Abstract
Background
There is limited data to guide the prevention and management of surgical site infections (SSI) in low- and middle-income countries. We prospectively studied aetiological agents associated with SSI and their corresponding antibiotic susceptibility patterns in a tertiary hospital in Ghana.
Methods
As part of a cohort study carried out at the surgical department of the Korle Bu Teaching Hospital (KBTH) from July 2017 to April 2019, wound swabs were collected from patients diagnosed with SSI. Isolates cultured from the wound swabs were identified by MALDI TOF and susceptibility testing was conducted according to EUCAST 2020 guidelines. Clinical data were monitored prospectively.
Results
Of 4577 patients, 438 developed an SSI and 352 microbial isolates were cultured. Isolates were predominantly Gram negative (286, 81%), a pattern seen for all kinds of surgery and all wound classes. The most common species included Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Staphylococcus aureus and Acinetobacter baumannii. The majority of organisms were multi-drug resistant including 86% of E. coli, 52% of A. baumannii and 86% of K. pneumoniae; and 65% (17/26) of the cefotaxime-resistant K. pneumoniae were extended spectrum β-lactamase producing. One of 139 E. coli, 15 of 49 P. aeruginosa, and 6 of 23 A. baumannii were meropenem resistant, but no clonal pattern was found. There was a 1% (5/428) prevalence of methicillin-resistant S. aureus.
Conclusions
The predominance of Gram-negative organisms and the high level of multi-drug resistance indicate a need to re-evaluate antibiotic prophylaxis and treatment protocols in surgical practice in low- and middle-income countries.
Keywords: Multidrug resistant, Surgical site infection, Gram-negative organisms, ESBL, Ghana
Background
Surgical site infections (SSI) is a major type of healthcare associated infections (HCAI), forming as much as 33% of all HCAI in sub-Saharan Africa [1, 2]. A major teaching hospital in Ghana also described an SSI incidence risk of 10% with an associated 9 extra days of stay and $1519 excess cost to the institution per patient [3, 4].
The global spread of antibiotic resistance further compounds the problem of SSI. Infections caused by resistant bacteria lead to poor treatment outcomes. Major drivers of antimicrobial resistance in low- and middle-income countries include inappropriate prescription practices and poor infection control measures [5]. Thus, the prevalence of antibiotic use among surgical patients in sub-Saharan Africa is 24–73% [6], and the majority of these antibiotics are prescribed for prophylaxis and usually beyond the recommended 24-h period [7]. Reasons for this prolonged antibiotic use include a fear of infections due to poor infection control [8].
In Ghana, as in most low- and middle-income countries, antimicrobial treatment of wound infections is mainly empirical due to limited laboratory services in most health facilities and the costs to patients of culture and susceptibility testing, when available [9]. The selection of prophylactic antibiotic therapy is based on the operation site and the normal resident flora. Thus, empirical choice of antibiotics relies on knowledge of the susceptibility patterns of common local pathogens. Absence of this data precludes rational use of antibiotics for treatment and prevention of SSI.
In this study, we prospectively identified the aetiological agents of SSI at the surgical department of a tertiary hospital in Ghana, with emphasis on their antibiotic susceptibility patterns, and related to the patient characteristics.
Methods
Study site, patient recruitment and collection of samples
Samples were collected as part of a cohort study at the surgical department of the Korle Bu Teaching Hospital from July 2017 to April 2019. The study design and results (for the period July 2017–December 2018) of the surveillance have been reported elsewhere [3]. All patients who underwent surgery in the unit were followed actively for the occurrence of SSI during admission and post-discharge for 30 days, and infection was defined according to Centers of Disease Control and Prevention (CDC) criteria [3, 10]. Wound contamination and SSI type were classified based on the CDC classification [10]. Implant surgery and surgery with wounds that could not be closed immediately were excluded.
Patients on admission had their change of dressing carried out on their bed in the wards. Post-discharge change of dressing was carried out in dressing rooms situated on each ward. Wound swabs were taken from consecutive patients diagnosed with SSI, either on admission or post discharge for microbiology analysis. The project staff had been trained to carefully collect exudate from the infected surgical site using sterile cotton- tipped applicators (Sterilin, U.K), which were transferred to the microbiology laboratory within an hour of sampling. Preliminary culture and susceptibility results were reported immediately to the physicians for management of patients. Isolates were then frozen at − 80 °C and transported to Copenhagen for confirmatory testing and additional analyses.
Laboratory analyses
Wound swabs were cultured on blood, chocolate and MacConkey agar plates. Isolates were identified using MALDI TOF Biotyper (Bruker Daltonics, Bremen, Germany). Here, susceptibility testing was conducted according to EUCAST 2020 guidelines [11]. The following discs were used; ampicillin (10 μg), amoxicillin-clavulanate (20/10 μg), gentamicin (10 μg), amikacin (30 μg), ciprofloxacin (5 μg), cefuroxime (30 μg), ceftazidime (10 μg), ceftriaxone (30 μg), cefotaxime (10 μg), tazobactam-piperracillin (30/6 μg), meropenem (10 μg), cefoxitin (30 μg), erythromycin (15 μg), clindamycin (2 μg), sulphamethoxazole-trimethoprim (25 μg), penicillin (1 unit), linezolid (10 μg), tetracycline (30 μg), (all from Oxoid Ltd., Basingstoke, United Kingdom (UK)). Isolates were categorized as “resistant” and “susceptible”, including those “susceptible at increased exposure” also classified as “susceptible” [12]. E. coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were used as controls for Gram negative and positive panels respectively. Klebsiella pneumoniae ATCC 700603 was used as quality control strains for ESBL screening.
Enterobacterales resistant to third generation cephalosporins were screened for production of extended spectrum beta lactamases (ESBL) using the double disc diffusion method [13]. Isolates with phenotypic resistance to meropenem were screened depending on the species. Pseudomonas spp. were screened for carbapenemases using GeneXpert (Xpert® Carba-R, Cepheid, France), Acinetobacter spp. were screened for OXA-23 using a rapid diagnostic test (OXA-23 K-SeT, Coris, Belgium), and Acinetobacter spp. and enterobacterales were screened for carbapenemases using NG-CARBA 5® (Hardy Diagnostics, CA, USA). All Staphylococcus aureus isolates were screened for methicillin resistance using cefoxitin. Multidrug resistance (MDR) was defined as resistance to ≥1 antibiotic in ≥3 antibiotic groups [14].
Data analysis
For each bacterial agent, the percentage frequency of resistant isolates was determined. The resistance patterns of isolates of the same species were screened visually for possible clonal distribution. Distribution of bacterial types by clinical groups were compared by chi-square test and p < 0.05 was considered significant. We used Stata /MP version 15.1 (Stata Corp., College Station, Tx, USA) for the analysis.
Results
Demographic and patient characteristics
A total of 4577 patients were included in the study. Of these, 438 (9.6%) patients, developed an SSI. The SSI risk was 9.3% (376/4054) in the general surgical department, 26.5% (31/117) in the department of urology and 7.6% (31/406) in paediatric surgery. The median age of the patients who developed an SSI was 45 years (interquartile range 31–60 years) and 239 (54%) patients were female.
From 382 (87%) of the 438 patients, a wound swab was taken for microbiological analysis. No wound swabs were taken in 13% (56/438) of the cases due to; surveillance team missing the opportunity to take a swab at change of wound dressing or at a relaparotomy for an SSI, missing the diagnosis of an SSI until the attention was drawn to the clinical signs and, wound swabs taken but accidentally sent to a private laboratory for analysis.
Characteristics of surgical wounds
The proportion of patients who developed an SSI was 5.2% (135/2589) for wounds classified as clean, 10.1% (66/655) for a clean contaminated wound, 12.1% (104/859) for contaminated and 27.4% (130/475) for dirty wounds.
We found 352 isolates in 327 (86%) of the 382 swabs. Most were monoculture, but two different species were cultured from each of 25 swabs. Isolates from clean wounds accounted for 29% (103/352) of isolates, clean contaminated wounds for 14% (48), contaminated wounds for 26% (91), and dirty wounds for 31% (110) of isolated microorganisms.
Aetiology of surgical site infections
Gram-negative microorganisms constituted 81% (286/352) of the isolates. The five most common microorganisms were Escherichia coli (139, 39%), Pseudomonas aeruginosa (49, 14%), Klebsiella pneumoniae (35, 10%), Staphylococcus aureus (33, 9%) and Acinetobacter baumannii (23, 6%), accounting for approximately 79% of the isolated organisms (Table 1).
Table 1.
Microbial isolates from infected surgical sites in a teaching hospital in Ghana
| Clinical isolates | N | % | Number of isolates with multidrug resistance (%) | Extended spectrum β-lactamase-producing isolates (%) | Meropenem resistant isolatesa (%) | Number of isolates with methicillin resistance (%) |
|---|---|---|---|---|---|---|
| Escherichia coli | 139 | 39.5 | 120 (86%) | 50 (36%) | 1 (1%) | – |
| Pseudomonas spp. | 49 | 13.9 | 17 (35%) | – | 15 (31%) | – |
| Klebsiella pneumonia | 35 | 9.9 | 30 (86%) | 17 (48%) | 0 | – |
| Staphylococcus aureus | 33 | 9.4 | 8 (24%) | – | – | 5 (15%) |
| Acinetobacter baumannii | 23 | 6.5 | 12 (52%) | – | 6 (26%) | – |
| Proteus spp. | 21 | 6.0 | 5 (24%) | 0 | 0 | – |
| Staphylococcus haemolyticus | 15 | 4.3 | 14 (93%) | – | – | 0 |
| Staphylococcus epidermidis | 11 | 3.1 | 10 (91%) | – | – | 0 |
| Enterobacter spp. | 11 | 3.1 | 5 (45%) | 2 (18%) | 0 | – |
| Corynebacterium spp. | 4 | 1.1 | 1 (25%) | – | – | – |
| Candida albicans | 2 | 0.6 | ND | – | – | – |
| Achromobacter spp. | 2 | 0.6 | 1 (50%) | – | 0 | – |
| Stenotrophomonas maltophilia | 2 | 0.6 | 0 | – | 0 | – |
| Providencia stuartii | 2 | 0.6 | 0 | – | 0 | – |
| Staphylococcus lugdunensis | 1 | 0.3 | 0 | – | – | 0 |
| Alcaligenes faecalis | 1 | 0.3 | 1 (100%) | – | – | – |
| Morganella morganii | 1 | 0.3 | 1 (100%) | – | 0 | – |
a Four P. aeruginosa had the vim (Verona integron-encoded metallo-β-lactamase) gene and one A. baumannii produced OXA-23
Table 2 describes the pathogens by type of surgery. At least one isolate was found in 187 patients (11%) of the 1646 gastrointestinal and other abdominal surgeries, in 12 (8%) of 140 genitourinary and prostate surgeries, 38 (4%) of 907 breast surgeries, 44 (5%) of 866 hernia and scrotal surgeries, 25 (16%) of 157 limb amputations, 9 (3%) of 307 thyroid surgeries and 12 (2%) of 554 other soft tissue surgeries. The ratio between Gram-negative and Gram-positive organisms differed by type of surgery (Table 3, p = 0.002). For gastro-intestinal and genito-urinary surgery, Staphylococcus spp. constituted < 16% of the isolates, whereas Staphylococcus spp. constituted > 25% of the isolates for hernia, breast, soft tissue and thyroid surgery. Despite this difference, Gram-negative organisms constituted ≥66% of isolates for all types of surgery and as much as 87% for gastro-intestinal surgery.
Table 2.
Distribution of microbial isolates in relation to type of surgical procedure
| Procedure performed | Total n = 4577 | SSI n = 438 |
E. coli n = 139 |
Pseudomonas spp. n = 49 |
Klebsiella spp. n = 35 |
A. baumannii n = 23 |
Proteus spp. n = 21 |
Enterobacter spp. n = 11 |
S. aureus n = 33 |
CoNS n = 27 |
Others n = 14 |
No growth n = 55 | No samples n = 56 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gastrointestinal + other abdominal surgery | 1646 | 253 | 117 | 15 | 21 | 18 | 4 | 4 | 6 | 16 | 5 | 33 | 33 |
| Hernia + scrotal surgery | 866 | 54 | 12 | 4 | 3 | 1 | 6 | 2 | 10 | 6 | 3 | 5 | 5 |
| Breast surgery | 907 | 49 | 3 | 13 | 3 | 2 | 6 | 0 | 8 | 3 | 2 | 5 | 6 |
| Limb amputation | 157 | 33 | 5 | 7 | 5 | 0 | 0 | 2 | 3 | 1 | 3 | 3 | 5 |
| Genitourinary tract + prostate surgery | 140 | 20 | 2 | 5 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 4 | 4 |
| Other soft tissue surgery | 554 | 16 | 0 | 2 | 2 | 1 | 2 | 1 | 3 | 0 | 0 | 2 | 2 |
| Thyroid surgery | 307 | 13 | 0 | 3 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 3 | 1 |
SSI surgical site infections
CoNS include S. haemolyticus and S. epidermidis and S. lugdunensis. Other microbial isolates include Corynebacterium spp., C. albicans, Achromobacter spp., S. maltophilia, P. stuartii, A. faecalis and M. morganii
Table 3.
Comparison of distribution of Gram negative: Gram positive for type of procedure performed and type of SSI
| Clinical characteristics | Total number of positive cultures n = 352 |
Total number of Gram-negative organisms n = 286 |
Gram-negative organisms % |
Total number of Staphylococcus spp.(60) + other Gram-positive organisms (4) n = 64 |
Staphylococcus spp. + Other Gram-positive organisms % |
Total number of other microbial isolates (fungi) n = 2 |
Other microbial isolates (fungi) % |
|---|---|---|---|---|---|---|---|
| Procedure performed | |||||||
| Gastrointestinal + other abdominal surgery | 207 | 181 | 87.4 | 24 | 11.6 | 2 | 1.0 |
| Hernia + scrotal surgery | 47 | 31 | 65.9 | 16 | 34.0 | 0 | 0.0 |
| Breast surgery | 40 | 29 | 72.5 | 11 | 27.5 | 0 | 0.0 |
| Limb amputation | 26 | 21 | 80.8 | 5 | 19.2 | 0 | 0.0 |
| Genitourinary tract + prostate surgery | 12 | 10 | 83.3 | 2 | 16.7 | 0 | 0.0 |
| Other soft tissue surgery | 10 | 7 | 70.0 | 3 | 30.0 | 0 | 0.0 |
| Thyroid surgery | 10 | 7 | 70.0 | 3 | 30.0 | 0 | 0.0 |
| Type of SSI | |||||||
| Superficial | 291 | 228 | 78.3 | 61 | 21.0 | 2 | 0.7 |
| Deep | 34 | 31 | 85.3 | 3 | 8.8 | 0 | 0.0 |
| Organ space | 27 | 27 | 100.0 | 0 | 0.0 | 0 | 0.0 |
There was a difference between type of wound and the ratio between Gram-negative and Gram-positive organisms (Table 3), and S. aureus was only cultured from superficial SSI (Table 4). Conversely, all isolates from organ-space SSI were Gram negative. A much higher proportion of organ/space infections than other types of infections were not cultured, mostly due to the surveillance team missing the opportunity to take a wound swab during a relaparotomy to drain abscesses.
Table 4.
Distribution of microbial isolates in relation to type of surgical site infection (SSI)
| Type of SSI | Number with surgical site infections |
E. coli n = 139 |
Pseudomonas spp. n = 49 |
Klebsiella spp. n = 35 |
A. baumannii n = 23 |
Proteus spp. n = 21 |
Enterobacter spp. n = 11 |
S. aureus n = 33 |
CoNS n = 27 |
Others n = 14 |
No growth n = 55 | No samples n = 56 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Superficial SSI | 366 (%) | 105 (29%) | 42 (11%) | 29 (8%) | 18 (5%) | 18 (5%) | 10 (3%) | 33 (9%) | 24 (6%) | 12 (3%) | 45 (12%) | 30 (8%) |
| Deep SSI | 49 (%) | 17 (34%) | 3 (6%) | 2 (4%) | 4 (8%) | 2 (4%) | 1 (2%) | 0 | 3 (6%) | 2 (4%) | 7 (14%) | 8 (16%) |
| Organ-space SSI | 48 (%) | 17 (35%) | 4 (8%) | 4 (8%) | 1 (2%) | 1 (2%) | 0 | 0 | 0 | 0 | 3 (6%) | 18 (37%) |
CoNS include S. haemolyticus and S. epidermidis and S. lugdunensis. Other microbial isolates include Corynebacterium spp., C. albicans, Achromobacter spp., S. maltophilia, P. stuartii, A. faecalis and M. morganii
Antimicrobial susceptibility patterns
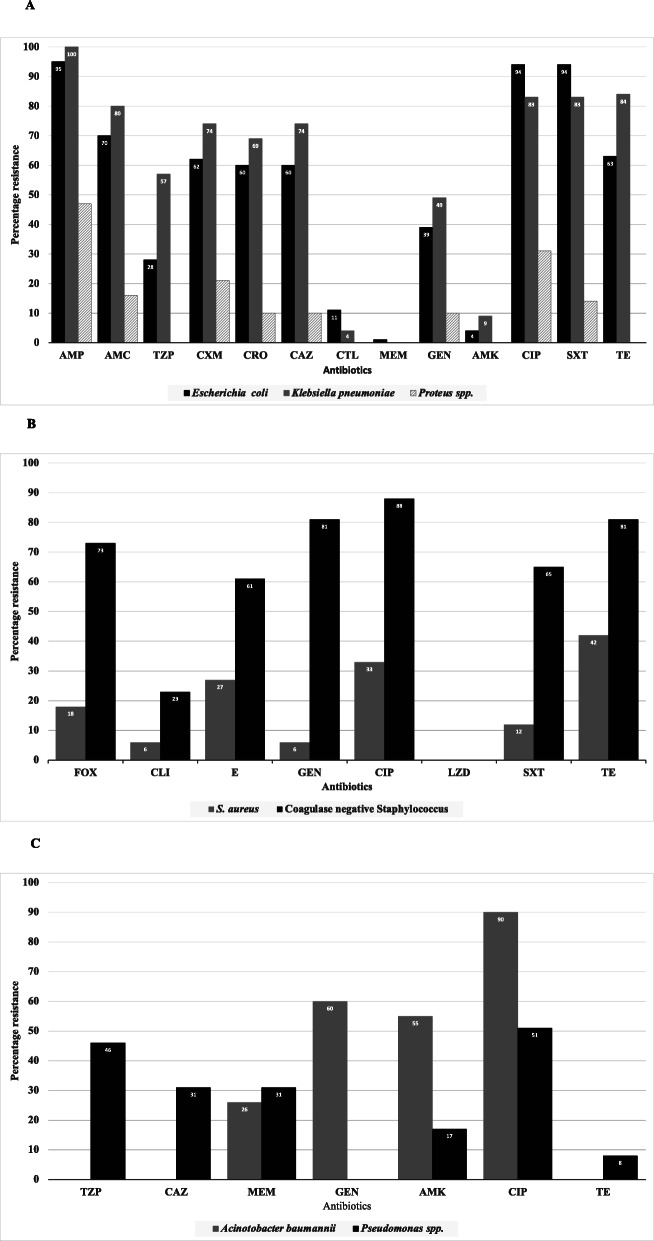
The majority of bacterial isolates were MDR, ranging from 23 to 86% for Proteus spp. and E. coli isolates, respectively (Table 1). Acquired resistance to commonly used antibiotics ranged from 1% to meropenem in E. coli to 95% to ampicillin, ciprofloxacin and trimethoprim-sulphamethoxazole in E. coli (Fig. 1a).
Fig. 1.
Antibiotic resistance pattern for bacteria isolated from infected surgical sites. a, Eschericia coli, Klebsiella spp. and Proteus spp., b, Pseudomonas aeruginosa and Acinetobacter baumannii, c. Staphylococcus spp. AMC – amoxicillin/clavulanic acid, AMK – amikacin, AMP – ampicillin, CAZ – ceftazidime, CIP – ciprofloxacin, CLI – clindamycin, CRO – ceftriazone, CTL – cefotaxime + clavulanic acid, CXM – cefuroxime, E – erythromycin, FOX – cefoxitin, GEN – gentamycin, LZD – linezolid, MEM – meropenem, SXT – trimethoprim-sulphamethoxazole, TE – tetracycline, TZP – piperacillin/tazobactam
Among E. coli and K. pneumoniae isolates, 60% (82/139) and 74% (26/35), respectively were resistant to third generation cephalosporins. Sixty-one percent (50/82) and 65% (17/26) of the cefotaxime-resistant E. coli and K. pneumoniae, respectively were ESBL producing.
Meropenem resistance in this study was mainly found in Pseudomonas spp. (15 of 49 isolates) and Acinetobacter baumannii (6 of 23 isolates) (Table 1, Fig. 1b). Four P. aeruginosa isolates harboured the vim gene, encoding Verona integron-encoded metallo-β-lactamase. One A. baumannii expressed OXA-23. K. pneumoniae showed no resistance to meropenem (Fig. 1a).
Fifteen percent (5/33) of S. aureus isolates were methicillin resistant (MRSA). Overall, the MRSA prevalence was 1% (5/438) in the cultured wounds. Resistance of S. aureus against other antibiotics ranged from 6% to gentamycin and clindamycin to 42% to tetracycline. Among coagulase-negative staphylococci, the percentage resistance ranged from 23% to clindamycin to 81% to tetracycline. Neither S. aureus nor coagulase negative staphylococci showed resistance to linezolid (Fig. 1c).
The sensitivity pattern of the MDR organisms, including those for which specific resistance mechanisms such as ESBL or carbapenemases were detected, did not give any indication of an outbreak of one or more clones (data not shown).
Discussion
In this study, from a surgical facility in Africa, Gram-negative rods were the primary aetiology of SSI. Despite differences in ratios between Gram-negative and Gram-positive organisms by types of surgery and SSI types, Gram-negatives dominated in all categories. There was a high level of MDR among isolated organisms, including carbapenem-resistant P. aeruginosa and A. baumannii and ESBL-producing E. coli and K. pneumoniae. There was no phenotypic indication of clonality of the MDR organisms, indicating that the findings could not be explained by an outbreak. These findings challenge the standard recommendations for empiric prevention and treatment of SSI in low- and middle-income countries.
Unlike earlier studies in Ghana, which recorded low rates of microbiology testing [9], this study recorded a high rate of testing. This can be explained by the fact that under the active study conditions, wound swab samples were taken by the surveillance team when an SSI was diagnosed, irrespective of the clinicians’ diagnosis, and the cost of the testing was also borne by the study. In low- and middle-income countries, low rates of microbiology testing are often reported, presumably due to the limited facilities for microbiology testing, the costs of testing, limited numbers of trained personnel, and the tendency for clinicians to underutilize existing microbiology facilities [9, 15–18] As reported previously, the active surveillance and access to microbiological testing in this study lead to immediate improvements in SSI rates in the course of the study [3].
E. coli was the commonest isolated organism, whereas, S. aureus and coagulase negative staphylococci constituted less than a fifth of the isolates. Some studies have also predominantly isolated E. coli from infected surgical sites post abdominal surgeries [19, 20] but in most studies in Africa, S. aureus was the most common pathogen isolated from SSI [21]. The large numbers of gastrointestinal surgeries performed in this study cannot fully explain the predominance of E. coli since Gram-negative rods were the predominant aetiology of SSI in all forms of surgery. A possible explanation of this could be a high rate of skin carriage of Gram-negative organisms as shown in a study from Tanzania [22]. In addition, the routine use of antibiotic prophylaxis with an effect on S. aureus in our department coupled with a relatively low rate of methicillin resistance may also have skewed the distribution of microorganisms toward Gram negatives. Finally we recently found high levels of antimicrobial air contamination in our surgical facility and demonstrated a causal relationship with SSI [23]. This study indicated that air contamination may have contributed to infections with environmental bacteria such as P. aeruginosa and A. baumannii, whereas enterobacterales were uncommon in the air samples.
Skin carriage of Gram-negative organisms may be associated with previous antibiotic use. In line with this, it is likely our patients had received antibiotic therapy at referring facilities before coming to our tertiary hospital, where the treatment may have been continued. We have recently documented long periods of administration of antibiotics in surgical units in Ghana, at all levels of health facilities [9]. Long periods of administration of antibiotics have also been reported in other health facilities in low- and middle-income countries [24, 25].
The high usage of antibiotics coupled with the low rate of microbiology testing to inform choice of antibiotic therapy, may explain the high levels of MDR. Hospital-based antimicrobial stewardship programs are said to decrease antibiotic use, though data on this is limited in low- and middle-income countries [26]. There is a need to develop and document the effect of functioning antibiotic stewardship programs based on longitudinal monitoring of microbiological test results from the surgical department.
The choice of antibiotics in the department is essentially based on the Ghana standard treatment guidelines, usually reviewed at intervals of 5 years or more [27, 28].
Antibiotics like ciprofloxacin are used routinely by surgeons as therapeutic and prophylactic treatment for gastrointestinal surgery, and in combination with clindamycin for limb amputation for dry and wet lower limb gangrene. The high prevalence of ciprofloxacin resistance in this study, ranging between 95 and 33% among E. coli and S. aureus respectively, shows the evolving pattern of resistance. In comparison, a 10% resistance to ciprofloxacin in fecal E. coli has been reported in the past [29]. Conversely, the low resistance to vancomycin, piperacillin/tazobactam and linezolid may reflect their low usage or unavailability in most Ghanaian facilities, based on the essential medicines list of the ministry of health [27].
The low prevalence of MRSA confirms previous findings from inpatients in our institution [30]. A high use of amoxicillin/clavulanic acid may explain the low frequency of S. aureus in our study, but the high prevalence of amoxicillin/clavulanic acid-resistant Gram-negative organisms suggests the need to re-evaluate the protocols.
The resistance to 3rd generation cephalosporins by K. pneumoniae and E. coli in our study was mainly caused by ESBL. This cannot be explained by a high usage of third generation cephalosporins as the Ghana treatment guidelines do not recommend this drug class. We have recently reported clonal outbreaks of MDR K. pneumoniae in neonatal intensive care units in Ghana [31], but in the present study we did not find indication of ongoing outbreaks, based on phenotypic analysis. The high ESBL rate may thus mimic antibiotic resistance in patients’ own flora indicating widespread carriage of resistant organisms in the society [32].
This study involves data from only one hospital, limiting the generalization of the results, though the hospital serves as a major referral center for a population of over 30 million.
Conclusion
Gram-negative organisms with a high level of MDR were the predominant organisms isolated from SSI. Antibiotic treatment protocols including prophylactic strategies need to be re-evaluated to improve outcomes and minimize the emergence of antimicrobial resistance.
Acknowledgements
We thank all the interns and nurses of the surgical department involved in taking wound swabs for microbiology analysis, Amos Akumwena of the department of Medical Microbiology of the University of Ghana Medical School, who did all the microbiology identification and susceptibility of isolates for clinical management in Ghana and Sonja Lekovic of the department of Microbiology, Copenhagen University hospital for the confirmation and analysis of the isolates.
Abbreviations
- CDC
Centers for Disease Control and Prevention
- Corp.
Corporation
- ESBL
Extended spectrum beta lactamases
- HCAI
Healthcare associated infections
- KBTH
Korle Bu Teaching Hospital
- MDR
Multidrug resistant
- MRSA
Methicillin resistant Staphylococcus aureus
- SSI
Surgical site infections
- Tx.
Texas
- UK
United Kingdom
- USA
United State of America
Authors’ contributions
ABB, JALK, KM, AKL, EO, and MJN conceptualized the study; participated in its design, coordination and helped to draft the manuscript. ABB collected data and performed the statistical analysis with help of KM. JALK and AKL contributed to microbiological testing. The manuscript was revised for intellectual content by KM, JALK and MJN. All authors have read and approved the final manuscript for submission.
Funding
This work was supported by DANIDA through the HAI-GHANA PROJECT, [grant number 16-PO1-GHA]. The funder had no role in study design, data collection, analysis and preparation of this manuscript.
Availability of data and materials
Data is available from corresponding author upon request.
Ethics approval and consent to participate
Ethical approval was granted by the Korle Bu Teaching Hospital Institutional Review Board, KBTH-STC/IRB/00022/2017 and the College of Health Sciences’ Ethical and Protocol review committee, CHS-Et/M.8-P4.5/2016–2017. All patients gave a written, informed consent.
Consent for publication
Not applicable.
Competing interests
None declared.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nejad SB, Allegranzi B, Syed SB, Ellis B, Pittet D. Health-care-associated infection in Africa: a systematic review. Bull World Heal Organ. 2011;89:757–765. doi: 10.2471/BLT.11.088179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labi A, Obeng-nkrumah N, Owusu E, Bjerrum S. Multi-Centre point-prevalence survey of hospital- acquired infections in Ghana. J Hosp Infect. 2019;101(1):60–68. doi: 10.1016/j.jhin.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 3.Bediako-Bowan A, Owusu E, Debrah S, Kjerulf A, Newman MJ, Kurtzhals JAL, et al. Surveillance of surgical site infection in a teaching hospital in Ghana: a prospective cohort study. J Hosp Infect. 2020;104(3):321–327. doi: 10.1016/j.jhin.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Fenny AP, Asante FA, Otieku E, Bediako-Bowan A, Enemark U. Attributable cost and extra length of stay of surgical site infection at a Ghanaian teaching hospital. Infect Prev Pract. 2020;2(2):100045. doi: 10.1016/j.infpip.2020.100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob Resist Infect Control. 2017;6 Available from: https://aricjournal.biomedcentral.com/track/pdf/10.1186/s13756-017-0208-x. [cited 2018 Sep 19]. [DOI] [PMC free article] [PubMed]
- 6.Atif M, Scahill S, Azeem M, Sarwar MR, Babar ZUD. Drug utilization patterns in the global context: a systematic review. Heal Policy Technol. 2017;6(4):457–470. [Google Scholar]
- 7.Machowska A, Sparrentoft J, Dhakaita SK, StålsbyLundborg C, Sharma M. Perioperative antibiotic prescribing in surgery departments of two private sector hospitals in Madhya Pradesh, India. Perioper Med. 2019;8(1):1–12. doi: 10.1186/s13741-019-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byarugaba DK. Antimicrobial resistance in developing countries and responsible risk factors. Int J Antimicrob Agents. 2004;24:105–110. doi: 10.1016/j.ijantimicag.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Bediako-Bowan AAA, Owusu E, Labi AK, Obeng-Nkrumah N, Sunkwa-Mills G, Bjerrum S, et al. Antibiotic use in surgical units of selected hospitals in Ghana: a multi-Centre point prevalence survey. BMC Public Health. 2019;19(1):797–807. doi: 10.1186/s12889-019-7162-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Center for Disease Prevention and Control . National Healthcare Safety Network (NHSN) Procedure-associated Module SSI. Surgical Site Infection (SSI ) Event. 2020. [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters. Version 10. http://www.eucast.org. 2020. 0–77 p. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf. Accessed on 5 Aug 2020.
- 12.European Committee on Antimicrobial Susceptibility Testing . EUCAST: Clinical breakpoints and dosing of antibiotics V 10.0. 2020. [Google Scholar]
- 13.European committee on antimicrobial susceptibility testing . EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. 2017. [Google Scholar]
- 14.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 15.Petti CA, Polage CR, Quinn TC, Ronald AR, Sande MA. Laboratory medicine in Africa: a barrier to effective health care. Clin Infect Dis. 2006;42(3):377–382. doi: 10.1086/499363. [DOI] [PubMed] [Google Scholar]
- 16.Labi AK, Obeng-Nkrumah N, Owusu E, Bjerrum S, Bediako-Bowan A, Sunkwa-Mills G, et al. Multi-centre point-prevalence survey of hospital-acquired infections in Ghana. J Hosp Infect. 2019;101(1):60–68. doi: 10.1016/j.jhin.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 17.Barbé B, Yansouni CPP, Affolabi D, Jacobs J. Implementation of quality management for clinical bacteriology in low-resource settings. Clin Microbiol Infect. 2017;23(7):426–433. doi: 10.1016/j.cmi.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Polage CR, Bedu-Addo G, Owusu-Ofori A, Frimpong E, Lloyd W, Zurcher E, et al. Laboratory use in Ghana: Physician perception and practice. Am J Trop Med Hyg. 2006;75(3):526–531. [PubMed] [Google Scholar]
- 19.Alkaaki A, Al-Radi OO, Khoja A, Alnawawi A, Alnawawi A, Maghrabi A, et al. Surgical site infection following abdominal surgery: a prospective cohort study. Can J Surg. 2019;62(2):111–117. doi: 10.1503/cjs.004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du M, Liu B, Li M, Cao J, Liu D, Wang Z, et al. Multicenter surveillance study of surgical site infection and its risk factors in radical resection of colon or rectal carcinoma. BMC Infect Dis. 2019;19(1):1–6. doi: 10.1186/s12879-019-4064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sisay M, Worku T, Edessa D. Microbial epidemiology and antimicrobial resistance patterns of wound infection in Ethiopia: a meta-analysis of laboratory-based cross-sectional studies. BMC Pharmacol Toxicol. 2019;20(1):1–19. doi: 10.1186/s40360-019-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moremi N, Claus H, Rutta L, Frosch M, Vogel U, Mshana SE. High carriage rate of extended-spectrum beta-lactamase-producing Enterobacteriaceae among patients admitted for surgery in Tanzanian hospitals with a low rate of endogenous surgical site infections. J Hosp Infect. 2018;100(1):47–53. doi: 10.1016/j.jhin.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Stauning MA, Bediako-Bowan A, Bjerrum S, Andersen LP, Andreu-Sánchez S, Labi AK, et al. Genetic relationship between bacteria isolated from intraoperative air samples and surgical site infections at a major teaching hospital in Ghana. J Hosp Infect. 2020;104(3):309–320. doi: 10.1016/j.jhin.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Talaat M, Saied T, Kandeel A, El-ata GAA, El-kholy A, Hafez S, et al. A point prevalence survey of antibiotic use in 18 hospitals in Egypt. Antibiotics. 2014;3:450–460. doi: 10.3390/antibiotics3030450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wattal C, Khanna S, Goel N, Oberoi JK, Rao BK. Antimicrobial prescribing patterns of surgical speciality in a tertiary care hospital in India: Role of persuasive intervention for changing antibiotic prescription behaviour. Indian J Med Microbiol. 2017;35(3):369–375. doi: 10.4103/ijmm.IJMM_17_273. [DOI] [PubMed] [Google Scholar]
- 26.Nathwani D, Varghese D, Stephens J, Ansari W, Martin S, Charbonneau C. Value of hospital antimicrobial stewardship programs [ASPs]: a systematic review. Antimicrob Resist Infect Control. 2019;8(1):1–13. doi: 10.1186/s13756-019-0471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ministry of Health . Standerd Treatment Guidelines. 7th ed. Standard Treatment Guidelines. Accra: Ghana National Drugs Programme (GNDP) Ministry of Health; 2017. pp. 1–1416. [Google Scholar]
- 28.Koduah A, Asare BA, Gavor E, Gyansa-Lutterodt M, Andrews Annan E, Ofei FW. Use of evidence and negotiation in the review of national standard treatment guidelines and essential medicines list: experience from Ghana. Health Policy Plan. 2019;34:II104–II120. doi: 10.1093/heapol/czz107. [DOI] [PubMed] [Google Scholar]
- 29.Namboodiri SS, Opintan JA, Lijek RS, Newman MJ, Okeke IN. Quinolone resistance in Escherichia coli from Accra, Ghana. BMC Microbiol. 2011;11. [DOI] [PMC free article] [PubMed]
- 30.Egyir B, Guardabassi L, Nielsen SS, Larsen J, Addo KK, Newman MJ, et al. Prevalence of nasal carriage and diversity of Staphylococcus aureus among inpatients and hospital staff at Korle Bu teaching hospital, Ghana. J Glob Antimicrob Resist. 2013;1(4):189–193. doi: 10.1016/j.jgar.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Labi A-K, Nielson K, Marvig R, Bjerrum S, Enweronu-Laryea C, Bennedbæk M, et al. Outbreak of carbapenemase producing Klebsiella pneumoniae in a neonatal intensive care unit in Ghana. Emerg Infect Dis. 2020; 26(9):2235-8. [DOI] [PMC free article] [PubMed]
- 32.Obeng-Nkrumah N, Molecular epidemiology of beta-lactamase producing Escherichia coli and Klebsiella pneumoniae in Ghana. Doctoral dissertation, University of Ghana; 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is available from corresponding author upon request.