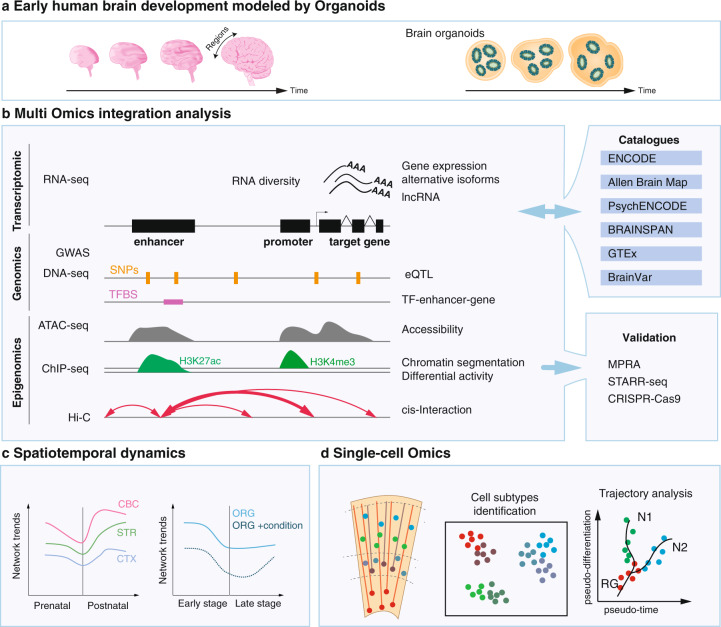
Fig. 1. Integrative approaches to study human brain development.
a Illustration of two complementary models, postmortem human brain tissue and iPSC-derived brain organoids. b Potential of multi-omics approaches to connect/integrate genomic, transcriptomic, and epigenomic information generated or available through datasets of in vivo/in vitro studies. Enhancer activity (identified from ATAC-seq, ChIP-seq) influences transcription (RNA-seq) of gene/s through DNA looping (identified through Hi-C). Similarly, eQTL connects risk variants (discovered via DNA-seq, SNPs, GWAS) to gene expression. These variants may disrupt transcription factor binding sites (TFBS) within enhancers. Human-specific gene regulatory mechanisms can be validated in live cell systems, i.e., iPSC-derived organoids, or human progenitor cell lines, by high throughput methods (e.g., MPRA, STARR-seq). c Studying gene-enhancer dynamics over time both in vivo and in organoids (ORG) can reveal biological insight into the formation and evolution of the human brain (e.g., the hourglass shape of interregional diversity over time between cortex (CTX), striatum (STR), cerebellum (CRB)). d Single-cell omics can define those dynamics at the cellular levels, for instance revealing how gene regulation evolves between radial glia (RG) and neuronal derivatives (N1, N2).