Abstract
The repeated failures of amyloid-targeting therapies have challenged our narrow understanding of Alzheimer’s disease (AD) pathogenesis and inspired wide-ranging investigations into the underlying mechanisms of disease. Increasing evidence indicates that AD develops from an intricate web of biochemical and cellular processes that extend far beyond amyloid and tau accumulation. This growing recognition surrounding the diversity of AD pathophysiology underscores the need for holistic systems-based approaches to explore AD pathogenesis. Here we describe how network-based proteomics has emerged as a powerful tool and how its application to the AD brain has provided an informative framework for the complex protein pathophysiology underlying the disease. Furthermore, we outline how the AD brain network proteome can be leveraged to advance additional scientific and translational efforts, including the discovery of novel protein biomarkers of disease.
Subject terms: Biochemistry, Neuroscience
Introduction
Alzheimer’s disease (AD), the most common cause of dementia worldwide, is characterized by progressive declines in cognition and everyday function [1]. An estimated 40 million people across the globe have AD and, due to extending lifespans, this number is only expected to increase [1, 2]. Therefore, without effective disease-modifying therapies, AD poses a uniquely devastating threat to the health and welfare of the elderly.
For the past several decades, AD research and biomarker development have centered around two proteins, amyloid-beta (Aβ) and tau. A wealth of evidence suggests these proteins play a pathogenic role in disease. Aβ, which begins accumulating over a decade prior to clinical symptoms, comprises the cortical extracellular plaques characteristic of AD [3]. In addition, gene mutations that directly enhance Aβ aggregation underlie rare familial forms of the disease [4, 5]. Meanwhile, tau accumulation as intracellular neurofibrillary tangles (NFTs) is another hallmark feature of AD and strongly associated with progressive neuronal loss and cognitive decline [3]. For these reasons, a definitive AD diagnosis requires the postmortem presence of both Aβ and tau pathologies in the cerebral cortex [6]. Antemortem diagnosis also relies heavily on the presence of Aβ and tau, either as measured in the spinal fluid or imaged using positron emission tomography (PET) [7–10].
However, mounting evidence indicates that Aβ and tau represent only a fraction of the complex and heterogeneous biology of AD. Large clinical trials of Aβ-targeting therapies have repeatedly failed, suggesting that amyloid-centric therapy is insufficient to quell disease [11]. Simultaneously, a growing number of genetic, cellular, and biochemical studies have linked AD pathogenesis to a more diverse array of biological mechanisms involving a variety of cell types. Thus, some have postulated that AD maintains two pathogenic phases, including a proteopathic “biochemical” phase characterized by the direct toxic effects of Aβ and tau aggregation and a “cellular” phase encompassing a web of complex feedback and feedforward cell-mediated mechanisms that promote irreversible degeneration [12]. This pathogenic framework argues that the dynamic interaction of both phases is critical for disease progression and cognitive decline.
The growing complexity of AD pathogenesis has highlighted the need for additional biomarkers that fully reflect underlying pathophysiology and effectively promote advancements in diagnostic profiling, disease monitoring, and treatment. In response to this urgent need for biomarker discovery, the Accelerating Medicines Partnership (AMP)-AD initiative was launched in 2014. This multidisciplinary effort between the National Institutes of Health (NIH), academia, and industry aims to leverage systems-based scientific strategies to better characterize AD pathophysiology and identify effective therapeutic targets [13]. Amidst this partnership, unbiased network-based proteomics has emerged as a powerful tool for unraveling the intricate biology underlying AD. In this review, we discuss recent large-scale, multidimensional, network proteomic analyses of the AD brain and the applications of these results to additional scientific and translational efforts.
Expanding AD pathogenesis beyond amyloid and tau
The “amyloid hypothesis” proposes that AD is caused by a linear series of events initiated by Aβ and culminating in progressive neuronal loss and cognitive decline [14]. Though never universally accepted, this hypothesis remained the most prominent theoretical framework for understanding AD for over two decades. However, the repeated failures of Aβ-targeting drug trials have recently bolstered critics of this hypothesis and broadened investigations of AD pathophysiology. Increasing evidence now suggests that AD features a diverse non-linear pathogenic landscape encompassing a variety of cellular mechanisms with equally prominent roles in disease progression [12].
The amyloid hypothesis controversy
Aβ peptide is the main component of extracellular plaque pathology in the AD brain [15]. Aberrant Aβ deposition begins over a decade before the onset of clinical symptoms and spreads diffusely throughout the cerebral cortex [16, 17]. According to the “amyloid hypothesis”, progressive Aβ accumulation triggers a cascade of detrimental processes, including intracellular NFT formation, synaptic dysfunction, neuronal death, and finally irreversible dementia [18, 19]. This hypothesis gained favor in the 1990s after several gene mutations directly responsible for aberrant Aβ accumulation (APP, PSEN1, PSEN2) were found to underlie rare familial forms of early-onset AD [20–22]. These genetic findings led to several transgenic mouse models that successfully recapitulated Aβ plaque formation, increasing support for the hypothesis (Table 1) [23–29]. Though familial AD cases account for <5% of the AD population, this amyloid-centric framework was promptly extended to sporadic late-onset AD (LOAD) given its identical hallmark pathology [3]. Subsequent genetic association studies of LOAD bolstered this framework, identifying risk factors such as APOE that modulate Aβ processing, trafficking, and clearance [19, 20, 30]. This was followed by evidence indicating that soluble oligomeric Aβ mediates synaptic dysfunction and neuronal toxicity [31–33]. For these reasons, enormous research efforts were expended toward Aβ-targeting drug therapies that either inhibited production or facilitated the removal of Aβ from the brain [11].
Table 1.
Representative transgenic mouse models commonly used to study Alzheimer’s disease (AD).
| Model name | Gene(s) | Mutation(s) | Promoter | Background | Availability | Ref. |
|---|---|---|---|---|---|---|
| J20 | huAPP770 | APP-Swe, Ind | PDGFB | C57BL/6 | The Jackson Laboratory: Stock No. 34836-JAX | [29] |
| TgCRND8 | huAPP695 | APP-Swe, Ind | Ham Prnp | Hybrid C3H/He-C57BL/6 | Peter St. George-Hyslop | [193] |
| Tg-SwDI | huAPP770 | APP-Swe, Dut, Iowa | Mo Thy1 | C57BL/6 | The Jackson Laboratory: Stock No. 34843-JAX | [194] |
| APP23 | huAPP751 | APP-Swe | Mo Thy1 | C57BL/6 | The Jackson Laboratory: Stock No. 030504 | [26] |
| Tg2576 | huAPP695 | APP-Swe | Ham Prnp | C57BL/6 x SJL; C57BL6 | Taconic: Stock No. 1349 | [195] |
| APPPS1 | huAPP, huPSEN1 | APP-Swe; PS1-L166P | Mo Thy1 | C57BL/6J | Mathias Jucker | [196] |
| APPswe/PSEN1dE9 (line 85) | mo/huAPP695, huPSEN1 | APP-Swe; PS1∆E9 | Mo Prnp | C57BL/6; C3H | The Jackson Laboratory: Stock No. 034829 | [197] |
| 5xFAD | huAPP, huPSEN1 | APP-Swe, Flo, Lon; PS1-M146L, L286V | Mo Thy1 | C57BL/6 x SJL; C57BL6 | The Jackson Laboratory: Stock No. 034840; Stock No. 034848 | [198] |
| JNPL3 | huMAPT 4R0N | P301L | Mo Prnp | C57BL/6, DBA/2, SW mixed | Taconic: Stock no. 2508 | [199] |
| hTau.P301S | huMAPT 4R0N | P301S | Mo Thy1.2 | CBAxC57BL/6 | Michel Goedert; LifeArc | [200] |
| PS19 | huMAPT 4R1N | P301S | Mo Prnp | C57BL/6 x C3H | The Jackson Laboratory: Stock No. 008169 | [201] |
| 3xTg-AD | huAPP, moPsen1, huMAPT 4R0N | APP-Swe; MAPT-P301L; Psen1 M146V knock-in | Mo Thy1.2 | C57BL/6;129 | The Jackson Laboratory: Stock No. 34830-JAX | [202] |
Although this is not a thorough list of mouse models, Jankowsky and Zheng [203] and www.alzforum.com/research-models provide detailed lists of transgenic and non-transgenic mouse models. Mutations: Swe = Swedish (M670/671NL); Flo = Florida (APP I716V); Lon = London (APP V717I), Ind = Indiana (V717F); Dut = Dutch (E693Q); Iowa (D694N).
APP amyloid precursor protein, PSEN1/PS1 presenilin 1, hu human, mo mouse, Ham hamster.
Yet, the repeated failures of these drugs have brought the amyloid hypothesis under fire [19, 31, 34]. While many argue that these trial failures are due to poor study design or patient selection, critics of an amyloid-centric framework maintain that Aβ is simply not the key to disease pathogenesis. Its tendency to accumulate for years without causing symptomatic disease (i.e., asymptomatic AD or AsymAD) supports this conclusion [35, 36]. In one notable case, even harboring an autosomal dominant PSEN1 mutation was insufficient to cause overt dementia [36]. Moreover, the growing recognition of overlapping pathologies (e.g., Lewy body inclusions, TDP-43 pathology, vascular lesions, etc.) among dementia patients indicates a wide range of biological heterogeneity within AD that necessitates a broader approach to scientific investigations of its pathophysiology [37–39].
The evolving role of tau
Hyperphosphorylated tau accumulates as intraneuronal NFTs, a hallmark feature of AD pathology [3]. Though the amyloid hypothesis places NFTs downstream of Aβ deposition, increasing evidence suggests that aberrant tau accumulation is a complex multifactorial process that can occur independent of amyloid. Such observations date back to the pathological descriptions of Heiko and Eva Braak in 1991, who found that AD-related tau deposition followed a stereotypical pattern of progression regardless of variations in Aβ distribution [40]. Subsequent studies identified the locus coeruleus as the first site of pathology in AD subjects, with NFTs preceding cortical tau or Aβ deposits [41–43]. More recently, Crary et al. coined the term “primary age-related tauopathy” (PART) after observing that the postmortem brains of many elderly individuals harbor NFTs indistinguishable from those of AD but in the absence of Aβ plaques or cognitive decline [44]. Furthermore, aggregated hyperphosphorylated tau is a well-described pathological hallmark of several other non-AD disorders, such as frontotemporal lobar degeneration (FTLD) [45]. Together, these findings argue against tau accumulation as a linear consequence of Aβ deposition.
As many critically question the amyloid hypothesis, tau has begun to receive more attention as a potential therapeutic target. Compared to amyloid burden, tau levels correlate much more strongly to cognitive symptoms, suggesting a more direct link to disease progression [46]. However, the efficacy of these tau-targeting strategies has yet to be demonstrated. In many instances, success in animal models has not translated into cognitive benefits in humans. Clinical trials of methylene blue-derived tau inhibitors in AD have generally yielded disappointing results [47–49]. Select tau immunotherapies currently employed in AD trials have previously failed to demonstrate any effect on disease progression in the primary tauopathy progressive supranuclear palsy (PSP), fueling reservations about their utility in other neurodegenerative disorders [49]. Indeed, many remain skeptical that targeting tau alone will be the key to successful AD modification, especially given the increasing evidence of other cell-mediated pathogenic processes. Nevertheless, numerous tau-targeting trials are currently on-going, tackling a variety of mechanisms that contribute to tauopathy development. The next several years should reveal much more about the utility of this approach and in turn, the role of tau in AD pathogenesis.
The cellular phase of AD
In a 2016 review, De Strooper and Karran discuss the overwhelming evidence for a “cellular phase” of AD, a decades-long period of complex feedback and feedforward mechanisms between neurons, glia, and the endothelium that ultimately precipitates the irreversible damage underlying cognitive decline [12]. In this framework, AD begins with a “biochemical phase”, characterized by aberrant amyloid precursor protein (APP) processing, Aβ accumulation, and tau hyperphosphorylation. These biochemical changes exert proteopathic or “aggregate” stress on surrounding tissues, sparking a cell-mediated phase of disease. Initially, this “cellular phase” maintains homeostasis amidst the growing proteopathic disruption. However, as the diseased individual ages, these compensatory cellular mechanisms evolve into increasingly dysfunctional and detrimental processes. Defective Aβ clearance, mediated by the brain’s perivascular circulation and glymphatic system, is considered one of the first events in this biochemical to cellular transition [12]. Dysfunction of the astrocytes and specialized endothelium comprising this clearance system not only contributes to increasing amyloid accumulation but also promotes irrevocable disruption of other neurovascular and glioneuronal functions, including lipid metabolism, myelin turnover, and immune regulation. Cell-mediated feedback and feedforward mechanisms allow this dysfunction to thrive independently of accumulating amyloid and tau, precipitating chronic inflammation and circuitry imbalances that result in cell failure and death.
It is this cellular transition, argue De Strooper and Karran, that propels the onset of cognitive symptoms. Thus, the hallmark proteinopathy of AD does not directly cause symptomatology but instead serves as an upstream trigger for destructive cellular processes. There is strong evidence to support the causative role of cell-mediated dysfunction in the pathogenesis of AD. Several genes encoding proteins intimately associated with the endothelium and Aβ clearance are highly validated AD risk factors, such as APOEε4, PICALM, and CLU [50, 51]. Likewise, genome-wide association studies (GWAS) have identified several single-nucleotide polymorphisms (SNPs) in immune genes as independent risk factors for LOAD [52–55], directly implicating microglia-mediated mechanisms in sporadic AD progression. In addition, two groups of investigators independently identified variants of the microglial TREM2 gene that increase susceptibility to LOAD with an odds ratio similar to that of APOEε4 [56–58]. These genetic findings are further supported by the common observation of aberrantly activated microglia in pathologically relevant AD brain regions, including within and surrounding Aβ plaques [59, 60].
The wealth of evidence indicating the causative role of these cellular processes in AD pathogenesis strongly supports a more holistic approach to AD investigation and biomarker discovery. In the remainder of the review, we discuss how global network-based proteomic approaches to the AD brain have helped characterize these cellular changes, their roles in disease, and associated protein biomarkers. This unbiased strategy does not ignore or discount the proteopathic biochemical phase, but instead puts this protein accumulation and its direct effects into the larger context of neurovascular and glioneuronal dysfunction.
A global network proteomic approach to the AD brain
The expanding framework of AD pathophysiology has necessitated more holistic network-based “-omic” approaches to scientific investigation. The multidisciplinary AMP-AD initiative has sprung to the forefront of these systems-based efforts, leveraging over 2000 brain tissues across its participating institutions to perform large-scale integration of network genomic, epigenomic, RNAseq, and proteomic data [13]. Under this initiative, network proteomic analysis has emerged as a valuable tool for assessing pathophysiological changes in both AsymAD and later stages of disease. This approach organizes complex proteomic data into unbiased groups or “modules” of protein co-expression that reflect various molecular, cellular, and circuit-level phenotypes.
This shift toward systems-level proteomic analysis mirrors efforts in AD transcriptomics, a field that has employed similar network strategies to contextualize the many genetic variants now associated with LOAD [61]. Yet, protein-level analyses have demonstrated disease-related alterations not readily measured in transcriptomic networks. Only 30–40% of modules in the AD brain network proteome overlap with those of the network transcriptome [62, 63]. While differential protein expression within these overlapping modules is reasonably concordant (R~0.5), we have repeatedly observed targets among these modules with highly discordant changes at the protein and RNA levels [62–64]. Furthermore, only half of the disease-related variance observed in the AD network proteome is reflected in gene expression at the transcriptome level, the remainder representing post-transcriptional and post-translational effects [64]. These findings, consistent with previous comparisons of protein and mRNA data [65], strongly support the utility of protein profiling in AD and its complementarity with transcriptomic studies. In the remainder of this section, we will discuss the mass spectrometry-based pipelines and computational methods used to conduct global network proteomic analyses of human AD brain tissue.
Strategies for mass spectrometry-based quantification
Network proteomic analyses of the AD brain have relied heavily on traditional “bottom-up” mass spectrometry (MS)-based techniques for protein identification and quantification (Fig. 1). This workflow typically comprises enzymatic protein digestion (e.g., trypsin) followed by liquid chromatography (LC) separation and tandem MS (MS/MS) measurement of peptides. Data-dependent acquisition (DDA) is commonly utilized, in which a limited number of precursor peptides are stochastically selected in the first stage of MS (MS1) to be fragmented and analyzed in the second tandem stage (MS2). These quantified peptides are subsequently identified by spectral matching and analyzed using well-validated statistical and bioinformatic strategies [66–71]. This workflow has proven robust and accurate in its protein profiling of complex mixtures, explaining its popularity among MS-based approaches.
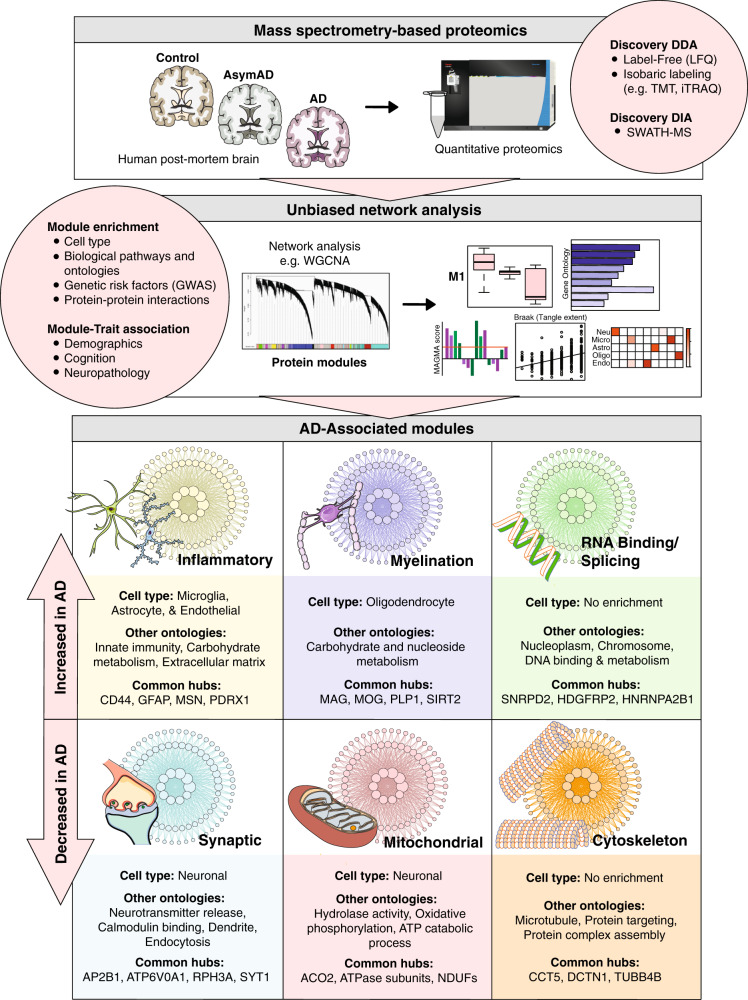
Fig. 1. Unbiased network-based proteomics to characterize the complex biochemical and cellular endophenotypes of Alzheimer’s disease (AD).
Traditional “bottom-up” mass spectrometry (MS)-based techniques, such as label-free quantitation (LFQ) or isobaric labeling, are used for protein identification and quantification in a large cohort of postmortem brain tissues from control, asymptomatic AD (AsymAD), and AD cases. Following fractionation, the deep discovery proteomic data generated from these large cohorts are organized into biologically meaningful groups, or modules, of proteins with sophisticated analytical methods, such as weighted gene correlation network analysis (WGCNA). The co-expression modules are assessed for enrichment with specific cell types, organelles, biological pathways, and genetic risk factors generated from genome-wide association studies (GWAS). In addition, abundance changes at the module level can be correlated with disease status, clinical features, and neuropathological measures. Using this framework, six core and highly conserved modules across AsymAD and AD cohorts with reproducible links to specific cell types, organelles, and biological functions have been identified. Three of the modules are consistently increased in the AD brain network proteome: inflammatory, myelination, and RNA binding/splicing, while the remaining three are consistently decreased: synaptic, mitochondrial, and cytoskeleton.
A variety of technological strategies have further enhanced this workflow, bolstering the quantification and depth of resulting proteomic datasets. For example, there has been an increasing transition from label-free to isobaric-labeling approaches. In label-free quantitation (LFQ), each sample is prepared and analyzed individually by LC-MS/MS. Though MS1 peptide selection is biased toward the most intense signals [72], the inherently stochastic nature of DDA impacts the consistency of peptides chosen and analyzed across samples. Variability in mass measurements and LC peptide retention times may also introduce inconsistencies in precursor peptide selection and quantification. This creates a well-described “missing value” problem that ultimately limits the number of quantifiable proteins in a given LFQ dataset [72–75]. Multiplex isobaric peptide labeling, as with tandem mass tags (TMTs) and isobaric tags for relative and absolute quantitation (iTRAQ), helps to mitigate the issue of missing values by enabling the analysis of multiple samples simultaneously within a single LC-MS/MS analysis, as many as 16 with TMT [76, 77]. This strategy, when coupled to off-line fractionation, can result in the quantification of thousands of additional proteins and has permitted remarkably deep proteomic analysis of AD brain tissue [74, 75, 78, 79]. Johnson et al. demonstrated this advantage in one of the first TMT-MS network proteomes of the AD brain [74], which quantified 6533 proteins across 47 brain tissues compared to just 2736 proteins quantified by LFQ-MS when applied to the same samples. However, despite these achievements in quantification, TMT-MS may still yield missing values across multiplexed batches when analyzing large numbers of samples [80].
To better address the problem of missing values, data-independent acquisition (DIA) has emerged as an alternative to the stochastic nature of DDA proteomics. Instead of selecting a limited number of MS1 peptides for analysis, DIA performs MS2 quantification on the entire MS1 spectrum. Therefore, DIA results in the identification of nearly all detectable peptides within a selected mass range, allowing for comprehensive and accurate quantification of identified proteins in the sample with minimal missing values [81, 82]. Due to complex chimeric mixtures of MS2 spectra, DIA data is more difficult to analyze compared to DDA spectra and typically requires more sophisticated computation. Yet, as DIA technology advances, such roadblocks to its widespread use in discovery-driven analyses will likely recede. For instance, sequential window acquisition of all theoretical mass spectra (SWATH-MS), a relatively user-friendly technology capable of deep proteome coverage and high quantitative accuracy [72, 83], stands to significantly advance the application of DIA to complex tissue analyses. Efforts to adapt this strategy for the quantification of bulk brain proteomes are already underway [84]. Meanwhile, targeted DIA approaches, such as selected or multiple reaction monitoring (SRM/MRM) and parallel reaction monitoring (PRM), have already become widely used in research settings for more robust quantification of pre-specified individual peptides and post-translational modifications (PTMs) [81]. The use of these targeted approaches in the validation and clinical translation of discovery-driven data is discussed later.
Principles of network building and module identification
Organizing such deep discovery proteomic datasets into modules of protein co-expression requires well-validated statistical algorithms. Widely validated in transcriptomic studies, weighted gene correlation network analysis (WGCNA) has been the statistical algorithm of choice among AD proteomic network analyses [61, 62, 85–87], though various alternatives are rapidly emerging [87, 88]. Using graph theory principles, these algorithms identify modules of proteins with highly correlated abundance levels across samples. The protein co-expression within each module may be driven by a variety of biological, physiological, and/or technical factors. Thus, it is important to comprehensively investigate individual module characteristics (Fig. 1). First, assessing connectivity within each module can identify module-specific hubs, or those proteins that are most central to module function [61]. In addition, co-expression modules can be analyzed for enrichment with markers of specific cell types, organelles, biological pathways, and genetic risk factors. Finally, abundance changes at the module level, often reported as eigenproteins or average expression levels, can be correlated with disease status, clinical features, and neuropathological measures. In this way, differential protein expression within a complex mixture can be effectively contextualized within the global biochemical and molecular pathways driving disease.
Network analyses of AD brain tissue demonstrate the most reproducibility at the module level, followed by the hub protein level, and then finally at the level of precise protein connectivity rankings and module protein membership [61, 89–94]. To obtain module-level reproducibility, 20 independent samples are typically considered adequate; however, systematic consistency at the more granular hub and module membership levels may require hundreds of samples [61, 93, 94]. In a typical proteomic pipeline, protein abundances are regressed for effects of age, sex, and postmortem interval (PMI) prior to building the network, but this decision will often depend on the goals of the analysis. Batch correction for technical variance can be performed using a variety of statistical approaches, each with its own strengths and weaknesses depending on the nature of the expression data [95, 96].
Module-level abundance profiles are often statistically correlated to various phenotypic traits of disease, such as amyloid burden, tangle deposition, and cognitive decline. These module-trait correlations indicate those protein groups with strong positive or inverse relationships to disease. Module-enrichment profiles can also offer important insights into proteomic composition. These analyses are achieved using well-validated reference databases and are designed to detect the over-representation of module proteins with known cell type, biological, or genetic risk factor associations. Cell type enrichment is typically performed by cross-referencing module proteins with marker lists derived from existing reference proteomes or transcriptomes of purified murine brain cells [62, 97, 98], although single-cell human datasets also hold promise in this regard [99]. Meanwhile, various resources exist for pathway and ontology analysis [100–102], such as GO-Elite. This flexible analytical tool allows users to incorporate a variety of reference and custom databases to examine ontological over-representation at the biological, molecular, and organellar levels [101]. Finally, genetic risk factor enrichment is performed using integrative algorithms like Multi-marker Analysis of GenoMic Annotation (MAGMA) [103], a powerful gene-set analysis tool that allows for module integration with AD GWAS data [62, 64, 74, 96]. Modules enriched with AD risk factors are considered reflective of causal, rather than reactive, disease mechanisms.
Limitations of proteomic analysis in human brain tissue
Despite a variety of technological advancements, proteomic analysis of bulk human brain tissue has its limitations. Perhaps the biggest challenge is the inter- and intra-regional variability found throughout brain tissue. Due to differences in neurodegenerative vulnerability, one cortical region rich in disease pathology may not reflect the protein pathophysiology of a relatively spared distant region. Intra-regional tissue heterogeneity in cell type densities or pathological burden may also yield poorly representative results. These challenges necessitate large-scale consensus proteomic comparisons involving multiple brain regions to draw confident conclusions regarding global findings. As discussed in the next section, collaborative efforts, such as AMP-AD, and advances in high-throughput proteomics have allowed the field to overcome many of these challenges and identify a systems-based proteomic organization conserved across brain samples from a variety of different AD cohorts and cortical regions.
While the network proteomic analysis of bulk tissue can provide an invaluable view of global cell type-specific changes in disease, this approach is limited in examination of the cellular microenvironment and its effects on protein levels. Cell populations display a wide dynamic range of protein concentrations driven by their immediate surroundings. Bulk tissue analysis averages these variations in microenvironment and their contributions to protein levels. In addition, reference data for cell type-specificity, derived from healthy brain tissues, may not fully reflect disease-related shifts in cellular protein expression. As detailed later, cell type-specific proteomics can combat many of these limitations by resolving individual communities of glial, endothelial, and neuronal cells and examining the protein expression of each cell population relative to disease state, brain region, and local microenvironment. Such studies promise to uncover the biological subtleties of the cellular phase not readily apparent in the global AD proteome.
Finally, one major limitation of discovery-driven bulk tissue proteomics is its inability to directly probe disease mechanisms. Indeed, additional molecular studies will be required to deduce the biological mechanisms underlying module changes and the precise roles of these protein communities in AD. Yet, a variety of analytical strategies commonly employed in network proteomics may offer valuable insights that effectively guide these additional studies. For example, genetic risk factor enrichment can identify those modules more likely to have causative, rather than reactive, roles in disease and support further investigations into the potential disease-related mechanisms associated with these protein communities. The identification of modules altered in AsymAD also provides a window into the earliest drivers of AD and may help guide the mechanistic evaluation of preclinical disease. Furthermore, as discussed below, the application of network approaches to protein interactomes has helped characterize key aggregation mechanisms underlying the biochemical phase of AD.
Organization of the AD brain network proteome
Nearly a dozen informative network-based analyses of the AD proteome have been performed in the human brain [62–64, 74, 78, 96, 104–108], including the dorsolateral prefrontal cortex (DLPFC), temporal cortex, and other cortical regions heavily affected in AD. Using high-throughput MS techniques, these studies have revealed highly reproducible modules across the AD proteome, several of which consistently demonstrate strong correlations to AD diagnosis, Aβ burden, and other phenotypic traits of disease. Alterations in these disease-associated modules are strongly conserved across cohorts and cortical regions, allowing for the recent construction of a large consensus AD brain network derived from hundreds of brain samples [96]. Cell type-specific perturbations appear to drive many of these disease-associated network changes with glia-enriched modules most increased in AD and neuronal modules most decreased in disease (Fig. 1). Yet, these network studies have also yielded AD-associated modules with no apparent links to specific cell types, suggesting these protein changes reflect the biochemical phase of AD. The following sub-sections discuss several of the most highly conserved modules of the AD brain network proteome and the insights they provide into disease. Of note, these consensus observations have been largely regressed for age, sex, and postmortem interval (PMI). Given the potential sex differences in AD, the AMP-AD consortium recently performed a large-scale analysis of sex influences on proteomic data and found no statistically significant contribution of sex to protein module alterations [96], though this remains an active area of study.
An early anti-inflammatory glial response
Microglia and astrocytes, glial cell populations that mediate the innate immunity and inflammatory responses of the central nervous system (CNS), have been linked in numerous studies to AD pathogenesis. Microglia are phagocytic immune cells that comprise ~10% of adult CNS cells with density varying across brain regions [109–112]. Variants of the microglial TREM2 gene have been found to increase the risk of sporadic AD by approximately 3- to 5-fold [56, 57]. Moreover, emerging evidence suggests that APOE, a potent immune modulator and strongest of genetic risk factors for LOAD [113–115], may regulate neurodegeneration in a TREM2-mediated fashion [116, 117]. Meanwhile, astrocytes are known components of many processes implicated in AD, including synaptogenesis, lipoprotein metabolism, and blood–brain barrier (BBB) regulation [12]. In the AD brain, astrocyte populations undergo a series of pathophysiological changes characterized by hypertrophy, proliferation, and increased expression of intermediate filaments. This “reactive” astrogliosis can be found throughout AD progression, including in early prodromal phases of disease.
Whether this heightened inflammation serves a primarily protective or detrimental purpose in AD remains a central question of disease pathogenesis. TREM2, APOE, and other microglial-expressed genes are thought to have protective, homeostatic roles in disease [118]. Yet, single-cell RNA sequencing, which enables the profiling of individual microglial cells with high-throughput datasets, has revealed significant functional heterogeneity among microglial subtypes and their inflammatory profiles in the neurodegenerative setting [109, 119]. Similarly, data examining the effects of microglia and inflammatory mediators on Aβ deposition in APP transgenic mice are mixed, with some studies demonstrating reduced and others exacerbated Aβ accumulation [120–122]. Indeed, it is becoming increasingly clear that glial-mediated inflammation is likely a complex process featuring a variety of disease-associated anti-inflammatory and pro-inflammatory components.
Network proteomics has provided insight into the complexity surrounding AD inflammation, supporting a predominantly protective, anti-inflammatory glial response in the earliest stages of disease. Multiple network proteomes have resolved a large module enriched with microglial and astrocytic proteins that demonstrates progressively increasing abundance levels throughout AsymAD and AD. This highly conserved module features hub proteins implicated in neuroprotection, such as the membrane receptor-cytoskeleton cross-linking protein moesin (MSN) [62–64, 96]. Its activation in association with proteins ezrin and radixin has been implicated in the neuroprotective non-amyloidogenic processing of APP [123, 124]. Peroxiredoxin 1 (PRDX1), another hub of this module, is a family member of antioxidant enzymes also implicated in protective cellular responses [125]. These module associations with neuroprotection are not only limited to hub proteins. In a recent network analysis comprising nearly 500 control, AsymAD, and AD brain tissues (DLPFC, precuneus, temporal cortex), Johnson et al. demonstrated enrichment of anti-inflammatory, neuroprotective markers throughout the MSN module [96]. In a comparison with prior proteomic studies, the authors also found several of these module proteins are decreased in rapidly progressive AD compared to sporadic disease, suggesting that a failure to activate this module may lead to more aggressive cognitive decline [96, 126]. Accordingly, this study and others have noted that the MSN module is enriched with AD genetic risk factors, suggesting it maintains a causal role in disease pathogenesis [62, 64, 74, 96]. The precise mechanisms employed by the MSN module are unclear, though many of its microglial proteins have known roles in phagocytosis, and module ontology analyses have demonstrated strong associations with glucose metabolism [96].
In addition to this early neuroprotective module, the AD network proteome features a second inflammatory module co-enriched with microglia, astrocytes, and endothelial cell type markers that is consistently elevated later in disease. This module contains extracellular matrix (ECM) proteins responsible for mediating cell–cell interactions, with its most preserved hubs including scaffolding (CAV1) and collagen proteins (COL6A1, COL6A3). The co-expression of glial and endothelial proteins supports the role of vascular-mediated inflammation in AD pathogenesis. The growing depth and complexity of proteomic networks may result in modules more specific to the endothelium and better define this vascular biology. We recently performed an unbiased network analysis on a remarkably deep AD brain proteome (DLPFC) of >8000 quantified proteins generated by TMT-MS coupled with off-line fractionation [63, 75]. Among the resulting 44 modules, we resolved an ECM-associated module with highly significant endothelial enrichment and relatively weak glial co-expression. This module was strongly associated with wound healing and other aspects of humoral immunity and featured modest upregulation in AD dementia. While these findings further imply a vascular contribution to AD pathogenesis, such disease-related alterations in vascular proteins could also reflect the impact of atherosclerosis or other cerebrovascular risk factors on the AD proteome [107].
A link between oligodendrocytes and AD risk
The AD brain network proteome also features conserved upregulation of oligodendrocyte-enriched modules associated with myelination (Fig. 1). Comprising ~75% of all glial cells in the brain, oligodendrocytes generate and maintain the lipid-rich myelin sheaths that insulate neuronal axons and facilitate signal transmission [12]. Despite their abundance, oligodendrocytes are perhaps the least studied of non-neuronal brain cells in the context of AD. Yet, a growing body of evidence suggests that myelin dysfunction is an early and essential component of AD pathogenesis. For instance, white matter lesions attributable to aberrant myelin breakdown have been widely reported in the brains of individuals with mild cognitive impairment (MCI) [127]. Furthermore, magnetic resonance imaging (MRI) data indicate that individuals with the risk allele APOEε4 have accentuated myelin breakdown in AD [128, 129]. This association with genetic risk and early disease indicates that myelin dysfunction may be an inciting event in the pathogenesis of disease.
Accordingly, proteomic network analyses of the AD brain have demonstrated modest upregulation of a highly conserved oligodendrocyte-enriched myelination module in both asymptomatic and symptomatic disease [62–64, 74, 96]. In addition, a handful of studies have found that like the MSN module, this myelin module features over-representation of AD GWAS candidates, suggesting a causal role for oligodendrocyte processes in disease [62, 96]. In an examination of APOE genotype on the brain network proteome, Johnson et al. found that the most significant effect was exerted by the APOEε2 allele on the oligodendrocyte module, suppressing its disease-related changes in those with AD [96]. Dai et al. also observed this APOEε2 effect on oligodendrocyte and other cell type markers in a separate network proteome of the AD brain [104]. These associations between AD risk and oligodendrocyte function mirror the results of a recent large-scale transcriptomic network analysis in the human AD brain (DLPFC, visual cortex, cerebellum), which demonstrated significant enrichment of AD GWAS candidates in conserved disease-associated oligodendrocyte-enriched modules, including BIN1, PICALM, and several others [130]. In a comparison of this transcriptome network to an independently constructed proteomic network from AD brain tissue, the authors found at least 50% overlap between oligodendrocyte modules of the two datasets. Additional transcriptomic network analyses have similarly found enrichment of genes associated with either AD risk or Aβ processing among oligodendrocyte-specific modules [131, 132].
The mechanisms by which oligodendrocyte dysfunction contribute to AD pathogenesis remain unclear. In an intriguing network proteomic analysis of over 400 brains (DLPFC), Wingo et al. found that cerebral atherosclerosis may be the missing link between oligodendrocytes and AD dementia [107]. In this study, cerebral atherosclerosis was pathologically assessed by visual inspection of Circle of Willis vessels and proximal branches and graded independently of gross infarcts, microinfarcts, or other white matter lesions. The oligodendrocyte-enriched myelin module was one of only two protein communities in the co-expression network associated with both cerebral atherosclerosis and AD dementia, independent of Aβ, tau tangles, infarct burden, and a variety of other pathologies. These associations also persisted after adjustment for hypertension, diabetes, and other cerebrovascular risk factors. In addition, the authors observed no similar statistical correlations between cerebral atherosclerosis and Aβ accumulation or tau burden. Overall, these findings suggest that cerebral atherosclerosis, a common occurrence among the elderly, contributes to AD through oligodendrocyte dysfunction and injury to the myelin sheath, regardless of hallmark proteinopathy, gross ischemic disease, or cerebrovascular risk factors.
A role for RNA binding in the biochemical phase
Modules enriched with RNA binding and splicing proteins are also consistently upregulated in the AD network proteome (Fig. 1). In contrast to the inflammatory and myelination modules, these RNA-binding/splicing modules are not enriched with glial or other cell type-specific markers. In a deep proteomic network analysis of control, AsymAD, and AD DLPFC tissues, Johnson et al. identified several AD-associated RNA-binding/splicing modules notably increased in asymptomatic and symptomatic AD. The disease-associated alterations in these protein modules were independent of cell type, persisting even after the dataset was regressed for cell type-specific effects [74]. The authors subsequently concluded that RNA-binding proteins may drive the proteopathic “biochemical” phase of AD.
These findings correlate well with previous analyses of the AD brain detergent-insoluble proteome, which have repeatedly demonstrated the association of U1 small nuclear ribonucleoproteins (U1 snRNPs) and other RNA-binding proteins with tau tangles [133–137]. These ribonucleoproteins, constituents of the spliceosome complex responsible for RNA processing, are mislocalized in AD to the cytoplasm of neuronal cell bodies where they aggregate with NFTs and contribute to widespread alterations in RNA processing [134]. Consistent with network proteomic findings, these aggregated U1 snRNPs have been observed in asymptomatic AD brains [135], providing further support for aberrant RNA processing in early disease. Interestingly, this aggregation of U1 snRNPs with tau appears to be unique to the AD brain and absent among other primary tauopathies, such as FTLD [134, 138].
In a recent study of U1-70K, an snRNP consistently associated with NFTs in the AD brain, Bishof et al. utilized a network-based proteomic approach to functionally characterize the co-immunoprecipitated interactome of U1-70K and identify those protein–protein interactions most dependent on U1-70K domains implicated in aggregation and AD pathogenesis [138]. This analysis successfully organized the U1-70K interactome into seven biologically meaningful modules, two of which demonstrated a strong association with pathogenic U1-70K domains. These two modules, both linked to RNA splicing ontologies, were enriched in proteins with structural and aggregation properties similar to those of U1-70K that could serve as additional AD biomarkers, such as pre-mRNA splicing factor LUC7L3. These findings underscore the utility of unbiased network-based analysis in the characterization of protein–protein interactions and their dependence on sub-molecular domains.
In contrast to cell type-enriched modules, RNA-binding/splicing modules demonstrate limited overlap with the transcriptomic network of the AD brain. This observation suggests that global protein-level analyses will maintain a uniquely valuable role in unraveling the proteopathic biochemical phase of AD. In addition to RNA-binding/splicing modules, network proteomics has revealed one other highly conserved AD-associated module with possible links to this phase of disease. Unlike the RNA-binding/splicing modules, this module is consistently downregulated throughout AD and strongly associated with the cytoskeleton (Fig. 1) [62, 63, 96]. Its hub proteins often include tubulin subunits or tubulin-associated proteins, such as dynactin subunit 1 (DCTN1). As microtubule stability is one emerging means of targeting tau pathology [49], this module may also yield promising disease-associated biochemical phase markers for therapeutic targeting.
A link between synaptic and mitochondrial dysfunction
It is well-known that synaptic loss is one of the strongest correlates of cognitive decline in AD, even more so than Aβ plaques and tau tangles [139]. While neuronal death contributes substantially to this synaptic decline, it is increasingly clear that other mechanisms may also play a role in AD synaptic dysfunction. A growing body of evidence in mouse models indicates alterations in synaptic networks, prior to the onset of gross neuronal loss in the cerebral cortex [140, 141]. This is reflected in the global AD network proteome, which consistently demonstrates the progressive downregulation of synaptic modules in asymptomatic and symptomatic AD brains, a finding that appears to be preserved across various cortical brain regions [62, 96, 105]. In addition, emerging cerebrospinal fluid (CSF) proteomic data has revealed alterations in a number of synapse-associated proteins in preclinical disease [63]. The mechanism underlying these early synaptic protein changes remains unclear, with possibilities including morphological remodeling, regional changes in distribution, and immune-mediated synaptic pruning, among others. Furthermore, it has yet to be determined whether these preclinical synapse changes represent a homeostatic, protective, or detrimental event in AD pathogenesis.
In a proteome-wide association study of nearly 150 subjects, Wingo et al. used co-expression network analysis to demonstrate the increased abundance of synaptic proteins in the postmortem brain tissues (DLPFC) of individuals with antemortem stable cognitive trajectories, regardless of the burden of Aβ plaques or tau tangles [106]. This suggests that the downregulation of synaptic proteins in AsymAD may ultimately prove detrimental to cognitive stability, promoting conversion to symptomatic disease. Interestingly, this study also found heightened levels of mitochondrial proteins among cases with preserved cognition, perhaps reflecting the close relationship between neuronal synapses and mitochondria. Synapses are markedly enriched with energy-producing mitochondria, which fuel both pre- and post-synaptic signaling processes. Therefore, mitochondria are critical for maintaining synaptic integrity and function, explaining why an over-abundance of both module types may be observed in individuals with high levels of cognitive resilience. In a more recent proteomic analysis, Yu et al. paralleled these network results, identifying associations between cognitive resilience and cortical proteins involved in synaptic, metabolic, and neurogenic functions [142].
Overall, the AD brain network proteome suggests close links between synaptic and mitochondrial modules. It is not uncommon for modules linked to synaptic ontologies to feature a variety of metabolic proteins [63]. As demonstrated in Fig. 1, ATPase subunits can be found among the preserved hubs of both synaptic and mitochondrial modules. As discussed later, this close link between synaptic and metabolic protein expression is also reflected in CSF, which features stark elevations in both types of proteins in the asymptomatic stages of disease [63]. These early CSF protein derangements suggest the aberrant exocytosis of neuronal components into the extracellular space, which could be explained by synaptic pruning. Emerging evidence in AD mouse models indicates this homeostatic, microglia-mediated synaptic phagocytosis may be one of the earliest events in AD progression and contribute to synaptic losses during asymptomatic stages of disease [143]. This mechanism would also explain, at least in part, the glial protein elevations observed in the early AD brain, providing a mechanistic framework for a variety of proteomic network changes during the cellular phase of disease.
Emerging technologies for cell type-specific proteomics
Proteomic profiling of human brain tissue has indirectly revealed potential cell type-specific mechanisms of AD. The next challenge for proteomic research is gaining the cellular and temporal resolution to further define the causative role of cell-mediated dysfunction in AD pathogenesis. In this section, we review several approaches for localizing the AD proteome to a brain region or cell type to advance our understanding of the cellular phase of disease (Fig. 2). Cell type-specific proteomics in the human brain is currently in its infancy, primarily due to the inability to isolate live, pure cell populations from frozen brain and limited access to fresh postmortem tissue. Thus, extending localized proteomic approaches to mouse models of AD pathology (Table 1) will be critical for confirming the network architecture and investigating causal molecular changes during the cellular phase of disease. Furthermore, cell type-specific proteomics in disease and aging mouse models can serve to de-convolute complex human brain data and provide cellular-level resolution to peripheral biomarkers. In addition, this work will significantly enhance our understanding of the aging-dependent course of cell type-specific perturbations, thus allowing us to pinpoint possible opportunities for therapeutic intervention and enhance biomarker development.
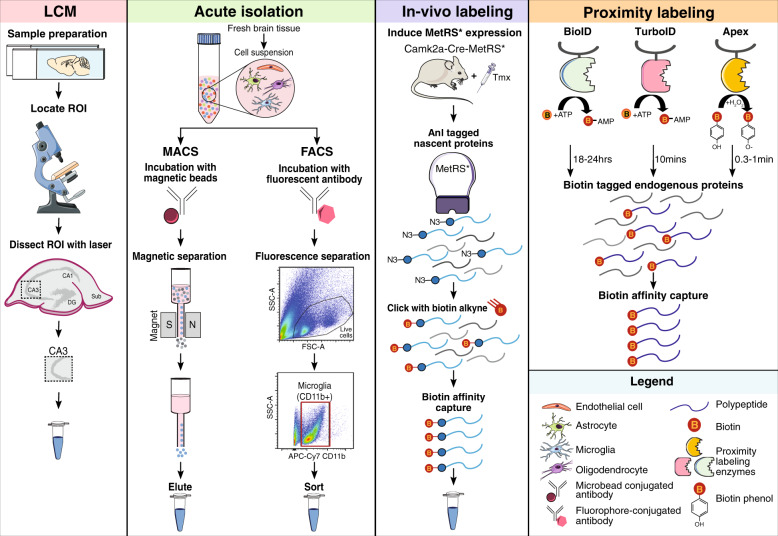
Fig. 2. Local proteomics approaches to define the cellular basis of Alzheimer’s disease.
Summary of approaches: Laser capture microdissection (LCM) is a method that can procure subpopulations of cells or very small regions of interest under direct microscopic visualization. LCM (pink panel) has been used to micro-dissect neurons from both fresh frozen human brain and formalin-fixed paraffin embedded tissue sections for proteomic analyses. Prior to dissection, immunohistochemical or histological staining of fixed tissue sections is performed to identify a specific cell type or population of cells in a region of interest without compromising protein quality. For acute isolation of brain cell types (green panel), a fresh brain sample needs to be processed to yield a single-cell suspension which is then subjected to magnetically-activated cell sorting (MACS) or fluorescent-activated cell sorting (FACS). For MACS, the desired cell type is labeled with a 50 nm magnetic microbead conjugated to an antibody specific to cell-surface receptor. After incubation, the sample is placed on a magnet to drain unbound cells and retain desired cell type within the column. Once the column is removed from the magnet, the bound cells are released and collected for downstream analyses. Analogous to MACS, in FACS, the single-cell suspension is incubated with a fluorophore-conjugated antibody specific to a cell-surface receptor. Subsequently, the desired cell type is sorted based on their size and fluorescent signal directly into a buffer amenable for downstream proteomic analyses. In vivo biorthogonal amino acid tagging (BONCAT) of proteins (purple panel) is achieved by expressing MetRS* under a cell type-specific promoter in a mouse (Camk2a-Cre-MetRS*). MetRS* harbors a mutation (L247G) in the amino acid binding site which preferentially tags nascent proteins with an azide-tagged methionine analog, azidonorleucine (Anl). After treating the mice with tamoxifen (Tmx) to facilitate Cre-mediated recombination, Camk2a cells express MetRS* and nascent proteins are tagged with Anl. The azide residue of Anl is amenable to copper-catalyzed azide-alkyne cycloaddition or “click” chemistry. Following protein extraction, Anl-tagged proteins residues are “clicked” with a PEG-biotin-alkyne and then purified using avidin beads or avidin resin for subsequent MS analyses. Proximity labeling (orange panel) is achieved by various enzymes that biotinylate proximal endogenous proteins. The BioID approach uses the Escherichia coli biotin ligase, BirA*, with a catalytic site mutation (R118G). The mutation destabilizes the enzyme, facilitating active biotin molecules (biotinoyl-5′-AMP) to dissociate and bind to primary amines of exposed lysine residues on adjacent proteins. APEX catalyzes the oxidation of biotin-phenol to the short-lived (<1 ms) biotin-phenoxyl radical in the presence of hydrogen peroxide, which then reacts with electron-rich amino acids, such as tyrosine, in neighboring proteins. TurboID was developed by taking advantage of yeast display-based directed evolution. TurboID retains the promiscuous biotinylation property of BioID, but rapidly labels proteins in 10 min compared to the 18–24 hrs by BioID. Subsequent to biotin labeling and protein extraction from a sample, proteins can be affinity captured by streptavidin beads or matrices for downstream proteomic analyses by MS.
MS-coupled laser capture microdissection
Laser capture microdissection (LCM) is a method that can procure subpopulations of cells or very small regions of interest under direct microscopic visualization (Fig. 2). LCM has been used to micro-dissect neurons from both fresh frozen human brain and formalin-fixed paraffin embedded tissue sections for proteomic analyses [144, 145]. Prior to dissection, immunohistochemical or histological staining of fixed tissue sections is performed to identify a specific cell type or population of cells in a region of interest. Protein yield for LC-MS/MS analysis is not impacted, suggesting that LCM-based approaches are very powerful and underutilized for cell type-specific proteomics of archival human brain samples. Drummond et al. [144] used LCM to comprehensively characterize the protein composition of Aβ plaques microdissected from the hippocampus of two AD subtypes: rapidly progressive AD and sporadic AD. LCM coupled to LFQ-MS of these samples revealed proteins increased in rapidly progressive AD plaques (e.g., α-synuclein) to be decreased or have no known involvement in sporadic AD [144]. This study is a powerful example of how an unbiased, localized proteomics approach can further our understanding of proteins involved in AD pathogenesis that would otherwise be diluted in a global proteomics analysis of bulk tissue. Due to recent technological advancements (e.g., optical resolution), LCM can capture areas or cells of interest with much higher precision and speed, though the time it takes to do the dissection is subjective (Table 2). In addition, LCM-based MS results in a lower proteome coverage compared to bulk brain proteomics, and the inability to confidently exclude contamination by cells that are not of interest poses a challenge (Table 2).
Table 2.
Summary of the advantages and disadvantages of local proteomics approaches.
| Approach | Advantages | Disadvantages |
|---|---|---|
| Laser capture microdissection (LCM) |
• Ability to image cell type and structure and acquire cell count • A wide range of tissues can be used: unfixed frozen postmortem brain tissue, formalin-fixed paraffin embedded brain tissue, hematoxylin & eosin stained or immunostained tissues • One tissue section can be dissected several times for different regions • Dissection does not disturb cells’ molecular state • Area size of 1.5 mm2 is sufficient for MS |
• Tissue drying during dissection • Dissection process can be time-consuming: 5 min to 8 hrs depending on the size, cell type, and number of areas or cells to be collected • Inability to confidently exclude cells that are not of interest • Protocol not optimized for smaller, non-neuronal cell types or single cells • Lower proteome coverage compared to bulk brain proteomics |
| Magnetic-activated cell sorting (MACS) |
• High throughput • High purity • Selective and rapid method • Enrichment can be scaled up or down to desired yield |
• Contamination by acellular debris or unbound cells • Immunomagnetic beads might cause mechanical shear • Sequential isolations significantly reduce yield • Cannot isolate intact neurons |
| Fluorescence-activated cell sorting (FACS) |
• High sensitivity, throughput, and purity • Isolate multiple cell types simultaneously based on immunopheno type alone • Sort complex cell types with multiple markers • Separate cells based on cell size, density, and morphology, cell cycle status, intracellular cytokine expression, and metabolic profile • Capture immunophenotyping data for 12 surface epitopes • Minimum of 12,000 cells are sufficient for MS |
• Long isolation procedure (3+ hrs) • Shear stress from the FACS instrument • Slow sorting process – depends on number of cell populations that need to be collect • Recovery is 50–70% on most sorters, need a high number of cells at the beginning • Fluorophore spillover into non-specific channels between cells with closely related immune phenotypes • Cannot isolate intact neurons |
| Bio-orthogonal non-canonical amino acid tagging (BONCAT) |
• Identification of low abundance, low copy number newly synthesized proteins with higher magnitude • Click chemistry procedure is modular and relatively simple • Lineage tracing of proteins • Established transgenic mouse line under the Cre/Lox system |
• Cost of Anl and special diet for mouse studies • Obtaining a good signal-to-noise ratio between endogenously biotinylated proteins and biotin clicked Anl-tagged proteins • Depth of proteome is lower than traditional proteomics • Reduced labeling efficiency due to competition between endogenous MetRS and MetRS* |
| Proximity labeling (BioID, APEX, TurboID) |
• Rapid kinetics of biotinylation without click chemistry • Detect weak or transient protein interactions as well as soluble and insoluble proteins • Ready bioavailability of biotin in the brain after peripheral administration • Acquire a more global proteome unlike the nascent proteins in BONCAT |
• Noise introduced by endogenous biotinylation • APEX approach is limited to in vitro experiments since biotin-phenol is toxic • TurboID can sequester endogenous biotin and cause toxicity • Saturation of proximal labeling sides with prolonged biotin supplementation • Lack of mouse models for BioID and TurboID approaches |
Acute isolation of brain cell types
Unlike transcriptomic studies that can be performed on intact nuclei from frozen brain (mouse or human), proteomic analyses of brain cell types require isolation of intact cells from the brain, for which fresh unfrozen brain is a pre-requisite. Several strategies exist to isolate the cell type(s) of interest in a highly pure form with minimal contamination by other cells or acellular elements.
Magnetic-activated cell sorting (MACS)
MACS aims to facilitate the rapid, high-throughput, immunomagnetic separation of a pure cell type population from a heterogeneous population (Fig. 2). The MACS approach has been successfully applied to enrich four brain cell types (neuronal progenitors, microglia, astrocytes, and oligodendrocytes) from adult mice for high-resolution MS-based proteomics [97]. This in-depth analysis resulted in the largest assortment of cell type-resolved proteomic data of the brain and is frequently used for cell type enrichment analyses [97]. Recently, quantitative TMT-MS was performed on acutely isolated CD11b+ MACS-enriched microglia from adult (6–7 mo) mice of normal, acute neuroinflammatory (lipopolysaccharide (LPS)-treatment), or chronic neurodegenerative (5xFAD model) states [146]. Of 4133 proteins identified, 187 microglial proteins were differentially expressed in 5xFAD mice, including proteins with known (e.g., Apoe) and novel (e.g., Cotl1) relevance to AD biology. Cotl1 was identified as a novel microglia-specific marker with increased expression and strong association with AD neuropathology. Apoe protein was also detected within Aβ plaque-associated microglia, suggesting a role for Apoe in phagocytic clearance of Aβ. Several proteins increased in human AD brain were also upregulated by 5xFAD microglia (e.g., Aβ peptide) [146]. This deep and comprehensive proteomic study of isolated mouse microglia revealed shared neuroinflammatory disease mechanisms between mouse models of AD pathology and human AD, emphasizing the value of state-of-the-art proteomics methods for resolving cell type-specific contributions to disease [146]. One key strength of the MACS is the ability to rapidly isolate cells of interest with relatively high purity without dependence on cell sorters; however, despite high cellular purity, MACS suffers from contamination by acellular debris or unbound cells (Table 2) [147].
Fluorescence-activated cell sorting (FACS)
FACS is another highly sensitive and high-throughput procedure for isolating cells from a heterogeneous population (Fig. 2). Fluorescent-conjugated antibodies against cell-surface epitopes are used to label cell type(s) of interest. The fluorescently labeled cells can be sorted directly into a buffer amenable for LC-MS/MS (Fig. 2). The FACS approach has been applied for transcriptomic studies [148–152] and proteomic applications are now gaining momentum. A recent study compared the proteomic profiles of MACS-enriched microglia and FACS-isolated microglia from adult mouse brain and found that the FACS-isolated microglia proteome was significantly enriched in canonical microglial proteins (e.g., Ctsd) and contained much lower levels of non-microglia proteins (e.g., Gfap) [153]. Also, FACS coupled to LC-MS/MS was employed to characterize neuronal and non-neuronal nuclear proteomes. Intact nuclei were purified from frozen human brain tissue and sorted based on the expression of a neuron-specific splicing factor, NeuN. Comparative analysis of NeuN-positive and NeuN-negative nuclear proteomes revealed a number of transcription and splicing factors not previously known to be expressed in a cell type-specific manner in human brain [154]. This method provides a unique opportunity to identify cell type-specific nuclear proteins, as well as histone modifications and regulation networks, that may be altered in AD. FACS is an ideal method for simultaneously sorting multiple cell types based solely on immunophenotype. Recently, a cell isolation methodology termed concurrent brain cell type acquisition (CoBrA) was used to isolate microglia, endothelial cells, astrocytes, and oligodendrocytes from mouse brain for RNAseq studies [152]. This FACS pipeline can be easily optimized for proteomic analyses of multiple cell types with >95% purity by confidently excluding debris. Moreover, utilizing FACS for high-throughput single-cell proteomics by MS (SCoPE-MS) [155], a fairly new method, holds great promise and potential for integration with single-cell RNAseq. Yet, FACS is not without limitations (Table 2). The prolonged handling of live cells for FACS and the shear stress imposed on them by the sorter can spuriously activate sensitive cells such as microglia. One inherent limitation of MACS and FACS is that live neurons cannot be sampled from adult brain, tremendously limiting our ability to understand neuron-specific proteomic changes occurring in vivo, as described below. Realistically, both approaches can be considered complementary and their relative strengths and weaknesses (Table 2) should guide rigorous study design with appropriate controls.
In vitro and in vivo protein labeling strategies
Bio-orthogonal non-canonical amino acid tagging (BONCAT)
Proteins within a specific cell type can be uniquely labeled in vivo via BONCAT (Fig. 2) and isolated from a complex tissue, such as the brain, for MS-based proteomics. BONCAT leverages the L274G mutant of mouse methionine-tRNA synthetase (MetRS*) to tag newly synthesized “nascent” proteins with an azide-tagged methionine analog, azidonorleucine (Anl) [156–158]. Anl residues are “clicked” with a PEG-biotin-alkyne and then purified using avidin beads or avidin resin for subsequent MS analyses. Recently, BONCAT was utilized to characterize excitatory and inhibitory neuronal proteomes in adult mice [158, 159]. MetRS* knock-in mice were crossed with Camk2a-Cre-ert2 mice followed by tamoxifen treatment to induce Cre-mediated recombination. Consequently, only Camk2a cells expressed MetRS* and could incorporate Anl into their proteome. Brain homogenate from these mice underwent click chemistry, affinity purification, and MS to obtain the proteomic profile of excitatory hippocampal neurons. The Camk2a hippocampal proteome was significantly enriched with proteins represented by neuronal components, such as synaptic transmission and synaptic plasticity. Several key proteins (e.g., APP, Grm5) linked to neurodevelopmental or neurodegenerative disorders were also identified [158]. Interesting data from this study showed that when mice were exposed to an environment with enriched sensory cues, there was a change in the neuronal proteome, which represented a response or adaptation to the external stimuli. The fidelity of in vivo BONCAT has thus far only been shown in mouse excitatory neurons; however, it represents a highly promising new strategy to derive nascent proteomes from heterogeneous brain tissue in a cell type-specific manner. Depth of proteome coverage, labeling efficiency, and obtaining a good signal-to-noise ratio between endogenously biotinylated proteins and biotin clicked Anl-tagged proteins are important factors to take into consideration when using this approach (Table 2).
Proximity labeling
Proximity labeling with enzymes is another strategy to achieve cell type-specific global proteomic labeling [160]. The enzymes catalyze the formation of reactive biotin species, which diffuse out of the active site to biotinylate proximal endogenous proteins (Fig. 2). Subsequently, biotinylated proteins can be enriched through affinity capture and characterized via MS. There are three proximity labeling technologies: BioID, APEX, and TurboID. The BioID approach uses the Escherichia coli biotin ligase, BirA*, to biotinylate proteins within ~10 angstroms [161]. BioID has been used extensively in vitro [162–166] to define interacting protein partners and proteins in subcellular compartments, but its application in vivo is limited. By fusing BirA* to a bait protein with a synaptic localization signal or nuclear pore signal, BirA* expression was specifically guided to the synapse or nuclear pore complex to achieve biotinylation of the respective proteomes [167, 168]. More recently, BioID was used to effectively label proteins within flies (Drosophila melanogaster) or worms (Caenorhabditis elegans) [169]. Similar to the BioID approach, an engineered ascorbate peroxidase, or APEX, could also be used for efficient proximity labeling of proteins [160] (Fig. 2). With APEX, researchers have been able to profile the mitochondrial matrix proteome and characterize the structure of a mitochondrial uniporter [170, 171]. A major advantage of APEX over BioID, is the rapid kinetics and high efficiency of labeling; however, APEX is not suitable for in vivo studies due to the toxicity of biotin-phenol. A rapid and non-toxic labeling technology, TurboID, was developed by taking advantage of yeast display-based directed evolution. TurboID retains the promiscuous biotinylation property of BioID, but rapidly labels proteins in 10 minutes compared to the 18–24 hours required by BioID (Fig. 2). TurboID-based proteome labeling has been successfully applied in flies and worms without much toxicity and with high labeling efficiency [169]. However, the levels of endogenous biotinylation in the rodent brain is a disadvantage of in vivo proximity-dependent biotinylation strategies (Table 2). This can be partly overcome by omitting regions with known elevated levels of endogenous biotinylation or subtracting out known endogenously biotinylated proteins from proteomic analyses. One significant advantage of these approaches (Table 2) is the ability to capture weak or transient protein interactions as well as soluble and insoluble proteins because of the high affinity of biotin for avidin, a challenge often faced with the use of classic antibody-based affinity purification/MS methods.
Translating the brain network proteome into AD biomarkers
We have outlined the proteomic evidence showcasing AD as a vastly complex, multi-system disorder that extends beyond “hallmark” pathology to include a variety of cellular mechanisms with potentially central roles in disease progression. Yet, the intricate pathology observed among meticulously categorized autopsy research cohorts likely reflects only a fraction of the vast biological complexity found in clinical AD populations. It is becoming increasingly clear that most demented elderly individuals harbor more than one “hallmark” dementia pathology [37]. Indeed, up to 90% of individuals with the classic amyloidosis of AD may feature concurrent vascular disease, TDP-43, or other degenerative pathologies [39]. These high rates of overlapping pathologies combined with additional biological heterogeneity introduced by individual genetic and environmental factors make for an overwhelmingly complex pathophysiological landscape among the over 40 million individuals thought to be living with AD worldwide.
Our current diagnostic framework for clinical AD fails in capturing this biological diversity. The limited number of clinically established AD biomarkers, such as amyloid PET imaging and CSF Aβ and tau levels, reflect only “hallmark” pathology. Such a reductionist approach to AD stands to hinder advancements in diagnostic subtyping, disease monitoring, and therapeutic development. Approaching such a biologically heterogenous disease as a single entity in clinical trials may account for many of the AD drug failures we have encountered to date. Thus, AD requires a new diagnostic framework, one reliant on more diverse biomarker assays reflective of a wide range of pathophysiology. A tool of this nature could not only allow for detailed biological subtyping, but also usher in a new age of patient-tailored therapeutics in neurodegeneration. For these reasons, systems-based approaches to biomarker discovery, as championed by the AMP-AD initiative, are growing in popularity among neurodegenerative disorders. In the following section, we describe early efforts to translate the network AD brain proteome into systems-based, physiologically diverse, multiplex biomarker assays capable of advancing our clinical framework of disease.
Integration of brain and biofluid proteomes
To increase the pathophysiological diversity among CSF biomarkers of AD, we have developed a novel approach that integrates the global AD brain network with proteomic analysis of CSF (Fig. 3). The direct proximity of CSF to the brain presents a strong rationale for discovery-driven integration of these two proteomes. Johnson et al. recently found that ~20 proteins from the highly conserved MSN module demonstrated significant elevations in AD CSF [96]. These CSF protein elevations were consistent across two different AD cohorts comprising a total of nearly 400 spinal fluid samples. As discussed previously, the anti-inflammatory microglia-enriched MSN module is elevated in the AsymAD brain and thought to play a causative role in disease pathogenesis. The reflection of such disease-associated cellular processes in the CSF could provide a means of detecting and monitoring treatment responses.
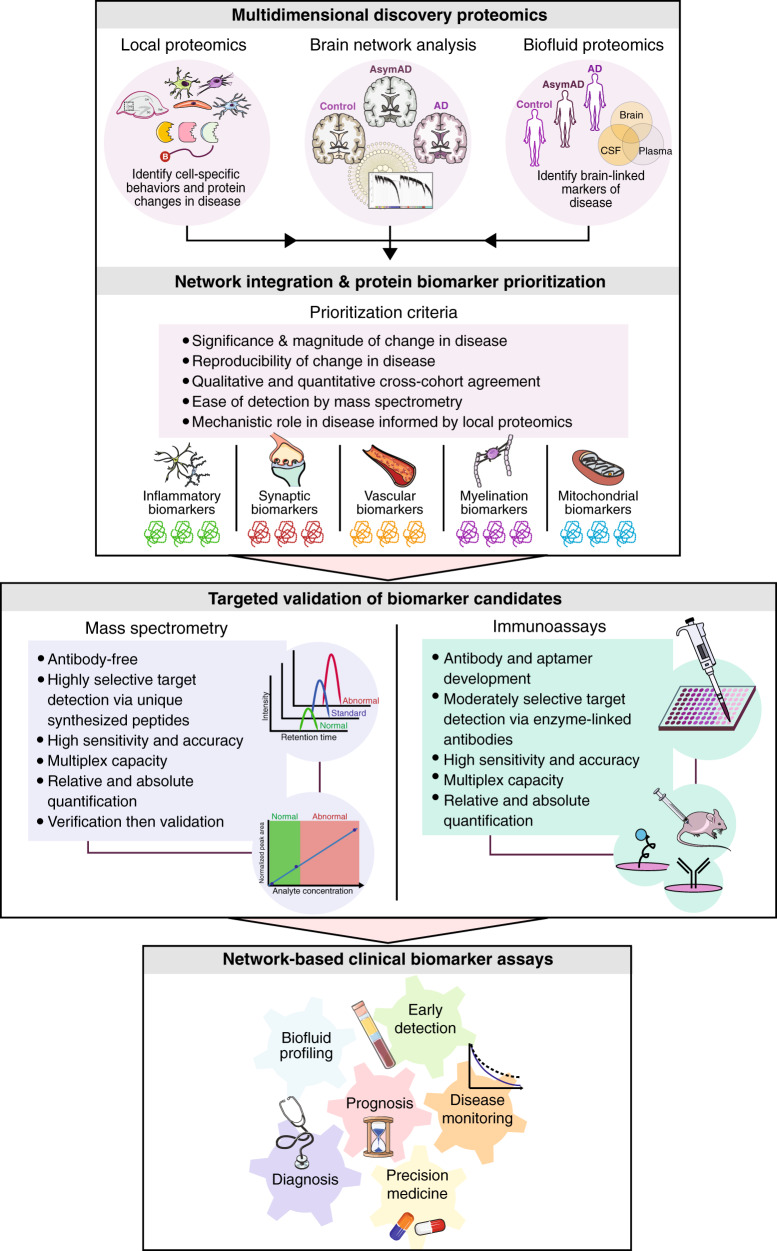
Fig. 3. Conceptual framework for translating brain protein networks into clinical biomarkers.
In this framework, multidimensional discovery-driven proteomics data collected from local cell type-specific approaches and antemortem biofluids will be integrated with the AD brain network proteome to identify systems-based panels of promising CSF and/or plasma biomarkers. Target prioritization will rest on the significance, magnitude, reproducibility, and ease of detection of the candidate biomarker in disease. Prioritized targets will also require links to disease mechanisms, informed in part by localized and cell type-specific proteomics. These network-based biomarker panels will then be validated using targeted quantitation approaches, including mass spectrometry (MS) and immunoassays. Both validation methods offer highly sensitive and accurate quantitation, though MS may offer certain advantages, such as highly selective target detection using unique peptides and the ability to cost-effectively analyze large panels of proteins in an initial verification phase prior to more expensive validation efforts. Validated biomarker panels representing a wide range of pathophysiologies could serve a variety of clinical uses, including preclinical profiling, disease monitoring, measuring therapeutic response, and confirming target engagement.
In another recent large-scale study, Higginbotham et al. used an integrative proteomic approach to examine the statistical overlap of the AD brain network proteome (DLPFC) with differential expression in the AD CSF proteome [63]. Fifteen of the 44 brain modules identified in this study strongly overlapped with the CSF proteome. These 15 brain modules were also high-yield sources of markers differentially expressed in AD CSF, collectively harboring nearly 300 proteins with significantly altered levels in AD spinal fluid compared to controls. Based on their corresponding brain modules, these ~300 CSF AD targets were then segregated into five systems-based biomarker panels representing a wide range of brain pathophysiology, including synaptic transmission, vascular biology, myelination, glial-mediated inflammation, and energy metabolism. Using high-throughput TMT proteomic analysis, proteins from these five panels were validated in multiple additional CSF cohorts, totaling >500 spinal fluid samples. The results of these validation studies were then used to narrow down these biomarker panels from roughly 300 to 60 proteins across the five panels. Target prioritization was based on a variety of criteria, including significance and magnitude of change in disease, reproducibility of these disease-related alterations, and the ease of mass spectrometry detection and quantification (Fig. 3). Interestingly, the final CSF marker panels demonstrated AD-specificity and altered levels in AsymAD spinal fluid, indicating a potential role for these panels in preclinical disease stages. In two separate smaller analyses of AD CSF, many of the same AD-associated synaptic and metabolic biomarkers were identified, further supporting the reproducibility these systems-based CSF marker panels [78, 172].
Plasma biomarker discovery represents another potential extension of AD brain network analysis. The rationale for using plasma biomarkers as proxies for brain processes [173] has been demonstrated for multiple neurodegenerative diseases [174, 175]. While targeted proteomic approaches have been primarily used to measure plasma Aβ [176, 177] and phosphorylated tau [178, 179], recent improvements in MS technology and chromatography have renewed interest in discovery plasma proteomics in AD [180]. For example, discovery TMT proteomics was recently applied to plasma samples of two independent cohorts to predict Aβ burden in preclinical disease with high accuracy [181]. Moving forward, integrative analyses that successfully correlate proteomic signatures across brain, CSF, and plasma will prove key for the advancement of ideal network-based biomarkers of disease (Fig. 3).
Targeted validation and assay development
Following discovery-driven proteomics, targeted assays are typically employed in large numbers of clinical samples to validate specific candidate biomarkers or multiplexed biomarker panels (Fig. 3). Antibody- or aptamer-based immunoassays are often used as a strategy for target validation [182, 183]. These assays are capable of high-fidelity validation, versatile in application, and easier to execute than MS-based assays. However, immunoassay validation can present certain challenges. First, the costs of commercially available antibody or aptamer reagents necessary for large-scale studies of hundreds or thousands of proteins in sizeable clinical research studies can be prohibitive, limiting validation and translation efforts. Second, in the absence of high-quality, commercially available antibodies or aptamers, assays must be generated de novo, optimized, and validated separately, a process that can also be costly and time-consuming. Finally, immunoassay approaches are often unable to specifically detect biologically or pathologically relevant isoforms or PTMs of a protein.
As an antibody-free platform with robust sensitivity, high accuracy, and an exceptional multiplex capacity, targeted MS technologies such as SRM/MRM and PRM offer a promising alternative to the challenges of immunoassay validation [184–187]. As Cilento et al. argue in a recent review of targeted MS in AD biomarker discovery, these approaches provide a cost-effective means of systematically verifying large panels of protein biomarker candidates prior to more expensive and large-scale validation testing [187]. This workflow typically begins with the synthesis of unique and protein-specific peptides to facilitate the direct detection of candidate biomarkers and development of multiplex assays. These targeted MS assays, which are capable of measuring hundreds of peptide biomarkers simultaneously, are then applied to a small set (~10–50) of patient samples and critically analyzed for reproducibility and assay adaptability. Once verified, a smaller panel of the most promising candidates can enter the validation stage, in which they are analyzed in several hundred (~100–500) samples to critically assess sensitivity and specificity. This validation analysis represents the final preclinical stage, after which chosen candidates can be further investigated for clinical application [187, 188].
Targeted MS approaches have been increasingly employed for the detection and quantification of protein biomarkers from biofluids [187]. Several studies have successfully used targeted MS strategies to quantify Aβ, tau, and APOE protein levels in CSF with similar disease-association efficacy to that of traditional ELISA assays [189–191]. More recently, targeted MS was applied to a more diverse panel of promising AD CSF biomarkers reflecting a much wider array of disease pathophysiologies [192]. In this study, Zhou and colleagues used PRM to quantify a panel of brain-derived proteins significantly altered in a discovery-driven proteomic analysis of AD CSF. Several proteins, including tau and neurofilaments, were significantly increased in AD CSF and capable of distinguishing AD cases from controls and non-AD dementia. Protein targets mapping to synaptic, metabolism, and neuroinflammation modules in brain [96] were quantified with high precision, showcasing their potential for future biomarker translation. SMOC1, YWHAZ, ALDOA, and MAP1B emerged as biomarkers that could best discriminate between individuals with AD and non-AD cognitive impairment, correlating well with Aβ and tau levels [192]. Overall, these results further illustrate the utility of targeted MS strategies to significantly advance AD biomarker discovery toward network-based protein assays with multiple clinical uses in disease (Fig. 3), including preclinical profiling, disease monitoring, measuring therapeutic response, and confirming target engagement.
Future directions
In summary, global network proteomics has revealed a highly reproducible and holistic window into the complex biochemical and cellular alterations in the brains of individuals with asymptomatic and symptomatic AD. As the network landscape of the AD proteome continues to emerge, future directions will include the (a) exploration of local cell type-specific proteomes to better elaborate the disease mechanisms implicated in these global studies, (b) integration of these brain networks and hubs with protein analyses of CSF and plasma, and (c) large-scale verification and validation of the most promising brain-linked biomarkers in additional human samples using targeted proteomic strategies. This validation will also include the longitudinal measurement of promising biomarkers in large cohorts, such as the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and the Dominantly Inherited Alzheimer Network (DIAN), to define their reactivity to disease progression and treatment. Together these efforts promise to not only expand our understanding of AD pathogenesis, but also fulfill many unmet clinical needs in AD diagnostics, disease monitoring, and therapeutics.
Funding and disclosure
Research reported in this publication was supported by the National Institute on Aging of the National Institutes of Health (F32AG064862, K08-NS099474-1, R01 NS114130-01A1, R01AG053960, R01AG057911, R01AG061800, RF1AG057471, RF1AG057470, RF1AG062181), the Accelerating Medicine Partnership for AD (U01AG046161 and U01AG061357), the Emory Alzheimer’s Disease Research Center (P50AG025688), and the NINDS Emory Neuroscience Core (P30NS055077). Additional support was provided by grants from the Alzheimer’s Association (Award no. 37102 to S. Rangaraju), Foundation for the National Institutes of Health (FNIH), Alzheimer’s Research UK (ARUK), and the Weston Brain Institute (11060). The authors declare no competing interests.
Acknowledgements
We would like to thank Dr. Eric B. Dammer and Dr. Erik Johnson for their critical review of this publication.
Author contributions
Writing—original draft preparation: S.Rayaprolu, LH, PB, CW, SZ, and S.Rangaraju, NTS; writing—review and editing: S.Rayaprolu, LH, PB, CW, SZ, AIL, S.Rangaraju, and NTS; visualization: S.Rayaprolu and LH; funding acquisition: S.Rayaprolu, AIL, S.Rangaraju, and NTS; project administration: PB and NTS. All authors have approved of the contents of this manuscript and provided consent for publication.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sruti Rayaprolu, Lenora Higginbotham
References
- 1.Association As. 2019 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2019;15:321–87. [Google Scholar]
- 2.Lane CA, Hardy J, Schott JM. Alzheimer’s disease. Eur J Neurol. 2018;25:59–70. doi: 10.1111/ene.13439. [DOI] [PubMed] [Google Scholar]
- 3.Duyckaerts C, Delatour B, Potier MC. Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 2009;118:5–36. doi: 10.1007/s00401-009-0532-1. [DOI] [PubMed] [Google Scholar]
- 4.Julia TCW, Goate AM. Genetics of beta-amyloid precursor protein in Alzheimer’s disease. Cold Spring Harb Perspect Med. 2017;7:a024539. [DOI] [PMC free article] [PubMed]
- 5.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–5. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 6.Jack CR, Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–62. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jack CR, Jr, Wiste HJ, Schwarz CG, Lowe VJ, Senjem ML, Vemuri P, et al. Longitudinal tau PET in ageing and Alzheimer’s disease. Brain. 2018;141:1517–28. doi: 10.1093/brain/awy059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe VJ, Lundt ES, Albertson SM, Przybelski SA, Senjem ML, Parisi JE, et al. Neuroimaging correlates with neuropathologic schemes in neurodegenerative disease. Alzheimers Dement. 2019;15:927–39. doi: 10.1016/j.jalz.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59:512–9. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 10.Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM. Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 2007;64:343–9. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- 11.Long JM, Holtzman DM. Alzheimer disease: an update on pathobiology and treatment strategies. Cell. 2019;179:312–39. doi: 10.1016/j.cell.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Strooper B, Karran E. The cellular phase of Alzheimer’s disease. Cell. 2016;164:603–15. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 13.Hodes RJ, Buckholtz N. Accelerating medicines partnership: Alzheimer’s disease (AMP-AD) knowledge portal aids Alzheimer’s drug discovery through open data sharing. Expert Opin Ther Targets. 2016;20:389–91. doi: 10.1517/14728222.2016.1135132. [DOI] [PubMed] [Google Scholar]
- 14.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–6. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 15.Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–90. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 16.Price JL, Morris JC. Tangles and plaques in nondemented aging and “preclinical” Alzheimer’s disease. Ann Neurol. 1999;45:358–68. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 17.Vermunt L, Sikkes SAM, van den Hout A, et al. Duration of preclinical, prodromal, and dementia stages of Alzheimer’s disease in relation to age, sex, and APOE genotype. Alzheimers Dement. 2019;15:888–98. doi: 10.1016/j.jalz.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy J. An ‘anatomical cascade hypothesis’ for Alzheimer’s disease. Trends Neurosci. 1992;15:200–1. doi: 10.1016/0166-2236(92)90033-5. [DOI] [PubMed] [Google Scholar]
- 19.Makin S. The amyloid hypothesis on trial. Nature. 2018;559:4–7. doi: 10.1038/d41586-018-05719-4. [DOI] [PubMed] [Google Scholar]
- 20.Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL. Alzheimer’s disease. Nat Rev Dis Prim. 2015;1:15056. doi: 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- 21.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–6. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 22.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2:864–70. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 23.Lamb BT, Call LM, Slunt HH, Bardel KA, Lawler AM, Eckman CB, et al. Altered metabolism of familial Alzheimer’s disease-linked amyloid precursor protein variants in yeast artificial chromosome transgenic mice. Hum Mol Genet. 1997;6:1535–41. doi: 10.1093/hmg/6.9.1535. [DOI] [PubMed] [Google Scholar]
- 24.Borchelt DR, Ratovitski T, van Lare J, Lee MK, Gonzales V, Jenkins NA, et al. Accelerated amyloid deposition in the brains of transgenic mice coexpressing mutant presenilin 1 and amyloid precursor proteins. Neuron. 1997;19:939–45. doi: 10.1016/s0896-6273(00)80974-5. [DOI] [PubMed] [Google Scholar]
- 25.Games D, Adams D, Alessandrini R, Barbour R, Berthelette P, Blackwell C, et al. Alzheimer-type neuropathology in transgenic mice overexpressing V717F beta-amyloid precursor protein. Nature. 1995;373:523–7. doi: 10.1038/373523a0. [DOI] [PubMed] [Google Scholar]
- 26.Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold KH, Mistl C, Rothacher S, et al. Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc Natl Acad Sci USA. 1997;94:13287–92. doi: 10.1073/pnas.94.24.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, et al. Early phenotypic changes in transgenic mice that overexpress different mutants of amyloid precursor protein in brain. J Biol Chem. 1999;274:6483–92. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]
- 28.Reaume AG, Howland DS, Trusko SP, Savage MJ, Lang DM, Greenberg BD, et al. Enhanced amyloidogenic processing of the beta-amyloid precursor protein in gene-targeted mice bearing the Swedish familial Alzheimer’s disease mutations and a “humanized” Abeta sequence. The. J Biol Chem. 1996;271:23380–8. doi: 10.1074/jbc.271.38.23380. [DOI] [PubMed] [Google Scholar]
- 29.Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, et al. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–8. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penke B, Paragi G, Gera J, Berkecz R, Kovacs Z, Crul T, et al. The role of lipids and membranes in the pathogenesis of Alzheimer’s disease: a comprehensive view. Curr Alzheimer Res. 2018;15:1191–212. doi: 10.2174/1567205015666180911151716. [DOI] [PubMed] [Google Scholar]
- 31.Benilova I, Karran E, De, Strooper B. The toxic Abeta oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat Neurosci. 2012;15:349–57. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 32.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–9. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 33.Glabe CG. Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol Aging. 2006;27:570–5. doi: 10.1016/j.neurobiolaging.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 34.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson PT, Alafuzoff I, Bigio EH, Bouras C, Braak H, Cairns NJ, et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J Neuropathol Exp Neurol. 2012;71:362–81. doi: 10.1097/NEN.0b013e31825018f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arboleda-Velasquez JF, Lopera F, O’Hare M, Delgado-Tirado S, Marino C, Chmielewska N, et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Nat Med. 2019;25:1680–83. doi: 10.1038/s41591-019-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armstrong RA, Lantos PL, Cairns NJ. Overlap between neurodegenerative disorders. Neuropathology. 2005;25:111–24.. doi: 10.1111/j.1440-1789.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- 38.Attems J, Jellinger KA. The overlap between vascular disease and Alzheimer’s disease-lessons from pathology. BMC Med. 2014;12:206. doi: 10.1186/s12916-014-0206-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapasi A, DeCarli C, Schneider JA. Impact of multiple pathologies on the threshold for clinically overt dementia. Acta Neuropathol. 2017;134:171–86. doi: 10.1007/s00401-017-1717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 41.Kelly SC, He B, Perez SE, Ginsberg SD, Mufson EJ, Counts SE. Locus coeruleus cellular and molecular pathology during the progression of Alzheimer’s disease. Acta Neuropathologica Commun. 2017;5:8. doi: 10.1186/s40478-017-0411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grudzien A, Shaw P, Weintraub S, Bigio E, Mash DC, Mesulam MM. Locus coeruleus neurofibrillary degeneration in aging, mild cognitive impairment and early Alzheimer’s disease. Neurobiol Aging. 2007;28:327–35. doi: 10.1016/j.neurobiolaging.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Bondareff W, Mountjoy CQ, Roth M, Rossor MN, Iversen LL, Reynolds GP, et al. Neuronal degeneration in locus ceruleus and cortical correlates of Alzheimer disease. Alzheimer Dis Assoc Disord. 1987;1:256–62. doi: 10.1097/00002093-198701040-00005. [DOI] [PubMed] [Google Scholar]
- 44.Crary JF, Trojanowski JQ, Schneider JA, Abisambra JF, Abner EL, Alafuzoff I, et al. Primary age-related tauopathy (PART): a common pathology associated with human aging. Acta Neuropathol. 2014;128:755–66. doi: 10.1007/s00401-014-1349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovacs GG. Invited review: Neuropathology of tauopathies: principles and practice. Neuropathol Appl Neurobiol. 2015;41:3–23. doi: 10.1111/nan.12208. [DOI] [PubMed] [Google Scholar]
- 46.Hanseeuw BJ, Betensky RA, Jacobs HIL, Schultz AP, Sepulcre J, Becker JA, et al. Association of amyloid and tau with cognition in preclinical Alzheimer disease: a longitudinal study. JAMA Neurol. 2019;76:915–24. doi: 10.1001/jamaneurol.2019.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gauthier S, Feldman HH, Schneider LS, Wilcock GK, Frisoni GB, et al. Efficacy and safety of tau-aggregation inhibitor therapy in patients with mild or moderate Alzheimer’s disease: a randomised, controlled, double-blind, parallel-arm, phase 3 trial. Lancet. 2016;388:2873–84. doi: 10.1016/S0140-6736(16)31275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seripa D, Solfrizzi V, Imbimbo BP, Daniele A, Santamato A, Lozupone M, et al. Tau-directed approaches for the treatment of Alzheimer’s disease: focus on leuco-methylthioninium. Expert Rev Neurother. 2016;16:259–77. doi: 10.1586/14737175.2016.1140039. [DOI] [PubMed] [Google Scholar]
- 49.Congdon EE, Sigurdsson EM. Tau-targeting therapies for Alzheimer disease. Nat Rev Neurol. 2018;14:399–415. doi: 10.1038/s41582-018-0013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, et al. ApoE influences amyloid-beta (Abeta) clearance despite minimal apoE/Abeta association in physiological conditions. Proc Natl Acad Sci USA. 2013;110:E1807–16. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Z, Sagare AP, Ma Q, Halliday MR, Kong P, Kisler K, et al. Central role for PICALM in amyloid-beta blood-brain barrier transcytosis and clearance. Nat Neurosci. 2015;18:978–87. doi: 10.1038/nn.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lambert JC, Grenier-Boley B, Chouraki V, Heath S, Zelenika D, Fievet N, et al. Implication of the immune system in Alzheimer’s disease: evidence from genome-wide pathway analysis. J Alzheimer’s Dis: JAD. 2010;20:1107–18. doi: 10.3233/JAD-2010-100018. [DOI] [PubMed] [Google Scholar]
- 54.Heneka MT, Golenbock DT, Latz E. Innate immunity in Alzheimer’s disease. Nat Immunol. 2015;16:229–36. doi: 10.1038/ni.3102. [DOI] [PubMed] [Google Scholar]
- 55.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, et al. TREM2 variants in Alzheimer’s disease. N. Engl J Med. 2013;368:117–27. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl J Med. 2013;368:107–16. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hickman SE, El Khoury J. TREM2 and the neuroimmunology of Alzheimer’s disease. Biochem Pharm. 2014;88:495–8. doi: 10.1016/j.bcp.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGeer PL, Itagaki S, Tago H, McGeer EG. Occurrence of HLA-DR reactive microglia in Alzheimer’s disease. Ann N. Y Acad Sci. 1988;540:319–23. doi: 10.1111/j.1749-6632.1988.tb27086.x. [DOI] [PubMed] [Google Scholar]
- 60.Heneka MT, Carson MJ, El Khoury J, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet. 2015;16:441–58. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin L, et al. A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer’s disease. Cell Syst. 2017;4:60–72. doi: 10.1016/j.cels.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higginbotham L, Ping L, Dammer EB, Duong DM, Zhou M, Gearing M, et al. Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer’s disease. bioRxiv. 2019, 10.1101/806752. [DOI] [PMC free article] [PubMed]
- 64.Swarup V, Chang TS, Duong DM, Dammer EB, Dai J, Lah JJ, et al. Identification of conserved proteomic networks in neurodegenerative dementia. Cell Rep. 2020;31:107807. doi: 10.1016/j.celrep.2020.107807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–26. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yates JR., 3rd Mass spectrometry and the age of the proteome. J Mass Spectrom. 1998;33:1–19. doi: 10.1002/(SICI)1096-9888(199801)33:1<1::AID-JMS624>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 67.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–90. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 68.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 69.Yates JR., 3rd Mass spectral analysis in proteomics. Annu Rev Biophys Biomol Struct. 2004;33:297–316. doi: 10.1146/annurev.biophys.33.111502.082538. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR., 3rd Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113:2343–94. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 72.Pappireddi N, Martin L, Wuhr M. A review on quantitative multiplexed proteomics. Chembiochem. 2019;20:1210–24. doi: 10.1002/cbic.201800650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gillet LC, Leitner A, Aebersold R. Mass spectrometry applied to bottom-up proteomics: entering the high-throughput era for hypothesis testing. Annu Rev Anal Chem. 2016;9:449–72. doi: 10.1146/annurev-anchem-071015-041535. [DOI] [PubMed] [Google Scholar]
- 74.Johnson ECB, Dammer EB, Duong DM, Yin L, Thambisetty M, Troncoso JC, et al. Deep proteomic network analysis of Alzheimer’s disease brain reveals alterations in RNA binding proteins and RNA splicing associated with disease. Mol Neurodegener. 2018;13:52. doi: 10.1186/s13024-018-0282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ping L, Duong DM, Yin L, Gearing M, Lah JJ, Levey AI, et al. Global quantitative analysis of the human brain proteome in Alzheimer’s and Parkinson’s disease. Sci Data. 2018;5:180036. doi: 10.1038/sdata.2018.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Van Vranken JG, Pontano Vaites L, Schweppe DK, Huttlin EL, Etienne C, et al. TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat Methods. 2020;17:399–404. doi: 10.1038/s41592-020-0781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rauniyar N, Yates JR., 3rd Isobaric labeling-based relative quantification in shotgun proteomics. J Proteome Res. 2014;13:5293–309. doi: 10.1021/pr500880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bai B, Wang X, Li Y, Chen PC, Yu K, Dey KK, et al. Deep multilayer brain proteomics identifies molecular networks in Alzheimer’s disease progression. Neuron. 2020;105:975–91. doi: 10.1016/j.neuron.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ping L, Kundinger SR, Duong DM, Yin L, Gearing M, Lah JJ, et al. Global quantitative analysis of the human brain proteome and phosphoproteome in Alzheimer’s disease. bioRxiv. 2020, 10.1101/2020.05.19.105197. [DOI] [PMC free article] [PubMed]
- 80.Brenes A, Hukelmann J, Bensaddek D, Lamond AI. Multibatch TMT reveals false positives, batch effects and missing values. Mol Cell Proteom. 2019;18:1967–80. doi: 10.1074/mcp.RA119.001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyer JG, Schilling B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Rev Proteom. 2017;14:419–29. doi: 10.1080/14789450.2017.1322904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crowgey EL, Matlock A, Venkatraman V, Fert-Bober J, Van Eyk JE. Mapping biological networks from quantitative data-independent acquisition mass spectrometry: data to knowledge pipelines. Methods Mol Biol. 2017;1558:395–413. doi: 10.1007/978-1-4939-6783-4_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ludwig C, Gillet L, Rosenberger G, Amon S, Collins BC, Aebersold R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol Syst Biol. 2018;14:e8126. doi: 10.15252/msb.20178126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Funk AJ, Labilloy G, Reigle J, Alnafisah R, Heaven MR, Roberts RC, et al. Region-specific PSD-95 interactomes contribute to functional diversity of excitatory synapses in human brain. bioRxiv. 2020, 10.1101/2020.05.04.076844.
- 85.Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci. 2008;28:1410–20. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–82. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prill RJ, Marbach D, Saez-Rodriguez J, Sorger PK, Alexopoulos LG, Xue X, et al. Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PLoS ONE. 2010;5:e9202. doi: 10.1371/journal.pone.0009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choobdar S, Ahsen ME, Crawford J, Tomasoni M, Fang T, Lamparter D, et al. Assessment of network module identification across complex diseases. Nat Methods. 2019;16:843–52. doi: 10.1038/s41592-019-0509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Song L, Langfelder P, Horvath S. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinforma. 2012;13:328. doi: 10.1186/1471-2105-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Allen JD, Xie Y, Chen M, Girard L, Xiao G. Comparing statistical methods for constructing large scale gene networks. PLoS ONE. 2012;7:e29348. doi: 10.1371/journal.pone.0029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2014;13:13–24. doi: 10.1111/gbb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ballouz S, Verleyen W, Gillis J. Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics. 2015;31:2123–30. doi: 10.1093/bioinformatics/btv118. [DOI] [PubMed] [Google Scholar]
- 94.Marbach D, Costello JC, Kuffner R, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nygaard V, Rodland EA, Hovig E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics. 2016;17:29–39. doi: 10.1093/biostatistics/kxv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson ECB, Dammer EB, Duong DM, Ping L, Zhou M, Yin L, et al. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med. 2020;26:769–80. doi: 10.1038/s41591-020-0815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18:1819–31. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–47. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat Methods. 2014;11:22–4. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 100.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zambon AC, Gaj S, Ho I, Hanspers K, Vranizan K, Evelo CT, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–10. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinforma. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dai J, Johnson ECB, Dammer EB, Duong DM, Gearing M, Lah JJ, et al. Effects of APOE genotype on brain proteomic network and cell type changes in Alzheimer’s disease. Front Mol Neurosci. 2018;11:454. doi: 10.3389/fnmol.2018.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Higginbotham L, Dammer EB, Duong DM, Modeste E, Montine TJ, Lah JJ, et al. Network analysis of a membrane-enriched brain proteome across stages of Alzheimer’s disease. Proteomes. 2019;7:30. doi: 10.3390/proteomes7030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wingo AP, Dammer EB, Breen MS, Logsdon BA, Duong DM, Troncosco JC, et al. Large-scale proteomic analysis of human brain identifies proteins associated with cognitive trajectory in advanced age. Nat Commun. 2019;10:1619. doi: 10.1038/s41467-019-09613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wingo AP, Fan W, Duong DM, Gerasimov ES, Dammer EB, Liu Y, et al. Shared proteomic effects of cerebral atherosclerosis and Alzheimer’s disease on the human brain. Nat Neurosci. 2020;23:696–700. doi: 10.1038/s41593-020-0635-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Q, Ma C, Gearing M, Wang PG, Chin LS, Li L. Integrated proteomics and network analysis identifies protein hubs and network alterations in Alzheimer’s disease. Acta Neuropathol Commun. 2018;6:19. doi: 10.1186/s40478-018-0524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Masuda T, Sankowski R, Staszewski O, Prinz M. Microglia heterogeneity in the single-cell era. Cell Rep. 2020;30:1271–81. doi: 10.1016/j.celrep.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 110.Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–70. doi: 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- 111.Lawson LJ, Perry VH, Gordon S. Turnover of resident microglia in the normal adult mouse brain. Neuroscience. 1992;48:405–15. doi: 10.1016/0306-4522(92)90500-2. [DOI] [PubMed] [Google Scholar]
- 112.Mittelbronn M, Dietz K, Schluesener HJ, Meyermann R. Local distribution of microglia in the normal adult human central nervous system differs by up to one order of magnitude. Acta Neuropathol. 2001;101:249–55. doi: 10.1007/s004010000284. [DOI] [PubMed] [Google Scholar]
- 113.Vitek MP, Brown CM, Colton CA. APOE genotype-specific differences in the innate immune response. Neurobiol Aging. 2009;30:1350–60. doi: 10.1016/j.neurobiolaging.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu Y, Nwabuisi-Heath E, Dumanis SB, Tai LM, Yu C, Rebeck GW, et al. APOE genotype alters glial activation and loss of synaptic markers in mice. Glia. 2012;60:559–69. doi: 10.1002/glia.22289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gale SC, Gao L, Mikacenic C, Coyle SM, Rafaels N, Murray Dudenkov T, et al. APOepsilon4 is associated with enhanced in vivo innate immune responses in human subjects. J Allergy Clin Immunol. 2014;134:127–34. doi: 10.1016/j.jaci.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shi Y, Manis M, Long J, Wang K, Sullivan PM, Remolina Serrano J, et al. Microglia drive APOE-dependent neurodegeneration in a tauopathy mouse model. J Exp Med. 2019;216:2546–61. doi: 10.1084/jem.20190980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47:566–81. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Galatro TF, Holtman IR, Lerario AM, Vainchtein ID, Brouwer N, Sola PR, et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat Neurosci. 2017;20:1162–71. doi: 10.1038/nn.4597. [DOI] [PubMed] [Google Scholar]
- 119.Gerrits E, Heng Y, Boddeke E, Eggen BJL. Transcriptional profiling of microglia; current state of the art and future perspectives. Glia. 2020;68:740–55. doi: 10.1002/glia.23767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chakrabarty P, Li A, Ceballos-Diaz C, Eddy JA, Funk CC, Moore B, et al. IL-10 alters immunoproteostasis in APP mice, increasing plaque burden and worsening cognitive behavior. Neuron. 2015;85:519–33. doi: 10.1016/j.neuron.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guillot-Sestier MV, Doty KR, Gate D, Rodriguez J, Jr, Leung BP, Rezai-Zadeh K, et al. Il10 deficiency rebalances innate immunity to mitigate Alzheimer-like pathology. Neuron. 2015;85:534–48. doi: 10.1016/j.neuron.2014.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Grathwohl SA, Kalin RE, Bolmont T, Prokop S, Winkelmann G, Kaeser SA, et al. Formation and maintenance of Alzheimer’s disease beta-amyloid plaques in the absence of microglia. Nat Neurosci. 2009;12:1361–3. doi: 10.1038/nn.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Darmellah A, Rayah A, Auger R, Cuif MH, Prigent M, Arpin M, et al. Ezrin/radixin/moesin are required for the purinergic P2X7 receptor (P2X7R)-dependent processing of the amyloid precursor protein. J Biol Chem. 2012;287:34583–95. doi: 10.1074/jbc.M112.400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ahmed ME, Selvakumar GP, Kempuraj D, Raikwar SP, Thangavel R, Bazley K, et al. Neuroinflammation mediated by GMF exacerbates neuronal injury in an in vitro model of traumatic brain injury. J Neurotrauma. 2020;37:1645–55. doi: 10.1089/neu.2019.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ding C, Fan X, Wu G. Peroxiredoxin 1—an antioxidant enzyme in cancer. J Cell Mol Med. 2017;21:193–202. doi: 10.1111/jcmm.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Drummond E, Nayak S, Faustin A, Pires G, Hickman RA, Askenazi M, et al. Proteomic differences in amyloid plaques in rapidly progressive and sporadic Alzheimer’s disease. Acta Neuropathol. 2017;133:933–54. doi: 10.1007/s00401-017-1691-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tosto G, Zimmerman ME, Hamilton JL, Carmichael OT, Brickman AM. Alzheimer’s Disease Neuroimaging I. The effect of white matter hyperintensities on neurodegeneration in mild cognitive impairment. Alzheimers Dement. 2015;11:1510–9. doi: 10.1016/j.jalz.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bartzokis G. Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiol Aging. 2011;32:1341–71. doi: 10.1016/j.neurobiolaging.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Scheltens P, Barkhof F, Valk J, Algra PR, van der Hoop RG, Nauta J, et al. White matter lesions on magnetic resonance imaging in clinically diagnosed Alzheimer’s disease. Evidence for heterogeneity. Brain. 1992;115:735–48. doi: 10.1093/brain/115.3.735. [DOI] [PubMed] [Google Scholar]
- 130.McKenzie AT, Moyon S, Wang M, Katsyv I, Song WM, Zhou X, et al. Multiscale network modeling of oligodendrocytes reveals molecular components of myelin dysregulation in Alzheimer’s disease. Mol Neurodegener. 2017;12:82. doi: 10.1186/s13024-017-0219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang M, Roussos P, McKenzie A, Zhou X, Kajiwara Y, Brennand KJ, et al. Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer’s disease. Genome Med. 2016;8:104. doi: 10.1186/s13073-016-0355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Allen M, Wang X, Burgess JD, Watzlawik J, Serie DJ, Younkin CS, et al. Conserved brain myelination networks are altered in Alzheimer’s and other neurodegenerative diseases. Alzheimers Dement. 2018;14:352–66. doi: 10.1016/j.jalz.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Diner I, Hales CM, Bishof I, Rabenold L, Duong DM, Yi H, et al. Aggregation properties of the small nuclear ribonucleoprotein U1-70K in Alzheimer disease. J Biol Chem. 2014;289:35296–313. doi: 10.1074/jbc.M114.562959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bai B, Hales CM, Chen PC, Gozal Y, Dammer EB, Fritz JJ, et al. U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer’s disease. Proc Natl Acad Sci USA. 2013;110:16562–7. doi: 10.1073/pnas.1310249110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hales CM, Dammer EB, Deng Q, Duong DM, Gearing M, Troncoso JC, et al. Changes in the detergent-insoluble brain proteome linked to amyloid and tau in Alzheimer’s disease progression. Proteomics. 2016;16:3042–53. doi: 10.1002/pmic.201600057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hales CM, Seyfried NT, Dammer EB, Duong D, Yi H, Gearing M, et al. U1 small nuclear ribonucleoproteins (snRNPs) aggregate in Alzheimer’s disease due to autosomal dominant genetic mutations and trisomy 21. Mol Neurodegener. 2014;9:15. doi: 10.1186/1750-1326-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maziuk BF, Apicco DJ, Cruz AL, Jiang L, Ash PEA, da Rocha EL, et al. RNA binding proteins co-localize with small tau inclusions in tauopathy. Acta Neuropathol Commun. 2018;6:71. doi: 10.1186/s40478-018-0574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bishof I, Dammer EB, Duong DM, Kundinger SR, Gearing M, Lah JJ, et al. RNA-binding proteins with basic-acidic dipeptide (BAD) domains self-assemble and aggregate in Alzheimer’s disease. J Biol Chem. 2018;293:11047–66. doi: 10.1074/jbc.RA118.001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Colom-Cadena M, Spires-Jones T, Zetterberg H, Blennow K, Caggiano A, DeKosky ST, et al. The clinical promise of biomarkers of synapse damage or loss in Alzheimer’s disease. Alzheimers Res Ther. 2020;12:21. doi: 10.1186/s13195-020-00588-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Palop JJ, Mucke L. Amyloid-beta-induced neuronal dysfunction in Alzheimer’s disease: from synapses toward neural networks. Nat Neurosci. 2010;13:812–8. doi: 10.1038/nn.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Menkes-Caspi N, Yamin HG, Kellner V, Spires-Jones TL, Cohen D, Stern EA. Pathological tau disrupts ongoing network activity. Neuron. 2015;85:959–66.. doi: 10.1016/j.neuron.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 142.Yu L, Tasaki S, Schneider JA, Arfanakis K, Duong DM, Wingo AP, et al. Cortical proteins associated with cognitive resilience in community-dwelling older persons. JAMA Psychiatry. 2020:e201807. [DOI] [PMC free article] [PubMed]
- 143.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–6. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Drummond ES, Nayak S, Ueberheide B, Wisniewski T. Proteomic analysis of neurons microdissected from formalin-fixed, paraffin-embedded Alzheimer’s disease brain tissue. Sci Rep. 2015;5:15456. doi: 10.1038/srep15456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hong I, Kang T, Yoo Y, Park R, Lee J, Lee S, et al. Quantitative proteomic analysis of the hippocampus in the 5XFAD mouse model at early stages of Alzheimer’s disease pathology. J Alzheimer’s Dis: JAD. 2013;36:321–34. doi: 10.3233/JAD-130311. [DOI] [PubMed] [Google Scholar]
- 146.Rangaraju S, Dammer EB, Raza SA, Gao T, Xiao H, Betarbet R, et al. Quantitative proteomics of acutely-isolated mouse microglia identifies novel immune Alzheimer’s disease-related proteins. Mol Neurodegeneration. 2018;13:34. doi: 10.1186/s13024-018-0266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rayaprolu S, Gao T, Xiao H, Ramesha S, Weinstock LD, Shah J, et al. Flow-cytometric microglial sorting coupled with quantitative proteomics identifies moesin as a highly-abundant microglial protein with relevance to Alzheimer’s disease. Mol Neurodegener. 2020;15:28. doi: 10.1186/s13024-020-00377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang Y, Sloan Steven A, Clarke Laura E, Caneda C, Plaza Colton A, Blumenthal Paul D, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–7. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016;113:E1738–46. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:eaal3222. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Swartzlander DB, Propson NE, Roy ER, Saito T, Saido T, Wang B, et al. Concurrent cell type-specific isolation and profiling of mouse brains in inflammation and Alzheimer's disease. JCI Insight. 2018;3:e121109. doi: 10.1172/jci.insight.121109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rayaprolu S, Gao T, Xiao H, Ramesha S, Duong DM, Dammer EB, et al. Quantitative multiplexed proteomics of mouse microglia by flow-cytometric sorting reveals a core set of highly-abundant microglial proteins. bioRxiv. 10.1101/802694.
- 154.Dammer EB, Duong DM, Diner I, Gearing M, Feng Y, Lah JJ, et al. Neuron enriched nuclear proteome isolated from human brain. J Proteome Res. 2013;12:3193–206. doi: 10.1021/pr400246t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Budnik B, Levy E, Harmange G, Slavov N. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018;19:161. doi: 10.1186/s13059-018-1547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Landgraf P, Antileo ER, Schuman EM, Dieterich DC. BONCAT: metabolic labeling, click chemistry, and affinity purification of newly synthesized proteomes. Methods Mol Biol. 2015;1266:199–215. doi: 10.1007/978-1-4939-2272-7_14. [DOI] [PubMed] [Google Scholar]
- 157.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103:9482–7. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alvarez-Castelao B, Schanzenbacher CT, Hanus C, Glock C, Tom Dieck S, Dorrbaum AR, et al. Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat Biotechnol. 2017;35:1196–201. doi: 10.1038/nbt.4016. [DOI] [PubMed] [Google Scholar]
- 159.Alvarez-Castelao B, Schanzenbacher CT, Langer JD, Schuman EM. Cell-type-specific metabolic labeling, detection and identification of nascent proteomes in vivo. Nat Protoc. 2019;14:556–75. doi: 10.1038/s41596-018-0106-6. [DOI] [PubMed] [Google Scholar]
- 160.Chen CL, Perrimon N. Proximity-dependent labeling methods for proteomic profiling in living cells. Wiley Interdiscip Rev Dev Biol. 2017;6:10.1002/wdev.272. doi: 10.1002/wdev.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–10. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen AL, Kim EW, Toh JY, Vashisht AA, Rashoff AQ, Van C, et al. Novel components of the Toxoplasma inner membrane complex revealed by BioID. mBio. 2015;6:e02357–14. doi: 10.1128/mBio.02357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, Wu Q, et al. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–99. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, et al. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell. 2013;12:356–67. doi: 10.1128/EC.00326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dingar D, Kalkat M, Chan PK, Srikumar T, Bailey SD, Tu WB, et al. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J Proteom. 2015;118:95–111. doi: 10.1016/j.jprot.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 166.Kehrer J, Frischknecht F, Mair GR. Proteomic analysis of the plasmodium berghei gametocyte egressome and vesicular bioID of osmiophilic body proteins identifies merozoite TRAP-like protein (MTRAP) as an essential factor for parasite transmission. Mol Cell Proteom. 2016;15:2852–62. doi: 10.1074/mcp.M116.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA. 2014;111:E2453–61. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Uezu A, Kanak DJ, Bradshaw TW, Soderblom EJ, Catavero CM, Burette AC, et al. Identification of an elaborate complex mediating postsynaptic inhibition. Science. 2016;353:1123–9. doi: 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol. 2018;36:880–7. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–8. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–31. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bader JM, Geyer PE, Muller JB, Strauss MT, Koch M, Leypoldt F, et al. Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer’s disease. Mol Syst Biol. 2020;16:e9356. doi: 10.15252/msb.20199356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:595–603. doi: 10.1002/ajmg.b.32150. [DOI] [PubMed] [Google Scholar]
- 174.Hamlett ED, Goetzl EJ, Ledreux A, Vasilevko V, Boger HA, LaRosa A, et al. Neuronal exosomes reveal Alzheimer’s disease biomarkers in Down syndrome. Alzheimers Dement. 2017;13:541–9. doi: 10.1016/j.jalz.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shi M, Kovac A, Korff A, Cook TJ, Ginghina C, Bullock KM, et al. CNS tau efflux via exosomes is likely increased in Parkinson’s disease but not in Alzheimer’s disease. Alzheimers Dement. 2016;12:1125–31. doi: 10.1016/j.jalz.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Verberk IMW, Slot RE, Verfaillie SCJ, Heijst H, Prins ND, van Berckel BNM, et al. Plasma amyloid as prescreener for the earliest alzheimer pathological changes. Ann Neurol. 2018;84:648–58. doi: 10.1002/ana.25334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schindler SE, Bollinger JG, Ovod V, Mawuenyega KG, Li Y, Gordon BA, et al. High-precision plasma beta-amyloid 42/40 predicts current and future brain amyloidosis. Neurology. 2019;93:e1647–59. doi: 10.1212/WNL.0000000000008081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Janelidze S, Mattsson N, Palmqvist S, Smith R, Beach TG, Serrano GE, et al. Plasma P-tau181 in Alzheimer’s disease: relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer’s dementia. Nat Med. 2020;26:379–86. doi: 10.1038/s41591-020-0755-1. [DOI] [PubMed] [Google Scholar]
- 179.Barthelemy NR, Li Y, Joseph-Mathurin N, Gordon BA, Hassenstab J, Benzinger TLS, et al. A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nat Med. 2020;26:398–407. doi: 10.1038/s41591-020-0781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Geyer PE, Kulak NA, Pichler G, Holdt LM, Teupser D, Mann M. Plasma proteome profiling to assess human health and disease. Cell Syst. 2016;2:185–95. doi: 10.1016/j.cels.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 181.Dey KK, Wang H, Niu M, Bai B, Wang X, Li Y, et al. Deep undepleted human serum proteome profiling toward biomarker discovery for Alzheimer’s disease. Clin Proteom. 2019;16:16. doi: 10.1186/s12014-019-9237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kim CH, Tworoger SS, Stampfer MJ, Dillon ST, Gu X, Sawyer SJ, et al. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci Rep. 2018;8:8382. doi: 10.1038/s41598-018-26640-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Giudice V, Biancotto A, Wu Z, Cheung F, Candia J, Fantoni G, et al. Aptamer-based proteomics of serum and plasma in acquired aplastic anemia. Exp Hematol. 2018;68:38–50. doi: 10.1016/j.exphem.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Carr SA, Abbatiello SE, Ackermann BL, Borchers C, Domon B, Deutsch EW, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteom. 2014;13:907–17. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ebhardt HA, Root A, Sander C, Aebersold R. Applications of targeted proteomics in systems biology and translational medicine. Proteomics. 2015;15:3193–208. doi: 10.1002/pmic.201500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–66. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 187.Cilento EM, Jin L, Stewart T, Shi M, Sheng L, Zhang J. Mass spectrometry: a platform for biomarker discovery and validation for Alzheimer’s and Parkinson’s diseases. J Neurochem. 2019;151:397–416. doi: 10.1111/jnc.14635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Parker CE, Borchers CH. Mass spectrometry based biomarker discovery, verification, and validation-quality assurance and control of protein biomarker assays. Mol Oncol. 2014;8:840–58. doi: 10.1016/j.molonc.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pannee J, Portelius E, Oppermann M, Atkins A, Hornshaw M, Zegers I, et al. A selected reaction monitoring (SRM)-based method for absolute quantification of Abeta38, Abeta40, and Abeta42 in cerebrospinal fluid of Alzheimer’s disease patients and healthy controls. J Alzheimers Dis. 2013;33:1021–32. doi: 10.3233/JAD-2012-121471. [DOI] [PubMed] [Google Scholar]
- 190.Pottiez G, Yang L, Stewart T, Song N, Aro P, Galasko DR, et al. Mass-spectrometry-based method to quantify in parallel tau and amyloid beta 1-42 in CSF for the diagnosis of Alzheimer’s disease. J Proteome Res. 2017;16:1228–38. doi: 10.1021/acs.jproteome.6b00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Han SH, Kim JS, Lee Y, Choi H, Kim JW, Na DL, et al. Both targeted mass spectrometry and flow sorting analysis methods detected the decreased serum apolipoprotein E level in Alzheimer’s disease patients. Mol Cell Proteom. 2014;13:407–19. doi: 10.1074/mcp.M113.028639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhou M, Haque RU, Dammer EB, Duong DM, Ping L, Johnson ECB, et al. Targeted mass spectrometry to quantify brain-derived cerebrospinal fluid biomarkers in Alzheimer’s disease. Clin Proteom. 2020;17:19. doi: 10.1186/s12014-020-09285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, et al. A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet. 1992;1:345–7. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 194.Davis J, Xu F, Deane R, Romanov G, Previti ML, Zeigler K, et al. Early-onset and robust cerebral microvascular accumulation of amyloid beta-protein in transgenic mice expressing low levels of a vasculotropic Dutch/Iowa mutant form of amyloid beta-protein precursor. J Biol Chem. 2004;279:20296–306. doi: 10.1074/jbc.M312946200. [DOI] [PubMed] [Google Scholar]
- 195.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, et al. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 196.Radde R, Bolmont T, Kaeser SA, Coomaraswamy J, Lindau D, Stoltze L, et al. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 2006;7:940–6. doi: 10.1038/sj.embor.7400784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13:159–70. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 198.Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–40. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lewis J, McGowan E, Rockwood J, Melrose H, Nacharaju P, Van Slegtenhorst M, et al. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat Genet. 2000;25:402–5. doi: 10.1038/78078. [DOI] [PubMed] [Google Scholar]
- 200.Allen B, Ingram E, Takao M, Smith MJ, Jakes R, Virdee K, et al. Abundant tau filaments and nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J Neurosci. 2002;22:9340–51. doi: 10.1523/JNEUROSCI.22-21-09340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yoshiyama Y, Higuchi M, Zhang B, Huang SM, Iwata N, Saido TC, et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–51. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 202.Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, et al. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–21. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 203.Jankowsky JL, Zheng H. Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol Neurodegeneration. 2017;12:89. doi: 10.1186/s13024-017-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]