Summary
Plastid‐encoded genes are coordinately transcribed by the nucleus‐encoded RNA polymerase (NEP) and the plastid‐encoded RNA polymerase (PEP). Resulting primary transcripts are frequently subject to RNA editing by cytidine‐to‐uridine conversions at specific sites. The physiological role of many editing events is largely unknown.
Here, we have used the CRISPR/Cas9 technique in rice to knock out a member of the PLS‐DYW subfamily of pentatricopeptide repeat (PPR) proteins.
We found that OsPPR16 is responsible for a single editing event at position 545 in the chloroplast rpoB messenger RNA (mRNA), resulting in an amino acid change from serine to leucine in the β‐subunit of the PEP. In striking contrast to loss‐of‐function mutations of the putative orthologue in Arabidopsis, which were reported to have no visible phenotype, knockout of OsPPR16 leads to impaired accumulation of RpoB, reduced expression of PEP‐dependent genes, and a pale phenotype during early plant development. Thus, by editing the rpoB mRNA, OsPPR16 is required for faithful plastid transcription, which in turn is required for Chl synthesis and efficient chloroplast development.
Our results provide new insights into the interconnection of the finely tuned regulatory mechanisms that operate at the transcriptional and post‐transcriptional levels of plastid gene expression.
Keywords: Chl biosynthesis, chloroplast development, plastid, RNA editing, RNA polymerase, transcription
Introduction
The chloroplast is the organelle where photosynthesis occurs, and its biogenesis and development are essential for plant development and crucial to crop productivity. Chloroplasts develop from proplastids, undifferentiated plastids that are present in meristems and embryonic tissues (Demarsy et al., 2006). In the absence of light, proplastids turn into etioplasts, a type of plastid that is arrested in chloroplast development. Upon illumination, proplastids and etioplasts develop into chloroplasts (Pogson et al., 2015; Armarego‐Marriott et al., 2019). The process of chloroplast development can be divided into three steps (Zhang et al., 2017). In the first step, plastid DNA synthesis and replication are activated. In the second step, genes encoding the plastid gene expression machinery are preferentially transcribed by nucleus‐encoded RNA polymerase (NEP) for rapid establishment of the plastid genetic system, leading to a strong increase in the transcription and translation levels of plastid‐encoded genes. In the final step, the activity of plastid‐encoded RNA polymerase (PEP) increases and is maintained at a high level, thus leading to strong expression of plastid‐encoded components of the photosynthetic apparatus. By contrast, NEP activity is reduced and kept at a low basal level. Impairments in the chloroplast biogenesis program result in a variety of abnormal phenotypes, such as embryo lethality, pigment‐deficient (e.g. albino) leaves, and retarded growth (Ruppel & Hangarter, 2007; Kadirjan‐Kalbach et al., 2012; Su et al., 2012; Tanoue et al., 2014; Lin et al., 2015; Lin et al., 2019; Wang et al., 2019; Xu et al., 2019). For example, in the pgp1pgp2 double mutant of Arabidopsis, the biosynthesis of phosphatidylglycerol (a major thylakoid membrane phospholipid) is blocked. This leads to impaired thylakoid membrane biogenesis and inhibition of chloroplast development, which in turn compromises seed germination (Tanoue et al., 2014). In a rice (Oryza sativa) mutant, a single base change in OsValRS2 (encoding an organellar Val‐tRNA synthetase) resulted in impaired chloroplast gene expression, in turn causing inefficient ribosome biogenesis, virescent to albino phenotypes in seedlings, and white panicles at heading (Wang et al., 2016). Although many genes involved directly or indirectly in chloroplast development have been identified, their complex interplay in the regulation of the biogenesis program is still far from being understood.
The chloroplast genome of seed plants contains approximately 130 genes, including 30 genes for transfer RNAs (tRNAs), approximately 30 genes related to plastid gene expression, and about 50 genes encoding photosynthesis‐associated proteins (Sugiura, 1992; Wakasugi et al., 2001; Kahlau et al., 2006; Daniell et al., 2016). The chloroplast has its own transcription and translation systems. Plastid‐encoded genes are coordinately transcribed by two RNA polymerases (RNAPs), dubbed NEP and PEP (Hedtke et al., 1997; Hess & Börner, 1999; Liere & Börner, 2007). NEP is encoded by RpoTp in the nuclear genome. PEP is composed of four core subunits (encoded by the plastid rpoA, rpoB, rpoC1, and rpoC2 genes), sigma factors, and a set of polymerase‐associated proteins encoded by nuclear genes. Plastid genes can be divided into three classes, depending on their promoter structures (Hajdukiewicz et al., 1997): PEP‐dependent genes (class I), PEP and NEP‐dependent genes (class II), and NEP‐dependent genes (class III). Photosynthesis‐related genes and most genes for tRNAs are PEP‐dependent genes. The four genes encoding the core subunits of PEP (rpoA, rpoB, rpoC1, and rpoC2) and the rpl23 gene (encoding ribosomal protein large subunit 23) were identified as NEP‐dependent genes (Zhelyazkova et al., 2012; Williams‐Carrier et al., 2014; Börner et al., 2015). When NEP or PEP functions are disturbed, the transcription of plastid genes becomes unbalanced, leading to abnormal chloroplast development and plant growth (Allison et al., 1996; Legen et al., 2002; Hricova et al., 2006; Courtois et al., 2007; Swiatecka‐Hagenbruch et al., 2008; Bock et al., 2014).
Pentatricopeptide repeat (PPR) proteins have been implicated in nearly all post‐transcriptional steps in plastid gene expression, including RNA editing (Cai et al., 2009; Toda et al., 2012; Hayes et al., 2015; Yap et al., 2015; Wang et al., 2017; Small et al., 2019), processing (Kazama & Toriyama, 2003; Hattori et al., 2007; Fujii et al., 2016; Wu et al., 2016), translation (Barkan et al., 2012; Zoschke et al., 2016), intron splicing (Hsieh et al., 2015; Aryamanesh et al., 2017; Ito et al., 2018; Sun et al., 2018; Wu et al., 2019), and messenger RNA (mRNA) stability (Beick et al., 2008; Hammani et al., 2016). In plastids of flowering plants, RNA editing results in alteration of the genetic information at the mRNA level through cytidine‐to‐uridine conversions at highly specific sites (Ichinose & Sugita, 2016; Lu, 2018). Several types of trans‐acting protein factors have been implicated in RNA editing (Chateigner‐Boutin et al., 2008; Tillich et al., 2009; Bentolila et al., 2012; Takenaka et al., 2012; Zhang et al., 2017; Sandoval et al., 2019; Small et al., 2019), with PPR proteins of the DYW type being the key factors that mediate the sequence‐specific recognition of editing sites (Okuda et al., 2009; Zhou et al., 2009; Barkan & Small, 2014; Jiang et al., 2018; Sun et al., 2018). A total of 491 PPR proteins have been found in rice (Chen et al., 2018), and 131 of these contain a DYW domain, suggesting that they could act as RNA editing factors (Salone et al., 2007). Of the rice DYW‐PPR proteins, 30 are predicted to be chloroplast localized. So far, 24 RNA editing sites have been identified in the rice plastid genome (Corneille et al., 2000), but which PPR proteins recognize these sites is not well understood. To date, only five rice PPR proteins (WSL5, DUA1, OsPPR4, OsPPR6 and OsPGL1) have been assigned to specific plastid RNA editing sites and implicated in the editing of altogether seven sites (Asano et al., 2013; Tang et al., 2017; Cui et al., 2018; Liu et al., 2018; Xiao et al., 2018). Also, although transplastomic studies in tobacco (Bock et al., 1994; Karcher et al., 2008; Loiacono et al., 2019) and mutant analyses in Arabidopsis (Robbins et al., 2009; Zhou et al., 2009; Bentolila et al., 2012; Ramos‐Vega et al., 2015) have revealed the importance of some RNA editing events in chloroplasts, the functional significance of many others remains unknown.
To investigate the effects of RNA editing on chloroplast development in rice, we have begun to use the CRISPR/Cas9 technique to knock out the 30 chloroplast‐localized PPR proteins containing a DYW motif. Here, we describe the generation and characterization of such knockout mutants for the OsPPR16 gene. We show that targeted inactivation of this DYW‐PPR gene results in a striking developmental phenotype, with leaves being pale during early plant development, whereas later leaves are largely unaffected. Analysis of all chloroplast editing sites revealed that editing of a single site in rpoB was defective in osppr16 mutants, resulting in decreased accumulation of the RpoB protein and low PEP activity. We provide evidence for low transcription of the trnE gene, resulting in strongly decreased Chl synthesis that in turn delays early chloroplast development.
Materials and Methods
Plant material and growth conditions
Rice cv Zhonghua 11 (Oryza sativa L. ssp. geng/japonica cv Zhonghua 11) was used in this study. OsPPR16 transgenic plants and Zhonghua 11 (as the wild‐type, WT) were planted in the experimental field of Huazhong Agricultural University, Wuhan, China (30.4°N, 114.2°E).
To analyze the phenotype of OsPPR16 loss of function, T1 plants were used. To this end, seeds were germinated on half‐strength MS medium (Murashige & Skoog, 1962) in a tissue culture room at 28°C under a 14 h : 10 h, light : dark, photoperiod until the five‐leaf stage. The seedlings were then transferred to the field and grown to maturity.
For norflurazon (NF; Sigma Aldrich) treatments, NF was added to the culture medium at 5 μM final concentration. Seeds were surface sterilized and then cultured with or without NF at 28°C under continuous illumination.
Knockout and knockdown of OsPPR16
The CRISPR/Cas9 technology was used to knock out OsPPR16. Two CRISPR/Cas9 vectors were constructed, as previously described (Ma et al., 2015). Three specific target sequences for OsPPR16 were designed using CRISPR‐P (Lei et al., 2014). Target1 and Target2 were cloned into the destination vector pYLCRISPR/Cas9‐MH, generating plasmid pYLCRISPR‐osppr16d, and Target3 was cloned into the destination vector pYLCRISPR/Cas9‐MH, generating pYLCRISPR‐osppr16s. The CRISPR vectors were introduced into rice cv Zhonghua 11 by Agrobacterium‐mediated transformation, as previously described (Hiei et al., 1994). The genotype of CRISPR/Cas9 plants was analyzed by PCR and direct sequencing of the amplification products. The sequences of targets and PCR primers are listed in Supporting Information Table S1.
RNA interference (RNAi) by double‐stranded RNA expression was used to knock down OsPPR16. An RNAi vector was constructed by amplifying a 275 bp fragment using Zhonghua 11 complementary DNA (cDNA) as a template and primers RNAi16‐F and RNAi16‐R (Table S1). The resulting amplification product was inserted into vector pDS1301 (Dai et al., 2007). The recombinant binary vector was transformed into Zhonghua 11 callus material. The relative expression of OsPPR16 in transgenic RNAi plants was analyzed by quantitative real‐time PCR (qRT‐PCR) using Real16‐F and Real16‐R as primers (Table S1).
Transmission electron microscopy
WT and osppr16d mutant leaves at the three and five‐leaf stages of plants were collected for transmission electron microscopy (TEM) processing as previously described (Leng et al., 2017).
RNA isolation and Northern blot analysis
Total RNA was extracted from leaf tissue using the Trizol reagent and following the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Northern blot analysis was performed as previously described (Kazama et al., 2008). Hybridization probes were labeled with digoxigenin (DIG) by PCR adding 0.01 μM DIG‐deoxyuridine triphosphate to the reaction mixture. The sequences of all primers are listed in Table S1.
Measurement of Chl contents
Samples of 0.015–0.035 g of fresh leaves were extracted with 1.8 ml of Chl extraction solution (volume ratio of ethanol : acetone : water 4.5 : 4.5 : 1). Chl content was measured using published methods (Mao et al., 2011).
Subcellular localization analyses
The coding region of OsPPR16 with the exception of the stop codon was amplified using genomic DNA as template and cloned into the pXCSG‐YFP vector (Feys et al., 2005). Rice protoplasts were transformed as described previously (Zhang et al., 2011). The yellow fluorescent protein (YFP) and the Chl fluorescence signal of the chloroplast were observed and photographed using a confocal microscope (FV 1200; Olympus, Toyko, Japan) after 16 h of dark culture of the transfected protoplasts at 28°C.
To further confirm the localization of OsPPR16 within the chloroplast, the coding region of OsPPR16 (without the stop codon) was cloned into the pAN580‐GFP vector (Yang et al., 2018) to generate pAN580‐OsPPR16GFP and co‐transformed with a PEND‐CFP construct (which expresses a fluorescent marker protein for plastid nucleoids; Terasawa & Sato, 2005) into rice protoplasts. The YFP, cyan fluorescent protein (CFP), and the Chl fluorescence signal of the chloroplast were subsequently observed by confocal laser‐scanning microscopy.
Sequence analyses and phylogenetic studies
The coding sequences (CDSs) of rpoB, ndhB, and ndhD in 79 land plants, whose plastid and nuclear genomes had been sequenced and annotated, were downloaded from the database of the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov). Homologous sequences of OsPPR16 in 79 land plants were identified using the NCBI’s Blastp search program. Multiple sequence alignments were conducted by Geneious 4.8.3 (www.geneious.com). The nucleotides corresponding to the editing sites of rpoB‐545, ndhB‐746, and ndhD‐887 were determined based on the aligned CDSs. A phylogenetic tree was constructed using Mega 5.0 (Tamura et al., 2011) and the neighbor‐joining method with 1000 bootstrap replicates. The accession numbers of RpoB, NdhB, NdhD, and the homologous of OsPPR16 are listed in Table S2.
Analysis of RNA editing, splicing, and relative gene expression
Total RNA was extracted with the Transzol reagent (TransGen Biotech Co. Ltd, Beijing, China). Thereafter, 2 μg of total RNA was reverse transcribed using M‐MLV reverse transcriptase (Invitrogen) and random primers to obtain cDNA according to the manufacturer’s instructions. Fragments containing editing sites were amplified by reverse transcription (RT)‐PCR using cDNA as template and previously described methods (Zhang et al., 2017). The RT‐PCR products were sequenced directly using the chain termination method. The sequences of all primers are listed in Table S1. RNA splicing was analyzed as described previously (Tan et al., 2014) using specific primers flanking the introns (Table S1). Relative gene expression was analyzed by qRT‐PCR using a FastStart Universal SYBR Green Master (Roche, Basel, Switzerland) on an ABI Wuantstudio 6 Flex Real‐Time PCR system. The amplification program was as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Relative gene expression was analyzed using the method (Livak & Schmittgen, 2001). Rice triosephosphate isomerase (OsTPI) was used as reference gene (Wang et al., 2016). Primer sequences are listed in Table S1.
Western blot analyses
Total protein samples of osppr16d and WT at the three and five‐leaf stages were isolated with an extraction buffer containing 50 mM Tris hydrochloride (pH 8.0), 150 mM sodium chloride, 1% NP40, and 2× protease inhibitor (COMPLETE; Roche). Protein concentrations were determined according to a previously described protocol (Bradford, 1976). Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, semi‐dry transferred onto polyvinylidene difluoride membranes, and incubated with antibodies. Signals were detected using the Clarity Western ECL Substrate kit (Bio‐Rad, Hercules, CA, USA) and visualized with an imaging system (ChemiDocTMXRS; Bio‐Rad). Heat‐shock protein 82 (HSP82) was used as an internal control. Antibodies against RpoB and HSP82 were obtained from Phytoab (www.phytoab.com) and BGI (Shenzhen, China), respectively. Antibodies against AtpB, PetB and PsaB were purchased from Agrisera (www.agrisera.com).
Results
Identification and characterization of the osppr16 mutant
To investigate the functional role of RNA editing in chloroplast biology in rice, we have begun to use the CRISPR/Cas9 technique to systematically knock out all members of the PLS‐DYW subfamily of PPR proteins, which are predicted to localize to the chloroplast. A striking developmental phenotype was observed in leaves of osppr16 mutants, which were pale at the early developmental stage, but normally green at later stages (Fig. 1). This interesting developmental gradient prompted us to investigate the function of OsPPR16 in detail.
Fig. 1.
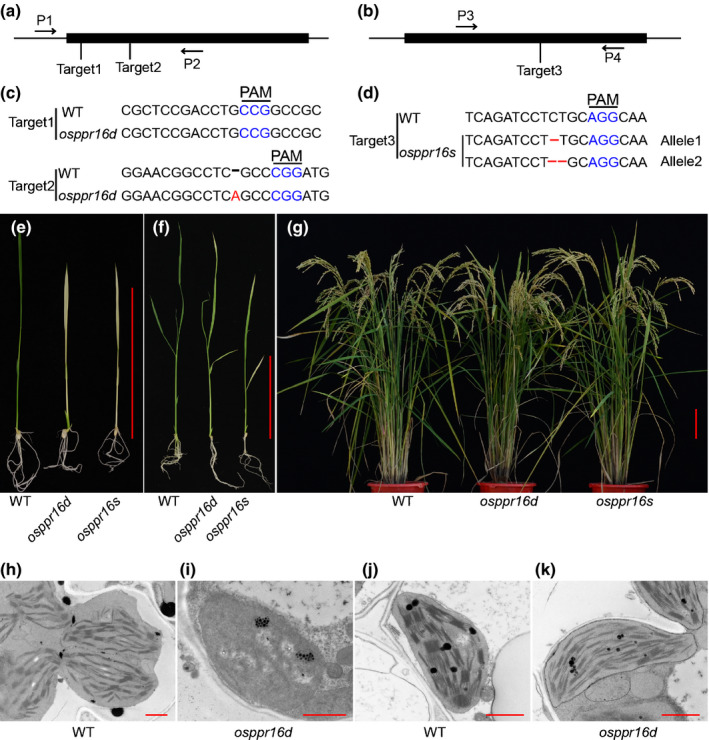
Generation and characterization of rice (Oryza sativa) osppr16 mutants. (a) Strategy I and (b) Strategy II for CRISPR/Cas9‐dependent gene editing of OsPPR16. Mutants obtained with Strategy I and Strategy II and selected for in‐depth characterization were named osppr16d and osppr16s, respectively. (c) Sequences around the two CRISPR targets (Target1 and Target2) in wild‐type (WT) and osppr16d. PAM, protospacer adjacent motif. (d) Sequence around Target3 in WT and osppr16s. (e) osppr16d, osppr16s, and WT plants at the three‐leaf stage. Bar, 10 cm. (f) osppr16d, osppr16s and WT plants at the five‐leaf stage. Bar, 10 cm. (g) osppr16d, osppr16s, and WT plants at seed set. Bar, 10 cm. (h, i) Chloroplast ultrastructure of (h) WT and (i) osppr16d at the three‐leaf stage. Bars, 1 μm. (j, k) Chloroplast ultrastructure of (j) WT and (k) osppr16d plants at the five‐leaf stage. Bars, 1 μm.
Two strategies were applied to knock out OsPPR16. One strategy involved a vector (pYLCRISPR‐osppr16d) that expresses two guide RNAs targeting two sites in the OsPPR16 gene (Fig. 1a). Thirty‐six T0 transgenic plants were obtained, 11 of which showed pale early leaves of tillers that gradually turned green (Fig. S1). At later developmental stages, no significant difference in leaf color was observed between transgenic and WT plants (Figs 1, S1). The other 25 transgenic lines were phenotypically indistinguishable from WT plants during the entire growth period. The genotype of the 36 T0 transgenic plants was assayed by direct sequencing of PCR products generated by amplification of the OsPPR16 locus (Fig. 1a). These analyses revealed that the gene was edited at Target1 and/or Target2 in the 11 lines with pale early leaves in tillers, whereas the 25 WT‐like lines had the WT OsPPR16 sequence or were heterozygous (Table S3).
Another CRISPR/Cas9 vector (pYLCRISPR‐osppr16s) was constructed that expresses a single guide RNA (Strategy II) that targets a site in the 3′ half of the OsPPR16 gene (Fig. 1b) and transformed into rice callus cells. This independent strategy was chosen first to provide additional evidence for the mutant phenotype being causally related to inactivation of the OsPPR16 gene, and second to rule out the possibility of off‐target genome editing by CRISPR/Cas9 (Fu et al., 2013; Lin et al., 2014; Jin et al., 2019). Thirty‐four T0 transgenic plants were obtained, 10 of which had the same phenotype as the mutant lines generated by Strategy I (Fig. S1c). The remaining 24 lines had a WT‐like phenotype, and their genotyping revealed that they were unmutated in OsPPR16 or heterozygous (Table S3). Taken together, these results strongly suggest that inactivation of OsPPR16 leads to pale early leaves of tillers in rice.
Two independently generated osppr16 lines were selected for further characterization: one line from Strategy I (named osppr16d) and one line from Strategy II (named osppr16s). In the osppr16d mutant, no base change was detected at Target1, whereas an additional adenine was inserted at Target2, resulting in a frameshift mutation (Fig. 1c). osppr16s represents a biallelic mutant, in which one nucleotide (C) was deleted in allele 1 and two nucleotides (CT) were deleted in allele 2 at Target3 (Fig. 1d). The leaves of both osppr16d and osppr16s T1 seedlings were pale during early leaf development (Fig. 1e) and turned green during the five‐leaf stage (Fig. 1f). A very similar phenotype of T1 plants was seen for T0 plants when they were transferred to the paddy field (Fig. S1). By contrast, leaf color of adult plants grown in the glasshouse or in the field was not different from the WT (Figs 1g, S1).
To investigate the effect of osppr16 mutation on chloroplasts biogenesis, chloroplast ultrastructure at the three‐leaf and five‐leaf stages was analyzed by TEM. Whereas chloroplasts in WT leaves showed well‐structured thylakoids at both the three‐leaf (Fig. 1h) and five‐leaf stages (Fig. 1j), the chloroplasts of osppr16d leaves were small and lacked organized thylakoid membranes at the three‐leaf stage (Fig. 1i). By contrast, chloroplasts containing well‐organized thylakoids (and stacked grana) were present at the five‐leaf stage (Fig. 1k). These results indicate that the OsPPR16 gene product plays an important role in the biogenesis of chloroplasts during early leaf development.
Chl synthesis in osppr16 is reduced during early leaf development
To further characterize the pigment‐deficient phenotype of the osppr16 mutants, the Chl content of leaves at different stages was analyzed (Fig. 2). The Chla contents in osppr16d and osppr16s leaves were 14.2% and 5.9%, respectively, of that in the WT at the three‐leaf stage and 71.1% and 70.9%, respectively, of the WT level at the five‐leaf stage. By contrast, no significant difference in Chla content was detectable between osppr16 and the WT at the time of seed set (Fig. 2a). The Chlb contents in osppr16d and osppr16s leaves were also much lower than in the WT at the three‐leaf stage, and, similar to Chla contents, partially recovered at the five‐leaf stage. However, in contrast to Chla contents, no full recovery to WT levels was seen in adult plants (Fig. 2b), although their total Chl content was not significantly different from that of the WT (Fig. 2c).
Fig. 2.
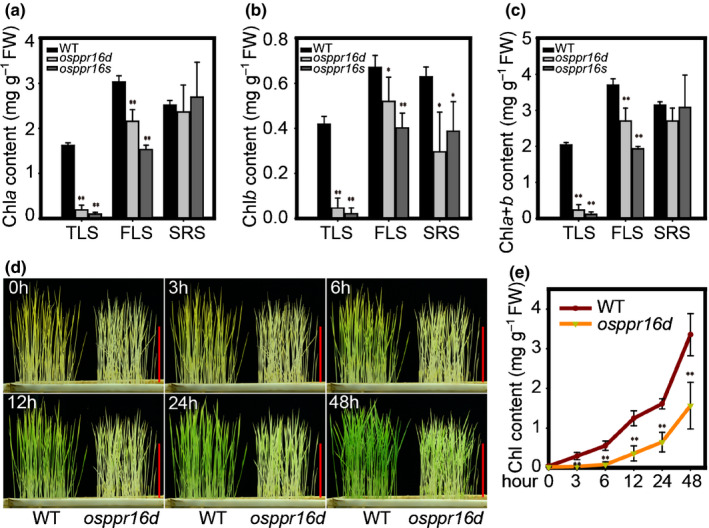
Analysis of Chl synthesis in rice (Oryza sativa) wild‐type (WT) and osppr16 mutant plants. (a–c) Chl content of leaves at different development stages in osppr16 and WT plants. The contents of (a) Chla, (b) Chlb, and (c) the total Chl are shown. TLS, three‐leaf stage; FLS, five‐leaf stage; SRS, seed ripening stage. Error bars indicate SD (n = 4), *, P < 0.05; **, P < 0.01 (Student’s t‐test). (d) Time course of de‐etiolation of WT and osppr16 seedlings. Plants were photographed 0, 3, 6, 12, 24, and 48 h after light treatment. Bars, 10 cm. (e) Chl contents in leaves of WT and osppr16d during the de‐etiolation time course. Error bars indicate SD (n = 4), **, P < 0.01 (Student’s t‐test).
To further investigate the effect of OsPPR16 on Chl synthesis, seeds of WT and osppr16d were germinated in soil in the dark at 28°C for 8 d and then treated with light. The Chl content of WT and osppr16d seedlings was analyzed at 0, 3, 6, 12, 24 and 48 h after lighting. After 3 h of lighting, the WT seedlings slightly turned green and the green color became obvious after 6 h of light treatment. By contrast, the greening of osppr16d seedlings was considerably delayed, and the seedlings did not turn visibly green until 12 h after lighting (Fig. 2d). After 48 h of de‐etiolation treatment, mutant seedlings were still only light green, whereas WT seedlings had virtually fully greened (Fig. 2d). The time course of Chl accumulation corresponded well with the visual observation of the greening process (Fig. 2e). These results suggest that inactivation of OsPPR16 reduces the rate of Chl synthesis or Chl stability.
OsPPR16 is a chloroplast‐localized PLS‐DYW subfamily PPR protein
Sequence analysis showed that the OsPPR16 gene contains a single exon (Fig. 1a). OsPPR16 is predicted to encode a PPR protein of 734 amino acids. Bioinformatics analysis of the sequence revealed that OsPPR16 consists of 14 PPR motifs, a typical DYW motif (representing the putative cytidine deaminase; Oldenkott et al., 2019), and a chloroplast transit peptide (Fig. 3a). Homologous proteins were identified by NCBI’s Blast to analyze the relationship between OsPPR16 and similar proteins in other species. The searches revealed that OsPPR16 shares 57.7% amino acid sequence identity with the PPR PLS‐DYW subfamily protein CRR22 in Arabidopsis thaliana (Okuda et al., 2009; Fig. S2). Phylogenetic tree analysis showed that proteins homologous to OsPPR16 are widely distributed across land plants (Fig. S3). CRR22 (encoded by At1g11290) was shown to be essential for the editing of the ndhB‐746, ndhD‐887 and rpoB‐551 sites in Arabidopsis (Okuda et al., 2009). The similarity of OsPPR16 to CRR22 suggested that OsPPR16 is a typical PLS‐DYW PPR protein and may also be involved in organellar RNA editing.
Fig. 3.
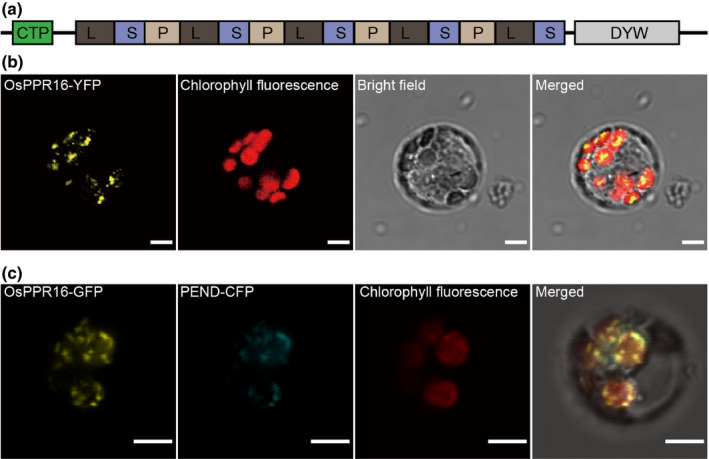
Domain structure and subcellular localization analysis of OsPPR16. (a) Structure of OsPPR16. CTP, chloroplast transit peptide. (b) Subcellular location of OsPPR16 in rice protoplasts. YFP, yellow fluorescent protein. Bars, 5 µm. (c) Co‐localization of OsPPR16‐GFP (yellow) and PEND‐CFP (cyan) in chloroplast nucleoids. Note that the GFP fluorescence was detected in the YFP channel, and therefore is represented as yellow color. GFP, green fluorescent protein; CFP, cyan fluorescent protein. Bars, 5 µm.
OsPPR16 was predicted to localize to the chloroplast by ChloroP (www.cbs.dtu.dk/services/ChloroP/) and Plant‐mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant‐multi/). To experimentally verify this prediction, a transformation vector expressing an OsPPR16‐YFP fusion protein was constructed and introduced into rice protoplasts. Analysis of transformed protoplasts by confocal laser‐scanning microscopy showed that the yellow fluorescence of OsPPR16‐YFP exclusively co‐localized with the Chl fluorescence of the chloroplasts (Fig. 3b). Interestingly, the YFP fluorescence was concentrated in small dot‐like structures within the transfected chloroplasts. Many chloroplast RNA‐binding proteins are associated with nucleoids, presumably because RNA processing initiates co‐transcriptionally (Majeran et al, 2012). To examine whether the dot‐like structures represent nucleoids, OsPPR16‐GFP was co‐expressed with PEND‐CFP, a fluorescent marker protein for plastid nucleoids (Terasawa & Sato, 2005). Indeed, the green fluorescent protein signals co‐localized with the CFP signals, indicating that OsPPR16 associates with chloroplast nucleoids (Fig. 3c).
Editing of rpoB‐545 is impaired in osppr16 mutant plants
Many PPR proteins, especially members of the PLS‐DYW subfamily, are involved in organellar RNA editing (Salone et al., 2007; Sun et al., 2016). To test whether this was the case for OsPPR16, the editing efficiency of all 24 editing sites in the rice chloroplast genome was examined in WT and osppr16d mutant leaves. cDNA sequencing revealed that editing of rpoB‐545 in osppr16d mutant plants was undetectable, whereas the other two editing sites in the rpoB mRNA (rpoB‐467 and rpoB‐560) and all other 21 editing sites in rice chloroplasts were edited normally and with similar efficiency as in the WT (Figs 4a, S4). RT‐PCR analysis also showed that splicing of all plastid introns occurred with similar efficiency as in the WT (Fig. S5). These results indicate that OsPPR16 is an editing factor specific for rpoB‐545.
Fig. 4.
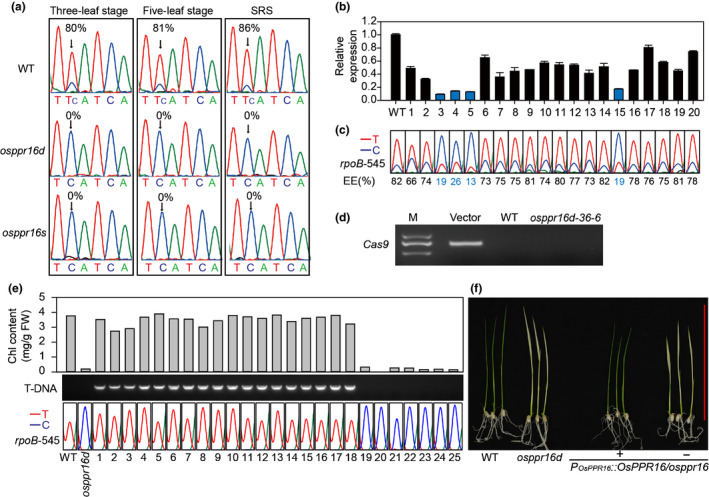
Editing analysis of rpoB‐545 in rice (Oryza sativa) wild‐type (WT), osppr16, RNA interference (RNAi), and complemented plants. (a) Editing efficiency of rpoB‐545 in WT and osppr16 leaves at three‐leaf, five‐leaf, and seed ripening stages. SRS, seed ripening stage. The arrows refer to the editing site. (b) Relative expression of OsPPR16 in RNAi transgenic plants, as determined by quantitative real‐time PCR. OsTPI was used as reference gene. (c) Editing efficiency of rpoB‐545 in OsPPR16 RNAi lines. EE, editing efficiency. (d) Assay for the Cas9 gene by PCR in osppr16d T1 plants. M, marker. Vector, pYLCRISPR‐osppr16d. The osppr16d‐36‐6 plant was confirmed as Cas9‐free line, and subsequently used to conduct complementation analysis. (e) Analysis of Chl content, transfer DNA (T‐DNA) presence, and RNA editing efficiency in WT, osppr16d, and T1 seedlings from a T0 complemented line at the three‐leaf stage. (f) Phenotypes of WT, osppr16d, and complemented T1 seedlings (POsPPR16::OsPPR16/osppr16). +, positive T1 seedlings segregated from a T0 complementation line; −, negative T1 seedlings segregated from a T0 complementation line. Bar, 10 cm.
RNAi was used to knock down OsPPR16 and determine the effect of reduced OsPPR16 expression on rpoB‐545 editing. Twenty T0 transgenic plants were generated, and the expression of OsPPR16 in lines 3, 4, 5 and 15 was determined to be less than 20% of that in the WT (Fig. 4b). When plastid RNA editing was assayed, the editing efficiency of rpoB‐545 in these RNAi lines was found to be more than 74% lower than that in the WT (Fig. 4c). In the other 16 lines, the expression level of OsPPR16 was only mildly decreased, correlating with an only slightly reduced editing efficiency of rpoB‐545 (Fig. 4b,c). These results confirm that OsPPR16 is required for the editing of rpoB‐545 in rice plastids.
Genetic complementation studies were also performed (according to Methods S1) to ultimately confirm that OsPPR16 was responsible for the mutant phenotype of osppr16. To this end, OsPPR16 fused to its native promoter was transformed into the osppr16d‐36‐6 mutant line, which was free of the CRISPR/Cas9 construct (Fig. 4d). Chl contents, presence of the transfer DNA, and RNA editing efficiency were analyzed in 25 T1 seedlings from a T0 complemented line. The Chl content and editing efficiency of rpoB‐545 were similar to that of the WT in all seedlings that tested positive for the complementation construct and had green leaves at the three‐leaf stage (Fig. 4e,f). Similar to the osppr16 mutant, seedlings not containing the complementation construct were Chl deficient and displayed impaired editing of rpoB‐545 (Fig. 4e,f). These results confirmed that OsPPR16 is required for the editing of rpoB‐545, and OsPPR16 disruption is the direct cause of the pale leaves of osppr16 during early leaf development.
Accumulation of the RpoB subunit of PEP is reduced in osppr16 mutant plants
To investigate the effect of impaired editing of rpoB‐545 on the RpoB protein, the catalytic subunit of the PEP, the accumulation of RpoB was determined by Western blot analysis in osppr16 and WT plants. At the three‐leaf stage, RpoB accumulation was much reduced in osppr16 compared with the WT (Fig. 5a). At the five‐leaf stage, the accumulation of RpoB was partly restored in osppr16 and only slightly lower than in WT plants at the same stage (Fig. 5a). Interestingly, accumulation of RpoB in the WT was considerably higher at the three‐leaf stage than at the five‐leaf stage (Fig. 5a). These results indicate that RpoB accumulation at an early developmental stage is severely affected in osppr16, presumably as a consequence of defective rpoB‐545 editing. In addition, the RpoB accumulation pattern in the WT suggests that more PEP is required for early chloroplast development than at later developmental stages.
Fig. 5.
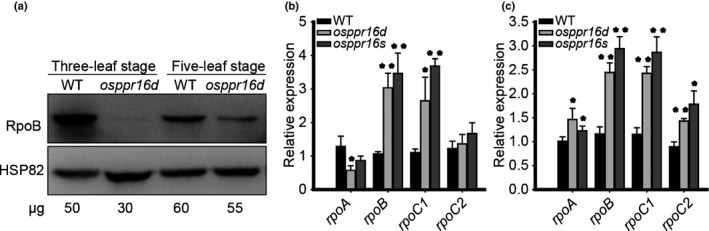
Protein and messenger RNA accumulation of plastid‐encoded RNA polymerase (PEP) core subunits in rice (Oryza sativa) wild‐type (WT) and osppr16 mutant plants. (a) Immunoblot analysis of RpoB in WT and osppr16 leaves at the three‐leaf and five‐leaf stages. An antibody against the heat‐shock protein HSP82 was used as an internal control. (b, c) Relative expression levels of the four genes encoding PEP core subunits in WT and osppr16 leaves at (b) the three‐leaf stage and (c) the five‐leaf stage, as determined by quantitative real‐time PCR. OsTPI was used as reference gene. Error bars indicate SD (n = 3); *, P < 0.05; **, P < 0.01 (Student’s t‐test).
PEP contains four core subunits encoded by the plastid genes rpoA, rpoB, rpoC1 and rpoC2. The expression levels of rpoA, rpoB, rpoC1 and rpoC2 were analyzed by qRT‐PCR. At the three‐leaf stage, expression of rpoB and rpoC1 in osppr16 was strongly upregulated compared with the WT (Fig. 5b). At the five‐leaf stage, expression of rpoB, rpoC1 and rpoC2 was increased in osppr16, whereas rpoA was only slightly upregulated compared with the WT (Fig. 5c). These results suggest that the mRNA levels of rpoB, rpoC1 and rpoC2 (which are NEP‐dependent genes) respond to the low PEP activity and may be regulated by a feedback mechanism that likely involves the NEP.
Structural basis of the PEP deficiency in the absence of editing at rpoB‐545
Impaired RNA editing of rpoB‐545 leads to reduced accumulation of the β‐subunit of PEP (Fig. 5a). To explore the possible molecular cause of the PEP deficiency in osppr16 mutant plants, we analyzed the structural consequences of the lack of rpoB RNA editing at the protein level.
Alignment of the amino acid sequences of the RNAP β‐subunits from five bacterial species (representing different phyla) shows a conserved leucine residue at the position corresponding to codon 182 of the rice rpoB gene (Fig. 6a). The crystal structure of the Thermus aquaticus RNAP‐promoter open complex shows that the conserved leucine (Leu197 in T. aquaticus) is buried inside the β‐lobe domain of the RNAP β‐subunit and is surrounded by hydrophobic residues (Fig. 6b,c; Bae et al., 2015), suggesting its importance in maintaining the hydrophobic core of the β‐lobe domain. In many plant species, the chloroplast gene for the β‐subunit of PEP contains a codon for a hydrophilic serine residue in the corresponding position (Fig. 6a), which is converted into a leucine codon by mRNA editing. Presence of a hydrophilic residue inside a hydrophobic domain potentially disrupts proper domain folding, thus decreasing protein stability and triggering proteolytic degradation (Dill & MacCallum, 2012). This explanation would be consistent with the reduced accumulation of the PEP β‐subunit in osppr16 mutant plants (Fig. 5a) and underscores the functional importance of RNA editing to restore the conserved leucine residue.
Fig. 6.
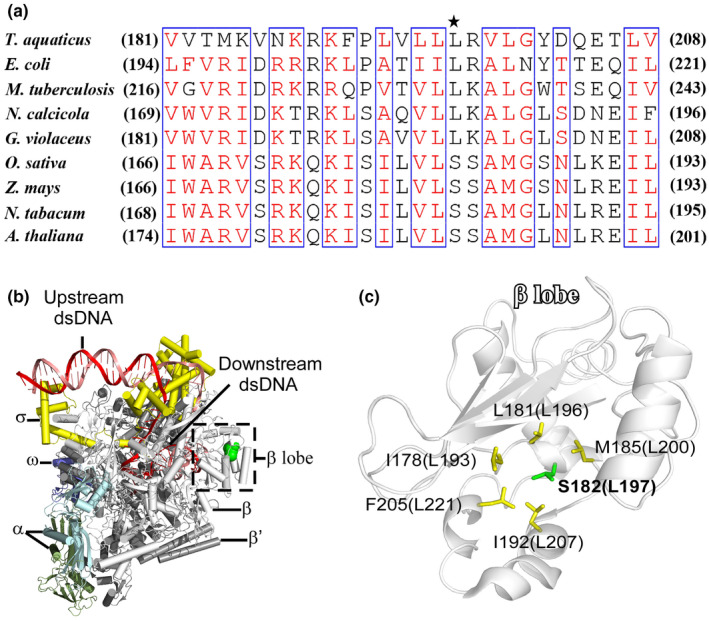
RNA editing at rpoB‐545 likely facilitates proper protein folding of the plastid‐encoded RNA polymerase (RNAP) β‐subunit. (a) Partial amino acid sequence alignment of RNAP β‐subunits of five bacterial species reveals a conserved leucine at the position corresponding to the edited codon (codon 182) of the rice chloroplast β‐subunit. By contrast, sequence alignment of chloroplast β‐subunits of four plant species reveals a serine at these positions (denoted by the asterisk). T. aquaticus, Thermus aquaticus; E. coli, Escherichia coli; M. tuberculosis, Mycobacterium tuberculosis; N. calcicola, Nostoc calcicola; G. violaceus, Gloeobacter violaceus; O. sativa, Oryza sativa; Z. mays, Zea mays; N. tabacum, Nicotiana tabacum; A. thaliana, Arabidopsis thaliana. (b) The crystal structure of T. aquaticus RNAP‐promoter open complex shows that L197 is located within the β‐lobe domain (dashed box) of the RNAP β‐subunit. The RNAP α, β, β′ and ω subunits are shown in cyan, gray, dark gray and blue, respectively, the promoter DNA is shown in red, and the σ‐factor is shown in yellow. (c) Amino acid L197 is buried inside the hydrophobic core of the β‐lobe domain. Numbers of amino acid residues correspond to the positions in the T. aquaticus RpoB protein. The equivalent positions in the O. sativa protein are given in parentheses.
Decreased PEP activity in osppr16 mutant plants during early leaf development
The accumulation of RpoB, the β‐subunit of PEP, was reduced in the absence of editing at rpoB‐545 in osppr16 mutants. To determine the consequences on PEP activity and transcription of the chloroplast genome, the expression of PEP‐dependent genes was analyzed by qRT‐PCR. In osppr16 mutant plants, the expression of PEP‐dependent genes such as atpB, psaA, psbB and rbcL was strongly reduced (to < 20% of the expression in the WT) at the three‐leaf stage (Fig. 7a). Interestingly, at the five‐leaf stage, the expression of most PEP‐dependent genes in osppr16 mutant plants had nearly fully recovered and was similar to that in the WT, with the exception that the expression of petN was slightly lower than in the WT (Fig. 7b). Immunoblot analysis of the accumulation of proteins encoded by PEP‐dependent genes confirmed the significance of the reduced mRNA levels at the level of the protein products (Fig. 7c,d). Together, these results indicate that PEP activity in osppr16 mutants is decreased during early leaf development but recovers at later developmental stages.
Fig. 7.
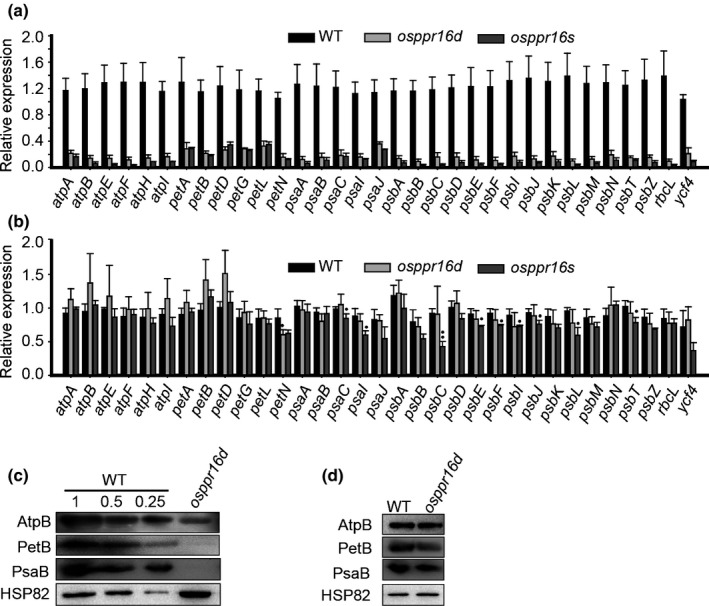
Transcript accumulation of plastid‐encoded RNA polymerase (PEP)‐dependent genes in rice (Oryza sativa) wild‐type (WT) and osppr16 mutant leaves. (a, b) Quantitative real‐time PCR analysis of the expression of PEP‐dependent genes in WT and osppr16 leaves at the (a) three‐leaf stage and (b) five‐leaf stage. OsTPI was used as reference gene. Error bars indicate SD (n = 3); *, P < 0.05; **, P < 0.01 (Student’s t‐test) in (b); all P < 0.01 in (a). (c) Immunoblot analysis of selected proteins encoded by PEP‐dependent genes in WT and osppr16 leaves at the three‐leaf stage. Samples of 25 μg total cellular protein extracted from osppr16d leaves were loaded and compared with a dilution series of WT protein extracts. The heat shock protein HSP82 was used as an internal control. (d) Immunoblot analysis of the same proteins at the five‐leaf stage. Samples of 25 μg total protein from WT and osppr16d leaves were analyzed by Western blot analysis. HSP82 was used as an internal control.
Low levels of tRNAGlu synthesis in osppr16 during early leaf development
tRNAGlu encoded by the chloroplast trnE gene is unique, in that it is not only required for chloroplast translation but also serves as substrate for Chl synthesis. In the committed step of the pathway, the enzyme glutamyl‐tRNA reductase (GluTR) utilizes charged tRNAGlu to synthesize the universal precursor for the biosynthesis of all tetrapyrroles: 5‐aminolevulinic acid (Beale, 1999). To explore the possibility that tRNAGlu provision limits Chl biosynthesis in osppr16 mutant plants, and thus is causally responsible for the pigment‐deficient phenotype, trnE gene expression was investigated by Northern blot analysis. Interestingly, at the three‐leaf stage, accumulation of tRNAGlu in osppr16 mutant plants was very low (Fig. 8a). By contrast, at the five‐leaf stage, tRNAGlu accumulation in the mutants had recovered and nearly reached WT levels (Fig. 8a). As controls, the accumulations of trnN and rpl23 were analyzed. trnN is a PEP and NEP‐dependent gene, according to the analysis of its promoter (Fig. S6a). The accumulation of trnN in osppr16 mutant plants was similar to that in the WT at the three‐leaf stage and even higher than in the WT at the five‐leaf stage (Fig. S6b). rpl23 is a NEP‐dependent gene (Zhelyazkova et al., 2012), and the accumulation of rpl23 transcripts in osppr16 mutant plants was higher than in the WT at the three‐leaf and five‐leaf stages (Fig. S6c). Together, these results suggest that tRNAGlu synthesis in osppr16 mutants is very strongly affected by the PEP deficiency in early leaf development.
Fig. 8.
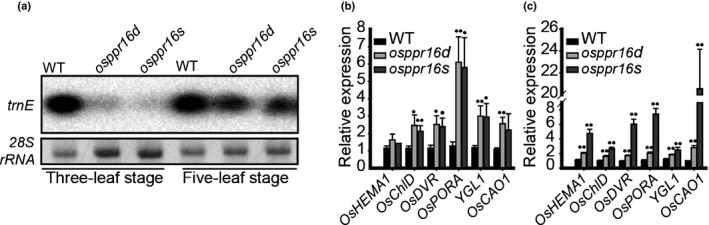
Expression of Chl biosynthesis‐related genes in rice (Oryza sativa) wild‐type (WT) and osppr16 mutant plants. (a) Northern blot analysis of accumulation of tRNAGlu in osppr16 mutants and WT plants at the three‐leaf stage and five‐leaf stage. 28S rRNA accumulation was used as internal loading control. (b, c) Quantitative real‐time PCR analysis of the expression of nuclear genes related to tetrapyrrole synthesis at the (b) three‐leaf stage and (c) five‐leaf stage. OsTPI was used as reference gene. Error bars indicate SD (n = 3); *, P < 0.05; **, P < 0.01 (Student’s t‐test).
To further explore the Chl deficiency in the mutants, the expression of key Chl synthesis genes (OsHEMA1, OsChlD, OsDVR, OsPORA, YGL1 and OsCAO1) was analyzed by qRT‐PCR. In the osppr16 mutants, all genes were found to be significantly upregulated at both the three‐leaf stage and the five‐leaf stage compared with WT plants (with the exception of OsHEMA1, which showed a significant increase only at the five‐leaf stage; Fig. 8b,c). These results suggest that, in osppr16 mutants, the expression of nuclear genes for components of the tetrapyrrole biosynthetic pathway responds to the Chl deficiency in the chloroplast.
osppr16 mutants display a gun phenotype
Expression of a large set of nuclear genes encoding plastid‐related functions is regulated by retrograde signals. Some retrograde signals emanate from developing chloroplasts (Singh et al., 2015) and control the expression of nuclear genes at the transcriptional and post‐transcriptional levels (Wu et al., 2019b, 2019). The genetic basis of retrograde signaling was revealed by the isolation of Arabidopsis mutants with disrupted retrograde signaling. These mutants were dubbed genomes uncoupled (gun) mutants, and most of them are related to genes involved in tetrapyrrole biosynthesis (Susek et al., 1993; Mochizuki et al., 2001; Larkin et al., 2003; Wu et al., 2018, 2019a). To explore whether retrograde signaling is affected in osppr16 mutants and determine whether the mutants display a gun phenotype at the molecular level, we analyzed the expression of OsLHCB2 and OsRBCS2, two classical target genes of GUN‐dependent retrograde signals. As a negative control, the expression of OsPPI2, a gene not regulated by plastid retrograde signals, was also analyzed. To visualize retrograde regulation, osppr16 and WT seedlings were grown in the presence or absence of NF at the three‐leaf stage. In the absence of NF, the expression levels of OsLHCB2 and OsRBCS2 were very similar in osppr16 mutants and the WT (Fig. S7a,b). By contrast, the expression of OsLHCB2 and OsRBCS2 in osppr16 mutants was higher (i.e. less repressed) than in the WT in the presence of NF (Fig. S7a,b). Expression of the control gene OsPPI2 showed no significant difference between osppr16 and the WT, neither in the absence nor in the presence of NF (Fig. S7c). Thus, osppr16 mutants appear to display a molecular gun phenotype that is suggestive of disturbed plastid retrograde signaling. Although the precise role of OsPPR16 in retrograde communication needs to be investigated further, it seems conceivable that OsPPR16 acts through its effects on the tetrapyrrole biosynthesis pathway and/or the plastid gene expression pathway of plastid retrograde signaling.
Discussion
In the course of this work, we have characterized a rice chloroplast RNA editing factor that acts in a striking developmental stage‐specific fashion. OsPPR16 is a chloroplast‐localized PLS‐DYW subfamily PPR protein (Fig. 3). We have shown that rpoB‐545 editing is undetectable in both pale and green leaves of osppr16 knockout mutants (Fig. 4a), whereas all other 23 chloroplast editing sites, including rpoB‐467 and rpoB‐560, are edited normally (Fig. S4). Complementation studies proved that rpoB‐545 editing in osppr16 can be restored by transgenic expression of OsPPR16 (Fig. 4e). Furthermore, knockdown of OsPPR16 leads to reduced editing efficiency at rpoB‐545 (Fig. 4b,c).
Previous studies have reported 21 PPR and PPR‐related proteins that are required in plastid RNA editing in Arabidopsis (Lu, 2018). Two PPR proteins (OsPPR4 and OsPPR6) have been shown to participate in editing of a single plastid RNA editing site in rice (Asano et al., 2013; Tang et al., 2017). In Arabidopsis, the expression of CRR22, which shares 57.7% amino acid identity with OsPPR16, is required for RNA editing of rpoB‐551, as well as of ndhB‐746 and ndhD‐887 (Okuda et al., 2009). In rice, the editing sites ndhB‐746 and ndhD‐887 are lost due to cytosine‐to‐thymine mutations at the DNA level. Thus, rpoB‐545 (corresponding to rpoB‐551 in Arabidopsis) remains as the only RNA editing target (Fig. S3). Phylogenetic analysis of OsPPR16 indicates that OsPPR16 homologues are conserved in many vascular plants but are absent from the moss Physcomitrella patens, in which none of the three editing sites (rpoB‐545, ndhB‐746 and ndhD‐887) is present (Fig. S3).
Although in Arabidopsis the editing of rpoB‐545, ndhB‐746 and ndhD‐887 is impaired in crr22 loss‐of‐function mutants, the mutant plants do not show an obvious phenotype under standard growth conditions (Okuda et al., 2009). In rice osppr16 mutants, only the editing at rpoB‐545 is defective, which, however, severely affects chloroplast development during early leaf development. The reason for this striking phenotypic difference between the Arabidopsis and rice mutants is currently unclear. A possible explanation could be that PEP activity plays a more important role in early chloroplast development in rice than in Arabidopsis. Alternatively, the edited event could be more important for the function of RpoB in rice than in Arabidopsis. Our findings reported here indicate that studies in a single species do not necessarily permit general conclusions about the functional relevance of RNA editing events in plant organelles.
In the Arabidopsis ys1 mutant, lack of editing at another site in the rpoB mRNA, rpoB‐338 (corresponding to position rpoB‐332 in rice, which does not undergo RNA editing, because a T is encoded at the DNA level) leads to a virescent phenotype at early leaf development that disappears 3 wk after germination (Zhou et al., 2009). This phenotype is similar to that of our osppr16 mutants and may suggest that high PEP activity is also crucial to early leaf development in Arabidopsis.
Although the rpoB mRNA level was increased in osppr16 mutants, the accumulation of the encoded β‐subunit of PEP was severely decreased at the three‐leaf stage (Fig. 5). Consequently, the expression of PEP‐dependent genes in osppr16 mutant plants was strongly affected at the three‐leaf stage (Fig. 7a), but virtually restored in the green leaves at the five‐leaf stage (Fig. 7b). Conversely, the expression of NEP‐dependent genes was upregulated (Fig. 5b,c) in the mutants, which is a known consequence of PEP deficiency (Allison et al., 1996; Hajdukiewicz et al., 1997). Taken together, our results indicate that the lack of rpoB editing in osppr16 mutants leads to low PEP activity, which limits chloroplast biogenesis in early leaf development, but is sufficient to sustain nearly normal levels of chloroplast gene expression at later stages of leaf development, when the biogenesis of the photosynthetic apparatus is completed and the demand for chloroplast RNA synthesis decreases. Our results do not entirely exclude the possibility that, through its RNA‐binding activity, OsPPR16 also directly regulates some other plastid genes, in addition to its function in RNA editing of rpoB‐545. Complementation of osppr16 with an ‘edited’ rpoB allele (encoding a leucine residue at the DNA level) would be a suitable experiment to ultimately confirm that the editing function of OsPPR16 is solely responsible for the greening defect observed in the osppr16 mutant. However, in the absence of a chloroplast transformation technology for rice, this experiment is currently not technically feasible.
In addition to participating in plastid protein synthesis, tRNAGlu is also the precursor for Chl synthesis (Beale, 1999). Thus, decreased accumulation of tRNAGlu can affect Chl synthesis and chloroplast biogenesis (Zhou et al., 2009; Börner et al., 2015). Consistent with such a limiting role, tRNAGlu accumulation in osppr16 mutants was found to be extremely low at the three‐leaf stage (Fig. 8a). Nearly full recovery at the five‐leaf stage corresponded well with the increase in Chl content and may suggest that the pattern of tRNAGlu accumulation in leaf development explains the developmental phenotype of the osppr16 mutant plants. We therefore propose a model (Fig. 9) in which active OsPPR16 expression in young leaves (Fig. S8) is required to edit rpoB transcripts, thus ensuring efficient RpoB protein accumulation. This leads to production of a sufficiently large and active pool of PEP that is capable of transcribing PEP‐dependent genes to levels that are sufficient to satisfy the high demands of the chloroplast biogenesis program. The accumulation of tRNAGlu is required to meet the large demand for Chl synthesis during the rapid development of proplastids and etioplasts to chloroplasts. At the same time, high PEP activity is also needed to synthesize Chl‐binding proteins (photosystem subunits), many of which are also encoded by PEP‐dependent genes. Loss of OsPPR16 leads to an extremely low level of tRNAGlu (and proteins encoded by other PEP‐dependent genes), thereby limiting Chl synthesis in the developing chloroplasts and leading to the pigment‐deficient phenotype during early leaf development. In the WT, the level of RpoB decreases at the five‐leaf stage, in response to the decreasing demands of plastid gene expression, now that chloroplast development is largely completed. By contrast, the accumulation of RpoB in osppr16 mutants increases over time to gradually overcome the PEP deficiency. Consequently, the accumulation of tRNAGlu and proteins encoded by other PEP‐dependent genes increases, ultimately reaching levels that are similar to those in the WT and sufficient to facilitate leaf greening.
Fig. 9.
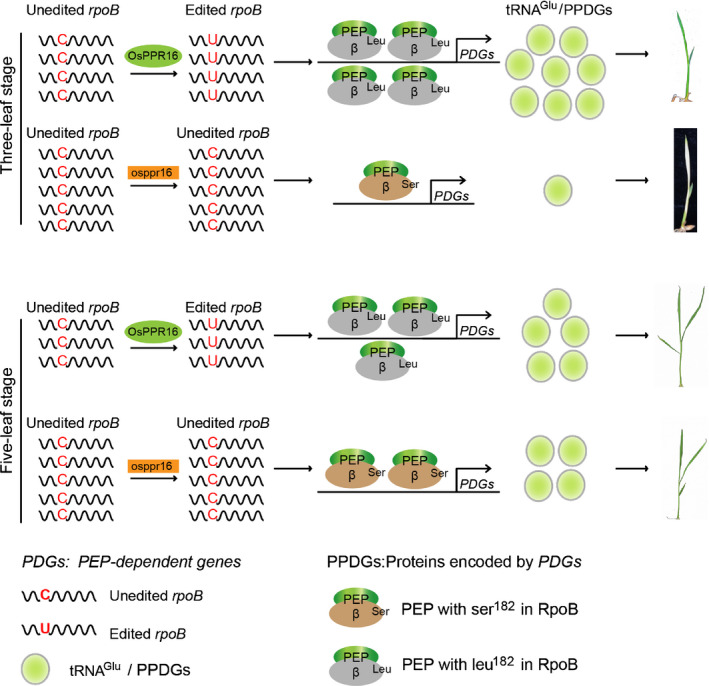
A working model for the role of OsPPR16 during chloroplast development in rice. During early chloroplast development, OsPPR16 is particularly important to edit rpoB transcripts to ensure maximum RpoB accumulation, thereby producing an active plastid‐encoded RNA polymerase (PEP) pool that is capable of efficiently transcribing PEP‐dependent genes (PDGs). The accumulation of tRNAGlu and proteins encoded by other PEP‐dependent genes is required to meet the demand for Chl and protein synthesis during the rapid development of proplastids into chloroplasts. Loss of OsPPR16 leads to an extremely low level of tRNAGlu (and proteins encoded by other PEP‐dependent genes, PPDGs), thereby limiting Chl biosynthesis and protein synthesis (and, potentially, also protein stability in the absence of sufficient pigments for incorporation into Chl‐binding proteins) in the developing chloroplasts, thus causing the pale phenotype during early leaf development. Lower demands for PEP activity at later developmental stages and gradually increasing PEP accumulation in the mutants alleviate the phenotype and facilitate leaf greening. See the Discussion section for more details.
In conclusion, our study suggests that OsPPR16 is needed for the efficient accumulation of RpoB by specifically editing site rpoB‐545. In particular, the high demand for Chl synthesis during early leaf development requires high PEP activity (for trnE gene expression) and provides a likely molecular explanation of the striking developmental phenotype of the rice osppr16 mutants. Our work highlights interesting differences in the function of editing PPRs in monocots vs dicots and links RNAP deficiency to a specific molecular process (tetrapyrrole biosynthesis) that becomes limiting in the chloroplast developmental program.
Author contributions
FZ, YL and WH conceived the project and designed the experiments. WH performed most of the experiments, assisted by Yongli Zhu, CZ and HC. Yang Zhang performed the qRT‐PCR analysis. LS and Yu Zhang performed the protein structural analysis. QF and CM performed the subcellular localization analyses. QL performed the splicing analyses. CG and Yong Zhou performed the bioinformatics analyses. JZ helped with the Northern blot. WH prepared all the figures and wrote the manuscript with contributions from FZ and RB. All the authors read and approved the final manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phenotype of rice (Oryza sativa) osppr16 mutant and WT plants at the tillering stage.
Fig. S2 Alignment of the amino acid sequences of OsPPR16 and its closest homologues from Arabidopsis thaliana (NP_172596.1, CRR22), Nicotiana tabacum (XP_016505961.1) and Zea mays (NP_001356569.1).
Fig. S3 Phylogenetic analysis of OsPPR16 and the nucleotide positions corresponding to the editing sites rpoB‐545, ndhB‐746 and ndhD‐887 in 79 land plants.
Fig. S4 Editing efficiency of 24 plastid RNA editing sites in rice (Oryza sativa) WT and osppr16 mutant plants.
Fig. S5 Splicing analyses of rice chloroplast transcripts in rice (Oryza sativa) WT and osppr16d mutant plants at the three‐leaf stage.
Fig. S6 Expression of trnN and rpl23 in rice (Oryza sativa) WT and osppr16 mutant plants.
Fig. S7 osppr16 mutants show a gun phenotype on norflurazon.
Fig. S8 Tissue‐specific expression profiles of OsPPR16 in rice (Oryza sativa) plants.
Methods S1 Complementation of the osppr16 mutant.
Table S1 List of oligonucleotides used in this study.
Table S2 Accession numbers of RpoB, NdhB, NdhD and homologous of OsPPR16 in 79 land plants.
Table S3 Genotype and phenotype of T0 transgenic plants.
Acknowledgements
We thank Professor Yaoguang Liu (South China Agricultural University) for providing the Cas9 vector. We are grateful to Professor Guozhang Wu (Shanghai Jiao Tong University) for helpful comments on the manuscript. We thank Dr Xia Li and the Core Facility Platform, Institute of Crop Sciences, Chinese Academy of Agricultural Science for their assistance with TEM analyses. This research was financially supported by the National Key Research and Development Program of China (2016YFD0100904), National Nature Science Foundation of China (31771752), and Fundamental Research Funds for the Central Universities (2662019PY083).
References
- Allison LA, Simon LD, Maliga P. 1996. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO Journal 15: 2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Armarego‐Marriott T, Kowalewska L, Burgos A, Fischer A, Thiele W, Erban A, Strand D, Kahlau S, Hertle A, Kopka J et al 2019. Highly resolved systems biology to dissect the etioplast‐to‐chloroplast transition in tobacco leaves. Plant Physiology 180: 654–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryamanesh N, Ruwe H, Sanglard LV, Eshraghi L, Bussell JD, Howell KA, Small I, des Francs‐Small CC. 2017. The pentatricopeptide repeat protein EMB2654 is essential for trans‐splicing of a chloroplast small ribosomal subunit transcript. Plant Physiology 173: 1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Miyao A, Hirochika H, Kikuchi S, Kadowaki K‐i. 2013. A pentatricopeptide repeat gene of rice is required for splicing of chloroplast transcripts and RNA editing of ndhA . Plant Biotechnology 30: 57–64. [Google Scholar]
- Bae B, Feklistov A, Lass‐Napiorkowska A, Landick R, Darst SA. 2015. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife 4: e08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. 2012. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genetics 8: e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Beale SI. 1999. Enzymes of chlorophyll biosynthesis. Photosynthesis Research 60: 43–73. [Google Scholar]
- Beick S, Schmitz‐Linneweber C, Williams‐Carrier R, Jensen B, Barkan A. 2008. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Molecular and Cellular Biology 28: 5337–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Heller WP, Sun T, Babina AM, Friso G, van Wijk KJ, Hanson MR. 2012. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proceedings of the National Academy of Sciences, USA 109: E1453–E1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Kossel H, Maliga P. 1994. Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO Journal 13: 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock S, Ortelt J, Link G. 2014. AtSIG6 and other members of the sigma gene family jointly but differentially determine plastid target gene expression in Arabidopsis thaliana . Frontiers in Plant Science 5: e667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV. 2015. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochimica et Biophysica Acta (BBA) – Bioenergetics 1847: 761–769. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Analytical Biochemistry 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Cai W, Ji D, Peng L, Guo J, Ma J, Zou M, Lu C, Zhang L. 2009. LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis . Plant Physiology 150: 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner‐Boutin AL, Ramos‐Vega M, Guevara‐García A, Andrés C, de la Luz Gutiérrez‐Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I et al 2008. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. The Plant Journal 56: 590–602. [DOI] [PubMed] [Google Scholar]
- Chen G, Zou Y, Hu J, Ding Y. 2018. Genome‐wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genomics 19: e720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneille S, Lutz K, Maliga P. 2000. Conservation of RNA editing between rice and maize plastids: are most editing events dispensable? Molecular and General Genetics 264: 419–424. [DOI] [PubMed] [Google Scholar]
- Courtois F, Merendino L, Demarsy E, Mache R, Lerbs‐Mache S. 2007. Phage‐type RNA polymerase RPOTmp transcribes the rrn operon from the PC promoter at early developmental stages in Arabidopsis . Plant Physiology 145: 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Wang Y, Wu J, Han X, Gu X, Lu T, Zhang Z. 2018. The RNA editing factor DUA1 is crucial to chloroplast development at low temperature in rice. New Phytologist 221: 834–849. [DOI] [PubMed] [Google Scholar]
- Dai M, Hu Y, Zhao Y, Liu H, Zhou DX. 2007. A WUSCHEL‐LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiology 144: 380–390.17351053 [Google Scholar]
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biology 17: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs‐Mache S. 2006. Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiology 142: 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill KA, MacCallum JL. 2012. The protein‐folding problem, 50 years on. Science 338: 1042–1046. [DOI] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina‐Escobar N, Neu C, Cabral A, Parker JE. 2005. Arabidopsis SENESCENCE‐ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. The Plant Cell 17: 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High‐frequency off‐target mutagenesis induced by CRISPR‐Cas nucleases in human cells. Nature Biotechnology 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Suzuki T, Giege P, Higashiyama T, Koizuka N, Shikanai T. 2016. The Restorer‐of‐fertility‐like 2 pentatricopeptide repeat protein and RNase P are required for the processing of mitochondrial orf291 RNA in Arabidopsis. The Plant Journal 86: 504–513. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P. 1997. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO Journal 16: 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Takenaka M, Miranda R, Barkan A. 2016. A PPR protein in the PLS subfamily stabilizes the 5′‐end of processed rpl16 mRNAs in maize chloroplasts. Nucleic Acids Research 44: 4278–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Miyake H, Sugita M. 2007. A pentatricopeptide repeat protein is required for RNA processing of clpP pre‐mRNA in moss chloroplasts. Journal of Biological Chemistry 282: 10773–10782. [DOI] [PubMed] [Google Scholar]
- Hayes ML, Dang KN, Diaz MF, Mulligan RM. 2015. A conserved glutamate residue in the C‐terminal deaminase domain of pentatricopeptide repeat proteins is required for RNA editing activity. Journal of Biological Chemistry 290: 10136–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A. 1997. Mitochondrial and chloroplast phage‐type RNA polymerases in Arabidopsis . Science 277: 809–811. [DOI] [PubMed] [Google Scholar]
- Hess WR, Börner T. 1999. Organellar RNA polymerases of higher plants. International Review of Cytology 190: 1–59. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. The Plant Journal 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Hricova A, Quesada V, Micol JL. 2006. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis . Plant Physiology 141: 942–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh WY, Liao JC, Chang CY, Harrison T, Boucher C, Hsieh MH. 2015. The SLOW GROWTH3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH Dehydrogenase Subunit7 intron 2 in Arabidopsis. Plant Physiology 168: 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose M, Sugita M. 2016. RNA editing and its molecular mechanism in plant organelles. Genes 8: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Sugita C, Ichinose M, Kato Y, Yamamoto H, Shikanai T, Sugita M. 2018. An evolutionarily conserved P‐subfamily pentatricopeptide repeat protein is required to splice the plastid ndhA transcript in the moss Physcomitrella patens and Arabidopsis thaliana . The Plant Journal 94: 638–648. [DOI] [PubMed] [Google Scholar]
- Jiang T, Zhang J, Rong L, Feng Y, Wang Q, Song Q, Zhang L, Ouyang M. 2018. ECD1 functions as an RNA editing trans‐factor of rps14‐149 in plastids and is required for early chloroplast development. Journal of Experimental Botany 69: 3037–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, Wang D, Qiu JL, Zhang F et al 2019. Cytosine, but not adenine, base editors induce genome‐wide off‐target mutations in rice. Science 364: 292–295. [DOI] [PubMed] [Google Scholar]
- Kadirjan‐Kalbach DK, Yoder DW, Ruckle ME, Larkin RM, Osteryoung KW. 2012. FtsHi1/ARC1 is an essential gene in Arabidopsis that links chloroplast biogenesis and division. The Plant Journal 72: 856–867. [DOI] [PubMed] [Google Scholar]
- Kahlau S, Aspinall S, Gray JC, Bock R. 2006. Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. Journal of Molecular Evolution 63: 194–207. [DOI] [PubMed] [Google Scholar]
- Karcher D, Kahlau S, Bock R. 2008. Faithful editing of a tomato‐specific mRNA editing site in transgenic tobacco chloroplasts. RNA 14: 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K. 2008. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT‐type cytoplasmic male sterile rice. The Plant Journal 55: 619–628. [DOI] [PubMed] [Google Scholar]
- Kazama T, Toriyama K. 2003. A pentatricopeptide repeat‐containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male‐sterile rice. FEBS Letters 544: 99–102. [DOI] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906. [DOI] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM. 2002. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild‐type and PEP‐deficient transcription machineries. The Plant Journal 31: 171–188. [DOI] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL. 2014. Crispr‐P: a web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Molecular Plant 7: 1494–1496. [DOI] [PubMed] [Google Scholar]
- Leng Y, Yang Y, Ren D, Huang L, Dai L, Wang Y, Chen L, Tu Z, Gao Y, Li X et al 2017. A rice PECTATE LYASE‐LIKE gene is required for plant growth and leaf senescence. Plant Physiology 174: 1151–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K, Börner T. 2007. Transcription and transcriptional regulation in plastids In: Bock R, ed. Cell and molecular biology of plastids. Berlin, Germany: Springer, 121–174. [Google Scholar]
- Lin D, Gong X, Jiang Q, Zheng K, Zhou H, Xu J, Teng S, Dong Y. 2015. The rice ALS3 encoding a novel pentatricopeptide repeat protein is required for chloroplast development and seedling growth. Rice 8: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Zhang L, Mei J, Chen J, Piao Z, Lee G, Dong Y. 2019. Mutation of the rice TCM12 gene encoding 2,3‐bisphosphoglycerate‐independent phosphoglycerate mutase affects chlorophyll synthesis, photosynthesis and chloroplast development at seedling stage at low temperatures. Plant Biology 21: 585–594. [DOI] [PubMed] [Google Scholar]
- Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, Wile BM, Vertino PM, Stewart FJ, Bao G. 2014. CRISPR/Cas9 systems have off‐target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Research 42: 7473–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lan J, Huang Y, Cao P, Zhou C, Ren Y, He N, Liu S, Tian Y, Nguyen T et al 2018. WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. Journal of Experimental Botany 69: 3949–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Loiacono FV, Thiele W, Schottler MA, Tillich M, Bock R. 2019. Establishment of a heterologous RNA editing event in chloroplasts. Plant Physiology 181: 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. 2018. RNA editing of plastid‐encoded genes. Photosynthetica 56: 48–61. [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y et al 2015. A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ. 2012. Nucleoid‐enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiology 158: 156–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Yu H, Liu T, Yang G, Xing Y. 2011. Two complementary recessive genes in duplicated segments control etiolation in rice. TAG. Theoretical and Applied Genetics 122: 373–383. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg‐chelatase H subunit in plastid‐to‐nucleus signal transduction. Proceedings of the National Academy of Sciences, USA 98: 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15: 473–497. [Google Scholar]
- Okuda K, Chateigner‐Boutin AL, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. 2009. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. The Plant Cell 21: 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenkott B, Yang Y, Lesch E, Knoop V, Schallenberg‐Rudinger M. 2019. Plant‐type pentatricopeptide repeat proteins with a DYW domain drive C‐to‐U RNA editing in Escherichia coli . Communications Biology 2: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht‐Borth V. 2015. Insights into chloroplast biogenesis and development. Biochimica et Biophysica Acta 1847: 1017–1024. [DOI] [PubMed] [Google Scholar]
- Ramos‐Vega M, Guevara‐Garcia A, Llamas E, Sanchez‐Leon N, Olmedo‐Monfil V, Vielle‐Calzada JP, Leon P. 2015. Functional analysis of the Arabidopsis thaliana CHLOROPLAST BIOGENESIS 19 pentatricopeptide repeat editing protein. New Phytologist 208: 430–441. [DOI] [PubMed] [Google Scholar]
- Robbins JC, Heller WP, Hanson MR. 2009. A comparative genomics approach identifies a PPR‐DYW protein that is essential for C‐to‐U editing of the Arabidopsis chloroplast accD transcript. RNA 15: 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppel NJ, Hangarter RP. 2007. Mutations in a plastid‐localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana . BMC Plant Biology 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salone V, Rudinger M, Polsakiewicz M, Hoffmann B, Groth‐Malonek M, Szurek B, Small I, Knoop V, Lurin C. 2007. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Letters 581: 4132–4138. [DOI] [PubMed] [Google Scholar]
- Sandoval R, Boyd RD, Kiszter AN, Mirzakhanyan Y, Santibańez P, Gershon PD, Hayes ML. 2019. Stable native RIP9 complexes associate with C‐to‐U RNA editing activity, PPRs, RIPs, OZ1, ORRM1 and ISE2. Plant Journal 99: 1116–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Singh S, Parihar P, Singh VP, Prasad SM. 2015. Retrograde signaling between plastid and nucleus: a review. Journal of Plant Physiology 181: 55–66. [DOI] [PubMed] [Google Scholar]
- Small ID, Schallenberg‐Rudinger M, Takenaka M, Mireau H, Ostersetzer‐Biran O. 2019. Plant organellar RNA editing: what 30 years of research has revealed. The Plant Journal 101: 1040–1056. [DOI] [PubMed] [Google Scholar]
- Su N, Hu ML, Wu DX, Wu FQ, Fei GL, Lan Y, Chen XL, Shu XL, Zhang X, Guo XP et al 2012. Disruption of a rice pentatricopeptide repeat protein causes a seedling‐specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiology 159: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M. 1992. The chloroplast genome. Plant Molecular Biology 19: 149–168. [DOI] [PubMed] [Google Scholar]
- Sun F, Zhang X, Shen Y, Wang H, Liu R, Wang X, Gao D, Yang YZ, Liu Y, Tan BC. 2018. The pentatricopeptide repeat protein EMPTY PERICARP 8 is required for the splicing of three mitochondrial introns and seed development in maize. The Plant Journal 95: 919–932. [DOI] [PubMed] [Google Scholar]
- Sun T, Bentolila S, Hanson MR. 2016. The unexpected diversity of plant organelle RNA editosomes. Trends in Plant Science 21: 962–973. [DOI] [PubMed] [Google Scholar]
- Sun YK, Gutmann B, Yap A, Kindgren P, Small I. 2018. Editing of chloroplast rps14 by PPR editing factor EMB2261 is essential for Arabidopsis development. Frontiers in Plant Science 9: e841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799. [DOI] [PubMed] [Google Scholar]
- Swiatecka‐Hagenbruch M, Emanuel C, Hedtke B, Liere K, Börner T. 2008. Impaired function of the phage‐type RNA polymerase RpoTp in transcription of chloroplast genes is compensated by a second phage‐type RNA polymerase. Nucleic Acids Research 36: 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Kugelmann M, Härtel B, Brennicke A. 2012. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proceedings of the National Academy of Sciences, USA 109: 5104–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. Mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Tan Z, Wu F, Sheng P, Heng Y, Wang X, Ren Y, Wang J, Guo X, Zhang X et al 2014. A novel chloroplast‐localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Molecular Plant 7: 1329–1349. [DOI] [PubMed] [Google Scholar]
- Tang J, Zhang W, Wen K, Chen G, Sun J, Tian Y, Tang W, Yu J, An H, Wu T. 2017. OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Molecular Biology 95: 345–357. [DOI] [PubMed] [Google Scholar]
- Tanoue R, Kobayashi M, Katayama K, Nagata N, Wada H. 2014. Phosphatidylglycerol biosynthesis is required for the development of embryos and normal membrane structures of chloroplasts and mitochondria in Arabidopsis . FEBS Letters 588: 1680–1685. [DOI] [PubMed] [Google Scholar]
- Terasawa K, Sato N. 2005. Visualization of plastid nucleoids in situ using the PEND–GFP fusion protein. Plant and Cell Physiology 46: 649–660. [DOI] [PubMed] [Google Scholar]
- Tillich M, Hardel SL, Kupsch C, Armbruster U, Delannoy E, Gualberto JM, Lehwark P, Leister D, Small ID, Schmitz‐Linneweber C. 2009. Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proceedings of the National Academy of Sciences, USA 106: 6002–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Fujii S, Noguchi K, Kazama T, Toriyama K. 2012. Rice MPR25 encodes a pentatricopeptide repeat protein and is essential for RNA editing of nad5 transcripts in mitochondria. The Plant Journal 72: 450–460. [DOI] [PubMed] [Google Scholar]
- Wakasugi T, Tsudzuki T, Sugiura M. 2001. The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynthesis Research 70: 107–118. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ren Y, Zhou K, Liu L, Wang J, Xu Y, Zhang H, Zhang L, Feng Z, Wang L. 2017. WHITE STRIPE LEAF4 encodes a novel P‐type PPR protein required for chloroplast biogenesis during early leaf development. Frontiers in Plant Science 8: e1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang C, Zheng M, Lyu J, Xu Y, Li X, Niu M, Long W, Wang D, Wang H et al 2016. WHITE PANICLE1, a Val‐tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiology 170: 2110–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhong P, Zhang X, Liu J, Zhang C, Yang X, Wan C, Liu C, Zhou H, Yang B et al 2019. GRA78 encoding a putative S‐sulfocysteine synthase is involved in chloroplast development at the early seedling stage of rice. Plant Science 280: 321–329. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang Y, Yang J, Hu K, An B, Deng X, Li Y. 2016. Reliable selection and holistic stability evaluation of reference genes for rice under 22 different experimental conditions. Applied Biochemistry and Biotechnology 179: 753–775. [DOI] [PubMed] [Google Scholar]
- Williams‐Carrier R, Zoschke R, Belcher S, Pfalz J, Barkan A. 2014. A major role for the plastid‐encoded RNA polymerase complex in the expression of plastid transfer RNAs. Plant Physiology 164: 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GZ, Chalvin C, Hoelscher M, Meyer EH, Wu XN, Bock R. 2018. Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiology 176: 2472–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GZ, Meyer EH, Richter AS, Schuster M, Ling Q, Schottler MA, Walther D, Zoschke R, Grimm B, Jarvis RP et al 2019a. Control of retrograde signalling by protein import and cytosolic folding stress. Nature Plants 5: 525–538. [DOI] [PubMed] [Google Scholar]
- Wu GZ, Meyer EH, Wu S, Bock R. 2019b. Extensive posttranscriptional regulation of nuclear gene expression by plastid retrograde signals. Plant Physiology 180: 2034–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Ren Y, Cai M, Wang Y, Zhu S, Zhu J, Hao Y, Teng X, Zhu X, Jing R et al 2019. Rice FLOURY ENDOSPERM10 encodes a pentatricopeptide repeat protein that is essential for the trans‐splicing of mitochondrial nad1 intron 1 and endosperm development. New Phytologist 223: 736–750. [DOI] [PubMed] [Google Scholar]
- Wu W, Liu S, Ruwe H, Zhang D, Melonek J, Zhu Y, Hu X, Gusewski S, Yin P, Small ID et al 2016. SOT1, a pentatricopeptide repeat protein with a small MutS‐related domain, is required for correct processing of plastid 23S–4.5S rRNA precursors in Arabidopsis thaliana . The Plant Journal 85: 607–621. [DOI] [PubMed] [Google Scholar]
- Xiao H, Xu Y, Ni C, Zhang Q, Zhong F, Huang J, Liu W, Peng L, Zhu Y, Hu J. 2018. A rice dual‐localized pentatricopeptide repeat protein is involved in organellar RNA editing together with OsMORFs. Journal of Experimental Botany 69: 2923–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Marino G, Klingl A, Enderle B, Monte E, Kurth J, Hiltbrunner A, Leister D, Kleine T. 2019. Extrachloroplastic PP7L functions in chloroplast development and abiotic stress tolerance. Plant Physiology 180: 323–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ren Y, Cai Y, Niu M, Feng Z, Jing R, Mou C, Liu X, Xiao L, Zhang X. 2018. Overexpression of OsbHLH107, a member of the basic helix–loop–helix transcription factor family, enhances grain size in rice (Oryza sativa L.). Rice 11: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap A, Kindgren P, Colas des Francs‐Small C, Kazama T, Tanz SK, Toriyama K, Small I. 2015. AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. The Plant Journal 81: 661–669. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D et al 2011. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast‐related processes. Plant Methods 7: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Cui X, Wang Y, Wu J, Gu X, Lu T. 2017. The RNA editing factor WSP1 is essential for chloroplast development in rice. Molecular Plant 10: 86–98. [DOI] [PubMed] [Google Scholar]
- Zhelyazkova P, Sharma CM, Forstner KU, Liere K, Vogel J, Börner T. 2012. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid‐encoded RNA polymerase. The Plant Cell 24: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Cheng Y, Yap A, Chateigner‐Boutin AL, Delannoy E, Hammani K, Small I, Huang J. 2009. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant Journal 58: 82–96. [DOI] [PubMed] [Google Scholar]
- Zoschke R, Watkins KP, Miranda RG, Barkan A. 2016. The PPR‐SMR protein PPR53 enhances the stability and translation of specific chloroplast RNA s in maize. The Plant Journal 85: 594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phenotype of rice (Oryza sativa) osppr16 mutant and WT plants at the tillering stage.
Fig. S2 Alignment of the amino acid sequences of OsPPR16 and its closest homologues from Arabidopsis thaliana (NP_172596.1, CRR22), Nicotiana tabacum (XP_016505961.1) and Zea mays (NP_001356569.1).
Fig. S3 Phylogenetic analysis of OsPPR16 and the nucleotide positions corresponding to the editing sites rpoB‐545, ndhB‐746 and ndhD‐887 in 79 land plants.
Fig. S4 Editing efficiency of 24 plastid RNA editing sites in rice (Oryza sativa) WT and osppr16 mutant plants.
Fig. S5 Splicing analyses of rice chloroplast transcripts in rice (Oryza sativa) WT and osppr16d mutant plants at the three‐leaf stage.
Fig. S6 Expression of trnN and rpl23 in rice (Oryza sativa) WT and osppr16 mutant plants.
Fig. S7 osppr16 mutants show a gun phenotype on norflurazon.
Fig. S8 Tissue‐specific expression profiles of OsPPR16 in rice (Oryza sativa) plants.
Methods S1 Complementation of the osppr16 mutant.
Table S1 List of oligonucleotides used in this study.
Table S2 Accession numbers of RpoB, NdhB, NdhD and homologous of OsPPR16 in 79 land plants.
Table S3 Genotype and phenotype of T0 transgenic plants.


