Abstract
Background
Patients with coeliac disease (CD) commonly report a variety of adverse symptoms to gluten, but descriptions of the symptomatic response in the literature may have been confounded by the presence of food components such as fermentable carbohydrates (FODMAPs) causing symptoms of irritable bowel syndrome independent of gluten. In recent unmasked and masked low FODMAP gluten challenge studies in small groups of treated CD patients, nausea and vomiting were shown to be the key symptoms associated with serum interleukin (IL)-2 release. Our objective was to utilise a large and diverse cohort of people with CD undertaking a standardised gluten food challenge to characterise the demographic, genetic and clinical factors influencing the severity and timing of acute gluten reactions and IL-2 production.
Methods
A total of 295 adults treated for CD were observed for 6 h after an unmasked food challenge consisting of 10 g vital wheat gluten (low in FODMAPs) in 100 ml water. Assessments included patient-reported outcomes, serum IL-2 and adverse events. Responses were analysed according to patient characteristics, HLA-DQ genotype, duodenal histology and response to a second gluten challenge.
Results
Peak symptom severity was at 3 h (median severity 5/10). Peak IL-2 was at 4 h (median 4 pg/ml, range undetectable to 1028 pg/ml). Older age, older age at diagnosis, HLA-DQ2.5 positivity and homozygosity for HLA-DQB1*02 were each significantly associated with IL-2 elevations after gluten. Patients positive for HLA-DQ2.5, DQ8, DQ2.2 or DQ7 showed elevated IL-2 after gluten. Patient factors were not significantly associated with severity of digestive symptoms, but symptoms were correlated to one another and serum IL-2. Gluten challenge after 5 months caused more vomiting and higher IL-2 levels, but responses correlated with the first.
Conclusions
Gluten-induced symptoms and cytokine release is common in adults with treated CD. Age, genetics and previous response to gluten predict these acute reactions to gluten challenge. Structured symptom assessment and serum IL-2 after standardised gluten challenge may inform on patient diagnosis, the role of gluten in symptomatology and the need for adjunctive treatment.
Trial registration
ClinicalTrials.gov, NCT03644069 Registered 21 May 2018.
Keywords: Coeliac disease, Gluten, Food challenge, Cytokine, Patient-reported outcome
Background
Coeliac disease (CD) is a prevalent, small intestinal immune-mediated enteropathy precipitated by exposure to dietary gluten in genetically predisposed people [1, 2]. Patients with CD commonly report adverse symptoms associated with gluten ingestion [3], but there are few studies that rigorously control for confounding effects to enable accurate gluten-related symptomatology to be recorded. These include nocebo effects, where patient’s negative expectations to potential gluten exposure influence their symptoms and the presence of non-gluten dietary components such as fermentable carbohydrates (FODMAPs) that can cause symptoms of irritable bowel syndrome (IBS) independent of gluten [4, 5]. In several recent gluten food challenge studies in small groups of patients with treated CD, unmasked and also double-blind, sham-controlled gluten challenges designed to be low in FODMAPs cause significant worsening of nausea, sometimes with vomiting, within 2-h and peak at 3 to 4 h, but rarely caused diarrhoea [6, 7]. This acute symptomatic reaction to gluten in patients with treated CD is linked to significant concomitant elevations in serum cytokines, which are not observed in individuals without CD or those with self-reported non-CD gluten sensitivity [7–9].
Serum levels of interleukin (IL)-2 increase earliest and most in the serum cytokine signature observed after gluten [10]. An almost identical set of digestive symptoms and systemic cytokine release occurs after patients with treated CD receive an intradermal injection of gluten peptides corresponding to immuno-dominant HLA-DQ2.5-restricted epitopes for gluten-specific CD4+ T cells [8, 11]. Collectively, these findings implicate an unexpected leading role for activated gluten-specific CD4+ T cells in the early symptoms after gluten. However, more detailed understanding requires studies in substantially larger cohorts that are not focused on patients positive for HLA-DQ2.5 alone.
The objective of the present study was to characterise demographic, genetic and clinical factors influencing the severity of symptoms and IL-2 release after gluten in a large cohort of patients having a standardised, low FODMAP gluten challenge. The data provide a roadmap outlining the potential clinical role for bolus gluten challenge in the care of patients with and without CD following a gluten-free diet.
Methods
Study design
The study design is shown in Fig. 1. Here, we report data collected from 295 CD individuals during the screening (pre-treatment) phase of the “RESET CeD Study”, a phase 2 clinical trial of an investigational treatment for CD (ClinicalTrials.gov identifier: NCT03644069). Full details of the RESET CeD Study design are described elsewhere [6]. There were 39 sites in the USA, Australia and New Zealand. All participants undertook an unmasked (i.e. open-label) gluten food challenge coupled with an assessment of IL-2 and symptom responses. A later component of the RESET CeD Study following the administration of the experimental drug/placebo involved CD participants completing a double-blind food challenge of gluten at the same dose and format as undertaken during screening or a matched non-gluten placebo, each low in FODMAPs and consumed 2 weeks apart. The data from the 36 CD patients who received placebo drug helps control for nocebo effects associated with open-label administration of gluten and is reported elsewhere [6].
Fig. 1.
Study outline
Participants and study procedures
All patients gave written, informed consent prior to undergoing any trial-related procedures. Full eligibility criteria are listed elsewhere [6]. To enter screening, patients’ ages were between 18 and 70 years, they had documentary evidence of duodenal villous atrophy and elevated CD-specific serology and had followed a gluten-free diet for at least 1 year. Patients were excluded from the analysis if their HLA-DQ genotype was subsequently found to be incompatible with CD (absence of HLA-DQA1*05, HLA-DQA1*03, HLA-DQB1*02 or HLA-DQB1*03), or no symptom data had been recorded after screening gluten challenge. Unmasked gluten challenge was as described by Tye-Din et al. [7]. The composition was as described by Daveson et al. [6]. The food challenge, taken as a bolus in 100 ml water over 2 min, comprised 10 g vital wheat gluten (Gem of the West® Vital Wheat Gluten, Manildra Group, Auburn, NSW, Australia). The protein content of the vital wheat gluten was 78 g per 100 g (Eurofins Microbiology Laboratories Inc). FODMAP content was low, with a total fructan of 0.074 g/serve and a total FODMAP of 0.089 g/serve using reported methods [12] (Monash University, Melbourne, Victoria, Australia).
Clinical assessments
Demographics, medical history and prior and concomitant medications were recorded. Patients completed the characteristics of CD survey described by Tye-Din et al. [7] and the Celiac Dietary Adherence Test (CDAT) [13]. As described elsewhere [7], self-reported symptom severity was recorded within an hour before, and then each hour up to 6 h after gluten challenge using a modified version of the “Celiac Disease Patient-Reported Outcome” (CeD PRO®) tool, and using a global gastrointestinal symptom survey (GloSS). Adverse events during the 24 h after gluten challenge were separately recorded and graded by investigators according to Common Terminology Criteria for Adverse Events, version 4.03.
Laboratory assessments
CD serology (QUANTA Lite® “R h-tTG IgA” and “gliadin IgG II (DGP)”, INOVA Diagnostics, San Diego, CA, USA) was performed by ARUP (Salt Lake City, UT). HLA-DQ genotype was evaluated by the UCLA Immunogenetics Center (Los Angeles, CA). The blood for IL-2 was allowed to clot for 30 min and, within 3 h, aliquots of the serum were frozen and stored at − 60 to − 80 °C. Serum IL-2 was measured by a commercial V-PLEX electrochemiluminescence assay as previously described [6]. The lower level of quantitation was 0.5 pg/ml.
Statistical analyses
We utilised non-parametric tests as we did not assume a normal distribution for the data. The Mann-Whitney test was used to compare two datasets of continuous variables that were unpaired, and for paired data, the Wilcoxon signed rank test and Spearman’s coefficient of correlation were used. Kruskal-Wallis testing was used to compare groups of three or more continuous variables in the same participant. Associations between categorical variables were tested by the Fisher exact test, chi-square test or Cochran-Armitage test depending on whether two variables, more than two variables or an order-dependent trend in variables was present, respectively. All statistical tests were 2-tailed, and a p value of 0.05 was taken as significant. Benjamini-Hochberg’s false discovery rate method was used to adjust p values for multiple comparisons where appropriate. Summary and significance statistics were computed using Graphpad Prism V7.0d. The overall clinical severity of a reaction to gluten for an individual from the least (“no symptoms”) to the worst (“very severe symptoms”) was deemed to be their worst recorded hourly GloSS descriptor over the 6-h interval after gluten.
Results
Patient characteristics
In total, data for 295 patients who had gluten challenges were analysed, which represented 77% of the 383 volunteers who were screened, and 95% of all the patients who were eligible and had a screening gluten challenge over the period from August 2018 to March 2019. Disposition of patients is shown in Additional file 1: Figure S1. The gluten challenge was on the first day of screening for 214 patients, or between 1 and 31 days after starting the screening for 96 patients. Demographics and characteristics of patients are shown in Table 1.
Table 1.
Patient characteristics
| Mean (SD) age in years | 43 (15) |
| Number (%) of females | 205 (69) |
| Mean (SD) height in centimetres | 169 (11) |
| Mean (SD) body mass in kilogrammes | 79 (19) |
| Mean (SD) body mass index | 28 (13) |
| Number (%) of White, not Hispanic or Latino | 277 (94) |
| Median (interquartile range) age in years at diagnosis of CD | 35 (25–46) |
| Median (interquartile range) years from coeliac diagnosis | 6 (3–10) |
| Number (%) of negative for both CD-specific serologies† | 241 (82) |
| Number (%) of positive for both CD-specific serologies† | 5 (2) |
| Number (%) of IgA deficient (< 7 mg/dl) | 2 (1) |
| Number (%) of positive for HLA-DQ2.5 genotype | 266 (90) |
| Number (%) of negative for HLA-DQ2.5 genotype | 29 (10) |
| Number (%) of recruited in the USA | 126 (43) |
| Number (%) of recruited in Australia | 123 (42) |
| Number (%) of recruited in New Zealand | 46 (16) |
†QUANTA Lite® R h-tTG IgA (normal range 3 U/ml or less) and gliadin IgG II (DGP), INOVA Diagnostics (normal range 19 U or less)
Patient-reported outcomes after gluten challenge
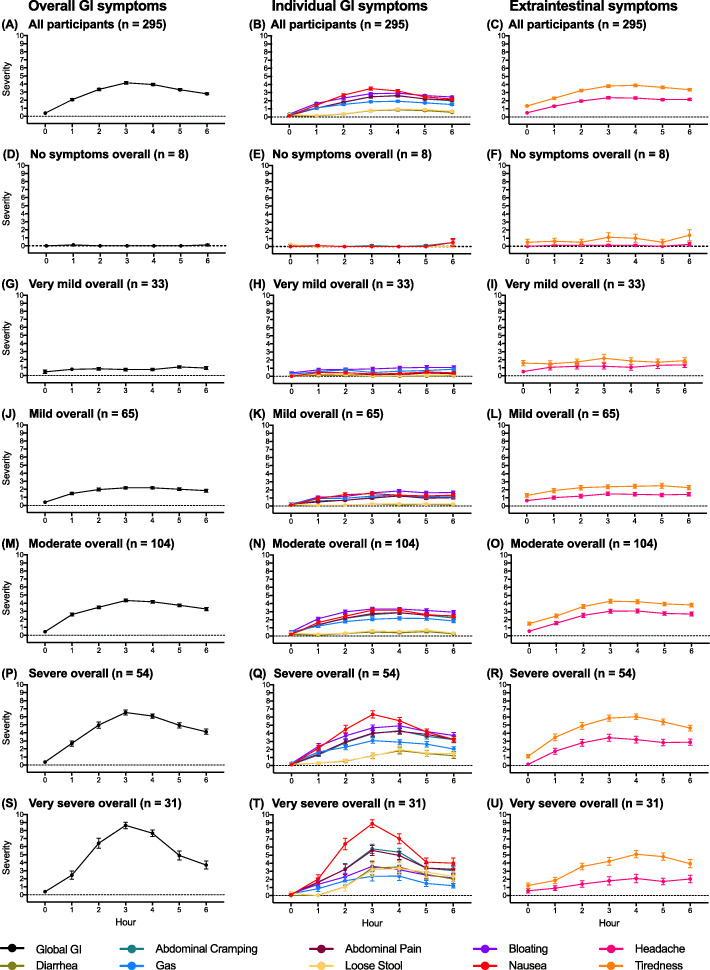
There was a good agreement between verbal descriptor and numerical score for peak hourly GloSS rating (Spearman r = 0.92, p < 10−15, n = 295) (Additional file 1: Table S1). Table 2 shows the distribution of peak GloSS severity and CeD PRO severity for individual symptoms. The overall severity of reactions based on peak GloSS descriptor was moderate and rated 5 on a scale from 0 to 10. Gluten reactions were graded moderate, severe or very severe by almost two thirds of patients. Figure 2 shows the time course for GloSS and CeD PRO ratings for global and individual symptoms, respectively. We again observed that patients’ expectations of symptoms were often not those experienced after gluten challenge (Additional file 1: Tables S2, S3) [6]. Onset was within 1 h, and peak overall severity of moderate, severe and very severe reactions was clearly at 3 h. Nausea, cramping and abdominal pain, bloating and tiredness steadily worsened as the overall severity of reactions increased, but diarrhoea and loose stool were only prominent in very severe reactions. Additional file 1: Table S4 shows that the peak severity of all individual symptoms assessed by the CeD PRO was correlated with one another. The strongest correlations were between the most prominent symptoms such as nausea, pain, cramping, bloating and tiredness. Table 3 shows how often patients rated individual symptoms among their three worst during the 6 h after gluten challenge. Nausea, tiredness and abdominal pain/cramping were each rated among the worst symptoms for almost half of the patients, but nausea and vomiting were clearly most often the worst symptoms for patients having very severe reactions.
Table 2.
Peak severity of symptoms up to 6 h after consuming gluten
| Gluten reaction severity‡ | All | ||||||
|---|---|---|---|---|---|---|---|
| None | Very mild | Mild | Moderate | Severe | Very severe | ||
| Number of patients (%) | 8 (3) | 33 (11) | 65 (22) | 104 (35) | 54 (18) | 31 (10) | 295 |
| Median peak hourly severity of digestive symptoms rated each hour in the Global Symptom Survey† (interquartile range) | |||||||
| Absolute value | 0 (0–0) | 2 (1–2) | 2 (1–2) | 6 (5–6) | 8 (8–8) | 10 (9–10) | 5 (3–8) |
| Change from baseline | 0 (0–0) | 1 (1–2) | 1 (1–2) | 5 (5–6) | 8 (7–8) | 10 (9–10) | 5 (3–7) |
| Median peak hourly symptom score in modified CeD PRO† (interquartile range) | |||||||
| Nausea | 0 (0–0) | 1 (0–2) | 2 (1–4) | 5 (3–6) | 8 (7–9) | 10 (10–10) | 4 (2–8) |
| Diarrhoea | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–1) | 1 (0–6) | 8 (1–10) | 0 (0–3) |
| Loose stool | 0 (0–0) | 0 (0–0) | 0 (0–1) | 0 (0–3) | 2 (0–8) | 8 (2–10) | 0 (0–3) |
| Cramping | 0 (0–1) | 1 (0–1) | 2 (1–3) | 4 (2–5) | 7 (4–8) | 8 (6–9) | 4 (1–6) |
| Pain | 0 (0–1) | 1 (0–1) | 2 (1–3) | 4 (3–5) | 6 (5–8) | 8 (6–9) | 4 (1–6) |
| Tiredness | 2 (0–3) | 2 (1–3) | 3 (2–5) | 6 (4–7) | 7 (7–8) | 6 (5–8) | 5 (3–7) |
| Bloating | 0 (0–0) | 2 (1–2) | 3 (2–3) | 5 (3–6) | 7 (5–7) | 5 (3–7) | 4 (2–6) |
| Gas | 0 (0–0) | 1 (1–2) | 2 (1–3) | 3 (2–4) | 4 (2–6) | 4 (1–7) | 2 (1–4) |
| Headache | 0 (0–0) | 1 (0–3) | 2 (0–4) | 4 (2–6) | 3 (1–8) | 2 (1–6) | 3 (1–5) |
‡Peak overall severity was the worst hourly descriptor rating reported by patients over 6 h after gluten challenge in the Global Symptom Survey for the question, “Overall, how would you rate the severity of your digestive symptoms in the past 1 h? (e.g. feeling or being sick, abdominal pain or cramps, feeling bloated, having loose stools or passing gas)”
†Whole number rating scale from 0 for no symptoms to 10 for worst possible symptoms
Fig. 2.
Six-hour time series after gluten for patient-reported outcome scores (modified CeD PRO and Global Symptom Survey). Profiles are for all patients (a–c) and for patients whose worst global gastrointestinal score over 6 h was “no symptoms” (d–f), “mild symptoms” (g–i), “very mild symptoms” (j–l), “moderate symptoms” (m–o) and “severe symptoms” (p–r), “very severe symptoms” (s–u). Points indicate mean ± standard error of the mean for scores rated 0 to 10
Table 3.
Number (%) of participants reporting a symptom as among their three most troubling symptoms after gluten challenge according to peak reaction severity
| Peak severity of global gastrointestinal symptoms (GloSS)‡ | Totalb | ||||||
|---|---|---|---|---|---|---|---|
| Nonea | Very milda | Milda | Moderatea | Severea | Very severea | ||
| Total patients | 8 (3) | 33 (11) | 65 (22) | 104 (35) | 54 (18) | 31 (11) | 295 (100) |
| Nausea | 1 (13) | 5 (15) | 29 (45) | 54 (52) | 32 (60) | 21 (72) | 142 (49) |
| Tiredness | 3 (38) | 16 (48) | 32 (50) | 50 (49) | 24 (45) | 3 (10) | 128 (44) |
| Abdominal pain/cramps | 1 (13) | 8 (24) | 24 (38) | 48 (47) | 24 (45) | 13 (45) | 118 (41) |
| Bloating | 0 | 15 (45) | 22 (52) | 43 (42) | 15 (28) | 1 (3) | 107 (37) |
| Headache | 1 (13) | 14 (42) | 24 (38) | 44 (43) | 16 (30) | 4 (14) | 103 (36) |
| Vomiting | 0 | 0 | 1 (2) | 13 (13) | 24 (45) | 26 (90) | 64 (22) |
| Gas | 0 | 8 (24) | 25 (39) | 21 (20) | 6 (11) | 0 | 60 (21) |
| Diarrhoea/loose stool | 0 | 1 (3) | 5 (8) | 12 (12) | 11 (21) | 14 (48) | 43 (15) |
| None | 5 (63) | 1 (3) | 0 | 1 (1) | 0 | 0 | 7 (2) |
| Others | 0 | 2 (6) | 3 (5) | 1 (1) | 0 | 0 | 6 (2) |
Participants’ responses to Q3 in the Global Symptom Survey, 6 h after gluten challenge, “What symptom/s overall have troubled you the most today, since having the food challenge?” Please select a maximum of 3 symptoms: A. none, B. abdominal pain/cramps, C. vomiting, D. nausea, E. diarrhoea/loose stool, F. gas (flatulence), G. bloating, H. tiredness, I. headache, J. others (please specify)
‡The peak overall severity was the worst hourly descriptor rating reported by patients over 6 h after gluten challenge in the Global Symptom Survey for the question, “Overall, how would you rate the severity of your digestive symptoms in the past 1 h? (e.g. feeling or being sick, abdominal pain or cramps, feeling bloated, having loose stools or passing gas)”
aPercentage of patients for each severity subgroup, e.g. none and very mild
bPercentage of all 295 patients
Adverse events after gluten challenge
The timing, prevalence, and severity of adverse events were generally consistent with patient-reported outcomes (Additional file 1: Table S5). Vomiting affected 62 (21%) patients and was the only common adverse event not among those included in the CeD PRO. There were no serious adverse events, but 37 (13%) patients experienced a severe adverse event. Nausea and vomiting accounted for the majority of adverse events graded severe. The timing of onset for adverse events indicated a consistent march of symptoms beginning with nausea.
Serum IL-2 after gluten challenge
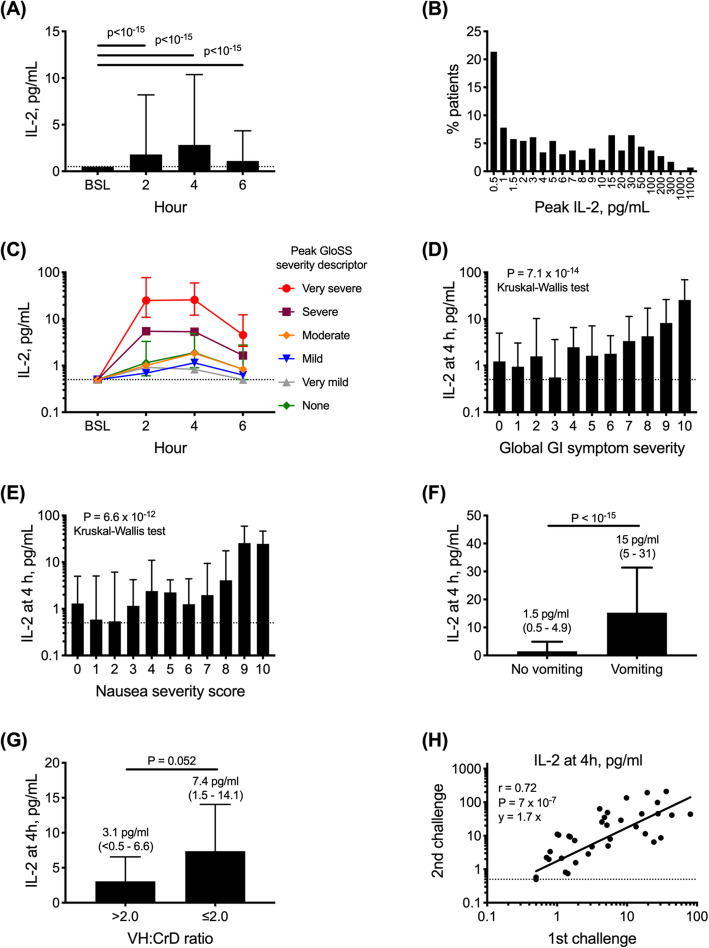
Serum IL-2 significantly increased by 2 h and typically peaked at 4 h (Fig. 3a). Altogether, serum IL-2 levels were elevated at 4 h in 216 (73%) patients who had baseline levels below 0.5 pg/ml. In addition, there were isolated elevations of IL-2 at 2 h in seven patients and at 6 h in four patients. Therefore, the total number of patients who elevated serum IL-2 from baseline to levels above the lower level of quantitation or at least 1.9 times more than the baseline was 229 (78%).
Fig. 3.
Serum IL-2 elevation in 295 patients over 6 h after gluten challenge. a Median and interquartile range of serum IL-2 concentrations at 2-h intervals after gluten; significance tested by Wilcoxon matched-pairs signed rank test. b The frequency distribution of peak serum IL-2 concentrations in patients after gluten. c Median and interquartile range of serum IL-2 concentrations at 4 h and peak hourly numerical rating for global severity of digestive symptoms (GloSS). d Median and interquartile range of serum IL-2 concentrations 4 h and peak hourly severity of nausea. e Median and interquartile range of serum IL-2 concentrations at 4 h and whether patients experienced vomiting after gluten challenge. f Mean and standard error of the mean of serum IL-2 concentrations after gluten according to gluten reaction severity (peak severity descriptor of digestive symptoms rated each hour in the GloSS). g Peak serum IL-2 concentrations after gluten according to villous height to crypt depth ratio (VH:CrD) in the second part duodenal biopsies indicating the presence of villous atrophy (VH:CrD ≤ 2) (n = 39 patients) or absence (n = 13). h Paired serum IL-2 concentrations at 4 h after two separate food challenges with 6-g gluten protein. Data for 36 patients who received placebo treatment during the RESET CeD Study and had both a screening (unmasked) and a second (masked) challenge 20 to 22 weeks later. The gluten challenges used the same format administering the equivalent of 6-g gluten protein in 10-g vital wheat gluten mixed in water
The median peak serum IL-2 concentration was 4 pg/ml (interquartile range 0.7–13), and the median peak fold change from baseline was 7 (interquartile range 1.2–25). The peak serum IL-2 levels were not normally distributed (Fig. 3b). The highest peak serum IL-2 concentration observed was 1075 pg/ml. Two (1%) patients had peak IL-2 levels above 1000 pg/ml, 15 (5%) had peak IL-2 levels above 100 pg/ml and 73 (25%) had peak IL-2 levels above 10 pg/ml, whereas serum IL-2 remained below 0.5 pg/ml in 63 (21%) patients. Serum IL-2 levels were above the lower level of quantitation (0.5 pg/ml) at baseline in five (2%) patients.
Correlation between symptoms and IL-2 after gluten challenge
The serum IL-2 concentration at 4 h was correlated most strongly with peak GloSS score (Additional file 1: Table S4, Fig. 3c), and serum IL-2 time profiles were elevated most and for longer in very severe reactions (Fig. 3d). Nausea was the symptom most correlated with peak serum IL-2 levels (Fig. 3e), but correlations were also found with the severity of abdominal pain, abdominal cramping, bloating, loose stool and diarrhoea (Additional file 1: Table S4). Patients who vomited after gluten had median serum IL-2 levels at 4 h that were ten times higher than other patients (Fig. 3f). Among the 65 patients who did not show any elevation in serum IL-2, their peak GloSS severity was rated very mild in 11 (17%), mild in 21 (31%), moderate in 27 (42%) and severe in 4 (6%). Comparing patients who had no detectable elevation of IL-2 with patients who did, only nausea (median 3 vs 5, p = 0.004), global symptoms (median 4 vs 6, p = 0.0009) and vomiting (1 vs 60, p = 0.000024) were different. Interestingly, all eight patients who reported no symptoms had elevations in serum IL-2 levels from below 0.5 pg/ml at baseline to between 0.8 and 6.9 pg/ml.
Associations between patient characteristics and gluten reactions
Additional file 1: Table S6 lists patient characteristics and their associations with peak global gastrointestinal symptom severity and with peak serum IL-2 after gluten challenge. None of the ten patient characteristics assessed was significantly associated with clinical severity. In contrast, patient age and age at diagnosis were associated with elevation in serum IL-2. In the subgroup of 52 patients described elsewhere who had quantitative histology performed on second part duodenal biopsies [14], patients with villous atrophy (Fig. 3g), or increased intra-epithelial lymphocyte (IEL) density, tended to have higher serum IL-2 levels at 4 h after gluten (IELs > 25 per 100 epithelial cells, n = 42, median 5.3 pg/ml, interquartile range 1.4–12.8 vs IELs ≤ 25, n = 10: 0.9, < 0.5–6.5; p = 0.083 by Mann-Whitney U test).
HLA-DQ genotype was associated with peak serum IL-2 concentration and the proportion of IL-2 responders, but not clinical severity of gluten reactions (Table 4, Additional file 1: Table S6 and S7). The median peak serum IL-2 concentration was more than seven times higher in patients positive for HLA-DQ2.5 than those who were negative (Table 4). Peak serum IL-2 concentrations were also significantly higher in homozygotes for HLA-DQB1*02 than heterozygotes. In the group of patients negative for HLA-DQ2.5, there were individuals positive for HLA-DQ8, HLA-DQ2.2 or HLA-DQ7 alone who elevated serum IL-2 levels and were symptomatic after gluten challenge (Additional file 1: Table S8). Most patients heterozygous for HLA-DQ8 who did not carry HLA-DQ2.2 or DQ7 did not elevate serum IL-2 after gluten. A very severe clinical reaction and strong serum IL-2 response was observed in one patient who was positive for HLA-DQ7 and HLA-DQ6.
Table 4.
HLA-DQ genotype and serum IL-2 after gluten
| Genotype group†, participants (n) | Serum IL-2 concentrations, median (interquartile range) pg/ml | Responders‡ participants, n (%) | |||
|---|---|---|---|---|---|
| 2 h | 4 h | 6 h | Maximum†† | ||
| 1. DQ2.5 positive (266) | 2.2 (< 0.5–8.6) | 3.2 (0.6–10.7) | 1.2 (< 0.5–4.5) | 4.6 (0.8–13.1) | 217 (82%) |
| 2. DQ2.5 negative‡‡ (29) | < 0.5 (< 0.5–1.2) | 0.6 (< 0.5–2.9) | < 0.5 (< 0.5–1.1) | 0.6 (< 0.5–3.6) | 17 (52%) |
| 3. DQ2.5, 2.5 (34) | 2.7 (0.8–8.0) | 3.7 (1.3–9.4) | 1.6 (0.6–4.9) | 5.9 (1.8–10.8) | 29 (85%) |
| 4. DQ2.5, 2.2 (56) | 5.5 (2.5–12.9) | 9.0 (2.6–22.9) | 3.7 (0.9–7.0) | 10.2 (4.7–24.5) | 52 (93%) |
| 5. DQ2.5, 7 (15) | 0.8 (< 0.5–7.4) | 1.8 (< 0.5–5.0) | 0.8 (< 0.5–1.4) | 1.8 (< 0.5–9.0) | 11 (73%) |
| 6. DQ2.5, 8 (36) | 1.1 (< 0.5–6.7) | 2.4 (< 0.5–5.7) | 0.9 (< 0.5–2.8) | 2.7 (0.6–8.5) | 28 (78%) |
| 7. DQ2.5trans (18) | 1.5 (0.7–8.2) | 3.3 (0.9–12.5) | 1.5 (0.6–6.9) | 3.3 (0.9–12.5) | 16 (89%) |
| 8. DQ2.5cis, X (107) | 1.4 (< 0.5–6.3) | 1.9 (< 0.5–8.1) | 0.8 (< 0.5–3.0) | 2.5 (0.6–10.4) | 81 (76%) |
†HLA-DQA and DQB alleles for the following genotype groups: 1. DQA1*05 and DQB1*02 both present; 2. absent either or both DQA1*05 and DQB1*02; 3; homozygous both DQA1*05 and DQB1*02; 4. DQA1*02,05, and DQB1*02 homozygous; 5. DQA1*05 homozygous and DQB1*02,03; 6. DQA1*03,05 and DQB1*02,03; 7. DQA1*02,05 and DQB1*02,03; 8. Other patients with DQA1*05 and DQB1*02
‡Responders defined by maximum serum IL-2 concentration at least 0.5 pg/ml
††Mann-Whitney test false discovery rate-adjusted p values for six comparisons were significant for the presence of DQ2.5 (group 1 vs 2), p = 0.004; DQB1*02 gene dose (groups 5, 6, 7 and 8 vs 3 and 4), p = 0.0009; p values were not significant for DQA1*05 gene dose (groups 4, 6, 7 and 8 vs 3 and 5), DQ2.5 homozygotes vs DQ2.5, 2.2 (group 3 vs 4), DQ2.5cis, X vs DQ2.5trans (group 7 vs 8) and for DQ2.5,8 vs DQ2.5cis, X (group 7 vs 8)
‡‡See Table S8 for genotypes, symptoms and IL-2 responses of HLA-DQ2.5-negative patients
Second gluten challenge after 5 months
A group of 36 placebo-treated patients had a masked gluten during the treatment period of the RESET CeD Study [6], which was between 20 and 22 weeks after their screening gluten challenge. Although the compositions of the two gluten challenges were identical, the second gluten challenge more often caused vomiting (16, 44% vs 8, 22%; p = 0.047, Fisher exact test), and serum IL-2 levels at 4 h were higher (median 10.1, 3.0–43 pg/ml vs 4.6, 1.3–17.8; p = 0.0013 Wilcoxon signed rank test). Individual patients’ serum IL-2 levels at 4 h after the first and second challenges were, however, closely correlated (Fig. 3h), and all but one patient who vomited after the first gluten challenge also vomited after the second (odds ratio [OR] 14.8; p = 0.012, Fisher exact test). A more detailed symptom analysis was not possible because patient-reported outcomes were assessed only after 1 day following the second gluten challenge [6].
Discussion
This is the largest study to characterise symptoms and cytokine release following acute gluten exposure in CD. Due to its large sample and participant diversity, this study provides novel insights into the relation between gluten-mediated symptoms and immune activation, and the influence of patient factors. For the first time, we provide functional evidence for the influence of HLA-DQB*02 gene dose on in vivo immune activation after oral gluten ingestion. We also provide functional data supporting population studies that HLA-DQ7 should be considered among genotypes conveying susceptibility to CD [15, 16].
The present report replicates and greatly extends findings from our earlier studies involving smaller, more selected patient cohorts [6–8]. Here, we show that the severity of individual digestive symptoms was strongly correlated with one another and with elevations in serum IL-2. The kinetics of serum IL-2 levels matched the changing severity of early digestive symptoms, especially nausea. Some symptoms in the CeD PRO including gas, headache and tiredness were not correlated with serum IL-2 levels and, therefore, may be more strongly influenced by non-gluten factors, such as a nocebo effect.
In this study, cytokine release was reproducible after successive gluten challenges, and responses were closely correlated for individual patients. Therefore, it is likely that in patients with treated CD who inadvertently ingest gluten, the small intestinal mucosa, the gut-associated lymphoid organs and extra-intestinal organs are regularly exposed to elevated concentrations of pro-inflammatory cytokines. Systemic cytokine release after gluten may offer an explanation for some of the extra-intestinal manifestations reported for CD.
Fever, hypotension and hypoxia are cardinal features of the cytokine release syndromes associated with systemically administered agents [17], but these adverse events were absent or rare after gluten. This study shows that a single one-off gluten exposure transiently elevates serum IL-2 levels to as high as 1000 pg/ml, which is higher than the peak levels after the first dose of an anti-CD3 biologic [18] or with CAR T cell immunotherapy [19].
This study had several limitations. Although the number of patients enrolled in this study was more than eight times larger and more diverse than in previous bolus gluten challenge studies [6–8], our findings may not be relevant to children and adolescents, patients regularly consuming gluten or those without CD who strictly avoid gluten. In particular, participants in this study had volunteered for an experimental immunotherapy trial and may not be representative of patients with CD in the communities where they lived. A potentially significant limitation of this study was also an important strength: the food challenge had a standardised format with a fixed amount of gluten from a single supplier and a confirmed low FODMAP content. Findings may not be the same for different amounts of gluten administered in other formats. Our previous studies, however, indicate that masked and unmasked food challenges with the same amount and format as the present study result in comparable elevations in serum IL-2 [6, 7]. The gluten challenge dose is likely to be much higher than the amount patients with CD following gluten-free diet might be exposed to with accidental or inadvertent exposure. For logistical reasons, we measured a single cytokine in serum to quantify immune activation after gluten. Other cytokines may be useful to complement IL-2, but data available at present do not clearly indicate which other cytokines could serve this role [10]. The present study included all 36 patients reported separately following placebo food challenge in the treatment period of the RESET CeD Study [6].
The relative size and responsiveness of the gluten-reactive CD4+ T cell population, and the level of antigenic stimulation it receives, are an attractive explanation for the severity of symptoms and peak levels of serum IL-2 after gluten challenge [8]. Previously, we reported significant correlations between peak IL-2 levels and the frequency of tetramer-stained gluten-specific effector memory CD4+ T cells in the blood [8]. Factors that were significantly associated with higher serum IL-2 elevation after gluten in the current study included higher IL-2 response after a previous gluten challenge and HLA-DQ genotype being positive for HLA-DQ2.5 and homozygous for HLA-DQB1*02. The data presented here is the first in vivo evidence supporting a HLA-DQ2.5 gene dose effect on the intensity of immune reactivation after patients ingest gluten. CD participants who were HLA-DQ8 (without HLA-DQB1*02) had much lower IL-2 responses after gluten, consistent with the weaker association between this genotype and CD [20, 21]. Future assessment of this uncommon CD subgroup is warranted. Here, we also observed that older patients and older patient age at diagnosis were associated with greater IL-2 elevations. Future studies including children as well as older patients could address this intriguing observation.
The current study showed that gluten challenge with assessments of symptom severity and serum IL-2 may inform the care of patients following a gluten-free diet. For example, in those on a gluten-free diet with an unclear diagnosis of CD, a positive IL-2 result would strongly support a CD diagnosis whilst a negative IL-2 could identify those who need a diagnostic re-evaluation of CD. In confirmed CD, the degree of symptoms and IL-2 rise following a standardised challenge may help stratify the patients who would benefit most from adjunctive pharmaceutical treatments currently under development. Prospective validation to define the clinical utility of this approach is warranted.
Conclusions
Most patients with treated CD experience a characteristic cytokine and symptom response after acute oral gluten challenge. Nausea and vomiting are hallmarks for moderate or more severe cytokine release reactions after gluten. Bolus gluten challenge with IL-2 assessments has the potential to be integrated into clinical practice to evaluate the diagnosis and management of patients on gluten-free diets.
Supplementary Information
Additional file 1: Table S1. Number (%) of participants using a qualitative descriptor and corresponding numerical score for peak global digestive symptom severity (GloSS) after gluten challenge. Table S2. Patients’ ability to predict their most troubling symptoms after gluten. Table S3. Indication for testing, worst symptoms at diagnosis, and worst symptom expected after gluten exposure on gluten free diet, number (%). Table S4. Correlations between peak severity of symptoms and IL-2 at 4 hours after gluten. Table S5. Adverse events within 24 h after gluten challenge. Table S6. Symptom severity and IL-2 after gluten according to patients’ characteristics. Table S7. Peak symptom severity after gluten according to HLA-DQ genotype. Table S8. Symptoms and serum IL-2 after gluten in 29 participants negative for HLA-DQ2.5 (DQA1*05 and DQB1*02). Table S9. Study Independent Ethics Committees and approvals. Figure S1. Patient disposition.
Acknowledgements
The authors thank the patients and their family members who supported this research. JAT-D is supported by a University of Melbourne Centenary Fellowship and an Australian National Health and Medical Research Council Investigator Grant.
‡The RESET CeD Study Group:
A. Adams (AB Clinical Trials, Las Vegas, NV, USA), J. Andrews (Royal Adelaide Hospital, Adelaide, SA, AUS), C. Behrend (Grand Teton Research Group, Idaho Falls, ID, USA), G. Brown (Alfred Hospital, Melbourne, VIC, AUS), S. Chen Yi Mei (Eastern Health-Box Hill Hospital, Box Hill, VIC, AUS), A. Coates (Gastroenterology Associates of Western Michigan, PLC d.b.a. West Michigan Clinical Research Center, Wyoming, MI, USA), A.J. Daveson (Coral Sea Clinical Research Institute, North Mackay, QLD, AUS, and The Wesley Hospital - The Wesley Research Institute, Auchenflower, QLD, AUS), A. DiMarino (Thomas Jefferson University Hospital, Philadelphia, PA, USA), H. Ee (Department of Gastroenterology, Sir Charles Gairdner Hospital, Nedlands, WA, AUS), D. Elliott (University Of Iowa, Iowa City, IA, USA), R. Epstein (ActivMed Practices & Research, Inc., Portsmouth, NH, USA), B. Feyen (PMG Research of McFarland Clinic, Ames, IA, USA), R. Fogel (Clinical Research Institute of Michigan, Chesterfield, MI, USA), K. Friedenberg (Great Lakes Gastroenterology Research, Mentor, OH, USA), R. Gearry (Department of Medicine, University of Otago, Christchurch, NZ), M. Gerdis (Drug Trials America, Hartsdale, NY, USA), M. Goldstein (Long Island Gastrointestinal Research Group, Great Neck, NY, USA), V. Gupta (Alliance Medical Research LLC, Lighthouse Point, FL, USA), R. Holmes (PMG Research of Winston-Salem, LLC, Winston-Salem, NC, USA), G. Holtmann (The University of Queensland - Princess Alexandra Hospital, Woolloongabba, QLD, AUS), S. Idarraga (Allegiance Research Specialists, Wauwatosa, WI, USA), G. James (Digestive Health Research, LLC, Hermitage, TN, USA), T. King (Auckland Clinical Studies Limited, Auckland, NZ), T. Klein (Heartland Research Associates, LLC, Wichita, KS, USA), S. Kupfer (The University of Chicago Medical Center, Chicago, IL, USA), B. Lebwohl (Celiac Disease Center at Columbia University, New York City, NY, USA), J. Lowe (Advanced Research Institute, Ogden, UT, USA), J. Murray (Mayo Clinic, Rochester, MN, USA), E. Newton (Omega Medical Research, Warwick, RI, USA), D. Quinn (P3 Research Limited, Wellington, NZ), D. Radin (Stamford Therapeutics Consortium, Stamford, CT, USA), T. Ritter (Texas Digestive Disease Consultants, Southlake, TX, USA), H. Stacey (Diablo Clinical Research, Inc., Walnut Creek, CA, USA), C. Strout (Coastal Carolina Research Center, Mount Pleasant, SC, USA), R. Stubbs (P3 Research Limited, Havelock North, NZ), S. Thackwray (Clinical Trials Centre - University of the Sunshine Coast, Sippy Downs, QLD, AUS), V. Trivedi (PMG Research, Inc., d/b/a PMG Research of Piedmont Healthcare, Statesville, NC, USA), J. A. Tye-Din (The Royal Melbourne Hospital - The Walter And Eliza Hall Institute of Medical Research, Parkville, VIC, AUS), J. Weber (Center for Digestive Health, Troy, MI, USA), and S. Wilson (Ocean State Clinical Research Partners, Lincoln, RI, USA)
Abbreviations
- CDAT
Celiac Dietary Adherence Test
- CeD PRO®
Celiac Disease Patient-Reported Outcome
- FODMAP
Fermentable oligo-, di-, monosaccharides and polyols
- GloSS
Global Gastrointestinal Symptom Survey
- HLA
Human leukocyte antigen
Authors’ contributions
RPA, KET and LJW designed the study. JAT-D, AJMD, KEG, HLH, KMN, KET and the RESET CeD Study Group conducted clinical studies. GG and RPA provided data integration and analysis. RPA wrote the first draft of the manuscript and prepared the tables and figures. All authors contributed to the manuscript revision and approved the text, tables and figures. The authors made the decision to submit the manuscript for publication and vouch for the accuracy of data and analyses and for the fidelity of this report to the trial protocol. RPA had full access to all data in the study and had final responsibility for the decision to submit for publication. The authors read and approved the final manuscript.
Funding
ImmusanT was the sponsor, funded and designed the RESET CeD Study that included gluten challenge during screening reported here. ImmusanT allowed full access to data from the RESET CeD Study, but did not play a role in the analysis of data reported here.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was conducted in accordance with the International Conference on Harmonisation for Good Clinical Practice. Local ethics committees approved the study (AUS: Melbourne Health Human Research Ethics Committee, Parkville VIC; Bellberry Limited, Eastwood SA; Uniting Care Health, Eastwood SA; NZ: Central Health and Disability Ethics Committee, Thorndon, Wellington; US: Copernicus Group IRB, Cary NC; Columbia University IRB, New York NY; Mayo Clinic IRB, Rochester MN and The University of Chicago IRB, Chicago IL) See Additional file 1: Table S9 for additional details. Written, informed consent was obtained from each patient included in the study.
Consent for publication
Not applicable
Competing interests
GG, KEG, HLH, KMN, KET, LJW and RPA were former employees of ImmusanT, Inc. JAT-D and AJMD served as advisors to ImmusanT, Inc. RPA is the inventor of patents, owned or licenced by ImmusanT, Inc., relating to the diagnostic application of gluten challenge.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jason A. Tye-Din and A. James M. Daveson contributed equally to this work.
Contributor Information
Robert P. Anderson, Email: banderson99133@gmail.com
on behalf of the RESET CeD Study Group:
A. Adams, J. Andrews, C. Behrend, G. Brown, S. Chen Yi Mei, A. Coates, A. J. Daveson, A. DiMarino, H. Ee, D. Elliott, R. Epstein, B. Feyen, R. Fogel, K. Friedenberg, R. Gearry, M. Gerdis, M. Goldstein, V. Gupta, R. Holmes, G. Holtmann, S. Idarraga, G. James, T. King, T. Klein, S. Kupfer, B. Lebwohl, J. Lowe, J. Murray, E. Newton, D. Quinn, D. Radin, T. Ritter, H. Stacey, C. Strout, R. Stubbs, S. Thackwray, V. Trivedi, J. A. Tye-Din, J. Weber, and S. Wilson
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12916-020-01828-y.
References
- 1.Ludvigsson JF, Leffler DA, Bai JC, Biagi F, Fasano A, Green PH, Hadjivassiliou M, Kaukinen K, Kelly CP, Leonard JN, et al. The Oslo definitions for coeliac disease and related terms. Gut. 2013;62(1):43–52. doi: 10.1136/gutjnl-2011-301346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caio G, Volta U, Sapone A, Leffler DA, De Giorgio R, Catassi C, Fasano A. Celiac disease: a comprehensive current review. BMC Med. 2019;17(1):142. doi: 10.1186/s12916-019-1380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silvester JA, Graff LA, Rigaux L, Walker JR, Duerksen DR. Symptomatic suspected gluten exposure is common among patients with coeliac disease on a gluten-free diet. Aliment Pharmacol Ther. 2016;44(6):612–619. doi: 10.1111/apt.13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skodje GI, Sarna VK, Minelle IH, Rolfsen KL, Muir JG, Gibson PR, Veierod MB, Henriksen C, Lundin KEA. Fructan, rather than gluten, induces symptoms in patients with self-reported non-celiac gluten sensitivity. Gastroenterology. 2018;154(3):529–539. doi: 10.1053/j.gastro.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 5.Colloca L, Barsky AJ. Placebo and nocebo effects. N Engl J Med. 2020;382(6):554–561. doi: 10.1056/NEJMra1907805. [DOI] [PubMed] [Google Scholar]
- 6.Daveson AJM, Tye-Din JA, Goel G, Goldstein KE, Hand HL, Neff KM, Williams LJ, Truitt KE, Anderson RP, Group RCS Masked bolus gluten challenge low in FODMAPs implicates nausea and vomiting as key symptoms associated with immune activation in treated coeliac disease. Aliment Pharmacol Ther. 2020;51(2):244–252. doi: 10.1111/apt.15551. [DOI] [PubMed] [Google Scholar]
- 7.Tye-Din JA, Daveson AJM, Ee HC, Goel G, MacDougall J, Acaster S, Goldstein KE, Dzuris JL, Neff KM, Truitt KE, et al. Elevated serum interleukin-2 after gluten correlates with symptoms and is a potential diagnostic biomarker for coeliac disease. Aliment Pharmacol Ther. 2019;50(8):901–910. doi: 10.1111/apt.15477. [DOI] [PubMed] [Google Scholar]
- 8.Goel G, Tye-Din JA, Qiao SW, Russell AK, Mayassi T, Ciszewski C, Sarna VK, Wang S, Goldstein KE, Dzuris JL, et al. Cytokine release and gastrointestinal symptoms after gluten challenge in celiac disease. Sci Adv. 2019;5(8):eaaw7756. doi: 10.1126/sciadv.aaw7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tye-Din JA, Skodje GI, Sarna VK, Dzuris JL, Russell AK, Goel G, Wang S, Goldstein KE, Williams LJ, Sollid LM, et al. Cytokine release after gluten ingestion differentiates coeliac disease from self-reported gluten sensitivity. United European Gastroenterol J. 2020;8(1):108–118. doi: 10.1177/2050640619874173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goel G, Daveson AJM, Hooi CE, Tye-Din JA, Wang S, Szymczak E, Williams LJ, Dzuris JL, Neff KM, Truitt KE, et al. Serum cytokines elevated during gluten-mediated cytokine release in coeliac disease. Clin Exp Immunol. 2020;199(1):68–78. doi: 10.1111/cei.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goel G, King T, Daveson AJ, Andrews JM, Krishnarajah J, Krause R, Brown GJE, Fogel R, Barish CF, Epstein R, et al. Epitope-specific immunotherapy targeting CD4-positive T cells in coeliac disease: two randomised, double-blind, placebo-controlled phase 1 studies. Lancet Gastroenterol Hepatol. 2017;2(7):479–493. doi: 10.1016/S2468-1253(17)30110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varney J, Barrett J, Scarlata K, Catsos P, Gibson PR, Muir JG. FODMAPs: food composition, defining cutoff values and international application. J Gastroenterol Hepatol. 2017;32(Suppl 1):53–61. doi: 10.1111/jgh.13698. [DOI] [PubMed] [Google Scholar]
- 13.Leffler DA, Dennis M, Edwards George JB, Jamma S, Magge S, Cook EF, Schuppan D, Kelly CP. A simple validated gluten-free diet adherence survey for adults with celiac disease. Clin Gastroenterol Hepatol. 2009;7(5):530–536. doi: 10.1016/j.cgh.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 14.Daveson AJM, Popp A, Taavela J, Goldstein KE, Isola J, Truitt KE, Maki M, Anderson RP, Group RCS Baseline quantitative histology in therapeutics trials reveals villus atrophy in most patients with coeliac disease who appear well controlled on gluten-free diet. GastroHep. 2020;2(1):22–30. [Google Scholar]
- 15.Tinto N, Cola A, Piscopo C, Capuano M, Galatola M, Greco L, Sacchetti L. High frequency of haplotype HLA-DQ7 in celiac disease patients from South Italy: retrospective evaluation of 5,535 subjects at risk of celiac disease. PLoS One. 2015;10(9):e0138324. doi: 10.1371/journal.pone.0138324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tye-Din JA, Cameron DJ, Daveson AJ, Day AS, Dellsperger P, Hogan C, Newnham ED, Shepherd SJ, Steele RH, Wienholt L, et al. Appropriate clinical use of human leukocyte antigen typing for coeliac disease: an Australasian perspective. Intern Med J. 2015;45(4):441–450. doi: 10.1111/imj.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee DW, Santomasso BD, Locke FL, Ghobadi A, Turtle CJ, Brudno JN, Maus MV, Park JH, Mead E, Pavletic S, et al. ASTCT Consensus Grading for Cytokine Release Syndrome and Neurologic Toxicity Associated with Immune Effector Cells. Biol Blood Marrow Transplant. 2019;25(4):625–638. doi: 10.1016/j.bbmt.2018.12.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumgart DC, Lowder JN, Targan SR, Sandborn WJ, Frankel MB. Transient cytokine-induced liver injury following administration of the humanized anti-CD3 antibody visilizumab (HuM291) in Crohn’s disease. Am J Gastroenterol. 2009;104(4):868–876. doi: 10.1038/ajg.2008.138. [DOI] [PubMed] [Google Scholar]
- 19.Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, Bartido S, Stefanski J, Taylor C, Olszewska M, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5(177):177ra138. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Toma A, Goerres MS, Meijer JW, Pena AS, Crusius JB, Mulder CJ. Human leukocyte antigen-DQ2 homozygosity and the development of refractory celiac disease and enteropathy-associated T-cell lymphoma. Clin Gastroenterol Hepatol. 2006;4(3):315–319. doi: 10.1016/j.cgh.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Karell K, Louka AS, Moodie SJ, Ascher H, Clot F, Greco L, Ciclitira PJ, Sollid LM, Partanen J. Hla types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease. Hum Immunol. 2003;64(4):469–477. doi: 10.1016/s0198-8859(03)00027-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Number (%) of participants using a qualitative descriptor and corresponding numerical score for peak global digestive symptom severity (GloSS) after gluten challenge. Table S2. Patients’ ability to predict their most troubling symptoms after gluten. Table S3. Indication for testing, worst symptoms at diagnosis, and worst symptom expected after gluten exposure on gluten free diet, number (%). Table S4. Correlations between peak severity of symptoms and IL-2 at 4 hours after gluten. Table S5. Adverse events within 24 h after gluten challenge. Table S6. Symptom severity and IL-2 after gluten according to patients’ characteristics. Table S7. Peak symptom severity after gluten according to HLA-DQ genotype. Table S8. Symptoms and serum IL-2 after gluten in 29 participants negative for HLA-DQ2.5 (DQA1*05 and DQB1*02). Table S9. Study Independent Ethics Committees and approvals. Figure S1. Patient disposition.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.