Abstract
Objective
To investigate the preoperative predictive value of non-invasive imaging biomarkers for programmed cell death protein 1/programmed cell death protein ligand 1 (PD-1/PD-L1) expression and outcome in intrahepatic cholangiocarcinoma (ICC) using machine learning.
Methods
PD-1/PD-L1 expression in 98 ICC patients was assessed by immunohistochemistry, and their prognostic effects were analysed using Cox regression and Kaplan-Meier analysis. Radiomic features were extracted from MRI in the arterial and portal vein phases, and three sets of Radiomics score (Radscore) with good performance were derived respectively as biomarkers for predicting PD-1, PD-L1 expression and overall survival (OS). PD-1 and PD-L1 expression models were developed using the Radscore (arterial phase), clinico-radiological factors and clinical factors, individually and in combination. The imaging-based OS predictive model was constructed by combining independent predictors among clinico-radiological, clinical factors and OS Radscore. Pathology-based OS model using pathological and clinical factors was also constructed and compared with imaging-based OS model.
Results
The highest area under the curves of the models predicting PD-1 and PD-L1 expression was 0.897 and 0.890, respectively. PD-1+ and PD-L1+ cases had worse outcomes than negative cases. The 5-year survival rates of PD-1+ and PD-1− cases were 12.5% and 48.3%, respectively (p<0.05), whereas the 5-year survival was 21.9% and 39.4% for PD-L1+ and PD-L1− cases, respectively (p<0.05). The imaging-based OS model involved predictors of clinico-radiological ‘imaging classification’, radiomics ‘Radscore’ from arterial phase and carcinoembryonic antigen (CEA) level (C-index:0.721). It performed better than pathology-based model (C-index: 0.698) constructed by PD-1/PD-L1 expression status and CEA level. The imaging-based OS model is potential for practice when the pathology assay is unavailable and could divide ICC patients into high-risk and low-risk groups, with 1-year, 3-year and 5-year survival rates of 57.1%, 14.3% and 12.4%, and 87.8%, 63.3% and 55.3%, respectively (p<0.001).
Conclusions
MRI radiomics could derive promising and non-invasive biomarker in evaluating PD-1/PD-L1 expression and prognosis of ICC patients.
Keywords: intrahepatic cholangiocarcinoma, PD-1, PD-L1, MRI, radiomics
Key questions.
What is already known about this subject?
Previous studies have reported that the programmed cell death protein 1/programmed cell death protein ligand 1 (PD-1/PD-L1) upregulation expression positively related to response to blocking the PD-1/PD-L1. However, there is still a lack of reports on non-invasive examination for PD-1/PD-L1 expression in intrahepatic cholangiocarcinoma (ICC).
What does this study add?
To our knowledge, our study is the first to preoperatively predict PD-1/PD-L1 expression in ICC using MRI radiomics based on machine learning.
How might this impact on clinical practice?
Our findings suggest that MRI radiomics could serve as a non-invasive biomarker in predicting PD-1/PD-L1 expression and prognosis of ICC patients. It may guide clinical decision-making in selecting ICC patients suitable for blocking PD-1/PD-L1 and prognostic evaluation.
Introduction
Intrahepatic cholangiocarcinoma (ICC) is the second most common primary liver malignancy after hepatocellular carcinoma.1 Currently, its morbidity and mortality rates have been steadily rising worldwide.2 Surgical resection cures a minority of ICC patients, yet most ICC patients are already in advanced clinical stages with unresectable tumours at the time of diagnosis, although great progress has been made in treatment modalities in recent decades.3 Due to local invasion, metastasis, and suboptimal treatments, the prognosis of ICC patients remains dismal,4 with a 5-year overall survival (OS) rate ranging from 14% to 40%.3 Consequently, new treatment strategies for ICC patients are urgently required.
Advances in tumour immunology have contributed to the clinical applications of new immunotherapeutic strategies. Immune checkpoint pathway members, such as programmed cell death protein 1 (PD-1) and programmed cell death protein ligand 1 (PD-L1), have attracted increasing attention in recent years. PD-1 presents on the membrane surface of T lymphocytes with various activation levels and has a negative regulatory effect in antigen responses.5 PD-L1 is broadly expressed on the membrane surface of antigen-presenting cells and tumour cells and induces the dysfunction or apoptosis of T cells after it binds to PD-1, thereby suppressing the antitumour immune response and escaping from it.
In recent years, drugs blocking the PD-1/PD-L1 pathway have shown potential in improving patient survival and in the treatment of a variety of malignant tumours.6–8 These agents suggest that improved treatment outcomes can be achieved for ICC patients. However, the main challenges in developing successful drugs targeting the immune checkpoint blockade are to select patient subgroups that would benefit most from them9 and to avoid ineffective treatments and potential side effects related to autoimmune effects resulting from blocking the PD-1/PD-L1 pathway.10 Because only 20%–50% of patients with advanced tumours respond to immune checkpoint inhibitors,11 discovering molecular markers to assess immunotherapy responses in ICC patients is important.12 Previous studies have demonstrated that the expression status of PD-L1 in tumours is associated with clinical outcomes and treatment responses of PD-1/PD-L1 pathway inhibition.13 Therefore, reliable preoperative prediction of the expression status of PD-1/PD-L1 has clinical implications in the immune checkpoint blockade for ICC patients.
Currently, immunohistochemical staining is a commonly used method to assess the expression status of PD-1/PD-L1 in tumours based on needle biopsies. However, outcomes are often confounded by the heterogeneous expression of these markers. This means that the value of measuring PD-1/PD-L1 expression in clinical applications is greatly limited. Molecular imaging can reveal the tumour microenvironment and enable visual monitoring of the expression of target molecules and cells in real-time using specific radionuclides or optical probes. Immune checkpoint target-associated molecular imaging has also been explored.14 However, the lack of validation for ICC and radiation concerns limit the rapid application of these probes.
Radiomics converts medical images into quantitative data15 to gain insight into the hidden information of tumour phenotypes based on the underlying hypothesis that cellular and molecular properties of tumours could be indirectly mirrored by medical imaging, and to produce image-driven biomarkers to better aid clinical decisions.16 This technique makes it possible to assess the microenvironment and spatial heterogeneity of tumours. Sun et al17 reported that radiomics can be used to assess lymphocytes and the response to immune checkpoint inhibitors. Tang et al18 showed that CT-radiomics correlate with CD3 infiltration and PD-L1 expression in non-small cell lung cancer. Hence, radiomics can be considered a noninvasive technology for predicting PD-1/PD-L1 expression in tumours. However, there is still a shortage of studies on the prediction of PD-1/PD-L1 in ICC patients.
In this study, we aimed to investigate the capability of MRI radiomics-based models for preoperatively predicting the expression of PD-1/PD-L1 and clinical outcome in ICC patients.
Materials and methods
Patients
The requirement for informed consent was waived.
We searched medical records at West China Hospital, Sichuan University (Chengdu, China) to identify ICC cases that had been pathologically diagnosed from January 2012 to December 2014. The inclusion and exclusion criteria are shown in online supplemental appendix 1. In total, 98 ICC patients were included in our study cohort during the above study period. The flow chart of patient enrolment is shown in online supplemental figure 1.
esmoopen-2020-000910supp001.pdf (49.8KB, pdf)
esmoopen-2020-000910supp002.pdf (396.7KB, pdf)
Imaging acquisition
The details are shown in online supplemental appendix 2.
esmoopen-2020-000910supp003.pdf (123.9KB, pdf)
Imaging evaluation
The details are shown in online supplemental appendix 3.
esmoopen-2020-000910supp004.pdf (49.8KB, pdf)
Histopathology
The details are shown in online supplemental appendix 4.
esmoopen-2020-000910supp005.pdf (47.9KB, pdf)
Follow-up
Patients were consistently followed up after surgery and monitored prospectively by chest X-ray, CT and/or MRI at intervals of 3–6 months. OS was defined as the interval between surgery and death or between surgery and the last follow-up. The data were censored at the last follow-up for surviving patients, or death was recorded.
Tumour segmentation and features extraction
The details of tumour segmentation are shown in online supplemental appendix 5. Radiomic features were extracted from each MRI modality and then normalised as described by the previous report.19
esmoopen-2020-000910supp006.pdf (54.1KB, pdf)
Prediction models of PD-1/PD-L1 expression
Briefly, the extracted radiomics features were selected separately to construct radiomics-based PD-1 and PD-L1 predicting model. For both models, the extracted radiomics features were initially assessed by Spearman rank correlation (r>0.7); the least absolute shrinkage and selection operator (LASSO) method was used to identify the most powerful predictive radiomics features (online supplemental figures 2 and 3). Logistic regression models were developed using radiomic features derived from the arterial phase (AP) and portal vein phase (PVP) MR images and the model with the best diagnostic performance was selected as the final radiomics model. The radiomic score (Radscore) for each patient was calculated using such determined radiomics model.
esmoopen-2020-000910supp007.pdf (953.5KB, pdf)
esmoopen-2020-000910supp008.pdf (964.4KB, pdf)
For MRI radiographic and clinical factors, univariate analysis with a generalised linear model was applied to identify predictors. Multivariate analysis, tests of the association with the Radscore, clinical factor evaluations and MRI were then performed to identify significant predictors based on a backward stepwise selection process with the Akaike information criterion. The predictive models including the individual or combination models were internally validated using fivefold cross-validation and 1000 times bootstrap internal validation were conducted for the radiomics models with the best performance.
Development of a clinical OS predictive model
The extracted radiomics features from AP and PVP MR images were selected separately to construct OS predictive model for ICC patients. And the model with better performance (C-index) was selected to be further combined with clinico-radiological and clinical factors, forming combined OS predictive model.
After using the Spearman correlation analysis to reduce redundant features, the univariate and multivariate LASSO Cox regression methods was applied separately and sequentially in AP and PVP MR images to identify the most relevant predictive radiomic feature set in a Cox proportional hazards model (online supplemental figure 4). The model performance was evaluated using C-index. The OS radiomics model (AP or PVP) with higher C-index was kept for further analysis. For each patient, the output of the radiomics model was evaluated using the Radscore, which was calculated as the linear combination of the selected radiomics features weighted by their individual coefficients.
esmoopen-2020-000910supp009.pdf (1,002.4KB, pdf)
We constructed two OS models including pathology-based model (pathological features and clinical factors) and imaging-based model (AP or PVP radiomics model with better performance +clinic-radiological factors+clinical factors), using multivariable Cox regression method.
Given a cut-off value of the median in the above model, ICC patients were divided into high-risk and low-risk groups. The log-rank test was applied to compare the two separate Kaplan-Meier (KM) survival curves. Furthermore, a nomogram was built for the imaging-based model to predict the probabilities of 1-year, 3-year and 5-year OS. A calibration curve was plotted to evaluate its predicting performance.
Statistical analysis
The t-test or the Mann-Whitney U test, and the χ2 test or Fisher’s exact test, were used for numerical variables and categorical variables, respectively. Interobserver agreement for assessing the reliability of MRI evaluation used the kappa test, the performance of each predictive model was quantified by the receiver operating characteristic) curve and the area under the curve (AUC), and the nomogram and its calibration were tested as previously reported.19 OS was evaluated based on the KM survival curves and the Cox proportional hazards model.
Statistical analysis was performed using R software (V.3.5.2). A p value of less than 0.05 was defined as significant in two-tailed analyses.
Results
Clinico-pathological characteristics and liver MRI
Out of 98 ICC patients included, 40 cases and 58 cases were PD-1+ and PD-1−, and 32 cases and 66 cases were PD-L1+ and PD-L1−, respectively. PD-1+ and PD-1− was statistically significant in pathological differentiation (OR 2.45; 95% CI 0.76 to 7.86), capsular retraction (OR 0.72; 95% CI 0.2 to 2.62), imaging classification (IC) (OR 0.32; 95% CI 0.07 to 1.59), intratumour vascularity (OR 4.11; 95% CI 0.69 to 24.3) and enhancement patterns (OR 0.18; 95% CI 0.04 to 0.96) based on multivariable analysis. However, no statistically significant clinico-pathological factors or MRI differences were found between the PD-L1+ and PD-L1− expression groups. The univariate analysis results are summarised in tables 1 and 2.
Table 1.
The patient’s demographics and clinicopathological characteristics
| PD-1 | PD-L1 | |||||||
| n | Negative | Positive | P value | n | Negative | Positive | P value | |
| Sex | 0.369 | 0.312 | ||||||
| Female | 47 | 30 (51.72) | 17 (42.50) | 47 | 34 (51.52) | 13 (40.62) | ||
| Male | 51 | 28 (48.28) | 23 (57.50) | 51 | 32 (48.48) | 19 (59.38) | ||
| Age | 98 | 57.47±11.86 | 56.65±10.79 | 0.729 | 98 | 56.71±11.92 | 58.00±10.32 | 0.602 |
| Adjacent organ invasion | 0.768 | 0.475 | ||||||
| Absent | 82 | 48 (82.76) | 34 (85.00) | 82 | 54 (81.82) | 28 (87.50) | ||
| Present | 16 | 10 (17.24) | 6 (15.00) | 16 | 12 (18.18) | 4 (12.50) | ||
| Nerve invasion | 0.349 | 0.802 | ||||||
| Absent | 78 | 48 (82.76) | 30 (75.00) | 78 | 53 (80.30) | 25 (78.12) | ||
| Present | 20 | 10 (17.24) | 10 (25.00) | 20 | 13 (19.70) | 7 (21.88) | ||
| Satellite nodule | 0.899 | 0.512 | ||||||
| Absent | 79 | 47 (81.03) | 32 (80.00) | 79 | 52 (78.79) | 27 (84.38) | ||
| Present | 19 | 11 (18.97) | 8 (20.00) | 19 | 14 (21.21) | 5 (15.62) | ||
| Necrosis | 0.973 | 0.147 | ||||||
| Absent | 81 | 48 (82.76) | 33 (82.50) | 81 | 52 (78.79) | 29 (90.62) | ||
| Present | 17 | 10 (17.24) | 7 (17.50) | 17 | 14 (21.21) | 3 (9.38) | ||
| Intravascular tumour thrombus | 0.86 | 0.097 | ||||||
| Absent | 90 | 53 (91.38) | 37 (92.50) | 90 | 58 (87.88) | 32 (100.00) | ||
| Present | 8 | 5 (8.62) | 3 (7.50) | 8 | 8 (12.12) | 0 (0.00) | ||
| Lymph node metastasis | 0.397 | 0.565 | ||||||
| Absent | 73 | 45 (77.59) | 28 (70.00) | 73 | 48 (72.73) | 25 (78.12) | ||
| Present | 25 | 13 (22.41) | 12 (30.00) | 25 | 18 (27.27) | 7 (21.88) | ||
| Surgical margin | 0.693 | 0.867 | ||||||
| R0 | 88 | 51 (87.93) | 37 (92.50) | 88 | 59 (89.39) | 29 (90.62) | ||
| R1 | 10 | 7 (12.07) | 3 (7.50) | 10 | 7 (10.61) | 3 (9.38) | ||
| ALT (IU/L) | 0.795 | 0.946 | ||||||
| <40 | 70 | 42 (72.41) | 28 (70.00) | 70 | 47 (71.21) | 23 (71.88) | ||
| ≥40 | 28 | 16 (27.59) | 12 (30.00) | 28 | 19 (28.79) | 9 (28.12) | ||
| AST (IU/L) | 0.395 | 0.476 | ||||||
| <35 | 66 | 41 (70.69) | 25 (62.50) | 66 | 46 (69.70) | 20 (62.50) | ||
| ≥35 | 32 | 17 (29.31) | 15 (37.50) | 32 | 20 (30.30) | 12 (37.50) | ||
| GGT (IU/L) | 0.106 | 0.603 | ||||||
| <45 | 31 | 22 (37.93) | 9 (22.50) | 31 | 22 (33.33) | 9 (28.12) | ||
| ≥45 | 67 | 36 (62.07) | 31 (77.50) | 67 | 44 (66.67) | 23 (71.88) | ||
| CEA (ng/mL) | 0.867 | 0.471 | ||||||
| <3.4 | 50 | 30 (51.72) | 20 (50.00) | 50 | 32 (48.48) | 18 (56.25) | ||
| ≥3.4 | 48 | 28 (48.28) | 20 (50.00) | 48 | 34 (51.52) | 14 (43.75) | ||
| CA-199 (U/mL) | 0.366 | 0.476 | ||||||
| <22 | 32 | 21 (36.21) | 11 (27.50) | 32 | 20 (30.30) | 12 (37.50) | ||
| ≥22 | 66 | 37 (63.79) | 29 (72.50) | 66 | 46 (69.70) | 20 (62.50) | ||
| AFP (ng/mL) | 0.853 | 0.871 | ||||||
| <8 | 85 | 50 (86.21) | 35 (87.50) | 85 | 57 (86.36) | 28 (87.50) | ||
| ≥8 | 13 | 8 (13.79) | 5 (12.50) | 13 | 9 (13.64) | 4 (12.50) | ||
| Cirrhosis | 0.61 | 0.147 | ||||||
| Absent | 81 | 47 (81.03) | 34 (85.00) | 81 | 52 (78.79) | 29 (90.62) | ||
| Present | 17 | 11 (18.97) | 6 (15.00) | 17 | 14 (21.21) | 3 (9.38) | ||
| Hepatitis B | 0.923 | 0.285 | ||||||
| Absent | 73 | 43 (74.14) | 30 (75.00) | 73 | 47 (71.21) | 26 (81.25) | ||
| Present | 25 | 15 (25.86) | 10 (25.00) | 25 | 19 (28.79) | 6 (18.75) | ||
| pathology | 0.04 | 0.665 | ||||||
| Well | 4 | 3 (5.17) | 1 (2.50) | 4 | 2 (3.03) | 2 (6.25) | ||
| Moderate | 68 | 44 (75.86) | 24 (60.00) | 68 | 48 (72.73) | 20 (62.50) | ||
| Poor | 26 | 11 (18.97) | 15 (37.50) | 26 | 16 (24.24) | 10 (31.25) | ||
| Diameter | 0.287 | 0.357 | ||||||
| ≤3 cm | 23 | 18 (31.03) | 5 (12.50) | 23 | 15 (22.73) | 8 (25.00) | ||
| >3 and ≤5 cm | 21 | 9 (15.52) | 12 (30.00) | 21 | 12 (18.18) | 9 (28.12) | ||
| >5 cm | 54 | 31 (53.45) | 23 (57.50) | 54 | 39 (59.09) | 15 (46.88) | ||
Data are the number of tumours, with percentages in parentheses.
AFP, a-fetoprotein; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CA-199, cancer antigen 199; CEA, carcinoembryonic antigen; GGT, γ-glutamyl transpeptidase; PD-1, programmed cell death protein 1; PD-L1, programmed cell death protein ligand 1.
Table 2.
The MRI
| PD-1 | PD-L1 | |||||||
| n | Negative | Positive | P value | n | Negative | Positive | P value | |
| Shape | 0.255 | 0.866 | ||||||
| Regular | 41 | 27 (46.55) | 14 (35.00) | 41 | 28 (42.42) | 13 (40.62) | ||
| Irregular | 57 | 31 (53.45) | 26 (65.00) | 57 | 38 (57.58) | 19 (59.38) | ||
| Margin | 0.165 | 0.464 | ||||||
| Well defined | 45 | 30 (51.72) | 15 (37.50) | 45 | 32 (48.48) | 13 (40.62) | ||
| Ill defined | 53 | 28 (48.28) | 25 (62.50) | 53 | 34 (51.52) | 19 (59.38) | ||
| Peritumoural bile duct dilatation | 0.749 | 0.672 | ||||||
| Absent | 46 | 28 (48.28) | 18 (45.00) | 46 | 30 (45.45) | 16 (50.00) | ||
| Present | 52 | 30 (51.72) | 22 (55.00) | 52 | 36 (54.55) | 16 (50.00) | ||
| Hepatic lobe atrophy | 0.899 | 0.328 | ||||||
| Absent | 79 | 47 (81.03) | 32 (80.00) | 79 | 55 (83.33) | 24 (75.00) | ||
| Present | 19 | 11 (18.97) | 8 (20.00) | 19 | 11 (16.67) | 8 (25.00) | ||
| Capsular retraction | 0.037 | 0.784 | ||||||
| Absent | 54 | 37 (63.79) | 17 (42.50) | 54 | 37 (56.06) | 17 (53.12) | ||
| Present | 44 | 21 (36.21) | 23 (57.50) | 44 | 29 (43.94) | 15 (46.88) | ||
| Imaging classification | 0.044 | 0.946 | ||||||
| Parenchymal | 28 | 21 (36.21) | 7 (17.50) | 28 | 19 (28.79) | 9 (28.12) | ||
| Ductal | 70 | 37 (63.79) | 33 (82.50) | 70 | 47 (71.21) | 23 (71.88) | ||
| Target sign in T2WI | 0.033 | 0.795 | ||||||
| Absent | 75 | 40 (68.97) | 35 (87.50) | 75 | 50 (75.76) | 25 (78.12) | ||
| Present | 23 | 18 (31.03) | 5 (12.50) | 23 | 16 (24.24) | 7 (21.88) | ||
| Target sign in DWI | 0.188 | 0.683 | ||||||
| Absent | 61 | 33 (56.90) | 28 (70.00) | 61 | 42 (63.64) | 19 (59.38) | ||
| Present | 37 | 25 (43.10) | 12 (30.00) | 37 | 24 (36.36) | 13 (40.62) | ||
| Enhancement patterns | 0.019 | 0.401 | ||||||
| Hypoarterial/mild-arterial enhancement | 68 | 35 (60.34) | 33 (82.50) | 68 | 44 (66.67) | 24 (75.00) | ||
| Hyperarterial enhancement | 30 | 23 (39.66) | 7 (17.50) | 30 | 22 (33.33) | 8 (25.00) | ||
| Intratumour vascularity | 0.004 | 0.871 | ||||||
| Absent | 85 | 55 (94.83) | 30 (75.00) | 85 | 57 (86.36) | 28 (87.50) | ||
| Present | 13 | 3 (5.17) | 10 (25.00) | 13 | 9 (13.64) | 4 (12.50) | ||
| Intratumoural separate structure | 0.88 | 0.764 | ||||||
| Absent | 45 | 27 (46.55) | 18 (45.00) | 45 | 31 (46.97) | 14 (43.75) | ||
| Present | 53 | 31 (53.45) | 22 (55.00) | 53 | 35 (53.03) | 18 (56.25) | ||
| Position | 0.173 | 0.897 | ||||||
| One lobe | 93 | 57 (98.28) | 36 (90.00) | 93 | 63 (95.45) | 30 (93.75) | ||
| More lobes | 5 | 1 (1.72) | 4 (10.00) | 5 | 3 (4.55) | 2 (6.25) | ||
Data are the number of tumours, with percentages in parentheses.
DWI, diffusion weighted imaging; PD-1, programmed cell death protein 1; PD-L1, programmed cell death protein ligand 1; T2WI, T2 weighted imaging.
Prediction of PD-1/PD-L1 expression
The ROIs size in different expression subgroups were no statistical difference between the AP and PVP (online supplemental table S1). The predictive model using the radiomics features derived from the AP showed better performance than that based on the PVP in the prediction of both PD-1 and PD-L1 expression.
esmoopen-2020-000910supp010.pdf (71.9KB, pdf)
For predicting PD-1 expression, the best diagnostic efficacy was achieved by the combination of the Radscore from AP images and clinico-radiological factors (‘pathology‘ ‘imaging classification’, ‘enhancement’, ‘intratumour vascularity’) (details in online supplemental appendix 6). For predicting PD-L1, only radiomics features in AP were used because no statistically significant clinico-radiological factors were found to be predictive. The predictive models showed high stability and reliability in intragroup fivefold cross-validation. The results were summarised in table 3 and shown in online supplemental figure 5). The selected radiomics features linked to PD-1 and PD-L1 expression are shown in online supplemental tables S2 and S3, respectively. As the radiomics model heavily relied on the multiple features, in order to check the reliability of the AP radiomics model for predicting PD-1 and PD-L1 expression status, they were internally validated using 1000 times bootstrap. For the AP radiomics model in predicting PD-1 expression, the overall accuracy is 0.703 compared with the accuracy (0.776) derived from the entire original dataset. For the AP radiomics model in predicting PD-L1 expression, the overall accuracy is 0.769 compared with the accuracy (0.878) derived from the entire original dataset. The adjusted C-index and the calibration curves for the two radiomics models (AP) were also summarised as online supplemental table S4) and (online supplemental figure 6.
Table 3.
Models performances of prediction PD-1/PD-L1 expression
| PD-1 | PD-L1 | |||||||||
| AUC (95% CI) | ACC | SPE | SEN | P value* | AUC (95% CI) | ACC | SPE | SEN | P value† | |
| AP | 0.840 (0.764 to 0.916) | 0.776 | 0.828 | 0.700 | 0.033 | 0.890 (0.812 to 0.967) | 0.878 | 0.924 | 0.781 | – |
| Mean folds | 0.740 | 0.707 | 0.800 | 0.680 | 0.750 | 0.745 | 0.877 | 0.622 | ||
| PVP | 0.773 (0.678 to 0.868) | 0.704 | 0.620 | 0.825 | 0.029 | 0.878 (0.805 to 0.950) | 0.836 | 0.894 | 0.719 | 0.8 |
| Mean folds | 0.604 | 0.672 | 0.649 | 0.600 | 0.793 | 0.776 | 0.848 | 0.739 | ||
| Radiological model | 0.694 (0.606 to 0.783) | 0.592 | 0.379 | 0.900 | <0.0001 | |||||
| Mean folds | 0.662 | 0.663 | 0.648 | 0.625 | ||||||
| Clinical radiologic | 0.724 (0.625 to 0.823) | 0.642 | 0.517 | 0.825 | <0.0001 | NA | ||||
| Mean folds | 0.689 | 0.663 | 0.506 | 0.775 | ||||||
| Radscore clinical | 0.866 (0.797 to 0.935) | 0.786 | 0.793 | 0.775 | 0.105 | NA | ||||
| Mean folds | 0.820 | 0.785 | 0.890 | 0.749 | ||||||
| Radscore radiologic | 0.883 (0.818 to 0.948) | 0.826 | 0.862 | 0.775 | 0.267 | NA | ||||
| Mean folds | 0.852 | 0.735 | 0.824 | 0.85 | ||||||
| Combined model | 0.897 (0.836 to 0.959) | 0.827 | 0.810 | 0.850 | – | NA | ||||
| Mean folds | 0.822 | 0.816 | 0.872 | 0.772 | ||||||
*P value of Delong’s test between every model and combined model in predicting PD-1.
†P value of Delong’s test between PVP and AP radiomics model in predicting PD-L1.
ACC, accuracy; AP, arterial phase; AUC, area under the curve; NA, not available; PD-1, programmed cell death protein 1; PD-L1, programmed cell death protein ligand 1; PVP, portal vein phase; SEN, sensitivity; SPE, specificity.
esmoopen-2020-000910supp011.pdf (252.6KB, pdf)
esmoopen-2020-000910supp012.pdf (1.1MB, pdf)
esmoopen-2020-000910supp013.pdf (68.2KB, pdf)
esmoopen-2020-000910supp014.pdf (70.3KB, pdf)
esmoopen-2020-000910supp015.pdf (39.5KB, pdf)
esmoopen-2020-000910supp016.pdf (177KB, pdf)
Correlation of PD-1/PD-L1 expression with prognosis
PD-1+ or PD-L1+ in ICC patients was more often associated with poor outcomes than PD-1− or PD-L1−, and the difference was statistically significant (p<0.05) (online supplemental figure 7). The median survival time in PD-1+ ICC patients was 16.5 months (IQR: 9–30 months); however, it was 44 months (IQR: 22–63.8 months) in PD-1− ICC patients. The 1-year, 3-year and 5-year survival rates of PD-1+ and PD-1− ICC patients were 55.0%, 22.5%, 12.5% and 84.5%, 50.0%, and 48.3%, respectively. The median survival time in PD-L1+ ICC patients was 26.1 months (IQR: 10.8–29.3 months) compared with 34.1 months (IQR: 13.3–63 months) in PD-L1− ICC patients. The 1-year, 3-year and 5-year survival rates of PD-L1+ and PD-L1− ICC patients were 65.6%, 21.9%, 21.9% and 75.8%, 47.0% and 39.4%, respectively.
esmoopen-2020-000910supp017.pdf (146.2KB, pdf)
Performance of the clinical OS predictive model
We used features extracted both from AP and PVP MR images to construct two sets of AP or PVP radiomics OS models. But we only retained one radiomics model (AP model) with better predictive performance (AP C-index: 0.673 vs PVP C-index: 0.594) for further study. By using univariate and multivariate LASSO Cox regression method, 11 radiomic features related to the OS of ICC patients were identified, and the corresponding Radscore was calculated (online supplemental table S5).
esmoopen-2020-000910supp018.pdf (62.8KB, pdf)
After the radiomics OS model (AP Radscore) determined, we constructed two OS models including pathology-based model and imaging-based model. The univariate and multivariate Cox regression results and the updated predictors in these two OS models were summarised in table 4.
Table 4.
Univariable and multivariable Cox analysis of factors associated with OS of intrahepatic cholangiocarcinoma
| Variable | Univariable analysis | Multivariable analysis Pathology OS model |
Multivariable analysis Imaging OS model |
|||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| PD-1 | 2.930 (1.793 to 4.789) | <0.001 | 3.347 (2.007 to 5.581) | <0.0001 | ||
| PD-L1 | 1.867 (1.134 to 3.074) | 0.017 | 1.868 (1.127 to 3.097) | 0.015 | ||
| Sex | 1.165 (0.715 to 1.898) | 0.539 | ||||
| Age | 0.997 (0.977 to 1.017) | 0.756 | ||||
| Pathology | 1.130 (0.693 to 1.844) | 0.625 | ||||
| Diameter | 1.134 (0.849 to 1.516) | 0.389 | ||||
| No | 2.203 (1.040 to 4.664) | 0.060 | ||||
| Adjacent organ invasion | 1.451 (0.776 to 2.714) | 0.263 | ||||
| Nerve invasion | 2.313 (1.334 to 4.010) | 0.005 | ||||
| Satellite nodule | 1.745 (0.990 to 3.076) | 0.067 | ||||
| Necrosis | 1.592 (0.867 to 2.923) | 0.153 | ||||
| Intravascular tumour thrombus | 1.715 (0.740 to 3.975) | 0.242 | ||||
| Lymph node metastasis | 1.827 (1.090 to 3.063) | 0.028 | ||||
| Surgical margin | 0.845 (0.365 to 1.956) | 0.687 | ||||
| ALT | 1.124 (0.665 to 1.901) | 0.664 | ||||
| AST | 1.285 (0.774 to 2.133) | 0.339 | ||||
| GGT | 1.952 (1.109 to 3.436) | 0.015 | ||||
| CEA | 1.918 (1.177 to 3.123) | 0.008 | 2.485 (1.502 to 4.111) | 0.0004 | 1.946 (1.189 to 3.186) | <0.001 |
| CA-199 | 1.573 (0.914 to 2.708) | 0.092 | ||||
| AFP | 0.606 (0.277 to 1.329) | 0.182 | ||||
| Cirrhosis | 1.169 (0.625 to 2.187) | 0.630 | ||||
| Hepatitis B | 0.642 (0.361 to 1.144) | 0.119 | ||||
| Position | 0.952 (0.641 to 1.413) | 0.806 | ||||
| Shape | 1.864 (1.115 to 3.117) | 0.015 | ||||
| Margin | 2.689 (1.599 to 4.523) | <0.001 | ||||
| Peritumoural bile duct dilatation | 2.400 (1.445 to 3.988) | <0.001 | ||||
| Hepatic lobe atrophy | 2.718 (1.544 to 4.784) | 0.001 | ||||
| Capsular retraction | 1.946 (1.192 to 3.179) | 0.008 | ||||
| Imaging classification | 3.583 (1.819 to 7.057) | <0.001 | 2.653 (1.317 to 5.343) | 0.006 | ||
| Target sign in T2WI | 0.376 (0.185,0.761) | 0.002 | ||||
| Target sign in DWI | 0.509 (0.298 to 0.870) | 0.010 | ||||
| Enhancement patterns | 0.246 (0.125 to 0.486) | <0.001 | ||||
| Intratumour vascularity | 1.051 (0.520 to 2.125) | 0.890 | ||||
| Intra-tumoural separate | 0.857 (0.528 to 1.390) | 0.532 | ||||
| Radscore | 4.419 (2.743 to 7.118) | <0.001 | 3.7213 (2.210 to 6.265) | <0.0001 | ||
Data in parentheses are 95% CIs.
The independent predictors in the pathology-based and imaging-based OS predicting models were summarised.
Bold values indicate that these are statistically significant in univariable analysis.
AFP, a-fetoprotein; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CA-199, cancer antigen 199; CEA, carcinoembryonic antigen; GGT, γ-glutamyl transpeptidase; OS, overall survival; PD-1, programmed cell death protein 1; PD-L1, programmed cell death protein ligand 1; Radscore, radiomics score.
The pathology-based OS model (C-index, 0.698; 95% CI 0.635 to 0.760) involved three predictors including PD-1, PD-L1 expression status and carcinoembryonic antigen (CEA) level, with a model cut value of 0.9104. The formula for the pathology-based OS model is:
Risk1=1.208×PD1 +0.625×PD-L1 +0.910× CEA.
This model could divide ICC patients into high-risk (median value of risk 1: 1.833, IQR: 1.29–2.118) and low-risk groups (median value of risk 1: 0.625, IQR: 0.00–0.9104) based on a cut-off value of 0.9104. The high-risk group included 46 patients and low-risk group included 52 patients. The median survival time in these two groups was 16.5 months (IQR: 9–36.75 months) and 44 months (IQR: 22.75–64 months), respectively. The 1-year, 3-year and 5 year survival rates of the high-risk and low-risk groups were 54.3%, 25.8% and 17.0% and 88.1%, 50.0% and 48.5%, respectively (p<0.0001) (online supplemental figure S8).
esmoopen-2020-000910supp019.pdf (192.1KB, pdf)
The imaging-based OS model (C-index, 0.721; 95% CI 0.658 to 0.783) also involved three predictors including IC, Radscore extracted from AP and CEA level, with a model cut value of 1.049. The formula for the imaging-based OS model is:
Risk2=0.975×IC+1.314×Radscore+0.666×CEA.
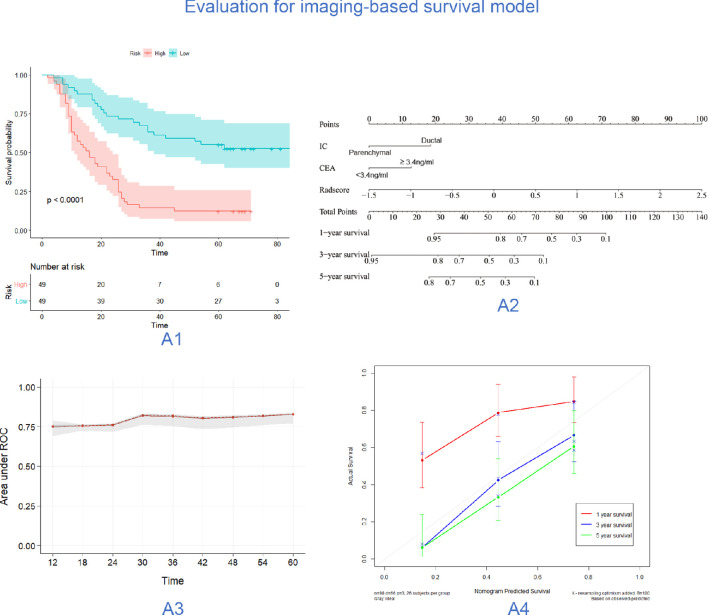
This model could divide ICC patients into high-risk (median value of risk 2: 1.641, IQR: 1.360–2.047) and low-risk groups (median value of risk 2: 0.424, IQR: −0.167–0.6589) based on a cut-off value of 1.049. There were both of 49 patients in each group. The median survival time in these two groups was 16 months (IQR: 9–26 months) and 61 months (IQR: 22–64 months), respectively. The 1-year, 3-year and 5-year survival rates of the high-risk and low-risk groups were 57.1%, 14.3%, and 12.4% and 87.8%, 63.3% and 55.3%, respectively (p<0.001). The calibration curve of the imaging-based OS model demonstrated good agreement between predictions and observations (figure 1).
Figure 1.
Evaluation of performance and clinical use of the imaging-based OS model. (A1) The results of Kaplan-Meier survival analyses based on imaging-based OS model for patients. (A2) A multiparameter radiomics nomogram. Locate the patient’s Radscore on the Radscore axis. Draw a line straight upward to the points’ axis to determine how many points toward the probability of OS the patient receives for his or her Radscore. Repeat the process for each variable. Sum the points achieved for each of the risk factors. Locate the final sum on the total point axis. Draw a line straight down to find the patients, probability of OS. (A3) The AUC of the model in different time periods, the red line shown the mean of AUC, and the grey areas was the IQR of AUC. (A4) The calibration curves for prediction model for 1 year, 3 years and 5 years OS. AUC, area under the curve; CEA, carcinoembryonic antigen; IC, imaging classification; OS, overall survival; ROC, receiver operating characteristic (ROC).
Discussion
With increasing blockage of the PD-1/PD-L1 pathway in cancer treatment, understanding their expression status in ICC patients could help identify those who might benefit most and achieve personalised treatment. In this study, we found that PD-1+ or PD-L1+ expression was linked to poor outcomes in ICC patients. Hence, the tumour MRI radiomic features were converted into a quantitative Radscore and further combined with clinico-radiological factors to preoperatively predict PD-1 and PD-L1 expression status in ICC patients, achieving an AUC of 0.897 and 0.890, respectively. Besides, an imaging-based OS predictive model integrating the Radscore, clinico-radiological factors, and clinical factors with a C-index of 0.721, was developed, accurately stratifying ICC patients into high-risk and low-risk groups. And such imaging-based OS model was potential for clinical practice and assist the pathology-based model (C-index: 0.698) using pathologically demonstrated PD-1, PD-L1 expression and CEA level. These results suggest that the findings of this study might provide helpful indications for the immunotherapy and prognosis of ICC patients.
In this study, we found that PD-1+ expression was linked to poor outcomes in ICC patients after receiving surgical treatment. This is consistent with a study conducted by Lu et al20 in which PD-1+ expression was a negative prognostic marker for ICC, indicating that PD-1 may be a potential therapeutic target.21 Furthermore, compare with PD-1− cases, we found that the PD-1+ expression group existed a significant difference in pathological differentiation, liver capsular retraction, enhancement patterns, IC and intratumour vascularity. The above factors may reflect the malignancy and poor prognosis of the tumour, as well as the presence of immune escape within the ICC. These results need to be confirmed by prospective studies.
However, there has been some controversy in previous studies regarding the effect of PD-L1+ expression on the prognosis of ICC.20–26 The present findings showed a significant relationship between PD-L1+ expression and poor outcomes in ICC patients since the PD-1/PD-L1 pathway contributes to malignant potential and immune tolerance in ICC.27 These contradictory results may be related to the use of different antibodies and cut-off values. Moreover, a meta-analysis showed that PD-L1+ expression is not associated with outcomes in cholangiocarcinoma.28 However, because that study included ICC and extrahepatic cholangiocarcinoma, this conclusion should be interpreted with caution and cannot be applied to ICC directly. Therefore, effort should be made in the future to clarify the significance of PD-L1+ expression in ICC patients.
Radiographical imaging contains invisible information on the differences in protein expression in tumours.29 Radiomics can provide insight into this information by quantitative analysis of medical imaging to infer protein expression in tumours.30 The present study found that the radiomics features provided increased power to predict PD-1 and PD-L1 expression in ICC, regardless of whether it was extracted from AP or PVP MR images. Overall, radiomics features extracted from AP were superior to those from PVP, a conclusion that was similar to a previous report that demonstrated that radiomics features derived from AP were better than those from PVP in predicting lymph node metastasis in ICC patients.31 It can be speculated that imaging from AP may contain more information than imaging from PVP in ICC, but the detailed reasons need further investigation. In addition, a prediction model combined with radiomics features extracted from AP and clinico-radiological factors was developed to predict PD-1 expression. Radiomics features extracted from AP were used to predict PD-L1 expression because no clinico-radiological factors were found for PD-L1. The AUCs of the above predictive models were 0.897 and 0.890, respectively. This has significant implications for clinical decision making for immune checkpoint inhibition in ICC.
This study found that imaging-based OS predicting model (C-index 0.721) could well assist the pathology-based model (C-index 0.698). PD-1 and PD-L1 were independent indicators for outcomes in ICC patients because activation of the PD-1/PD-L1 pathway inhibits the host’s antitumour response21 and enhances tumour cell invasion and migration in ICC. But considering the fluctuations in the assay condition, IHCs antibody, it might be encouraging to find out some easy method to assist such pathology-based assessment. The imaging-based model in the current study involved the easy-to-get radiomics features, traditional MRI features and clinical factor CEA as predictors, which might be potential in clinical practice. CEA is a serum biomarker that reflects tumour burden in ICC. In accordance with a previous study,32 it was found that CEA was an independent factor that affected ICC patients’ OS and simultaneously involved into pathology-based and imaging-based OS models in the current study. For clinic-radiological features, IC serves to classify ICC into parenchymal and ductal types, reflecting the origin of the small duct and large ducts, according to whether there is intrahepatic bile duct dilatation or abnormality on the liver MRI.33 It was found that the ductal type of ICC has a worse outcome than the parenchymal type, which is in line with previous studies.33 The ductal type of ICC more frequently shows perineural invasion and lymph node metastasis,34 which are poor prognostic factors. The Radscore, which was calculated from radiomics features, was also found to be an independent prognostic indicator. Radiomics extracts additional information that can reveal intratumoural spatial heterogeneity and can be used to infer gene or protein expression of the tumour, which in turn can predict tumour prognosis.29 Wang et al35 reported that MRI radiomics can preoperatively predict the survival of patients with hepatocellular carcinoma. Therefore, radiomics can potentially be used as a quantitative and noninvasive prognostic biomarker.
In our PD-1 and PD-L1 predictive model, we found that the GLCMEnergy and GLCMEntropy present similar predictive tendency. The higher the GLCMEnergy and lower GLCMEntropy seemed more likely to appear PD-1 or PD-L1 expression. While in the OS model, it seems that GLCMEnergy and GLCMEntropy are good prognosis predictors. Tang et al18 found that the higher the intensity SD, the lower mean intensity and higher texture inhomogeneity might indicate a lower tumour PD-L1 expression and higher frequency of CD3+ cells (immune activated) which indicate a favourable prognosis. In our result, we found that in the PD-L1 prediction model, the larger RelativeDeviation is more likely not to appear PD-L1 expression and in the OS model the features reflecting homogeneity (InverseDifferenceMoment, LongRunEmphasis) were poor prognosis predictors might have some similar reflection with Tang’s findings. But we could not make any suspect on this as we have no further biological assays. We will try to make more investigations on these aspects in the future works.
This study suffered from several limitations. First, many ICC patients without surgery or MRI scanning were excluded since this study had a retrospective design, and therefore a potential selection bias might have existed. Second, the study was drawn from a single centre with small sample size, and the results will need to be externally validated at other centres. Third, the antibody for PD-L1 in our study is not validated in the clinic and the results may be greatly affected. Finally, a tracer was not used in the scanning procedure, and therefore the data may be slightly biased.
In conclusion, our study demonstrated that machine learning consistently achieved desirable results in the prediction of PD1/PD-L1 expression and outcome based on MRI radiomics. The clinical OS predictive model integrating Radscore, clinico-radiological and clinical factors also showed excellent performance in dividing ICC patients into high-risk and low-risk groups. These findings suggested that the MRI radiomics could provide promising and non-invasive indicative biomarkers for the evaluation of PD-1/PD-L1 expression and prognosis of ICC patients.
esmoopen-2020-000910supp020.pdf (141.3KB, pdf)
Footnotes
JZ and ZW contributed equally.
Contributors: All authors have full access to all data used in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. JZ, ZW, YS and BS designed the study. JZ, ZW, JZ and FY acquired data. JZ, ZW, XZ, SL, JZ, FY, YS and BS analysed, and interpreted the data. XZ and SL did the statistical analysis. JZ and ZW drafted the manuscript. YS and BS critically revised the manuscript. BS provided administrative and material support. All authors approved the final version of the manuscript for submission.
Funding: This project was supported by the National Natural Science Foundations of China (81771797 and 81971571) and the 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYJC18008).
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: This retrospective study was approved by our institutional review board. All procedures were performed in accordance with the 1975 Declaration of Helsinki and its later amendments.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available in a public, open access repository. This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC4.0) licence, which permits others to distribute, remix, adapt, build on this work non-commercially, and licence their derivative works on different terms, provided the original work is properly cited, any changes made are indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Torre LA, Bray F, Siegel RL, et al. . Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Razumilava N, Gores GJ. Cholangiocarcinoma. The Lancet 2014;383:2168–79. 10.1016/S0140-6736(13)61903-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridgewater J, Galle PR, Khan SA, et al. . Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J Hepatol 2014;60:1268–89. 10.1016/j.jhep.2014.01.021 [DOI] [PubMed] [Google Scholar]
- 4.de Jong MC, Nathan H, Sotiropoulos GC, et al. . Intrahepatic cholangiocarcinoma: an international multi-institutional analysis of prognostic factors and lymph node assessment. J Clin Oncol 2011;29:3140–5. 10.1200/JCO.2011.35.6519 [DOI] [PubMed] [Google Scholar]
- 5.Wei SC, Levine JH, Cogdill AP, et al. . Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 2017;170:1120–33. 10.1016/j.cell.2017.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Migden MR, Rischin D, Schmults CD, et al. . Pd-1 blockade with Cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med 2018;379:341–51. 10.1056/NEJMoa1805131 [DOI] [PubMed] [Google Scholar]
- 7.Chowell D, Morris LGT, Grigg CM, et al. . Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359:582–7. 10.1126/science.aao4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid P, Adams S, Rugo HS, et al. . Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med 2018;379:2108–21. 10.1056/NEJMoa1809615 [DOI] [PubMed] [Google Scholar]
- 9.Herbst RS, Soria J-C, Kowanetz M, et al. . Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563–7. 10.1038/nature14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamid O, Robert C, Daud A, et al. . Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 2013;369:134–44. 10.1056/NEJMoa1305133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 12.Sirica AE, Gores GJ, Groopman JD, et al. . Intrahepatic cholangiocarcinoma: continuing challenges and translational advances. Hepatology 2019;69:1803–15. 10.1002/hep.30289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanmamed MF, Chen L. A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell 2018;175:313–26. 10.1016/j.cell.2018.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larimer BM, Wehrenberg-Klee E, Dubois F, et al. . Granzyme B PET imaging as a predictive biomarker of immunotherapy response. Cancer Res 2017;77:2318–27. 10.1158/0008-5472.CAN-16-3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limkin EJ, Sun R, Dercle L, et al. . Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 2017;28:1191–206. 10.1093/annonc/mdx034 [DOI] [PubMed] [Google Scholar]
- 16.Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. . Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun R, Limkin EJ, Vakalopoulou M, et al. . A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180–91. 10.1016/S1470-2045(18)30413-3 [DOI] [PubMed] [Google Scholar]
- 18.Tang C, Hobbs B, Amer A, et al. . Development of an Immune-Pathology informed Radiomics model for non-small cell lung cancer. Sci Rep 2018;8:1922. 10.1038/s41598-018-20471-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Huang Z, Cao L, et al. . Differentiation combined hepatocellular and cholangiocarcinoma from intrahepatic cholangiocarcinoma based on radiomics machine learning. Ann Transl Med 2020;8:119. 10.21037/atm.2020.01.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J-C, Zeng H-Y, Sun Q-M, et al. . Distinct PD-L1/PD1 profiles and clinical implications in intrahepatic cholangiocarcinoma patients with different risk factors. Theranostics 2019;9:4678–87. 10.7150/thno.36276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gani F, Nagarajan N, Kim Y, et al. . Program death 1 immune checkpoint and tumor microenvironment: implications for patients with intrahepatic cholangiocarcinoma. Ann Surg Oncol 2016;23:2610–7. 10.1245/s10434-016-5101-y [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Wang X-Y, Zhang Y, et al. . Programmed death ligand 1 expression in human intrahepatic cholangiocarcinoma and its association with prognosis and CD8+ T-cell immune responses. Cancer Manag Res 2018;10:4113–23. 10.2147/CMAR.S172719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jing C-Y, Fu Y-P, Yi Y, et al. . HHLA2 in intrahepatic cholangiocarcinoma: an immune checkpoint with prognostic significance and wider expression compared with PD-L1. J Immunother Cancer 2019;7:77. 10.1186/s40425-019-0554-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asahi Y, Hatanaka KC, Hatanaka Y, et al. . Prognostic impact of CD8+ T cell distribution and its association with the HLA class I expression in intrahepatic cholangiocarcinoma. Surg Today 2020;50:931–40. 10.1007/s00595-020-01967-y [DOI] [PubMed] [Google Scholar]
- 25.Dong Z, Liao B, Shen W, et al. . Expression of programmed death ligand 1 is associated with the prognosis of intrahepatic cholangiocarcinoma. Dig Dis Sci 2020;65:480–8. 10.1007/s10620-019-05787-0 [DOI] [PubMed] [Google Scholar]
- 26.Sabbatino F, Villani V, Yearley JH, et al. . Pd-L1 and HLA class I antigen expression and clinical course of the disease in intrahepatic cholangiocarcinoma. Clin Cancer Res 2016;22:470–8. 10.1158/1078-0432.CCR-15-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Y, Zhou L, Xie X, et al. . Interaction of B7-H1 on intrahepatic cholangiocarcinoma cells with PD-1 on tumor-infiltrating T cells as a mechanism of immune evasion. J Surg Oncol 2009;100:500–4. 10.1002/jso.21376 [DOI] [PubMed] [Google Scholar]
- 28.Xu G, Sun L, Li Y, et al. . The clinicopathological and prognostic value of PD-L1 expression in cholangiocarcinoma: a meta-analysis. Front Oncol 2019;9:897. 10.3389/fonc.2019.00897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbs SK, Shi G, Homer R, et al. . Magnetic resonance image-guided proteomics of human glioblastoma multiforme. J Magn Reson Imaging 2003;18:530–6. 10.1002/jmri.10395 [DOI] [PubMed] [Google Scholar]
- 30.Lambin P, Rios-Velazquez E, Leijenaar R, et al. . Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441–6. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji G-W, Zhu F-P, Zhang Y-D, et al. . A radiomics approach to predict lymph node metastasis and clinical outcome of intrahepatic cholangiocarcinoma. Eur Radiol 2019;29:3725–35. 10.1007/s00330-019-06142-7 [DOI] [PubMed] [Google Scholar]
- 32.Tian M, Liu W, Tao C, et al. . Prediction of overall survival in resectable intrahepatic cholangiocarcinoma: ISICC -applied prediction model. Cancer Sci 2020;111:1084–92. 10.1111/cas.14315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhee H, Kim M-J, Park YN, et al. . A proposal of imaging classification of intrahepatic mass-forming cholangiocarcinoma into ductal and parenchymal types: clinicopathologic significance. Eur Radiol 2019;29:3111–21. 10.1007/s00330-018-5898-9 [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Li J, Xia Y, et al. . Prognostic nomogram for intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin Oncol 2013;31:1188–95. 10.1200/JCO.2012.41.5984 [DOI] [PubMed] [Google Scholar]
- 35.Wang X-H, Long L-H, Cui Y, et al. . Mri-Based radiomics model for preoperative prediction of 5-year survival in patients with hepatocellular carcinoma. Br J Cancer 2020;122:978–85. 10.1038/s41416-019-0706-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2020-000910supp001.pdf (49.8KB, pdf)
esmoopen-2020-000910supp002.pdf (396.7KB, pdf)
esmoopen-2020-000910supp003.pdf (123.9KB, pdf)
esmoopen-2020-000910supp004.pdf (49.8KB, pdf)
esmoopen-2020-000910supp005.pdf (47.9KB, pdf)
esmoopen-2020-000910supp006.pdf (54.1KB, pdf)
esmoopen-2020-000910supp007.pdf (953.5KB, pdf)
esmoopen-2020-000910supp008.pdf (964.4KB, pdf)
esmoopen-2020-000910supp009.pdf (1,002.4KB, pdf)
esmoopen-2020-000910supp010.pdf (71.9KB, pdf)
esmoopen-2020-000910supp011.pdf (252.6KB, pdf)
esmoopen-2020-000910supp012.pdf (1.1MB, pdf)
esmoopen-2020-000910supp013.pdf (68.2KB, pdf)
esmoopen-2020-000910supp014.pdf (70.3KB, pdf)
esmoopen-2020-000910supp015.pdf (39.5KB, pdf)
esmoopen-2020-000910supp016.pdf (177KB, pdf)
esmoopen-2020-000910supp017.pdf (146.2KB, pdf)
esmoopen-2020-000910supp018.pdf (62.8KB, pdf)
esmoopen-2020-000910supp019.pdf (192.1KB, pdf)
esmoopen-2020-000910supp020.pdf (141.3KB, pdf)