Summary
Abscisic acid (ABA) and jasmonic acid (JA) both inhibit seed germination, but their interactions during this process remain elusive. Here, we report the identification of a ‘SAPK10‐bZIP72‐AOC’ pathway, through which ABA promotes JA biosynthesis to synergistically inhibit rice seed germination.
Using biochemical interaction and phosphorylation assays, we show that SAPK10 exhibits autophosphorylation activity on the 177th serine, which enables it to phosphorylate bZIP72 majorly on 71st serine. The SAPK10‐dependent phosphorylation enhances bZIP72 protein stability as well as the DNA‐binding ability to the G‐box cis‐element of AOC promoter, thereby elevating the AOC transcription and the endogenous concentration of JA.
Blocking of JA biosynthesis significantly alleviated the ABA sensitivity on seed germination, suggesting that ABA‐imposed inhibition partially relied on the elevated concentration of JA.
Our findings shed a novel insight into the molecular networks of ABA–JA synergistic interaction during rice seed germination.
Keywords: abscisic acid (ABA), bZIP, jasmonic acid (JA), phosphorylation, rice, seed germination, SnRK2
Introduction
Seeds of cereals such as rice and wheat acquire dormancy during the seed maturation, which enables the offspring to survive from the harsh conditions and initiate the new life cycle after sensing optimal environment cues (Bentsink & Koornneef, 2008). In the agricultural production, seeds acquiring precocious germination are predisposed to cause preharvest sprouting, ultimately leading to losses of grain yield and quality (Gubler et al., 2005).
Abscisic acid (ABA) is a primary phytohormone repressing seed germination majorly by inhibiting cell wall loosening and expansion, which is the key step to starting germination (Gimeno‐Gilles et al., 2009). The inhibitory effect of ABA on germination has been supported by much genetic evidence. Seeds of typical ABA‐deficient or ABA‐insensitive mutants have faster germination than the wild‐type (WT), while ABA catabolism mutants with high ABA accumulation levels exhibit lower germination rate and speed (Shu et al., 2016). About a decade ago, a ‘PYR/PYL/RCAR‐PP2C‐SnRK2’ cascade model for ABA signaling was constituted in Arabidopsis (Umezawa et al., 2010). In this model, PYR/PYL/RCAR receptors perceive the ABA signal to form a favorable configuration for the binding of PP2C phosphatases, thus releasing their dephosphorylation activity to autophosphorylate SnRK2s. Phosphorylation‐activated SnRK2s further pass the signals to AREB/ABF transcription factors (TFs) through protein phosphorylation, and finally activate the downstream genes to respond to the ABA signals. As the core element in ABA signaling pathway, SnRK2 protein kinases SnRK2.2 and SnRK2.3 have been reported to play essential roles in ABA‐mediated seed germination inhibition, and the snrk2.2 snrk2.3 snrk2.6 triple mutant became almost completely insensitive to ABA in seed germination in Arabidopsis (Fujii & Zhu, 2009). Rice SnRK2 members are termed SAPK1‐10 (osmotic stress/ABA‐activated protein kinase), of which SAPK6, SAPK8, SAPK9 and SAPK10 are reported to be functionally related to ABA signaling, and SAPK10 shows the highest homology to SnRK2.2, SnRK2.3 and SnRK2.6 in Arabidopsis, while its role in seed germination remains unclear (Kobayashi et al., 2004; Chae et al., 2007; Yuhko et al., 2010). To date, only a few ABFs have been identified as SnRK2 substrates, including ABI5 and AtDPBF2 which are implicated in seed maturation and germination in Arabidopsis (Lopezmolina et al., 2001; Kulik et al., 2011). Using a quantitative phosphoproteome strategy, Wang et al (2013) identified 58 potential substrate proteins of Arabidopsis SnRK2s, but their kinase‐substrate relationships need more verification by biochemical analysis (Wang et al., 2013).
Jasmonic acid (JA) and its derivates have been found to inhibit seed germination by disrupting the peroxisomal Adenosine triphosphate (ATP) binding cassette transporter or core β‐oxidation process (Dave et al., 2011; Liu et al., 2015). Owing to the fact that exogenous ABA or stress‐induced endogenous ABA usually stimulates JA biosynthesis (Adie et al., 2007; Avramova, 2017), JA is generally believed to work synergistically with ABA in most of the biological processes, including seed germination (Liu et al., 2015; Tang et al., 2020), despite the fact that a few exceptional cases in plant biotic stress have also been reported to show antagonistic effects of ABA‐JA (Garcia‐Andrade et al., 2011; Xie et al., 2018). Previous efforts have provided clues that JAM1, as well as MYC2, may serve as the linkers of ABA and JA signaling (Nakata et al., 2013; Aleman et al., 2016). Nevertheless, the mechanisms underlying ABA‐mediated JA biosynthesis are still largely elusive, particularly in the seed germination process. In this study, we attempted to address this question by identifying a regulatory pathway ‘SAPK10‐bZIP72‐AOC’, through which ABA promotes the JA biosynthesis to synergistically inhibit seed germination. Moreover, we revealed that the inhibitory effect of ABA on seed germination is partially based on the elevated concentration of JA. This study provides a novel insight into the molecular mechanism of ABA and JA synergy in rice seed germination, which would deepen our understanding of the hormone‐mediated fine‐tuning network of seed germination in crops.
Materials and Methods
Vector construction and plant transformation
For overexpression of SAPK10 or bZIP72, the coding sequence (CDS) of SAPK10 or bZIP72 was amplified using the cDNA of the WT (Oryza sativa L. ssp. japonica cv Nipponbare) as the template and cloned into the binary vector PU1301 driven by the maize ubiquitin promoter, by using the KpnI and BclI sites for SAPK10 and the KpnI and SpeI sites for bZIP72. The S177A mutation in SAPK10 and S71A mutation in bZIP72 were amplified using overlap PCR (Higuchi et al., 1988) and inserted into PU1301 vector to generate PU1301‐SAPK10S177A or PU1301‐bZIP72S71A, respectively. Mutants of SAPK10, bZIP72 and bZIP72/TRAB1 were generated by CRISPR/Cas9 system as described previously (Ma et al., 2015). Briefly, annealed double‐strand oligos of the gDNA sequences were ligated into the pYLCRISPR/Cas9‐MH using BsaI site (Thermo, Waltham, MA, USA). All the binary vectors were transformed into Nipponbare callus using the Agrobacterium‐mediated transformation method as described previously (Hiei et al., 1994). The primers used were listed in Supporting Information Table S1.
Seed germination and seedling growth assays
Seed germination assay was performed as previously reported (Lin et al., 2015) with a few modifications. Briefly, dehulled rice seeds from WT and transgenic lines in Nipponbare (O. sativa L. ssp. japonica) background were immersed in 75% ethanol for 1 min. Then the seeds were sterilized in 2.5% (v/v) NaClO for 30 min and subsequently washed five times with sterilized water (each time for 5 min). Rinsed seeds were planted on half‐strength Murashige & Skoog medium (½MS; pH 5.8) with 0.3% plant agar. Then the seeds were placed in a growth chamber (28 ± 2°C, 12 h : 12 h photoperiod with 60% relative humidity) for 4 d to reach the maximum germination rate of the WT seeds under mock treatment. Complete germination was defined as the growth of the coleoptile to 5 mm in length (Lin et al., 2015). The seed germination rates were recorded as a percentage every 12 h. Triple biological repeats (each replicate containing 50 seeds) were performed for each sample. Postgermination growth assay was performed as follows: the seedlings were grown using rice nutrient solution as previously described (W. Wang et al., 2014; X.F. Wang et al., 2014) under different treatments for 10 days, then the shoot height was recorded. The relative germination of the seeds under ABA treatments were determined at 4 d after germination (DAG) and expressed as a percentage of ABA treatment germination rate vs mock treatment germination rate. The sensitivity to ABA treatment was evaluated by analysis of variance and multiple comparisons using R functions ‘aov’ and ‘emmeans’.
Yeast two‐hybrid assays
Yeast two‐hybrid (Y2H) assays were performed using the matchmaker GAL4 two‐hybrid system (Clontech, CA, USA) according to the manufacturer's instructions. The coding sequences of SAPK10 were digested with EcoRI and XhoI and cloned into the pGADT7 vector, the coding sequences of SAPK10 were digested with EcoRI and PstI and cloned into the pGBKT7 vector, and the coding sequences of bZIP72 were digested with NdeI and EcoRI and cloned into the pGBKT7 vector, respectively. Primers used were listed in Table S1. The resulting constructs were cotransformed into the yeast strain Y2H Gold (Clontech) and selected on synthetic medium lacking leucine, tryptophan and histidine with 0.04 mg ml−1 X‐α‐Gal and 100 ng ml−1 Aureobasidin A applied.
Pull‐down assays
Full‐length CDSs of bZIP72 and bZIP72S71A were digested with EcoRI and SmaI and cloned into the pGEX‐4T‐1 vector (GE Healthcare, Chicago, IL, USA), and the full‐length CDS of SAPK10 was cloned into pGEX‐4T‐1 using the EcoRI and SalI sites, and full‐length CDSs of SAPK10 and SAPK10S177A were cloned into pET28a (Thermo), both by using the EcoRI and XhoI sites. Primers used were listed in Table S1. These recombinant vectors were transformed into Transetta (DE3) chemically competent cell (Transgen, Beijing, China), and purified using the glutathione S‐transferase (GST)‐Sefinose™ Kit (Sangon Biotech, Shanghai, China) and 6 × His‐Tagged Protein Purification Kit (CWBIO, Beijing, China) according to the manufacturer's protocols, respectively. The detected interactive proteins were incubated with glutathione high‐capacity magnetic agarose beads (Sigma‐Aldrich) in pull‐down buffer (50 mM Tris‐HCl, pH 7.5, 5% glycerol, 1 mM EDTA, 1 mM DL‐Dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.01% Nonidet P‐40, and 150 mM KCl) at 4°C for 2 h. After washing five times with pull‐down buffer, the beads were suspended in 50 μl 1 × PBS and 10 μl 6 × sodium dodecyl sulfate (SDS) protein loading buffer and boiled for 5 min for immunoblot analysis on 10% SDS‐polyacrylamide gel electrophoresis. Individual bands were detected using Supersignal West Pico Chemiluminescent Substrate (Thermo) and the ChemDoc Touch Imaging system (Bio‐Rad). The dilution for anti‐GST (Yeasen Ltd, Shanghai, China) and anti‐His (Yeasen) was 1 : 5000.
Coimmunoprecipitation (Co‐IP) assays
The full‐length CDS of bZIP72 was digested with KpnI and SpeI and cloned into the proUbi–FLAG vector, and the full‐length CDS of SAPK10 was digested with KpnI and XbaI and cloned into pCAMBIA1300‐35S‐GFP vector, respectively. Primers used were listed in Table S1. bZIP72‐FLAG was transiently coexpressed with empty green fluorescent protein (GFP) or SAPK10‐GFP in tobacco leaves by Agrobacterium infiltration. The cotransformed tobacco leaves were then ground into fine powders in liquid nitrogen and resuspended in protein extraction buffer (25 mM Tris‐HCl, pH 7.4, 150 mM NaCl,1 mM EDTA, 1% NonidetP‐40, and 5% glycerol, 1 mM PMSF, 20 μM MG132, and 1× Roche protease inhibitor cocktail (Roche)). After brief centrifugation twice (12 000 g for 10 min each time), the resulting supernatant was incubated with anti‐FLAG M2 magnetic beads (Sigma‐Aldrich) at 4°C for 2 h. Beads were washed five times with washing buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.2% Triton X‐100, 1 mM PMSF, and 1× Roche protease inhibitor cocktail). The immunoprecipitated proteins were suspended in 50 μl of 1 × PBS and 10 μl of 6 × SDS loading buffer, boiled for 5 min, and resolved on 10% acrylamide gels. Individual bands were detected using Supersignal West Pico Chemiluminescent Substrate (Thermo) and the ChemDoc™ Touch Imaging system (Bio‐Rad). The dilution for anti‐FLAG (Sigma‐Aldrich) and anti‐GFP (Yeasen) antibodies was 1 : 5000.
In vitro phosphorylation assays
In vitro kinase assays were performed as previously described (Hou et al., 2019). Briefly, for the SAPK10 autophosphorylation assay, GST, GST‐SAPK10 or different mutated versions of GST‐SAPK10 (GST‐SAPK10S177A, GST‐SAPK10T178A and GST‐SAPK10S177A/T178A) was expressed in Transetta (DE3) chemically competent cell (Transgen), and purified using the GST‐Sefinose™ Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions, respectively. The purified proteins (100 ng) were incubated with 1 µg calf intestinal phosphatase (CIAP; Takara, Dalian, China) at 37°C for 30 min, and then separated by electrophoresis on 10% acrylamide gels. Phosphorylated bands were detected using biotinylated Phos‐tag™ zinc complex BTL111 purchased from Wako (Osaka, Japan) according to the manufacturer’s instructions. For the kinase assay of SAPK10 and bZIP72, GST‐bZIP72 or GST‐bZIP72S71A was coexpressed with His‐SAPK10 in Transetta (DE3) chemically competent cell, and purified using the GST‐Sefinose™ Kit (Sangon Biotech) and 6 × His‐Tagged Protein Purification Kit (CWBIO), respectively. The detection of bZIP72 phosphorylated bands was the same as that of SAPK10 autophosphorylation bands.
In vivo phosphorylation assays
The experiments were performed as described previously (Zhou et al., 2018) with a few modifications. SAPK10‐FLAG and bZIP72‐FLAG overexpression lines were hydroponically cultured for 2 wk, then transferred to the solutions containing different concentrations of ABA (0, 25, 50, 100 µM) for 6 h, respectively. Then the seedlings of SAPK10‐FLAG or bZIP72‐FLAG were harvested and ground into fine powders in liquid nitrogen and resuspended in protein extraction buffer (25 mM Tris‐HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NonidetP‐40, and 5% glycerol, 1 mM PMSF, 20 μM MG132, and 1 × Roche protease inhibitor cocktail (Roche). After brief centrifugation twice (12 000 g for 10 min each time), the resulting supernatant was incubated with anti‐FLAG M2 magnetic beads (Sigma‐Aldrich) at 4°C for 1 h. The beads were washed five times with washing buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.2 % Triton X‐100, 1 mM PMSF, and 1 × Roche protease inhibitor cocktail). Bound proteins were eluted with 50 μl of 1 × PBS and 10 μl of 6 × SDS loading buffer, boiled for 5 min, and resolved on 10% acrylamide gels. The serine phosphorylation of SAPK10‐FLAG and bZIP72‐FLAG were detected using antiphosphoserine antibody. The dilution for antiphosphoserine antibody (Abcam, Cambridge, UK) and anti‐FLAG (Sigma‐Aldrich) antibodies was 1 : 5000.
Cell‐free degradation assays
The experiments were conducted as described previously (Lv et al., 2014). Briefly, the wild‐type (japonica cv Nipponbare) were hydroponically cultured under mock or +ABA (2 µM ABA) nutrition solutions for 2 wk. Then the seedlings were harvested and ground into fine powders in liquid nitrogen, then resuspended in the degradation buffer (25 mM Tris‐HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM PMSF, 5 mM DTT and 10 mM ATP). After brief centrifugation twice (12 000 g for 10 min each time), the resulting total proteins (30 µg of each) were incubated with recombinant GST‐bZIP72 or GST‐bZIP72S71D (100 ng of each) at 28°C for the individual assays. The reactions were taken at indicated intervals for determination of bZIP72 abundance by immunoblotting. The protein intensities were quantified using imagej software. The dissociation‐one phase exponential decay curve was plotted on a semilog graph using graphpad prism (5.0) software to calculate the half‐life of GST‐bZIP72. The dilution for anti‐GST (Yeasen) was 1 : 5000.
Electrophoresis mobility shift assay (EMSA)
Recombinant p‐GST‐bZIP72 (phosphorylated GST‐bZIP72) was purified from Transetta (DE3) chemically competent cell (Transgen) coexpressing GST‐ bZIP72 and His‐SAPK10. The EMSA probes in a length of 59 nt in the AOC promoter were commercially synthesized by Tsingke Co. (Tsingke Biotech, Hangzhou, China) and labeled using an EMSA Probe Biotin Labeling Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. Probes used were listed in Table S1. An equal amount of purified recombinant proteins were preincubated with EMSA/gel‐shift binding buffer (Beyotime) at 25°C for 20 min, then incubated with 20 fmol labeled probes with or without nonlabeled competitive DNA probes for another 20 min. Then the incubated mixed samples were separated by electrophoresis on 6% acrylamide gels and transferred to the Nylon membrane (Beyotime, Shanghai, China). The labeled DNA probes was detected using the LightShift Chemiluminescent EMSA kit (Thermo) according to the manufacturer’s instructions. Migration of biotin‐labeled probes was detected using Supersignal West Pico Chemiluminescent Substrate (Thermo) and the ChemDoc™ Touch Imaging system (Bio‐Rad).
Chromatin immunoprecipitation‐quantitative PCR (ChIP‐qPCR)
The experiments were conducted as described previously (Hou et al., 2015). Briefly, chromatin was isolated from 2 g crosslinked leaves of the wild‐type plants. Isolated chromatin was sonicated for DNA fragmentation ranging from 200 to 700 bp. Subsequently, the DNA/protein complex was immune‐precipitated with polyclonal rabbit bZIP72 antibody against the amino acid residues 258 to 271 (C‐ DSGDKGNSDLSSPT ‐COOH) of bZIP72, which was commercially synthesized and affinity‐purified by Genescript company (Genescript, Shanghai, China). Then the immunoprecipitated DNA was purified with phenol/chloroform after reverse cross‐linking and proteinase K treatment. The immunoprecipitated and input DNA were used for quantitative reverse transcription polymerase chain reaction (qRT‐PCR) with gene‐specific primers, respectively. The gene‐specific primers used were listed in Table S1. The quantitative PCR results were analyzed according to the manual of the Magna ChIP™ HiSens kit (Millipore, Darmstadt, Germany). The quantitative ChIP‐qPCR was performed with triple biological replicates.
Luciferase transient transcriptional activity assay
The CDS of bZIP72 and bZIP72S71A were cloned into ‘None’ vectors as effectors by using the BamHI and EcoRI sites, and the promoter region of AOC was cloned into 190fLUC vector as reporter by using the HindIII and BglII sites. The primers used were listed in Table S1. Then the plasmids were transformed into rice protoplasts as reported previously (Xie & Yang, 2013). Then the transformed protoplasts were resuspended in 50 μl lysis buffer, and 30–50 μl lysate was used to measure the luciferase activity. Firefly luciferase assay substrate buffer (100 μl) was added into the lysate and the firefly luciferase (fLUC) activity was measured. Subsequently, Stop & Renilla luciferase substrate buffer (100 μl) was added to the reaction and the Renilla luciferase (rLUC) activity was determined. All the luciferase activities were detected by the Dual Luciferase Reporter Gene Assay Kit (Beyotime) using the Tecan infinite M200 system (Tecan, Mannedorf, Switzerland). The relative luciferase activity was calculated as the ratio between fLUC and rLUC (fLUC/rLUC). AtUbi3:rLUC was used as an internal control. The luciferase activity for each sample was detected with triple biological replicates.
Quantification of JA concentrations in rice
Jasmonic acid extraction and quantification were performed as described previously (Zhang et al., 2018). Briefly, c. 0.1 g seedlings grown on ½MS medium for 4 d were ground into fine powers and extracted with 1 ml plant hormone extraction buffer (isopropanol : H2O : hydrochloric acid, 2 : 1 : 0.002 v/v/v). The extract was gently agitated at 4°C for 30 min. Then 20 ml of dichloromethane was added to the samples and gently agitated at 4°C for 30 min. Subsequently, the samples were centrifuged at 8000 g for 10 min and the organic phase was extracted and dried with liquid nitrogen. Ultimately, the pellets were dissolved in 150 ml of 0.1% methanol and filtered with 0.22 µm filter membranes. The JA contents of the purified products were quantified using high‐performance liquid chromatography‐tandem MS (HPLC‐MS/MS). The MS conditions were performed as reported previously (Zhang et al., 2018).
Results
SAPK10 is autophosphorylated on serine 177
As the closest ortholog of OST1 in Arabidopsis and AAPK in Vicia faba, SAPK10, which is an ABA‐inducible SnRK2 type kinase in rice, has been reported to be a core component in ABA signaling (Kobayashi et al., 2004, 2005). It is demonstrated that the function of SnRK2s, such as OST1, is activated by autophosphorylation within the activation T‐loop domain (Belin et al., 2006; Ng et al., 2011). Sequence similarity analysis of SAPK10 identified two putative phosphorylation sites, Serine 177 and Threonine 178 (S177 and T178, respectively) within the T‐loop region, which were conserved with those of OST1 in Arabidopsis (Figs 1a, S1). Autophosphorylation assay of SAPK10 showed that a phosphorylation band of purified GST‐SAPK10 was detected by phos‐tag, and it disappeared after CIAP treatment, while GST as a negative control was not phosphorylated, indicating that SAPK10 has an autophosphorylation activity on its own (Fig. 1b). When Ser177 and Thr178 of SAPK10 were replaced by alanines (SAPK10S177A and SAPK10T178A, respectively), the autophosphorylation band was not detected in SAPK10S177A and SAPK10S177A/T178A (Fig. 1b). However, the phosphorylation intensity of SAPK10T178A was also decreased to nearly half that of the native SAPK10 (Fig. 1b). Therefore, Ser177 might be the major autophosphorylation site of SAPK10. Moreover, we treated proUbi:SAPK10‐FLAG seedlings with exogenous ABA at various concentrations, and purified the SAPK10‐FLAG proteins to examine their phosphorylation intensities by immunoblot. Obviously, the application of ABA significantly increased the SAPK10 phosphorylation intensity in a dosage‐dependent manner, and the phosphorylation bands were effectively removed by CIAP treatment, suggesting that the autophosphorylation on SAPK10 is induced by the ABA concentration in vivo (Fig. 1c). The autophosphorylation activity of SAPK10 intrigued us to test the protein self‐binding ability. The Y2H and in vitro pull‐down assays clearly demonstrated that SAPK10 could physically bind itself to form a homodimer. The autophosphorylation on SAPK10 may not affect its protein‐binding ability, as SAPK10 was found to physically interact with SAPK10S177A in yeast and in vitro (Fig. 1d–e).
Fig. 1.
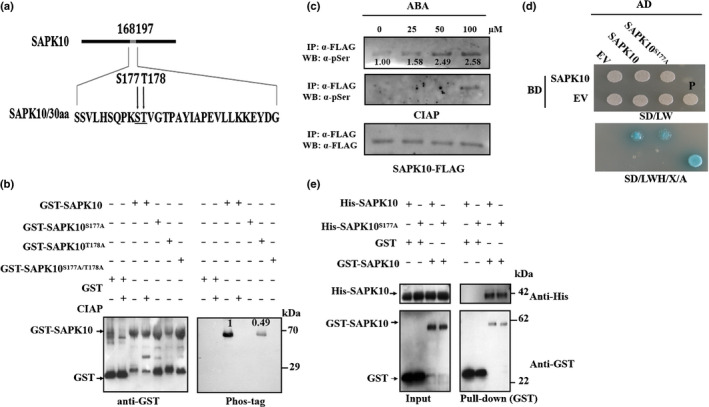
SAPK10 is autophosphorylated on serine 177. (a) Schematic representation of the putative autophosphorylation sites of SAPK10. Ser‐177 and Thr‐178 in the T‐ loop region of SAPK10 (amino acids 168–197) are indicated in bold. (b) Serine 177 is the key phosphosite of SAPK10. Equal amounts of GST‐SAPK10 and other mutated versions (GST‐SAPK10S177A, GST‐SAPK10T178A, GST‐SAPK10S177A/T178A) were detected with anti‐GST antibody (left panel) and the phosphorylated proteins were detected with biotinylated Phos‐tag™ zinc BTL111 complex (right panel). The GST protein was used as negative control. GST, glutathione S‐transferase. (c) SAPK10 phosphorylation was enhanced under abscisic acid (ABA) treatment. Two‐week‐old pro35S:SAPK10‐FLAG seedlings (Nipponbare, Oryza sativa ssp. japonica) were treated with ABA in different concentrations (0, 25, 50 and 100 μM) for 6 h. Equal amounts of immunoprecipitated SAPK10‐FLAG proteins were immunoblotted against antiphosphoserine (a‐pSer) antibody (top panel), treated with calf intestinal phosphatase (CIAP) at 37°C for 30 min and against a‐pSer antibody (middle panel) and anti‐FLAG antibody (bottom panel). The relative intensity of SAPK10‐FLAG under no‐ABA treatment was set to 1.00. (d) Yeast two‐hybrid assay. Yeast cells cotransformed with SAPK10 or SAPK10S177A fused to the GAL4 activation domain (SAPK10‐AD or SAPK10S177A‐AD) and SAPK10 fused to the GAL4‐binding domain (SAPK10‐BD) were grown on selective media. BD, pGBKT7; AD, pGADT7; EV, empty vector; SD/LW, ‐Leu‐Trp; SD/LWH, ‐Leu‐Trp‐His; P, positive control using pGADT7‐T + pGBKT7‐53; X: X‐α‐Gal in 0.04 mg ml−1; a, Aureobasidin A in 100 ng mI−1. (e) Pull‐down assay. Purified His‐SAPK10, His‐SAPK10S177A, GST and GST‐SAPK10 were subjected to pull‐down assays and detected with anti‐His and anti‐GST antibodies, respectively. Molecular mass markers are shown (kDa).
Overexpression of SAPK10 confers rice with hypersensitivity to ABA
SAPK10 overexpression lines (OxSAPK10) as well as CRISPR/Cas9‐mediated knockout mutants (crsapk10) were generated to dissect the biological roles of SAPK10 in rice. The seeds of T2 homozygous crsapk10 lines, which harbored an A insertion or an A/G insertion in the first exon and shifted the open reading frame of SAPK10, showed similar germination rate as the WT under the conditions with or without the additions of ABA (Fig. S2). This may be ascribed to the functional redundancy of SAPK10 with other ABA‐responsive SAPK members like SAPK6, SAPK8 and SAPK9, as the transcription levels of SAPK8 and SAPK9 were elevated when SAPK10 was knocked out (Fig. S3). Further developing higher‐order mutants of SAPK6, 8 and 9 with SAPK10 would be helpful to clarify this issue more clearly. Two representative OxSAPK10 lines with substantially increased transcriptional level were also picked for seed germination assays (Fig. S4). Under no‐ABA conditions, the seed germination and seedling growth of OxSAPK10 lines were significantly retarded in comparison with the WT (Fig. 2a–c). Similarly, OxSAPK10 displayed more severe ABA‐mediated repression on seed germination and seedling growth than did the WT (Fig. 2a–d). To evaluate the relative ABA sensitivity of the seeds, we compared the germination rates at 4 DAG under mock and ABA treatments. As a result, the WT relative germination rates of 2 μM ABA/mock and 5 μM ABA/mock were 93% and 84%, respectively. However, for the OxSAPK10 seeds, these numbers decreased to 61% and 56%, respectively. Thus, OxSAPK10 is hypersensitive to ABA when compared with the WT (P < 0.01) (Fig. 2b). Subsequently, SAPK10S177A overexpression lines (OxSAPK10S177A) were generated to explore the effects of autophosphorylation of SAPK10 on its own biological functions (Fig. S4). The lines OxSAPK10S177A‐2 and OxSAPK10S177A‐3, which have comparable SAPK10 transcriptional level with OxSAPK10‐3, was selected for phenotyping (Fig. S4). Interestingly, in comparison with the delayed seed germination and seedling growth in OxSAPK10‐3, the OxSAPK10S177A line lost its hypersensitivity to ABA, and showed similar germination rate and seedling growth as that of the WT (Fig. 2). Thus, these results indicated that blocking of the autophosphorylation site impairs the SAPK10 function in ABA signaling and inhibiting seed germination.
Fig. 2.
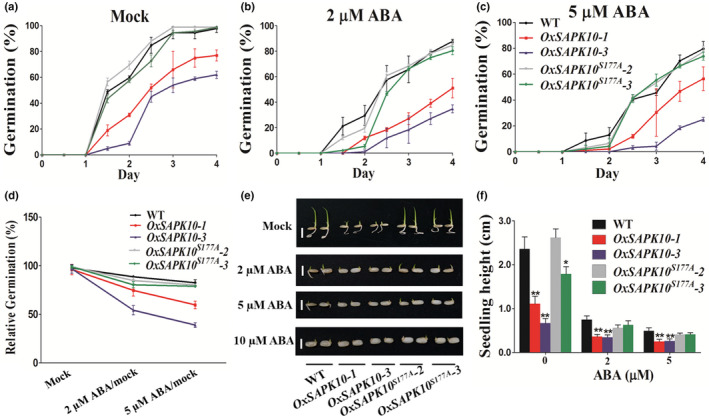
Overexpression of SAPK10 confers rice with hypersensitivity to abscisic acid (ABA). (a) Germination time courses of wild‐type (WT), OxSAPK10 and OxSAPK10S177A seeds grown on half‐strength Murashige & Skoog (½MS) medium containing different concentrations of ABA (0, 2 and 5 μM), respectively. (b) Relative germination of the WT, OxSAPK10 and OxSAPK10S177A seeds under ABA treatments were determined after 4 d and expressed as a percentage of those grown under the ‘mock’ condition. (c) Germination phenotypes of the WT, OxSAPK10 and OxSAPK10S177A treated with 0, 2 and 5 μM ABA, respectively. Photographs were taken on day 7. Bars, 1cm. (d) Seedling heights of the WT, OxSAPK10 and OxSAPK10S177A in accordance with (c). Error bars indicate SD with biological triplicates (n = 3, each replicates containing 50 seeds) in (a) and 50 biological replicates (n = 50) in (d). Asterisks indicate the significance of differences between the WT and transgenic lines as determined by Student's t‐test analysis: *, P < 0.05; **, P < 0.01.
SAPK10 phosphorylates and stabilizes bZIP72
After screening over two million colonies from a rice seed‐derived cDNA library using Y2H system, we found that transcription factor bZIP72 could interact with SAPK10. Interestingly, bZIP72 and SAPK10 both have relatively high transcriptional levels in developing seeds and could be significantly induced by exogenous ABA during seed germination (Fig. S5a,b). Their expression patterns in other various tissues and growth stages, as well as their subcellular localizations, are largely overlapped (Fig. S5c–e). The interaction between SAPK10 and bZIP72 was further validated by Y2H, in vitro pull‐down and in vivo Co‐IP assays, though bZIP72 somehow displayed weak autoactivation activity in Y2H (Fig. 3a–c). In support of our conclusion that autophosphorylation on SAPK10 does not affect its protein–protein binding ability, SAPK10S177A could also physically bind with bZIP72 in yeast and in vitro, just as SAPK10 did (Fig. 3a,b). Meanwhile, almost equal amounts of the His‐SAPK10 and His‐SAPK10S177A proteins were coimmunoprecipitated by GST‐bZIP72 when the pull‐down assays were performed in parallel (Fig. 3b), suggesting that ABA‐induced phosphorylation on SAPK10 has little or no impact on its binding intensity to bZIP72 either.
Fig. 3.
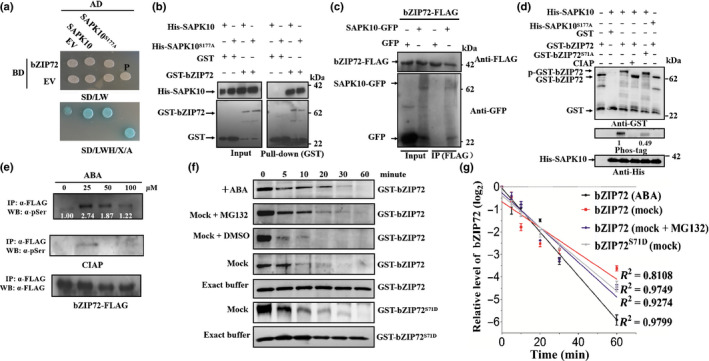
SAPK10 phosphorylates and stabilizes bZIP72. (a) Yeast two‐hybrid assays. Yeast cells cotransformed with SAPK10‐AD or SAPK10S177A‐AD and bZIP72 fused to the GAL4‐binding domain (bZIP72‐BD) were grown on selective media. BD, pGBKT7; AD, pGADT7; EV, empty vector; SD/LW, ‐Leu‐Trp; SD/LWH, ‐Leu‐Trp‐His; P, positive control using pGADT7‐T + pGBKT7‐53; X: X‐α‐Gal in 0.04 mg ml−1. (a) Aureobasidin A in 100 ng ml−1. (b) Pull‐down assay. Purified His‐SAPK10, His‐SAPK10S177A, GST and GST‐bZIP72 were subjected to pull‐down assays and detected with anti‐His and anti‐GST antibodies, respectively. (c) Coimmunoprecipitation (Co‐IP) assay. GFP, SAPK10‐GFP and bZIP72‐FLAG extracted from infiltrated tobacco (Nicotiana benthamiana) leaves were used in a Co‐IP assay. Precipitates were detected with anti‐GFP and anti‐FLAG antibodies, respectively. (d) bZIP72 was phosphorylated by SAPK10 at Ser71. Equal amounts of the recombinant proteins were detected with anti‐GST (top panel) and anti‐His (bottom panel) antibodies, respectively. The phosphorylated proteins were detected with biotinylated Phos‐tag™ zinc BTL111 complex (middle panel). GST‐bZIP72 protein alone or GST purified from coexpressed forms with His‐SAPK10 was used as the negative control. p‐GST‐bZIP72, GST‐bZIP72 phosphorylated band. The relative intensity of GST‐bZIP72 phosphorylated band was set to 1.00 in the middle panel. (e) In vivo phosphorylation of bZIP72 was enhanced under abscisic acid (ABA) treatment. Two‐week‐old pro35S:bZIP72‐FLAG seedlings (Nipponbare, Oryza sativa ssp. japonica) were treated with ABA at different concentrations (0, 25, 50 and 100 μM) for 6 h. Equal amount of immunoprecipitated bZIP72‐FLAG proteins were immunoblotted against antiphosphoserine (α‐pSer) antibody (top panel), treated with calf intestinal phosphatase (CIAP) at 37°C for 30 min and against a‐pSer antibody (middle panel), and anti‐FLAG antibody (bottom panel). The relative intensity of bZIP72‐FLAG phosphorylated band under no‐ABA treatment was set to 1.00 in the top panel. (f) Cell‐free degradation assay of GST‐bZIP72 in the absence or presence of ABA. Equal amounts of total plant (Nipponbare) proteins (30 μg) were incubated with GST‐bZIP72 or GST‐bZIP72S71D (mimicking the phosphorylation form of bZIP72) and detected with anti‐GST antibody. For control, 100 ng GST‐bZIP72 or GST‐bZIP72S71D was incubated with 30 μl of extract buffer. (g) The relative remaining amount of bZIP72 protein was calculated and plotted on a semilog graph in accordance with (f). A ratio of protein signal at the corresponding time to the signal at the start (0 min) was shown as a dissociation‐one phase exponential decay curve. Error bars indicate SD with biological triplicates (n = 3).
Given the protein kinase feature of SAPK10, the kinase‐substrate relationship between SAPK10 and bZIP72 was tested by kinase assay in E.coli. As shown in Fig. 3(d), the GST‐bZIP72 was only phosphorylated by His‐SAPK10, but not by SAPK10S177A, indicating that the autophosphorylation on SAPK10 is a key switch controlling its kinase activity (Fig. 3d). Our previous phosphoproteomic study of ABA‐induced protein phosphorylation revealed that exogenous ABA enhanced the bZIP72 phosphorylation intensity on Serine 71 (Qiu et al., 2017). Thus, we attempted to specify the residues responsible for SAPK10‐mediated phosphorylation on bZIP72 by using bZIP72S71A as the substrate for the kinase assay. The phosphorylation intensity of bZIP72S71A is only half that of the native bZIP72, suggesting that Ser71 is a major site of the SAPK10‐mediated phosphorylation (Fig. 3d). Moreover, we found that the in vivo phosphorylation intensity on bZIP72 also displayed an ABA dosage‐dependent pattern, which is quite a similar pattern to SAPK10 (Fig. 3e).
Protein phosphorylation has been linked to targeting protein turnover and stability (Chen et al., 2015). Therefore, we attempted to check the influences of SAPK10‐mediated phosphorylation on the stability of bZIP7 by cell‐free protein degradation assay. As shown in Fig. 3(f) and (g), treatment of the seedlings with ABA significantly extended the half‐life of GST‐bZIP72 to 10 min, whereas GST‐bZIP72 was degraded to half amount in only 6 min when the mock protein extracts were used, suggesting that ABA enhanced the protein stability of bZIP72 (Fig. 3f,g). On the other hand, by using GST‐bZIP72S71D as the substrate, in which the Ser71 was replaced by aspartic acid to mimic constitutive phosphorylation status of bZIP72, we found that the mimicked phosphorylation significantly stabilized the protein with a half‐life of 9 min when incubated with the mock total protein extracts (Fig. 3f‐g). The addition of MG132, a 26S proteasome inhibitor, drastically delayed the in vitro degradation of GST‐bZIP72, which implied that the protein is under the degradation of the Ubiquitin/26S proteasome pathway (Fig. 3f‐g). Taken together, these results strongly suggested that the stability of bZIP72 was enhanced by ABA and SAPK10‐mediated phosphorylation on Ser71.
bZIP72 and TRAB1 functions redundantly in ABA signaling
The kinase‐substrate relationship between SAPK10 and bZIP72 motivated us to investigate the function of bZIP72 in ABA signaling by generating various types of mutants and overexpression lines. Interestingly, the CRISPR/Cas9‐mediated bzip72 homozygous mutants (crbzip72) did not show any significant differences with the WT in seed germination, although we have confirmed the indel mutations in bZIP72 (Figs 4a,b,e,g, S6a,b). It was assumed that the loss‐of‐function of bZIP72 was compensated by its closest homolog, TRAB1/OsbZIP66 (LOC_Os08g36790), because transcription of TRAB1 was significantly upregulated when bZIP72 was mutated (Fig. S7). Subsequently, we knocked out both genes simultaneously (Fig. S6c,d), and found that the bzip72/trab1 double mutants (crbzip72/crtrab1) became less sensitive to ABA during seed germination, while the trab1 single mutants (crtrab1) were very similar to the WT and bzip72 in phenotype (Fig. 4a,b,e,g). The result suggested that bZIP72 and TRAB1 functions redundantly in ABA signaling. Despite the reduced sensitivity of bzip72/trab1 in seed germination, it should be noted that the seedling growth of bzip72/trab1 and the WT did not show significant differences (P > 0.05), implying that ABA‐mediated repression on germination and shoot growth may depend on different pathways or mechanisms. Despite the reduced sensitivity of bzip72/trab1 in seed germination, it should be noted that the seedling growth of bzip72/trab1 and the WT did not show significant differences (P > 0.05), implying that ABA‐mediated repression on germination and shoot growth may depend on different pathways or mechanisms. Similar to the phenotypes observed in OxSAPK10, bZIP72 overexpression lines showed retarded seed germination and seedling growth under no‐ABA application, and enhanced sensitivity to ABA treatment, as 2 μM ABA/mock and 5 μM ABA/mock relative germination rates were 79% and 66% for OxbZIP72, respectively, and 88% and 85% for the WT (Figs 4c,d,f,g, S8). To further investigate the effect of SAPK10‐mediated phosphorylation on the biological function of bZIP72, the lines OxbZIPS71A‐15 and OxbZIP72S71A‐17, which have comparable bZIP72 transcriptional level with OxbZIP72, were selected for phenotyping (Fig. S8). Nevertheless, OxbZIP72S71A lost the hypersensitivity to ABA with restored seed germination rates and seedling growth (Fig. 4c,d,f,h), indicating that SAPK10‐mediated phosphorylation on serine 71 is a key switch turning on the bZIP72 function in ABA signaling.
Fig. 4.
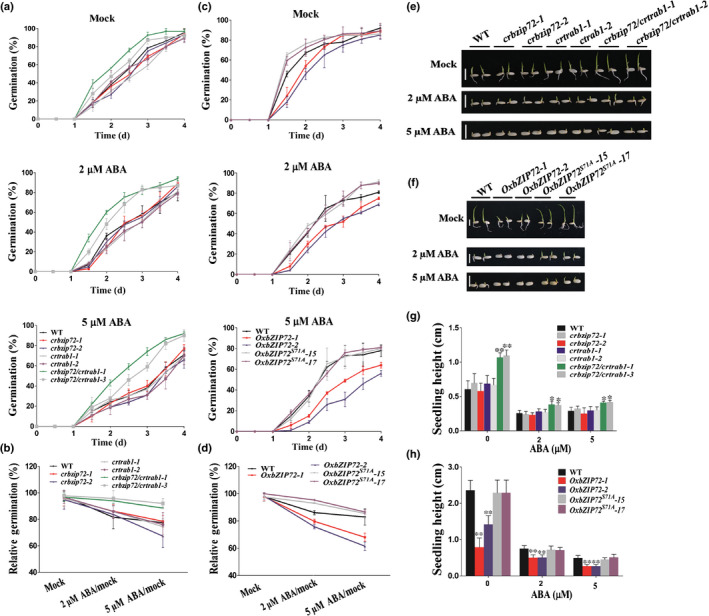
bZIP72 and TRAB1 function redundantly in abscisic acid (ABA) signaling in Nipponbare. (a) Germination time courses of the wild‐type (WT), crbzip72, crtrab1 and crbzip72/crtrab1 seeds grown on half‐strength Murashige & Skoog (½MS) medium containing different concentrations of ABA (0, 2 and 5 μM). (b) The relative germination of the WT, crbzip72, crtrab1 and crbzip72/crtrab1 seeds under ABA treatments were determined after 4 d and expressed as a percentage of those grown under ‘mock’ conditions. (c) Germination time courses of the WT, OxbZIP72 and OxbZIP72S71A seeds grown on ½MS medium containing different concentrations of ABA (0, 2 and 5 μM). (d) The relative germination of the WT, OxbZIP72 and OxbZIP72S71A seeds under ABA treatments were determined after 4 d and expressed as a percentage of those grown under ‘mock’ conditions. (e) Germination phenotypes of the WT, crbzip72, crtrab1 and crbzip72/crtrab1 treated with 0, 2 and 5 μM ABA. Photographs were taken on day 4. Bars, 1 cm. (f) Germination phenotypes of the WT, OxbZIP72 and OxbZIP72S71A treated with 0, 2 and 5 μM ABA. Photographs were taken on day 7. Bars, 1cm. (g) Seedling heights of the WT, crbzip72, crtrab1 and crbzip72/crtrab1 in accordance with (e). (h) Seedling heights of the WT, OxbZIP72 and OxbZIP72S71A in accordance with (f). (a, c, g, h) Error bars indicate SD with biological triplicates (n = 3, each replicate containing 50 seeds) (a, c) and 50 biological replicates (n = 50) (g, h). Asterisks indicate the significance of differences between the WT and transgenic lines as determined by Student's t‐test analysis: *, P < 0.05; **, P < 0.01.
Phosphorylated bZIP72 directly activates AOC transcription by binding to the G‐box in the promoter
As Gibberellins (GAs) and JA are actively involved in seed germination (Nambara et al., 2010), we examined the transcription level of some crucial GA or JA pathway genes in the germinating embryos of OxbZIP72 and the WT. Intriguingly, a couple of JA biosynthesis step‐limiting genes, such as LOX1, LOX2, AOS1, AOS2, AOC and OPR7 (Agrawal et al., 2003), were significantly upregulated in OxbZIP72, but remained unchanged in OxbZIP72S71A, while GA metabolism genes such as CYP714B‐1 and GA2ox6 displayed the opposite tendency (Fig. S9). As predicted by the online tool PlantCARE (Plant Cis‐Acting Regulatory Elements) (Lescot et al., 2002), promoters of LOX1, AOS1 and AOC harbored G‐box cis‐elements, which are the conserved binding sites for bZIPs. Thus, we performed EMSA to test the DNA‐binding ability of bZIP72 on the promoter of these targets. As a result, no positive interactions were detected when unphosphorylated GST‐bZIP72 was used (Fig. S10). Nevertheless, we found that phosphorylated p‐GST‐bZIP72 could specifically bind to the probe 1 (P1) region of AOC, which is a master regulator of JA biosynthesis in the production of JA precursor 12‐oxo‐phytodienoic acid (12‐OPDA) (Fig. 5a,b). The EMSA binding was substantially weakened by nonlabeled, competitive probes in a dosage‐dependent manner (Fig. 5c). This result implied that the SAPK10‐mediated phosphorylation confers bZIP72 DNA‐binding ability to the promoter of AOC, in addition to enhancing bZIP72 protein stability. The protein‐DNA binding was compromised when some of the nucleotides in the conserved G‐box in the P1 region were mutated, demonstrating that the G‐box acts as a core site for recognition by bZIP72 (Fig. 5a,d). Subsequently, ChIP‐qPCR was performed to validate such binding in vivo. A total of four fragments representing the promoter, UTR and CDS regions were used for the examination (Fig. 5a). In support of the results of EMSA, bZIP72 was significantly enriched in the P1 region of the AOC promoter under ABA treatment, while there was no significant enrichment in the other fragments under the mock condition, except that the P3 region, which is located in the CDS region of AOC, exhibited slight bZIP72 enrichment (Fig. 5e). Finally, a dual‐luciferase (LUC) transient transcriptional activity assay was performed to determine the regulatory effect of bZIP72 on AOC transcription (Fig. 5f). In comparison with the empty effector, pro35S:bZIP72:tNOS drastically elevated the level of fLUC reporter, but such induction was blocked when the effector bZIP72 was replaced by bZIP72S71A, which is in accordance with the transcription pattern of AOC in OxbZIP72 and OxbZIP72S71A transgenic lines, respectively (Fig. 5g). Notably, the upregulation of AOC by bZIP72 was significantly enhanced when exogenous ABA was applied (Fig. 5g), suggesting that ABA might induce SAPK10‐mediated phosphorylation on bZIP72 to promote the transcription level of AOC. Previous studies have clarified that a rice AOC mutant cpm2 has lower endogenous JA concentration and unchanged ABA contents when compared with the WT (O. sativa L. ssp. japonica cv Nihonmasari) (Hazman et al., 2015). Seed germination assay revealed that cpm2 also displayed reduced sensitivity to ABA, because the relative germination rates of cpm2 of 2 μM/mock and 5 μM/mock were 76% and 64%, respectively, whereas these of the WT were 64% and 45% (Fig. 6a,b). cpm2 also showed a significantly higher relative seedling growth than the WT (Fig. 6c–e). Taken together, these experiments strongly supported the possibility that AOC positively participates in ABA‐mediated seed germination and postgermination growth processes.
Fig. 5.
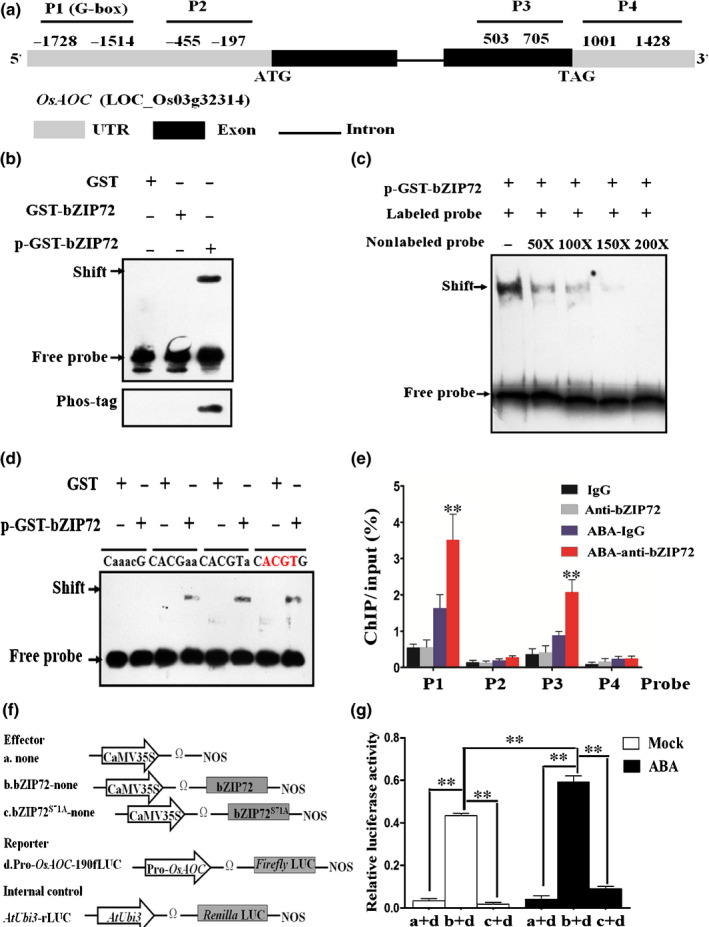
Phosphorylated bZIP72 directly activates AOC transcription by binding to the G‐box in the promoter. (a) Probe positions on AOC promoter and genome. Gray boxes, untranslated regions; black boxes, exons; black line, intron. Transcription starting site ATG was set as 0. Numbers indicate the distances (bps) of probes to the ATG. P1–P4, probes 1–4. (b) Electrophoresis mobility shift assay (EMSA) showed that p‐GST‐bZIP72 (phosphorylated GST‐bZIP72 band) binds with the P1 on the promoter of AOC in (a). Purified GST, GST‐bZIP72 and p‐GST‐bZIP72 were detected with anti‐GST antibody. p‐GST‐bZIP72 was detected with biotinylated Phos‐tag™ zinc BTL111 complex. GST, glutathione S‐transferase.(c) EMSA showed that p‐GST‐bZIP72 specifically bound to the G‐box in the P1 region of AOC promoter. The 50‐, 100‐, 150‐ and 200‐fold excess nonlabeled probes were applied as competitiors. (d) EMSA assay showed that CACGTG in the P1 region of AOC promoter was required for p‐GST‐bZIP72 to bind AOC. Probe 1 sequence (60 bp) contains G‐box (CACGTG) in (a), with the core element sequences ACGT in this motif marked by red letters. CACGTG was substituted by CaaacG, CACGaa and CACGTa in the mutant probe and the substitution nucleotide acids are marked in lowercase. (e) Chromatin immunoprecipitation‐quantitative PCR (ChIP‐qPCR) assay showed that abscisic acid (ABA) enhances bZIP72 binding to the promoter and coding sequence (CDS) regions of AOC. P1–P4 represents the regions shown in (a) detected by ChIP‐qPCR. The enrichment values were normalized to input. Immunoglobulin G (IgG) immunoprecipitated DNA was used as a control. Error bars indicate SD with biological triplicates. (f, g) Luciferase transient transcriptional activity assay in rice protoplast. Effectors: 35S:tNOS, 35S:bZIP72:tNOS and 35S:bZIP72S71A:tNOS; Reporter: proAOC:LUC. Error bars represent means ± SD (n = 3) in (e) and (g). **, P < 0.01 by Student's t‐test analysis.
Fig. 6.
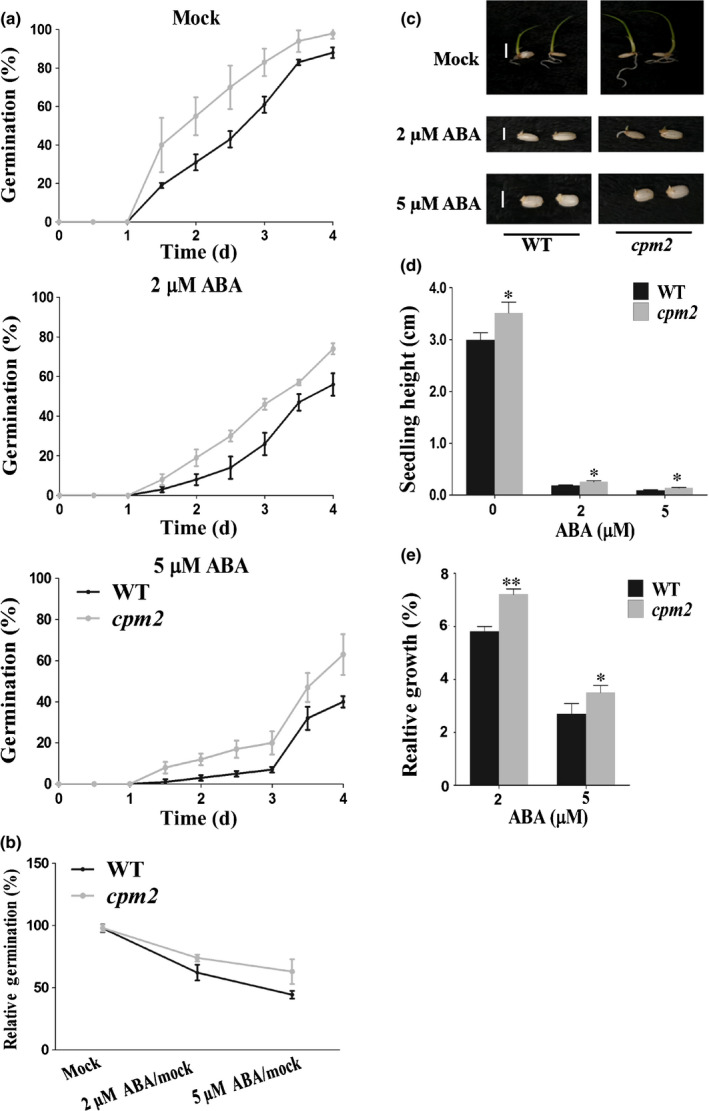
cpm2, a rice AOC mutant, displayed hyposensitivity to abscisic acid (ABA). (a) Germination time courses of the wild‐type (WT) and cpm2 mutants grown on half‐strength Murashige & Skoog (½MS) medium containing different concentrations of ABA (0, 2, 5 μM). (b) The relative germination of the WT and cpm2 seeds under ABA treatments were determined after 4 d and expressed as a percentage of those grown under the ‘mock’ condition. (c) Germination phenotypes of the WT and cpm2 treated with 0, 2, 5 μM ABA. Photographs were taken on day 4. Bars, 1cm. (d) Seedling heights of the WT and cpm2 in accordance with (c). (e) Growth inhibition of seedling height by ABA. The relative growth of the WT and cpm2 seeds under ABA treatments were determined after 4 d and expressed as a percentage of those grown under the ‘mock’ condition. (a, d) Error bars indicate SD with biological triplicates (n = 3, each replicate containing 50 seeds) (a) and 50 biological replicates (n = 50) (d). Asterisks indicate the significance of differences between the WT and transgenic lines as determined by Student’s t‐test analysis in (d) and (e): *, P < 0.05; **, P < 0.01. Oryza sativa ssp. japonica cv Nihonmasari was used as the wild‐type.
Additionally, we tested the binding ability of GST‐bZIP72 on the promoters of some rice GA pathway genes such as GA3ox1, GA2ox5 and GA20ox2 with G‐box motifs (Itoh et al., 2001; Spielmeyer et al., 2002; Lo et al., 2008), and found that GST‐bZIP72 was able to bind to the promoter of GA20ox2, indicating the involvement of bZIP72 in GA homeostasis as well (Fig. S10).
ABA partially inhibits seed germination by inducing JA accumulation
The ‘SAPK10‐bZIP72‐AOC’ regulatory pathway identified in the previous section hinted a positive regulatory pathway in the interplay between ABA and JA during seed germination. To address this issue, exogenous ABA was applied to germinating seeds of the WT. The JA accumulation level was significantly elevated when the WT germinating embryos were treated by ABA (Fig. 7a). Consistent with this result, a set of ABA signaling genes and JA pathway genes, including AOC, were drastically upregulated (Fig. S11). As both ABA and JA have been known as inhibitors for seed germination (Liu et al., 2015; Shu et al., 2016), it is likely that the final inhibition effects of ABA on seed germination may be partially based on the ABA‐promoted JA accumulation.
Fig. 7.
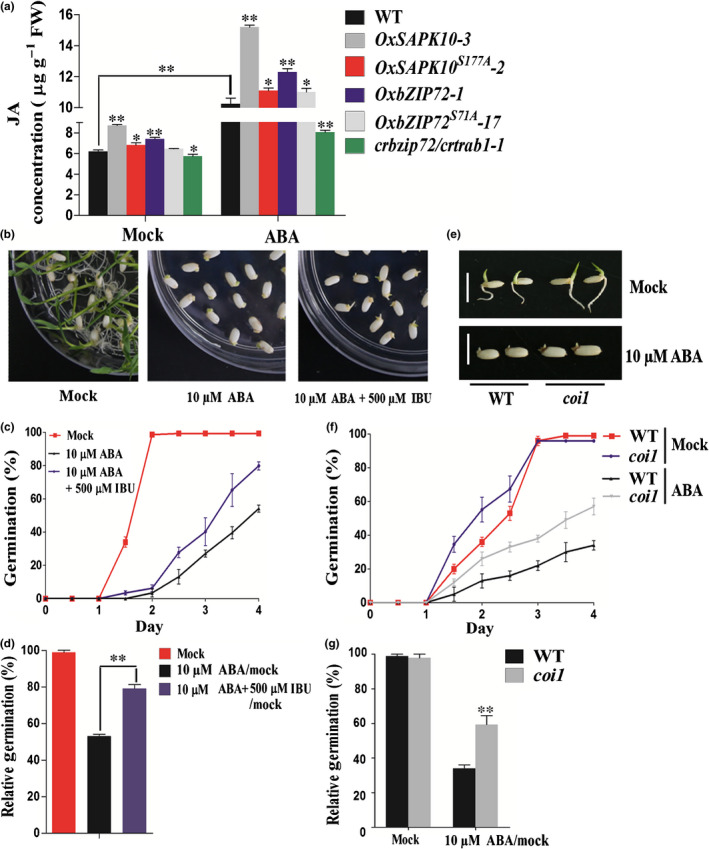
Abscisic acid (ABA) inhibits seed germination by partially inducing jasmonic acid (JA) accumulation in Nipponbare. (a) Measurement of JA concentrations in the seeds of the wild‐type (WT), OxSAPK10, OxSAPK10S177A, OxbZIP72, OxbZIP72S71A and crbzip72/crtrab1 after being grown on half‐strength Murashige & Skoog (½MS) medium under mock or 10 μM ABA treatment for 4 d. Error bars indicate SD with three biological replicates (n = 3). Asterisks indicate the significance of differences between the WT and transgenic lines as determined by Student's t‐test analysis: *, P < 0.05; **, P < 0.01. (b) Germination phenotypes of the WT under mock, 10 μM ABA or 10 μM ABA + 500 μM Ibuprofen (IBU) treatment. Photographs were taken on day 10. (c) Germination time courses of the WT grown on ½MS medium under mock, 10 μM ABA or 10 μM ABA + 500 μM IBU treatment, respectively. (d) The relative germination of the WT seeds under 10 μM ABA or 10 μM ABA + 500 μM IBU treatment were determined after 4 d and expressed as a percentage of those grown under the ‘mock’ condition. (e) Germination phenotypes of the WT and coi1 mutants under mock or 10 μM ABA treatment, respectively. Photographs were taken on day 4. Bars, 1 cm. (f) Germination time courses of WT and coi1 grown on ½MS medium containing 0 or 10 μM ABA. (g) The relative germination of the WT and coi1 seeds under 10 μM ABA treatment was determined after 4 d and expressed as a percentage of those grown under the ‘mock’ condition. (c, f) Error bars indicate SD with biological triplicates (n = 3, each replicates containing 50 seeds).
By blocking lipoxygenase, which is the key enzyme catalyzing the first step of JA biosynthesis, Ibuprofen (IBU) has been widely used as a JA inhibitor (Vick & Zimmerman, 1984; Staswick et al., 1991; Schaller & Stintzi, 2009). We found that the seed germination and post‐germination growth restricted by ABA were obviously relieved when IBU was applied (Figs 7b–d, S12). Additionally, coi1, a JA receptor mutant in rice (Yang et al., 2012), also showed reduced sensitivity to ABA in germination, as the relative 10 μM ABA/mock germination rate was 59% for coi1 and only 34% for the WT. Seedling growth of coi1 showed a similar reduced ABA sensitivity as the germination rate (Fig. 7e–g).
The endogenous JA contents were also quantified in OxSAPK10 and OxbZIP72 germinating embryos with or without ABA treatment, respectively. Under nonABA conditions, the JA concentrations in OxSAPK10 and OxbZIP72 were significantly higher than the WT (Fig. 7a). However, the JA contents in OxSAPK10S177A and OxbZIP72S71A dropped drastically when compared with those in OxSAPK10 and OxbZIP72, respectively (Fig. 7a). Moreover, the endogenous JA concentration in the crbzip72/crtrab1 double mutant was significantly reduced under either mock or ABA treatment conditions, and a series of JA biosynthesis genes, including AOC, were downregulated in crbzip72/crtrab1 (Figs 7a, 8i). Application of exogenous ABA showed similar inclinations as the nonABA conditions, but in a more severe manner (Fig. 8i). In accordance with the JA quantification assays, IBU relieved the inhibition on seed germination and seedling growth of OxSAPK10 and OxbZIP72, while OxSAPK10S177A and OxbZIP72S71A showed no significant difference between the mock and IBU treatments, which is similar to what was seen in the WT (Figs 8a–h, S13). To this end, we concluded that ABA hinders seed germination partially through inducing JA accumulation.
Fig. 8.
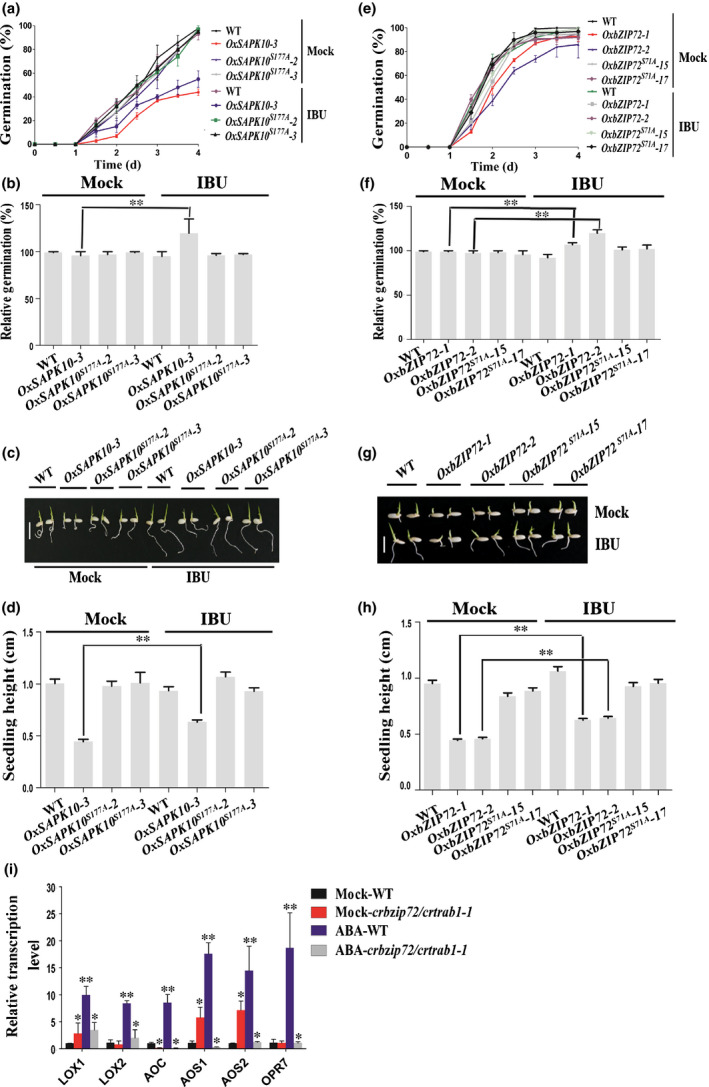
Ibuprofen (IBU) relieved the inhibition of OxSAPK10 and OxbZIP72 on seed germination in Nipponbare. (a) Germination time courses of the wild‐type (WT), OxSAPK10 and OxSAPK10S177A grown on half‐strength Murashige & Skoog (½MS) medium under mock or 100 μM IBU treatment. (b) The relative germination of the WT, OxSAPK10 and OxSAPK10S177A seeds under 100 μM IBU treatment were determined after 4 d and expressed as a percentage of those grown under ‘mock’ conditions. (c) Germination phenotypes of the WT, OxSAPK10 and OxSAPK10S177A treated with 0 or 100 μM IBU. Photographs were taken on day 4. Bar , 1cm. (d) Seedling heights of the WT, OxSAPK10 and OxSAPK10S177A in accordance with (c). (e) Germination time courses of the WT, OxbZIP72 and OxbZIP72S71A grown on ½MS medium under mock or 100 μM IBU treatments. (f) The relative germination of the WT, OxbZIP72 and OxbZIP72S71A seeds under 100 μM IBU treatment were determined after 3 d and expressed as a percentage of those grown under ‘mock’ conditions. (g) Germination phenotypes of the WT, OxbZIP72 and OxbZIP72S71A treated with 0 or 100 μM IBU. Photographs were taken on day 4. Bar, 1cm. (h) Seedling heights of the WT, OxbZIP72 and OxbZIP72S71A in accordance with (g). (i) Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis for transcript accumulation of abscisic acid (ABA) and jasmonic acid (JA) pathway genes in the seeds of the WT and crbzip72/crtrab1. Seeds were grown on mock or ½MS medium treated with 0 or 10 μM ABA for 6 h before harvest for total RNA isolation. (a, d, e, h, i) Error bars indicate SD with biological triplicates (n = 3, each replicate containing 50 seeds) (a, e), 50 biological replicates (n = 50) (d, h) and biological triplicates (n = 3) (i). (b, d, f, h) Asterisks indicate the significance of differences between the mock and IBU treatment (Student's t‐test analysis; **, P < 0.01). (i) Asterisks indicate the significance of differences between the WT and transgenic lines (Student's t‐test analysis; *, P < 0.05; **, P < 0.01).
Discussion
‘SAPK10‐bZIP72‐AOC’ is a key pathway of ABA signaling in seed germination
The current study identified a key ABA signaling pathway, ‘SAPK10‐bZIP72‐AOC’, in rice seed germination. This pathway is supported by several layers of genetics and biochemical evidence: genetically, overexpression of SAPK10 or bZIP72 conferred plants with hypersensitivity to ABA during seed germination, whereas the double knockout of bZIP72/TRAB1 or single knockout of AOC compromised the ABA sensitivity of seeds, suggesting that the three genes had similar positive functions in ABA signaling and seed germination; ABA‐inducible SAPK10 physically bound to and phosphorylated bZIP72, and the phosphorylation on bZIP72 could be enhanced by exogenous ABA; and SAPK10‐mediated phosphorylation strengthened the binding ability of bZIP72 on the AOC promoter and activated the transcription of AOC. Likewise, ABA also enhanced the bZIP72‐dependent activation on AOC transcription.
SAPK10 is the closest rice ortholog of OST1, which has been characterized as a key player in ABA signaling in Arabidopsis (Nakashima et al., 2009). Nevertheless, except for the discovery of its kinase activity on TRAB1 (Kobayashi et al., 2005), little has been known about the biological functions of SAPK10 until very recently. It is now clear that, together with SAPK8 and 9, SAPK10 could phosphorylate Tiller enhancer to block the GA signaling with elevated endogenous ABA concentration and retarded seedling growth, which is implicated in the ABA‐mediated inhibition of plant growth (Lin et al., 2015). SAPK10 is also involved in the ABA‐regulated rice flowering via phosphorylating bZIP77 in the FAC‐MADS15 pathway (D. Liu et al., 2019; X. Liu et al., 2019).
bZIPs are typical ABF TFs that can be phosphorylated by SnRK2 in ABA signaling (D. Liu et al., 2019; X. Liu et al., 2019). The current study showed that ABA‐inducible bZIP72 is subject to the protein phosphorylation regulation by SAPK10. bZIP72 works redundantly with TRAB1 to positively transmit ABA signal, as the ABA sensitivity in seed germination was attenuated only when both genes were mutated (Fig. 4a,b,e,g). Meanwhile, OxbZIP72 became hypersensitive to ABA in seed germination (Fig. 4c,d,f,h), which is also consistent with its reported function in drought resistance (Lu et al., 2009). Interestingly, we found that AOC encoding an allene oxide cyclase in JA biosynthesis is directly transcribed by bZIP72. AOC has been implicated in coleoptile photomorphogenesis, abiotic stress resistance to salinity, as well as biotic stress resistance to piercing‐sucking insects and Magnaporthe oryzae (Riemann et al., 2013; Guo et al., 2014; Hazman et al., 2015). The ABA insensitivity of cpm2 seeds hinted at an essential role of AOC in ABA signaling and crosstalk between ABA and JA in rice (Fig. 6).
Comprehensive effects of SAPK10‐mediated phosphorylation on the ABA signaling elements
Protein phosphorylation represents a significant mechanism of the signal transduction of phytohormones such as ABA and brassinosteroids (BR) (W. Wang et al., 2014; Yang et al., 2017). The introduction or removal of a charged, hydrophilic phosphate group in the sidechain of amino acids could possibly change the structure of target protein by altering interactions with nearby amino acids, which finally imposes profound effects on the protein functions (Reinders & Sickmann, 2005). Under cold stress, phosphorylation facilitates the shuttling of 14‐3‐3 proteins from cytosol to the nucleus to interact with CBFs, while OST1‐dependent phosphorylation enhances the protein–protein interaction of BTF3 with their partners (Liu et al., 2017; Ding et al., 2018). Effects of phosphorylation on WRKYs are somehow elusive. For example, MPK3/MPK6 phosphorylates WRKY53 to enhance its transcriptional activity on defense‐related genes (Chujo et al., 2014). By contrast, WRKY72 had weakened trans‐activation on JA synthesis genes when phosphorylation occurred on its 129th threonine (Hou et al., 2019). In this study, we revealed the comprehensive effects of SAPK10‐mediated phosphorylation on the target proteins. Biochemical evidence revealed that the autophosphorylation on Ser 177 of SAPK10 is crucial to maintaining its kinase activity on both itself and the substrate protein bZIP72, but this modification does not alter its protein–protein binding ability (Figs 1, 3, S1). Artificial blocking of the major autophosphorylation site Ser 177 of SAPK10 made the protein dysfunctional, because it was observed that overexpression of SAPK10S177A did not exhibit hyper sensitivity to ABA as the native SAPK10 did (Fig. 2). A similar phenomenon was observed on OxbZIP72S71A, the seed sensitivity to ABA of which remained at the same level as that of the WT (Fig. 4). However, the underlying molecular mechanisms were distinct; as indicated in the cell‐free protein degradation assay, constitutive phosphoryaltion on Ser 71 of bZIP72 made the protein more resistant to the degradation by 26S proteosome (Fig. 3f,g). Application of exogenous ABA achieved the same effect, and this is in agreement with the logic that ABA could enhance the SAPK10‐mediated phosphorylation on bZIP72 (Fig. 3). In addition to the protein stability, the phosphorylation strengthened the DNA‐binding ability of bZIP72 on the G‐box region of AOC promoter, and eventually elevated the transcriptional level of AOC (Fig. 5). Notably, despite the fact that we functionally characterized the major phosphosite on bZIP72, the minor phosphosites mediated by SAPK10 as well as phosphorylations catalyzed by other kinases are still in the dark. Identification and characterization of the other unknown phosphorylation events on bZIP72 will certainly be helpful to us in understanding the regulatory mechanism of bZIP72 in ABA signaling.
ABA promotes JA accumulation to synergistically inhibit seed germination
Although their synergism relationship has been recognized for a long time, little is known about the molecular network between ABA and JA, especially in seed germination (Nambara et al., 2010). Recent work on Arabidopsis and wheat seed germination demonstrated that exogenous ABA elevated the endogenous JA concentration with the upregulation of a couple of JA biosynthesis genes (Ju et al., 2019). Unfortunately, because the research was more focused on how JA affected the ABA signaling and response, the possible mechanism of ABA‐regulated JA accumulation was untouched. Wang et al (2018) suggested that ABA may induce the expression of several plastid phospholipase such as PLIP2 and PLIP3 to promote the JA biosynthesis in chloroplast of Arabidopsis (Wang et al., 2018). In rice, the ABA‐responsive gene OsbZIP82 positively regulates JA content, primarily through direct regulation of the JA metabolism genes, instead of biosynthesis genes (D. Liu et al., 2019; X. Liu et al., 2019). In this study, we linked the ABA signaling with JA production through a novel pathway, ‘SAPK10‐bZIP72‐AOC’, in which ABA promoted the JA accumulation in germinating seeds. Similarly, higher JA concentration was also detected in OxSAPK10, which has elevated endogenous ABA concentration as well as augmented ABA signaling. It appeared that a set of important JA biosynthesis genes, including AOS1, AOS2 and AOC, were upregulated in the germinating embryos of OxbZIP72 (Fig. S9). In particular, we found that AOC could be directly activated by bZIP72 after receiving the phosphorylation signal from the ABA signaling core element SAPK10 (Fig. 5). As a rate‐limiting gene of JA biosynthesis, the AOC mutant cpm2 was found to be defective in producing JA precursor 12‐oxo‐phytodienoic acid (12‐OPDA), which ultimately led to lower JA concentration (Hazman et al., 2015). By conrast, overexpression of AOC resulted in greater JA accumulation in the plants (Guo et al., 2014). The reduced ABA sensitivity in cpm2 suggested that AOC serves as a key node in the ABA signaling and ABA–JA interaction (Fig. 6).
Considering the inhibition effect of JA in seed germination (Wilen et al., 1991; Yang et al., 2012), it is rational to speculate that the final ABA‐imposed inhibition on seed germination might be the additive effects from both ABA and ABA‐induced JA. Indeed, this was attested by three independent germination assays in which the JA biosynthesis was blocked by using IBU or by knocking out AOC, or JA signaling was repressed by knocking down COI1. Under all the three conditions described, ABA sensitivities were significantly compromised (Figs 6, 7). Additionally, in OxSAPK10 and OxbZIP72 seeds, the inhibition caused by augmented ABA signaling could also be neutralized by IBU (Fig. 8). Therefore, we concluded that the ABA inhibition is partially based on the activated JA concentration.
As a few previous studies have shown that JA‐isoleucine (JA‐Ile), instead of JA, works as the active molecule triggering JA signaling (Thines et al., 2007; Katsir et al., 2008), it was concerned that the elevated endogenous JA concentration may not necessarily represent a higher JA response in case that JA‐JA‐Ile conversion is blocked. However, in the current study, the application of IBU relieved the ABA‐induced JA repression on OxSAPK10 and OxbZIP72 seeds (Fig. 8a,b,e,f), which means JA signaling is effectively working, and JA to JA‐Ile conversion should not be affected. Given the reported strong positive correlation between JA and JA‐Ile in plants (Suza & Staswick, 2008; Shimizu et al., 2013), we propose that JA‐Ile concentration should also be elevated with the increase of JA in this case. Additionally, JA can also be converted to 12‐OH‐JA without being converted to JA‐Ile, and the JA‐Ile peak might not be visible as it could be converted to its oxidated form quickly (Heitz et al., 2019). Hence the role of JA meta‐ and catabolism in seed germination of rice warrants further studies by using jar1 mutant.
A model summarizing the role of the ‘SAPK10‐bZIP72‐AOC’ pathway in rice seed germination is depicted in Fig. 9. Exogenous ABA confers autophosphorylation on the 177th serine of SAPK10, which enables it to phosphorylate bZIP72 to a great extent on the 71st serine (Fig. 9). The SAPK10‐dependent phosphorylation enhances bZIP72 protein stability against 26S proteasome‐mediated degradation as well as the DNA‐binding ability to the G‐box cis‐element of the AOC promoter, which elevated the AOC transcription, and finally increased the endogenous JA concentration to synergistically inhibit seed germination (Fig. 9).
Fig. 9.
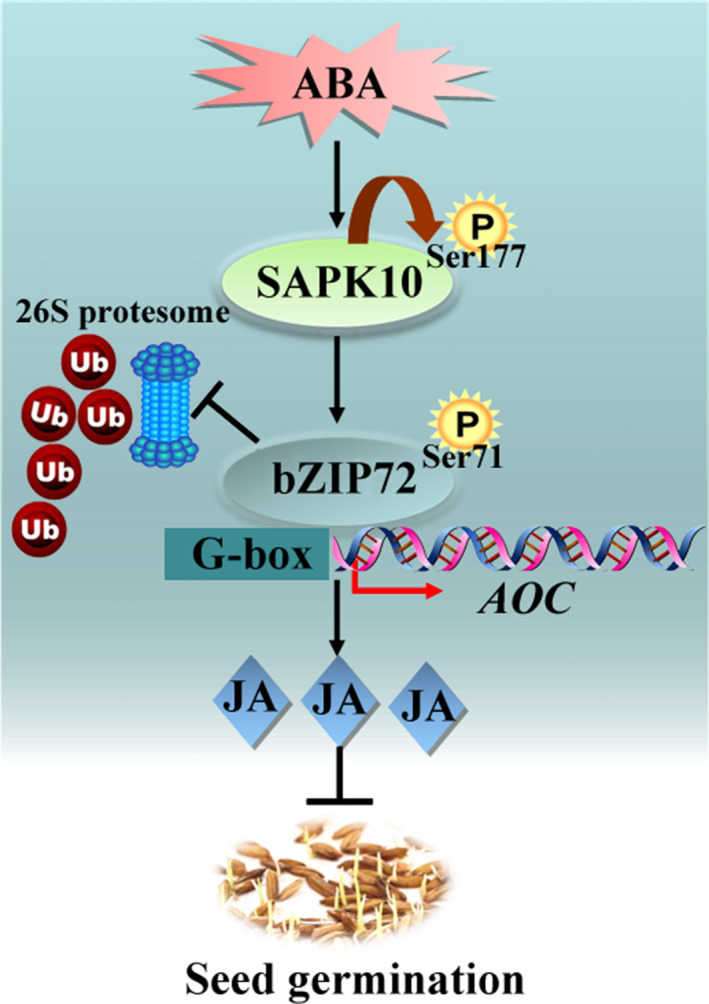
Working model for ‘SAPK10‐bZIP72‐AOC’ pathway in rice seed germination. P, phosphorylation group; Ub, ubiquitin; Ser, serine. Arrowheads show positive regulation, flat‐ended lines show negative regulation and the brown bent arrow represents phosphorylation.
Author contributions
YW and JZ conceived and designed the experiments; YW, YH, JQ, HW, SW, LT and XT conducted the experiments; YW, YH, and JZ analyzed the data; and YW and JZ wrote the manuscript. YW and YH contributed equally to this work.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Sequence alignment of T‐ loop region of 10 Arabidopsis and 10 rice SnRK2 family members.
Fig. S2 Molecular characterization of sapk10 mutant and seed germination in response to ABA.
Fig. S3 qRT‐PCR analysis for transcript accumulation of nine SnRK2 family members (SAPK1‐SAPK9) in the seeds of WT or crsapk10 lines, respectively.
Fig. S4 Molecular characterization of SAPK10, SAPK10S177A overexpressing transgenic lines.
Fig. S5 Expression pattern of SAPK10 and bZIP72.
Fig. S6 Molecular characterization of crbzip72, crtrab1 and crbzip72/crtrab1 mutants.
Fig. S7 qRT‐PCR analysis for transcript accumulation of TRAB1 in the seeds of crbzip72 transgenic line.
Fig. S8 Molecular characterization of bZIP72 and bZIP72S71A overexpressing transgenic lines.
Fig. S9 qRT‐PCR analysis for transcript accumulation of JA and GA pathway genes in the seeds of the WT, OxbZIP72 and OxbZIP72S71A transgenic lines.
Fig. S10 EMSA of bZIP72 on JA pathway genes AOC, AOS1 and LOX1 promoter regions and GA pathway genes GA3ox1, GA2ox5 and GA20ox2 promoter regions.
Fig. S11 qRT‐PCR analysis for transcript accumulation of ABA and JA pathway genes in the seeds of the WT.
Fig. S12 IBU relieved the inhibition of ABA on post‐germination growth.
Fig. S13 SAPK10‐mediated retarded seed germination was relieved by exogenous application of IBU.
Table S1 Sequences of primers used in this study.
Acknowledgements
We thank Professors Moritoshi Iino (Osaka City University) and Donglei Yang (Nanjing Agricultural University) for cpm2 and coi1 seeds, respectively, and Dr Hongyu Zhao (Institute of Agricultural Resources and Regional Planning) for assistance with the subcellular localization experiment. The manuscript was critically reviewed by Prof. Yong Xiang, Xingliang Hou, Wenqiang Tang, Honghong Hu and Dr Abolore Adijat Ajadi. This work was supported by the National Natural Science Foundation of China (grant nos 31401366, 31701395 and 31871229), the ASTIP program of CAAS, and the Chinese High‐yielding Rice Transgenic Program (grant no. 2016ZX08001004‐001). The authors declare there are no competing interests.
References
- Adie BAT, Perez‐Perez J, Perez‐Perez MM, Godoy M, Sanchez‐Serrano J‐J, Schmelz EA, Solano R. 2007. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis . Plant Cell 19: 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal GK, Jwa NS, Agrawal SK, Tamogami S, Iwahashi H, Rakwal R. 2003. Cloning of novel rice allene oxide cyclase (OsAOC): mRNA expression and comparative analysis with allene oxide synthase (OsAOS) gene provides insight into the transcriptional regulation of octadecanoid pathway biosynthetic genes in rice. Plant Science 164: 979–992. [Google Scholar]
- Aleman F, Yazaki J, Lee M, Takahashi Y, Kim AY, Li Z, Kinoshita T, Ecker JR, Schroeder JI. 2016. An ABA‐increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: a putative link of ABA and JA signaling. Scientific Reports 6: 28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova Z. 2017. The jasmonic acid‐signalling and abscisic acid‐signalling pathways cross talk during one, but not repeated, dehydration stress: a non‐specific 'panicky' or a meaningful response? Plant, Cell & Environment 40: 1704–1710. [DOI] [PubMed] [Google Scholar]
- Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier‐Brygoo H, Thomine S. 2006. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiology 141: 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Koornneef M. 2008. Seed dormancy and germination. Arabidopsis Book 6: e0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae MJ, Lee JS, Nam MH, Cho K, Hong JY, Yi SA, Suh SC, Yoon IS. 2007. A rice dehydration‐inducible SNF1‐related protein kinase 2 phosphorylates an abscisic acid responsive element‐binding factor and associates with ABA signaling. Plant Molecular Biology 63: 151–169. [DOI] [PubMed] [Google Scholar]
- Chen JY, Wang YF, Wang F, Yang J, Gao MX, Li CY, Liu YY, Liu Y, Yamaji N, Ma JF et al 2015. The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 27: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Miyamoto K, Ogawa S, Masuda Y, Shimizu T, Kishi‐Kaboshi M, Takahashi A, Nishizawa Y, Minami E, Nojiri H. 2014. Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS ONE 9: e98737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A, Hernandez ML, He ZS, Andriotis VME, Vaistij FE, Larson TR, Graham IA. 2011. 12‐Oxo‐phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis . Plant Cell 23: 583–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Jia Y, Shi Y, Zhang X, Song C, Gong Z, Yang S. 2018. OST1‐mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO Journal 37: e98228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. 2009. Arabidopsis mutant deficient in 3 abscisic acid‐activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceedings of the National Academy of Sciences, USA 106: 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Andrade J, Ramirez V, Flors V, Vera P. 2011. Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen‐induced callose deposition. The Plant Journal 67: 783–794. [DOI] [PubMed] [Google Scholar]
- Gimeno‐Gilles C, Lelievre E, Viau L, Malik‐Ghulam M, Ricoult C, Niebel A, Leduc N, Limami AM. 2009. ABA‐mediated inhibition of germination is related to the inhibition of genes encoding cell‐wall biosynthetic and architecture: modifying enzymes and structural proteins in Medicago truncatula embryo axis. Molecular Plant 2: 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV. 2005. Dormancy release, ABA and pre‐harvest sprouting. Current Opinion in Plant Biology 8: 183–187. [DOI] [PubMed] [Google Scholar]
- Guo HM, Li HC, Zhou SR, Xue HW, Miao XX. 2014. Cis‐12‐oxo‐phytodienoic acid stimulates rice defense response to a piercing‐sucking insect. Molecular Plant 7: 1683–1692. [DOI] [PubMed] [Google Scholar]
- Hazman M, Hause B, Eiche E, Nick P, Riemann M. 2015. Increased tolerance to salt stress in OPDA‐deficient rice ALLENE OXIDE CYCLASE mutants is linked to an increased ROS‐scavenging activity. Journal of Experimental Botany 66: 3339–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz T, Smirnova E, Marquis V, Poirier L. 2019. Metabolic control within the jasmonate biochemical pathway. Plant and Cell Physiology 60: 2621–2628. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. The Plant Journal 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Research 16: 7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Wang L, Liu L, Li L, Sun L, Rao Q, Zhang J, Huang S. 2015. JMJ704 positively regulates rice defense response against Xanthomonas oryzae pv. oryzae infection via reducing H3K4me2/3 associated with negative disease resistance regulators. BMC Plant Biology 15: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Wang Y, Tang L, Tong X, Wang L, Liu L, Huang S, Zhang J. 2019. SAPK10‐mediated phosphorylation on WRKY72 releases its suppression on jasmonic acid biosynthesis and bacterial blight resistance. iScience 16: 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi‐Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. 2001. Cloning and functional analysis of two gibberellin 3 beta‐hydroxylase genes that are differently expressed during the growth of rice. Proceedings of the National Academy of Sciences, USA 98: 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Jing Y, Shi P, Liu J, Chen J, Yan J, Chu J, Chen KM, Sun J. 2019. JAZ proteins modulate seed germination through interaction with ABI 5 in bread wheat and Arabidopsis . New Phytologist 223: 246–260. [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. 2008. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proceedings of the National Academy of Sciences, USA 105: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. 2005. Abscisic acid‐activated SNRK2 protein kinases function in the gene‐regulation pathway of ABA signal transduction by phosphorylating ABA response element‐binding factors. The Plant Journal 44: 939–949. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. 2004. Differential activation of the rice sucrose nonfermenting1‐related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16: 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik A, Wawer I, Krzywinska E, Bucholc M, Dobrowolska G. 2011. SnRK2 protein kinases–key regulators of plant response to abiotic stresses. OMICS: A Journal of Integrative Biology 15: 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. 2002. PlantCARE, a database of plant cis‐acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wu F, Sheng P, Zhang Z, Zhang X, Guo X, Wang J, Cheng Z, Wang H, Wan J. 2015. The SnRK2‐APC/C(TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nature Communications 6: 7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Shi S, Hao Z, Xiong W, Luo M. 2019. OsbZIP81, A Homologue of Arabidopsis VIP1, may positively regulate JA levels by directly targetting the genes in JA signaling and metabolism pathway in rice. International Journal of Molecular Sciences 20: 2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li Z, Hou Y, Wang Y, Wang H, Tong X, Ao H, Zhang J. 2019. Protein interactomic analysis of SAPKs and ABA‐inducible bZIPs revealed key roles of SAPK10 in rice flowering. International Journal of Molecular Sciences 20: 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S. 2017. Plasma membrane CRPK1‐mediated phosphorylation of 14‐3‐3 proteins induces their nuclear import to fine‐tune CBF signaling during cold response. Molecular Cell 66: 117–128. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang S, Sun N, Liu H, Zhao Y, Liang Y, Zhang L, Han Y. 2015. Functional diversity of jasmonates in rice. Rice 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SF, Yang SY, Chen KT, Hsing YL, Zeevaart JAD, Chen LJ, Yu SM. 2008. A novel class of gibberellin 2‐oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopezmolina L, Mongrand S, Chua NH. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis . Proceedings of the National Academy of Sciences, USA 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Gao CX, Zheng XN, Han B. 2009. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229: 605–615. [DOI] [PubMed] [Google Scholar]
- Lv QD, Zhong YJ, Wang YG, Zhang L, Shi J, Wu ZC, Liu Y, Mao CZ, Yi KK, Wu P. 2014. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26: 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR et al 2015. A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M et al 2009. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant and Cell Physiology 50: 1345–1363. [DOI] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJK, Moreno JE, Suzuki K, Howe GA, Ohme‐Takagi M. 2013. A bHLH‐type transcription factor, ABA‐INDUCIBLE BHLH‐TYPE TRANSCRIPTION FACTOR/JA‐ASSOCIATED MYC2‐LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis . Plant Cell 25: 1641–1656.23673982 [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y. 2010. Abscisic acid and the control of seed dormancy and germination. Seed Science Research 20: 55–67. [Google Scholar]
- Ng LM, Soon FF, Zhou XE, West GM, Kovach A, Suino‐Powell KM, Chalmers MJ, Li J, Yong EL, Zhu JK et al 2011. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proceedings of the National Academy of Sciences, USA 108: 21259–21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Hou Y, Wang Y, Li Z, Zhao J, Tong X, Lin H, Wei X, Ao H, Zhang J. 2017. A comprehensive proteomic survey of ABA‐induced protein phosphorylation in rice (Oryza sativa L.). International Journal of Molecular Sciences 18: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Sickmann A. 2005. State‐of‐the‐art in phosphoproteomics. Proteomics 5: 4052–4061. [DOI] [PubMed] [Google Scholar]
- Riemann M, Haga K, Shimizu T, Okada K, Ando S, Mochizuki S, Nishizawa Y, Yamanouchi U, Nick P, Yano M et al 2013. Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae . The Plant Journal 74: 226–238. [DOI] [PubMed] [Google Scholar]
- Schaller A, Stintzi A. 2009. Enzymes in jasmonate biosynthesis – structure, function, regulation. Phytochemistry 70: 1532–1538. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Miyamoto K, Miyamoto K, Minami E, Nishizawa Y, Iino M, Nojiri H, Yamane H, Okada K. 2013. OsJAR1 contributes mainly to biosynthesis of the stress‐induced jasmonoyl‐isoleucine involved in defense responses in rice. Bioscience, Biotechnology, and Biochemistry 77: 1556–1564. [DOI] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH. 2016. Two faces of one seed: hormonal regulation of dormancy and germination. Molecular Plant 9: 34–45. [DOI] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM. 2002. Semidwarf (sd‐1), "green revolution" rice, contains a defective gibberellin 20‐oxidase gene. Proceedings of the National Academy of Sciences, USA 99: 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Huang JF, Rhee Y. 1991. Nitrogen and methyl jasmonate induction of soybean vegetative storage protein genes. Plant Physiology 96: 130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suza WP, Staswick PE. 2008. The role of JAR1 in Jasmonoyl‐L‐isoleucine production during Arabidopsis wound response. Planta 227: 1221–1232. [DOI] [PubMed] [Google Scholar]
- Tang G, Ma J, Hause B, Nick P, Riemann M. 2020. Jasmonate is required for the response to osmotic stress in rice. Environmental and Experimental Botany 175: 104047. [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi‐Shinozaki K. 2010. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant and Cell Physiology 51: 1821–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC. 1984. Biosynthesis of jasmonic Acid by several plant species. Plant Physiology 75: 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Guo Q, Froehlich JE, Hersh HL, Zienkiewicz A, Howe GA, Benning C. 2018. Two abscisic acid‐responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana . Plant Cell 30: 1006–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou Y‐J, Van MJO, Zhang H, Tao AW, Zhu J‐K. 2013. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proceedings of the National Academy of Sciences, USA 110: 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Bai MY, Wang ZY. 2014. The brassinosteroid signaling network‐a paradigm of signal integration. Current Opinion in Plant Biology 21: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Wang YF, Pineros MA, Wang ZY, Wang WX, Li CY, Wu ZC, Kochian LV, Wu P. 2014. Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant, Cell & Environment 37: 1159–1170. [DOI] [PubMed] [Google Scholar]
- Wilen RW, van Rooijen GJ, Pearce DW, Pharis RP, Holbrook LA, Moloney MM. 1991. Effects of jasmonic acid on embryo‐specific processes in Brassica and Linum oilseeds. Plant Physiology 95: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie KL, Li LL, Zhang HH, Wang R, Tan XX, He YQ, Hong GJ, Li JM, Ming F, Yao XF et al 2018. Abscisic acid negatively modulates plant defence against rice black‐streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant, Cell & Environment 41: 2504–2514. [DOI] [PubMed] [Google Scholar]
- Xie KB, Yang YN. 2013. RNA‐guided genome editing in plants using a CRISPRCas system. Molecular Plant 6: 1975–1983. [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li JG, Deng XW et al 2012. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences, USA 109: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zhang W, Wang X. 2017. Post‐translational control of ABA signaling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnology Journal 15: 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuhko K, Michiharu M, Hideyuki M, Shuhei Y, Yasuaki K, Tokunori H, Akiko Y, Tsukaho H. 2010. Abscisic acid‐activated SNRK2 protein kinases function in the gene‐regulation pathway of ABA signal transduction by phosphorylating ABA response element‐binding factors. The Plant Journal 44: 939–949. [DOI] [PubMed] [Google Scholar]
- Zhang XB, Feng BH, Wang HM, Xu X, Shi YF, He Y, Chen Z, Sathe AP, Shi L, Wu JL. 2018. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. Journal of Integrative Plant Biology 60: 160–172. [DOI] [PubMed] [Google Scholar]
- Zhou YB, Liu C, Tang DY, Yan L, Wang D, Yang YZ, Gui JS, Zhao XY, Li LG, Tang XD et al 2018. The receptor‐like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 30: 1100–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Sequence alignment of T‐ loop region of 10 Arabidopsis and 10 rice SnRK2 family members.
Fig. S2 Molecular characterization of sapk10 mutant and seed germination in response to ABA.
Fig. S3 qRT‐PCR analysis for transcript accumulation of nine SnRK2 family members (SAPK1‐SAPK9) in the seeds of WT or crsapk10 lines, respectively.
Fig. S4 Molecular characterization of SAPK10, SAPK10S177A overexpressing transgenic lines.
Fig. S5 Expression pattern of SAPK10 and bZIP72.
Fig. S6 Molecular characterization of crbzip72, crtrab1 and crbzip72/crtrab1 mutants.
Fig. S7 qRT‐PCR analysis for transcript accumulation of TRAB1 in the seeds of crbzip72 transgenic line.
Fig. S8 Molecular characterization of bZIP72 and bZIP72S71A overexpressing transgenic lines.
Fig. S9 qRT‐PCR analysis for transcript accumulation of JA and GA pathway genes in the seeds of the WT, OxbZIP72 and OxbZIP72S71A transgenic lines.
Fig. S10 EMSA of bZIP72 on JA pathway genes AOC, AOS1 and LOX1 promoter regions and GA pathway genes GA3ox1, GA2ox5 and GA20ox2 promoter regions.
Fig. S11 qRT‐PCR analysis for transcript accumulation of ABA and JA pathway genes in the seeds of the WT.
Fig. S12 IBU relieved the inhibition of ABA on post‐germination growth.
Fig. S13 SAPK10‐mediated retarded seed germination was relieved by exogenous application of IBU.
Table S1 Sequences of primers used in this study.


