Abstract
Vitamin B12, cobalamin, is a cobalt-containing ring-contracted modified tetrapyrrole that represents one of the most complex small molecules made by nature. In prokaryotes it is utilised as a cofactor, coenzyme, light sensor and gene regulator yet has a restricted role in assisting only two enzymes within specific eukaryotes including mammals. This deployment disparity is reflected in another unique attribute of vitamin B12 in that its biosynthesis is limited to only certain prokaryotes, with synthesisers pivotal in establishing mutualistic microbial communities. The core component of cobalamin is the corrin macrocycle that acts as the main ligand for the cobalt. Within this review we investigate why cobalt is paired specifically with the corrin ring, how cobalt is inserted during the biosynthetic process, how cobalt is made available within the cell and explore the cellular control of cobalt and cobalamin levels. The partitioning of cobalt for cobalamin biosynthesis exemplifies how cells assist metalation.
Abbreviations: CoA, co-enzyme A; GDP, guanosine diphosphate; TCA, tricarboxylic acid; GC–MS, gas chromatography–mass spectrometry; ROS, reactive oxygen species; ABC, ATP-binding cassette; NiCoT, nickel/cobalt transporter; OM, outer membrane; ECF, Electron-coupled factor; SBP, substrate-binding protein; ATP, Adenosine triphosphate; MFS, major facilitator superfamily; RND, resistance nodulation division; AdoCbl, adenosylcobalamin; AqCbl, aquacobalamin; MeCbl, methylcobalamin; RBS, ribosome binding site; GTP, guanosine triphosphate; MCM, methylmalonyl-CoA mutase
Keywords: Cobalamin, Cobamide, Metals, Chelation, Homeostasis, sensors
Highlights
-
•
The corrin-cobalt couple is ideal for forming metal-carbon bonds
-
•
A fraction of bacteria make vitamin B12 and supply other organisms including humans
-
•
The biogenesis of cobalamin: Cobalt chelatases and chaperones
-
•
A thermodynamic framework for metalation has been established
-
•
The partitioning of cobalt into cobalamin exemplifies how cells assist metalation
1. Vitamin B12 - its structure and biological roles
Vitamin B12 boasts a complex façade yet mediates an abundance of intricate chemistries that ultimately derive from the properties of a central cobalt ion. This essential dietary component first came to prominence a century ago when it was identified as the anti-pernicious anaemia factor that is present in raw liver [1,2]. The isolation of the nutrient from liver extracts was enhanced by the development of a microbial bioassay [3], using a bacterial vitamin B12 auxotroph, and culminated in the generation of purified bright red crystals [4,5], that were shown to contain cobalt [6,7], and which famously made their way into the hands of Dorothy Hodgkin for X-ray diffraction studies [8,9]. Through her pioneering work she was able to deduce the structure of vitamin B12 to reveal it as the most complex structure known at that time. It is an inauspicious coincidence that the name cobalt originates from German miners who when extracting the metal for its ability to colour glass believed their ores were contaminated by a pernicious goblin, or kobold, since when heated they emitted poisonous arsenic and sulphur containing gases – and that the main human cobalt deficiency in humans is related to vitamin B12 which is also associated with a pernicious ailment.
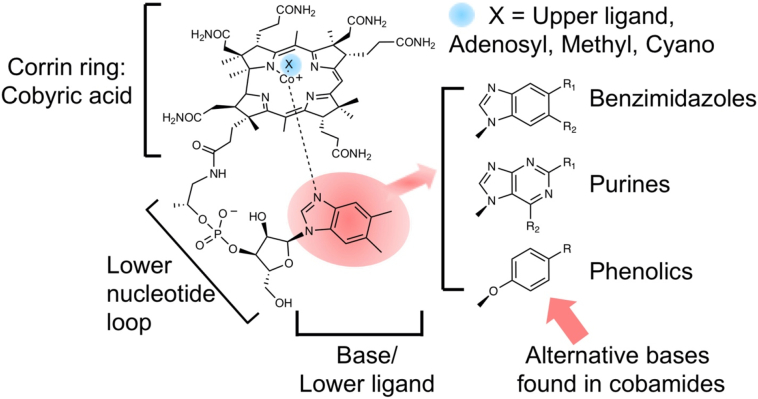
The structure of vitamin B12 can be considered in three parts (Fig. 1). Firstly, there is a modified tetrapyrrole that ligands a central cobalt ion. This tetrapyrrole-derived ring is unusual in that it has undergone a ring-contraction process meaning that one of the bridging (meso) carbon atoms that are used to connect the four pyrrole rings has been eliminated, thereby generating a macrocycle that is both contracted and lop-sided in comparison to the tetrapyrrole-frameworks that are associated with heme and chlorophylls [10]. This contracted ring structure is called a corrin. Secondly, the molecule contains a nucleotide loop that houses an unusual base, which in vitamin B12 is called dimethylbenzimidazole. The nucleotide loop is attached to one of the propionate side chains of the corrin ring through an aminopropanol linker, extending underneath the plane of the corrin ring such that the dimethylbenzimidazole base is able to act as a lower ligand for the cobalt. Finally, the third component of vitamin is the upper ligand to the cobalt ion, which in vitamin B12 is cyanide. The corrin ring and the lower nucleotide loop with its dimethylbenzimidazole base represents a molecule that is called cobalamin. Technically, vitamin B12 is therefore cyanocobalamin but vitamin B12 is quite often used loosely to refer to cobalamin. The cyano group in vitamin B12 is a consequence of the way the molecule is isolated, where cyanide is added to help its extraction and purification [11].
Fig. 1.
Structure of cobalamin (vitamin B12).
The core component of cobalamin is the corrin ring, which houses a central cobalt ion. The corrin ring together with the cobalt is called cobyric acid. Attached to the propionic side chain of cobyric acid is a lower nucleotide loop, that contains an unusual base called dimethylbenzimidazole and which acts as a lower ligand to the cobalt ion. The upper ligand in vitamin B12, marked as a X in the diagram is a cyano group. In the biological forms of cobalamin, the upper ligand is usually a methyl or an adenosyl group. Some bacteria make variant forms of cobalamin where the dimethylbenzimidazole base is replaced with other bases such as other benzimidazoles (with variations around R1 and R2), purines (with variations around R1 and R2) and phenolics (with variations around R).
Some bacteria make alternative forms of cobalamin that differ in the nature of the lower nucleotide loop (Fig. 1). Here, the differences largely relate to the base that is incorporated into the loop so, for instance, some bacteria incorporate adenine rather than dimethylbenzimidazole to give rise to a cobalamin analogue that is referred to as pseudocobalamin [12]. However, across different bacterial species around 15 different variants of cobalamin are made and these are collectively referred to as cobamides or corrinoids [13,14]. Cobalamin therefore is just one member of a broader cobamide family that all contain the same corrin ring with a liganded cobalt ion. It is the relationship between the cobalt ion and the corrin ring that is important with respect to its biological function [15,16].
The main active biological forms of cobalamin are where the upper ligand is either a methyl or an adenosyl group, which are present in methylcobalamin and adenosylcobalamin, respectively [17,18]. Methylcobalamin acts as a cofactor in a number of methyl transferase reactions, including in methionine synthase. Adenosylcobalamin acts as a coenzyme in rearrangement/isomerase reactions such as with methylmalonyl CoA mutase. These two enzymes catalyse probably the two best-known B12 -dependent reactions which also represent the only two known B12 -dependent processes in mammals. However, in prokaryotes there are a range of other cobamide-dependent enzymes including the diol dehydratases, ethanolamine ammonia lyase, ribonucleotide reductase, reductive dehalogenases and a host of radical SAM enzymes [13,19]. Adenosylcobalamin has also been shown to act as a light sensor in the CarH transcription factor [20]. Finally, cobamides can also directly influence levels of transcription and translation through interacting with riboswitches, where binding of specific cobamide forms to regulatory regions within the mRNA controls the production of encoded proteins (Section 5.3.1) [21,22]. Most cobamide riboswitches regulate genes associated with cobalt transport and cobalamin metabolism.
1.1. Chemistry of cobalt and the corrin ring
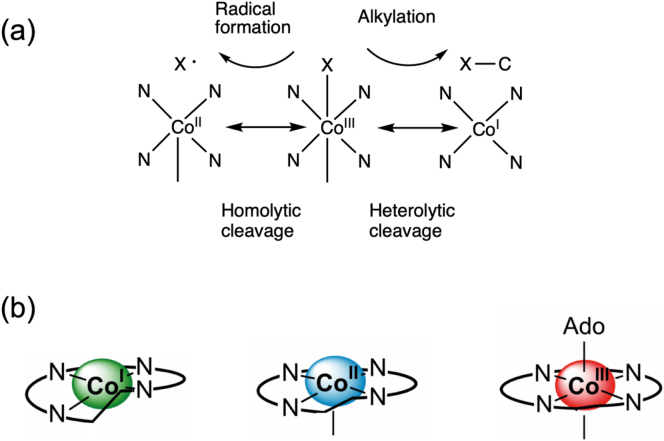
One of the long-standing questions concerning cobamides relates to why nature has selected the combination of cobalt and the corrin ring as a metalloprosthetic group partnership. What is the advantage of a corrin ring over the versatile porphyrin macrocycle of heme, and what is it about cobalt that so suits it for B12-dependent processes? This is particularly relevant given that (i) the availability of cobalt as a trace element varies significantly on both land and in water [[23], [24], [25]], and (ii) that the corrin ring is so difficult to make [[26], [27], [28]]. The main driving force for utilising cobalt is the chemistry that is mediated by the metal. Fundamental to this is the ability of cobalt to form metal-carbon bonds, the formation of which is facilitated by the powerful nucleophilicity of the Co(I) species [17,18,29]. The ability to control the nature of the Co-carbon complex, especially with control over homo- or heterolytic cleavage of this bond is crucial to why nature has selected the cobalt-corrin couple (Fig. 2a) [30,31].
Fig. 2.
Cobalt coordination and chemistry associated with cobamides.
(a) Co(III) corrinoids such as adenosylcobalamin (where X = deoxyadenosyl) can undergo homolytic cleavage to generate a Co(II) species and an adenosyl radical (X•), whereas methylcobalamin undergoes heterolytic cleavage to generate a Co(I) species and a methylated product. (b) The corrin ring naturally adopts a helical arrangement and this fits well with Co(I). More increasingly planar forms of the corrin ring are generated with Co(II) and Co(I) respectively. The ability to transition between planar and helical conformations means that the corrin ring acts as an entatic state module.
For instance, in the methyltransfer reaction associated with methionine synthase the cobalt in methylcobamide is in the Co(III) state and the methyl group is transferred after heterolytic cleavage of the Co-CH3 bond to generate the nucleophilic Co(I) species, which rapidly acquires another methyl group from Me-tetrahydrofolate [17,18]. In this process the cobamide cycles between Co(III) and Co(I) allowing for efficient transfer of the methyl group from methyl-tetrahydrofolate to homocysteine in the generation of methionine with the cobamide acting as a recycling cofactor. With isomerisation reactions the Co-carbon bond of adenosylcobamide undergoes homolytic cleavage, converting the Co(III) of adenosylcobamide to a Co(II) species and generating an adenosyl radical. The adenosyl radical is then able to abstract a hydrogen from the substrate, thereby inducing substrate radical formation and rearrangement, prior to hydrogen abstraction from the adenosyl group and reformation of the Co(III) adenosylcobamide [17,18]. The chemistry of cobamide-dependent reactions therefore relates to the controlled change in the oxidation state of the cobalt ion, changes that also reflect preferences for specific co-ordination of the metal ion. Thus, within the corrin macrocycle Co(III) prefers to bind in a 6 coordinate fashion, Co(II) in 5 coordinate fashion and Co(I) in a 4 coordinate manner. These changes in the oxidation state of the cobalt ion are, in part, facilitated by the corrin ring, which unlike like the planar ring of the porphyrin adopts a helical conformation [15]. The liganding of the cobalt ion into the corrin ring induces changes in the level of helicity within the corrin structure depending upon the oxidation state of the metal such that the corrin ring acts as an entatic state module that facilitates metal-oxidative state conversion (Fig. 2b) [15,32].
2. Vitamin B12 in prokaryotes and eukaryotes
The inclusion of both upper and lower ligands for cobalt within the corrin framework make cobamides much more complex structures than observed with other modified tetrapyrroles [10]. This complexity is reflected in equally convoluted biosynthetic pathways, involving upwards of 30 enzymatic steps for their complete de novo biogenesis [33]. This molecular intricacy is the likely explanation as to why the biosynthesis of cobamides has remained a prokaryotic prerogative, with the synthesis never making the transition to eukaryotes [26]. Even within the prokaryotic world there is a significant amount of variation as to which bacteria make the nutrient and those that cannot make it but require it. It appears that somewhere around 90% of bacteria contain a B12-dependent enzyme, yet only around a third of bacteria have the ability to make the nutrient [34]. This discrepancy between the synthesisers and utilisers likely reflects an attempt to share metabolic burden between microbes that occupy similar environmental niches and participate in mutualistic interactions [34,35].
Cobamides are very efficient cofactors and coenzymes, and coupled with the fact that they are very stable entities means that, in general, only very small quantities of the nutrient are required within a cell. For instance, in humans the dietary requirement for cobalamin is around 2 μg per day and hence a small amount of vitamin B12 goes a long way [36]. However, many systems have evolved to live in a B12-less setting. Higher plants, fungi and yeasts all exist in the absence of cobamides, with cobamide-dependent enzymes replaced with less efficient alternatives. This has implications for the food chain since crop-grazing mammals have to acquire their dietary requirement for cobalamin from alternative sources. For ruminants this demand for the nutrient is met by the ruminal microbiota that produce the necessary cobalamin, which is then absorbed into the circulatory system. Mono-gastric crop-feeding animals acquire their cobalamin either from surface bacteria on their food or by coprophagy. Humans acquire their cobalamin generally from dairy and meat produce. Although the human gastro-intestinal microbiome produces a range of cobamides, cobalamin only represents a minor component of this total [37]. Moreover, the human cobalamin uptake mechanism is located within the small intestine [38] whereas the gut microbiota is largely located within the large intestine. The take home message from this is that humans on vegetarian and more specifically vegan diets are prone to B12-deficiency [36].
3. Biosynthesis of vitamin B12
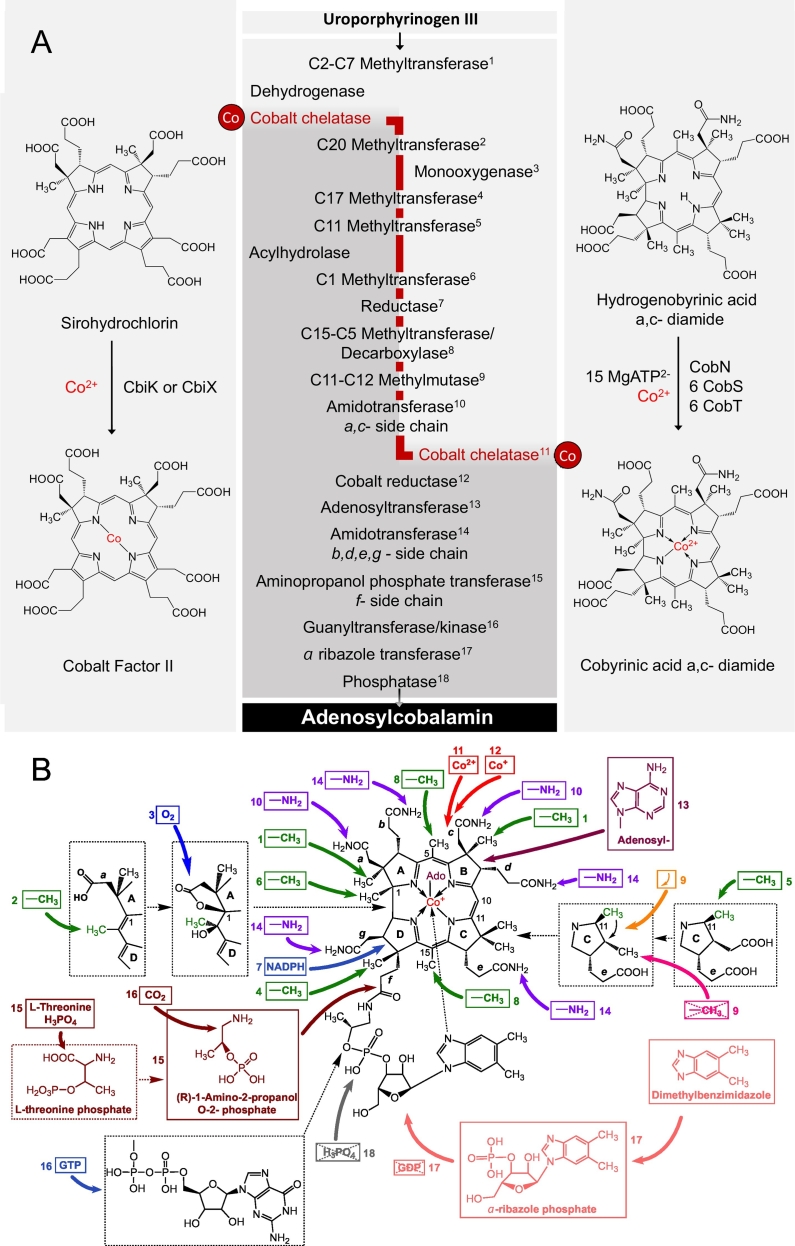
As alluded to earlier, the biogenesis of cobamides such as cobalamin is a complex process, and as with all modified tetrapyrroles the construction is based on the primogenitor blueprint of uroporphyrinogen III [10]. The transformation of uroporphyrinogen III into the corrin ring component of cobalamin, a molecule called cobyric acid, requires 8 methylations, 6 amidations, extrusion of the C20 meso position, decarboxylation of an acetic acid side chain and the insertion of cobalt (Fig. 3). The conversion of cobyric acid into cobalamin requires the attachment of a threonine-derived aminopropanol linker to the remaining propionic acid side chain to give cobinamide, activation of cobinamide with GDP and then substitution of the GDP moiety with α-ribazole-5′-P. The α-ribazole-5′-P is synthesised by attachment of the ribosyl-5′-phosphate moiety of nicotinate mononucleotide to dimethylbenzimidazole (Fig. 3b). More detailed reviews of cobalamin biosynthesis can be found elsewhere [13,33,39,40].
Fig. 3.
The biogenesis of cobalamin.
(a) The biosynthesis of cobalamin from the common tetrapyrrole primogenitor uroporphyrinogen III involves in excess of twenty steps. Although there are two pathways for the synthesis of cobalamin, referred to as the aerobic (or cobalt-late) and anaerobic (cobalt early) routes, the series of methylation, amidation and rearrangement reactions are broadly similar and these steps are highlighted. The cobalt-insertion steps are also shown, with the cobalt-early insertion shown on the left hand side and the cobalt-late insertion stage shown on the right hand side. (b) The modifications that are associated with the cobalt-late pathway are highlighted and cross-referenced with (a). The Figure highlights the extensive modification that take place during the biogenesis of cobalamin.
The biosynthesis of cobamides is further confounded by the presence of two genetically distinct but similar biosynthetic pathways for the construction of the corrin ring component of the molecule [10,41]. These pathways differ in their requirement for molecular oxygen and the timing of cobalt insertion (Fig. 3a). In the anaerobic pathway cobalt is inserted at an early stage, prior to ring contraction, and the pathway is independent of molecular oxygen. By way of contrast the aerobic pathway requires molecular oxygen to help promote the contraction process and inserts cobalt after all the methylations have taken place. Although these two routes are referred to as the anaerobic and aerobic pathways they should more accurately be called the early or late cobalt insertion pathways, respectively [27]. Despite the differences in the timing of cobalt insertion and the mechanism for ring contraction many of the other enzymes that are associated with methylations and amidations are very similar and operate in the same order along the pathway. The two pathways start from uroporphyrinogen III and progress via an ordered attachment of methylation and amidation reactions interspersed with ring contraction, decarboxylation, methyl migration and adenosylation reactions. The two pathways diverge after the first two methylation events, facilitated by the uroporphyrinogen III methyltransferase, and re-join around the point of the final amidations.
4. Cobalt toxicity
Cobalt deficiency is a challenge for those organisms which synthesise B12, while all organisms must handle this metal carefully since cobalt can mediate a range of adverse effects. Metallostasis for cobalt, like other transition metals, is necessary at the upper-end of the tolerable range to avoid toxicity arising from aberrant reactivity including mismetalation. Cobalt is a well-known cause of contact dermatitis, leading to it being named ‘Allergen of the Year’ in 2016 by the American Contact Dermatitis Society. Severe cases of cobalt toxicity in humans can cause neurotoxicity, pneumonia and increase lung cancer risk when inhaled [42]. In bacteria, cobalt toxicity arises due to the disruption of iron-sulphur cluster formation, mismetalation of metallocomplexes and the formation of reactive oxygen species.
4.1. Limitation of iron availability
Many of the mechanisms of cobalt toxicity are due to competition between iron and cobalt. Iron and cobalt are both similar in size and charge. One of the effects of cobalt stress is the reduction of intracellular iron content by 50% [43]. The addition of exogenous iron has been shown to restore bacterial growth at high concentrations of cobalt, suggesting that cobalt can out compete iron at metal binding sites. Cobalt has been shown to compete with the metal binding site of Fur, the iron regulatory protein, and significantly impair its activity [44]. Fur binds iron as a corepressor, so iron availability mediates the activity of Fur. In E. coli Fur controls the iron-dependent expression over 90 genes, including genes encoding iron uptake and enzymes involved in glycolysis and the TCA cycle [45]. The notion of iron and cobalt competition is best shown when Fur is deleted from E. coli. These strains have unregulated iron import and are far less sensitive to cobalt than wildtype strains [46,47].
4.2. Iron-sulphur clusters
Fe—S clusters are most commonly found in either the [4Fe—4S], [3Fe—4S] or [2Fe—2S] forms. So far there have been three biosynthetic pathways identified in E. coli: the ISC, SUF and CSD. These pathways are induced at different times. The ISC pathway is thought to be used under normal growth whereas the SUF is believed to be used under periods of cell stress that result in Fe—S cluster degradation [48,49]. The biological role and timing of the CSD pathway is still unknown. The formation of Fe—S clusters in the ISC and SUF pathways requires protein scaffolds IscU, IscA and SufA [[50], [51], [52]]. These are important in the formation and delivery of clusters to the target apoprotein. The exposure of E. coli to 200 μM CoCl2 resulted in the cobalt and iron competing with scaffold proteins IscA and SufA and the insertion of mixed [Fe/Co-S] clusters into proteins [43]. Mature iron-sulphur clusters have been shown not to be affected by cobalt. MiaB, a [4Fe—4S] radical SAM enzyme involved in TRNA modification; FhuF, a [2Fe—2S] enzyme involved in ferrisiderophore reduction and aconitase were incubated with CoCl2 in vitro and had little effect upon activity [43].
Many Fe—S proteins are found within the tricarboxylic acid (TCA) cycle. Aconitase catalyses the conversion of citrate to isocitrate in the TCA cycle and an Fe—S cluster. It has been previously shown that presence of toxic levels of cobalt during the growth of E. coli caused the activity of aconitase to drop 70–80% [43]. Succinic dehydrogenase and fumarate reductase also contain Fe—S clusters and likely cease functioning due to toxic levels of cobalt, causing metabolic dead ends. Majtan et al. showed that E. coli adapts to oxidative stress by utilising modified mixed acid fermentation under aerobic conditions [53]. High levels of citrate, 2-hydroxyglutarate, succinate, lactate, fumarate and malate were found in cobalt-containing growth media through GC/MS analysis [53]. These intermediates correlate with TCA enzymes aconitase, succinic dehydrogenase and fumarate reductases placement within the cycle.
4.3. Oxidative stress and DNA damage
Cobalt-induced formation of reactive oxygen species (ROS) has been well documented in both eukaryotic and prokaryotic cells due to its ability to form different oxidative states. It has been shown in vitro that cobalt in reduced states can react with O2 and H2O2 through Fenton reactions to generate ROS, superoxides and hydroxy radicals that can go on to damage DNA and inhibit DNA repair mechanisms [54].
It has been observed that E. coli grown in the presence of 250 μM CoCl2 has a 1.4-fold increase in ROS when measured using an oxidative stress-sensitive fluorescent probe [46]. Conversely to other studies, Kumar et al. found that the addition of 500 μM CoCl2 to E. coli generated no ROS species and that DNA was damaged through an oxidative stress-independent pathway [47]. The addition of nickel and cobalt was demonstrated to inhibit the rate of DNA replication up to 50%. RecBCD, a helicase that initiates the SOS response, was inhibited, leading to DNA being damaged without an SOS response. Further to these findings, it was also noted that the generation of mixed acids, as described earlier, in combination with an acidic environment only enhanced cobalt toxicity.
4.4. Interference in sulphur assimilation
Thorgersen and Downs found that cobalt may affect sulphur assimilation in Salmonella enterica serovar Typhimurium (hereafter Salmonella) [44]. The sulphur assimilation pathway uses CysJI to convert sulphites to sulphides, which are then converted to cysteine. CysJI contains a [4Fe—4S] site and requires siroheme as a cofactor. It was shown there was a correlation between high exogenous cobalt concentrations and minimal CysJI activity. Both siroheme and [4Fe—4S] require iron – an element whose availability is reduced during cobalt toxicity. Cysteine is also the source of sulphur during [Fe—S] cluster formation, meaning cobalt affects both components of the complex.
4.5. Mismetalation of tetrapyrroles
Cobalt and iron have been shown to compete again during the formation of heme in E. coli, where protoporphyrin IX is mismetalated with cobalt [53]. Cobalt-protoprophyrin IX was found inside membrane-bound cytochromes, leading to the inhibition of the electron transport chain and decreased respiration. This is not the first account of the ferrochelatase inserting incorrect metals. The formation of zinc-protoporphyrin is well document and arises during periods of iron limitation [55].
5. The components of cobalt homeostasis
Metallostasis for cobalt, in common with other metals, involves the combined actions of, for example, importers and exporters (Section 5.1), delivery proteins (Section 5.4) and sensors that match supply with demand while avoiding toxic excess (Section 5.2) [56,57]. Peculiar to cobalt metallostasis in some bacteria is the use of B12-riboswitches to coordinate the substantial contribution of vitamin B12 biosynthesis to cobalt demand. Riboswitches also integrate B12 uptake with demand when the cofactor itself is available (Section 5.3).
5.1. Transporters
5.1.1. Cobalt uptake
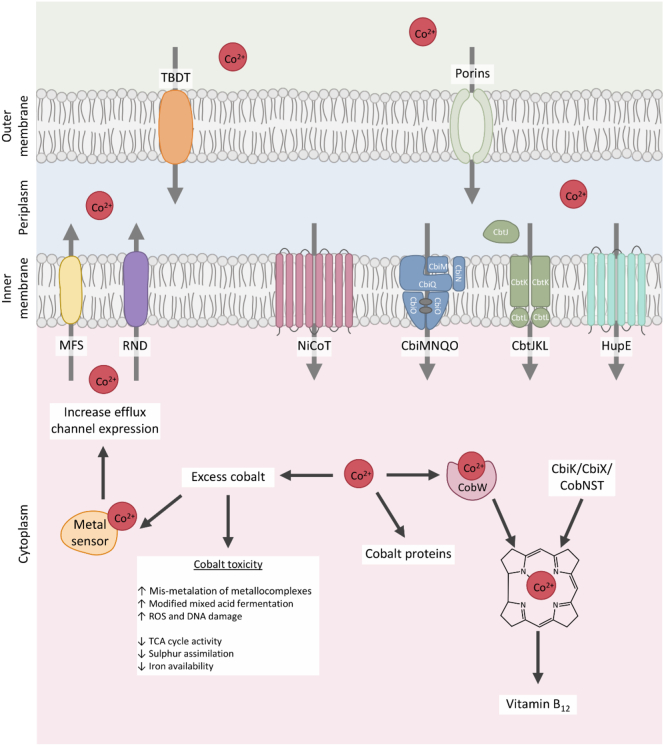
As there are only a handful of non-corrin cobalt-containing enzymes, cobalt uptake and requirement is generally distributed among organisms that can synthesise vitamin B12 de novo. Bacteria have developed many uptake systems for cobalt that sustain growth without inducing cobalt toxicity (Fig. 4). These transporters are largely highly specific but low capacity due to the toxic nature of cobalt. Many now-known cobalt transporters were identified in silica due to their location within B12 biosynthetic operons or their presence elsewhere in the genome under the control of an adenosylcobalamin riboswitch. Cobalt is primarily transported by ATP-binding cassette (ABC) transport systems with the addition of secondary transport systems such as nickel-cobalt (NiCoT) permeases (Table 1) .
Fig. 4.
Cobalt transporters.
Cobalt is transported through the outer membrane by porins or by TonB-dependent transporters (TBDT). From the periplasm, cobalt is transported through the inner membrane by a range of primary and secondary transporters. When cobalt is in excess, cobalt is transported out of the cytoplasm by MFS (major facilitator) permeases and RND (resistance nodulation division pumps) whose expression is controlled by a variety of metal-sensing proteins.
Table 1.
Transport proteins involved in cobalt homeostasis.
| Transporter | Transporter family | Organism | Metal specificity | Reference |
|---|---|---|---|---|
| Cobalt uptake | ||||
| CbtJKL | ABC | Sinorhizobium meliloti | Co(II) | [61] |
| CbiMNQO | ECF | Salmonella | Co(II), Ni(II) | [22] |
| NhlF | NiCoT | Rhodococcus rhodochorus | Co(II), Ni(II) | [70,71] |
| HupE | HupE/UreJ | Synechocystis sp. PCC 6803 | Co(II) | [73] |
| CorA | CorA | Escherichia coli | Mg(II), Co(II), Ni(II) | [238,239] |
| Thermotoga maritima | Co(II), Mg(II) | [77] | ||
| Cobalt sensing and efflux | ||||
| RcnA | MFS | Escherichia coli | Ni(II), Co(II) | [240,241] |
| CoaT | Synechocystis sp. PCC 6803 | Co(II) | [94] | |
| CzcABC | RND | Cupriavidus mtallidurans CH34 | Zn(II), Co(II), CdII | [242] |
| CnrABC | Co(II), Ni(II) | [243] | ||
ABC – ATP-binding cassette, ECF – electron-coupled factor, NiCoT – nickel/cobalt transporter, MFS – major facilitator factor, RND – resistance nodulation division.
5.1.1.1. Transport across the outer membrane
In Gram-negative bacteria, cobalt must cross both the inner and outer membranes. Large molecules, such as vitamin B12, are transported across the membrane by TonB-dependent transport. In the case of vitamin B12, the outer membrane receptor BtuB transports B12 into the periplasm [58]. Some TonB OM receptors have been identified and predicted to be cobalt-specific. In the case of Novosphinogobium aromaticivorans, the gene encoding a TonB outer membrane receptor is preceded by an adenosylcobalamin riboswitch and then followed by genes encoding a NiCoT permease and cobaltochelatase, CobW (Section 5.4) [22]. No cobalt-specific outer membrane receptors have been confirmed so far but some have been identified for nickel scavenging in Helicobacter species. In H. pylori, the outer membrane receptor FrpB4 and the ExbB/ExbD/TonB complex were required for efficient nickel scavenging [59]. A similar protein, NikH, was identified in H. mustelae [60].
5.1.1.2. Canonical ABC transporters
An ABC-type cobalt importer, CbtJKL, has been reported in Sinorhizobium meliloti [61]. Originally thought to encode a cobalamin transporter due to sequence similarity between CbtJ and BtuF, the CbtJKL complex transports Co(II) through the inner membrane into the cytoplasm. The complex was found to be responsible for cobalt transport when S. meliloti strains with a cbtJKL knock-out could only be grown in the presence of 1–10 μM CoCl2. It has not been reported on whether this system can also transport nickel in addition to cobalt. cbtJKL is found on the pSymB megaplasmid of S. meliloti and is unsurprisingly under the control of an adenosylcobalamin riboswitch [61].
5.1.1.3. ECF transporters
Electron-coupled factor (ECF) transporters are a subfamily of the ABC transporter family that lack the cytoplasmic substrate-binding protein (SBP) found in ABC transporters. Cobalt-specific ECF transporters were first identified in Salmonella where a cluster of four genes, cbiMNQO, were found in the B12 biosynthetic operon [62]. This was later shown to be a cobalt-specific transport system [22]. ECF transporters consist of a substrate-specific transmembrane domain (S unit), a transmembrane protein (T unit) and two ABC ATPase domains (A unit) [63].
CbiMNQO contains a T unit (CbiQ), an ABC ATPase (CbiO) and two S unit transmembrane proteins (CbiMN). CbiMQO form a stable complex with a stoichiometry of 1:1:2 and a crystal structure of the complex was published in 2017 [64]. CbiM lies across the periplasmic face of the inner membrane with CbiQ wrapping around in a ‘C' shape. Two CbiO units are found beneath CbiQ and CbiM, forming a cone shape and the substrate binding site is a large cleft formed by CbiQ and CbiO. The interactions of CbiN and the CbiMQO complex are weak: it has been frequently demonstrated that CbiN cannot be co-purified with the rest of the complex and only interacts transiently, but CbiM and CbiN are the minimal required proteins to transport cobalt across the membrane [22,[64], [65], [66]]. CbiN has a short flexible extracytoplasmic loop between its two transmembrane helices that lies on top of the metal binding pocket of CbiM. It has been shown that truncations or modifications to this loop severely impair cobalt transport though the mechanism behind their interactions is still not well understood [67]. It is understood that CbiM provides ATPase activity to the complex without the presence of CbiN, and that CbiO units undergo ATP binding and product release-induced conformational change, but the full transport cycle is yet to be elucidated [65].
5.1.1.4. NiCoT permeases
The most diverse group of cobalt transporters are the nickel/cobalt permeases (NiCoT; TC2.A52). NiCoTs are found across many bacteria and some archaea and urease-producing fungi [22,68]. NiCoTs have 6–8 transmembrane helices containing 4 characteristic domains, show in Table 2, of which Domain 2 has been demonstrated to be responsible for metal binding [69]. The N- and C-terminal helices are separated by a large charged and hydrophilic loop. The first cobalt-specific NiCoT identified was NhlF from Rhodococcus rhodochorus, but later shown to also transport nickel [70,71]. NiCoTs can be divided into three classes based on their substrate specificity: (i) Class I only transport nickel; (ii) Class II transport nickel and cobalt with preference for cobalt, e.g. NhlF; and (iii) Class III transport nickel and cobalt with a preference for nickel. The substrate specificity of this family cannot be predicted by sequence identity. The best predictor of substrate specificity is genomic localisation. Many genes encoding nickel-specific NiCoTs are located downstream of [NiFe] hydrogenase operons or under transcriptional control by NikR, whereas genes encoding predicted cobalt-transporting NiCoTs are located within B12 biosynthetic operons, or close to cobalt-containing nitrile hydratase genes [72].
Table 2.
Signature domains of nickel-cobalt permeases. From [244].
| Domain | Motif |
|---|---|
| TM2 | (R/K)HAXDADH(I/L) |
| TM3 | FXXGHS(T/S)(V/I)V |
| TM5 | LGX(D/E)T(A/S)(T/S)E |
| TM6 | GMXXXD(T/S)XD |
5.1.1.5. HupE/UreJ
Members of the HupE/UreJ transporter family are distributed throughout proteobacteria and cyanobacteria. Many are found within [NiFe] hydrogenase and urease gene clusters, leading to their assumed function in nickel uptake. However, some homologs in cyanobacteria have been identified as cobalt transporters as they are regulated by an adenosylcobalamin riboswitch [21,22]. HupE cobalt transport has been demonstrated experimentally. hupE mutants of Synechocystis PCC 6803 could only be rescued by the addition of cobalt or methionine to the cultures [73]. HupE/UreJ proteins are thought to contain 6 transmembrane helices with a signature motif sequence (HPXXGXDH) in the first transmembrane helix [74].
5.1.1.6. CorA as a cobalt transporter
CorA is a family of divalent cation transporters that primarily transport magnesium, with nickel and cobalt as secondary metals, the binding affinities of which are 15–20 μM, 200–400 μM and 20–40 μM respectively [75,76]. As these concentrations of nickel and cobalt are not typically present physiologically, CorA is primarily a magnesium transporter. However, one CorA transporter from Thermotoga maritima has been found to select for cobalt over magnesium when present at 100-fold lower concentrations [77]. CorA itself is a homopentameric protein with each subunit formed of 8 helices that come together to form a funnel shape [78]. In organisms such as E. coli which have no dedicated cobalt transport system, CorA is likely to be the main source of cobalt entry into the cytoplasm.
5.1.2. Cobalt efflux
Whereas cobalt import systems are regulated by a cobalt-dependent product such as adenosylcobalamin, cobalt efflux systems are generally regulated by the cobalt ion itself. Cobalt efflux proteins can be divided into two groups: the major facilitator superfamily (MFS) permeases and resistance nodulation division (RND) pumps (Table 1).
5.1.2.1. MFS permeases
As described in Section 5.2.1, RcnR regulates the expression of RcnA, a Co(II)/Ni(II) efflux pump, that is responsible for detoxification of cobalt and nickel. Other members of this group are P1B-4-ATPases, a family for transmembrane metal transporters involved in transition metal homeostasis. Such transporters include CoaT as well as CtpD and CtpJ from Mycobacterium smegmatis and Mycobacterium tuberculosis respectively. These proteins have been demonstrated to be involved in cobalt homeostasis through cobalt export [79,80]. Both proteins contain six transmembrane domains with signature motifs SPC and HEG(S/G)T found in TM4 and TM6 respectively.
PmtA has been found to function as a Fe(II) and Co(II) efflux pump in Streptococcus suis whose expression is regulated by Fe(II), Co(II) and Ni(II) [81]. Deletion of pmtA resulted in increased Fe(II) and Co(II) sensitivity and accumulation of these metals. PmtA bears similarity to other P1B-4-ATPases such as PfeT from B. subtilis that is responsible for Fe(II) and Co(II) efflux [82].
5.1.2.2. RND pumps
Cuprivadius metallidurans contains two RND pumps to mediate cobalt efflux: the cobalt, zinc and cadmium resistance system (CzcABC) and the cobalt-nickel resistance system (CnrABC): their mechanisms of regulation are described in 5.2.4, 5.2.5, respectively.
5.2. Protein-based cobalt sensors
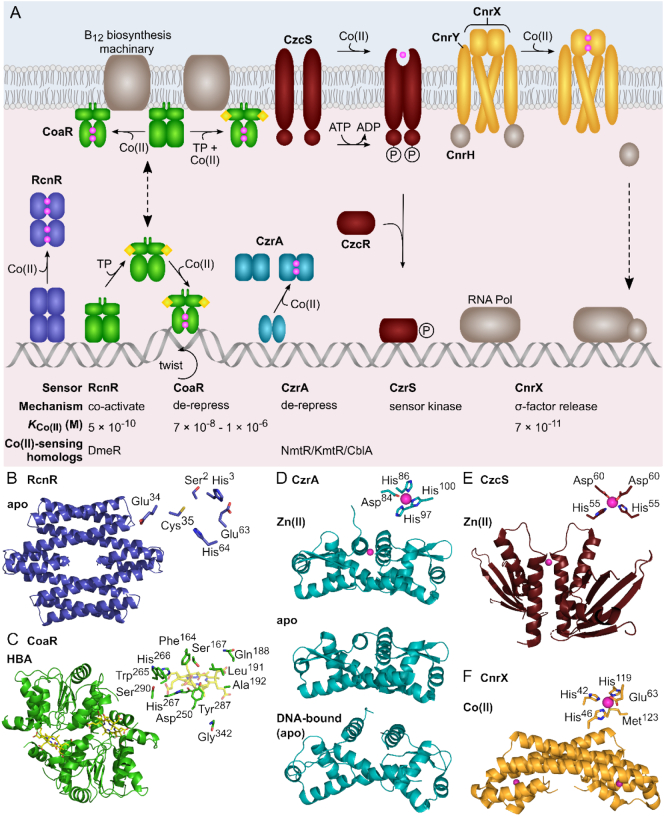
Protein-based cobalt sensors are typically cytoplasmic proteins which both sense intracellular cobalt and regulate gene expression, although transmembrane proteins which can sense periplasmic cobalt have also been described (Fig. 5). These sensors modulate the expression of cobalt-efflux systems (or in one case a cobalt-requiring enzyme) as cobalt availability increases. No protein-based cobalt sensor has, as yet, been identified that regulates expression of a cobalt importer, with import commonly regulated via negative feedback from a cobalt-containing biosynthetic product such as vitamin B12.
Fig. 5.
Protein-based cobalt sensors.
A, RcnR represses expression of rcnRAB (in E. coli and Salmonella, two RcnR tetramers bind the target site). RcnR can bind four Co(II) ions per tetramer, which weakens the affinity for DNA, de-repressing expression [83]. CoaR binds the coaT promoter in the absence of effector. Co(II) binding is predicted to induce a conformational change which distorts the operator-promoter, enabling recruitment of RNA polymerase and activation of expression. CoaR also harbours a tetrapyrrole (TP) binding domain including a hydrophobic patch which is capable of interacting with membranes [96]. Tetrapyrrole binding might tighten the affinity of CoaR for Co(II), and/or association with membrane-associated B12 biosynthetic machinery could confer a kinetic advantage for Co(II) and/or tetrapyrrole acquisition. CzrA represses expression of czrAB, and Co(II) binding weakens the DNA affinity of the CzrA dimer, alleviating repression. The structures of apo- and Zn(II)-CzrA are similar in the absence of DNA and an entropic mechanism for allosteric regulation has been demonstrated [103]. CzcS sensor histidine kinase detects periplasmic Co(II), and metal-binding induces autophosphorylation at a conserved histidine by Mg(II)-GTP. Transphosphorylation of the response regulator CzcR induces a conformational change promoting DNA-binding and activation of czcCBA [113]. CnrX is a transmembrane protein which forms a complex with CnrY and cytoplasmic CnrH. Co(II) binding to the periplasmic domain of CnrX, induces a conformational change which is transduced through the complex, releasing the CnrH extracytoplasmic function sigma factor. CnrH is recruited by RNA polymerase enabling expression of cnrCBA [112,113]. B–F, structural models of Co(II)-sensing proteins. B, Structural representation of RcnR tetramer (dimer of dimers) generated from PDB 5LYC. The ligands identified for co-ordination of Co(II) are shown. C, Dimeric representation of the deduced tetrapyrrole binding domain of CoaR bound to hydrogenobyrinic acid (HBA), modelled on the structure of CobH precorrin isomerase (PDB 1I1H) as described in [96] (main chain of Gly342 shown). The CoaR ligands which spatially overlay with the HBA-binding site of CobH are shown. D, The structures of Zn(II)- (PDB 2M30) and apo-CzrA (PDB 1R1U) reveal similar ‘open’ conformations which contrasts with the ‘closed’ conformation of the DNA-bound form (PDB 2KJB) [103]. The Zn(II) (and Co(II)) binding site at the dimer interface is shown. E, The sensor domain of P. aeruginosa Zn(II)-CzcS (PDB 5GPO) suggests Zn(II) binds to reciprocal residues at the dimer interface. His55, but not Asp60, is conserved in the Co(II)-responsive CzcS homolog from C. metallidurans [114]. F, The soluble periplasmic domain of CnrX (PDB 2Y3B) displays a similar structural fold to MerR-family sensors such as RcnR. The Co(II)-binding site is shown: a sulphur donor ligand is provided by a conserved methionine in contrast to the invariant cysteine in RcnR [83,120].
5.2.1. CsoR/RcnR-family: RcnR/DmeR
The best characterised cytoplasmic Co(II)-sensor belongs to the CsoR/RcnR-family of metal-dependent de-repressors [83]. These sensors typically repress the expression of transporters required for metal-export from the cell. DNA binding is inhibited upon metal co-ordination, relieving repression and enabling the removal of excess metal. RcnR was initially identified in E. coli as a regulator of the rcnR-rcnAB divergon, encoding the Co(II)/Ni(II)-efflux pump, RcnA (Section 5.1.2.1), and periplasmic protein, RcnB, plus autoregulation of its own expression [84]. RcnR is a tetrameric α-helical dimer of dimers that responds to both Co(II) and Ni(II) in the cell (Fig. 5b) [85]. Binding of the cognate metals is via a six-coordinate site and requires a cysteine (invariant in the CsoR/RcnR-family) and 5 O/N donor ligands (Fig. 5b) [86]. Co(II)-binding recruits, and orders, the amino terminus enabling a cross-link between subunits to drive allostery [86,87].
RcnR, and the homologous protein, DmeR, have also been functionally characterised in organisms which can synthesise B12 de novo such as the human pathogen Salmonella, the leguminous plant symbionts Rhizobium leguminosarum and Sinorhizobium meliloti, and plant pathogen Agrobacterium tumefaciens [[88], [89], [90], [91], [92]]. In each case the genetic architecture differs from E. coli (which does not synthesise B12): in Salmonella, rcnB is not part of the rcnR operon, and in the plant microbes, DmeR regulates expression of dmeF, encoding a cation diffusion facilitator Ni(II)/Co(II)-efflux pump, distinct from RcnA [88,89,91]. The binding affinity, or stability of the Co(II)-RcnR complex as a KD, for Salmonella RcnR is 5 × 10−10 M. Autoregulation by Salmonella RcnR confers hysteresis: This dampens the response to elevated cobalt and shifts the dynamic range away from what might be predicted from metal-affinity alone [93]. How Co(II)-sensing by RcnR/DmeR relates to B12-production in these organisms remains to be investigated.
5.2.2. MerR-family: CoaR
CoaR is a Co(II)-sensing MerR-family regulator from the cyanobacterium Synechocystis PCC 6803 [94] (Fig. 5a). Regulation by MerR-family sensors is characterised by sub-optimal spacing of the −10 and −35 promoter elements upstream of the target gene. These sensors bind their target DNA sequence with only a modest change in affinity between the apo- and holo-forms, and metal-binding causes a conformational change which under-winds the DNA allowing recruitment of RNA polymerase [95]. During exposure of Synechocystis to increased Co(II) (but not other metals), CoaR activates expression of coaT encoding a Co(II)-effluxing P1-type ATPase (Section 5.1.2.1) [94]. A Cys-His-Cys motif at the carboxy terminus of CoaR is implicated in Co(II)-coordination. CoaR also contains a putative tetrapyrrole-binding domain with homology to CbiC/CobH, the enzyme which catalyses the 1, 5-sigmatropic migration of a methyl group of precorrin-8 to generate hydrogenobyrinic acid during the aerobic biosynthesis of vitamin B12 (Fig. 5c) [96]. Synechocystis synthesises B12 via the anaerobic pathway and cbiE mutants, which will accumulate B12-precursors prior to cobalt-precorrin 7, demonstrate altered expression of coaT [94]. When expressed in E. coli, Co(II)-dependent activation of CoaR also requires residues predicted for tetrapyrrole binding, in addition to those which form the cobalt site [96]. One model is that CoaR has dual effectors in order to somehow coordinate both the levels of Co(II) and B12 (or precursor) in the regulation of Co(II)-export.
Thermodynamic characterisation of multiple metal sensors in Synechocystis reveals that Zn(II)-sensors ZiaR and Zur bind Co(II) at least 100-fold more tightly than does CoaR (CoaR KCo(II) = 7 × 10−8–1 × 10−6 M), and Co(II) is able to activate the allosteric response of both Zn(II) sensors [96]. This raises the question: how is CoaR more competent than ZiaR and Zur at responding to Co(II) in vivo? Notably, metal-sensitivity is not solely a function of metal-affinity but also access, allostery, affinity for DNA, and abundance [57]. Sequence analysis of CoaR reveals a carboxy-terminal hydrophobic region predicted to interact with membranes [94,96]. Membrane association with CoaR could confer a kinetic advantage to Co(II)-sensing by CoaR over ZiaR and Zur in effect providing preferential access to cobalt [96]. Several components of the cobalamin biosynthetic pathway are thought to be membrane associated and could therefore encourage formation of a membrane-based metabolon [97,98], limiting the release of precursors during synthesis. The possibility that CoaR co-localises with this machinery, perhaps via its precorrin isomerase-like domain, and that this promotes channelling of Co(II) and/or a B12 precursor to CoaR, requires further investigation. In future, the magnitude of such kinetic effects could be quantified by reference to a thermodynamic framework such as that set out in Section 6.5.4. Progress in understanding the metalation of CobW (Section 6.6) highlights the merits of such an approach: By further analogy to CobW, the possibility that the Co(II) affinity of CoaR is enhanced by interaction of its precorrin isomerase domain with a tetrapyrrole or other partner, should also be considered.
5.2.3. ArsR-SmtB family: NmtR/KmtR/CzrA/CblA
The ArsR-SmtB family of metal sensors are particularly widespread in bacteria and two examples which sense Co(II) (and Ni(II)) have been identified in Mycobacterium tuberculosis: NmtR and KmtR [99,100]. NmtR and KmtR repress expression of a Co(II)/Ni(II)-exporting P1-type ATPase and a cation diffuser facilitator metal-efflux pump, respectively. Upon metal co-ordination (which is distinct for each sensor [99,100]), the affinity for their respective DNA binding site is weakened, resulting in de-repression of the downstream gene [101]. Characterisation of NmtR and KmtR has revealed facets of metal-sensing that highlight the difficulties in designating the cognate metal(s) to metal sensor proteins and the vital contribution of metal-access, that is intracellular metal-bioavailability, to metal-specificity.
Repression by both NmtR and KmtR is alleviated by Co(II) and Ni(II) in mycobacterial cells. However, during heterologous expression in cyanobacteria, NmtR responds solely to Co(II) and not Ni(II). This suggests that the different cell types have distinct metal availabilities equating to differences in available free energies, see Section 6.6, and/or altered access due to metal channelling [99].
When mycobacterial cells are grown in minimal medium, both KmtR and NmtR are responsive to the cognate metals. However, when cultivated in a complex growth medium, expression of the gene regulated by KmtR is already elevated and unaffected by metals. In contrast, NmtR retains Co(II) and Ni(II)-responsiveness [99]. The prediction that KmtR is more sensitive than NmtR to Co(II) and Ni(II) was tested by competition between the two proteins in vitro: KmtR was able to out-compete NmtR for both Co(II) and Ni(II) consistent with a tighter metal affinity and increased sensitivity in vivo [99]. This could enable a graded stress response to increasing Co(II) (and Ni(II)) in mycobacteria reflecting discrete functions of the two transporters.
Staphylococcus aureus CzrA demonstrates selectivity towards Zn(II) and Co(II) in vitro and in vivo [100,102]. Allosteric regulation requires a tetrahedrally co-ordinated metal ion afforded by Co(II) and Zn(II), but not Ni(II) (Fig. 5a,d) [100]. The contribution of allostery to the selectivity of metal-sensor proteins (even within the same family) is highlighted by comparing this with the allosteric site of NmtR: here a six-coordinate site is required to trigger the allosteric mechanism, discriminating against Zn(II) (despite NmtR having a tight Zn(II) affinity) [100]. Structural determinations of apo- and holo-CzrA reveal similar “open” conformations in contrast to the “closed” conformation of the DNA-bound apo-structure [103]. Rather than a major structural reorganisation, cognate metal ion binding to CzrA is entropically driven and alters the internal protein dynamics [103]. A contribution from solvent entropy to allostery has also been established suggesting a model where dynamic redistribution between CzrA and surface water molecules occurs without significant perturbations in the protein structure [104]. This may be a general model for metal-sensing by members of the ArsR/SmtB family which undergo only small structural changes upon metal-binding.
Recently, a cobalt-sensing member of the ArsR-SmtB family has been characterised in Rhodococcus rhodochrous [105]. Atypically, rather than regulating metal-export machinery, CblA de-represses expression of a cobalt-dependant nitrile hydratase (NHase) in response to cobalt and nickel [106]. NHases are of interest to industrial biotechnology for the large-scale production of acrylamide and nicotinamide in addition to their relevance for plant-microbial interactions [107]. This is the first description of a NHase being regulated by cobalt in addition to substrate-dependant activation of transcription. Cobalt NHases have a non-corrin catalytic Co(III) at their active site [108]. A role for trapping kinetically stable Co(III) ions in the detection of cobalt by CblA is an intriguing possibility.
5.2.4. Two-component regulatory system: CzcRS
The plasmid-borne resistance to cadmium-zinc-cobalt (czc) determinant originally identified in Cupriavidus metallidurans encodes genes for the CzcABC resistance nodulation and cell division (RND) exporter (Section 5.1.2.2) [109]. Expression is induced by Co(II), Zn(II), and Cd(II) in vivo [110,111]. The regulatory mechanism includes CzcR (response regulator) and CzcS (sensor histidine kinase) as a two-component regulatory system [112,113] (Fig. 5a). In contrast to cytoplasmic metal sensors, metal detection is by the periplasmic sensing domain of CzcS which initiates a signalling cascade, and phosphorylation of CzcR. Activation of CzcR leads to transcriptional activation of czcABC. The structure of the Zn(II)-bound CzcS sensor domain from Pseudomonas aeruginosa has been determined revealing that a His2Asp2 tetrahedral site could form at the dimer interface (Fig. 5e). However, CzcS from P. aeruginosa appears to be distinct from that of C. metallidurans, as the former does not respond to Co(II) in vivo [114], despite CzcA providing resistance to Co(II) in both organisms. Mutation of the metal-binding site in P. aeruginosa CzcS reduces Zn(II) sensing and substitution of the Asp ligands with Cys enables Co(II) detection in vivo [109,115]. Sequence differences between CzcS protein, and/or different metal availabilities in the periplasm, may explain the distinct metal-specificities of CzcS sensors in each organism.
5.2.5. Transmembrane anti-sigma factor: CnrYXH
In addition to CzcS, C. metallidurans senses periplasmic Co(II) (and Ni(II)) by the transmembrane signal transduction system CnrYXH (Fig. 5a) [116]. CnrX is a transmembrane protein with a metal-sensing domain that extends into the periplasm [117]. CnrX interacts with the transmembrane protein CnrY, an anti-sigma factor which sequesters the extra-cytoplasmatic function sigma factor CnrH at the membrane [118,119]. Binding of Co(II) or Ni(II) to CnrX induces a conformational change which is transduced to CnrY, weakening the interaction with CnrH. Cytoplasmic release of CnrH allows recruitment by RNA polymerase, promoter recognition and expression of cnrABC [119]. CnrABC, a RND efflux pump confers Co(II) and Ni(II) resistance to C. metallidurans (Section 5.1.2.2) [119]. The structure of the holo-CnrX soluble periplasmic domain reveals a dimer with an octahedrally co-ordinated Co(II) or Ni(II) ion [117,120]. Signal transduction by CnrX requires a conserved methionine (Met123) which is also recruited (via the thioether sulphur) to the metal-binding site by Co(II) and Ni(II) (Fig. 5f) [117,120]. Zn(II) adopts a trigonal bipyramidal binding site not involving Met123 and Zn(II)-CnrX is non-functional [117]. Intriguingly, the helical bundle of CnrX displays structural similarity to RcnR/CsoR-family cytoplasmic metal sensors (Fig. 5a, f). These sensors use an invariant cysteine as a metal ligand [83]. Met123 is conserved among CnrX homologs and although an uncommon metal ligand, is considered to be more suited to the redox environment of the periplasm. Methionine has been observed in the periplasmic metal-binding site of Cu(I)-exporting RND pump CusABC, divalent metal transporter NRAMP and periplasmic chaperone CopC [[121], [122], [123]]. Substitution of Met123 with cysteine in CnrX abolishes Ni(II)-responsive expression and decreases the Co(II)-affinity by ~2000-fold [120]. Spectroscopic analysis suggests Cys123 is not involved in Co(II) co-ordination. Interestingly, the Zn(II) affinity of wild type CnrX is ~3-fold weaker than the affinity for Co(II) and this remains unchanged in the Met to Cys substitution [120]. The use of methionine in metal-coordination may contribute to allosteric metal-selectivity [117,120].
While the metal affinities of many cytosolic metal sensors have been determined, few are known for extra-cytoplasmic metal sensors likely due to the analytical challenge of working with membrane-bound proteins. For cytoplasmic sensors, these metal-affinities can be used to infer intracellular metal availabilities (Section 6.5), but it is less clear whether the same relationship exists for extra-cytoplasmic metal sensors and metal availabilities. One option for estimating Co(II) availability in the extra-cytoplasmic space is to measure transport rates of Co(II)-importers (Section 5.1.1). Co(II)-uptake assays with CbiMNQO and NhlF suggest Km values in the range of 10−7 to 10−8 M [124]. In Gram-negative bacteria, controlling metal availability in the periplasm presents a greater challenge than for the cytoplasm: the periplasm is more vulnerable to external fluctuations than the cytosol and metal ions may diffuse through outer membrane porins [125]. The outer membrane does afford some protection against the extracellular environment but there may be regions which are contiguous with the extracellular environment. Despite this, the Co(II) and Ni(II) affinities of the CnrX soluble periplasmic domain have been determined to be 6.5 × 10−11 M and 1.7 × 10−12 M, respectively [120,126]. These affinities are notably tight and suggest that the availability of metal ions in the periplasm may be somewhat analogous to the cytoplasm (Section 6.5). Metal speciation in the extra-cytoplasmic space is not well defined although there is a suggestion that copper could be more available than in the cytosol (Section 6.3), perhaps relating to a predominance of cupric rather than cuprous ions. Low molecular weight molecules (e.g. amino acids, glutathione, citrate), and adventitious sites on proteins and membrane components are candidates for chelating and buffering metals in this compartment. Glutathione is exported into the periplasm of Gram-negative bacteria, and can buffer Cu(I) below 10−15 M when in excess [127,128]. The substrate for NikA-mediated Ni(II)-import in E. coli and Helicobacter pylori is a Ni(II)-histidine2 complex which might suggest the presence of histidine in a bacterial periplasm [129]. Histidine is implicated in the speciation of Ni(II) and Zn(II) in labile metal pools [130,131]. By analogy to cytoplasmic metal sensors, knowledge of the metal availabilities to which sensors are tuned (Section 6.5), should advance understanding of how metals, including Co(II), are discerned by extra-cytoplasmic proteins.
5.2.6. Metal sensors are fine-tuned to a narrow dynamic range
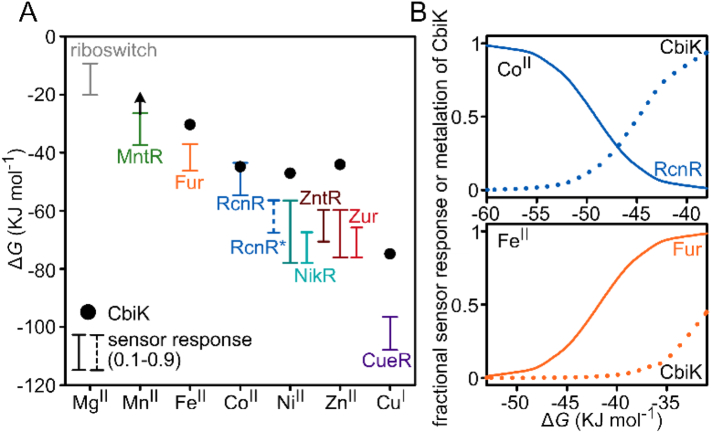
The Co(II)-sensor RcnR can bind Zn(II) in vitro and DNA-binding is impaired by Zn(II) [90]. By measuring affinities for both Co(II) and Zn(II), along with affinities for DNA of the apo- and metal-bound forms of the protein, sensitivities to both metals were calculated [90] (also see Section 6.5.1). Counterintuitively, RcnR is about two-orders of magnitude more sensitive to Zn(II) than to Co(II), yet in vivo RcnR does not respond to non-inhibitory levels of Zn(II). A sensor that does respond to Zn(II) in vivo, Zur, is reciprocally competent to respond to Co(II) in vitro but fails to do so under normal conditions in living cells. Importantly, the bona fide sensors are only about an order of magnitude more sensitive to the cognate metal compared to the non-cognate sensors [90]. Thus, the sensors have evolved to show perfect discrimination only within a narrow range of metal concentrations.
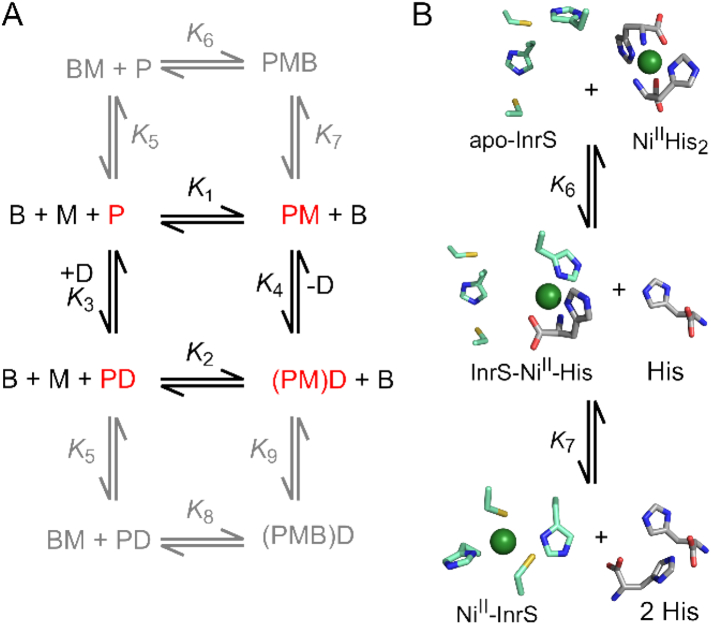
Variants of the Ni(II) sensor InrS with weakened Ni(II) affinities retained the ability to dissociate from DNA in vitro but lost the ability to detect sub-inhibitory elevated Ni(II) in vivo [132]. Again, only a modest (about ten-fold) change in sensitivity was enough to lose detection of the cognate metal. Subsequent experiments showed that the dynamic range of InrS was poised to track with the degree of Ni(II)-saturation of a cytosolic-like buffer (Section 6.5.2). Under such a regime, modest changes in metal concentration can reflect large changes in the total number of atoms bound within the buffer. In this manner, sensors can be set to detect when the buffer approaches metal-saturation or -depletion to trigger, for example, export or uptake respectively.
5.3. Riboswitches for B12 and for cobalt
Riboswitches are allosteric RNA molecules, present in the upstream region of bacterial mRNAs, and whose conformation is modified upon selective binding of a specific inducer. Riboswitches are generally split into two domains: an aptamer which recognises the effector molecule and an expression platform which modulates the expression of the downstream gene(s). Effector binding to the aptamer domain induces a conformational change in the expression platform leading to transcription termination, translation initiation, message stability, or alternative splicing of the associated transcript [133]. Characterised riboswitches present a diverse repertoire of regulatory mechanisms and effectors: the first effector described being B12 and now more recently the identification of riboswitches for metal ions.
5.3.1. The B12 riboswitch
To test the credibility of an “RNA World” experiments were performed to establish whether the catalytic repertoire of RNA enzymes could readily be extended beyond the limited hydrolytic activities known in nature, for example of self-splicing introns [134]. In 1990, the Systematic Evolution of Ligands by EXponential enrichment (SELEX) approach was established which identified, from a random pool of oligonucleotides, single stranded nucleic acids that could fold into structures (aptamers) with high affinity and specificity for small biological molecules [135,136]. Despite the presence of only four nucleotides, single-stranded nucleic acid aptamers have the versatility to adopt structures with comparable ligand binding affinities and specificities to proteins. A B12-binding aptamer was identified from a pool of 1015 molecules in 1994 and bound cyanocobalamin with tight affinity (KD ~ 9 × 10−8 M) but with negligible affinity for adenosylcobalamin (AdoCbl), highlighting the degree of specificity that can be afforded by an RNA aptamer [137]. These data supported the notion that B12 could have been an early evolutionary cofactor with a role during the transition from the RNA-dominated biological world to the present, where DNA and proteins prevail [26].
The verification that RNA molecules could bind biological targets led to the pursuit of naturally occurring RNAs as regulatory elements, now known as riboswitches. The first riboswitch was identified in the 5′-untranslated region of btuB mRNA, encoding the outer membrane TonB-dependent B12-importer in E. coli and other bacteria [138]. Expression of btuB is down-regulated in the presence of B12 by inhibiting association of the ribosome with btuB mRNA [139]. No protein-based regulatory sensor which recognised B12 could be identified and the unusually long 5′-leader sequence, which forms secondary structure elements, is now known to be responsible for the allosteric regulation of btuB translation by B12 [138]. The btuB riboswitch is formed of a ~ 200 nucleotide B12-binding aptamer (receptor domain) and a ~ 40 nucleotide expression platform (regulatory domain) [140].
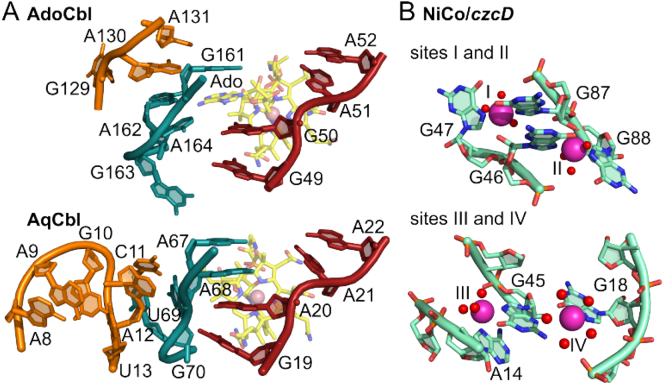
The complex secondary structure of the B12 riboswitch involves twelve helical domains and formation of a tertiary interaction with a kissing loop close to the B12 binding site [141]. Substrate binding by the receptor domain relies on shape complementarity, primarily through van der Waals contacts, with relatively few hydrogen bond interactions, and the kissing-loop is required for the allosteric conformational change in the regulatory domain to alter gene-expression [141,142]. The ligand binding pocket is formed by a four-way junction and specific nucleotide interactions to the 5′-deoxyadenosyl moiety of AdoCbl (Fig. 6a) [142]. Riboswitches with selectivity for different B12 derivatives have peripheral extensions which discriminate against AdoCbl by occluding the bulky adenosyl group in favour of smaller upper ligands as in aquacobalamin (AqCbl) and methylcobalamin (MeCbl) (Fig. 6b) [142]. It has recently been shown that Mg(II) can pre-organise the btuB riboswitch into a binding-competent conformation that recognises AdoCbl [143]. Binding affinities for B12 riboswitches range from 2.5 × 10−7 M for AdoCbl (btuB riboswitch) to 7.5 × 10−9 M for MeCbl (env8AqCbl riboswitch) [142]. However, the degree to which co-transcriptional folding might affect the affinity and selectivity of these riboswitches remains unknown.
Fig. 6.
Riboswitches for B12 and Co(II).
A, The binding site of AdoCbl and AqCbl in the AdoCbl riboswitch (Thermoanaerobacter tengcongensis; PDB 4GMA) and env8AqCbl riboswitch (marine metagenome; PDB 4FRN), respectively (adapted from [236]) . Although similar orientations of the J3/4 strand (red) are observed, the differential positioning of the central J6/3 strand (teal) enables accommodation of the deoxyadenosyl moiety (Ado) of AdoCbl which base pairs with A162 in the AdoCbl riboswitch, but is occluded by A20 and A68 in env8AqCbl [148]. B, The Co(II)-binding sites in the NiCo riboswitch from Erysipelotrichaceae bacterium (PDB 4RUM). The four Co(II) ions bind with octahedral co-ordination geometry and Co(II) ions I and II are co-ordinated by four conserved guanine nucleotides (G46, G47, G87, G88). Site III requires G45 which also forms the outer co-ordination sphere of site II implicating an allosteric linkage between sites I-III [148].
When B12 levels are low, the btuB ribosome binding site (RBS) is available for ribosome binding, and translation proceeds. In the presence of AdoCbl, the ribosome binding site (RBS) is sequestered by an anti-RBS complementary sequence forming an intrinsic terminator which precludes translation initiation [142]. Selective interaction of the anti-RBS with the RBS or an anti-anti-RBS sequence, when bound or unbound by AdoCbl, respectively, enables the conformational switch necessary for B12-responsive translation of btuB [133]. Folding of the riboswitch into these alternate structures is aided by transcriptional pausing by RNA polymerase [144].
The B12 riboswitch is widespread in both Gram-negative and Gram-positive bacteria, associated with genes that are involved in B12-biosynthesis, −uptake, −utilisation and also cobalt-supply [21]. The sequence and secondary structure of the B12 aptamer domain is evolutionarily conserved. Genomic analysis using this sequence has identified genes encoding high-affinity uptake systems for Co(II) (versus for Ni(II) which have a NikR binding site) that are required for cobalt supply to B12 biosynthesis (Section 5.1.1), and B12-independent isoenzymes that can be utilised by prokaryotes when B12 is scarce [21,22]. In Listeria monocytogenes hierarchal regulation mediated by the B12 riboswitch has been observed [145]. The negative feedback regulation of B12 biosynthesis, mediated by the B12 riboswitch, maintains appropriate B12 levels in the cell. However, this might also limit the commercial production of B12 industrially using organisms for which limited genetic tools exist [146]. The possibility of engineering B12 biosynthetic pathways in heterologous, genetically tractable hosts where regulation can be easily modified (such as E. coli), present attractive alternatives.
5.3.2. Riboswitches for cobalt
In addition to transcriptional regulating proteins (Section 5.2), cis-acting metal-responsive RNA molecules, “metalloriboswitches”, have also been proposed to selectively alter gene expression in response to Co(II), Ni(II), Mg(II), Mn(II) and Fe(II) [147]. These metalloriboswitches are typically located in the 5′-untranslated region of transcripts encoding metal transporters and in some cases act in concert with the protein-based metal sensors.
The czcD (NiCo) riboswitch was identified upstream of czcD, encoding a CDF-family transporter forming part of the cadmium-zinc-cobalt resistance determinant [148]. This riboswitch promotes expression of czcD in the presence of Co(II) and Ni(II), but not Mn(II) or Zn(II) in Clostridium species, suggesting a selective role in metal resistance in vivo [148]. Metal sensor proteins which selectively respond to Co(II) in the cell bind Zn(II) and Cu(I) more tightly than the cognate metal (in accordance with the Irving Williams series, (Section 6.2)), and in many cases, these noncognate metals also trigger allostery (for example RcnR and CoaR) [90,96]. Selectivity in vivo is enabled because the protein's affinity for Co(II) matches (is tuned to) the cellular availability of Co(II), but the affinities for Zn(II) and Cu(I) (although tighter than the affinity for Co(II)) are too weak to compete with the availability of these metals in the cell (Section 6.5). In contrast, affinities of the czcD riboswitch from Clostridium botulinum (Cbo) indicate that it binds Ni(II) and Co(II) more tightly than other metals (KD values = 6.5 μM and 13 μM, respectively), with Mn(II) also binding weakly (KD = 220 μM) with no binding of iron, copper or zinc detected [148]. Affinities were determined aerobically using in-line probing which monitors ligand induced structural changes in the RNA molecule, therefore values might represent a combination of affinity and allostery. It is possible that a metal ion can bind tightly but not induce the allosteric conformational change, as has been observed for some metal sensor proteins [149]. Furthermore, aerobic conditions are problematic when working with some metal ions, for example those which are sensitive to oxidation, such as Cu(I) and Fe(II). Compared to metal sensor proteins, the affinities of the czcD riboswitch for Co(II) and Ni(II) appear too weak to explain the selective response in vivo (Section 6.5).
The mechanism of regulation by the czcD riboswitch is via an intrinsic transcriptional terminator hairpin which prevents synthesis of the full-length mRNA in the absence of metal ions. Upon metal binding, the RNA is stabilised in a structure which promotes read-through of the full length transcript by RNA polymerase, and expression of czcD is enabled [148].
The structure of the czcD riboswitch from Erysipelotrichaceae bacterium (Eba) bound to Co(II) has been determined and reveals two pairs of coaxially stacked helices with no tertiary interactions [148]. Four Co(II) binding sites were identified (Fig. 6b). Two Co(II) ions (sites 1 and 2) are six-co-ordinate, each with three co-ordinating water molecules. Four highly conserved nucleotides (G46, G47, G87, G88) directly co-ordinate the two Co(II) ions. In each case co-ordination involves the N7 position (as well as 2′-OH for G46 and G87), the most nucleophilic RNA functional group which has also been implicated in metal ion co-ordination in other metalloriboswitches (Fig. 6b) [150]. Co-operative metal binding is observed for both Eba-czcD and Cbo-czcD riboswitches such that binding of the first Co(II) ion stabilises the second Co(II) site, with G46 and G47 providing a link in the primary co-ordination sphere of the two sites [148]. Two additional Co(II) sites are observed in the crystal structure, each co-coordinated by at least four water molecules: site three is linked to an outer co-ordination solvent molecule of site 2 via G45, with the fourth site possibly an artefact of the crystallisation conditions bound by only a single non-conserved nucleotide [148]. As such, the lability of the czcD riboswitch Co(II) sites is much greater than for metal sites of metal sensor proteins [151].
Recently, the Eba-czcD riboswitch has been biochemically characterised anaerobically using a fluorescent variant, and affinities determined under in vitro conditions where the metal ions have been buffered [151]. In this experimental setup, affinities of the czcD riboswitch were determined to be 120 nM for Co(II), 61 nM for Ni(II) and 11 μM for Mn(II). In vitro responses were also observed for Fe(II) and Zn(II) with affinities of 400 and 93 nM, respectively [151]. Here, there is closer agreement with the Irving Williams series than previously observed, and the results highlight the technical challenges of measuring metal affinities (Section 6.1). It is interesting to note that unlike metal sensor proteins, the metal binding sites formed by nucleic acids lack a sulphur donor, likely influencing metal binding affinities. When expressed in E. coli, the Eba-czcD riboswitch is activated by manganese, iron, cobalt, nickel and zinc, but when the in vitro affinities are compared with recent estimates of bacterial cellular metal availability, only Fe(II) and Mn(II) would be able to bind czcD in the cell [93,151]. Furthermore, the czcD riboswitch of Listeria monocytogenes appears to regulate a P1B-ATPase metal exporter (LMO3448) with homology to transporter PfeT, implicated in Fe(II)-efflux. Heterologous expression of LMO3448 in Bacillus subtilis lacking pfeT provides resistance to Fe(II), and to some degree Mn(II) and Zn(II), but not to Co(II) or Zn(II) [151].
The cognate metal(s) detected by the czcD riboswitch remain unclear with conflicting data in different organisms and affinities for Co(II) being weaker than corresponding protein-based Co(II)-sensors (Section 5.2) [148,151]. That being said, sensors with relatively weak Co(II) affinities (~10−7 M) have been reported to respond to cobalt in the cell under certain conditions [92,96]. It is possible that the czcD riboswitch presents a mechanism of ‘last-resort’ which starts to respond as the cellular buffer for Co(II) (or other metal) becomes saturated. Conversely, it might respond to metal ions lower down the Irving Williams series where availability in the cell is in the nano- to micromolar range (Section 6.5). Robust characterisation of the czcD riboswitch in different organisms, as has been done for metal sensor proteins, is necessary to fully understand the role of the czcD riboswitch and which metal(s) it senses. This requires analyses of metal responsiveness in vivo under conditions which do not overwhelm cellular metal homeostasis, and adapting in vitro experimental designs which have been developed to biochemically characterise metalloproteins. It is possible that in different organisms, the czcD riboswitch responds to different metals depending on the metal availability and/or specific nucleotide differences which alter metal selectivity. In this way, the same RNA scaffold could have a different specificity for metal ions afforded by relatively small sequence changes (analogous to the selectivity of the B12 riboswitches for different B12 derivatives, and metal sensor proteins of the same family which detect different metals). Is it possible that metal-sites in RNA molecules are more rigid than those created by the side-chains of amino-acids, offering greater steric selection and hence departing from the constraints of the Irving-Williams series? As a relatively new area in the cell biology of metals, metalloriboswitches present novel questions.
5.4. Chaperones and chelatases for cobalt and B12
The selective advantages of metal delivery proteins versus metal insertases, metallochaperones versus metal-chelatases, overlap and distinctions blur. Viewed simplistically, chelatases distort their tetrapyrrole substrates to catalyse the insertion of cognate metals [152], while metallochaperones supply cognate metals to the active sites of proteins and cofactors and are necessary in vivo because of limited metal availabilities inside cells [153,154]: But chelatases can also confer a metal supply advantage. Reciprocally, some metallochaperones modify their partners to aid metal insertion. Vitamin B12 biosynthesis pathways utilise two remarkably different chelatases, the class II (CbiX/CbiK) and class I (CobNST) enzymes, for early- and late-stage cobalt insertion, respectively (detailed descriptions in 5.4.1, 5.4.2). The class II enzyme CbiK catalyses cobalt insertion into sirohydrochlorin at in vivo cobalt availabilities [93] and thus carries out both metal supply and insertion functions. The class I enzyme CobNST catalyses metal insertion into hydrogenobyrinic acid a,c-diamide when high concentrations of cobalt are supplied in vitro [155] but an additional aerobic B12 synthesis enzyme, CobW (Section 5.4.2.1), is necessary for cobalt supply in vivo, at least when cobalt is not in surplus [156].
5.4.1. The early cobalt chelatases and related enzymes associated with metal insertion
In the early cobalt insertion, or anaerobic, pathway cobalt is inserted into a macrocycle called sirohydrochlorin (Fig. 3a), an intermediate that represents a branch-point with a number of other modified tetrapyrroles including siroheme, coenzyme F430 and heme [10]. Indeed, metal insertion at this stage directs the intermediate to the corresponding final product. Thus, cobalt insertion directs the intermediate towards cobalamin biogenesis, whereas nickel insertion directs the intermediate towards coenzyme F430, whilst insertion of iron dictates the molecule is destined for either siroheme or heme synthesis. Metal insertion in all these cases is mediated by separate class II chelatase enzymes [157], all of which share a common structure and are clearly homologous. For cobalt insertion three separate enzymes have been identified and are known as CbiK [[158], [159], [160]], CbiXL [160,161] and CbiXS [157,162]. These enzymes insert Co2+ into sirohydrochlorin to generate Co(II)-sirohydrochlorin but are structurally related to the iron-inserting enzyme SirB that converts sirohydrochlorin into siroheme [157,163,164], as well as to CbfA, which is the nickel chelatase that inserts Ni(II) into sirohydrochlorin for coenzyme F430 biosynthesis [165,166].
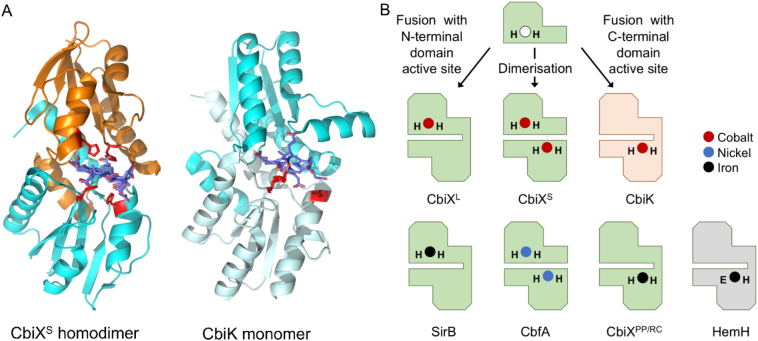
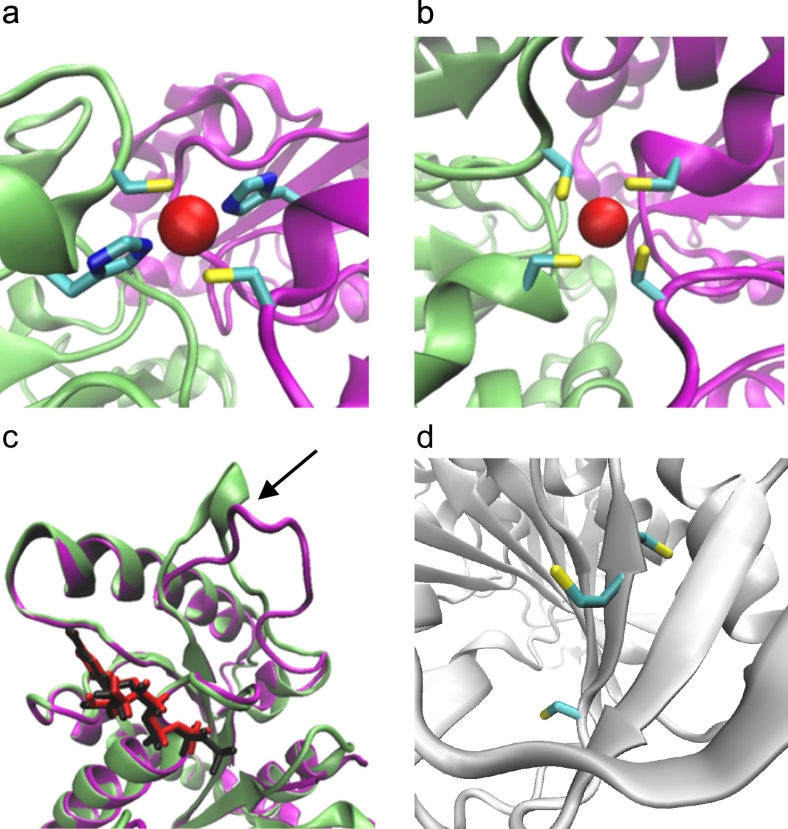
CbiXS refers to a “small” CbiX enzyme, composed of around 120–145 amino acids and is generally found in the Archaea [162]. CbiXS is active as a homo-dimer (Fig. 7a), with the active site formed at the junction between the two proteins. Consequently, the active site is symmetrical. Interestingly, the other class II chelatases CbiK and CbiXL (the large form of CbiX) are about twice the size of CbiXS and have N-terminal and C-terminal regions that separately align with the smaller protein. This indicates that both CbiK and CbiXL have arisen following cbiXS duplication [157]. Indeed, the structures of these larger enzymes confirm this finding, revealing a bilobal architecture composed of two α/β domains (Fig. 7a). The two lobes of the enzyme not only relate to one another by a pseudo-2-fold symmetry but also to the primordial CbiXS. The small CbiX is therefore considered as the “ancestral” class II chelatase [157].
Fig. 7.
The chelatases associated with the cobalt-early pathway.
(a) The type II chelatases are associated with the insertion of Co(II) into cobalamin, Fe(II) into siroheme and heme and Ni(II) into coenzyme F430. They all have a common fold despite often having limited sequence similarity. The CbiXS cobaltochelatase from Archaeoglobus fulgidus is a homo-dimer composed of two identical subunits (coloured orange and cyan). The active site is formed at the junction of the two subunits and is symmetrical. Because of the symmetry imposed by having two identical subunits, sirohydrochlorin is sandwiched by two catalytic histidines from each subunit. In other type II chelatases, such as CbiK from Salmonella, the two subunits have fused together to give a single polypeptide chain and only two active site histidines (His 145 and 207) are found here – in this case located below the sirohydrochlorin substrate. The pseudo two-fold symmetry of CbiK, reflecting the ancestral CbiXS heritage, is shown by the light and dark cyan colouring. (b) The evolution of the different type II chelatases is reflected in where the active site histidine residues are located. The cartoon highlights that the enzymes have the same basic architecture and are homologous, with those with a higher level of homology shown in green. For the small chelatases such as CbiXS and CfbA the enzyme has a symmetrical active site with a total of 4 histidines, with two from each subunit. In the larger single subunit enzymes the two histidines are located either in the N-terminal domain of the protein or the C-terminal domain. Thus CbiXL and CbiK (shown in pink) have evolved by retaining the active site histidine residues in different domains. Similarly, CbiXPP/RC and SirB, which are both ferrochelatases associated with siroheme biogenesis, have also evolved with differential retention of the active site histidines. HemH (shown in grey), which is a ferrochelatase associated with heme biogenesis has changed one of the histidine residues for a glutamate and retained these in its C-terminal domain.
The chelation reaction promoted by the sirohydrochlorin chelatases is mediated by a pair of conserved histidine residues. In the Archaeoglobus fulgidus CbiXS enzyme these two conserved histidine residues are located at His10 and His74 [162]. In its dimeric form the CbiXS complex therefore has a symmetrical active site that contains a total of four histidines. Two analogous histidines, His145 and His207 (Fig. 7a), are found only in the C-terminal domain of Salmonella CbiK and have been shown to be important for cobalt ion insertion indicating an asymmetrical active site [159,162,167]. In Bacillus megaterium CbiXL, the catalytically important histidine residues are found in the N-terminal domain of the protein (His14 and His79) indicating that this protein has evolved from a fusion of two CbiXS in a different way to CbiK. Thus, the evolution of CbiK and CbiXL can be followed by retention of the essential active site histidines within either the N- or C- terminal domain of these bilobal enzymes (Fig. 7b) [167]. Unexpectedly, CbiXL was also found to contain an extended C-terminal extension that harbours both a 4Fe—4S centre as well as a long histidine-rich region. Deletion of this region does not affect the cobalt insertion reaction but it has been proposed that the region may be involved in metal ion storage and delivery [161]. However, it is clear that several types of chelatases contain Fe—S centres although it is not clear why this should be so as the centres are not required for catalytic activity [168,169]. Finally, another CbiXL-like protein from Paracoccus pantotrophus has been characterised and shown to be involved in siroheme synthesis – so technically this is a SirB. In this case the two catalytic histidine residues within CbiXL-PP are found in the C-terminal region of the protein, which is also the closest ortholog to the Rhodobacter capsulatus CbiX that was shown to play a role in defending against photooxidative stress, possibly by altering flux through the tetrapyrrole pathway [170].
Duplication of the ancestral small chelatase CbiXS has therefore led to the appearance in nature of a suite of enzymes including CbiXL, CbiK, CfbA and SirB that are able to mediate the insertion of divalent transition metal ions including Co(II), Fe(II) and Ni(II) (Fig. 7b). Indeed, most class II chelatases that insert metals into sirohydrochlorin appear to lack metal specificity and hence these enzymes will insert Fe(II), Ni(II) and Co(II) into the tetrapyrrole substrate [163]. It is not possible to predict which metal ion they are specific for from their tertiary structures and hence metal specificity is likely to be imposed within the cell depending on their binding affinity (see 6.2, 6.4, 6.5.4).
The type II chelatases are thought to function mechanistically by binding the tetrapyrrole substrate in a distorted fashion, thereby exposing the pyrrole nitrogens to allow them to deprotonate [152,167]. The deprotonation event may involve the active site histidine residues, which would then allow for direct metal insertion. However, it may be that the histidines are also associated with metal ion delivery to the active site to help promote catalysis through proximity. The distortion of the substrate coupled with the immediacy of the metal ion is all that is needed to promote the liganding event, which also occurs spontaneously at a slower rate in the absence of enzyme. Indeed, the insertion of various metal ions into sirohydrochlorin follows the Irving-Williams series in terms of relative rate, indicating that metal availability to the chelatase is a key determinant of metal selection [171].
The early insertion of cobalt into sirohydrochlorin generates cobalt-sirohydrochlorin (also known as cobalt-Factor II), and dictates that the molecule is destined for life as a cobamide. Cobalt-sirohydrochlorin next undergoes a series of methylation events that promote ring contraction prior to decarboxylation and methyl group migration and the amidation of the side chains, generating cobyric acid. The attachment of the lower nucleotide loop then produces the final cobamide [10].
5.4.2. The late cobalt insertion pathway and the type I chelatases
The late cobalt insertion pathway proceeds from uroporphyrinogen III through the same series of methylation events as with the early insertion pathway, but obviously with cobalt-free intermediates (Fig. 3b) [10]. Cobalt is only added after the main corrin-ring has been generated, at an intermediate called hydrogenobyrinic acid a,c-diamide, which is the substrate for the late cobalt insertion chelatase (Fig. 3a) [172]. Unlike the monomeric ATP-independent chelatase of the early cobalt insertion pathway, this chelatase requires three proteins, CobN, CobS and CobT and consumes ATP. CobN is a large monomeric protein (120–150 kDa) that contains the main active site responsible for the transfer of the cobalt into the corrin substrate.
Hydrogenobyrinic acid a,c-diamide binds to CobN and the protein co-purifies with the substrate. CobS and CobT form a 450 kDa complex, which is required along with CobN to form a functional cobaltochelatase [172]. CobS and CobT share some similarity to each other and appear to form a two-tiered hexameric ring [173]. Whilst the CobS subunit contains a characteristic ATP binding Walker A and Walker B motifs belonging to the AAA+ superfamily of proteins, the CobT subunit has an integrin I domain, or von Willebrand factor, at its C-terminus, a motif that is usually associated with protein interactions associated with cell adhesion. CobS is thought to be the engine of the cobaltochelatase whereas CobT is more likely to be involved in the stabilisation of the motor complex platform [173].
The CobNST cobaltochelatase system is homologous to the magnesium chelatase system associated with the insertion of magnesium into protoporphyrin IX during the formation of bacteriochlorophyll (Bch) and chlorophyll (Chl) [174], which is catalysed by a very similar ATP-dependent heterotrimeric system consuming 15 ATP per round of catalysis [175]. The three proteins involved in the magnesium chelatase complex are named BchH/ChlH, a large monomeric subunit similar to CobN, BchD/ChlD that are the counterparts of CobT and finally BchI/ChlI subunits that belongs to the AAA+ (ATPases associated with diverse cellular activities) and are similar to CobS. This magnesium chelatase together with the cobalt chelatases form the Class I metal chelatase group [157]. In contrast to the sparse amount of information concerning the cobalt chelatase mechanism, many more studies have been undertaken on the magnesium chelatase.
The structure of Synechocystis PCC 6803 ChlH was solved in 2015 although no substrate complex was determined [176]. The protein is formed of six domains, including N-terminal ‘head’ and ‘neck’ regions and a central cage-like assembly. The ChlI subunits can self-assemble into a hexameric ring structure without ATP to resemble a 6-tooth rotor motor with an L-shaped helix to form the base and the outer ring for the motor [177]. The N-terminal domain of BchD/ChlD is similar to BchI/ChlI and its C-terminal domain contains a proline-rich region followed by a C-terminal integrin I domain [178]. This subunit also forms a hexameric ring but does not possess ATPase activity. As with CobS and CobT, the ChlD and ChlI form a double hexameric complex, which can be purified together and it was shown that the N-terminal domain of ChlD interacts with ChlI [179]. The 6-tooth rotor motor of BchI/ChlI is thought to twist upon ATP hydrolysis, mediating the movement via ChlD and its integrin I domain to bridge the ATPase activity through to the ChlH metal ion active site [180]. It is likely that similar conformational changes are also associated with the CobNST complex.
It is not fully understood why ATP hydrolysis is required for metal chelation within the class I chelatases. For cobalt insertion, the energy may be required to help promote insertion of the metal into the limited confines of a ring-contracted macrocycle. Moreover, the structure of the corrin substrate indicates that two of the opposite-facing pyrrole nitrogens remain protonated, thereby providing a steric block to the metal [15]. Hence, the cobalt chelation process into the corrin ring may require energy to distort the corrin macrocycle, promote proton abstraction and then encourage metal insertion. Unlike the type II chelatase, which can accept a range of metal substrates, CobNST is highly specific for cobalt and no other metals appear to function with the enzyme. Moreover, the type I cobaltochelatase requires a specific cobalt delivery system, a function performed by a protein called CobW (see Section 5.4.2.1).
5.4.2.1. CobW and the COG0523 family of G3E GTPases
CobW was first identified as a component of the aerobic B12 pathway nearly three decades ago, when it was discovered that disruptions to the cobW gene impaired cobalamin biosynthesis in Pseudomonas denitrificans [181]. The functional role of CobW in Co(II) supply has finally been elucidated [156], with ambiguity about its metal-specificity resolved (Section 6.6).
CobW belongs to the COG0523 family, a subset of P-loop G3E GTPases whose common role is to support metallocentre assembly [182,183]. There are four branches of the G3E superfamily: MeaB, UreG, HypB and COG0523. MeaB, UreG and HypB have well-established functions: MeaB gates the insertion of B12 into methylmalonyl-CoA mutase (MCM) [184] while UreG and HypB support Ni(II)-acquisition by urease and hydrogenase, respectively [[185], [186], [187], [188]]. The genes encoding these proteins are generally found only once in a genome, in MCM-encoding, urease maturation and hydrogenase maturation gene clusters, respectively [183]. In contrast, diverse functions have been attributed to the COG0523 family which are found in numerous gene contexts and it is common to find more than one homolog within the same genome. This has led to the identification of at least 15 subgroups of the COG0523 family which differ in both function and metal-selectivity [183]. These include Nha3 which activates the Fe(III)-type nitrile hydratase (NHase) [189,190], numerous subgroups (including YeiR, YjiA, ZigA and ZagA) connected to Zn(II) homeostasis [130,[191], [192], [193], [194]], and CobW connected to vitamin B12 [181]. An estimated 12.5% of COG0523 genes are cobW, usually located adjacent to cobN in aerobic cobalamin biosynthesis gene clusters [183]. Given its homology to known metallochaperones, and genomic co-localisation with the cobaltochelatase gene cobN, a potential role as cobalt chaperone was suggested for CobW [195]. However, alternative functions (such as chaperone for the iron-requiring B12 enzyme CobG) were possible, particularly considering the roles of related COG0523 proteins in iron homeostasis. The mystery was solved when, by reference to intracellular metal-availabilities, CobW was shown to acquire Co(II) in vivo and its functional role in Co(II) supply was revealed [156] (Section 6.6).
COG0523 proteins are the largest of the G3E family. In addition to the highly conserved N-terminal domain which retains key structural features common to all GTPases [182,196], COG0523s contain a second, highly variable, C-terminal domain [183]. This is thought to interact specifically with client proteins with the high sequence variability suggesting a broad range of functional targets, as first indicated by gene context [183], and increasingly substantiated with experimental evidence [130,156,189,194]. The cobaltochelatase CobNST is the anticipated target for CobW, although no specific protein interaction has yet been demonstrated. Interestingly, while the Co(II)-acquisition site of CobW is likely localised to a conserved CxCC motif in the N-terminal domain [156] (Section 6.6), the C-terminal domain contains a conserved His/Asp-rich region [130] which could play a role in metal-handover.
6. A paradigm for metalation inside cells: CbiK and CobW
6.1. The determination of metal-protein affinities: A cautionary note
Widely used methods for determining the metal affinities of proteins can produce erroneous values [197]. Indeed, a substantial fraction of reported metal-protein affinities are incorrect [197,198]. As researchers follow published experimental precedents, so wrong values propagate and the ‘noise-to-signal’ ratio within the literature increasingly confounds the development and testing of hypotheses. Care will be needed to exclude unreliable values if metal affinities are interrogated by machine learning routines. Common issues include unrecognised upper or lower limits of experimental methods, inadvertent inclusion of competing ligands (eg buffers, salts), presence of metal-binding protein tags, poor pH control, the formation of adducts, non-equilibrium assays, oxidation of metal or ligand, contaminating metals, and misinterpretation of experimental data. To circumvent these pitfalls, robust methods for the determination of metal-protein affinities (usually via competition with well-characterised ligand standards) have been described [[197], [198], [199], [200]].
6.2. The metal affinities of CbiK do not match its requirement for Co(II)
The metal affinities of the CbiK cobalt chelatase of Salmonella were measured by competition against probes of known affinities: fura-2 for Co(II) and mag-fura-2 for Fe(II), Ni(II) and Zn(II), while the affinity for Mn(II) was too weak for either competitor [93]. An affinity for Cu(I) was obtained by competition against bicinchoninic acid. The affinity for Co(II) is similar to that for Zn(II) and slightly weaker than that for Ni(II), but Cu(I) forms substantially the most stable complexes. Viewed in isolation, these data indicate that CbiK might be vulnerable to mismetalation with other ions and perhaps preferentially Cu(I).
Because proteins are flexible, they offer negligible steric selection, at least in their nascent forms and in the absence of interacting partners (Section 6.5). Under these conditions the order of metal binding follows the Irving-Williams series [201]. Notably this was co-discovered by R.J.P. Williams who also proposed the entatic state (Section 1.1).
The Irving–Williams series for divalent metals [201], plus Cu(I) and Mg(II). Note reversal of the less-than sign after copper. All first row metals can exist in the divalent form and Cu(I) ions, thought to predominate in the cytosol, also bind tightly, while Mg(II) binds weakly.
The order of metal binding to CbiK exemplifies the Irving-Williams series and highlights a universal challenge for metalation.
Mismetalation can involve a sub-set of ligands from the bona fide metal site, a different coordination geometry or even recruit additional ligands. However, the biological challenge is to populate each locus within metallo-proteins with the correct ions regardless of the coordination chemistries of aberrant binding. Inclusion of a metal within a cofactor such as a tetrapyrrole largely resolves these issues for the final metalloenzyme which can now contain a specific binding pocket that recognises the complex molecule. However, this still requires that the correct metal has previously partitioned into the tetrapyrrole (Section 6.4). A metal delivery protein or metallochaperone can resolve the specificity of metal insertion via specific protein-protein recognition between delivery protein and chelatase, but ultimately the challenge still remains: How does the correct metal, in this case Co(II), partition onto the delivery protein if other metals bind more tightly (6.4, 6.6)?
6.3. Bioavailability influences metal-protein speciation in vivo
A pair of proteins in the periplasm of a cyanobacterium share the same cupin fold, the same metal-binding ligands yet bind different metals from near opposing ends of the Irving-Williams series in vivo, manganese and copper [202]. Refolding of either apo-protein (MncA or CucA) in vitro in the presence of equimolar amounts of manganese and copper (either cuprous or cupric) led to binding of copper which inactivates oxalate oxidase activity of the manganese-protein [202]. A ten-thousand times excess of manganese is required at folding in order to correctly metalate the manganese-protein. This implies that manganese is at least ten thousand times more available than copper at the site where the nascent manganese-protein folds. Notably, while the cupro-protein is secreted in an unfolded state via the sec-machinery, the manganese-protein is secreted pre-folded via the Tat-machinery [[202], [203], [204]]. These data imply that the cytosol is a metal-protected environment where the bioavailability of the more competitive metals is controlled to prevent mismetalation of proteins that require less competitive ions.
The metal ions in MncA and CucA are buried with no, or negligible, access to solvent [202,205]. Within folded manganese-MncA metals become kinetically trapped. This implies that at some step along the folding pathway there is a nascent metal-binding site which requires a 10,000 times excess of manganese over copper to avoid entrapment of the incorrect ion. The MncA-related oxalate decarboxylase enzyme (OxdC) from Bacillus subtilis has been expressed in the cytosol of E. coli and found to entrap a range of inactivating metals [206]. For example, growth of cells in excess cobalt leads to the accumulation of cobaltous-MncA. The proteins of metallostasis must normally act to sustain the correct availabilities of the different elements to enable correct metalation of proteins and delivery pathways, including those that supply metals to tetrapyrroles and vitamin B12. But what are these vital availabilities: What defines intracellular metal-bioavailability and how can this parameter be measured?
6.4. CbiK as a paradigm for in vivo metalation
The cytosol is over-supplied with binding sites for essential metals [207,208]. This is also anticipated to be true in other biological compartments. Under these conditions proteins and other molecules compete for a limited supply of metals, rather than metals competing with each other for a limited supply of binding sites. To understand how Co(II) partitions onto CbiK, this competition was defined [93]. Reference values for cytosolic metal availabilities were generated (Section 6.5) against which the binding preferences of CbiK for different metals (Section 6.2) could be related.
6.5. Metal availability in the cell determined from the sensitivities of metal-sensors
It is straight-forward to determine the total amount of metal in a cell. By digesting a known number of cells in acid and then determining the concentration of each metal in the extract, for example by ICP-MS, the number of atoms per cell is readily calculated. Using the average volume of a cell, this can be converted into a pseudo concentration: Pseudo because many of these atoms will not be available in solution but sequestered in unavailable sites, kinetically trapped and/or compartmentalised. The crucial question is how much of each metal is accessible to nascent metalloproteins and how tightly must a protein (such as CbiK) bind in order to compete for each of these ions?
Metals such as Cu(I) and Zn(II) are at much lower availabilities than less-competitive (for binding to ligands) metals such as Co(II), Mn(II) and Fe(II) (Section 6.4) [93]. More than 20 years ago it was concluded that the cytosol contains essentially no ‘free’ Cu(I) or Zn(II) [207,209]. We take the term free to refer to the aquo-form (or any comparably weak ligands). To estimate the bioavailability of each metal a number of methodologies have been used including chemical probes, genetically encoded probes, and measurements of the metal affinities of cognate metal sensors [57,210].
DNA-binding metal-sensing transcriptional regulators have evolved to respond to the availability of a specific metal ion [90]. Metal co-ordination by the metal sensor induces a conformational change which allosterically activates or inhibits the interaction of the protein with its target DNA binding site to modulate downstream gene expression. Together, these systems enable the cell to mount an adaptive response to metals: achieving metal sufficiency whilst avoiding metal toxicity (5.1, 5). Each metal sensor is tuned to the cellular availability of its cognate metal and operates within a relatively narrow range (Section 5.2.6) [90]. If the sensitivity (which is determined by the allosteric mechanism and protein-abundance as well as metal-affinity, Section 6.5.1) of each metal sensor can be determined, it should become possible to determine the bioavailability of each metal in a cell (Section 6.5.3). To this end, a complete set of metal sensors was thermodynamically characterised in Salmonella (Fig. 8a) [93].
Fig. 8.
Thermodynamic characterisation of a set of sensors.
A, The set of cytosolic protein-based metal sensors from Salmonella with cognate metal ion(s) and allosteric mechanism of regulation shown, as described in [93]. B, Thermodynamic cycle depicting the allosteric coupling of metal (M) binding and DNA (D) binding by metal sensor proteins (P) [211]. C, Fractional occupancy of target DNA binding site(s) with respective metal sensor protein as a function of buffered metal concentration, following determination of K1–4, and number of sensor molecules and target DNA-sites per cell (B), sensor protein abundance and number of target DNA binding sites as described in [93]. D, the ranges of total (triangles) and the available (buffered; circles) metal concentrations in Salmonella. Total metal was determined in cells grown in minimal media with (closed triangles) and without (open triangles) metal supplementation [93]. Open triangles obscured for Mn(II) and Zn(II). Available metal reflects the mid-point of each sensor's dynamic range in (c). The concentration equating to one hydrated atom per cell is shown by a dashed line.
6.5.1. Thermodynamic characterisation of a set of metal sensors
Metal binding to a metal sensor protein is thermodynamically coupled to DNA binding (Fig. 8b) [211]. As a consequence, the metal affinity of the DNA-bound form can be tighter or weaker than that of the sensor when free from DNA: in the case of the Salmonella Co(II)-sensor, the DNA affinity of Co(II)-RcnR is ~100-fold weaker than that of apo-RcnR [212], and this will be reciprocated by a ~100-fold weaker Co(II)-affinity of DNA-bound RcnR (K2 = (K1K4)/K3) (Fig. 8b). The opposite will be true for sensors whose DNA-affinity is tightened upon metal binding, for example co-repressors such as Zn(II)-sensing Zur. Ultimately, the sensitivity of a metal sensor depends on the population which associates with DNA (or activates the promoter in the case of co-activators) to alter gene expression [90]. Thus, the metal-affinity of the sensor when not bound to DNA (K1) is an imperfect approximation of sensitivity.
Salmonella metal sensors were identified for Mn(II), Fe(II), Co(II), Ni(II), Zn(II) and Cu(I) Fig. 8a [90,213,214]. For each sensor, the cognate metal-affinity (K1), and the DNA-affinities of the apo- and metalated-protein (K3 and K4) were measured in vitro (Fig. 8b) [93]. Notably, accurate metal-affinity measurements can be challenging (Section 6.1), due to the tight affinities exhibited by many metal-protein complexes (such as 10−19 M for Cu(I)-CueR) [93]. Informatic analyses and multiple reaction monitoring-mass spectrometry was used to determine the number of target DNA-binding sites and the number of sensor molecules per cell, respectively [93]. The thermodynamic cycle in Fig. 8b was solved predicting the DNA occupancy of each sensor as a function of buffered metal concentration [93]. This revealed that the buffered metal concentrations at which each sensor exhibited 0.5 of its response spanned twelve orders of magnitude from Mn(II) to Cu(I) (Fig. 8c) [93]. The cellular buffered metal concentration inferred from the mid-point of the dynamic range for each sensor is much lower than the total metal in a Salmonella cell when expressed as a concentration per cell volume. For Cu(I), Zn(II), Ni(II), and possibly Co(II), the buffered available metal concentration equates to less than one hydrated metal ion per cell (Fig. 8d) (Section 6.5.2). This is consistent with the hypothesis that the cell has an over-capacity to chelate metal [208].
6.5.2. An associative cell biology of metals: Metal availabilities as free energy changes
The volume of an E. coli cell is about one femto litre. A single atom or molecule dissolved in this volume equates to a concentration of almost one nano molar (10−8 to 10−9 M). Estimates described in Section 6.5.1, and elsewhere [207,209,215,216], indicate that the available concentrations of many essential metals are sufficiently low as to equate to just a handful of atoms, or less than one hydrated atom, per cell (Fig. 8a). This has been a source of confusion: Perhaps metal availability must be considered at a population level with pico molar available Zn(II) equating to one atom in every thousandth cell; or perhaps the system must be time-resolved with one atom being available one thousandth of the time. Tremendous advances in technical abilities to observe molecular events in single live cells, make these explanations attractive. But the above arguments assume a dissociative cell biology of metals. An alternative hypothesis is that metals are widely delivered by specific protein-protein interactions via rapid ligand exchange reactions [57]. Again, progress in live cell imaging makes this explanation appealing. However, with such a large number of metalloproteins this implies an unfeasibly large number of undiscovered delivery systems. Moreover, this amplifies rather than solves the challenge associated with correct metalation since now the correct metals must somehow partition onto a multitude of delivery pathways.
An associative cell biology of metals via buffered metal-pools provides an explanation (Fig. 9a) [57,93]. For example, a simplified cytosolic solution matching the intracellular concentrations of a dozen free amino acids and glutathione, was shown to buffer available Ni(II) such that the affinity of the Ni(II) sensor InrS was poised to track with metal saturation of the buffer, elaborated in Section 5.2.6 [132]. Histidine was the main competing species, and as noted in Section 5.2.5, some Ni(II)-importers such as NikABC, translocate Ni(II)-histidine complexes [129]. Fig. 9b shows atoms of Ni(II) in complex with histidine binding to InrS via associative ligand exchange. Such exchange reactions are rapid because they bypass the otherwise rate limiting step of release to the hydrated state. With such tight metal-protein affinities, off-rates to the hydrated state would be so slow as to exceed the lifetimes of proteins before thermodynamic equilibrium is approached: Associative ligand exchange overcomes this limitation.
Fig. 9.
An associative cell biology of metals.
A, The four allosteric end states of a metal sensor protein: apo-protein (P), metal-bound protein (PM), DNA-bound apo-protein (PD) and DNA-bound metal-protein ((PM)D). Metal (M) exchange occurs between the metal sensor protein and cellular buffer (B) via K5-K9 by an associative mechanism [57,93,211]. B, Proposed metal-acquisition by a metal-sensor protein (InrS, PDB 5FMN) from a cellular metal-buffer complex (Ni(II)His2, LHISNI01) via formation of a heterocomplex (modelled on PDB 4XKN). The Ni(II)-InrS model was generated using PDB 2HH7 plus free His (HIS_LFOH). InrS side chains are light blue, histidine from the cellular buffer is grey, nickel is green.
It seems perverse to describe metal availabilities as the concentrations of almost non-existent hydrated species if the cell biology of metals is associative and the hydrated species irrelevant: But these values also provide information on the state of the relevant bound metal-species. Specifically, they indicate how tightly the metal is bound and hence which proteins can out-compete the buffer to acquire the metal by associative ligand-exchange. The terms chemical activity or chemical potential encompass this concept. To avoid confusion, cellular metal availabilities have been expressed as free energy changes, ΔG, associated with in vivo metal complex formation: the values for metal concentrations can be used to calculate the ΔG using the standard equation RTlnKA, where KA is the affinity of a molecule that would be 50% saturated with metal at the available buffered concentration [93]. These values provide a point of reference for understanding intracellular metalation.
6.5.3. Intracellular metal availability follows the Irving-Williams series
Fig. 10a shows the intracellular metal availabilities (as ΔG) at the dynamic response ranges (0.1 to 0.9 fractional DNA occupancy) of the DNA-binding metal-sensors of Salmonella [93]. The sensors are tuned to the opposite of the Irving Williams series (Section 6.2) [93,201]: That is the most competitive metals are at the lowest ΔG, hence tuned to the lowest availabilities, while the less competitive ones are at less negative ΔG and hence tuned to the highest availabilities (Fig. 10a). As suggested in Section 6.3, intracellular metal availabilities compensate for the inherent metal-binding preferences of proteins. Under these conditions different proteins can acquire different metals determined by their ‘relative’ (with reference to the competing species within the intracellular milieu) metal-binding preferences, rather than absolute affinities.
Fig. 10.
Metalation of CbiK.
A, Metal availabilities (expressed as free-energy changes, ΔG) in the bacterial cytosol determined from the thermodynamic characterisation of a set of metal-sensor proteins and related riboswitch (Fig. 8) [93]. Bars show the ΔG range as each metal sensor shifts from 10 to 90% of its transcriptional response (i.e. 0.1–0.9 fractional DNA occupancy). In cases where there are two sensors for one metal, the combined range is also shown. An estimated Ni(II) availability from thermodynamic values for RcnR (RcnR*) is shown, using determined RcnR DNA-binding affinities and the Ni(II)-affinity of InrS, a related Ni(II)-sensor (as only a limit Ni(II) affinity is known for RcnR) [132,237]. The ΔG associated with metal binding to CbiK is also shown. The arrow for Mn(II) reflects a lower limiting value. B, Fractional occupancy of CbiK (dotted lines) with Co(II) (upper) or Fe(II) (lower) at the intracellular availabilities where RcnR and Fur (sold lines) shift from 1 to 99% of their responses, respectively.
The hypothesis that intracellular metal availabilities will be set to the opposite of the Irving-Williams series is not new [217]. A diversity of estimates of metal availabilities, based on the metal-affinities of proteins of metallostasis [207,209], or by using metal-responsive probes for example [218], are consistent with this notion. Moreover, the complex mixture of intracellular ligands (free amino acids, other small molecules, adventitious binding sites on the surface of macromolecules), that can be assembled into a diversity of coordination sites with limited geometric constraint, provides an ideal mixture which will inevitably buffer according to the Irving-Williams series [201]. It is anticipated that over the course of evolution metal-sensors and other proteins of metallostasis have been selected to limit saturation or depletion of this buffer. Similarly, metalloenzymes and proteins that acquire metals to supply cofactors will have evolved to obtain the correct metal in the context of these competing species.
6.5.4. A thermodynamic framework for metalation predicts that CbiK acquires Co(II)
By calculating the ΔG associated with forming metal-CbiK complexes from the affinities described in Section 6.2, in comparison with the metal availabilities described in Section 6.5.3, it becomes evident that cobalt is poised to enable formation of Co(II)-CbiK (Fig. 10a). Other metals are estimated to bind CbiK more weakly than the competing intracellular species [93]. In vitro assays establish that sirohydrochlorin can spontaneously acquire cobalt in vitro. However non-catalysed insertion is negligible when Co(II) is buffered to an availability approximating the mid-point of the dynamic range for RcnR [93]. In contrast, under these buffered conditions insertion is sustained by CbiK, revealing the essential role of the chelatase for B12 biosynthesis in the context of competing intracellular species is noted in Section 6.4 [93]. By reference to this thermodynamic framework it becomes possible to understand why and how CbiK acquires the correct metal for the synthesis of vitamin B12.
Within the dynamic range of RcnR, Co(II) occupancy of CbiK is estimated to switch from entirely apo to fully saturated (Fig. 10b). This suggests that under some growth conditions Co(II) supply for vitamin B12 is limiting. Moreover, at the upper end of the dynamic range for Fur partial metalation of CbiK with iron becomes possible (Fig. 10a), albeit such mismetalation would be overcome by coincident supplementation with cobalt (Section 6.6). Notably, in mutants deficient in the chelatase for siroheme, CbiK can complement the deficiency and support iron-supply to siroheme [158]. Importantly, the data in Fig. 10a provides a thermodynamic framework within which it becomes possible to understand, and potentially optimise, the metalation of vitamin B12 and indeed other metal-dependent proteins and pathways.
6.6. Metalation of CobW
6.6.1. Identification of the cognate metal
CobW contains a CxCC sequence motif which is highly conserved in the COG0523 family [183] (Section 5.4.2.1). Mutational analysis has confirmed that the CxCC sequence is necessary for metal-handling activities of various COG0523 proteins including in vivo activation of Fe(III)-type NHase by Nha3 [190], the protective effect of YeiR under conditions of Zn(II) depletion [191], and for high-affinity Zn(II) binding to ZigA [193]. It was recently discovered that, upon binding Mg(II) and GTP, CobW assembles a thiol-rich, metal-binding site that is likely localised to this same motif [156]. The site binds Co(II), and the Co(II) affinity for the GTP-complex is more than one thousand times stronger than for the GDP-complex (KCo(II) ~ 30 pM versus ~100 nM, respectively). Notably, with reference to the availability of Co(II) in cells [93] (see Section 6.5), only the GTP-complex has a sufficiently tight affinity to acquire Co(II) in vivo [156].
Similarly to CbiK (Section 6.2), the metal affinities of CobW (in the GTP-bound ‘metal acquisition’ form) for Cu(I) and Zn(II) are tighter than for Co(II) [156]. In vivo Cu(I) binding can be ruled out, since intracellular Cu(I) availability in a cell is too low to enable its acquisition [93]. However, the remarkably high Zn(II) affinity (~190 fM) is sufficient for in vivo Zn(II) acquisition [156]. Since both Co(II) and Zn(II) bind at the same (or similar) sites, it was necessary to account for competition between these two metals to determine in vivo metal-occupancy of CobW. Thermodynamics favours Co(II) binding because the free energy difference between metal-binding to CobW and metal-binding to the cytosolic buffer (termed ΔΔG), is greater (ie more negative) for Co(II) than for Zn(II) [156]. Thus, Co(II) is the cognate metal.
6.6.2. Allosteric coupling of nucleotide and metal binding
In all G3E GTPases (Section 5.4.2.1) nucleotide binding and substrate binding are closely interconnected. UreG and HypB both form square-planar Ni(II) binding sites at dimerization interfaces which are induced by nucleotide binding (Fig. 11a,b) [[219], [220], [221]]. Moreover, the Ni(II) affinities of these sites vary significantly depending on the nature of the bound nucleotide (ie GDP versus GTP or a non-hydrolysable analogue) [[222], [223], [224]]. MeaB binding affinity for its substrate MCM is also affected by nucleotides [225]. X-ray crystallography has revealed nucleotide-dependent conformational changes in proteins from the MeaB [226], HypB [221,222] and UreG [219,220] families which rationalise these observations (Fig. 11a-c). However, structural determinations of CobW in complex with nucleotides are still needed to identify the configuration of the Co(II) site and to explain the substantial impact of nucleotide-binding on Co(II) affinity. Structural information is currently only available for one member of the COG0523 family (E. coli YjiA), and in the absence of bound nucleotides [192,227]. In these structures, the CxCC putative metal-binding motif is located along a beta strand in a conformation that would not likely support metal coordination (Fig. 11d). It is tantalising to note that this motif lies between the canonical switch I and switch II regions which change conformation as a function of GTP-binding and hydrolysis [196,228].
Fig. 11.
Allosteric relationship between nucleotide and substrate binding in G3E GTPases.
a–b Square-planar Ni(II) coordination sites at dimeric interfaces in (a) Klebsiella pneumoniae UreG (in complex with GMPPNP; PDB 5XKT [220]) and (b) Helicobacter pylori HypB (in complex with GDP; PDB 4LPS [222]); the two protomers of each homodimer are coloured green and magenta, respectively. c) Structure of Methylorubrum extorquens MeaB in complex with GMPPNP (green; PDB 4JYB [226]) or GDP (magenta; PDB 4LC1 [226]); bound nucleotides are shown in black and red, respectively. Arrow points to the ‘Switch III’ loop, which changes conformation upon nucleotide hydrolysis and is crucial for chaperone function [226]. d) The CXCC putative metal-binding motif in Escherichia coli YjiA (of the COG0523 family; PDB 1NIJ [227]), crystallised in the absence of nucleotides.
UV–visible spectroscopy of nucleotide-bound CobW is characteristic of tetrahedral Co(II) coordination geometry with three ligating thiols (originating, at least in part, from the CxCC motif) [156]. It is possible that GTP-binding induces a conformational change in CobW that imposes a tetrahedral geometry on the coordination site. This could explain why, in contrast to CbiK (and the Irving-Williams series generally, which applies in the absence of steric selection; see Section 6), CobW binds Co(II) more tightly than Ni(II) [156] because Ni(II) favours square planar coordination geometry (see Fig. 11a,b) due to ligand-field stabilisation for the d8 metal ion. Structural differences which alter the metal selectivity of different COG0523 proteins for Fe(II), Co(II) or Zn(II) (see Section 5.4.2.1) also remain to be discovered.
6.6.3. Function and mismetalation
Thermodynamic calculations which established that CobW is specific for binding Co(II) in vivo (Section 6.6.1) assume ‘idealised’ cellular metal availabilities, where sensors are at the mid-points of their responses (Section 6.5.1). If metal availabilities diverge from idealised values, calculations of CobW metalation change. Indeed, when engineered B12-producing E. coli strains (which, notably, lack an identified dedicated cobalt import system [22]) are cultured in standard media, intracellular Co(II) availability is limited and mismetalation of CobW by Zn(II) is predicted [156]. Only upon Co(II) supplementation does the intracellular Co(II) availability increase enough to elicit a response from the Co(II) sensor RcnR and in turn to lead to predictions that Co(II) will displace Zn(II) from CobW.
In vivo studies show that CobW significantly enhances B12-production at limiting intracellular Co(II) availabilities and CobNST can only acquire Co(II) directly from the cellular milieu if intracellular Co(II) levels are high [156]. In common with the contributions of copper-metallochaperones to metal-supply [207], it is predicted that CobW is needed to deliver Co(II) for insertion into the corrin ring when intracellular buffering molecules are not metal-saturated but competing. Since GDP-bound CobW has a drastically weaker Co(II) affinity than the GTP-bound form, GTP-hydrolysis is predicted to trigger Co(II) release [156]. An appealing possibility is that CobNST may act as a guanine nucleotide activating protein (GAP) to target metal delivery to the chelatase complex. This would be consistent with mechanisms of related G3E GTPases: GTPase activity of MeaB is enhanced 100-fold in the presence of its protein partner MCM [225]; while enhanced (albeit to a lesser extent) NTPase activities of UreG and HypB have also been reported in the presence of their respective interaction partners UreE [224] and HypA [229]. It is not known whether the GTPase activity of CobW provides an additional ‘check-point’ for metal fidelity, or whether the chelatase CobNST, whose in vitro insertase activity appears remarkably specific for Co(II) [155], is solely responsible for metal selectivity at the insertion step.
7. Future prospects: food security, biotechnology and the metalation of vitamin B12
As increasing numbers of individuals adopt vegan or ‘flexitarian’ (with reduced consumption of animal products) diets, so B12 insufficiency becomes more prevalent. Complete synthesis of B12 is possible [230], but it is not an economically viable option for mass production. Native strains that make B12 are generally less well suited to large scale industrial fermentations than, for example, E. coli or yeast, making this the most expensive vitamin on the market with prices having risen significantly over the past few years in a volatile market. Commercial production of vitamin B12 is made more challenging by the need to add cobalt to the growth media and the need to dispose of this carefully after growth given the health and environmental concerns associated with the metal. Similar concerns are associated with the addition of cyanide during the extraction and purification of the nutrient. It is likely that changes associated with improvements to the safer production of vitamin B12 have been one of the drivers associated with the increase in price of the nutrient. Therefore, there exists an exciting prospect of improving B12 manufacture coordinated with precise cobalt addition by engineering the biosynthesis and uptake pathways into heterologous strains to produce cobalamin in a controlled manner to help facilitate a simpler purification.
Fig. 10a provides a thermodynamic framework which may be used to inform the optimisation of vitamin B12 metalation, either by manipulating media cobalt and/or competing ions (6.5.4, 6.6), or by engineering the metallostasis of cobalt and/or competing ions. More broadly, with half of the reactions of life requiring metals, this framework may be applied to the improved manufacture of a wide range of products through sustainable bio-transformations. However, this is also relevant to areas of human health, especially in the human gut microbiome, where around a third of bacteria produce cobamides that help sustain many of the other cobamide-dependent bacteria [231]. Corrinoids are known to play a key role as modulators of the microbiome and the availability and integration of cobalt into the corrin-framework is a key step in the health of this large and complex ecosystem [232]. Indeed, it is interesting to note that human cobalamin-deficiency is often associated with inflammatory bowel disease and bacterial dysbiosis [233].
Another area of developing interest is the synthesis of metal variants of cobalamin, where the central cobalt ion is replaced with an alternative transition metal ion. The properties of the different metal ions within the corrin ring means that most of these metal-corrin analogues tend to act as anti-vitamins. For instance, the inclusion of rhodium in place of cobalt produces rhodibalamin, which forms a stronger and more stable metal-carbon bond, which prevents catalytic turnover [16]. Replacement of cobalt with other metals such as zinc or nickel generate structures that more closely resemble the Co(II) or Co(I) corrins, respectively, and have significant potential in the study of B12-dependent processes [234,235]. Excitingly, the capacity to make metal-free forms of cobalamin linked with the ability to insert a range of metal isotopes also paves the way for the development cobalamin as an imaging agent [15].
Significant advances have been made from using cobalt and the corrin ring to gain new knowledge about how metal availability, binding and delivery are coordinated to produce metallo-products. There has been ongoing debate about the contribution of kinetic factors [208], mediated by specific protein interactions and metal-niches, to metalation. To what extent is metal supply ‘hard-wired’ from import to metalation via specific protein-interactions, or thermodynamically equilibrated with buffered intracellular pools. Probably some combination applies to differing degrees for different metalloenzymes and cofactors. The framework in Fig. 10a offers the exciting prospect of uncovering the answers to such questions.
CRediT authorship contribution statement
Deenah Osman: Writing - review & editing. Anastasia Cooke: Writing - review & editing. Tessa R. Young: Writing - review & editing. Evelyne Deery: Writing - review & editing. Nigel J. Robinson: Conceptualization, Writing - review & editing. Martin J. Warren: Conceptualization, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We acknowledge the help of Arthur Glasfeld (Reed College, USA) with Fig. 9b. This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) grant awards BB/S014020/1, BB/S002197/1 and BB/R002118/1, and the Royal Commission for the Exhibition of 1851.
Footnotes
This article is part of a Special Issue entitled: Cell Biology of Metals III edited by Roland Lill and Mick Petris.
Contributor Information
Deenah Osman, Email: deenah.osman@durham.ac.uk.
Anastasia Cooke, Email: ac948@kent.ac.uk.
Tessa R. Young, Email: tessa.r.young@durham.ac.uk.
Evelyne Deery, Email: E.Deery@kent.ac.uk.
Nigel J. Robinson, Email: nigel.robinson@durham.ac.uk.
Martin J. Warren, Email: martin.warren@quadram.ac.uk.
References
- 1.Whipple G.H., Hooper C.W., Robscheit F.S. Blood regeneration following anemia, iv: in- fluence of meat, liver and various extrac- tives, alone or combined with standard diets. JAMA. 1920;53:236–262. [Google Scholar]
- 2.Minot G.R., Murphy W.P. Treatment of perni- cious anemia by a special diet. JAMA. 1926;87:470–476. [Google Scholar]
- 3.Shorb M.S. Activity of vitamin B12 for the growth of Lactobacillus lactis. Science. 1948;107(2781):397–398. doi: 10.1126/science.107.2781.397. [DOI] [PubMed] [Google Scholar]
- 4.Rickes E.L., Brink N.G., Koniuszy F.R., Wood T.R., Folkers K. Crystalline vitamin B12. Science. 1948;107(2781):396–397. doi: 10.1126/science.107.2781.396. [DOI] [PubMed] [Google Scholar]
- 5.Smith E.L. Purification of anti-pernicious anaemia factors from liver. Nature. 1948;161(4095):638. doi: 10.1038/161638a0. [DOI] [PubMed] [Google Scholar]
- 6.Rickes E.L., Brink N.G., Koniuszy F.R., Wood T.R., Folkers K. Vitamin B12, a cobalt complex. Science. 1948;108(2797):134. doi: 10.1126/science.108.2797.134. [DOI] [PubMed] [Google Scholar]
- 7.Smith E.L. Presence of cobalt in the anti-pernicious anaemia factor. Nature. 1948;162(4108):144. doi: 10.1038/162144b0. [DOI] [PubMed] [Google Scholar]
- 8.Brink C., Hodgkin D.C., Lindsey J., Pickworth J., Robertson J.R., White J.G. X-ray crystallographic evidence on the structure of vitamin B12. Nature. 1954;174(4443):1169–1171. doi: 10.1038/1741169a0. [DOI] [PubMed] [Google Scholar]
- 9.Hodgkin D.C., Kamper J., Mackay M., Pickworth J., Trueblood K.N., White J.G. Structure of vitamin B12. Nature. 1956;178(4524):64–66. doi: 10.1038/178064a0. [DOI] [PubMed] [Google Scholar]
- 10.Bryant D.A., Hunter C.N., Warren M.J. Biosynthesis of the modified tetrapyrroles-the pigments of life. J. Biol. Chem. 2020;295(20):6888–6925. doi: 10.1074/jbc.REV120.006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martens J.H., Barg H., Warren M.J., Jahn D. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 2002;58(3):275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 12.Barker H.A., Weissbach H., Smyth R.D. A coenzyme containing Pseudovitamin B(12) Proc. Natl. Acad. Sci. U. S. A. 1958;44(11):1093–1097. doi: 10.1073/pnas.44.11.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.F.G. Costa, E. Deery, M.J. Warren, J.C. Escalante-Semerena, New insights into the biosynthesis of cobamides and their use, in: H.-W. Lui, T.P. Begley (Eds.), Comprehensive Natural Products III, Elsevier 2020, p. (In press).
- 14.Kennedy K.J., Taga M.E. Cobamides. Curr. Biol. 2020;30(2):R55–R56. doi: 10.1016/j.cub.2019.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieninger C., Deery E., Lawrence A., Podewitz M., Wurst K., Nemoto-Smith E., Widner F.J., Baker J.A., Jockusch S., Kreutz C.R., Liedl K.R., Gruber K., Warren M.J., Krautler B. The Hydrogenobyric acid structure reveals the Corrin ligand as an Entatic state module empowering B12-cofactors for catalysis. Angew. Chem. Int. Ed. Engl. 2019;58(31):10756–10760. doi: 10.1002/anie.201904713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Widner F.J., Lawrence A.D., Deery E., Heldt D., Frank S., Gruber K., Wurst K., Warren M.J., Krautler B. Total synthesis, structure, and biological activity of Adenosylrhodibalamin, the non-natural rhodium homologue of coenzyme B12. Angew Chem Int Ed Engl. 2016;55(37):11281–11286. doi: 10.1002/anie.201603738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee R., Ragsdale S.W. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 18.Gruber K., Puffer B., Krautler B. Vitamin B12-derivatives-enzyme cofactors and ligands of proteins and nucleic acids. Chem. Soc. Rev. 2011;40(8):4346–4363. doi: 10.1039/c1cs15118e. [DOI] [PubMed] [Google Scholar]
- 19.Bridwell-Rabb J., Drennan C.L. Vitamin B12 in the spotlight again. Curr. Opin. Chem. Biol. 2017;37:63–70. doi: 10.1016/j.cbpa.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jost M., Fernandez-Zapata J., Polanco M.C., Ortiz-Guerrero J.M., Chen P.Y., Kang G., Padmanabhan S., Elias-Arnanz M., Drennan C.L. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature. 2015;526(7574):536–541. doi: 10.1038/nature14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodionov D.A., Vitreschak A.G., Mironov A.A., Gelfand M.S. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 22.Rodionov D.A., Hebbeln P., Gelfand M.S., Eitinger T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 2006;188(1):317–327. doi: 10.1128/JB.188.1.317-327.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawco N.J., McIlvin M.M., Bundy R.M., Tagliabue A., Goepfert T.J., Moran D.M., Valentin-Alvarado L., DiTullio G.R., Saito M.A. Minimal cobalt metabolism in the marine cyanobacterium Prochlorococcus. Proc. Natl. Acad. Sci. U. S. A. 2020;117(27):15740–15747. doi: 10.1073/pnas.2001393117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tagliabue A., Hawco N.J., Bundy R.M., Landing W.M., Milne A., Morton P.L., Saito M.A. The role of external inputs and internal cycling in shaping the Global Ocean cobalt distribution: insights from the first cobalt biogeochemical model. Glob. Biogeochem. Cycles. 2018;32(4):594–616. doi: 10.1002/2017GB005830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsay D., Kerr W. Cobalt close-up. Nat. Chem. 2011;3(6):494. doi: 10.1038/nchem.1053. [DOI] [PubMed] [Google Scholar]
- 26.Roth J.R., Lawrence J.G., Bobik T.A. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 1996;50(1):137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 27.Raux E., Schubert H.L., Roper J.M., Wilson K.S., Warren M.J. Vitamin B-12: insights into biosynthesis’s mount improbable. Bioorg. Chem. 1999;27(2):100–118. [Google Scholar]
- 28.Raux E., Schubert H.L., Warren M.J. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell. Mol. Life Sci. 2000;57(13–14):1880–1893. doi: 10.1007/PL00000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krautler B. Vitamin B12: chemistry and biochemistry. Biochem. Soc. Trans. 2005;33(Pt 4):806–810. doi: 10.1042/BST0330806. [DOI] [PubMed] [Google Scholar]
- 30.Ando N., Kung Y., Can M., Bender G., Ragsdale S.W., Drennan C.L. Transient B12-dependent methyltransferase complexes revealed by small-angle X-ray scattering. J. Am. Chem. Soc. 2012;134(43):17945–17954. doi: 10.1021/ja3055782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bridwell-Rabb J., Grell T.A.J., Drennan C.L. A rich man, poor man story of S-Adenosylmethionine and Cobalamin revisited. Annu. Rev. Biochem. 2018;87:555–584. doi: 10.1146/annurev-biochem-062917-012500. [DOI] [PubMed] [Google Scholar]
- 32.Williams R.J. The entatic state. Cold Spring Harb. Symp. Quant. Biol. 1972;36:53–62. doi: 10.1101/sqb.1972.036.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Warren M.J., Raux E., Schubert H.L., Escalante-Semerena J.C. The biosynthesis of adenosylcobalamin (vitamin B12) Nat. Prod. Rep. 2002;19(4):390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 34.Shelton A.N., Seth E.C., Mok K.C., Han A.W., Jackson S.N., Haft D.R., Taga M.E. Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 2019;13(3):789–804. doi: 10.1038/s41396-018-0304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Croft M.T., Lawrence A.D., Raux-Deery E., Warren M.J., Smith A.G. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438(7064):90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 36.Stabler S.P., Allen R.H. Vitamin B12 deficiency as a worldwide problem. Annu. Rev. Nutr. 2004;24:299–326. doi: 10.1146/annurev.nutr.24.012003.132440. [DOI] [PubMed] [Google Scholar]
- 37.Allen R.H., Stabler S.P. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am. J. Clin. Nutr. 2008;87(5):1324–1335. doi: 10.1093/ajcn/87.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gherasim C., Lofgren M., Banerjee R. Navigating the B(12) road: assimilation, delivery, and disorders of cobalamin. J. Biol. Chem. 2013;288(19):13186–13193. doi: 10.1074/jbc.R113.458810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battersby A.R. How nature builds the pigments of life: the conquest of vitamin B12. Science. 1994;264(5165):1551–1557. doi: 10.1126/science.8202709. [DOI] [PubMed] [Google Scholar]
- 40.Blanche F., Cameron B., Crouzet J., Debussche L., Thibaut D., Vuilhorgne M., Leeper F.J., Battersby A.R. Vitamin-B12 - how the problem of its biosynthesis was solved. Angew. Chem. Int. Edit. 1995;34(4):383–411. [Google Scholar]
- 41.Blanche F., Thibaut D., Debussche L., Hertle R., Zipfel F., Muller G. Parallels and decisive differences in vitamin-B12 biosyntheses. Angew. Chem. Int. Edit. 1993;32(11):1651–1653. [Google Scholar]
- 42.Leyssens L., Vinck B., Van Der Straeten C., Wuyts F., Maes L. Cobalt toxicity in humans-a review of the potential sources and systemic health effects. Toxicology. 2017;387:43–56. doi: 10.1016/j.tox.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 43.Ranquet C., Ollagnier-de-Choudens S., Loiseau L., Barras F., Fontecave M. Cobalt stress in Escherichia coli. The effect on the iron-sulfur proteins, J Biol Chem. 2007;282(42):30442–30451. doi: 10.1074/jbc.M702519200. [DOI] [PubMed] [Google Scholar]
- 44.Thorgersen M.P., Downs D.M. Cobalt targets multiple metabolic processes in Salmonella enterica. J. Bacteriol. 2007;189(21):7774–7781. doi: 10.1128/JB.00962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrews S.C., Robinson A.K., Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003;27(2–3):215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 46.Fantino J.R., Py B., Fontecave M., Barras F. A genetic analysis of the response of Escherichia coli to cobalt stress. Environ. Microbiol. 2010;12(10):2846–2857. doi: 10.1111/j.1462-2920.2010.02265.x. [DOI] [PubMed] [Google Scholar]
- 47.Kumar V., Mishra R.K., Kaur G., Dutta D. Cobalt and nickel impair DNA metabolism by the oxidative stress independent pathway. Metallomics. 2017;9(11):1596–1609. doi: 10.1039/c7mt00231a. [DOI] [PubMed] [Google Scholar]
- 48.Nachin L., Loiseau L., Expert D., Barras F. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 2003;22(3):427–437. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Outten F.W., Djaman O., Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 2004;52(3):861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 50.Agar J.N., Krebs C., Frazzon J., Huynh B.H., Dean D.R., Johnson M.K. IscU as a scaffold for iron-sulfur cluster biosynthesis: sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry. 2000;39(27):7856–7862. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- 51.Krebs C., Agar J.N., Smith A.D., Frazzon J., Dean D.R., Huynh B.H., Johnson M.K. IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry. 2001;40(46):14069–14080. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- 52.Lu J., Yang J., Tan G., Ding H. Complementary roles of SufA and IscA in the biogenesis of iron-sulfur clusters in Escherichia coli. Biochem. J. 2008;409(2):535–543. doi: 10.1042/BJ20071166. [DOI] [PubMed] [Google Scholar]
- 53.Majtan T., Frerman F.E., Kraus J.P. Effect of cobalt on Escherichia coli metabolism and metalloporphyrin formation. Biometals. 2011;24(2):335–347. doi: 10.1007/s10534-010-9400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jomova K., Valko M. Advances in metal-induced oxidative stress and human disease. Toxicology. 2011;283(2–3):65–87. doi: 10.1016/j.tox.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 55.Labbe R.F., Vreman H.J., Stevenson D.K. Zinc protoporphyrin: a metabolite with a mission. Clin. Chem. 1999;45(12):2060–2072. [PubMed] [Google Scholar]
- 56.Chandrangsu P., Rensing C., Helmann J.D. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017;15(6):338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foster A.W., Osman D., Robinson N.J. Metal preferences and metallation. J. Biol. Chem. 2014;289(41):28095–28103. doi: 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borths E.L., Poolman B., Hvorup R.N., Locher K.P., Rees D.C. In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry. 2005;44(49):16301–16309. doi: 10.1021/bi0513103. [DOI] [PubMed] [Google Scholar]
- 59.Schauer K., Gouget B., Carriere M., Labigne A., de Reuse H. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 2007;63(4):1054–1068. doi: 10.1111/j.1365-2958.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- 60.Stoof J., Kuipers E.J., Klaver G., van Vliet A.H. An ABC transporter and a TonB ortholog contribute to helicobacter mustelae nickel and cobalt acquisition. Infect. Immun. 2010;78(10):4261–4267. doi: 10.1128/IAI.00365-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng J., Poduska B., Morton R.A., Finan T.M. An ABC-type cobalt transport system is essential for growth of Sinorhizobium meliloti at trace metal concentrations. J. Bacteriol. 2011;193(17):4405–4416. doi: 10.1128/JB.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roth J.R., Lawrence J.G., Rubenfield M., Kieffer-Higgins S., Church G.M. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 1993;175(11):3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rempel S., Stanek W.K., Slotboom D.J. ECF-type ATP-binding cassette transporters. Annu. Rev. Biochem. 2019;88:551–576. doi: 10.1146/annurev-biochem-013118-111705. [DOI] [PubMed] [Google Scholar]
- 64.Kirsch F., Eitinger T. Transport of nickel and cobalt ions into bacterial cells by S components of ECF transporters. Biometals. 2014;27(4):653–660. doi: 10.1007/s10534-014-9738-3. [DOI] [PubMed] [Google Scholar]
- 65.Bao Z., Qi X., Hong S., Xu K., He F., Zhang M., Chen J., Chao D., Zhao W., Li D., Wang J., Zhang P. Structure and mechanism of a group-I cobalt energy coupling factor transporter. Cell Res. 2017;27(5):675–687. doi: 10.1038/cr.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siche S., Neubauer O., Hebbeln P., Eitinger T. A bipartite S unit of an ECF-type cobalt transporter. Res. Microbiol. 2010;161(10):824–829. doi: 10.1016/j.resmic.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 67.Finkenwirth F., Sippach M., Pecina S.N., Gade M., Ruta J., Ricke A., Bondarenko E., Klare J.P., Zinke M., Lange S., Lange A., Steinhoff H.J., Eitinger T. Dynamic interactions of CbiN and CbiM trigger activity of a cobalt energy-coupling-factor transporter. Biochim. Biophys. Acta Biomembr. 1862;2020(2):183114. doi: 10.1016/j.bbamem.2019.183114. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y., Rodionov D.A., Gelfand M.S., Gladyshev V.N. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics. 2009;10:78. doi: 10.1186/1471-2164-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Degen O., Eitinger T. Substrate specificity of nickel/cobalt permeases: insights from mutants altered in transmembrane domains I and II. J. Bacteriol. 2002;184(13):3569–3577. doi: 10.1128/JB.184.13.3569-3577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Degen O., Kobayashi M., Shimizu S., Eitinger T. Selective transport of divalent cations by transition metal permeases: the Alcaligenes eutrophus HoxN and the Rhodococcus rhodochrous NhlF. Arch. Microbiol. 1999;171(3):139–145. doi: 10.1007/s002030050691. [DOI] [PubMed] [Google Scholar]
- 71.Komeda H., Kobayashi M., Shimizu S. A novel transporter involved in cobalt uptake. Proc. Natl. Acad. Sci. U. S. A. 1997;94(1):36–41. doi: 10.1073/pnas.94.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hebbeln P., Eitinger T. Heterologous production and characterization of bacterial nickel/cobalt permeases. FEMS Microbiol. Lett. 2004;230(1):129–135. doi: 10.1016/S0378-1097(03)00885-1. [DOI] [PubMed] [Google Scholar]
- 73.Hoffmann D., Gutekunst K., Klissenbauer M., Schulz-Friedrich R., Appel J. Mutagenesis of hydrogenase accessory genes of Synechocystis sp. PCC 6803. Additional homologues of hypA and hypB are not active in hydrogenase maturation. FEBS J. 2006;273(19):4516–4527. doi: 10.1111/j.1742-4658.2006.05460.x. [DOI] [PubMed] [Google Scholar]
- 74.T. Eitinger, Nickel, cobalt transport in prokaryotes, in: R.A. Scott (Ed.), Encycl. Inorg. Bioinorg. Chem., American Cancer Society2013, pp. 1–10.
- 75.Hmiel S.P., Snavely M.D., Miller C.G., Maguire M.E. Magnesium transport in Salmonella typhimurium: characterization of magnesium influx and cloning of a transport gene. J. Bacteriol. 1986;168(3):1444–1450. doi: 10.1128/jb.168.3.1444-1450.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Snavely M.D., Florer J.B., Miller C.G., Maguire M.E. Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA. MgtA, and MgtB systems, J Bacteriol. 1989;171(9):4761–4766. doi: 10.1128/jb.171.9.4761-4766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xia Y., Lundback A.K., Sahaf N., Nordlund G., Brzezinski P., Eshaghi S. Co2+ selectivity of Thermotoga maritima CorA and its inability to regulate Mg2+ homeostasis present a new class of CorA proteins. J. Biol. Chem. 2011;286(18):16525–16532. doi: 10.1074/jbc.M111.222166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maguire M.E. The structure of CorA: a Mg(2+)-selective channel. Curr. Opin. Struct. Biol. 2006;16(4):432–438. doi: 10.1016/j.sbi.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 79.Raimunda D., Long J.E., Padilla-Benavides T., Sassetti C.M., Arguello J.M. Differential roles for the Co(2+)/Ni(2+) transporting ATPases. CtpD and CtpJ, in Mycobacterium tuberculosis virulence, Mol Microbiol. 2014;91(1):185–197. doi: 10.1111/mmi.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raimunda D., Long J.E., Sassetti C.M., Arguello J.M. Role in metal homeostasis of CtpD, a Co(2)(+) transporting P(1B4)-ATPase of Mycobacterium smegmatis. Mol. Microbiol. 2012;84(6):1139–1149. doi: 10.1111/j.1365-2958.2012.08082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng C., Jia M., Gao M., Lu T., Li L., Zhou P. PmtA functions as a ferrous iron and cobalt efflux pump in Streptococcus suis. Emerg Microbes Infect. 2019;8(1):1254–1264. doi: 10.1080/22221751.2019.1660233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guan G., Pinochet-Barros A., Gaballa A., Patel S.J., Arguello J.M., Helmann J.D. PfeT, a P1B4 -type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Mol. Microbiol. 2015;98(4):787–803. doi: 10.1111/mmi.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Higgins K.A., Giedroc D. Insights into protein allostery in the CsoR/RcnR family of transcriptional repressors. Chem. Lett. 2014;43(1):20–25. doi: 10.1246/cl.130965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iwig J.S., Rowe J.L., Chivers P.T. Nickel homeostasis in Escherichia coli – the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol. Microbiol. 2006;62(1):252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 85.Iwig J.S., Chivers P.T. DNA recognition and wrapping by Escherichia coli RcnR. J. Mol. Biol. 2009;393(2):514–526. doi: 10.1016/j.jmb.2009.08.038. [DOI] [PubMed] [Google Scholar]
- 86.Huang H.-T., Maroney M.J. Ni(II) sensing by RcnR does not require an FrmR-like intersubunit linkage. Inorg. Chem. 2019;58(20):13639–13653. doi: 10.1021/acs.inorgchem.9b01096. [DOI] [PubMed] [Google Scholar]
- 87.Huang H.-T., Bobst C.E., Iwig J.S., Chivers P.T., Kaltashov I.A., Maroney M.J. Co(II) and Ni(II) binding of the Escherichia coli transcriptional repressor RcnR orders its N terminus, alters helix dynamics, and reduces DNA affinity. J. Biol. Chem. 2018;293(1):324–332. doi: 10.1074/jbc.RA117.000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dokpikul T., Chaoprasid P., Saninjuk K., Sirirakphaisarn S., Johnrod J., Nookabkaew S., Sukchawalit R., Mongkolsuk S. Regulation of the cobalt/nickel efflux operon dmeRF in Agrobacterium tumefaciens and a link between the iron-sensing regulator RirA and cobalt/nickel resistance. Appl. Environ. Microbiol. 2016;82(15):4732–4742. doi: 10.1128/AEM.01262-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rubio-Sanz L., Prieto R.I., Imperial J., Palacios J.M., Brito B. Functional and expression analysis of the metal-inducible dmeRF system from Rhizobium leguminosarum bv. viciae. Appl. Environ. Microbiol. 2013;79(20):6414–6422. doi: 10.1128/AEM.01954-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Osman D., Foster A.W., Chen J., Svedaite K., Steed J.W., Lurie-Luke E., Huggins T.G., Robinson N.J. Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat. Commun. 2017;8(1):1884. doi: 10.1038/s41467-017-02085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z., Song X., Wang J., Bai X., Gao E., Wei G. Nickel and cobalt resistance properties of Sinorhizobium meliloti isolated from Medicago lupulina growing in gold mine tailing. PeerJ. 2018;6 doi: 10.7717/peerj.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Osman D., Piergentili C., Chen J., Chakrabarti B., Foster A.W., Lurie-Luke E., Huggins T.G., Robinson N.J. Generating a metal-responsive transcriptional regulator to test what confers metal sensing in cells. J. Biol. Chem. 2015;290(32):19806–19822. doi: 10.1074/jbc.M115.663427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Osman D., Martini M.A., Foster A.W., Chen J., Scott A.J., Morton R.J., Steed J.W., Lurie-Luke E., Huggins T.G., Lawrence A.D. Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat. Chem. Biol. 2019;15(3):241–249. doi: 10.1038/s41589-018-0211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rutherford J.C., Cavet J.S., Robinson N.J. Cobalt-dependent transcriptional switching by a dual-effector MerR-like protein regulates a cobalt-exporting variant CPx-type ATPase. J. Biol. Chem. 1999;274(36):25827–25832. doi: 10.1074/jbc.274.36.25827. [DOI] [PubMed] [Google Scholar]
- 95.Philips S.J., Canalizo-Hernandez M., Yildirim I., Schatz G.C., Mondragón A., O'Halloran T.V. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science. 2015;349(6250):877–881. doi: 10.1126/science.aaa9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Patterson C.J., Pernil R., Dainty S.J., Chakrabarti B., Henry C.E., Money V.A., Foster A.W., Robinson N.J. Co(II)-detection does not follow KCo(II) gradient: channelling in Co(II)-sensing. Metallomics. 2013;5(4):352–362. doi: 10.1039/c3mt20241k. [DOI] [PubMed] [Google Scholar]
- 97.Maggio-Hall L.A., Claas K.R., Escalante-Semerena J.C. The last step in coenzyme B(12) synthesis is localized to the cell membrane in bacteria and archaea. Microbiology. 2004;150(Pt 5):1385–1395. doi: 10.1099/mic.0.26952-0. [DOI] [PubMed] [Google Scholar]
- 98.Zayas C.L., Claas K., Escalante-Semerena J.C. The CbiB protein of Salmonella enterica is an integral membrane protein involved in the last step of the de novo corrin ring biosynthetic pathway. J. Bacteriol. 2007;189(21):7697–7708. doi: 10.1128/JB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Campbell D.R., Chapman K.E., Waldron K.J., Tottey S., Kendall S., Cavallaro G., Andreini C., Hinds J., Stoker N.G., Robinson N.J., Cavet J.S. Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent. J. Biol. Chem. 2007;282(44):32298–32310. doi: 10.1074/jbc.M703451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pennella M.A., Shokes J.E., Cosper N.J., Scott R.A., Giedroc D.P. Structural elements of metal selectivity in metal sensor proteins. Proc. Nat. Acad. Sci. 2003;100(7):3713–3718. doi: 10.1073/pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Osman D., Cavet J.S. Bacterial metal-sensing proteins exemplified by ArsR–SmtB family repressors. Nat. Prod. Rep. 2010;27(5):668–680. doi: 10.1039/b906682a. [DOI] [PubMed] [Google Scholar]
- 102.Singh V.K., Xiong A., Usgaard T.R., Chakrabarti S., Deora R., Misra T.K., Jayaswal R.K. ZntR is an autoregulatory protein and negatively regulates the chromosomal zinc resistance operon znt of Staphylococcus aureus. Mol. Microbiol. 1999;33(1):200–207. doi: 10.1046/j.1365-2958.1999.01466.x. [DOI] [PubMed] [Google Scholar]
- 103.Capdevila D.A., Braymer J.J., Edmonds K.A., Wu H., Giedroc D.P. Entropy redistribution controls allostery in a metalloregulatory protein. Proc. Natl. Acad. Sci. U. S. A. 2017;114(17):4424–4429. doi: 10.1073/pnas.1620665114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Capdevila D.A., Edmonds K.A., Campanello G.C., Wu H., Gonzalez-Gutierrez G., Giedroc D.P. Functional role of solvent entropy and conformational entropy of metal binding in a dynamically driven allosteric system. J. Am. Chem. Soc. 2018;140(29):9108–9119. doi: 10.1021/jacs.8b02129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lavrov K.V., Shemyakina A.O., Grechishnikova E.G., Novikov A.D., Derbikov D.D., Kalinina T.I., Yanenko A.S. New cblA gene participates in regulation of cobalt-dependent transcription of nitrile hydratase genes in Rhodococcus rhodochrous. Res. Microbiol. 2018;169(4):227–236. doi: 10.1016/j.resmic.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 106.Lavrov K.V., Shemyakina A.O., Grechishnikova E.G., Novikov A.D., Kalinina T.I., Yanenko A.S. In vivo metal selectivity of metal-dependent biosynthesis of cobalt-type nitrile hydratase in Rhodococcus bacteria: a new look at the nitrile hydratase maturation mechanism? Metallomics. 2019;11(6):1162–1171. doi: 10.1039/c8mt00129d. [DOI] [PubMed] [Google Scholar]
- 107.Prasad S., Bhalla T.C. Nitrile hydratases (NHases): at the interface of academia and industry. Biotechnol. Adv. 2010;28(6):725–741. doi: 10.1016/j.biotechadv.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 108.Miyanaga A., Fushinobu S., Ito K., Wakagi T. Crystal structure of cobalt-containing nitrile hydratase. Biochem. Biophys. Res. Commun. 2001;288(5):1169–1174. doi: 10.1006/bbrc.2001.5897. [DOI] [PubMed] [Google Scholar]
- 109.Goldberg M., Pribyl T., Juhnke S., Nies D.H. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 1999;274(37):26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 110.Nies D.H. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J. Bacteriol. 1992;174(24):8102. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grosse C., Grass G., Anton A., Franke S., Santos A.N., Lawley B., Brown N.L., Nies D.H. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J. Bacteriol. 1999;181(8):2385–2393. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Große C., Anton A., Hoffmann T., Franke S., Schleuder G., Nies D.H. Identification of a regulatory pathway that controls the heavy-metal resistance system Czc via promoter czcNp in Ralstonia metallidurans. Arch. Microbiol. 2004;182(2):109–118. doi: 10.1007/s00203-004-0670-8. [DOI] [PubMed] [Google Scholar]
- 113.van der Lelie D., Schwuchow T., Schwidetzky U., Wuertz S., Baeyens W., Mergeay M., Nies D.H. Two-component regulatory system involved in transcriptional control of heavy-metal homoeostasis in Alcaligenes eutrophus. Mol. Microbiol. 1997;23(3):493–503. doi: 10.1046/j.1365-2958.1997.d01-1866.x. [DOI] [PubMed] [Google Scholar]
- 114.Wang D., Chen W., Huang S., He Y., Liu X., Hu Q., Wei T., Sang H., Gan J., Chen H. Structural basis of Zn(II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa. PLoS Pathog. 2017;13(7) doi: 10.1371/journal.ppat.1006533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Perron K., Caille O., Rossier C., van Delden C., Dumas J.-L., Köhler T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279(10):8761–8768. doi: 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- 116.Grass G., Große C., Nies D.H. Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. strain CH34. J. Bacteriol. 2000;182(5):1390–1398. doi: 10.1128/jb.182.5.1390-1398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Trepreau J., Girard E., Maillard A.P., de Rosny E., Petit-Haertlein I., Kahn R., Covès J. Structural basis for metal sensing by CnrX. J. Mol. Biol. 2011;408(4):766–779. doi: 10.1016/j.jmb.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 118.Grosse C., Friedrich S., Nies D.H. Contribution of extracytoplasmic function sigma factors to transition metal homeostasis in Cupriavidus metallidurans strain CH34. J. Mol. Microbiol. Biotechnol. 2007;12(3–4):227–240. doi: 10.1159/000099644. [DOI] [PubMed] [Google Scholar]
- 119.Grass G., Fricke B., Nies D.H. Control of expression of a periplasmic nickel efflux pump by periplasmic nickel concentrations. Biometals. 2005;18(4):437–448. doi: 10.1007/s10534-005-3718-6. [DOI] [PubMed] [Google Scholar]
- 120.Trepreau J., Grosse C., Mouesca J.-M., Sarret G., Girard E., Petit-Haertlein I., Kuennemann S., Desbourdes C., de Rosny E., Maillard A.P., Nies D.H., Covès J. Metal sensing and signal transduction by CnrX from Cupriavidus metallidurans CH34: role of the only methionine assessed by a functional, spectroscopic, and theoretical study. Metallomics. 2014;6(2):263–273. doi: 10.1039/c3mt00248a. [DOI] [PubMed] [Google Scholar]
- 121.Long F., Su C.C., Zimmermann M.T., Boyken S.E., Rajashankar K.R., Jernigan R.L., Yu E.W. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467(7314):484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bozzi A.T., Bane L.B., Weihofen W.A., McCabe A.L., Singharoy A., Chipot C.J., Schulten K., Gaudet R. Conserved methionine dictates substrate preference in Nramp-family divalent metal transporters, Proc. Natl. Acad. Sci. U. S. A. 2016;201607734 doi: 10.1073/pnas.1607734113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lawton T.J., Kenney G.E., Hurley J.D., Rosenzweig A.C. The CopC family: structural and bioinformatic insights into a diverse group of periplasmic copper binding proteins. Biochemistry. 2016;55(15):2278–2290. doi: 10.1021/acs.biochem.6b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chivers P.T. The Royal Society of Chemistry. 2014. Cobalt and Nickel, binding, transport and storage of metal ions in biological cells; pp. 381–428. [Google Scholar]
- 125.Vergalli J., Bodrenko I.V., Masi M., Moynié L., Acosta-Gutiérrez S., Naismith J.H., Davin-Regli A., Ceccarelli M., van den Berg B., Winterhalter M., Pagès J.-M. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 2020;18(3):164–176. doi: 10.1038/s41579-019-0294-2. [DOI] [PubMed] [Google Scholar]
- 126.Maillard A.P., Künnemann S., Große C., Volbeda A., Schleuder G., Petit-Härtlein I., de Rosny E., Nies D.H., Covès J. Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding. Metallomics. 2015;7(4):622–631. doi: 10.1039/c4mt00293h. [DOI] [PubMed] [Google Scholar]
- 127.Morgan M.T., Nguyen L.A.H., Hancock H.L., Fahrni C.J. Glutathione limits aquacopper(I) to sub-femtomolar concentrations through cooperative assembly of a tetranuclear cluster. J. Biol. Chem. 2017;292(52):21558–21567. doi: 10.1074/jbc.M117.817452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pittman M.S., Robinson H.C., Poole R.K. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 2005;280(37):32254–32261. doi: 10.1074/jbc.M503075200. [DOI] [PubMed] [Google Scholar]
- 129.Chivers P.T., Benanti E.L., Heil-Chapdelaine V., Iwig J.S., Rowe J.L. Identification of Ni-(L-His) 2 as a substrate for NikABCDE-dependent nickel uptake in Escherichia coli. Metallomics. 2012;4(10):1043–1050. doi: 10.1039/c2mt20139a. [DOI] [PubMed] [Google Scholar]
- 130.Nairn Brittany L., Lonergan Zachery R., Wang J., Braymer Joseph J., Zhang Y., Calcutt M.W., Lisher John P., Gilston Benjamin A., Chazin Walter J., de Crécy-Lagard V., Giedroc David P., Skaar Eric P. The response of acinetobacter baumannii to zinc starvation, cell host & microbe. 2016;19(6):826–836. doi: 10.1016/j.chom.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Murphy J.T., Bruinsma J.J., Schneider D.L., Collier S., Guthrie J., Chinwalla A., Robertson J.D., Mardis E.R., Kornfeld K. Histidine protects against zinc and nickel toxicity in Caenorhabditis elegans. PLoS Genet. 2011;7(3) doi: 10.1371/journal.pgen.1002013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Foster A.W., Pernil R., Patterson C.J., Scott A.J.P., Pålsson L.-O., Pal R., Cummins I., Chivers P.T., Pohl E., Robinson N.J. A tight tunable range for Ni(II) sensing and buffering in cells. Nat. Chem. Biol. 2017;13(4):409–414. doi: 10.1038/nchembio.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mandal M., Breaker R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell. Biol. 2004;5(6):451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 134.Gold L. SELEX: how it happened and where it will go. J. Mol. Evol. 2015;81(5):140–143. doi: 10.1007/s00239-015-9705-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 136.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 137.Lorsch J.R., Szostak J.W. In vitro selection of RNA aptamers specific for cyanocobalamin. Biochemistry. 1994;33:973–982. doi: 10.1021/bi00170a016. [DOI] [PubMed] [Google Scholar]
- 138.Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9(9):1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 139.Nou X., Kadner R.J. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc. Natl. Acad. Sci. U. S. A. 2000;97(13):7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nahvi A., Barrick J.E., Breaker R.R. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lussier A., Bastet L., Chauvier A., Lafontaine D.A. A kissing loop is important for btuB riboswitch ligand sensing and regulatory control. J. Biol. Chem. 2015;290(44):26739–26751. doi: 10.1074/jbc.M115.684134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johnson J.E., Jr., Reyes F.E., Polaski J.T., Batey R.T. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492(7427):133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Choudhary P.K., Gallo S., Sigel R.K.O. Tb3+-cleavage assays reveal specific Mg2+ binding sites necessary to pre-fold the btuB riboswitch for AdoCbl binding. Front Chem. 2017;5:10. doi: 10.3389/fchem.2017.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Perdrizet G.A., 2nd, Artsimovitch I., Furman R., Sosnick T.R., Pan T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Nat. Acad. Sci. 2012;109(9):3323–3328. doi: 10.1073/pnas.1113086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mellin J.R., Koutero M., Dar D., Nahori M.-A., Sorek R., Cossart P. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science. 2014;345:940–943. doi: 10.1126/science.1255083. [DOI] [PubMed] [Google Scholar]
- 146.Biedendieck R., Malten M., Barg H., Bunk B., Martens J.-H., Deery E., Leech H., Warren M.J., Jahn D. Metabolic engineering of cobalamin (vitamin B12) production in Bacillus megaterium. Microb. Biotechnol. 2010;3:24–37. doi: 10.1111/j.1751-7915.2009.00125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wedekind J.E., Dutta D., Belashov I.A., Jenkins J.L. Metalloriboswitches: RNA-based inorganic ion sensors that regulate genes. J. Biol. Chem. 2017;292:9441–9450. doi: 10.1074/jbc.R117.787713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Furukawa K., Ramesh A., Zhou Z., Weinberg Z., Vallery T., Winkler W.C., Breaker R.R. Bacterial riboswitches cooperatively bind Ni2+ or Co2+ ions and control expression of heavy metal transporters. Mol. Cell. 2015;57:1088–1098. doi: 10.1016/j.molcel.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cavet J.S., Meng W., Pennella M.A., Appelhoff R.J., Giedroc D.P., Robinson N.J. A nickel-cobalt-sensing ArsR-SmtB family repressor: contributions of cytosol and effector binding sites to metal selectivity. J. Biol. Chem. 2002;277(41):38441–38448. doi: 10.1074/jbc.M207677200. [DOI] [PubMed] [Google Scholar]
- 150.Price I.R., Gaballa A., Ding F., Helmann J.D., Ke A. Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol. Cell. 2015;57:1110–1123. doi: 10.1016/j.molcel.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu J., Cotruvo J.A., Jr. The czcD. NiCo) riboswitch responds to iron(IIBiochemistry. 2020;59:1508–1516. doi: 10.1021/acs.biochem.0c00074. [DOI] [PubMed] [Google Scholar]
- 152.Al-Karadaghi S., Franco R., Hansson M., Shelnutt J.A., Isaya G., Ferreira G.C. Chelatases: distort to select? Trends Biochem. Sci. 2006;31(3):135–142. doi: 10.1016/j.tibs.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.O’Halloran T.V., Culotta V.C. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 2000;275(33):25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 154.Capdevila Daiana A., Edmonds Katherine A., Giedroc David P. Metallochaperones and metalloregulation in bacteria. Essays Biochem. 2017;61(2):177–200. doi: 10.1042/EBC20160076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Debussche L., Couder M., Thibaut D., Cameron B., Crouzet J., Blanche F. Assay, purification, and characterization of cobaltochelatase, a unique complex enzyme catalyzing cobalt insertion in hydrogenobyrinic acid a,c-diamide during coenzyme B12 biosynthesis in Pseudomonas denitrificans. J. Bacteriol. 1992;174(22):7445–7451. doi: 10.1128/jb.174.22.7445-7451.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Young T.R., Martini M.A., Osman D., Morton R.J., Deery E., Warren M.J., Robinson N.J. 2020. Calculating Metalation in Cells Reveals CobW Acquires CoII for Vitamin B12 Biosynthesis upon Binding Nucleotide, bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Brindley A.A., Raux E., Leech H.K., Schubert H.L., Warren M.J. A story of chelatase evolution: identification and characterization of a small 13-15-kDa “ancestral” cobaltochelatase (CbiXS) in the archaea. J. Biol. Chem. 2003;278(25):22388–22395. doi: 10.1074/jbc.M302468200. [DOI] [PubMed] [Google Scholar]
- 158.Raux E., Thermes C., Heathcote P., Rambach A., Warren M.J. A role for Salmonella typhimurium cbiK in cobalamin (vitamin B12) and siroheme biosynthesis. J. Bacteriol. 1997;179(10):3202–3212. doi: 10.1128/jb.179.10.3202-3212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schubert H.L., Raux E., Wilson K.S., Warren M.J. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry. 1999;38(33):10660–10669. doi: 10.1021/bi9906773. [DOI] [PubMed] [Google Scholar]
- 160.Beck R., Raux E., Thermes C., Rambach A., Warren M. CbiX: a novel metal-binding protein involved in sirohaem biosynthesis in Bacillus megaterium. Biochem. Soc. Trans. 1997;25(1):77S. doi: 10.1042/bst025077s. [DOI] [PubMed] [Google Scholar]
- 161.Leech H.K., Raux E., McLean K.J., Munro A.W., Robinson N.J., Borrelly G.P., Malten M., Jahn D., Rigby S.E., Heathcote P., Warren M.J. Characterization of the cobaltochelatase CbiXL: evidence for a 4Fe-4S center housed within an MXCXXC motif. J. Biol. Chem. 2003;278(43):41900–41907. doi: 10.1074/jbc.M306112200. [DOI] [PubMed] [Google Scholar]
- 162.Yin J., Xu L.X., Cherney M.M., Raux-Deery E., Bindley A.A., Savchenko A., Walker J.R., Cuff M.E., Warren M.J., James M.N. Crystal structure of the vitamin B12 biosynthetic cobaltochelatase. CbiXS, from Archaeoglobus fulgidus, J Struct Funct Genomics. 2006;7(1):37–50. doi: 10.1007/s10969-006-9008-x. [DOI] [PubMed] [Google Scholar]
- 163.Leech H.K., Raux-Deery E., Heathcote P., Warren M.J. Production of cobalamin and sirohaem in Bacillus megaterium: an investigation into the role of the branchpoint chelatases sirohydrochlorin ferrochelatase (SirB) and sirohydrochlorin cobalt chelatase (CbiX) Biochem. Soc. Trans. 2002;30(4):610–613. doi: 10.1042/bst0300610. [DOI] [PubMed] [Google Scholar]
- 164.Raux E., Leech H.K., Beck R., Schubert H.L., Santander P.J., Roessner C.A., Scott A.I., Martens J.H., Jahn D., Thermes C., Rambach A., Warren M.J. Identification and functional analysis of enzymes required for precorrin-2 dehydrogenation and metal ion insertion in the biosynthesis of sirohaem and cobalamin in Bacillus megaterium. Biochem. J. 2003;370(Pt 2):505–516. doi: 10.1042/BJ20021443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moore S.J., Sowa S.T., Schuchardt C., Deery E., Lawrence A.D., Ramos J.V., Billig S., Birkemeyer C., Chivers P.T., Howard M.J., Rigby S.E., Layer G., Warren M.J. Elucidation of the biosynthesis of the methane catalyst coenzyme F430. Nature. 2017;543(7643):78–82. doi: 10.1038/nature21427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zheng K., Ngo P.D., Owens V.L., Yang X.P., Mansoorabadi S.O. The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science. 2016;354(6310):339–342. doi: 10.1126/science.aag2947. [DOI] [PubMed] [Google Scholar]
- 167.Romao C.V., Ladakis D., Lobo S.A., Carrondo M.A., Brindley A.A., Deery E., Matias P.M., Pickersgill R.W., Saraiva L.M., Warren M.J. Evolution in a family of chelatases facilitated by the introduction of active site asymmetry and protein oligomerization. Proc. Natl. Acad. Sci. U. S. A. 2011;108(1):97–102. doi: 10.1073/pnas.1014298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shepherd M., Dailey T.A., Dailey H.A. A new class of [2Fe-2S]-cluster-containing protoporphyrin (IX) ferrochelatases. Biochem. J. 2006;397(1):47–52. doi: 10.1042/BJ20051967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sirijovski N., Mamedov F., Olsson U., Styring S., Hansson M. Rhodobacter capsulatus magnesium chelatase subunit BchH contains an oxygen sensitive iron-sulfur cluster. Arch. Microbiol. 2007;188(6):599–608. doi: 10.1007/s00203-007-0282-1. [DOI] [PubMed] [Google Scholar]
- 170.Bali S., Rollauer S., Roversi P., Raux-Deery E., Lea S.M., Warren M.J., Ferguson S.J. Identification and characterization of the ‘missing’ terminal enzyme for siroheme biosynthesis in alpha-proteobacteria. Mol. Microbiol. 2014;92(1):153–163. doi: 10.1111/mmi.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Leech H.K. Queen Mary University of London; 2004. The Biosynthesis of Sirohydrochlorin and its Transformation into Sirohaem and Vitamin B12. [Google Scholar]
- 172.Debussche L., Couder M., Thibaut D., Cameron B., Crouzet J., Blanche F. Assay, purification, and characterization of cobaltochelatase, a unique complex enzyme catalyzing cobalt insertion in hydrogenobyrinic acid a,C-diamide during coenzyme B-12 biosynthesis in pseudomonas-denitrificans. J. Bacteriol. 1992;174(22):7445–7451. doi: 10.1128/jb.174.22.7445-7451.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lundqvist J., Elmlund D., Heldt D., Deery E., Soderberg C.A., Hansson M., Warren M., Al-Karadaghi S. The AAA(+) motor complex of subunits CobS and CobT of cobaltochelatase visualized by single particle electron microscopy. J. Struct. Biol. 2009;167(3):227–234. doi: 10.1016/j.jsb.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 174.Gibson L.C., Willows R.D., Kannangara C.G., von Wettstein D., Hunter C.N. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1995;92(6):1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Reid J.D., Hunter C.N. Magnesium-dependent ATPase activity and cooperativity of magnesium chelatase from Synechocystis sp. PCC6803. J Biol Chem. 2004;279(26):26893–26899. doi: 10.1074/jbc.M400958200. [DOI] [PubMed] [Google Scholar]
- 176.Chen X., Pu H., Fang Y., Wang X., Zhao S., Lin Y., Zhang M., Dai H.E., Gong W., Liu L. Crystal structure of the catalytic subunit of magnesium chelatase. Nat Plants. 2015;1:15125. doi: 10.1038/nplants.2015.125. [DOI] [PubMed] [Google Scholar]
- 177.Gao Y.S., Wang Y.L., Wang X., Liu L. Hexameric structure of the ATPase motor subunit of magnesium chelatase in chlorophyll biosynthesis. Protein Sci. 2020;29(4):1040–1046. doi: 10.1002/pro.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fodje M.N., Hansson A., Hansson M., Olsen J.G., Gough S., Willows R.D., Al-Karadaghi S. Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase. J. Mol. Biol. 2001;311(1):111–122. doi: 10.1006/jmbi.2001.4834. [DOI] [PubMed] [Google Scholar]
- 179.Adams N.B., Vasilev C., Brindley A.A., Hunter C.N. Nanomechanical and thermophoretic analyses of the nucleotide-dependent interactions between the AAA(+) subunits of magnesium chelatase. J. Am. Chem. Soc. 2016;138(20):6591–6597. doi: 10.1021/jacs.6b02827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Farmer D.A., Brindley A.A., Hitchcock A., Jackson P.J., Johnson B., Dickman M.J., Hunter C.N., Reid J.D., Adams N.B.P. The ChlD subunit links the motor and porphyrin binding subunits of magnesium chelatase. Biochem. J. 2019;476(13):1875–1887. doi: 10.1042/BCJ20190095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Crouzet J., Levy-Schil S., Cameron B., Cauchois L., Rigault S., Rouyez M.C., Blanche F., Debussche L., Thibaut D. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. Journal of Bacteriology. 1991;173(19):6074–6087. doi: 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Leipe D.D., Wolf Y.I., Koonin E.V., Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 183.Haas C.E., Rodionov D.A., Kropat J., Malasarn D., Merchant S.S., de Crécy-Lagard V. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics. 2009;10(1):470. doi: 10.1186/1471-2164-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Padovani D., Banerjee R. A G-protein editor gates coenzyme B12 loading and is corrupted in methylmalonic aciduria. Proc. Natl. Acad. Sci. 2009;106(51):21567–21572. doi: 10.1073/pnas.0908106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Maier T., Jacobi A., Sauter M., Böck A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J. Bacteriol. 1993;175(3):630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lee M.H., Mulrooney S.B., Renner M.J., Markowicz Y., Hausinger R.P. Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J. Bacteriol. 1992;174(13):4324–4330. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Maier R.J., Benoit S.L., Seshadri S. Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori. Biometals. 2007;20(3–4):655–664. doi: 10.1007/s10534-006-9061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li Y., Zamble D.B. Nickel homeostasis and nickel regulation: an overview. Chem. Rev. 2009;109(10):4617–4643. doi: 10.1021/cr900010n. [DOI] [PubMed] [Google Scholar]
- 189.Nojiri M., Yohda M., Odaka M., Matsushita Y., Tsujimura M., Yoshida T., Dohmae N., Takio K., Endo I. Functional expression of nitrile hydratase in Escherichia coli: requirement of a nitrile hydratase activator and post-translational modification of a ligand cysteine1. The Journal of Biochemistry. 1999;125(4):696–704. doi: 10.1093/oxfordjournals.jbchem.a022339. [DOI] [PubMed] [Google Scholar]
- 190.Lu J., Zheng Y., Yamagishi H., Odaka M., Tsujimura M., Maeda M., Endo I. Motif CXCC in nitrile hydratase activator is critical for NHase biogenesis in vivo. FEBS Lett. 2003;553(3):391–396. doi: 10.1016/s0014-5793(03)01070-6. [DOI] [PubMed] [Google Scholar]
- 191.Blaby-Haas C.E., Flood J.A., Crécy-Lagard V.D., Zamble D.B. YeiR: a metal-binding GTPase from Escherichia coli involved in metal homeostasis. Metallomics. 2012;4(5):488–497. doi: 10.1039/c2mt20012k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sydor A.M., Jost M., Ryan K.S., Turo K.E., Douglas C.D., Drennan C.L., Zamble D.B. Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases. Biochemistry. 2013;52(10):1788–1801. doi: 10.1021/bi301600z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Jordan M.R., Wang J., Weiss A., Skaar E.P., Capdevila D.A., Giedroc D.P. Mechanistic insights into the metal-dependent activation of ZnII-dependent metallochaperones. Inorg. Chem. 2019;58(20):13661–13672. doi: 10.1021/acs.inorgchem.9b01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chandrangsu P., Huang X., Gaballa A., Helmann J.D. Bacillus subtilis FolE is sustained by the ZagA zinc metallochaperone and the alarmone ZTP under conditions of zinc deficiency. Mol. Microbiol. 2019;112(3):751–765. doi: 10.1111/mmi.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Heldt D., Lawrence A.D., Lindenmeyer M., Deery E., Heathcote P., Rigby S.E., Warren M.J. Aerobic synthesis of vitamin B12: ring contraction and cobalt chelation. Biochem. Soc. Trans. 2005;33(4):815–819. doi: 10.1042/BST0330815. [DOI] [PubMed] [Google Scholar]
- 196.Bourne H.R., Sanders D.A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 197.Xiao Z., Wedd A.G. The challenges of determining metal–protein affinities. Nat. Prod. Rep. 2010;27(5):768–789. doi: 10.1039/b906690j. [DOI] [PubMed] [Google Scholar]
- 198.Xiao Z., Brose J., Schimo S., Ackland S.M., La Fontaine S., Wedd A.G. Unification of the copper(I) binding affinities of the Metallo-chaperones Atx1, Atox1, and related proteins: Detection Probes And Affinity Standards. J. Biol. Chem. 2011;286(13):11047–11055. doi: 10.1074/jbc.M110.213074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wijekoon C.J.K., Ukuwela A.A., Wedd A.G., Xiao Z. Evaluation of employing poly-lysine tags versus poly-histidine tags for purification and characterization of recombinant copper-binding proteins. J. Inorg. Biochem. 2016;162:286–294. doi: 10.1016/j.jinorgbio.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 200.Young T.R., Wedd A.G., Xiao Z. Evaluation of cu(i) binding to the E2 domain of the amyloid precursor protein – a lesson in quantification of metal binding to proteins via ligand competition. Metallomics. 2018;10(1):108–119. doi: 10.1039/c7mt00291b. [DOI] [PubMed] [Google Scholar]
- 201.Irving H., Williams R. Order of stability of metal complexes. Nature. 1948;162(4123):746–747. [Google Scholar]
- 202.Tottey S., Waldron K.J., Firbank S.J., Reale B., Bessant C., Sato K., Cheek T.R., Gray J., Banfield M.J., Dennison C., Robinson N.J. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455(7216):1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 203.Fulda S., Huang F., Nilsson F., Hagemann M., Norling B. Proteomics of synechocystis sp. strain PCC 6803. Eur. J. Biochem. 2000;267(19):5900–5907. doi: 10.1046/j.1432-1327.2000.01642.x. [DOI] [PubMed] [Google Scholar]
- 204.Robinson C., Bolhuis A. Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2004;1694(1):135–147. doi: 10.1016/j.bbamcr.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 205.Waldron K.J., Firbank S.J., Dainty S.J., Pérez-Rama M., Tottey S., Robinson N.J. Structure and metal loading of a soluble periplasm cuproprotein. J. Biol. Chem. 2010;285(42):32504–32511. doi: 10.1074/jbc.M110.153080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Moomaw E.W., Angerhofer A., Moussatche P., Ozarowski A., García-Rubio I., Richards N.G. Metal dependence of oxalate decarboxylase activity. Biochemistry. 2009;48(26):6116–6125. doi: 10.1021/bi801856k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rae T.D., Schmidt P.J., Pufahl R.A., Culotta V.C., Halloran O.V.T. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284(5415):805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 208.Finney L.A., O’Halloran T.V. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300(5621):931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 209.Outten C.E., Thomas Halloran V. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292(5526):2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 210.Carter K.P., Young A.M., Palmer A.E. Fluorescent sensors for measuring metal ions in living systems. Chem. Rev. 2014;114(8):4564–4601. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Giedroc D.P., Arunkumar A.I. Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Transactions. 2007;29:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 212.Osman D., Piergentili C., Chen J., Sayer L.N., Usón I., Huggins T.G., Robinson N.J., Pohl E. The effectors and sensory sites of formaldehyde-responsive regulator FrmR and metal-sensing variant. J. Biol. Chem. 2016;291(37):19502–19516. doi: 10.1074/jbc.M116.745174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ikeda J.S., Janakiraman A., Kehres D.G., Maguire M.E., Slauch J.M. Transcriptional regulation of sitABCD of Salmonella enterica serovar typhimurium by MntR and Fur. J. Bacteriol. 2005;187(3):912–922. doi: 10.1128/JB.187.3.912-922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Osman D., Waldron K.J., Denton H., Taylor C.M., Grant A.J., Mastroeni P., Robinson N.J., Cavet J.S. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 2010;285(33):25259–25268. doi: 10.1074/jbc.M110.145953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wegner S.V., Arslan H., Sunbul M., Yin J., He C. Dynamic copper(I) imaging in mammalian cells with a genetically encoded fluorescent copper(I) sensor. J. Am. Chem. Soc. 2010;132(8):2567–2569. doi: 10.1021/ja9097324. [DOI] [PubMed] [Google Scholar]
- 216.Qiao W., Mooney M., Bird A.J., Winge D.R., Eide D.J. Zinc binding to a regulatory zinc-sensing domain monitored <em>in vivo</em> by using FRET. Proc. Natl. Acad. Sci. 2006;103(23):8674–8679. doi: 10.1073/pnas.0600928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Da Silva J.F., Williams R.J.P. Oxford University Press; 2001. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. [Google Scholar]
- 218.Morgan M.T., Yang B., Harankhedkar S., Nabatilan A., Bourassa D., McCallum A.M., Sun F., Wu R., Forest C.R., Fahrni C.J. Stabilization of aliphatic phosphines by auxiliary phosphine sulfides offers zeptomolar affinity and unprecedented selectivity for probing biological CUI. Angew. Chem. Int. Ed. 2018;57(31):9711–9715. doi: 10.1002/anie.201804072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yuen M.H., Fong Y.H., Nim Y.S., Lau P.H., Wong K.-B. Structural insights into how GTP-dependent conformational changes in a metallochaperone UreG facilitate urease maturation. Proc. Natl. Acad. Sci. 2017;114(51):E10890–E10898. doi: 10.1073/pnas.1712658114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fong Y.H., Wong H.C., Yuen M.H., Lau P.H., Chen Y.W., Wong K.-B. Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of helicobacter pylori urease. PLoS Biol. 2013;11(10) doi: 10.1371/journal.pbio.1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gasper R., Scrima A., Wittinghofer A. Structural insights into HypB, a GTP-binding protein that regulates metal binding. J. Biol. Chem. 2006;281(37):27492–27502. doi: 10.1074/jbc.M600809200. [DOI] [PubMed] [Google Scholar]
- 222.Sydor A.M., Lebrette H., Ariyakumaran R., Cavazza C., Zamble D.B. Relationship between Ni(II) and Zn(II) coordination and nucleotide binding by the helicobacter pylori [NiFe]-Hydrogenase and urease maturation factor HypB. J. Biol. Chem. 2014;289(7):3828–3841. doi: 10.1074/jbc.M113.502781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lacasse M.J., Douglas C.D., Zamble D.B. Mechanism of selective nickel transfer from HypB to HypA, Escherichia coli [NiFe]-Hydrogenase accessory proteins. Biochemistry. 2016;55(49):6821–6831. doi: 10.1021/acs.biochem.6b00706. [DOI] [PubMed] [Google Scholar]
- 224.Yang X., Li H., Lai T.-P., Sun H. UreE-UreG complex facilitates nickel transfer and Preactivates GTPase of UreG in helicobacter pylori. J. Biol. Chem. 2015;290(20):12474–12485. doi: 10.1074/jbc.M114.632364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Padovani D., Labunska T., Banerjee R. Energetics of interaction between the G-protein chaperone, MeaB, and B12-dependent Methylmalonyl-CoA Mutase. J. Biol. Chem. 2006;281(26):17838–17844. doi: 10.1074/jbc.M600047200. [DOI] [PubMed] [Google Scholar]
- 226.Lofgren M., Padovani D., Koutmos M., Banerjee R. A switch III motif relays signaling between a B12 enzyme and its G-protein chaperone. Nat. Chem. Biol. 2013;9(9):535–539. doi: 10.1038/nchembio.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Khil P.P., Obmolova G., Teplyakov A., Howard A.J., Gilliland G.L., Camerini-Otero R.D. Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function. Proteins: Structure, Function, and Bioinformatics. 2004;54(2):371–374. doi: 10.1002/prot.10430. [DOI] [PubMed] [Google Scholar]
- 228.Verstraeten N., Fauvart M., Versées W., Michiels J. The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 2011;75(3):507–542. doi: 10.1128/MMBR.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Watanabe S., Kawashima T., Nishitani Y., Kanai T., Wada T., Inaba K., Atomi H., Imanaka T., Miki K. Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer. Proc. Natl. Acad. Sci. 2015;112(25):7701–7706. doi: 10.1073/pnas.1503102112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Woodward R.B. The total synthesis of vitamin B12. Pure Appl. Chem. 1973;33(1):145–178. doi: 10.1351/pac197333010145. [DOI] [PubMed] [Google Scholar]
- 231.Magnusdottir S., Ravcheev D., de Crecy-Lagard V., Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 2015;6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Degnan P.H., Taga M.E., Goodman A.L. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014;20(5):769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ward M.G., Kariyawasam V.C., Mogan S.B., Patel K.V., Pantelidou M., Sobczynska-Malefora A., Porte F., Griffin N., Anderson S.H., Sanderson J.D., Harrington D.J., Irving P.M. Prevalence and risk factors for functional vitamin B12 deficiency in patients with Crohn’s disease. Inflamm. Bowel Dis. 2015;21(12):2839–2847. doi: 10.1097/MIB.0000000000000559. [DOI] [PubMed] [Google Scholar]
- 234.Kieninger C., Baker J.A., Podewitz M., Wurst K., Jockusch S., Lawrence A.D., Deery E., Gruber K., Liedl K.R., Warren M.J., Krautler B. Zinc substitution of cobalt in vitamin B12: zincobyric acid and zincobalamin as luminescent structural B12 -mimics. Angew Chem Int Ed Engl. 2019;58(41):14568–14572. doi: 10.1002/anie.201908428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kieninger C., Wurst K., Podewitz M., Stanley M., Deery E., Lawrence A., Liedl K.R., Warren M.J., Krautler B. Replacement of the cobalt-center of vitamin B12 by nickel - nibalamin and nibyric acid prepared from metal-free B12-ligands hydrogenobalamin and hydrogenobyric acid. Angew Chem Int Ed Engl. 2020 doi: 10.1002/anie.202008407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Polaski J.T., Webster S.M., Johnson J.E., Batey R.T. Cobalamin riboswitches exhibit a broad range of ability to discriminate between methylcobalamin and adenosylcobalamin. J. Biol. Chem. 2017;292(28):11650–11658. doi: 10.1074/jbc.M117.787176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Iwig J.S., Leitch S., Herbst R.W., Maroney M.J., Chivers P.T. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J. Am. Chem. Soc. 2008;130(24):7592–7606. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nelson D.L., Kennedy E.P. Magnesium transport in Escherichia coli. Inhibition by cobaltous ion, J Biol Chem. 1971;246(9):3042–3049. [PubMed] [Google Scholar]
- 239.Silver S. Active transport of magnesium in escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1969;62(3):764–771. doi: 10.1073/pnas.62.3.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Iwig J.S., Rowe J.L., Chivers P.T. Nickel homeostasis in Escherichia coli - the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol. Microbiol. 2006;62(1):252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 241.Rodrigue A., Effantin G., Mandrand-Berthelot M.A. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J. Bacteriol. 2005;187(8):2912–2916. doi: 10.1128/JB.187.8.2912-2916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Nies D.H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J. Bacteriol. 1995;177(10):2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Liesegang H., Lemke K., Siddiqui R.A., Schlegel H.G. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J. Bacteriol. 1993;175(3):767–778. doi: 10.1128/jb.175.3.767-778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Eitinger T. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, FRG: 2003. Transport of Nickel and Cobalt in Prokaryotes, Microb. Transp. Syst. pp. 397–417. [Google Scholar]