Abstract
The endocytic pathway is a common strategy that several highly pathogenic viruses use to enter into the cell. To demonstrate the usefulness of this pathway as a common target for the development of broad-spectrum antivirals, the inhibitory effect of drug compounds targeting endosomal membrane proteins were investigated. This study entailed direct comparison of drug effectiveness against animal and human pathogenic viruses, namely Ebola (EBOV), African swine fever virus (ASFV), and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
A panel of experimental and FDA-approved compounds targeting calcium channels and PIKfyve at the endosomal membrane caused potent reductions of entry up to 90% in SARS-CoV-2 S-protein pseudotyped retrovirus. Similar inhibition was observed against transduced EBOV glycoprotein pseudovirus and ASFV. SARS-CoV-2 infection was potently inhibited by selective estrogen receptor modulators in cells transduced with pseudovirus, among them Raloxifen inhibited ASFV with very low 50% inhibitory concentration. Finally, the mechanism of the inhibition caused by the latter in ASFV infection was analyzed.
Overall, this work shows that cellular proteins related to the endocytic pathway can constitute suitable cellular targets for broad range antiviral compounds.
Keywords: SARS-CoV-2, EBOV, ASFV, Antivirals, PIKfyve, Calcium channels
Graphical abstract
1. Introduction
The worldwide spread of COVID-19 highlights the need for developing efficient therapeutics against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2 (Gil et al., 2020)). The disease was declared a public health emergency and it is having an enormous impact on human health and deep social and economic consequences. There are several international efforts to develop vaccines in many vaccine platforms, some of them undergoing clinical trials (Dagotto et al., 2020). However, the therapeutic arsenal to treat COVID-19 is limited to date, despite the research efforts to develop effective and safe compounds (Gil et al., 2020). Antiviral Remdesivir, first developed against Ebola virus (EBOV), is a consistent therapeutic option in severe COVID-19 patients approved by the FDA (Yin et al., 2020). Although several large drug screens have described antiviral activity for a number of apparently unrelated FDA-approved compounds, it is increasingly evident that a deeper understanding of the inhibitory mechanism of antiviral compounds is needed to find useful and broad-spectrum antivirals (V’Kovski et al., 2020).
SARS-CoV-2 is a positive sense, single-stranded RNA betacoronavirus of a 30 kb genome. It is an enveloped virus, decorated with the surface glycoprotein, the Spike (S) protein. Other structural proteins are envelope protein (E), membrane protein (M), and nucleocapsid protein (N). Protein S interacts with the cellular receptor ACE2 for cell entry, facilitated by S protein processing by human proteases. S-protein priming can be performed by surface transmembrane serine protease 2 (TMPRSS2) (Hoffmann et al., 2020) and endosomal cathepsins (Shang et al., 2020). Unlike SARS-CoV, cell entry of SARS-CoV-2 is preactivated by furin, reducing its dependence on target cell proteases for entry. Furin preactivation of the spike and the high binding affinity of the processed receptor-binding domain (RBD) when exposed, are proposed contributors to the wide spread of the pandemic virus (Shang et al., 2020). New host co-factors are being identified, such as neuropilin (NRP) (Daly et al., 2020) or heparan sulfate (Clausen et al., 2020). Furin cleavage of S-protein generates a sequence in S1 fragment that binds surface receptor NRP and allows entry. SARS-CoV-2 infects most human and some animal cell lines provided that they express SARS-CoV-2 entry limiting factors such as ACE2 (Hoffmann et al., 2020). COVID-19 may start with fever, malaise, and acute pneumonia. Some patients suddenly deteriorate into severe respiratory failure and a multisystemic inflammatory syndrome is associated with immune dysfunction or macrophage activation state characterized by hyper-cytokinemia lymphopenia, and frequent fatalities (Guan et al., 2020).
Ebola virus (EBOV) is another highly pathogenic virus that spread to western countries during the largest epidemics of this disease. Declared by WHO as a public health emergency, it started in 2013 in West Africa and lasted until 2016. Re-emergences in the Democratic Republic of Congo in 2017–2020 evidenced the continuous risk of this disease (Ilunga Kalenga et al., 2019). EBOV infects virtually all organs and cell types except lymphocytes causing an acute clinical syndrome with fever, multiorgan failure, cytokine storm, coagulation alterations, and vascular leakage with very high fatality rates (Hunt et al., 2015). EBOV is a Filovirus, with a negative-sense RNA genome surrounded by a nucleocapsid decorated with the virus glycoprotein (GP). The largest EBOV epidemics boosted research in vaccine platforms and therapeutic compounds. Finally, an approved vaccine (VSV-GP) is being currently used and over 280 thousand people have been immunized so far (Ollmann Saphire, 2020).
On another note, the recent spread of an animal virus, the African Swine Fever virus (ASFV) through Europe and Asia is causing massive economic losses (Karger et al., 2019). In Europe, it has been spreading from east to west and the first cases of ASFV in Germany have been declared since September 2020 near the polish border (FLI web page) (www.fli.de, 2020). ASFV is a double-stranded DNA virus with icosahedral morphology that belongs to the Asfarviridae family (Alonso et al., 2018). This is another deadly virus with a high impact on animal health, causing over 95% mortality in its swine host. It develops a severe acute disease characterized by fever and a hyperinflammatory state. Despite its high socio-economic and environmental burden, a commercial vaccine against ASFV is under development, but not yet available. The search for antiviral drugs should continue until an effective vaccine is developed (Alonso et al., 2013).
An entry route for several viruses, including coronavirus is the endocytic pathway. It is commonly accepted that SARS-CoV infection requires the acidic endosomal environment and is also dependent on endosomal cathepsin L (Mingo et al., 2015, Ou et al., 2020, Wang et al., 2008) but subsequent steps are not yet fully characterized. EBOV enters the cells by clathrin-mediated endocytosis and macropinocytosis (Aleksandrowicz et al., 2011). At endosomes, the EBOV GP undergoes conformational changes due to the action of cathepsins and other endosomal proteases to prime the viral GP for fusion and endosomal exit (Chandran et al., 2005).
ASFV also infects macrophages and Vero cells by endocytosis and its transit through the endocytic pathway has been well characterized. ASFV viral particles undergo disassembly in endosomal compartments that the virus traffics en route to the site of replication. This disassembly relies on the acid pH of late endosomes (Cuesta-Geijo et al., 2012). After the fusion of the viral internal membrane with the endosomal membrane, naked cores are released to the cytoplasm to start replication (Hernaez et al., 2016).
Here, we examined the antiviral activity of a panel of experimental as well as FDA-approved compounds acting on the endosomal membrane that could constitute a potential target against SARS-CoV-2, EBOV, and ASFV because although distant, all these viruses share similar entry pathways. Specifically, tested compounds are known to act as PIKfyve inhibitors or as modifiers of intracellular calcium levels with different degrees of knowledge with respect to their antiviral activity. With the aim to decipher the specific inhibitory mechanisms of some of these compounds, we further analyzed and compared their inhibitory potential.
2. Materials and methods
2.1. Cell culture and viruses
Vero cells (ATCC CCL-81; renal fibroblasts) and Vero E6 (ATCC CRL-1586) were obtained from the American Type Culture Collection and cultured at 37 °C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, and 5–10% heat-inactivated fetal bovine serum (FBS) which was reduced to 2% during viral infection. Baby hamster kidney cells (BHK-21, 12-14-17 MAW, Kerafast, Boston, MA) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 25 μg/ml gentamycin, 2 mM L-glutamine and 10% heat-inactivated fetal bovine serum (FBS). Swine alveolar macrophages were cultured in RPMI medium with 10% swine serum, 2 mM L-glutamine, 50 μM 2-mercaptoethanol, 20 mM Hepes and 30 μg/ml gentamycin.
For compound screening, we used a pseudotyped retrovirus system expressing SARS-CoV-2 S protein or vesicular stomatitis virus (VSV) expressing the Ebola virus (EBOV) GP protein, as it will be below explained.
The first selection of compounds was performed on an infectious recombinant version of the common cold coronavirus 229E, which expresses the green fluorescent protein (GFP) gene (229E-GFP; kindly given by V. Thiel, University of Bern, Switzerland), in Huh-7 Lunet C3 cells, a gift from T. Pietschman, Twincore, Germany. Then, we examined infectivity values by determination of GFP expression by fluorimetry using the SpectraMax iD3 from Molecular Devices.
For ASFV, we used the Vero-adapted ASFV isolate BA71V (Enjuanes et al., 1976) in Vero cells and primary alveolar macrophages. For flow cytometry (FACS) analyses, we used the infectious recombinant ASFV, Bpp30GFP, which expresses the green fluorescent protein (GFP) gene, fused to the promoter of the early viral p30 protein (Barrado-Gil et al., 2017) and the infectious recombinant ASFV, B54ChFP expressing the Cherry fluorescence protein (ChFP) as a fusion protein of viral p54 (Alonso et al., 2013). Preparation of viral stocks, titrations, and infection experiments were carried out as previously described (Enjuanes et al., 1976).
2.2. SARS-2-CoV spike protein-pseudotyped retroviral system
Retroviral particles pseudotyped with SARS-2-CoV spike envelope protein (SARS-2-Spp) were produced in HEK293T cells following a previously described procedure (Mingorance et al., 2014) with materials kindly provided by Dr. F. L. Cosset (INSERM, Lyon) and J. M. Casanovas and J. G. Arriaza (CNB–CSIC) for the S protein cDNA. Particles devoid of envelope glycoproteins were produced in parallel as controls.
For SARS-2-Spp entry experiments, Vero-E6 cells (104 cells/well) were seeded onto 96-well plates the day before. Compounds were diluted in complete media [(DMEM supplemented with 10 mM HEPES, 1× non-essential amino acids (Gibco), 100 U/ml penicillin-streptomycin (Gibco) and 10% Fetal Bovine Serum (heat-inactivated at 56 °C for 30 min)] to achieve the desired final concentration. Compound and vehicle dilutions were applied to the cell cultures 1 h before virus inoculation. Pre-treatment was removed and fresh compound dilutions containing the SARS-2-Spp were used to inoculate the cultures for 24 h, time after which the mixture was replaced with fresh media devoid of inhibitors. Forty-eight hours post-inoculation, cells were lysed for luciferase activity determination using Luciferase Assay System (Promega) and a luminometer. Relative infection values were determined by normalizing the data to the average relative light units detected in the vehicle control cells.
2.3. Ebola glycoprotein-pseudotyped recombinant vesicular stomatitis virus (rVSV-luc system)
rVSV-luc pseudotypes were generated following a published protocol (Whitt, 2010). BHK-21 were transfected to express the Ebola virus Makona glycoprotein (EBOV-GP) (KM233102.1) synthesized and cloned into pcDNA3.1 by GeneArt AG technology (Life Technologies, Regensburg, Germany) or VSV-G with Lipofectamine 3000 following the manufacturer's instructions (Fisher Scientific).
After 24 h, transfected cells were inoculated with a replication-deficient rVSV-luc pseudotype (MOI 1–5 pfu/cell) that contain firefly luciferase instead of the VSV-G open reading frame, rVSVΔG-luciferase (G*ΔG-luciferase, Kerafast) during 1 h at 37 °C.
Next, the inoculum was removed, cells were washed 3 times with phosphate buffered saline (PBS) and finally the medium added. Pseudotyped particles were harvested 24 h and 48 h postinoculation, clarified from cellular debris by centrifugation for 10 min at 1200 r.p.m. and stored at −80 °C. Infectious titers were estimated as tissue culture infectious dose per ml by limiting dilution of each rVSV-luc virus-containing supernatants on Vero E6 cells (African green monkey cells). Luciferase activity was determined by luciferase assay (Luciferase Assay System, Promega, Madison, WI).
2.4. Compounds tested
We tested the inhibitory effect of the following compounds at the doses indicated except as otherwise stated, Tetrandrine (TETR 10 μM), Verapamil (VER 100 μM), Curcumin (CUR 75 μM), Bafilomycin (BAF 200 nM), Apilimod (APL 25 μM), Tamoxifen citrate (TMX 10 μM), Raloxifene hydrochloride (RLX 10 μM), Fulvestrant (ICI 182,780; FUL 100 μM) and β-Estradiol (EST 100 μM) were purchased from Sigma-Aldrich. Bapta-AM (BAP 10 μM) and YM201636 (YM 1 μM) and also controls Hydroxycloroquine (HCL 10 μM) endosomal acidification inhibitor and Teicoplanin (TEI 10 μM), an inhibitor of cathepsin L (Zhou et al., 2016) were purchased from Abcam. Stock solutions were dissolved in DMSO and working solutions were freshly prepared in DMEM 2% fetal bovine serum (FBS) at indicated concentrations. First, we pursued cell viability and cytotoxicity tests of all reagents using the CellTiter 96 Non-radioactive Cell Proliferation Assay (Promega) following the Manufacturer's instructions (Supplementary Fig. S2, Supplementary Fig. S3, Supplementary Fig. S4). We also studied the cytotoxic activity of the organic solvent DMSO. Based on these experiments we selected optimal non-toxic working concentrations for infection assays.
2.5. Flow cytometry analysis
Detection of ASFV infected cells was performed by flow cytometry. Vero cells or swine alveolar macrophages were pretreated with compounds at the indicated concentrations in growth medium for 1 h at 37 °C, followed by infection with recombinants ASFV Bpp30GFP at a moi of 1 pfu/cell for 16 h. Cells were washed with PBS, harvested by trypsinization, and then washed and collected with flow cytometry buffer (PBS, 0.01% sodium azide, and 0.1% bovine serum albumin. In order to determine the percentage of infected cells per condition, 10,000 cells/time point were scored and analyzed in a FACS Canto II flow cytometer (BDSciences). Untreated control infected cultures yielded 80–90% of infected cells from the total cells examined. Infected cell percentages obtained after compounds treatments were normalized to values found in control samples.
2.6. Quantitative real time PCR
DNA from Vero cells treated with the indicated concentrations of inhibitors and infected with ASFV at a moi of 1 pfu/cell for 16 hpi, was purified using the DNAeasy blood and tissue kit (Qiagen). DNA concentration was measured using a Nanodrop spectrophotometer. Untreated- and DMSO-treated cells were used as a control. The qPCR assay used fluorescent hybridisation probes to amplify a region of the p72 viral gene, as described previously (King et al., 2003). The amplification mixture was 300 ng of DNA template added to a final reaction mixture of 20 μl containing 50 pmol sense primers, 50 pmol anti-sense primer, 5 pmol of probe and 10 μl of Premix Ex Taq (2×) (Takara). Positive amplification controls were DNA purified from ASFV virions at different concentrations used as standards. Each sample was included in triplicates and values were normalized to standard positive controls. Reactions were performed using the ABI 7500 Fast Real-Time PCR System (Applied Biosystems) with the following parameters: 94 °C 10 min and 45 cycles of 94 °C for 10 s and 58 °C for 60 s.
2.7. Western blotting
Vero cells were seeded in 6-well plates and infected with ASFV at a moi of 1 pfu/cell in the presence or absence of compounds. At 24 hpi, cells were lysed in Laemmli sample buffer and equivalent amounts of sample were electrophoresed in sodium dodecyl sulfate polyacrylamide gels and transferred to a nitrocellulose membrane (Bio-Rad). Non-specific antibody binding sites were blocked with skimmed milk in phosphate-buffered saline (PBS) and then incubated with anti p30 monoclonal antibody (kindly given by Dr. Escribano, INIA) 1/1000, anti p72 monoclonal antibody, clone 1BC11 (Ingenasa) 1:1000, anti-estrogen receptor α clone 60C (Millipore) 1:2000 or anti-tubulin (Sigma) 1:2000. Bound antibodies were detected with HRP (Horseradish peroxidase)-conjugated secondary antibody and the ECL system (Amersham) using a ChemidocXRS imaging system (Bio-Rad). Band densitometry was performed with Image Lab software (BioRad) and data were normalized to control values.
2.8. Filipin staining
To detect cholesterol accumulation, we used fluorescent filipin (Sigma). Filipin is a naturally fluorescent polyene antibiotic that binds to cholesterol but not to esterified sterols. Thus, it is useful for detecting free cholesterol. Vero cells plated on glass coverslips were treated with inhibitors at the indicated concentrations for 16 h. After fixation with 4% paraformaldehyde, cells were washed with PBS, incubated with 50 mg/ml filipin in PBS for 1 h, and washed again with PBS, after which coverslips were mounted and imaged on a fluorescence microscope.
2.9. Endosome acidification assays
Endosomal acidification was assessed with LysoTracker Red (Molecular Probes) as a probe for low-pH organelles. Cells were pretreated with DMSO or inhibitors for 2 h at 37 °C and then incubated with 75 nM LysoTracker Red (Molecular Probes) for an additional 1 h. Cells were fixed with 4% paraformaldehyde and analyzed by confocal fluorescence microscopy. Also, HCL (10 μM) and BAF (200 nM) were used as controls when indicated.
2.10. Indirect immunofluorescence
Vero cells were grown on coverslips at 60% confluence and fixed with Methanol at −20 °C for 5 min or with 4% paraformaldehyde (PFA) in PBS for 12 min. When PFA was used, after washing in PBS, cells were permeabilized for 15 min with PBS 0.1% Triton X-100. For staining, fixed cells were incubated with anti-tubulin (clon 6-11B-1 Sigma) 1:2000, anti-acetylated tubulin (Sigma) 1:1000, or monoclonal anti-body against major ASFV capsid protein p72 (Ingenasa) 1:1000 1 h a 37 °C. A secondary antibody conjugated to Alexa fluor-488 was purchased from Molecular Probes. Coverslips were mounted onto glass slides using ProLong Gold (Invitrogen) and examined in a TCS SPE confocal or a conventional vertical DRB microscope (Leica).
2.11. Endolysosomal calcium assay
Vero cells grown on coverslips were pre-incubated with 10 μM RLX, 100 μM VER or DMSO as a control for 16 h, and then loaded with 5 μM membrane-permeable calcium indicators Fluo8-AM (Abcam) for 1 h at 37 °C and fixed in 4% paraformaldehyde in PBS for 12 min. Coverslips were mounted onto glass slides using ProLong Gold (Invitrogen) and examined in a TCS SPE confocal microscope (Leica). In experiments to examine fluorescence intensity, Vero cells were grown in wells of MW 96, pre-incubated with 10 μM RLX, 100 μM VER, or DMSO as a control for 16 h, loaded with 5 μM Fluo8-AM and monitored hourly in a TECAN GENios Microplate Reader. When cells were infected, ASFV was added after 1 h treatment with the inhibitors.
2.12. Statistical analysis
The experimental data were analyzed by one-way ANOVA by Graph Pad Prism 6 Software. Bonferroni's correction was applied for multiple comparisons. Values were expressed in graph bars as mean ± SD of at least three independent experiments and a p-value <0.05 was considered statistically significant.
3. Results
3.1. Endosomal calcium channels inhibitors
We screened a collection of experimental as well as FDA-approved compounds acting at the endosomal membrane searching for an antiviral effect. With this aim, we included Tetrandrine (TETR), a specific TPC-1 and -2 endosomal calcium channels inhibitor and also, Verapamil (VER) and other compounds, which are not specific for endosomal channels but modify intracellular calcium levels. Experimental drugs included as controls were Curcumin (CUR) that is a regulator of intracellular calcium release from ER to endosomes (Bardak et al., 2017), and Bapta-AM (BAP), a cell-permeable calcium chelator that modifies intracellular calcium (Tang et al., 2020). In this inhibitor panel, we included some drugs known to inhibit EBOV infection (Kang et al., 2020). We analyzed the effect of Apilimod (APL) and YM201636, as FDA-approved and experimental PIKfyve inhibitors respectively, and finally, the selective estrogen receptor modulators (SERMs) Raloxifen (RLX) and Tamoxifen (TMX) between others. Endosomal phosphatydil-inositols (PtdIns) are crucial molecules in endosomal maturation. PtdIns (3,5)-biphosphate (PtdIns 3,5-P2) is an agonist of TPC channels at the late endosome. Hence, inhibition of the enzyme PIKfyve that is responsible for the biphosphate synthesis could have an impact on the infection. SERMs are molecules with tertiary structures that permit binding to the estrogen receptor (ESR) with an unclear mechanism of action at other cellular levels. Fulvestrant (FUL) was used as it is an antagonist of ESR receptor.
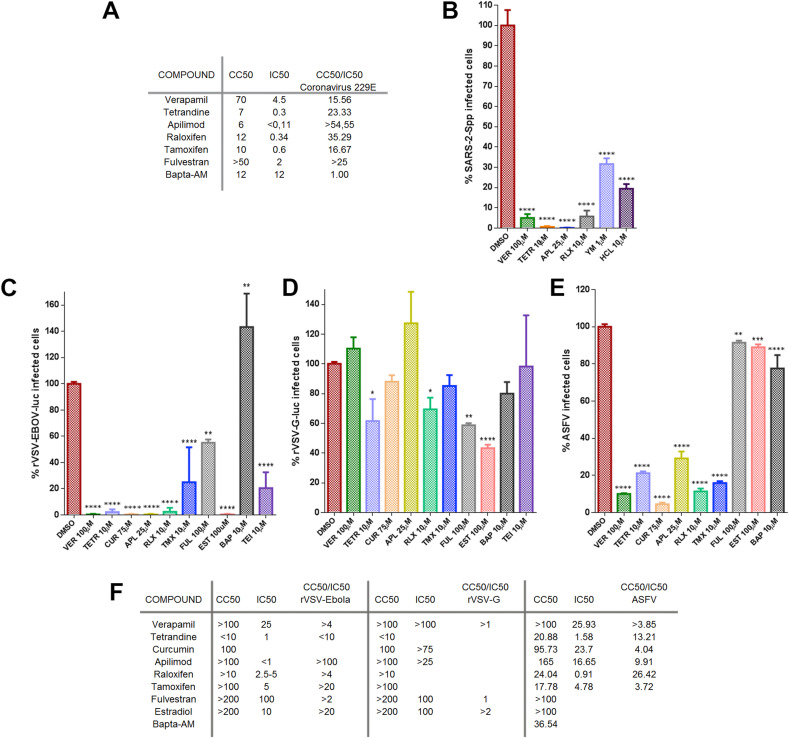
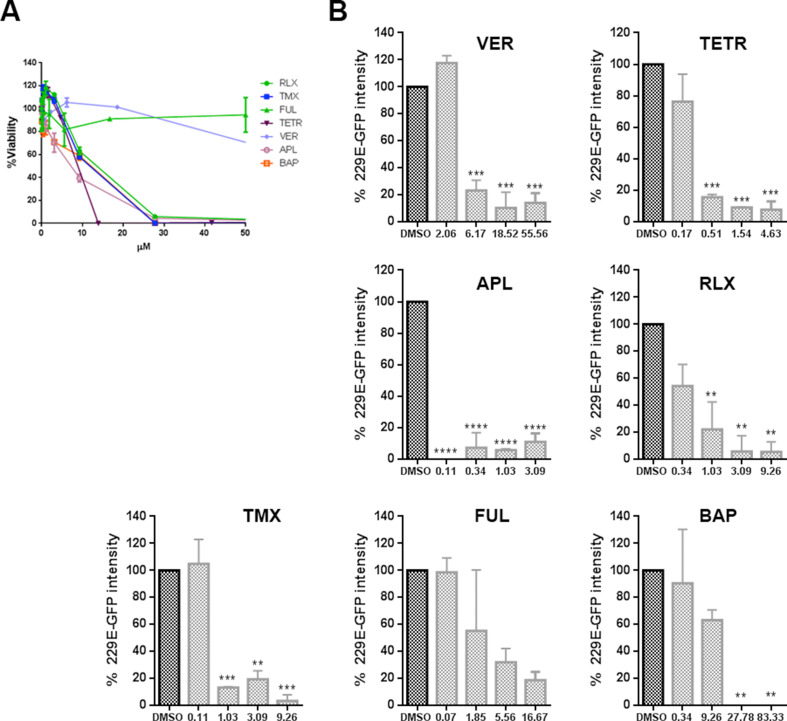
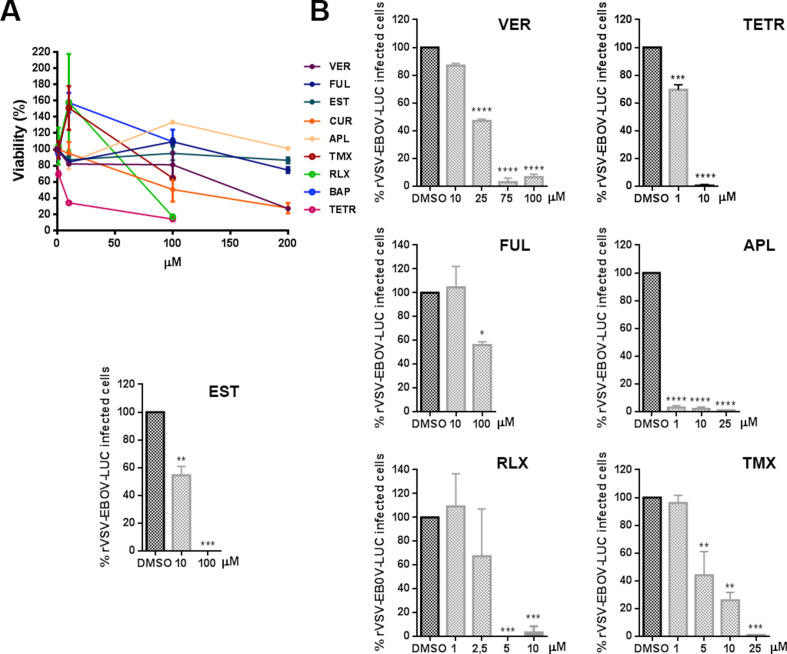
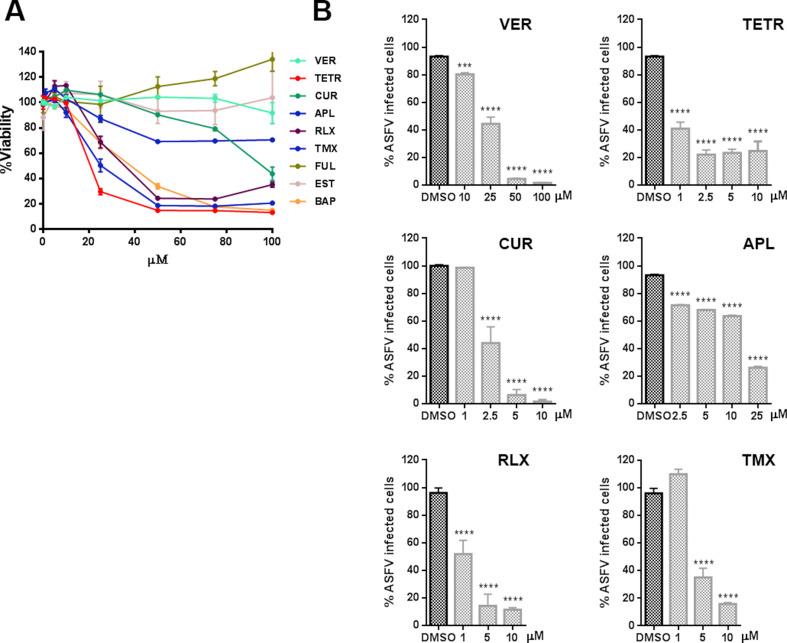
A first initial screen of the selected inhibitor panel was performed to investigate their impact on the infection of coronavirus 229E (CoV-229E-GFP) in Huh-7 cells by fluorimetry. With the best-selected compounds, we proceeded to the evaluation of the minimal concentration of compound that induced a 50% reduction of cell viability or cytotoxicity concentration 50% (CC50), the inhibitory concentration 50% (IC50), and ratio (Fig. 1 A). All tested compounds except Bapta-AM showed an IC50 < 4.5 μM. Considering also CC50, Tamoxifen was discarded for the next study in this virus because its selectivity index.
Fig. 1.
Inhibition of endosomal calcium channels impaired SARS-CoV-2, EBOV, and ASFV infection. A) Screening and selection of compounds that inhibited CoV229E infection in Huh-7 Lunet C3 cells. B) Intracellular luciferase activity of Vero E6 cells pretreated with DMSO or endosomal targeting compounds at the indicated concentrations, and transduced with the SARS-CoV-2 protein-pseudotyped virus (SARS-2-Spp). C) Infection percentages of vesicular stomatitis virus (VSV) pseudovirions with EBOV GP (rVSV-EBOV-luc) or D) VSV-G protein in Vero cells (rVSV-G-luc) pretreated with DMSO or drug compounds. E) ASFV Bapp30GFP infection percentages at 16 h in DMSO controls or Vero cells pretreated with the indicated concentrations of compounds and detected by flow cytometry. Percentages were normalized to values in DMSO treated cells. (F) Table of inhibitory activities of several compounds on rVSV-EBOV or rVSVG or ASFV Bapp30GFP on Vero cells. (B–E) Error bars indicate S.D. from three independent experiments. Statistically significant differences are indicated by asterisks (****p < 0.0001).
Once compounds have been tested against CoV-229E, cells were treated with the compounds and transduced with the SARS-2-CoV S protein-pseudotyped virus (SARS-2-Spp) in the presence of freshly added compounds (or DMSO as control) and then incubated for 48 h. At that point, the intracellular luciferase activity was measured. Hydroxicloroquine (HCL), a drug that inhibits endocytosis by blocking endosomal acidification (Shang et al., 2020) was included as positive control (Fig. 1B). All compounds tested reduced pseudotyped SARS-2-Spp entry in Vero cells to different degrees but most drugs reduced viral entry over HCL treatment. The most active inhibition was achieved with Apilimod and Tetrandrine (over 99% reduction), followed by Verapamil and Raloxifen that reduced the luciferase levels over 90% compared to vehicle-treated conditions. Treatment with YM201636 (1 μM) produced a 65% reduction in the luciferase activity levels.
Then, we compared the entry reduction caused by those drugs on vesicular stomatitis pseudovirus with EBOV glycoprotein GP (rVSV-EBOV-luc) or control rVSV-G-luc (Fig. 1C–D). Cathepsin L inhibitor TEI inhibited virus entry to similar levels. The effect of these compounds on cell viability and the non-toxic working concentrations were calculated and DMSO, used as vehicle, was taken as control. Vero cells were pretreated for 1 h with the compounds as indicated in the Materials and Methods section. These drugs also showed a potent inhibitory effect of EBOV GP pseudovirus. Almost complete inhibition of EBOV GP pseudotyped virions entry was achieved with RLX, APL, and TETR. TETR presented low IC50 of below 1.5 in the three viruses, the lower for 229E CoV that presented IC50 0.3 (Fig. 1A and F). Also, VER, CUR and TEI presented a strong inhibition (Fig. 1C) while control VSV G entry was not significantly affected in most cases (Fig. 1 D). Teicoplanin (TEI) a cathepsin L inhibitor (Zhou et al., 2016) that would affect viral protein processing was included as positive control.
When testing compounds, we used pseudotyped retrovirus or vesicular stomatitis pseudovirus as these systems are well-suited for viral entry studies avoiding unnecessary risks of working with infectious virus EBOV or SARS-CoV-2. However, infectious attenuated ASFV Ba71V as is a well-known model, easy to handle that infects primary alveolar macrophages and Vero cells. Moreover, its infectious cycle and transit through the endosomal route have been thoroughly documented (Cuesta-Geijo et al., 2012; Hernaez et al., 2016).
The inhibitory effect of these drugs was also found in distant virus ASFV. This virus undergoes acid-dependent decapsidation and fusion at the late endosome (Cuesta-Geijo et al., 2012). Vero cells were pre-treated with the compounds before inoculation with ASFV Bpp30GFP at a MOI of 1 pfu/cell for 16 h and early GFP expression was measured by flow cytometry. Calcium channel inhibitors reduced ASFV infectivity over 80–90%, especially TETR and VER (Fig. 1E). Since PIKfyve and calcium channel inhibitors target the endocytic pathway, which is a common route for several viruses, these may act as wide spectrum antivirals. Apilimod exerted a potent inhibitory activity in pseudotyped systems for SARS-CoV-2 spike (IC50 < 1), EBOV-GP, and against attenuated ASFV (Fig. 1E–F). However, also RLX and TMX caused a potent inhibitory effect by an unknown mechanism. Remarkably, RLX exhibited an IC50 of 0.91 for ASFV (Fig. 1F).
The corresponding viability and dose/response curves are shown in Supplementary Fig. S2, Supplementary Fig. S3, Supplementary Fig. S4 for Vero cells and for primary swine macrophages in Supplementary Fig. S5.
Then, we found that SERMs were potent antivirals, especially for ASFV, but while the mechanism has been recently characterized for the other viruses, the mechanism underlying the inhibitory effect of SERMs is not yet known for ASFV. Then, we further studied their effect on ASFV infection.
3.2. SERMs entry inhibition is not related to the estrogen receptor pathway
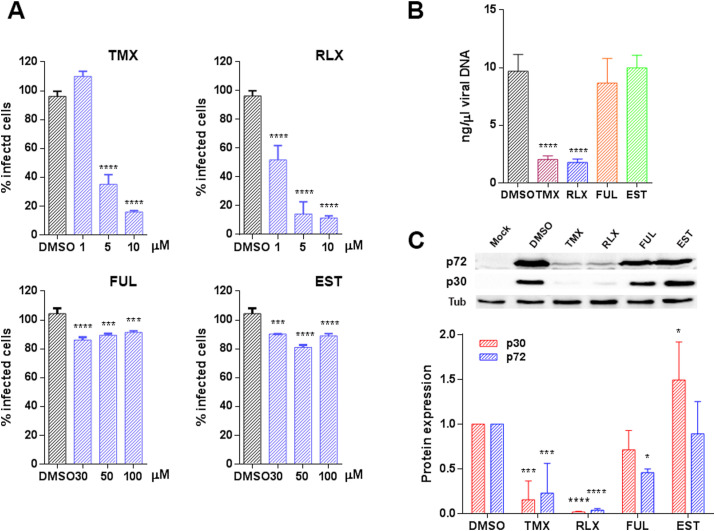
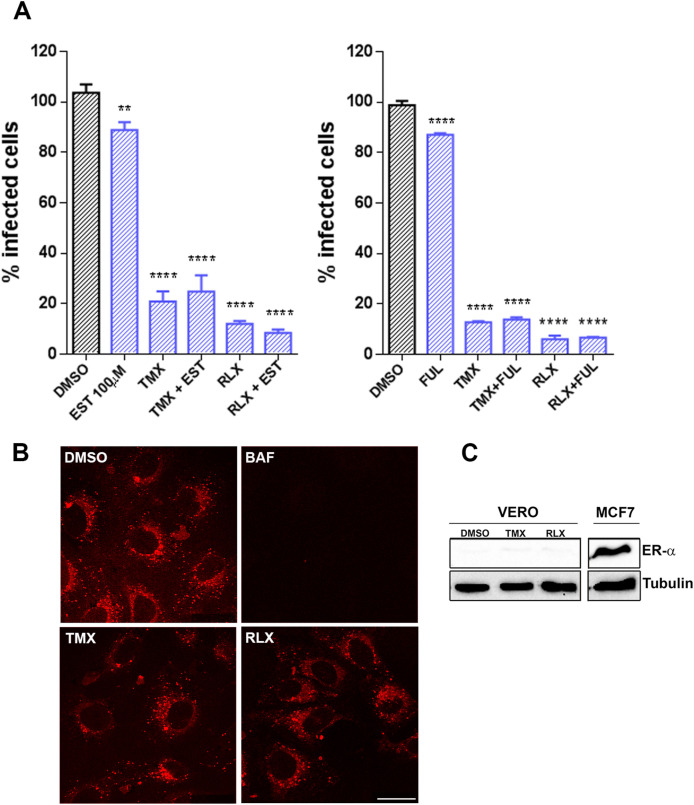
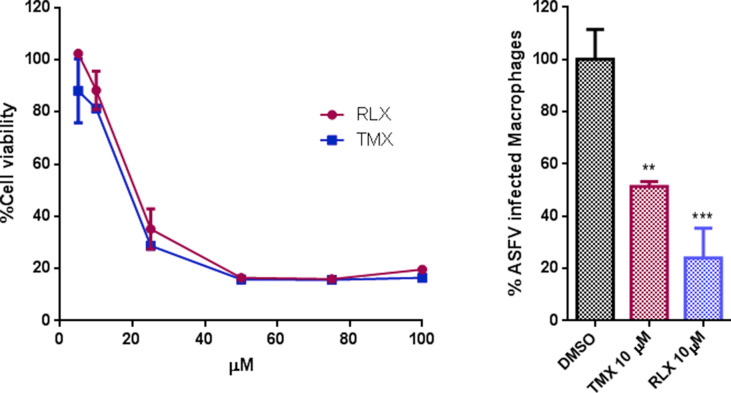
Then, we further investigated the mechanism of inhibition caused by SERMs. To investigate whether this inhibition was achieved through the estrogen receptor (ESR), we included both ESR agonist and antagonist, the natural ligand of ESR 17β-estradiol (EST), and Fulvestrant (FUL) respectively, in infectivity assays (Fig. 2 A). Vero cells were infected with ASFV Bpp30GFP, a recombinant virus that expresses the GFP protein under the early viral protein promoter p30. Raloxifen (RLX) and Tamoxifen (TMX) caused a dose-dependent reduction of infectivity compared to untreated cells with a reduction of 80% at a 10 μM concentration of TMX or RLX. However, treatment with the natural ligand of ESR 17β -estradiol Estradiol (EST) or the Estrogen Receptor antagonist Fulvestrant (FUL) had a low impact on virus infectivity under 20% at 100 μM concentrations of FUL.
Fig. 2.
TMX and RLX potently inhibited ASFV infection. (A) Reduction of ASFV entry in Vero cells pretreated with TMX, RLX, FUL, and EST on ASFV infectivity evaluated by flow cytometry. Cells were infected with Bapp30GFP for 16 h and percentages were normalized to values in DMSO treated cells. (B) ASFV genome copy number in drug-treated and ASFV infected cells analyzed by real-time PCR. (C) Early p30 and late p72 protein expression were detected by WB at 16 h. Quantification of the bands were corrected to tubulin data and then normalized to control values. Error bars indicate S.D. from three independent experiments. Statistically significant differences are indicated by asterisks (****p < 0.0001, ***p < 0.001, *p < 0.05).
To further confirm these results, we evaluated other infection parameters. Inhibition of viral DNA replication was analyzed by real-time PCR at 16 h. The presence of RLX and TMX upon ASFV the infection reduced viral DNA synthesis (Fig. 2B). Moreover, the decrease in infectivity correlated with a reduction of the expression of early viral protein p30 and late viral protein p72 (Fig. 2C) as detected by western blotting of infected Vero cell extracts.
Again, we ruled out the possibility that SERMs could be acting through the estrogen receptor (ESR) itself as shown by combinations of drugs with EST or FUL (Fig. 3 A). The agonist did not modify substantially ASFV infection but neither did the estrogen receptor antagonist FUL (Fig. 3A). RLX and TMX strongly reduced ASFV entry and neither the addition of EST nor FUL could prevent this inhibitory effect. This result suggests that RLX and TMX would not act through competition, neither with the receptor agonist EST nor with FUL for the target molecules involved in viral entry.
Fig. 3.
SERMs inhibition of ASFV infection was independent of estrogen receptor (ESR) α or endosomal acidification. (A) The addition of ESR agonist EST or ESR antagonist FUL with TMX or RLX did not rescue the inhibitory effect of the latter. After treatment, infected cells were detected by flow cytometry and percentages normalized to values in DMSO treated cells. (B) Endosome acidification was assessed with LysoTracker Red, a probe for low-pH organelles in Vero cells. TMX, RLX, BAF, or DMSO treatment did not induce changes while BAF eliminated acidic endosomes staining. This compound was included as a positive control for inhibition of acidification. (C) Absence of ESR expression in Vero cells. There was no detectable expression of ESR in Vero cells by Western blotting while it was evident in MCF-7 cell lysates used as a positive control for ESR. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Then, we proceeded to investigate other possible targets for SERMs antiviral effect. We evaluated whether RLX and TMX could affect endosome acidification, which is a pre-requisite for ASFV entry (Fig. 3B). Endosome acidification was evaluated with a probe for low-pH organelles, in cells treated with vehicle, RLX, TMX, or Bafilomycin (BAF), the latter as a positive control for inhibition of acidification. As expected, we observed a strong reduction of acidic endosomes with BAF but remained unaltered in the presence of RLX or TMX (Fig. 3B). These experiments were performed in the Vero cell line. In these cells, the estrogen receptor was not detected as previously described (Johansen et al., 2013) compared to MCF-7 cells in a western blot probed for estrogen receptor alpha (Fig. 3C).
3.3. SERMs induce endosomal cholesterol accumulation
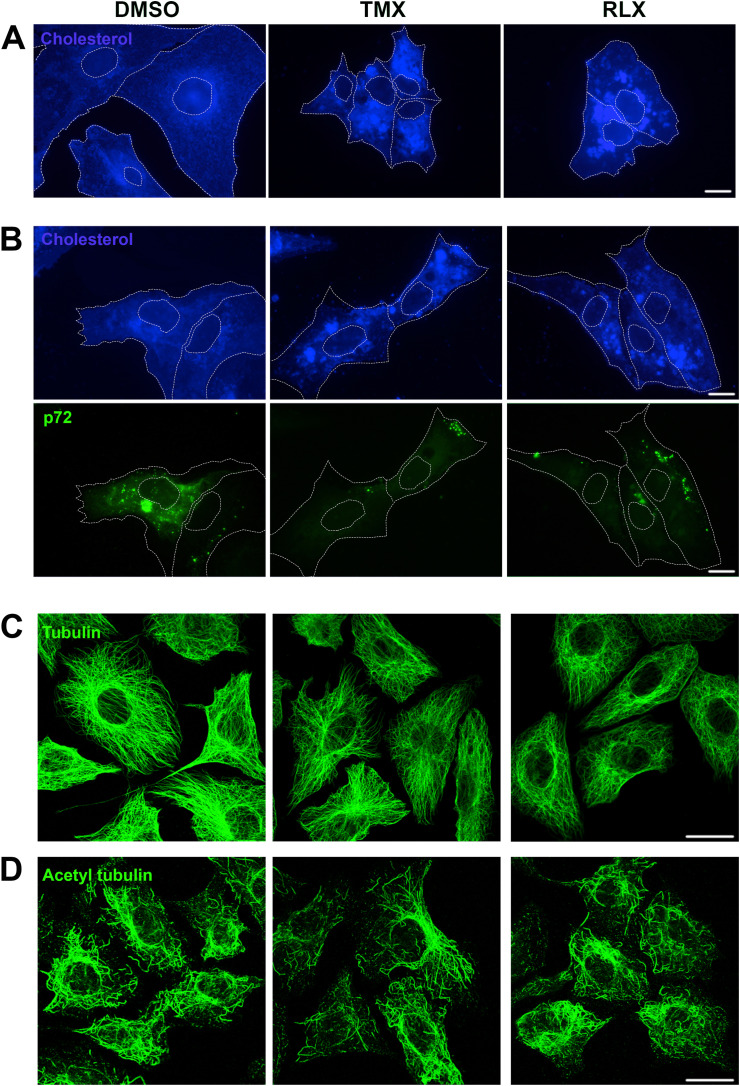
Then, to further understand the mechanism of SERMs antiviral effect, we evaluated the potential activity of these compounds on subcellular cholesterol distribution. In controls, cholesterol-positive structures displayed a diffuse distribution in the cytoplasm, with distinguishable small vesicles (Fig. 4 A). The addition of RLX and TMX changed this pattern and caused the appearance of larger filipin-positive vesicles in the cytoplasm, indicative of the accumulation of free cholesterol. We also studied the cholesterol distribution in ASFV infected Vero cells in the presence of these drugs (Fig. 4B). RLX and TLX also caused a marked accumulation of free cholesterol in large vesicles and the absence of the characteristic viral factories.
Fig. 4.
TMX and RLX induced alterations of cholesterol distribution. (A) Fluorescence microscopy of Vero cells treated or not with compounds and stained with Filipin. Treated cells presented large accumulations of cholesterol-laden vesicles that were absent in untreated cells. (B) Absence of viral factories in Vero cells infected for 16 hpi and treated or not with compounds. Filipin was detected in blue and viral protein p72 in green. Bar 25 μm. (C) Conserved microtubule cytoskeleton upon TMX and RLX treatment. Confocal fluorescence microscopy of Vero cells treated with indicated drugs or solvent DMSO and immunostained for alpha-tubulin or (D) acetylated tubulin, both presented a conserved microtubule pattern. Bar 25 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Like many other viruses, ASFV interacts with the microtubule cytoskeleton during infection (Hernaez et al., 2010). It has been described that SERMs can modulate microtubule assembly causing microtubule defects (Lo et al., 2019). Then, we studied microtubule organization in the presence or absence of SERMs. Vero cells were treated with compounds during 3 h, fixed and stained for tubulin or acetylated tubulin, and observed at the confocal microscope. Cells treated with Tubacin (TUB) were used as a positive control of acetyl-tubulin (not shown). As shown in Fig. 4B, no significant differences were detected between treated and untreated cells at the concentrations used in this study.
3.4. Raloxifen induced alterations in the calcium flux in cells
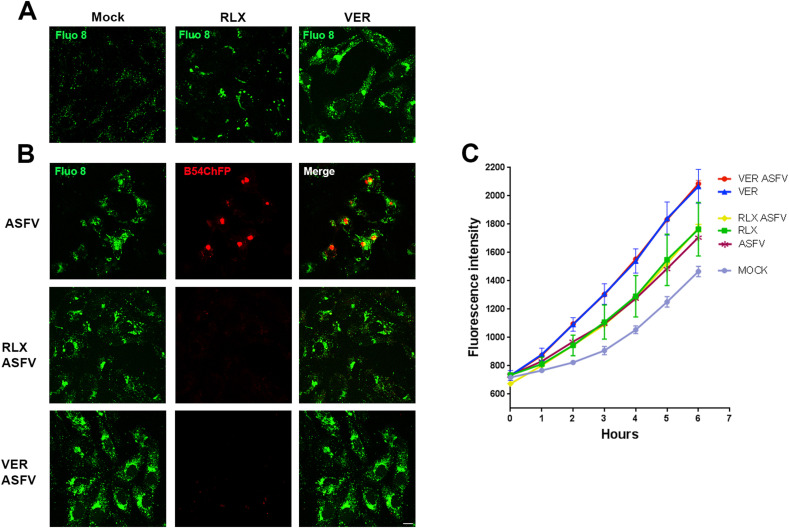
Finally, we investigated another possible mechanism of action of SERMs in ASFV infection, by studying endolysosomal calcium. The fluorescent calcium indicator fluo8 generally displayed a homogeneous diffuse distribution in control Vero cells indicating a conserved calcium flux from endosomes. However, in cells treated with RLX, calcium indicator markedly accumulated in fluorescent vesicles similar to those produced in the presence of Verapamil, a calcium channel inhibitor (Fig. 5 A). We also studied calcium distribution in infected cells in absence or presence of these compounds that produced an accumulation of fluorescent vesicles in the cytoplasm distributed around the viral replication sites (Fig. 5B merge panel). RLX and VER caused a marked accumulation of fluorescent vesicles and the absence of viral factories because of infection inhibition (Fig. 5B).
Fig. 5.
Intracellular calcium levels upon SERMs treatment. (A) Representative confocal micrographs of calcium imaging with Fluo8 in Vero cells pretreated with Verapamil, Raloxifene, or DMSO. (B) Representative images of ASFV B54-cherryFP infected cells after 16 h harboring the characteristic viral replication factories in controls. ASFV factories were absent in drug-treated cells. Bar 25 μm. (C) Time-response of intracellular Ca2+ detected with Fluo8. Each graph represented the average Ca2+ response to individual compounds in uninfected or infected cells. VER was included as a known positive responder that elevated intracellular Ca2+ levels. The graph shows fluorescence intensities in cells shown in (A) and (B), at the time points indicated. Error bars indicate S.D. from three independent experiments.
Next, we performed a time course of fluorescence intensity during 6 h after fluo8 incubation, the highest fluorescence intensity was quantified in infected or uninfected VER treated cells (Fig. 5C). The fluorescence intensity curve displayed high yields in cells treated with RLX at similar rates in uninfected or infected cells. Untreated infected cells enhanced fluorescence intensity over the controls. Taken together these data regarding calcium channel inhibitors, suggest that ASFV could not only rely on but also trigger the activation of calcium-signaling pathways.
4. Discussion
Emerging viral diseases continuously impact human and animal health as occurred with SARS-CoV-2 that caused a human pandemic of unpredictable consequences (Guan et al., 2020). EBOV is another emerging human pathogen causing a fatal hemorrhagic fever. Other viruses that affect mammals and other species pose a continuous threat to human health, food availability, or environmental and natural resources. Currently, a major problem in animal health is caused by ASFV, an animal virus that has spread to three continents with a high economic and social burden.
The diseases caused by those unrelated viruses are coincident in their potential to produce a multisystemic inflammatory syndrome (MIS) with cytokine storm, hyperinflammation and coagulation alterations that can be severe and frequently lethal (Hunt et al., 2015). Each disease has its characteristic features but there are some similarities in the clinical syndrome caused by these highly pathogenic viruses.
Currently, there are increasing number of studies on potential therapeutics against these viruses (Riva et al., 2020) but several of them are lacking insights on the mechanism of action of inhibitory compounds that could help understanding the molecular pathogenesis of these diseases and pave the way for further antiviral discovery (Jeon et al., 2020; Weston et al., 2020).
These pathological agents have evolved targets for virus entry at endosomes through adaptation to the mammalian cell shared by different viruses (Helenius, 2018). The endocytic pathway is essentially dynamic and highly organized in several maturation compartments, characterized by molecular cues that confer identity to each vesicle (Huotari and Helenius, 2011). One of them is the endosome luminal pH controlled by the ATP-dependent H+ pump at the endosomal membrane (Toei et al., 2010), which is also dependent on ion flux (Marshansky and Futai, 2008). Then, drugs targeting this pump or calcium channels are potential broad-spectrum antiviral candidates to include in drug screening. Maturation of these vesicles characteristically implies luminal acidification, and this is required for infectious entry of several virus families, including all three viruses in this study. Moreover, EBOV find in endosomes the necessary enzymes for their entry, namely the cathepsins required for the cleavage and activation of the GP protein. SARS-CoV-2 S protein priming is performed by proteases like TMPRSS2 or by cathepsins (Chandran et al., 2005; Ou et al., 2020).
Endosomal maturation also requires membrane proteins that are specific for each compartment (Huotari and Helenius, 2011). Early endosomes express the small GTPases Rab 5 on their surface and their lumen is alkaline. In contrast, late endosomes express Rab7 on their surface and have an acidic luminal pH. Importantly, unconventional lipids named phosphatidyl inositols or inositol phosphates confer organelle identity to endosomes and allow docking of Rab proteins at the membrane (Wallroth and Haucke, 2018). Inositol phosphates undergo rapid interconversions by phosphorylation of the inositol ring mediated through kinases, one of them being PIKfyve (Ikonomov et al., 2002).
The objective of this work was to study experimental and FDA-approved drugs targeting functional proteins at the endosomal membrane as antiviral candidates. These could inhibit a wide range of viruses, among them viruses of humans, SARS-CoV-2 and EBOV, or animals, like ASFV. Our results highlight the potential of endosomes in viral entry. However, it might be relevant to consider other aspects of the vesicular transport machinery in which this organelle participates such as, innate immunity, sorting, recycling, transport, exit, metabolism, autophagy, chaperone-mediated degradation, and a handful of other cellular processes. In general, endosomes function as major cellular signaling hubs that are required for viral infections (Helenius, 2018).
Compounds targeting endosomal kinase PIKfyve, the kinase responsible for the synthesis of phosphatidyl inositol-(3,5)-biphosphate (PtdIns 3,5-P2) at the late endosome, are active against EBOV and SARS-CoV-2 (Kang et al., 2020; Nelson et al., 2017; Ou et al., 2020). We first described the antiviral effect of PIKfyve inhibition for a virus, against ASFV (Cuesta-Geijo et al., 2012). ASFV seems to be dependent on this kinase as ASFV upregulates PIKfyve expression in infected cells (Quetglas et al., 2012). Then, we included drugs targeting PIKfyve in this comparative study to check our hypothesis, finding that Apilimod potently inhibited the infection of the three viruses to similar levels over 75% infectivity reductions (Fig. 1E). These compounds induce deep changes in the entire endocytic pathway blocking endosomal maturation. We have previously described the profound alteration of endosomes undergoing enlargement and vacuolization (Cuesta-Geijo et al., 2012).
All endosome-targeting compounds screened in this study, significantly reduced the infectivity of SARS-CoV-2 S protein-pseudotyped retrovirus infection efficiency in Vero E6 cells to a high degree. The most active compounds were Apilimod and Tetrandrine over 99%, followed by Verapamil and Raloxifen over 90% (Fig. 1B). The same drugs strongly reduced rVSV-EBOV-GP infectivity compared with much less effect on rVSV-G (Fig. 1C–D). Finally, we observed entry reductions in ASFV infectivity over 80% using the following set of compounds, APL, TETR, VER, and RLX (Fig. 1E).
Due to the importance of steroids in viral infections and because SERMs are Food and Drug Administration (FDA)-approved compounds with oral availability, safety, and a long history of use, which were recently found in drug screens against SARS-CoV-2 (Weston et al., 2020), we found them suitable candidates to study their effect against ASFV infection. Also, the relevance of cholesterol in ASFV infection, suggested that SERMs could be potentially effective antivirals against it.
The impact of these compounds was high on the three-virus screened, SARS-2-Spp retrovirus system, rVSV-EBOV-GP, rVSV-G and ASFV. However, TMX and RLX caused a dose-dependent reduction of ASFV infectivity ca. 80–90% with the lowest IC50 value of 0.98. Hence, we further studied the potential mechanism of action of SERMs on this virus. The decrease in ASFV infectivity correlated with reductions on the expression of early viral protein p30 that indicates that these drugs affected infection at very early stages. Viral replication in the presence of these two components was also strongly inhibited.
Although the efficacy of SERMs as antivirals has been documented against the Human Immunodeficiency Virus (HIV), Hepatitis C Virus (HCV), Herpes Simplex Virus 1 (HSV-1), and Vesicular Stomatitis Virus (VSV) (Montoya and Krysan, 2018), their mechanism of action is not fully understood. In fact, these compounds were reported to inhibit EBOV by a mechanism independent of the estrogen-receptor (Johansen et al., 2013) and for SARS-CoV-2 (Weston et al., 2020). A mechanism suggested for this inhibition is by destabilizing GP for fusion (Zhao et al., 2016) or by inhibition of TPCs (Penny et al., 2019), but this is still under discussion.
Then, we described first hat SERMs were effective antivirals against ASFV and explored possible mechanisms underlying their inhibitory action against this virus. We ruled out the possibility that SERMs could be acting through the estrogen receptor given the lack of effect of the natural ligand EST, or the antagonist FUL (even at high concentrations of 100 μM). The absence of recovery supplied at the time of infection reinforces the independence of the receptor for the antiviral effect. Estrogen receptor expression was absent as previously reported in Vero cells both untreated or treated with TMX or RLX (Johansen et al., 2013). These results indicate that these compounds should be mediating their antiviral effects through cell-based mechanisms unrelated to the classical estrogen signaling pathway.
Endosomal acidification was not altered in Vero cells treated with TMX or RLX. This fact would have altered the entry of any of the virus tested as it is a prerequisite for endosomal maturation. We also found that these compounds modified cholesterol efflux from endosomes as treated cells presented coarse accumulations of cholesterol-laden vesicles that were absent in untreated cells. A similar interference of SERMs with cholesterol was found in macrophages or human umbilical vein endothelial cells (HUVECs), indicating a mode of action of SERMs related to cholesterol distribution (Fernandez-Suarez et al., 2016, Shim et al., 2015). In fact, cholesterol is required for successful ASFV entry and blocking cholesterol flux at the endosomal membrane causes retention of virions inside endosomes, inhibiting infection progression (Cuesta-Geijo et al., 2016). Despite previous reports of microtubule acetylation (Lo et al., 2019), neither RLX nor TMX induced significant changes in the distribution of microtubules or the degree of acetylation at the concentrations used in this study (Fig. 4).
As shown here (Fig. 5), SERMs strongly modified endosomal calcium flux. This could impact the ionic equilibrium of the endosome in such way that would affect virus transit through these organelles. In fact, another drug unrelated to SERMs, Tetrandrine, has been one of the most potent antivirals against the three viruses analyzed. TETR is an inhibitor of two-pore channel 2 (TPC2), an ion transporter protein that mediates calcium efflux out of late endosomes (Wang et al., 2012). A minimum local concentration of calcium may be required for viral fusion as it was previously shown for EBOV (Sakurai et al., 2015), Rubella virus (Dube et al., 2014) and importantly, coronavirus SARS-CoV (Lai et al., 2017) and MERS (Tang et al., 2020). It is to consider that SERMs could also act as cationic amphiphilic drugs (CADs) causing alterations in acidification and vesicular transport (Chen et al., 1999). Moreover, this mechanism of action would explain their inhibitory potential in the viruses under study indicating that estrogen inhibitors could join some FDA-approved drugs potentially useful against several viruses sharing this pathway, including SARS-CoV-2.
This work suggested a common mechanism underlying the inhibitory action of these compounds on endosomal proteins that could constitute an important target when developing broad-spectrum antivirals against several important viruses. Also, the characterization of such common features for distant virus infections help to understand the molecular mechanisms underlying severe viral infections in which endosomes contain important host factors.
The results obtained with clinically-approved drugs could lead to a quick repurposing for clinical use. Also, in vivo experiments in animals (pigs) are ongoing in our laboratories and compounds presented an optimal tolerance and lack toxicity by the oral route in an ASFV infection model.
Finally, these drugs could be also considered as lead compounds to develop optimized candidates focusing in the antiviral properties and deciphered mechanism of action. These results pave the way for medicinal chemistry investigation of structural features directly responsible for activity and makes possible the optimization of these structures to develop novel compounds.
5. Conclusion
Several viruses require relevant proteins and receptors located on the endosomal membrane upon cell entry for cleavage or activation of functional virus proteins that should occur at this stage. Therefore, those proteins constitute suitable cellular targets for a wide range of antiviral compounds. Based on this premise, we have described the inhibitory effect of experimental and FDA-approved drugs acting on the endosomal membrane against viruses of high relevance for human and animal health, namely SARS-CoV-2, EBOV, or ASFV. Among the tested compounds, we have corroborated the antiviral properties of inhibitors of endosomal calcium channels and PIKfyve kinase. Moreover, we have partially characterized the inhibitory mechanisms of selective estrogen receptor modulators what makes them attractive candidates to be repurposed as antiviral drugs.
Acknowledgments
We are thankful to François-Loïc Cosset (Ecole Normale Superieure, Lyon, France) and to Jose María Casanovas and Juan García Arriaza (CNB–CSIC) for providing the retroviral pseudotyped plasmid system and the S protein cDNA, respectively. We thank Volker Thiel from the University of Bern, Switzerland for CoV 229E-GFP and Thomas Pietschman, Twincore, Germany for Huh-7 Lunet C3 cells. This research was partially supported through “La Caixa” Banking Foundation (HR18-00469), Spain; Instituto de Salud Carlos III, Spain (FIS PI181007 and ISCIII–COV20/01007); CSIC, Spain (201980E024 and 202020E079); Spanish Ministry of Science and Innovation, Spain (RTI2018-097305-R-I00); the European Commission, European Union Horizon 2020 Framework Programme VACDIVA-SFS-12-2019-1-862874; and Comunidad de Madrid, Spain PEJD-2019-PRE/BMD-16764 for PB.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104990.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Structures of the antiviral compounds tested.
Supplementary Fig. S2.
A. Evaluation of the cytotoxicity at increasing concentrations of compounds in Huh-7 cells. B. CoV-229E-GFP signal intensity percentages at increasing doses of compounds. Error bars indicate S.D. from two independent experiments. Statistically significant differences are indicated by asterisks (*p < 0.5; **p < 0.01).
Supplementary Fig. S3.
A. Evaluation of the cytotoxicity at increasing concentrations of the compounds in Vero E6 cells. B. EBOV infectivity percentages at increasing doses of compounds. Error bars indicate S.D. from three independent experiments. Statistically significant differences are indicated by asterisks (****p < 0.0001).
Supplementary Fig. S4.
A. Evaluation of the cytotoxicity at increasing concentrations of the compounds in Vero cells. B. ASFV infectivity percentages at increasing doses of compounds. Error bars indicate S.D. from three independent experiments. Statistically significant differences are indicated by asterisks (****p < 0.0001).
Supplementary Fig. S5.
A. Cell viability of primary lung macrophages from swine, treated with increasing concentrations of compounds for 1 h. B. ASFV Ba71V infectivity percentages at increasing doses of compounds in macrophages pretreated for 1 h, and then infected with fluorescent virus Bpp30GFP for 24 hpi. Drug compounds are maintained in the cultures for 24 h. Error bars indicate S.D. from three independent experiments. Statistically significant differences are indicated by asterisks (***p < 0.001).
References
- Aleksandrowicz P., Marzi A., Biedenkopf N., Beimforde N., Becker S., Hoenen T., Feldmann H., Schnittler H.J. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J. Infect. Dis. 2011;204(Suppl. 3):S957–S967. doi: 10.1093/infdis/jir326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C., Borca M., Dixon L., Revilla Y., Rodriguez F., Escribano J.M., Ictv Report, C. ICTV virus taxonomy profile: Asfarviridae. J. Gen. Virol. 2018;99:613–614. doi: 10.1099/jgv.0.001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C., Galindo I., Cuesta-Geijo M.A., Cabezas M., Hernaez B., Munoz-Moreno R. African swine fever virus-cell interactions: from virus entry to cell survival. Virus Res. 2013;173:42–57. doi: 10.1016/j.virusres.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardak H., Uguz A.C., Bardak Y. Curcumin regulates intracellular calcium release and inhibits oxidative stress parameters, VEGF, and caspase-3/-9 levels in human retinal pigment epithelium cells. Phys. Int. 2017;104:301–315. doi: 10.1556/2060.104.2017.4.3. [DOI] [PubMed] [Google Scholar]
- Barrado-Gil L., Galindo I., Martinez-Alonso D., Viedma S., Alonso C. The ubiquitin-proteasome system is required for African swine fever replication. PLoS One. 2017;12 doi: 10.1371/journal.pone.0189741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., Thacker B.E., Glass C.A., Yang Z., Torres J.L., Golden G.J., Bartels P.L., Porell R.N., Garretson A.F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B.M., Caradonna T.M., Kellman B.P., Martino C., Gordts P., Chanda S.K., Schmidt A.G., Godula K., Leibel S.L., Jose J., Corbett K.D., Ward A.B., Carlin A.F., Esko J.D. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057 e1015. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Geijo M.A., Chiappi M., Galindo I., Barrado-Gil L., Munoz-Moreno R., Carrascosa J.L., Alonso C. Cholesterol flux is required for endosomal progression of African swine fever virions during the initial establishment of infection. J. Virol. 2016;90:1534–1543. doi: 10.1128/JVI.02694-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Geijo M.A., Galindo I., Hernaez B., Quetglas J.I., Dalmau-Mena I., Alonso C. Endosomal maturation, Rab7 GTPase and phosphoinositides in African swine fever virus entry. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Schindler M., Simon S.M. A mechanism for tamoxifen-mediated inhibition of acidification. J. Biol. Chem. 1999;274:18364–18373. doi: 10.1074/jbc.274.26.18364. [DOI] [PubMed] [Google Scholar]
- Dagotto G., Yu J., Barouch D.H. Approaches and challenges in SARS-CoV-2 vaccine development. Cell Host Microbe. 2020;28:364–370. doi: 10.1016/j.chom.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Anton-Plagaro C., Shoemark D.K., Simon-Gracia L., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Collins B.M., Cullen P.J., Yamauchi Y. Neuropilin-1 is a host factor for SARS-788 CoV-2 infection. Science. 2020 doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M., Rey F.A., Kielian M. Rubella virus: first calcium-requiring viral fusion protein. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Carrascosa A.L., Moreno M.A., Vinuela E. Titration of African swine fever (ASF) virus. J. Gen. Virol. 1976;32:471–477. doi: 10.1099/0022-1317-32-3-471. [DOI] [PubMed] [Google Scholar]
- Fernandez-Suarez M.E., Escola-Gil J.C., Pastor O., Davalos A., Blanco-Vaca F., Lasuncion M.A., Martinez-Botas J., Gomez-Coronado D. Clinically used selective estrogen receptor modulators affect different steps of macrophage-specific reverse cholesterol transport. Sci. Rep. 2016;6:32105. doi: 10.1038/srep32105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.A., Urquiza J., Ramirez D., Alonso C., Campillo N.E., Martinez A. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020;60(21):12359–12389. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S., China Medical Treatment Expert Group for, C. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A. Virus entry: looking back and moving forward. J. Mol. Biol. 2018;430:1853–1862. doi: 10.1016/j.jmb.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernaez B., Guerra M., Salas M.L., Andres G. African swine fever virus undergoes outer envelope disruption, capsid disassembly and inner envelope fusion before core release from multivesicular endosomes. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernaez B., Tarrago T., Giralt E., Escribano J.M., Alonso C. Small peptide inhibitors disrupt a high-affinity interaction between cytoplasmic dynein and a viral cargo protein. J. Virol. 2010;84:10792–10801. doi: 10.1128/JVI.01168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L., Gupta-Wright A., Simms V., Tamba F., Knott V., Tamba K., Heisenberg-Mansaray S., Tamba E., Sheriff A., Conteh S., Smith T., Tobin S., Brooks T., Houlihan C., Cummings R., Fletcher T. Clinical presentation, biochemical, and haematological parameters and their association with outcome in patients with Ebola virus disease: an observational cohort study. Lancet Infect. Dis. 2015;15:1292–1299. doi: 10.1016/S1473-3099(15)00144-9. [DOI] [PubMed] [Google Scholar]
- Huotari J., Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O.C., Sbrissa D., Mlak K., Kanzaki M., Pessin J., Shisheva A. Functional dissection of lipid and protein kinase signals of PIKfyve reveals the role of PtdIns 3,5-P2 production for endomembrane integrity. J. Biol. Chem. 2002;277:9206–9211. doi: 10.1074/jbc.M108750200. [DOI] [PubMed] [Google Scholar]
- Ilunga Kalenga O., Moeti M., Sparrow A., Nguyen V.K., Lucey D., Ghebreyesus T.A. The ongoing Ebola epidemic in the Democratic Republic of Congo, 2018-2019. N. Engl. J. Med. 2019;381:373–383. doi: 10.1056/NEJMsr1904253. [DOI] [PubMed] [Google Scholar]
- Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.M., Brannan J.M., Delos S.E., Shoemaker C.J., Stossel A., Lear C., Hoffstrom B.G., Dewald L.E., Schornberg K.L., Scully C., Lehar J., Hensley L.E., White J.M., Olinger G.G. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.L., Chou Y.Y., Rothlauf P.W., Liu Z., Soh T.K., Cureton D., Case J.B., Chen R.E., Diamond M.S., Whelan S.P.J., Kirchhausen T. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karger A., Perez-Nunez D., Urquiza J., Hinojar P., Alonso C., Freitas F.B., Revilla Y., Le Potier M.F., Montoya M. 2019. An Update on African Swine Fever Virology. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.P., Reid S.M., Hutchings G.H., Grierson S.S., Wilkinson P.J., Dixon L.K., Bastos A.D., Drew T.W. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol Methods. 2003;107:53–61. doi: 10.1016/s0166-0934(02)00189-1. [DOI] [PubMed] [Google Scholar]
- Lai A.L., Millet J.K., Daniel S., Freed J.H., Whittaker G.R. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium-dependent manner. J. Mol. Biol. 2017;429:3875–3892. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.C., Cormier O., Liu T., Nettles K.W., Katzenellenbogen J.A., Stearns T., Altman R.B. Pocket similarity identifies selective estrogen receptor modulators as microtubule modulators at the taxane site. Nat. Commun. 2019;10:1033. doi: 10.1038/s41467-019-08965-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshansky V., Futai M. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr. Opin. Cell Biol. 2008;20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingo R.M., Simmons J.A., Shoemaker C.J., Nelson E.A., Schornberg K.L., D'Souza R.S., Casanova J.E., White J.M. Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J. Virol. 2015;89:2931–2943. doi: 10.1128/JVI.03398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingorance L., Friesland M., Coto-Llerena M., Perez-del-Pulgar S., Boix L., Lopez-Oliva J.M., Bruix J., Forns X., Gastaminza P. Selective inhibition of hepatitis C virus infection by hydroxyzine and benztropine. Antimicrob. Agents Chemother. 2014;58:3451–3460. doi: 10.1128/AAC.02619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya M.C., Krysan D.J. Repurposing estrogen receptor antagonists for the treatment of infectious disease. mBio. 2018;9 doi: 10.1128/mBio.02272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson E.A., Dyall J., Hoenen T., Barnes A.B., Zhou H., Liang J.Y., Michelotti J., Dewey W.H., DeWald L.E., Bennett R.S., Morris P.J., Guha R., Klumpp-Thomas C., McKnight C., Chen Y.C., Xu X., Wang A., Hughes E., Martin S., Thomas C., Jahrling P.B., Hensley L.E., Olinger G.G., Jr., White J.M. The phosphatidylinositol-3-phosphate 5-kinase inhibitor apilimod blocks filoviral entry and infection. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollmann Saphire E. A vaccine against Ebola virus. Cell. 2020;181:6. doi: 10.1016/j.cell.2020.03.011. [DOI] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny C.J., Vassileva K., Jha A., Yuan Y., Chee X., Yates E., Mazzon M., Kilpatrick B.S., Muallem S., Marsh M., Rahman T., Patel S. Mining of Ebola virus entry inhibitors identifies approved drugs as two-pore channel pore blockers. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:1151–1161. doi: 10.1016/j.bbamcr.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quetglas J.I., Hernaez B., Galindo I., Munoz-Moreno R., Cuesta-Geijo M.A., Alonso C. Small rho GTPases and cholesterol biosynthetic pathway intermediates in African swine fever virus infection. J. Virol. 2012;86:1758–1767. doi: 10.1128/JVI.05666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., Chang M.W., Chan J.F., Cao J., Poon V.K., Herbert K.M., Cheng K., Nguyen T.H., Rubanov A., Pu Y., Nguyen C., Choi A., Rathnasinghe R., Schotsaert M., Miorin L., Dejosez M., Zwaka T.P., Sit K.Y., Martinez-Sobrido L., Liu W.C., White K.M., Chapman M.E., Lendy E.K., Glynne R.J., Albrecht R., Ruppin E., Mesecar A.D., Johnson J.R., Benner C., Sun R., Schultz P.G., Su A.I., Garcia-Sastre A., Chatterjee A.K., Yuen K.Y., Chanda S.K. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai Y., Kolokoltsov A.A., Chen C.C., Tidwell M.W., Bauta W.E., Klugbauer N., Grimm C., Wahl-Schott C., Biel M., Davey R.A. Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science. 2015;347:995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.S., Li R.J., Lv J., Head S.A., Yang E.J., Liu J.O. Inhibition of angiogenesis by selective estrogen receptor modulators through blockade of cholesterol trafficking rather than estrogen receptor antagonism. Canc. Lett. 2015;362:106–115. doi: 10.1016/j.canlet.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toei M., Saum R., Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49:4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020:1–16. doi: 10.1038/s41579-020-00468-6. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallroth A., Haucke V. Phosphoinositide conversion in endocytosis and the endolysosomal system. J. Biol. Chem. 2018;293:1526–1535. doi: 10.1074/jbc.R117.000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang X., Dong X.P., Samie M., Li X., Cheng X., Goschka A., Shen D., Zhou Y., Harlow J., Zhu M.X., Clapham D.E., Ren D., Xu H. TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell. 2012;151:372–383. doi: 10.1016/j.cell.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S., Coleman C.M., Haupt R., Logue J., Matthews K., Li Y., Reyes H.M., Weiss S.R., Frieman M.B. Broad anti-coronavirus activity of food and drug administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. J. Virol. 2020;94 doi: 10.1128/JVI.01218-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt M.A. Generation of VSV pseudotypes using recombinant DeltaG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J. Virol Methods. 2010;169:365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- www.fli.de, 2020. https://www.fli.de/en/news/short-messages/short-message/wild-boar-with-african-swine-fever-in-the-district-of-maerkisch-oderland/ 09/30/2020.
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ren J., Harlos K., Jones D.M., Zeltina A., Bowden T.A., Padilla-Parra S., Fry E.E., Stuart D.I. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Pan T., Zhang J., Li Q., Zhang X., Bai C., Huang F., Peng T., Zhang J., Liu C., Tao L., Zhang H. Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) J. Biol. Chem. 2016;291:9218–9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structures of the antiviral compounds tested.