Abstract
Background
Allergic rhinitis (AR) symptoms exhibit prominent 24‐hour variations associated with the biological clock. Although endogenous glucocorticoids synchronize circadian oscillator in the nasal mucosa, the precise mechanism of AR remains unclear. Therefore, using a mouse model, we investigated the association between circadian‐clock genes and AR symptoms at various time‐points.
Methods
Based on the rhythmic secretion of corticosterone levels, we chose 2 time‐points, ZT4 (10:00 AM) and ZT16 (10:00 PM), to observe dynamic changes of nasal symptoms, immunologic responses, and circadian‐clock gene period (Per) expressions.
Results
In the AR group, nasal symptom scores at ZT4 were significantly higher than at ZT16, with a greater increase in eosinophils, mast cells, and total immunoglobulin E levels at ZT4. The scores had a negative correlation with fluctuation of corticosterone levels. T‐helper 1 (Th1) cell counts and interferon‐γ levels decreased significantly at ZT4 compared with ZT16 in the AR group, whereas Th2 cells; Th17 cells; and interleukin (IL)‐4, ‐13, and ‐17A levels increased significantly at ZT4 compared with ZT16. Furthermore, Per2 gene expression levels were attenuated at ZT4 and elevated at ZT16, but correlated negatively with Th2 and Th17 responses associated with G ata3 and Rorγt expression levels that were enhanced at ZT4 and reduced at ZT16 in the AR group.
Conclusion
Our results suggest that the Per2 gene may influence diurnal variations of AR symptom severity, partially through its possible anti‐inflammatory effect on the circadian regulation of GATA3 and RORγt levels in immune cells. This further demonstrates the neural‐immune‐endocrinal mechanism of circadian rhythm in AR and sheds new light on chronotherapeutic approaches to AR.
Keywords: allergic rhinitis, immune response, endogenous glucocorticoid, circadian clock, Per2, mouse
Many allergic diseases exhibit time‐of‐day‒dependent variations associated with the endogenous circadian system. 1 , 2 For example, in most allergic rhinitis (AR) patients, clinical symptoms often worsen during the nighttime or early morning, compromising nighttime sleep and resulting in poor daytime performance. 3 , 4 Actually, the clock genes in the central and peripheral oscillators may affect maintenance of the nasal cycle and play a critical role in the daily changes of nasal symptomology 5 ; however, the precise mechanisms involved in AR remain to be further investigated.
The circadian clock drives ≈24‐hour rhythms in many physiologic and behavioral processes, such as sleep‐wake cycles, hormonal secretions, and food intake. 6 In mammals, the master clock located in the hypothalamic suprachiasmatic nucleus (SCN) is activated to synchronize peripheral clocks distributed throughout the body via neural and endocrine pathways. 6 , 7 The molecular circadian clock is generated by autoregulatory transcriptional‐translational feedback loops comprising many clock genes, such as Clock, Bmal1, Period (Per), cryptochrome (Cry), and their proteins. How these clock genes work in relation to human physiology and pathology has been described in detail elsewhere. 3 , 8 , 9 Interestingly, this circadian oscillation lasts ≈24 hours for one cycle of the feedback loop. 6 , 7 , 10 , 11
Some immune responses and allergic reactions are under the control of the circadian clock. 12 , 13 , 14 , 15 , 16 The clock genes Per (Per1, Per2, and Per3) are the core negative regulators of the circadian timing system that mediate rhythmic variability in immune‐inflammatory processes. 7 , 15 In particular, endogenous glucocorticoids have been considered Zeitgebers (time‐givers), which receive signals from SCN to synchronize peripheral clocks, such as nasal mucosa tissues. 4 Glucocorticoid hormones are secreted by the adrenal gland, which is regulated by the hypothalamus‐pituitary‐adrenal (HPA) axis in a circadian manner. These hormones are potent phase‐shifting agents for circadian gene expression in peripheral mouse tissues. 4 , 17 In nocturnal animals, serum glucocorticoids reach peak levels around or just before the onset of the activity phase (between Zeitgeber time 12 [ZT12] and ZT16), with a nadir of a short duration between ZT0 and ZT4.17 A recent study confirmed a circadian rhythm in PER2 level in the mouse nasal mucosa, with peaks in PER2 expression observed at the phase corresponding to the early dark period (ZT12), consistent with peak levels of serum glucocorticoids. The data suggest that endogenous glucocorticoids are important timing signals for peripheral oscillators and can reset the nasal cyclic activities via Per2 expression in the nasal mucosa. 4 However, the regulatory mechanism of the circadian clock on AR at the molecular level has yet to be determined.
It is well known that AR is an immunoglobulin E (IgE)‐mediated allergic inflammation characterized by a predominant T‐helper 2 (Th2)‐ and Th17‐type response, and involves eosinophil (EOS) infiltration, goblet cell hyperplasia, and mast cell (MC) degranulation in the local nasal mucosa. 18 , 19 Nevertheless, the periodic changes in AR symptoms may be partly explained by the circadian rhythms in the neural‐immune‐humoral mechanism of AR. 4 , 20 , 21 New evidence has indicated that the circadian clock regulates the adaptive immune system, mainly by influencing CD4+ T‐cell lineage immigration and differentiation. 22 , 23 , 24 Some T‐cell‒associated cytokines and transcription factors, such as nuclear factor/interleukin‐3‒regulated protein (E4BP4), and retinoic acid receptor‒related orphan receptor (ROR)γt synthesis, are circadian in nature. 9 , 22 Furthermore, one recent study showed that E4BP4 is a basically negative transcription factor of Th2 and Th17 cells that is directly controlled by clock genes Rev‐erbα and linked to the clock‐regulatory network. 22 Therefore, we hypothesized that the daily changes of nasal symptoms in the development of AR may be regulated by circadian‐clock genes through alteration of different Th‐specific transcription factors under the rhythmic secretion of endogenous glucocorticoids.
In our previous study, we measured plasma corticosterone levels at various time‐points in mice and found that peak plasma corticosterone levels were observed at ZT16 (10:00 PM, during the active phase) along with a clear nadir at ZT4 (10:00 AM, during the resting phase). 25 In the present study, we chose ZT4 and ZT16 as suitable time‐points for observation, as indicated in previous work. 6 , 14
In this study we investigated circadian variability of AR symptoms and circadian expression of the core clock genes Per1, Per2, and Per3 in nasal mucosal tissues and peripheral blood mononuclear cells (PBMCs) of an ovalbumin (OVA)‐sensitized mouse AR model. We sought to determine whether endogenous glucocorticoids influence the diurnal severity of AR symptoms via regulation of clock genes and explored the possible molecular mechanism and potential chronotherapeutic strategy in the pathophysiology of AR.
Materials and methods
Animals
Eight‐week‐old male BALB/c mice (15 mice per group; body weight, 20‐22 gm) were obtained from the Shanxi Medical University Affiliated Animal Centre (Taiyuan, China). Mice were housed in a specific pathogen‐free facility with ad libitum access to regular chow and water. All animals were reared under these conditions with a 12‐hour light/dark cycle for at least 2 weeks before experimental use. The lights were turned on at 6:00 AM (defined as ZT0 and turned off at 6:00 AM [ZT12]). Our experimental design was reviewed and approved by the institutional animal care and use committee at Shanxi Medical University (Approval No. 15857), and all experiments were performed according to the regulations on the administration of experimental animals of the State Science and Technology Commission and the Guide for the Care and Use of Laboratory Animals (institutional animal care and use committee of Shanxi Medical University).
Establishment of an AR mouse model
Mice were immunized and challenged with OVA (Sigma‐Aldrich, St Louis, MO) according to our previously described method 18 , 25 (Fig. 1A). Briefly, mice were intraperitoneally injected (50 µg/mouse) with OVA in phosphate‐buffered saline (PBS) mixed with an equal volume of 4 mg/mL aluminum hydroxide gel (Sigma‐Aldrich) as an adjuvant in a total volume of 1 mL at the indicated time‐points on days 1, 7, and 14, respectively. This was followed by intranasal administration of 3% OVA solution in PBS into the bilateral nasal cavity, either in the morning (ZT4) or evening (ZT16), daily for 12 consecutive days from days 21 to 32. PBS alone was administered as a control (Fig. 1B).
FIGURE 1.
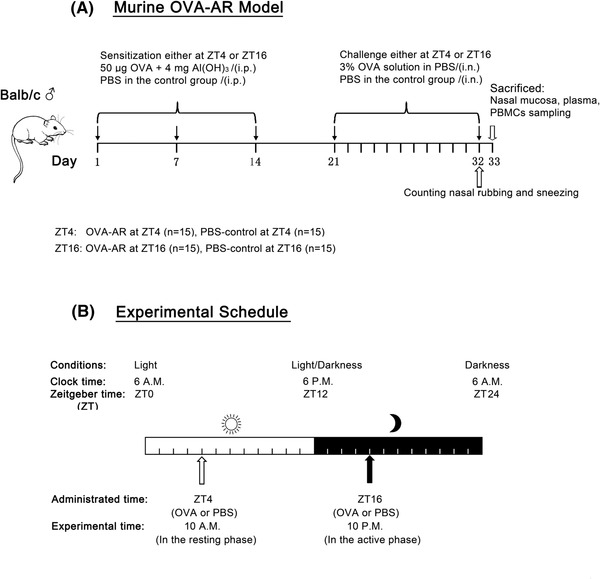
(A) Experimental protocol. Protocol used for the OVA‐AR mouse model. Mice in the AR group were sensitized with 50 µg OVA + 4 mg Al(OH)3 on days 1, 7, and 14. On days 21 through 32, the animals were challenged intranasally each day with a 3% OVA solution in PBS (50 µL per nostril), either at ZT4 or ZT16. PBS was used instead of OVA in the control group. (B) Experimental schedule. White and black bars represent the light and dark periods, respectively. Mice were administered OVA or PBS at ZT4 or ZT16. Behavioral tests or sample collections were conducted 24 hours after the final challenge. AR = allergic rhinitus; OVA = ovalbumin; PBS = phosphate‐buffered saline; ZT = Zeitgeber time.
Evaluation of AR symptoms in mice
After the final OVA challenge, nasal rubbing and sneezing were counted and recorded in a 10‐minute interval by using a video camera under an infrared lamp to avoid light‐induced circadian phase shifting. AR‐like symptoms were recorded at ZT4 (10:00 AM) and ZT16 (10:00 PM), respectively. The AR mouse model was successfully established and evaluated according to a behavioral symptoms scoring system. 18 , 19 AR scores were determined as follows: for nasal rubbing, slightly scratching the nose one time corresponded to a score of 1; continuously scratching the nose and face corresponded to a score of 2; and scratching everywhere corresponded to a score of 3. For nasal sneezing, sneezing 1 to 3 times corresponded to a score of 1; sneezing 4 to 10 times corresponded to a score of 2; and sneezing >11 times corresponded to a score of 3. For rhinorrhea, nasal mucus flowing to the anterior naris corresponded to a score of 1; nasal mucus flowing to the lower anterior naris corresponded to a score of 2; and a runny nose corresponded to a score of 3. A superposition method was used to obtain total scores (scores for each term were added together for each mouse to obtain 8 scores for each group); a mean score >5 was considered to indicate that the mouse had AR.
Nasal mucosa tissue histology
Mice were exsanguinated at ZT4 or ZT16, and nasal septal mucosa tissues were completely excised from each mouse to control standard quality and reduce experimental staining error. After fixing with 4% paraformaldehyde for 48 hours at room temperature, the specimens were embedded in paraffin wax and dehydrated in gradient concentrations of ethanol. The sections were cut to 4‐μm thickness, and each section was stained with hematoxylin and eosin (H&E) or toluidine blue. EOS and MC numbers in the sections were counted in 5 randomly selected fields at high magnification (400×). EOS and MC frequencies were presented as cells/mm2.
Measurement of corticosterone and IgE
Blood samples at different time‐points were collected in ethylene‐diamine tetraacetic acid (EDTA)‐containing tubes, and plasma was obtained by centrifugation for 15 minutes at 1000g and stored at −80°C until use. Plasma levels of corticosterone, total IgE, and OVA‐specific IgE were measured using a 96‐well microplate and enzyme‐linked immunosorbent assay (ELISA) kits (corticosterone, total IgE [Abcam, Beijing, China]; OVA‐specific IgE [BlueGene, Shanghai, China]) according to the manufacturer's instructions. Absorbance at 450 nm was measured using an enzyme‐linked immunoassay (ELISA) reader (Cytation3; BioTek, Winooski, VT).
Measurement of cytokine concentration
Plasma levels of cytokines, including interferon (IFN)‐γ and interleukin (IL)‐4, IL‐6, IL‐13, and IL‐17A, were determined using ELISA kits (Abcam, Beijing, China) according to the manufacturer's instructions. Absorbance at 450 nm was measured using the ELISA reader (Cytation3).
Isolation of PBMCs
Venous blood samples were collected through cardiac puncture under general anesthesia. The blood was immediately put into sterile EDTA tubes. PBMCs were isolated using a mouse peripheral blood lymphocyte separator kit (Solarbio, Beijing, China). Briefly, diluted blood samples in PBS (1:1 [v/v]) were layered onto a low‐gradient lymphocyte separator solution (1:3 [v/v]) and centrifuged at 800g for 20 minutes. The interface between the plasma and transparent separator solution containing the PBMCs was then collected, centrifuged at 100g for 5 minutes, and washed twice with sterile PBS. A total of 2 × 106 PBMCs were resuspended and cultured in RPMI‐1640 medium (Gibco, Gaithersburg, MD) supplemented with 10% fetal bovine serum (Gibco), 2 mmol/L L‐glutamine (Sigma‐Aldrich), 1% nonessential amino acids (Gibco), 1 mmol/L sodium pyruvate (Sigma‐Aldrich), 100 U/mL penicillin (Gibco), and 100 µg/mL streptomycin (Gibco) at 37°C in a humidified atmosphere of 5% CO2.
Flow cytometric analysis
After isolation, 1 × 106 PBMCs were stimulated with 50 µg/L phorbol 12‐myristate, 100 µg/L ionomycin mixture, 3 mg/L brefeldin A, and 1.4 mg/L monensin mixture, respectively, for 18 hours. For cell‐surface staining, the cells were stained with phycoerythrin (PE)‒cyanine 5‒labeled anti‐mouse CD4 antibody (eBioscience, San Diego, CA). The stained cells were then incubated with a PE‐labeled anti‐mouse IL‐4 antibody (eBioscience), a flourescein isothiocyanate‒labeled anti‐mouse IFN‐γ antibody (eBioscience), and an allophycocyanin‐labeled anti‐mouse IL‐17A antibody (eBioscience), after fixation and permeabilization using a fixation/permeabilization solution kit (eBioscience) according to manufacturer's instructions. Flow cytometric data for Th1, Th2, and Th17 cells were acquired using a flow cytometer and analyzed using FlowJo software version 7.6 (FlowJo, LLC, Ashland, OR).
RNA extraction and quantitative real‐time polymerase chain reaction
Total RNA was extracted from mouse nasal mucosa tissue and PBMCs using an RNAiso Plus kit (TaKaRa, Shiga, Japan), and RNA concentrations for each sample were determined using the Cytation3 system (BioTek). Approximately 500 ng of total RNA was reverse transcribed to obtain cDNA in a 10‐μL reaction using 5× PrimeScript RT Master Mix (TaKaRa), an ABI‐PRISM 7500 system (Applied Biosystems, Foster City, CA) with ROX reference dye II (50×) and SYBR Premix Ex Taq II (Tli RNaseH Plus), and a quantitative PCR kit (TaKaRa). The housekeeping gene β‐actin was used as an internal standard to obtain relative expressions of Per1, Per2, Per3, T‐bet, Gata3, E4bp4, and Rorγt. mRNA levels were quantified using specific primers (Table S1).
PCR was performed as follows: 1 cycle of 30 seconds at 95°C, followed by 40 cycles of 5 seconds at 95°C and 34 seconds at 60°C. Cycle threshold (Ct) values were used to analyze target and internal reference cDNA levels, and target cDNA levels were normalized to that of nonoscillated β‐actin using the 2−∆∆Ct method.
Statistical analysis
All results are expressed as mean ± standard deviation (SD). GraphPad Prism software version 6.0 (GraphPad Software, La Jolla, CA) was used for all data analyses. To determine whether the data were normally distributed and showed equal or biased variance, we chose the frequency distribution and used the Kolmogorov‐Smirnov test. Parametric analysis was conducted using an unpaired Student t test for 2‐group comparisons. Nonparametric analysis was performed using a Mann‐Whitney U test and a Wilcoxon signed rank test. p < 0.05 was considered statistically significant.
Results
AR‐like symptoms are severe during the early resting phase
To demonstrate the effects of the circadian clock on the allergic response in vivo, we investigated whether the AR symptoms differed in severity at the indicated time‐points (ZT4 and ZT16). As shown in Figure 2A and C, the frequency of nasal rubbing was significantly higher at ZT4 (resting phase) than at ZT16 (active phase) (tnasal rubbing, AR = −3.391, p < 0.01) in the AR group and accompanied by increased sneezing (tnasal sneezing, AR = −1.779, p < 0.05), although the frequencies of these activities in the AR group were significantly higher than those in the control group (tnasal rubbing, ZT4 = −5.175, tnasal rubbing, ZT16 = −5.197, p < 0.01; tnasal sneezing, ZT4 = −3.792, tnasal sneezing, ZT16 = −3.646, p < 0.01) at both time‐points. In addition, relative nasal rubbing and sneezing are presented in Figure 2B and D and the results show that nasal symptoms were more severe at ZT4 compared with ZT16 in the AR group (t2B = −2.515, t2D = −2.109, p < 0.05). In the control group, symptom comparison between ZT4 and ZT16 showed a relative difference in nasal rubbing and sneezing, with no significant difference. Furthermore, nasal symptom scores were calculated to be 8.63 ± 0.52 at ZT4 and 6.50 ± 0.53 at ZT16, respectively, in the AR group, with scores of 3.13 ± 0.64 at ZT4 and 2.75 ± 0.71 at ZT16, respectively, in the control group (Table S2A). The data show that AR mouse models were successfully established and there were significant differences at both time‐points between the AR and control groups (tscores, ZT4 = −3.478, tscores, ZT16 = −3.448, p < 0.01). Nasal symptom scores were significantly higher at ZT4 than at ZT16 in the AR group (tscores, AR = −3.475, p < 0.01), with slightly higher scores at ZT4 compared with ZT16 in the control group, indicating no significant difference (Fig. 2E and F).
FIGURE 2.
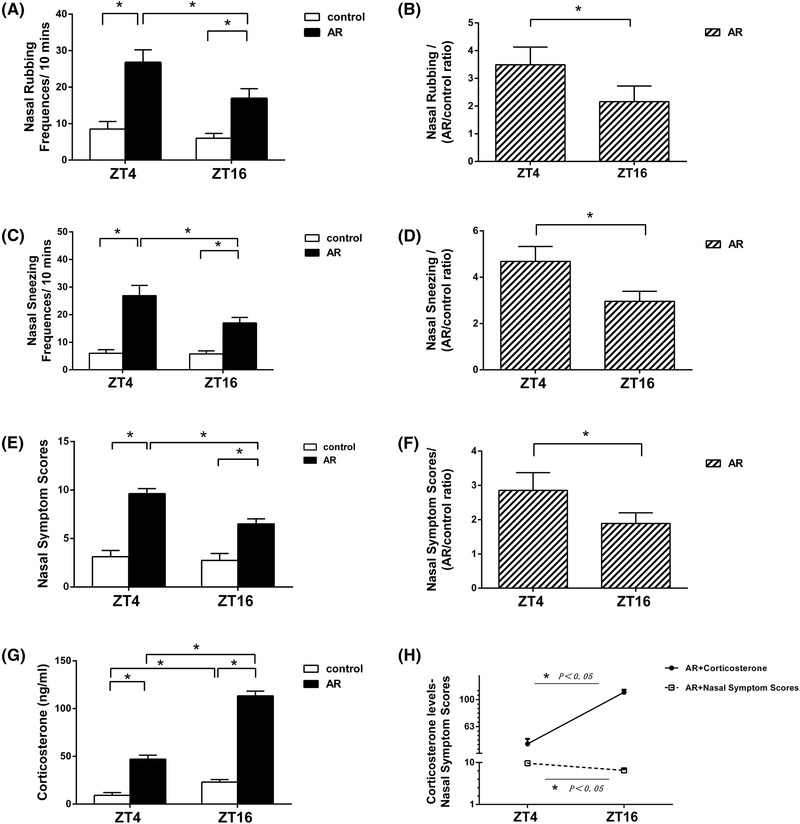
Nasal symptom scores over 10 minutes at ZT4 and ZT16 at 1 day after final OVA challenge, and the correlation with plasma corticosterone levels at the indicated time‐points. Nasal rubbing and sneezing increased significantly at ZT4 compared with ZT16 in the AR mice (A‐D). AR mice exhibited significantly higher allergic symptom scores than the control mice (E, F). Plasma levels of corticosterone (G) and a correlative analysis with nasal symptom scores in the AR group (H) are presented. Data represent the mean ± standard deviation (n = 15 per group). * p < 0.05. AR = allergic rhinitus; OVA = ovalbumin; ZT = Zeitgeber time.
Nasal symptom scores show negative correlation with corticosterone levels
Figure 2G shows the plasma corticosterone levels of rhythmic secretion in the AR and control groups. Plasma corticosterone levels were increased significantly at ZT16 compared with ZT4 in both the AR and control groups (tcorticosterone, AR = −3.477, tcorticosterone, control = −3.099, p < 0.01), whereas plasma corticosterone levels were increased significantly at ZT4 and ZT16 in the AR group when compared with the control group (tcorticosterone, ZT4 = −2.014, tcorticosterone, ZT16 = −2.570, p < 0.01). Interestingly, as shown in Figure 2H, the fluctuation of plasma corticosterone levels correlated negatively with nasal symptom scores, suggesting a possible anti‐inflammatory effect on the diurnal changes in the AR process.
EOS infiltration and MC degranulation increases in early resting phase
To analyze histologic changes in the nasal mucosa, samples obtained at ZT4 and ZT16 were stained with H&E and toluidine blue. As shown in Figure 3, EOS infiltration and MC degranulation in the nasal mucosa of the AR group were more severe when compared with the control group (tEOS, ZT4 = −3.908, tEOS, ZT16 = −3.868, p < 0.01; tMC, ZT4 = −3.963, tMC, ZT16 = −3.966, p < 0.01). In addition, in the AR group we observed a significant increase in numbers of EOS (22.80 ± 3.27) and MC (9.67 ± 2.02) at ZT4, as compared with ZT16 (EOS, 14.76 ± 2.93; MC, 7.34 ± 1.27) (tEOS, AR = 1.143, tMC, AR = 0.999, p < 0.05), but there was no significant difference between ZT4 and ZT16 in the control group (Table S2B).
FIGURE 3.
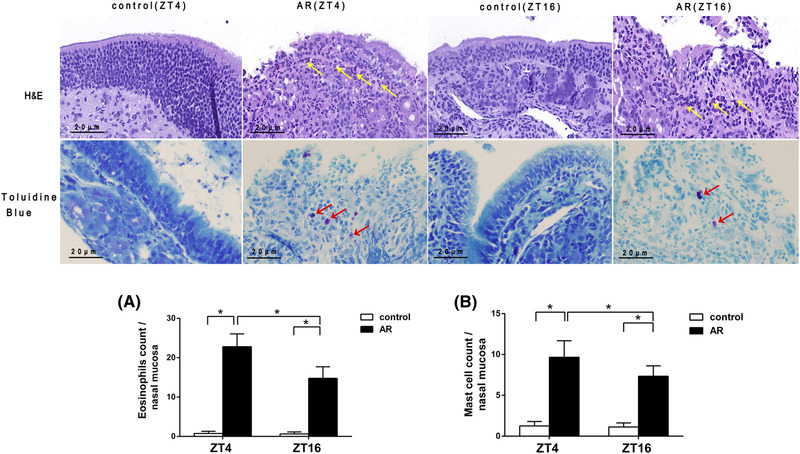
Histopathologic examination of mouse nasal mucosa tissue. EOS and MC numbers increased significantly at ZT4 as compared with ZT16 in the AR group. H&E and toluidine blue staining was performed on nasal mucosa tissue collected from both the AR and control groups at ZT4 and ZT16. Yellow and red arrows indicate EOSs and MCs, respectively. Statistical analysis was done for EOS infiltration (A) and MC degranulation (B) of nasal mucosa at ZT4 and ZT16. Original magnification: 400×. Scale bars = 20 µm. Data represent the mean ± standard deviation (n = 15 per group). * p < 0.05. AR = allergic rhinitus; EOS = eosinophil; H&E = hematoxylin and eosin; MC = mast cell; ZT = Zeitgeber time.
Total IgE and OVA‐specific IgE increase after OVA administration
To explore the diurnal rhythm of immunologic changes in vivo, total IgE and OVA‐specific IgE levels in both the AR and control groups were evaluated at ZT4 and ZT16. As shown in Figure 4A and B, plasma levels of both total IgE and OVA‐specific IgE in the AR group at ZT4 and ZT16 were higher than in the control group (ttotal IgE, ZT4 = 0.449, ttotal IgE, ZT16 = −1.025, p < 0.05; tOVA‐sIgE, ZT4 = −8.924, tOVA‐sIgE, ZT16 = −3.782, p < 0.01), suggesting involvement of IgE in AR pathogenesis. Moreover, plasma levels of total IgE at ZT4 (529.51 ± 27.74) ng/mL in the AR group were significantly higher than those at ZT16 (414.79 ± 24.09) ng/mL (ttotal IgE, AR = 0.734, p < 0.05), but there was no significant difference between ZT4 (298.23 ± 19.34) ng/mL and ZT16 (237.51 ± 17.33)ng/mL in the control group. In addition, we found that plasma levels of OVA‐specific IgE at ZT4 were relatively higher than those at ZT16, but the difference was not statistically significant between groups.
FIGURE 4.
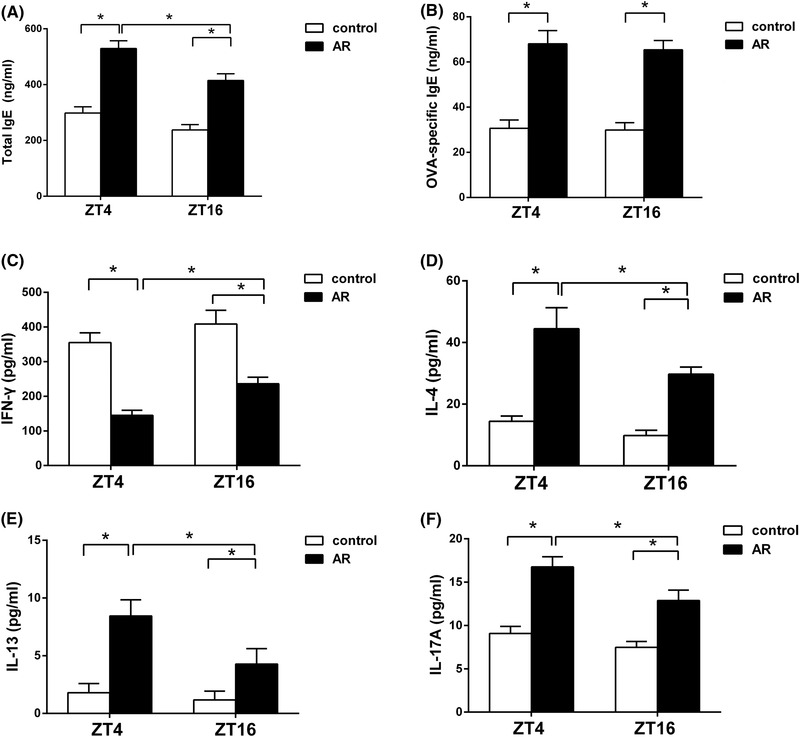
Temporal variations in plasma levels of total IgE, OVA‐specific IgE, and Th‐specific cytokine levels. Plasma levels of total IgE, IL‐4, IL‐13, and IL‐17A at ZT4 were significantly higher than those at ZT16 in the AR group, whereas plasma IFN‐γ levels at ZT4 were significantly lower than those at ZT16 in the AR group. Plasma levels of total IgE (A), OVA‐specific IgE (B), IFN‐γ (C), IL‐4 (D), IL‐13 (E), and IL‐17A (F), according to ELISA. Data represent mean ± standard deviation (n = 15/per group). * p < 0.05. AR = allergic rhinitus; ELISA = enzyme‐linked immunoassay; OVA = ovalbumin; IFN‐γ = interferon‐gamma; IgE = immunoglobulin E; IL = interleukin; Th = T helper; ZT = Zeitgeber time.
Th‐specific cytokine expression shows marked day‐night changes
To evaluate the diurnal rhythm of OVA‐induced immune responses, plasma cytokine levels for Th1 (IFN‐γ), Th2 (IL‐4 and IL‐13), and Th17 (IL‐17A) were measured by ELISA. As shown in Figure 4C‐4F, IFN‐γ levels decreased significantly, whereas IL‐4, IL‐13, and IL‐17A levels increased significantly at both time‐points in the AR group relative the control group (tIFN‐γ, ZT4 = −3.728, tIFN‐γ, ZT16 = 13.827, p < 0.05; tIL‐4, ZT4 = −13.485, tIL‐4, ZT16 = −22.023, p < 0.05; tIL‐13, ZT4 = −5.582, tIL‐13, ZT16 = −6.355, p < 0.05; tIL‐17A, ZT4 = −13.116, tIL‐17A, ZT16 = −9.530, p < 0.05). In addition, plasma IFN‐γ levels at ZT4 were significantly lower than those at ZT16 in the AR group (tIFN‐γ, AR = −7.449, p < 0.01), whereas plasma IL‐4, IL‐13, and IL‐17A levels at ZT4 were significantly higher than those at ZT16 in the AR group (tIL‐4, AR = 6.446, tIL‐13, AR = 3.363, tIL‐17A, AR = 5.630, p < 0.01). Moreover, plasma IFN‐γ levels at ZT4 were lower than those at ZT16 in the control group, but plasma IL‐4, IL‐13, and IL‐17A levels at ZT4 were higher than those at ZT16 in the control group; however, we observed no significant difference in these cytokines in the control group.
Th1, Th2, and Th17 cell numbers show temporal variations in vivo
We then investigated the circadian rhythm of altered proportions of CD4+IFN‐γ+ Th1, CD4+IL‐4+ Th2, and CD4+IL‐17A+ Th17 cells in PBMCs by flow cytometry. As shown in Figure 5, there was a decreased number of Th1 cells in the AR group compared with the control group, yet the numbers of Th2 and Th17 cells increased significantly in the AR group compared with the control group (tTh1, ZT4 = −3.026, tTh1, ZT16 = −4.296; tTh2, ZT4 = −6.383, tTh2, ZT16 = −3.825; tTh17, ZT4 = −6.363, t Th17, ZT16 = −2.402, p < 0.01). In addition, proportions of Th1, Th2, and Th17 cells showed diurnal changes in both groups, with significant differences between ZT4 and ZT16 in the AR group (tTh1, AR = −2.306, p < 0.05; tTh2, AR = 3.201, p < 0.01; tTh17, AR = 3.756, p < 0.01). However, we observed no significant difference in these proportions between ZT4 and ZT16 in the control group.
FIGURE 5.
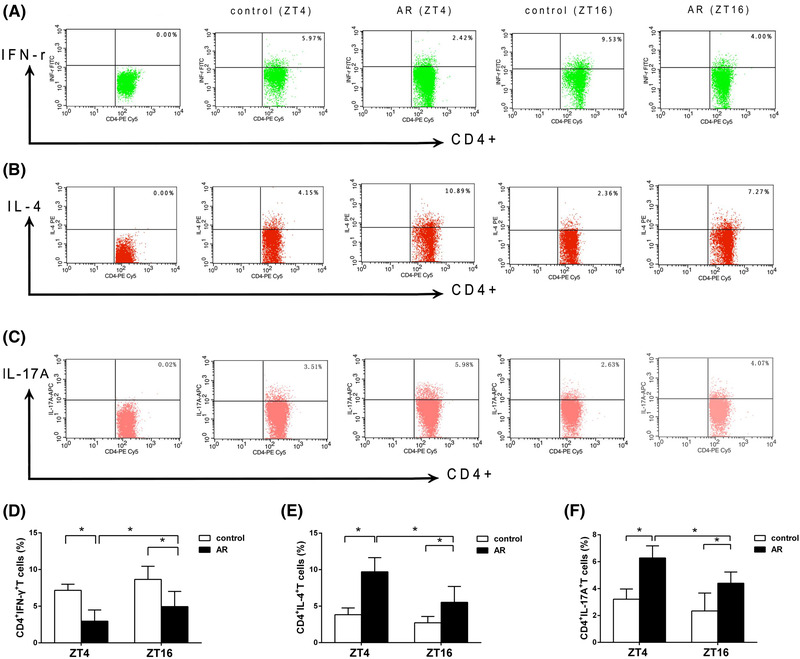
Temporal variations in T‐lymphocyte populations. Th1, Th2, and Th17 from PBMCs in both AR and control groups at ZT4 and ZT16 were quantified by flow cytometry. The proportion of Th1 cells decreased significantly at ZT4 as compared with ZT16 in the AR group, whereas the proportions of Th2 and Th17 cells increased significantly at ZT4 as compared with ZT16 in the AR group. Representative results are shown for Th1 (A), Th2 (B), and Th17 cells (C). Comparisons of the percentages of Th1 (D), Th2 (E), and Th17 cells (F) between the AR and control groups. Data represent mean ± standard deviation (n = 15 per group). * p < 0.05. AR = allergic rhinitus; PMBC = peripheral blood mononuclear cell; Th = T helper; ZT = Zeitgeber time.
Regulation of allergic reactions by circadian‐clock genes
To determine transcriptional activation of circadian‐clock genes according to time of day in AR, we performed quantitative reverse transcript polymerase chain reaction (qRT‐PCR) analysis of messenger RNA (mRNA) levels of Per1, Per2, and Per3 in both nasal mucosa tissues and PBMCs. In addition, we evaluated mRNA levels of Th‐specific transcription factors (Th1: T‐bet; Th2: Gata3 and E4bp4; Th17: Rorγt) in PBMCs.
As shown in Figure 6, circadian‐clock genes were expressed in the nasal mucosa tissues (Fig. 6A) and PBMCs (Fig. 6B) across the day‐night cycle, with no significant differences observed between ZT4 and ZT16 in Per1 levels in both groups, Per2 levels in the control group, and Per3 levels in the AR group. In the nasal mucosa tissues, Per1 levels at ZT4 in the control group were significantly higher than those in the AR group (tPer1 , ZT4 = −1.229, p < 0.05); Per2 levels at ZT4 were significantly lower than those at ZT16 in the AR group (tPer2 , AR = −2.970, p < 0.01); and Per3 levels at ZT4 were significantly higher than those at ZT16 in the control group (tPer3 , control = −2.872, p < 0.01). For PBMCs, Per2 and Per3 levels at ZT4 in the control group were significantly higher than those in the AR group (tPer2 ,PBMCs, ZT4 = 2.365, p < 0.05; tPer3 ,PBMCs, ZT4 = 3.856, p < 0.01); Per2 levels at ZT4 were significantly lower than those at ZT16 in the AR group (tPer2 ,PBMCs, AR = −4.204, p < 0.05); and Per3 levels at ZT4 were significantly higher than those at ZT16 in the control group (tPer3 ,PBMCs, control = 2.324, p < 0.05).
FIGURE 6.
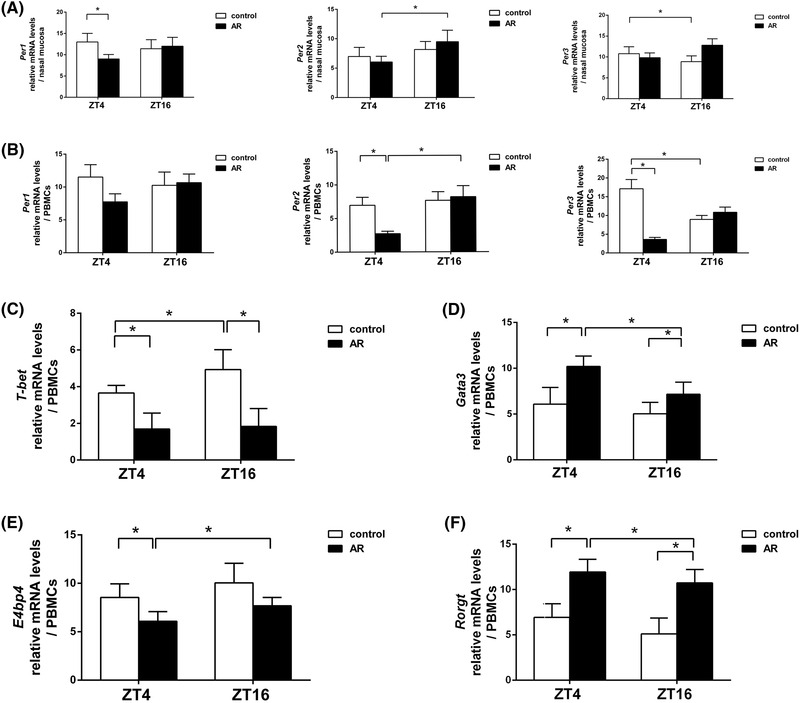
Messenger RNA levels of circadian‐clock genes and transcription factors at ZT4 and ZT16. qPCR analysis of Per1, Per2, Per3 mRNA levels in nasal mucosa tissue (A) and PBMCs (B), and T‐bet (C), Gata3 (D), E4bp4 (E), and Rorγt (F) in PBMCs. Data show that Per2 levels from nasal mucosa tissues and PBMCs at ZT4 were significantly lower than those at ZT16 in the AR group. The levels of Gata3 and Rorγt from PBMCs at ZT4 were significantly higher than those at ZT16 in the AR group, whereas E4bp4 levels in PBMCs at ZT4 were significantly lower than those at ZT16 in the AR group. Data represent mean ± standard deviation (n = 15 per group). * p < 0.05. AR = allergic rhinitus; PMBC = peripheral blood mononuclear cell; qPCR = quantitative polymerase chain reaction; ZT = Zeitgeber time.
Figure 6C and F shows that the expression of T‐cell−associated transcription factors changed across the day‐night cycle, although differences between ZT4 and ZT16 regarding T‐bet levels in the AR group and Gata3, E4bp4, and Rorγt levels in the control group were not statistically significant. Compared with data at ZT16, T‐bet levels at ZT4 decreased in the control group (tT‐bet , control = −0.694, p < 0.05), Gata3 and Rorγt levels at ZT4 increased in the AR group (tGata3 , AR = 1.096, tRorγt , AR = 1.437, p < 0.05), and E4bp4 levels at ZT4 decreased in the AR group (tE4bp4 , AR = 0.733, p < 0.05), although E4bp4 levels at ZT4 in the control group increased compared with the AR group (tE4bp4 , ZT4 = −2.082, p < 0.05). Furthermore, T‐bet levels at both ZT4 and ZT16 in the control group were significantly higher than those in the AR group (tT‐bet , ZT4 = 1.629, tT‐bet , ZT16 = 0.824, p < 0.05), whereas Gata3 and Rorγt levels at both ZT4 and ZT16 in the control group were significantly lower than those in the AR group (tGata3 , ZT4 = −1.303, tGata3 , ZT16 = −0.968, p < 0.05; tRorγt , ZT4 = −7.390, tRorγt , ZT16 = −4.392, p < 0.01). These data suggest that expression of circadian‐clock genes, especially Per2, are significantly altered during AR pathogenesis, indicating a possible role in AR and regulation of Th2‐ and Th17‐related responses.
Fluctuation of plasma corticosterone and Per2 and Th‐specific transcription factor levels from PBMCs between ZT4 and ZT16 are compared and analyzed in Figure 7. As shown in Figure 7A, plasma corticosterone levels and Per2 expression at ZT16 increased significantly compared with ZT4 in the AR group, indicating a similar pattern and rhythmicity. However, the fluctuation of nasal symptom scores and Per2 expression levels between ZT4 and ZT16 appeared to change in an opposite way, as shown in Figure 7B. Figure 7C shows that Per2 expression levels and Th1‐specific T‐bet levels demonstrated no significant correlation between ZT4 and ZT16 due to the loss of T‐bet rhythmicity. As shown in Figure 7D and F, Per2 expression levels and Th2‐specific Gata3 and Th17‐specific Rorγt levels changed differently and showed a negative interaction between ZT4 and ZT16 in the AR group. Due to the negative transcription factor of Th2‐ and Th17‐related E4bp4, Per2 expressions showed a similar fluctuation of E4bp4 levels at both time‐points in the AR group, with a nadir value at ZT4 and a maximal value at ZT16 (Fig. 7E).
FIGURE 7.
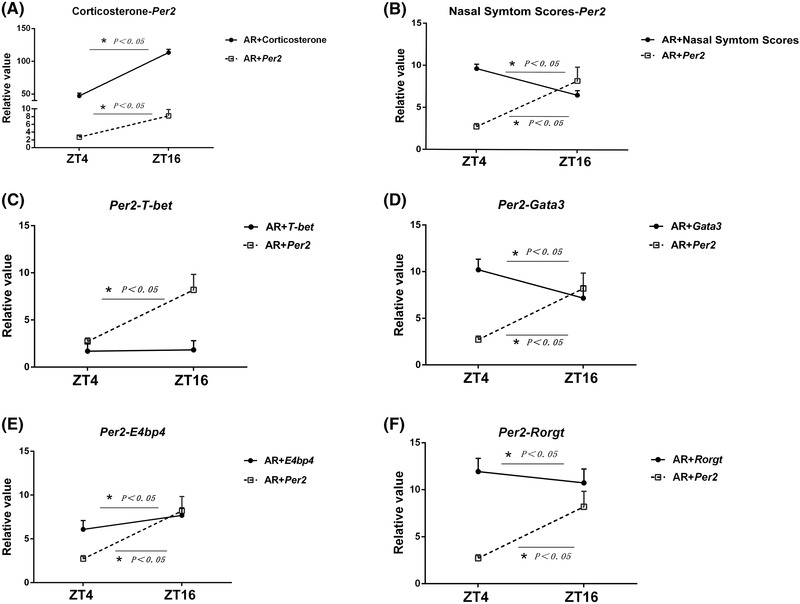
Correlative analysis among plasma corticosterone levels, nasal symptom scores, Per2 levels, and some T‐helper‒specific transcription factors from PBMCs. Data show a positive correlation between plasma corticosterone and Per2 expression (A), a negative correlation between nasal symptom scores and Per2 expression (B), no significant correlation between Per2 and T‐bet expression (C), a negative correlation between Per2 and Gata3 expression (D), a positive correlation between Per2 and E4bp4 expression (E), and a negative correlation between Per2 and Rorγt expression (F) at ZT4 and ZT16 in the AR groups, respectively. Data represent mean ± standard deviation (n = 15 per group). * p < 0.05. AR = allergic rhinitis; PMBC = peripheral blood mononuclear cell; ZT = Zeitgeber time.
Discussion
The role of the circadian rhythm in allergic diseases has been recognized 1 , 2 , 26 ; however, little is known about the immunomodulatory mechanisms associated with the circadian clock in AR. In this study, we have demonstrated that endogenous glucocorticoids could synchronize circadian expression of the clock genes, especially Per2, from both nasal mucosa tissues and PBMCs, which showed a marked day‐night cycle that was negatively correlated with diurnal changes in nasal symptoms (nasal rubbing and sneezing), total IgE, proportions of Th2 and Th17 cells, and levels of Th‐specific cytokines (IL‐4, IL‐13, and IL‐17A) and Th‐specific transcription factors (eg, GATA‐binding protein 3 [GATA3], and RORγt) in an AR mouse model. Our results indicate that the core circadian‐clock gene Per2 may contribute to temporal symptomatic variations in AR by its possible anti‐inflammatory effect on controlling rhythmic changes in levels of Th2‐ and Th17‐associated transcription factors. This prompted us to further investigate the cellular mechanisms of circadian rhythm to determine the clinically optimal “chronotherapy” for AR.
Based on the preliminary findings, we chose ZT4 and ZT16 as suitable time‐points to observe circadian expressions of the clock genes and the possible AR‐related immunologic mechanism. We found that nasal symptom scores, with allergic symptoms (nasal rubbing and sneezing), increased significantly at ZT4 relative to those at ZT16, as shown in Figure 2. Moreover, we observed significantly enhanced EOS infiltration and MC degranulation in the nasal mucosa at ZT4, as compared with that at ZT16 in AR mice, as shown in Figure 3. Similar to our results, other researchers have reported that the IgE/MC‐mediated passive cutaneous anaphylactic reaction showed a time‐of‐day–dependent variation in wild‐type mice, with a maximum at ZT4 (10:00 AM) and a minimum at ZT16 (10:00 AM), whereas the reaction was absent in mice with a loss‐of‐function mutation of Per2 (mPer2 m/m mice). 14 In most patients with AR, the severity of allergic symptoms with nasal congestion, sneezing, rhinorrhea, and nasal pruritus follows a circadian rhythm, being worst at night and in the early morning, between midnight and 6:00 AM during sleep. 27 Importantly in this context, the circadian variation in nasal reactivity is associated with changes in activity of EOS and basophils, which are found at significantly higher levels in nasal secretion at 6:00 AM. 28 Clinically, when drugs such as antihistamine mequitazine or leukotriene receptor antagonists are administered in the evening, they can significantly reduce allergic symptoms and improve nighttime rhinitis symptoms. 27 Given that mice are nocturnal animals, ZT4 corresponds to the resting phase and ZT16 corresponds to the active phase. Sublingual immunotherapy for AR in mice may vary with the time of day and be more effective at ZT4 (ie, nighttime in humans) than at ZT16 through a larger decrease in total and OVA‐specific IgE levels, as well as in IL‐4, IL‐10, and IL‐13 production. 29 To some extent, the amount of day‐night variation in symptom severity is probably related to circadian shifts in cortisol levels. 20
The current data demonstrate that Per2 levels in the nasal mucosa tissues and PBMCs and plasma corticosterone levels displayed similar circadian patterns in mice, with both peaking around the onset of night (ZT16) during the active phase. The fluctuation of plasma corticosterone levels and Per2 levels in the PBMCs correlated negatively with nasal symptom scores, with higher scores at ZT4 and lower scores at ZT16, as shown in Figures 2H and 7B. Therefore, we speculate that Per2 may exert an anti‐inflammatory effect on allergic responses, including those associated with AR. In line with this hypothesis, one recent study showed that corticosterone (or dexamethasone) suppressed IgE‐mediated allergic reactions in mouse bone marrow–derived MCs or basophils and PCA reactions in mice in association with increased PER2 levels in MC or basophils. 30 Moreover, a recent study confirmed that endogenous glucocorticoids control the peripheral clock via daily expression of clock gene Per2 in epithelial cells, vascular endothelial cells, and the fascicules of submucosal nerve termini in the mouse nasal mucosa. 4 In addition, So et al demonstrated that glucocorticoid‐response elements (GREs) located in the Per2 promoter are continuously occupied by the glucocorticoid receptor (GR), which acts as a transcription factor to promote rhythmic Per2 expression and is essential for glucocorticoid regulation of this gene in vivo. 31 In another study, Nakamura et al suggested that Per2 expression in MCs exhibits a time‐of‐day‒dependent variation in vivo. 14 It is possible that Per2 functions in MCs to inhibit CLOCK/BMAL1 activity, which regulates expression of the β subunit of FcεRIβ, an amplifier of FcεRIβ expression and signaling, 32 in a circadian manner. Thus, pharmacologically resetting the Per2 gene in MCs to reduce FcεRI signaling may inhibit IgE‐mediated allergic reactions. 33
Our findings demonstrate that levels of Th1, Th2, and Th17 cells exhibited periodic variations in mouse PBMCs, which corresponded to the circadian variations observed in plasma cytokine levels, with a major decrease in Th1 cell count and IFN‐γ levels at ZT4 and a major increase in Th2 and Th17 cell counts and IL‐4, IL‐13, and IL‐17A levels at ZT4, when compared with those at ZT16 in the AR group (Figs. 4 and 5). AR is an immune disorder associated with imbalances in Th1/Th2 and Th17/regulatory T‐cell ratios accompanied by an abnormal increase in numbers of Th2 and Th17 cells. 25 Th2 and Th17 cells produce type 2–like cytokines, such as IL‐4, IL‐13, and IL‐17A, whereas Th1 cells produce type 1 cytokines, such as IFN‐γ. In addition, Th2 and Th17 cytokines are known to play pivotal roles in the induction and maintenance of the allergic inflammatory cascade. 19 It has been reported that Th1‐specific IFN‐γ inhibits the synthesis of IgE and the differentiation of precursor cells to Th2 cells, whereas Th2‐specific IL‐4 and IL‐13 cause class switching in B cells to synthesize IgE production and induce MC degranulation in AR response. 18 , 34 Although Th‐cell development and their respective cytokine profiles are under the control of the circadian clock, 2 it remains unclear how Per2 regulates Th‐specific responses in AR. Previous studies suggested that circadian‐clock genes act directly on Th‐specific transcription factors associated with cytokine expression, thereby resulting in rhythmic cytokine secretion. 9 , 22 In the present study, we found that levels of T‐cell‒associated transcription factors varied significantly during the day‐night cycle and were consistent with observed patterns of Th‐specific cytokine expression.
On the other hand, we found significant circadian rhythms of expression for Per2, E4bp4, Gata3, and Rorγt in the local tissues in the AR group, as shown in Figure 7. Interestingly, Per2 expression levels and Th2‐specific Gata3, Th17‐specific Rorγt levels changed in the opposite way, with a negative correlation between ZT4 and ZT16 in the AR group. Due to the negative transcription factor of Th2‐ and Th17‐related E4bp4, Per2 expression levels exhibited similar fluctuation of E4bp4 levels at both time‐points in the AR group, with a lowest value at ZT4 and a highest value at ZT16. Some studies showed that production of type 2 cytokines is controlled by the transcription factor E4BP4, which negatively regulates Th2‐ and Th17‐cell differentiation. 22 Importantly, E4bp4 activity is regulated by Rev‐Erba, and its expression in human CD4+ T cells follows a circadian rhythm. 9 , 23 In addition, E4BP4 suppresses Th17‐cell development by binding and repressing the promoter region of RORγt, a Th17‐specific transcription factor. 22 In the present study, the findings coincide with those of a previous study showing that E4bp4 levels were lower during the resting phase (at ZT4) and were higher during the active phase (at ZT16), whereas RORγt levels were just the opposite―that is, higher during the resting phase and lower during the active phase in mice. 9 , 22 Moreover, one study reported that an OVA‐induced mouse food allergy model displayed lower E4BP4 levels during the light period (resting phase), which resulted in elevated IL‐13 and IL‐5 production and lower IL‐4 levels relative to those observed in the group observed during the dark period (active phase). 9 In the present study, we found that IL‐4 and IL‐13 levels in OVA‐induced AR mice were elevated at ZT4 relative to those at ZT16, suggesting its possible regulation by other transcription factors, including GATA3, signal transducer and activator of transcription (STAT)3, STAT5, and nuclear factor‐kappaB. 2 , 9 Furthermore, we found that AR‐related Th1 cells and IFN‐γ levels varied in a circadian manner, although this was not the case with Th1‐type transcription factor T‐bet levels. This finding coincides with that of Graham et al, and was explained by the rhythmic expression of killer‐cell lectin‐like receptor subfamily C member 2 in Th1 cells, indicating the involvement of the circadian rhythm in the Th1 response. 35 In our study, plasma levels of total IgE exhibited temporal variations, but plasma levels of OVA‐specific IgE did not differ between ZT4 and ZT16, possibly due to antigen administration and inflammation, both of which influence the circadian clock. Despite these findings, the molecular pathways linking systemic cues (eg, antigen‐uptake time) and specific allergic outputs (eg, MC degranulation) remain to be elucidated.
The findings of the preliminary study add to our knowledge of the possible neural‐immune‐endocrinal mechanism of clock gene activities involved in AR. In addition, we intend to use a PER2 inhibitor or Per2‐mutant mice to test our hypothesis in the near future. Accordingly, further studies are required to explore the molecular mechanisms through which PER2 regulates MC degranulation in vitro during AR development and investigate possible upstream circadian‐clock‒related activity using SCN‐disturbed, adrenalectomized, or aged mice in vivo.
Conclusion
Taken together, these results demonstrate that the Per2 gene may regulate daily variations of nasal symptoms severity in an AR mouse model, partially through its possible anti‐inflammatory function as well as by circadian regulation of Th2 and Th17 allergic responses. Our results suggest that the circadian clock is involved in the pathogenesis of AR, which establishes the theoretical foundation of a neural‐immune‐endocrinal mechanism in AR while prompting us to further explore the clinically appropriate chronotherapy for AR.
Supporting information
Supporting Material
Supporting Material
Supporting Material
Acknowledgements
The authors thank Dr Jimin Cao, Shanxi Medical University, Taiyuan, China, for review of the manuscript.
How to Cite this Article:Cheng F‐L, An Y‐F, Han Z‐Q, et al. Period2 gene regulates diurnal changes of nasal symptoms in an allergic rhinitis mouse model. Int Forum Allergy Rhinol. 2020;10:1236–1248.
Funding sources for the study: National Natural Science Foundation of China (81670914 and 81870707); Key R&D Project of Shanxi Province (201703D121003); Innovation Project of Postgraduate Education in Shanxi Province (2019BY076); junior travel grant to RhinoWorld 2019 (to FL.C.).
Potential conflict of interest: None provided.
Presented orally in abstract form at the RhinoWorld Conference on June 5‐9, 2019, in Chicago, IL, and published in a supplement issue of the International Forum of Allergy and Rhinology.
References
- 1. Nakamura Y, Nakano N, Ishimaru K, et al. Circadian regulation of allergic reactions by the mast cell clock in mice. J Allergy Clin Immunol. 2014;133:568‐575. [DOI] [PubMed] [Google Scholar]
- 2. Nakao A. Temporal regulation of cytokines by the circadian clock. J Immunol Res. 2014;2014:614529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ando N, Nakamura Y, Ishimaru K, et al. Allergen‐specific basophil reactivity exhibits daily variations in seasonal allergic rhinitis. Allergy. 2015;70:319‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Honma A, Yamada Y, Nakamaru Y, Fukuda S, Honma K, Honma S. Glucocorticoids reset the nasal circadian clock in mice. Endocrinology. 2015;156:4302‐4311. [DOI] [PubMed] [Google Scholar]
- 5. Kim HK, Kim HJ, Kim JH, Kim TH, Lee SH. Asymmetric expression level of clock genes in left vs. right nasal mucosa in humans with and without allergies and in rats: circadian characteristics and possible contribution to nasal cycle. PLoS One. 2018;13:e0194018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakao A, Nakamura Y, Shibata S. The circadian clock functions as a potent regulator of allergic reaction. Allergy. 2015;70:467‐473. [DOI] [PubMed] [Google Scholar]
- 7. Nakamura Y, Ishimaru K, Shibata S, Nakao A. Regulation of plasma histamine levels by the mast cell clock and its modulation by stress. Sci Rep. 2017;7:39934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sundar IK, Yao H, Sellix MT, Rahman I. Circadian molecular clock in lung pathophysiology. Am J Physiol Lung Cell Mol Physiol. 2015;309:L1056‐1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanabe K, Kitagawa E, Wada M, et al. Antigen exposure in the late light period induces severe symptoms of food allergy in an OVA‐allergic mouse model. Sci Rep. 2015;5:14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawauchi T, Ishimaru K, Nakamura Y, et al. Clock‐dependent temporal regulation of IL‐33/ST2‐mediated mast cell response. Allergol Int. 2017;66:472‐478. [DOI] [PubMed] [Google Scholar]
- 11. Koritala BSC, Cakmakli S. The human circadian clock from health to economics. Psych J. 2018;7:176‐196. [DOI] [PubMed] [Google Scholar]
- 12. Baumann A, Gonnenwein S, Bischoff SC, et al. The circadian clock is functional in eosinophils and mast cells. Immunology. 2013;140:465‐474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dumbell R, Matveeva O, Oster H. Circadian clocks, stress, and immunity. Front Endocrinol. 2016;7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakamura Y, Harama D, Shimokawa N, et al. Circadian clock gene Period2 regulates a time‐of‐day‐dependent variation in cutaneous anaphylactic reaction. J Allergy Clin Immunol. 2011;127:1038‐1045.e1‐3. [DOI] [PubMed] [Google Scholar]
- 15. Silver AC, Arjona A, Hughes ME, Nitabach MN, Fikrig E. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immunol. 2012;26:407‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Reece SP, Van Scott MR, Brown JM. A circadian clock in murine bone marrow‐derived mast cells modulates IgE‐dependent activation in vitro. Brain Behav Immun. 2011;25:127‐134. [DOI] [PubMed] [Google Scholar]
- 17. Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food‐induced phase‐shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128‐7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qi X, Suo L, Zhao C, Yang P. Investigation on the role of TIM4 in the pathogenesis of allergic rhinitis in mice (in Chinese). Zhonghua er bi yan hou tou jing wai ke za zhi [Chinese J Otolaryngol Head Neck]. 2014;49:283‐287. [PubMed] [Google Scholar]
- 19. Tang Z, Wang Y, Lv L, Li L, Zhang H. Mice with double knockout of H2‐Eb1 and H2‐Ab1 exhibit reduced susceptibility to allergic rhinitis. PLoS One. 2018;13:e0206122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fidan V, Alp HH, Gozeler M, Karaaslan O, Binay O, Cingi C. Variance of melatonin and cortisol rhythm in patients with allergic rhinitis. Am J Otolaryngol. 2013;34:416‐419. [DOI] [PubMed] [Google Scholar]
- 21. Zhao CQ, Li C. Advancement on the mechanism of mast cell degranulation in relation to neural‐endocrine regulation in allergic rhinitis (in Chinese). Zhonghua er bi yan hou tou jing wai ke za zhi [Chinese J Otolaryngol Head Neck]. 2018;53:730‐732. [DOI] [PubMed] [Google Scholar]
- 22. Yu X, Rollins D, Ruhn KA, et al. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342:727‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bollinger T, Leutz A, Leliavski A, et al. Circadian clocks in mouse and human CD4+ T cells. PLoS One. 2011;6:e29801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang G, Zhang H, Liu Y, et al. Alternation of circadian clock modulates forkhead box protein‐3 gene transcription in CD4(+) T cells in the intestine. J Allergy Clin Immunol. 2016;138:1446‐1449.e10. [DOI] [PubMed] [Google Scholar]
- 25. Cheng FL, Qi XP, Zhao CQ, An YF, Ren JJ, Li C. Effects of endogenous glucocorticoid deprivation on immune response of allergic rhinitis in mice (in Chinese). Zhonghua er bi yan hou tou jing wai ke za zhi [Chinese J Otolaryngol Head Neck]. 2018;53:757‐764. [DOI] [PubMed] [Google Scholar]
- 26. Nakao A. Circadian regulation of urticaria and anaphylaxis (in Japanese). Nihon Rinsho Jpn J Clin Med. 2013;71:2153‐2157. [PubMed] [Google Scholar]
- 27. Storms WW. Pharmacologic approaches to daytime and nighttime symptoms of allergic rhinitis. J Allergy Clin Immunol. 2004;114(suppl):S146‐153. [DOI] [PubMed] [Google Scholar]
- 28. Aoyagi M, Watanabe H, Sekine K, et al. Circadian variation in nasal reactivity in children with allergic rhinitis: correlation with the activity of eosinophils and basophilic cells. Int Arch Allergy Immunol. 1999;120(suppl 1):95‐99. [DOI] [PubMed] [Google Scholar]
- 29. Igarashi S, Suzuki K, Nakamura Y, et al. The efficacy of Sublingual Immunotherapy for allergic rhinitis may vary with the time of day. Int Arch Allergy Immunol. 2016;171:111‐118. [DOI] [PubMed] [Google Scholar]
- 30. Nakamura Y, Nakano N, Ishimaru K, et al. Inhibition of IgE‐mediated allergic reactions by pharmacologically targeting the circadian clock. J Allergy Clin Immunol. 2016;137:1226‐1235. [DOI] [PubMed] [Google Scholar]
- 31. So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci. 2009;106:17582‐17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ra C, Nunomura S, Okayama Y. Fine‐tuning of mast cell activation by FcepsilonRIbeta chain. Front Immunol. 2012;3:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Orihara K, Saito H. Controlling the peripheral clock might be a new treatment strategy in allergy and immunology. J Allergy Clin Immunol. 2016;137:1236‐1237. [DOI] [PubMed] [Google Scholar]
- 34. Coomes SM, Kannan Y, Pelly VS, et al. CD4(+) Th2 cells are directly regulated by IL‐10 during allergic airway inflammation. Mucosal Immunol. 2017;10:150‐161. [DOI] [PubMed] [Google Scholar]
- 35. Graham CM, Christensen JR, Thomas DB. Differential induction of CD94 and NKG2 in CD4 helper T cells. A consequence of influenza virus infection and interferon‐gamma? Immunology. 2007;121:238‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Material
Supporting Material
Supporting Material


