Abstract
The global spread of antimicrobial resistance (AMR) and lack of novel alternative treatments have been declared a global public health emergency by WHO. The greatest impact of AMR is experienced in resource-poor settings, because of lack of access to alternative antibiotics and because the prevalence of multidrug-resistant bacterial strains may be higher in low-income and middle-income countries (LMICs). Intelligent surveillance of AMR infections is key to informed policy decisions and public health interventions to counter AMR. Molecular surveillance using whole-genome sequencing (WGS) can be a valuable addition to phenotypic surveillance of AMR. WGS provides insights into the genetic basis of resistance mechanisms, as well as pathogen evolution and population dynamics at different spatial and temporal scales. Due to its high cost and complexity, WGS is currently mainly carried out in high-income countries. However, given its potential to inform national and international action plans against AMR, establishing WGS as a surveillance tool in LMICs will be important in order to produce a truly global picture. Here, we describe a roadmap for incorporating WGS into existing AMR surveillance frameworks, including WHO Global Antimicrobial Resistance Surveillance System, informed by our ongoing, practical experiences developing WGS surveillance systems in national reference laboratories in Colombia, India, Nigeria and the Philippines. Challenges and barriers to WGS in LMICs will be discussed together with a roadmap to possible solutions.
Keywords: epidemiology, other diagnostic or tool, public health
Summary box.
Antimicrobial resistance (AMR) has been declared a global public health emergency by WHO.
Low-income and middle-income countries (LMICs) bear the greatest burden of AMR infections.
Whole-genome sequencing (WGS) has the potential to greatly enhance AMR surveillance at both the national and international level.
A roadmap with examples is presented on how obstacles to the implementation of WGS for AMR surveillance in LMICs can be overcome.
Collaborations between high-income countries and LMICs at various stages of implementing WGS for AMR surveillance are learning opportunities for all partners and can form cornerstones for global surveillance in future.
The NIHR Global Health Research Unit on Genomic Surveillance of AMR (GHRU) aims to make WGS tools for AMR surveillance available in LMICs through equitable partnerships and to provide actionable data for public health policy.
Introduction: what is the value of whole-genome sequencing for antimicrobial resistance surveillance?
Antimicrobial resistance (AMR) is the ability of bacteria to resist antimicrobial treatment. As a result, bacterial infections cannot be cleared and there is a high risk of onward transmission. A related group of AMR bacteria that share a common ancestor and genetic background is known as an AMR strain, which is frequently used synonymously with AMR clone. The global spread of AMR pathogens has been declared a global public health emergency.1
AMR imposes a substantial cost on societies that endangers economic growth and balanced access to resources (table 1).2 AMR disproportionately affects low-income and middle-income countries (LMICs) and is a risk to several United Nations sustainable development goals.3 4 For example, AMR endangers the health and well-being of individuals and their livestock, the functioning of health systems and clean water supplies.5 In high-income countries (HICs), the infrastructure to ensure clean water supplies and to regulate the use of antimicrobials among both the human and animal populations is more readily available leading to a reduced impact of AMR strains.
Table 1.
Summary of how AMR adversely affects a number of UN sustainable development goals
| UN global development goal (no) | Impact of AMR |
| No poverty (1) | AMR strikes hardest on the poor2 87 |
| Zero hunger (2) | Negative impact of AMR on animal husbandry88 |
| Good health and well-being (3) | AMR endangers health, well-being and functioning of health systems89 |
| Clean water and sanitation (6) | Antimicrobials and AMR organisms are released in wastewater90 |
| Decent work and economic growth (8) | AMR causes a cost to societies that endangers growth and employment91 |
| Responsible consumption and production (12) | AMR endangers balanced access to medical and environmental resources92 |
AMR, antimicrobial resistance.
Applied as part of a one-health approach, whole-genome sequencing (WGS) can be used to infer transmission events between humans and animals and trace the origin of foodborne diseases. For example, a WGS study of Salmonella typhimurium DT104 suggests that several transmission events occurred between human and cattle populations over the past 50 years.6 As antimicrobial use in farm animals and release of antibiotics in the environment are suspected to affect AMR in human diseases, one health has become an important component of action plans against AMR.7 In addition, the transmission of pathogens through the food chain from farm to fork can also be tracked. In this way, when a putative source has been identified, investigations can be carried out and transmission routes eliminated by cooperation of organisations with different realms of responsibility within the food chain. This methodology has been demonstrated, for example, in public health investigations of Salmonella in eggs8 and in colistin-resistant Escherichia coli in pigs and raw meat in China.9 It is important that the chain from farm to fork be monitored as part of National Action Plans (NAPs).10
Reducing AMR and preventing the spread of AMR organisms requires understanding the mechanisms of resistance, transmission routes and the epidemiology of AMR organisms.11–17 To collect this information, WHO has built the Global Antimicrobial Resistance Surveillance System (GLASS) that collects data on eight priority pathogens at high risk of developing resistance to all available antimicrobial treatment options.1 GLASS collects national-level phenotypic AMR data using WHONET, a free database software developed by the WHO Collaborating Centre for Surveillance of Antimicrobial Resistance at Brigham and Women’s Hospital and Harvard Medical School supported in over 2300 laboratories in over 120 countries.18
Phenotypic antimicrobial susceptibility testing (AST) determines if cultured bacterial isolates grow in vitro in the presence of a specified concentration of a given antimicrobial agent. Phenotypic data include the minimum inhibitory concentration of an antimicrobial that prevents growth of the tested bacterial isolate or zones of growth inhibition collected from disk diffusion tests. AST results provide important information for clinical management and surveillance but no direct information about resistance mechanism, transmission routes or pathogen evolution.
Molecular surveillance data based on bacterial DNA sequence information can be a valuable addition to a national surveillance system, and a complement to phenotypic surveillance by providing more detailed insights into the epidemiology of pathogens, including AMR strains. Using WGS achieves superior reproducibility and resolution compared with other molecular surveillance methods allowing not only for the possible origin of the host bacteria to be determined but also the genetics of the loci responsible for resistance to be investigated. WGS has become a key technology for understanding pathogen evolution and population dynamics on different spatial and temporal scales.19 Additionally, WGS can determine other pathogen characteristics of public health importance, such as virulence and transmissibility.20 Knowledge of these characteristics can improve the management of disease outbreaks and epidemics and have a direct impact on the health of individuals within a region.
WGS captures neutral evolution as well as the evolution of AMR determinants and can therefore be used to infer the origins and transmission routes of AMR strains. The ability to infer bacterial evolution and transmission routes from WGS data is important because frequently only a fraction of infections is captured and sequenced rather than the entire transmission chain. For example, a WGS study of early MRSA isolates revealed that the mecA gene that renders Staphylococcus aureus resistant to the second-generation β-lactam methicillin originally arose 14 years prior to the first clinical use of methicillin, presumably as an adaptation against the use of first-generation β-lactams.21 Another WGS study on MRSA proved that transmission of MRSA is not restricted to events within the same hospitals but increases proportionally with the number of patients referred between different hospitals.22
The same methods used to construct phylogenetic trees of patient isolates can be used to estimate important epidemiological parameters, such as the basic reproductive number R0 that describes how many secondary infections can arise from one primary case. R0 indicates how fast an infection spreads through a population (transmissibility) and how difficult it is to contain.6 23 Even though it can be difficult to determine R0 from genetic information from fast-growing bacteria, phylogenetic analysis of WGS data from Mycobacterium tuberculosis isolates has been used to estimate R0 in a tuberculosis (TB) outbreak in a low-burden environment in Canada.7
AMR emerges and spreads within observable timescales. Therefore, constant monitoring of the population structure of known pathogens facilitates a targeted response to emerging high-risk clones. A high-risk clone is a genetically uniform group of bacteria that by common ancestry share the same critical resistance mutations and genes making them resistant to one or more standard treatments. High-risk clones can be identified from WGS data based on clonal relatedness and abundance and inferring virulence and resistance profiles from gene content.8
It has been argued that continuous surveillance is more cost-effective for preventing disease outbreaks than attempts at predicting potential new pathogenic strains from available sequence and epidemiological data.9 In combination with phenotypic surveillance and epidemiological data, evidence from WGS data can be used to strengthen programmes for infection prevention and control, inform emergency responses and refine clinical decision making by lending further evidence on the origin of resistant clones that are dominant within a geographical region or globally.
In some circumstances, WGS data may allow for pre-emptive actions against AMR spread. by aiding the development of rapid and sensitive molecular diagnostics that can detect AMR and be employed as point-of-care tests (POCTs). For example, quinolone-resistance in Neisseria gonorrhoeae is generally associated with known mutations in the gyrA and parC genes, which could be exploited to design a molecular POCT for quinolone resistance.10 Another example for the development of POCTs with the help of WGS is TB.24
What are the benefits of incorporating WGS into AMR surveillance programmes in LMICs?
LMICs are disproportionately impacted by AMR. An important driver of AMR in LMICs is unregulated antimicrobial use and the sale of counterfeit products.25–27 Sanitation is frequently poor and waste water enters the environment untreated.28 A recent study in New Delhi found that the concentration of 28 tested antimicrobials exceeded 0.1 µg/L in some of the city’s aquifers. Most water bodies had antimicrobial concentrations greater than 0.01 µg/L.29 The majority of LMICs are located in tropical/subtropical regions and have above average biodiversity levels.30 Therefore, the risk of AMR arising through the mixing of different species and bacterial strains is potentially high. For instance, a study from Vietnam found that rats and shrews captured on livestock farms harboured eight times more multidrug-resistant (MDR) E. coli strains than mammals caught in the wild.31 For these reasons, the evolution of AMR in LMICs should be carefully monitored.
Agriculture and livestock farming play a major role in the economies of most LMICs. The interactions between humans and farm animals in LMICs are complex and the use of antimicrobials in farming is largely uncontrolled.26 Consequently, the risk of AMR epidemics in food animals and transmission between humans and livestock along various points in the food production chain is high, but poorly understood.10 WGS studies can untangle the processes that lead to the emergence and spread of AMR organisms in human-livestock interactions and identify novel resistance mechanisms in animal pathogens.32 The benefits of comprehensive AMR surveillance in LMICs will extend to the animal health and agricultural sector. Hence, AMR surveillance can suggest measures to maintain and boost economic growth and resource preservation in affected countries. One example of an integrated food safety surveillance programme that can be expanded by incorporating WGS is the AGROSAVIA (the Colombian Institute for Farming and Livestock Research, previously under the name of COIPARS).33
WGS has been successfully applied to identify drivers of AMR burden, including in LMICs. For example, WGS data from E. coli isolates from children in South Asia and sub-Saharan Africa found that 65% of isolates were resistant against three or more antimicrobial classes and that resistance correlated with geography and antimicrobial usage, rather than lineage. AMR genes were frequently colocated which could facilitate the acquisition of MDR by as yet susceptible bacterial strains.34 This implies that antimicrobial use drives high rates of AMR and that acquired AMR genes can be lost again in bacterial lineages if antimicrobial exposure ceases for long enough.
Coinfections with other pathogens, especially immunosuppressive agents, such as HIV and undernutrition, contribute to the disease burden imposed by AMR in some LMICs. For example, invasive nontyphoidal Salmonellosis has been shown to be a common cause of febrile illness in HIV patients in Kenya.35 The majority of sequenced Salmonella isolates in this study were found to be MDR.
Figure 1 illustrates how building local partnerships among LMICs and between LMICs and HICs can lead to a ‘virtuous circle’ in which improved local capacity for AMR surveillance using WGS, enhanced reference databases and scientific research all feed into each other to produce added value locally for public health policy and AMR control.
Figure 1.
Virtuous circle of improved local capacity for WGS for AMR surveillance, improved reference databases and scientific research. Expanded capacity for AMR surveillance locally in LMICs (and HICs), including systematic sampling of resistant isolates, quality control and collaborative networks will improve and extend reference databases of AMR organisms which will drive scientific research and lead to new science-driven engineering solutions that in turn can improve the use of WGS for AMR surveillance. Together these three interlocking systems can lead to improved public health policy and AMR control and technological innovation. AMR, antimicrobial resistance; HICs, high-income countries; LMICs, low-income and middle-income countries; WGS, whole-genome sequencing.
Although LMICs are the most affected by AMR, efforts to lower the burden of AMR in LMICs will have global benefits. High rates of international movement of people and livestock facilitate the spread of AMR organisms across borders. For example, Cohen et al used WGS data to identify movement of MDR M. tuberculosis between high-incidence and low-incidence countries, although the WGS data were not dense enough to infer directionality.36 Designing impactful strategies to counter AMR at the regional and global level requires an adequate assessment of AMR burden, identifying hotspots of AMR emergence and tracing dominant transmission routes.
Improved surveillance can reduce unnecessary use of standard and reserve antimicrobials and allow the use of narrow spectrum drugs, all of which reduce selective pressure for resistance. Collecting this information requires a potent surveillance system that generates high-quality, standardised data. WGS is one of the most data-rich surveillance technologies and can be a valuable supplement to existing national surveillance. As the costs for WGS continue to fall, an increasing number of countries are expected to incorporate WGS into national AMR surveillance programmes.37–39 Eventually, these national surveillance programmes will contribute to building a global AMR surveillance network that, in addition to quantifying the extent of AMR in various parts of the world, will trace the emergence and spread of AMR within and across countries.40 These data also feed into building and improving global tools that enable LMICs to access data in real time and contribute to global surveillance in the long term. To start building a global AMR surveillance system, the most highly affected regions need experienced and trained personnel. Therefore, investing in expanding AMR surveillance and in improved technologies, such as WGS, in LMICs should form an integral part of any long-term global strategy for AMR control.
Developing a roadmap to establish AMR surveillance in LMICs
WGS contributes an important new dimension to surveillance systems, including for AMR. Policy-makers are starting to explore big data for precision public health, combining traditional medical data with novel data and technologies from fields including genomics, enabling a better understanding of disease pathogenesis and more targeted diagnoses and treatments.41 With new guidelines and publicly accessible tools available giving countries real-time access to global pathogen information, countries have an opportunity to consider how WGS can be implemented as part of their broader surveillance systems.
Every country has a different context and organisational model for surveillance, including for AMR. Based on our experience, we recommend that there are four steps that are important to establishing WGS within a surveillance system. These steps can be regarded as a ‘roadmap’ for countries looking to adopt WGS and can be flexibly adapted to each country’s surveillance system.
First step: commitment
Most countries have commitments under the International Health Regulations, and to global strategies for AMR, including through WHO GLASS. It is important to have local and national commitment to establishing WGS as a surveillance tool in (LMICs) within these broader systems as a first step. In our experience, commitment by the hosting organisation was an important first feature to establish WGS as a feature within each surveillance system. The experiences of other countries currently implementing WGS within their surveillance systems was an important aspect of making the business case for implementation.
Second step: assessment
An important second step in all countries with whom we have worked has been a system assessment of how WGS could be developed for use within existing surveillance systems. System assessments of laboratories, bioinformatics and supporting management systems proved a useful second step for the countries with which we worked, identifying how systems could work together, as well as gaps and needs. For example, WGS can be developed for use in a complementary way with existing phenotypic AST to contextualise bacterial isolates within the background population obtained from retrospective sequencing and the cumulative data from ongoing sequencing.
Third step: technical development
This step establishes the technical capacity within a country to sustain WGS for the long term. Across our work, this has included capacity building for locally, led research projects by providing resources and training, the development of sampling frameworks to address national public health and research needs, data collection to identify high-risk bacterial lineages and AMR genes, provision and integrated use of open-access tools for genomics analysis and data visualisation to understand the distribution of bacterial lineages.
Technical guidance and tools are globally available to support the development of WGS for AMR surveillance. WHO technical guidance32 is available to support the introduction of molecular methods in LMICs. In addition, open-access tools enable health professionals and policy-makers real time access to, and sharing of, genomic pathogen data for national AMR surveillance. Tools such as Pathogenwatch33 and Microreact42 43 enable epidemiological data to be combined with genomics to inform pathogen control strategies and interventions on a local, national and international scale.
A starting point for technical development is to establish a reference database including WHO priority pathogens and species of local importance. This can be achieved by retrospectively sequencing stored isolates that have ideally been collected in a structured way, including those which are susceptible to antibiotics. This is important to discern whether AMR strains originate from pre-existing locally circulating susceptible strains or are imported.
An appropriate prospective sampling strategy can be developed based on identified public health and research needs. Sampling strategies should include both passive surveillance and targeted surveillance of outbreaks and unusual isolates. Passive surveillance can be based on structured surveys in which susceptible and resistant isolates are sequenced to obtain an overview of the population genetic background of circulating bacterial strains. The combination of passive and targeted surveillance can identify putative high-risk clones through the prediction of resistance, virulence and global spread of bacterial lineages.
Capacity building is an essential step at this stage to support WGS within the emerging surveillance system. In our group of countries, workshops and training that included other countries’ experiences were highly beneficial for developing and maintaining a solutions perspective as challenges were experienced. This challenges-and-solutions approach is detailed further in the following sections.
Fourth step: create a business plan and standard operating practice for WGS to ensure long-term sustainability
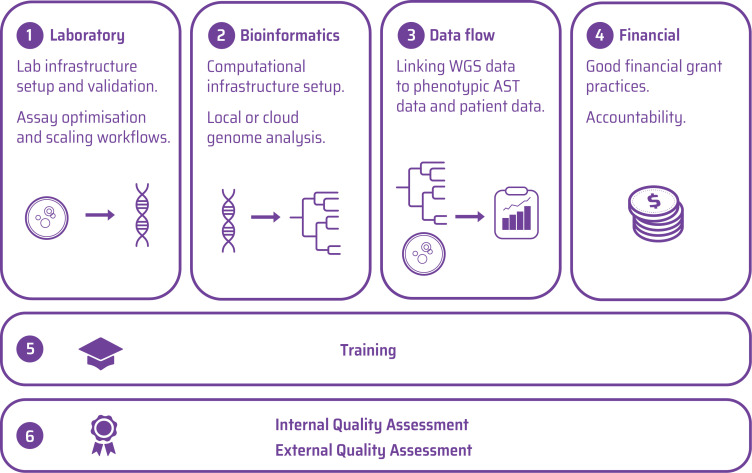
A business plan covering laboratory, bioinformatics, data flow and financial management is useful to integrate WGS as part of the broader surveillance system. At a practical level, partner countries created a range of sustainable business practices including standard operating and supply chain procedures, quality assurance (QA) and reporting processes, as well as ongoing capacity development, and processes for ongoing governance and financial sustainability. The workflow components that need to be put into place, the associated training and quality assessment that are necessary to efficiently use WGS for AMR surveillance are suggested as a practical business planning framework in figure 2.
Figure 2.
Workflow components required to implement WGS for AMR surveillance. AMR, antimicrobial resistance; AST, antimicrobial susceptibility testing; WGS, whole-genome sequencing.
Integrating WGS within national surveillance systems is an opportunity underpinned by global technical standards and open-access tools. However, it is not without challenges. In the following sections, we review both the challenges and solutions of integrating WGS as part of AMR surveillance, based on our ongoing experiences in developing such surveillance in National Reference Laboratories in Colombia, India, Nigeria and the Philippines.
What are the main challenges to WGS surveillance in general?
Despite its promises, WGS surveillance for AMR in bacterial pathogens poses a number of challenges. In this section, we discuss overarching challenges for the application of WGS for AMR surveillance in any setting. In the following section, we cover the logistic and operational challenges faced by LMICs wishing to incorporate WGS in their AMR surveillance system.
WGS in itself does not test for AMR phenotypes. Consequently, it can only detect known resistance genotypes, or genetic variants that are very similar to known resistance genotypes. However, for species with a well-characterised genome, such as Salmonella enterica, in silico prediction of AMR phenotypes from WGS data with very few discrepant results is possible and can be an effective replacement for phenotypic testing.44 A comparison of different bioinformatic methods has shown that there is high concordance in predicting AMR and virulence factors in S. aureus, Salmonella and other pathogens.45–49
Given the variety of WGS technologies available and differences in laboratory settings, external QA (EQA) or performance testing of laboratory procedures is important to ensure that results generated by different laboratories are comparable. EQA usually has laboratories analyse the same set of known, well-characterised bacterial strains. The results produced by different laboratories are compared with predefined quality markers assessing the laboratories’ ability for DNA preparation, library construction and eventually sequencing and the performance in identifying epidemiological markers.50 EQA should be done for all bacterial species for which WGS surveillance data is to be collected. The number of different subsequent steps in WGS and the variety of available WGS platforms makes EQA for WGS non-trivial.38 For global surveillance projects, EQA protocols with agreed common and standardised quality markers will have to be developed that are applicable in different countries with varying laboratory capacities and varying levels of previous experience with WGS analysis. EQA should ultimately test a laboratory’s ability to predict antimicrobial susceptibility from genomic data given a set of strains with known phenotypic AMR profiles. An additional level of assessment could be to test if the laboratory can determine which resistance loci and alleles are present. This requires EQA strains with high-quality closed genome sequences where the ‘true’ sequence of all loci has been confirmed through high-depth, long-read sequencing technologies. In addition, EQA strains also need a reliable, high-quality AST profile. The Global Microbial Identifier Consortium and GenomeTrakr have piloted proficiency testing protocols for DNA extraction, library preparation, WGS, assembly, phylogenetic analysis and detection of AMR genes.51 52 A full roll-out of the protocols is currently underway.
A related problem in the implementation of WGS for AMR surveillance is contamination of samples, equipment or reagents with DNA that does not belong to the isolate to be tested. WGS relies on highly sensitive assays that could amplify signals from contaminating DNA and hence lead to erroneous results.53 The risk of contamination can be reduced by partitioning the laboratory space into separate areas for pre-sequencing, sequencing and postsequencing work steps and establishing a unidirectional workflow. Moreover, the use of negative controls can give an indication of whether contamination has occurred.54 More recently, various software tools have become available to detect and remove suspected contaminating sequences from microbial WGS data.55–57
Most WGS studies so far have focused on reconstructing transmission chains from past events.58 Applying WGS prospectively to predict emerging trends in AMR organisms is more challenging, not least because of the difficulty to discern an evolutionary signal in the data from genetic background noise in the microbiological population.59 Nevertheless, a recent study at a tertiary care hospital in Germany showed that WGS surveillance of MDR isolates of S. aureus, E. coli, Enterococcus faecium and Pseudomonas aeruginosa was not only feasible but also cost-effective. WGS surveillance allowed for changes in the isolation policy of infected patients that led to cost savings of more than €200 000 over a 6-month time frame.15
Another challenge to implementing WGS as a surveillance tool for AMR is data accessibility. While WGS data is digital and can easily be shared, metadata that is crucial for interpreting WGS results is mostly distributed across multiple unlinked databases and not standardised.60 Metadata in this context comprises patient data (eg, age, gender, symptoms, geographical location), information on the microbiological sample (eg, sample type, date, anatomical location) and information on the laboratory techniques used to extract and sequence DNA together with quality indicators. Frequently, a trade-off between data accessibility and concerns for patient privacy exists.61 There is a need for efficient and secure platforms that facilitate the integrated analysis and interpretation of WGS data with standardised and anonymised metadata. An example for such a platform is the NIAID TB Portals Programme database repository (TB DEPOT) that contains clinical, socioeconomic, laboratory and genomic data.62
What are the main challenges to implementing WGS surveillance in LMICs?
Besides the inherent technological challenges described in the previous paragraph, LMICs face their own set of practical difficulties in implementing WGS for AMR surveillance. Although the technology is available, policy-makers may be reluctant to invest in it because they do not recognise the added value that WGS can bring to health systems. These issues need to be addressed, but we are not going to discuss them here. The problems around political buy-in and societal trust for WGS in public health and possible solutions have been discussed for TB surveillance in Jackson et al 63, for human genome sequencing in Tekola-Ayele and Rotimi,64 and for rabies surveillance in Brunker et al.65 Instead, we focus on the logistic and operational aspects illustrated in figure 2.
Laboratory
The first major obstacle faced by many LMICs is the need for a high-performing microbiology laboratory which is not always a given.66 For example, sterility of samples and workspaces can be difficult to maintain. Moreover, resource constraints may hamper a laboratory’s ability to set up microbiological cultures from which the substrate for many sequencing technologies is grown.67
The second major issue is the cost of laboratory equipment and machines. Next-generation sequencing machines are expensive in HICs, but paradoxically, because of supplier-based pricing models that weigh product prices against demand, they can be even more expensive in LMICs. Country-specific regulatory and administrative procedures have to be considered when planning to implement WGS surveillance. For example, laboratory equipment can only be purchased from state-approved suppliers which may further limit available options for WGS technologies in LMICs.
In addition to the initial purchase costs, laboratory equipment and machines need reagents, maintenance and infrastructure that is frequently absent in LMIC settings. For example, sequencing machines need continuous electricity supply, chemical reactions need to be prepared with laboratory-quality clean water and occur at specified optimal temperatures, and many reagents and DNA need to be cooled while stored. As an example, the total cost of installing GeneXpert MTB/RIF for TB diagnostics at several sites in Nigeria was more than twice as much the basic price for installation of the machine alone in a high-income setting.68
In some cases, LMICs acquire laboratory equipment, such as sequencing devices, with the help of international collaborations that provide funding and logistic support. For example, the Philippines purchased a sequencing device for AMR surveillance as part of a UK National Institute for Health Research (NIHR)-funded collaboration with the Centre for Genomic Pathogen Surveillance (CGPS).13 Apart from the high purchasing costs (US$215 000 for the sequencing device plus US$160 000 for presequencing and postsequencing equipment and US$6000 for renovations that were necessary to set up a sequencing laboratory), the Philippines initially faced the problem of finding a local supplier. Some reagents could not be purchased locally and had to be bought and shipped by UK collaborators. The problem with this solution is that it is not sustainable, as long-term financing for laboratory and device maintenance will have to be found and local supply chains ought to be established.
Bioinformatics
Analysing the outputs from sequencing requires significant computing resources. A laboratory setting up a WGS service is faced with two options: either build and maintain local infrastructure that has sufficient computing power for the likely sample throughput or upload the data to computing resources hosted elsewhere. The cost for local setup will be between the equivalent of a minimum of ten and many tens of thousands of dollars, and there are both known (power and cooling) and unknown (component replacement) ongoing costs which are often overlooked, yet significant. As the infrastructure in many LMICs cannot provide all of these requirements, laboratories may have to spend additional resources and space on generators and uninterruptible power supplies. Data upload to online resources relies on continuously available broadband internet connection which may not be available, either. In particular, the relative cost for the same type of connection compared with the cost of living can vary significantly among countries. However, once the data are remotely available, the full economic cost of running routine genome analysis is lower than on premise solutions.
Data flow
Currently, there is little standardisation of laboratory information management systems among different laboratories. In LMICs, data management systems are often paper based, which is error-prone and can hamper integration with digital sequence data. As most LMICs are still to implement WGS for AMR surveillance, there is currently an opportunity for developing standardised digital data management systems. At the same time, these systems must be flexible enough to be applicable in a range of different laboratory settings and pathogens. Data management systems should be based on a documented data standard and provide a user-friendly interface for linking sequence data with patient data and AST data. Moreover, they have to comply with regulations on data protection and privacy by providing options to restrict data sharing to authorised individuals and to define for how long sensitive data will be stored. Open-source free software can save costs and is also more transparent than proprietary software.
Financial
Building capacity for WGS surveillance of AMR in LMICs requires sustainable and flexible funding. In addition to one-off purchase costs for preparation and sequencing machines and the IT infrastructure to process and analyse sequence information, funding also needs to be secured for maintenance and servicing of laboratory machines and computational servers. Ordering machines and equipment, hiring staff and satisfying regulatory procedures all take time, especially when a WGS surveillance laboratory is set up for the first time. Therefore, grants will have to be flexible enough to accommodate delays in timelines and work packages. At the same time, grant approval and management must follow ethical and accountability standards. Formal procedures to ensure these standards may have to be put into place.
Training
Another major difficulty for the implementation of WGS surveillance in LMICs is the lack of trained personnel. Within-country expertise may be lacking both for laboratory scientists with genomics experience and bioinformaticians. Even if qualified bioinformaticians can be found, they often lack desirable epidemiology and microbiology training. Conversely, epidemiologists frequently lack the necessary bioinformatics skills to implement WGS surveillance. An associated concern is the lack of funded permanent positions in many LMICs which hinders the recruitment and retention of trained staff. In addition, even given the availability of trained staff, developing standard protocols for sequencing and genome analysis is a non-trivial task. If the regulations on handling biological specimens are complex, trained administrative staff may have to be hired in addition to specialist scientific staff. Similarly, administrative staff trained in accounting and ethical grant management will be required.
Quality assessment
Many LMICs do not yet have an NAP for AMR surveillance or a functional surveillance system. Consequently, implementing WGS surveillance may not be a priority. However, laboratories that do conduct WGS surveillance should be required to pass an EQA exercise before they form part of a national surveillance system to ensure the collection of high-quality, reliable data. Several national and international AMR surveillance networks and public health institutions have established standards that can be used for proficiency testing, for example, PulseNet, Gen-FS, CDC, FDA, GMI.50 69–71 EQA standards need to be stringent enough to ensure good-quality sequence data and reliable analysis results. However, there is a risk that strict standards at the laboratory level will exclude LMIC laboratories from participating in surveillance networks. An alternative solution could be to initially perform quality testing at the sequence level, while simultaneously providing support to improve laboratory standards. Optimising assays in new WGS laboratories to meet accreditation criteria will take time that needs to be taken into account when setting up surveillance systems.
Where local laboratories cannot be accredited as sequencing centres, another solution could be to ship samples to a centralised laboratory for sequencing. Finding a suitable courier service may be another hurdle. For example, not all couriers accept biological specimens. Moreover, DNA samples should ideally be shipped on dry ice to maintain low temperatures and thus to minimise damage to the samples. However, this method of shipping is expensive and may not be available in all countries. Thus, alternative storage and transport protocols may have to be designed and tested. Shipping samples abroad may further be subject to regulations on biodiversity conservation. In the future, this may also affect the sharing of DNA sequence information.72
How can barriers to implementing WGS surveillance in LMICs be overcome? Driving down time to implementation
Having outlined the main challenges for implementing WGS for AMR surveillance in LMICs, we propose practical solutions for overcoming the challenges associated with each of the workflow components (figure 2).
Laboratory
In LMICs, the main challenge beyond the initial setup of a laboratory is keeping it up and running. To guarantee continuous operations in surveillance laboratories, stable supply chains for reagents and single-use laboratory items should be established locally. Commercial suppliers are more likely to offer stable goods deliveries if several laboratories within a country or region purchase from them. A sufficient amount of initial investment with a funding horizon of several years is required to kick-start the implementation of WGS surveillance in LMICs. Large-scale funders like the NIHR or philanthropic organisations may be able to exert pressure on suppliers to lower prices or offer alternative and more transparent pricing models to laboratories in LMICs.
Given resource and staff constraints, public health bodies and laboratories in LMICs will have to prioritise which bacterial isolates to sequence. However, prioritisation of isolates also occurs in HICs. For example, in the UK all Salmonella isolates are routinely sequenced, but not all S. aureus isolates.73 74 Prioritisation should be guided by what are the most urgent local public health concerns and associated research questions. Cost-effectiveness also plays a role in the decision on what isolates to sequence.75 Both passive surveillance based on random sampling of susceptible and resistant isolates to establish the population genetic context of circulating bacterial strains and targeted surveillance of highly resistant and outbreak isolates are important. Background surveillance does not need to be continuous, but could be based on structured surveys as in the case for TB surveillance in many countries.76 This may require a change in laboratory outlook from a diagnostic to a surveillance perspective.
Bioinformatics
If the computational infrastructure to conduct bioinformatic analysis is to be set up locally, establishing continuous supply chains for component replacement and upgrades will be important. Where technology has to be imported, custom arrangements should be simplified to guarantee timely delivery. With modern technologies such as workflow managers to describe analytical steps and containers to bundle software dependencies, the technical complexity of setting up analytical pipelines can be greatly simplified.77 This allows for reproducible analyses that can be run on a diverse range of infrastructures with little configuration. For example, the same bioinformatics pipelines for genome assembly, phylogenetic tree construction, Multilocus Sequence Typing (MLST) calling and AMR prediction have been implemented in partner laboratories in Colombia, India, Nigeria, the Philippines and the UK, using Nextflow78 as a workflow manager and Docker79 or Singularity80 containers to ensure that pipelines are compatible with local operating systems. All bioinformatics tools that are part of each workflow are open access and open source. A recent review of bioinformatics tools suitable for workflows in both LMICs and HICs has been published by Hendriksen et al.81
Where building local computing infrastructure is currently too expensive, alternatives are now available using genome analysis pipeline software and cloud-based infrastructure. Examples for cloud-based bacterial genomic analysis tools are the Bacterial Analysis Platform82 and Pathogenwatch.33 However, this requires financial systems within host organisations to allow payment for resource-usage-based subscription services in the cloud, such as Amazon Web Services, Google Compute Platform or Microsoft Azure. It also requires that data transfer via a stable internet connection is reliable and not subject to capped and expensive data plans.
Data flow
Reference databases and software need to be developed to make genomic information accessible across clinical, public health, environmental and agricultural sectors. Such databases already exist but need to be expanded and a single standard for data entry should be agreed. Genomic and associated metadata should be open access, for example, via a data portal on a website. Some information will have to be kept confidential, for example, metadata from hospital outbreaks that could be used to identify patients. WGS data should be stored alongside phenotypic AST data and metadata linked to the isolate sequences. To facilitate the curation of joint databases, the commonalities in reporting these different data types can be assembled and pipelined. The combined analysis of connected data types will give insights into behavioural patterns or events that drive the emergence and spread of resistant bacterial lineages and high-risk clones. Existing application programming interfaces and metadata ontologies are available, for example, the Minimal Data for Matching by the Global Microbial Identifier and System for Enteric Disease Response, Investigation and Coordination by the US CDC.83 84 WHONET is the most widely used example of a database software for the curation of phenotypic AST data. It could be extended with an option to include WGS surveillance data. For example, WGS data could be stored in one of the already existing databases, and the WHONET file could contain a variable that links to the relevant database entry. This separation could also guarantee that sequence data which on their own do not contain sensitive information are easily accessible, whereas metadata which are sensitive can only be accessed by approved public health officials and scientific collaborators. A web interface based on a WHONET configuration file could be produced with a flexible option for WGS data collection. Ideally, the interface should work on portable tablets that do not need constant power.
Financial
Various schemes exist to standardise and simplify grant management practices and to provide accreditation for good financial grant practices, for example, the Global Grant Community.85 The initiative sets out standard guidelines that need to be followed by both grantors (institutions awarding a grant) and grantees (institutions receiving a grant). As part of the assessment process, grantees are rated according to their financial capacity, that is, how large and complex a grant they are able to handle. Accreditation reassures grantors that grantees can responsibly manage received funding. Another advantage for grantees is that once accredited they will not have to repeat the assessment process for each new grant they apply for.
Training
Joint initiatives between HICs and LMICs are great opportunities for training and knowledge transfer. To maximise utility, training programmes should be tailored to the specific needs of participating countries. The curricula should be developed in discussion with local researchers to fill skills gaps and support local research agendas. Importantly, training documents should be accessible via online services such as Google Docs so that they are both easy to share and keep up to date. Keeping the information up to date could be the responsibility of Regional Reference Laboratories in collaboration with WHO or international surveillance networks. In addition, practical exercises should not require specialist IT equipment and should run on a relatively modest local computer. The training material and course structure should be streamed and modular so that they can be adapted for scientists to become trainers of local colleagues (train-the-trainer schemes). Other programmes have emphasised the need to provide incentives for staff retention once training is completed, as failure to retain staff can lead to interruption of surveillance, for instance, the Collaborative African Genomics Network provides funding for staff faculty positions at their home universities.86 To decrease the risk that WGS surveillance will be disrupted when a staff member leaves, training initiatives could be targeted at teams rather than individuals.
Quality assessment
EQA procedures should be standardised among collaborating centres to ensure that results are comparable and of high quality. The genomic-based EQA should be developed in collaboration with QA providers. Samples with known phenotypic and genotypic characteristics can be used as a standard against which the performance of a participating institute in laboratory and bioinformatics techniques can be measured.
Even though few LMICs will have the capacity for implementing WGS surveillance immediately, many more will have AMR surveillance programmes based on phenotypic testing. These programmes can be leveraged and built on to incorporate WGS surveillance later. NAPs can be developed or extended with this goal in mind. In order for WGS surveillance to become part of NAPs, standard operating procedures (SOPs) for sample collection, sample preparation, sequencing of isolates and regulatory procedures concerning access to and handling of samples and DNA sequence information will have to be developed. SOPs should take into account challenges experienced in LMIC settings and offer workable solutions. Once they are tested and approved, SOPs should be published in peer-reviewed journals to make them available to other researchers in LMICs and encourage a broader uptake of standardised protocols.
Conclusion: a way forward for implementing WGS surveillance for AMR in LMICs
WGS is a promising technology for AMR surveillance that can provide actionable data for public health decision making. Nevertheless, the use of WGS for public health surveillance poses a number of challenges. LMICs in particular face high hurdles in the implementation of WGS surveillance for AMR. However, since LMICs are disproportionately affected by AMR infections and are hotspots for the emergence of high-risk clones, strong surveillance networks with the capacity for WGS in LMICs will have local and global benefits. A possible roadmap to the implementation of WGS surveillance in LMICs and building global surveillance networks is being developed, with the aim of achieving the virtuous circle sketched out in figure 1. This virtuous circle can be boosted by the development of open source tools that help countries to strengthen their surveillance systems and access critical data in real time. The NIHR GHRU Consortium is a partnership between the CGPS (at the UK-based University of Oxford and Sanger Institute) and strategically important sites in Asia (India, The Philippines), Africa (Nigeria) and South America (Colombia) with the aim to provide actionable data for public health policy to control high-risk bacterial pathogens and to make WGS tools for AMR surveillance available in LMICs. The Philippines have already successfully implemented WGS as part of their national AMR surveillance programme and can serve as a template for other LMICs that plan to strengthen their surveillance capacity with WGS.13
The most important lessons learnt so far with regard to WGS surveillance are: (1) decision making based on surveillance requires good-quality and timely data that is representative for the whole country and sustained over time, (2) surveillance data need to be disseminated, preferably via the web (short time and low cost), (3) surveillance methods should be updated regularly and (4) high-quality surveillance has a cost. These lessons are not exclusive to WGS surveillance in LMICs but apply to surveillance programmes in general. International research collaborations between HICs and LMICs at various stages of developing and implementing NAPs for AMR surveillance are learning opportunities for all participating partners and can form cornerstones for global surveillance and research networks in the future.
Acknowledgments
We acknowledge John Stelling for advice on the use of WHONET, Neil Woodford and Matthew Ellington for helpful discussions about genomic EQAs for AMR.
Footnotes
Handling editor: Seye Abimbola
Collaborators: Carolin Vegyari (Infectious Disease Epidemiology, Imperial College London, London, UK), Anthony Underwood (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK; Centre for Genomic Pathogen Surveillance, Big Data Institute, University of Oxford, Oxford, UK), Mihir Kekre (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK; Centre for Genomic Pathogen Surveillance, Big Data Institute, University of Oxford, Oxford, UK), Silvia Argimon (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK; Centre for Genomic Pathogen Surveillance, Big Data Institute, University of Oxford, Oxford, UK), Dawn Muddyman (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK; Centre for Genomic Pathogen Surveillance, Big Data Institute, University of Oxford, Oxford, UK), Monica Abrudan (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK; Centre for Genomic Pathogen Surveillance, Big Data Institute, University of Oxford, Oxford, UK), Celia Carlos (Research Institute for Tropical Medicine, Muntinlupa City, Philippines), Pilar Donado-Godoy (AGROSAVIA, Bogota, Colombia), Iruka N. Okeke (University of Ibadan, Ibadan, Nigeria), K L Ravikumar (KIMS Hospital and Research Centre, Bangalore, India), Khalil Abudahab (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK), Ayorinde Afolayan (University of Ibadan, Ibadan, Nigeria), Alejandra Arevalo (AGROSAVIA, Bogota, Colombia), Johan Bernal-Morales (AGROSAVIA, Bogota, Colombia), Erik Christopher Castro (AGROSAVIA, Bogota, Colombia), June Gayeta (Research Institute for Tropical Medicine, Muntinlupa City, Philippines), Vandan Govindan (KIMS Hospital and Research Centre, Bangalore, India), Maria Fernanda Guerrero (AGROSAVIA, Bogota, Colombia), Harry Harste (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK; Centre for Genomic Pathogen Surveillance, Big Data Institute, University of Oxford, Oxford, UK), Elmer Herrera (Research Institute for Tropical Medicine, Muntinlupa City, Philippines), Jolaade Janet Ajiboye (University of Ibadan, Ibadan, Nigeria), Polle Krystle Valdenarro (Research Institute for Tropical Medicine, Muntinlupa City, Philippines), Marietta Lagrada (Research Institute for Tropical Medicine, Muntinlupa City, Philippines), Melissa Ana Masim (Research Institute for Tropical Medicine, Muntinlupa City, Philippines), Ali Molloy (London, UK), Geeta Nagaraj (KIMS Hospital and Research Centre, Bangalore, India), Anderson Osemahu Oaikhena (University of Ibadan, Ibadan, Nigeria), Agnettah Olorosa (Research Institute for Tropical Medicine, Muntinlupa City, Philippines), Akshata Prabhu (KIMS Hospital and Research Centre, Bangalore, India), Varun Shammana (KIMS Hospital and Research Centre, Bangalore, India), M.R. Shincy (KIMS Hospital and Research Centre, Bangalore, India), Darmavaram Sravani (KIMS Hospital and Research Centre, Bangalore, India), John Stelling (Brigham and Women's Hospital, Boston, Massachusetts, USA), Ben Taylor (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK), Nicole Wheeler (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK; Centre for Genomic Pathogen Surveillance, Big Data Institute, University of Oxford, Oxford, UK), Anneke Schmider (Centre for Genomic Pathogen Surveillance, Big Data Institute, University of Oxford and Wellcome Genome Campus, Oxford, UK), David Aanensen (Centre for Genomic Pathogen Surveillance, Wellcome Trust Sanger Institute, Cambridge, UK; Big Data Institute, University of Oxford, Oxford, UK)
Contributors: AA: Bioinformatician. AA: Laboratory scientist/microbiologist. AM: Graphics Designer. AOO: Laboratory scientist/ microbiologist. AP: Research Associate. AS: Consultant. AU: Bioinformatics Implementation Manager. BT: Software Developer. CC: Lead PI, Philippines National Surveillance Unit. CV: Scientific Writer. DM: GHRU Head of Programme Management. DMA: PI from UK NIHR Global Health Research Unit. DS: Research Associate. ECDOC: Administrative professional. EH: Financial and Management Officer. GN: Senior lab manager / Project Coordinator. HH: Finance Implementation Manager. INO: Lead PI, Nigeria National Surveillance Unit. JFB-M: Bioinformatician. JG: Bioinformatician. JJA: Project Coordinator / Admin SupportPilar. JS: WHONET Software Consultant. KA: Software Developer. KLR: Lead PI, India National Surveillance Unit. MA: Postdoctoral researcher in Bioinformatics. MAM: Bioinformatician. MFVG: Laboratory scientist/microbiologist. MK: NGS Operations Lead. ML: Senior laboratory scientist / microbiologist. MRS: Research Associate. NW: Data Scientist. PD-G: Lead PI, Colombia National Surveillance Unit. PKVM: Laboratory scientist/ microbiologist. SA: Genomic Epidemiologist. VG: Bioinformatician/WHONET. VS: Bioinformatician. All authors contributed to the conceptualisation and editing of the manuscript.
Funding: This research was funded by the National Institute for Health Research (NIHR) (award reference 16_136_111) using UK aid from the UK Government to support global health research. Further funding provided by Li Ka Shing Foundation (DMA)
Disclaimer: The views expressed in this publication are those of the author(s) and not necessarily those of the NIHR or the UK Department of Health and Social Care.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Contributor Information
NIHR Global Health Research Unit on Genomic Surveillance of AMR:
Celia Carlos, Marietta Lagrada, Polle K. Macaranas, Agnettah M. Olorosa, June Gayeta, Melissa Ana Masim, Elmer M. Herrera, David M Aanensen, Khalil Abudahab, Monica Abrudan, Silvia Argimon, Harry Harste, Mihir Kekre, Ali Molloy, Dawn Muddyman, Anneke Schmider, Ben Taylor, Anthony Underwood, Nicole Wheeler, María del Pilar Donado Godoy, Johan Fabian Bernal Morales, Alejandra Arevalo, Maria Fernanda Valencia Guerrero, Erik Cristopher Dustin Osma Castro, Iruka N Okeke, Anderson O. Oaikhena, Ayorinde Oluwatobiloba Afolayan, Jolaade J Ajiboye, Ravikumar K. L., Geetha Nagaraj, Varun Shammana, Vandana Govindan, Akshata Prabhu, Darmavaram Sravani, Shincy M. R., Rajitha G. V., Carolin Vegvari, and John Stelling
References
- 1. WHO Global antimicrobial resistance surveillance system: manual for early implementation. Geneva, Switzerland, 2015. [Google Scholar]
- 2. Alividza V, Mariano V, Ahmad R, et al. Investigating the impact of poverty on colonization and infection with drug-resistant organisms in humans: a systematic review. Infect Dis Poverty 2018;7:76. 10.1186/s40249-018-0459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jasovský D, Littmann J, Zorzet A, et al. Antimicrobial resistance-a threat to the world's sustainable development. Ups J Med Sci 2016;121:159–64. 10.1080/03009734.2016.1195900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bloom G, Merrett GB, Wilkinson A, et al. Antimicrobial resistance and universal health coverage. BMJ Glob Health 2017;2:e000518. 10.1136/bmjgh-2017-000518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pokharel S, Raut S, Adhikari B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob Health 2019;4:e002104. 10.1136/bmjgh-2019-002104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li LM, Grassly NC, Fraser C. Genomic analysis of emerging pathogens: methods, application and future trends. Genome Biol 2014;15:541. 10.1186/s13059-014-0541-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Didelot X, Gardy J, Colijn C. Bayesian inference of infectious disease transmission from whole-genome sequence data. Mol Biol Evol 2014;31:1869–79. 10.1093/molbev/msu121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aanensen DM, Feil EJ, Holden MTG, et al. Whole-Genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. mBio 2016;7. 10.1128/mBio.00444-16. [Epub ahead of print: 05 May 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holmes EC, Rambaut A, Andersen KG. Pandemics: spend on surveillance, not prediction. Nature 2018;558:180–2. 10.1038/d41586-018-05373-w [DOI] [PubMed] [Google Scholar]
- 10. Grad YH, Harris SR, Kirkcaldy RD, et al. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000-2013. J Infect Dis 2016;214:1579–87. 10.1093/infdis/jiw420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collineau L, Boerlin P, Carson CA, et al. Integrating whole-genome sequencing data into quantitative risk assessment of foodborne antimicrobial resistance: a review of opportunities and challenges. Front Microbiol 2019;10:1107. 10.3389/fmicb.2019.01107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 2019;4:1919–29. 10.1038/s41564-019-0492-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Argimón S, Masim MAL, Gayeta JM, et al. See and sequence: integrating whole-genome sequencing within the National antimicrobial resistance surveillance program in the Philippines. bioRxiv 2019;808378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tong SYC, Holden MTG, Nickerson EK, et al. Genome sequencing defines phylogeny and spread of methicillin-resistant Staphylococcus aureus in a high transmission setting. Genome Res 2015;25:111–8. 10.1101/gr.174730.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mellmann A, Bletz S, Böking T, et al. Real-Time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol 2016;54:2874–81. 10.1128/JCM.00790-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kwong JC, Chow EPF, Stevens K, et al. Whole-genome sequencing reveals transmission of gonococcal antibiotic resistance among men who have sex with men: an observational study. Sex Transm Infect 2018;94:151–7. 10.1136/sextrans-2017-053287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerner-Smidt P, Besser J, Concepción-Acevedo J, et al. Whole genome sequencing: bridging One-Health surveillance of foodborne diseases. Front Public Health 2019;7:172. 10.3389/fpubh.2019.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Brien TF, Stelling JM. WHONET: an information system for monitoring antimicrobial resistance. Emerg Infect Dis 1995;1:66. 10.3201/eid0102.950209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baker S, Thomson N, Weill F-X, et al. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 2018;360:733–8. 10.1126/science.aar3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bentley SD, Parkhill J. Genomic perspectives on the evolution and spread of bacterial pathogens. Proc Biol Sci 2015;282:20150488. 10.1098/rspb.2015.0488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harkins CP, Pichon B, Doumith M, et al. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol 2017;18:130. 10.1186/s13059-017-1252-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donker T, Reuter S, Scriberras J, et al. Population genetic structuring of methicillin-resistant Staphylococcus aureus clone EMRSA-15 within UK reflects patient referral patterns. Microb Genom 2017;3:e000113. 10.1099/mgen.0.000113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grad YH, Lipsitch M. Epidemiologic data and pathogen genome sequences: a powerful synergy for public health. Genome Biol 2014;15:538. 10.1186/s13059-014-0538-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Votintseva AA, Bradley P, Pankhurst L, et al. Same-Day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol 2017;55:1285–98. 10.1128/JCM.02483-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kelesidis T, Falagas ME. Substandard/counterfeit antimicrobial drugs. Clin Microbiol Rev 2015;28:443–64. 10.1128/CMR.00072-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morgan DJ, Okeke IN, Laxminarayan R, et al. Non-prescription antimicrobial use worldwide: a systematic review. Lancet Infect Dis 2011;11:692–701. 10.1016/S1473-3099(11)70054-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okeke IN, Lamikanra A, Edelman R. Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerg Infect Dis 1999;5:18–27. 10.3201/eid0501.990103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mara D, Evans B. Sanitation and water supply in low- income countries;149. [Google Scholar]
- 29. Velpandian T, Halder N, Nath M, et al. Un-segregated waste disposal: an alarming threat of antimicrobials in surface and ground water sources in Delhi. Environ Sci Pollut Res Int 2018;25:29518–28. 10.1007/s11356-018-2927-9 [DOI] [PubMed] [Google Scholar]
- 30. Brown J, Lomolino M. Biogeography. Sinauer associates, Sunderland 1998.
- 31. Nhung NT, Cuong NV, Campbell J, et al. High levels of antimicrobial resistance among Escherichia coli isolates from livestock farms and synanthropic rats and shrews in the Mekong delta of Vietnam. Appl Environ Microbiol 2015;81:812–20. 10.1128/AEM.03366-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. GLASS, WHO . Molecular methods for antimicrobial resistance (AMR) diagnostics to enhance the global antimicrobial resistance surveillance system.. Available: http://www.who.int/glass/resources/publications/molecular-methods-for-amr-diagnostics/en/ [Accessed 3 Mar 2020].
- 33. Pathogenwatch A global platform for genomic surveillance. Available: https://pathogen.watch/ [Accessed 24 Feb 2020].
- 34. Ingle DJ, Levine MM, Kotloff KL, et al. Dynamics of antimicrobial resistance in intestinal Escherichia coli from children in community settings in South Asia and sub-Saharan Africa. Nat Microbiol 2018;3:1063–73. 10.1038/s41564-018-0217-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kariuki S, Onsare RS. Epidemiology and genomics of invasive nontyphoidal Salmonella infections in Kenya. Clin Infect Dis 2015;61 Suppl 4:S317–24. 10.1093/cid/civ711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cohen KA, Manson AL, Abeel T, et al. Extensive global movement of multidrug-resistantM. tuberculosis strains revealed by whole-genome analysis. Thorax 2019;74:882–9. 10.1136/thoraxjnl-2018-211616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grundmann H. Towards a global antibiotic resistance surveillance system: a primer for a roadmap. Ups J Med Sci 2014;119:87–95. 10.3109/03009734.2014.904458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rossen JWA, Friedrich AW, Moran-Gilad J, et al. Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect 2018;24:355–60. 10.1016/j.cmi.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 39. Schürch AC, van Schaik W. Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann N Y Acad Sci 2017;1388:108–20. 10.1111/nyas.13310 [DOI] [PubMed] [Google Scholar]
- 40. Seale AC, Gordon NC, Islam J, et al. AMR Surveillance in low and middle-income settings - A roadmap for participation in the Global Antimicrobial Surveillance System (GLASS). Wellcome Open Res 2017;2:92. 10.12688/wellcomeopenres.12527.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. OECD iLibrary Health in the 21st Century : Putting Data to Work for Stronger Health Systems [Accessed 3 Mar 2020].
- 42. Microbiology Society Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Available: https://www.microbiologyresearch.org/content/journal/mgen/ [Accessed 24 Feb 2020]. [DOI] [PMC free article] [PubMed]
- 43. Argimón S, Abudahab K, Goater RJE, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2016;2:e000093. 10.1099/mgen.0.000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neuert S, Nair S, Day MR, et al. Prediction of Phenotypic Antimicrobial Resistance Profiles From Whole Genome Sequences of Non-typhoidal Salmonella enterica. Front Microbiol 2018;9:592. 10.3389/fmicb.2018.00592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Day MR, Doumith M, Do Nascimento V, et al. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Salmonella enterica serovars typhi and Paratyphi. J Antimicrob Chemother 2018;73:365–72. 10.1093/jac/dkx379 [DOI] [PubMed] [Google Scholar]
- 46. Mason A, Foster D, Bradley P, et al. Accuracy of different bioinformatics methods in detecting antibiotic resistance and virulence factors from Staphylococcus aureus whole-genome sequences. J Clin Microbiol 2018;56. 10.1128/JCM.01815-17. [Epub ahead of print: 27 Aug 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Monk JM. Predicting antimicrobial resistance and associated genomic features from whole-genome sequencing. J Clin Microbiol 2019;57. 10.1128/JCM.01610-18. [Epub ahead of print: 30 Jan 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nguyen M, Long SW, McDermott PF, et al. Using Machine Learning To Predict Antimicrobial MICs and Associated Genomic Features for Nontyphoidal Salmonella. J Clin Microbiol 2019;57:e01260–18. 10.1128/JCM.01260-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shelburne SA, Kim J, Munita JM, et al. Whole-Genome sequencing accurately identifies resistance to extended-spectrum β-lactams for major gram-negative bacterial pathogens. Clin Infect Dis 2017;65:738–45. 10.1093/cid/cix417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moran-Gilad J, Sintchenko V, Pedersen SK, et al. Proficiency testing for bacterial whole genome sequencing: an end-user survey of current capabilities, requirements and priorities. BMC Infect Dis 2015;15:174. 10.1186/s12879-015-0902-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. GMI The proficiency test (pilot) report of the global microbial identifier (GMI) initiative, year 2014., 2016. Available: http://www.globalmicrobialidentifier.org/-/media/Sites/gmi/Work-groups/GMI_pilot-PT_report_2014_v4d_final.ashx?la¼da [Accessed 24 Feb 2020].
- 52. Timme RE, Rand H, Sanchez Leon M, et al. GenomeTrakr proficiency testing for foodborne pathogen surveillance: an exercise from 2015. Microb Genom 2018;4. 10.1099/mgen.0.000185. [Epub ahead of print: 15 Jun 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014;12:87. 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol 2019;14:319–38. 10.1146/annurev-pathmechdis-012418-012751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Low AJ, Koziol AG, Manninger PA, et al. ConFindr: rapid detection of intraspecies and cross-species contamination in bacterial whole-genome sequence data. PeerJ 2019;7:e6995. 10.7717/peerj.6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xi W, Gao Y, Cheng Z, et al. Using QC-Blind for quality control and contamination screening of bacteria DNA sequencing data without reference genome. Front Microbiol 2019;10:1560. 10.3389/fmicb.2019.01560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Breitwieser FP, Pertea M, Zimin AV, et al. Human contamination in bacterial genomes has created thousands of spurious proteins. Genome Res 2019;29:gr.245373:118–960. 10.1101/gr.245373.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Köser CU, Ellington MJ, Peacock SJ. Whole-genome sequencing to control antimicrobial resistance. Trends Genet 2014;30:401–7. 10.1016/j.tig.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palmer AC, Kishony R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat Rev Genet 2013;14:243–8. 10.1038/nrg3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gardy JL, Loman NJ. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat Rev Genet 2018;19:9–20. 10.1038/nrg.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vandenberg O, Kozlakidis Z, Schrenzel J, et al. Control of infectious diseases in the era of European clinical microbiology laboratory consolidation: new challenges and opportunities for the patient and for public health surveillance. Front Med 2018;5:15. 10.3389/fmed.2018.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gabrielian A, Engle E, Harris M, et al. TB DEPOT (data exploration portal): a multi-domain tuberculosis data analysis resource. PLoS One 2019;14:e0217410. 10.1371/journal.pone.0217410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jackson C, Gardy JL, Shadiloo HC, et al. Trust and the ethical challenges in the use of whole genome sequencing for tuberculosis surveillance: a qualitative study of stakeholder perspectives. BMC Med Ethics 2019;20:43. 10.1186/s12910-019-0380-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tekola-Ayele F, Rotimi CN. Translational genomics in low- and middle-income countries: opportunities and challenges. Public Health Genomics 2015;18:242–7. 10.1159/000433518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brunker K, Jaswant G, Thumbi SM, et al. Rapid in-country sequencing of whole virus genomes to inform rabies elimination programmes. Wellcome Open Res 2020;5:3. 10.12688/wellcomeopenres.15518.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ahmed SS, Alp E, Ulu-Kilic A, et al. Establishing molecular microbiology facilities in developing countries. J Infect Public Health 2015;8:513–25. 10.1016/j.jiph.2015.04.029 [DOI] [PubMed] [Google Scholar]
- 67. Dailey PJ, Osborn J, Ashley EA, et al. Defining system requirements for simplified blood culture to enable widespread use in resource-limited settings. Diagnostics 2019;9:10. 10.3390/diagnostics9010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Abdurrahman ST, Emenyonu N, Obasanya OJ, et al. The hidden costs of installing Xpert machines in a tuberculosis high-burden country: experiences from Nigeria. Pan Afr Med J 2014;18:277. 10.11604/pamj.2014.18.277.3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nadon C, Van Walle I, Gerner-Smidt P, et al. Pulsenet international: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill 2017;22:30544. 10.2807/1560-7917.ES.2017.22.23.30544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tolar B, Joseph LA, Schroeder MN, et al. An overview of PulseNet USA databases. Foodborne Pathog Dis 2019;16:457–62. 10.1089/fpd.2019.2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Timme RE, Rand H, Shumway M, et al. Benchmark datasets for phylogenomic pipeline validation, applications for foodborne pathogen surveillance. PeerJ 2017;5:e3893. 10.7717/peerj.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Neumann D, Borisenko AV, Coddington JA, et al. Global biodiversity research tied up by juridical interpretations of access and benefit sharing. Org Divers Evol 2018;18:1–12. 10.1007/s13127-017-0347-1 [DOI] [Google Scholar]
- 73. Chattaway MA, Dallman TJ, Larkin L, et al. The Transformation of Reference Microbiology Methods and Surveillance for Salmonella With the Use of Whole Genome Sequencing in England and Wales. Front Public Health 2019;7:317. 10.3389/fpubh.2019.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Toleman MS, Reuter S, Jamrozy D, et al. Prospective genomic surveillance of methicillin-resistant Staphylococcus aureus (MRSA) associated with bloodstream infection, England, 1 October 2012 to 30 September 2013. Euro Surveill 2019;24:24. 10.2807/1560-7917.ES.2019.24.4.1800215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dymond A, Davies H, Mealing S, et al. Genomic surveillance of methicillin-resistant Staphylococcus aureus: a mathematical early modelling study of cost effectiveness. Clin Infect Dis 2019. 10.1093/cid/ciz480. [Epub ahead of print: 18 Jun 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. World Health Organization Global tuberculosis report 2019. World Health Organization, 2019. [Google Scholar]
- 77. Perkel JM. Workflow systems turn raw data into scientific knowledge. Nature 2019;573:149–50. 10.1038/d41586-019-02619-z [DOI] [PubMed] [Google Scholar]
- 78. Nextflow A DSL for parallel and scalable computational pipelines. Available: https://www.nextflow.io/ [Accessed 3 Mar 2020].
- 79. Docker Empowering app development for developers. Available: https://www.docker.com/ [Accessed 3 Mar 2020].
- 80. Singularity Sylabs.io. Available: https://sylabs.io/singularity/ [Accessed 3 Mar 2020].
- 81. Hendriksen RS, Bortolaia V, Tate H, et al. Using genomics to track global antimicrobial resistance. Front Public Health 2019;7:242. 10.3389/fpubh.2019.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Thomsen MCF, Ahrenfeldt J, Cisneros JLB, et al. A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One 2016;11:e0157718. 10.1371/journal.pone.0157718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Taboada EN, Graham MR, Carriço JA, et al. Food safety in the age of next generation sequencing, bioinformatics, and open data access. Front Microbiol 2017;8:909. 10.3389/fmicb.2017.00909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Centers for Disease Control and Prevention SEDRIC: system for enteric disease response, investigation, and coordination, 2018. Available: https://www.cdc.gov/foodsafety/outbreaks/investigating-outbreaks/sedric.html [Accessed 24 Feb 2020].
- 85. Global grant community. Available: https://www.globalgrantcommunity.org/ [Accessed 10 Nov 2019].
- 86. Mlotshwa BC, Mwesigwa S, Mboowa G, et al. The Collaborative African genomics network training program: a trainee perspective on training the next generation of African scientists. Genet Med 2017;19:826–33. 10.1038/gim.2016.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Alvarez-Uria G, Gandra S, Laxminarayan R. Poverty and prevalence of antimicrobial resistance in invasive isolates. Int J Infect Dis 2016;52:59–61. 10.1016/j.ijid.2016.09.026 [DOI] [PubMed] [Google Scholar]
- 88. McEwen SA, Collignon PJ. Antimicrobial resistance: a one health perspective : Antimicrobial resistance in bacteria from livestock and companion animals. John Wiley & Sons, Ltd, 2018: 521–47. [Google Scholar]
- 89. Ferri M, Ranucci E, Romagnoli P, et al. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr 2017;57:2857–76. 10.1080/10408398.2015.1077192 [DOI] [PubMed] [Google Scholar]
- 90. Barancheshme F, Munir M. Strategies to combat antibiotic resistance in the wastewater treatment plants. Front Microbiol 2017;8:2603. 10.3389/fmicb.2017.02603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. World Bank Drug-resistant infections: a threat to our economic future. Washington, DC: World Bank, 2017. http://documents.worldbank.org/curated/en/323311493396993758/pdf/final-report.pdf [Google Scholar]
- 92. Littmann J, Viens AM. The ethical significance of antimicrobial resistance. Public Health Ethics 2015;8:phv025:209–24. 10.1093/phe/phv025 [DOI] [PMC free article] [PubMed] [Google Scholar]