Abstract
Crucial decisions involving cell fate and connectivity that shape the distinctive development of the human brain occur in the embryonic and fetal stages—stages that are difficult to access and investigate in humans. The last decade has seen an impressive increase in resources—from atlases and databases to biological models—that is progressively lifting the curtain on this critical period. In this review, we describe the current state of genomic, transcriptomic, and epigenomic datasets charting the development of normal human brain with a particular focus on recent single-cell technologies. We discuss the emergence of brain organoids generated from pluripotent stem cells as a model to compensate for the limited availability of fetal tissue. Indeed, comparisons of neural lineages, transcriptional dynamics, and noncoding element activity between fetal brain and organoids have helped identify gene regulatory networks functioning at early stages of brain development. Altogether, we argue that large multi-omics investigations have pushed brain development into the “big data” era, and that current and future transversal approaches needed to leverage both fetal brain and organoid resources promise to answer major questions of brain biology and psychiatry.
Subject terms: Neural stem cells, Neural patterning
Introduction: human-specific features of brain development
Understanding the development of the human central nervous system is fundamental for unraveling its complex functions, evolutionary innovations, and pathophysiology of neuropsychiatric disorders. The central nervous system originates from the neural tube, which forms within the ectoderm in the dorsal part of the embryo. In early development, succeeding orthogonal waves of diffusible molecules, called morphogens, control the specification and differentiation of its broad domains (i.e., telencephalon, diencephalon, mesencephalon, and spinal cord), their subsequent subdivisions into regions (for instance, the distinction of hippocampus and neocortex in the dorsal telencephalon or the lateral, medial, and caudal ganglionic eminences in the basal telencephalon), and further refined suborganizations (such as the neuronal layers of the neocortex) [1]. This tissue organization is accompanied on the cellular scale by several transitions, starting from a transition of symmetrically dividing neuroepithelial cells into neurogenic radial glia (RG), the stem cells of the developing brain. RG generate over time and space sequential waves of neuronal progenitors, neurons, astrocytes, and oligodendrocytes. In parallel, cells migrate across different regions and form reciprocal interconnections through axonogenesis, finally resulting in neurons and glia developing network electrical activities and completing the process of brain development.
Compared with other mammals, the human brain has more neurons, larger neuronal diversity and pronounced morphological differences—with a complex pattern of gyri and sulci—and a greater brain lateralization compared with nonhuman primates. These features are the result of an increased duration of neurogenesis, an increased number and diversity of progenitors, and an increased complexity of cell fate programs. This divergence is supported by a longer gestational period and a postnatal maturation finishing only in the third decade of life (reviewed in [2]). The cerebral cortex is the region that has undergone the most remarkable number of changes at the anatomical, cellular, and molecular levels over the course of evolution. The human cerebral cortex has a six-layered laminar architecture, which is relatively conserved in mammals, but with a surface area 1000 times larger than that of a mouse [3]. Cortical areas, with their inherent neuronal architecture and connections, are much more diversified in humans, with many novel areas associated with cognitive functions [4, 5]. The cortex is an anatomical and functional map of our interactions with the external world, and, together with its interconnected regions, forms a biological entity responsible for higher human cognitive abilities and associated psychopathology.
The emerging discipline of functional genomics is crucial to understand the establishment of this cellular and functional architecture (see Box 1 for a glossary of important terms and technologies). Gene regulatory networks, composed by cascades of transcription factors (TFs) and epigenetic modifications, are central to the correct differentiation of every neural cell as the brain develops [6]. TFs bind to genomic noncoding regions called enhancers and the 3D chromatin architecture enables their interactions with gene promoters and other regulatory proteins, even over long distances [7]. These transcriptional machineries control the expression of their target genes in space and time (Fig. 1b). Enhancer activity is regulated in part through epigenetic modification of nearby histones (i.e., acetylation, methylation, and hydroxylation). Altogether, enhancer activities are used as an integrative mechanism for the control of gene expression and are central to the establishment and maintenance of cell identity, affecting the fate of multiple classes of neural cells [8, 9]. As such, they are controlled on a more refined level than the expression of TFs [10, 11]. This fundamental role highlights the importance of identifying TF-enhancer-gene relationships as well as their epigenetic regulation to understand how external information is integrated by the cell. Enhancers, along with other regulatory regions, therefore constitute the main “data” necessary to understand the control of brain development. Finally, mutations within those regions, such as single-nucleotide polymorphisms (SNPs), can affect transcription by, for instance, modifying TF binding affinity or chromatin conformation. Therefore, mutations in noncoding regions can have a more subtle, precise, and context-dependent impact than when affecting coding regions, making them an important subject to understand complex genetic diseases, including many neuropsychiatric diseases [12, 13].
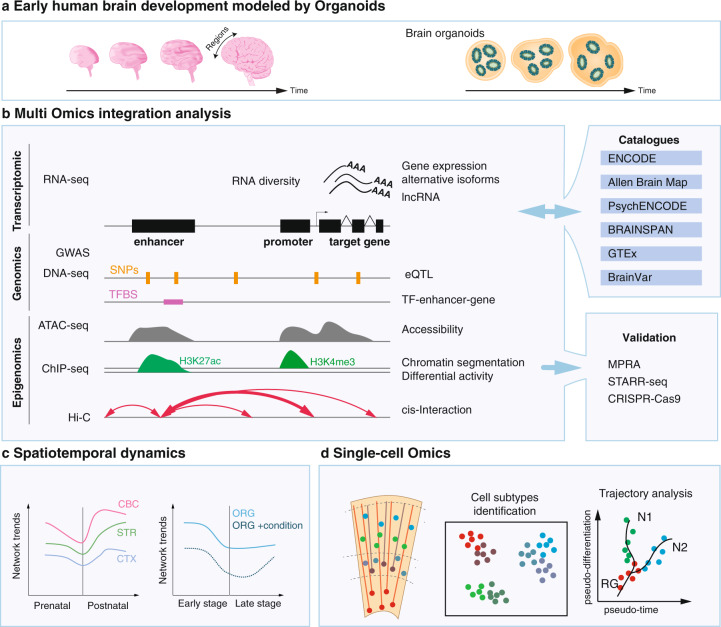
Fig. 1. Integrative approaches to study human brain development.
a Illustration of two complementary models, postmortem human brain tissue and iPSC-derived brain organoids. b Potential of multi-omics approaches to connect/integrate genomic, transcriptomic, and epigenomic information generated or available through datasets of in vivo/in vitro studies. Enhancer activity (identified from ATAC-seq, ChIP-seq) influences transcription (RNA-seq) of gene/s through DNA looping (identified through Hi-C). Similarly, eQTL connects risk variants (discovered via DNA-seq, SNPs, GWAS) to gene expression. These variants may disrupt transcription factor binding sites (TFBS) within enhancers. Human-specific gene regulatory mechanisms can be validated in live cell systems, i.e., iPSC-derived organoids, or human progenitor cell lines, by high throughput methods (e.g., MPRA, STARR-seq). c Studying gene-enhancer dynamics over time both in vivo and in organoids (ORG) can reveal biological insight into the formation and evolution of the human brain (e.g., the hourglass shape of interregional diversity over time between cortex (CTX), striatum (STR), cerebellum (CRB)). d Single-cell omics can define those dynamics at the cellular levels, for instance revealing how gene regulation evolves between radial glia (RG) and neuronal derivatives (N1, N2).
Studying functional genomics of the developing human brain is challenging due to the limited availability of fetal tissue for science. The generation of embryonic stem cell (ESC) and human induced pluripotent stem cell (iPSC) lines have recently offered an alternative. Using those cell lines, several labs have developed 3D in vitro models of brain development generally referred to as brain organoids [14–16]. By mimicking the morphology of the embryonic nervous system—especially the apico-basal polarity of the RG in the ventricular zone (VZ) and the generation of the outer VZ typical of the human brain — organoids have the inherent capability of generating the multiple cellular lineages of the brain while reproducing in vivo cell-to-cell communications and organization [17]. The organoid model presents multiple advantages, from the ability of studying individual variations using iPSCs from different human genetic backgrounds to the ability of conducting longitudinal analyses in the same genomic context. Brain organoids can be easily perturbed to model the effects of environmental and genetic factors on neural development in a controlled setting. While holding invaluable promises, the brain organoid field is still emerging and many developments remain necessary to translate its discoveries into the clinical world (see below, “Exploiting scRNA-seq to study organoids cellular composition and gene networks” section).
In this review, we will describe how the application of functional genomics to the human brain has led to the development of invaluable resources and datasets. We will then focus on recent studies that leveraged those resources to characterize the unique features of the developing human brain, and how the boundaries have been pushed by the emergence of single-cell technologies. The applications of the brain organoid model are presented and we discuss how they compare and can be integrated with their in vivo counterpart, illustrating this by our recent work and the plethora of single-cell studies recently published (Table 1). Overall, we present a blueprint of integrative studies and techniques at the crossroad of developmental biology, stem cell biology, neuroscience, genomics, and system biology to understand brain development and its disorders (Fig. 1).
Table 1.
List of scRNA-seq and scATAC-seq studies in human brain organoids.
| Authors | Reference | Main focus of study | Type of organoid protocol | Single-cell technology | Number of PSC line | Stages | Biological samples | Total cells | Main analytical toolkits: visualization, clustering, batch effect, trajectory, gene network | Main cluster annotations | Brain tissue reference (human scRNA-seq/RNA-seq) | Main findings and observations related to the scRNA-seq of human organoids | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | D | E | M | L | ||||||||||||
| Camp et al. | [85] | Organoid model validation | Undirected | Fluidigm C1 | 1 iPSC, 1 ESC | X | 11 | 508 | Seurat, BackSPIN, t-SNE, Monocle | early/ventral NPC, dorsal NPC, dorsal neur., ventral neur., mesenchymal cells, RSPO + cells | 226 cells neocortex (12–13 pcw) | First study to demonstrate a similar single-cell expression profile in organoid and cortex. Yet, organoids presented less basal progenitor than apical progenitors compare with brain. | ||||
| Quadrato et al. | [145] | Organoid model validation | Undirected | Drop-Seq | 1 iPSC, 1 ESC | X | X | 31 | ##### | Seurat, t-SNE | NEC, div., Forebrain (RG, IPC, Callosal and Corticofugal neur., IN), Ast., dopa neur., retina, mesodermal prog. | Pollen et al. [93] | Organoid mimick brain neurogenesis with coherent cell subtypes progression. Presence of mesodermal cells. Bioreactors batch influences cell composition. | |||
| Birey et al. | [134] | Regional specification and interaction | Directed: cortical and subpallial + assembloids | BD Resolve/ Smart-Seq2 | 1 iPSC (?) +DLX-lineage | X | 4 | ##### | Seurat, t-SNE | hCS: RG, IPC, Glut. Neur., Choroid, hSS: GABA neur., ventral prog., Ast., OPC | Specified forebrain subregions. Assembloids reproduce regional interactions, with migration of interneurons expressing subpallial and cortical IN markers (e.g., DLXs) | |||||
| Sloan et al. | [123] | Cell-subtype specification | Directed: cortical spheroid | Smart-Seq2 | 2 iPSC | X | X | 8 | 710 | PAGODA, t-SNE, Monocle | Neur., div. glial, intermediate glial, mature Ast. | Pollen et al. [93], Darmanis et al. [77] | Described the maturation of astrocytes over ~20 months in cortical spheroids, with transcriptional and functional similarities to primary tissue. | |||
| Xiang et al. | [135] | Regional specification and interaction | Directed: cortical and MGE | 10X Chromium | 1 ESC, 1 iPSC | X | X | 8 | ##### | Seurat, t-SNE, random-forest | NPC, IPC, cortical neur., IN, Non-committed neur., Subplate, Oligo., Ast., Glia, OPC, Ependymocyte | Darmanis et al. [77] | MGE organoids generate NKX2-1+ IN lineage and oligodendrocytes. Cortical organoids generate interneurons (NKX2-1 negative). | |||
| Madhavan et al. | [125] | Cell-subtype specification | Directed: oligocortical | 10X Chromium | 1 ESC | X | 1? | ##### | CellRanger, t-SNE | Glial and neuronal clusters | Nowakowski et al. [86] | Ability to promote oligodendrocyte lineage while keeping overall cortical fate in spheroids. | ||||
| Amiri et al. | [70] | Organoid model validation | Directed: dorsal forebrain | 10X Chromium | 1 iPSC | X | X | 3 | ##### | Seurat, t-SNE, Monocle | early RG, oRG, tRG, MGE-RG, divRG, IPC, early EN, late EN, Glycolysis, Choroid, U | Nowakowski et al. [86]; Liu et al. [104] | Organoids model cortical development equivalent to 5 and 16 pcw brain tissue, reproducing radial glial and early neuronal diversity. Mapping of enhancer elements and target genes active over organoid development. | |||
| Yoon et al. | [144] | Reproducibility | Directed: cortical | BD Rhapsody system | 3 iPSC | X | 3 | ##### | BD Rhapsody, t-SNE | RG, IPC, ventral prog., Glut. Neur., GABA neur., Ast., OPC, choroid | Previous data | Reliability of cell composition over multiple IPS lines, culture conditions, and differentiation. | ||||
| Marton et al. | [124] | Cell-subtype specification | Directed: oligodendrocyte | Smart-Seq2 | 2 iPSC | X | 2 | 295 | k-mean, t-SNE, Monocle | Neurons, Astrocytes, Microglia, Endothelial, OPC, div. prog., myelinating Oligo. | Darmanis et al. [77]; Sloan et al. [123] | Induced oligodendrocytes myelinated neurons with transcriptional similarity with in vivo counterparts. Transcriptional variability of cells decreased over time. | ||||
| Pollen et al. | [151] | Evolution | Directed: telencephalic | Fluidigm C1 | 10 PSC | X | X | 32 | 1223 | NN-Louvain, CCA, t-SNE, WGCNA | RG, div (G1/S/G2/M), IPC, Reelin+ EN, Maturing EN, IN, Glycolysis, hindbrain, Choroid, retina, mesenchymal cells, outliers | Gene networks are preserved in organoids. Some lines generate off-target cells (mesenchymal, hindbrain, retina). Noted a variability in cortical cells composition, presence of glycolysis related gene modules and less interneurons than in primary samples. | ||||
| Field et al. | [150] | Evolution and lncRNA | Directed: cortical | 10X Chromium | 1 ESC | X | X | X | 4 | ##### | k-mean, t-SNE | RG, Neuroepithelium, Cajal–Retzius cells | Described the cell type-specific presence of evolutionary conserved transiently-expressed lncRNA in human organoids. | |||
| Xiang et al. | [116] | Regional specification and interaction | Directed: Thalamic | 10X Chromium | 1 ESC | X | X | 2 | ##### | Seurat, CCA, Destiny? | NPC, IPC, immature neur., mature neur., glial prog., Ast., BMP-response, Cilium, proteoglycan, UPR | Darmanis et al. [77]; Thalamus (Allen Brain Atlas) | Thalamic organoids mostly produced glutamatergic neurons and beared more similarity with cortical organoids than MGE organoids, while exhibiting specific gene expression. | |||
| Giandomenico et al. | [120] | Regional interaction | Undirected | 10X Chromium | 2 ESC | X | 6 | ##### | Seurat, t-SNE, Monocle | VZ-SVZ RG, div. prog., deep-layer neur., immature upper layer neur., mature neur., IN | Camp et al. [85] | Air–liquid interface promotes survival and axon outgrowth with intracortical and subcortical identities. | ||||
| Velasco et al. | [143] | Reproducibility | Directed: Dorsal forebrain | 10X Chromium | 4 iPSC, 1 ESC | X | X | 21 | ##### | Seurat, CCA, t-SNE, Monocle | RG, oRG, div. prog., IPC, Cajal–Retzius cells, immat./mat. projection neur. (callosal, corticofugal), div. IN prog., immat.IN, U, Ast. | Nowakowski et al. [86]; Habib et al. [179], Fan et al. [89] | Dorsally patterned forebrain organoids generate similar cell type composition with low intra- and inter-line variabilities, similarly to brain tissue datasets. | |||
| Trujillo et al. | [119] | Organoid model validation | Directed: cortical spheroid | 10X Chromium | Not explicited (?) | X | X | X | 4 | ##### | Seurat, UMAP | Prog., div. Prog., IPC, Glut. neur., GABA neur., Glia, U | Functional GABAergic interneurons develop after 6 months in cortical organoids. Glial cells become more abundant than progenitors after 3 months. | |||
| Cakir et al. | [127] | Cell-subtype specification | Directed: cortical and vascular cells | 10X Chromium | 1 ESC | X | 2 | ##### | Seurat, CCA, t-SNE, Monocle | RG, IPC, cortical neur., IN, Ast., glial prog., endothelial cells and progenitors, cilium, BMP-response, proteoglycan, EMT, UPR | Zhong et al. [88] | Induction of proto-vascularization in organoids resulted in acceleration of neuronal maturation. Endothelial-like cells expressed vasculogenesis, pericytes, collagen and cell adhesion markers. | ||||
| Kanton et al. | [152] | Evolution | Undirected | 10X Chromium | 6 IPSC, 1 ESC | X | X | X | X | 36 | ##### | Seurat, CCA, t-SNE, RSS, SPRING Diffusion map, RNA velocity | Stem cells, NPC forebrain (dorsal and ventral telencephalon, diencephalon), midbrain, hindbrain and retina, EN, IN, Ast. | Nowakowski et al. [86]; Pollen et al. [151]; Miller et al. [6]; BrainSpan; Camp et al. [85]; Mora-Bermúdez et al. [149] | Excitatory and inhibitory lineages are distinguished after 2 months, and astrocytes at 4 months. iPSC lines varied in cell composition but lineage-specific signatures remained highly correlated. Human organoids present a delayed maturation compare with chimpanzee and macaque organoids. | |
| scATAC-seq C1 Fluidigm | 9 | 521 | MACS2, Cicero, t-SNE, Diffusion map | Pluripotency, Neuroectoderm, Neuroepithelium, NPC, Neuron | Human VISTA enhancer | Differentially accessible peaks with chimpanzee with cell specific pattern are enriched for single-nucleotide evolutionary change. | ||||||||||
| Bhaduri et al. | [87] | Organoid model validation | 2 directed + 1 undirected | 10X Chromium | 3 iPSC, 1 ESC | X | X | X | X | 36 | ##### | Seurat, t-SNE, WGCNA, variance partition | RG, mixed prog./neur., EN, IN, endothelial, Ast., U | 5 cortical samples (6-22 GW), 7 regions, 189,409 cells | Stress pathway is activated in organoid models and impaired cell diversity (low IPC, oRG, and upper layer neurons) and specification compare with brain samples. Yet organoids display diverse regional signatures. | |
| Qian et al. | [122] | Organoid model validation | Directed: cortical (sliced) | SPLiT-seq (nuclei) | 1 iPSC | X | 1 | ##### | Seurat, t-SNE | RG, div, IPC, Neurons Layer I, upper layer, deep layer | Zhong et al. [88]; Zhang et al. [180] | Presence of layer specific neocortical neurons and mature astrocytes. | ||||
The information aggregated in this table focuses solely on human pluripotent stem cell (PSC) i.e., ESC- or iPSC-derived brain organoid scRNA-seq data published as of April 2020. The table highlights the historic progression of scRNA-seq usage and reflects the evolution of the technology (from low number of cells at high coverage to high number at lower coverage) and methods. All study metrics (col. F-M) were reported from the published “Method” section, main text, figures, supplementary information, or tables to the best of our comprehension, as no standard for scRNA-seq report exists. Annotations of the main cell types (col. O) have been harmonized when judged equivalent. The difference in type of clusters depends on the protocol used (col D). For instance, undirected protocol generates multiple regional identities (forebrain, midbrain, hindbrain) and mesenchymal fates and regional-directed protocols focus the annotation on relevant cell types. Difficulties and inconsistencies in annotation can partially be explained by the fact that accurate unsupervised clustering (col N) is difficult and its level of resolution is highly dependent on the question central to the study. This highlights the importance of validating annotations though some form of correlation analysis with scRNA-seq or RNA-seq from human fetal brain references (col P).
Abbreviations for stages (col G-K): P pluripotent stem cell stage, D differentiation stage (below 1 month from the beginning of the protocol), E Early (1–3 months), M Middle (3–6 months), and L Late stage organoids (>6 months).
Abbreviation for methods (col N): NN-Louvain Nearest-neighbor graph combined with Louvain clustering method (also used in Seurat), CCA canonical correlation analysis for batch effect, WGCNA weighted correlation network analysis, RSS reference similarity spectrum.
Abbreviations for cluster annotations (col O): NPC neural precursor cells, NEC neuropithelial cell, neur. neurons, div. dividing/cycling cells, RG radial glia, IPC intermediate precursor cell, IN interneuron, Ast. astrocyte, dopa dopaminergic, prog. progenitor, Glut. glutamatergic, OPC oligodendrocyte precursor cell, Choroid Choroid Plexus cells, Oligo. oligodendrocyte, U unknown/undetermined/unlabeled, EN excitatory neurons, UPR unfolded protein response related, VZ ventricular zone, SVZ subventricular zone, (im)mat. (im)mature, EMT epithelial-mesenchymal transition cells, int. intermediate.
Box 1 Glossary.
| Transcription factors (TFs) | Proteins that bind specific target DNA sequences, located within enhancers or promoters, contributing to the regulation of RNA transcription of specific gene(s). |
| Promoter | DNA regulatory region typically present upstream of the transcription start site (TSS) of a gene. RNA polymerase recruitment at those loci triggers the transcription of the gene(s). |
| Enhancer | DNA element that modulates the transcription by binding TFs and bringing them in physical interaction with the cognate promoter(s) of gene(s). In contrast to promoters, enhancers act independently of the distance (up to hundreds of kilobases or even megabases) and orientation to the respective target gene(s). They are characterized by accessible chromatin devoid of nucleosomes and flanked by nucleosomes with specific histone modifications (H3K27ac and/or H3K4me1). |
| ChIP-seq | Chromatin immunoprecipitation followed by sequencing is a method for finding DNA–protein interactions by combining immunoprecipitation and DNA-sequencing. Can be applied to TF to validate their binding sites or to covalent histone modifications (e.g., H3K27 acetylation, H3K4 methylation) to identify putative regulatory regions—a process referred to as chromatin segmentation—and their differential activity. |
| RNA-seq | RNA-sequencing is a next-generation sequencing method that provides an overall unbiased quantification of RNA content, which can be refined by ribosomal RNA depletion or mRNA enrichment before cDNA synthesis, library preparation, and sequencing. The relative abundance of transcripts obtained constitute the input to evaluate differential gene expression between samples or conditions. |
| scRNA-seq | Single-cell RNA-sequencing; a technology that allows the gene expression analysis at single-cell resolution. Isolating individual cells, this technique highlights transcriptional differences between cells of the same biological sample, otherwise obscured using only the bulk RNA-seq analysis. |
| Hi-C |
Hi-C is based on the chromosome conformation capture (3C) technology and it is used to detect, in a genome-wide manner, the chromatin interactions inside the nucleus. Chromatin is crosslinked with formaldehyde; the spatially close genomic fragments are ligated generating chimeric DNA fragments which are captured and identified by deep sequencing. |
| TAD | Topologically associate domains (TADs) are 3D chromosome structures, whose boundaries are relatively conserved while their internal conformation can change depending upon epigenetic marks and nucleosome structures. Within TADs, chromatin looping brings together regulatory element in close proximity (i.e., enhancers with promoters). |
| MPRA | Massively parallel reporter assay; method that provides a quantitative measurement of the activity of thousand potential DNA regulatory sequences in parallel. A library of candidate sequences cloned upstream of a promoter/reporter construct are transfected into a cellular model system and identified by deep sequencing. |
| STARR-seq | Self-transcribing active regulatory region sequencing; like MPRA, this technology assesses the activity of thousand potential regulatory regions, in a cellular model. Candidate sequences are cloned downstream of a promoter/reporter construct. Potential regulatory regions are self-transcribed and identified by RNA-seq to measure candidate regions’ activity. |
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats. Cas9 is an endonuclease that can be directed to the target region by a synthetic guide RNA (sgRNA) complementary to the target causing a DNA strand break. A mutated version of Cas9 without endonuclease activity (dead or dCas9) if fused with activator or repressor domains can induce chromatin changes leading to inactivation (CRISPRi) or activation (CRISPRa) of the target regions. |
| GWAS | Genome-wide association study. It is an observational study of a set of genetic variants (genome-wide) in different individuals to see if any variant is associated with a specific trait. Variants are detected by microarray or whole-genome sequencing technology. |
| eQTL | Expression quantitative trait loci. Regions of DNA in which genetic variation is associated with variability in the expression of one or more genes. |
| DNase-seq | A method based on DNaseI hypersensitivity for identifying accessible regions of the genome. |
| ATAC-seq | Assay for transposase accessible chromatin; a method for identifying accessible regions of the genome, based on transposase activity. |
| WGCNA | Weighted gene co-expression network analysis identifies modules of genes which exhibit correlated patterns of gene expression across samples and often represent similar cell types. Trends in the network can be summarized by eigengenes or hub genes that are central to the network structure. |
Omics and big datasets
Genomics and epigenomics
The development of cost-efficient sequencing in the last 10 years has drastically accelerated the reproducible generation of high throughput nucleic acid sequence data (hereafter referred generally as Omics). These include genomic variant discovery, like SNPs, by whole-exome or whole-genome sequencing (WGS). Transcriptome analysis by RNA-sequencing (RNA-seq) profiles global expression patterns of distinct RNA species, including messenger RNA (mRNA), microRNA or long noncoding RNA (lncRNA) and their eventual posttranscriptional modification, like alternative splicing or RNA editing. Finally, ChIP-seq directed at histone modifications and Hi-C or ATAC-seq that inform about chromatin structure, can be used to evaluate epigenomic changes (Fig. 1b) (Glossary in Box 1).
Large-scale Omics datasets are key to establish exhaustive repertories of cellular and molecular features of normal and diseased-affected brain development, estimate the validity and fidelity of in vitro models and feed in silico drug discovery. Several public–private research initiatives, national or international (e.g., NIH, EU, Simons Foundation, Allen Institute for Brain Science), have funded collaborative efforts to catalog and analyze genomic, transcriptomic and epigenomic data of cells and tissue in human, nonhuman primates, and model organisms. Data have been uploaded in open repositories such as Gene expression Omnibus or in user-friendly databases to allow further analysis by different groups and enable cross comparisons.
It is now known that many neurodevelopmental and neuropsychiatric disorders, such as autism spectrum disorders (ASD), bipolar disorder, or schizophrenia, are genetically heterogeneous and do not always follow a Mendelian inheritance [13, 18, 19]. Instead, such polygenetic disorders arise from multiple causative variants, both common and rare, and complex environmental factors. Genome and exome sequencing studies in patient cohorts and normal individuals have identified potential causative genetic variations of all sizes, from SNPs to large structural variation in the DNA [13, 20, 21], however the major challenge is to interpret their functional impact. The importance of sharing genetic and phenotypic data has fostered global networks since 2015, like Matchmaker Exchange, Decipher and GeneMatcher [22, 23], in which patients with similar genetic variants and/or phenotypes were matched. Other essential resources have been developed and implemented, including databases of diseases-associated genes, like OMIM and ORPHANET [24], or clinical interpretation of variation, like ClinVar or ClinGen [25, 26], initiating a move towards precision medicine and research. Population-wide cataloging of genetic variation, like gnomAD, has allowed the exclusion of variants that were too common at the population level to be plausible causes of diseases [27].
For complex diseases, numerous sequencing-based gene association studies have been done to link phenotype differences with variant allele frequencies. Genome-wide association studies (GWAS)—the first dating back to early 2000s—have been charting common genomic variants across the genome in individuals with or without the disease, initially using genome-wide SNPs arrays and more recently WGS. GWAS studies have demonstrated that most of such variants are found in noncoding regions of the genome, 60% associated to enhancer and super-enhancer, and so are more likely to be involved in gene regulation [28]. This approach has led to the recent identification of common risk variants for schizophrenia, albeit it required more than 36,000 subjects and 110,000 controls [29]. A GWAS effort focused upon neuropsychiatric disorders is the Psychiatric Genomics Consortium that covered 11 psychiatric disorders including attention-deficit/hyperactivity disorder, Alzheimer’s disease, ASD, bipolar disorder, eating disorders, major depressive disorder, obsessive-compulsive disorder/Tourette syndrome, post-traumatic stress disorder, schizophrenia, substance use disorders, and all other anxiety disorders [30]. Of note, the NHGRI-EBI published a catalog of GWAS (ebi.ac.uk/gwas). More focused consortia subsequently developed, for instance the Brain Somatic Mosaicism Network (BSMN), aimed at studying somatic mosaicism both in neurotypical postmortem human brains and in schizophrenia, ASD, bipolar disorder, Tourette syndrome, and epilepsy patient populations. The BSMN aims at cataloging the frequency and pattern of somatic mutations, which are not inherited but occur during the life. To overcome the challenges inherent to discovering somatic mutations, which are present at low frequency in a subset of an individual’s cells, members integrate a variety of complementary approaches which include clonal analyses, deep coverage DNA-sequencing, single-cell genomics, and cutting-edge bioinformatics, while the BSMN enables a cross-platform integrated analysis with other omic-datasets [31–33].
Several brain transcriptomic studies have been performed in both human and nonhuman samples offering the opportunity to probe the molecular basis of neuronal function, understanding its developmental process, and discovering conserved evolutionary mechanisms or diversity between species (reviewed in [34]). The analysis of transcripts has proved challenging, as RNA is more unstable than DNA, especially for postmortem samples. Yet, modification of transcriptional activity remains the core way to link genomic variants to alteration in gene expression through identification of expression quantitative trait loci (eQTL) and other analyses (Fig. 1). Pioneer transcriptome studies were performed in human, macaque, and mouse brain regions across lifespan by microarrays, highlighting time-dependent, layer-, region-, and species-specific features in cortical gene expression profiles [6, 35–41]. Larger transcriptomic datasets were soon generated using RNA-seq by several consortia (e.g., Allen brain Map, BRAINSPAN) (see ALLEN BRAIN ATLAS data portal: https://portal.brain-map.org/) from multiple cell lines, human brain regions at mid-fetal and adult stages, and from embryonic and adult mouse cortex. The GTEx project [42, 43] generated transcriptome datasets across tissues in many “normal”, non-diseased tissues and each donor was genotyped for common SNPs, creating one of the biggest eQTL studies. Such studies have offered not only the characterization of variation in gene expression levels, but also its link to genetic variants and the basic process of gene regulation. With its extension in 2017, Enhancing GTEx proposes to integrate previous data with telomere length, DNA accessibility, histone modifications, DNA and RNA methylation, somatic mutation, allele-specific expression, and protein quantification across individuals [44].
Another important role of these multicentric studies was to investigate the exquisite gene regulatory mechanisms upstream to the transcriptome, through epigenomic studies. The Encyclopedia of DNA Elements (ENCODE) consortium offered the first functional annotation of regulatory elements in the genome, both coding and noncoding and systematically in human, mouse, fly and worm. To date, the Encode includes 10,868 projects and several bio-sample types. Through many assays (DNA binding, accessibility, methylation, transcription, RNA binding, replication timing, and 3D chromatin structure) the ENCODE performed the first segmentation of the human genome into different categories of functional elements [45]. These include active enhancers, which are typically enriched in H3K27ac-labeled histones; poised enhancers, which exhibit H3K4me1; promoters, which are associated with H3K4me3; and repressed chromatin, associated with H3K27me3.
Finally, our group has taken part in an integrative omics analysis initiative called the Psychiatric Encyclopedia of DNA Elements (PsychENCODE), a consortium focused on understanding gene regulatory mechanisms in the human brain [46]. This is in contrast to the ENCODE, that focused largely on human cell lines. The PsychENCODE consortium has generated a comprehensive online resource (http://www.psychencode.org/) of transcriptomic, epigenomic, and genomic data from postmortem developing and adult human brains, both normal and diseased (schizophrenia, ASD, and bipolar disorder), and human cellular model systems. Three main research areas were pursued: dissecting human brain development, studying disease transcriptomes and its regulation, and finally integrating bulk tissue and single-cell data with deep learning approaches to deconvolute the unique features of the human brain. In the following sections, we present some of the main findings of the consortium and related research.
Emergence of single-cell omics
The recent years have seen an explosion of technologies to study genome, transcriptome, and epigenome at the single-cell level [47]. This advancement was enabled by improvements in single-cell isolation and barcoding techniques, coupled to a reduction in sequencing costs. Many single-cell isolation methods now exist, each having different advantages and caveats [48–51]. Alternative methods that rely on combinatorial barcoding to identify single cells without requiring physical isolation have also been applied successfully to neural tissues [52].
Single-cell RNA-sequencing (scRNA-seq) generates transcriptomic signatures of hundreds to millions of single cells, revealing both cellular composition (Fig. 1d) and cell-type-specific gene networks in pluricellular structures such as brains or organoids (Table 1) [47]. The quality of scRNA-seq is highly dependent on correct cell isolation, to avoid doublets (two cells instead of one), and unbiased transcript capture and amplification from each cell, to avoid representation artefacts. Indeed, most techniques capture only a limited fraction of the cell’s RNA content, leading to transcripts dropout. This unavoidable stochastic loss of transcripts requires the aggregated analysis of multiple cells to recover statistically significant information. Consequently, scRNA-seq output describes the state of cell subpopulations and not of single cells per se [47]. Although sequencing coverage is an important parameter, it has been shown that low coverage scRNA-seq (i.e., 50,000 reads per cell) is enough to identify and reconstitute cell diversity in the developing cortex [48]. While most single-cell platforms analyze only the three-prime end of mRNA, some library preparations allow to sequence the full-length mRNA transcript, improving sensitivity in isoforms detection [53].
Bioinformatic analyses of scRNA-seq have become highly complex and are still under active development (reviewed in [47, 54]). For best practice recommendations and a workflow in scRNA-seq data analysis, see [55]. While an extensive overview of current analytical steps is beyond the scope of this review, we wanted to highlight some commonly used tools. The Seurat package (developed in R by the Satija Lab) [56] has become the most commonly used in our field (referenced in 13 out of 19 scRNA organoid studies listed in Table 1) largely due to its centralized handling of the scRNA-seq analytical pipeline, including normalization, batch-effect correction, clustering, visualization, and multifeature integration [57]. Visualization is an important part of scRNA-seq interpretation and mainly relies on nonlinear dimensionality reduction (e.g., t-SNE or UMAP) to reduce the data to a 2D plot of single cells. In developmental datasets, such as fetal brain or organoids, cells are evolving along lineages (e.g., from RG to neurons). This hidden dimension can be revealed along a pseudo-time or pseudo-differentiation axis projection through trajectory analysis [58], appropriate visualization tools [59, 60] (see below, Fig. 1d and Table 1) and further verified by elegant methods such as RNA velocity analysis [61].
Improvements in scRNA-seq analytical tools are still required, especially to merge together the increasing number of datasets generated across multiple technologies and studies, including integration with multiple other cellular features. Indeed, similar to bulk methods presented in the previous section, single-cell studies are progressively becoming multimodal, capturing multiple information from the same sample or even from the same cell, including spatial, epigenomic, morphological, immunophenotype, DNA sequence or mutations, or even electrophysiological [62, 63]. Spatial transcriptomics is a recent development that allows capturing transcriptomic data from a given location in a tissue slice while retaining spatial information close to single-cell resolution [64]. Similarly, profiling single-cell epigenetic information, such as open chromatin state through scATAC-seq or DNA methylation, opens a new feature of classification of cell diversity [65]. Finally, the simultaneous collection of electrophysiological (e.g., patch-clamp or calcium imaging) and transcriptomic data from the same neural cells constitutes an important innovation for neuroscience [66, 67].
Genomics trajectories of the developing brain
Reconstituting human neural development from postmortem human tissues though transcriptomic, epigenomic, and integrative analyses: the PsychENCODE Consortium
The PsychENCODE project is aimed at defining a comprehensive map of functional regulatory genomic elements active in the human brain, differently than the ENCODE project, that mainly focused on peripheral and/or transformed cell lines [45]. The main success of PsychENCODE [46] has been the multi-omic approach that allowed a systematic characterization of noncoding elements, along with the transcriptome, in neurotypical developing and adult brains, in individuals with neuropsychiatric disorders and in human cellular models [12, 68–73]. Coupled with other notable recent studies in developmental genomics [74, 75] and single-cell studies (discussed below) this resource provides new insights into the biology of brain development and its diseases.
Among the main findings, it was observed that the overall transcriptomic signature of all brain regions undergoes a sharp transition phase between mid-fetal and late-fetal stages, suggesting that major changes occur around birth [68]. This temporal trend was accompanied by a transient drop in interregional variability. This suggests that cortical regions become more similar around birth and that adult region-specific signatures arise mainly after the late infancy stages. Part of these dynamic changes could be explained by a regional variability in cell type composition, including differences in progenitor populations during the prenatal stages and differences in mature cell types and functional diversification during later postnatal stages [68, 69]. Different levels of alternative splicing contributed to the overall transcriptional variability over time and space. As splicing dysregulation has been shown to be involved in neurodevelopmental diseases, including ASD, schizophrenia, and bipolar disorder [71], this highlights the importance of studying alternative splicing in early development.
Spatiotemporal variability was also described at the epigenomic level. Major changes in chromatin accessibility as assessed by ATAC-seq between the germinal zone and the cortical plate in fetal cortical samples reflected the transcriptomic changes that happen during neurogenesis [74]. The study also associated putative enhancers to their corresponding TF using binding sites enrichment analysis, confirming that the germinal zone accessible regions are enriched in binding sites for TF implicated in neural progenitor specification (i.e., PAX6, SOX2, ARX, EMX1/2, LHX2, etc.), although the study stopped short of comprehensively defining actual enhancers. This is important because many studies, including large-scale chromatin conformation analysis in mid-gestation brain samples, have revealed that most enhancer–promoter interactions within topologically associating domains (TAD) were long range and not with the adjacent genes [75].
Overall, there is a good concordance between DNA methylation, histone marks, and gene expression over brain development [68]. For instance, enhancers active during the fetal period were associated with genes linked to neural development functions, and became hypermethylated over the postnatal period, heralding the expected decrease in target gene activity. Genome-wide, chromatin accessibility correlated relatively well with gene expression, both at transcription start sites (TSS, r = 0.417) and at regulatory regions (putative enhancers), especially when the latter were defined using Hi-C chromatin interactions (r = 0.456) [74, 75].
All those studies constituted the bases of an integrative model for the discovery and interpretation of functional genomics of the adult human brain within the PsychEncode consortium [69]. This included adult brain bulk transcriptome, chromatin, genotype, and Hi-C and single-cell datasets from major human brain regions and merged these datasets with others available through GTEx, ENCODE, and Roadmap Epigenomics (see Fig. 1). All the datasets were uniformly processed to create many fundamental resources, including a list of brain-expressed genes, co-expression modules, 79,000 brain-active enhancers, and their putative targets; more than 2.5 million eQTLs, including relationship with splice isoforms, cell fractions, and chromatin activity. The study generated a brain gene regulatory network where TFs, enhancers, and target genes are linked to each other, based on QTLs, element-activity correlation and Hi-C data. Disease genes were linked to GWAS variants for psychiatric disorders. The regulatory network was used as an input for a machine-learning model to predict psychiatric phenotypes, giving back a threefold increase in prediction compare with other models, highlighting the value of having both epigenome and transcriptome data. This integration remains to be extended to the developmental brain and functionally validated. Interestingly, the integrative model revealed that cell composition is the major contributor to the overall developmental trajectory signature of the human brain, a central result that was only possible to obtain by using single-cell resources.
A parallel longitudinal in vitro study of iPSC-derived organoids and fetal brains (described in more detail below) generated a dataset of roughly 96,000 enhancer elements active in early brain development and linked to genes by chromatin conformation analyses, and demonstrated a good correlation between enhancer activity and gene expression along neural differentiation (see below, Fig. 1 and [70]). Altogether, this integration between transcriptome and epigenomic studies demonstrated the validity of multi-omic approaches to deconvolute complex processes like neurodevelopment.
Single-cell transcriptomic of postmortem developing brain samples
Single-cell nucleus analysis of adult human cortex have been successfully applied to decipher human-specific diversity and organization (e.g., [76, 77]), such as transcriptomic signature of multiple cortical areas [78] and integrated analyses between transcriptomic and epigenomic signatures [79]. In addition, scRNA-seq has revealed cell-type-specific alterations in multiple human brain disorders, like ASD [80], glioblastoma [81], multiple sclerosis [82, 83], or Alzheimer’s disease [84].
Single-nucleus analysis of fetal neocortex brain tissue from different post-conceptional weeks (PCW) and gestational weeks (GW) were described in a seminal series of publications by the Kriegstein group and others (GW 16 in Pollen et al. [48]; PCW12-13 in Camp et al. [85]; PCW 6-37, in Nowakowski et al. [86]; GW6-22 in Bhaduri et al. [87]). Other groups have refined and extended the single-cell spatiotemporal dynamics (GW 16-18, Darmanis et al. [77]; GW8-26 in Zhong et al. [88]; PCW 22-23 in Fan et al. [89]; 5-20 PCW in Li et al. [68]; GW17-18 in Polioudakis et al. [90]). Although the neocortex captures most of the attention, similar valuable datasets have been produced for other regions, such as the hippocampus (GW 16-27, [91]) or the embryonic ventral midbrain (PCW 6-11, La Manno et al. [92]). The quality, number of cells, and depth of analysis in those studies have followed the advances in single-cell technology. Many of these studies confirm decades of neurodevelopmental research: from the organization into ventricular, subventricular, subplate, and mantle zones, the order in which neurons of each cortical layer are specified over time, to the switch towards gliogenesis in the later stage. The full diversities of cell type transcriptomes are being characterized, covering not only RG, intermediate progenitor cells, inhibitory, and excitatory neurons, but also astrocytes and oligodendrocyte precursor cells, microglia, choroid plexus cells, mural cells, and endothelial cells [86].
In addition to cataloging the transcriptomic signatures of every single population, scRNA-seq brings the promise of refining or giving a new take on important unresolved questions of forebrain development biology in human brain, and specifically the most evolved regions such as the cerebral cortex.
Identifications of the outer- and truncated-RG subtypes (oRG, tRG) have received a particular attention for their hypothesized impact on human-specific brain size and morphology. Multiple studies have now established their molecular profiles, such as the oRG marker HOPX [93–96]. Interestingly, among the RG subpopulation, oRG presents an enrichment of the mTOR pathway, which has been implicated in pathological conditions related to dysplastic growth and defective cortical migration with co-morbid epilepsy such as focal cortical dysplasia type 2 and hemimegalencephaly [86, 97–99]. Another interesting aspect highlighted by these recent studies is that humans gained enhancers (referred to as HGE) which increase in activity over the course of human evolution, seem to preferentially target genes expressed in the oRG and regulate their growth during neurogenesis [74].
Another important question that has been debated in the mouse literature is the establishment of cortical patterning following either a premitotic model (specialized progenitors) or a postmitotic model (common progenitor but specialized neurons). By comparing visual and prefrontal cortex areas, Nowakowski et al. [86] noted that RG do not present regional transcriptional signatures but show evidence of a progressive divergence in gene networks during neurogenesis. They also noted that those two areas seem to mature at different speeds. This asynchronicity was also observed in another study analyzing 20 different areas, where the authors also observed different proportions of interneurons across the cortical regions [89]. This is in apparent contrast to a more recent study that found cortical areal signatures already present in RG progenitor cells [87].
The development of cortical layers and subpopulations of excitatory neurons seems to go through a phase where immature neurons expressed combination of genes, in particular TFs, that are known to be expressed by distinct cortical layers in adult neurons [68]. An example is the co-expression in embryonic and mid-fetal excitatory cells of BCL11B (CTIP2) and FEZF2 (both known markers of layers V/VI) with CUX2, an upper layer marker (layers II/IV). Another example is the co-expression of RELN and PCP4, specific for layer I and deep layers, respectively. This suggests that human neuronal cell types could be very malleable during early postmitotic differentiation and their molecular identities not completely resolved before the end of mid-fetal development.
The origin and establishment of interneurons diversity in the neocortex have received major attention and there is still a controversy regarding the capacity of dorsal cortical RG to generate interneurons [100–103]. It seems that similarly to excitatory neurons, different proportion of interneurons subtypes populate different cortical areas [89]. There is an early presence of GABAergic progenitors and SST and CALB2+ interneurons in the cortex [88, 89] and one study suggested that some SST-expressing cells could originate from the cortex around GW7 although without entering the cell cycle [88], but there is no evidence of interneurons progenitors later in the neocortex at GW17 [90]. Hence, generation of interneurons from the human neocortical neuroepithelium remains controversial.
Using GW13 to 23 samples, Liu et al. [104] cataloged the expression of lncRNAs in human neocortex development and applied it to detect 1400 lncRNA in single cells from four neocortex samples (19–23 GW) and in previously published samples (Pollen et al. [93]). They resolved cell-type-specific expression (e.g., higher expression of MEG3 or DLX6-AS lncRNA in interneurons compared with excitatory neurons), showed that some lncRNA that are barely detectable in bulk tissue are enriched only in certain cell types, and showed examples of a lncRNA regulating proliferation in RG.
The availability of scATAC-seq allows the mapping of regulatory elements in a cell-type-specific fashion. This is particularly important since regulatory elements exhibit far more cell-type specificity than genes [105, 106]. Open DNA (i.e., DNA accessible to transcriptional regulation) and the transcriptome can then be intersected across single cells [107] and used to infer gene regulatory network at the cell level [90]. This information can be integrated with cell-type enrichment of TFs and co-factors, and intersected with published epigenomic datasets [74]. Using disease variants datasets and scATAC-seq datasets, it is now possible to identify cell type enrichment not only in neurodevelopmental disorder risk genes, but also in SNPs within enhancers active in particular cell types [90, 108].
Finally, it is now possible to link transcriptomic information to functional heterogeneity, as Mayer et al. [67] demonstrated by coupling scRNA-seq with calcium imaging in dissociated cells from mid-gestational (PCW14-22) cortical plate, subplate, and germinal zone.
Altogether, multi-omics and single-cell information of developing brain samples constitute an invaluable resource. In addition to allowing to reconstitute the dynamics of brain development, they represent a necessary reference to validate and improve results obtained using human in vitro models, such as brain organoids.
Brain organoid, an in vitro model to validate features of human brain development
Although a diversity of in vitro models exists relying on human cells, we will focus on recent developments in the organoid field, presenting the incremental improvements of protocols, the multi-omics characterization and integration with brain data and the main conclusions from 5 years of single-cell study on many aspects of organoids biology (Table 1). Finally, we review innovative approaches for characterization of noncoding elements which, in our opinion, could leverage the power of organoids to answer long-standing questions of human genetics posed in the first part of this review.
Diversity of organoid protocols and applications
Organoid protocols can be separated in three major types—undirected, directed, and patterned—which reflects the extent to which molecular cues are used to guide neural differentiation. Each protocol has benefits and limitations to model different aspects of brain development. Undirected protocols rely on the capacity of neural progenitors to self-organize and yield multiple regional fates [16]. On the contrary, directed organoids take advantage of morphogen agonists or antagonists to mimic developmental cues, guiding cell fates during neural tube patterning in vivo [15, 109–112]. This encompasses canonical signaling pathways such as BMPs, TGFß, Wnts, or SHH. Although at first many directed protocols focused on obtaining telencephalon and neocortex in particular [17, 110, 111], there now exist a full repertoire of protocols to generate regional organoids, including hippocampus [113], cerebellum [114], midbrain [115], thalamus [116], and others [117]. Finally, spatial patterning of organoids is a recent addition to the field where a local molecular signal, for instance SHH, allows long distance spatial organization inside the organoid, a process mimicking the morphogen gradients fundamental to establishing positional identity during development [118].
Maintenance of healthy organoids over a long period of time is crucial for the emergence of spontaneous neuronal activity and network oscillation patterns [119]. This time requirement and the aspiration to mimic other in vivo aspects has led to new inventive methodologies: moving from static towards dynamic culture systems (e.g., spinning bioreactors [112] or SpinΩ [117], growing organoids at the air–liquid interface [120] or even incorporating engineered microfilaments [121]). Recently, Qian et al. demonstrated that cultivating thick organoid sections—instead of culturing whole organoids—improves nutrient access, decreases necrosis in the organoid core and results in an extended formation of most human neocortical layers [122]. In parallel, other groups have focused their efforts on obtaining all neural cell types, including astrocytes [123] or oligodendrocytes [124, 125]. Important for favoring neuronal maturation and energy exchanges, vascularization has been modeled by transplanting organoids in mouse brains [126] or by incorporating external mesodermal sources [127, 128]. Proper angiogenesis and reproducing vascular cues will certainly improve both longevity and fidelity of neural organoids since in mouse they strongly influence neurogenesis dynamics in a region-dependent manner [129]. Microglial cells are another mesodermal cell type that is a key player of neural development and are difficult to obtain in brain organoids. While it has been reported that an undirected protocol can yield some microglia [130], others proposed to rely on an external source [131–133]. Finally, to study interregional migration and connectivity over development, several studies work on a fusion of different regionally directed organoids, baptized assembloids [116, 134, 135]. Interestingly, human cells within organoids can form electrophysiologically active connections with mouse cells in xenografts, as nerve tracts have been modeled using co-culture with mouse spinal cord-muscle explants [120, 126]. Altogether, these studies show that the organoid research field is innovating new approaches at a rapid pace to explore what can be done using this model system.
In addition to studying normal development, the organoid model has also been successfully used to characterize the neurological impact of many conditions, including the Zika virus [117], drug exposure [136], syndromic mendelian gene mutations (e.g., in Rett syndrome [137] or Timothy syndrome [138]), and common idiopathic neuropsychiatric disorders such as ASD [110, 139] and schizophrenia [140, 141]. For this, organoids have the unprecedented advantage to be able to model and manipulate pathological mechanisms in a controlled human neural system, deciphering their impact on a large range of features in a dynamic manner (Fig. 1a, c), which is difficult to achieve in other relevant models such as postmortem human brain.
Multi-omics integration of organoids with the developing human brain
The correspondence between organoids and normal brain development has been difficult to investigate, in part due to scarcity of human brain samples, especially at the early stage of development, as well as genetic heterogeneity. The analysis of gene regulation in organoids has been pioneered by Amiri et al., who performed an integrative analysis of enhancer activity and gene expression in organoids derived from fetal skull fibroblasts of three individuals, and compared it to the isogenic fetal human brains [70]. Gene expression dynamics were assessed by RNA-seq during the transition from stem cell to neuronal progenitor cells and from progenitor to neurons, and reflected cell cycle exit, increase in neuronal differentiation, transcriptional regulation in cortical precursor cells and increase in synaptic transmission, cell adhesion, and axon guidance. Transcriptome comparison between organoids, isogenic brains and the PsychEncode developmental dataset showed that organoids mapped more closely to human cortex before 15 PCW.
The study also included cortical brain tissue from the same subjects (15–17 PCW), as internal reference tissue. In comparison with organoids, brain samples were enriched in more mature neuronal transcripts while depleted in transcript related to RG and cell division. Noncoding elements (enhancers, promoters, and repressed chromatin) were mapped by ChIP-seq for several histone posttranslational modifications. Comparison in enhancer number and activity between organoids and brains revealed 1.8 more enhancers in organoids, as 59% of the enhancers still active in organoids were already inactivated in the mid-fetal brain samples.
Overall, this study identified over 300,000 putative enhancers active in organoids and fetal brains during development [70]. Proximity and chromatin conformation analyses were used to link these putative enhancers with their target genes. About 30% (96,375) of the enhancers, among which 10% were novel, could be associated with protein coding genes. Among the ~96,000 gene-linked enhancers, 35% were shared with the isogenic human cortex. Based on correlations between enhancer activity and the expression level of their associated genes, enhancers were cataloged into potentially activating (A-reg) or repressing (R-reg) regulators of gene expression. This classification was reflected in A-reg and R-reg being significantly enriched in genes respectively upregulated or downregulated over time.
Gene expression and enhancer activities were then modeled into a weighted gene correlation network (WGCNA) encompassing 54 co-expressed gene modules, and 29 co-active enhancer modules with specific activity profiles and biological annotations consistent with the organoid’s developmental trajectory [70]. Over 24% of the SFARI ASD-associated genes were differentially expressed in the organoids over time, and 80% were associated to enhancers active in both organoids and fetal brain. Genes associated with ASD by postmortem transcriptome analyses were significantly overrepresented in three gene modules related to synapse development and the regulation of cell proliferation. Similar enrichment was observed in the corresponding gene-associated enhancer modules, most of them showing an upregulated activity across development.
Organoids have also been shown to be a promising system to study the genetic mechanisms driving human brain evolution. Over 60% of the human gained enhancers [142], those set of enhancers that possess increased activity in early human brain development compared with rhesus macaque and mouse brains, were active in organoids, particularly at the earliest stages, and showed decreasing activity along differentiation. This evidence suggests that organoids can capture dynamic gene regulatory events, pointing them out as potentially involved in brain neurodevelopmental disorders.
The findings by Amiri et al. provides the most comprehensive integrative analysis of gene-enhancer interactions in human brain organoids so far, where enhancers were identified by a combination of peak calling (ChIP-seq) and chromatin segmentation, followed by identification of interacting gene-enhancer pairs achieved by cross-reference with Hi-C data of human fetal brain. One of the most notable results was the definition of a convergent gene and enhancer network defining global pattern of expression and activity along trajectories of neural cell differentiation and maturation. These enhancers harbored mutations found in ASD probands from external datasets, suggesting that organoids may provide a system to better understand the functional impact of disease-associated risk variants located in noncoding regions of the human genome and their potential to disrupt certain TF binding sites [70].
Exploiting scRNA-seq to study organoids cellular composition and gene networks
Brain organoids are a multi-cellular 3D model by definition. Therefore, the characterization of organoid models depends on identification of cell types and structures obtained, often by immunohistochemistry using antibodies against known markers of neural development and regional identities. The scRNA-seq technology was used early on to unbiasedly identify cellular diversity and lineage in organoid models [85] and has now become a standard for the field. This has led to a recent increase in the organoid single-cell transcriptomic data available. In the 19 studies that we have reviewed and listed in Table 1, the field has accumulated transcriptomic information over close to 800,000 cells, encompassing multiple relevant stages and types of human organoids.
Overall, organoid models seem to reproduce the mechanisms and temporal dynamics of neural system development, which can be observed both in cellular composition over time and gene expression networks (Table 1). Notably, the systematic presence of cell clusters with expression signatures typical of RG (marked by PAX6, NESTIN, SOX2, etc.), intermediate precursor cells (marked by EOMES/TBR2) and different subtypes of excitatory neurons (marked by TBR1, BCL11B, SATB2, CUX1, etc.) confirm the presence of cortical neurogenesis in most models. Interestingly, the ability to clearly distinguish between multiple subtypes of cortical neurons, including Cajal–Retzius cells/layer I neurons and lower/upper layer projection neurons, seems to be dependent on a sufficient period of culture [122]. As in vivo, astrocytes and oligodendrocytes require an extensive period of development to be generated (at least 8 months for “mature astrocytes”) [123–125, 135]. Despite their subpallial origin, it is also surprising to note that a GABAergic lineage of interneuron is observed in cortically driven organoids [110, 119, 135, 143, 144], although they become more abundant when specifying ganglionic eminence fate. Finally, there seems to be other lineages emerging in many models, such as choroid plexus cells and ependymocytes of the VZ, although there is less clear consensus on their annotations (Table 1). Since many studies specifically use cortical organoids, a consensus on annotation strategies and markers of reference could be established in future work.
Despite this overall fidelity in reproducing brain cellular lineages and gene networks, Bhaduri et al. [87] recently exposed a divergence in the specification and maturation of different cell subtypes between organoids and brain samples (GW6-22). They link decreased neuronal maturation with an aberrant activation of oxidative and stress pathways in vitro, including glycolysis and endoplasmic reticulum stress, which could suggest improvements of organoid culture conditions (e.g., modifying glucose concentration or oxygen level).
Variability remains a major limitation of organoid models. Undirected brain organoids seem to present a higher intra- and inter-organoid regional variability, perhaps due to stochastic organization, with not only forebrain identity, but also regions such as retina, spinal cord, and others [16, 145]. Study of cortical development in those models often requires micro-dissection of cortex-like areas that can be visually identified from the rest of the organoid, although extracted regions can end up having non-cortical identity [85]. Dorsal forebrain patterned organoids seem to present less organoid to organoid and line to line variability [143, 144], although some variation, for instance in cortical areal identity pattern, have been reported [87]. The assessment of organoid cell composition variability, both between lines and between batches, is an important step relevant to statistical modeling and differential gene expression analysis. Notably, many published scRNA-seq studies are generated from 1 or 2 biologically distinct ESC or iPSC lines (14 out of 19 datasets in Table 1), and with rare intra-line variability estimation.
Integration efforts, especially across the diversity of existing protocols, should generate meaningful transversal conclusions. One limitation of the integration is the batch correction step when dealing with diverse datasets in terms of isolation technology, library preparation, and read depth level per cell, with data origin often driving the clustering [87]. Using canonical correlation analysis, Tanaka et al. recently integrated eight datasets from different studies containing both directed and undirected protocols and found similar cellular composition and gene expression per lineage [146]. With recent improvements in batch correction and integration methods [65, 147, 148], such transversal analysis should become more accessible, and lead to establishing a common single-cell transcriptomics organoid atlas defined through common marker genes and reference datasets. Such integration will lead to a clear transcriptomic definition of the cellular space of in vitro brain organoids, leading to a protocol-independent definition of artefactual cell types and structuring the diverse lineage trajectories. Such integration is vitally important to determine inter- and intra-protocol variability in cell fate.
Finally, evolution has received a particular interest in single-cell organoid studies, owing to the capacity of generating iPSCs and brain organoids from multiple primate species, including chimpanzee, orangutan, and rhesus macaque [149–152]. It was demonstrated that compared with other apes, human organoids present a delayed maturation, with less mature neuronal signatures and astrocytes presence at equivalent stages, which agrees with the longer gestational period in humans [152]. Interestingly, transcriptomic divergence in the telencephalic lineage seems to consist mainly of gains in new gene expression in humans, with related functions spanning proliferation of RG, neuron migration, and neurite formation [152]. Using scATAC-seq in complement to scRNA-seq, Kanton et al. also showed that differentially accessible peaks between human and chimpanzee have a cell-type-specific pattern and are enriched in single-nucleotide evolutionary changes. Coherently, evolution seems to affect gene networks differently in different neural lineages, with major changes occurring in astrocytes [152].
Functional validation of noncoding elements in vitro
Identifying the physical location of putative gene regulatory elements does not represent definitive proof of their functional activity in regulating gene expression. This is true even if the degree of open chromatin, quantified by ATAC-seq or ChIP-seq signals, correlates well with gene expression. There is, therefore, a need to combine the biochemical annotation-based techniques, aimed at assessing both the accessibility of chromatin (DNaseI-seq and ATAC-seq) and its interactions (Hi-C and variant technologies such as capture Hi-C [153], PLAC-seq [154], and HiChIP [155]) with orthogonal assays more directed on demonstrating regulatory element activity and their effect on target genes.
In this regard, massively parallel reporter assay (MPRA) [156, 157] (Box 1) allows testing thousands of regulatory elements in a single experiment. A synthetized library of candidate sequences is cloned into a vector, wherein each candidate element is upstream of a basal promoter and a reporter gene. Each reporter gene is associated with a unique barcode providing a quantitative readout of the cognate candidate enhancer activity. Permitting the artificial introduction of SNPs [158], eQTL [159] or potential TF binding sites disrupting variants [160], MPRA is a powerful system for studying the effect of traits or disease-associated genomic variations on the functionality of regulatory regions. The shortcoming of this technique is the limited size of the tested fragments, that might not fully represent the entire enhancer’s region. Furthermore, both the use of episomal and lentiviral (lentiMPRA [161, 162]) reporter vectors do not reflect the enhancers functionality in their endogenous genomic context. Finally, the MPRA is a good tool to measure enhancers’ activity but fails to identify their endogenous target gene/s.
An alternative method for the enhancers’ validation is STARR-seq (self-transcribing active regulatory region sequencing [163]) (Box 1). Differently than MPRA, the candidate regulatory sequences are placed downstream of the minimal promoter and reporter gene, and therefore they will be transcribed as “enhancer RNA” such that each enhancer sequence works as its own barcode. Like MPRA, STARR-seq allows to investigate the functional activity of regions of interest selected by other predictive analyses (CapSTARR-seq [164], ChIP STARR-seq [165]) or to test the functional effect of disease-associated SNPs but removes any epigenetic contextualization.
Using the CRISPR/Cas9 technology [166] (Box 1) is complementary to MPRA and STARR-seq as it allows to perturb the sequences of interest in their native context and can also reveal the cognate gene(s) of enhancer regulation. Many studies use the active Cas9 and sgRNA libraries to “destroy” specific noncoding regions (up to hundreds of kilobases) in order to affect the expression of target genes [167–171]. An alternative approach alters the epigenomic landscape rather than the genomic sequence of the target region (CRISPR-epigenome editing). A catalytically inactive Cas9 (dCas9), fused with functional repressor or activator domains, triggers, respectively, repressive (CRISPR interference, or CRISPRi) or activating (CRISPR activation, or CRISPRa) chromatin modifications [172, 173]. Alongside CRISPR-mediated strategies based on detectable features (expression of the genetic reporter [171], drug resistance [169], or growth assays [174, 175]) recent works have combined CRISPRi-based enhancers perturbation with scRNA-seq in order to evaluate the variation of the transcriptome across the genome at single-cell resolution [176, 177]. However, all the CRISPR-based screens are not exempt from technical issues such as the potential presence of false positives and false negatives, or inefficiency of the Cas9-fused repressors/activators on certain enhancers [178].
The still unexplored combination of the above-mentioned methods (MPRA, STARR-seq, and CRISPR) with the powerful system of the cerebral organoids can open new avenues for a deeper understanding of the regulatory network involved in brain development and disorders.
Future research directions
There is an impressive amount of data available for the developmental neuroscience field, both from postmortem brain samples and in vitro models, which are largely derived from genome-scale sequencing efforts at both DNA and RNA levels. As demonstrated by some of the most recent studies presented here, we argue that the future lies in integration of these different levels of analyses. Impactful results can come from the intersections between transcriptomic information, epigenomic context, and genomic variation, which could be made even more compelling by integration of imaging and electrophysiological results. Targets relevant to the clinic can be obtained by intersecting those results with GWAS, WGS, exome and other databases to begin to understand disease pathophysiology during development of the brain. We tentatively summarize this multimodal approach in Fig. 1.
As all new technology, brain organoids come with obstacles and challenges. Although organoids resemble the embryonic to early fetal human brain at the molecular level, along with the capacity to generate most of the neural lineages found in humans, they are still less mature compared with adult neurons. Moreover, this system lacks the capacity to fully recapitulate features of human brain development like gyrification, full distinct cortical neuronal layers, gliogenesis, and complex neuronal circuitry formation. Features that undoubtedly will be explored in future years are how to promote vascularization and proper morphogenetic patterning to acquire better fidelity to in vivo development and try to recapitulate later stages of fetal development. Meanwhile the human iPSC-derived brain organoids have opened new ways to analyze brain development for a specific individual and in a longitudinal fashion. They promise to help us gain better understanding of the field of functional genomics, including defining enhancer–gene relationships and other regulatory mechanisms that govern brain development (Fig. 1b).
Multimodal studies, incorporating genome-scale analyses with other biological features of both brain tissue and organoids, have become more complex to understand. Developmental neurobiology is undergoing a progressive transformation from traditional one gene-one function studies to integrative “big data” studies, and the shift towards single-cell analysis has exponentially increased the amount of information available. We predict that transversal and meta-analysis will reveal more than originally meet the eyes, and that computational biology and machine-learning techniques will allow attaining a deeper understanding of brain development.
Funding and disclosure
The authors declare no competing financial interests related to this article. FMV, DC, and SS are members of the PsychEncode consortium. This work was funded by NIH grants #R01 MH109648, #R56 MH114911, #U01 MH103365, and Simons Foundation grant# 632742.
Acknowledgements
We thank Jeremy Schreiner for proof editing this paper.
Author contributions
Paper conception/outline: FMV and AJ. Paper writing: FMV, AJ, SS, DC, and AA. Display item preparation (Fig. 1): AJ, SS, and FMV. (Table 1) curated by AJ. (Box 1) curated by AJ, SS, DC, FMV. All authors participated in paper editing and proofreading.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Alexandre Jourdon, Soraya Scuderi
References
- 1.Sansom SN, Livesey FJ. Gradients in the brain: the control of the development of form and function in the cerebral cortex. Cold Spring Harb Perspect Biol. 2009;1:a002519. doi: 10.1101/cshperspect.a002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molnar Z, Clowry GJ, Sestan N, Alzu’bi A, Bakken T, Hevner RF, et al. New insights into the development of the human cerebral cortex. J Anat. 2019;235:432–51. doi: 10.1111/joa.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–8. doi: 10.1016/0166-2236(95)93934-P. [DOI] [PubMed] [Google Scholar]
- 4.Shine JM, Breakspear M, Bell PT, Ehgoetz Martens KA, Shine R, Koyejo O, et al. Human cognition involves the dynamic integration of neural activity and neuromodulatory systems. Nat Neurosci. 2019;22:289–96. doi: 10.1038/s41593-018-0312-0. [DOI] [PubMed] [Google Scholar]
- 5.Helfrich RF, Knight RT. Cognitive neurophysiology of the prefrontal cortex. Handb Clin Neurol. 2019;163:35–59. doi: 10.1016/B978-0-12-804281-6.00003-3. [DOI] [PubMed] [Google Scholar]
- 6.Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Shendure J. Mechanisms of interplay between transcription factors and the 3D genome. Mol Cell. 2019;76:306–19. doi: 10.1016/j.molcel.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Kim TK, Shiekhattar R. Architectural and functional commonalities between enhancers and promoters. Cell. 2015;162:948–59. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167:1170–87. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickel DE, Ypsilanti AR, Pla R, Zhu Y, Barozzi I, Mannion BJ, et al. Ultraconserved enhancers are required for normal development. Cell. 2018;172:491–9. doi: 10.1016/j.cell.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casino C, et al. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science. 2018;362. 10.1126/science.aat4311. [DOI] [PMC free article] [PubMed]
- 13.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–44. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–32. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12770–5. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, et al. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell. 2017;20:435–49. doi: 10.1016/j.stem.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH. The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci. 2016;19:1397–407. doi: 10.1038/nn.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–37. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coe BP, Stessman HAF, Sulovari A, Geisheker MR, Bakken TE, Lake AM, et al. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet. 2019;51:106–16. doi: 10.1038/s41588-018-0288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–84. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am J Hum Genet. 2009;84:524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat. 2015;36:915–21. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–98. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. ClinGen-the clinical genome resource. N Engl J Med. 2015;372:2235–42. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart J, et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48:D835–D44. doi: 10.1093/nar/gkz972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Psychiatric GCSC. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–7. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- 31.McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, et al. Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science. 2017;356. 10.1126/science.aal1641. [DOI] [PMC free article] [PubMed]
- 32.Abyzov A, Vaccarino FM, Urban AE, Sarangi V. Approaches and methods for variant analysis in the genome of a single cell. In: Moskalev A, editor. Biomarkers of Human Aging and Longevity: Springer, Cham; 2019. p. 203–28.
- 33.D’Gama AM, Walsh CA. Somatic mosaicism and neurodevelopmental disease. Nat Neurosci. 2018;21:1504–14. doi: 10.1038/s41593-018-0257-3. [DOI] [PubMed] [Google Scholar]
- 34.Keil JM, Qalieh A, Kwan KY. Brain transcriptome databases: a user’s guide. J Neurosci. 2018;38:2399–412. doi: 10.1523/JNEUROSCI.1930-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, et al. A comprehensive transcriptional map of primate brain development. Nature. 2016;535:367–75. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–23. doi: 10.1038/nature10524nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–9. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayoub AE, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, et al. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci USA. 2011;108:14950–5. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–16. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Consortium GT. Erratum: Genetic effects on gene expression across human tissues. Nature. 2018;553:530. doi: 10.1038/nature25160. [DOI] [PubMed] [Google Scholar]
- 44.e GP. Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat Genet. 2017;49:1664–70. doi: 10.1038/ng.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Psych EC, Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–12. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lahnemann D, Koster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020;21:31. doi: 10.1186/s13059-020-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014. 10.1038/nbt.2967 [DOI] [PMC free article] [PubMed]
- 49.Fan HC, Fu GK, Fodor SP. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 50.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360:176–82. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–8. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 54.Amezquita RA, Lun ATL, Becht E, Carey VJ, Carpp LN, Geistlinger L, et al. Orchestrating single-cell analysis with bioconductor. Nat Methods. 2020;17:137–45. doi: 10.1038/s41592-019-0654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saelens W, Cannoodt R, Todorov H, Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol. 2019;37:547–54. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- 59.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018. 10.1038/nbt.4314. [DOI] [PubMed]
- 60.Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB, Chen WS, et al. Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol. 2019;37:1482–92. doi: 10.1038/s41587-019-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. 2018;560:494–8. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van den Hurk M, Bardy C. Single-cell multimodal transcriptomics to study neuronal diversity in human stem cell-derived brain tissue and organoid models. J Neurosci Methods. 2019;325:108350. doi: 10.1016/j.jneumeth.2019.108350. [DOI] [PubMed] [Google Scholar]
- 63.Macaulay IC, Ponting CP, Voet T. Single-cell multiomics: multiple measurements from single cells. Trends Genet. 2017;33:155–68. doi: 10.1016/j.tig.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–7. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol. 2016;34:199–203. doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayer S, Chen J, Velmeshev D, Mayer A, Eze UC, Bhaduri A, et al. Multimodal single-cell analysis reveals physiological maturation in the developing human neocortex. Neuron. 2019;102:143–58.e7. doi: 10.1016/j.neuron.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362. 10.1126/science.aat7615. [DOI] [PMC free article] [PubMed]
- 69.Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362. 10.1126/science.aat8464. [DOI] [PMC free article] [PubMed]
- 70.Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, et al. Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 2018;362. 10.1126/science.aat6720. [DOI] [PMC free article] [PubMed]
- 71.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362. 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed]
- 72.Rhie SK, Schreiner S, Witt H, Armoskus C, Lay FD, Camarena A, et al. Using 3D epigenomic maps of primary olfactory neuronal cells from living individuals to understand gene regulation. Sci Adv. 2018;4:eaav8550. doi: 10.1126/sciadv.aav8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.An JY, Lin K, Zhu L, Werling DM, Dong S, Brand H, et al. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science. 2018;362. 10.1126/science.aat6576. [DOI] [PMC free article] [PubMed]
- 74.de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, et al. The dynamic landscape of open chromatin during human cortical neurogenesis. Cell. 2018;172:289–304. doi: 10.1016/j.cell.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–7. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019. 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed]
- 77.Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA. 2015;112:7285–90. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–90. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 2018;36:70–80. doi: 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science. 2019;364:685–9. doi: 10.1126/science.aav8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhaduri A, Di Lullo E, Jung D, Muller S, Crouch EE, Espinosa CS, et al. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell. 2020;26:48–63. doi: 10.1016/j.stem.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jakel S, Agirre E, Mendanha Falcao A, van Bruggen D, Lee KW, Knuesel I, et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature. 2019;566:543–7. doi: 10.1038/s41586-019-0903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schirmer L, Velmeshev D, Holmqvist S, Kaufmann M, Werneburg S, Jung D, et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature. 2019;573:75–82. doi: 10.1038/s41586-019-1404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–7. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci USA. 2015;112:15672–7. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–23. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–8. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–8. doi: 10.1038/nature25980. [DOI] [PubMed] [Google Scholar]
- 89.Fan X, Dong J, Zhong S, Wei Y, Wu Q, Yan L, et al. Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis. Cell Res. 2018;28:730–45. doi: 10.1038/s41422-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Polioudakis D, de la Torre-Ubieta L, Langerman J, Elkins AG, Shi X, Stein JL, et al. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron. 2019;103:785–801.e8. doi: 10.1016/j.neuron.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhong S, Ding W, Sun L, Lu Y, Dong H, Fan X, et al. Decoding the development of the human hippocampus. Nature. 2020;577:531–6. doi: 10.1038/s41586-019-1917-5. [DOI] [PubMed] [Google Scholar]
- 92.La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell. 2016;167:566–80.e19. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, et al. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat Methods. 2016;13:87–93. doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci. 2015;18:637–46. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, Kriegstein AR. Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron. 2016;91:1219–27. doi: 10.1016/j.neuron.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941–5. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–8. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marin-Valencia I, Guerrini R, Gleeson JG. Pathogenetic mechanisms of focal cortical dysplasia. Epilepsia. 2014;55:970–8. doi: 10.1111/epi.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu X, Zecevic N. Dorsal radial glial cells have the potential to generate cortical interneurons in human but not in mouse brain. J Neurosci. 2011;31:2413–20. doi: 10.1523/JNEUROSCI.5249-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, Alvarez-Buylla A, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16:1576–87. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–9. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- 103.Ma T, Wang C, Wang L, Zhou X, Tian M, Zhang Q, et al. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16:1588–97. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 104.Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nott A, Holtman IR, Coufal NG, Schlachetzki JCM, Yu M, Hu R, et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science. 2019;366:1134–9. doi: 10.1126/science.aay0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci USA. 2013;110:20284–9. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–90. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015. 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed]
- 112.Rigamonti A, Repetti GG, Sun C, Price FD, Reny DC, Rapino F, et al. Large-scale production of mature neurons from human pluripotent stem cells in a three-dimensional suspension culture system. Stem cell Rep. 2016;6:993–1008. doi: 10.1016/j.stemcr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, et al. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10:537–50. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 115.Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem cell Rep. 2017;8:1144–54. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell. 2019;24:487–97. doi: 10.1016/j.stem.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–54. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cederquist GY, Asciolla JJ, Tchieu J, Walsh RM, Cornacchia D, Resh MD, et al. Specification of positional identity in forebrain organoids. Nat Biotechnol. 2019;37:436–44. doi: 10.1038/s41587-019-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019;25:558–69. doi: 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 2019;22:669–79. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35:659–66. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Qian X, Su Y, Adam CD, Deutschmann AU, Pather SR, Goldberg EM, et al. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell. 2020. 10.1016/j.stem.2020.02.002. [DOI] [PMC free article] [PubMed]
- 123.Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron. 2017;95:779–90. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marton RM, Miura Y, Sloan SA, Li Q, Revah O, Levy RJ, et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci. 2019;22:484–91. doi: 10.1038/s41593-018-0316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Madhavan M, Nevin ZS, Shick HE, Garrison E, Clarkson-Paredes C, Karl M, et al. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat Methods. 2018;15:700–6. doi: 10.1038/s41592-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mansour AA, Goncalves JT, Bloyd CW, Li H, Fernandes S, Quang D, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol. 2018;36:432–41. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang YJ, et al. Engineering of human brain organoids with a functional vascular-like system. Nat Methods. 2019;16:1169–75. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Worsdorfer P, Dalda N, Kern A, Kruger S, Wagner N, Kwok CK, et al. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci Rep. 2019;9:15663. doi: 10.1038/s41598-019-52204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Paredes I, Himmels P, Ruiz de Almodovar C. Neurovascular communication during CNS development. Dev Cell. 2018;45:10–32. doi: 10.1016/j.devcel.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 130.Ormel PR, Vieira de Sa R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. Microglia innately develop within cerebral organoids. Nat Commun. 2018;9:4167. doi: 10.1038/s41467-018-06684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Song L, Yuan X, Jones Z, Vied C, Miao Y, Marzano M, et al. Functionalization of brain region-specific spheroids with isogenic microglia-like cells. Sci Rep. 2019;9:11055. doi: 10.1038/s41598-019-47444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 2017;94:278–93. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Abreu CM, Gama L, Krasemann S, Chesnut M, Odwin-Dacosta S, Hogberg HT, et al. Microglia increase inflammatory responses in iPSC-derived human BrainSpheres. Front Microbiol. 2018;9:2766. doi: 10.3389/fmicb.2018.02766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–9. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21:383–98. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dang J, Tiwari SK, Agrawal K, Hui H, Qin Y, Rana TM. Glial cell diversity and methamphetamine-induced neuroinflammation in human cerebral organoids. Mol Psychiatry. 2020. 10.1038/s41380-020-0676-x [DOI] [PMC free article] [PubMed]
- 137.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–39. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–62. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marchetto MC, Belinson H, Tian Y, Freitas BC, Fu C, Vadodaria K, et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol Psychiatry. 2017;22:820–35. doi: 10.1038/mp.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–5. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, et al. Evolutionary genomics evolutionary changes in promoter and enhancer activity during human corticogenesis. Science. 2015;347:1155–9. doi: 10.1126/science.1260943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–7. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yoon SJ, Elahi LS, Pasca AM, Marton RM, Gordon A, Revah O, et al. Reliability of human cortical organoid generation. Nat Methods. 2019;16:75–8. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tanaka Y, Cakir B, Xiang Y, Sullivan GJ, Park IH. Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep. 2020;30:1682–9. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36:421–7. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mora-Bermudez F, Badsha F, Kanton S, Camp JG, Vernot B, Kohler K, et al. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. eLife. 2016;5. 10.7554/eLife.18683. [DOI] [PMC free article] [PubMed]
- 150.Field AR, Jacobs FMJ, Fiddes IT, Phillips APR, Reyes-Ortiz AM, LaMontagne E, et al. Structurally conserved primate LncRNAs are transiently expressed during human cortical differentiation and influence cell-type-specific genes. Stem cell Rep. 2019;12:245–57. doi: 10.1016/j.stemcr.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell. 2019;176:743–56.e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Sanchis-Calleja F, et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–22. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 153.Dryden NH, Broome LR, Dudbridge F, Johnson N, Orr N, Schoenfelder S, et al. Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res. 2014;24:1854–68. doi: 10.1101/gr.175034.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fang R, Yu M, Li G, Chee S, Liu T, Schmitt AD, et al. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 2016;26:1345–8. doi: 10.1038/cr.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13:919–22. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Patwardhan RP, Lee C, Litvin O, Young DL, Pe’er D, Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–5. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–7. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ulirsch JC, Nandakumar SK, Wang L, Giani FC, Zhang X, Rogov P, et al. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell. 2016;165:1530–45. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tewhey R, Kotliar D, Park DS, Liu B, Winnicki S, Reilly SK, et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell. 2016;165:1519–29. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grossman SR, Zhang X, Wang L, Engreitz J, Melnikov A, Rogov P, et al. Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc Natl Acad Sci USA. 2017;114:E1291–E300. doi: 10.1073/pnas.1621150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Inoue F, Kircher M, Martin B, Cooper GM, Witten DM, McManus MT, et al. A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res. 2017;27:38–52. doi: 10.1101/gr.212092.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Inoue F, Kreimer A, Ashuach T, Ahituv N, Yosef N. Identification and massively parallel characterization of regulatory elements driving neural induction. Cell Stem Cell. 2019;25:713–27. doi: 10.1016/j.stem.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–7. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 164.Vanhille L, Griffon A, Maqbool MA, Zacarias-Cabeza J, Dao LT, Fernandez N, et al. High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq. Nat Commun. 2015;6:6905. doi: 10.1038/ncomms7905. [DOI] [PubMed] [Google Scholar]
- 165.Vockley CM, D’Ippolito AM, McDowell IC, Majoros WH, Safi A, Song L, et al. Direct GR binding sites potentiate clusters of TF binding across the human Genome. Cell. 2016;166:1269–81. doi: 10.1016/j.cell.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–7. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gasperini M, Findlay GM, McKenna A, Milbank JH, Lee C, Zhang MD, et al. CRISPR/Cas9-mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am J Hum Genet. 2017;101:192–205. doi: 10.1016/j.ajhg.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–9. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rajagopal N, Srinivasan S, Kooshesh K, Guo Y, Edwards MD, Banerjee B, et al. High-throughput mapping of regulatory DNA. Nat Biotechnol. 2016;34:167–74. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Diao Y, Li B, Meng Z, Jung I, Lee AY, Dixon J, et al. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 2016;26:397–405. doi: 10.1101/gr.197152.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–7. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–9. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–73. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34:192–8. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 176.Xie S, Duan J, Li B, Zhou P, Hon GC. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol Cell. 2017;66:285–99. doi: 10.1016/j.molcel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 177.Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, et al. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell. 2019;176:377–90. doi: 10.1016/j.cell.2018.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Klein JC, Chen W, Gasperini M, Shendure J. Identifying novel enhancer elements with CRISPR-based screens. ACS Chem Biol. 2018;13:326–32. doi: 10.1021/acschembio.7b00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–8. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]