Abstract
The thickening of Zizania latifolia shoots, referred to as gall formation, depends on infection with the fungal endophyte Ustilago esculenta. The swollen and juicy shoots are a popular vegetable in Asia. A key role for cytokinin action in this process was postulated. Here, trans-zeatin stimulated swelling in fungi-infected Z. latifolia. A two-component system (TCS) linked cytokinin binding to receptors with transcriptional regulation in the nucleus and played important roles in diverse biological processes. We characterized 69 TCS genes encoding for 25 histidine kinase/histidine-kinase-like (HK(L)) (21 HKs and 4 HKLs), 8 histidine phosphotransfer proteins (HP) (5 authentic and 3 pseudo), and 36 response regulators (RR; 14 type A, 14 type B, 2 type C, and 6 pseudo) in the genome of Z. latifolia. These TCS genes have a close phylogenetic relationship with their rice counterparts. Nineteen duplicated TCS gene pairs were found and the ratio of nonsynonymous to synonymous mutations indicated that a strong purifying selection acted on these duplicated genes, leading to few mutations during evolution. Finally, ZlCHK1, ZlRRA5, ZIRRA9, ZlRRA10, ZlPRR1, and ZlPHYA expression was associated with gall formation. Among them, ARR5, ARR9, and ZlPHYA are quickly induced by trans-zeatin, suggesting a role for cytokinin signaling in shoot swelling of Z. latifolia.
Keywords: two-component system, Z. latifolia, shoot swelling, cytokinin signals
1. Introduction
Zizania latifolia belongs to the wild rice genus Zizania and was an important grain crop in ancient China. Around 2000 years ago, Z. latifolia was infected by the fungal endophyte Ustilago esculenta which causes swelling in shoot apical tissues, resulting in juicy galls. Nowadays, the swollen shoots of Z. latifolia are a popular vegetable in China (called “Jiaobai”), East Asian countries, and Japan [1,2].
In nature, plant tumors or “galls” can be induced by different pathogens such as bacteria, viruses, fungi, protists, and insects. Despite different pathogenic mechanism to induce galls, most of these plant pathogens demonstrate the ability to change the phytohormone level in the host plants, especially cytokinins. For example, the well-studied gall-forming Agrobacterium tumefaciens carries the Ti plasmid, which can integrate a DNA segment called T-DNA into the host’s genome [3]. The T-DNA carries several genes including isopentenyl transferase (IPT), a key enzyme of cytokinin biosynthesis, which instigates increased cytokinin production in the transformed cells of the host [4].
Increased production of cytokinins in the infected plant leads to uncontrolled cell proliferation and gall formation [5,6]. Chan and Thrower [7] isolated zeatin in Ustilago esculenta liquid culture, suggesting this fungi may also produce cytokinin in vivo. Another study revealed that the content of cis-zeatin riboside and cytokinin activity in terms of kinetin equivalents from fungi-infected gall tissue was elevated compared to non-fungi-infected shoots in Z. latifolia [8]. These results indicate that cytokinins play an important role in the swelling of Z. latifolia shoots.
Cytokinins are a class of key phytohormones that mediate several processes including apical dominance, lateral branching, germination, flower and fruit development, chloroplast differentiation, plant–pathogen interactions, senescence, cell division in shoots, and spatial control of differentiation in roots [9,10,11,12,13]. Cytokinin signal transduction is through a phospho-relay derived from the two-component system (TCS) consisting of membrane-associated histidine kinase receptor proteins (HK), histidine-containing phosphotransfer proteins (HPs), and the response regulators (RR) gene. The TCS genes in the Arabidopsis thaliana model have been identified and functionally characterized. Three histidine kinase proteins of AHK2, AHK3, and AHK4 have been characterized as cytokinin receptors [14]. All cytokinin receptors were found to share a domain in the predicted extracytoplasmic region, designated as cyclase/histidine kinase-associated sensory extracellular (CHASE) domain which is the putative binding site for cytokinin molecules [15]. Upon perceiving the stimuli, the receptors are autophosphorylated. The phosphate group is then transferred to the AHPs (Arabidopsis histidine-containing phosphotransfer proteins), which are able to enter the nucleus and transfer the phosphate group to type B RRs. In the nucleus, phosphorylated type B RRs can, amongst other targets, transcriptionally activate type A RRs. Type A RRs act as repressors of the primary cytokinin response, providing a negative feedback regulation [16,17,18]. This TCS-mediated histidine-aspartate signaling has been demonstrated to be able to control diverse biological processes, such as osmosensing, responses to environmental stimuli, cell growth, and proliferation [14,19,20,21,22,23,24,25,26,27,28,29,30]
Capitalizing on the progress in genome sequencing, TCS genes have been identified and characterized in many other different plant species such as rice [21], maize [25], soybean [24], Chinese cabbage [26], tomato [27], and cucumber [28]. In this study, the effect of exogenous trans-zeatin in Z. latifolia shoots swelling was investigated. Exogenous cytokinin stimulated the gall formation in fungi-infected shoots, in line with previous reports. As a first step towards understanding this response to cytokinins, genome-wide identification of TCS genes in Z. latifolia was conducted, which included sequences and functional domains analysis, phylogenetic relationship prediction, genes and protein structure analysis, and the analysis of the evolution of these members. Secondly, the expression of the TCS genes during shoot swelling was investigated by transcriptomics. Thirdly, the response of a subset of TCS genes to exogenous cytokinin was analyzed by quantitative real-time PCR (qRT-PCR). The obtained results establish a base for future functional analysis of TCS genes in cytokinin signal transduction in Z. latifolia, especially during gall formation.
2. Results and Discussion
2.1. Effect of Exogenous Cytokinin Treatment on Z. latifolia Gall Formation
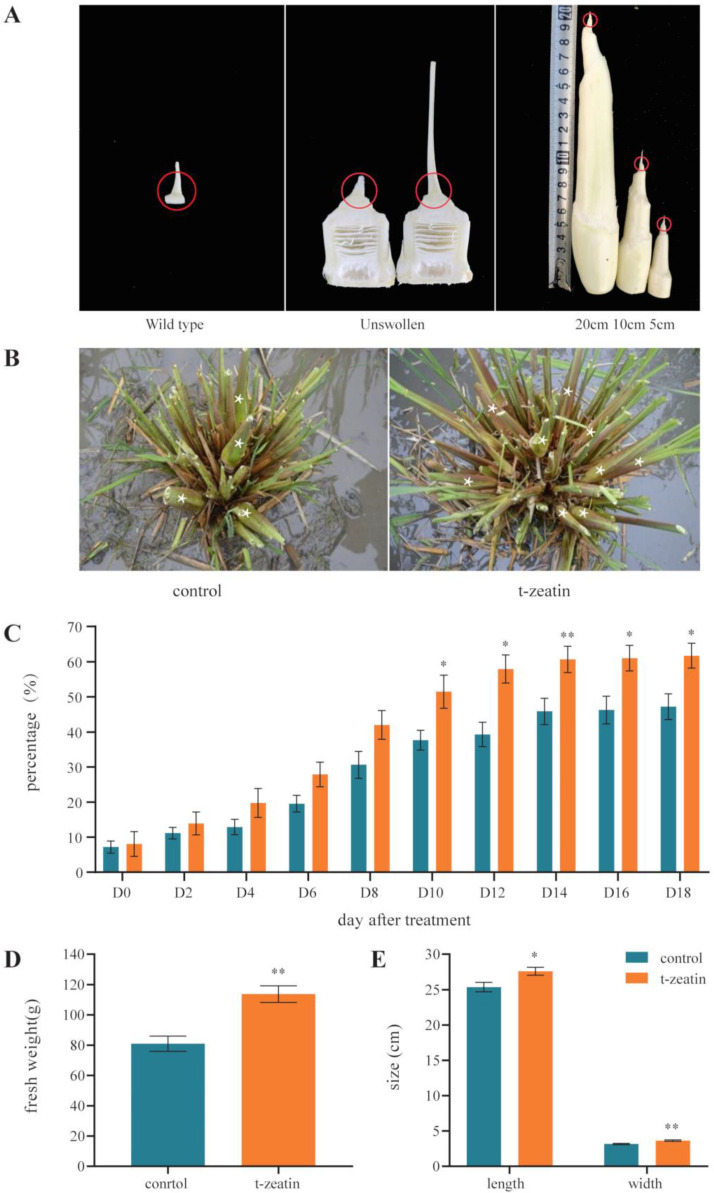
In contrast to non-infected wild-type Z. latifolia shoots, Ustilago esculenta-infected plants develop galls which become up to 20 cm long over two weeks (Figure 1A). Trans-zeatin treatment could significantly increase the frequency of shoot swelling by about 15% (p < 0.05) (Figure 1B,C). Further measurements suggested that cytokinin treatment also significantly stimulated the shoots swelling in size and weight (p < 0.05) (Figure 1D,E). Chan and Thrower [7] suggested that three kinds of cytokinins including zeatin and zeatin riboside were isolated in Z. latifolia gall tissue, and Ustilago esculenta could produce zeatin in culture. However, no structural confirmation on the cytokinin identity was available at time. A higher level of cis-zeatin riboside was detected in the Z. latifolia gall compared to the uninfected shoot [8]. Furthermore, ZlIPT, the most important enzyme to catalyze cytokinins synthesis, was more highly expressed in gall tissue compared to the uninfected shoot [31]. Collectively, cytokinins may play a key role in Z. latifolia gall formation and the cytokinins produced by Z. latifolia and/or Ustilago esculenta could stimulate cell division and enlargement, facilitating gall formation. This study also demonstrated that exogenous cytokinin could stimulate and promote the gall formation; therefore, cytokinins can be used as a plant growth regulator to improve the horticultural production of Z. latifolia galls.
Figure 1.
Development of and effects of cytokinin on Zizania latifolia galls. (A) In contrast to uninfected wild-type (WT) shoots (left panel), infected shoots develop large galls under their apices up to 20 cm length, over a time course of 2 weeks (middle, initial shape of apex, right panel stages of gall formation). The red circles indicate the size of the collected samples collected for expression analysis. (B) Morphology (stars label the swollen shoots) and (C) frequency of gall formation of 150 mg/L t-zeatin treated and untreated Z. latifolia plants (n = 12). (D) Fresh weight and size (E) of treated and untreated galls (n > 100). Each histogram represents the mean value and the bar indicates the means ± standard error of biological replicates. * Significant difference at p < 0.05. ** Significant difference at p < 0.01.
2.2. Identification of TCS Genes in Z. latifolia
To identify members of the TCS family in Z. latifolia, HMM searches and BLASTP analysis were performed by employing 415 TCS protein sequences from Arabidopsis [14], rice [21], maize [25], soybean [24], Chinese cabbage [26], and tomato [27] as queries. After repetitive sequences were removed, all identified sequences were reserved and submitted to NCBI CDD, Pfam, and SMART to confirm their typical domains. A total of 69 non-redundant sequences, including 25 HK(L)s, 8 HPs, and 36 RRs, were identified in the Z. latifolia genome. All TCS members in Z. latifolia were named according to the homologous genes in Arabidopsis [27,28,32]. This nomenclature has been used in tomato [27], cucumber, and watermelon [28]. The TCS genes have been intensively studied in model plants and some important crops. The number of TCS genes in Z. latifolia (n = 69) is similar to that in tomato (n = 65) but larger than that of all reported species except soybean (n = 98) and Chinese cabbage (n = 85; Table S1).
2.3. HK Proteins in Z. latifolia
Sequence analysis of the entire Z. latifolia genome revealed 25 distinct putative histidine protein kinases genes, including 21 histidine protein kinases (ZlHKs) and 4 related proteins (ZlHKLs), based on whether their HK domains were conserved or divergent (Table 1). These histidine protein kinase homologs can be further divided into distinct subfamilies: Eight cytokinin receptors (CHKs), one CKI-like protein, seven ethylene receptors (ERSs and ETRs), five phytochrome photoreceptors (PHYs), and four pyruvate dehydrogenase kinases (PDKs; Table 1). A typical HK domain (corresponding to motifs 1 and 7 in Figure S2) has five conserved signature motifs—H, N, G1, F, and G2—with the conserved His as the key feature in the H motif. The other four motifs were defined as the nucleotide-binding cleft [33]. Through multiple-sequence alignment, all HKs and HKLs proteins were found to be conserved in the H motif with His residue, except ZlCHK2 and ZlCHK3 (Figure S1A). All proteins had complete N motifs. However, the G1, F, and G2 motifs occurred only in ZlCHK3 and ZlCHK4, suggesting that other HK/HKL proteins may not have full HK function or that these motifs may not be necessary for HK function (Figure S1A). Notably, only ZlCHK4 had a full HK domain, suggesting that it has a critical function in Z. latifolia. Table 1 and Figure S1B also show that both CHASE domains (motif 11 in Figure S2) and 1–4 transmembrane sequences (TM) occurred in these cytokinin receptors, except for ZlCHK2 and ZlCHK3. Both CHASE and TM were demonstrated to be crucial for membrane-associated cytokinin recognizing and binding [17]. In addition, the receiver domain (motifs 9 and 13 in Figure S2) with Asp residue was conserved in all ZlCHKs except for ZlCHK7 (Figure S1B). Two AHK2-like genes (ZlCHK1 and ZlCHK2) were 65% and 46% similar, respectively, to their Arabidopsis counterparts of AHK2, whereas two AHK3-like members (ZlCHK3 and ZlCHK4) were 61% and 63% similar, respectively, to Arabidopsis AHK3. Another four AHK4-like cytokinin receptor genes (ZlCHK5, ZlCHK6, ZlCHK7, and ZlCHK8) were highly similar (55%–72%) to Arabidopsis CRE1/AHK4/WOL (Table 1). AHK2, AHK3, and CRE1/AHK4/WOL are cytokinin receptors and can regulate cell division in Arabidopsis [15,34,35,36,37,38,39,40,41]. Some of the cell cycle and meristem control master genes, such as CYCD3s and STM1, respond to cytokinins and function downstream of CRE1/AHK4/WOL to regulate shoot and root development [42].
Table 1.
Histidine kinase/histidine-kinase-like (HK(L)) proteins in Z. Latifolia.
| Gene ID | Gene Name | Character a | Gene Family | Counterpart | Score (bits) | Identity (%) | Length (aa) | Molecular Weight (kDa) | Isoelectric Point (PI) | Number of TM | Subcellular localization b |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Zlat_10043954 | ZlCHK1 | HisKA, HATPase_c, CHASE, REC, TM | Cytokinin Receptor | AHK2 | 532 | 65 | 609 | 67.56 | 7.26 | 4 | Endomembrane system |
| Zlat_10043955 | ZlCHK2 | HisKA, HATPase_c, REC | Cytokinin Receptor | AHK2 | 207 | 46 | 445 | 49.66 | 6.65 | 0 | Nucleus |
| Zlat_10033100 | ZlCHK3 | HisKA, HATPase_c, REC | Cytokinin Receptor | AHK3 | 673 | 61 | 570 | 63.2 | 6.72 | 0 | Nucleus |
| Zlat_10033475 | ZlCHK4 | HisKA, HATPase_c, CHASE, REC, TM | Cytokinin Receptor | AHK3 | 1045 | 63 | 861 | 95.89 | 8.55 | 1 | Nucleus |
| Zlat_10002572 | ZlCHK5 | HisKA, HATPase_c, CHASE, REC, TM | Cytokinin Receptor | AHK4 | 746 | 68 | 907 | 99.75 | 6.02 | 2 | Endomembrane system |
| Zlat_10008874 | ZlCHK6 | HisKA, HATPase_c, CHASE, REC, TM | Cytokinin Receptor | AHK4 | 202 | 72 | 430 | 48.69 | 5.58 | 1 | Organelle membrane |
| Zlat_10009428 | ZlCHK7 | HisKA, HATPase_c, CHASE, REC, TM | Cytokinin Receptor | AHK4 | 620 | 69 | 618 | 69.42 | 5.27 | 1 | Nucleus |
| Zlat_10007870 | ZlCHK8 | HisKA, HATPase_c, CHASE, REC, TM | Cytokinin Receptor | AHK4 | 335 | 55 | 716 | 78.44 | 5.41 | 1 | Nucleus |
| Zlat_10005586 | ZlCKI1 | HisKA, HATPase_c, REC | CKI1 | 330 | 36 | 810 | 89.62 | 5.81 | 0 | Nucleus | |
| Zlat_10007295 | ZlERS1 | HisKA, HATPase_c, GAF, TM | Ethylene Receptor | ETR1 | 900 | 74 | 636 | 70.93 | 6.86 | 3 | Endomembrane system |
| Zlat_10019946 | ZlERS2 | HisKA, HATPase_c, GAF | Ethylene Receptor | ETR1 | 380 | 65 | 314 | 34.48 | 5.29 | 0 | Nucleus |
| Zlat_10028651 | ZlERS3 | HisKA, HATPase_c, GAF, TM | Ethylene Receptor | ETR1 | 879 | 75 | 635 | 70.79 | 6.82 | 3 | Endomembrane system |
| Zlat_10043320 | ZlETR1 | HisKA, HATPase_c, GAF, REC, TM | Ethylene Receptor | ETR2 | 549 | 47 | 842 | 93.21 | 6.89 | 4 | Plasma membrane |
| Zlat_10010084 | ZlETR2 | HisKA, HATPase_c, GAF, REC, TM | Ethylene Receptor | EIN4 | 669 | 51 | 759 | 85.01 | 6.21 | 3 | Endomembrane system |
| Zlat_10043113 | ZlETR3 | HisKA, HATPase_c, GAF, REC, TM | Ethylene Receptor | EIN4 | 662 | 51 | 758 | 84.99 | 6.18 | 3 | Plasma membrane |
| Zlat_10044950 | ZlETR4 | HisKA, HATPase_c, GAF, REC | Ethylene Receptor | EIN4 | 302 | 39 | 507 | 54.79 | 7.52 | 0 | Nucleus |
| Zlat_10007862 | ZlPHYA | HisKA, HATPase_c, GAF, PAS | Phytochrome | PHYA | 1456 | 64 | 1128 | 125.1 | 5.78 | 0 | Nucleus |
| Zlat_10002747 | ZlPHYB | HisKA, HATPase_c, GAF, PAS | Phytochrome | PHYA | 1439 | 63 | 1129 | 125.24 | 5.83 | 0 | Nucleus |
| Zlat_10036932 | ZlPHYC | HisKA, HATPase_c, GAF, PAS | Phytochrome | PHYB | 1736 | 75 | 1190 | 130.73 | 5.72 | 0 | Organelle membrane |
| Zlat_10005292 | ZlPHYD | HisKA, HATPase_c, GAF, PAS | Phytochrome | PHYB | 1710 | 75 | 1178 | 123.76 | 5.69 | 0 | Nucleus |
| Zlat_10007618 | ZlPHYE | HisKA, HATPase_c, GAF, PAS | Phytochrome | PHYC | 1384 | 59 | 1137 | 125.65 | 5.62 | 0 | Chloroplast |
| Zlat_10046362 | ZlPDK1 | HATPase_c | Pyruvate dehydrogenase kinase | PDK | 530 | 74 | 363 | 40.77 | 6.08 | 0 | Chloroplast |
| Zlat_10041425 | ZlPDK2 | HATPase_c | Pyruvate dehydrogenase kinase | PDK | 521 | 72 | 363 | 40.73 | 6.33 | 0 | Chloroplast |
| Zlat_10030552 | ZlPDK3 | HATPase_c | Pyruvate dehydrogenase kinase | PDK | 551 | 76 | 363 | 40.91 | 6.68 | 0 | Nucleus |
| Zlat_10018898 | ZlPDK4 | HATPase_c | Pyruvate dehydrogenase kinase | PDK | 555 | 73 | 405 | 44.76 | 6.84 | 0 | Chloroplast |
a Character indicates conserved histidine–kinase domain (HK), receiver domain (Rec), CHASE domain for cytokinin binding (CHASE), and chromophore-binding domain (PHY). b Subcellular localization predicted with BUSCA (Bologna Unified Subcellular Component Annotator).
The ethylene receptors, including ZlETR1–4 and ZlERS1–3, contain HK, HATPase, and cyclic GMP adenylyl cyclase FhlA (GAF) domains (Table 1). Although histidine–kinase activity can subtly modulate the ethylene response, no major role has yet been identified in ethylene signal transduction [43,44,45]. However, histidine kinase activity could allow cross-talk between ethylene perception and other TCS pathways such as cytokinin signal transduction. All five PHY subfamily members (ZlPHYA, B, C, D, E) possess PHY (chromophore-binding), GAF, and PAS (signal sensor) domains (Table 1). These domains were found to be essential for responding to red and far-red light signals during plant development in Arabidopsis [46]. The HATPase domain was identified in all ZlPDKs proteins (ZlPDK1–4). A newly published paper has revealed that PDK1 could regulate auxin transport and vascular development through phosphorylation of AGC1 kinase PAX in Arabidopsis [47].
To further understand the structural diversity of Z. latifolia HK genes, we analyzed the exon–intron organization and conserved protein motifs (Figure S2). These analyses revealed that HK genes in the same group usually had a similar gene structure and motif composition, which strongly support the reliability of the phylogenetic classification.
2.4. HP Proteins in Z. latifolia
Eight ZlHP genes were identified: Five authentic HPs and three pseudo-HPs (Table 2). All ZlHPs were small proteins with 149–276 amino acids and had two conserved motifs (motifs 1 and 2 in Figure S3) that encode the Hpt domain. Five authentic HPs (ZlHP1, ZlHP2, ZlHP4, ZlHP7, ZlHP8) contained conserved His (H) residues in the HP signature sequence of XHQXKGSSXS, whereas in the other three pseudo-HPs (ZlHP3, ZlHP5, and ZlHP6), the histidine phosphorylation site was replaced by Gln (Q) residue (Figure S4). Notably, all HPs except ZlHP1 and ZlHP3 were localized in the nucleus (Table 2), which might be essential for their phosphorelay, during which HPs translocate from the cytoplasm to the nucleus [48,49]. In addition, all ZlHPs had a very similar gene structure containing 5–7 introns and motif compositions (Figure S3).
Table 2.
Histidine phosphotransfer proteins (HP) proteins in Z. latifolia.
| Gene ID | Gene Name | Character a | Gene Family | Counterpart | Score(bits) | Identity (%) | Length (aa) | Molecular Weight(kDa) | Isoelectric Point (PI) | Subcellular localization b |
|---|---|---|---|---|---|---|---|---|---|---|
| Zlat_10020936 | ZlHP1 | HPt | HPt | AHP1 | 117 | 20 | 210 | 23.93 | 8.57 | Organelle membrane |
| Zlat_10031305 | ZlHP2 | HPt | HPt | AHP1 | 115 | 50 | 146 | 16.63 | 5.28 | Nucleus |
| Zlat_10002667 | ZlHP3 | pseudo-HPt | pseudo-HPt | AHP4 | 162 | 61 | 276 | 30.89 | 7.11 | Extracellular space |
| Zlat_10013094 | ZlHP4 | HPt | HPt | AHP4 | 184 | 59 | 151 | 17.84 | 8.33 | Nucleus |
| Zlat_10015279 | ZlHP5 | pseudo-HPt | pseudo-HPt | AHP4 | 187 | 59 | 158 | 18.03 | 8.2 | Nucleus |
| Zlat_10032102 | ZlHP6 | pseudo-HPt | pseudo-HPt | AHP4 | 197 | 63 | 151 | 17.42 | 7.55 | Nucleus |
| Zlat_10034774 | ZlHP7 | HPt | HPt | AHP5 | 125 | 46 | 149 | 16.78 | 4.71 | Nucleus |
| Zlat_10034845 | ZlHP8 | HPt | HPt | AHP5 | 125 | 46 | 149 | 16.79 | 4.66 | Nucleus |
a Character indicates whether the proteins possess a conserved His-containing phosphotransfer domain (HPt) or a pseudo-HPt lacking the His phosphorylation site. b Subcellular localization predicted with BUSCA (Bologna Unified Subcellular Component Annotator).
Two AHP1-like members of ZlHP1 and ZlHP2 were 20% and 50% similar to their Arabidopsis homolog AHP1, respectively (Table 2). AHP1 could interact with CRE1/AHK4/WOL and B-type ARRs such as ARR1, suggesting that it may function as a phosphotransfer intermediate [50,51,52,53]. Four AHP4-like members (ZlHP3, ZlHP4, ZlHP5, ZlHP6) were found to be highly similar (59%–63%) to Arabidopsis AHP4 (Table 2). Interestingly, three pseudo-HPs of (ZlHP3, ZlHP5, ZlHP6) had a higher similarity to the authentic AHP4 than to the pseudo APHP1 in Arabidopsis. This was also observed in rice and cucumber [21,28]. Both ZlHP7 and ZlHP8 were 46% similar to their Arabidopsis counterpart AHP5 (Table 2). The high sequence similarity between ZlHP7 and ZlHP8 suggests possible functional redundancy between these two genes.
2.5. RR Proteins in Z. latifolia
Thirty-six genes were identified as ZlRRs: 14 type A RRs, 14 type B RRs, 2 type C RRs, and 6 pseudo-RRs (Table 3). The type A RRs (ZlRRA1–14) were a group of small proteins with 76–269 amino acids. All type A RRs contained a receiver domain corresponding to three motifs (motifs 1, 3, and 4 in Figure S5) along with short N- and C-terminal extensions (Table 3 and Figure S5). Most type A RRs contained four introns. The 14 ZlRRAs were highly similar (52%–81%) to their Arabidopsis homologs (Table 3).
Table 3.
Response regulator (RR) proteins in Z. latifolia.
| Gene ID | Gene Name | Character a | Gene Family | Counterpart | Score(bits) | Identity (%) | Length (aa) | Molecular Weight(kDa) | Isoelectric Point (PI) | Subcellular Localization b |
|---|---|---|---|---|---|---|---|---|---|---|
| Zlat_10022933 | ZlRRA1 | REC | Type A | ARR3 | 103 | 61 | 167 | 16.23 | 5.71 | Nucleus |
| Zlat_10026415 | ZlRRA2 | REC | Type A | ARR3 | 110 | 65 | 175 | 16.16 | 6.84 | Chloroplast |
| Zlat_10028329 | ZlRRA3 | REC | Type A | ARR3 | 129 | 64 | 143 | 15.11 | 9.46 | Nucleus |
| Zlat_10028974 | ZlRRA4 | REC | Type A | ARR3 | 129 | 64 | 137 | 14.75 | 6.59 | Nucleus |
| Zlat_10030869 | ZlRRA5 | REC | Type A | ARR3 | 124 | 54 | 258 | 28.13 | 5.99 | Extracellular space |
| Zlat_10032021 | ZlRRA6 | REC | Type A | ARR3 | 121 | 52 | 269 | 29.31 | 7.6 | Plasma membrane |
| Zlat_10033406 | ZlRRA7 | REC | Type A | ARR6 | 152 | 64 | 165 | 17.89 | 9.1 | Chloroplast outer membrane |
| Zlat_10001454 | ZlRRA8 | REC | Type A | ARR8 | 200 | 72 | 234 | 25.56 | 4.83 | Nucleus |
| Zlat_10017478 | ZlRRA9 | REC | Type A | ARR8 | 99.4 | 59 | 187 | 21.08 | 6.81 | Nucleus |
| Zlat_10003241 | ZlRRA10 | REC | Type A | ARR9 | 112 | 72 | 76 | 8.31 | 8.71 | Extracellular space |
| Zlat_10016677 | ZlRRA11 | REC | Type A | ARR9 | 204 | 57 | 215 | 24.01 | 6.06 | Nucleus |
| Zlat_10020468 | ZlRRA12 | REC | Type A | ARR9 | 137 | 81 | 176 | 19.42 | 6.74 | Nucleus |
| Zlat_10029578 | ZlRRA13 | REC | Type A | ARR9 | 134 | 57 | 231 | 25.92 | 4.07 | Extracellular space |
| Zlat_10044807 | ZlRRA14 | REC | Type A | ARR9 | 220 | 70 | 196 | 22.08 | 6.05 | Nucleus |
| Zlat_10024295 | ZlRRB1 | REC, Myb | Type B | ARR1 | 352 | 57 | 689 | 73.95 | 5.99 | Nucleus |
| Zlat_10041210 | ZlRRB2 | REC, Myb | Type B | ARR1 | 356 | 57 | 688 | 73.83 | 6.13 | Nucleus |
| Zlat_10019278 | ZlRRB3 | REC, Myb | Type B | ARR2 | 186 | 39 | 353 | 40.14 | 7.07 | Nucleus |
| Zlat_10010976 | ZlRRB4 | REC, Myb | Type B | ARR10 | 278 | 42 | 627 | 68.56 | 5.95 | Nucleus |
| Zlat_10006425 | ZlRRB5 | REC, Myb | Type B | ARR11 | 128 | 62 | 179 | 20.47 | 4.81 | Nucleus |
| Zlat_10007685 | ZlRRB6 | REC, Myb | Type B | ARR11 | 47.4 | 30 | 340 | 36.56 | 5.61 | Cytoplasm |
| Zlat_10010099 | ZlRRB7 | REC, Myb | Type B | ARR11 | 328 | 50 | 583 | 65.13 | 5.04 | Nucleus |
| Zlat_10033001 | ZlRRB8 | REC, Myb | Type B | ARR11 | 348 | 54 | 580 | 65.07 | 5.16 | Nucleus |
| Zlat_10007027 | ZlRRB9 | REC, Myb | Type B | ARR12 | 321 | 49 | 707 | 76.01 | 6.25 | Nucleus |
| Zlat_10018529 | ZlRRB10 | REC, Myb | Type B | ARR12 | 343 | 55 | 694 | 76.02 | 5.87 | Nucleus |
| Zlat_10018563 | ZlRRB11 | REC, Myb | Type B | ARR12 | 322 | 66 | 693 | 75.34 | 6.2 | Nucleus |
| Zlat_10022449 | ZlRRB12 | REC, Myb | Type B | ARR12 | 336 | 59 | 621 | 68.29 | 5.88 | Nucleus |
| Zlat_10028769 | ZlRRB13 | REC, Myb | Type B | ARR12 | 348 | 60 | 626 | 68.84 | 5.8 | Nucleus |
| Zlat_10041971 | ZlRRB14 | REC, Myb | Type B | ARR14 | 106 | 46 | 299 | 31.15 | 6.61 | Nucleus |
| Zlat_10027036 | ZlRRC1 | REC | Type C | ARR24 | 66.6 | 40 | 128 | 13.85 | 5.69 | Nucleus |
| Zlat_10029496 | ZlRRC2 | REC | Type C | ARR24 | 67.8 | 41 | 129 | 13.8 | 5.68 | Nucleus |
| Zlat_10006383 | ZlPRR1 | Pseudo-REC, CCT | Pseudo | APRR1 | 306 | 39 | 518 | 57.68 | 6.34 | Nucleus |
| Zlat_10008561 | ZlPRR2 | Pseudo-REC, CCT | Pseudo | APRR7 | 318 | 41 | 764 | 82.49 | 8.52 | Chloroplast outer membrane |
| Zlat_10024918 | ZlPRR3 | Pseudo-REC, CCT | Pseudo | APRR7 | 347 | 40 | 742 | 81.17 | 6.23 | Nucleus |
| Zlat_10017908 | ZlPRR4 | Pseudo-REC, CCT | Pseudo | APRR5 | 228 | 39 | 629 | 70.06 | 6.78 | Nucleus |
| Zlat_10023948 | ZlPRR5 | Pseudo-REC, CCT | Pseudo | APRR5 | 212 | 53 | 684 | 74.99 | 8.21 | Nucleus |
| Zlat_10035513 | ZlPRR6 | Pseudo-REC, CCT | Pseudo | APRR5 | 215 | 54 | 682 | 75.18 | 8.44 | Nucleus |
a Character indicates receiver domain (REC), pseudo-receiver domain (pseudo-REC) lacking the conserved D, Myb-like domain (Myb), plant-specific CCT motif found in clock proteins. b Subcellular localization predicted with BUSCA (Bologna Unified Subcellular Component Annotator).
In addition to having a conserved receiver domain, all type B RRs (ZlRRB1–14) contained long C-terminal extensions with a Myb-like DNA binding domain (motif 2 in Figure S5), suggesting their function as transcription factors. Except for ZlRRB6, all type B RRs were predicted to be localized in the nucleus (Table 3). Most type B RRs had four or five introns (Figure S5). Type B RRs were 30%–66% similar to their counterparts in Arabidopsis.
Different from type A RRs, two identified type C RRs (ZlRRC1, 2) contained a receiver domain (motifs 1 and 4 in Figure S5) but lacked a long C-terminal extension. Both proteins had three introns and were 40% and 41% similar to their Arabidopsis counterpart, ARR24, respectively. In addition, six proteins identified as pseudo-RRs (ZlPRR1–6) all had a conserved receiver domain (motifs 1 and 4 in Figure S5) and a CCT domain (motif 5), which was found to play an important role in regulating circadian rhythms [54]. These pseudo-RRs were 39%–54% similar to their counterparts in Arabidopsis. Except ZlPRR2, all these type C and pseudo-RRs were localized in the nucleus (Table 3, Figure S5). Figure S5 also shows that the ZlRRs genes in the same category usually had very similar gene structure and motif composition, thereby supporting the evolutionary conservation and reliability of the phylogenetic classification.
2.6. Phylogenetic Relationship of TCS Members in Z. latifolia
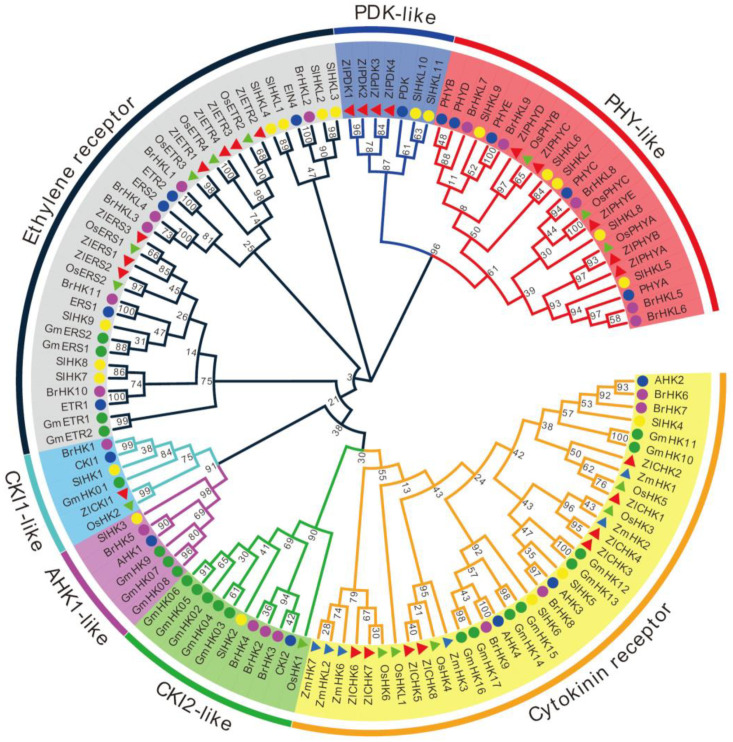
To investigate the evolutionary relationships of TCS genes, we used all amino acid sequences of 124 HK(L)s, 53 HPs, and 275 RRs from Arabidopsis [14], rice [21], maize [25], soybean [24], Chinese cabbage [26], tomato [27], and Z. latifolia to construct maximum likelihood (ML) trees [55]. On the basis of the bootstrap values and the topology of the tree, all HK(L)s proteins in the seven species were divided into seven distinct subfamilies, including cytokinin receptor, ethylene receptor, PHY-like, CKI1-like, CKI2/AHK5-like, AHK1-like, and PDK-like subfamilies (Figure 2). Similar phylogenetic structures were viewed in previous studies [14,17,21]. The HK(L)s in Z. latifolia usually have much closer relationships to those of rice and maize than other species, as they are all members of the monocotyledon. This indicates that the HK(L)s gene expansion occurred after the divergence of monocot and dicot plants. In addition, HK(L)s genes of monocots and eudicots showed an alternative distribution pattern in each subfamily, but no monocots occurred in the AHK1-like subgroup.
Figure 2.
Phylogenetic relationship of HK(L) proteins in Arabidopsis, rice, maize, soybean, Chinese cabbage, tomato, and Z. latifolia. A maximum likelihood phylogenetic tree indicates the different subgroups of HK(L)s in different colors.
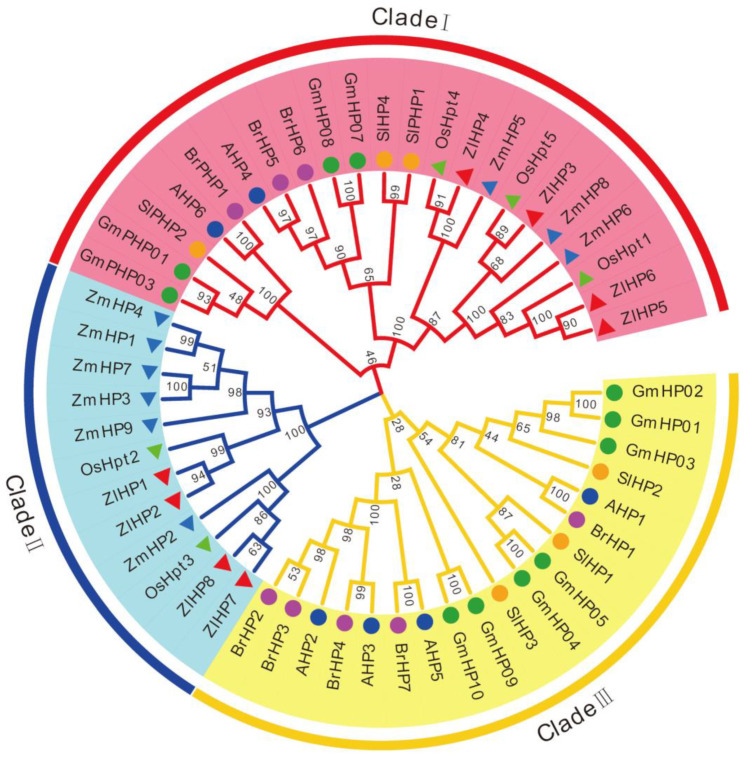
All HP proteins from the seven species were divided into three clades—clades I, II, and III (Figure 3). Clade II contained HPs only from monocots, whereas all HPs in clade III were from dicots. These results suggest that the gene expansion events for HPs occurred after the divergence of monocots and dicots. Four authentic ZlHPs except ZlHP4 were grouped into clade II, whereas clade I mainly contained pseudo-HPs from both monocots and dicots. All ZlHPs showed the closest phylogenetic relationship to rice OsHpts (Figure 3).
Figure 3.
Phylogenetic relationship of phosphotransfer proteins (HP) and related proteins in Arabidopsis, rice, maize, soybean, Chinese cabbage, tomato, and Z. latifolia.
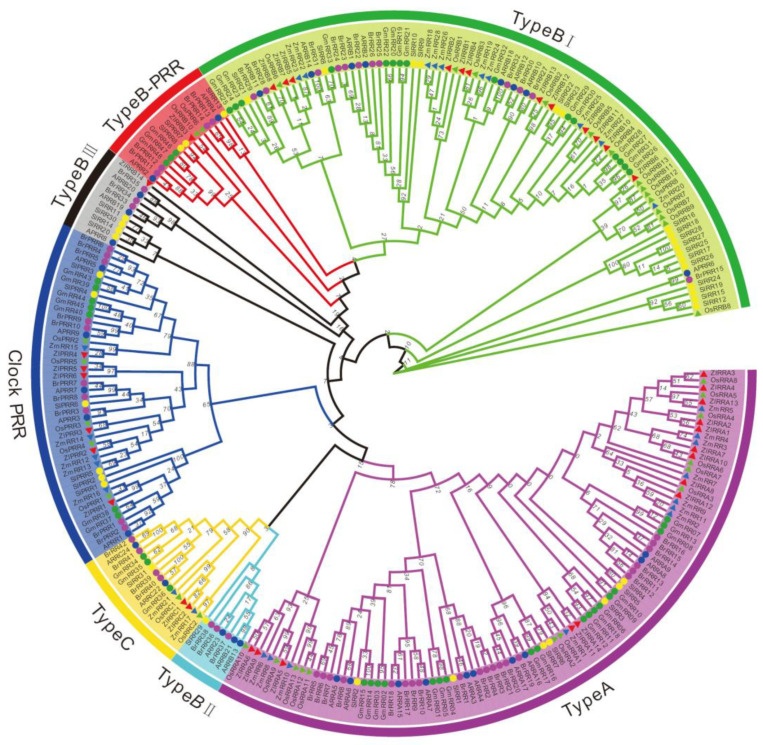
The 275 RR proteins from the seven plant species were classified into type A, type B, type C, and pseudo-RR groups (Figure 4). Generally, the ZlRRs in Z. latifolia were phylogenetically closer to rice OsRRs in all these groups. Phylogenetic analyses indicated that all type A RRs shared a close evolutionary relationship and an alternating distribution between monocots and dicots plants, suggesting that type A RR genes might already have expanded before the monocot–dicot divergence. Type B RRs from these species could be divided into three subgroups (I, II, and III), consistent with previous studies [17,26]. In detail, type B II subgroups contain RRs only from dicots of Arabidopsis, Chinese cabbage, and tomato; the RRs from monocots might have been lost during evolution. All type B ZlRRs from Z. latifolia except for ZlRRB3 and ZlRRB14 were classified into the type B I subfamily. A similar phenomenon was also viewed in soybean and cucumber [24,28]. All pseudo-RRs from seven plant species were subclassified into type B PRRs or clock PRRs (Figure 4).
Figure 4.
Phylogenetic relationship of response regulator (RR) proteins and related proteins in Arabidopsis, rice, maize, soybean, Chinese cabbage, tomato, and Z. latifolia.
2.7. Strong Purifying Selection for TCS Genes in Z. latifolia
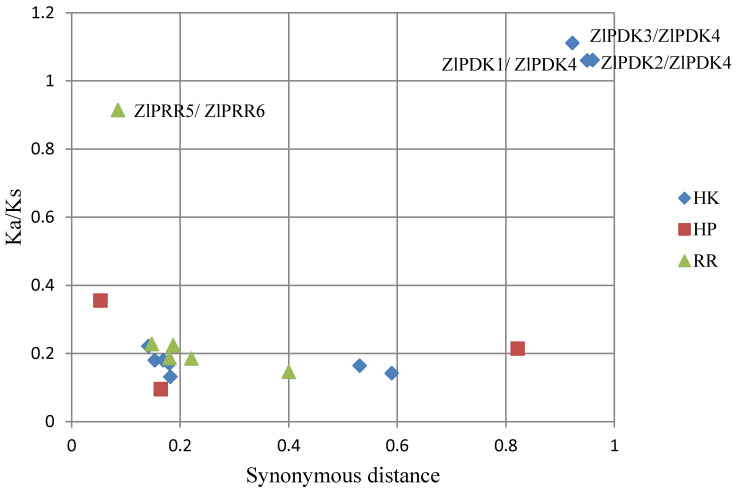
To further understand how the TCS gene family in Z. latifolia expanded during evolution, the gene duplication events were investigated. Nineteen duplicated gene pairs in Z. latifolia TCS genes were identified including 10 ZlHK pairs, 3 ZlHP pairs, and 6 ZlRR pairs (Table S2). To study the selection pressures on the TCS gene family, nonsynonymous (Ka), synonymous (Ks), and Ka/Ks ratios were calculated. In this study, the Ka/Ks values from the 19 pairs of Z. latifolia TCS genes were much less than 1, except ZlPDK1/ZlPDK4, ZlPDK2/ZlPDK4, and ZlPDK3/ZlPDK4 (Table S2, Figure 5). This suggests that the TCS gene family in Z. latifolia underwent strong purifying selection, with a slow rate of accumulation of missense mutations during evolution.
Figure 5.
Scatter plots of the Ka/Ks ratios of duplicated two-component system (TCS) genes in Z. latifolia. The x-axis represents the synonymous distance and the y-axis represents the Ka/Ks ratio for each pair.
2.8. TCS Gene Expression During Z. latifolia Shoots Swelling
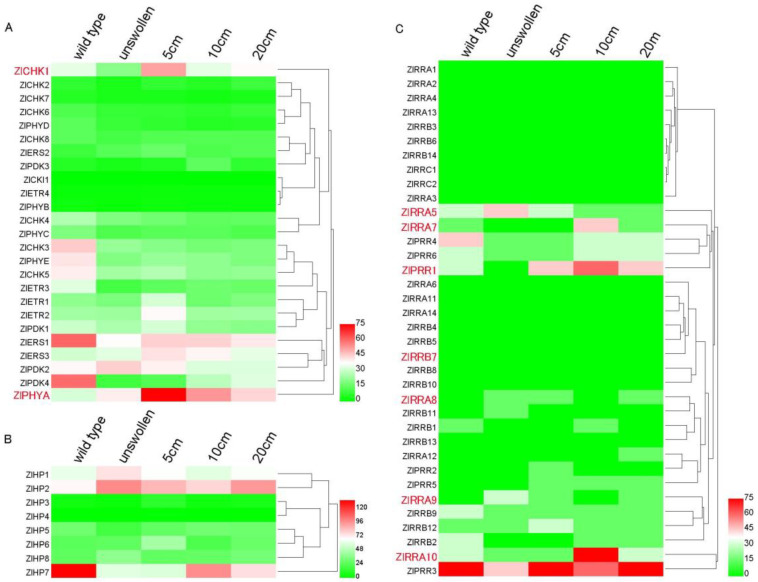
To gain more insight into the TCS gene function in cytokinin-regulated gall formation, the transcriptome measured by RNA sequencing was mined to investigate the expression of TCS genes in shoot apices during shoot swelling in Z. latifolia. Levels of transcripts of TCS genes in wild-type (collected from uninfected shoots) were compared to samples from young but infected unswollen shoots (collected from fungi-infected plant with eight leaves) and samples from developing galls (Figure 1A).
We noted the transient kinetics of the ZICHK1 and ZIETR2 receptor transcripts, the transcripts of the gene encoding for the red-light receptor phytochrome A (ZIPHA) transiently accumulated during gall formation and the activation of ZIPDK2 in young infected shoots (Figure 6A). Elevated expression of the phosphotransfer protein ZIAHP1, ZIAHP2, and ZIAHP6 was associated with infected shoots (Figure 6B). We noted that the homologous gene pairs of ZlHP2/ZlHP7 showed different expression patterns, suggesting their functional divergence after the duplication event (Figure 6B). The response regulator encoding ZlRRA5 and ZIARR9 was activated early, whereas ZIAAR7 and ZlRRA10 type A and ZlPRR1 response regulator genes were activated later in more developed galls. Overall, levels of ARRB type encoding transcripts were lower, with ZIRRB12 displaying a higher level of transient expression kinetics (Figure 6C). To validate the transcriptome data, the transcript levels of five genes with distinct kinetics including an HK, HPs, and an RR and PHYA were monitored by qPCR. The expression trends agreed with the RNA seq data, suggesting the RNA seq results were reliable (Figure S6).
Figure 6.
Heatmap showing the expression of ZlHK(L)s (A), ZlHPs (B), and ZlRRs (C) in Z. latifolia. The expression levels of genes are presented using transcripts Per Million reads (TPM) values and the color scale are shown at the top of heat map. Green represents low expression and red represents strong expression.
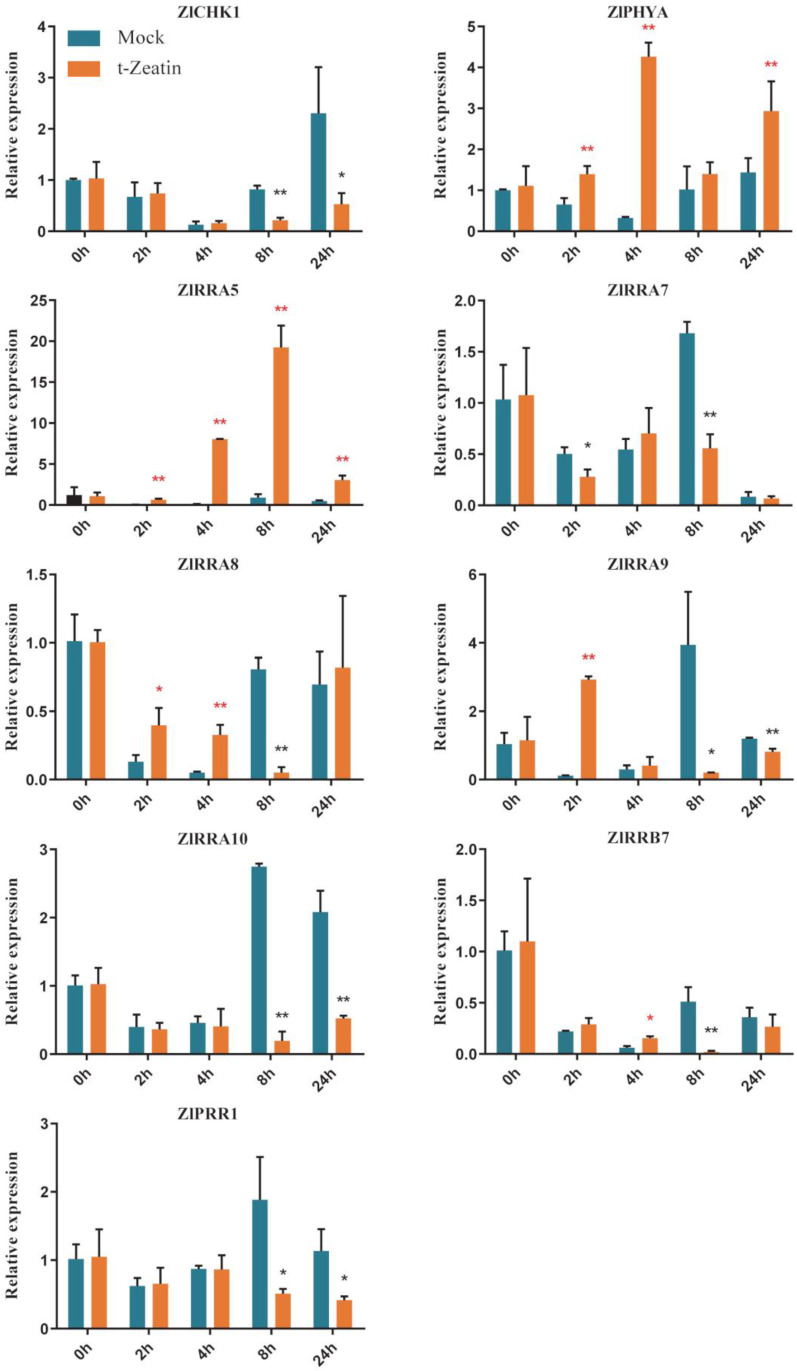
2.9. TCS Gene Response to Exogenous Cytokinin
Our observations indicated that exogenous cytokinin can increase the frequency of gall formation and promote the growth of galls in Z. latifolia shoots. To verify whether the gall-associated TCS genes were responsive to exogenous cytokinin, the expression of 9 selected genes was measured through real-time PCR upon trans-zeatin treatment (Figure 7). Transcript levels of ZlCHK1, ZIRRA5, ZlRRA7, ZIRRA8, ZIRRA9, ZlRRA10, ZIPHYA, ZIRRB7, and pseudo-RR gene of ZlPRR1 in samples of 150 mg/L trans-zeatin sprayed shoots were compared with mock-sprayed samples after 0 (9:00 am in the morning), 2, 4, 8, and 24 h. The mock-sprayed samples displayed a similar trend for all genes measured. In these mock samples, we observed a transient decline in transcript levels followed by an increase after 8 h. In contrast, transcript levels of ZIRRA5, ZIRRA9, and ZIPHYA were elevated by zeatin treatment after two to four hours (Figure 7), and this response was transient. These results indicate that the expression of only a subset of A-type RR genes is triggered quite rapidly upon cytokinin treatment, indicating the complexity of cytokinin signaling-regulated Z. latifolia gall formation. Interestingly, the expression of red and far-red light sensor genes of ZlPHYA was quickly induced after 4 h trans-zeatin treatment (Figure 7). In Arabidopsis, PHYA, the counterpart gene of ZlPHYA played various roles in light-regulated plant development such as seed germination [56,57], internode elongation [58], and root hair growth [59]. The function of ZlPHYA in cytokinin-regulated gall swelling in Z. latifolia needs further elucidation.
Figure 7.
Expression kinetics of nine selected TCS genes in response to trans-zeatin treatment in Z. latifolia. Data are the means ± standard error (n = 3). Two percent EtOH was used as mock treatment. * Indicates the significant differences (p < 0.05) and ** indicates significant differences (p < 0.01).
3. Materials and Methods
3.1. Plant Growth and Treatments
Z. latifolia cv. Ivory, was provided by Zibing Ge, Shucheng Agricultural Science Research Institute, Demonstration Park of Agricultural Science Institute of Taoxi Town, Shucheng, China. The plants were grown in the field of the Anhui Agricultural University Farm (Hefei, China). To investigate the morphological changes of Z. latifolia in response to cytokinin, a 150 mg/L trans-zeatin (HENUO WEIYE Co., China) solution in 2% EtOH was sprayed onto the leaves of fungi-infected plants two weeks before their shoot swelling began. Two percent EtOH was used as mock treatment. The frequency of swelling was recorded daily over a period of 18 days. As the swelling ceased, shoots were harvested, and weight and size were recorded.
To examine the expression profiles of TCS genes responding to cytokinin, 150 mg/L trans-zeatin solution was sprayed until the fully expanded leaves were all covered with the solution. The shoots’ apical tissues were then collected at 0, 1, 2, 4, 8, and 24 h after treatments. All samples were collected in three biological replicates with 3 shoots each and then stored at −80 °C until RNA extraction.
3.2. Identification of TCS Genes in Z. latifolia
The genome sequences of Z. latifolia were downloaded from the Ricerelativesb GD (http://ibi.zju.edu.cn/ricerelativesgd/download.php). The protein sequences of other TCS genes from Arabidopsis, rice, maize, tomato, soybean, and Chinese cabbage were downloaded from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html) or NCBI (www.ncbi.nlm.nih.gov). All TCS sequences were used to build a local blast database and then used as queries to perform BLASTP (Basic Local Alignment Search Tool: Protein BLAST) searches with an E-value of 1e−5 as the threshold [24,60]. However, the putative TCS proteins of Z. latifolia were searched with the hidden Markov model (HMM) of TCS characteristic domains from Pfam (http://pfam.janelia.org/). On the basis of the above two independent methods, the redundancy removal sequences were obtained. To confirm the reliability of the searched results, all candidate protein sequences were then confirmed with SMART (http://smart.embl-heidelberg.de/) [61], Pfam (http://pfam.xfam.org/) [62,63], and NCBI Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/cdd/) [64] according to whether these sequences possess the specific structural characteristics and conserved domains of TCS elements, including HisKA, HATPase, CHASE, His-containing phosphotransfer (HPt), and receiver (Rec) domains.
3.3. Genes Structure Prediction, Protein Sequence Analysis, and Phylogenetic Analysis
Analysis of exons and introns were conducted using Gene Structure Display Server 2.04 (http://gsds.cbi.pku.edu.cn/) [65] by comparing the coding sequences with their corresponding gene sequences. The isoelectric point (PI) and protein molecular weight in kDa were obtained using ExPASy proteomics server (http://www.expasy.org/tools/) [66]. Protein subcellular localizations were predicted using Target P online software (http://www.cbs.dtu.dk/services/TargetP/). Multiple Expectation Maximization for Motif Elicitation (MEME; http://meme.sdsc.edu/meme4_3_0/intro.html) [67] was used for motif analysis to confirm the presence of the conserved motifs in these TCS proteins. Multiple-sequence alignment for the predicted peptide sequences of conserved domains was generated using default parameters [68]. Next, phylogenetic analysis was performed with the MEGA 7 program by using the maximum likelihood (ML) method with 1000 replicates of the bootstrap based on the full-length protein sequences [69].
3.4. Calculation of Nonsynonymous (Ka) to Synonymous (Ks) Substitutions
The TCS paralogous genes in Z. latifolia were subjected to a manual BLAST search, and two genes with a similarity of >75% were considered paralogous gene pairs. However, the tandem or segmental duplication genes could not be determined due to the lack of chromosome information. The selection pressure of paralogs was then determined by analyzing the synonymous and nonsynonymous nucleotide substitutions rates (Ka/Ks ratio) of each paralogous gene pair. In theory, Ka/Ks < 1 indicates purifying or negative selection, Ka/Ks = 1 indicates neutral selection, and Ka/Ks > 1 indicates positive selection [70]. The Ka/Ks ratio was calculated using DnaSP v 5.0 software [71].
3.5. Transcriptome Analysis and Expression of TCS Genes
To obtain a better understanding of the functions of the TCS genes, we investigated their transcriptional levels in the different gall formation stages in Z. latifolia. Samples from shoots apical tissue (0.5 cm in length) were collected from wild-type Z. latifolia (no fungi infected and never swollen), and galls in different size including non-swollen, 5, 10, and 20 cm, respectively. All samples were collected in three biological replicates with 3 shoots each, frozen immediately in liquid nitrogen, and then stored at −80 °C until used. RNA was extracted and the cDNA library was constructed using Illumina TruseqTM RNA sample prep Kit. The Illumina Novaseq 6000 platform was used for transcriptome sequencing, which generated a total of 254.5 Gb of sequence data. Upon filtering, the sequencing data to eliminate poor quality sequences, the sequences were mapped to the Z. latifolia genome using TopHat2 (http;//tophat.cbcb.umd.edu/) [72]. The Transcripts Per Million reads (TPM) algorithm was used for quantification of gene expression. The heatmap of the expression patterns of TCS genes was generated by Heml 1.0 (Heatmap Illustrator) software (CUCKOO, Wuhan, China). All the transcriptomic data have been deposited at GenBank under accession PRJNA664353.
3.6. RNA Isolation and Real-Time PCR
Total RNA was extracted using an RNAprep Pure Plant Plus Kit (polysaccharides and polyphenolics rich; TIANGEN, Beijing, China). The first cDNA strand was synthesized with 2 μg of total RNA by using a FastKing RT Kit (with gDNase) (TIANGEN, Beijing, China), as per the manufacturer’s instructions. qRT-PCR was performed with the CFX96 Touch C1000 Real-Time PCR System (Bio-Rad, Hercules, CA, USA) by using specific primers designed by Primer 5 Software (Table S3). Three biological and three technical replicates for each sample were performed with 20 µL of reaction volume using the SYBR® Green Realtime PCR Master Mix (Toyobo, Osaka, Japan). The ACTIN gene was used as the housekeeping loading reference. The relative gene expression was calculated using the 2−ΔΔCt method [73].
4. Conclusions
A role for cytokinin in gall formation was postulated. Here we observed a positive effect of cytokinin treatment on the gall formation in Z. latifolia in line with earlier reports [7,8], which raised the question of how cytokinin signaling regulates the gall formation. To address this problem, 69 two-component system genes, including 25 HK(L)s (21 HKs and 4 HKLs), 8 HPs (5 authentic and 3 pseudo-HPs), and 36 RRs (14 type A, 14 type B, 2 type C, and 6 pseudo-RRs), were identified in the Z. latifolia genome; the classification was supported by conserved motifs and domains, exon and intron structure, and phylogeny. Our phylogenetic analysis revealed the closest relationships between rice and Z. latifolia in seven species comparisons. Gene duplication events contributed to the TCS gene expansion in the Z. latifolia genome, and Ka/Ks analysis suggested that these duplicated gene pairs experienced strong positive selection during evolution.
We observed the activation of the expression of several of these TCS genes during gall formation, either in the very early stages or during the development of the gall. Among these, two genes encoding for RRA-type response regulators were also rapidly induced by cytokinin treatment. This transient upregulation of these RRA type was in line with RRA genes being a direct target of cytokinin signaling in other plant systems. Furthermore, the far-red light receptor phytochrome A was revealed to be a putative target.
In conclusion, these observations suggest an important role for cytokinin signaling in the process of gall formation and they highlight an important role for cytokinins in the ancient relationship between the endophyte Ustilago esculenta and its host Z. latifolia.
Acknowledgments
We would like to thank Da Li and Jiwang Fan for technical assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/11/1409/s1, Figure S1: Amino acid sequence alignment of ZlHK(L)s in Z. Latifolia; Figure S2: Phylogenetic relationship, gene structures, and conserved motif of all HK(L) genes in Z. latifolia; Figure S3: Phylogenetic relationship, gene structures, and conserved motif of the HP family members in Z. Latifolia; Figure S4: Amino acid sequence alignment of ZlHP proteins in Z. latifolia; Figure S5: Phylogenetic relationship, gene structures, and conserved motif of RR genes in Z. latifolia; Figure S6: The validation of TCS gene expression by qPCR; Table S1: Summary of the two-component system (TCS) gene numbers identified in plants; Table S2: The Ka/Ks ratios of duplicated TCS genes in Z. latifolia; Table S3: The primer sequences.
Author Contributions
B.W. and W.D. conceived and designed the experiments; L.H., F.Z., and Y.H. conducted the experiments; L.H., F.Z., Y.H., and X.W. collected and analyzed the data; L.H., L.D., and F.Z. carried out the experiments and performed software; F.Z. analyzed the transcriptome data; B.W., W.D., and L.H. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31672167) and Key Research Development Program of Anhui Province (201904b11020039).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guo L., Qiu J., Han Z., Ye Z., Chen C., Liu C., Xin X., Ye C.Y., Wang Y.Y., Xie H., et al. A host plant genome (Zizania latifolia) after a century-long endophyte infection. Plant J. Cell Mol. Biol. 2015;83:600–609. doi: 10.1111/tpj.12912. [DOI] [PubMed] [Google Scholar]
- 2.Guo H.B., Li S.M., Peng J., Ke W.D. Zizania latifolia Turcz. cultivated in China. Genet. Resour. Crop Evol. 2007;54:1211–1217. doi: 10.1007/s10722-006-9102-8. [DOI] [Google Scholar]
- 3.Chilton M.D., Drummond M.H., Merio D.J., Sciaky D., Montoya A.L., Gordon M.P., Nester E.W. Stable incorporation of plasmid DNA into higher plant cells: The molecular basis of crown gall tumorigenesis. Cell. 1977;11:263–271. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- 4.Barry G.F., Rogers S.G., Fraley R.T., Brand L. Identification of a cloned cytokinin biosynthetic gene. Proc. Natl. Acad. Sci. USA. 1984;81:4776–4780. doi: 10.1073/pnas.81.15.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crespi M., Messens E., Caplan A.B., van Montagu M., Desomer J. Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 1992;11:795–804. doi: 10.1002/j.1460-2075.1992.tb05116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandeputte O., Oden S., Mol A., Vereecke D., Goethals K., El Jaziri M., Prinsen E. Biosynthesis of auxin by the gram-positive phytopathogen Rhodococcus fascians is controlled by compounds specific to infected plant tissues. Appl. Environ. Microbiol. 2005;71:1169–1177. doi: 10.1128/AEM.71.3.1169-1177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan Y.S., Thrower L. The host-parasite relationship between Zizania caduciflora Turcz. and Ustilago esculebta p. Heen. IV. Growth substances in the host-parasite combination. New Phytol. 1980;85:225–233. doi: 10.1111/j.1469-8137.1980.tb04464.x. [DOI] [Google Scholar]
- 8.Lin Y.L., Lin C.H. Involvement of tRNA bound cytokinin on the gall formation in Zizania. J. Exp. Bot. 1990;41:277–281. doi: 10.1093/jxb/41.3.277. [DOI] [Google Scholar]
- 9.Werner T., Schmulling T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009;12:527–538. doi: 10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Han Y., Zhang C., Yang H., Jiao Y. Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:6840–6845. doi: 10.1073/pnas.1318532111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards K.D., Takata N., Johansson M., Jurca M., Novak O., Henykova E., Liverani S., Kozarewa I., Strnad M., Millar A.J., et al. Circadian clock components control daily growth activities by modulating cytokinin levels and cell division-associated gene expression in Populus trees. Plant Cell Environ. 2018;41:1468–1482. doi: 10.1111/pce.13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieber J.J., Schaller G.E. Cytokinin signaling in plant development. Development. 2018;145:dev149344. doi: 10.1242/dev.149344. [DOI] [PubMed] [Google Scholar]
- 13.Hwang I., Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 14.Schaller G.E., Kieber J.J., Shiu S.H. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arab. Book. 2008;6:e0112. doi: 10.1199/tab.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., Miwa K., Yamashino T., Mizuno T. The Arabidopsis AHK4 Histidine Kinase is a Cytokinin-Binding Receptor that Transduces Cytokinin Signals Across the Membrane. Plant Cell Physiol. 2001;42:1017–1023. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]
- 16.Wurgler-Murphy S.M., Saito H. Two-component signal transducers and MAPK cascades. Trends Biochem. Sci. 1997;22:172–176. doi: 10.1016/S0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 17.Hwang I. Two-Component Signal Transduction Pathways in Arabidopsis. Plant Physiol. 2002;129:500–515. doi: 10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grefen C., Harter K. Plant two-component systems: Principles, functions, complexity and cross talk. Planta. 2004;219:733–742. doi: 10.1007/s00425-004-1316-4. [DOI] [PubMed] [Google Scholar]
- 19.Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., De Peer Y.V., Rouze P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuno T. Two-component phosphorelay signal transduction systems in plants: From hormone responses to circadian rhythms. Biosci. Biotechnol. Biochem. 2005;69:2263–2276. doi: 10.1271/bbb.69.2263. [DOI] [PubMed] [Google Scholar]
- 21.Pareek A., Singh A., Kumar M., Kushwaha H.R., Lynn A.M., Singla-Pareek S.L. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 2006;142:380–397. doi: 10.1104/pp.106.086371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishida K., Niwa Y., Yamashino T., Mizuno T. A genome-wide compilation of the two-component systems in Lotus japonicus. DNA Res. 2009;16:237–247. doi: 10.1093/dnares/dsp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishida K., Yamashino T., Nakanishi H., Mizuno T. Classification of the Genes Involved in the Two-Component System of the MossPhyscomitrella patens. Biosci. Biotechnol. Biochem. 2010;74:2542–2545. doi: 10.1271/bbb.100623. [DOI] [PubMed] [Google Scholar]
- 24.Mochida K., Yoshida T., Sakurai T., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 2010;17:303–324. doi: 10.1093/dnares/dsq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu Z.X., Ma Q., Lin Y.X., Tang X.L., Zhou Y.Q., Zhu S.W., Fan J., Cheng B.J. Genome-wide identification, classification, and analysis of two-component signal system genes in maize. Genet. Mol. Res. 2011;10:3316–3330. doi: 10.4238/2011.December.8.3. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z., Zhang M., Kong L., Lv Y., Zou M., Lu G., Cao J., Yu X. Genome-Wide Identification, Phylogeny, Duplication, and Expression Analyses of Two-Component System Genes in Chinese Cabbage (Brassica rapa ssp. pekinensis) DNA Res. 2014;21:379–396. doi: 10.1093/dnares/dsu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y., Liu X., Ye L., Pan C., Chen L., Zou T., Lu G. Genome-Wide Identification and Expression Analysis of Two-Component System Genes in Tomato. Int. J. Mol. Sci. 2016;17:1204. doi: 10.3390/ijms17081204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y., Liu X., Zou T., Pan C., Qin L., Chen L., Lu G. Genome-Wide Identification of Two-Component System Genes in Cucurbitaceae Crops and Expression Profiling Analyses in Cucumber. Front. Plant Sci. 2016;7:899. doi: 10.3389/fpls.2016.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P., Yang X., Zhang Y., Wang S., Ge Q., Li Q., Wang C., Shi Q., Ren Z., Wang L. Genome-Wide Identification of Two-Component Signal Transduction System Genes in Melon (Cucumis melon L.) Agric. Sci. 2018;9:469–479. doi: 10.4236/as.2018.94032. [DOI] [Google Scholar]
- 30.Liu P., Wang S., Wang X., Yang X., Li Q., Wang C., Chen C., Shi Q., Ren Z., Wang L. Genome-wide characterization of two-component system (TCS) genes in melon (Cucumis melo L.) Plant Physiol. Biochem. 2020;151:197–213. doi: 10.1016/j.plaphy.2020.03.017. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z.D., Yan N., Wang Z.H., Zhang X.H., Zhang J.Z., Xue H.M., Wang L.X., Zhan Q., Xu Y.P., Guo D.P. RNA-seq analysis provides insight into reprogramming of culm development in Zizania latifolia induced by Ustilago esculenta. Plant Mol. Biol. 2017;95:533–547. doi: 10.1007/s11103-017-0658-9. [DOI] [PubMed] [Google Scholar]
- 32.Heyl A., Brault M., Frugier F., Kuderova A., Lindner A.C., Motyka V., Rashotte A.M., Schwartzenberg K.V., Vankova R., Schaller G.E. Nomenclature for members of the two-component signaling pathway of plants. Plant Physiol. 2013;161:1063–1065. doi: 10.1104/pp.112.213207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock A.M., Robinson V.L., Goudreau P.N. Two-Component Signal Transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 34.Ueguchi C., Sato S., Kato T., Tabata S. The AHK4 Gene Involved in the Cytokinin-Signaling Pathway as a Direct Receptor Molecule in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:751–755. doi: 10.1093/pcp/pce094. [DOI] [PubMed] [Google Scholar]
- 35.Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki T., Miwa K., Ishikawa K., Yamada H., Aiba H., Mizuno T. The Arabidopsis Sensor His-kinase, AHK4, Can Respond to Cytokinins. Plant Cell Physiol. 2001;42:107–113. doi: 10.1093/pcp/pce037. [DOI] [PubMed] [Google Scholar]
- 37.Spíchal L., Rakova N.Y., Riefler M., Mizuno T., Romanov G.A., Strnad M., Schmülling T. Two Cytokinin Receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, Differ in their Ligand Specificity in a Bacterial Assay. Plant Cell Physiol. 2004;45:1299–1305. doi: 10.1093/pcp/pch132. [DOI] [PubMed] [Google Scholar]
- 38.Romanov G.A., Lomin S.N., Schmulling T. Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J. Exp. Botany. 2006;57:4051–4058. doi: 10.1093/jxb/erl179. [DOI] [PubMed] [Google Scholar]
- 39.Stolz A., Riefler M., Lomin S.N., Achazi K., Romanov G.A., Schmülling T. The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 2011;67:157–168. doi: 10.1111/j.1365-313X.2011.04584.x. [DOI] [PubMed] [Google Scholar]
- 40.Lomin S.N., Krivosheev D.M., Steklov M.Y., Arkhipov D.V., Osolodkin D.I., Schmulling T., Romanov G.A. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J. Exp. Botany. 2015;66:1851–1863. doi: 10.1093/jxb/eru522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartrina I., Jensen H., Novak O., Strnad M., Werner T., Schmulling T. Gain-of-Function Mutants of the Cytokinin Receptors AHK2 and AHK3 Regulate Plant Organ Size, Flowering Time and Plant Longevity. Plant Physiol. 2017;173:1783–1797. doi: 10.1104/pp.16.01903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rashotte A.M., Carson S.D.B., To J.P.C., Kieber J.J. Expression Profiling of Cytokinin Action in Arabidopsis. Plant Physiol. 2003;132:1998–2011. doi: 10.1104/pp.103.021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W., Hall A.E., O’Malley R., Bleecker A.B. Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc. Natl. Acad. Sci. USA. 2003;100:352–357. doi: 10.1073/pnas.0237085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Binder B.M., O’Malley R.C., Wang W., Moore J.M., Parks B.M., Spalding E.P., Bleecker A.B. Arabidopsis seedling growth response and recovery to ethylene. A kinetic analysis. Plant Physiol. 2004;136:2913–2920. doi: 10.1104/pp.104.050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu X., Schaller G.E. Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol. 2004;136:2961–2970. doi: 10.1104/pp.104.047126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rockwell N.C., Su Y.-S., Lagarias J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao Y., Offringa R. PDK1 regulates auxin transport and Arabidopsis vascular development through AGC1 kinase PAX. Nat. Plants. 2020;6:544–555. doi: 10.1038/s41477-020-0650-2. [DOI] [PubMed] [Google Scholar]
- 48.Sun L., Zhang Q., Wu J., Zhang L., Jiao X., Zhang S., Zhang Z., Sun D., Lu T., Sun Y. Two rice authentic histidine phosphotransfer proteins, OsAHP1 and OsAHP2, mediate cytokinin signaling and stress responses in rice. Plant Physiol. 2014;165:335–345. doi: 10.1104/pp.113.232629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fassler J.S., West A.H. Histidine phosphotransfer proteins in fungal two-component signal transduction pathways. Eukaryot. Cell. 2013;12:1052–1060. doi: 10.1128/EC.00083-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyata S.-i., Urao T., Yamaguchi-Shinozaki K., Shinozaki K. Characterization of genes for two-component phosphorelay mediators with a single HPt domain in Arabidopsis thaliana. FEBS Lett. 1998;437:11–14. doi: 10.1016/S0014-5793(98)01188-0. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki T., Imamura A., Ueguchi C., Mizuno T. Histidine-Containing Phosphotransfer (HPt) Signal Transducers Implicated in His-to-Asp Phosphorelay in Arabidopsis. Plant Cell Physiol. 1999;39:1258–1268. doi: 10.1093/oxfordjournals.pcp.a029329. [DOI] [PubMed] [Google Scholar]
- 52.Urao T., Miyata S., Yamaguchi-Shinozaki K., Shinozaki K. Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 2000;478:227–232. doi: 10.1016/S0014-5793(00)01860-3. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki T., Sakurai K., Ueguchi C., Mizuno T. Two Types of Putative Nuclear Factors that Physically Interactwith Histidine-Containing Phosphotransfer (Hpt) Domains, Signaling Mediators in His-to-Asp Phosphorelay, in Arabidopsisthaliana. Plant Cell Physiol. 2001;42:37–45. doi: 10.1093/pcp/pce011. [DOI] [PubMed] [Google Scholar]
- 54.Niwa Y., Ito S., Nakamichi N., Mizoguchi T., Niinuma K., Yamashino T., Mizuno T. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:925–937. doi: 10.1093/pcp/pcm067. [DOI] [PubMed] [Google Scholar]
- 55.Swapan K.S., Alak C.D., Raju B. Morphogenesis of edible gall in Zizania latifolia (Griseb.) Turcz. ex Stapf due to Ustilago esculenta Henn. infection in India. Afr. J. Microbiol. Res. 2016;10:1215–1223. doi: 10.5897/AJMR2016.8207. [DOI] [Google Scholar]
- 56.Franklin K.A., Allen T., Whitelam G.C. Phytochrome A is an irradiance-dependent red light sensor. Plant J. 2007;50:108–117. doi: 10.1111/j.1365-313X.2007.03036.x. [DOI] [PubMed] [Google Scholar]
- 57.Monte E., Alonso J.M., Ecker J.R., Zhang Y., Li X., Young J., Austin-Phillips S., Quail P.H. Isolation and Characterization of phyC Mutants in Arabidopsis Reveals Complex Crosstalk between Phytochrome Signaling Pathways. Plant Cell. 2003;15:1962–1980. doi: 10.1105/tpc.012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Devlin P.F., Patel S.R., Whitelam G.C. Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gahlaut V., Mathur S., Dhariwal R., Khurana J.P., Tyagi A.K., Balyan H.S., Gupta P.K. A multi-step phosphorelay two-component system impacts on tolerance against dehydration stress in common wheat. Funct. Integr. Genom. 2014;14:707–716. doi: 10.1007/s10142-014-0398-8. [DOI] [PubMed] [Google Scholar]
- 61.Letunic I., Doerks T., Bork P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Punta M., Coggill P., Eberhardt R., Mistry J., Tate J., Boursnell C., Pang N., Forslund S., Ceric G., Clements J., et al. The Pfam protein families database. Nucleic Acids Res. 2011;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo A., Zhu Q., Chen X., Luo J. GSDS: A gene structure display server. Yi Chuan. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- 66.Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E., et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The MEME Suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 69.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hurst L.D. The K a/K s ratio:diagnosing the form of sequence evolution. Trends Genet. 2002;18:486. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 71.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 72.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.