Abstract
The novel coronavirus SARS-CoV-2 enters into the human body mainly through the ACE2 + TMPRSS2+ nasal epithelial cells. The initial host response to this pathogen occurs in a peculiar immune microenvironment that, starting from the Nasopharynx-Associated Lymphoid Tissue (NALT) system, is the product of a long evolutionary process that is aimed to first recognize exogenous airborne agents. In the present work, we want to critically review the latest molecular and cellular findings on the mucosal response to SARS-CoV-2 in the nasal cavity and in NALT, and to analyze its impact in the subsequent course of COVID-19. Finally, we want to explore the possibility that the regulation of the systemic inflammatory network against the virus can be modulated starting from the initial phases of the nasal and nasopharyngeal response and this may have several clinical and epidemiological implications starting from a mucosal vaccine development.
Introduction
The nose represents an important component of the mucosal immunity in upper airways (UA), and it is involved both in host protection and immune homeostasis between the commensal microbiota and invading pathogens.1 Phylogenetically, the nasal cavity and the nasopharynx-associated lymphoid tissue (NALT) constitute an ancient barrier system that can be found even in early bony vertebrates.1, 2 While in rodents, this immune compartment is found in the floor of the nasal cavity (hence the name nasal-associated lymphoid tissue), in humans it is more properly referred to as NALT since it is located in the most cranial pharyngeal mucosa.2 Strikingly, this tissue has coevolved along with the olfactory system under the same evolutionary forces, both in terrestrial and in aquatic animals.3 In addition, being in direct physical contact with the external environment, the nasal mucosa routinely filters, moistens, and warms the inhaled air to minimize the irritative effects on lower airways, to maintain the mucociliary clearance, and to favor gaseous exchanges.4, 5
Through both innate and acquired immunity, the nose and NALT play a central role in the induction of mucosal immune responses, including the generation of Th1- and Th2-polarized lymphocytes, and IgA-committed B cells.1, 3 Together with other cellular elements, such as dendritic cells (DCs), microfold (M) cells, and macrophages, nasal epithelial ciliated and goblet cells form a unique gateway involved in the induction of the local and systemic response to a wide range of pathogens and allergens.1, 6 Accordingly, intranasal immunization is able to activate an antigen-specific protective immunity in both the mucosal and systemic immune compartments.7
The Severe Acute Respiratory Syndrome-CoronaVirus-2 (SARS-CoV-2) is a novel RNA beta-coronavirus of probable zoonotic origin that is currently causing an unprecedented pandemic.8 The disease originating from SARS-CoV-2 is known as COronaVIrus Disease 19 (COVID-19) and it develops initially as a respiratory infection but, in some individuals, it may progress to systemic involvement. Like many other airborne viral diseases, penetration into the UA is the first step of the infection, as a higher viral load was found in nasal swabs when compared to throat swabs.9 By single-cell RNA sequencing, nasal epithelial cells were shown to express the highest levels of angiotensin-converting enzyme 2 (ACE2) and of the cellular serine protease TMPRSS2, both working as the main entry receptors for SARS-CoV-2 following interaction with viral (S)pike protein.10, 11, 12 Given that the nose is not only the mere entry site but also the main target of SARS-CoV-2, understanding the initial host–viral interaction in the nasal and NALT microenvironments may be one of the keys to understand and modulate the systemic inflammatory response.11 In this paper, we want to critically review the currently available cellular, experimental, and clinical data regarding the central role of the nose and the NALT in the transmission, modulation, and progression of COVID-19.
SARS-CoV-2 transmission and entry
SARS-CoV-2 emerged in Wuhan, China, in December 2019. Thanks to the human-to-human airborne transmission, it rapidly became a pandemic within few months and, as of 24 October, about 42 million cases and 1.1 million deaths have been reported worldwide by the World Health Organization (WHO).13
A general consensus exists on the possibility of SARS-CoV-2 diffusion by respiratory droplets, as it was already demonstrated for other pathogens like SARS-CoV, Middle East respiratory syndrome coronavirus (MERS), and influenza viruses.11, 14, 15 In agreement, in COVID-19 symptomatic patients, Zou et al showed that the viral load increases after symptoms onset, with higher viral loads detected in the nose than in the oral cavity.9 Notably, viral shedding was confirmed also in asymptomatic patients that can represent up to 60% of a fixed cohort.16 As a matter of fact, viable viral particles were also found in the aerosol generated under experimental conditions.17, 18 By definition, an aerosol is defined as fluid particles <5 μm that can spread over distances >1 m, and the distinction between aerosols and droplets has also a practical implication.19 For instance, a physical distance of 1–2 m may not protect enough given that a review of the literature has found that “droplets” can deposit as far away as 6–8 m (Fig. 1 ).20 In the hospital environment, high infection rates were registered for several “aerosol-generating procedures” (invasive and noninvasive ventilation, intubation, tracheostomy, etc.); again, it has not yet been proved that aerosols are the culprits for the increased transmissibility but healthcare workers are advised to wear N95/FFP2 or FFP3 in such a context.21, 22
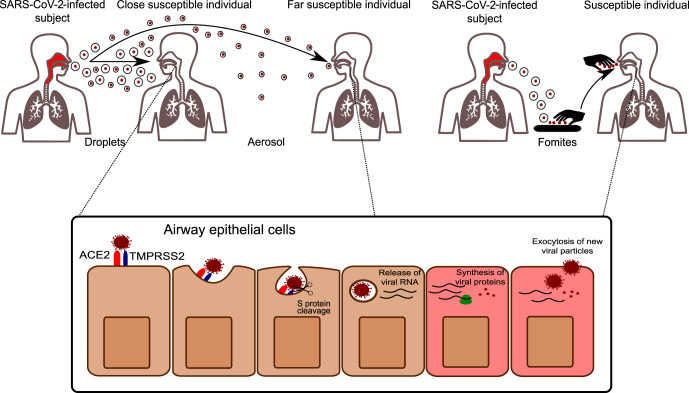
Fig. 1.
SARS-CoV-2 transmission and infection. SARS-CoV-2 can be transmitted by respiratory particles released from an infected subject. Droplets are large particles that commonly deposit within a few meters away from the emitting subject and are responsible for the infection of close individuals. Aerosol instead has a smaller diameter (<5 μm) and can infect subjects at higher distances. In addition to these routes, also contaminated objects (fomites) can be a source of transmission, especially if not practicing regular hand hygiene. Once entered in the airways, SARS-CoV-2 interacts with S protein to its receptor ACE2. The proteolytic cleavage of S, mediated by the cellular protease TMPRSS2, facilitates SARS-CoV-2 infection, which is followed by the release of viral nucleic acid, protein synthesis, and assembly of new viral particles.
The velocity of the airflow exhaled by a sneeze is much higher than that of breathing or coughing, and the corresponding turbulence is about 18 times larger than that of a cough.23, 24 However, it must be remembered that droplet ejection at high speed may also generate an aerosol.23 These physical differences have a crucial role in terms of site of deposition in the host: large droplets can stop in UA where they are trapped in nasal secretions and carried by the mucociliary clearance, to be expelled, inhaled, or swallowed; on the other hand, inhaled aerosolized particles can penetrate more deeply into the lower airways.25 At variance, other studies have suggested that the nose may be the dominant initial site for SARS-CoV-2 infection also in presence of infected aerosol. In fact, SARS-CoV-2 was detected in aerosol particles in the range of aerodynamic sizes exhaled during normal tidal breathing, and it can also deposit at the highest density into the nose according to a mechanical delivery deposition model in naive subjects.26, 27 Actually, the dichotomy in large versus small particles is simplistic, since the emission of pathogen-laden fluids is best described as a multiphase turbulent gas cloud and large droplets themselves can rapidly transform into an aerosol by evaporation.28 Several human activities might be able to generate aerosols (coughing, talking, sneezing, vomiting, and even toilet flushing) but no clear evidence linking them to actual infection exists outside the laboratory setting.19, 22
As a matter of fact, indirect transmission of SARS-CoV-2 may take place after the droplets are dispersed upon several objects and surfaces (fomites) where the virus can survive for several days (Fig. 1).17, 23 However, subsequent papers have criticized these findings because the concentration of the inoculum implemented in the simulations was far higher than what is measured in real life and the chance of transmission through fomites is believed to be very small.29 Whatever the estimated risks, at least in the hospital setting, periodically disinfecting all the touchable surfaces remains a key recommendation from the WHO.29, 30
Once the airborne particles reach the airways, many biological and physicochemical factors determine the capability to invade the mucosal barrier, including the molecular tropism for a specific receptor, the presence of activating proteases, tissue temperature, pH, and the resident microbiota.31, 32 Commensal bacteria are classically regarded as protective against other pathogens.33 However, they can also favor some viral infections, as it was demonstrated for the colonization by S. pneumoniae and H. influenza, which increases the binding of the respiratory syncytial virus to UA epithelium thanks to the upregulation of adhesion receptors, such as intercellular adhesion molecule 1.32
The two main entry factors for SARS-CoV-2, ACE2, and TMPRSS2, have been identified in many human epithelial and non-epithelial tissues. For example, the finding of high expression of ACE2 in the oral cavity mucosa has raised the issue of oro-fecal transmission.34 However, the co-expression of ACE2 and TMPRSS2 that is necessary for efficient viral entry was demonstrated only in some subtypes of respiratory, corneal, and intestinal cells (Fig. 1).10 In detail, subsequent research has shown that only type II pneumocytes, nasal secretory cells, and absorptive enterocytes are the principal host targets and, by a quantitative analysis using GFP chromophore, a remarkable gradient of infection was demonstrated.11, 12 The UA and the nasal ciliated cells, in particular, are the main site of infection while the goblet secretory ACE2 + TMPRSS2 + MUC5B+ cells were not infected in vitro or in vivo, ultimately suggesting that other unknown variables mediate tropism.11 Furthermore, inter-individual variations of ACE2 and TMPRSS2 expression may account for different susceptibility to the infection. In this regard, three subpopulations need to be separately considered. First, children (<9 years old, in particular) have fewer and less severe symptoms compared with adults and a significantly reduced nasal ACE2 expression was demonstrated.35 Second, ACE2 is also less expressed in people with allergic respiratory disease and especially those with a clinical/laboratory profile of type 2 inflammation.36, 37 Since the ACE2 receptor is an INF-regulated gene whose expression is reduced in allergic patients with T2 cytokine pattern, a possible reduction of infection risk has been postulated.38 These molecular findings represent some of the foundations for the potential clinical protective role of asthma in COVID-19 infection.36 Finally, nasal SARS-CoV-2 was identified also in patients who underwent a total laryngectomy and whose UA are iatrogenically separated from the lungs.39, 40 Despite the lack of airflow induces progressive squamous metaplasia and submucosal fibrosis, viral cell tropism is maintained and the hematogenous route or inoculation by contact with a fomite is the supposed route of transmission.39, 40
Other possible human receptors for SARS-CoV-2 are the non-tyrosine kinase neuropilin-1 (NRP1) and neuropilin-2 (NRP2).41 There is preliminary evidence that the cleavage of S protein can generate a polybasic Arg-Arg-Ala-Arg C-terminal sequence on the S1 subunit that can efficiently bind to NRP1 and NRP2.41 These single-pass transmembrane receptors are widely expressed in several immune (macrophages, T lymphocytes, etc.) and nonimmune cells, with many documented physiological roles (e.g., cardiac and neural development, etc.) and future studies are anticipated in this regard.42
The preeminent role of nasal mucosa in viral transmission has also some important practical implications. The diagnostic gold standard for COVID-19 is the reverse‐transcriptase polymerase chain reaction on secretions taken from nasopharyngeal or oropharyngeal swabs.43 However, less invasive and self-collected swabs from the nasal fossa/middle turbinate were shown to have comparable diagnostic accuracy and they would represent a more cost-effective strategy for massive testing.44 All procedures that involve the nasal mucosa, on the other hand, must be possibly regarded as aerosol-generating and therefore they should be performed only using the appropriate protective measures.45, 46
Mucosal and systemic immune response to SARS-CoV-2
Nasal defense mechanisms and the primary phase of COVID-19
There are two major types of nasal mucosal defenses.47, 48 A physicochemical barrier is formed by the tightly bound ciliated, goblet, and basal epithelial cells, by the double mucus layer, and by the basement membrane.49 In addition, several extracellular molecules such as the beta-defensins and galectins show proinflammatory and antimicrobial properties.50
Airway mucus is the first double-layer (gel and periciliary/sol) barrier that pathogens encounter and it is mainly composed of water (95%) and different other molecules produced by the goblet cells and submucosal nasal glands.47, 51 Mucins are the most represented glycoproteins in the gel layer and they contribute to innate immunity through the interaction with other components, such as IgA, collectins, or defensins.51 Among the 11 genes so far described in human airways, MUC5AC and MUC5B are the predominant gel-forming mucins in the nasal mucosa.47, 51 Most importantly, many cytokines (IL-1, IL-4, IL-6, IL-9, IL-13, IFNs) were shown to upregulate the expression of mucins genes and, for influenza virus, there is experimental evidence that the production of MUC1 limits the binding of the virion to other sialic acid-expressing glycoproteins in the mucus layer, thereby limiting viral entry and the inflammatory damage.47, 52 Pathogens can disrupt the mucus barrier by the production of several degradative enzymes, by exploiting the uptake by M cells or, when present, by the aid of a flagellum.51 Regarding SARS-CoV-2, a Chinese study has shown that COVID-19 patients present higher concentrations of MUC5AC and MUC1 in respiratory secretions compared to healthy controls but its clinical relevance is unclear.53 With about 200–300 cilia in every single respiratory epithelial cell, the constant microtubule-based clearance process is another fundamental first-line defense against microbes.54 Again, despite the shedding of these structures was demonstrated in both SARS-CoV and SARS-CoV-2, its significance in the pathogenesis of coronavirus infection remains elusive.55, 56
The second nasal immunological barrier is made of a network of cells and molecules of the innate and adaptive immune system, such as resident microfold M cells, macrophages, innate lymphoid cells, dendritic cells, and B and T lymphocytes.57, 58
Furthermore, the UA is characterized by MALT of the Waldeyer's ring including NALT, tubal, palatine, and lingual tonsils.59, 60, 61, 62 Notably, NALT is underrepresented in germ-free mice compared with wild-type mice, suggesting that its development comes from the interaction with the microbiota after birth.1, 59 In addition, its formation depends on several unique factors such as ID2 (inhibitor of DNA binding 2) at variance with other secondary lymphoid tissues such as Peyer's patches.1 NALT is constantly challenged by potentially pathogenic agents that merge with a complex resident microbiota.1, 59
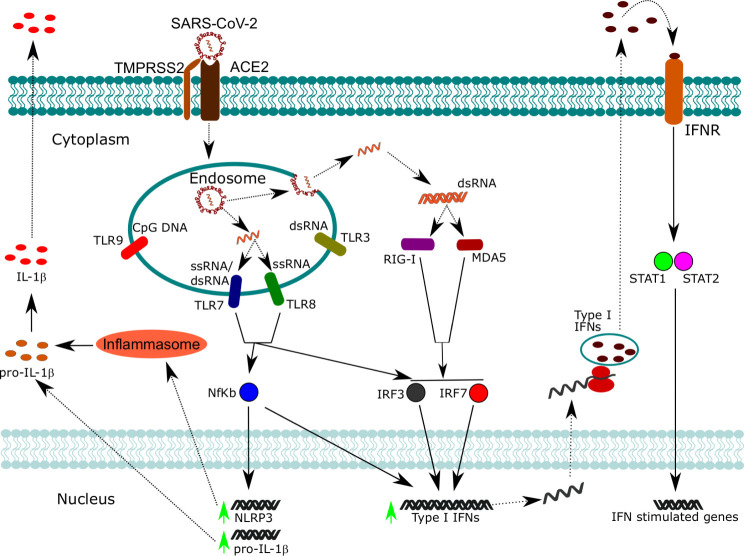
In this scenario, NALT represents the first lymphoepithelial barrier exerting a “gate control” on airborne antigens including respiratory viruses such as SARS-CoV-2.63, 64 At the mucosal level, SARS-CoV-2 is a cytopathic virus able to induce death and injury to infected tissues by eliciting a highly inflammatory form of programmed cell death termed pyroptosis.65 This process involves an increased secretion of several cytokines and chemokines (IL-6, IFN-gamma, MCP1, and IP-10), which are released into the blood.65 During pyroptosis, the release of damage-associated molecular patterns rapidly activates the cells of innate immunity. Moreover, innate immune cell activation is based on a complex system of pattern recognition receptors (PRRs), that are able to recognize a large number of pathogen-associated molecular patterns, shared by an entire class of pathogens but not expressed on host cells.47 At least two families of PRRs are involved in the recognition of viral structures: Toll-like receptors (TLRs) and retinoic-acid-inducible gene I (RIG-I)-like receptors (RLRs). Among TLRs, TLR3, TLR7, TLR8, and TLR9 are involved in the recognition of viral structures, and they are all predominantly located in endosomes.66 RLRs instead are ubiquitously expressed in the cytoplasm.66 Collectively, these receptors can recognize the different types of viral nucleic acids (dsDNA, dsRNA, ssRNA) and converge toward the activation of transcription factors such as interferon-regulatory factor 3 (IRF3), IRF7, and NF-kB, which ultimately lead to the expression of type 1 INF, such as IFN-β, and pro-inflammatory cytokines as IL-1β, thus starting the pro-inflammatory antiviral response mechanisms (Fig. 2 ).66
Fig. 2.
SARS-CoV-2 recognition by nucleic acid sensors and activation of pro-inflammatory and antiviral responses. Following SARS-CoV-2 infection, viral particles are contained within endosomal compartments. Here, viral ssRNA is released and can be recognized by TLR7 and TLR8, which activate downstream signaling pathways converging to the activation of NF-kB, IRF3, and IRF7. NF-kB promotes the transcription of pro-inflammatory cytokines such as pro-IL-1β and the inflammasome component NLRP3. The inflammasome promotes the maturation of pro-IL-1β in mature IL-1β, which can be secreted. NF-kB cooperates also with IRF3 and IRF7 for the expression of type I IFNs. When the viral ssRNA is released in the cytoplasm, it is replicated by an RNA-dependent RNA polymerase which forms dsRNA intermediates. dsRNA can be recognized by cytoplasmic sensors such as RIG-I and MDA5, which converge to the activation of IRF3 and IRF7. Type I IFNs can act in an autocrine manner, activating STAT1 and STAT2 and thus promoting the expression of interferon-stimulated genes involved in anti-viral defense.
Components of innate immunity involved in patrolling the mucosal surfaces also include many humoral factors: a role for soluble PRR has been proposed in SARS-CoV-2 infection.67, 68 Mannose-binding lectin (MBL) is a member of the collectin family and it is composed of multiple carbohydrate recognition domains that bind to repetitive mannose molecules, highly expressed on both bacteria and viruses. Following the attachment to the pathogen's surface, MBL can activate either the complement cascade or act as an opsonin. The concentration of MBL varies significantly among individuals and depends on polymorphisms of the gene.69 The deficiency of MBL has been associated with increased susceptibility to various infectious diseases, including the original SARS-CoV infection.70, 71, 72 In agreement, it has been demonstrated that MBL can selectively bind to SARS-S protein, leading to a reduced viral infectious capacity.73 This latter phenomenon does not occur by interfering with the interaction between S and its target ACE2, but presumably via the inhibition of post-binding structural S rearrangements that are crucial for optimal infection.73 MBL expression progressively declines with age, suggesting that the elderly may be more susceptible to the development of severe COVID-19 because of a lack of efficacious innate response.74 In addition to soluble PRRs, natural antibodies (nAbs) have also an important antimicrobial role.68 nAbs are predominantly of IgM and IgA classes and are produced by B cells in absence of any antigenic stimulation.75 nAbs bind to both exogenous and autologous epitopes, providing the first line of defense when pathogen-specific T cell-dependent antibodies have not been produced yet.75 A subset of nAbs can recognize the antigens of the AB0 blood group, which results from the post-translational glycosylation of the H antigen expressed on red blood cells (RBCs). A-group individuals can enzymatically add N-acetylgalactosamine to the H antigen, while in B-group subjects is added galactose. These two enzymatic activities are both present in AB individuals, while they are both absent in 0 individuals. In addition to RBC, AB0 antigens are also present in many epithelial cell types, including in the lungs.76 Given that enveloped viruses (including coronaviruses) are highly glycosylated, it has been proposed that SARS-CoV and SARS-CoV-2 virions generating from lung alveolar epithelial cells of individuals of A or B blood group may harbor these antigens on their envelope. For this reason, natural anti-A or anti-B antibodies may confer protection, as it was demonstrated for other viruses.77 In agreement, it has been shown that anti-A antibodies can prevent the interaction between S protein and ACE2 when Spike is produced in a cell type expressing the A antigen.78 Further confirming this hypothesis, individuals with 0 blood group displayed a reduced risk of infection compared to non-0 individuals during the 2003 SARS-CoV outbreak in Hong Kong.79 Given the similarity between SARS-CoV and SARS-CoV-2, a protective role for nAbs is expected also in the current pandemic and it has already been shown that A blood group patients seem to have an increased risk to develop severe disease, while individuals with 0 group are relatively protected.80, 81
In addition to alterations in soluble components, it has been proposed that also a dysregulated cellular innate response occurs in SARS-CoV-2 infection. Most of the data produced so far has been obtained from peripheral blood of COVID-19 patients, given the difficulty to obtain lung samples. Monocytes are large mononuclear cells with a key role in inflammation and pathogen eradication. Three subsets of monocytes can be distinguished, based on their activation status and the combinatorial expression of CD14 and CD16. Immature classical (c) monocytes are CD14 + + CD16−, while more differentiated and inflammatory monocytes are defined as transitional (t) CD14 + CD16+ and, non-classical (nc) CD14-CD16++. The proportions of these three subsets in the circulation of healthy individuals are well defined, although an expansion of inflammatory monocytes has been described in autoimmunity and chronic inflammation.82 Accumulating data have shown that proportions of monocyte subsets are altered in COVID-19,83, 84 together with a significant downregulation of monocyte HLA-DR expression.83, 85, 86 In addition to monocytes, DCs are another innate cell population with a crucial role in nasal immunity.87 Two subsets of DCs have been described: plasmacytoid DCs (pDCs), which rapidly respond to viral infections secreting high amounts of type I and III IFNs, and myeloid DC (mDC), which act as sentinels ready to stimulate T cells. Myeloid DCs can be further divided into two subsets, cDC1 and cDC2, depending on the expression of CD141 and CD1c, respectively. A significant decrease of peripheral pDCs and mDCs in COVID-19 patients has been demonstrated.83, 84, 85, 86 Moreover, Sànchez-Cerrillo et al. demonstrated that dense secretions obtained from bronchoscopies of COVID-19 patients are enriched of t- and nc-monocytes, as well as cDC2, suggesting that these subsets are selectively recruited from the circulation.84
As already stated, type I and III IFNs have an important role in early phases of viral infections, but it has been shown that they are both secreted at lower levels in SARS-CoV-2 infection, compared to other respiratory viruses.88 Moreover, at least 10% of patients with life-threatening pneumonia have serum neutralizing autoantibodies targeting type I IFNs. These individuals are generally old persons, notably males. Neutralizing anti-type I IFN are clinically silent before SARS-CoV-2 infection, but then predispose patients to severe COVID-19.89 Further supporting this concept, loss of function mutations at TLR3 and IRF7, both involved in type I IFN immunity, were identified in patients with severe COVID-19.90
Finally, increased proportions of circulating neutrophils have been demonstrated in COVID-19, with increased expression of the activation marker CD66b.83, 86, 91 In agreement, increased percentages of granulocytes were observed in pulmonary secretions of COVID-19 individuals,84 and neutrophils extracellular traps were identified in postmortem lung specimens from subjects died of COVID-19.92
In addition to providing the first line of defense, innate immunity allows the development of both cell-mediated and humoral adaptive response. In particular, it has been shown that anti-SARS-CoV-2 IgG seroconversion occurs within 19 days from symptoms onset, with a median of 13 days.93 Regarding T cell response, accumulating studies in the literature show that SARS-CoV-2 specific CD4+ and CD8+ cells can be found in nearly all convalescent patients.94, 95, 96, 97, 98, 99, 100, 101, 102 Of note, it was observed that the frequency of virus-specific CD4 + T cells correlates with specific antibody titers.94, 102 This is of great importance, suggesting that memory T cells efficiently develop in convalescent patients, thus providing the rationale for vaccine research. Moreover, it was shown that in some exposed individuals, SARS-CoV-2 specific T cell responses can be observed even without detectable specific antibodies.98 In most cases, immune activation is able to resolve the infection and to confine it into the UA.65, 68 It has been demonstrated in mice that robust activation of the immune response in the UA to a murine coronavirus infection can prepare and modulate the activation status of immune cells in lower airways, preventing a dysregulated and self-destructing response in the lungs, but this remains still unclear for SARS-CoV-2 in humans (Fig. 2).103 So far, it has been shown that the transcription of proteins belonging to the nuclear factor κB (NF-κB) and tumor necrosis factor α (TNF-α) pathways (e.g., IL-18, NFKB2, NFKBIA, TNFA, etc.) is significantly higher in nasal versus bronchial epithelium, thus confirming a distinctive immune response between the upper and lower airways and future studies are needed to explore this difference.104
A comparison with other coronaviruses: mucosal immunity against SARS-CoV and MERS-CoV
The 2002 bat-derived SARS and the 2012 dromedary camels-derived MERS constitute the two best-studied immunopathological and clinical models of coronavirus infection in humans.105, 106, 107, 108, 109 SARS-CoV enters the human cells thanks to the S1 domain of the trimeric S glycoprotein which efficiently binds the ACE2 receptor.110 MERS, on the other hand, exploits the CD26 (also known as dipeptidyl peptidase 4, DPP4) receptor.111 Interestingly, dromedaries mainly develop UA symptoms because DPP4 is highly expressed in the camel nasal turbinates and larynx.106 In humans, DPP4 is primarily expressed in type I and II pneumocytes and, thus, lower respiratory tract infection is more common. However, non-ciliated bronchial epithelial cells, endothelial cells, and epithelial cells of many other organs (kidney, intestine, liver, thymus and bone marrow) are also susceptible to MERS-CoV.109
Once in the low-pH environment of the endosome, the cysteine protease cathepsin L is the principal responsible for the cleavage of S protein of SARS-CoV, a required event to activate the membrane fusion domain.112 Other proteases expressed in the respiratory tract, capable of processing SARS-CoV S, include the human airway trypsin-like protease (HAT or TMPRSS11d), TMPRSS11a, and TMPRSS2.113, 114 Regarding MERS-CoV, furin protease is the major activator in the Golgi apparatus but TMPRSS2 can also play a role.113, 115 In 2019, an elegant work has revealed the complexity of the CoV-membrane-fusion activation, using human neutralizing anti-S antibodies derived from SARS and MERS survivors: many trimers cooperate to form the fusion pore while the S1 subunit should be viewed as a molecular chaperone. Moreover, at least for SARS-CoV, host antibodies can even trigger S fusogenic conformational changes in a marvelous example of functional mimicry.116
The mucosal response against SARS-CoV begins in nasal and bronchial ciliated respiratory cells which express ACE-2.55 Notably, binding to ACE2 downregulates the expression of the host receptor which is a key element in the protection against acute lung injury.108, 112 Analogously to SARS-CoV-2, an early type I IFN-driven inflammatory response (IFNα, IFNγ, CXCL10, CCL2, and other related proteins), an in vitro direct cytopathic effect, and the induction of apoptosis via a caspase-dependent pathway were demonstrated.108, 112 There are different mechanisms through which SARS-CoV counteracts the immune response: for example, at least eight viral proteins were shown to block the signaling cascades downstream of PRRs (e.g., M protein blocks E3 ubiquitin kinase, nonstructural proteins nsp3b and nsp6 blocks the IFN-regulatory factor 3 IRF3, etc.).112 Furthermore, a membrane protein can inhibit the formation of a molecular complex that contains IKKε and, ultimately, it represses the induction of type I IFN genes.112
There is much less data available on MERS-CoV immunopathogenesis because of the few cases registered worldwide (<2500, as of 31, December 2019) and of the fact that autopsies were almost never performed for healthcare workers protection and cultural reasons.109 The virus can be isolated from nasal secretions and it is capable of inducing robust virus-specific CD8 T-cell responses that were detected in most patients with severe or moderate disease.109, 117 MERS-CoV ORF4a has an analogous type I IFN-repressing function thanks to the inhibition of melanoma differentiation-associated protein 5; again, viral ORF4a, ORF4b, ORF5 are able to inhibit the nuclear trafficking of IRF3 and activation of the IFNB promoter.112
Systemic diffusion and immune escape of SARS-CoV-2
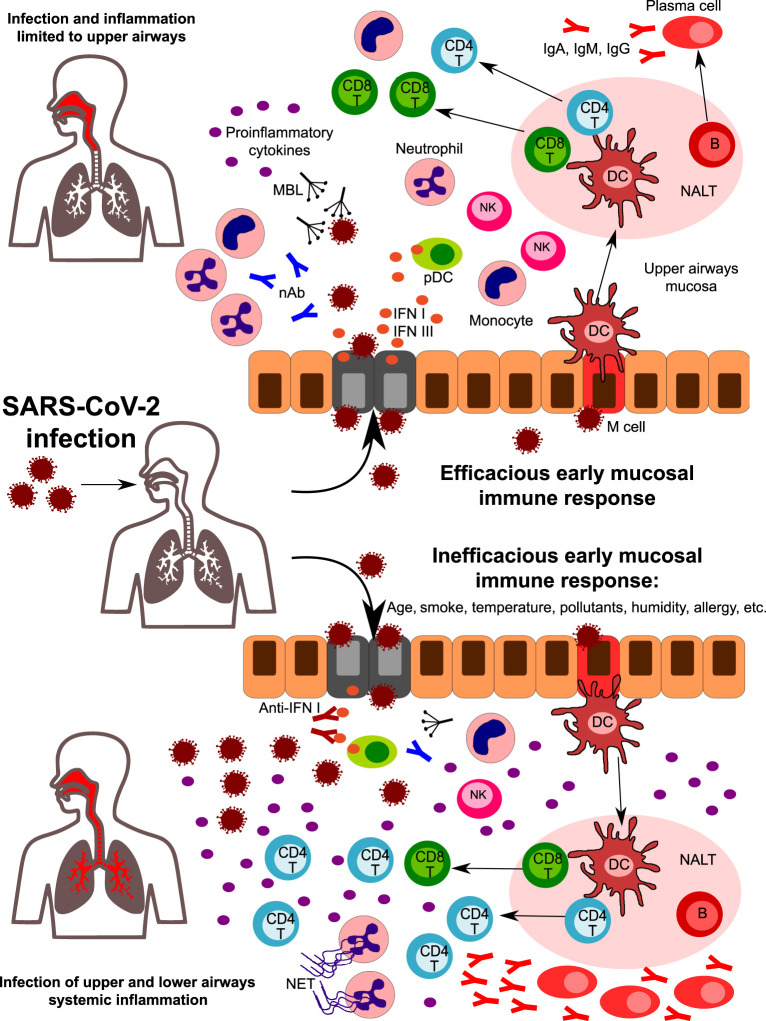
SARS-CoV-2 infection results in no apparent symptoms in about 80% of patients (Fig. 3 ).118, 119 These subjects were shown to have a weaker immune response and a longer viral shedding from the UA compared to symptomatic cases.93 Asymptomatic individuals display a reduced frequency of circulating SARS-CoV-2 specific T cells and lower antibody titers than symptomatic ones.95, 102 In agreement with this observation, the frequency of SARS-CoV-2 specific CD4 + T cells is higher in patients with severe COVID-19 compared to those with moderate disease, although with reduced proportions of IFN-γ secreting cells.120 Increased frequencies of SARS-CoV-2 specific CD8 + T cells were observed in mild cases, suggesting that impaired cytotoxic T cell response may predispose to severe COVID-19.96 On the contrary, in almost 10% of cases, an inefficient response in the UA leads to an uncontrolled viral replication and an exaggerated systemic response (Fig. 3).121 The subsequent elevation of serum cytokine levels (i.e., the so-called “cytokine storm”) constitutes a potential source of damage for many organs, eventually leading to multiorgan failure.65 In agreement, infected patients show elevated serum levels of IL1B, IFNγ, IP10, and MCP1, while GCSF, IP10, MCP1, MIP1A, IL-6, and TNFα levels were significantly higher in severe than in non-severe cases.122 IL-6 levels continue to increase in non-survivors compared to survivor patients,123, 124 and a recent meta-analysis of mean IL-6 concentrations reported 2.9-fold higher levels in patients with complicated COVID-19 infection when compared with patients without complications.125 Cytokine storm is also responsible for a general inhibition of immune cell functions because T and NK lymphocytes from COVID-19 patients display impaired effector functions.91 The neutralization of IL-6 via the drug Tocilizumab or the generalized desensitization of cytokine network via the use of the JAK1-2 inhibitor Ruxolitinib allow restoration of immune cell functionality.91, 126, 127 Moreover, post-mortem examination of thoracic lymph nodes and spleens from COVID-19 deceased because of the cytokine storm shows how TNF-a may severely affect T follicular helper cells and germinal centers development, thus leading to impaired humoral immunity.128 This observation is in agreement with data showing that humoral immunity to SARS-CoV-2 is often short-lived.95
Fig. 3.
A hypothetical role for the early mucosal response in the upper airways in SARS-CoV-2 infection progression. Following SARS-CoV-2 entrance in the nasal or oral cavity, epithelial cells of the upper airways are the primary target site of infection. Innate immunity components in the upper airway mucosa are responsible for the first line of defense. Humoral components such as natural antibodies (nAb) and lectins (including mannose-binding lectin, MBL) can recognize glycoside structures of the virus, while epithelial infected cells and plasmacytoid DC release high amounts of type I IFNs, that are crucial in the initial antiviral response. Cells of innate immunity are activated by viral PAMPS and by the release of DAMPS from infected cells. An efficacious early mucosal innate response allows the development of adaptive immunity, with the expansion of CD4 + helper and CD8 + cytotoxic T cells and the differentiation of Ig-secreting plasma cells. Coordinated innate and adaptive responses contribute to the final elimination of the pathogen (upper part). However, factors such as age, smoke, pollutants, temperature, humidity, and genetics can affect the early response, leading to an inefficacious control of viral replication (lower part). Reduced levels of nAb and/or MBL, the presence of autoantibodies neutralizing type I IFN activity, and of impaired development of antigen-specific CD8 + T cells are some of the factors that can predispose to uncontrolled viral propagation and infection of lower airways. Viral escape is accompanied by a huge release of pro-inflammatory cytokines, with the recruitment and expansion of several subsets of innate and adaptive immunity. Neutrophils accumulate in the lungs and are massively activated, leading to NETs formation. All together, these mechanisms amplify the inflammatory response, leading to uncontrolled systemic inflammation.
Among the risk factors that can predict the development of severe disease (ACE2 expression, smoking history, comorbidities), age is one of the most prominent.123 It is well known that the functionality of the immune system progressively declines with age, leading to a higher susceptibility to infections. This condition is known as “immune-senescence” and is characterized by a progressive dysregulation of innate and adaptive cell functions in the elderly.129 Indeed, aged innate immune cells display reduced antimicrobial functions, with a parallel, sustained secretion of pro-inflammatory cytokines. The latter phenomenon is responsible for the occurrence of chronic, low-grade inflammation known as “inflamm-aging” that significantly affects adaptive immune cells.129 It can be hypothesized that low-grade inflammation, together with the accumulation of dysfunctional immune cells in older individuals, may predispose to the development of uncontrolled hyper-inflammation following SARS-CoV-2 infection. Older people also display reduced proportions of naïve T cells, with an accumulation of terminally differentiated cells. This phenomenon depends on a reorganization of the bone marrow that is skewed to myeloid cells generation in the elderly, and to the involution of the thymus that occurs with age.130 Supporting this hypothesis, it has been demonstrated that the low frequency of naïve CD8 + T cells in the elderly is a risk factor for severe COVID-19.131 Moreover, it has been suggested that age-related alterations may occur earlier in the mucosal than in the system immune cells.132 This phenomenon has been demonstrated to occur initially in the gut133 and then in the nasal mucosa.134 Altogether, these observations suggest that the impairment of the nasal and NALT immune response that commonly occurs with aging may predispose to uncontrolled SARS-CoV-2 infection, thus resulting in systemic involvement.
Summing up, the central role of the immune response in patients with severe COVID-19 infection is highlighted by the immunological characteristics of the patients who have died. Given that SARS-CoV-2 is a new virus for the human species, upon infection, there are no components of adaptive immunity ready to fight it and an efficacious early innate response in the nasal mucosa might contribute to controlling viral replication and spreading, allowing the development of adaptive immune responses (Fig. 2).68
COVID-19 associated olfactory dysfunction and clinical implications
The importance of the interaction between the virus and the UA is also suggested by the discovery that loss of smell represents one of the early symptoms of COVID-19 infection: it is frequently reported at the onset, sometimes as the only symptom in the nonhospitalized population.135 Although the loss of smell can arise from several other sinonasal conditions, such as head trauma, chronic rhinosinusitis, exposure to toxic agents, and many neurological disorders, viral infections are the principal cause of both acute and chronic olfactory dysfunction.136 For these viruses, both a direct injury of the olfactory neuroepithelium and indirect mechanisms (swelling of the mucosa in the olfactory cleft, alteration in mucus production and composition, and changes in olfactory signaling by local cytokines) were demonstrated.137
An observational study that involved over two million subjects showed that isolated anosmia (i.e., without rhinorrhea or nasal congestion) is more predictive than all other COVID-19 symptoms, including fever or cough.138 Although it depends on the way anosmia is measured, it seems that this symptom clinically resolves in a significant proportion of cases: in a recent study, 48.7% of patients reported complete resolution of chemosensory impairment, 40.7% reported at least an improvement in the severity, and only 10.6% reported no modification.139
The pathophysiological foundations for olfactory dysfunction are yet unclear: while early studies had suggested, analogously to other coronaviruses, a direct invasion of olfactory neurons (ONs),140 this mechanism was not confirmed in post-mortem studies.141 The olfactory epithelium shows a complex cytological architecture where ONs are supported by multiple non-neural cell types (sustentacular, Bowman's gland, and microvillar cells) plus the basal globose and horizontal cells which are a reservoir of ONs stem cells.137 Using single-cell RNA sequencing, it was shown that only supporting cells and horizontal stem cells in the olfactory cleft plus vascular pericytes in the olfactory bulb coexpress ACE2 and TMPRSS2.137, 142 Brann et al. have therefore proposed that local infection of these cells may cause an “inflammatory” block of odor conduction where the global disruption of the mucosal architecture can hamper the processing and signal transmission to the brain.142 In the subset of patients showing an extensive involvement of stem cells, epithelial regeneration cannot occur and this would explain the small proportion of patients that do not recover.142 Regarding the prognosis, most of COVID-19 patients with olfactory dysfunction reported an improvement within a few weeks, and intranasal glucocorticosteroids along with olfactory rehabilitation can be useful for those who do not fully recover.143, 144
As previously stated, NRP1 and NRP2 are two other possible receptors for viral entry: an international group has recently shown that, in mice, SARS-CoV-2 infection may occur in neuronal cells of the olfactory bulb. Remarkably, also in five out of six COVID-19 patients, infection in the nasal olfactory neuroepithelium was demonstrated at autopsy, thus suggesting the existence of an NRP-dependent intranasal brain entry pathway that will be the focus of future inquiry.145
Towards a COVID-19 intranasal vaccine?
A global race is underway to identify an effective vaccine against COVID-19 and, thanks to previous research on SARS-CoV and MERS-CoV,146, 147 the timeline from the first discovery of the novel pathogen and the first phase I trial of a potential vaccine has been shortened from the usual 3–9 years to just 6 months.148, 149 The S protein with its receptor-binding domain (RBD) represents an ideal candidate to elicit both cellular and humoral responses and this is supported by the preliminary clinical experience.146, 147, 150 For vaccine development purposes, mouse ACE2 does not bind viral RBD but researchers have shown that mice transduced with adenoviruses encoding human ACE2 are efficiently infected and develop SARS-CoV-2 pneumonia.151 In detail, the specific epitopes of RBD and their structural interaction with neutralizing antibodies have been described, thus providing a framework for the rational development of an effective vaccine.152
Besides the many technical aspects regarding SARS-CoV-2 vaccine development (reviewed in153), the observation that SARS-CoV-2 receptors are highly expressed by ciliated nasal cells and the central role of NALT in generating an immune response to inhaled environmental agents supports the central role of the nasal route in this regard. From experimental evidence, NALT appears to be both a target and a key factor in the regulation of the in vivo local immune response, with effects on distant sites including the lungs.103, 154 Since the nose is exposed to different concentrations of environmental agents,59 it is likely that variable nasal infectivity might influence host immune response and, in turn, the variability of the clinical syndrome of COVID-19.
The pathogenetic role of the nose and the relationship between upper and lower airways in SARS-CoV-2 infection is also supported by clinical observations that suggest an early infection in the UA followed by subsequent aspiration and infection of the lungs.11 The role of the nasal cavity in the “viral seeding” of the lower airways is also supported by studies of the influenza virus in the ferret model where it was recently shown that the nasal cavity is the main site for viral replication and shedding.155 Together, a cross-talk between upper and lower respiratory tract seems to be the key factor in the transmission of infection from the early site of virus replication to the lungs and this transmission seems to be regulated by the activation of innate immunity by NALT, with a modulation of the systemic immune response.156 Such a modulation of infectivity, replication, and activation of the immune response at the mucosal level might be able to drive the evolution/progression of the disease. However, the precise mechanisms by which UA infection or inflammation modulates the lung environment remain poorly understood. For example, the low expression of ACE2 receptors in alveolar cells as well as its lower infectivity in vitro might support this hypothesis although no definitive evidence exists.10, 11 In contrast, given the contamination of the lower airways because of the aspiration of infected nasal secretions, we could hypothesize that the inflammatory lung damage may be a function of the quantity of inhaled virions.11
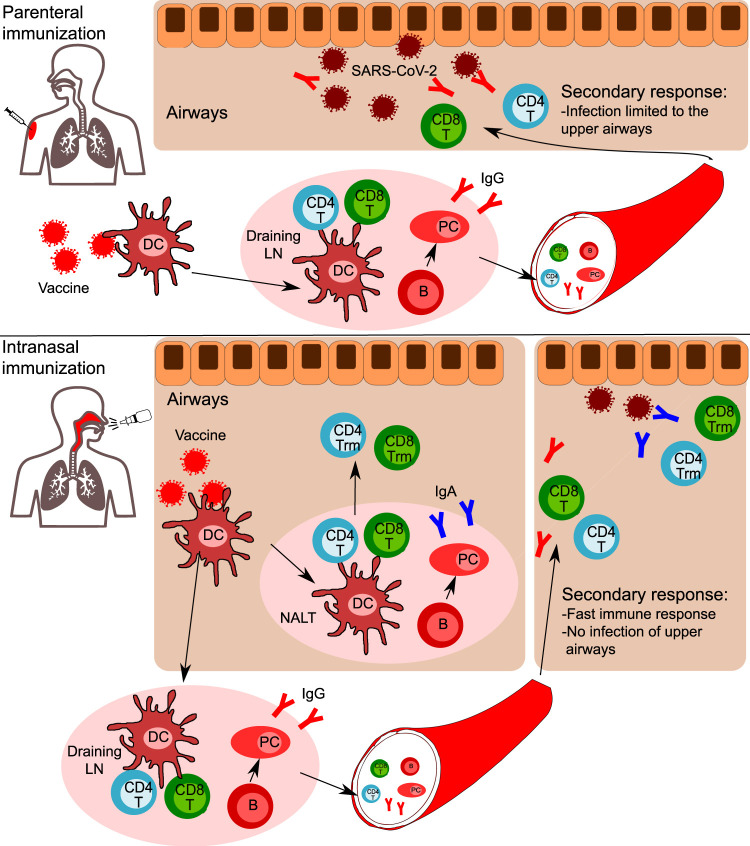
The host-viral initial interactions in the nasal cavity and in the UA are crucial to modulate the subsequent systemic immune response to SARS-CoV-2 as well as many other airborne pathogens. In this view, the nasal administration of a future vaccine may be helpful. Intranasal vaccines, when compared to the conventional parenteral administration, are able to induce efficient protective immunity in both the mucosal (by secretory IgA) and systemic immune systems (Fig. 4 ).1, 7, 157 In the last years, some specific mechanisms showing how this immunity is induced and maintained have been revealed. In mice intranasally exposed to pneumococcal surface protein A, epithelium-derived thymic stromal lymphopoietin (TSLP) was shown to be a key factor in the differentiation of CD11b+ DCs, which, in turn, favor mucosal defenses by producing IgA-inducible cytokines such as APRIL, BAFF, IL-6, and TGF-b. Interestingly, TSLP-mediated stimulation of IgA production was heavily dependent on the IL-6 levels in the mucosal microenvironment.157
Fig. 4.
Comparison of parenteral and intranasal vaccination strategies. Following parenteral immunization, dendritic cells (DC) rapidly uptake the vaccine's antigens and migrate to draining lymph nodes. Here, DC prime antigen-specific CD4 + and CD8 + T cells. Contemporary, antigen-specific B cells activate and differentiate in memory B cells and Ig-secreting plasma cells. CD4 + T cells, CD8 + T cells, B cells, and plasma cells egress the lymph node into the bloodstream. CD4 + T cells, CD8 + T cells, and B cells enter the pool of recirculating lymphocytes, while plasma cells will home to bone marrow niches, where they will continue to secrete antigen-specific IgG. In case of a secondary response (SARS-CoV-2 encounter), in addition to the presence of specific IgG, CD4 + and CD8 + T cells will be recruited to the airways. The infection is limited to upper airways, with no involvement of lower airways and no systemic inflammation. In the case of intranasal vaccination, in addition to the response occurring in draining lymph nodes, a mucosal response occurs in the NALT. B cells differentiate in plasma cells secreting IgA, while CD4 + and CD8 + T cells migrating in the airway mucosa develop a tissue-resident phenotype, thus do not recirculate but reside in tissues. In the case of SARS-CoV-2 encounter, specific T cells and IgA are immediately available in the upper airways to fight the virus and are subsequently assisted by IgG and the recruitment of T cells from the bloodstream. Thus, a faster immune response occurs, with rapid elimination of the virus and no infection of upper airways.
Regarding coronaviruses, we already knew that following intranasal vaccination to SARS-CoV, virus-specific Trm cells develop in the lungs, while if the vaccine is administered subcutaneously virus-specific cells accumulate in the spleen.158 Of note, mice are completely protected from lethal SARS-CoV infection only after intranasal, not subcutaneous, immunization.158 Indeed, the nasal immunization route may promote the development and accumulation of SARS-CoV-2 specific CD4 and CD8 T cells with a tissue-resident memory phenotype (Trm) in the UA. Trm cells are memory lymphocytes that, following the resolution of the infection, do not recirculate in the blood and secondary lymphoid organs but reside in the originally infected tissue, providing a fast response in case of secondary infection by the same pathogen.159 Supporting this hypothesis, it has been shown that Trm cells in the UA could prevent the transmission of influenza to the lower airways and the development of severe lung inflammation.160 Moreover, intranasal vaccination with live attenuated influenza virus could guarantee the development of CD4 and CD8 memory T cells in the UA and lungs.161 Interestingly, O'Hara et al. showed that parenteral administration of killed pneumococci is able to generate IL-17A+CD4+ TRM in the nasal mucosa of mice, and this prevents subsequent colonization with living pneumococci.162
An intramuscular chimpanzee adenovirus-vectored vaccine against SAR-CoV-2 (ChAdOx1 nCoV-19) is currently tested on humans after encouraging phase 1/2 trials results.150 However, in rhesus macaques models, ChAdOx1 nCoV-19 reduced the incidence of pneumonia but it did not generate a UA mucosal response and it did not prevent viral shedding and, ultimately, transmission.163 The authors themselves observe, in their conclusions, that a change in terms of the route of vaccination by exposing respiratory surfaces may induce a more potent mucosal immunity, which is a key to stop nasal shedding and transmission.163
Another group of researchers has thus exploited the intranasal administration of a chimpanzee Ad (simian Ad-36)-based SARS-CoV-2 vaccine (ChAd-SARS-CoV-2-S) in the aforementioned mouse model.151 Strikingly, intranasal ChAd-SARS-CoV-2-S induced mucosal immunity, provided superior protection compared to the parenteral route, and it could even promote “sterilizing” immunity that can block interhuman transmission.164
Concluding remarks
In conclusion, promoting the accumulation of SARS-CoV-2 specific T cells in the UA thanks to a nasal vaccination may guarantee a fast cellular adaptive response in case of exposure, thereby allowing to control viral spreading in the UA and preventing its dissemination to the lungs and other organs. Understanding the complex network of the many cellular and molecular elements involved is a necessary step to develop efficient therapeutic agents and to induce a successful long-lasting mucosal vaccination. If the nose is indeed the initial site of infection, therapeutic strategies that can modulate the mucosal immune response in the UA may prove very useful against COVID-19.
ADDITIONAL INFORMATION
Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Published online: 26 November 2020
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
These authors contributed equally: Oreste Gallo, Luca Giovanni Locatello, Alessio Mazzoni
Author contributions
OG: conceptualization, data curation, investigation, methodology, visualization, writing—original draft. LGL: conceptualization, data curation, writing—original draft. AM: conceptualization, methodology, investigation, writing—original draft. LN: data curation. FA: conceptualization, data curation, methodology, investigation, visualization, writing—original draft.
Competing interests
The authors declare no competing interests.
References
- 1.Kiyono H, Fukuyama S. NALT-versus Peyer's-patch-mediated mucosal immunity. Nat. Rev. Immunol. 2004;4:699–710. doi: 10.1038/nri1439. 1:CAS:528:DC%2BD2cXntFCmtL0%3D, 7097243, 15343369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandtzaeg P, Kiyono H, Pabst R, Russell MW. Terminology: nomenclature of mucosa-associated lymphoid tissue. Mucosal. Immunol. 2008;1:31–37. doi: 10.1038/mi.2007.9. 1:CAS:528:DC%2BD1cXhtlWhuw%3D%3D, 19079158. [DOI] [PubMed] [Google Scholar]
- 3.Tacchi L, et al. Nasal immunity is an ancient arm of the mucosal immune system of vertebrates. Nat. Commun. 2014;5:1–11. doi: 10.1038/ncomms6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elad D, Wolf M, Keck T. Air-conditioning in the human nasal cavity. Respir. Physiol. Neurobiol. 2008;163:121–127. doi: 10.1016/j.resp.2008.05.002. 18565805. [DOI] [PubMed] [Google Scholar]
- 5.Newsome H, et al. Clinical importance of nasal air conditioning: a review of the literature. Am. J. Rhinol. Allergy. 2019;33:763–769. doi: 10.1177/1945892419863033. 31291132. [DOI] [PubMed] [Google Scholar]
- 6.Takeda K, et al. Allergic conversion of protective mucosal immunity against nasal bacteria in patients with chronic rhinosinusitis with nasal polyposis. J. Allergy Clin. Immunol. 2019;143:1163–1175. doi: 10.1016/j.jaci.2018.07.006. 1:CAS:528:DC%2BC1cXhsFCjsbnN, 30053529. [DOI] [PubMed] [Google Scholar]
- 7.Bernocchi B, Carpentier R, Betbeder D. Nasal nanovaccines. Int J. Pharm. 2017;530:128–138. doi: 10.1016/j.ijpharm.2017.07.012. 1:CAS:528:DC%2BC2sXht1GhsrjJ, 28698066. [DOI] [PubMed] [Google Scholar]
- 8.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. 1:CAS:528:DC%2BB3cXltFCjtbY%3D, 32284615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou, L. et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med 10.1056/NEJMc2001737 (2020). [DOI] [PMC free article] [PubMed]
- 10.Sungnak W, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. 1:CAS:528:DC%2BB3cXotVCjurY%3D, 8637938, 32327758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou, Y. J. et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell (2020). [DOI] [PMC free article] [PubMed]
- 12.Ziegler, C. G. et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell (2020). [DOI] [PMC free article] [PubMed]
- 13.WHO Coronavirus Disease (COVID-19) Dashboard, data consulted on 2020/10/24), https://covid19.who.int/.
- 14.Lauer, S. A. et al. The Incubation Period of Coronavirus Disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 10.7326/M20-0504 (2020). [DOI] [PMC free article] [PubMed]
- 15.Chao CYH, Wan MP, Sze To GN. Transport and removal of expiratory droplets in hospital ward environment. Aerosol Sci. Technol. 2008;42:377–394. 1:CAS:528:DC%2BD1cXnsFGgt7w%3D. [Google Scholar]
- 16.Sakurai, A. et al. Natural history of asymptomatic SARS-CoV-2 Infection. N. Engl. J. Med. 10.1056/NEJMc2013020 (2020). [DOI] [PMC free article] [PubMed]
- 17.Van Doremalen N, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. 32182409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asadi, S., Bouvier, N., Wexler, A. S. & Ristenpart, W. D. The coronavirus pandemic and aerosols: does COVID-19 transmit via expiratory particles? Aerosol Sci. Technol. 10.1080/02786826.2020.1749229 (2020). [DOI] [PMC free article] [PubMed]
- 19.Fernstrom, A. & Goldblatt, M. Aerobiology and its role in the transmission of infectious diseases. J. Pathog. 493960 (2013). [DOI] [PMC free article] [PubMed]
- 20.Bahl, P. et al. Airborne or droplet precautions for health workers treating COVID-19? J. Infect. Dis. (2020). [DOI] [PMC free article] [PubMed]
- 21.Sommerstein R, et al. Risk of SARS-CoV-2 transmission by aerosols, the rational use of masks, and protection of healthcare workers from COVID-19. Antimicrobial Resistance Infect. Control. 2020;9:1–8. doi: 10.1186/s13756-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO Scientific Brief: Transmission of SARS-CoV-2: implications for infection prevention precautions. Geneva: World Health Organization; 2020. https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions. Accessed 24 Oct 2020.
- 23.Meselon, M. Droplet and aereosol in the transmission of SARS-CoV-2. N. Engl. J. Med.382, 2063 (2020). 10.1056/NEJMc2009324. [DOI] [PMC free article] [PubMed]
- 24.Han, Z. Y., Weng, W. G. & Huang, Q. Y. Characterizations of particle size distribution of the droplets exhaled by sneeze. J. R. Soc. Interface10, 20130560 (2013). 10.1098/rsif.2013.0560. [DOI] [PMC free article] [PubMed]
- 25.Farzal Z, et al. Comparative study of simulated nebulized and spray particle deposition in chronic rhinosinusitis patients. Int. Forum Allergy Rhinol. 2019;9:746–758. doi: 10.1002/alr.22324. 7457377, 30821929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Booth TF, et al. Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J. Infect. Dis. 2005;191:1472–1477. doi: 10.1086/429634. 15809906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Y. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature 10.1038/s41586-020-2271-3 (2020). [DOI] [PubMed]
- 28.Bourouiba L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID-19. JAMA. 2020;323:1837–1838. doi: 10.1001/jama.2020.4756. 32215590. [DOI] [PubMed] [Google Scholar]
- 29.Goldman E. Exaggerated risk of transmission of COVID-19 by fomites. Lancet Infect. Dis. 2020;20:892–893. doi: 10.1016/S1473-3099(20)30561-2. 1:CAS:528:DC%2BB3cXhtlegu7rF, 7333993, 32628907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cleaning and disinfection of environmental surfaces in the context of COVID-19. Geneva: World Health Organization; 2020. https://www.who.int/publications/i/item/cleaning-and-disinfection-of-environmental-surfaces-inthe-context-of-covid-19. Accessed 24 Oct 2020.
- 31.Herfst S, et al. Drivers of airborne human-to-human pathogen transmission. Curr. Opin. Virol. 2017;22:22–29. doi: 10.1016/j.coviro.2016.11.006. 27918958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. 1:CAS:528:DC%2BC2sXks1GlsbY%3D, 7097736, 28316330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown RL, Sequeira RP, Clarke TB. The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 2017;8:1–11. doi: 10.1038/s41467-017-01803-x. 1:CAS:528:DC%2BC1cXovFygt7g%3D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral. Sci. 2020;12:1–5. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunyavanich, S., Do, A. & Vicencio, A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 10.1001/jama.2020.8707 (2020). [DOI] [PMC free article] [PubMed]
- 36.Carli, G., Cecchi, L., Stebbing, J., Parronchi, P. & Farsi, A. Is asthma protective against COVID‐19? Allergy (2020). [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 37.Jackson, D. et al. Association of respiratory allergy, asthma and expression of the SARS-CoV-2 receptor, ACE2. J. Allergy Clin. Immunol. (2020). [DOI] [PMC free article] [PubMed]
- 38.Jian, L. et al. Perspective: COVID-19, implications of nasal diseases and consequences for their management. J. Allergy Clin. Immunol. (2020). [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 39.Patel, T. R., Teitcher, J. E., Tajudeen, B. A. & Revenaugh, P. C. Disparate nasopharyngeal and tracheal COVID-19 diagnostic test results in a patient with a total laryngectomy. Otolaryngol.—Head Neck Surg. (2020). [DOI] [PubMed]
- 40.Gallo, O. et al. SARS-CoV-2 in upper and lower airway samples of a laryngectomized patient: new insights and many lessons. Oral Oncol. 107, 104841 (2020). [DOI] [PMC free article] [PubMed]
- 41.Daly, J. L. et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. bioRxiv. (2020). [DOI] [PMC free article] [PubMed]
- 42.Roy S, et al. Multifaceted role of neuropilins in the immune system: potential targets for immunotherapy. Front. Immunol. 2017;8:1228. doi: 10.3389/fimmu.2017.01228. 5641316, 29067024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. 1:CAS:528:DC%2BB3cXps1Srurs%3D, 7066521, 32159775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tu Y, et al. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N. Engl. J. Med. 2020;383:494–496. doi: 10.1056/NEJMc2016321. 32492294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vukkadala, N., Qian, Z. J., Holsinger, F. C., Patel, Z. M., & Rosenthal, E. COVID‐19 and the otolaryngologist: preliminary evidence‐based review. Laryngoscope130, 2537–2543 (2020). 10.1002/lary.28672. [DOI] [PubMed]
- 46.Workman, A. D. et al. Endonasal instrumentation and aerosolization risk in the era of COVID‐19: simulation, literature review, and proposed mitigation strategies. Int. Forum Allergy Rhinol. 10, 798–805 (2020). [DOI] [PubMed]
- 47.Zhang N, Van Crombruggen K, Gevaert E, Bachert C. Barrier function of the nasal mucosa in health and type‐2 biased airway diseases. Allergy. 2016;71:295–307. doi: 10.1111/all.12809. 1:CAS:528:DC%2BC28XitF2ksbg%3D, 26606240. [DOI] [PubMed] [Google Scholar]
- 48.Weitnauer M, Mijošek V, Dalpke AH. Control of local immunity by airway epithelial cells. Mucosal. Immunol. 2016;9:287–298. doi: 10.1038/mi.2015.126. 1:CAS:528:DC%2BC2MXhvFeqsbfF, 26627458. [DOI] [PubMed] [Google Scholar]
- 49.Hellings PW, Steelant B. Epithelial barriers in allergy and asthma. J. Allergy Clin. Immunol. 2020;145:1499–1509. doi: 10.1016/j.jaci.2020.04.010. 7270816, 32507228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ooi EH, Wormald PJ, Tan LW. Innate immunity in the paranasal sinuses: a review of nasal host defenses. Am. J. Rhinol. 2008;22:13–19. doi: 10.2500/ajr.2008.22.3127. 18284853. [DOI] [PubMed] [Google Scholar]
- 51.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. 1:CAS:528:DC%2BD1cXksFygurs%3D, 7100821, 19079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McAuley JL, et al. The cell surface mucin MUC1 limits the severity of influenza A virus infection. Mucosal Immunol. 2017;10:1581–1593. doi: 10.1038/mi.2017.16. 1:CAS:528:DC%2BC2sXkvFans7Y%3D, 28327617. [DOI] [PubMed] [Google Scholar]
- 53.Lu, W. et al. Elevated MUC1 and MUC5AC mucin protein levels in airway mucus of critical ill COVID‐19 patients. J. Med. Virol. (2020). [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 54.Kuek, L. E. & Lee, R. J. First contact: the role of respiratory cilia in host-pathogen interactions in the airways. Am. J. Physiol-Lung Cell Mol. Physiol. (2020). [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 55.Sims AC, et al. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 2005;79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. 1:CAS:528:DC%2BD28XhslOntA%3D%3D, 1316022, 16306622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang Y, et al. Distinct stem/progenitor cells proliferate to regenerate the trachea, intrapulmonary airways and alveoli in COVID-19 patients. Cell Res. 2020;30:705–707. doi: 10.1038/s41422-020-0367-9. 1:CAS:528:DC%2BB3cXht1yqsLbP, 7325636, 32606347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jochems SP, et al. Novel analysis of immune cells from nasal microbiopsy demonstrates reliable, reproducible data for immune populations, and superior cytokine detection compared to nasal wash. PloS One. 2017;12:e0169805. doi: 10.1371/journal.pone.0169805. 5249128, 28107457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panda SK, Colonna M. Innate lymphoid cells in mucosal immunity. Front. Immunol. 2019;10:861. doi: 10.3389/fimmu.2019.00861. 1:CAS:528:DC%2BC1MXhvVSlsrrF, 6515929, 31134050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu HY, Nguyen HH, Russell MW. Nasal lymphoid tissue (NALT) as a mucosal inductive site. Scand. J. Immunol. 1997;46:506–513. doi: 10.1046/j.1365-3083.1997.d01-159.x. 1:STN:280:DyaK1c%2FlsFeksA%3D%3D, 9393634. [DOI] [PubMed] [Google Scholar]
- 60.Gallo O, Bani D, Rucci L, Fini Storchi O. Does the epithelium play a central role in the immune function of rhinopharyngeal tonsils? An immunohistochemical and ultrastructural study. Int J. Ped. Otolaryngol. 1991;22:219–222. doi: 10.1016/0165-5876(91)90076-n. 1:STN:280:DyaK38%2FptFWgsA%3D%3D. [DOI] [PubMed] [Google Scholar]
- 61.Gallo O, Bani D, Fini Storchi O. Intraepithelial lymphocyte subpopulations and dendritic accessory cells in normal and hypertrophic adenoids. Laryngoscope. 1994;104:869–873. doi: 10.1288/00005537-199407000-00017. 7517484. [DOI] [PubMed] [Google Scholar]
- 62.Heritage PL, Underdown BJ, Arsenault AL, Sinder DP, Mc Dermott MR. Comparison of the murine nasal-associated lymphoid tissue and Peyer's patches. Am. J. Respir. Crit. Care Med. 1997;156:1256–1262. doi: 10.1164/ajrccm.156.4.97-03017. 1:STN:280:DyaK1c%2FhtF2qtw%3D%3D, 9351630. [DOI] [PubMed] [Google Scholar]
- 63.Lehtinen MJ, et al. Nasal microbiota clusters associate with inflammatory response, viral load, and symptom severity in experimental rhinovirus challenge. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-29793-w. 6065324, 30061588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rose MA, Zielen S, Baumann U. Mucosal immunity and nasal influenza vaccination. Exp. Rev. Vaccines. 2012;11:595–607. doi: 10.1586/erv.12.31. 1:CAS:528:DC%2BC38Xos1Sltbs%3D. [DOI] [PubMed] [Google Scholar]
- 65.Tay, M.Z., Poh, C.M., Rénia, L. et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 20, 363–374 (2020). 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed]
- 66.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008;8:911–922. doi: 10.1038/nri2436. 1:CAS:528:DC%2BD1cXhsVWlsb7P, 7097711, 18989317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. 1:CAS:528:DC%2BB3cXjsValtbk%3D, 7164771, 32085846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matricardi, P. M., Dal Negro, R. W. & Nisini, R. The first, holistic immunological model of COVID‐19: implications for prevention, diagnosis, and public health measures. Pediatr. Allergy Immunol. 10.1111/pai.13271 (2020). [DOI] [PMC free article] [PubMed]
- 69.Madsen HO, et al. Interplay between promoter and structural gene variants control basal serum level of mannan-binding protein. J. Immunol. 1995;155:3013–3020. 1:CAS:528:DyaK2MXnvFCit7s%3D, 7673719. [PubMed] [Google Scholar]
- 70.Eisen DP, Minchinton RM. Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin. Infect. Dis. 2003;37:1496–1505. doi: 10.1086/379324. 1:CAS:528:DC%2BD2cXpslGm, 14614673. [DOI] [PubMed] [Google Scholar]
- 71.Zhang H, et al. Association between mannose-binding lectin gene polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;192:1355–1361. doi: 10.1086/491479. 1:CAS:528:DC%2BD2MXhtFKktbvJ, 16170752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ip WE, et al. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;191:1697–1704. doi: 10.1086/429631. 1:CAS:528:DC%2BD2MXkvVWisLo%3D, 15838797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, et al. A single asparagine-linked glycosylation site of the severe acute respiratory syndrome coronavirus spike glycoprotein facilitates inhibition by mannose-binding lectin through multiple mechanisms. J. Virol. 2010;84:8753–8764. doi: 10.1128/JVI.00554-10. 1:CAS:528:DC%2BC3cXht1CmsLbK, 2919028, 20573835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tomaiuolo R, et al. Activity of mannose‐binding lectin in centenarians. Aging Cell. 2012;11:394–400. doi: 10.1111/j.1474-9726.2012.00793.x. 1:CAS:528:DC%2BC38XosFentLg%3D, 22239660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holodick NE, Rodríguez-Zhurbenko N, Hernández AM. Defining natural antibodies. Front Immunol. 2017;8:872. doi: 10.3389/fimmu.2017.00872. 5526850, 28798747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oriol R, Mollicone R, Couillin P, Dalix AM, Candelier JJ. Genetic regulation of the expression of ABH and Lewis antigens in tissues. APMIS. 1992;100(Supp 27):28–38. [PubMed] [Google Scholar]
- 77.Galili U. Evolution in primates by “Catastrophic-selection” interplay between enveloped virus epidemics, mutated genes of enzymes synthesizing carbohydrate antigens, and natural anti-carbohydrate antibodies. Am. J. Phys. Anthropol. 2019;168:352–363. doi: 10.1002/ajpa.23745. 30578545. [DOI] [PubMed] [Google Scholar]
- 78.Guillon P, et al. Inhibition of the interaction between the SARS-CoV spike protein and its cellular receptor by anti-histo-blood group antibodies. Glycobiology. 2008;18:1085–1093. doi: 10.1093/glycob/cwn093. 1:CAS:528:DC%2BD1cXhsVChtLrJ, 18818423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng Y, Cheng G, Chui CH, Lau FY. ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA. 2005;293:1450–1451. doi: 10.1001/jama.293.12.1450-c. 1:CAS:528:DC%2BD2MXis1OhsrY%3D, 15784866. [DOI] [PubMed] [Google Scholar]
- 80.Gérard C, Maggipinto G, Minon JM. COVID‐19 & ABO blood group: another viewpoint. Br. J. Haematol. 2020;190:e93–e94. doi: 10.1111/bjh.16884. 32453863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ellinghaus, D. et al. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 10.1056/NEJMoa2020283 (2020). [DOI] [PMC free article] [PubMed]
- 82.Narasimhan PB, Marcovecchio P, Hamers AAJ, Hedrick CC. Nonclassical monocytes in health and disease. Annu. Rev. Immunol. 2019;37:439–456. doi: 10.1146/annurev-immunol-042617-053119. 1:CAS:528:DC%2BC1MXosVSrtbc%3D, 31026415. [DOI] [PubMed] [Google Scholar]
- 83.Peruzzi, B. et al. Quantitative and qualitative alterations of circulating myeloid cells and plasmacytoid DC in SARS-CoV-2 infection. Immunology 10.1111/imm.13254 (2020). [DOI] [PMC free article] [PubMed]
- 84.Sánchez-Cerrillo, I. et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DC and inflammatory transitional and nonclassical monocytes. J. Clin. Investig. 10.1172/JCI140335 (2020). [DOI] [PMC free article] [PubMed]
- 85.Giamarellos-Bourboulis EJ, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. 1:CAS:528:DC%2BB3cXnvFehsb4%3D, 7172841, 32320677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuri-Cervantes L, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. 1:CAS:528:DC%2BB3cXhs1anu7zN, 7402634, 32669287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee H, et al. Phenotype and function of nasal dendritic cells. Mucosal Immunol. 2015;8:1083–1098. doi: 10.1038/mi.2014.135. 1:CAS:528:DC%2BC2MXisVyis7o%3D, 4532662, 25669151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. 1:CAS:528:DC%2BB3cXps1yjtrk%3D, 7227586, 32416070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bastard, P. et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science 10.1126/science.abd4585 (2020). [DOI] [PMC free article] [PubMed]
- 90.Zhang, Q. et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 10.1126/science.abd4570 (2020). [DOI] [PMC free article] [PubMed]
- 91.Mazzoni, A. et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Investig. 138554. 10.1172/JCI138554 (2020). [DOI] [PMC free article] [PubMed]
- 92.Radermecker C, et al. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 2020;217:e20201012. doi: 10.1084/jem.20201012. 1:CAS:528:DC%2BB3cXhvFKrsb3E, 7488867, 32926097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Long QX, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. 1:CAS:528:DC%2BB3cXot1aktLk%3D, 32350462. [DOI] [PubMed] [Google Scholar]
- 94.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. 1:CAS:528:DC%2BB3cXhtVOmu73N, 7237901, 32473127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Long QX, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. 1:CAS:528:DC%2BB3cXhtF2qtbrM, 32555424. [DOI] [PubMed] [Google Scholar]
- 96.Peng, Y. et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 10.1038/s41590-020-0782-6 (2020). [DOI] [PMC free article] [PubMed]
- 97.Ni L, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. 1:CAS:528:DC%2BB3cXptlCjtb4%3D, 7196424, 32413330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sekine, T. et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 183, 158–168 (2020). 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed]
- 99.Le Bert N, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. 32668444. [DOI] [PubMed] [Google Scholar]
- 100.Braun, J. et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 10.1038/s41586-020-2598-9 (2020). [DOI] [PubMed]
- 101.Weiskopf D, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. 1:CAS:528:DC%2BB3cXhsVCmtbjP, 7319493, 32591408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mazzoni, A. et al. Cell-mediated and humoral adaptive immune responses to SARS-CoV-2 are lower in asymptomatic than symptomatic COVID-19 patients. Eur. J. Immunol. 10.1002/eji.202048915 (2020). [DOI] [PubMed]
- 103.Hua X, et al. Nasal priming by a murine coronavirus provides protective immunity against lethal heterologous virus pneumonia. JCI Insight. 2018;3:e99025. doi: 10.1172/jci.insight.99025. 6124400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pizzorno A, et al. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep. Med. 2020;1:100059. doi: 10.1016/j.xcrm.2020.100059. 7373044, 32835306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morens DM, Daszak P, Taubenberger JK. Escaping Pandora's box—another novel coronavirus. N. Engl. J. Med. 2020;382:1293–1295. doi: 10.1056/NEJMp2002106. 1:CAS:528:DC%2BB3cXmslOnsLw%3D, 32101660. [DOI] [PubMed] [Google Scholar]
- 106.De Wit E, Van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523. doi: 10.1038/nrmicro.2016.81. 7097822, 27344959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fehr, A. R., Channappanavar, R. & Perlman, S. Middle East respiratory syndrome: emergence of a pathogenic human coronavirus. Annu. Rev. Med.68, 387–399 (2017). [DOI] [PMC free article] [PubMed]
- 108.Gu J, Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 2007;170:1136–1147. doi: 10.2353/ajpath.2007.061088. 1:CAS:528:DC%2BD2sXkvFymsLw%3D, 1829448, 17392154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Memish, Z. A., Perlman, S., Van Kerkhove, M. D. & Zumla, A. Middle East respiratory syndrome. Lancet (2020). [DOI] [PMC free article] [PubMed]
- 110.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. 1:CAS:528:DC%2BD3sXpt1GlsLs%3D, 7095016, 14647384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu G, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. 1:CAS:528:DC%2BC3sXhtVKhtbfK, 7095341, 23831647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Du L, et al. The spike protein of SARS-CoV-a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. 1:CAS:528:DC%2BD1MXhsFeqs7Y%3D, 2750777, 19198616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lu G, Wang Q, Gao GF. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. 1:CAS:528:DC%2BC2MXhtV2iu73E, 7125587, 26206723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Glowacka I, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. 3126222, 21325420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shirato K. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87:12552–12561. doi: 10.1128/JVI.01890-13. 1:CAS:528:DC%2BC3sXhvVygsrjM, 3838146, 24027332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Walls AC, et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. 1:CAS:528:DC%2BC1MXisVWgu74%3D, 6751136, 30712865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mahallawi WH. Case report: detection of the Middle East respiratory syndrome coronavirus (MERS-CoV) in nasal secretions of a dead human. J. Taibah Univ. Med. Sci. 2018;13:302–304. doi: 10.1016/j.jtumed.2017.07.004. 31435338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li R, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. 1:CAS:528:DC%2BB3cXosVSjur0%3D, 7164387, 32179701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hoxha, A. et al. Asymptomatic SARS-CoV-2 infection in Belgian long-term care facilities. Lancet Infect. Dis. 10.1016/S1473-3099(20)30560-0 (2020). [DOI] [PMC free article] [PubMed]
- 120.Sattler A. et al. SARS-CoV-2 specific T-cell responses and correlations with COVID-19 patient predisposition. J. Clin. Investig. 10.1172/JCI140965 (2020). [DOI] [PMC free article] [PubMed]
- 121.Maggi E, Canonica GW, Moretta L. COVID-19: unanswered questions on immune response and pathogenesis. J. Allergy Clin. Immunol. 2020;146:18–22. doi: 10.1016/j.jaci.2020.05.001. 1:CAS:528:DC%2BB3cXpvFegsLs%3D, 7205667, 32389590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China [published correction appears in Lancet. 2020 Jan 30] Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. 1:CAS:528:DC%2BB3cXhs1Kqu7c%3D, 7159299, 31986264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou, F. et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet395, 1038 (2020). [DOI] [PMC free article] [PubMed]
- 124.Vultaggio A, et al. Prompt predicting of early clinical deterioration of moderate-to-severe COVID-19 patients: usefulness of a combined score using IL-6 in a preliminary study. J. Allergy Clin. Immunol. Pract. 2020;S2213-2198:30611–30615. doi: 10.1016/j.jaip.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aziz, M., Fatima, R. & Assaly, R. Elevated interleukin-6 and severe COVID-19: a meta-analysis. J. Med. Virol. 10.1002/jmv.25948 (2020). [DOI] [PMC free article] [PubMed]
- 126.Xu X, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl Acad. Sci. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. 1:CAS:528:DC%2BB3cXhtFCit7zM, 7245089, 32350134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vannucchi, A. M. et al. Compassionate use of JAK1/2 inhibitor ruxolitinib for severe COVID-19: a prospective observational study. Leukemia 1–13 (2020). [DOI] [PMC free article] [PubMed]
- 128.Kaneko, N. et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell S0092-S8674:31067-9. 10.1016/j.cell.2020.08.025 (2020). [DOI] [PMC free article] [PubMed]
- 129.Ray D, Yung R. Immune senescence, epigenetics and autoimmunity. Clin. Immunol. 2018;196:59–63. doi: 10.1016/j.clim.2018.04.002. 1:CAS:528:DC%2BC1cXns1Whurk%3D, 6548177, 29654845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Masters AR, Haynes L, Su DM, Palmer DB. Immune senescence: significance of the stromal microenvironment. Clin. Exp. Immunol. 2017;187:6–15. doi: 10.1111/cei.12851. 1:STN:280:DC%2BC2szhtVemuw%3D%3D, 27529161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moderbacher, C. R. et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 10.1016/j.cell.2020.09.038 (2020). [DOI] [PMC free article] [PubMed]
- 132.Fujihashi K, Kiyono H. Mucosal immunosenescence: new developments and vaccines to control infectious diseases. Trends Immunol. 2009;30:334–343. doi: 10.1016/j.it.2009.04.004. 1:CAS:528:DC%2BD1MXotF2gtrY%3D, 19540811. [DOI] [PubMed] [Google Scholar]
- 133.Koga T, et al. Evidence for early aging in the mucosal immune system. J. Immunol. 2000;165:5352–5359. doi: 10.4049/jimmunol.165.9.5352. 1:CAS:528:DC%2BD3cXnvVOntLk%3D, 11046071. [DOI] [PubMed] [Google Scholar]
- 134.Hagiwara Y, et al. Protective mucosal immunity in aging is associated with functional CD4+ T cells in nasopharyngeal-associated lymphoreticular tissue. J. Immunol. 2003;170:1754–1762. doi: 10.4049/jimmunol.170.4.1754. 1:CAS:528:DC%2BD3sXht12it7c%3D, 12574339. [DOI] [PubMed] [Google Scholar]
- 135.Lechien JR, et al. Loss of smell and taste in 2013 European patients with mild to moderate COVID-19. Ann. Intern. Med. 2020 doi: 10.7326/M20-2428. 7505100, 10.7326/M20-2428. 32449883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Whitcroft KL, Hummel T. Olfactory dysfunction in COVID-19: diagnosis and management. JAMA. 2020;323:2512–2514. doi: 10.1001/jama.2020.8391. 1:CAS:528:DC%2BB3cXhtlSnt7%2FE, 32432682. [DOI] [PubMed] [Google Scholar]
- 137.Cooper KW, et al. COVID-19 and the chemical senses: supporting players take center stage. Neuron. 2020;107:219–233. doi: 10.1016/j.neuron.2020.06.032. 1:CAS:528:DC%2BB3cXhtlKgu7%2FM, 7328585, 32640192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Menni C, et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020;26:1037–1040. doi: 10.1038/s41591-020-0916-2. 1:CAS:528:DC%2BB3cXptVKhsrg%3D, 7751267, 32393804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Spinato G, et al. Alterations in smell or taste in mildly symptomatic outpatients with SARS-CoV-2 infection. JAMA. 2020;323:2089–2090. doi: 10.1001/jama.2020.6771. 1:CAS:528:DC%2BB3cXhtVGrs7bI, 7177631, 32320008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li YC, Bai WZ, Hashikawa T. The neuroinvasive potential of SARS‐CoV2 may play a role in the respiratory failure of COVID‐19 patients. J. Med. Virol. 2020;92:552–555. doi: 10.1002/jmv.25728. 1:CAS:528:DC%2BB3cXntlelsr4%3D, 32104915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Solomon, I. H. et al. Neuropathological features of Covid-19. N. Engl. J. Med. 10.1056/NEJMc2019373 (2020). [DOI] [PMC free article] [PubMed]
- 142.Brann, D. H. et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfaory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. eabc5801 (2020). [DOI] [PMC free article] [PubMed]
- 143.Boscolo-Rizzo P, et al. Evolution of altered sense of smell or taste in patients with mildly symptomatic COVID-19. JAMA Otolaryngol.—Head. Neck Surg. 2020;146:729–732. doi: 10.1001/jamaoto.2020.1379. 7333173, 32614442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Locatello, L. G., Maggiore, G., Bruno, C., Trotta, M. & Gallo, O. An integrated care strategy for the follow-up of patients with COVID-19–associated chemosensory dysfunction. Otolaryngol.—Head Neck Surg. 0194599820950716 (2020). [DOI] [PubMed]
- 145.Cantuti-Castelvetri L. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science eabd2985. 10.1126/science.abd2985 (2020). [DOI] [PMC free article] [PubMed]
- 146.Martin JE, et al. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine. 2008;26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. 1:CAS:528:DC%2BD1cXhtlyhtr7N, 2612543, 18824060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Folegatti, P. M. et al. Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect. Dis.20, 816–826 (2020). [DOI] [PMC free article] [PubMed]
- 148.Krammer, F. SARS-CoV-2 vaccines in development. Nature 10.1038/s41586-020-2798-3 (2020). [DOI] [PubMed]
- 149.Heaton, P. M. The Covid-19 vaccine-development multiverse. NEJM (2020). [DOI] [PMC free article] [PubMed]
- 150.Folegatti PM, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. 1:CAS:528:DC%2BB3cXhsVaku7fE, 7445431, 32702298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hassan AO, et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753. doi: 10.1016/j.cell.2020.06.011. 1:CAS:528:DC%2BB3cXhtFyktLzJ, 7284254, 32553273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zost, S. J., Gilchuk, P., Case, J. B. et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020). 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed]
- 153.Shin MD, et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020;15:646–655. doi: 10.1038/s41565-020-0737-y. 1:CAS:528:DC%2BB3cXhsVSju73L, 32669664. [DOI] [PubMed] [Google Scholar]
- 154.Zhang S, Caldeira-Dantas S, Smith CJ, Snyder CM. Persistent viral replication and the development of T-cell responses after intranasal infection by MCMV. Med. Microbiol. Immunol. 2019;208:457–468. doi: 10.1007/s00430-019-00589-7. 1:CAS:528:DC%2BB3cXltl2qsb8%3D, 6635021, 30848361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Richard M, et al. Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-14626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Giavina-Bianchi P, Aun MV, Takejima P, Kalil J, Agondi RC. United airway disease: current perspectives. J. Asthma Allergy. 2016;9:93. doi: 10.2147/JAA.S81541. 1:CAS:528:DC%2BC1cXmsFGhsbY%3D, 4872272, 27257389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Joo S, et al. Critical role of TSLP-responsive mucosal dendritic cells in the induction of nasal antigen-specific IgA response. Mucosal. Immunol. 2017;10:901–911. doi: 10.1038/mi.2016.103. 1:CAS:528:DC%2BC28XitVajs7nK, 27924821. [DOI] [PubMed] [Google Scholar]
- 158.Zhao J, et al. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. 1:CAS:528:DC%2BC28XpsVGltLw%3D, 4917442, 27287409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mueller SN, Mackay LK. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 2016;16:79–89. doi: 10.1038/nri.2015.3. 1:CAS:528:DC%2BC2MXitVCqsLbE, 26688350. [DOI] [PubMed] [Google Scholar]
- 160.Pizzolla A, et al. Resident memory CD8+ T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci. Immunol. 2017;2:eaam6970. doi: 10.1126/sciimmunol.aam6970. 28783656. [DOI] [PubMed] [Google Scholar]
- 161.Zens KD, Chen JK, Farber DL. Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight. 2016;1:e85832. doi: 10.1172/jci.insight.85832. 4959801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.O'Hara JM, et al. Generation of protective pneumococcal-specific nasal resident memory CD4+ T cells via parenteral immunization. Mucosal. Immunol. 2020;13:172–182. doi: 10.1038/s41385-019-0218-5. 31659300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.van Doremalen, N., Lambe, T., Spencer, A. et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586, 578–582 (2020). 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed]
- 164.Hassan, A. O. et al. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell183 169–184.e13 (2020). 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed]