Abstract
Background
To develop a deep learning model to classify primary bone tumors from preoperative radiographs and compare performance with radiologists.
Methods
A total of 1356 patients (2899 images) with histologically confirmed primary bone tumors and pre-operative radiographs were identified from five institutions’ pathology databases. Manual cropping was performed by radiologists to label the lesions. Binary discriminatory capacity (benign versus not-benign and malignant versus not-malignant) and three-way classification (benign versus intermediate versus malignant) performance of our model were evaluated. The generalizability of our model was investigated on data from external test set. Final model performance was compared with interpretation from five radiologists of varying level of experience using the Permutations tests.
Findings
For benign vs. not benign, model achieved area under curve (AUC) of 0•894 and 0•877 on cross-validation and external testing, respectively. For malignant vs. not malignant, model achieved AUC of 0•907 and 0•916 on cross-validation and external testing, respectively. For three-way classification, model achieved 72•1% accuracy vs. 74•6% and 72•1% for the two subspecialists on cross-validation (p = 0•03 and p = 0•52, respectively). On external testing, model achieved 73•4% accuracy vs. 69•3%, 73•4%, 73•1%, 67•9%, and 63•4% for the two subspecialists and three junior radiologists (p = 0•14, p = 0•89, p = 0•93, p = 0•02, p < 0•01 for radiologists 1–5, respectively).
Interpretation
Deep learning can classify primary bone tumors using conventional radiographs in a multi-institutional dataset with similar accuracy compared to subspecialists, and better performance than junior radiologists.
Funding
The project described was supported by RSNA Research & Education Foundation, through grant number RSCH2004 to Harrison X. Bai.
Keywords: Deep learning, Convolutional neural network, Primary bone tumor, Plain radiograph
Research in Context section.
Evidence before this study
Primary malignancy of the bone and joints is ranked as the third leading cause of death in patients with cancer who are younger than 20 years. The plain radiograph remains the most useful examination for differentiating benign from aggressive lesions. Because of the low incidence and variety of uncommon feature of primary bone tumors, few radiologists develop sufficient expertise to make a definite diagnosis. For general radiologists and those working in resource limited regions, radiographic interpretation can be less accurate, leading to misdiagnosis and unnecessary biopsies. Classification of primary bone tumors correctly via radiography is a challenging problem even for subspecialists. The aim of this project was to raise the level of plain radiography analysis through deep learning to the level of the musculoskeletal subspecialist. Artificial intelligence, especially deep learning with convolutional neural networks has shown great promise in classifying two-dimensional images of some common diseases. With the use of PubMed and Google Scholar, a systematic literature search was performed to identify original research papers in English from inception to October, 2019, using the terms (“bone tumors” OR “bone cancer”) AND (“DCNN” OR “deep learning” OR “machine learning”) AND (“radiographs” OR “plain film”). No previously published report was found.
Added value of this study
Our study is the first to establish a deep learning algorithm for classifying primary bone tumors on conventional radiographs using a multi-institutional dataset with similar accuracy to subspecialists and higher accuracy than junior radiologists. The performance is expected to improve further in the future with larger datasets.
Implications of all the available evidence
Correctly classifying bone tumors on plain radiograph is important for clinical decision making as it can guide subsequent management. Our algorithm has the potential to improve primary bone tumor radiographs interpretation to the level of the subspecialists. If further validated, the algorithm can prevent patients from undergoing unnecessary invasive biopsies and help guide clinical management, especially in areas without subspecialty expertise.
Alt-text: Unlabelled box
Introduction
Although primary bone tumors are uncommon with incidence rates of 4–7% among children and adolescents in the United States [1], primary malignancies of the bone and joints are ranked as the third leading cause of death in patients with cancer who are younger than 20 years of age [2]. Bone tumors vary widely in their biological behavior and require different management depending on their classification as benign, intermediate, or malignant, by the World Health Organization (WHO) [3]. Benign bone tumors (e.g. osteochondroma, osteoid osteoma, etc.) have a limited capacity for local recurrence, and are almost always readily cured by complete local excision/curettage [3]. Tumors in the intermediate group (e.g. giant cell tumor, chondroblastoma, etc.) have the potential to be locally aggressive or metastasize in rare cases. Therefore, bone tumors classified as intermediate often require wide excision margins inclusive of normal tissue, and/or the use of adjuvant therapy in order to ensure local control [3]. Malignant bone tumors (e.g. chondrosarcoma, osteosarcoma, etc.) not only have the potential for locally destructive growth and recurrence, but also carry significant risk for distant metastases [3].
Differential diagnoses of primary bone tumor mostly depend on the review of the conventional radiographs and the age of the patient. The plain radiograph remains the most useful examination for differentiating these cases, while CT and MRI are only helpful in selected cases. Besides demographic information such as the patient's age, radiographic appearance of the tumor including size, location, margin, type of matrix, presence of periosteal reaction and cortical destruction are other key clues in helping the radiologist differentiate indolent from aggressive bone tumors [3]. Because bone tumors have a variety of appearances and are relatively uncommon, few radiologists develop sufficient expertise to make a definite diagnosis. Among general radiologists, accuracy in interpretation of bone lesions can be low, leading to misdiagnosis which can be detrimental to patient outcome [4]. Many patients with benign tumors are referred to bone biopsy, which has the issue of increased morbidity and cost, and is subject to sampling error [5] or evaluated with advanced imaging modalities which increase health care costs.
Artificial intelligence, especially deep learning with convolutional neural networks has shown great promise in classifying two-dimensional images of some common diseases and relies on databases of thousands of annotated or unannotated images [6], [7], [8], [9]. Deep learning models can recognize predictive features directly from images by utilizing a back-propagation algorithm which recalibrates the model's internal parameters after each round of training [10]. Recent studies have shown the potential of deep learning in the assessment of solid liver lesions on ultrasonography [11], renal lesions [12,13] and glioma on MR Imaging [10,[14], [15], [16], [17]] and abnormal chest radiographs [18].
An algorithm that can distinguish benign from malignant bone tumors on routine radiographs with high accuracy can facilitate triage, guide patient management, and save patients from unnecessary procedures. In this study, we trained a deep learning algorithm to classify primary bone tumors on plain film and compare performance with radiologists of varying level of experience.
Materials and methods
Patient cohort
Patients with primary bone tumor confirmed by histology according to the 2013 World Health Organization (WHO) classification were retrospectively identified from five large academic centers from July 2008 to July 2019. Plain radiograph and clinical variables including patient demographics (i.e., age and sex) were collected. The study was conducted in accordance with Declaration of Helsinki and approved by the Institutional Review Boards at all five institutions. The inclusion criteria for the study were (i) histopathologically confirmed (biopsy or surgery) primary bone tumor according to current WHO criteria, (ii) available pre-procedure plain radiograph including all the projections it had which can show the lesion clearly, and years of experience reading musculoskeletal (MSK) plain film. Our final dataset consisted of 2,899 images from 1,356 patients (institution 1: 410 images from 160 patients, institution 2: 745 images from 333 patients, institution 3: 1,105 images from 572 patients, institution 4: 390 images from 186 patients, institution 5: 249 images from 105 patients). Each patient contributed 1 lesion to the dataset. Of the 1,356 lesions, 679 (1,523 images) were benign based on histopathology, 317 (635 images) were intermediate, and 360 (741 images) were malignant (see Supplementary Figure S1, which demonstrates inclusion and exclusion criteria). In respect of patient confidentiality and consent, the radiographs and clinical information datasets analyzed in this study are not available for download but are available upon reasonable request to the corresponding author."?> years of experience reading musculoskeletal (MSK) plain film. Our final dataset consisted of 2,899 images from 1,356 patients (institution 1: 410 images from 160 patients, institution 2: 745 images from 333 patients, institution 3: 1,105 images from 572 patients, institution 4: 390 images from 186 patients, institution 5: 249 images from 105 patients). Each patient contributed 1 lesion to the dataset. Of the 1,356 lesions, 679 (1,523 images) were benign based on histopathology, 317 (635 images) were intermediate, and 360 (741 images) were malignant (see Supplementary Figure S1, which demonstrates inclusion and exclusion criteria). In respect of patient confidentiality and consent, the radiographs and clinical information datasets analyzed in this study are not available for download but are available upon reasonable request to the corresponding author."?> (iii) quality of the images was adequate for analysis, without motion or artifacts. The images were screened by a radiologist (Y.H.) with 7 years of experience reading musculoskeletal (MSK) plain film. Our final dataset consisted of 2899 images from 1356 patients (institution 1: 410 images from 160 patients, institution 2: 745 images from 333 patients, institution 3: 1105 images from 572 patients, institution 4: 390 images from 186 patients, institution 5: 249 images from 105 patients). Each patient contributed 1 lesion to the dataset. Of the 1356 lesions, 679 (1523 images) were benign based on histopathology, 317 (635 images) were intermediate, and 360 (741 images) were malignant (see Supplementary Figure S1, which demonstrates inclusion and exclusion criteria). In respect of patient confidentiality and consent, the radiographs and clinical information datasets analyzed in this study are not available for download but are available upon reasonable request to the corresponding author.
Preprocessing
All images were downloaded in DICOM format at their original dimensions and resolution. Images were converted from DICOM to 8-bit JPEG. Then the images were loaded into Click 2 Crop software (v5.2.2), and regions of interest containing the whole tumor were manually cropped from the original image to include some surrounding while capturing the margin of the lesion, by a radiologist (Y.H.) with 7 years of experience reading musculoskeletal (MSK) plain film. Images were padded and resized to 512 by 512 pixels. Single-channel images were converted to 3-channel images by repeating the single channel 3 times [12,19,20]. Pixel values were normalized by scaling values into the range [0, 1], then subtracting (0•485, 0•456, 0•406) and dividing by (0•229, 0•224, 0•225) channel-wise.
Training and inference
Model training was performed in Python 3.7 and PyTorch 1.6 using a NVIDIA GV100 32GB graphics processing unit. Models were based on the EfficientNet-B0 convolutional neural network architecture [21]. Model weights were initialized with weights pretrained on the ImageNet database. Training was performed using a batch size of 96, dropout probability of 0.2 before the final fully-connected layer, and data augmentation consisting of horizontal flips, affine transformations, and contrast adjustments. Models were trained for 3-way classification (benign, intermediate, and malignant) and binary classification (benign versus not-benign and malignant versus not-malignant). The RAdam optimizer was used with a categorical cross-entropy loss and a cosine annealing learning rate schedule with an initial learning rate of 3 × 10e−4. Models were trained for 20 epochs. The selected model for each training episode was selected based on the Cohen's kappa score on the validation set. For each test fold, 3 training episodes were performed to form a 3-model ensemble. Predictions were averaged across all models and all radiographic views to produce a final prediction for each case. During each training epoch, 1 image from 1 patient is sampled so that the model is exposed to the same number of images per patient over the course of the entire training period. An external test set comprised of images from two of our five institutions (institutions 4 and 5) was used to evaluate the generalization performance of the model. The external test set consisted of 639 images from 291 patients. Each patient contributed 1 lesion to the dataset. Of the 291 lesions, 162 (368 images) were benign based on histopathology, 61 (126 images) were intermediate, and 68 (145 images) were malignant. Please see Supplementary Figure S2 for schematic of our pipeline.
Evaluation
Radiologist evaluation
Two board-certified musculoskeletal subspecialists (H.L. and S.P.), who see more than 100 bone tumors per year, with 25 and 23 years of experience, and three junior radiologists, who with 6, 1, and 7 years of experience reading MSK plain film respectively, blind to histopathologic data, evaluated conventional radiographs of the bone lesions, and labeled each case as benign, intermediate, or malignant with their own interpretations. They were given clinical information of age and sex of each patient. The 2 musculoskeletal subspecialists interpreted the uncropped images of entire cohort (data from all five institutions) and the cropped images of external test set (data from institutions 4 and 5), while the 2 junior radiologists evaluated only the uncropped and cropped images of external test set. One junior radiologist (J.G.) only evaluated the uncropped images of the external set, because she was exposed to the gold standard during the recropping process. Ground truth labels were obtained using the final pathology results. The model’ results were compared with radiologists’ interpretations and final pathology results to assess model performance. Information on the five radiologists is shown in Supplemental Table S1.
Model evaluation
The model performance was evaluated using several metrics. Receiver operating characteristic (ROC) curves and area under curve (AUC) for benign versus not-benign and malignant versus not-malignant were used to evaluate binary discriminatory capacity. Cohen's kappa scores and categorical accuracy were used to evaluate the three-way classification performance of the model and radiologists. Five-fold cross-validation was used to analyze model performance, ensuring no patient overlap across different folds. First, the model was divided into 5 disjoint partitions based on patient ID, each approximately 20% of the overall dataset, which comprise the test folds. Next, the remaining 80% of the dataset was used for training (70%) and validation (10%). A separate model was trained for each fold, and the out-of-fold predictions were obtained for the test fold. The cross-validation scheme is illustrated in Supplemental Figure S3 and Supplemental Table S2. This cross-validation procedure allowed us to obtain an out-of-fold prediction for each sample in the dataset to maximize the sample size on which the model performance was evaluated without data leakage. Model performance was also evaluated on an external test set to evaluate generalizability beyond the institutions present in the internal cohort. To evaluate the impact of manual lesion cropping on model performance, a second radiologist (J.G.) with 1 years of experience reading MSK plain film independently recropped the images in the external test set, and model performance was evaluated on the external test set using this set of recropped images.
Statistical analysis
Statistical analysis was performed using the R statistical computing language, as well as non-parametric methods implemented in Python 3.7. 95% confidence intervals for AUCs were obtained via the DeLong method. For Cohen's kappa scores and categorical accuracy, 95% confidence intervals were generated using 10,000 bootstrap samples. Permutation tests with 10,000 iterations were used to calculate p-values. p < 0•05 was considered to indicate a statistically significant difference in performance. Comparison with radiologists was performed only for 3-way classification. Subgroup analysis based on age was also performed.
Results
Table 1 summarizes the 5 datasets used in this study. Overall, the mean age was 24•7 ± 18•1 years with 50•1% benign tumors (average age 22•8 ± 16•9), 23•4% intermediate tumors (average age 23•5 ± 15•7), and 26•5% malignant tumors (average age 27•7 ± 21•1), as indicated by the final pathology results. There was a slight male predominance (58•2%). Differences in the distributions of age (One-way analysis of variance, p < 0•01), sex (Chi-square test, p = 0•013) and pathology (Chi-square test, p < 0•01) were statistically significant among the 5 institutions. Please see Supplementary Figure S4 for examples of benign, intermediate and malignant bone tumors.
Table 1.
Demographics for each of the 5 institutions.
| Institution 1 (PENN) |
Institution 2 (CHOP) |
Institution 3 (China 2) |
Institution 4 (China 1) |
Institution 5 (China 3) |
|
|---|---|---|---|---|---|
| Hospital type | Adult & Pediatric | Pediatric | Adult & Pediatric | Adult & Pediatric | Pediatric |
| Number of patients | 160 | 333 | 572 | 186 | 105 |
| Age, mean, years (SD) | 40•3 (17•8) | 12•6 (4•8) | 28•4 (17•8) | 31•3 (19•3) | 7•5 (3•9) |
| Sex (% male) | 82 (51•2) | 188 (56•5) | 328(57•3) | 116 (62•4) | 75 (71•4) |
| Pathology (%) | |||||
| Benign | 69 (43•1) | 112 (33•6) | 336 (58•7) | 78 (41•9) | 84 (80•0) |
| Intermediate | 35 (21•9) | 78 (23•4) | 143 (25•0) | 45 (24•2) | 16 (15•2) |
| Malignant | 56 (35•0) | 143 (42•9) | 93 (16•3) | 63 (33•9) | 5 (4•8) |
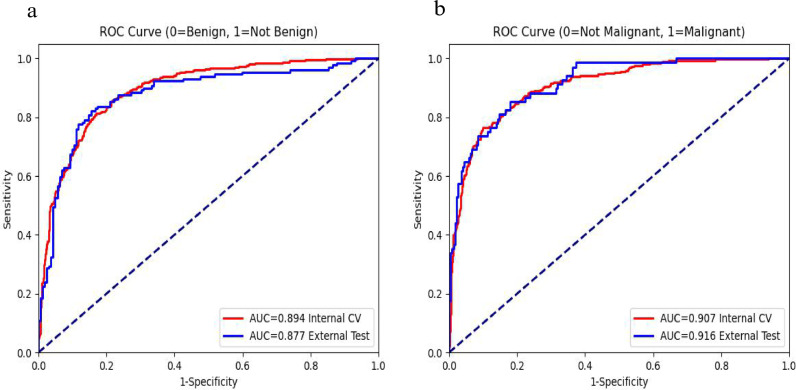
On cross-validation, the AUCs for the two classifications were 0•894 and 0•907, respectively. For benign vs. not benign, at a naive threshold of 0•5, the model achieved 82•7% sensitivity and 81•8% specificity. Sensitivity and specificity for the model can be adjusted along the ROC curve by calibrating the model threshold. For malignant vs. not malignant, at a naive threshold of 0•5, the model achieved 77•7% sensitivity and 89•6% specificity. On external testing, the AUCs for these 2 classifications were 0•877 and 0•916, respectively. The data were divided into quartiles by age for subgroup analysis: younger than 12 years old, 12–18 years old, 19–36 years old, and older than 36 years old. Performance of 2 formulated binary classification problems: benign vs. not benign and malignant vs. not malignant for the deep learning model are summarized in Table 2. Fig. 1 depicts the ROC curves for the 2 formulated binary classification problems: benign vs. not benign and malignant vs. not malignant on cross-validation and external testing.
Table 2.
Model performance of 2 formulated binary classification problems: benign vs. not benign and malignant vs. not malignant. 95% confidence intervals for AUCs were obtained via the DeLong method.
| AUC | ||
|---|---|---|
| Cross-validation (Institution 1, 2 and 3, n=1065) |
Not Benign | 0•894 (0•874, 0•912) |
| Not Malignant | 0•907 (0•886, 0•926) | |
| Divided into quartiles by age for subgroup | ||
| Age (<12, n=268) | Not Benign | 0•891 (0•849, 0•928) |
| Not Malignant | 0•915 (0•870, 0•953) | |
| Age (12-18, n=277) | Not Benign | 0•933 (0•903, 0•960) |
| Not Malignant | 0•933 (0•900, 0•962) | |
| Age (19-36, n= 263) | Not Benign | 0•897 (0•858, 0•933) |
| Not Malignant | 0•946 (0•910, 0•975) | |
| Age (>36, n= 257) | Not Benign | 0•844 (0•849, 0•928) |
| Not Malignant | 0•819 (0•870, 0•953) | |
| External testing (Institution 4 and 5, n=291) |
Not Benign | 0•877 (0•833, 0•918) |
| Not Malignant | 0•916 (0•877, 0•949) | |
AUC: area under curve
Fig. 1.
Receiver operating characteristic curves for the 2 formulated binary classification problems. benign vs. not-benign (a) and malignant vs. not-malignant (b). Area under curve (AUC) of internal cross-validation (CV, red) and external testing (blue) are also included.
Three-way classification results for the deep learning model and two subspecialists are shown in Table 3. For three-way classification, Cohen's kappa scores for the model and subspecialists were 0•548, 0•605, and 0•565, respectively. On cross validation, differences between model predictions and subspecialist 1’s rating was found to be statistically significant (Permutation tests, p = 0•03). Differences between model predictions and subspecialist 2’s ratings were not found to be statistically significant (Permutation tests, p = 0•52). In addition, the data were divided into age quartiles, and detailed stratified model performance by age is summarized in Table 3. Whereas class distributions for both subspecialists were similar, the model predicted a higher number of benign tumors (50•9% vs. 43•2% and 43•5%) and fewer intermediate tumors (18•1% vs. 23•6% and 24•5%). Malignant tumor predictions were more similar across model and subspecialists (31•0% vs. 33•1% and 32•0%).
Table 3.
Comparison of model performance with subspecialists on cross-validation. For Cohen's kappa scores and categorical accuracy, 95% confidence intervals were generated using 10,000 bootstrap samples. Permutation tests with 10,000 iterations were used to calculate p-values.
| Accuracy | Cohen's κ | Difference in κ | p-value | ||
|---|---|---|---|---|---|
| Total (n=1065) |
Model | 72•1% | 0•548 (0•504, 0•590) | ||
| Rater 1 | 74•6% | 0•605 (0•564, 0•644) | 0•057 (0•007, 0•107) | 0•03 | |
| Rater 2 | 72•1% | 0•565 (0•523, 0•607) | 0•017 (-0•034, 0•068) | 0•52 | |
| Age (<12, n=268) | Model | 73•9% | 0•557 (0•473, 0•641) | ||
| Rater 1 | 71•3% | 0•544 (0•464, 0•625) | -0•013 (-0•106, 0•079) | 0•77 | |
| Rater 2 | 73•9% | 0•587 (0•506, 0•666) | 0•030 (-0•069, 0•128) | 0•56 | |
| Age (12-18, n=277) | Model | 76•7% | 0•617 (0•537, 0•693) | ||
| Rater 1 | 77•4% | 0•646 (0•570, 0•721) | 0•029 (-0•065, 0•126) | 0•55 | |
| Rater 2 | 75•6% | 0•615 (0•534, 0•689) | -0•002 (-0•098, 0•094) | 0•96 | |
| Age (19-36, n= 263) | Model | 75•8% | 0•610 (0•523, 0•692) | ||
| Rater 1 | 77•8% | 0•653 (0•571, 0•731) | 0•043 (-0•062, 0•148) | 0•43 | |
| Rater 2 | 70•6% | 0•541 (0•451, 0•628) | -0•069 (-0•174, 0•036) | 0•22 | |
| Age (>36, n= 257) | Model | 62•2% | 0•384 (0•291, 0•473) | ||
| Rater 1 | 72•1% | 0•558 (0•472, 0•641) | 0•174 (0•065, 0•284) | 0•003 | |
| Rater 2 | 68•3% | 0•499 (0•413, 0•583) | 0•115 (0•004, 0•227) | 0•05 |
Rater 1 and 2 are subspecialists.
Three-way classification results for the deep learning model and five radiologists on uncropped images of external testing data are shown in Table 4. Cohen's kappa scores for the model and five radiologists were 0•560, 0•483 0•553, 0•555, 0•430, and 0•367, respectively. Differences between model predictions and 1-3 radiologist's ratings were not found to be statistically significant (Permutation tests, p = 0•14, p = 0•89 and p = 0•93). Differences between model predictions and 4-5 radiologist's ratings were found to be statistically significant (Permutation tests, p = 0•02 and p < 0•05). In addition, the data were divided into three equally sized age groups, and detailed stratified model performance by age is shown in Table 4.
Table 4.
Comparison of model performance with subspecialists and junior radiologists evaluating uncropped images of the external testing data and stratified by age group. For Cohen's kappa scores and categorical accuracy, 95% confidence intervals were generated using 10,000 bootstrap samples. Permutation tests with 10,000 iterations were used to calculate p-values.
| Accuracy | Cohen's κ | Difference in κ | p-value | ||
|---|---|---|---|---|---|
| Total (n = 291) |
Model | 73•4% | 0•560 (0•481, 0•639) | ||
| Rater 1 | 69•3% | 0•483 (0•394, 0•567) | -0•077 (-0•180, 0•021) | 0•14 | |
| Rater 2 | 73•4% | 0•553 (0•468, 0•634) | -0•007 (-0•112, 0•096) | 0•89 | |
| Rater 3 | 73•1% | 0•555 (0•472, 0•633) | -0•005 (-0•115, 0•103) | 0•93 | |
| Rater 4 | 67•9% | 0•430 (0•340, 0•519) | -0•130 (-0•240, -0•020) | 0•02 | |
| Rater 5 | 63•4% | 0•367 (0•285, 0•449) | -0•193 (-0•293, -0•093) | 0•0005 | |
| Age (<10, n = 97) | Model | 74•2% | 0•383 (0•210, 0•542) | ||
| Rater 1 | 79•4% | 0•478 (0•278, 0•655) | 0•095 (-0•128, 0•314) | 0•41 | |
| Rater 2 | 79•4% | 0•515 (0•334, 0•678) | 0•132 (-0•080, 0•343) | 0•23 | |
| Rater 3 | 79•4% | 0•535 (0•367, 0•695) | 0•152 (-0•080, 0•393) | 0•25 | |
| Rater 4 | 80•4% | 0•448 (0•239, 0•637) | 0•065 (-0•177, 0•314) | 0•61 | |
| Rater 5 | 69•1% | 0•229 (0•064, 0•390) | -0•154 (-0•341, 0•017) | 0•11 | |
| Age (10-24, n = 97) | Model | 77•3% | 0•630 (0•498, 0•755) | ||
| Rater 1 | 70•1% | 0•496 (0•336, 0•640) | -0•134 (-0•311, 0•038) | 0•13 | |
| Rater 2 | 72•2% | 0•538 (0•392, 0•676) | -0•092 (-0•261, 0•075) | 0•28 | |
| Rater 3 | 77•3% | 0•618 (0•473, 0•749) | -0•012 (-0•183, 0•156) | 0•88 | |
| Rater 4 | 69•1% | 0•450 (0•291, 0•596) | -0•180 (-0•352, -0•011) | 0•045 | |
| Rater 5 | 52•6% | 0•217 (0•085, 0•354) | -0•413 (-0•576, -0•246) | <1•0e-6 | |
| Age (>24, n = 97) | Model | 68•8% | 0•514 (0•366, 0•648) | ||
| Rater 1 | 58•3% | 0•386 (0•250, 0•521) | -0•128 (-0•304, 0•047) | 0•15 | |
| Rater 2 | 68•8% | 0•526 (0•385, 0•660) | 0•012 (-0•178, 0•200) | 0•89 | |
| Rater 3 | 62•5% | 0•413 (0•263, 0•556) | -0•101 (-0•294, 0•093) | 0•31 | |
| Rater 4 | 54•2% | 0•282 (0•132, 0•429) | -0•232 (-0•426, -0•033) | 0•025 | |
| Rater 5 | 68•8% | 0•479 (0•345, 0•608) | -0•035 (-0•198, 0•137) | 0•71 |
Rater 1 and 2 are subspecialists, while rater 3-5 are junior radiologists.
Comparison of model performance on the images in the external test set cropped by the original radiologist (Y.H.) and the second radiologist (J. G.) demonstrated no significant difference (Cohen's kappa score of 0.560 versus 0.549, p = 0.67).
Three-way classification results for the deep learning model and five radiologists on cropped images of external testing data are shown in Supplementary Table S3. Intra-rater reliability for evaluation using cropped versus uncropped images on the external test data showed that radiologist 1-3’s ratings were moderate while radiologist 5’s rating was fair. Cohen's kappa scores of intra-rater reliability for the four radiologists were 0•544, 0•560, 0•509, and 0•385, respectively.
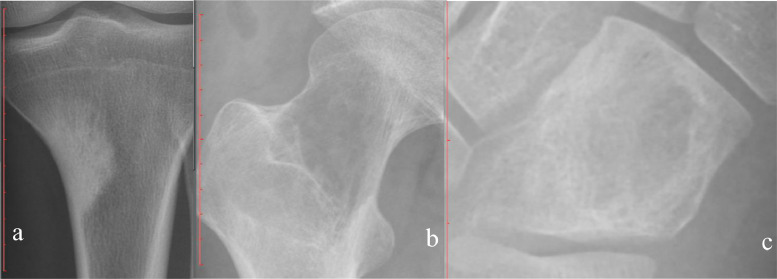
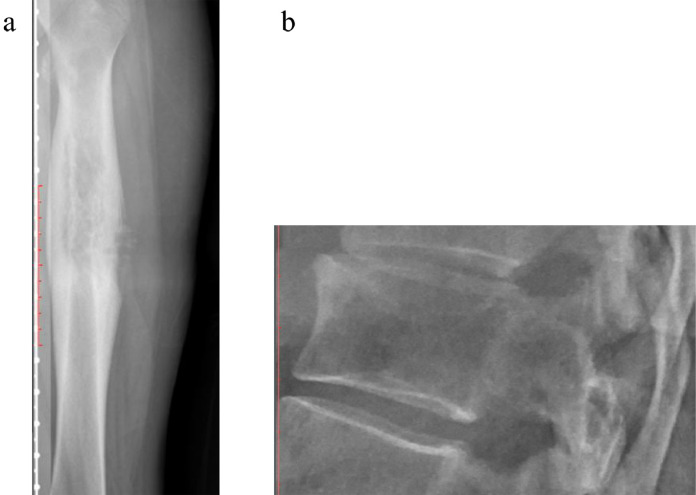
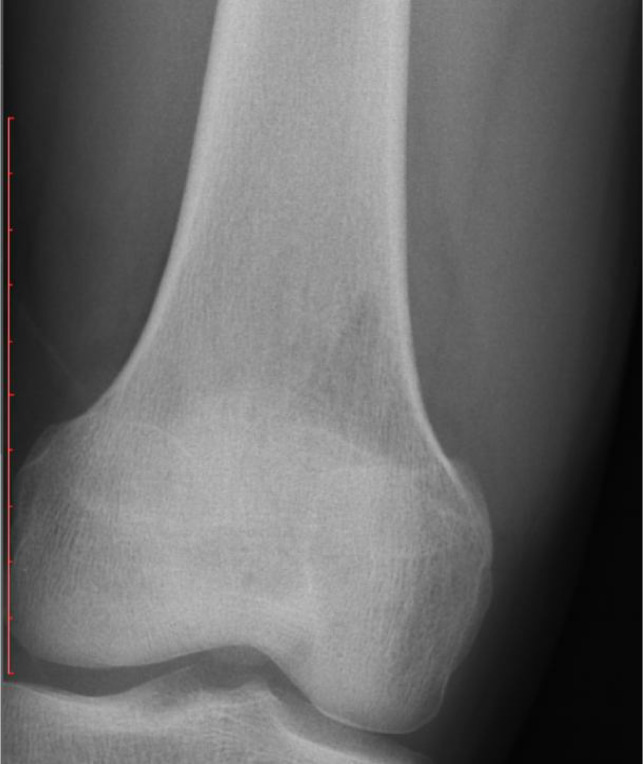
Figs. 2–4 depicts examples of model-subspecialist disagreement under 3 scenarios for prediction of malignancy. Fig. 2 depicts 3 examples of malignant tumors that were predicted to be not malignant by both deep learning model and subspecialists, selected from total number of 15 lesions in the first scenario. These cases either had uncharacteristic appearances (n = 8) or were located in unusual locations (e.g., vertebral body or coccyx) (n = 7). Examples include an osteosarcoma that is completely sclerotic (Fig 2a), a chondrosarcoma that has no calcification of cartilage matrix (Fig 2b), and an Ewing sarcoma that has no cortical destruction or periosteal reaction (Fig 2c). Fig. 3 depicts one example of malignant tumor that was predicted to be malignant by the deep learning model and otherwise by the subspecialists, selected from total number of 9 lesions in the second scenario. Almost all these cases were ill-defined lytic lesions without aggressive periostitis (Fig 3). Fig. 4 demonstrates 2 instances of the opposite of Fig. 3, selected from total number of 22 lesions in the third scenario. These cases all have an aggressive type of periosteal reaction (lamellated, amorphous or sunburst) (Fig 4a), or have a permeative or moth-eaten appearance (Fig 4b).
Fig. 2.
Three examples of malignant tumors that were predicted to be not malignant by both deep learning model and subspecialists. a, Osteosarcoma in upper left tibia predicted to be benign by the deep learning model (67•1%) and benign by 2 subspecialists. b, Chondrosarcoma in upper right femur predicted to be intermediate by the deep learning model (80•5%) and benign and intermediate by 2 subspecialists. c, Ewing sarcoma in right cuboid bone predicted to be benign by the deep learning model (77•2%) and intermediate by 2 subspecialists.
Fig. 4.
Two examples of malignant tumor predicted to be malignant by the subspecialists and otherwise by the deep learning model. a, Ewing sarcoma in left femur diaphysis, predicted to be benign by the deep learning model (95•0%). b, Plasma cell myeloma in T12 vertebral body, predicted to be benign by the deep learning model (81•5%).
Fig. 3.
Examples of malignant tumor that was predicted to be malignant by the deep learning model and otherwise by subspecialists. Osteosarcoma in distal right femur, predicted to be malignant by the deep learning model (99•9%) and intermediate by 2 subspecialists.
Discussion
In this study, we constructed and evaluated a deep learning model for lesion classification on a collection of 2899 images from 1356 patients with histologically confirmed primary bone tumors and pre-operative radiographs. The model achieved similar grouping ability in three-way classification when compared to subspecialists, and better performance than the junior radiologists.
Correctly classifying bone tumors on plain radiograph is important for clinical decision making as it can guide subsequent management [3]. This is especially true in locales where there is a relative lack of subspecialty radiology expertise. Because many bone lesions are uncommon or rare, few radiologists develop sufficient expertise to diagnose them accurately. In clinical practice, one relies on learning and recalling characteristic imaging features of various lesions, both of which are subject to bias. Inappropriate classification of benign bone tumor can lead to unnecessary biopsy and subsequently increased morbidity and cost. In fact, a study utilizing questionnaires revealed that biopsy wounds yielded complications in 17•3% of patients with malignant primary tumors of bone or soft tissue who underwent biopsy, and that biopsy was detrimental to these patients’ prognosis and overall outcome 8•5% of the time [22]. Biopsy of malignant bone tumor without appropriate planning can increase the risk of tumor seeding along the biopsy tract, with the incidence of seeding reported as up to 19•2% following osteosarcoma biopsy [23]. Sampling error presents as another problem for bone biopsy. A diagnosis was not obtained successfully in 7•9% of cases reported with CT image-guided core biopsies of musculoskeletal tumors [24], as well as in 4•7% of open biopsy cases [25,26]. Incorrect diagnosis from tertiary cancer centers also range from 6% to 12% for image-guided core needle biopsies [27]. When the referring center is accounted for, this rate increases to 23% [28]. In addition, CT-guided core biopsy is associated with re-biopsy rate up to 20% of cases [26]. There are also a host of other factors that can prevent providers from obtaining adequate tissue for diagnosis. For instance, many bone tumors are extremely vascular and often yield what appears to be blood only. For lesions that have massive bony sclerosis, such as osteosarcomas, the material obtained is often of poor quality and non-diagnostic. Specimens of benign or malignant cystic lesions or tumors with necrosis are also difficult to obtain for biopsy. The aim of this project was to raise the level of plain radiography analysis through deep learning to the level of the musculoskeletal subspecialists.
Most radiologists rely on “pattern recognition” to differentiate benign from malignant lesions on plain radiograph, which can often lead to erroneous conclusion. Some common radiologic criteria used for this distinction include cortical destruction [29], periostitis [29], orientation or axis of the lesion [30], and zone of transition [30]. However, all have limitations. Cortical bone can be replaced by part of the noncalcified matrix (fibrous matrix or chondroid matrix) of benign fibro-osseous lesions and cartilaginous lesions, giving the false impression of cortical destruction on plain film [31]. Periostitis and orientation of the lesion can be nonspecific [30]. Although the zone of transition is arguably the most useful indicator of whether a lesion is benign or malignant (i.e. a narrow zone of transition indicates a benign lesion and vice versa), it only applies to lytic lesions—a blastic or sclerotic lesion will always appear to have a narrow zone of transition and may erroneously be diagnosed as benign even if it is malignant [31]. Despite the challenges, we have identified no study in the literature which applies deep learning to differentiate benign from malignant bone lesions on plain radiograph. Past studies had the limitations of small cohort size, focus on specific differential diagnoses, and use of advanced imaging modalities [7,8,32,33].
Our model demonstrated good binary discriminatory capacity on cases from different hospitals stratified by age. For the older than 36 years old group, the model's binary discriminatory capacity was slightly lower than that of the younger age group. This can be explained by the smaller sample size on which the deep learning algorithm was trained since most bone tumors were diagnosed in pediatric patients. The good model performance on external testing supports generalizability of our algorithm.
On cross-validation and external testing, our model achieved similar categorical accuracy to the subspecialists for the 3-category classification. This demonstrates that classification of primary bone tumors on radiographs is a challenging problem even for experienced radiologists subspecialized in MSK. Our model performed better than the junior radiologists for all the different age groups, except the younger than 10 years old group. That may be caused by our external testing data containing excessively high proportion of benign bone tumor, such as osteochondroma and osteoid osteoma, which is easy to recognize even for less experienced radiologists.
Deep learning is often considered black box. To understand the choices and mistakes that the model and subspecialists made, we investigated specific cases of model-subspecialist disagreement under 3 scenarios for prediction of malignancy. We also concentrated on wrong prediction of malignant tumors because this would have impacted management and outcome if our algorithm were used in lieu of biopsy. In the first scenario where both model and subspecialists were wrong, we found that the tumors either had uncharacteristic appearances or were located in unusual locations, such as vertebral bodies or the coccyx, where the characterization on plain film was poor. In the second scenario where the model was right but the subspecialists were wrong, we found that almost all the cases were ill-defined lytic lesions without aggressive periostitis. It appears that our deep learning model was better than the subspecialists at evaluating the zone of transition, which is often considered the most reliable plain film indicator for benign versus malignant lesions as discussed above. In the third scenario where the model was wrong and the subspecialists were right, there are some common findings. These cases all had a permeative, moth-eaten appearance or an aggressive type of periosteal reaction (lamellated, amorphous, or sunburst). Although many benign lesions can cause aggressive periostitis such as infection, eosinophilic granuloma, and trauma, our study only included primary bone tumors so aggressive periostitis helped the subspecialist recognize them as malignant. It seems that our deep learning model was not good at recognizing a permeative appearance or aggressive periostitis and associating it with malignancy. This can be explained by either a lack in number or variety of these patterns or both in training. It is also important to note that the difference in class distribution between model and subspecialist predictions. The deep learning model predicted a greater number of benign tumors (50•9% vs. 43•2–43•5%) than subspecialists, largely at the expense of intermediate lesions (18•1% vs. 23•6–24•5%). This is most likely due to the class distribution in the training set (48•7% benign, 24•0% intermediate, and 27•2% malignant), as deep learning model predictions will tend toward the training distribution. This may also suggest that benign and intermediate lesions share similar features learned by the model, causing confusion between these 2 classes.
There are several limitations to our work. First, this is a retrospective study with cases identified from a search of pathology databases at five institutions. In the general population, benign bone tumors are far more common than malignant ones. But due to the tertiary care center character of the five including centers, most typical benign bone tumors are diagnosed directly, without biopsy and pathology. Therefore, our data contained a smaller number of benign bone tumor and large number of intermediate and malignant bone tumor, indicating selection bias. Second, we included only primary bone tumors, but did not consider other situations (e.g. osteomyelitis, metastasis, bone-tumor mimickers, etc.) commonly encountered in clinical practice that often cause diagnostic difficulty. It is also well known that benign processes such as infection and eosinophilic granuloma can mimic malignant tumors [5]. However, a lot of cases were without pathology or were not diagnosed with confidence on pathology. Future studies will include these cases. Third, the images were cropped by a MSK radiologist to highlight the tumor before being inputted into the network. To evaluate the impact of this manual cropped on model performance, a junior radiologist was asked to recrop the images in the external set and model performance was evaluated on this recropped set to compare with the original radiologist. Although we believe that manual cropping keeps the radiologist in the loop who are already interpreting the study, is easy to implement clinically and requires only seconds to complete, it is important to emphasize that the current pipeline is not ready for real-time clinical use. Future study will incorporate deep learning based lesion localization before classification to achieve a fully automated pipeline for clinical integration. Finally, our cohort size is still small compared to the millions of images on ImageNet used to train deep neural network models. Algorithm development can benefit from incorporation of more data from additional institutions, which will result in better performance.
In conclusion, our study shows that deep learning with DCNN can classify primary bone tumors on conventional radiographs using a multi-institutional dataset with similar accuracy to subspecialists, and better performance than the junior radiologists. Our algorithm has the potential to improve primary bone tumor radiographs interpretation to the level of the subspecialists. Future study will focus on development of a fully automatic pipeline including lesion localization, incorporation of studies such as CT or MRI through deep learning and inclusion of bone tumor mimic pathologies.
Author Contributions
HXB and ZZ conceived the study; YH, JW, BB, MC,XP and PJZ collected the data; YH and JG preprocessed the images; HL, RAS, SP and JG evaluated the bone lesions; IP analyzed the data; HXB and KH helped in the analyses and discussion of the results; YH and IP wrote the manuscript; TY, ATD, EF, LJS helped in manuscript editing and revision. All authors contributed to the review, edit, and approvaled the final version of the manuscript.
Ethics approval and consent to participate
The study was conducted in accordance with Declaration of Helsinki and approved by the Institutional Review Boards at Hospital of University of Pennsylvania, Children's Hospital of Philadelphia, Hunan Children's Hospital, Xiangya Hospital and Second Xiangya Hospital.
Consent for publication
The need for informed consent was waived by the institutional review board for this retrospective study.
Availability of data and materials
In respect of patient confidentiality and consent, the radiographs and clinical information datasets analyzed in this study are not available for download but are available upon reasonable request to the corresponding author.
Data sharing
Source code have been uploaded to a public GitHub repository at: https://github.com/i-pan/bone-tumor.
Declaration of Competing Interest
The authors declare that they have no conflicts of interests.
Acknowledgments
Acknowledgments
We acknowledged the help of Ke Jin (K.J.) in data collection.
Funding sources
The project described was supported by RSNA Research & Education Foundation, through grant number RSCH2004 to H. Bai. The content is solely the responsibility of the authors and does not necessarily represent the official views of the RSNA R&E Foundation. The funders had no role in study design, data collection, data analysis, interpretation, writing of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103121.
Contributor Information
Jing Wu, Email: wujing622@csu.edu.cn.
Xianjing Peng, Email: pengxianjing@csu.edu.cn.
Harrison X. Bai, Email: Harrison_Bai@Brown.edu.
Appendix. Supplementary materials
Reference
- 1.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher CDM. International Agency for research on cancer. WHO classification of tumours of soft tissue and bone. 4th ed. IARC Press; Lyon: 2013. World Health Organization; p. 468. p. p. [Google Scholar]
- 4.Do BH, Langlotz C, Beaulieu CF. Bone Tumor Diagnosis Using a Naive Bayesian Model of Demographic and Radiographic Features. J Digit Imaging. 2017;30:640–647. doi: 10.1007/s10278-017-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helms . Wolters Kluwer Health; Philadelphia: 2012. WEBCA. Fundamentals of diagnostic radiology; p. 1420. [Google Scholar]
- 6.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284:574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 7.Bradshaw T, Perk T, Chen S. Deep learning for classification of benign and malignant bone lesions in [F-18]NaF PET/CT images. J Nucl Med. 2018;59:327. [Google Scholar]
- 8.Cheng CT, Ho TY, Lee TY. Application of a deep learning algorithm for detection and visualization of hip fractures on plain pelvic radiographs. Eur Radiol. 2019;29:5469–5477. doi: 10.1007/s00330-019-06167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Fauw J, Ledsam JR, Romera-Paredes B. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 10.Chang K, Bai HX, Zhou H. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24:1073–1081. doi: 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xi IL, Wu J, Guan J. Deep learning for differentiation of benign and malignant solid liver lesions on ultrasonography. Abdom Radiol. 2020 doi: 10.1007/s00261-020-2564-w. [DOI] [PubMed] [Google Scholar]
- 12.Xi IL, Zhao Y, Wang R. Deep learning to distinguish benign from malignant renal lesions based on routine MR imaging. Clin Cancer Res. 2020;26:1944–1952. doi: 10.1158/1078-0432.CCR-19-0374. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y, Chang M, Wang R. Deep learning based on MRI for differentiation of low- and high-grade in low-stage renal cell carcinoma. J Magn Reson Imaging. 2020 doi: 10.1002/jmri.27153. [DOI] [PubMed] [Google Scholar]
- 14.Chang K, Beers AL, Bai HX. Automatic assessment of glioma burden: a deep learning algorithm for fully automated volumetric and bidimensional measurement. NeuroOncol. 2019;21:1412–1422. doi: 10.1093/neuonc/noz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H, Chang K, Bai HX. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J Neurooncol. 2019;142:299–307. doi: 10.1007/s11060-019-03096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuthuru S, Deaderick W, Bai H. A visually interpretable, dictionary-based approach to imaging-genomic modeling, with low-grade glioma as a case study. Cancer Inform. 2018;17 doi: 10.1177/1176935118802796. DOI: 1176935118802796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Vallieres M, Bai HX. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19:862–870. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan I, Agarwal S, Merck D. Generalizable Inter-Institutional Classification of Abnormal Chest Radiographs Using Efficient Convolutional Neural Networks. J Digit Imaging. 2019;32:888–896. doi: 10.1007/s10278-019-00180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo YH, Xi IL, Wang R. Deep learning based on mr imaging for predicting outcome of uterine fibroid embolization. J Vasc Interv Radiol. 2020;31 doi: 10.1016/j.jvir.2019.11.032. 1010-7.e3. [DOI] [PubMed] [Google Scholar]
- 20.Bai HX, Wang R, Xiong Z. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. 2020;296:E156–EE65. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mingxing T, Quoc V L. EfficientNet: rethinking model scaling for convolutional neural networks: ICML 2019 [Available from: https://arxiv.org/pdf/1905.11946.pdf.
- 22.Mankin HJ, Lange TA, Spanier SS. THE CLASSIC: The hazards of biopsy in patients with malignant primary bone and soft-tissue tumors. The Journal of Bone and Joint Surgery, 1982;64:1121-1127. Clin Orthop Relat Res. 2006;450:4–10. doi: 10.1097/01.blo.0000229299.36969.b5. [DOI] [PubMed] [Google Scholar]
- 23.Seeger LL. Revisiting tract seeding and compartmental anatomy for percutaneous image-guided musculoskeletal biopsies. Skeletal Radiol. 2019;48:499–501. doi: 10.1007/s00256-018-3127-x. [DOI] [PubMed] [Google Scholar]
- 24.Altuntas AO, Slavin J, Smith PJ. Accuracy of computed tomography guided core needle biopsy of musculoskeletal tumours. ANZ J Surg. 2005;75:187–191. doi: 10.1111/j.1445-2197.2005.03332.x. [DOI] [PubMed] [Google Scholar]
- 25.Wallace MT, Lin PP, Bird JE, Moon BS, Satcher RL, Lewis VO. The accuracy and clinical utility of intraoperative frozen section analysis in open biopsy of bone. J Am Acad Orthop Surg. 2019;27:410–417. doi: 10.5435/JAAOS-D-18-00071. [DOI] [PubMed] [Google Scholar]
- 26.Ashford RU, McCarthy SW, Scolyer RA, Bonar SF, Karim RZ, Stalley PD. Surgical biopsy with intra-operative frozen section. An accurate and cost-effective method for diagnosis of musculoskeletal sarcomas. J Bone Jt Surg Br. 2006;88:1207–1211. doi: 10.1302/0301-620X.88B9.17680. [DOI] [PubMed] [Google Scholar]
- 27.Jelinek JS, Murphey MD, Welker JA. Diagnosis of primary bone tumors with image-guided percutaneous biopsy: experience with 110 tumors. Radiology. 2002;223:731–737. doi: 10.1148/radiol.2233011050. [DOI] [PubMed] [Google Scholar]
- 28.Saifuddin A, Mitchell R, Burnett SJ, Sandison A, Pringle JA. Ultrasound-guided needle biopsy of primary bone tumours. J Bone Jt Surg Br. 2000;82:50–54. doi: 10.1302/0301-620x.82b1.10141. [DOI] [PubMed] [Google Scholar]
- 29.Teo HE, Peh WC. Primary bone tumors of adulthood. Cancer Imaging. 2004;4:74–83. doi: 10.1102/1470-7330.2004.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umer M, Hasan OHA, Khan D, Uddin N, Noordin S. Systematic approach to musculoskeletal benign tumors. Int J Surg Oncol. 2017;2:e46. doi: 10.1097/IJ9.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remotti F, Feldman F. Nonneoplastic lesions that simulate primary tumors of bone. Arch Pathol Lab Med. 2012;136:772–788. doi: 10.5858/arpa.2011-0557-RA. [DOI] [PubMed] [Google Scholar]
- 32.Filograna L, Lenkowicz J, Cellini F. Identification of the most significant magnetic resonance imaging (MRI) radiomic features in oncological patients with vertebral bone marrow metastatic disease: a feasibility study. Radiol Med. 2019;124:50–57. doi: 10.1007/s11547-018-0935-y. [DOI] [PubMed] [Google Scholar]
- 33.Yin P, Mao N, Zhao C. Comparison of radiomics machine-learning classifiers and feature selection for differentiation of sacral chordoma and sacral giant cell tumour based on 3D computed tomography features. Eur Radiol. 2019;29:1841–1847. doi: 10.1007/s00330-018-5730-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In respect of patient confidentiality and consent, the radiographs and clinical information datasets analyzed in this study are not available for download but are available upon reasonable request to the corresponding author.