Abstract
Coptis chinensis has been long used as the potential herbal remedy for the treatment of influenza A infection. The six isoquinolone alkaloids extracted from C. chinensis rhizomes are reported to have good inhibition activity on neuraminidase (NA) of Clostridium perfringens, A/H1N1/1918, and recombinant NA-1; however, the study of the effect of these candidates on other NAs of threatening influenza A causing pandemic and seasonal flu recently has not considered yet. The purpose of this study is to investigate the interaction between these compounds and NAs of different wild and mutant subtypes of influenza A. This process involved the molecular docking of 3D structures of those compounds (ligand) into target proteins NA of A/H1N1/1918, A/H1N1/2009pdm, H3N2/2010 wild type, H3N2/2010 D151G mutant, H5N1 wild type, and H5N1 H274Y mutant. Then, the Protein-Ligand Interaction Profiler (PLIP) was utilized to demonstrate the bond formed between the ligand and the binding pocket of receptors of interest. The results showed that six candidates including palmatine, berberine, jatrorrhizine, epiberberine, columbamine, and coptisine have a higher affinity to all six selected proteins than commercial drugs such as oseltamivir, zanamivir, and natural binding ligand sialic acid. The results could be explained via the 2D picture, which showed the hydrophobic interaction and hydrogen bonding forming between the oxygen molecules of the ligand with the free residue of proteins.
1. Introduction
Influenza A virus affects directly the human lower respiratory and immune system. In the last two centuries, there are several influenza A subtypes that threatened human life by causing seasonal and pandemic flu around the world with high mobility and mortality, such as the “Spanish flu” in 1918,1 the pandemic in 2009 by the A/H1N1 strain,2 and the “Avian flu” in 2004–2007 via the A/H5N1 strain. Up to now, most of the anti-influenza viral drugs target inhibition action on neuraminidase (NA) and M2 channel (M2), two of the three kinds of surface glycoprotein antigens of this virus. However, two commercial M2 inhibitor drugs amantadine (Symmetrel) and rimantadine (Flumadine) were not recommended by the WHO in influenza A infection treatment as viral resistance has been shown in 100% of seasonal H3N2 and H1N1pdm09 samples tested. Neuraminidase (NA) inhibitors, oseltamivir (Tamiflu) and zanamivir (Relenza), are still effective drugs recently. Despite the fact that no influenza has shown any resistance signs to zanamivir in the United States, the inhalation administration is still inconvenient for patients. The use of oseltamivir is more comfortable by oral intake, but the efficacy is lower as the presence of some influenza A has shown resistance to it. Moreover, in Vietnam, the supply of these drugs is still limited and depends on the manufacturers. Therefore, it is necessary to develop new drug candidates that originated in Vietnam not only to avoid the viral resistance but also affordable for Vietnamese to handle once the pandemic or seasonal flu occurs.
Vietnam carries a highly diverse practice of traditional medicine in which various combinations of herbs have been widely used as remedies for many types of diseases.3 Research on metabolites of herbs has provided fundamental information and multidimensional perspectives from herbal medicines to modern drug discovery.4Coptis chinensis is a potential herbal medicine that is found growing freely at the highland of Lao Cai Province, Sapa, and Hoang Lien Son Mountain Range.5 From previous studies, this herb was recorded having strong antibacterial activity and used to treat bacterial diarrheas, acute conjunctivitis, and cutaneous and visceral leishmaniasis.6,7 Additionally, its rhizome is a good source of the isoquinoline alkaloids berberine, palmatine, coptisine, columbamine, jatrorrhizine, and epiberberine.5 These compounds were also reported showing inhibition activity on the NAs of Clostridium perfringens and H1N1 (NA-1 recombinant).8
In this study, we focus on investigating the inhibition of main bioactive compounds contained in the C. chinensis rhizome on the NAs of different subtypes of influenza A causing pandemics and common seasonal flu such as A/H1N1, A/H5N1, and A/H3N2. From our prior study on neuraminidase,9 to generalize the effects of these compounds, the NAs of H1N1/1918, H1N1/2009pdm, H3N2/2010 wild type, H3N2/2010 D151G mutant, H5N1 wild type, and H5N1 H274Y mutant were selected as target receptors.
2. Materials and Method
2.1. Receptor
The 3D structures of NA protein from different subtypes of influenza A were taken from Protein Data Bank, namely, NA-1 with mutant H169AY (PDB code 2HTY),10 NA-1 wild type (PDB code 3B7E),11 NA-1 with H274Y mutant (PDB code 3CKZ),12 NA-1 pandemic influenza virus resistance of the I223R mutant (PDB code 4B7R),13 NA-2 with the D151G mutant (PDB code 4GZQ),14 and NA-2 neuraminidase with the D151G mutant (PDB code 4GZT) (Table 1).14 These proteins were tested again at the binding pocket to verify the capacity of the model in experiment with the new ligand. These structures were tested again with molecules oseltamivir and sialic acid, which served as control docking models. This work was performed using Autodock Vina15 and visualized using MD.16
Table 1. All Receptors (Both Wild-Type and Mutant) Used on the Docking Simulations.
| PDB ID | name | length (amino acids) | mutated position |
|---|---|---|---|
| 3B7E | wild-type NA on the H1N1 strain | 385 | |
| 4B7R | mutant NA on the H1N1 strain | 387 | 269 |
| 4GZQ | wild-type NA on the H3N2 strain | 393 | |
| 4GZT | mutant NA on the H3N2 strain | 393 | 75 |
| 2HTY | wild-type NA on the H5N1 strain | 387 | |
| 3CKZ | mutant NA on the H5N1 strain | 385 | 95 |
2.2. Bioactive Compounds in C. chinensis
Most of the 3D structures of drug molecules in C. chinensis were downloaded from the PubChem Compound section of the National Center for Biotechnology Information (NCBI). Optimization was performed by Gaussian with a semiempirical AM1 method. The 2D structures of ligands and the bioactive compound are illustrated in Table 2.
Table 2. Bioactive Compounds and Ligand Control of the Influenza Virus.
2.3. Docking Simulations
Autodock Vina15 was used for performing the docking process. One section in Molecular Graphic Laboratory (MGL tools) was applied to build a complete pdbqt file name of ligands and receptors. Receptor preparation was initial fix with the SWISS PDB viewer17 to add the lost molecule such as hydrogen, nitrogen, or oxygen. Then MGL tools were carried out with four major substeps: adding polar hydrogen, removing a water molecule, computation of Gasteiger charges, and location of a grid box. The site of the grid box is illustrated in Table 3. From our prior study on the drug binding pathway, the grid box should be big enough to cover the path in which the drug will enter the substrate binding site.18 All of the ligands were prepared by the code package of Autodock Vina. These codes will check Torsion root (the amide bonds were treated as nonrotatable) and then convert it to .pdbqt.
Table 3. Position of the Grid Box Center.
| PDB ID | center of X | center of Y | center of Z |
|---|---|---|---|
| 2HTY | 7.9074 | 3.4235 | 8.52545 |
| 2HU4 | 6.7047 | 4.3269 | 8.434 |
| 3B7E | 7.2891 | 4.5236 | 8.867 |
| 3CKZ | –30.9278 | –56.3483 | 8.6463 |
| 4B7R | 7.277 | 4.47395 | 9.1283 |
| 4GZQ | 6.7346 | 4.43535 | 8.3664 |
| 4GZT | 6.8653 | 4.279 | 8.2432 |
Before running the Autodock Vina, a multiconfigure file was built to encode information that included the position of receptors file, ligand file, data of the grid-box’s coordinate, the size of the grid box, which was set up in 30 × 30 × 30 points, an exhaustiveness of 1000, and the number of modes, which was 10.
2.4. Pharmacophore Modeling
Protein-Ligand Interaction Profiler (PLIP)19 helps to virtualize the 3D structure with the position and interaction of the ligand in the binding pocket of the receptor. From the 3D observation, some types of bonds were identified by color and symbol, such as the hydrogen bond and hydrophobic bond, which were demonstrated as a blue solid line and gray dashed line, respectively.
3. Results and Discussion
3.1. Free Energy Binding of Isoquinolone Alkaloid Compounds to Targert NA Antigens
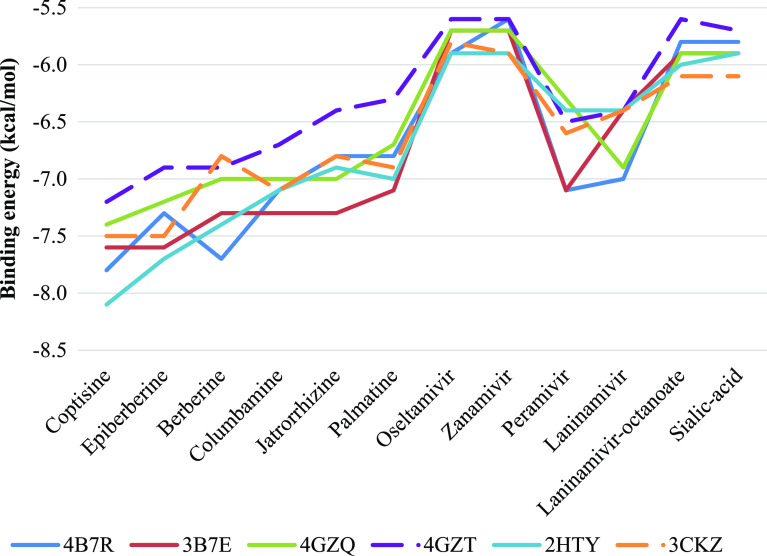
In order to investigate binding capacity of bioactive compounds in C. chinensis on NA of influenza A, molecular docking was ultilized. Among docking results, the absolute value of binding energy ranged from 5.6 to 8.1 kcal/mol, which are presented in Table 4. Bioactive compounds in C. chinensis had best binding on H1N1 (3B7E) and H5N1(2HTY) wild-type proteins, whereas the lowest effect belonged to H3N2 the mutant strain (4GZT). Despite fluctuations, all compounds had greater binding affinities to all NA-1 and NA-2 than the two commercial antiviral drugs approved by the FDA, oseltamivir and zanamivir, as well as the natural binding ligand sialic acid, as well demonstrated in Figure 1. The high binding energy could be explained by the high number of 1-benzylisoquinoline backbone that helps the candidates to fit into the binding pockets. The advantage of this steric effect is avoiding the binding of natural compounds after the drug entrance. Thus, the results provided strong evidence that bioactive compounds derived from C. chinensis have potential in development of influenza A infection treatment. Additionally, all selected compounds shown in Table 2 indicate that these molecules meet the requirement of Lipinski’s rule of five, which consequent to these compounds could be considered as orally active drugs.
Table 4. Binding Energies (kcal/mol) Collected after Running Molecular Docking on Six Protein Receptors (3B7E, 4B7R, 4GZQ, 4GZT, 2HTY, 3CKZ).
| H1N1 |
H3N2 |
H5N1 |
||||
|---|---|---|---|---|---|---|
| ligand | wild-type 3B7E | mutant 4B7R | wild-type 4GZQ | mutant 4GZT | wild-type 2HTY | mutant 3CKZ |
| coptisine | –7.6 | –7.8 | –7.4 | –7.2 | –8.1 | –7.5 |
| epiberberine | –7.6 | –7.3 | –7.2 | –6.9 | –7.7 | –7.5 |
| berberine | –7.3 | –7.7 | –7.0 | –6.9 | –7.4 | –6.8 |
| columbamine | –7.3 | –7.1 | –7.0 | –6.7 | –7.1 | –7.1 |
| jatrorrhizine | –7.3 | –6.8 | –7.0 | –6.4 | –6.9 | –6.8 |
| palmatine | –7.1 | –6.8 | –6.7 | –6.3 | –7.0 | –6.9 |
| oseltamivir | –5.9 | –5.7 | –5.7 | –5.6 | –5.9 | –5.8 |
| zanamivir | –5.6 | –5.7 | –5.7 | –5.6 | –5.9 | –5.9 |
| peramivir | –7.1 | –7.1 | –6.3 | –6.5 | –6.4 | –6.6 |
| laninamivir | –7.0 | –6.4 | –6.9 | –6.4 | –6.4 | –6.4 |
| laninamivir-octanoate | –5.8 | –5.9 | –5.9 | –5.6 | –6.0 | –6.1 |
| sialic acid | –5.8 | –5.9 | –5.9 | –5.7 | –5.9 | –6.1 |
Figure 1.
Binding energies of 12 ligands to 6 receptors, including six tested compounds (coptisine, epiberberine, berberine, columbamine, jatrorrhizine, palmatine), five protein inhibitors (oseltamivir, zanamivir, laninamivir, laninamivir octanoate), and sialic acid.
3.2. Pharmacophore Modeling
After the ligand test, the interaction analysis was conducted and four chemical constituents extracted from C. chinensis were chosen based on the high number of hydrogen interactions with receptors.
3.3. Wild-Type and I223R Mutant of H1N1
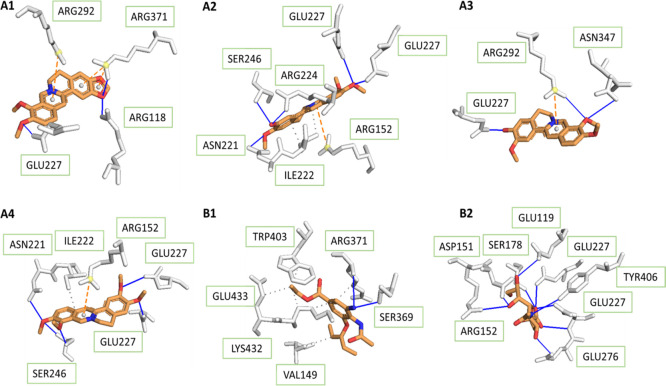
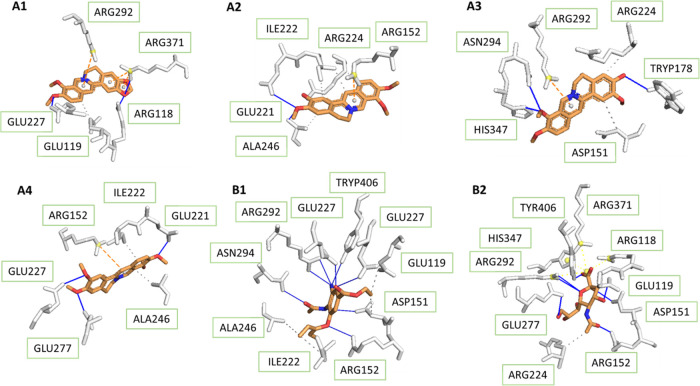
For the wild-type of H1N1 shown in Figure 2, Ser-246 formed hydrogen bonds with columbamine and palmatine in the binding pocket; amino acid residue Glu-227 bound strongly to all four tested compounds and sialic acid. For this reason, the compounds could be potential inhibitors for the binding of sialic acid to the influenza protein. The Arg-371 residue not only was observed to make hydrogen interaction with berberine but also set up hydrophobic contact with hydrophobic parts of the ligands. Because of the frequent presence within hydrogen contact, the three residues seemed to be important substrates in recognition of the 3B7E protein. In addition, the structure of protein contains Ile-222 and Arg-292, both of which are involved frequently in hydrogen and hydrophobic binding to tested ligands.
Figure 2.
Interactions between H1N1 NA protein (PDB ID: 3B7E) and its ligands, including four tested compounds ((A1) berberine, (A2) columbamine, (A3) jatrorrhizine, (A4) palmatine), oseltamivir (B1), and sialic acid (B2).
As indicated in Figure 2B1, the binding conformation of oseltamivir showed direct interaction via hydrogen bonds with residues Arg-371 and Ser-369 and through hydrophobic contacts with Val-149, Lys-432, Glu-433, and Trp-403, which contributed to inhibitory capacity at the catalytic site. Similar to oseltamivir, berberine also created hydrogen-binding with NA at the position Arg-371. Compared to all four selected compounds, oseltamivir had fewer hydrogen bonds while that number of sialic acid was much higher, which was responsible for the inhibition capacity of these substances.
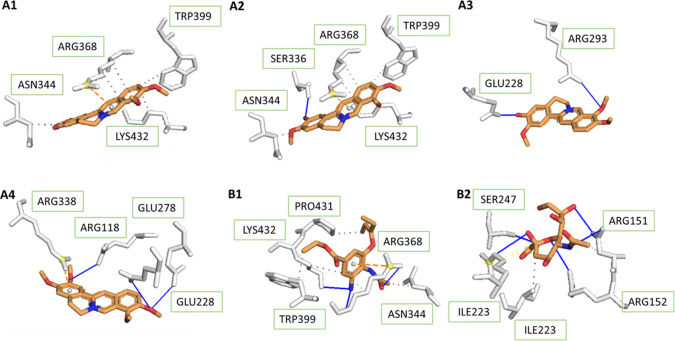
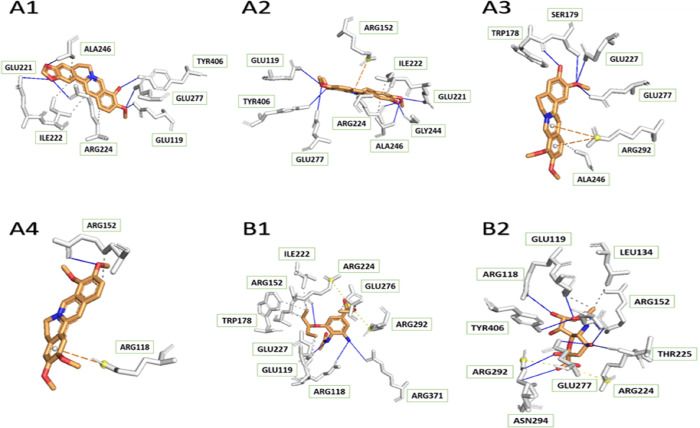
On the other hand, for H1N1 with the I223R mutant (4B7R), Figure 3 illustrates that there was an important function of Glu-228 in the binding of two compounds, jatrorrhizine and palmatine, to the viral protein. The same kind of interaction happened between experimental ligands and amino acid residues, which were Ser-336, Arg-293, Arg-118, and Glu-278. There was no hydrogen interaction reported in the complex of NA–berberine; thus, berberine may not be a good drug for H1N1 mutation. In comparison with the wild-type receptor, four tested medicinal constituents in contact with the mutant one formed a lower number of hydrogen bonds. This was a good explanation for the greater binding affinities in complex with 3B7E, except for berberine.
Figure 3.
Interactions between H1N1 NA protein (PDB ID: 4B7R) and its ligands, including four tested compounds ((A1) berberine, (A2) columbamine, (A3) jatrorrhizine, (A4) palmatine), oseltamivir (B1), and sialic acid (B2).
According to Figure 3, while Arg-368, Lys-432, and Asn-344 were the binding sites of oseltamivir on the NA protein, these also contacted with berberine and columbamine through hydrophobic bonds. Especially, Lys-432 and oseltamivir shared a hydrogen interaction. Therefore, those compounds could be a replacement for the commercial drug.
3.4. Wild-Type and D151G Mutant of H3N2
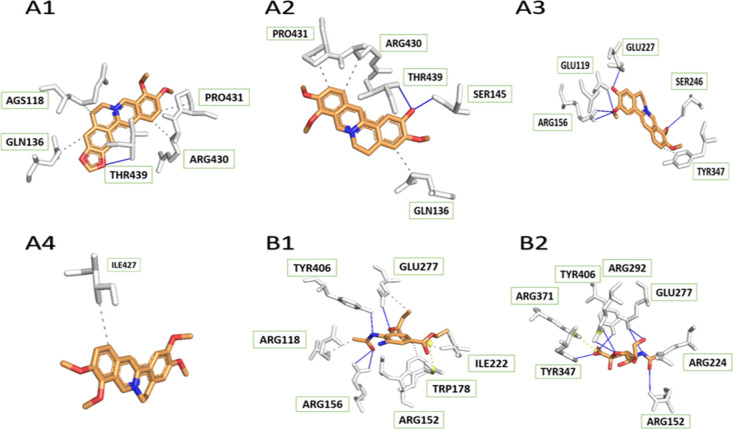
In the 4GZQ protein, though the NA–berberine complex showed fewer hydrogen interactions than the others, its binding affinities were quite low, which was due to the many hydrophobic components of coptisine (six benzene rings). It is obvious from Figure 4 that amino acid residues Glu-227 and Glu-221 were reported to interact directly with berberine, columbamine, and palmatine. Especially, Glu-227 could also be found in strong contact with oseltamivir and sialic acid at higher binding affinity. Meanwhile, Asn-294 was the residue forming hydrogen-binding to both jatrorrhizine and oseltamivir, but that of jatrorrhizine had lower binding energy. Residue 294 was identified to be an important one for drug binding in our prior study.18 Whereas Ala-246 was a residue in hydrogen interaction of columbamine, it shared a hydrophobic part to palmatine and oseltamivir. Not only contacts via hydrophobic interaction to columbamine and the approved drug, but Ile-222 also had both kinds of bonds in complex with palmatine. Consequently, all four medicinal constituents of the herb could be potential influenza inhibitors.
Figure 4.
Interactions between H3N2 NA protein (PDB ID: 4GZQ) and its ligands, including four tested compounds ((A1) berberine, (A2) columbamine, (A3) jatrorrhizine, (A4) palmatine), oseltamivir (B1), and sialic acid (B2).
Unlike binding to sialic acid, His-347 interacted with jatrorrhizine through a hydrogen bond. The same circumstance also happened to Arg-371 and Arg-118 in interaction with berberine. Therefore, berberine and jatrorrhizine could inhibit binding of sialic acid to the NA protein.
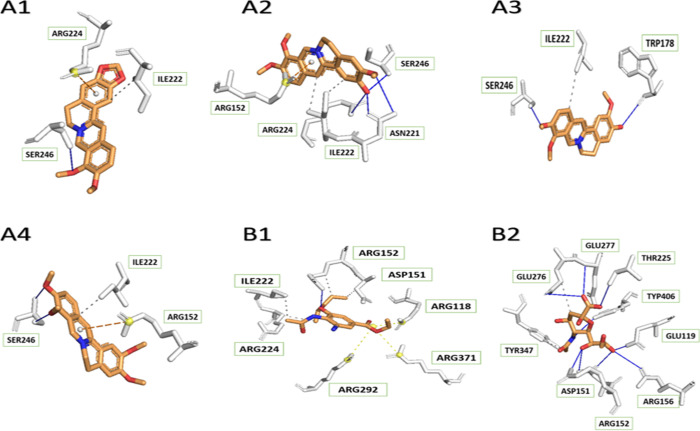
According to the molecular interaction, there was a significant difference between 4GZQ and 4GZT proteins of H3N2 NA in contact with all four tested ligands. There were high numbers of hydrogen bonds in complexes of berberine, columbamine, and jatrorrhizine with 4GZT compared to 4GZQ. In contrast, palmatine linked to 4GZT by a lower number of hydrogen bonds (Figure 5).
Figure 5.
Interactions between H3N2 NA protein (PDB ID: 4GZT) and its ligands, including four tested compounds ((A1) berberine, (A2) columbamine, (A3) jatrorrhizine, (A4) palmatine), oseltamivir (B1), and sialic acid (B2).
3.5. Wild-Type and H274Y Mutant of H5N1
Although berberine represented the lowest binding energy in complex with 2HTY and 3CKZ, it shared a low number of hydrogen bonds compared to other compounds. The reason is that berberine contains many hydrophobic components, which are aromatic rings, with their ability to construct hydrophobic interaction with the free residues of the receptors. For wild-type H5N1 influenza neuraminidase, Figure 6 illustrates that Thr-439 residues played a vital role in hydrogen bond formation to berberine and columbamine while Arg-156 interacted with jatrorrhizine and oseltamivir through the same kind of interaction. Moreover, both berberine and oseltamivir created hydrophobic interaction at Arg-118 with the 2HTY NA protein. In contrast, there was no hydrogen-binding between palmatine and NA. Because the tested ligands rarely shared strong interaction with the protein, compared to both oseltamivir and sialic acid, there was no doubt that these compounds could not be protein inhibitors, except for jatrorrhizine, which was able to form four hydrogen bonds as well as have one similar reception site as indicated.
Figure 6.
Interactions between H5N1 NA protein (PDB ID: 2HTY) and its ligands, including four tested compounds ((A1) berberine, (A2) columbamine, (A3) jatrorrhizine, (A4) palmatine), oseltamivir (B1), and sialic acid (B2).
In the case of N1 neuraminidase H274Y (3CKZ), hydrogen bonds were frequently shared between the amino acid residue Ser-246 and selective compounds. All tested compounds shared hydrophobic interactions with the receptor at the positions of Ile-222, Arg-224, and Arg-152, but the mechanisms are different among the positions. Ile-222 and Arg-224 residues of the hydrophobic receptor developed hydrophobic bonds with all four tested ligands from methyl groups. Meanwhile, the link between benzene rings and parts of interest of the receptor was a decisive tendency for Arg-152. Similar to medicinal compounds, the three residues were in contact with oseltamivir via hydrophobic interactions. Although the number of hydrogen bonds from sialic acid was dominant compared to the others, that of tested medicines was more than that of oseltamivir, and thus, they absolutely had the capability of becoming potential drugs (Figure 7).
Figure 7.
Interactions between H5N1 NA protein (PDB ID: 3CKZ) and its ligands, including four tested compounds ((A1) berberine, (A2) columbamine, (A3) jatrorrhizine, (A4) palmatine), oseltamivir (B1), and sialic acid (B2).
4. Conclusions
The docking study of six isoquinoline alkaloid compounds extracted from C. chinensis showed the good inhibitor action on the NA-1 and NA-2 as having higher binding affinity to all receptors of interest compared with available commercial drugs and other phase III clinical candidates. These binding results were presented by PLIP to show the high number of hydrogen bonds and hydrophobic interactions. Among the selected candidates, berberine, jatrorrhizine, and palmatine were consistent with the previous study.
This in vivo study has partially investigated the inhibitor activity on different NA antigens of a wide subtype range of the influenza A virus. By binding energy calculation and pharmacophore analysis, all six compounds are shown to be promising in anti-influenza A drug development. However, further in silico studies to understand well their bioavailability, pharmacokinetics, and pharmacodynamics are highly recommended.
Acknowledgments
This work was supported by the NAFOSTED (The National Foundation for Science and Technology) under Grant No. 108.06-2017.332.
The authors declare no competing financial interest.
References
- Crosby A. W., America’s Forgotten Pandemic: The Influenza of 1918. 2 ed.; Cambridge University Press: Cambridge, 2003, 10.1017/CBO9780511586576. [DOI] [Google Scholar]
- Dawood F. S.; Jain S.; Finelli L.; Shaw M. W.; Lindstrom S.; Garten R. J.; Gubareva L. V.; Xu X.; Bridges C. B.; Uyeki T. M. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. The New England journal of medicine 2009, 360, 2605–2615. 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- Nguyen-Vo T. H.; Le T.; Pham D.; Nguyen T.; Le P.; Nguyen A.; Nguyen T.; Nguyen T. N.; Nguyen V.; Do H.; Trinh K.; Duong H. T.; Le L. VIETHERB: A Database for Vietnamese Herbal Species. J. Chem. Inf. Model. 2019, 59, 1–9. 10.1021/acs.jcim.8b00399. [DOI] [PubMed] [Google Scholar]
- Nguyen-Vo T. H.; Nguyen L.; Do N.; Nguyen T. N.; Trinh K.; Cao H.; Le L. Plant Metabolite Databases: From Herbal Medicines to Modern Drug Discovery. J. Chem. Inf. Model. 2020, 60, 1101–1110. 10.1021/acs.jcim.9b00826. [DOI] [PubMed] [Google Scholar]
- Lọi D̵. T., Nhũng Cây Thuốc Và Vị Thuốc Viột Nam. NXB Y Hộc: Viột Nam, 2006. [Google Scholar]
- Xiang K. L.; Wu S. D.; Yu S. X.; Liu Y.; Jabbour F.; Erst A. S.; Zhao L.; Wang W.; Chen Z. D. The First Comprehensive Phylogeny of Coptis (Ranunculaceae) and Its Implications for Character Evolution and Classification. PloSone 2016, 11, e0153127 10.1371/journal.pone.0153127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D.; Jin C.; Xiao X. H.; Dong X. P. Antimicrobial properties of berberines alkaloids in Coptis chinensis Franch by microcalorimetry. J. Biochem. Biophys. Methods 2008, 70, 845–849. 10.1016/j.jbbm.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Li H.; Shi Z.; Gao S.; Wei S.; Li K.; Wang J.; Li J.; Wang R.; Gong M.; Zhao Y.; Xiao X. Inhibition activity of a traditional Chinese herbal formula Huang-Lian-Jie-Du-Tang and its major components found in its plasma profile on neuraminidase-1. Sci. Rep. 2017, 7, 15549. 10.1038/s41598-017-15733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. T.; Le L.; Truong T. N.. Top-hits for H1N1pdm Identified by Virtual Screening Using Ensemble-based Docking. PLoS currents 2009, 1, 10.1371/currents.RRN1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. J.; Haire L. F.; Stevens D. J.; Collins P. J.; Lin Y. P.; Blackburn G. M.; Hay A. J.; Gamblin S. J.; Skehel J. J. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 2006, 443, 45–49. 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- Xu X.; Zhu X.; Dwek R. A.; Stevens J.; Wilson I. A. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J. Virol. 2008, 82, 10493–10501. 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P. J.; Haire L. F.; Lin Y. P.; Liu J.; Russell R. J.; Walker P. A.; Skehel J. J.; Martin S. R.; Hay A. J.; Gamblin S. J. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 2008, 453, 1258–1261. 10.1038/nature06956. [DOI] [PubMed] [Google Scholar]
- Van Der Vries E.; Collins P. J.; Vachieri S. G.; Xiong X.; Liu J.; Walker P. A.; Haire L. F.; Hay A. J.; Schutten M.; Osterhaus A. D. M. E.; Martin S. R.; Boucher C. A. B.; Skehel J. J.; Gamblin S. J. H1N1 2009 pandemic influenza virus: resistance of the I223R neuraminidase mutant explained by kinetic and structural analysis. PLoS Pathog. 2012, 8, e1002914 10.1371/journal.ppat.1002914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; McBride R.; Nycholat C. M.; Yu W.; Paulson J. C.; Wilson I. A. Influenza Virus Neuraminidases with Reduced Enzymatic Activity That Avidly Bind Sialic Acid Receptors. J. Virol. 2012, 86, 13371–13383. 10.1128/JVI.01426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Guex N.; Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Nguyen H.; Tran T.; Fukunishi Y.; Higo J.; Nakamura H.; Le L. Computational Study of Drug Binding Affinity to Influenza A Neuraminidase Using Smooth Reaction Path Generation (SRPG) Method. J. Chem. Inf. Model. 2015, 55, 1936–1943. 10.1021/acs.jcim.5b00319. [DOI] [PubMed] [Google Scholar]
- Salentin S.; Schreiber S.; Haupt V. J.; Adasme M. F.; Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, 443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]