSPEAR-AS2 promoter region in GC cells (Figure 1H). Furthermore, downregulation or overexpression of BTEB2 decreased or increased BTEB2 enrichment within the TSPEAR-AS2 promoter, respectively (Figure 1I). Taken together, our data demonstrated that increased TSPEAR-AS2 expression could be transcriptionally activated by the key transcription factor BTEB2 in GC.
TSPEAR-AS2 Boosted the Oncogenic Activities of GC Cells In Vitro and In Vivo
To illuminate the function of TSPEAR-AS2 in GC progression, we performed loss- and gain-of function assays to effectively alter TSPEAR-AS2 expression in GC cells (Figures 2A and 2B). Knockdown of TSPEAR-AS2 obviously inhibited GC cell proliferation and impaired colony-formation ability (Figures 2C and 2E), while TSPEAR-AS2 overexpression displayed the reverse effects (Figures 2D and 2F). Moreover, Ethynyldeoxyuridineanaly (EdU) assays verified that TSPEAR-AS2 knockdown obviously decreased the proliferative capacities of GC cells (Figure 2G). Flow cytometry assays demonstrated that inhibition of TSPEAR-AS2 significantly increased the proportion of apoptosis in GC cells (Figure 2H). Our data verified that the inhibition of cell proliferation induced by TSPEAR-AS2 knockdown could be partly attributed to activated apoptosis in GC cells. In addition, TSPEAR-AS2 repression markedly weakened the migratory and invasive capacities of GC cells (Figures 2I and S2A), and overexpression of TSPEAR-AS2 elicited the opposite impacts (Figures 4K and S2B).
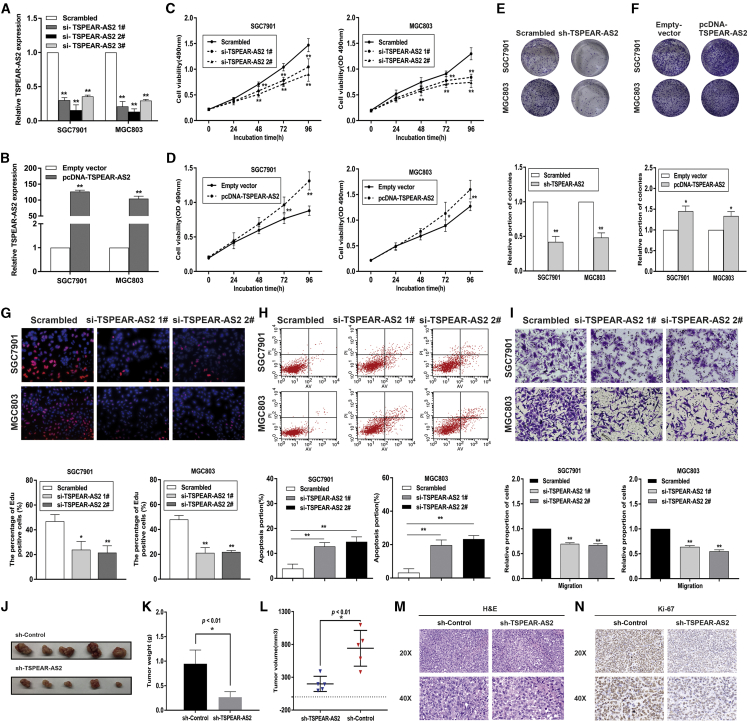
Figure 2.
The Biological Role of TSPEAR-AS2 in GC Progression
(A) TSPEAR-AS2 knockdown efficiency was analyzed by qRT-PCR in GC cells. (B) TSPEAR-AS2 overexpression efficiency was analyzed by qRT-PCR in GC cells. (C) Cell viability examinations of GC cells with TSPEAR-AS2 knockdown. (D) Cell viability examinations of GC cells with TSPEAR-AS2 overexpression. (E) Colony-forming assays were conducted to determine the proliferation of GC cells with TSPEAR-AS2 knockdown. (F) Colony-forming assays were conducted to determine the proliferation of GC cells with TSPEAR-AS2 overexpression. (G) Cell proliferation of GC cells was evaluated 48h after transfection with TSPEAR-AS2 siRNAs or Scrambled using EdU assays. (H) The apoptosis of GC cells transfected with TSPEAR-AS2 siRNAs or Scrambled was analyzed by flow cytometry assays. (I) Transwell assays were performed in GC cells transfected with TSPEAR-AS2 siRNAs or Scrambled. (J) The dissected tumors bearing from MGC803 cells transfection of sh-Control or shRNA groups. (K) The weight of tumors obtained from sh-TSPEAR-AS2 group or control group. (L) The volume of tumors obtained from sh-TSPEAR-AS2 group or control group. (M) H&E staining of the tumors isolated from mice. 20×images and 40×images were shown. (N) Ki-67 staining of the tumors isolated from mice. 20×images and 40×images were shown. *p < 0.05, **p < 0.01.
Figure 4.
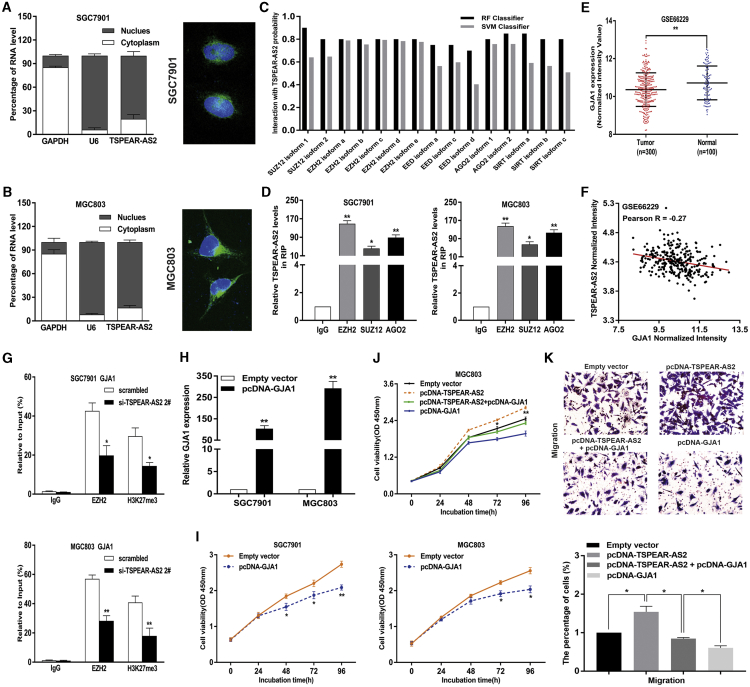
lncRNA TSPEAR-AS2 Silenced GJA1 Transcription through Binding to EZH2 in GC
(A) The subcellular localization analysis of the location of TSPEAR-AS2 in the cytoplasm and nuclear fractions in GC cells. (B) The FISH analysis of the location of TSPEAR-AS2 in the cytoplasm and nuclear fractions in GC cells. (C) Prediction of the interaction probability between TSPEAR-AS2 and RNA binding proteins by bioinformatics (http://pridb.gdcb.iastate.edu/RPISeq/). (D) The interaction of TSPEAR-AS2 with EZH2, SUZ12, and AGO2 was verified by RIP assay in GC cells. (E) The expression of GJA1 in GC tissues compared with normal tissues in GSE66229 datasets. (F) The correlation between the level of TSPEAR-AS2 and GJA1 in GC tissues was detected using data from GSE66229 datasets. (G) ChIP-qPCR assay of EZH2 and H3K27me3 occupancy in the GJA1 promoter in GC cells transfected with TSPEAR-AS2 siRNA or Scrambled. (H) The expression level of GJA1 is detected in GC cells transfected with pcDNA-GJA1 through qRT-PCR. (I) The effects of GJA1 overexpression on GC cell viability were detected using CCK8 assays. (J) Overexpression of GJA1 can partly reverse the promotion effects of GC cell proliferation mediated by TSPEAR-AS2 overexpression. (K) Overexpression of GJA1 can partly reverse the promotion effects of GC cell migration mediated by TSPEAR-AS2 overexpression. *p < 0.05, **p < 0.01.
To determine TSPEAR-AS2 impact on tumorigenic capacities in vivo, a xenograft tumor model was constructed. TSPEAR-AS2-stable-knockdown MGC803 cells or control cells were subcutaneously injected into male nude mice. We found that the tumors derived from the control group were markedly larger than tumors obtained from the TSPEAR-AS2-stable-knockdown group (Figure 2J). Additionally, the volume and weight of the tumors formed from TSPEAR-AS2-stable-knockdown group were significantly decreased compared with those derived from the control group, indicating the promotion effects of TSPEAR-AS2 on the tumorigenic abilities of GC cells (Figures 2K and 2L). As shown in Figures 2M and 2N, tumors derived from the control group revealed stronger staining of Ki-67 compared with tumors obtained from the TSPEAR-AS2-stable-knockdown group (Figures 2M and 2N).
Silencing of GJA1 Was Regulated by Interaction of TSPEAR-AS2 and Polycomb Repressive Complex 2
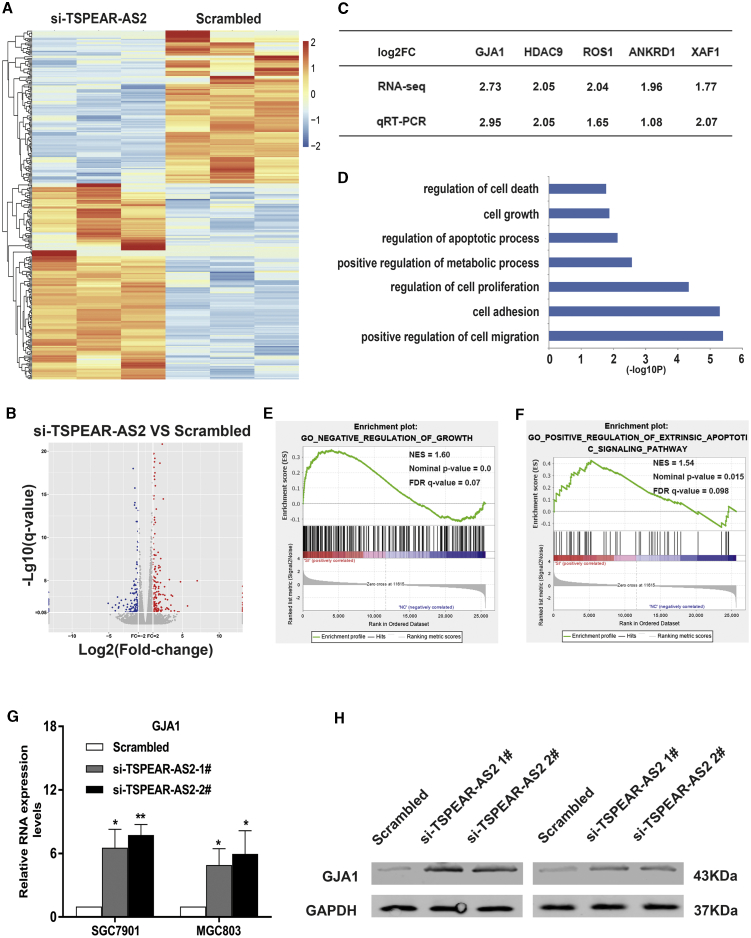
To uncover the underlying mechanism about how lncRNA TSPEAR-AS2 contribute to the malignant phenotype of GC, we evaluated the gene expression profiles of a control group and TSPEAR-AS2 knockdown group via RNA-seq (Figures 3A and 3B). Gene Ontology (GO) analysis of RNA-seq assays of TSPEAR-AS2 knockdown demonstrated that the alteration in gene set was closely associated with cell migration, cell proliferation, apoptotic process, and cell growth (Figure 3D). As shown in Figures 3E and 3F, Gene Set Enrichment Analysis (GSEA) was applied to further explore the pathways involved in GC pathogenesis. Enrichment plots of GSEA highlighted that the gene signatures of negative regulation of growth and positive regulative extrinsic apoptotic signaling pathway were much more involved in TSPEAR-AS2-depleted cells compared with the control group (Figures 3E and 3F). Then, qRT-PCR assays were used to identify key regulators mediated by TSPEAR-AS2 knockdown and thus further the understanding of TSPEAR-AS2-mediated GC progression. Among these aberrantly altered key genes, GJA1 displayed the highest level in TSPEAR-AS2-depleted SGC7901 cells relative to the control group (Figure 3C). Current evidence has demonstrated that GJA1 exhibits close association with cancer development, distant metastasis, and survival condition.23,24 Moreover, GJA1 was dramatically upregulated in TSPEAR-AS2-depleted GC cells, as demonstrated by qRT-PCR and western blot experiments (Figures 3G and 3H).
Figure 3.
GJA1 Acted as a Key Downstream Target of TSPEAR-AS2 in GC Progression
(A) Heatmap of altered genes in GC cells transfected with TSPEAR-AS2 siRNA or Scrambled. (B) Hierarchical clustering gene transcription altered in GC cells after knockdown of TSPEAR-AS2. (C) The qRT-PCR assays were conducted to validate the level of key genes in GC cells with TSPEAR-AS2 knockdown. (D) Gene Ontology analysis for all genes with altered expressions after knockdown of TSPEAR-AS2. (E) GSEA explored the gene sets enriched by genes in response to TSPEAR-AS2 knockdown (Negative regulation of growth). (F) GSEA explored the gene sets enriched by genes in response to TSPEAR-AS2 knockdown (Positive regulation of extrinsic apoptotic signaling pathway). (G) The expression of GJA1 was determined in GC cells treated with TSPEAR-AS2 siRNA or scrambled using qRT-PCR assays. (H) The expression of GJA1 was determined in GC cells treated with TSPEAR-AS2 siRNA or scrambled using western blot assays. *p < 0.05, **p < 0.01.
To further clarify TSPEAR-AS2-involved regulatory mechanisms in GC progression, we performed subcellular fractionation and RNA-fluorescence in situ hybridization (FISH) assays in GC cells. Our findings elucidated that TSPEAR-AS2 was more prevalent in the nucleus than in the cytoplasm in SGC7901 and MGC803 cells (Figures 4A and 4B). These findings may support the potential transcriptional regulation of TSPEAR-AS2 in GC progression. Currently, researchers demonstrated that lncRNAs could modulate epigenetic modification or gene silencing through binding with RBPs.25,26 To further determine the regulatory mechanism of TSPEAR-AS2-induced GJA1 silencing, bioinformatics analysis was first conducted to predict interaction possibilities of RBPs and TSPEAR-AS2. The analysis data indicated that TSPEAR-AS2 was predicted to potentially bind with EZH2, SUZ12, EED, STAU1, and Ago2, with the score of RF or SVM greater than 0.5 (Figure 4C). To verify this prediction, radioimmunoprecipitation (RIP) assays testified the binding of TSPEAR-AS2 with EZH2, SUZ12, and AGO2 in GC cells (Figure 4D).
Methyltransferase EZH2, a critical member of PRC2, enhances methylation of H3K27, leading to silencing of tumor suppressors.27,28 The amplification of EZH2 was observed in various types of cancers.29, 30, 31 Previous research also highlighted the oncogenic role of EZH2 in GC, indicating its emerging role in this active field.32 In this study, the level of EZH2 was effectively impaired in GC cells with siRNAs against EZH2 (Figure S3A). Intriguingly, knockdown of EZH2 can dramatically upregulate GJA1 level in GC (Figure S3B). Together, GJA1 may be coregulated by TSPEAR-AS2 and EZH2 in GC.
To elucidate whether TSPEAR-AS2 was involved in regulating gene transcription by recruiting PRC2, we performed ChIP assays in SGC7901 and MGC803 cells. It was demonstrated that TSPEAR-AS2 knockdown significantly reduced the binding of EZH2 and H3K27me3 across the GJA1 promoter region (Figure 4G). Together, our data revealed that TSPEAR-AS2 participate in the tumorigenesis of GC through the transcriptional regulation of GJA1 via binding to PRC2 in GC cells.
Oncogenic Function of TSPEAR-AS2 by Repressing GJA1 Expression
Given the potential regulatory role of GJA1 in TSPEAR-AS2-involved GC progression, bioinformatics analysis was implicated to verify the expression pattern of TSPEAR-AS2 in specimens from patients with GC. It was shown that GJA1 was prominently downregulated in GC tissue samples (n = 300) compared with non-tumor samples (n = 100) (Figure 4E). Meanwhile, it is interesting to note the negative relationship between GJA1 level and TSPEAR-AS2 level in GC tumor specimens (n = 300) (Figure 4F). In addition, the level of GJA1 was verified in GC cells treated with empty vector or pcDNA-GJA1. It can be observed that GC cells with pcDNA-GJA1 displayed remarkable upregulation compared with the control group (Figure 4H). Cell viability tests verified the inhibition impact of GJA1 overexpression on GC cell proliferation (Figure 4I).
To investigate the function of GJA1 in TSPEAR-AS2-induced promotion of GC proliferation, invasion, and migration, rescue assays were conducted in GC cells, which were cotransfected with pcDNA-TSPEAR-AS2 and pcDNA-GJA1. Of note, ectopic expression of TSPEAR-AS2 remarkably activated GC cell proliferation, migration, and invasion, and overexpression of GJA1 was capable of reversing the influence mediated by TSPEAR-AS2 (Figures 4J, 4K, and S2B). These data elucidated that the effects of TSPEAR-AS2 on GC progression may partially depend on the regulation of GJA1.
lncRNA TSPEAR-AS2 Elevated CLDN4 Expression through Competing for miR-1207-5p
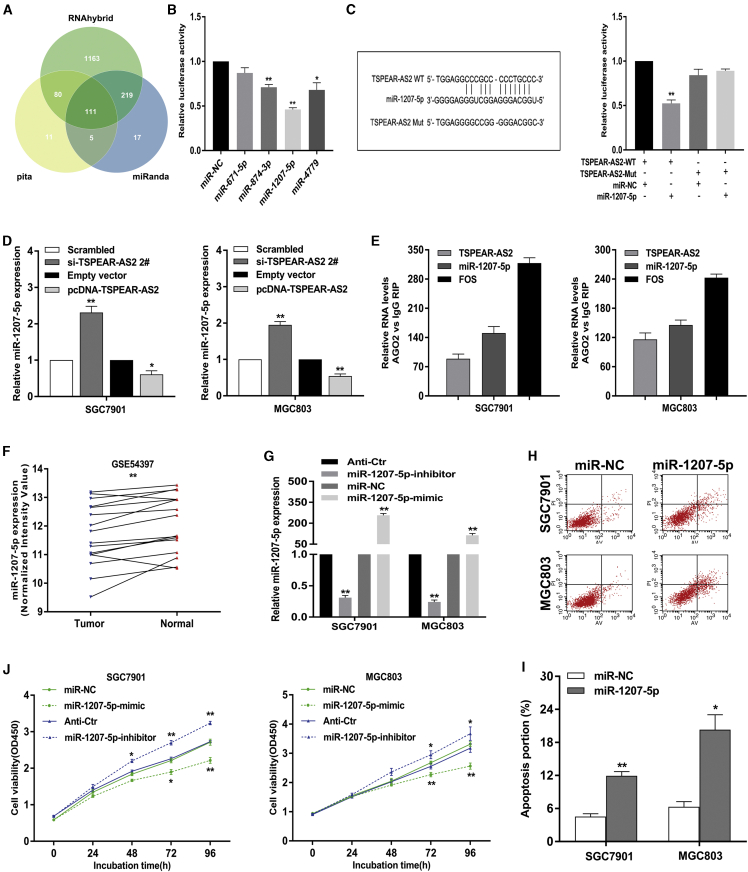
Importantly, TSPEAR-AS2 may also play a post-transcriptional role in gene regulation. It is known that lncRNAs have been implicated in post-transcription regulation, such as sponging activity for miRNAs.33,34 To investigate whether TSPEAR-AS2 played such a role, we used miRanda, pita, and RNAhybrid to make a prediction of possible miRNAs targeting sites on TSPEAR-AS2 (Figure 5A). According to the prediction result and current evidence, we filtered out a number of miRNAs, which have been shown to inhibit the malignant phenotype of tumor.35, 36, 37 Therefore, the implementation of dual-luciferase reporter assays testfied the interacted correlation between TSPEAR-AS2 and these miRNAs. We observed that the luciferase activity of pmir-GLO-TSPEAR-AS2 that contained full-length TSPEAR-AS2 can be significantly repressed by the transfection of miR-874-3p, miR-1207-5p, and miR-4779 (Figure 5B). Moreover, miRNA-1207-5p showed the strongest inhibition effect (Figure 5B). Thus, miR-1207-5p was selected for further analysis.
Figure 5.
Regulation Relationship between lncRNA TSPEAR-AS2 and miR-1207-5p in GC
(A) Bioinformatics databases (miRanda, pita, and RNAhybrid) were analyzed to predict potential miRNAs binding with TSPEAR-AS2. (B) Luciferase activity of HEK293T cells cotransfected with 4 various miRNA-coding plasmids and the luciferase reporter plasmids (pmirGLO-TSPEAR-AS2-WT). Data are presented as the ratio of firefly luciferase to Renilla luciferase activity. (C) Luciferase activity in HEK293T cells cotransfected with miR-1207-5p or negative control and pmirGLO-TSPEAR-AS2-WT or pmirGLO-TSPEAR-AS2-Mut. (D) The level of miR-1207-5p was examined in GC cells with TSPEAR-AS2 knockdown or overexpression through qRT-PCR assays. (E) RNA levels in immunoprecipitates were presented as fold change in Ago2 relative to IgG immunoprecipitates. (F) Relative expression of miR-1207-5p in GC tissues and paired normal tissues was analyzed in the GSE54397 database. (G) The level of miR-1207-5p was detected in GC cells transfected with mimic or inhibitor against miR-1207-5p using qRT-PCR assays. (H and I) The effects of miR-1207-5p on GC cell apoptosis were analyzed by flow cytometry assays. (J) The effects of miR-1207-5p knockdown or overexpression on GC cell viability. ∗p < 0.05, ∗∗p < 0.01.
Thereafter, site-targeted mutagenesis was constructed within the speculative miR-1207-5p-binding site in the lncRNA TSPEAR-AS2 sequence (Figure 5C). It was shown that pmir-GLO-TSPEAR-AS2-mut (TSPEAR-AS2-Mut) failed to respond to miR-1207-5p, clarifying that TSPEAR-AS2 acts to sponge miR-1207-5p (Figure 5C). In addition, knockdown of TSPEAR-AS2 gave rise to an elevated level of miR-1207-5p, whereas the boosted TSPEAR-AS2 level reflected contrary impact on miR-1207-5p level in GC cells (Figure 5D). However, overexpression of miR-1207-5p displayed no significant difference on TSPEAR-AS2 expression (Figure S3C). More importantly, RIP assays were performed in GC cells to verify whether TSPEAR-AS2 and miR-1207-5p were involved in the RNA-induced silencing complex (RISC). It was shown that both TSPEAR-AS2 and miR-1207-5p are drastically enriched in AGO2 immunoprecipitates compared with those in the immunoglobulin G (IgG) pellet in SGC7901 and MGC803 cells (Figure 5E). These results suggested that TSPEAR-AS2 physically existed in the AGO2-based miRNA-modulated repression complex and exhibited close association with miR-1207-5p, but miR-1207-5p did not induce TSPEAR-AS2 degradation.
Previous evidence has indicated that miR-1207-5p is of great significance in many fetal malignancies.38,39 Nevertheless, the functional biology of miR-1207-5p was not comprehensively elucidated in GC. Furthermore, the mechanistic model of miR-1207-5p in TSPEAR-AS2-mediated GC progression remains unclear. Therefore, the profile of the miR-1207-5p level was verified in paired tissue specimens from patients with GC (GSE54397) (Figure 5F). The significant downregulation of miR-1207-5p can be observed in GC tissue samples relative to matched normal tissue samples (Figure 5F). Then, significant overexpression or knockdown of miR-1207-5p level was made in GC cells using mimic or inhibitor against miR-1207-5p (Figure 5G). We gave the first evidence that the ectopic level of miR-1207-5p dramatically improve the apoptotic proportion of GC cells (Figures 5H and 5I). Consistently, knockdown of miR-1207-5p could obviously activate GC cell proliferation (Figure 5J). By contrast, elevation of miR-1207-5p impacted contrary effects on GC cell proliferation (Figure 5J). These findings highlight the critical role of miR-1207-5p in TSPEAR-AS2-correlated GC progression
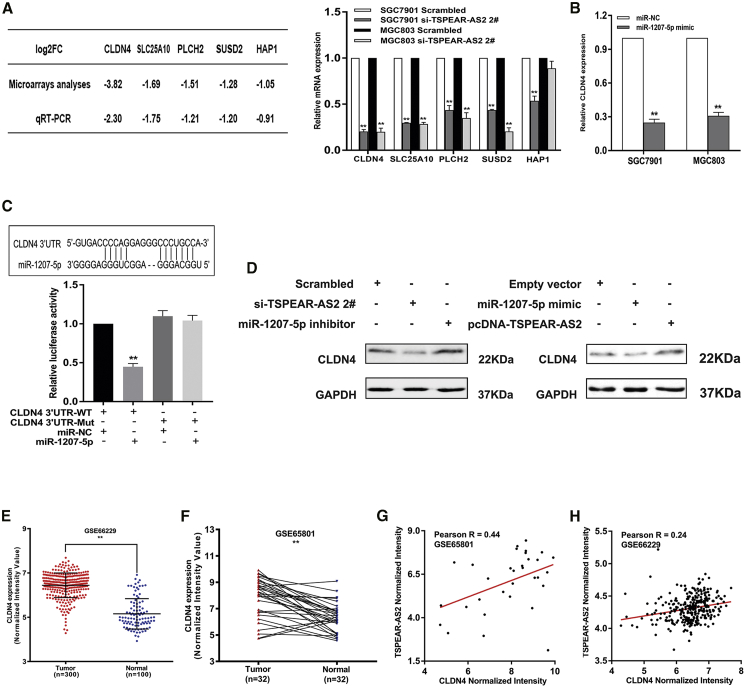
To detail the TSPEAR-AS2-involved mechanism of post-transcriptional regulation, data mining was processed in RNA-seq analysis of a control group and the TSPEAR-AS2-knockdown group. We observed that a number of key genes displayed obvious downregulation in abundance of log2FC (fold change) ≤−1 (Figure 3B). Then, a series of verification experiments were applied to test the alteration after TSPEAR-AS2 knockdown. It was found that a number of key genes were dramatically decreased in TSPEAR-AS2-depleted GC cells, including CLDN4, solute carrier family 25 member 10 (SLC25A10), phospholipase C eta 2 (PLCH2), and sushi domain containing 2 (SUSD2), in both SGC7901 and MGC803 cells. Among these potential genes, TSPEAR-AS2 knockdown displayed the strongest inhibition effects on CLDN4 level (Figure 6A). It is known that the alteration of CLDN4 is tightly linked with the malignant progression of various malignancies and therapeutic resistance.40, 41, 42 Here, we found the obvious downregulation of CLDN4 in TSPEAR-AS2-depleted GC cells and significant upregulation of CLDN4 in TSPEAR-AS2-overexpressed GC cells, identifying that CLDN4 may be a key downstream effector of TSPEAR-AS2 in GC progression (Figure 6D).
Figure 6.
lncRNA TSPEAR-AS2 Modulates CLDN4 Expression by Competing for miR-1207-5p in GC
(A) The qRT-PCR assays were conducted to validate the alteration of key genes in GC cells with TSPEAR-AS2 knockdown. (B) The effects of miR-1207-5p overexpression on CLDN4 expression in GC cells. (C) Luciferase activity in HEK293T cells cotransfected with miR-1207-5p or negative control and pmirGLO-CLDN4 3′UTR-WT or pmirGLO-CLDN4 3′UTR-Mut. (D) Western blot assay was used to detect CLDN4 protein level in GC cells upon miR-1207-5p mimic or pcDNA-TSPEAR-AS2 treatment (right), and in the absence of TSPEAR-AS2 expression or miR-1207-5p inhibitor (left). (E) The level of CLDN4 in gastric cancer tissues and normal tissues was measured in the data provided from GEO (GSE66229) datasets. (F) The level of CLDN4 in gastric cancer tissues and normal tissues was measured in the data provided from GEO (GSE65801) datasets. (G) The relationship between CLDN4 and TSPEAR-AS2 in GC tissues was determined based on the data from GSE65801 datasets. (H) The relationship between CLDN4 and TSPEAR-AS2 in GC tissues was determined based on the data from GSE66229 datasets.
Subsequently, we assumed that TSPEAR-AS2, miR-1207-5p, and CLDN4 were involved in a competing endogenous RNA (ceRNA) regulatory network. To verify our hypothesis, we first performed miR-1207-5p overexpression assays in GC cells and found that the elevation of miR-1207-5p resulted in the obvious decrease of CLDN4 at the mRNA level (Figure 6B). Moreover, we further revealed that the depletion or increase of miR-1207-5p can significantly upregulate or downregulate CLDN4 expression in GC cells (Figure 6D). Interestingly, the prediction analysis from miRanda database showed that miR-1207-5p can possibly bind to CLDN4 (Figure 6C). As shown in Figure 6C, a luciferase activity assay further demonstrated that miR-1207-5p elevation induced the effective suppression of luciferase activity of CLDN4-3′ UTR-wild-type (WT) but not CLDN4-3′ UTR-Mut (Figure 6C). The analysis of GSE66229 database highlighted the abundance of CLDN4 in GC tissue specimens (n = 300) compared with normal specimens (n = 100) (Figure 6E). We also detected the CLDN4 level in paired specimens from patients with GC and indicated CLDN4 upregulation in GC (Figure 6F). Interestingly, the positive correlation can be observed between TSPEAR-AS2 and CLDN4 in GC tumor samples, highlighting the tight regulatory correlation between TSPEAR-AS2 and CLDN4 in GC progression based on data from GEO datasets (GEO: GSE65801 and GSE66229) (Figures 6G and 6H). Taken together, our findings elucidated that lncRNA TSPEAR-AS2 worked as a ceRNA for miR-1207-5p, consequently leading to a boosted level of CLDN4 in GC progression.
Discussion
Mounting evidence has reported that lncRNAs are effective biological regulators rather than “transcriptional noise.”11,43, 44, 45 Despite that lncRNAs have been considered to play important roles during cancer progression, a variety of mechanisms need to be developed and clarified in various types of cancers, especially GC. To detect lncRNAs potentially involved in GC progression, we first explored the publicly available profiling data of GC from TCGA and GEO datasets. A novel lncRNA, TSPEAR-AS2, was screened out as a candidate gene associated with GC progression. The ectopic expression of TSPEAR-AS2 exhibited close correlation with the survival condition of patients with GC. Through gain- and loss-of function assays, TSPEAR-AS2 could induce GC cell apoptosis and promote GC proliferation, migration, and invasion. Together, TSPEAR-AS2 may exhibit an oncogenic role in GC progression. To the best of our knowledge, this is the first study to systematically evaluate the role of TSPEAR-AS2 in GC initiation and development.
Current studies indicate that many transcription factors have been revealed to be highly expressed in various malignancies, contributing to the activities of transcriptional activation of lncRNAs.46,47 In this study, a high level of transcription factor BTEB2 was observed in GC specimens and potentially correlated with TSPEAR-AS2 abundance, contributing to TSPEAR-AS2 overexpression in GC. Both FISH and subcellular fractionation assays demonstrated that TSPEAR-AS2 was more prevalent in the nucleus of GC cells, suggesting that TSPEAR-AS2 may mediate GC progression at the transcriptional level. RNA-seq found that inhibition of TSPEAR-AS2 affected key regulators correlated with cancer, such as GJA1, HDAC9, ROS1, ANKRD1, and XAF1. The expression level of GJA1 exhibited significant upregulation in GC cells with knockdown of EZH2, which is a critical member of PRC2. Interestingly, GJA1 can be coregulated by TSPEAR-AS2/EZH2. Mechanistic assays showed that TSPEAR-AS2 may participate in the tumorigenesis of GC via transcription repression of key regulators through interaction with EZH2. Additionally, TSPEAR-AS2-induced GC proliferation, migration, and invasion can be significantly reversed by overexpression of GJA1 in GC.
Recently, a novel regulatory mechanism has been illuminated to exist between lncRNAs and miRNAs in many malignant diseases.48, 49, 50 Both lncRNAs and miRNAs exert dynamic function in transcriptional and translational regulation.51,52 lncRNAs can serve as ceRNAs to protect mRNAs through competing for their targeting miRNAs.49, 50, 51 In the present study, bioinformatics databases (miRanda, pita, and RNAhybrid) were analyzed to predict miRNAs, which may potentially bind with TSPEAR-AS2. Among these miRNAs, miR-1207-5p showed the strongest repressive abilities of TSPEAR-AS2-mediated luciferase activity. Furthermore, RNA-seq assays and verified assays confirmed that CLDN4 is among the most downregulated gene in GC cells with TSPEAR-AS2 knockdown. Subsequently, a series of assays were designed to determine the novel ceRNA network formed by TSPEAR-AS2, miR-1207-5p, and CLDN4. TSPEAR-AS2 knockdown or overexpression could obviously boost or impair the level of miR-1207-5p, while miR-1207-5p elevation had no impact on TSPEAR-AS2 regulation. RIP assays revealed that both TSPEAR-AS2 and miR-1207-5p were involved in the same RISC. Dual-luciferase-reporter assays clarified the direct binding ability of the predicted miR-1207-5p binding site on TSPEAR-AS2 and further demonstrated that miR-1207-5p could directly target CLDN4 in GC. These data strongly indicated that TSPEAR-AS2 could serve as a ceRNA for miR-1207-5p to regulate CLDN4 expression at the post-transcriptional level in GC. Rescue assays further revealed that CLDN4 knockdown can, at least in part, reverse the promotion of GC progression caused by overexpressing TSPEAR-AS2.
In summary, this is the first report documenting the clinical value, biological role, and mechanism of TSPEAR-AS2 in GC. The BTEB2-activated TSPEAR-AS2 model was first elucidated in this study, leading to TSPEAR-AS2 transcription promotion in GC. Meanwhile, TSPEAR-AS2 could serve as a ceRNA for miRNAs or interact with PRC2 in GC. Our data highlight the key involvements of TSPEAR-AS2 in GC progression, implicating the axis of TSPEAR-AS2/EZH2/GJA1 and TSPEAR-AS2/miR-1207-5p/CLDN4 as novel targets for GC therapeutics. Importantly, the investigation of the expression and mechanistic model of TSPEAR-AS2 in other GC cells is urgently needed in future research. More explorations are also required to detect other upstream effectors or downstream effectors of TSPEAR-AS2 in GC progression.
Materials and Methods
Cell Culture
Human gastric adenocarcinoma cancer cell lines and normal gastric epithelium cell line (GES-1) were maintained as previously reported.53
RNA Immunoprecipitation
We used EZMagna RIP Kit (Millipore, Billerica, MA, USA) to perform RIP assays in GC cell lines. The detailed information is summarized in the Supplemental Information. The details for antibodies, primers, and siRNA oligonucleotides are listed in Table S1.
Luciferase Assays
Luciferase assays were performed as previously described.53
Chromatin Immunoprecipitation Assays
EZ-ChIP Kit (Millipore, Billerica, MA, USA) was used to conduct ChIP assays in accordance with the manufacturer’s instructions. The details of ChIP procedures can be found in the Supplemental Information.
Statistical Analysis
SPSS version 25.0 software was used to assess statistical differences. The significance between groups was assessed using a paired, two-tailed Student’s t test, Wilcoxon test, or χ2 test. The curves of OS were obtained using the Kaplan-Meier method. p <0.05 was indicative of significant difference.
Acknowledgments
This study was funded by the Innovation Fund for Outstanding Doctoral Candidates of Peking University Health Science Center (grant number: 71013Y2029); the Cancer Hospital Foundation of Jiangsu (grant number: ZM201907); the National Natural Science Foundation of China (81772603); the Science Foundation of Peking University Cancer Hospital (2017-23, A001538); and the Peking University Clinical Scientists Program (BMU2019LCKXJ011), supported by the Fundamental Research Funds for the Central Universities.
Author Contributions
Z.H.M., Y.S., K.M.W., X.Z.W., and J.F.J. designed the whole study. Z.H.M. drafted the manuscript and prepared the figures. Z.H.M., Y.S., X.Y.G., and Y.Y. performed in vitro assays and data analysis. Y.S. performed in vivo assays and bioinformatics analysis. K.M.W., X.Z.W., and J.F.J. provided full guidance. All authors have read and approved the final version.
Declaration of Interest
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.022.
Contributor Information
Ke-Ming Wang, Email: kemingwang@njmu.edu.cn.
Xian-Zi Wen, Email: wenxz@bjmu.edu.cn.
Jia-Fu Ji, Email: jijiafu@hsc.pku.edu.cn.
Supplemental Information
References
- 1.Arnold M., Park J.Y., Camargo M.C., Lunet N., Forman D., Soerjomataram I. Is gastric cancer becoming a rare disease? A global assessment of predicted incidence trends to 2035. Gut. 2020;69:823–829. doi: 10.1136/gutjnl-2019-320234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Uno Y. Prevention of gastric cancer by Helicobacter pylori eradication: A review from Japan. Cancer Med. 2019;8:3992–4000. doi: 10.1002/cam4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun W., Yan L. Gastric cancer: current and evolving treatment landscape. Chin. J. Cancer. 2016;35:83. doi: 10.1186/s40880-016-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zong L., Abe M., Seto Y., Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388:2606. doi: 10.1016/S0140-6736(16)32226-7. [DOI] [PubMed] [Google Scholar]
- 6.Ransohoff J.D., Wei Y., Khavari P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uszczynska-Ratajczak B., Lagarde J., Frankish A., Guigó R., Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018;19:535–548. doi: 10.1038/s41576-018-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan C., Cao J., Chen L., Xi X., Wang S., Zhu Y., Yang L., Ma L., Wang D., Yin J. Noncoding RNAs Serve as Diagnosis and Prognosis Biomarkers for Hepatocellular Carcinoma. Clin. Chem. 2019;65:905–915. doi: 10.1373/clinchem.2018.301150. [DOI] [PubMed] [Google Scholar]
- 10.Hoefel G., Tay H., Foster P. MicroRNAs in Lung Diseases. Chest. 2019;156:991–1000. doi: 10.1016/j.chest.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Pichler M., Rodriguez-Aguayo C., Nam S.Y., Dragomir M.P., Bayraktar R., Anfossi S., Knutsen E., Ivan C., Fuentes-Mattei E., Lee S.K. Therapeutic potential of FLANC, a novel primate-specific long non-coding RNA in colorectal cancer. Gut. 2020;69:1818–1831. doi: 10.1136/gutjnl-2019-318903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Zuo H., Jin J., Lv W., Xu Z., Fan Y., Zhang J., Zuo B. Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis. 2019;10:505. doi: 10.1038/s41419-019-1742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klingenberg M., Groß M., Goyal A., Polycarpou-Schwarz M., Miersch T., Ernst A.S., Leupold J., Patil N., Warnken U., Allgayer H. The Long Noncoding RNA Cancer Susceptibility 9 and RNA Binding Protein Heterogeneous Nuclear Ribonucleoprotein L Form a Complex and Coregulate Genes Linked to AKT Signaling. Hepatology. 2018;68:1817–1832. doi: 10.1002/hep.30102. [DOI] [PubMed] [Google Scholar]
- 14.Qu L., Wang Z.L., Chen Q., Li Y.M., He H.W., Hsieh J.J., Xue S., Wu Z.J., Liu B., Tang H. Prognostic Value of a Long Non-coding RNA Signature in Localized Clear Cell Renal Cell Carcinoma. Eur. Urol. 2018;74:756–763. doi: 10.1016/j.eururo.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 15.Chen S., Xu H., Hu F., Wang T. Identification of Key Players Involved in CoCl2 Hypoxia Induced Pulmonary Artery Hypertension in vitro. Front. Genet. 2020;11:232. doi: 10.3389/fgene.2020.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang Y.J., Yang D.C., Kong L., Hou M., Meng Y.Q., Wei L., Gao G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi F., Liu X., Wu H., Yu X., Wei C., Huang X., Ji G., Nie F., Wang K. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J. Hematol. Oncol. 2017;10:48. doi: 10.1186/s13045-017-0420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak M.K., Lee H.J., Hur K., Park D.J., Lee H.S., Kim W.H., Lee K.U., Choe K.J., Guilford P., Yang H.K. Expression of Krüppel-like factor 5 in human gastric carcinomas. J. Cancer Res. Clin. Oncol. 2008;134:163–167. doi: 10.1007/s00432-007-0265-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L., Wu Y., Wu J., Zhou M., Li D., Wan X., Jin F., Wang Y., Lin W., Zha X., Liu Y. KLF5-mediated COX2 upregulation contributes to tumorigenesis driven by PTEN deficiency. Cell. Signal. 2020;75:109767. doi: 10.1016/j.cellsig.2020.109767. [DOI] [PubMed] [Google Scholar]
- 22.Xu T.P., Ma P., Wang W.Y., Shuai Y., Wang Y.F., Yu T., Xia R., Shu Y.Q. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell Death Differ. 2019;26:2179–2193. doi: 10.1038/s41418-018-0236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busby M., Hallett M.T., Plante I. The Complex Subtype-Dependent Role of Connexin 43 (GJA1) in Breast Cancer. Int. J. Mol. Sci. 2018;19:693. doi: 10.3390/ijms19030693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonacquisti E.E., Nguyen J. Connexin 43 (Cx43) in cancer: Implications for therapeutic approaches via gap junctions. Cancer Lett. 2019;442:439–444. doi: 10.1016/j.canlet.2018.10.043. [DOI] [PubMed] [Google Scholar]
- 25.Zhang E., Han L., Yin D., He X., Hong L., Si X., Qiu M., Xu T., De W., Xu L. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086–3101. doi: 10.1093/nar/gkw1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z., Yang B., Zhang M., Guo W., Wu Z., Wang Y., Jia L., Li S., Xie W., Yang D., Cancer Genome Atlas Research Network lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell. 2018;33:706–720.e9. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat. Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emran A.A., Chatterjee A., Rodger E.J., Tiffen J.C., Gallagher S.J., Eccles M.R., Hersey P. Targeting DNA Methylation and EZH2 Activity to Overcome Melanoma Resistance to Immunotherapy. Trends Immunol. 2019;40:328–344. doi: 10.1016/j.it.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Ler L.D., Ghosh S., Chai X., Thike A.A., Heng H.L., Siew E.Y., Dey S., Koh L.K., Lim J.Q., Lim W.K. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci. Transl. Med. 2017;9:eaai8312. doi: 10.1126/scitranslmed.aai8312. [DOI] [PubMed] [Google Scholar]
- 31.Wan L., Xu K., Wei Y., Zhang J., Han T., Fry C., Zhang Z., Wang Y.V., Huang L., Yuan M. Phosphorylation of EZH2 by AMPK Suppresses PRC2 Methyltransferase Activity and Oncogenic Function. Mol. Cell. 2018;69:279–291.e5. doi: 10.1016/j.molcel.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan L., Xu M., Hua R., Tan C., Zhang J., Gong Y., Wu Z., Weng W., Sheng W., Guo W. The polycomb group protein EZH2 induces epithelial-mesenchymal transition and pluripotent phenotype of gastric cancer cells by binding to PTEN promoter. J. Hematol. Oncol. 2018;11:9. doi: 10.1186/s13045-017-0547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J., Meng Q., Li X., Yang H., Xu J., Gao N., Sun H., Wu S., Familiari G., Relucenti M. Long Noncoding RNA MIR17HG Promotes Colorectal Cancer Progression via miR-17-5p. Cancer Res. 2019;79:4882–4895. doi: 10.1158/0008-5472.CAN-18-3880. [DOI] [PubMed] [Google Scholar]
- 34.Li N., Yang G., Luo L., Ling L., Wang X., Shi L., Lan J., Jia X., Zhang Q., Long Z. LncRNA THAP9-AS1 promotes pancreatic ductal adenocarcinoma growth and leads to a poor clinical outcome via sponging miR-484 and interacting with YAP. Clin. Cancer Res. 2020;26:1736–1748. doi: 10.1158/1078-0432.CCR-19-0674. [DOI] [PubMed] [Google Scholar]
- 35.Xia B., Lin M., Dong W., Chen H., Li B., Zhang X., Hou Y., Lou G. Upregulation of miR-874-3p and miR-874-5p inhibits epithelial ovarian cancer malignancy via SIK2. J. Biochem. Mol. Toxicol. 2018;32:e22168. doi: 10.1002/jbt.22168. [DOI] [PubMed] [Google Scholar]
- 36.Wang X., Li L., Xiao W., Xu Q. Plasma microRNA-1207-5p as a Potential Biomarker for Diagnosis and Prognosis of Colorectal Cancer. Clin. Lab. 2020;66 doi: 10.7754/Clin.Lab.2020.191269. [DOI] [PubMed] [Google Scholar]
- 37.Koo K.H., Kwon H. MicroRNA miR-4779 suppresses tumor growth by inducing apoptosis and cell cycle arrest through direct targeting of PAK2 and CCND3. Cell Death Dis. 2018;9:77. doi: 10.1038/s41419-017-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L., Lü M.H., Zhang D., Hao N.B., Fan Y.H., Wu Y.Y., Wang S.M., Xie R., Fang D.C., Zhang H. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014;5:e1034. doi: 10.1038/cddis.2013.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You L., Wang H., Yang G., Zhao F., Zhang J., Liu Z., Zhang T., Liang Z., Liu C., Zhao Y. Gemcitabine exhibits a suppressive effect on pancreatic cancer cell growth by regulating processing of PVT1 to miR1207. Mol. Oncol. 2018;12:2147–2164. doi: 10.1002/1878-0261.12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y., Kishi S., Sasaki T., Ohmori H., Fujiwara-Tani R., Mori S., Goto K., Nishiguchi Y., Mori T., Kawahara I. Targeting claudin-4 enhances chemosensitivity in breast cancer. Cancer Sci. 2020;111:1840–1850. doi: 10.1111/cas.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin C.H., Li H.Y., Liu Y.P., Kuo P.F., Wang W.C., Lin F.C., Chang W.L., Sheu B.S., Wang Y.C., Hung W.C. High-CLDN4 ESCC cells harbor stem-like properties and indicate for poor concurrent chemoradiation therapy response in esophageal squamous cell carcinoma. Ther. Adv. Med. Oncol. 2019;11 doi: 10.1177/1758835919875324. 1758835919875324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki T., Fujiwara-Tani R., Kishi S., Mori S., Luo Y., Ohmori H., Kawahara I., Goto K., Nishiguchi Y., Mori T. Targeting claudin-4 enhances chemosensitivity of pancreatic ductal carcinomas. Cancer Med. 2019;8:6700–6708. doi: 10.1002/cam4.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kopp F., Mendell J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu H., Guo J., Shen Y., Dong W., Gao H., Miao Y., Li C., Zhang Y. Functions and Mechanisms of Tumor Necrosis Factor-α and Noncoding RNAs in Bone-Invasive Pituitary Adenomas. Clin. Cancer Res. 2018;24:5757–5766. doi: 10.1158/1078-0432.CCR-18-0472. [DOI] [PubMed] [Google Scholar]
- 45.Dragomir M.P., Kopetz S., Ajani J.A., Calin G.A. Non-coding RNAs in GI cancers: from cancer hallmarks to clinical utility. Gut. 2020;69:748–763. doi: 10.1136/gutjnl-2019-318279. [DOI] [PubMed] [Google Scholar]
- 46.Liu H.T., Liu S., Liu L., Ma R.R., Gao P. EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer. Cancer Res. 2018;78:5877–5890. doi: 10.1158/0008-5472.CAN-18-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C., Tan C., Wen Y., Zhang D., Li G., Chang L., Su J., Wang X. FOXP1-induced lncRNA CLRN1-AS1 acts as a tumor suppressor in pituitary prolactinoma by repressing the autophagy via inactivating Wnt/β-catenin signaling pathway. Cell Death Dis. 2019;10:499. doi: 10.1038/s41419-019-1694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang M., Mao C., Ouyang L., Liu Y., Lai W., Liu N., Shi Y., Chen L., Xiao D., Yu F. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26:2329–2343. doi: 10.1038/s41418-019-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X.S., Wang F., Li H.F., Hu Y.P., Jiang L., Zhang F., Li M.L., Wang X.A., Jin Y.P., Zhang Y.J. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18:1837–1853. doi: 10.15252/embr.201744147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J., Yu Y., Li H., Hu Q., Chen X., He Y., Xue C., Ren F., Ren Z., Li J. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer. 2019;18:33. doi: 10.1186/s12943-019-0947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang P., Huang F.Z., Liu H.Z., Zhang T.Y., Yang M.S., Sun C.Z. LncRNA MEG3 functions as a ceRNA in regulating hepatic lipogenesis by competitively binding to miR-21 with LRP6. Metabolism. 2019;94:1–8. doi: 10.1016/j.metabol.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 52.Humphries B., Wang Z., Yang C. MicroRNA Regulation of Epigenetic Modifiers in Breast Cancer. Cancers (Basel) 2019;11:897. doi: 10.3390/cancers11070897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Liang Q., Zhang L., Gou H., Li Z., Chen H., Dong Y., Ji J., Yu J. C8orf76 Promotes Gastric Tumorigenicity and Metastasis by Directly Inducing lncRNA DUSP5P1 and Associates with Patient Outcomes. Clin. Cancer Res. 2019;25:3128–3140. doi: 10.1158/1078-0432.CCR-18-2804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.