Summary
Primary sclerosing cholangitis (PSC) is a common indication for liver transplantation (LT). Up to 25% of patients experience recurrence of PSC (rPSC) after LT, which is associated with significant morbidity and mortality. To date, it is not possible to predict which patients are at risk for rPSC. The aetiology of PSC is complex and is speculated to involve translocation of intestinal bacteria to the liver, because of its frequent co‐occurrence with inflammatory bowel diseases (IBD). Here, we investigate whether the mucosal intestinal microbiome of PSC patients (n = 97) at time of first LT can identify those patients who will develop rPSC. 16S gene sequencing of bacterial DNA isolated from formalin‐fixed paraffin‐embedded biopsies showed that PSC patients with Crohn’s disease (n = 15) have a reduced microbial diversity and that inflammation of the mucosa is associated with beta‐diversity changes and feature differences. No differences in alpha‐ or beta diversity were observed between patients with rPSC (n = 14) and without rPSC (n = 83). However, many over‐represented bacterial features were detected in patients with rPSC, while surprisingly, those without recurrence of disease were characterized by an increased presence of the Gammaproteobacteria Shigella. This pilot study warrants further investigation into bacterial differences between rPSC and non‐rPSC patients.
Keywords: liver transplantation, microbiome, primary sclerosing cholangitis, recurrent disease
Introduction
Primary sclerosing cholangitis (PSC) is a liver disease characterized by chronic inflammation, distributed across both the intra‐ and extrahepatic biliary tree. PSC is progressive and results in cholestasis, fibrosis, liver cirrhosis and in some cases cholangiocarcinoma. In the absence of liver transplantation (LT), PSC eventually leads to an early death as there is no medical cure [1, 2, 3, 4]. Outcome after LT is excellent with 87.2%, 78.2% and 70.3% survival after 1, 5 and 10 years, respectively [5]. Unfortunately, PSC recurs in 8.6–27% of patients after an average of 4.7 years after LT [6, 7, 8, 9]. Recurrence of PSC (rPSC) is associated with significant morbidity and mortality and may necessitate retransplantation. Similar to the primary disease, the aetiopathogenesis of recurrent PSC is poorly understood [10]. Therefore, it is as of yet not possible to accurately predict, let alone prevent, recurrence.
PSC pathogenesis is thought to be multifactorial combining genetic predisposition with immunologic and environmental factors [10]. Interestingly, PSC is prominently associated with inflammatory bowel disease (IBD), with up to 80% of patients affected by PSC developing IBD [3]. This striking coexistence is partially explained by genetics [11]. However, genetic predisposition is only responsible for 10% of the disease aetiopathogenesis, implicating a large number of other factors involved in the development of PSC [12]. The gut microbiota is an eminent factor of importance in both IBD and PSC; a reservoir of bacteria, archaea, eukaryotes and viruses providing endless interaction with the human immune system [13, 14]. Impaired epithelial lining of the gut (i.e. leaky gut syndrome) is a phenomenon in IBD, resulting in bacterial translocation from gut to liver. This enhanced bacterial translocation in IBD is speculated to be one of the driving forces of PSC, and it has been hypothesized that bacteria (or other microbes) carried into the liver via the portal vein may induce local inflammatory responses [15]. Microbial alterations generally observed in IBD include a decreased bacterial diversity, in particular in Crohn’s disease (CD) [16]. Recent publications have shown alterations in gut microbiome and mycobiome in PSC patients as well, suggesting the microbiota as a potential biomarker or treatment target [17, 18, 19, 20, 21, 22, 23, 24, 25, 26].
Interestingly, a recent review with meta‐analysis showed that colectomy before LT is associated with a decreased risk of rPSC [6, 27], with the commentary that patients receiving an end ileostomy seem to achieve the best graft survival after colectomy [28]. The association between colectomy and rPSC development is pointing at a potential role for gut microbes in rPSC. Although routine colectomy for rPSC prevention is not advocated, some authors suggest further exploration of the option and timing of colectomy before or during LT in selected PSC patients [29]. Better understanding of the involvement of the gut microbiome in disease progression might help to define clinical management.
The development of rPSC could be driven by similar factors as PSC. Discoveries made in the pathogenesis of rPSC may be valuable for identification of patients at risk of recurrence, developing preventive therapies, and directing future PSC research in general. LT patients are monitored prospectively according to a follow‐up protocol offering a well‐documented cohort of patients with a common starting point after the surgical procedure. With the availability of these data, we initiated this study to determine whether the gut microbiome differs between liver transplant recipients who do or do not develop rPSC.
Materials and methods
Subjects and sample collection
All consecutive patients with PSC who received a liver transplant between 1987 and 2015 at our transplant centre were considered eligible for study enrolment if they had undergone a screening colonoscopy with surveillance biopsies pre‐LT. Up until 2015, colonoscopy was a routine practice in our liver transplant screening procedure. In all PSC patients, either with or without IBD, random sampling at 10 sites was regularly performed for pathological assessment of underlying dysplasia. Biopsy material was stored at stable conditions as formalin‐fixed paraffin‐embedded (FFPE) blocks. Transplant cases were identified in our prospective database, and the biobank was searched for availability of FFPE colon samples. Only patients with available colon biopsies were enrolled in our study. Samples were preferentially taken from the right hemicolon.
Definition and diagnosis of rPSC
Recurrence PSC was defined according to the Mayo criteria, which includes classic PSC‐like disease after liver transplant and excludes secondary causes of post‐LT biliary disease [30]. Inclusion criteria were the following: confirmed diagnosis of PSC before LT; endoscopic or magnetic resonance cholangiography showing intrahepatic and/or extrahepatic biliary strictures, beading, and irregularities at least> 90 days after LT; and histological findings of fibrous cholangitis and/or fibro‐obliterative lesions with or without ductopenia, biliary fibrosis, or biliary cirrhosis on liver biopsy or explant histology at time of transplant. Exclusion criteria were the following: hepatic artery thrombosis/stenosis; established chronic ductopenic rejection; anastomotic biliary strictures alone; nonanastomotic strictures before post‐LT day 90; and ABO incompatibility between donor and recipient.
Data on rPSC were collected prospectively, and the diagnosis was confirmed by an expert hepatologist, radiologist and pathologist.
Definition of IBD
Inflammatory bowel disease status was derived from patient charts. A positive IBD diagnosis was based on a known history of IBD and pathological assessment of biopsies taken prior to LT by our expert pathologist. Crohn’s disease, ulcerative colitis (UC) and IBD‐unclassified (IBD‐U) were identified.
Antibiotic use
As the use of antibiotics may influence the colon microbiome, we recorded all antibiotic treatments within 3 months before the date of the colon biopsy and considered it as a potential confounder. The use of antibiotics leads to rapid luminal microbial changes, and after cessation, subsequent restoration of the composition occurs within 1.5 months [31, 32]. Three categories were defined: (i) no antibiotics, (ii) short‐term antibiotics and (iii) prolonged antibiotic treatment. Short‐term antibiotics usage was defined as the incidental use of antibiotics for an (suspected) infection, within 3 months prior to colon sampling. Prolonged antibiotic usage was defined as a prolonged maintenance treatment to prevent recurrent (biliary) infections within 3 months prior to colon sampling.
Ethical considerations
All transplant candidates have given informed consent for the use of both biopsy specimens and clinical data collected during the process of LT for scientific purposes. All samples used in this study were primarily collected for routine clinical care. All clinical data were collected from medical charts and stored anonymously.
DNA extraction and 16S sequencing
The FFPE samples were sliced. For each sample, we used 14 slices of five micron; seven superficial slices and seven slices halfway the biopsy, to cover most of the biopsy. The slices were rinsed with Xylene to remove all paraffin. Subsequently, bacterial DNA was extracted from the colon biopsies using the RTP Bacteria DNA Mini Kit (Stratec®) as per manufacturer’s protocol [33]. An empty paraffin block not containing human tissue but undergoing the same DNA extraction protocol was used as a ‘kitome’ control to assess contamination. The DNA isolates were sent to Macrogen® for 16S rRNA gene sequencing (V3 ‐ V4 regions), using Illumina MiSeq.
Data analysis
Data were analysed and displayed using QIIME V 1.9.1 [34] and Emperor [35].
Operational taxonomic units representing more than 1% of the total amount of reads as determined by kitome control were considered to be contaminant features and were removed from all samples. All samples were rarefied to a depth of 1200 reads. To investigate whether selection bias had occurred, the prevalence of rPSC was determined in all patients who had undergone a transplant for PSC. Patient characteristics were described using mean and standard deviation or median and range (based on normal distribution), or count and proportion, and compared between patients with and without recurrence using chi‐squared tests (for categorical variables) and t‐tests (for approximately normally distributed variables). The relative frequency of the phyla and orders found in the samples was visualized using bar plots. Alpha diversity of each sample, providing information on the number of species (richness) and distribution of those species (evenness) within a given sample, was calculated using Shannon and Chao alpha‐diversity indices. Alpha‐diversity means were compared with pairwise T‐tests. Alpha diversity was visualized using box plots, showing mean (plus sign), median, interquartile range, minimum and maximum, and outliers. Beta diversity, indicating the diversity of species between samples and providing information on how samples compare to each other (similarity), given how different the species within said samples are, was assessed using the unweighted and weighted UniFrac distance metrics, displayed as principal coordinates analysis (PCoA) plots. Mann–Whitney U‐tests on alpha diversity and analysis of similarities (ANOSIM) tests on beta diversity were used to assess differences in diversity between patients with and without IBD, active inflammation, antibiotics use and rPSC; and for differences between sub‐groups of IBD (UC, CD, IBD‐U, and no IBD) and differences with regard to the length of antibiotics use. Linear discriminant analysis Effect Size (LEfSe) analyses were used to compare and visualize differences in microbiome between groups, both univariable and multivariable, on all available taxonomic levels (phylum, class, order, family and genus) [36].
Results
Study population
A total of 169 patients were transplanted for PSC between 1987 and 2015. Colon biopsy samples were available from 98 (58%) of these patients. In the remaining 71 (42%) cases, the FFPE blocks could not be provided as too little material was left in the biobank. One patient sample was excluded during analysis due to a low read count, leaving a total of 97 (57%) patients for all characteristics and analyses. The PSC recurrence rate was 12% (n = 21) in the total group of 169 patients and 14% (n = 14) for the patients for whom samples were available (P = 0.83), suggesting that no severe selection bias had occurred.
Study cohort characteristics are presented in Table 1. Median time between colon sampling and LT was 7.2 months (i.e. median time on waiting list, range 1.7–32.8 months). After LT, 14 patients were diagnosed with rPSC, after a mean of 4.7 years (SD 3.5). Mean follow‐up time post‐LT was 10.7 years (SD 6.0).
Table 1.
Characteristics of the study population.
| Total cohort | no rPSC | rPSC | P‐value | |
|---|---|---|---|---|
| n = 97 | n = 83 | n = 14 | ||
| Age at Dx PSC (years), mean (SD) | 34.7 (12) | 34.6 (12.3) | 35.1 (10.7) | 0.87 |
| Age at LT (years), mean (SD) | 44.4 (11) | 44.7 (11.1) | 42.7 (10.8) | 0.53 |
| CIT (minutes), mean (SD) | 470.9 (186.5) | 466 (193.1) | 500.1 (143.8) | 0.53 |
| WIT (minutes), mean (SD) | 35.9 (20.8) | 36.6 (21.7) | 31.6 (14.2) | 0.40 |
| MELD at listing, mean (SD) | 15.8 (5.7) | 15.8 (5.4) | 15.4 (7.2) | 0.81 |
| Gender | 0.32 | |||
| Male (%) | 65 (67) | 54 (65) | 11 (79) | |
| Female (%) | 32 (33) | 29 (35) | 3 (21) | |
| AB | 0.51 | |||
| No (%) | 63 (65) | 55 (66) | 8 (57) | |
| Yes (%) | 34 (35) | 28 (34) | 6 (43) | |
| AB category | ||||
| None (%) | 63 (65) | 55 (66) | 8 (57) | 0.10 |
| Short (%) | 11 (11) | 11 (13) | 0 (0) | |
| Prolonged (%) | 23 (24) | 17 (21) | 6 (43) | |
| Active colonic inflammation | 0.48 | |||
| No (%) | 50 (52) | 44 (53) | 6 (43) | |
| Yes (%) | 47 (48) | 39 (47) | 8 (57) | |
| Indication listing | 0.003 | |||
| Cirrhosis (%) | 77 (79) | 70 (84) | 7 (50) | |
| Recurrent cholangitis (%) | 20 (21) | 13 (16) | 7 (50) | |
| Biopsy location | 0.81 | |||
| Right hemicolon (%) | 51 (53) | 43 (52) | 8 (57) | |
| Left hemicolon (%) | 34 (35) | 29 (35) | 5 (36) | |
| Unspecified colon (%) | 12 (12) | 11 (13) | 1 (7) | |
| IBD yes/no | 0.25 | |||
| No (%) | 26 (27) | 24 (29) | 2 (14) | |
| Yes (%) | 71 (73) | 59 (71) | 12 (86) | |
| IBD type | 0.53 | |||
| No IBD (%) | 26 (27) | 24 (29) | 2 (14) | |
| UC (%) | 53 (55) | 45 (54) | 8 (57) | |
| m. Crohn (%) | 15 (16) | 12 (15) | 3 (21) | |
| IBD‐U (%) | 3 (3) | 2 (2) | 1 (7) |
Bold P‐values indicate statistically significant differences.
rPSC, recurrence of primary sclerosing cholangitis; Dx, diagnosis; PSC, primary sclerosing cholangitis; LT, liver transplantation; CIT, cold ischaemia time; WIT, warm ischaemia time; MELD, model for end‐stage liver disease; AB, antibiotics; IBD, inflammatory bowel disease; UC, ulcerative colitis; IBD‐U, inflammatory bowel disease – unclassified; SD, standard deviation.
Mucosal microbiome is dominated by Proteobacteria in PSC patients
First, we investigated the taxonomy of all samples (Fig. 1a for phylum level, Figure S1 for order level). On average, a relatively high abundancy of the phylum Proteobacteria is noticeable (41.9%). The commonly dominating phyla Bacteriodetes (27.3%) and Firmicutes (16.7%), and in lesser amount Actinobacteria (8%), are the next most abundant phyla. Microbiomes of three patients (indicated by arrows in Fig. 1a,b) were dominated by one single genus of bacteria: Shigella, Staphylococcus and Actinomycetales, respectively. This also resulted in a drastic reduction in the alpha diversity of the microbial composition in these patients (Fig. 1b). In one of these cases, the patient was diagnosed with an IBD flare at time of colon biopsy, which in retrospect could have been an acute Shigella infection mimicking a flare of IBD. In the other two cases, there is no clear clinical explanation for the domination of one single genus, as no clinical problems were reported at that time. Both the Staphylococcus and Actinomycetales genus are commonly found in gut microbiome samples, but not in these amounts. All downstream analyses were performed with and without exclusion of these three patients, with similar results (data not shown).
Figure 1.
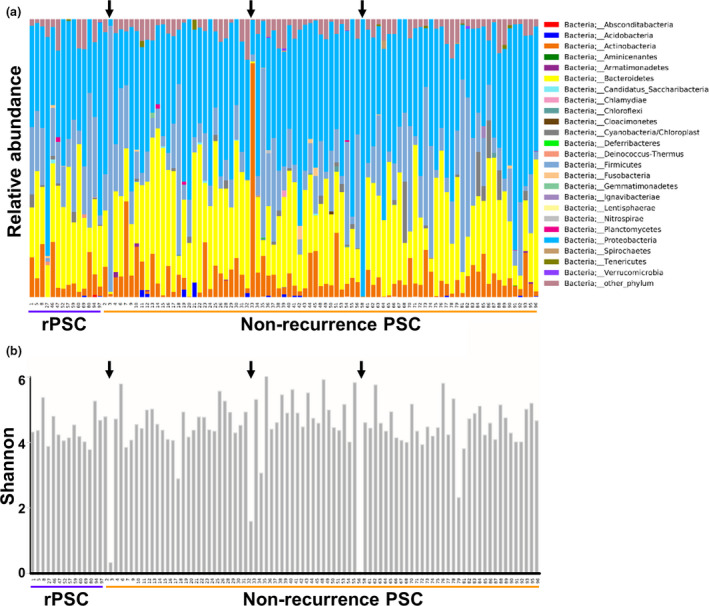
Mucosal microbiome of PSC patients. FFPE samples from PSC patients were collected pre‐LT, and microbial composition was determined. (a) Taxonomy plots at the phylum level. (b) Alpha diversity using the Shannon index. The black arrows point to patients with overgrowth of specific bacteria, corresponding to a reduced alpha diversity.
Low microbial diversity in mucosal samples from patients with CD‐PSC
To assess the validity of our set to investigate PSC and its recurrence, we first investigated whether known microbial changes seen in IBD patients were also present in our cohort. Fig. 2a,b shows that the colon mucosal microbiome of PSC patients with and without IBD shows no major differences for either alpha diversity (Chao P = 0.41; Shannon P = 0.69) or beta diversity (unweighted UniFrac P = 0.64; weighted UniFrac P = 0.90). However, when sub‐classifying IBD entities (Fig. 2c), a significantly lower alpha diversity was seen in CD patients as determined by the Shannon index (4.63 vs. 3.83, P = 0.03). Investigation of individual feature differences (Fig. 2d) showed a significantly increased abundance of the phylum Bacteroidetes in the samples of patients without IBD. Rhodobacteraceae (order) of the Rhodobacterales family was also more abundant in the non‐IBD group. On the other hand, PSC patients with IBD showed increased levels of the Proteobacterium Moraxellaceae. The corresponding genus, Acinetobacter, was also significantly more abundant in IBD‐PSC patients in our cohort as compared to PSC patients without IBD.
Figure 2.
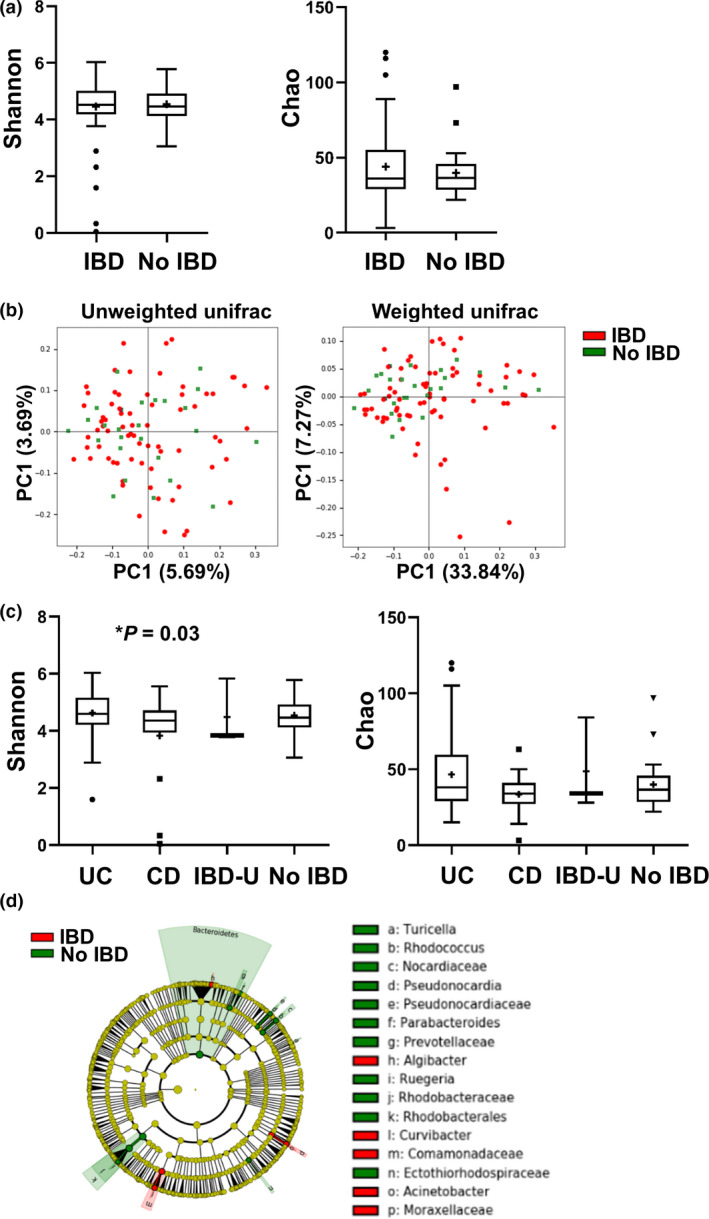
The microbiome of PSC‐CD patients is less diverse as compared to PSC‐UC and PSC. PSC patients were divided according to co‐occurring IBD disease (n = 71) or no IBD (n = 26). (a) Alpha diversity using the Shannon index (left panel) and Chao index (right panel). (b) Beta diversity using principal coordinate analysis (PCoA) of unweighted (left) and weighted (right) UniFrac distances. (c) PSC patients were subdivided according to IBD disease type: UC (n = 53), CD (n = 15), IBD‐U (n = 3) and no IBD (n = 26). Alpha diversity using the Shannon index (left panel) and Chao index (right panel). Significant changes are indicated with an asterisk. (d) Cladogram of significant differentially abundant microbial taxa obtained using LEfSe of PSC patients subdivided according to IBD disease.
Active colonic inflammation versus no colonic inflammation
As inflammation has been shown to affect the microbial composition, we next investigated the differences in microbial composition between samples demonstrating active inflammation versus inactive disease (Fig. 3). No significant differences were observed in alpha diversity between samples with and without inflammation. However, there was a significant difference in beta diversity between inflamed and noninflamed states using unweighted (P = 0.024), but not in weighted UniFrac (P = 0.62). LEfSe analysis showed several features that were significantly different between inflamed and noninflamed samples, which are summarized in Fig. 3c. Of note, an increased abundance of Fusobacteriaceae, Stenotrophomonas and Micrococcus was seen in inflamed tissues. Furthermore, an increased presence of Deinococcus–thermus was seen in inflamed tissues in our study. In inflamed mucosae, reduced levels of Bacteroidetes (phylum) and Caulobacterales (order) were seen.
Figure 3.
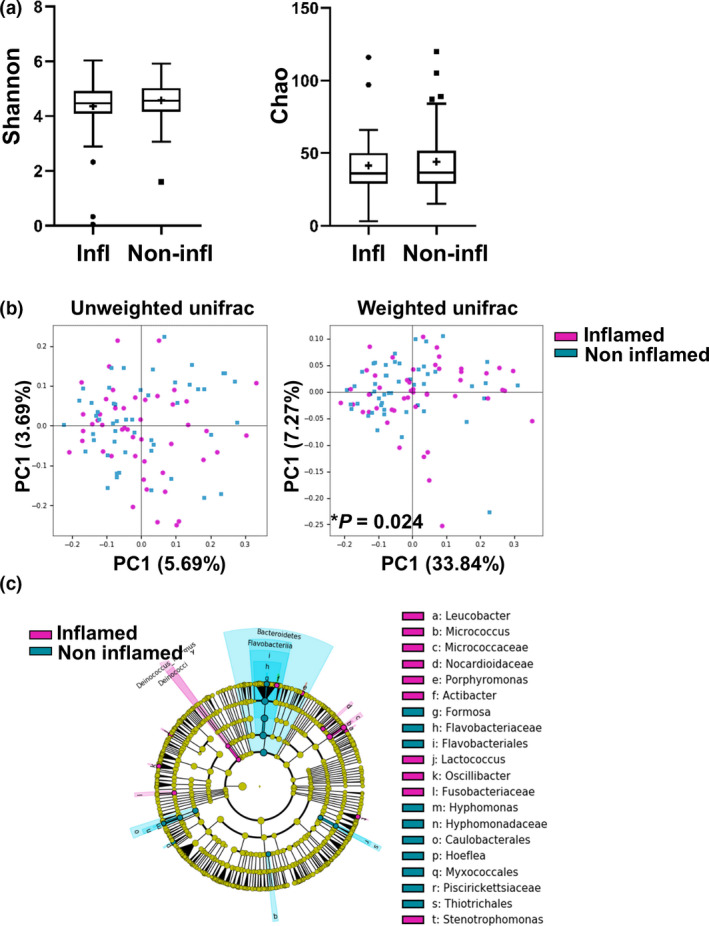
Mucosal inflammation in PSC patients is associated with microbial alterations. Mucosal biopsy specimens were grouped into those showing active inflammation at time of sampling (n = 47) versus having no active disease (n = 50). (a) Alpha diversity using the Shannon index (left panel) and Chao index (right panel). (b) Beta diversity using principal coordinate analysis (PCoA) of unweighted (left) and weighted (right) UniFrac distances. Significant changes are indicated with an asterisk. (c) Cladogram of significant differentially abundant microbial taxa obtained using LEfSe of samples according to inflammation stage.
Antibiotics use
In total, 34 (35%) patients used antibiotics within 3 months before colon sampling was performed. No significant differences were noted for both alpha diversity (Shannon P = 0.15; Chao P = 0.076, Figure S2A) and beta diversity (unweighted UniFrac P = 0.15; weighted UniFrac P = 0.69, Figure S2B) for patients who did or did not take antibiotics. Investigation of individual feature differences showed only minor changes, with an increase of mainly Gram‐negative bacteria in the group of patients who were on antibiotics (Figure S2C). To further explore the effect of antibiotics on mucosal microbiome composition, we took into account the length of antibiotics treatment. The usage of 11 (32%) patients was qualified as short‐term antibiotics treatment, whereas in 23 (68%) cases the usage qualified as prolonged. All patients with prolonged treatment were taking antibiotics on the day of sampling. When comparing these categories, no significant differences were noted either for alpha diversity or beta diversity (data not shown). The agents used were predominantly fluorquinoles and penicillins. Further analyses by antibiotic group were not feasible due to limited numbers. In total, these data suggest that use of antibiotics at the time of sampling did not result in major microbial alterations in the mucosa and is hence not a confounding factor in our study.
PSC versus rPSC
We next investigated whether differences in microbial signatures existed between rPSC patients and patients who did not develop rPSC. The alpha‐ and beta‐diversity indices showed no significant differences, indicating that there are no major differences in mucosal microbiome composition in PSC patients who do or do not develop rPSC (Fig. 4a,b). LEfSe analysis however shows extensive feature differences between the two groups, as shown in Fig. 4c and Figure S3. Gammaproteobacteria (class) and Shigella (genus) were significantly more abundant in patients who do not develop rPSC (Fig. 4d). An increased presence of several other microbial features was noted in rPSC, many of which were unclassified. Of note, Deinococcus_Thermus and Fusobacteraceae were increased in rPSC patients. Other pathogenic features increased in rPSC include Listeriaceae, Desulfosarcina [37] and Fastidiosipila [38].
Figure 4.
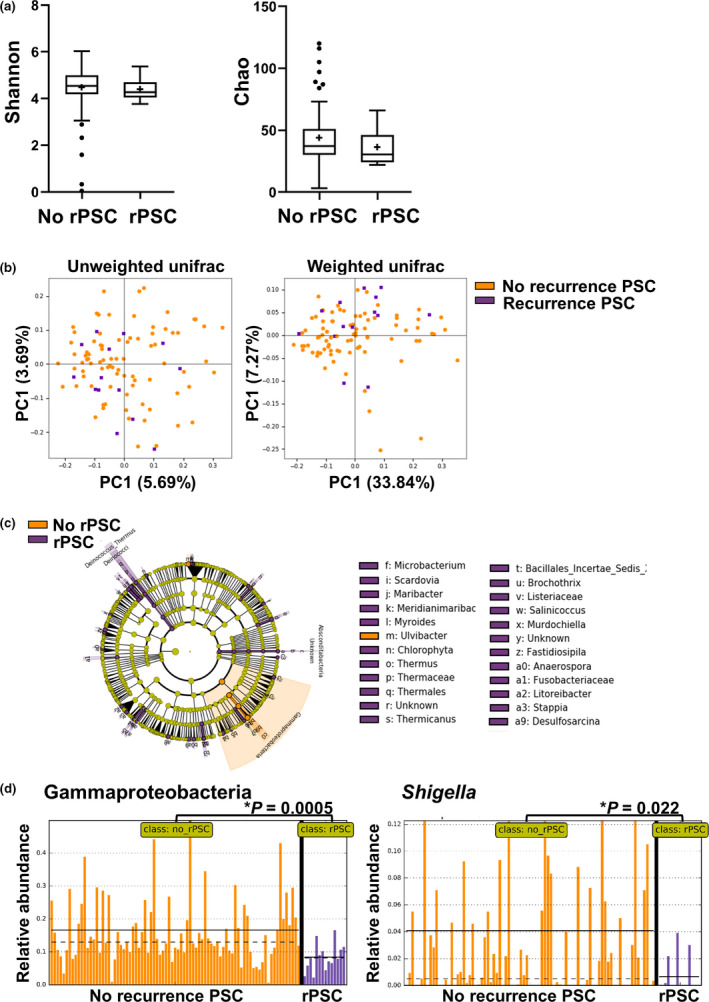
Specific changes in microbiome according to PSC recurrence status. Patients were grouped into those showing no recurrence of PSC after LT (no rPSC) (n = 83) and those showing recurrence during follow‐up (rPSC, n = 14). Significant changes are indicated with an asterisk. (a) Alpha diversity using the Shannon index (left panel) and Chao index (right panel). (b) Beta diversity using principal coordinate analysis (PCoA) of unweighted (left) and weighted (right) UniFrac distances. (c) Cladogram of significant differentially abundant microbial taxa obtained using LEfSe of samples separated according to recurrence or no recurrence of PSC. Legend is abbreviated – for full list of feature differences, see Figure S3. (d) Relative abundance plots for Gammaproteobacteria (left) and Shigella (right) according to PSC recurrence status. Mean and median are indicated by solid and dotted lines, respectively. Mean relative abundance of Gammaproteobacteria was 0.167 vs. 0.084 for no rPSC versus rPSC (P = 0.0005), and Mean relative abundance of Shigella was 0.0409 vs. 0.0066 for no rPSC versus rPSC (P = 0.022).
To investigate whether these feature differences were affected by confounding factors, we next performed multiple multivariable LEfSe analyses with a second variable, consecutively including the use of antibiotics, IBD, indication for listing, inflammation or site of biopsy. In all these analyses, the phylum Gammaproteobacteria remained significantly increased in non‐rPSC patients (Figure S4). The genus Shigella remained significant after adding all variables, except for IBD. All other feature differences were not significant after adding the variables, which are probably related to the limited number of samples in which certain bacteria were detected. When the one sample dominated by Shigella was removed from the analysis, both the phylum Gammaproteobacteria and the genus Shigella remained significantly higher in patients without rPSC (data not shown).
Discussion
In this study, we analysed the association between the gut microbiome before LT and development of rPSC after LT. Our approach is unprecedented in rPSC research with a large group of PSC patients after LT with biomaterials available. While rPSC has not been studied in this context, the colonic microbiome of PSC has been described before [19, 39, 40, 41, 42, 43, 44]. Various studies have indicated different microbial alterations in PSC patients. Some showed an overrepresentation of the genera Enterococcus, Lactobaccillus and Fusobacterium, others demonstrated enhanced presence of Proteobacteria and Parabacteroides [39], and yet others demonstrated enhanced levels of Rothia, Enterococcus, Streptococcus and Veillonella in PSC patients [41, 43]. However, most studies agree that richness and diversity of both the mucosal and faecal gut microbiome are considerably lower in PSC patients than in non‐PSC patients [19, 22].
Additionally, two previous studies found that the alterations in PSC microbiomes are independent of IBD status, with less IBD‐specific microbial changes in PSC patients [39, 43], suggesting that the end‐stage PSC microbiome blueprint may possibly be dominant over the IBD blueprint [45]. This may explain why in our study not all microbial signatures associated with IBD only seen in literature are reproduced. However, we did observe reduced alpha diversity in PSC patients with concomitant CD compared to UC. Furthermore, a markedly increased abundance of the phylum Proteobacteria was found in PSC‐IBD patients, which has been described in relation to both IBD and PSC before, and has been related to epithelial dysfunction and intestinal inflammation [46, 47]. In addition, the genus Acinetobacter, one of the main genera of the Proteobacteria phyla to contribute to IBD disease activity [48], was significantly more abundant in IBD‐PSC patients as compared to PSC patients without IBD. An increased abundance of Fusobacteriaceae, Stenotrophomonas and Micrococcus was seen in inflamed tissues, which is in accordance with previous investigations of IBD disease activity [49, 50, 51], while the found reduced levels of Bacteroidetes (phylum) and Caulobacterales (order) in inflamed tissue are consistent with mouse studies of colitis [52]. Thus, these findings are in line with previously published results for IBD and inflammation [53], indicating that the microbial signature present in the biopsies accurately represent disease state and that our sample is therefore useful for further exploration regarding rPSC.
In this study, we showed that mucosal presence of Gammaproteobacteria Shigella was significantly increased in patients who were not diagnosed with rPSC. The class Gammaproteobacteria is a large heterogenic group of Gram‐negative bacteria that are able to oxidize chemical bonds, hydrogen and sulphur to obtain their energy. This class contains several important pathogens: Salmonella spp., Yersinia pestis, Vibrio cholerae, Pseudomonas aeruginosa, Klebsiella, Escherichia coli and Shigella. Shigella is capable of invading into the colonic epithelium and the lamina propria, followed by a cytokine‐mediated inflammation of the colon and consecutive necrosis of the colonic epithelium. This results in ulcers in the colonic mucosa, causing bloody stools with mucus with or without febrile diarrhoea, which could mimic IBD flares [54, 55]. An overrepresentation of pathogenic bacteria in the gut of patients who are seemingly at lower risk of rPSC, seems counterintuitive. Shigella, however, has been linked to multiple immunomodulatory mechanisms which are believed to facilitate and prolong its colonization [55, 56]. These mechanisms could, directly or indirectly, have an effect on the development of rPSC as well. It has been suggested that Shigella releases mucolytic molecules to effectuate progression through the colonic mucus layer, a protective layer preventing the colonic epithelial cells from making contact with microbes. In reaction, probably to restore the mucus layer, induced mucin expression levels (MUC2, MUC5AC, MUC4 and MUC15) were detected after Shigella infection in both in vitro and in vivo models, the latter in a rat ileal loop model [57, 58, 59, 60]. Thus, it might be speculated that an improved mucus layer in the presence of Shigella prevents contact with and invasion of other microbes, resulting in less activation of the immune system and a reduced chance of rPSC. In addition, MUC2 and MUC5AC are both weakly expressed in cholangiocytes [61]. If Shigella would also activate mucin production in bile ducts, one could theoretically reason that this may be beneficial in protecting these cells against, thus far, unknown harmful insults involved in rPSC development. However, these are theoretical possibilities that need confirming data in future studies.
We acknowledge several strengths and weaknesses in our study. First, in most studies investigating PSC microbiome, faecal samples have been used. Here, we chose to use FFPE material, as prospective biobanking of fresh samples to address rPSC is logistically challenging, and would require long follow‐up times. While we took sections from multiple depths of the biopsy in order to capture the full spectrum of mucosal microbes as best we could, an underrepresentation as compared to whole biopsies may still have occurred. However, while the faecal microbiome is altered by everyday changes (e.g. food intake, alcohol consumption, exercise), the mucosal microbiome is more consistent over time [62]. Furthermore, the mucosal microbiome may be more relevant to disease aetiology as these bacteria have the ability to invade the colonic epithelium, thereby releasing toxins and metabolites into the portal bloodstream and eventually translocate to the liver [47]. Second, while in our centre 169 liver transplants were performed for PSC up to 2015, and all patients underwent a screening colonoscopy, samples were only available for 98 (58%) of these. While this is a relatively large cohort in PSC research, only 14 patients were diagnosed with rPSC. However, given our long follow‐up (median 10.7 years) and the fact that the majority of rPSC patients are diagnosed in the first 5 years after LT, it is unlikely that prolonging the follow‐up would have yielded more patients [9]. To verify the associations found in this pilot study, larger studies should be performed, ideally including both an IBD control group and patients undergoing LT for other liver diseases like primary biliary cholangitis or autoimmune hepatitis. Third, the diagnosis of rPSC is notoriously challenging, with other post‐LT complications such as nonanastomotic strictures mimicking the disease. However, we have used combined radiology, pathology and hepatology expert opinion to determine whether patients qualified for the diagnosis of rPSC, while applying the strict Mayo criteria. Lastly, all samples were taken just prior to the first LT, at which time all patients are affected by end‐stage PSC. Although this created a rather homogenous sample, it is conceivable that not all alterations in microbiomes are yet visible at this point in time. Post‐LT biopsies might have given additional insights, but unfortunately, these were not available as all biopsies were performed for clinical reasons and not protocoled in a research setting. It would be interesting to analyse changes in microbiome over time and correlate this to the onset of rPSC pathogenesis in future studies.
In conclusion, we demonstrate for the first time that the pre‐LT gut microbiome of patients undergoing LT for PSC and eventually developing rPSC post‐LT is associated with specific compositional alterations as compared to those not developing rPSC. It would be interesting to investigate to what extent the protective effect of colectomy on rPSC is associated with microbial load. As altering the gut microbiome could be an alternative to colectomy, further explorations in extending the criteria for colectomy before or during LT should be done with extensive caution. While it is too early for the development of effective and harmless ways to manipulate the microbiome for the prevention or treatment of rPSC, it is likely that further investigations in this field may eventually lead to considerable advancements in the foreseeable future [63, 64, 65].
Authorship
TV: designed research/study, performed research/study, collected data, analysed data and wrote paper. GMF: designed research/study, analysed data and wrote paper. NSE: designed research/study, analysed data and revised paper. YRAN: analysed data and revised paper. HJM, JNMI, SDM and MPP: designed research/study and revised paper.
Funding
The authors have declared no funding.
Conflicts of interest
The authors have no conflicts of interest to disclose.
Supporting information
Figure S1 Mucosal microbiome of PSC patients.
Figure S2 Antibiotics have minimal effect on mucosal microbiome from PSC patients.
Figure S3 Specific changes in microbiome according to PSC recurrence status.
Figure S4 Relative abundance plots for Gammaproteobacteria according to PSC recurrence status, using IBD disease status as confounder.
Acknowledgements
We would like to thank several colleagues for their time and expertise who helped us in performing this research. Auke Verhaar is acknowledged for his supervision during the experiments. Michael Doukas (pathologist) and Roy Dwarkasing (radiologist) are acknowledged for their expertise during the revision of rPSC diagnoses. Marith van Citteren, Kim de Groot ‐ Kreefft and Blerdi Blakaj are thanked for their assistance in the laboratory, and both Marcel Kap and Stef Luijmes are thanked for their help during the construction of the database.
References
- 1. Chapman RW, Arborgh BA, Rhodes JM, et al Primary sclerosing cholangitis: a review of its clinical features, cholangiography, and hepatic histology. Gut 1980; 21: 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ponsioen CY, Vrouenraets SM, Prawirodirdjo W, et al Natural history of primary sclerosing cholangitis and prognostic value of cholangiography in a Dutch population. Gut 2002; 51: 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hirschfield GM, Karlsen TH, Lindor KD, Adams DH. Primary sclerosing cholangitis. Lancet 2013; 382: 1587. [DOI] [PubMed] [Google Scholar]
- 4. Boonstra K, Weersma RK, van Erpecum KJ, et al Population‐based epidemiology, malignancy risk, and outcome of primary sclerosing cholangitis. Hepatology 2013; 58: 2045. [DOI] [PubMed] [Google Scholar]
- 5. Visseren T, Polak WG, Adam R, et al Recurrence of primary sclerosing cholangitis is associated with lower survival after liver transplantation: an analysis of the European liver transplant registry data base. J Hepatol 2017; 66: S200. [Google Scholar]
- 6. Lindstrom L, Jorgensen KK, Boberg KM, et al Risk factors and prognosis for recurrent primary sclerosing cholangitis after liver transplantation: a Nordic Multicentre Study. Scand J Gastroenterol. 2018; 53: 297. [DOI] [PubMed] [Google Scholar]
- 7. Ravikumar R, Tsochatzis E, Jose S, et al Risk factors for recurrent primary sclerosing cholangitis after liver transplantation. J Hepatol. 2015; 63: 1139. [DOI] [PubMed] [Google Scholar]
- 8. Trivedi PJ, Scalera I, Slaney E, et al Clinical outcomes of donation after circulatory death liver transplantation in primary sclerosing cholangitis. J Hepatol. 2017; 67: 957. [DOI] [PubMed] [Google Scholar]
- 9. Visseren T, Darwish Murad S. Recurrence of primary sclerosing cholangitis, primary biliary cholangitis and auto‐immune hepatitis after liver transplantation. Best Pract Res Clin Gastroenterol. 2017; 31: 187. [DOI] [PubMed] [Google Scholar]
- 10. Karlsen TH, Folseraas T, Thorburn D, Vesterhus M. Primary sclerosing cholangitis – a comprehensive review. J Hepatol. 2017; 67: 1298. [DOI] [PubMed] [Google Scholar]
- 11. Ji SG, Juran BD, Mucha S, et al Genome‐wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat Genet. 2017; 49: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang X, Karlsen TH. Genetics of primary sclerosing cholangitis and pathophysiological implications. Nat Rev Gastroenterol Hepatol. 2017; 14: 279. [DOI] [PubMed] [Google Scholar]
- 13. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 2012; 489: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Turnbaugh PJ, Ley RE, Hamady M, Fraser‐Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature 2007; 449: 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tabibian JH, Varghese C, LaRusso NF, O'Hara SP. The enteric microbiome in hepatobiliary health and disease. Liver Int 2016; 36: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pascal V, Pozuelo M, Borruel N, et al A microbial signature for Crohn's disease. Gut 2017; 66: 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hov JR, Kummen M. Intestinal microbiota in primary sclerosing cholangitis. Curr Opin Gastroenterol. 2017; 33: 85. [DOI] [PubMed] [Google Scholar]
- 18. Kevans D, Tyler AD, Holm K, et al Characterization of intestinal microbiota in ulcerative colitis patients with and without primary sclerosing cholangitis. J Crohns Colitis 2016; 10: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rossen NG, Fuentes S, Boonstra K, et al The mucosa‐associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J Crohns Colitis 2015; 9: 342. [DOI] [PubMed] [Google Scholar]
- 20. Torres J, Bao X, Goel A, et al The features of mucosa‐associated microbiota in primary sclerosing cholangitis. Aliment Pharmacol Ther 2016; 43: 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kummen M, Holm K, Anmarkrud JA, et al The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 2017; 66: 611. [DOI] [PubMed] [Google Scholar]
- 22. Sabino J, Vieira‐Silva S, Machiels K, et al Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 2016; 65: 1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quraishi MN, Sergeant M, Kay G, et al The gut‐adherent microbiota of PSC‐IBD is distinct to that of IBD. Gut 2017; 66: 386. [DOI] [PubMed] [Google Scholar]
- 24. Ruhlemann MC, Heinsen FA, Zenouzi R, Lieb W, Franke A, Schramm C. Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut 2017; 66: 753. [DOI] [PubMed] [Google Scholar]
- 25. Ruhlemann MC, Solovjeva MEL, Zenouzi R, et al Gut mycobiome of primary sclerosing cholangitis patients is characterised by an increase of Trichocladium griseum and Candida species. Gut 2019: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemoinne S, Kemgang A, Ben Belkacem K, et al Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 2020; 69: 92. [DOI] [PubMed] [Google Scholar]
- 27. Steenstraten IC, Sebib Korkmaz K, Trivedi PJ, et al Systematic review with meta‐analysis: risk factors for recurrent primary sclerosing cholangitis after liver transplantation. Aliment Pharmacol Ther. 2019; 49: 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trivedi PJ, Reece J, Laing RW, et al The impact of ileal pouch‐anal anastomosis on graft survival following liver transplantation for primary sclerosing cholangitis. Aliment Pharmacol Ther. 2018; 48: 322. [DOI] [PubMed] [Google Scholar]
- 29. Buchholz BM, Lykoudis PM, Ravikumar R, Pollok JM, Fusai GK. Role of colectomy in preventing recurrent primary sclerosing cholangitis in liver transplant recipients. World J Gastroenterol. 2018; 24: 3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Graziadei IW, Wiesner RH, Batts KP, et al Recurrence of primary sclerosing cholangitis following liver transplantation. Hepatology 1999; 29: 1050. [DOI] [PubMed] [Google Scholar]
- 31. Palleja A, Mikkelsen KH, Forslund SK, et al Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. 2018; 3: 1255. [DOI] [PubMed] [Google Scholar]
- 32. Dubinsky V, Reshef L, Bar N, et al Predominantly antibiotic‐resistant intestinal microbiome persists in patients with pouchitis who respond to antibiotic therapy. Gastroenterology 2019; 158: 610–624.e13. [DOI] [PubMed] [Google Scholar]
- 33. STRATEC.Molecular . RTP© Bacteria DNA Mini Kit 0515.
- 34. Caporaso JG, Kuczynski J, Stombaugh J, et al QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 2010; 7: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vázquez‐Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high‐throughput microbial community data. Gigascience 2013; 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Segata N, Izard J, Waldron L, et al Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp Biol Med (Maywood). 2004; 229: 586. [DOI] [PubMed] [Google Scholar]
- 38. Bay V, Griffiths B, Carter S, et al 16S rRNA amplicon sequencing reveals a polymicrobial nature of complicated claw horn disruption lesions and interdigital phlegmon in dairy cattle. Sci Rep 2018; 8: 15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ruhlemann M, Liwinski T, Heinsen FA, et al Consistent alterations in faecal microbiomes of patients with primary sclerosing cholangitis independent of associated colitis. Aliment Pharmacol Ther 2019; 50: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vaughn BP, Kaiser T, Staley C, et al A pilot study of fecal bile acid and microbiota profiles in inflammatory bowel disease and primary sclerosing cholangitis. Clin Exp Gastroenterol 2019; 12: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vieira‐Silva S, Sabino J, Valles‐Colomer M, et al Quantitative microbiome profiling disentangles inflammation‐ and bile duct obstruction‐associated microbiota alterations across PSC/IBD diagnoses. Nat Microbiol 2019; 4: 1826. [DOI] [PubMed] [Google Scholar]
- 42. Hov JR, Karlsen TH. The microbiome in primary sclerosing cholangitis: current evidence and potential concepts. Semin Liver Dis 2017; 37: 314. [DOI] [PubMed] [Google Scholar]
- 43. Bajer L, Kverka M, Kostovcik M, et al Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol 2017; 23: 4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tabibian JH, O'Hara SP, Lindor KD. Primary sclerosing cholangitis and the microbiota: current knowledge and perspectives on etiopathogenesis and emerging therapies. Scand J Gastroenterol 2014; 49: 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Halfvarson J, Brislawn CJ, Lamendella R, et al Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017; 2: 17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol 2015; 37: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Altomare A, Putignani L, Del Chierico F, et al Gut mucosal‐associated microbiota better discloses inflammatory bowel disease differential patterns than faecal microbiota. Dig Liver Dis 2019; 51: 648. [DOI] [PubMed] [Google Scholar]
- 48. Leung JM, Davenport M, Wolff MJ, et al IL‐22‐producing CD4+ cells are depleted in actively inflamed colitis tissue. Mucosal Immunol 2014; 7: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strauss J, Kaplan GG, Beck PL, et al Invasive potential of gut mucosa‐derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis 2011; 17: 1971. [DOI] [PubMed] [Google Scholar]
- 50. Bharadwaj RS. Role of bacteria in Inflammatory bowel disease (IBD). Int J Infect Dis 2016; 45: 134. [Google Scholar]
- 51. Dinakaran V, Mandape SN, Shuba K, et al Identification of specific oral and gut pathogens in full thickness colon of colitis patients: implications for colon motility. Front Microbiol 2018; 9: 3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martinez‐Herrero S, Larrayoz IM, Narro‐Iniguez J, et al Lack of adrenomedullin results in microbiota changes and aggravates azoxymethane and dextran sulfate sodium‐induced colitis in mice. Front Physiol 2016; 7: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang W, Jovel J, Halloran B, et al Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria. Inflamm Bowel Dis. 2015; 21: 1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hale TL, Keusch GT. Shigella In: Baron S, ed. Medical Microbiology. 4th edn. Galveston, TX: University of Texas Medical Branch at Galveston; 1996. [PubMed] [Google Scholar]
- 55. Anderson M, Sansonetti PJ, Marteyn BS. Shigella diversity and changing landscape: insights for the twenty‐first century. Front Cell Infect Microbiol 2016; 6: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cascales E, Buchanan SK, Duche D, et al Colicin biology. Microbiol Mol Biol Rev 2007; 71: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gopal A, Iyer SC, Gopal U, Devaraj N, Halagowder D. Shigella dysenteriae modulates BMP pathway to induce mucin gene expression in vivo and in vitro. PLoS One 2014; 9: e111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Prakash R, Bharathi Raja S, Devaraj H, Devaraj SN. Up‐regulation of MUC2 and IL‐1beta expression in human colonic epithelial cells by Shigella and its interaction with mucins. PLoS One 2011; 6: e27046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Raja SB, Murali MR, Devaraj H, Devaraj SN. Differential expression of gastric MUC5AC in colonic epithelial cells: TFF3‐wired IL1 beta/Akt crosstalk‐induced mucosal immune response against Shigella dysenteriae infection. J Cell Sci 2012; 125(Pt 3): 703. [DOI] [PubMed] [Google Scholar]
- 60. Sperandio B, Fischer N, Joncquel Chevalier‐Curt M, et al Virulent Shigella flexneri affects secretion, expression, and glycosylation of gel‐forming mucins in mucus‐producing cells. Infect Immun 2013; 81: 3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kasprzak A, Adamek A. Mucins: the old, the new and the promising factors in hepatobiliary carcinogenesis. Int J Mol Sci 2019; 20: 1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gevers D, Kugathasan S, Denson LA, et al The treatment‐naive microbiome in new‐onset Crohn's disease. Cell Host Microbe 2014; 15: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mooser C, Ganal‐Vonarburg SC. Microbiota as a cornerstone in the development of primary sclerosing cholangitis: paving the path for translational diagnostic and therapeutic approaches. Gut 2019; 68: 1353. [DOI] [PubMed] [Google Scholar]
- 64. Allegretti JR, Kassam Z, Carrellas M, et al Fecal microbiota transplantation in patients with primary sclerosing cholangitis: a pilot clinical trial. Am J Gastroenterol 2019; 114: 1071. [DOI] [PubMed] [Google Scholar]
- 65. Torres J, Palmela C, Brito H, et al The gut microbiota, bile acids and their correlation in primary sclerosing cholangitis associated with inflammatory bowel disease. United Eur Gastroenterol J 2018; 6: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Mucosal microbiome of PSC patients.
Figure S2 Antibiotics have minimal effect on mucosal microbiome from PSC patients.
Figure S3 Specific changes in microbiome according to PSC recurrence status.
Figure S4 Relative abundance plots for Gammaproteobacteria according to PSC recurrence status, using IBD disease status as confounder.


