Abstract
The Rac1-specific GTPase activating protein Slit-Robo GAP2 (Srgap2) is dramatically up regulated during RANKL-induced osteoclastogenesis. Srgap2 interacts with the cell membrane to locally inhibit activity of Rac1. In this study, we determined the role of Srgap2 in the myeloid lineage on bone homeostasis and the osteoclastic response to TNFα treatment. The bone phenotype of mice specifically lacking Srgap2 in the myeloid lineage (Srgap2f/f:LysM-Cre; Srgap2 cKO) was investigated using histomorphomeric analysis, in vitro cultures and western blot analysis. Similar methods were used to determine the impact of TNFα challenge on osteoclast formation in Srgap2 cKO mice. Bone parameters in male Srgap2 cKO mice were unaffected. However, female cKO mice displayed higher trabecular bone volume due to increased osteoblast surface and bone formation rate, while osteoclastic parameters were unaltered. In vitro, cells from Srgap2 cKO had strongly enhanced Rac1 activation, but RANKL-induced osteoclast formation was unaffected. In contrast, conditioned medium from Srgap2 cKO osteoclasts promoted osteoblast differentiation and had increased levels of the bone anabolic clastokine SLIT3, providing a possible mechanism for increased bone formation in vivo. Rac1 is rapidly activated by the inflammatory cytokine TNFα. Supracalvarial injection of TNFα caused an augmented osteoclastic response in Srgap2 cKO mice. In vitro, cells from Srgap2 cKO mice displayed increased osteoclast formation in response to TNFα.
We conclude that Srgap2 plays a prominent role in limiting osteoclastogenesis during inflammation through Rac1, and restricts expression of the paracrine clastokine SLIT3, a positive regulator of bone formation.
Keywords: inflammatory osteolysis, Rac1, tumor necrosis factor alpha, clastokine, SLIT3
INTRODUCTION
Osteoclasts are specialized multinucleated cells that resorb mineralized bone matrix. Their activity is essential for calcium homeostasis, for maintaining bone quality and for returning repaired bone to its normal shape post-fracture. However, excess osteoclast activity contributes to the development of prevalent bone diseases such as osteoporosis and to the pathology associated with osteolytic bone metastases and multiple myeloma (1–3).
Osteoclasts arise by the fusion of macrophage/monocyte progenitors, which is induced by macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL), both of which can be synthesized by bone-forming osteoblasts (4, 5). Moreover, the potent pro-inflammatory cytokine TNFα is known to stimulate osteoclastogenesis with or without involvement of RANKL (6–8). In vivo, TNFα increases osteoclast precursors (9–12), promotes RANK expression and stimulates the synthesis of M-CSF and RANKL by mesenchymal cells (13–15). This contributes to both localized bone loss at sites of inflammation as well as a generalized increase in osteoclast formation due to increased circulating TNFα.
In addition to resorbing bone, mature osteoclasts regulate the differentiation of bone forming osteoblasts. The term “clastokine” was coined for osteoclast-derived factors that positively or negatively regulate osteoblast differentiation. Recently identified clastokines include bone morphogenetic protein 6 (BMP6), EphrinB2 (EFNB2), sphingosine 1-phosphate (S1P), collagen triple helix repeat containing 1 (CTHRC1), and SLIT guidance ligand 3 (SLIT3) (16–19). The interplay of these paracrine factors between osteoclasts and osteoblasts contributes to the tight regulation of bone homeostasis.
Multiple signaling pathways regulate the differentiation and function of osteoclasts. Of these, the Rho family of GTPases, which includes RhoA, Cdc42 and Rac1, are responsible for cytoskeletal reorganization critical for cell motility and for the development of the specialized structures needed to facilitate bone resorption. During osteoclast maturation, actin polymerizes and forms podosomal rings at sealing zones on bone resorbing surfaces. At the ruffled border within the sealing zone, protons and lysosomal enzymes are released into resorption lacuna to dissolve bone mineral and digest organic matrix (20). Whereas all Rho GTPases have an impact on bone mass regulation and osteoclast function, their individual biological properties are subtle and distinct (21–23).
In osteoclasts, Rac1 regulates survival, migration and actin organization, promoting resorption capacity, but not necessarily differentiation (23–26). Although the conformational change of Rac1 between inactive GDP-bound and active GTP-bound forms is facilitated by multiple guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) respectively, only a small subset of these factors are established to function in osteoclasts. Rac1 deficiency or the deficiency of Rac1-specific GEFs (e.g., Vav3 or Dock5), or the failure to control Rac1 activity, causes bone disorders due to abnormal osteoclast activity (26–29). However, the impact of Rac1-specific GAPs on osteoclasts and bone is unknown.
In non-biased screen, we found SLIT-ROBO GTPase activating protein (Srgap2) RNA is dramatically up-regulated in myeloid cells undergoing M-CSF plus RANKL-induced osteoclastogenesis, compared with cells treated with M-CSF alone. The expression of this Rac1-specific GAP in the osteoclast lineage has not been previously reported. In this study, we demonstrate that Srgap2 strongly limits Rac1 activation and attenuates osteoclast-mediated bone resorption in vitro. However, mice lacking Srgap2 specifically in the monocyte lineage (Srgap2f/f:LysM-Cre; Srgap2 cKO mice) displayed increased bone formation and enhanced production of the bone-anabolic clastokine SLIT3. In contrast, Srgap2 cKO mice exhibited augmented osteoclastogenic response to the pro-inflammatory cytokine TNFα in vivo and in vitro, suggesting that Srgap2 plays a more prominent role in regulating osteoclastogenesis during inflammation.
MATERIALS AND METHODS
Animals
Floxed Srgap2 mice were generated using homologous recombination in C57BL/6J ES cells (see Figure 1B for design). They were then crossed with LysMCre mice [Lyz2tm1(cre)Ifo, Jackson Laboratory]. Wild type and floxed Srgap2 alleles were differentiated using PCR of genomic DNA (Figure 1D). LysMCre+/−: Srgap2fl/fl mice were crossed with Srgap2fl/fl to generate littermate controls (LysMCre−/−: Srgap2fl/fl) and mice with monocyte lineage-restricted deletion of Srgap2 (LysMCre+/−: Srgap2fl/fl; Srgap2-cKO). All controls and Srgap2-cKO mice used for experiments were 7–9 weeks-old.
Mice were maintained and handled in accordance with the guidelines of the Institutional Animal Care and Use Committee at UConn Health. Littermates were housed in a temperature and humidity controlled specific pathogen free facility in micro-isolator cages on a 12 hour light/dark schedule. Mice had access to standard mouse chow and water ad libitum, and were provided with species-appropriate enrichment. Recombinant mouse TNFα (50 μg/kg) was injected supracalvarially daily for 4 days; 24 hours later, calvarial tissues were removed for histology. Mice of each genotype were randomly assigned to vehicle or TNFα treatment groups.
Micro-CT and histomorphometric analyses
Right femurs from Srgap2-cKO and littermate control mice were fixed in 70% ethanol. Metaphyseal region of the distal femur was analyzed at 8 μm voxel size resolution using micro-CT (μCT40, Scanco Medical AG, Bassersdorf, Switzerland) to quantify trabecular and cortical morphometry (30). The sample sizes used are sufficient to detect a 20–25% difference in bone parameters with 80% power, p<0.05 with 14% coefficient of variation. Left femurs were fixed in 4% paraformaldehyde, decalcified in 14% EDTA, embedded in paraffin blocks, sectioned at 7 μm thickness, and subjected to TRAP and hematoxylin staining.
For dynamic bone histomorphometry, 10 mg/kg calcein (Sigma-Aldrich) and 30 mg/kg alizarin-3-methyliminodiacetic acid (Sigma-Aldrich) were injected into mice 7 and 2 days prior to sacrifice. Femurs were fixed in 4% paraformaldehyde for 6 days, incubated in 30% sucrose overnight and embedded in OCT compound (Thermo Fischer Scientific). Femurs were sectioned at 7 μm thickness using a cryostat (Leica) and tape transfer system (Section-lab). Sections were analyzed using Osteomeasure software (OsteoMetrics) and quantified 0.2 mm from growth plates and cortices, as recommended by the Nomenclature Committee of the American Society for Bone and Mineral Research (31).
ELISA
Serum was harvested at time of euthanasia from animals at 8 weeks of age and subjected to ELISA as per manufacturer’s protocol. Bone resorption was quantitated with CTX (Mouse CTX ELISA, MyBioSource), and bone formation was quantitated with P1NP (Mouse P1NP ELISA, MyBioSource) ELISAs. SLIT3 ELISA (Biomatik, Cambridge, ON, Canada) was performed using serum from control and Srgap2-cKO mice, as well as conditioned media from BMM cultures that were treated with M-CSF and RANKL for 5 days.
RANKL- and TNF-induced osteoclast formation assay
Bone marrow cells were flushed from tibias and femurs with αMEM and cultured overnight on tissue culture plastic to remove stromal cells. Nonadherent cells were subjected to Ficoll-Hypaque (GE Healthcare) density gradient to prepare bone marrow monocytes (BMMs) from the buffy coat layer. BMMs (2X104 cells/well in 96 well plate) were cultured with recombinant human M-CSF (30 ng/ml) and human RANKL (30 ng/ml) or mouse TNFα (30 ng/ml) for up to 5 days. Culture medium and cytokines were refreshed at day 3. Multinucleated osteoclasts were stained for TRAP and cells with 3 or more nuclei were counted.
Recombinant human M-CSF and human RANKL were expressed in our laboratory using the constructs kindly provided by Dr. D. Fremont from Washington University and Dr. Y. Choi from University of Pennsylvania, respectively. Recombinant murine TNF was prepared in our laboratory as follows: a murine TNF-α cDNA fragment encoding amino acid residues 83–235 was cloned by PCR using primers 5’-CCCCATATGCTCAGATCATCTTCTCAA-3’ and 5’-CCCCTCGAGTCACAGAGCAATGACTCC-3’. The PCR product was digested with NdeI and XhoI, and cloned into a pET28a expression vector (EMD Biosciences, Billerica, MA) to generate a HIS-fusion protein. HIS-TNFα was expressed in Escherichia coli BL21 cells (Stratagene, La Jolla, CA).
Resorption pit formation assay
BMMs were cultured on collagen type I-coated plates with hM-CSF (30 ng/ml) and hRANKL (30 ng/ml) for 5 days. Multinucleated osteoclasts were collected by digestion using 0.1% collagenase P (Roche), and equal numbers of osteoclasts were cultured on bovine cortical bone slices with hM-CSF (30 ng/ml) and hRANKL (30 ng/ml) for an additional 24 hours. Cells were removed by sonication, and bone slices were stained with 1% toluidine blue. The resorption pits were viewed via light microscopy (Olympus BX53) and measured using Olympus CellSens imaging software.
Flow cytometry
BMMs were incubated in ACK lysing buffer (Thermo Fisher Scientific) for 5 minutes and washed with Hanks’ balanced salt solution containing 10 mM HEPES (pH 7.4) and 2% FBS. Cells were incubated with fluorochrome-conjugated antibodies: anti-CD3, anti-CD45R (B220), anti-CD11b (Mac-1), anti-CD117 (c-kit) and anti-CD115 (c-Fms) for 30 min on ice. All antibodies were purchased from eBioscience. After washing, cells were analyzed using a LSR flow cytometer and FlowJo software (Tree Star).
Osteoblast differentiation assay
To prepare primary osteoblasts, cells were liberated from neonatal C57BL/6 mouse calvaria by 5 sequential 15 minute incubations with 0.1% bacterial collagenase (Collagenase P, Roche), and 0.1% trypsin. Cells were collected by centrifugation after each digestion and washed with DMEM and 10% heat inactivated fetal calf serum (HIFCS, HyClone). Cells obtained from digestions 4–5 were pooled and used as primary osteoblasts (32).
Osteoclast conditioned medium was collected from cells on day 5 of culture and concentrated 100 fold using 100 kDa-pore size Amicon® Ultra-4 Centrifugal Filter Unit (Millipore), to eliminate low molecular weight proteins. Concentrated osteoclast conditioned medium was added at 1:50 ratio onto osteoblasts cultured with 8 mM β-glycerophosphate and 50 μg/ml ascorbic acid. Recombinant SLIT3 0.5 μg/ml (R & D Systems) was used as a positive control. Osteoblast differentiation was induced for up to 12 days. Culture medium was replaced every 3 days. In some experiments, neutralizing antibody to SLIT3 (5 μg/ml, R & D Systems) was added to cultures along with the osteoclast conditioned medium.
Osteoblastic cells were stained for alkaline phosphatase activity using Leukocyte Alkaline Phosphatase Kit (Sigma-Aldrich). Alkaline phosphatase activity per ug protein was evaluated using the substrate 2 mg/ml p-nitrophenyl phosphate as previously described, and absorbance was measured at 415 nm (33). For calcium deposition assay, cells were fixed in 4 % paraformaldehyde for 5 minutes and stained with 1 % alizarin red S (ARS, pH 5.5). The ARS stains were eluted in 10 % acetic acid and absorbance was measured at 415 nm.
RNA extraction and RT-PCR
TRI reagent (Molecular Research Center Cincinnati, OH) was used to extract total RNA from either control or Srgap2-cKO BMM cultures that had been treated with M-CSF and RANKL for 4 days. Total RNA was converted to cDNA by reverse transcriptase (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems, Carlsbad, CA) using random hexamer primers, and aliquots of RT mixtures were used for PCR amplification. PCR amplification was done using gene-specific real time PCR primers (Applied Biosystems) and expressed as fold differences compared WT controls.
Western blot antibodies
Rabbit polyclonal anti-Srgap2 antibody generated in-house by Dr. Franck Polleux was previously described (34). The following commercial antibodies were used for Western blotting: mouse anti-NFATc1 (BD Biosciences); rabbit anti-c-Src, rabbit anti-p-IκB, rabbit anti-p-ERK, rabbit anti-ERK, rabbit anti-p-JNK, rabbit anti-JNK, rabbit anti-p-p38, rabbit anti-p38, and rabbit anti-β-actin antibodies (Cell Signaling Technology); mouse anti-Myc (Millipore); anti-SLIT3 antibody (Abnova).
Rho GTPase activity assay
BMMs were cultured with hM-CSF and hRANKL (both at 30 ng/ml) or hM-CSF and mTNFα for 3 days and serum starved for 2 hours. Whole cell extracts were incubated with GST-fused PAK1 PBD for overnight. GTP bound Rac1 or Cdc42 were pulled down by GST pull down assay (Cell BioLabs, Inc).
Retroviral transduction
Myc-tagged constitutively active form of Rac1V12 and Cdc42V12 in pMX-puro vectors were transfected into the retrovirus packaging cell line, Plat-E (Cell Biolabs) using Lipofectamine 2000 (Invitrogen). Retroviral vectors and Plat-E cells were kindly provided by H. Kim (University of Pennsylvania, Philadelphia, USA) (25). After 48 hours, retroviral supernatants plus 8 μg/ml polybrene were used to transduce BMMs expanded for 2 days in medium containing hM-CSF (150 ng/ml). Transduced BMMs were detached with Trypsin/EDTA and re-plated with hM-CSF (30 ng/ml) and puromycin (2 μg/ml) for additional 2 days. Puromycin-resistant cells were used and expression of the constructs was confirmed by Western blotting using anti-Myc antibody.
Statistical analysis
All data are expressed as mean ± SEM. In vitro data were obtained from at least 3 independent experiments. Statistical analysis was evaluated by paired Student’s t-test to compare two groups. Results were considered significant when p < 0.05.
RESULTS
Srgap2 deficiency increases Rac1 activity and resorption activity
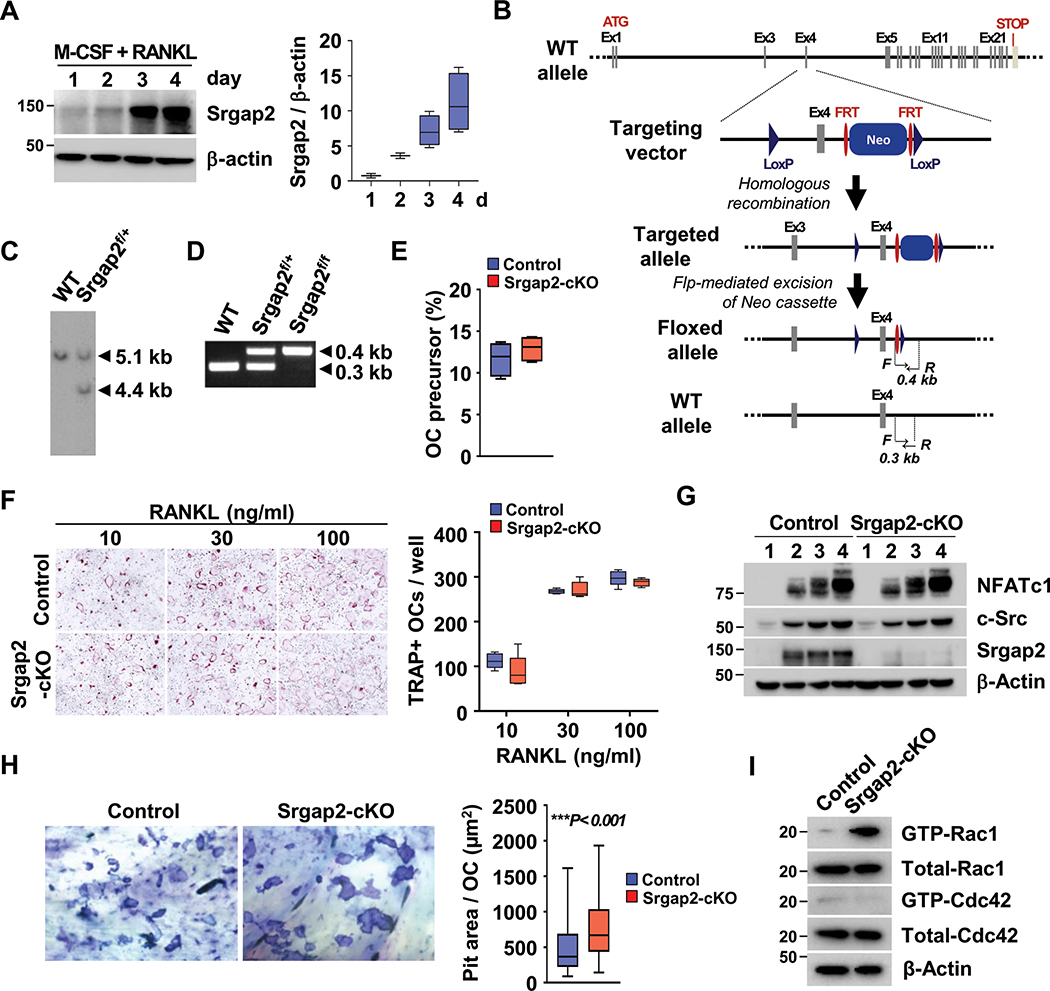
As previously reported, a CD11b−/lowCD45R−CD3−CD115+ population from mouse bone marrow contains a highly efficient population of osteoclast precursors (35). Microarray analysis performed on this FACS sorted population cultured with M-CSF or M-CSF and RANKL showed a 22 fold increase in Srgap2 expression in cells cultured with RANKL (Suppl. Figure 1A). We therefore determined Srgap2 protein levels during osteoclastogenesis by culturing bone marrow monocytes (BMMs) with M-CSF and RANKL. Srgap2 protein was progressively induced and highly expressed in pre- and multinucleated osteoclasts on day 3 and 4 of differentiation, imply that Srgap2 may play a role in osteoclast differentiation and/or function (Figure 1A). For comparison, we also determined whether cultured neonatal mouse calvarial osteoblasts express Srgap2 (Suppl. Figure 1B). We found that these primary osteoblasts expressed similar levels of Srgap2 protein when cultured in standard growth medium and when cultured in osteogenic medium, suggesting constitutive expression.
Figure 1.
Deficiency of Srgap2 affects function of osteoclasts. (A) Srgap2 expression during osteoclast differentiation with M-CSF and RANKL for up to 4 days. (B, C, D) Schematic representation of Srgap2 genomic locus and targeting vector used to induce homologous recombination in mouse embryonic stem (ES) cells. Flp-mediated recombination of the FRT sites deleted the neomycin cassette. The LoxP sites flanking Exon 4 will allow Cre-recombinase mediated excision of Exon 4 and efficient interruption of open reading frame. (C) Southern blot analysis of AflII-digested genomic DNA using a probe distinguishing the targeted allele (4.4 kb) from the wild-type allele (5.1 kb). (D) The wild type and Srgap2f/f alleles can be genotyped by PCR of genomic DNA using primers indicated in bottom two panels of (B). (E) Flow cytometric analysis of a CD11b−/lowCD45R−CD3−CD115+ population for osteoclast precursors was performed. (F) Osteoclast formation assay with 30 ng/ml M-CSF and increasing RANKL concentrations using BMMs from Srgap2-cKO and littermate control mice. (G) Western blot analysis of the osteoclastogenic factors NFATc1 and c-Src during days 1 to 4 of RANKL-induced osteoclastogenesis. Deletion of Srgap2 was confirmed by Srgap2 Western blotting; β-actin was used as control. (H) Bone resorption assay. Equal numbers of osteoclasts differentiated on collagen type I coated plates were cultured on bone slices for 24 hours and pit area per osteoclast was measured. A representative of the pit formation stained by 1% toluidine blue is shown. (I) BMMs were cultured with M-CSF and RANKL for 3 days and starved serum for 2 hours. Active forms of Rac1 and Cdc42 were detected by pull-down assays. Total Rac1 and Cdc42 were detected in whole cell lysate and β-Actin was used as an internal control. ***, p<0.001. Data represent mean ± SEM. Cells were isolated from female mice.
To determine the role of Srgap2 in osteoclast differentiation and function, we generated Srgap2 conditional knockout mice carrying LoxP sites flanking Exon 4 (Srgapf/f; Figure 1B–D), and induced monocyte/macrophage lineage-restricted deletion by crossing floxed mice with mice expressing Cre under the control of the lysozyme 2 promoter (LysMCre) [B6N.129P2(B6)-Lyz2tm1(cre)Ifo/J, Jackson Laboratory]. To determine whether deletion of Srgap2 in LysM-expressing cells impacted the abundance of osteoclast precursors in bone marrow, we performed flow cytometric analysis (Figure 1E). Both control (Srgap2f/f) and Srgap2-cKO (Srgap2f/f:LysM-Cre) mice had a similar number of the CD11b−/lowCD45R−CD3−CD115+ osteoclast precursor population. The ability of Srgap2 to regulate osteoclast formation was evaluated using BMMs from Srgap2-cKO and control mice cultured with M-CSF and RANKL at various concentrations. Srgap2 deficient BMMs formed multinucleated osteoclasts similar to control cells, and normally expressed osteoclastogenic marker proteins NFATc1 and c-Src (Figure 1F and G). However, resorption pit assays performed using equal numbers of purified mature osteoclasts cultured on bone slices showed that Srgap2-cKO cells generated increased pit area compared to controls, although osteoclasts from control and Srgap2-cKO mice express similar level of cathepsin K (Figure 1H and Suppl. Figure 5A).
Since it is known that the GAP protein domain of Srgap2 specifically inhibits Rac1 activity in neuronal cells (36), we assessed Rac1 activity in osteoclast precursor cells. BMMs were cultured with M-CSF and RANKL for 3 days, then serum and cytokines were depleted to determine basal level of Rac1 activity. The active GTP-bound form of Rac1 was greater in Srgap2-cKO cells compared to control, while there was no difference in Cdc42 activation, suggesting specificity for Rac1 in the osteoclast lineage as well as in neuronal cells (Figure 1I). These results indicate that Srgap2 modulates Rac1 activity and osteoclast function without altering RANKL-induced osteoclast differentiation.
Deletion of Srgap2 in myeloid cells increases bone mass due to enhanced bone forming activity
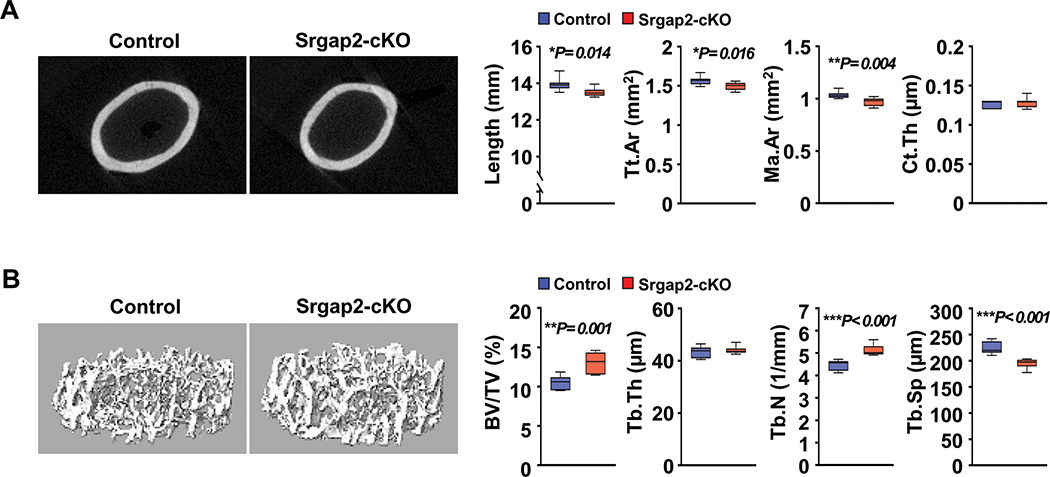
To determine the physiological role of Srgap2 in myeloid lineage cells in the skeleton, we performed μCT analysis of femurs from 8 week old control and Srgap2-cKO mice. In males, significant differences in cortical bone parameters were not observed between genotypes (Suppl. Figure 2A). In contrast, female Srgap2-cKO mice displayed significantly reduced femur length, cortical area and marrow area compared to littermate controls, while cortical thickness was similar to control mice (Figure 2A). Although osteoclasts from Srgap2-cKO mice showed increased bone resorption activity in vitro, trabecular bone volume (BV/TV) was normal in Srgap2-cKO male mice and significantly increased (29%) in Srgap2-cKO female mice compared to controls (Suppl. Figure 2B and Figure 2B). In the females, this increased trabecular bone volume was driven by an increase in trabecular number and decreased trabecular spacing, without alterations in trabecular thickness. Due to the sex-specific impact of Srgap2 deletion in the monocyte lineage, we focused our subsequent analyses on bones and cells from females.
Figure 2.
Micro-CT analysis of femurs from Srgap2-cKO female mice. (A) μCT analysis of cortical bone of femurs from 8-week-old Srgap2-cKO and littermate controls. Total cortical area (Tt.Ar); Marrow area (Ma.Ar); Cortical thickness (Ct.Th). (B) μCT analysis of trabecular bone of femurs from 8-week-old Srgap2-cKO and littermate control mice. Bone volume/total volume (BV/TV); Trabecular thickness (Tb.Th); Trabecular number (Tb.N); Trabecular spacing (Tb.Sp). N=7–8. *, p<0.05, **, p<0.01, ***, p<0.001. Data represent mean ±SEM.
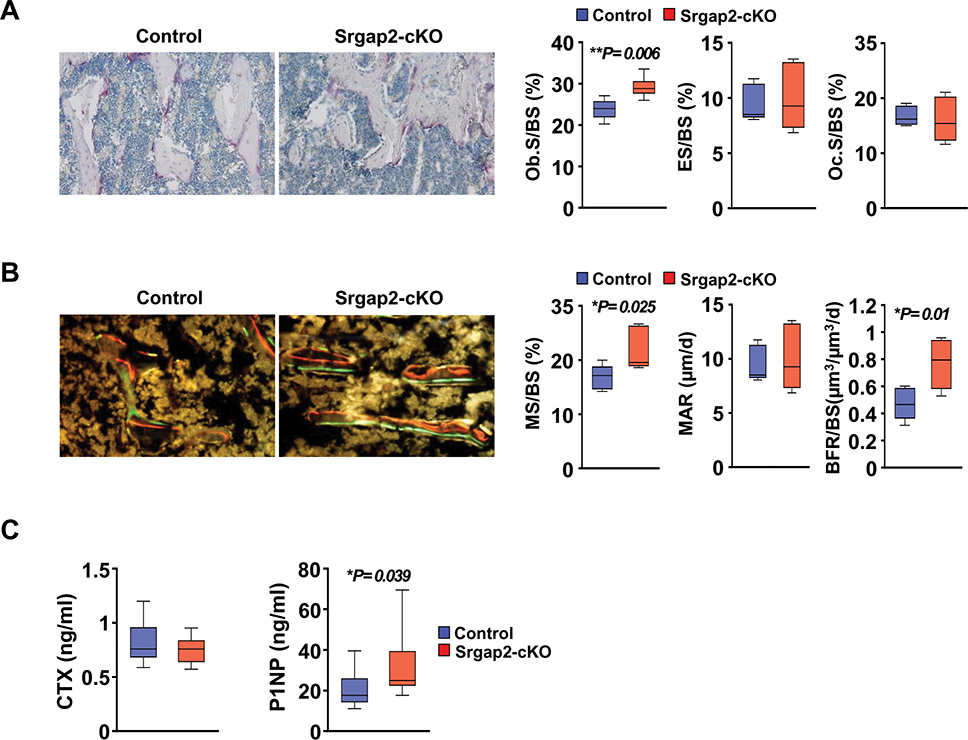
Static and dynamic histomorphometric analysis showed a significant increase in osteoblast surface (Ob.S/BS, 22%), mineralizing surface (MS/BS, 30%) and bone formation rate (BFR/BS, 68%) in Srgap2-cKO female mice, without alterations in eroded surface (ES/BS) and osteoclast surface (Oc.S/BS) (Figure 3A and B). Evaluation of serum markers for bone formation and resorption corroborated these histomorphometry data. Specifically, P1NP level were significantly higher in serum from Srgap2-cKO mice when compared to control mice, while no significant differences were observed in the CTX levels between control and Srgap2-cKO mice (Figure 3C). Together, these results suggest that Srgap2 deficiency in the myeloid lineage promotes osteoblast activity, resulting in increased bone formation and trabecular bone volume.
Figure 3.
Histomorphometric analysis of femurs from Srgap2-cKO female mice. (A) Static histomorphometric analysis of femurs from 8-week-old female Srgap2-cKO and littermate control mice. Sections were stained for TRAP activity and counterstained with hematoxylin. Osteoblast/bone surface (Ob.S/BS); Eroded surface/bone surface (ES/BS); Osteoclast surface/bone surface (Oc.S/BS). (B) Dynamic histomorphometric analysis of femurs double-labeled with calcein and alizarin complexone from 8-week-old Srgap2-cKO and littermate control mice. Mineralized surface/bone surface (MS/BS); Mineral apposition rate (MAR); bone formation rate/bone surface (BFR/BS). N=5–6. (C) Serum CTX and P1NP were measured from control and Srgap2-cKO mice. N=12–13. *, p<0.05, **, p<0.01. Data represent mean ±SEM.
Rac1 activation by Srgap2 deficiency in myeloid cells enhances SLIT3
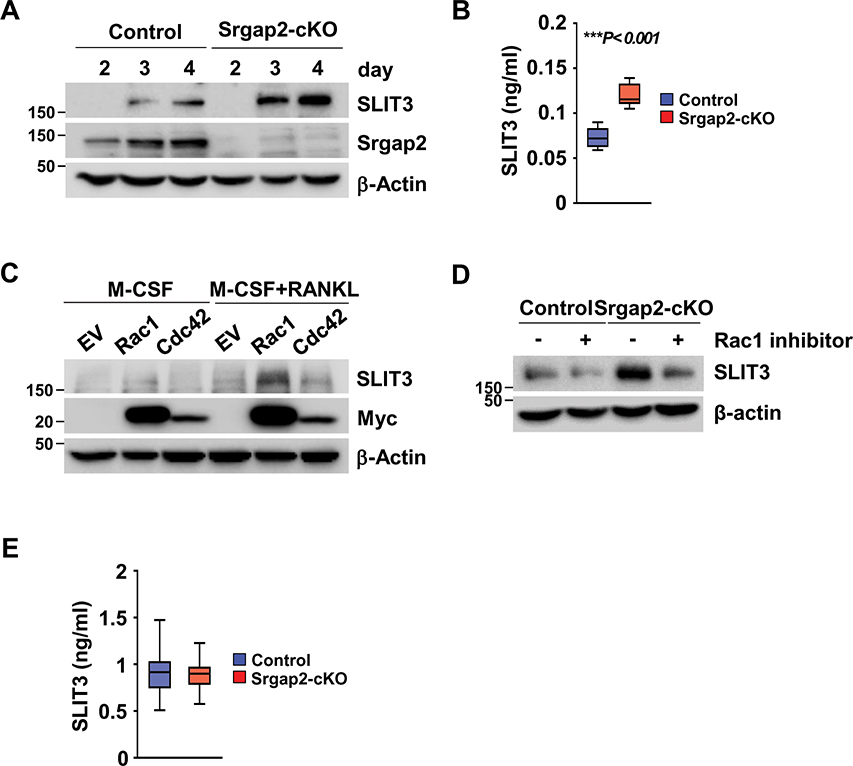
Given that deletion of Srgap2 in the myeloid lineage caused an increase in bone formation, we considered potential clastokines that may mediate such a response. Recently, SLIT3 was identified by two different groups as an osteoclast- and osteoblast-derived bone anabolic factor that acts in a paracrine fashion (19, 37). Western blot analysis confirmed that SLIT3 protein was induced in BMMs cultured in the presence of M-CSF and RANKL; moreover, expression of SLIT3 was greater in differentiated osteoclasts from Srgap2-cKO mice compared to control (Figure 4A). ELISA confirmed the increased levels of SLIT3 secreted into conditioned medium from Srgap2-cKO osteoclasts (Figure 4B).
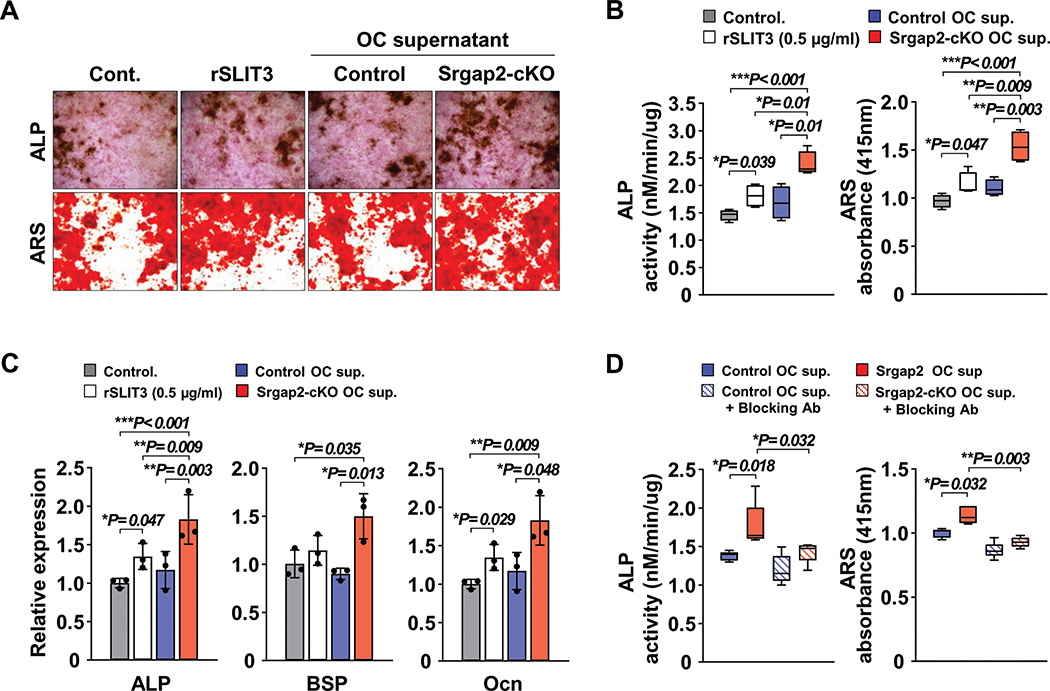
Figure 4.
Protein levels of SLIT3 in Srgap2 deficient and Rac1-activated osteoclasts. (A) SLIT3 was detected by Western blotting of lysates from Srgap2-cKO and control BMMs cultured with M-CSF and RANKL for up to 4 days. Srgap2 was detected to confirm the genotype and β-Actin was used as an internal control. (B) SLIT3 levels were measured in the conditioned medium from control and Srgap2-cKO cells that were treated with M-CSF and RANKL for 5 days. (C) SLIT3 proteins were detected by Western blotting in wild type BMMs transduced with empty vector (EV), Myc-tagged constitutively active Rac1 or Myc-tagged constitutively active Cdc42. Transduced BMMs were cultured with M-CSF and RANKL for 4 days. Transduced genes were confirmed by Western blotting with anti-Myc antibody and β-Actin was used as a loading control. (D) BMM cells from control and Srgap2-cKO mice were cultured with or without NSC23766, a specific Rac1 inhibitor, for 4 days in the presence of M-CSF and RANKL. (E) SLIT3 levels in the serum from control and Srgap2-cKO mice. N=12–13. ***, p<0.001.
Srgap2-cKO cells display increased Rac1 activation (Figure 1I); to establish if up-regulation of SLIT3 could be mediated by Rac1 activation, we cultured BMMs expressing constitutively active Rac1 or Cdc42 with M-CSF and RANKL and examined SLIT3 levels. Western blot analysis revealed that SLIT3 was greater in constitutively active Rac1-expressing BMMs compared to empty vector or BMMs expressing constitutively active Cdc42 (Figure 4C). To further confirm the activation of Rac1 caused enhanced SLIT3 expression, we used NSC23766, a specific inhibitor of Rac1 activation (38). As shown in Figure 4D, SLIT3 expression was reduced in cultures that were treated with NSC23766 in both control and Srgap2-cKO cells. Since it is possible to detect bone-derived peptides in serum, we measured SLIT3 in serum from WT and cKO mice (Figure 4E). However, significant differences in serum SLIT3 levels between WT and Srgap2-cKO mice were not detected. This was not wholly unexpected, as Slit3 can be expressed in other tissues such as neuronal cells, heart and kidney. Therefore, the increased levels SLIT3 derived from Srgap2-deficient monocyte lineage cells is not likely to have a systemic effect in vivo. Overall, these results indicate that SLIT3 is highly expressed in Srgap2 deficient osteoclastic cells, an effect that can be driven by Rac1 activation.
Conditioned medium from Srgap2 deficient osteoclastic cells promotes osteoblast differentiation
Although SLIT3 is a bone anabolic clastokine, other osteoclast derived-coupling factors have been published (e.g., BMP6, EphrinB2, CTHR1, Wnt10b and SPHK1). To determine whether the expression of these might be altered in Srgap2-cKO cells, we examined their mRNA expression in BMM cultures by real time PCR. As shown in Suppl. Figure 3, none of these factors were significantly altered in Srgap2-cKO osteoclasts. To confirm that SLIT3 secreted from Srgap2 deficient myeloid cells enhances osteoblast differentiation, we collected conditioned medium from Srgap2-cKO and control osteoclast cultures and examined its impact on osteoblast differentiation. Most osteoclast derived-coupling factors published thus far are 27–56 kDa proteins, while SLIT3 is a much larger ~168 kDa protein. Therefore, we used a 100K NMWL (normal molecular weight limit) filter to first exclude molecules smaller than 100kDa and to concentrate osteoclast culture supernatants. Conditioned medium concentrates were used to treat primary calvarial osteoblast cells cultured in the presence of β-glycerol phosphate and ascorbic acid (osteogenic medium). Alkaline phosphatase (ALP) activity is critical for collagen crosslinking and is an early osteogenic marker. Whereas recombinant SLIT3 increased both ALP staining and activity assays, culture supernatant from Srgap2-cKO osteoclasts further enhanced osteoblast differentiation compared to culture supernatant from control (Figure 5A and B). Deposition of mineralized matrix is a hallmark of differentiated osteoblasts. Alizarin red S (ARS) staining and quantification revealed increased calcium deposition in the cell layer of cultures treated with supernatant from Srgap2-cKO osteoclasts (Figure 5A and B). Likewise, mRNA expression of osteoblastic marker genes such as ALP, bone sialoprotein (BSP) and osteocalcin (OCN) were also significantly increased in cultures that received supernatant from Srgap2-cKO osteoclasts (Figure 5C). To confirm this enhancement in osteoblast differentiation in Srgap2-cKO conditioned medium treated cultures was due to increased SLIT3 expression, we used neutralizing antibody specific to SLIT3 in similar conditioned medium transfer experiments. As shown in Figure 5D, conditioned medium from Srgap2-cKO cultures enhanced both ALP activity and ARS, this effect was ablated in cultures co-treated with SLIT3 specific neutralizing antibody. These data indicate that increased SLIT3 secretion by Srgap2 osteoclasts promotes osteoblastic differentiation.
Figure 5.
Conditioned medium from Srgap2 deficient osteoclastic cells promotes osteoblastic differentiation in vitro. Recombinant SLIT3 (rSLIT3), conditioned medium of osteoclast cultures from Srgap2-cKO or control mice were added to osteoblasts cultured with β-glycerophosphate and ascorbic acid. (A) Representative ALP and ARS staining of osteoblastic cultures. (B) Alkaline phosphatase (ALP) activity assay and Alizarin red S (ARS) eluted from stained wells was measured at 415nm absorbance. (C) Osteoblastic gene expression levels, alkaline phosphatase (ALP), bone sialoprotein (BSP), osteocalcin (Ocn) relative to Gapdh were analyzed by quantitative real-time PCR. (D) ALP activity and ARS measurement after treating the cells with control or Srgap2-cKO osteoclast conditioned medium in the presence or absence of SLIT3 neutralizing antibody, to block SLIT3 activity. *, p<0.05, **, p<0.01, ***, p<0.001. Data represent mean ±SEM.
Deficiency of Srgap2 in myeloid cells enhanced TNFα-induced osteoclastogenesis
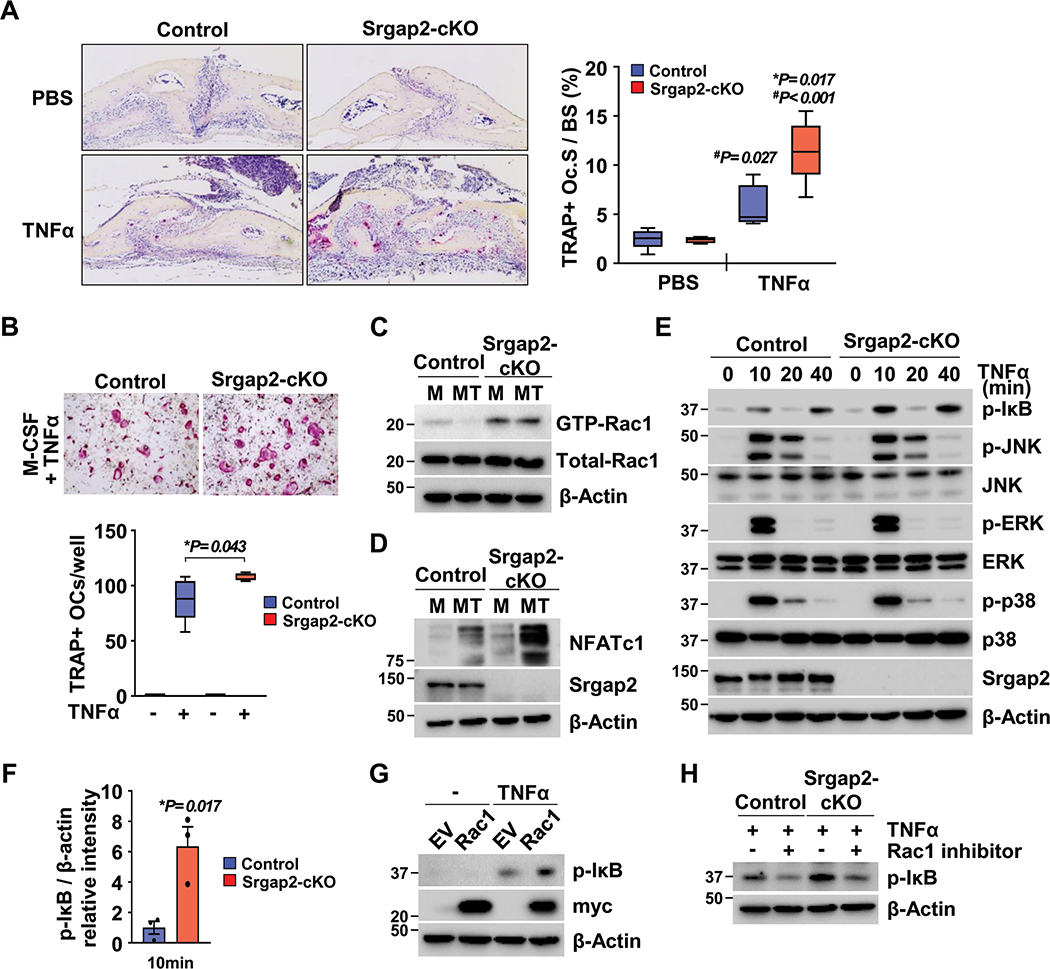
Since Rac1 is rapidly activated by TNFα and required for TNFα-induced MAP kinase signaling (39), we investigated if Srgap2 deficiency in the myeloid lineage affects TNFα-induced osteoclastogenesis in vivo and in vitro. Recombinant TNFα was injected into mice over the calvaria once a day for 4 days, and TRAP+ osteoclast surface area was evaluated. TRAP stained calvarial sections showed that TNFα challenge led to a significant increase in TRAP+ osteoclast surface area in Srgap2-cKO mice compared to TNFα-treated littermate controls (Figure 6A). In vitro studies confirmed that Srgap2-cKO BMMs were more responsive to TNFα-induced osteoclast formation in vitro (Figure 6B). This is in contrast to RANKL-induced osteoclast formation, which was comparable between Srgap2-cKO and control (Figure 1F). Further, TNFα treatment more dramatically increased NFATc1 protein level in Srgap2-cKO cells (Figure 6D).
Figure 6.
Regulation of Rac1 activity by Srgap2 is critical for TNFα-induced osteoclastogenesis in vivo and in vitro. (A) Srgap2-cKO and littermate control mice were injected with PBS or TNFα daily for 4 days over the calvaria. Calvarial sections were stained for TRAP with hematoxylin counterstain. TRAP(+) osteoclast surface area was quantified. (B) BMMs from Srgap2-cKO and littermate control mice were cultured with M-CSF and TNFα for 5 days. A representative image of osteoclasts stained for TRAP is shown and TRAP(+) osteoclasts were counted. (C) BMMs were cultured with M-CSF with or without TNFα for 3 days and starved serum for 2 hours. Active form of Rac1 was detected by pull-down assays. Total Rac1 was detected from whole cell lysate and β-Actin was used as an internal control. (D) Western blot analysis of NFATc1 in cultures of Srgap2-cKO and control BMMs cultured in M-CSF alone (M) or M-CSF and TNFα (MT) for 3 days. (E) BMMs from Srgap2-cKO and littermate mice cultured with M-CSF for 3 days were starved serum for 2 hours and then exposed to TNFα for up to 40 minutes. Effect of Srgap2 deficiency on TNFα signaling were assessed by Western blotting of phosphorylated of IκB, JNK, ERK, and p38. Blots were reprobed for total JNK, ERK, and p38. Srgap2 was detected to confirm genotype and β-Actin was used an internal control. (F) Fold induction of phospho-IκB in Srgap2-cKO cells compared to control cells in response to TNFα for 10 minutes. (G) BMMs transduced empty vector (EV) or Myc-tagged constitutively active Rac1 were starved serum for 2 hours and then exposed to TNFα for 15 minutes. Phosphorylation of IκB, was assessed by Western blotting. Transduced genes were confirmed by Western blotting with anti-Myc antibody and β-actin was used an internal control. (H) BMMs from control and Srgap2-cKO mice were cultured with M-CSF for 3 days, serum starved and pretreated with the Rac1 inhibitor (NSC23766), for 2 hours, then treated with TNF for 10 minutes. *, Significant effect of Srgap2 deficiency, p<0.05; significant effect of TNFα treatment, #, p<0.05,. Data represent mean ±SEM.
To examine the impact of TNFα on basal Rac1 activation, BMMs were cultured with M-CSF in the presence or absence of TNFα for 3 days, then serum and cytokines were depleted. The active GTP-bound form of Rac1 level was greater in Srgap2-cKO cells compared to control, both in the presence or absence of TNFα (Figure 6C). Although TNF-induced osteoclast formation was enhanced in Srgap2 cultures and these cells displayed increased basal level of Rac1 activation, TNF-induced osteoclastic differentiation was not accompanied by an increase in SLIT3 expression (Suppl. Figure 4). This is in contrast to RANKL-induced osteoclastogenesis, which causes a dramatic increase in SLIT3 expression (Figure 1A).
To investigate the mechanism which leads to increased osteoclast formation in Srgap2-cKO in response to TNFα treatment, we examined whether Srgap2 deficiency affected TNFα-induced NF-κB and MAP kinase activation. IκB phosphorylation, an indicator of NF-κB activation, was highly induced by TNFα stimulation in Srgap2-cKO BMMs (Figure 6E and F). In contrast, TNFα activation of MAP kinase pathway components (JNK, ERK and p38) was similar in control and Srgap2-cKO cells. To confirm that up-regulation of NF-κB by TNFα can be mediated by Rac1 activation, we treated BMMs expressing constitutively active Rac1 with TNFα. We found that IκB phosphorylation was increased by TNFα in constitutively active Rac1-expressing BMMs (Figure 6G). Further, treatment with the Rac1 inhibitor NSC23766 dramatically downregulated IκB phosphorylation in both control and Srgap2-cKO cells cultured in TNFα (Figure 6H). Taken together, these results demonstrate that Rac1 is highly activated in TNFα treated Srgap2-cKO cells, with a corresponding activation of NF-κB and enhanced osteoclastogenesis, suggesting Srgap2 control of Rac1 activity is an important anti-inflammatory mechanism.
DISCUSSION
This study connects Rac1 signaling activity in osteoclasts with clastokine-mediated bone anabolism and enhanced osteoclastogenesis in response to inflammatory cues. We demonstrated that expression of the Rac1 specific GTPase activating protein Srgap2 is progressively increased during the course of RANKL-induced osteoclastogenesis in vitro. Deletion of Srgap2 resulted in high levels of Rac1 activation. While this modestly increased osteoclastic activity, it caused a dramatic increase in expression of the bone anabolic clastokine SLIT3. Thus, in vivo, Srgap2 deletion in the myeloid lineage promoted bone formation without altering osteoclastic parameters. Lastly, Srgap2-cKO mice displayed enhanced osteoclast formation in response to the inflammatory cytokine TNFα, an effect mediated by accentuated NF-kB signaling. These data suggest that Srgap2 activity in the osteoclast lineage restrains osteoclast-osteoblast coupling and tempers osteoclastogenesis in response to inflammatory mediators.
Osteoclasts express both Rac1 and Rac2, which appear to have overlapping and compensatory functions in this cell type. Previous studies exploring the role of Rac in osteoclasts have been primarily limited to “loss of function” studies performed using mice deficient for Rac1 and/or Rac2 or the Rac-specific GTP exchange factors Vav3 and Dock5 (23, 28, 29). These studies indicate that Rac activity is more important for RANKL-induced osteoclast function than formation; our in vitro studies support this concept (Figure 1). Although Vav3 appears to be constitutively expressed during osteoclastogenesis in vitro, Dock5 expression is low in committed cells and increases during maturation (28, 29). In this regard, the expression pattern of Srgap2 is most similar to that of Dock5 (Figure 1A). It is likely that based on their localization within the cell, these GAP/GEFs fine tune Rac1 activation in osteoclasts.
Our observation that Srgap2 is expressed in the osteoclast lineage is novel; most previous work on Srgap2 function was performed in neuronal cells. There, Srgap2 decreases migration and promotes filopodia-like membrane protrusions needed for neurite outgrowth and branching, a process requiring Srgap2 to recognize concave membrane curvatures (40). Recently, Srgap2 was shown to be critical for the polarization of neutrophils, stabilizing plasma membrane curvature and activating signaling cascades needed for polarization (41). Future studies could investigate localization of Srgap2, Rac1 and Rac-specific GEFs in osteoclasts cultured on bone slices, to examine distribution in basal vs apical surfaces.
Rac1 activation was strongly increased in Srgap2-cKO osteoclasts, and constitutive activation of Rac1 in osteoclastic cells promoted the expression of the clastokine SLIT3 (Figures 1I and 4C). Other investigators demonstrated that, in BMMs treated with M-CSF and RANKL, the addition of exogenous SLIT3 inhibited Rac1 activation. This effect of SLIT3 on osteoclastic cells was mediated by the ROBO1 receptor, which interacts with the SH3 domain of Srgap2 (19, 42). These data suggest a feedback loop in which Rac activity stimulates the expression of SLIT3, which in turn acts in an autocrine manner to limit Rac activation in osteoclastic cells. However, our studies did not reveal decreased osteoclast parameters in Srgap2-cKO mice in vivo, nor was RANKL-induced osteoclastogenesis in vitro affected, suggesting perhaps other compensatory mechanisms.
In addition to autocrine activities, SLIT3 synthesized by osteoclasts has paracrine effects on osteoblasts. To our knowledge, SLIT3 is the only high molecular weight osteoclast-derived coupling factor described to date. SLIT3 synthesis is induced in Srgap2-cKO osteoclasts, which promoted osteoblastic differentiation in vitro (Figure 5). Importantly, mice with myeloid lineage-restricted deletion of Srgap2 displayed increased trabecular bone volume and increased bone formation rate, while osteoclast surface and eroded surface remained unchanged (Figures 2 and 3). These results complement published studies demonstrating that deletion of Slit3 in cathepsin K expressing cells decreased bone formation rate and trabecular bone volume, although osteoclast number and eroded surface were also increased in this mouse model (19). While the impact of osteoblast lineage-derived SLIT3 on the skeleton remains somewhat controversial (19, 37), our data firmly support the concept that osteoclast-derived coupling factors, including SLIT3, play an important role in skeletal homeostasis.
BMMs from male and female mice express similar levels of SRGAP2, and Srgap2-cKO cells from both sexes display dramatically increased Rac1 activation (Suppl. Figure 5). However, at 8 weeks of age, we observed the increased trabecular bone phenotype only in female in Srgap2-cKO mice. Sex-restricted phenotypes in genetic mouse models are not uncommon (43–48). Skeletal sexual dimorphism depends not only on androgen action in males and estrogen action in females, but also on complex sex and time-specific interactions between sex hormones, the growth hormone (GH) / insulin-like growth factor-1 (IGF-1) axis, and mechanical loading. It is worth noting that 8 week/2 month old C57BL/6 mice, particularly the males, are still growing (49). Examination of more skeletally mature mice may reveal differences between control and Srgap2-cKO males, especially since C57BL/6 lose trabecular bone volume with maturity. The bone environment in vivo is complex, and studies are underway to determine the impact of Srgap2 deletion in the monocyte lineage on bone quantity and quality in mature and aging mice of both sexes. Impact of this was not apparent in the complex in vivo environment at 8 weeks of age.
Although inactivation of Srgap2 had no impact on RANKL-induced osteoclast differentiation, the absence of Srgap2 had a profound stimulatory effect on TNFα-induced osteoclastogenesis, in vivo and in vitro (compare Figures 1 and 6). In BMMs, Rac is rapidly activated by TNFα stimulation, through Src-mediated phosphorylation of the VAV family of Rho GEFs. Moreover, this Rac activation is thought to play a critical role in initiating the MAPK signaling cascade in TNFα treated cells (39). While NF-κB signaling was significantly augmented by TNFα in Srgap2-cKO BMMs, the p38, JNK and ERK MAPK signaling pathways were not (Figure 6). These data suggest that the increased osteoclastogenesis in TNFα treated Srgap2-cKO mice was mediated primarily by a boost in NF-κB signaling, and indicate that Srgap2 activity plays a prominent role in limiting inflammatory osteolysis.
While little known about the molecular mechanisms regulating Srgap2 expression, intriguingly, only ~3.6 kb of intergenic DNA separates the Srgap2 gene and its nearest neighbor Fam72a, and this genomic organization is conserved across species. Srgap2 and Fam72a are transcribed in opposite directions and appear to share this short intergenic region as a promoter. In neuronal cells, this intergenic region can mediate co-transcription of Srgap2 and Fam72a or their independent transcription, depending on cellular context (50). However, transcription factors regulating the expression of Srgap2 in osteoclasts or neuronal cells are not yet identified, and the expression of Fam72a in myeloid cells has not been reported. In contrast, we previously demonstrated that miR-29 family members target Srgap2 mRNA. miR-29 positively regulates osteoclastogenesis and expression of the miR-29 family increases during osteoclast differentiation in vitro (51). It is likely that miR-29 tempers Srgap2 expression in maturing osteoclasts. While other miRNAs have been proposed to target Srgap2, these miRNA-target interactions have not yet been validated (52).
Whereas mice have only one Srgap2 gene, humans have three paralogs due to partial human-specific gene duplication events. Human SRGAP2A is orthologous to murine Srgap2, while human SRGAP2C and SRGAP2B are truncated paralogs containing only a partial F-BAR domain (53). SRGAP2C can dimerize with SRGAP2A and inhibit its function (34, 54). The expression of SRGAP2 paralogs in human osteoclastic cells has yet to be determined. However, since deletion of Srgap2 activity in mouse myeloid cells resulted in a bone-anabolic effect in vivo, it is tempting to speculate that interfering with Srgap2 function using small molecules targeted to bone resorbing surfaces or by increasing the expression of SRGAP2B/C may constitute a novel approach for treating osteoporosis.
In summary, up-regulation of Rac1 activation due to Srgap2 deficiency in osteoclasts revealed novel roles for Rac1 in bone homeostasis and in the inflammatory response. Our studies highlight the importance of osteoblast-osteoclast crosstalk in the maintenance of bone mass, and the potential role of SLIT3 in this process. Further, our work suggests that Rac1 activity plays a critical role in the control of inflammatory osteoclastogenesis.
Supplementary Material
Supplemental Figure 1. A) Srgap2 mRNA expression in a population of bone marrow osteoclast precursor cells (CD45R− CD3− CD11b−/lo CD115+), which was FACS sorted and cultured for 4 days with M-CSF or M-CSF + RANKL. Microarray analysis was performed using Illumina Mouse WG-6 Beadchip. N=3. B) Primary osteoblasts express Srgap2. Western blot analysis of Srgap2 in primary osteoblasts prepared from neonatal mouse calvaria and cultured before or after osteogenic medium (with ascorbic acid {AA} and ß-glycerophosphate {BGP}) for 12 days.
Supplemental Figure 2. Micro-CT analysis of femurs from Srgap2-cKO male mice. (A) μCT analysis of cortical bone of femurs from 8-week-old Srgap2-cKO and littermate controls. Total cortical area (Tt.Ar); Marrow area (Ma.Ar); Cortical thickness (Ct.Th). (B) μCT analysis of trabecular bone of femurs from 8-week-old Srgap2-cKO and littermate control mice. Bone volume/total volume (BV/TV); Trabecular thickness (Tb.Th); Trabecular number (Tb.N); Trabecular spacing (Tb.Sp). N=7–8. Data represent mean ±SEM. Statistically significant differences were not detected.
Supplemental Figure 3. Real time PCR analysis of various clastokines in BMM cultures. Bone marrow cells from control and Srgap2 cKO mice were cultured with M-CSF and RANKL for 4 days and real time PCR analysis using gene specific Taqman assay (Applied Biosystems). N=4. Significant differences between control and Srgap2-cKO cultures were not detected.
Supplemental Figure 4. Western blot analysis of SLIT3 expression in BMM cells treated with M-CSF and TNFα for 5 days. M-CSF and RANKL treated WT BMM cells were used as positive control for SLIT3 expression.
Supplemental Figure 5. (A) Comparable levels of Srgap2 and cathepsin K in BMM cultures from male control mice and female control and Srgap2-cKO mice detected by western blot analysis. Female BMMs from Srgap2-cKO mice are shown as negative control for Srgap2 expression. β-Actin was used as an internal control. (B) Increased Rac1 activation in BMM cells from male Srgap2-cKO mice. BMMs were cultured with M-CSF and RANKL for 3 days and starved serum for 2 hours. Active forms of Rac1 was detected by pull-down assays. Total Rac1 was detected in whole cell lysate and β-Actin was used as an internal control.
Acknowledgements
The authors thank the UConn Health MicroCT Core facility for assistance with MicroCT data acquisition.
Funding Sources: Supported by grants from NIH (AR064867 to AD and SKL; NS067557 to FP; NS100323 to ES), The Roger De Spoelberch Foundation (FP), Netherlands Organization for Scientific Research (NOW Rubicon 825.14.017 to ES) and The EMBO (Long Term Fellowship ALTF 1055-2014 to ES)
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Waning DL, Guise TA. Molecular mechanisms of bone metastasis and associated muscle weakness. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(12):3071–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silbermann R, Roodman GD. Myeloma bone disease: Pathophysiology and management. Journal of bone oncology. 2013;2(2):59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorenzo J, Horowitz M, Choi Y. Osteoimmunology: interactions of the bone and immune system. Endocrine reviews. 2008;29(4):403–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takami M, Woo JT, Nagai K. Osteoblastic cells induce fusion and activation of osteoclasts through a mechanism independent of macrophage-colony-stimulating factor production. Cell Tissue Res 1999;298(2):327–34. [DOI] [PubMed] [Google Scholar]
- 5.Chambers TJ. Regulation of the differentiation and function of osteoclasts. J Pathol 2000;192(1):4–13. [DOI] [PubMed] [Google Scholar]
- 6.Abu-Amer Y, Ross FP, Edwards J, Teitelbaum SL. Lipopolysaccharide-stimulated osteoclastogenesis is mediated by tumor necrosis factor via its P55 receptor. J Clin Invest 1997;100(6):1557–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam J, Takeshita S, Barker JE, Kanagawa O, Ross FP, Teitelbaum SL. TNF-alpha induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J Clin Invest 2000;106(12):1481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi K, Takahashi N, Jimi E, Udagawa N, Takami M, Kotake S, et al. Tumor necrosis factor alpha stimulates osteoclast differentiation by a mechanism independent of the ODF/RANKL-RANK interaction. J Exp Med. 2000;191(2):275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anandarajah AP, Schwarz EM, Totterman S, Monu J, Feng CY, Shao T, et al. The effect of etanercept on osteoclast precursor frequency and enhancing bone marrow oedema in patients with psoriatic arthritis. Ann Rheum Dis 2008;67(3):296–301. [DOI] [PubMed] [Google Scholar]
- 10.Yao Z, Li P, Zhang Q, Schwarz EM, Keng P, Arbini A, et al. Tumor necrosis factor-alpha increases circulating osteoclast precursor numbers by promoting their proliferation and differentiation in the bone marrow through up-regulation of c-Fms expression. J Biol Chem 2006;281(17):11846–55. [DOI] [PubMed] [Google Scholar]
- 11.Li P, Schwarz EM, O’Keefe RJ, Ma L, Looney RJ, Ritchlin CT, et al. Systemic tumor necrosis factor alpha mediates an increase in peripheral CD11bhigh osteoclast precursors in tumor necrosis factor alpha-transgenic mice. Arthritis Rheum 2004;50(1):265–76. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Guo R, Schwarz EM, Boyce BF, Xing L. TNF inhibits production of stromal cell-derived factor 1 by bone stromal cells and increases osteoclast precursor mobilization from bone marrow to peripheral blood. Arthritis Res Ther 2008;10(2):R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teitelbaum SL. Osteoclasts; culprits in inflammatory osteolysis. Arthritis Res Ther 2006;8(1):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitaura H, Kimura K, Ishida M, Kohara H, Yoshimatsu M, Takano-Yamamoto T. Immunological reaction in TNF-alpha-mediated osteoclast formation and bone resorption in vitro and in vivo. Clin Dev Immunol 2013;2013:181849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh MC, Choi Y. Biology of the RANKL-RANK-OPG System in Immunity, Bone, and Beyond. Front Immunol 2014;5:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pederson L, Ruan M, Westendorf JJ, Khosla S, Oursler MJ. Regulation of bone formation by osteoclasts involves Wnt/BMP signaling and the chemokine sphingosine-1-phosphate. Proc Natl Acad Sci U S A. 2008;105(52):20764–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao C, Irie N, Takada Y, Shimoda K, Miyamoto T, Nishiwaki T, et al. Bidirectional ephrinB2-EphB4 signaling controls bone homeostasis. Cell Metab 2006;4(2):111–21. [DOI] [PubMed] [Google Scholar]
- 18.Ryu J, Kim HJ, Chang EJ, Huang H, Banno Y, Kim HH. Sphingosine 1-phosphate as a regulator of osteoclast differentiation and osteoclast-osteoblast coupling. EMBO J 2006;25(24):5840–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim BJ, Lee YS, Lee SY, Baek WY, Choi YJ, Moon SA, et al. Osteoclast-secreted SLIT3 coordinates bone resorption and formation. J Clin Invest 2018;128(4):1429–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaananen HK, Zhao H, Mulari M, Halleen JM. The cell biology of osteoclast function. J Cell Sci 2000;113 ( Pt 3):377–81. [DOI] [PubMed] [Google Scholar]
- 21.Chellaiah MA, Soga N, Swanson S, McAllister S, Alvarez U, Wang D, et al. Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J Biol Chem. 2000;275(16):11993–2002. [DOI] [PubMed] [Google Scholar]
- 22.Ito Y, Teitelbaum SL, Zou W, Zheng Y, Johnson JF, Chappel J, et al. Cdc42 regulates bone modeling and remodeling in mice by modulating RANKL/M-CSF signaling and osteoclast polarization. J Clin Invest 2010;120(6):1981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croke M, Ross FP, Korhonen M, Williams DA, Zou W, Teitelbaum SL. Rac deletion in osteoclasts causes severe osteopetrosis. J Cell Sci. 2011;124(Pt 22):3811–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukuda A, Hikita A, Wakeyama H, Akiyama T, Oda H, Nakamura K, et al. Regulation of osteoclast apoptosis and motility by small GTPase binding protein Rac1. J Bone Miner Res 2005;20(12):2245–53. [DOI] [PubMed] [Google Scholar]
- 25.Kim H, Choi HK, Shin JH, Kim KH, Huh JY, Lee SA, et al. Selective inhibition of RANK blocks osteoclast maturation and function and prevents bone loss in mice. J Clin Invest 2009;119(4):813–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan J, Chen S, Zhang Y, Li X, Li Y, Wu X, et al. Rac1 mediates the osteoclast gains-in-function induced by haploinsufficiency of Nf1. Hum Mol Genet 2008;17(7):936–48. [DOI] [PubMed] [Google Scholar]
- 27.Brazier H, Stephens S, Ory S, Fort P, Morrison N, Blangy A. Expression profile of RhoGTPases and RhoGEFs during RANKL-stimulated osteoclastogenesis: identification of essential genes in osteoclasts. J Bone Miner Res 2006;21(9):1387–98. [DOI] [PubMed] [Google Scholar]
- 28.Faccio R, Teitelbaum SL, Fujikawa K, Chappel J, Zallone A, Tybulewicz VL, et al. Vav3 regulates osteoclast function and bone mass. Nat Med 2005;11(3):284–90. [DOI] [PubMed] [Google Scholar]
- 29.Vives V, Laurin M, Cres G, Larrousse P, Morichaud Z, Noel D, et al. The Rac1 exchange factor Dock5 is essential for bone resorption by osteoclasts. J Bone Miner Res 2011;26(5):1099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding M, Danielsen CC, Hvid I, Overgaard S. Three-dimensional microarchitecture of adolescent cancellous bone. Bone. 2012;51(5):953–60. [DOI] [PubMed] [Google Scholar]
- 31.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 2013;28(1):2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakker AD, Klein-Nulend J. Osteoblast isolation from murine calvaria and long bones. Methods Mol Biol 2012;816:19–29. [DOI] [PubMed] [Google Scholar]
- 33.Sabokbar A, Millett PJ, Myer B, Rushton N. A rapid, quantitative assay for measuring alkaline phosphatase activity in osteoblastic cells in vitro. Bone Miner 1994;27(1):57–67. [DOI] [PubMed] [Google Scholar]
- 34.Charrier C, Joshi K, Coutinho-Budd J, Kim JE, Lambert N, de Marchena J, et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149(4):923–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacquin C, Gran DE, Lee SK, Lorenzo JA, Aguila HL. Identification of multiple osteoclast precursor populations in murine bone marrow. J Bone Miner Res 2006;21(1):67–77. [DOI] [PubMed] [Google Scholar]
- 36.Mason FM, Heimsath EG, Higgs HN, Soderling SH. Bi-modal regulation of a formin by srGAP2. J Biol Chem 2011;286(8):6577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu R, Yallowitz A, Qin A, Wu Z, Shin DY, Kim JM, et al. Targeting skeletal endothelium to ameliorate bone loss. Nat Med 2018;24(6):823–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levay M, Krobert K, Wittig K, Voigt N, Bermudez M, Wolber G, et al. NSC23766, a widely used inhibitor of Rac1 activation, additionally acts as a competitive antagonist at muscarinic acetylcholine receptors. J Pharmacol Exp Ther 2013;347(1):69–79. [DOI] [PubMed] [Google Scholar]
- 39.Kant S, Swat W, Zhang S, Zhang ZY, Neel BG, Flavell RA, et al. TNF-stimulated MAP kinase activation mediated by a Rho family GTPase signaling pathway. Genes Dev 2011;25(19):2069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, et al. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138(5):990–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren C, Yuan Q, Braun M, Zhang X, Petri B, Zhang J, et al. Leukocyte Cytoskeleton Polarization Is Initiated by Plasma Membrane Curvature from Cell Attachment. Dev Cell. 2019;49(2):206–19 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas B, Hardin J. Mind the (sr)GAP - roles of Slit-Robo GAPs in neurons, brains and beyond. J Cell Sci 2017;130(23):3965–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Govoni KE, Baylink DJ, Chen J, Mohan S. Disruption of four-and-a-half LIM 2 decreases bone mineral content and bone mineral density in femur and tibia bones of female mice. Calcif Tissue Int 2006;79(2):112–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi C, Iura A, Terajima M, Liu F, Lyons K, Pan H, et al. Deletion of BMP receptor type IB decreased bone mass in association with compromised osteoblastic differentiation of bone marrow mesenchymal progenitors. Sci Rep 2016;6:24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Rundle CH, Wergedal JE, Srivastava AK, Mohan S, Lau KH. Loss of sex-specific difference in femoral bone parameters in male leptin knockout mice. Calcif Tissue Int 2007;80(6):374–82. [DOI] [PubMed] [Google Scholar]
- 46.Callewaert F, Sinnesael M, Gielen E, Boonen S, Vanderschueren D. Skeletal sexual dimorphism: relative contribution of sex steroids, GH-IGF1, and mechanical loading. J Endocrinol 2010;207(2):127–34. [DOI] [PubMed] [Google Scholar]
- 47.Won H, Mun S, Shin B, Lee S. Contradictory role of CD97 in basal and tumor necrosis factor–induced osteoclastogenesis in vivo. Arthritis & Rheumatology. 2016;68(5):1301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloom A, Collins F, Hof RVt, Ryan E, Jones E, Hughes T, et al. Deletion of the membrane complement inhibitor CD59a drives age and gender-dependent alterations to bone phenotype in mice. Bone. 2016;84:253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glatt V E C, Stadmeyer L, Bouxsein M. Age-related changes in trabecular architecture differ in female and male C57BL/6J mice. J Bone Miner Res 2007;22(8):1197–207. [DOI] [PubMed] [Google Scholar]
- 50.Ho NTT, Kutzner A, Heese K. A Novel Divergent Gene Transcription Paradigm-the Decisive, Brain-Specific, Neural |-Srgap2-Fam72a-| Master Gene Paradigm. Molecular neurobiology. 2019. [DOI] [PubMed] [Google Scholar]
- 51.Franceschetti T, Kessler CB, Lee SK, Delany AM. miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J Biol Chem 2013;288(46):33347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J, et al. Effects of miR-335–5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res 2011;26(8):1953–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149(4):912–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fossati M, Pizzarelli R, Schmidt ER, Kupferman JV, Stroebel D, Polleux F, et al. SRGAP2 and Its Human-Specific Paralog Co-Regulate the Development of Excitatory and Inhibitory Synapses. Neuron 2016;91(2):356–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. A) Srgap2 mRNA expression in a population of bone marrow osteoclast precursor cells (CD45R− CD3− CD11b−/lo CD115+), which was FACS sorted and cultured for 4 days with M-CSF or M-CSF + RANKL. Microarray analysis was performed using Illumina Mouse WG-6 Beadchip. N=3. B) Primary osteoblasts express Srgap2. Western blot analysis of Srgap2 in primary osteoblasts prepared from neonatal mouse calvaria and cultured before or after osteogenic medium (with ascorbic acid {AA} and ß-glycerophosphate {BGP}) for 12 days.
Supplemental Figure 2. Micro-CT analysis of femurs from Srgap2-cKO male mice. (A) μCT analysis of cortical bone of femurs from 8-week-old Srgap2-cKO and littermate controls. Total cortical area (Tt.Ar); Marrow area (Ma.Ar); Cortical thickness (Ct.Th). (B) μCT analysis of trabecular bone of femurs from 8-week-old Srgap2-cKO and littermate control mice. Bone volume/total volume (BV/TV); Trabecular thickness (Tb.Th); Trabecular number (Tb.N); Trabecular spacing (Tb.Sp). N=7–8. Data represent mean ±SEM. Statistically significant differences were not detected.
Supplemental Figure 3. Real time PCR analysis of various clastokines in BMM cultures. Bone marrow cells from control and Srgap2 cKO mice were cultured with M-CSF and RANKL for 4 days and real time PCR analysis using gene specific Taqman assay (Applied Biosystems). N=4. Significant differences between control and Srgap2-cKO cultures were not detected.
Supplemental Figure 4. Western blot analysis of SLIT3 expression in BMM cells treated with M-CSF and TNFα for 5 days. M-CSF and RANKL treated WT BMM cells were used as positive control for SLIT3 expression.
Supplemental Figure 5. (A) Comparable levels of Srgap2 and cathepsin K in BMM cultures from male control mice and female control and Srgap2-cKO mice detected by western blot analysis. Female BMMs from Srgap2-cKO mice are shown as negative control for Srgap2 expression. β-Actin was used as an internal control. (B) Increased Rac1 activation in BMM cells from male Srgap2-cKO mice. BMMs were cultured with M-CSF and RANKL for 3 days and starved serum for 2 hours. Active forms of Rac1 was detected by pull-down assays. Total Rac1 was detected in whole cell lysate and β-Actin was used as an internal control.