Abstract
Background
Actionable fibroblast growth factor receptor 3 (FGFR3)–transforming acidic coiled-coil protein 3 fusions (F3T3) are found in approximately 3% of gliomas, but their characteristics and prognostic significance are still poorly defined. Our goal was to characterize the clinical, radiological, and molecular profile of F3T3 positive diffuse gliomas.
Methods
We screened F3T3 fusion by real-time (RT)-PCR and FGFR3 immunohistochemistry in a large series of gliomas, characterized for main genetic alterations, histology, and clinical evolution. We performed a radiological and radiomic case control study, using an exploratory and a validation cohort.
Results
We screened 1162 diffuse gliomas (951 unselected cases and 211 preselected for FGFR3 protein immunopositivity), identifying 80 F3T3 positive gliomas. F3T3 was mutually exclusive with IDH mutation (P < 0.001) and EGFR amplification (P = 0.01), defining a distinct molecular cluster associated with CDK4 (P = 0.04) and MDM2 amplification (P = 0.03). F3T3 fusion was associated with longer survival for the whole series and for glioblastomas (median overall survival was 31.1 vs 19.9 mo, P = 0.02) and was an independent predictor of better outcome on multivariate analysis.
F3T3 positive gliomas had specific MRI features, affecting preferentially insula and temporal lobe, and with poorly defined tumor margins. F3T3 fusion was correctly predicted by radiomics analysis on both the exploratory (area under the curve [AUC] = 0.87) and the validation MRI (AUC = 0.75) cohort. Using Cox proportional hazards models, radiomics predicted survival with a high C-index (0.75, SD 0.04), while the model combining clinical, genetic, and radiomic data showed the highest C-index (0.81, SD 0.04).
Conclusion
F3T3 positive gliomas have distinct molecular and radiological features, and better outcome.
Keywords: diffuse gliomas, F3T3 gene fusions, lesion to symptom mapping, VASARI features
Key Point.
1. We show that gliomas with FGFR3-TACC3 fusion represent, whatever the grade, a unique histo-prognostic entity, with specific molecular and radiological characteristics and a less aggressive clinical evolution.
Oncogenic chromosomal translocations of fibroblast growth factor receptor 3 (FGFR3)–transforming acidic coiled-coil protein 3 (TACC3) that fuse the tyrosine kinase coding domain of the FGFR3 gene to the coiled-coil domain of transforming acidic coiled-coil domain containing protein 3 (F3T3) are driver events occurring in 3% of gliomas and a large number of other cancers.1–6 Inhibition of FGFR kinase showed clinical benefit in patients harboring the F3T3 fusion,1 and different FGFR inhibitors are being tested in clinical trials (NCT02824133; NCT02052778) with encouraging preliminary results.6
The F3T3 fusion protein activates noncanonical pathways to implement mitotic and chromosomal instability,1 and promotes oxidative phosphorylation and dependency upon mitochondrial metabolism.7 This last property is consistent with the strikingly high vessel density that characterizes these tumors,8 suggesting that F3T3 gliomas represent a unique entity among gliomas.
Here, we characterized a large series of F3T3-positive gliomas, defined the repertoire of structural variants of the fusion gene and unraveled the unique molecular, radiological and clinical features of gliomas harboring F3T3 fusions. We used a subset of comprehensive feature set schema, known as Visually AcceSAble Rembrandt Images (VASARI9) to describe morphology on MR images and a lesion-to-symptom mapping (LSM) for spatial distribution. Our aim was to use radiomic features to improve survival prediction and classify F3T3 fusion non-invasively.
Patients and Methods
Patient Cohort
The patient cohort includes retrospective6 and prospective cases of gliomas identified (since January 2014) at Pitié-Salpêtrière Hospital, 10 other French institutions in the setting of the Association of French-Speaking Neuro-Oncologists (ANOCEF)10 and Management of Anaplastic Oligodendrogliomas (POLA) networks,11,12 and the C. Besta Institute in Milan, Italy (Supplementary Table 1).
Cases from external institutions and cases diagnosed at Pitié-Salpêtrière after February 2018 were preselected according to FGFR3 immunopositivity6,8 and underwent confirmation of F3T3-fusion by real-time (RT)-PCR using frozen material (Supplementary Table 1).
Tumor specimens, blood samples, and clinico-pathological information were collected with informed consent and relevant ethical board approval in accordance with the tenets of the Declaration of Helsinki.
For the samples from the Pitié-Salpêtrière Hospital, clinical data and follow-up are available in the neuro-oncology database (OncoNeuroTek, GH Pitié-Salpêtrière, Paris).
Identification of Fusion Transcripts and Molecular Characterization
Performed as previously described6,13–16 were RNA extraction, RT-PCR for detection of F3T3 fusion transcript, Sanger sequencing of isocitrate dehydrogenase 1 (IDH1)R132, IDH2R172, TERT promoter, histones H3B and H3F3A, PTEN, BRAFV600, and epidermal growth factor receptor variant III (EGFRvIII). Promoter methylation status of O6-methylguanine-DNA methyltransferase (MGMT) was determined by pyrosequencing.10
Copy number variations were detected by comparative genomic hybridization arrays.6 In a subset of cases, a custom next-generation sequencing panel was used, covering IDH1R132, IDH2R172, TERT promoter, H3B and H3F3A, BRAFV600 mutations, EGFR, cyclin-dependent kinase 4 (CDK4), murine double minute 2 (MDM2) amplifications, CDK inhibitor 2A (CDKN2A) deletions, and chromosomal gain and losses on 1p, 19q, 9p, and 10.
Immunohistochemistry
Integrated histological diagnosis was reviewed according to World Health Organization (WHO) 2016 classification17 by 2 independent neuropathologists (K.M. and F.B.). Immunohistochemistry for mutant IDH1R132H, ATRX, p53, Ki67, and FGFR3 was performed as previously described.8,11 F.B. and K.M. reviewed FGFR3 immunolabeling.
Radiological Analysis
F3T3-positive cases and F3T3-negative controls were selected within the cohort of patients tested by RT-PCR, based on the availability of MRI at diagnosis (before surgery), including at least T1 pre- and post-contrast and fluid-attenuated inversion recovery (FLAIR) sequences. Controls negative for F3T3 fusion (2 for each F3T3-positive case) were paired by sex, histological diagnosis, and age at diagnosis ±2 years (when no patients available, range was extended to ±6 years). Further details are provided in the supplementary methods, available at https://sansonlab.gitbook.io/workspace/. For each patient, 3 independent investigators (J.S., E.S., A.P.), including 2 board-certified neuroradiologists (J.S., E.S.), blinded for F3T3 status, reviewed 3D T1-weighted MR images before and after gadolinium administration, axial T2-weighted FLAIR image, and, when available, T2, T2*, and diffusion sequences according to the standardized VASARI feature set.9,18 Contrast-enhanced tumor, non-enhanced tumor, necrosis, and edema were visually estimated (0–5%, 6–33%, 34–67%, 68–95%, 95–100%) according to the VASARI guidelines.19 Tumor volumes were calculated by semi-automatic segmentation using ITK-SNAP v3.6.0 software.20 The distribution of radiological features was validated using an additional independent cohort of F3T3-positive cases with paired F3T3-negative controls, selected according to the same design. Further details are provided in the Supplementary Material and supplementary methods (https://sansonlab.gitbook.io/workspace/).
Image Postprocessing and Radiomic Feature Extraction
Image registration was performed using ANTsR v0.4.21 FLAIR images were co-registered to T1 gadolinium weighted 3D imaging for each patient. Binary masks were produced using semi-automatic segmentation using ITK-SNAP 3.8.20 These images were registered to the brain extracted T1 enhanced volume using affine registration. To normalize image intensity we used the WhiteStripe R package.22 Tumor segmentation was performed semi-automatically for the contrast-enhancing portion and separately for the peritumoral edema (as previously described) using a region-growing segmentation algorithm implemented in ITK-SNAP.20 The segmented volumes were all reviewed by one of us (A.P., A.L.D.S., A.A.) and corrected manually when necessary. The tumor distribution of binary masks was compared (F3T3-positive vs F3T3-negative) using Sparse Canonical Correlation Analysis for Neuroimaging (SCCAN). This multivariate lesion-to-symptom mapping (LSM) method improves robustness and reliability of mapping and was performed using the LESYMAP R package.23 Radiomic feature extraction was performed with a PyRadiomics (v2.0.0) pipeline.24 Within each volume of interest, we extracted 2616 radiomic features, including first-order features, volume and shape features, and texture features, and we performed different transformations of these features (Laplacian of Gaussian (log) filter, logarithm transformation, square transformation, Squareroot, exponential transformation, wavelet transformation). Further details are provided in the Supplementary Material and supplementary methods (https://sansonlab.gitbook.io/workspace/). To consolidate our findings, we split a first cohort of patients into a training set (70% of the sample size) and a validation set (30% of the sample size). We then completed our analysis retrieving a completely independent second cohort which was entirely used for validation.
Statistical Analysis
Differences in the distribution on categorical variables were analyzed using the Fisher exact test and chi-square test according to sample size and data distribution. Differences in quantitative variables were assessed by Student’s t-test, in case of equal variance and normal distribution, or the Mann‒Whitney test in the other cases. Correlations between non-parametric features were analyzed by the Spearman method. When indicated, P-values were adjusted for multiple testing according to the Benjamini‒Hochberg false discovery rate.25 A q-value of 0.05 (2-sided) was considered to be statistically significant.
Overall survival (OS) was defined as the time between diagnosis and death. Patients who were still alive at the last follow-up were considered censored. Survival curves were calculated using the Kaplan‒Meier method. Statistically significant differences between survivals were assessed using the log-rank test. The Cox model was used to evaluate the effect of quantitative variables on survival, and for multivariate survival analysis.
Using the plsRcox package, several Cox models were constructed with either clinical, genetics, and radiomic features as well as the different combinations of these 3 individual Cox models. The goodness of fit or the performance of the different Cox proportional hazards models was assessed with Harrell’s concordance index (C-index)26 using a stratified 5-fold cross-validation with the same number of deaths per fold. Further details are provided in the Supplementary Material and supplementary methods (https://sansonlab.gitbook.io/workspace/).
The ability of radiomic features to classify F3T3-positive gliomas was assessed using random forest and the R caret package v6.0–80.27 In order to better represent the real distribution of F3T3 samples within high-grade gliomas (~3%), we weighted the classification model with this proportion to correct the imbalance between F3T3 groups. We divided the first dataset into a training cohort (composed of 70% of the samples) and a validation cohort including the rest of samples. We then used a second, independent dataset to validate the model. The classification was assessed using receiver operating characteristic (ROC) area under the curve (AUC), obtained with pROC R package v1.12.1.28 The 95% confidence interval (CI) was computed with 2000 stratified bootstrapped replicates.
Analyses were performed using R software v3.5.
Results
Identification of FGFR-TACC3 Fusions
We screened by RT-PCR6 a cohort of 1162 diffuse gliomas (826 WHO grade IV, 156 grade III, 113 grade II, and 67 gliomas with no grading information [unclassified]), including 193 IDH mutant (42 grade IV, 74 grade III, and 77 grade II).
We tested unselected 951 cases from Pitié-Salpêtrière Hospital tumor bank (including 591 previously reported; 6 711 grade IV, 129 grade III, 104 grade II, 7 unclassified). We screened an additional 211 specimens from Pitié-Salpêtrière and external institutions, preselected for FGFR3 immunopositivity8 (115 grade IV, 27 grade III, 9 grade II, 60 unclassified).
Overall, we found 80 F3T3-positive gliomas, 24 among unselected cases (2.5%) and 56 among FGFR3-immunopositive gliomas (26.5%), all IDH wildtype (except 4 with unknown status), mostly grade IV (84%, 67/80) (Table 1, Supplementary Table 1).
Table 1.
Distribution of F3T3 diffuse gliomas according to WHO grading and IDH status
| Histological Entity | FGFR3-TACC3 Fusions Identified/Samples Tested | |
|---|---|---|
| Without Prescreening for FGFR3 Immunopositivity | Prescreened for FGFR3 Immunopositivity | |
| Diffuse glioma (grade II-IV), IDH mutant | 0/187 | 0/6 |
| Grade II glioma, IDH wildtype | 2/24 (8.3%) | 4/6 (66.7%) |
| Grade III glioma, IDH wildtype | 1/56 (1.8%) | 5/19 (26.3%) |
| Glioblastoma, IDH wildtype | 21/588 (3.6%) | 42/84 (50%) |
| Diffuse glioma NOS, IDH wildtype | 0/7 | 1/60 (1.7%) |
| Diffuse glioma (grade II-III), IDH NOS | 0/7 | 0/6 |
| Glioblastoma, IDH NOS | 0/82 | 4/30 (13.3%) |
| Total | 24/951 (2.5%) | 56/211 (26.5%) |
NOS = not otherwise specified. Note. Twenty-four F3T3 cases were identified from a systematic screening of 951 gliomas from Pitié-Salpêtrière tumor bank without FGFR3 IHC pre-screening (frequency of F3T3 fusions among unselected diffuse gliomas, grades II–IV: 2.5%), while 56 F3T3 cases were detected from an additional series of 211 preselected gliomas with FGFR3 protein immunopositivity (frequency of F3T3 fusions among FGFR3-immunopositive tumors: 26.5%).
F3T3 fusion mRNAs invariably included the intact FGFR3 tyrosine kinase (TK) domain fused in-frame with variable sequences from the TACC3 gene that constantly included the coiled-coil domain of TACC3. F3T3 RT-PCR amplicon ranged from 805 bp (for FGFR3-ex18-TACC3-ex13) to 1706 bp (for FGFR3-ex18-TACC3-ex4), corresponding to notably variable F3T3 fusion isoforms. However, the fusions FGFR3-ex17-TACC3-ex11, FGFR3-ex17-TACC3-ex10, and FGFR3-ex17-TACC3-ex8 were the most recurrent variants that together accounted for 79% (63/80) of the entire F3T3-positive glioma cohort. Notably, 3 F3T3 fusion variants had not been reported before, and 1 variant was reported only in bladder cancer (https://cancer.sanger.ac.uk/cosmic) (Fig. 1A).
Fig.1.
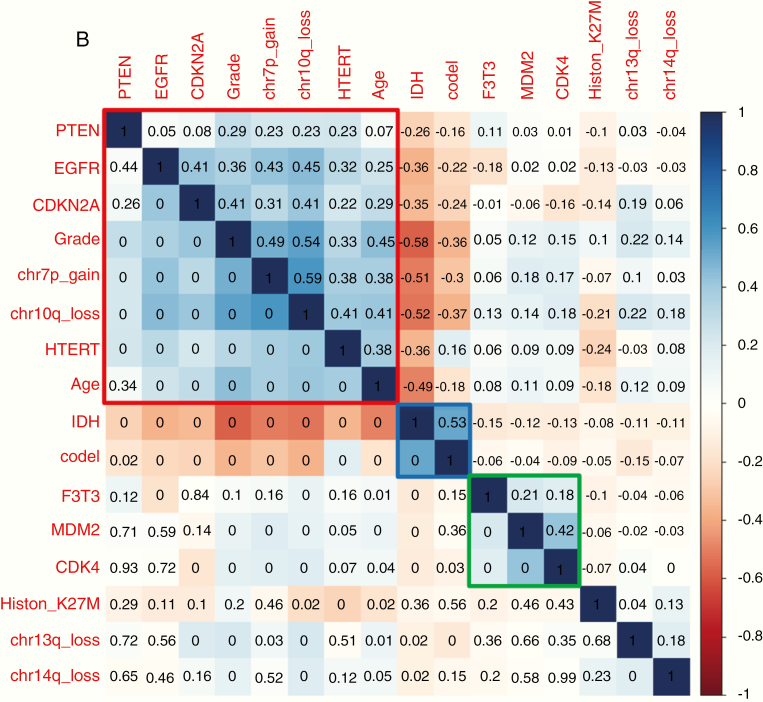
(A) Structure of F3T3 fusions identified by RT-PCR Sanger sequencing. Predicted F3T3 fusion protein encoded by the transcripts identified by RT-PCR. Regions corresponding to FGFR3 or TACC3 are shown in red or blue, respectively. On the left are indicated FGFR3 and TACC3 exons joined in the fused mRNA; the presence of TACC3 introns is also reported when they are spliced in the fusion cDNA. In the second column, the number of patients harboring the corresponding fusion variant is indicated. The novel transcripts discovered in this study are highlighted in red. Black arrows indicate the position of the primers used for the F3T3 fusion screening. (B) Correlation matrix of the main genetic alterations in the whole data set of 1162 diffuse gliomas. The variables are indicated in blue when associated, in red when inversely associated. Upper triangle: strength of the correlation (rho values). Lower triangle: statistical significance (P-values; 0 means P < 0.01). Three main clusters were identified: EGFR amplification with 7p gain, 10q loss and older age at diagnosis (cluster 1, in red), IDH mutation with 1p/19q codeletion (cluster 2, in blue), and F3T3 fusion with MDM2 and CDK4 amplifications (cluster 3, in green).
Clinical Features and FGFR3 Staining
The clinical and prognostic features of F3T3-positive and F3T3-negative IDH wildtype gliomas were similar, except for a lower rate of MGMT methylation status in F3T3-positive patients (Table 2). All F3T3-positive gliomas were immunopositive for FGFR3.
Table 2.
Distribution of clinical and prognostic features according to F3T3 status
| FGFR3-TACC3+ | FGFR3-TACC3- | P value | |
|---|---|---|---|
| Clinical features of the IDH wildtype glioma cohort (grades II‒IV), n = 969 | |||
| Sex ratio (M/F) | 1.19 | 1.51 | 0.33 |
| Median age at diagnosis, y (range) | 57 (25–87) | 61 (15–90) | 0.20 |
| Median KPS score (range) | 80 (40–100) | 80 (40–100) | 0.68 |
| Surgery at diagnosis (available records), n (%) | |||
| • Surgery | 54 (72) | 411 (79) | 0.20 |
| • Biopsy | 21 (28) | 112 (21) | |
| MGMT promoter methylation/tested | 15/47 (32) | 129/268 (48) | 0.04 |
| Clinical features of the IDH wildtype and IDH NOS GBM subgroup, n = 784 | |||
| Sex ratio (M/F) | 1.20 | 1.56 | 0.31 |
| Median age at diagnosis, y (range) | 58 (35–87) | 63 (21–90) | 0.05 |
| Median KPS score, range | 80 (40–100) | 80 (40–100) | 0.55 |
| Surgery at diagnosis (available records), n (%) | |||
| • Surgery | 47 (76) | 359 (80) | 0.42 |
| • Biopsy | 15 (24) | 89 (20) | |
| First line treatment after surgery, n (%) | 0.20 | ||
| • RT plus concomitant and adjuvant TMZ | 51 (84) | 333 (76) | |
| • Other treatments | 10 (16) | 103 (24) | |
| MGMT promoter methylation/tested (%) | 13/40 (33) | 112/239 (47) | 0.09 |
Note. In the whole cohort of 969 IDH wildtype diffuse gliomas (upper part) and in the subgroup of 581 IDH wildtype/IDH NOS glioblastoma patients (lower part).
In a subset of 495 unselected gliomas tested for both F3T3 fusion and FGFR3 immunostaining, FGFR3 immunostaining showed 100% sensitivity, 86% specificity, 28% positive predictive value, and 100% negative predictive value for F3T3 status (25 true positive, 0 false negative, 65 false positive, 405 true negative cases). This finding indicates that positive staining for FGFR3 is a very useful biomarker to preselect potential F3T3-positive tumors.
Genomic Profile of F3T3-Positive Glioma
Correlation analysis on available molecular features for the 1162 gliomas (Fig. 1B) showed that F3T3 fusions clustered with amplification of MDM2 (r = 0.211, P < 0.001) and CDK4 (r = 0.180, P < 0.001), both located on chromosome (Chr) 12q13.15, and were mutually exclusive with both IDH mutation (r = −0.147, P < 0.001) and EGFR amplification (r = −0.183, P < 0.001). This analysis identified 3 main clusters: (i) EGFR amplification with Chr 7p gain, Chr 10q loss, and older age at diagnosis, (ii) IDH mutation with 1p/19q codeletion, and (iii) F3T3 fusions with frequent MDM2 and CDK4 amplifications (Fig. 1B and Supplementary Table 2).
F3T3 Gene Fusion Is an Independent Predictor of Favorable Outcome
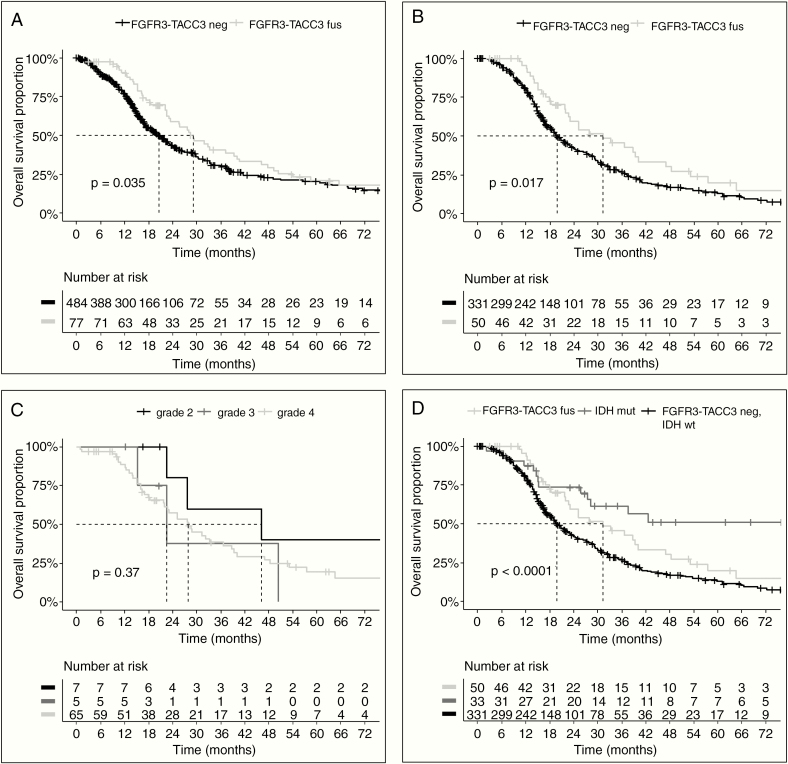
The clinical follow-up of patients with F3T3-positive gliomas revealed that they have a better survival than patients with IDH wildtype tumors lacking F3T3 fusions (median OS 29.1 vs 20.5 mo, P = 0.04; Fig. 2A). This was not due to differences in grading as we observed a similar difference in survival when the analysis was restricted to IDH wildtype glioblastoma (GBM) patients, who all received standard radiochemotherapy with temozolomide after surgery or biopsy29 (median OS, 31.1 mo for F3T3-positive vs 19.9 mo for F3T3-negative, P = 0.02; Fig. 2B). In contrast, grading of F3T3-positive tumors seemed to have a limited impact on survival (Fig. 2C). No difference in survival was associated with the 3 most frequent F3T3 isoforms (FGFR3-ex17-TACC3-ex11, FGFR3-ex17-TACC3-ex10, and FGFR3-ex17-TACC3-ex8) (P = 0.2). While F3T3 fusion and EGFR amplification were mutually exclusive, EGFR amplification per se had no impact on survival in the GBM cohort. Strikingly, CDK4 amplification was associated with longer survival (median OS, 57.5 vs 25.1 mo, P = 0.04) in F3T3 positive GBM, and a similar trend was observed for MDM2 amplification (median OS, 47.0 mo vs 28.6, P = 0.22) (Supplementary Figure 1). Conversely, no differences in survival associated to CDK4 and MDM2 amplifications were seen in F3T3-negative GBM (Supplementary Figure 2A, B). A marked benefit in survival was seen in CDK4/MDM2 amplified GBM harboring the F3T3 fusion compared with CDK4/MDM2 amplified, F3T3-negative cases (Supplementary Figure 2C, D). The distribution of the main known prognostic factors of F3T3-positive versus F3T3-negative gliomas (grades II–IV) and GBM is reported in Table 2. The multivariate analysis showed that F3T3 fusion predicts longer survival in IDH wildtype GBM (hazard ratio [HR] = 0.47; 95% CI: 0.27–0.80), independently of age, Karnofsky performance status, extent of resection, MGMT status, and treatment (Table 3). When analysis was restricted to IDH wildtype patients treated by standard radiochemotherapy, the F3T3 fusion remained an independent predictor of better outcome (HR = 0.33; 95% CI: 0.17–0.63). By comparing the 3 prognostic subcategories of GBM (IDH mutant, IDH wildtype/F3T3-positive, and IDH wildtype/F3T3-negative) after standard radiochemotherapy, we found that IDH wildtype/F3T3-positive GBM patients had an intermediate survival between IDH mutant patients (median OS = not reached, 5-y OS = 51.2%) and IDH wildtype/F3T3-negative patients (Fig. 2D). Therefore, F3T3-positive GBM represents a distinct group of IDH wildtype gliomas with unique molecular and clinical features.
Fig. 2.
Survival analysis of patients in the study. (A) Survival of IDH wildtype diffuse glioma (grades II‒IV) patients according to the F3T3 status (median OS = 29.1 mo for F3T3-positive vs 20.5 mo for F3T3-negative; P = 0.04). (B) Survival of IDH wildtype glioblastoma (GBM) patients treated with standard radiation therapy plus concomitant and adjuvant temozolomide after surgery or biopsy, according to the F3T3 status (median OS, 31.1 mo for F3T3-positive vs. 19.9 mo for F3T3-negative, P = 0.02). (C) Survival of F3T3-positive patients according to grade (P = 0.37). (D) Survival of GBM patients according to F3T3 and IDH status (median OS, 29.1 mo for F3T3-positive/IDH wildtype GBM vs 19.9 mo for F3T3-negative/IDH wildtype GBM, P = 0.04; median OS not reached for F3T3-negative/IDH mutant, P < 0.001). Tick marks represent censored patients.
Table 3.
Multivariate analysis of clinical and pathological factors influencing survival in IDH wildtype glioblastoma patients
| Variables | Hazard Ratio | 95% CI | P-value |
|---|---|---|---|
| FGFR3-TACC3 fusion | 0.47 | 0.27–0.80 | 0.005 |
| Age at diagnosis | 1.04 | 1.02–1.06 | <0.001 |
| Karnofsky performance status | 0.96 | 0.94–0.98 | <0.001 |
| No resection at diagnosis (biopsy only) | 2.00 | 1.04–3.86 | 0.039 |
| MGMT promoter methylated | 0.44 | 0.28–0.68 | <0.001 |
| Standard radiochemotherapy with temozolomide | 0.39 | 0.25–0.59 | <0.001 |
Radiogenomic Features and Spatial Distribution
We first performed a blinded analysis of qualitative and quantitative features from the VASARI feature set on preoperative MRIs of 22 F3T3-positive and 44 matched (see Methods) F3T3-negative GBM. We found no differences between F3T3-positive and F3T3-negative patients in terms of tumor volumes (P = 0.16) and subvolumes (contrast-enhancing, P = 0.6; non-contrast-enhancing, P = 0.16; and necrosis volumes, P = 0.7). However, F3T3-positive GBM involved less frequently eloquent areas (r = −0.35, P < 0.01), had more frequently poorly-defined enhancing margins (r = 0.46, P < 0.01), non-enhancing margins (r = 0.62, P < 0.01), and increased edema (r = 0.38, P < 0.01). A trend towards reduced contrast enhancement quality was also noted (r = −0.24, P = 0.05) (Supplementary Figure 3).
The same analysis was then performed on an independent validation cohort of 26 F3T3-positive and 52 matched F3T3-negative GBM. We confirmed the association of F3T3-positive status with reduced contrast enhancement quality (r = -0.36, P < 0.01), poorly-defined enhancing margins(r = 0.27, P = 0.02), and a trend of association with poorly-defined non-enhancing margins (r = 0.21, P = 0.08) and reduced eloquent area involvement (r = −0.21, P = 0.06) (Supplementary Figure 3).
We then extended the study to non GBM and analyzed an additional group of 6 F3T3-positive (5 grade II, 1 grade III) and 12 paired F3T3-negative/IDH wildtype lower-grade gliomas (10 grade II, 2 grade III): again, in this smaller group, F3T3 positive gliomas were associated with poorly defined enhancing and non-enhancing tumor borders (r = −0.45 and −0.42, respectively, P < 0.05).
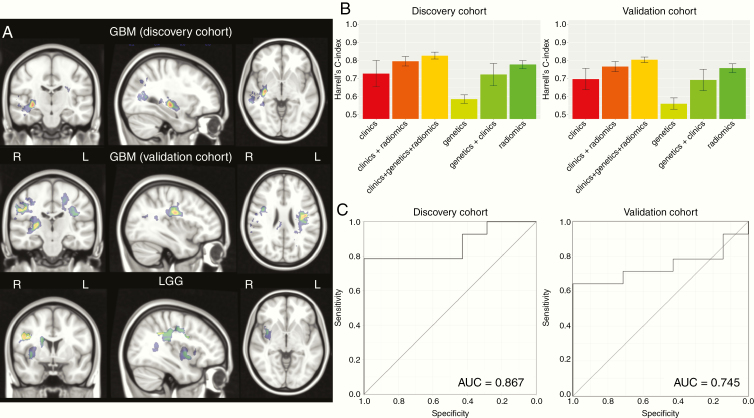
Spatial localization of the analyzed gliomas according to the enhanced tumor Region of interest is reported in Supplementary Figure 4. Multivariate SCCAN based on contrast enhancement and FLAIR sequences showed more frequent involvement of cortical and subcortical regions, especially the insula, in F3T3 gliomas, irrespective of tumor grade (Fig. 3A).
Fig. 3.
(A) Heatmap of brain regions with positive significant correlation with F3T3 fusion (adjusted P < 0.05) using a SCCAN (sparse canonical correlation analysis). Color scale represents the value r of correlation (from black = 0 to red = 1). R = right hemisphere; L = left hemisphere. This analysis shows a more frequent involvement of insula and cortico-subcortical regions. (B) Barplots representing the mean value of Harrell’s C-index of 5 cross-validations using different types of data (genetics, clinics, and radiomics) in the 2 independent exploratory and validation cohorts of GBM. Error bars indicate the SD of 5 cross-validations. (C) Performance of the F3T3 status classification in the 2 independent exploratory and validation cohorts of GBM, assessed by ROC curve. Further details are provided in the supplementary methods, available at https://sansonlab.gitbook.io/workspace/.
Radiomics Analysis
We extracted 2616 radiomic features per patient. The distribution of the 200 most variable radiomic features according to the medium absolute distance is presented as a heatmap in Supplementary Figure 5.
To further evaluate the value of radiomic features to predict OS, we compared different Cox models using Harrell’s C-index. The Cox model using radiomic features reached a C-index of 0.75 ± 0.04, versus 0.59 ± 0.05 using genetic characteristics only, and 0.7 ± 0.15 for clinical variables. Finally, the model using the combination of the different types of data (clinical, genetic, and radiomics) showed the highest C-index (0.81 ± 0.04) (Fig. 3B, left). In the same light, in the validation dataset, radiomic features also reached a C-index of 0.75 ± 0.05 versus 0.56 ± 0.05 using genetic characteristics only and 0.68 ± 0.16 for clinical variables, while the combination model again showed the highest C-index (0.8 ± 0.04) (Fig. 3B, right). The 50 most relevant radiomic features to predict OS according to their weight are presented in Supplementary Fig. 6.
Using a random forest model to classify F3T3 status in the first set of GBM (22 F3T3-positive and 44 F3T3-negative) with training cohort (70% of the sample size) and validation cohort (30% of the sample size), the performance of classification assessed by AUC was 0.87 (95% CI: 0.66–0.99). We confirmed this result on the second independent set of GBM (26 F3T3-positive and 52 F3T3-negative) (AUC = 0.745; 95% CI: 0.52–0.93) (Fig. 3C). The 25 most relevant radiomic features in this model are provided in Supplementary Figure 7. The vast majority of the radiomic features in both the Cox model (Supplementary Figure 6) and the random forest model that predict the F3T3 status (Supplementary Figure 7) were highly enriched with T1-weighed gadolinium post-contrast images. Together, these data suggest that F3T3 gene fusions confer a specific radiomic signature.
Discussion
F3T3-positive gliomas comprise a specific entity among the group of IDH wildtype gliomas. The unique characteristics of F3T3-positive gliomas include morphological features (monomorphous ovoid nuclei, nuclear palisading and thin parallel cytoplasmic processes, endocrinoid network of thin capillaries, frequent microcalcifications, and desmoplasia) associated with a rich vascularization.8F3T3 is a novel oncogenic driver resulting in the activation of mitochondrial metabolism. This diverges from the majority of GBM that depend mostly on glycolysis.7 Here we show that beside other diffuse glioma entities (ie, IDH mutant gliomas‒1p/19q codeleted; IDH mutant gliomas/non-codeleted; IDH wildtype gliomas with EGFR amplifications/7p gain and 10 q loss), F3T3 is associated with Chr 12q13.15 amplicons involving CDK4 and MDM2 oncogenes. Given the limited number of cases and the lack of complete testing in all patients, we may have missed some other genetic associations. We also found that among IDH wildtype tumors, F3T3 gliomas have a more favorable clinical outcome, especially when associated with 12q13.15 amplicons. The association between F3T3 and patient survival is independent of tumor grading, thus indicating that F3T3-positive gliomas are a distinct tumor subgroup within IDH wildtype tumors. The unique clinical course of F3T3-positive gliomas may be related to the oxidative phosphorylation phenotype of these tumors. F3T3 activates at the same time macromolecule synthesis—necessary to tumor growth—and mitochondrial metabolism, triggering oxidative phosphorylation.7 It is still unknown whether, as for other cancer types such as lung cancer,30,31 the oxidative phosphorylation phenotype is associated with better outcome in GBM.
In order to better characterize the radiological features of F3T3-positive gliomas, we collected preoperative MRIs from 2 independent sets of F3T3-positive and paired F3T3-negative patients and performed and validated a case-control retrospective study for both morphological features from the VASARI set and spatial distribution of tumor locations. This analysis identified recurrent features in F3T3-positive GBM: involvement of non-eloquent areas, poorly defined margins for contrast-enhancing and non-contrast-enhancing tumors and a reduced enhancement intensity. By contrast, GBM with EGFR amplification had a higher T2/FLAIR hyperintensity and contrast-enhancing tumor volume,32,33 and a higher contrast-enhancing volume to necrotic tumor volume ratio.34 We also observed an important enrichment for T1-weighted post-contrast features in the prediction of both survival and F3T3 status, which could be explained by the high vessel density found in F3T3 samples.8 One of our main findings—validated on both the exploratory and the validation cohort—is that radiomic data identified F3T3-positive patients with good accuracy, and might be used to predict F3T3-positive gliomas non-invasively. We also found that radiomics predicted OS with higher accuracy than clinical or genetic data, comparable to a model including genetics and clinical data, while the model combining genetics, clinical, and radiomic data had the best prediction accuracy. Radiomics has the advantage of capturing a large number of features. We assume that these features reflect a broad range of biological features, which are not explored by the limited genetic testing done in this study. Taken together, quantitative assessment of MRI data complements genetic and clinical biomarkers and improves survival prediction.
Finally, we investigated a specific tropism of F3T3-positive gliomas for specific areas of the brain by multivariate optimization technique (SCCAN). We report an association between F3T3-positive gliomas (both lower grade and GBM) and cortical-subcortical areas of the brain, insula, and temporal lobe location. FGFs are required for early stages of CNS development, and specific roles of the different FGFR signaling are emerging35,36 with potential connections with oncogene-based tumor locations36: FGFR3 controls the development of the cortex by regulating proliferation and apoptosis of cortical progenitors, the Asn540Lys FGFR3 germline mutation results in bilateral medial temporal lobe anomalies,37,38 and different FGF receptors are required for the development of the medial prefrontal cortex and its connections with limbic circuits, including the insula.39 These parallelisms open new fields of research in F3T3-positive glioma ontogenesis.
In conclusion, the present study reports the comprehensive characterization of F3T3-positive gliomas and reveals that these tumors exhibit molecular, radiological, and clinical features that are unique within IDH wildtype gliomas. Together with the recent histological and metabolic characterization of these tumors,7,8 we suggest that F3T3-positive gliomas should be managed as an independent subgroup of brain tumors requiring specific approaches for diagnosis, prognosis, and therapy.
Supplementary Material
Acknowledgments
ALDS received support from a donation to Fondation Foch in the memory of Mr Roland Lemetre and Mr Vincent Clausse. This work benefited from the facilities and expertise of the NeuroBioTec Collection (Groupement Hospitalier Est, Bron France).
Funding
This work was supported by grant INCa-DGOS-Inserm_12560 of the SiRIC CURAMUS, by the Ligue Nationale Contre le Cancer (équipe labellisée), and by a grant from Investissements d’avenir.
Conflict of interest statement
Anna Lasorella and Antonio Iavarone received research grants from AstraZeneca and Taiho Pharmaceuticals; Marc Sanson received financial support from AstraZeneca for the TARGET Trial, and is investigator for the TAS-120 trial. No other authors have financial disclosures.
>Authorship statement
A.L.D.S., J.S., A.A., and M.S. designed the study. A.L.D.S., A.P., J.S., A.A., A.L., A.I, and M.S. wrote the manuscript. A.L.D., F.D., M.E., B.M., L.C., V.B., C.V., M.S. and the TARGET study group (attached) provided the cases. A.L. and A.I. designed and helped to validate the RT-PCR screen for F3T3. A.L.D.S., Y.S., R.P., and J.L. performed the genes analysis. F.B., C.V., J.L., and K.M. performed the histological analysis and immunohistochemistry. E.S., A.P., J.S., and A.A. analyzed MRIs. A.L.D.S., A.A., A.P. performed tumor 3D segmentation and spatial distribution analysis. A.A., A.G., C.P., V.F., performed texture analysis. Clinical and imaging data were collected and analyzed by A.L.D.S., A.P., L.B., and M.S. All contributed to the data analysis and interpretation. All read and approved the manuscript.
TARGET study group non-author contributors
Paule Augerau,1 Antoine Carpentier,2 Isabelle Catry-Thomas,3 Olivier Chinot,4 Caroline Dehais,5 Jean-Yves Delattre,5 Dominique Figarella-Branger,4 Stephan Gaillard,6 David Guyon,6 Khe Hoang-Xuan,5 Caroline Houillier,5 Ahmed Idbaih,5 Florence Laigle-Donadey,5 Emilie Le Rhun,7 David Meyronet,8 Elisabeth Moyal,9 Dimitri Psimaras,5 Luc Taillandier,10 Mehdi Touat,5 Nadia Younan.5
1. CHU d’Angers, Angers, France
2. APHP, Hôpital St-Louis, France
3. CHU de Bordeaux, Bordeaux, France
4. APHM, Hopital de la Timone, Marseille, France
5. APHP, Hôpital de la Pitié-Salpêtrière, France
6. Hôpital Foch, Suresnes, France
7. CHU de Lille, Lille, France
8. Hospices Civils de Lyon, Université Claude Bernard Lyon, France
9. Oncopole, Department of Radiotherapy, Toulouse, France
10. CHU de Nancy, Nancy, France
References
- 1. Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carneiro BA, Elvin JA, Kamath SD, et al. FGFR3-TACC3: A novel gene fusion in cervical cancer. Gynecol Oncol Rep. 2015;13:53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Capelletti M, Dodge ME, Ercan D, et al. Identification of recurrent FGFR3-TACC3 fusion oncogenes from lung adenocarcinoma. Clin Cancer Res. 2014;20(24):6551–6558. [DOI] [PubMed] [Google Scholar]
- 4. Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22(4):795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yuan L, Liu ZH, Lin ZR, Xu LH, Zhong Q, Zeng MS. Recurrent FGFR3-TACC3 fusion gene in nasopharyngeal carcinoma. Cancer Biol Ther. 2014;15(12):1613–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Stefano AL, Fucci A, Frattini V, et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res. 2015;21(14):3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frattini V, Pagnotta SM, Tala, et al. A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature. 2018;553(7687):222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bielle F, Di Stefano AL, Meyronet D, et al. Diffuse gliomas with FGFR3-TACC3 fusion have characteristic histopathological and molecular features. Brain Pathol. 2018;28(5):674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273(1):168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer. 2012;118(17):4201–4211. [DOI] [PubMed] [Google Scholar]
- 11. Bielle F, Ducray F, Mokhtari K, et al. ; Pola Network Tumor cells with neuronal intermediate progenitor features define a subgroup of 1p/19q co-deleted anaplastic gliomas. Brain Pathol. 2017;27(5):567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. [DOI] [PubMed] [Google Scholar]
- 13. Labussière M, Boisselier B, Mokhtari K, et al. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology. 2014;83(13):1200–1206. [DOI] [PubMed] [Google Scholar]
- 14. Labussière M, Di Stefano AL, Gleize V, et al. TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations. Br J Cancer. 2014;111(10):2024–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Picca A, Berzero G, Bielle F, et al. FGFR1 actionable mutations, molecular specificities, and outcome of adult midline gliomas. Neurology. 2018;90(23):e2086–e2094. [DOI] [PubMed] [Google Scholar]
- 16. Idbaih A, Aimard J, Boisselier B, et al. Epidermal growth factor receptor extracellular domain mutations in primary glioblastoma. Neuropathol Appl Neurobiol. 2009;35(2):208–213. [DOI] [PubMed] [Google Scholar]
- 17. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 18. Rios Velazquez E, Meier R, Dunn WD Jr, et al. Fully automatic GBM segmentation in the TCGA-GBM dataset: Prognosis and correlation with VASARI features. Sci Rep. 2015;5:16822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. VASARI Research Project. The Cancer Imaging Archive (TCIA) Public Access, Cancer Imaging Archive Wiki https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project. Accessed September 27, 2019.
- 20. Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- 21. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shinohara RT, Sweeney EM, Goldsmith J, et al. ; Australian Imaging Biomarkers Lifestyle Flagship Study of Ageing; Alzheimer’s Disease Neuroimaging Initiative Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 2014;6:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pustina D, Avants B, Faseyitan OK, Medaglia JD, Coslett HB. Improved accuracy of lesion to symptom mapping with multivariate sparse canonical correlations. Neuropsychologia. 2018;115:154–166. [DOI] [PubMed] [Google Scholar]
- 24. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 26. Harrell F. Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis. New York, NY: Springer-Verlag; 2001. //www.springer.com/gb/book/9781441929181. Accessed January 30, 2019. [Google Scholar]
- 27. Tsiliki G, Munteanu CR, Seoane JA, Fernandez-Lozano C, Sarimveis H, Willighagen EL. RRegrs: an R package for computer-aided model selection with multiple regression models. J Cheminform. 2015;7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 30. Shackelford DB, Abt E, Gerken L, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23(2):143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cruz-Bermúdez A, Vicente-Blanco RJ, Laza-Briviesca R, et al. PGC-1alpha levels correlate with survival in patients with stage III NSCLC and may define a new biomarker to metabolism-targeted therapy. Sci Rep. 2017;7(1):16661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ellingson BM, Lai A, Harris RJ, et al. Probabilistic radiographic atlas of glioblastoma phenotypes. AJNR Am J Neuroradiol. 2013;34(3):533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ellingson BM. Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr Neurol Neurosci Rep. 2015;15(1):506. [DOI] [PubMed] [Google Scholar]
- 34. Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105(13):5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Inglis-Broadgate SL, Thomson RE, Pellicano F, et al. FGFR3 regulates brain size by controlling progenitor cell proliferation and apoptosis during embryonic development. Dev Biol. 2005;279(1):73–85. [DOI] [PubMed] [Google Scholar]
- 36. Smith KM, Ohkubo Y, Maragnoli ME, et al. Midline radial glia translocation and corpus callosum formation require FGF signaling. Nat Neurosci. 2006;9(6):787–797. [DOI] [PubMed] [Google Scholar]
- 37. Romeo A, Lodi M, Viri M, et al. Does the co-occurrence of FGFR3 gene mutation in hypochondroplasia, medial temporal lobe dysgenesis, and focal epilepsy suggest a syndrome? Pediatr Neurol. 2014;50(4):427–430. [DOI] [PubMed] [Google Scholar]
- 38. Okazaki T, Saito Y, Ueda R, et al. Epileptic phenotype of FGFR3-related bilateral medial temporal lobe dysgenesis. Brain Dev. 2017;39(1):67–71. [DOI] [PubMed] [Google Scholar]
- 39. Stevens HE, Smith KM, Maragnoli ME, et al. Fgfr2 is required for the development of the medial prefrontal cortex and its connections with limbic circuits. J Neurosci. 2010;30(16):5590–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.