Abstract
Latest advancement of omics technologies allows in-depth characterization of venom compositions. In the present work we present a proteomic study of two snake venoms of the genus Naja i.e., Naja naja (black cobra) and Naja oxiana (brown cobra) of Pakistani origin. The present study has shown that these snake venoms consist of a highly diversified proteome. Furthermore, the data also revealed variation among closely related species. High throughput mass spectrometric analysis of the venom proteome allowed to identify for the N. naja venom 34 protein families and for the N. oxiana 24 protein families. The comparative evaluation of the two venoms showed that N. naja consists of a more complex venom proteome than N. oxiana venom. Analysis also showed N-terminal acetylation (N-ace) of a few proteins in both venoms. To the best of our knowledge, this is the first study revealing this posttranslational modification in snake venom. N-ace can shed light on the mechanism of regulation of venom proteins inside the venom gland. Furthermore, our data showed the presence of other body proteins, e.g., ankyrin repeats, leucine repeats, zinc finger, cobra serum albumin, transferrin, insulin, deoxyribonuclease-2-alpha, and other regulatory proteins in these venoms. Interestingly, our data identified Ras-GTpase type of proteins, which indicate the presence of extracellular vesicles in the venom. The data can support the production of distinct and specific anti-venoms and also allow a better understanding of the envenomation and mechanism of distribution of toxins. Data are available via ProteomeXchange with identifier PXD018726.
Keywords: Naja naja, Naja oxiana, venom proteome, Ras-GTPase, ankyrin repeat, N-terminal acetylation, extracellular vesicles
1. Introduction
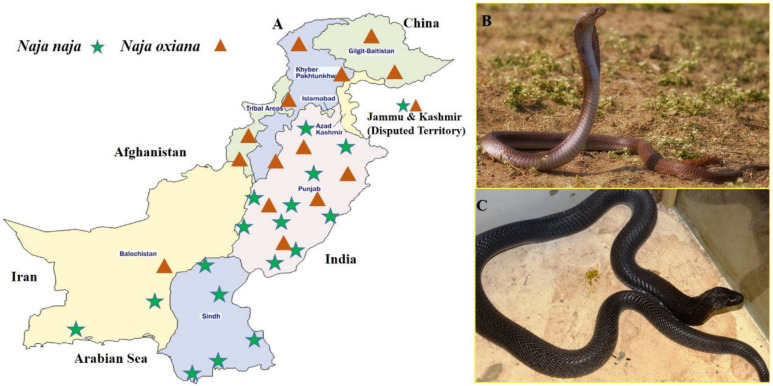
Pakistan has a particular geographical location and hosts an array of habitats such a, mountains, glaciers, coastal areas, swamps, plane areas, fresh water, and sandy areas [1]. The country is located between two zoogeographical regions (Palearctic and Oriental) and hosts a diverse venomous fauna. Nine habitat zones are recognized according to the distribution of snakes in Pakistan [2]. Seventy-two snake species are known to Pakistan, among which 14 marine and 12 terrestrial are venomous [1]. According to ITIS (Integrated Taxonomic Information System) database there are 29 snake species belonging to the genus Naja [3]. Among these two are found in Pakistan, i.e., Naja naja and Naja oxiana [2]. Both of these snakes are non-spitting cobras [4]. These snakes are shy of humans. However, upon assessing threat they lift the anterior part of their body, display a hood, and if provoked, hiss loudly and sway their hood to frighten their adversary. These snakes attack very furiously, chewing the bitten part. They usually feed on rodents, birds, frogs, lizards, and snakes. They are found in rocky, stony foothills, forests and around the villages [2]. N. naja (black cobra) is known to have variable color and pattern. However, in Pakistan juveniles and young adults tend to be grey with hood marks, but the adult specimens are usually uniformly black. In addition, the throat pattern is obscured in adult snakes, due to pigmentation [4,5]. N. naja is distributed in North West Pakistan, south and desert areas, except most of Baluchistan. N. oxiana (brown cobra) occurs sympatrically in the Northern half of Pakistan with N. naja. Adult N. oxiana is normally brown in color [5,6]. These snakes and their geographical distribution are shown in Figure 1.
Figure 1.
(A) Geographical distribution of the genus Naja snakes in Pakistan. (B) Naja oxiana (Brown cobra) (C) Naja naja (black cobra).
Only a few reliable data exist reporting the frequency of morbidity because of snakebites in developing countries. However, it is predicted that snakebite is responsible for a substantial amount of morbidity and mortality in remote areas [7]. The hidden toll of suffering continues to affect the families of the deceased, and patients who survived with crippling deformity [8]. World Health Organization (WHO), included snakebite in its list of “Neglected Tropical disease” in 2007 [9]. Recently, WHO also added snakebite envenoming at high preference in the list of Neglected Tropical disease, in 2017, upon request of some member states of United Nations. The supply of antivenom and snakebite management was declared as a global public health emergency. WHO has included snake antivenom immunoglobulins in the “WHO Model List for Essential Medicines” WHO has also encouraged countries to ensure their national antivenom stocks [10] Despite these efforts, snakebite has not gained attention on international public health agendas [11]. The snakes commonly responsible for clinically significant bites in Pakistan are Bungarus caeruleus (common krait), N. naja (cobra), Daboia russelii (Russel’s viper), and Echis carinatus (Saw-scaled viper) [12]. National Institute of Health, Pakistan, produces around 30,000 vials of polyvalent anti-venom per year. However, the amount of this antivenom is not sufficient and can only treat a fraction of snakebite cases in the country (https://www.nih.org.pk/1255-2/) [13]. To meet the requirement of antivenoms, snake antivenom sera are presently also imported from India. However, studies have shown that Indian antivenoms provide partial neutralization, particularly for N. naja venom [14,15,16]. Although, N. naja and N. oxiana are also prevalent in India, but the venom composition is known to vary within the same species, due to change in geographical and ecological factors [17,18,19,20]. A study reported that Pakistani N. naja is more neurotoxic with lower LD50, then that prevalent in India [8]. Gender, diet, and age of the snake is also known to influence the composition of venoms [21,22,23,24].
Depending on the amount of venom injected, paralysis following cobra bites can occur within several hours, with death ensuing if breathing is not assisted [8]. On average cobras can inject 60 mg of venom in a bite [25]. Cobra venom is a postsynaptic neurotoxin and presents a variety of symptoms like pain, edema, necrosis, respiratory paralysis, headache, cardiac arrest, hypotension, and bleeding wounds [26]. The use of anticholinesterase, such as neostigmine, has been suggested to compensate a cobra bite, in addition to the administration of antivenom [25,26].
Recent scientific advances have paved the way to explore venomous snake composition in detail and various strategies have been evolved to better understand venom components, their function and immunological properties [27]. Genomic and transcriptomic studies have proved to be an invaluable tool in the discovery of the snake venom evolution and proteoform [28,29,30,31,32,33,34]. Consequently, investigations are directed towards the discovery of pharmacologically active snake venom compounds [35,36,37,38]. For example, a recent study reported Mambaquaretin-1 (peptide from green mamba venom), as a promising candidate for the treatment of polycystic kidney disease [39]. Another study described Nubein6.8, a peptide from the venom of N. nubiae, as a promising template for the treatment of human melanoma and ovarian cancer [40].
In the present study, we describe an in-depth comparative proteomic study of two Pakistani snake species of the family elapid and genus Naja, i.e., N. oxiana (brown cobra/Caspian cobra/Central Asian cobra) and N. naja (black cobra/Indian cobra/Spectacled cobra). In Pakistani region both species of adult cobras are melatonic and N. oxiana is commonly known as brown, while N. naja is known as black cobra. These snakes were previously known as Naja n. oxiana and Naja n. karachienis respectively, but now they are named according to the ITIS database [41]. Till now only a few studies have been reported about the proteomics of Pakistani N. naja [42,43,44]. The N. naja venom samples in these studies were collected from Southern Punjab and Sindh Province of Pakistan. These research groups performed pre fractionation of the venom sample either by reverse phase chromatography, 1-dimensional gel electrophoresis (1D gel) or 2-dimensional gel electrophoresis (2 D gel) or a combination of these methods. Further mass spectrometric analysis of peptide fragments obtained from in gel trypsin digestion, was carried out by MALDI TOF/TOF, ion trap or ESI MS. Chanda et al. also reported the venom proteomics of N. naja, from Western and Eastern parts of India [45,46]. In their study of the venom sample from East India, they pre fractionated the crude venom by 1D gel prior to LTQ orbitrap analysis. However, the proteomic analysis of the venom sample from Western India was performed by a combination of fractionation methods and LC- MS/MS was done by QTOF mass spectrometer. Analysis of the comparative statement of the research group showed that pre fractionation of the crude venom by gel filtration chromatography followed by gel electrophoresis, worked best in their hands. The same group reported the proteomic study of South Indian N. naja venom, recently [47].
In this work, they separated the crude venom components by 1D gel electrophoresis. The mass spectrometric analysis of the tryptic peptide was performed on QTOF. The results of this study derive a comparison of common and unique toxins in N. naja venom obtained from all the three different Indian regions. Our results revealed remarkable differences in the relative abundance of the venom components, as compared to the previous studies. In addition, our investigations unveiled new venom components, not reported before in these venoms. The variation in the results could be different geographical of the snakes from which we collected the venom samples. Further, our workflow did not involve any pre fractionation of the venom. Pre fractionation by gel electrophoresis or liquid chromatography might lead to the loss of some low abundant venom components. Also, we used a modern version of the orbitrap mass spectrometer in this work which is very sensitive equipment.
To the best of our knowledge, this is the first report on the proteomic study of Naja oxiana venom. The abbreviations used for proteins and peptides are given in Table 1.
Table 1.
Comparative evaluation of snake venom protein families in the venom of N. naja and N. oxiana.
| Protein Family | First Report in Nn Venom | Abbreviation Used | NN (No of Peptides) | %Age | NO(No. of Peptides) | %Age |
|---|---|---|---|---|---|---|
| Three-Finger toxin | 3FTX | 157 | 21 | 41 | 16 | |
| Snake venom metalloprotease family | SVMP | 72 | 10 | 39 | 15 | |
| Cobra venom factor | CVF | 62 | 9 | 22 | 8.7 | |
| Cysteine-rich secretory protein | CRISP | 53 | 7 | 7 | 2.8 | |
| Phospholipase A2 | PLA2 | 46 | 6 | 32 | 12.6 | |
| Phospholipase B | PLB | 1 | 0.1 | 4 | 1.6 | |
| Phospholipase inhibitor | ✓ | CNF-I | 3 | 0.4 | - | - |
| L-amino-acid oxidase | LAAO | 31 | 4 | 14 | 5.5 | |
| Snake Venom Serine proteinase | SP | 15 | 2 | 11 | 4.3 | |
| Ohanin | Oh | 11 | 1.5 | 2 | 0.8 | |
| Kunitz type serine protease inhibitor | KSPI | 14 | 2 | 4 | 1.6 | |
| Nerve Growth Factor | NGF | 12 | 1.7 | 11 | 4.3 | |
| 5′-nucleotidase | 5-Ntd | 10 | 1.4 | 1 | 0.4 | |
| Serum Albumin | ✓ | SA | 10 | 1.4 | - | |
| Glutathione peroxidase | ✓ | GP | 9 | 1.2 | 3 | 1.2 |
| Phosphodiesterase | Pde | 8 | 1.1 | 8 | 3.1 | |
| Aminopeptidase | - | 7 | 1 | 4 | 1.6 | |
| TNF receptor family | ✓ | TNF | 2 | 0.3 | 3 | 1.2 |
| Lectin | ✓ | - | 3 | 0.4 | 1 | 0.4 |
| Natriuretic peptide family | NP | 4 | 0.54 | 1 | 0.4 | |
| Cystatin | - | 4 | 0.54 | - | ||
| Cathelicidin | ✓ | cath | 1 | 0.1 | - | |
| N-acetylcholinesterase | N-Ache | 1 | 0.1 | 1 | 0.4 | |
| Vascular endothelial growth factor | VEGF | 1 | 0.1 | - | ||
| Transforming growth factor | ✓ | TGF | 2 | 0.3 | - | |
| Zinc finger protein | ✓ | ZFP | 6 | 0.8 | 4 | 1.6 |
| Insulin | ✓ | In | 2 | 0.3 | - | |
| Transferrin | ✓ | TF | 2 | 0.3 | - | |
| Ankyrin repeat | ✓ | AR | 2 | 0.3 | 1 | 0.4 |
| Leucine repeat | ✓ | LR | 1 | 0.1 | 1 | 0.4 |
| Endonuclease | ✓ | - | 3 | 0.4 | - | |
| SLRP family | ✓ | SLRP | 2 | 0.3 | 1 | 0.4 |
| Ras-like protein | ✓ | Ras | 5 | 0.7 | - | |
| Serpin | - | - | 1 | 0.4 | ||
| Others | - | 158 | 37 | |||
| Total | 735 | 254 |
Bold text in the first column indicates protein families exclusively identified in N. naja venom. Blue coloured text indicates protein family identified only in N. oxiana. Check mark (✓) in the second column, indicates that this work is the first report of the identification of the corresponding protein families in N.naja venom. The dash sign indicate that the protein family was not identified in the venom.
2. Results
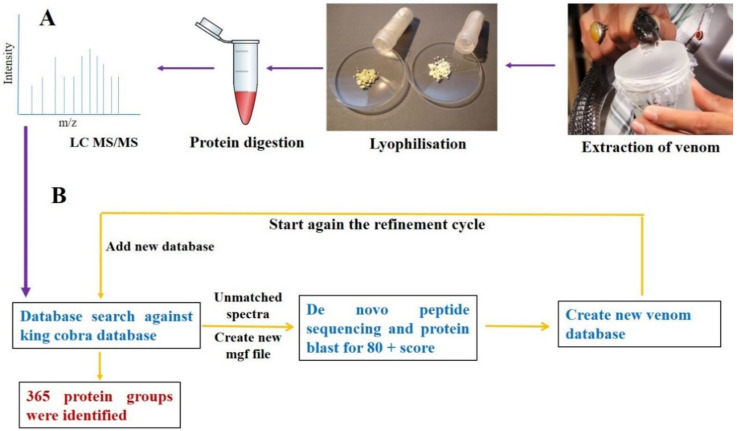
The venom proteome of N. naja (NN) and N. oxiana (NO) snakes was investigated by mass spectrometric analysis, using a shotgun proteomic approach. We were able to provide an extensive overview of various protein families present in both venoms, based on data base searches and BLAST analysis of the de novo sequenced tandem mass spectra. A total of 735 peptides from NN and 254 peptides from NO were sequenced (Supplementary Table S1 and S2). Subsequently 365 proteins in NN venom (Table 2) and 140 proteins were identified in NO venom (Table 3). The sequences of the protein fragments are listed in Supplementary Tables S1 and S2. The results obtained allowed us to cluster the venom protein content into 34 protein families for N. naja and in 24 protein families for the N. oxiana venom. Figure 2A illustrates the preparation for MS acquisition and Figure 2B represents the strategy applied for data base searches. In the present work, we performed data base search against Serpents, King cobra utilizing Uniprot data base. The venom of Ophiophagus hannah has been well studied and genomic and proteomic data are available in the database [28,48,49]. A recent study showed similarity between the genome of Indian cobra and King cobra [50]. This group analyzed 139 N. naja venom gland toxin genes to identify orthologs in the King cobra. It was determined that 96 genes matched while 43 did not. It was suggested that, although some genes are likely to be unique to Indian cobra, the majority were not annotated in King cobra genome. The possible reason could be its highly fragment assembly. Based on this similarity, we searched our data against King cobra database also. Further, in the data base complete proteome of only King cobra is available. The details of our search against Serpent database are presented in Supplementary Table S3 and S5 while that against King cobra are compiled in Supplementary Table S4 and S6. The results presented and discussed are a conclusion of both data base searches.
Table 2.
Summary of the venom proteome of Naja naja.
| S. No | Protein Family | Protein | Accession Code | Number of Matched Peptides | Homology with a Protein from the Venom of Snake Species |
|---|---|---|---|---|---|
| 1 | 3FTXs (Neurotoxin) | Long neurotoxin | AHZ08824 | 9 | Micropechis ikaheca |
| 2 | P01389 | 1 | Naja anchietae | ||
| 3 | P01390 | 2 | Naja nivea | ||
| 4 | Long neurotoxin homolog | O93422 | 5 | Naja atra | |
| 5 | Long neurotoxin 1 | P25668 | 4 | Naja naja | |
| 6 | Long neurotoxin 1 | P01380 | 1 | Hydrophis stokesii | |
| 7 | Long neurotoxin 1 | P25674 | 3 | Naja haje haje | |
| 8 | Long neurotoxin 4 | P25672 | 3 | Naja naja | |
| 9 | Long neurotoxin 7 | O42257 | 3 | Naja sputatrix | |
| 10 | putative long neurotoxin | ABX58151 | 1 | Austrelaps labialis | |
| 11 | putative long neurotoxin | ABX58163 | 1 | Austrelaps labialis | |
| 12 | Alpha-neurotoxin NTX-3 | O57326 | 1 | Naja sputatrix | |
| 13 | Short neurotoxin 3 | P01420 | 1 | Naja annulifera | |
| 14 | Short neurotoxin III | P59275 | 1 | Naja kaouthia | |
| 15 | Neurotoxin II | P01427 | 6 | Naja oxiana | |
| 16 | cobrotoxin b | CAA73829 | 3 | Naja atra | |
| 17 | Cobrotoxin-b | P80958 | 4 | Naja atra | |
| 18 | Alpha-cobratoxin | P01391 | 4 | Naja kaouthia | |
| 19 | kappa-cobrotoxin | CAA76846 | 1 | Naja atra | |
| 20 | Weak toxin 2 | Q8AY50 | 2 | Bungarus candidus | |
| 21 | Weak neurotoxin 7 | P29181 | 7 | Naja naja | |
| 22 | Weak neurotoxin 10 | Q802B2 | 1 | Naja sputatrix | |
| 23 | Weak toxin CM-11 | P01401 | 4 | Naja haje haje | |
| 24 | Weak toxin S4C11 | P01400 | 5 | Naja melanoleuca | |
| 25 | three-finger toxin precursor, partial | ADN67572 | 4 | Bungarus multicinctus | |
| 26 | three-finger toxin precursor, partial | ADN67582 | 9 | Naja atra | |
| 27 | three-finger toxin precursor, partial | ADN67583 | 1 | Naja atra | |
| 28 | three-finger toxin precursor | ADN67579 | 1 | Naja atra | |
| 29 | Muscarinic toxin-like protein 3 | P82464 | 3 | Naja kaouthia | |
| 30 | Muscarinic toxin-like protein | Q9W727 | 4 | Bungarus multicinctus | |
| 31 | Muscarinic toxin-like protein 2 | P82463 | 6 | Naja kaouthia | |
| 32 | Muscarinic toxin-like protein 1 | P82462 | 2 | Naja kaouthia | |
| 33 | Muscarinic toxin 38 | Q2VBN0 | 1 | Ophiophagus hannah | |
| 34 | Alpha-elapitoxin-Nk2a | P01391 | 4 | Naja kaouthia | |
| 36 | three finger toxin V | ABX82866 | 1 | Walterinnesia aegyptia | |
| 37 | Three finger toxin W-V | C1IC49 | 3 | Walterinnesia aegyptia | |
| 38 | Chain A, Putative Ancestral Mamba Toxin 1 | 5MG9_A | 1 | Dendroaspis angusticeps | |
| 39 | 3FTXs (cytotoxins) | cytotoxin 17, partial | BAU24676 | 13 | Naja naja |
| 40 | Cytotoxin Vc-5 | Q9PS34 | 6 | Naja oxiana | |
| 41 | Cytotoxin 3a | P86539 | 4 | Naja naja | |
| 42 | Cytotoxin SP15c | P60308 | 13 | Naja atra | |
| 43 | cardiotoxin 7a | AAB36929 | 2 | Naja atra | |
| 44 | cardiotoxin 7a | Q91126 | 3 | Naja atra | |
| 46 | Cytotoxin 8 | P86540 | 2 | Naja naja | |
| 47 | Cytotoxin 1 | P01447 | 1 | Naja naja | |
| 48 | Cytotoxin II | P01441 | 1 | Naja oxiana | |
| 49 | Cytotoxin 5 | P25517 | 2 | Naja mossambica | |
| 50 | Cardiotoxin-6 | Q98965 | 1 | Naja atra | |
| 51 | Cytotoxin 10 | P86541 | 1 | Naja naja | |
| 52 | Cytotoxin homolog 3 | P01473 | 1 | Naja melanoleuca | |
| 53 | Cardiotoxin-like basic polypeptide ah | P0C547 | 2 | Naja atra | |
| 54 | cardiotoxin 1e | AAA90960 | 4 | Naja atra | |
| 55 | Venom complement C3-like | Venom factor | AAX86641 | 5 | Austrelaps superbus |
| 56 | Cobra venom factor | Q91132 | 31 | Naja kaouthia | |
| 57 | Cobra venom factor gamma chain | Q91132 | 2 | Naja kaouthia | |
| 58 | Cobra venom factor alpha chain | Q91132 | 2 | Naja kaouthia | |
| 59 | cobra venom factor precursor | AAA68989 | 1 | Naja kaouthia | |
| 60 | venom factor-like, partial | XP_025025833 | 2 | Python bivittatus | |
| 61 | cobra venom factor 1, partial | AXL96620 | 13 | Ahaetulla prasina | |
| 62 | cobra venom factor, partial | AXL95279 | 1 | Spilotes sulphureus | |
| 63 | cobra venom factor, partial | AWX67646 | 1 | Boiga irregularis | |
| 64 | Ophiophagus venom factor | I2C090 | 3 | Ophiophagus hannah | |
| 66 | Venom Kunitz-type family | Kunitz-type serine protease inhibitor | P19859 | 1 | Naja naja |
| 67 | Kunitz-type serine protease inhibitor | P20229 | 6 | Naja naja | |
| 68 | Kunitz-type serine protease inhibitor isoform 7 | ACY68703 | 1 | Parasuta nigriceps | |
| 69 | Kunitz inhibitor b, partial | AAL30069 | 1 | Bungarus candidus | |
| 70 | protease inhibitor | AFA90080 | 1 | Daboia siamensis | |
| 71 | Venom basic protease inhibitor 2 | P00986 | 1 | Naja nivea | |
| 72 | Kunitz-type protease inhibitor, partial | AWX67660 | 1 | Boiga irregularis | |
| 73 | papilin-like, partial | XP_025032351 | 1 | Python bivittatus | |
| 74 | Kunitz inhibitor I | ABX82867 | 1 | Walterinnesia aegyptia | |
| 75 | natriuretic peptide family | Natriuretic peptide Na-NP | D9IX97 | 2 | Naja atra |
| 76 | natriuretic peptide | ADK12001 | 1 | Naja atra | |
| 77 | natriuretic peptide | ADK12001 | 1 | Naja atra | |
| 78 | cystatin | Cystatin | E3P6P4 | 4 | Naja kaouthia |
| 79 | NGF-beta family | Venom nerve growth factor 2 | Q5YF89 | 2 | Naja sputatrix |
| 80 | Venom nerve growth factor 3 | Q3HXY1 | 7 | Pseudechis australis | |
| 81 | nerve growth factor, partial | AAR24530 | 1 | Bitis gabonica | |
| 82 | nerve growth factor | BAN82142 | 4 | Ovophis okinavensis | |
| 83 | nerve growth factor beta chain precursor | A59218 | 1 | Naja kaouthia | |
| 84 | Ohanin/vespryn family. | Ohanin | P83234 | 4 | Ophiophagus hannah |
| 85 | Thaicobrin | P82885 | 2 | Naja kaouthia | |
| 86 | Venom PRY-SPRY domain-containing protein, partial | AHZ08803 | 4 | Micropechis ikaheca | |
| 87 | Vespryn | AEJ32004 | 1 | Crotalus adamanteus | |
| 88 | Insulin family | Insulin-like growth factor-binding protein 3, partial | XP_025032248 | 1 | Python bivittatus |
| 89 | Insulin enhancer protein ISL-1, partial | ETE72105 | 1 | Ophiophagus hannah | |
| 90 | Snake venom VEGF subfamily | Snake venom vascular endothelial growth factor toxin barietin | C0K3N1 | 1 | Bitis arietans |
| 91 | CRISP | Cysteine-rich venom protein 25 | P84806 | 6 | Naja haje haje |
| 92 | cysteine-rich seceretory protein Ts-CRPM | ACE73574 | 2 | Trimeresurus stejnegeri | |
| 93 | Cysteine-rich venom protein mossambin | P0DL16 | 2 | Naja mossambica | |
| 94 | Cysteine-rich venom protein natrin-1 | Q7T1K6 | 16 | Naja atra | |
| 95 | Cysteine-rich venom protein ophanin | Q7ZT98 | 3 | Ophiophagus hannah | |
| 96 | cysteine-rich venom protein, partial | BAP39957 | 1 | Protobothrops flavoviridis | |
| 97 | Cysteine-rich venom protein natrin-2 | Q7ZZN8 | 3 | Naja atra | |
| 98 | Cysteine-rich seceretory protein Ts-CRPM | N-ACE73574 | 1 | Trimeresurus stejnegeri | |
| 99 | Cysteine-rich venom protein 25-A | P84807 | 1 | Naja haje haje | |
| 100 | Helicopsin | P0DJG8 | 2 | Helicops angulatus | |
| 101 | Cysteine-rich venom protein bucarin | P81993 | 1 | Bungarus candidus | |
| 102 | Cysteine-rich venom protein latisemin | Q8JI38 | 1 | Laticauda semifasciata | |
| 103 | Cysteine-rich venom protein ophanin | AAO62996 | 1 | Ophiophagus hannah | |
| 104 | cysteine-rich secretory protein 4, partial | AXL96584 | 2 | Borikenophis portoricensis | |
| 105 | Cysteine-rich venom protein kaouthin-1 | P84805 | 1 | Naja kaouthia | |
| 106 | Cysteine-rich venom protein annuliferin-b | P0DL15 | 1 | Naja annulifera | |
| 107 | Cysteine-rich venom protein | AAP20603 | 2 | Naja atra | |
| 108 | Cysteine-rich secretory protein | AJB84505 | 1 | Philodryas chamissonis | |
| 109 | Opharin precursor | AAP81292 | 1 | Ophiophagus hannah | |
| 110 | Cysteine rich secretory protein 2, partial | AXL96629 | 4 | Ahaetulla prasina | |
| 111 | Cathelicidin family | Cathelicidin-related protein precursor | ACF21000 | 1 | Naja atra |
| 112 | TGF-beta family | Transforming growth factor beta-3, partial | ETE71774 | 1 | Ophiophagus hannah |
| 113 | Glial cell line-derived neurotrophic factor, partial | ETE67324 | 1 | Ophiophagus hannah | |
| 114 | Phospholipase A2 | Acidic phospholipase A2 3 | P60045 | 4 | Naja sagittifera |
| 115 | 85 kDa calcium-independent phospholipase A2, partial | ETE71158 | 2 | Ophiophagus hannah | |
| 116 | Acidic phospholipase A2 1 | P00596 | 4 | Naja kaouthia | |
| 117 | Acidic phospholipase A2 1 | Q9W7J4 | 6 | Pseudonaja textilis | |
| 118 | Basic phospholipase A2 T1-2 A chain | P84472 | 2 | Bungarus candidus | |
| 119 | Acidic phospholipase A2 C | Q92086 | 5 | Naja sputatrix | |
| 120 | Acidic phospholipase A2 1 | P00598 | 3 | Naja naja | |
| 121 | Acidic phospholipase A2 2 | P60044 | 1 | Naja sagittifera | |
| 122 | Acidic phospholipase A2 1 | P00596 | 4 | Naja kaouthia | |
| 123 | Phospholipase A2 | BAA36403 | 1 | Naja kaouthia | |
| 124 | Acidic phospholipase A2 beta-bungarotoxin A4 chain | P17934 | 2 | Bungarus multicinctus | |
| 125 | Phospholipase A2-III | ABD24038 | 1 | Daboia russelii russelii | |
| 126 | Basic phospholipase A2 homolog 1 | P10117 | 1 | Laticauda colubrina | |
| 127 | Phospholipase A2 | AAL55555 | 1 | Hydrophis hardwickii | |
| 128 | Phospholipase A2 | P15445 (2WQ5) | 1 | Naja naja | |
| 129 | Phospholipase A2 3 | P21792 | 3 | Micrurus nigrocinctus | |
| 130 | Phospholipase A2I precursor | BAC77655 | 1 | Bungarus flaviceps | |
| 131 | Phospholipase a2 | CAA45372 | 1 | Naja naja | |
| 132 | Phospholipase A2 | AAA66029 | 1 | Naja naja | |
| 133 | Phosphatidylcholine 2-acylhydrolase T1-2 A | P84472 | 2 | Bungarus candidus | |
| 134 | Phospholipase B-like family | Phospholipase B-like 1, partial | ETE59578 | 1 | Ophiophagus hannah |
| 135 | CNF-like-inhibitor family | Phospholipase A2 inhibitor subunit gamma A | Q9PWI4 | 1 | Elaphe quadrivirgata |
| 136 | Phospholipase A2 inhibitor beta subunit isoform OMI-2B | AAF21049 | 1 | Oxyuranus microlepidotus | |
| 137 | Phospholipase A2 inhibitor 31 kDa subunit | Q7LZI1 | 1 | Naja kaouthia | |
| 138 | SVMP (PIII) | Acutolysin e precursor | AAD27891 | 1 | Deinagkistrodon acutus |
| 139 | Snake venom metalloproteinase | D5LMJ3 | 12 | Naja atra | |
| 140 | Snake venom metalloproteinase | D3TTC1 | 20 | Naja atra | |
| 141 | Snake venom metalloproteinase | D3TTC2 | 8 | Naja atra | |
| 142 | Snake venom metalloproteinase-disintegrin-like mocarhagin | Q10749 | 7 | Naja mossambica | |
| 143 | Snake venom metalloproteinase | Q9PVK7 | 5 | Naja kaouthia | |
| 144 | Snake venom metalloproteinase | A8QL49 | 2 | Bungarus multicinctus | |
| 145 | Snake venom metalloproteinase | P82942 | 8 | Naja kaouthia | |
| 146 | Snake venom metalloprotease(ADAM) | ACS74986 | 1 | Philodryas olfersii | |
| 147 | Snake venom metalloproteinase 27, partial | AXL96577 | 1 | Borikenophis portoricensis | |
| 148 | Disintegrin and metalloproteinase domain-containing protein 21, partial | ETE71596 | 2 | Ophiophagus hannah | |
| 149 | Microlepidotease-1 | ABQ01137 | 1 | Oxyuranus microlepidotus | |
| 150 | Metalloproteinase atrase B, partial | ADD14036 | 1 | Naja atra | |
| 151 | Metalloproteinase 7, partial | AXL96626 | 1 | Ahaetulla prasina | |
| 152 | Snake venom metalloproteinase | P0DM46 | 1 | Micrurus corallinus | |
| 153 | K-like metalloprotease precursor, partial | ACN50005 | 1 | Naja atra | |
| 154 | Snake venom serine proteinase (peptidase S1 family) |
Tissue-type plasminogen activator, partial | ETE66683 | 3 | Ophiophagus hannah |
| 155 | Tissue-type plasminogen activator-like, partial | XP_025033187 | 3 | Python bivittatus | |
| 156 | Complement factor B precursor | AAR21601 | 1 | Naja kaouthia | |
| 157 | Thrombin-like enzyme TLP | P86545 | 2 | Naja naja | |
| 158 | Serine endopeptidase | AUS82567 | 1 | Crotalus tigris | |
| 159 | Snake venom serine protease NaSP | A8QL53 | 1 | Naja atra | |
| 160 | Snake venom serine protease catroxase-1 | Q8QHK3 | 1 | Crotalus atrox | |
| 161 | Anionic trypsin-1-like | XP_007434941 | 1 | Python bivittatus | |
| 162 | Coagulation factor X isoform 1, partial | ETE73401 | 1 | Ophiophagus hannah | |
| 163 | Serine endopeptidase | AUS82552 | 1 | Crotalus scutulatus | |
| 164 | 5’-nucleotidase family | 5-nucleotidase | BAP39972 | 5 | Protobothrops flavoviridis |
| 165 | Venom 5’-nucleotidase | A0A2I4HXH5 | 3 | Naja atra | |
| 166 | 5’-nucleotidase, partial | ETE67245 | 1 | Ophiophagus hannah | |
| 167 | Snake venom 5’-nucleotidase | B6EWW8 | 1 | Gloydius brevicaudus | |
| 168 | Aminopeptidase | Aminopeptidase N, partial | ETE61021 | 1 | Ophiophagus hannah |
| 169 | Aminopeptidase N | BAG82599 | 6 | Gloydius brevicaudus | |
| 170 | Type-B carboxylesterase/lipase | N-acetylcholinesterase | AAC59905 | 1 | Bungarus fasciatus |
| 171 | Phosphodiesterase | Snake venom Phosphodiesterase | A0A2D0TC04 | 3 | Naja atra |
| 172 | Phosphodiesterase | AHJ80885 | 1 | Macrovipera lebetina | |
| 173 | Phosphodiesterase, partial | AXL96599 | 2 | Borikenophis portoricensis | |
| 174 | Phosphodiesterase | BAN89425 | 2 | Ovophis okinavensis | |
| 175 | Flavin monoamine oxidase family | L-amino acid oxidase, partial | AAZ08620 | 1 | Daboia siamensis |
| 176 | L-amino acid oxidase, partial | AVX27607 | 4 | Naja atra | |
| 177 | L-amino-acid oxidase | Q4JHE1 | 5 | Pseudechis australis | |
| 178 | L-amino-acid oxidase | P0C2D5 | 1 | Protobothrops flavoviridis | |
| 179 | L-amino-acid oxidase | A8QL51 | 1 | Bungarus multicinctus | |
| 180 | L-amino-acid oxidase | Q4JHE3 | 3 | Oxyuranus scutellatus scutellatus | |
| 181 | L-amino acid oxidase, partial | AVX27607 | 4 | Naja atra | |
| 182 | L-amino-acid oxidase | A8QL58 | 6 | Naja atra | |
| 183 | L-amino-acid oxidase | Q4JHE3 | 3 | Oxyuranus scutellatus scutellatus | |
| 184 | L-amino acid oxidase precursor | AAY89682 | 2 | Pseudechis australis | |
| 185 | L-amino-acid oxidase | CAQ72894 | 1 | Echis ocellatus | |
| 186 | True venom lectin family | C-type lectin galactose-binding isoform | D2YVK1 | 2 | Hoplocephalus stephensii |
| 187 | BJcuL precursor | AAQ92957 | 1 | Bothrops jararacussu | |
| 188 | Ankyrin SOCS box (ASB) family | Ankyrin repeat and SOCS box protein 7, partial | ETE63895 | 1 | Ophiophagus hannah |
| 189 | Ankyrin repeat domain-containing protein 50, partial | ETE61041 | 1 | Ophiophagus hannah | |
| 190 | Transferrin | Transferrin | CAK18221 | 2 | Natrix natrix |
| 191 | Cobra serum albumin | Cobra serum albumin | S59517 | 1 | Naja kaouthia |
| 192 | Serum albumin precursor | S59517 | 3 | Naja naja | |
| 193 | Cobra serum albumin | CAA55333 | 3 | Naja naja | |
| 194 | Serum albumin/Alpha-fetoprotein/Afamin | Alpha-fetoprotein, partial | ETE59846 | 3 | Ophiophagus hannah |
| 195 | Leucine repeat | Leucine-rich repeat neuronal protein 4 | XP_007424790 | 1 | Python bivittatus |
| 196 | Small leucine-rich proteoglycan (SLRP) family | Decorin, partial | ETE60606 | 1 | Ophiophagus hannah |
| 197 | Leucine-rich repeat and WD repeat-containing protein, partial | ETE61323 | 1 | Ophiophagus hannah | |
| 198 | XPG/RAD2 endonuclease family | Endonuclease domain-containing 1 protein, partial | ETE59939 | 2 | Ophiophagus hannah |
| 199 | Deoxyribonuclease-2-alpha, partial | ETE73206 | 1 | Ophiophagus hannah | |
| 200 | NHS Family | NHS-like protein 1, partial | ETE71282 | 1 | Ophiophagus hannah |
| 201 | G-protein coupled receptor | G-protein coupled receptor 161 | XP_007428215 | 1 | Python bivittatus |
| 202 | Putative G-protein coupled receptor | ETE61591 | 2 | Ophiophagus hannah | |
| 203 | Melanocyte-stimulating hormone receptor, partial | ETE69163 | 1 | Ophiophagus hannah | |
| 204 | Latrophilin-2, partial | ETE73569 | 1 | Ophiophagus hannah | |
| 205 | Cadherin EGF LAG seven-pass G-type receptor 2, partial | ETE72621 | 1 | Ophiophagus hannah | |
| 206 | Putative G-protein coupled receptor, partial | ETE70400 | 1 | Ophiophagus hannah | |
| 207 | Zinc finger protein | Thioredoxin domain-containing protein 11, partial | ETE72118 | 1 | Ophiophagus hannah |
| 208 | Zinc finger protein 91-like isoform X2 | XP_007443313 | 1 | Python bivittatus | |
| 209 | Zinc finger protein 687 isoform X1 | XP_025027118 | 1 | Python bivittatus | |
| 210 | Zinc finger FYVE domain-containing protein 16, partial | ETE66135 | 1 | Ophiophagus hannah | |
| 211 | Zinc finger and BTB domain-containing protein 14, partial | XP_026555390 | 1 | Pseudonaja textilis | |
| 212 | Zinc finger protein 609 isoform X1 | XP_007426825 | 1 | Python bivittatus | |
| 213 | Ras-like protein | Ras GTPase-activating protein 3, partial | ETE71570 | 1 | Ophiophagus hannah |
| 214 | Rac GTPase-activating protein 1, partial | ETE61861 | 1 | Ophiophagus hannah | |
| 215 | Ras-related protein Rap-2a, partial | ETE66602 | 1 | Ophiophagus hannah | |
| 216 | RalA-binding protein 1, partial | ETE67818 | 1 | Ophiophagus hannah | |
| 217 | Guanylate-binding protein 1-like | XP_007444632 | 1 | Python bivittatus | |
| 218 | Glutathione peroxidase family | Glutathione peroxidase 3, partial | ETE68810 | 9 | Ophiophagus hannah |
| 219 | Protein family not assigned | Octapeptide-repeat protein T2, partial | ETE65834 | 1 | Ophiophagus hannah |
| 220 | Atrial natriuretic peptide receptor 2, partial | ETE58463 | 1 | Ophiophagus hannah | |
| 221 | Octapeptide-repeat protein T2, partial | ETE61441 | 1 | Ophiophagus hannah | |
| 222 | GAS2-like protein 2, partial | ETE67730 | 1 | Ophiophagus hannah | |
| 223 | Exocyst complex component 3, partial | ETE60130 | 1 | Ophiophagus hannah | |
| 224 | Vacuolar protein sorting-associated protein 54 | ETE70627 | 1 | Ophiophagus hannah | |
| 225 | Cohesin subunit SA-2, partial | ETE63002 | Ophiophagus hannah | ||
| 226 | Zona pellucida sperm-binding protein 3 receptor, partial | ETE59512 | 1 | Ophiophagus hannah | |
| 227 | Ubiquitin carboxyl-terminal hydrolase 32, partial | ETE63263 | 1 | Ophiophagus hannah | |
| 228 | Putative E3 ubiquitin-protein ligase UBR7 | ETE67503 | 1 | Ophiophagus hannah | |
| 229 | Mdm2-binding protein, partial | ETE64533 | 1 | Ophiophagus hannah | |
| 230 | E3 ubiquitin-protein ligase TTC3, partial | ETE73451 | 1 | Ophiophagus hannah | |
| 231 | Protocadherin-23 | XP_007425673 | 1 | Python bivittatus | |
| 232 | Nucleolar complex protein 4-like protein, partial | ETE59886 | 1 | Ophiophagus hannah | |
| 233 | Low molecular weight phosphotyrosine protein phosphatase, partial | ETE66708 | 1 | Ophiophagus hannah | |
| 234 | Major histocompatibility complex class I-related protein, partial | ETE56816 | 1 | Ophiophagus hannah | |
| 235 | Beta-2-microglobulin, partial | ETE58426 | 1 | Ophiophagus hannah | |
| 236 | GRAM domain-containing protein 1B, partial | ETE59875 | 1 | Ophiophagus hannah | |
| 237 | von Willebrand factor A domain-containing protein 3B, partial | ETE71898 | 1 | Ophiophagus hannah | |
| 238 | Homeobox protein PKNOX1, partial | XP_007435014 | 1 | Python bivittatus | |
| 239 | Homeobox protein prophet of Pit-1, partial | ETE69018 | 1 | Ophiophagus hannah | |
| 240 | Homeobox protein cut-like 2, partial | ETE71612 | 1 | ||
| 241 | Inosine-uridine preferring nucleoside hydrolase, partial | ETE68936 | 1 | Ophiophagus hannah | |
| 242 | Signal recognition particle receptor subunit beta | ETE61181 | 1 | Ophiophagus hannah | |
| 243 | Sodium channel protein type 1 subunit alpha | XP_025024892 | 1 | Python bivittatus | |
| 244 | Small serum protein-4 | BAJ14709 | 1 | Gloydius blomhoffii blomhoffii | |
| 245 | Clathrin heavy chain 1, partial | ETE68739 | 1 | Ophiophagus hannah | |
| 246 | Neutral amino acid transporter A, partial | ETE71889 | 1 | Ophiophagus hannah | |
| 247 | Bystin | ETE67512 | 1 | Ophiophagus hannah | |
| 248 | Peroxisome biogenesis factor 1-like isoform X1 | XP_025032182 | 1 | Python bivittatus | |
| 249 | Dapper-like 1, partial | ETE59781 | 1 | Ophiophagus hannah | |
| 250 | Protein patched-like 2, partial | ETE72035 | 1 | Ophiophagus hannah | |
| 251 | Keratin, type II cytoskeletal 1, partial | ETE67131 | 1 | Ophiophagus hannah | |
| 252 | Keratin, type II cytoskeletal 6A-like | XP_007441333 | 1 | Python bivittatus | |
| 253 | Cytosolic carboxypeptidase 2, partial | ETE72716 | 1 | Ophiophagus hannah | |
| 254 | NADH dehydrogenase subunit 4 | YP_003540795 | 1 | Hypsiglena ochrorhyncha klauberi | |
| 255 | Olfactory receptor 2D2-like | XP_007442854 | 1 | Python bivittatus | |
| 256 | Histone-lysine N-methyltransferase SETD1B, partial | ETE63606 | 1 | Ophiophagus hannah | |
| 257 | Helicase SRCAP, partial | ETE66458 | 1 | Ophiophagus hannah | |
| 258 | Tyrosine-protein phosphatase non-receptor type 11-like | XP_015743235 | 1 | Python bivittatus | |
| 259 | Glycerol-3-phosphate acyltransferase 4 | ETE64295 | 1 | Ophiophagus hannah | |
| 260 | NEDD4-binding protein 1, partial | ETE71789 | 1 | Ophiophagus hannah | |
| 261 | Nuclear pore complex protein, partial | ETE72717 | 1 | Ophiophagus hannah | |
| 262 | G1/S-specific cyclin-E1, partial | ETE69419 | 1 | Ophiophagus hannah | |
| 263 | Copine-3 | ETE62235 | 1 | Ophiophagus hannah | |
| 264 | Disks large-like 1, partial | ETE60775 | 1 | Ophiophagus hannah | |
| 265 | Tumor necrosis factor receptor superfamily member 11B | ETE67452 | 1 | Ophiophagus hannah | |
| 266 | Extracellular matrix protein 1, partial | ETE63009 | 3 | Ophiophagus hannah | |
| 267 | Protein PRRC2C isoform X7 | XP_025025988 | 1 | Python bivittatus | |
| 268 | Protein dispatched-like 2, partial | ETE65280 | 1 | Ophiophagus hannah | |
| 269 | Cytoplasmic FMR1-interacting protein 1 | ETE70074 | 1 | Ophiophagus hannah | |
| 270 | Sushi domain-containing protein 2 isoform X1 | XP_007439094 | 1 | Python bivittatus | |
| 271 | POU domain, class 2, transcription factor 1, partial | ETE68887 | 1 | Ophiophagus hannah | |
| 272 | Vomeronasal type-2 receptor 26-like | XP_015746172 | 1 | Python bivittatus | |
| 273 | snRNA-activating protein complex subunit 4, partial | ETE66257 | 1 | Ophiophagus hannah | |
| 274 | Small subunit processome component 20-like protein, partia | ETE62675 | 1 | Ophiophagus hannah | |
| 275 | Retrotransposon-derived protein PEG10, partial | ETE60414 | 1 | Ophiophagus hannah | |
| 276 | Heterogeneous nuclear ribonucleoprotein R | ETE70095 | 1 | Ophiophagus hannah | |
| 277 | Sacsin, partial | ETE73074 | 1 | Ophiophagus hannah | |
| 278 | Trafficking protein particle complex subunit 3 | XP_007439119 | 1 | Python bivittatus | |
| 279 | Putative protein C4orf34 | ETE61848 | 1 | Ophiophagus hannah | |
| 280 | Sulfate transporter, partial | ETE72250 | 1 | Ophiophagus hannah | |
| 281 | Solute carrier family 2, facilitated glucose transporter member 11, partial | ETE65979 | 1 | Ophiophagus hannah | |
| 282 | Solute carrier family 25 member 47, partial | ETE64737 | 1 | Ophiophagus hannah | |
| 283 | Citrate synthase, mitochondrial | ETE71902 | 1 | Ophiophagus hannah | |
| 284 | Separin, partial | ETE71706 | 1 | Ophiophagus hannah | |
| 285 | 5,6-dihydroxyindole-2-carboxylic acid oxidase, partial | ETE63759 | 1 | Ophiophagus hannah | |
| 286 | Protocadherin-15, partial | ETE73122 | 1 | Ophiophagus hannah | |
| 287 | Tumor necrosis factor receptor superfamily member 11B isoform X2 | XP_025019261 | 1 | Python bivittatus | |
| 288 | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, partial | ETE72267 | 1 | Ophiophagus hannah | |
| 289 | Ubiquitin carboxyl-terminal hydrolase CYLD | XP_015680147 | 1 | Protobothrops mucrosquamatus | |
| 290 | Peroxidasin, partial | ETE57820 | 1 | Ophiophagus hannah | |
| 291 | Serine palmitoyltransferase small subunit B | XP_025028624 | 1 | Python bivittatus | |
| 292 | C-terminal-binding protein 1, partial | ETE64323 | 1 | Ophiophagus hannah | |
| 293 | StAR-related lipid transfer protein 13 | ETE69978 | 1 | Ophiophagus hannah | |
| 294 | Ty3b-i, partial | ETE59080 | 1 | Ophiophagus hannah | |
| 295 | E3 ubiquitin-protein ligase RNF19B, partial | ETE68153 | 1 | Ophiophagus hannah | |
| 296 | PDZ domain-containing protein 6, partial | ETE69093 | 1 | Ophiophagus hannah | |
| 297 | Nebulin, partial | ETE70906 | 2 | Ophiophagus hannah | |
| 298 | Myoferlin, partial | ETE66870 | 1 | Ophiophagus hannah | |
| 299 | Protein mago nashi-like 2 | ETE70612 | 1 | Ophiophagus hannah | |
| 300 | H(+)/Cl(-) exchange transporter 7, partial | ETE72134 | 1 | Ophiophagus hannah | |
| 301 | Membrane cofactor protein-like | XP_025021316 | 2 | Python bivittatus | |
| 302 | Holliday junction recognition protein isoform X1 | XP_025025001 | 1 | Python bivittatus | |
| 303 | Adenylate cyclase type 2, partial | ETE62750 | 1 | Ophiophagus hannah | |
| 304 | Transmembrane protein, partial | ETE59610 | 1 | Ophiophagus hannah | |
| 305 | Transmembrane protein, partial | ETE58244 | 1 | Ophiophagus hannah | |
| 306 | Type I inositol 3,4-bisphosphate 4-phosphatase | XP_015686159 | 1 | Protobothrops mucrosquamatus | |
| 307 | Complement decay-accelerating factor transmembrane isoform, partial | ETE63384 | 8 | Ophiophagus hannah | |
| 308 | NACHT, LRR and PYD domains-containing protein 6(Belongs to NLRP family) | XP_015679160 | 1 | Protobothrops mucrosquamatus | |
| 309 | Ubiquitin carboxyl-terminal hydrolase 24 | ETE67725 | 1 | Ophiophagus hannah | |
| 310 | Epiplakin, partial | ETE58258 | 1 | Ophiophagus hannah | |
| 311 | 5’ nucleotidase, partial | AXL95273 | 1 | Spilotes sulphureus | |
| 312 | GTP-binding protein 2, partial | ETE70473 | 1 | Ophiophagus hannah | |
| 313 | Transmembrane protein 41A | XP_007420693 | 1 | Python bivittatus | |
| 314 | Serine/threonine-protein kinase TAO2, partial | ETE67077 | 1 | Ophiophagus hannah | |
| 315 | Serine/threonine-protein kinase WNK1, partial | ETE61641 | 1 | Ophiophagus hannah | |
| 316 | cilia- and flagella-associated protein 57-like, partial | XP_007436852 | 1 | Python bivittatus | |
| 317 | Lymphocyte antigen 6 complex locus protein G6d | ETE61452 | 1 | Ophiophagus hannah | |
| 318 | Histamine H3 receptor, partial | ETE72972 | 1 | Ophiophagus hannah | |
| 319 | Glycerol-3-phosphate acyltransferase 1, mitochondrial, partial | ETE59719 | 1 | Ophiophagus hannah | |
| 320 | Cleft lip and palate transmembrane protein 1-like protein, partial | ETE61569 | 1 | Ophiophagus hannah | |
| 321 | Complement factor B precursor | AAR21601 | 1 | Naja kaouthia | |
| 322 | Selenocysteine lyase | XP_015669194 | 1 | Protobothrops mucrosquamatus | |
| 323 | Serine/threonine-protein kinase Nek1, partial | ETE68306 | 1 | Ophiophagus hannah | |
| 324 | Collagen alpha-1(IV) chain, partial | ETE60834 | 1 | Ophiophagus hannah | |
| 325 | DmX-like protein 2, partial | ETE63888 | 1 | Ophiophagus hannah | |
| 326 | Aldehyde dehydrogenase family 3 member B1, partial | ETE72723 | 1 | Ophiophagus hannah | |
| 327 | Putative ATP-dependent RNA helicase DHX40, partial | ETE68740 | 1 | Ophiophagus hannah | |
| 328 | Immunoglobulin Y2 heavy chain, partial | AFR33766 | 1 | Python bivittatus | |
| 329 | Myomesin-1, partial | ETE65385 | 1 | Ophiophagus hannah | |
| 330 | Cyclic AMP-dependent transcription factor ATF-1, partial | ETE65149 | 1 | Ophiophagus hannah | |
| 331 | Toll-like receptor 4, partial | ETE72495 | 1 | Ophiophagus hannah | |
| 332 | Serine palmitoyltransferase small subunit B | XP_025028624 | 1 | Python bivittatus | |
| 333 | Histone-lysine N-methyltransferase, H3 lysine-79 specific, partial | ETE65559 | 1 | Ophiophagus hannah | |
| 334 | Creatine kinase B-type, partial | ETE69249 | 1 | Ophiophagus hannah | |
| 335 | Fibroblast growth factor 3, partial | ETE69378 | 1 | Ophiophagus hannah | |
| 336 | RB1-inducible coiled-coil protein 1, partial | ETE67067 | 1 | Ophiophagus hannah | |
| 337 | Phosphoinositide 3-kinase regulatory subunit 5, partial | ETE74144 | 1 | Ophiophagus hannah | |
| 338 | Cadherin EGF LAG seven-pass G-type receptor 2, partial | ETE72621 | 1 | Ophiophagus hannah | |
| 339 | Trafficking kinesin-binding protein 1, partial | ETE68220 | 1 | Ophiophagus hannah | |
| 340 | YTH domain family protein 2 | ETE65464 | 1 | Ophiophagus hannah | |
| 341 | Vigilin, partial | ETE61946 | 1 | Ophiophagus hannah | |
| 342 | 39S ribosomal protein L44, mitochondrial, partial | ETE68399 | 1 | Ophiophagus hannah | |
| 343 | Pseudouridine-5’-monophosphatase, partial | ETE71697 | 1 | Ophiophagus hannah | |
| 344 | Kelch-like protein 13, partial | ETE71947 | 1 | Ophiophagus hannah | |
| 345 | Maleylacetoacetate isomerase | ETE68752 | 1 | Ophiophagus hannah | |
| 346 | Neurexophilin-2, partial | ETE71784 | 1 | Ophiophagus hannah | |
| 347 | Myocyte-specific enhancer factor 2A isoform X1 | XP_007425135 | 1 | Python bivittatus | |
| 348 | Membrane cofactor protein-like isoform X1 | XP_015743425 | 1 | Python bivittatus | |
| 349 | Ninein-like protein, partial | ETE70166 | 1 | Ophiophagus hannah | |
| 350 | Keratin, type I cytoskeletal 19, partial | ETE70217 | 1 | Ophiophagus hannah | |
| 351 | Intraflagellar transport protein 88-like protein | ETE73657 | 1 | Ophiophagus hannah | |
| 352 | Complement receptor type 2, partial | ETE63383 | 1 | Ophiophagus hannah | |
| 353 | Complement decay-accelerating factor, partial | ETE59511 | 1 | Ophiophagus hannah | |
| 354 | Keratin, type II cytoskeletal 5-like | XP_025030548 | 1 | Python bivittatus | |
| 355 | 7-dehydrocholesterol reductase, partial | ETE67784 | 1 | Ophiophagus hannah | |
| 356 | La-related protein 4B | ETE62671 | 1 | Ophiophagus hannah | |
| 357 | Intelectin-1a, partial | ETE57886 | 1 | Ophiophagus hannah | |
| 358 | Cation-independent mannose-6-phosphate receptor | ETE64374 | 2 | Ophiophagus hannah | |
| 359 | Cerebellin-4 | ETE65277 | 1 | Ophiophagus hannah | |
| 360 | C3 and PZP-like alpha-2-macroglobulin domain-containing protein 8, partial | ASU45032 | 1 | Ophiophagus hannah | |
| 361 | Neuronal PAS domain-containing protein 2, partial | ETE63668 | 1 | Ophiophagus hannah | |
| 362 | Interferon-induced transmembrane protein 10, partial | ETE66904 | 1 | Ophiophagus hannah | |
| 363 | Myotubularin-related protein 11, partial | ETE72068 | 1 | Ophiophagus hannah | |
| 364 | Tyrosyl-DNA phosphodiesterase 2 | XP_026525751 | 1 | Notechis scutatus | |
| 365 | Phosphoinositide 3-kinase regulatory subunit 5, partial | ETE74144 | 1 | Ophiophagus hannah |
The bold text indicates the proteins identified to have N-terminal acetylation.
Table 3.
Summary of the venom proteome of N. oxiana.
| S. No. | Protein Family | Protein | Accession Code | Number of Matched Peptides | Homology with Protein from the Venom of Snake Species | |
|---|---|---|---|---|---|---|
| 1 | 3FTX (Neurotoxin | Neurotoxin homolog NL1 | Q9DEQ3 | 1 | Naja atra | |
| 2 | Short neurotoxin SNTX-1 | A6MFK6 | 1 | Demansia vestigiata | ||
| 3 | Neurotoxin II | P01427 | 1 | Naja oxiana | ||
| 4 | Cobrotoxin-b | P80958 | 1 | Naja atra | ||
| 5 | Alpha-cobratoxin | P01391 | 3 | Naja kaouthia | ||
| 6 | Weak toxin 2 | Q8AY50 | 2 | Bungarus candidus | ||
| 7 | Weak neurotoxin 6 | O42256 | 1 | Naja sputatrix | ||
| 8 | Weak neurotoxin 7 | P29181 | 2 | Naja naja | ||
| 9 | Weak toxin S4C11 | P01400 | 1 | Naja melanoleuca | ||
| 10 | Muscarinic toxin-like protein 3 | P82464 | 4 | Naja kaouthia | ||
| 11 | Muscarinic toxin-like protein 2 | P82463 | 4 | Naja kaouthia | ||
| 12 | Muscarinic toxin-like protein | Q9W727 | 1 | Bungarus multicinctus | ||
| 13 | Three-finger toxin precursor, partial | ADN67582 | 1 | Naja atra | ||
| 14 | Three-finger toxin precursor, partial | ADN67582 | 1 | Naja atra | ||
| 15 | 3FTXs (cytotoxins) | Cytotoxin Vc-5 | Q9PS34 | 2 | Naja oxiana | |
| 16 | Cytotoxin homolog | P14541 | 1 | Naja kaouthia | ||
| 17 | Cytotoxin homolog 5V | Q9W716 | 1 | Naja atra | ||
| 18 | Cytotoxin SP15c | P60308 | 1 | Naja atra | ||
| 19 | Cytotoxin 8 | P86540 | 2 | Naja naja | ||
| 20 | Cytotoxin 1 | P01447 | 2 | Naja naja | ||
| 21 | Cardiotoxin 7a | Q91126 | 6 | Naja atra | ||
| 22 | Cardiotoxin 1e | AAA90960 | 2 | Naja atra | ||
| 23 | Venom Complement C3-like | Venom factor | AAX86641 | 1 | Austrelaps superbus | |
| 24 | Cobra venom factor | Q91132 | 10 | Naja kaouthia | ||
| 25 | A.superbus venom factor 1 | Q0ZZJ6 | 1 | Austrelaps superbus | ||
| 26 | Cobra venom factor alpha chain | Q91132 | 1 | Naja kaouthia | ||
| 27 | Cobra venom factor 1, partial | AXL96620 | 6 | Ahaetulla prasina | ||
| 28 | Cobra venom factor, partial | AWX67646 | 2 | Boiga irregularis | ||
| 29 | Ophiophagus venom factor | I2C090 | 1 | Ophiophagus hannah | ||
| 30 | Venom Kunitz-type family | Kunitz-type serine protease inhibitor | P20229 | 2 | Naja naja | |
| 31 | BPTI/Kunitz domain-containing protein-like | XP_026546510 | 1 | Notechis scutatus | ||
| 32 | Kunitz/BPTI-like toxin | XP_026579467 | 1 | Pseudonaja textilis | ||
| 33 | natriuretic peptide family | Natriuretic peptide PaNP-c precursor, partial | AAZ82822 | 1 | Pseudechis australis | |
| 34 | NGF-beta family | Venom nerve growth factor 2 | Q5YF89 | 5 | Naja sputatrix | |
| 35 | Nerve growth factor, partial | AAR24530 | 1 | Bitis gabonica | ||
| 36 | Nerve growth factor | BAN82142 | 4 | Ovophis okinavensis | ||
| 37 | Venom nerve growth factor 2 | Q3HXX9 | 1 | Hoplocephalus stephensii | ||
| 38 | ohanin/vespryn family. | Thaicobrin | P82885 | 1 | Naja kaouthia | |
| 39 | venom PRY-SPRY domain-containing protein, partial | AHZ08803 | 1 | Micropechis ikaheca | ||
| 40 | CRISP | Cysteine-rich venom protein natrin-1 | Q7T1K6 | 3 | Naja atra | |
| 41 | Cysteine-rich secretory protein 1, partial | AXL96607 | 1 | Ahaetulla prasina | ||
| 42 | Cysteine-rich venom protein ophanin | Q7ZT98 | 1 | Ophiophagus hannah | ||
| 43 | Cysteine-rich venom protein, partial | BAP39957 | 1 | Protobothrops flavoviridis | ||
| 44 | Cysteine-rich venom protein 2 | Q7ZZN8 | 1 | Naja atra | ||
| 45 | Phosoholipase A2 | Acidic phospholipase A2 3 | P60045 | 1 | Naja sagittifera | |
| 46 | Acidic phospholipase A2 2 | P00597 | 1 | Naja kaouthia | ||
| 47 | Phospholipase a2 | CAA45372 | 3 | Naja naja | ||
| 48 | Neutral phospholipase A2 paradoxin-like beta chain | Q45Z46 | 2 | Oxyuranus microlepidotus | ||
| 49 | Phospholipase A2 | AHZ08810 | 1 | Micropechis ikaheca | ||
| 50 | Phospholipase A2 | AAA66029.1 | 1 | Naja naja | ||
| 51 | Acidic phospholipase A2 2 | P15445 | 1 | Naja naja | ||
| 52 | Acidic phospholipase A2 1 | P00596 | 6 | Naja kaouthia | ||
| 53 | Acidic phospholipase A2 1 | Q9W7J4 | 1 | Pseudonaja textilis | ||
| 54 | Basic phospholipase A2 T1-2 A chain | P84472 | 1 | Bungarus candidus | ||
| 55 | Acidic phospholipase A2 C | Q92086 | 11 | Naja sputatrix | ||
| 56 | Acidic phospholipase A2 1 | P00598 | 1 | Naja naja | ||
| 57 | Acidic phospholipase A2 beta-bungarotoxin A4 chain | P17934 | 1 | Bungarus multicinctus | ||
| 58 | Phospholipase A2 3 | P21792 | 1 | Micrurus nigrocinctus | ||
| 59 | Phospholipase B | Phospholipase B, partial | AXL95274 | 1 | Spilotes sulphureus | |
| 60 | Phospholipase B1, partial | AXL96606 | 2 | Ahaetulla prasina | ||
| 61 | Phospholipase B1, membrane-associated | XP_02653746 | 1 | Notechis scutatus | ||
| 62 | SVMP | Snake venom metalloproteinase | D3TTC2 | 4 | Naja atra | |
| 63 | Snake venom metalloproteinase | F8RKW1 | 1 | Drysdalia coronoides | ||
| 64 | Snake venom metalloproteinase | Q9PVK7 | 1 | Naja kaouthia | ||
| 65 | Disintegrin and metalloproteinase domain-containing protein 20, partial | ETE72945 | 1 | Ophiophagus hannah | ||
| 66 | Disintegrin and metalloproteinase domain-containing protein 21, partial | ETE71596 | 1 | Ophiophagus hannah | ||
| 67 | disintegrin and metalloproteinase domain-containing protein 10-like, partial | XP_026580760 | 1 | Pseudonaja textilis | ||
| 68 | P-III snake venom metalloprotease, partial | AHZ08819 | 1 | Micropechis ikaheca | ||
| 69 | Zinc metalloproteinase-disintegrin-like kaouthiagin-like | D3TTC1 | 7 | Naja atra | ||
| 70 | Zinc metalloproteinase-disintegrin-like atrase-A | D5LMJ3 | 14 | Naja atra | ||
| 71 | Hemorrhagic metalloproteinase-disintegrin-like kaouthiagin | P82942 | 2 | Naja kaouthia | ||
| 72 | metalloproteinase 7, partial | AXL96626 | 1 | Ahaetulla prasina | ||
| 73 | metalloproteinase, partial | AWX67576 | 1 | Boiga irregularis | ||
| 74 | Snake venom metalloproteinase-disintegrin-like mocarhagin | Q10749 | 3 | Naja mossambica | ||
| 75 | Snake venom metalloproteinase | Q9W6M5 | 1 | Deinagkistrodon acutus | ||
| 76 | Snake venom serine proteinase (peptidase S1 family) |
Tissue-type plasminogen activator, partial | ETE66683 | 3 | Ophiophagus hannah | |
| 77 | tissue-type plasminogen activator, partial | XP_026544671 | 2 | Notechis scutatus | ||
| 78 | Snake venom serine protease 3 | O13058 | 1 | Protobothrops flavoviridis | ||
| 79 | Serine protease 27, partial | ETE64653 | 1 | Ophiophagus hannah | ||
| 80 | Thrombin-like enzyme TLP | P86545 | 1 | Naja naja | ||
| 81 | Snake venom serine protease 3 | AAG10790 | 1 | Protobothrops jerdonii | ||
| 82 | Snake venom serine protease Dav-PA | Q9I8X1 | 1 | Deinagkistrodon acutus | ||
| 83 | serine protease 53 | XP_026576912 | 1 | Pseudonaja textilis | ||
| 84 | 5’-nucleotidase family | 5’ nucleotidase, partial | AXL95273 | 1 | Spilotes sulphureus | |
| 85 | Aminopeptidase | aminopeptidase N isoform X2 | XP_026565037 | 4 | Pseudonaja textilis | |
| 86 | type-B carboxylesterase/lipase | acetylcholinesterase | XP_026549820 | 1 | Notechis scutatus | |
| 87 | Phosphodiesterase | Phosphodiesterase | BAN89425 | 2 | Ovophis okinavensis | |
| 88 | Phosphodiesterase partial | ALA20853 | 1 | Macropisthodon rudis | ||
| 89 | Phosphodiesterase partial | AXL96599 | 1 | Borikenophis portoricensis | ||
| 90 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 isoform X2 | XP_026561286 | 2 | Pseudonaja textilis | ||
| 91 | Snake venom Phosphodiesterase | A0A2D0TC04 | 2 | Naja atra | ||
| 92 | Flavin monoamine oxidase family | L-amino acid oxidase, partial | AVX27607 | 7 | Naja atra | |
| 93 | L-amino-acid oxidase | XP_026538830 | 4 | Notechis scutatus | ||
| 94 | L-amino-acid oxidase | Q4JHE3 | 1 | Oxyuranus scutellatus scutellatus | ||
| 95 | L-amino-acid oxidase | Q4JHE1 | 1 | Pseudechis australis | ||
| 96 | L-amino-acid oxidase | A8QL58 | 1 | Naja atra | ||
| 97 | True venom lectin family | C-type lectin Cal | P21963 | 1 | Crotalus atrox | |
| 98 | Glutathione peroxidase | Glutathione peroxidase 3, partial | ETE68810 | 1 | Ophiophagus hannah | |
| 99 | Glutathione peroxidase 3 isoform X1 | XP_026541908 | 1 | Notechis scutatus | ||
| 100 | Glutathione peroxidase 3 isoform X1 | XP_026552406 | 1 | Pseudonaja textilis | ||
| 101 | Leucine repeat | Leucine-rich repeat and death domain-containing protein 1 | XP_026543987 | 1 | Notechis scutatus | |
| 102 | TNF receptor superfamily | Tumor necrosis factor receptor superfamily member 11B | XP_026545353 | 1 | Notechis scutatus | |
| 103 | Tumor necrosis factor receptor superfamily member 11B | XP_026559377 | 1 | Pseudonaja textilis | ||
| 104 | Tumor necrosis factor receptor superfamily member 11B | ETE67452 | 1 | Ophiophagus hannah | ||
| 105 | Intermediate filament family | Keratin, type II cytoskeletal 1, partia | ETE67131 | 1 | Ophiophagus hannah | |
| 106 | Keratin, type II cytoskeletal 4-like | XP_026539658 | 1 | Notechis scutatus | ||
| 107 | Keratin, type II cytoskeletal 5, partial | ETE59039 | 1 | Ophiophagus hannah | ||
| 108 | Keratin, type II cytoskeletal 5, partial | ETE59038 | 2 | Ophiophagus hannah | ||
| 109 | Keratin, type II cytoskeletal 1-like | XP_026573193 | 1 | Pseudonaja textilis | ||
| 110 | Keratin, type I cytoskeletal 19, partial | ETE70217 | 2 | Ophiophagus hannah | ||
| 111 | Keratin, type I cytoskeletal 18-like isoform X1 | XP_026521302 | 1 | Notechis scutatus | ||
| 112 | Serpin Family | Serpin B5, partial | ETE65002 | 1 | Ophiophagus hannah | |
| 113 | Ankyrin repeat domain | M-phase phosphoprotein 8, partial | ETE73652 | 1 | Ophiophagus hannah | |
| 114 | Zinc finger containing proteins | Zinc finger protein, partial | ETE62318 | 1 | Ophiophagus hannah | |
| 115 | Zinc finger protein, partial | ETE62303 | 1 | Ophiophagus hannah | ||
| 116 | Zinc finger protein 804A | XP_026552505 | 1 | Pseudonaja textilis | ||
| 117 | Zinc finger SWIM domain-containing protein 6 | XP_026572863 | 1 | Pseudonaja textilis | ||
| 118 | Zinc finger MYM-type protein 2 isoform X1 | XP_026564670 | 1 | Pseudonaja textilis | ||
| 119 | Zinc finger BED domain-containing protein 1 | XP_026522663 | 1 | Notechis scutatus | ||
| 120 | NHS Family | NHS-like protein 1 isoform X1 | XP_026561348 | 1 | Pseudonaja textilis | |
| 121 | Protein family not assigned | Holliday junction recognition protein | XP_026519764 | 1 | Notechis scutatus | |
| 122 | N-acetylgalactosaminyltransferase 7 isoform X1 | XP_026555474 | 1 | Pseudonaja textilis | ||
| 123 | PHD finger protein 3 | XP_026520899 | 1 | Notechis scutatus | ||
| 124 | Sulfhydryl oxidase 1(contains FAD binding domain) | ETE70041 | 1 | Ophiophagus hannah | ||
| 125 | C-C chemokine receptor type 10, partial | ETE65216 | 1 | Ophiophagus hannah | ||
| 126 | Cytosolic carboxypeptidase 2 | XP_026521145 | 1 | Notechis scutatus | ||
| 127 | SUMO-specific isopeptidase USPL1 isoform X1 | XP_026564646 | 1 | Pseudonaja textilis | ||
| 128 | Protein VPRBP | ETE70381 | 1 | Ophiophagus hannah | ||
| 129 | Cilia- and flagella-associated protein 97 | XP_026553667 | 1 | Pseudonaja textilis | ||
| 130 | lpxK, partial | ETE68446 | 1 | Ophiophagus hannah | ||
| 131 | Zinc phosphodiesterase ELAC protein 2, partial | ETE70777 | 1 | Ophiophagus hannah | ||
| 132 | NHS-like protein 1 isoform X1 | XP_026561348 | 1 | Pseudonaja textilis | ||
| 133 | Pro-cathepsin H | XP_026565144 | 1 | Pseudonaja textilis | ||
| 134 | C4b-binding protein alpha chain-like isoform X1 | XP_026571379 | 2 | Pseudonaja textilis | ||
| 135 | Janus kinase and microtubule-interacting protein 3 isoform X1 | XP_026566312 | 1 | Pseudonaja textilis | ||
| 136 | WD and tetratricopeptide repeats protein 1 | XP_026558310 | 1 | Pseudonaja textilis | ||
| 137 | Pro-cathepsin H | XP_026565144 | 1 | Pseudonaja textilis | ||
| 138 | C4b-binding protein alpha chain-like isoform X1 | XP_026571379 | 2 | Pseudonaja textilis | ||
| 139 | Janus kinase and microtubule-interacting protein 3 isoform X1 | XP_026566312 | 1 | Pseudonaja textilis | ||
| 140 | WD and tetratricopeptide repeats protein 1 | XP_026558310 | 1 | Pseudonaja textilis | ||
The bold text indicates the proteins identified to have N-terminal acetylation.
Figure 2.
(A) Milking of N. naja venom and sample preparation for LC-MS/MS analysis (B) Data base search cycle.
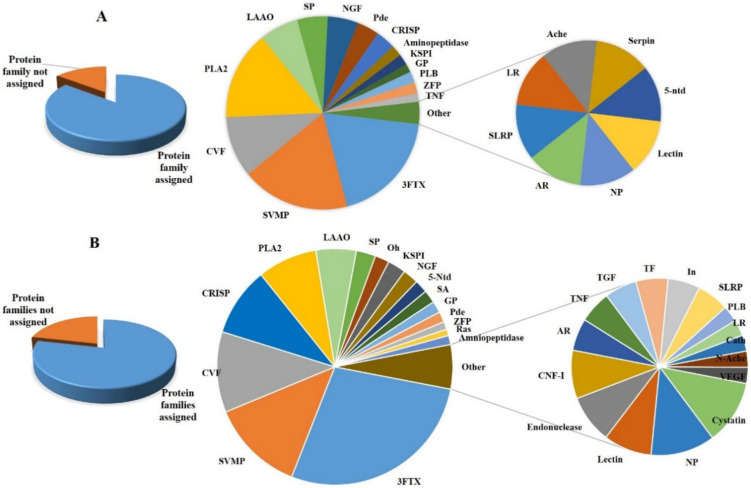
A comparative summary of the protein families of the two venoms is presented in Table 1. Figure 3, presents a comparison of the relative abundance of different venom protein families as pie charts. From the pie charts, it can be observed that there are significant differences in the proteome of two snake venoms. In the venom of N. naja, three-finger toxins (3FTx) are more abundant, while in N. oxiana venom, both 3FTXs and snake venom metalloproteinase (SVMPs) are almost equally abundant. In NO, snake venom serine proteases (SVSPs) and phospholipase A2 (PLA2s) are much more abundant than in NN. There are other subtle variations in the relative abundance of protein families between the two venoms. For example, Cysteine-rich Secretory Protein (CRISP) family is much more abundant in NN as compared to NO. Further, NN venom contains 11 protein families, which could not be found in NO venom, listed in Table 1 and highlighted in red color. Whereas NO venom contains serpins, which are absent in NN venom. Figure 3 shows that NN venom is much more versatile and contains a number of different proteins (Table 1). Data base searches revealed that our data provide a deeper insight of the NN and NO venom proteomes. There are several protein families, which have not been reported earlier in NN venom, including western and eastern Indian N. naja. In Table 1, the protein families discovered and reported for the first time in terms of our investigations are shown with check mark (✓). Interestingly previous studies reported PLA2 as the second most abundant protein family found in N. naja venom, and that SVMP was present in relatively low abundance [8,42,43,44,45,46]. In contrast, our data showed SVMP as the second most abundant protein family in N. naja. The venom proteome of N. oxiana displays that, both 3FTXs and SVMP are equally abundant like that of king cobra (Ophiophagus hannah) [51], as illustrated in Figure 3A.
Figure 3.
Pie chart illustrations highlighting the relative abundance of various protein families in the two venoms. (A) N. oxiana (B) N. naja.
In the present work, three types of posttranslational modification were also observed, i.e., N-terminus pyro-glutamate, methionine oxidation and N-terminal acetylation (N-ace). Pyro-glutamate posttranslational modifications of the venom proteins has been described before and are known to confer stability to the proteins and peptides [52,53,54,55]. However, modification of methionine residues and pyro-glutamate cannot be excluded during sample preparation. Therefore, keeping in view this possibility we have not discussed the observed methionine and pyro-glutamate modifications. The current study is the first description of N-terminal acetylation of venom proteins. In N. naja venom we were able to identify three peptide fragments (Muscarinic toxin-like protein 3, Acid phospholipase A2 and weak neurotoxin 7) containing N-ace modification. Whereas in N. oxiana one peptide (Muscarinic toxin-like protein 3) was identified with N-ace. These sequences have been highlighted with green colour in Supplementary Tables S1 and S2.
In the present work, we have identified a number of proteins like cobra serum albumin, leucine repeat, zinc finger containing protein, venom lectin protein, Ras-like protein. The presence of Ras-like protein demonstrates the presence of extracellular vesicles in the venom of Naja naja. The comparison of our proteomic data with that of N. naja snake both from western and eastern India, reveals that such proteins were not identified in Indian N. naja, Further in Pakistani Naja naja snake we not could identify cholinesterase, butyrylcholinesterase, hyalurinidase and snaclec proteins which were previously reported in Indian N. naja venom [45,46].
Below we briefly describe and discuss the different venom protein families identified.
3. Discussion
3.1. Major Protein Components (Relative Abundance >2%)
3.1.1. Three-Finger Toxins
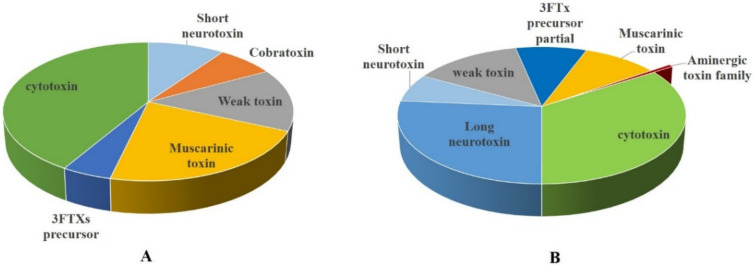
The detailed proteomic investigations of the, NN and NO snake venoms identified two main types of three-finger toxins, i.e., neurotoxins and cytotoxins. The venom of NN consists of an overall higher abundance and a greater diversity of 3FTXs, as compared to NO (Table 1 and Table 2, Figure 3). Neurotoxins are predominant in both venoms, as compared to cytotoxins, Figure 4. Among the neurotoxins, long, muscarinic, weak, 3FTxs precursor and aminergic toxin families are present in both venoms. In case of NO, a rather low amount of long neurotoxin is present, represented by one neurotoxin, cobratoxin. Whereas, long neurotoxins constitute a major proportion of neurotoxins found in the NN venom. Figure 4 indicates that in NO venom, muscarinic toxins are present in relatively higher amounts as compared to NN venom. It is interesting to note that NN venom contains an aminergic neurotoxin with homology to Dendroaspis angusticeps venom toxin AdTx1. This toxin is known to impair G-protein-coupled receptors [56,57].
Figure 4.
Pie chart illustrating a comparative profile of the three-finger toxins present in the two snake venoms. (A) N. oxiana (B) N. naja.
Previous studies have shown that 3FTXs make up approximately 56–84% of venom proteome in various species of Naja [58]. However, our results of Pakistani Naja venom samples show a much lower percentage of 3FTXS as compared to other investigations, which is 21% in NN and 16% in NO of the total venom proteins. In contrast to Pakistani N. naja venom proteome, the eastern Indian N. naja venom comprises of 61% 3FTXs and western Indian N. naja contains 68% 3FTXs. Interestingly eastern Indian N. naja lacks short neurotoxins, which are present in both western Indian and Pakistani N. naja snake venom [45,46].
Investigating 3FTXs are not only of interest to characterize their toxicity, but also of great significance for structural studies, as well as for biotechnological, biomedical and evolutionary studies [59,60,61,62,63]. Already, 3FTXs have proven to be an efficient tool to analyze various receptor types, and to study diseases like Parkinson’s disease, myasthenia gravis and cancer [64,65,66,67,68,69,70,71]. The aminergic toxins from mamba venom served as good candidates for protein resurrection methodology [72].
3.1.2. Phospholipase A2
Both Naja snake venom contain PLA2. The percentage abundance of PLA2 enzymes (12.6%) is higher in NO as compared to NN. PLA2s make up 6% of the venom of NN (Table 1). A recent study reported the comparative enzymatic activity of PLA2s in ten different Naja species, with highest activity in N. siamensis and lowest in N. nivea [73]. The venom proteome study of Indian N. naja venom carried out by A. K. Mukherjee research group reported that Indian N. naja contains 20–27% PLA2s [45,46]. This is much higher than the amount of PLA2s present in Pakistani N. naja. A proteomic study of N. kaouthia venom reported PLA2s as one of the most abundant venom proteins [74]. While another study on the venom proteome of N. annulifera did not detect PLA2s [75]. In the venom of N. philippinensis PLA2s made up 22.88% of venom proteome [76]. Another study showed distinct distribution of PLA2s in Afro-Asian cobra venom. Asian spitting cobras showed highest PLA2 activity. Asian non-spitting and African spitting cobras showed moderate activity and low activity was shown by African non-spiting cobras [77].
Table 3 shows that both venom comprise of acidic and basic PLA2s. However, acidic PLA2s are more abundant in the two venoms. Two fragments of phospholipases from NO bear homology to neutral PLA2s paradoxin-like beta chain from Oxyuranus microlepidotus. This protein was found to be one of the most potent presynaptic neurotoxins [78]. Eleven peptide fragments bearing homology to acidic phospholipase in the venom of Naja sputatrix were identified (Table 3). In the Naja naja peptide fragments having homology to acidic PLA2s from the venom of other Naja species were determined (Table 2). Six peptide fragments showed homology to acidic PLA2 from the venom of Pseudonaja textilis. A previous study reported this molecule to have moderate enzymatic activity and procoagulant property and was found to be non-lethal [79]. In the NN venom two peptide fragments matching Basic phospholipase A2 from Bungarus candidus venom and one matching with basic PLA2 with sea krait was identified. While in NO venom only one peptide fragment having homology to a basic PLA2 from Bungarus candidus was found. The activity and specificity of basic phospholipases from Agkistrodon h. blomhiffii and Pakistani N. naja was studied on intact human erythrocytes. Although belonging to two different snake families, similar response was reported for these molecules, from both venoms. Basic PLA2 induced the hydrolysis of membrane phospholipids and total cell hemolysis [80]. Despite the fact that acidic PLA2s are found abundantly in the snake venom, their role is poorly understood [81]. In spite of having high catalytic activity as compared to basic PLA2s, they do not induce toxicity [82]. Studies have suggested acidic PLA2s to participate in prey digestion [83]. Other studies have suggested that acidic PLA2s work synergistically with other venom toxins, as PLA2s, metalloprotease and cytotoxins [84,85,86].
PLA2 is ubiquitously found in nature [87,88]. In mammals, they are known to play important and vital role in many life processes [89,90,91]. On the other hand, snake venom PLA2s are toxic and interfere with a number of physiological processes, upon envenomation [87]. Phospholipase A2 are also known to be responsible for the hepatic injury, inflammation and anticoagulation in a victim [26].
3.1.3. Snake Venom Metalloproteinase
The present study shows that N. naja and N. oxiana snake venom contain significant amounts of metalloproteinases, which are the second most abundant protein family. Proteomics study of other Naja species shows the presence of SVMPs in varying amounts ranging from as low as 0.9% to 16% [74,92,93,94,95,96,97,98,99,100]. Previous proteomic studies reported a lower abundance of SVMP in Pakistani N. naja venom [42,43,44]. Three SVMPs bearing relatively higher homology with snake venom metalloproteinase from N. atra were determined in each of the two venoms. Twenty fragments of SVMPs were detected in N. naja venom, which are homologous to K-like SVMPs from N. atra. 13 Peptide fragments were found to match with SVMPs from N. kaouthia (Table 2). The data shows that in case of N. oxiana venom higher number of peptide fragments match with SVMPs from N. atra venom (Table 3). The eastern Indian N. naja contains only 6% SVMPs in contrast to Pakistani N. naja, which contains 10% of SVMPS. It is interesting to note that western Indian N. naja contains 16% SVMPs as determined by A. K. Mukherjee and his research group [45,46].
SVMPs are found in all advanced snakes and make up the major component of the venom of Crotalid and Viperid snakes [101,102,103,104]. SVMPs are structurally versatile and act on different hemostatic targets of prey upon envenomation [105]. These toxins provoke many systemic changes, such as hemorrhage, acute renal failure, coagulopathy, and/or platelet aggregation inhibition [106]. The SVMPs identified in terms of our investigations, in both of the venoms, belong to subfamily P-III. The P-III SVMPs possess gelatinolytic and hemorrhagic activities [105]. A previous study reported the hemorrhagic response of Pakistani N. naja venom in chicken egg [107]. The determination of a higher amount of SVMPs in both NO and NN venom indicates that there is potential for hemorrhage as a response of NO and NN snakebite envenomation.
3.1.4. L-Amino Acid Oxidase
Snake venom L-Amino acid oxidase (LAAOs) belong to the Flavin monoamine oxidase family and are dependent on FAD group for their activity. These proteins are present in both venoms studied and constitute approximately 4–5% of the venom proteome (Table 1). Peptide fragments bearing sequence similarity to LAAO from different snake venoms were detected and summarized (Table 2 and Table 3). In contrast to our results, studies of western Indian N. naja venom report only 0.31% LAAO. However, Indian N. naja venom contains 3% LAAO, which is similar to that of Pakistani N. naja [45,46]. In terms of our investigations we identified that LAAO from both, NN and NO venom, have homology with LAAO from N. atra venom. LAAO is known to be prevalent in many snake venoms [108] but its role in snake venom envenomation pathology is not clear. A recent study reported that LAAO might contribute to severe tissue disruption. This study suggested that LAAO might elicit its toxicity by catalyzing the intracellular substrates [108]. Different biological activities of the isolated LAAO have been reported like, edema, cytotoxic, antibacterial, antiparasitic, and/or platelet aggregation effects [109,110]. Also some investigations reported antitumor effects of LAAO [111]. LAAO is a glycoprotein and glycosylation is also considered to play a significant role in the toxicity of LAAO, and cause cell death by interacting with the cell surface [112,113].
3.1.5. Cobra Venom Factor
Cobra venom factor (CVF) belong to the venom complement C3 homologue family. CVF constitutes approximately 9% of the total proteins identified in both venoms. The identified CVF peptides bear homology mainly to the CVFs from N. kaouthia. Fragments matching to CVF alpha chain and gamma chain were also analyzed. Peptide fragments showing sequence similarity to CVF proteins from other Elapidae and Colubridae have also been identified (Table 2 and Table 3). Proteomic study of Indian N. naja venom showed that it contains only 0.03–1.7% CVF [45,46], which is significantly less compared to our results obtained for the Pakistani N. naja. A venom proteome study of Naja philippinensis showed that it contains less than 4% [76]. The venom of N. ashei contains only 0.12% CVF [99]. Cobra venom factor is a complement activating protein and is functionally and structurally similar to complement component C3b. It is a glycoprotein and herein glycosylation contributes in the immunogenicity of CVF [114,115]. In vivo studies have shown that CVF causes an acute inflammatory injury in the lungs [116]. CVF serves as a gold standard molecule for the evaluation of drugs for trials, to control diseases involving the complement system [117]. A recent study reported CVF as a promising candidate for the treatment of IRI-induced hepatic injury [118]. Our data reveals that CVF is one of the abundant proteins in the venom of Pakistani Naja naja and Naja oxiana (Figure 3). Therefore, these venom can be a good source of isolating CVF for use in biomedical research.
3.1.6. Cysteine-Rich Secretory Protein
Cysteine-rich secretory proteins (CRISPs) have been identified in many animal venoms. These proteins have two domains, a pathogenesis related domain at the N-terminal region and a cysteine rich domain at the C-terminus. Based on sequence homology the CRISP family is classified into four classes, and snake venom CRISPs belong to the group III [119]. CRISPs were found in much higher abundance in N. naja (7%) as compared to N. oxiana (2.8%) and peptide fragments showing similarities to CRISPs from different snake venoms were found in both venoms. However, highest similarity was found with the cysteine-rich venom protein natrin-1(NA-CRVP1) from N. atra. Investigations indicated that NA-CRVP1 could act as inflammatory modulator that could perturb the wound-healing process of a bitten victim by regulating the expression of adhesion molecules in endothelial cells. This study also showed that natrin contains a zinc-binding domain at the N-terminus and elicits its proinflammatory activity in a zinc and heparan-sulfate dependent manner [119]. Natrin has also been reported as a potassium channel blocker and in this context can weakly block muscle contraction [120,121,122,123]. In our study six peptide fragments matching CRISP from N. haje. A study reported this CRISP was found to be non-toxic when administered to crickets [124]. The venom proteome of N. haje contain 10% CRISP [92]. Different species of Naja contain varying amounts of CRISP, from as low as 0.2% to 10% of the total venom proteome. The Indian N. naja contains 1.14–3% CRISPs [45,46].
3.1.7. Snake Venom Serine Proteinase
Snake Venom Serine Proteinase (SVSPs) belong to the peptidase S1 family. N. oxiana venom proteome shows relatively higher abundance of serine proteinases (4%) as compared to N. naja venom, which contains only 2% (Table 1; Figure 3). Both of the venoms contain peptide fragments, which bear homology to tissue-type plasminogen activators from Ophiophagus hannah and the thrombin-like enzyme TLP from Indian N. naja. In addition to this, peptide fragments having sequence similarity to SVSP have also been identified (Table 2 and Table 3). SVSPs have been identified in only few Naja species venom. In western Indian N. naja the SVSPs contributed only 0.69% to venom proteome [46]. N. philippinensis venom proteome consists of 0.35% SVSPs [76]. Previous studies showed that SVSPs are absent in Eastern Indian N.naja venom, while a small percentage (0.03%) was reported for the western Indian snake [45,46].
SVSPs have high specificity towards their substrates. Based on their biological roles, they have also been classified as activators of the fibrinolytic system, procoagulant, anticoagulant and platelet-aggregating enzymes [125]. A few SVSPs, like ancrod and batroxobin have already applications in the treatment of cardiovascular problems, while reptilase serves today as a diagnostic probe for dysfibrinogenemia [126].
3.1.8. Snake Venom Nerve Growth Factor
Snake venom Nerve Growth Factor (NGF) were identified in both venoms but were relatively more abundant in the venom of N. oxiana (4%) as compared to N. naja (2%), Table 3. In both venoms, peptides sharing homology with Ovophis okinavensis, N. sputatrix, and Bitis gabonica NGF were identified (Table 1 and Table 2). In N. naja seven peptides bearing homology with Pseudechis australis were also identified. Further, additional peptide fragments of NGF were also identified in terms of our investigations (Table 2 and Table 3). A previous proteomic study also showed Pakistani N. naja venom to contain 2% NGF [42]. N. naja snake venom from east India contained 3.1% and 1% in western India sample. In the same study N. kaouthia from eastern India was shown to contain 1% NGF [45,46]. N. philippinensis contain only 0.06% NGF [76]. Proteomic analysis of other Naja species venom have also shown them to contain NGF but their relative abundance was not calculated [93,100]. Moroccan cobra venom contains 5% NGF of total venom proteome [92].
Till now not much is known about the contribution and function of NGFs in envenomation. Various bioassays have shown that NGFs have neurotropic activity. Snake venom NGFs have been suggested as a pharmacological tool to study the structure function relationship of human trkA receptor [127]. Studies show that NGFs assert venom toxicity indirectly, either by acting as a carrier of other neurotoxins, which do not have specific recognition sites, like phospholipase or by inducing plasma extravasation at the site of snakebite. NGF is known to coexist with CVF in cobra snake venom, and might be responsible for enhancing the toxic mechanism of CVF in an indirect manner [128]. In 1986, two scientists were awarded a Nobel Prize for their pioneering work, which allowed to explain cell growth regulation. And in context of this investigations Cohen serendipitously discovered NGF from snake venom of Agkistrodon piscivorus [129].
3.1.9. Snake Venom Phosphodiesterase
A lower abundance of snake venom phosphodieterases (PDEs) was determined in both venoms, although relatively higher in N. oxiana, i.e., 3.1%. N. naja venom contains only 1.1% of PDEs. Peptides fragments matching with PDEs from the venom of N. atra, Ovophis okinavensis, and Borikenophis portoricensis were identified in both venom. A recent study determined PDE activity in the venom of ten different species of Naja. All the species showed PDE activity with minor variation [73]. The Indian N. naja venom was reported to contain less than 1 % PDEs, which is similar to Pakistani N. naja.
PDEs are ubiquitously present in snake venom but their activity is higher in Viperidae venom as compared to Elapidae family [130]. In recent years, there has been considerable interest in snake venom PDEs due to their potential applications as pharmacological tool and drug lead. The endonuclease activity of PDEs rendered their use in sequencing of polynucleotides and oligonucleotides [130]. Phosphoribosylation, a protein modification, can also be processed by PDEs [131]. Recent innovative approaches, have utilized snake venom PDEs to digest genomic DNA into single nucleosides to study modifications of DNA [132,133,134].
3.2. Minor Protein Components (Relative Abundance ≤2%)
A large number of low abundant proteins were found in both venoms, particularly in N. naja (Table 2 and Table 3). Ras-like proteins, identified in the venom of N. naja were of particular interest, as they indicate the presence of extra0cellular vesicles in the venom. Snake venom extracellular vesicles (SVEVs) have previously been isolated from the venom of Agkistrodon contortrix contortrix, Crotalus atrox, Crotalus viridis, and Crotalus cerberus oreganus. The size distribution of SVEVs was found to be between approximately 50–500 nm. Proteomic investigations revealed that SVEVs could be assigned to eight different protein classes, such as SVMP, SVSP, and disintegrins [135].
Exosome-like vesicles have also been reported in the venom of Gloydius blomhoffii blomhoffii [136]. In this context extracellular vesicles (EV) are known to carry a diverse cargo of molecules as proteins, DNA, RNA, and/or lipids [137]. Further, EVs are involved in cell-to-cell communication, immune response and apoptotic rescue [138,139] and participate in the maintenance of normal as well as pathophysiological conditions, like cancer [140,141,142]. The proteomic study of extracellular vesicles released from cancer cells have shown the presence of Ras proteins functioning as biomarkers for extracellular vesicles [137,143,144,145]. Studies have shown that Ras proteins are involved in the regulation and assembly of extracellular vesicles cargo [145,146,147,148]. Therefore, the identification of Ras-like proteins indicates the presence of extracellular vesicles in the venom of N. naja. However direct experimental work needs to be done to confirm the presence of such vesicles in the venom. SVEVs in the venom may be involved in another mechanism to secrete membrane proteins like aminopeptidase A and G coupled receptors. SVEVs may also offer an additional route for the envenomation process, thereby facilitating toxins to translocate inside the prey cells.
In the present work, a number of proteins have been identified for the first time in the proteome of these venoms, like G-protein coupled receptors, zinc finger proteins, ankyrin repeat, leucine repeat, Ubiquitin carboxyl-terminal hydrolase and a number of other protein. It can be assumed that these proteins have also a function in the venom. Ankyrin repeats and zinc finger proteins were also identified recently, in the venom of King cobra, Naja annulifera and Micrurus pyrrhocryptus [51,100,149]. A rather old publication reported cobra serum albumin in the venom of cobra snakes [150]. Our data also revealed the presence of cobra serum albumin in the venom of N. naja. It is possible that upon envenomation cobra serum albumin is responsible or supporting the transportation of other venom proteins in the prey serum. Previous studies have reported Cobra blood serum albumin as an antitoxic protein, having the potential to sequester endogenous toxins [151,152]. Cobra serum albumin was also reported in the venom proteome of N. sumatrana [93]. Further, we identified glutathione peroxidase in both venoms. A recent proteomic study also reported the presence of glutathione peroxidase in the venom of Micrurus pyrrhocryptus and N. annulifera [100,149]. It can be speculated that this protein might be involved in protecting the venom gland from oxidative damage. Phospholipase A2 inhibitors, bearing similarity to PLA2 inhibitor isolated from the serum of Elaphe quadrivirgata and Naja kaouthia snakes, were identified in the venom of N. naja. This inhibitor was shown to inhibit the enzymatic activity of all till now known PLA2 enzymes [153,154]. Phospholipase B was also identified in both venom. Only 0.1% constituted the venom proteome of NN while that of NO contained 1.6% of the total venom proteome. Studies have shown that PLBs make up approximately 0.34% of venom components, and in Viperidae venom it varies between 0.23% to 2.5% [155]. Insulin and Transferrin proteins were also identified in the venom of N. naja. Transferrin is a metal binding proteins. Transferrin was also reported before in the venom of P. australis, utilizing two dimensional gel electrophoresis [156]. Snake venom VEGF bearing similarities to that isolated also in Bitis arietans venom, identified in N. naja venom as well. Studies have shown that different variants of snake venom VEGF induce angiogenesis and vascular permeability through different mechanisms [157,158]. Snake venom VEGF are potential candidates for therapeutic angiogenesis [159]. A low abundance of Kunitz type serine protease inhibitors (KSPI) was identified in the venom of both snakes. Snake venom KSPI have the potential to selective inhibit distinct serine proteases [35]. Some of the snake venom KSPI have evolved as potassium channel blockers [160]. BF9 a snake venom KSPI, which act as potassium channel blockers and retain the serine protease inhibitory activity. This bifunctional KSPI was reported in the venom of Bungarus fasciatus [161]. Interestingly another type of serine protease inhibitor, i.e., serpin, was identified in the venom of N. oxiana. Proteins belonging to Ohanin/Vespryn family were found in both venoms. They are small proteins with an average mass of approx. 12 kDa, and are neurotoxic in nature [162]. Further, we could identify 5′-nucleotidase in both venoms. This family of protein is found in different snake venoms [163]. These enzymes play a major role in the release of adenosine upon envenomation, which facilitates prey immobilization. In vivo studies have shown that 5-nucleotidases act synergistically with other venom components like phospholipases, disintegrins to exert a pronounced anticoagulant effect [164]. Aminopeptidase was identified in both N. naja and N. oxiana venoms. Aminopeptidase A activity has been found in the venoms of snakes belonging to Elapidae and Viperidae families. Till now not much is known about the contribution of this enzymes within envenomation pathology [165]. Cystatin, having similarity to cystatin from the venom of N. kaouthia, was identified for the first time in the venom of N. naja in the present study. Cystatin is a cysteine protease inhibitor [166]. Natriuretic peptides were identified in both N. naja and N. oxiana venom. These peptides are known to induce hypotension upon envenomation [167,168]. Cathelicidin was identified in the venom of N. naja, and previous studies have shown it to be potent antimicrobial peptide [169].
3.3. Posttranslational Modifications
In terms of our investigations, we were able to identify N-terminal acetylation (N-ace) for the first time in the snake venom. This posttranslational modification is known to carry and support out different functions in the cell. A most analyzed function of N-ace is the regulation of protein half-life, by labelling proteins for polyubiquitation and thus degradation by the proteasome [170,171]. N-ace modification plays a role in protein folding and protein complex formation [172,173]. Furthermore, studies have shown that N-ace modification mediates the interaction of proteins with membrane and subcellular localization [173]. A probable role of this modification in snake venom proteins could be to stabilize them against proteolytic cleavage, and to assist in distinct protein–protein interactions upon envenomation. In both venoms a peptide fragment bearing homology to muscarinic toxin like-protein 3, from the venom of Naja kaouthia was found to be N- terminal acetylated. Whereas in Naja naja two other peptide fragments were identified to be N-terminal acetylated. One bearing homology to phospholipase A2 and other to a weak neurotoxin 7 (Supplementary Table S1 and S2).
4. Conclusions
Using the MS shotgun approach we could provide a holistic view of the venom profile of the two Pakistani cobra snakes N. naja and N. oxiana. Our data shows for the first time the venom proteome of N. oxiana. The comparative evaluation of the venom proteome of the two snakes reveals differences, as well as similarities in their venom composition. Both snake venoms contain different types of three-finger toxins in their venom, although they exit sympatrically. There are a few toxin families, which were only found in the venom of N. naja, like cystatin, VEGF, TGF, BPP, and Cathelicidin. Therefore, we can suggest, that venom samples from both species should be utilized for the production of effective antivenoms. Also, applying state-of-the-art mass spectrometric tools allowed to identify a number of proteins not known before to be in these venoms, like Ras-GTPase, Ankyrin repeats, leucine repeat, G-protein coupled receptor, zinc finger protein, holiday junction protein, and endonuclease. In this context, identification of Ras-like proteins provided a clue about the presence of extracellular vesicles. These vesicles might function as an additional carrier to transport venom components in the prey upon envenomation. Further, our data highlight N-terminal acetylation of venom proteins for the first time and the results delineate the unique complexity of snake venoms, and open routes for further research to understand function of these molecules upon envenomation.
5. Materials and Methods
5.1. Venom Collection
Venom was milked manually from adult snake species of N. naja (black cobra/Indian cobra/Spectacled cobra) and N. oxiana (brown cobra/Caspian cobra/Central Asian cobra). For the proteomic studies of each species the venom was collected from three adult healthy snakes and pooled. The sex of the snakes was not determined. N. naja snakes were captured from the rural surroundings of Mianwali district, while N. oxiana snakes were caught from Lahore, Punjab province, Pakistan. The venom was freeze dried and kept at −20 °C till further use.
5.2. Sample Preparation for LC-MS/MS
For LC-MS/MS analysis the lyophilized crude venom from N. naja (black cobra) and N. oxiana (brown cobra) was dissolved in 10 mM Triethylamonium bicarbonate (TEAB, Thermo Fisher Scientific), 1% v/w Sodium deoxycholate (SDC, Sigma) buffer. Protein concentration was determined using a bicinchoninic acid protein assay (Pierce™ BCA Protein Assay Kit, Thermo Fisher Scientific) and 20 µg of venom protein was tryptically digested. In brief, cysteine residues were reduced for 30 min. in the presence of 10 mM dithiothreitol (DTT, Sigma) at 60 °C and alkylated for 30 min. with 20 mM iodoacetamide (IAA, Sigma) at 37 °C in the dark. Thereafter, sequencing grade trypsin (Promega) was added in a protease/protein ratio of 1:100 at 37 °C to hydrolyze venom proteins overnight. Enzymatic activity was quenched by addition of 1% v/v formic acid (FA, Fluka) and the SDC was precipitated by centrifugation at 16000 g for 5 min. The peptide containing supernatant was vacuum dried and reconstituted in 0.1% FA for LC-MS/MS analysis.
5.3. LC-MS/MS Analysis of the Digested Venom
LC-MS/MS analysis of the venom samples was performed using a nano ACQUITY UPLC® System (Waters, Manchester, UK) coupled to a Hybrid-Quadrupole-Orbitrap mass spectrometer (Q Exactive™, Thermo Fisher Scientific). The LC system was equipped with a reversed phase chromatography (RPC) columns [ACQUITY UPLC® Symmetry C 18 (180 µm i.d × 20 cm, 5 µm particle size, 100 Å pore size, Waters, Manchester, UK) as trap column and a RPC separation column (ACQUITY UPLC® Peptide BEH C-18 (75µm i.d × 20 cm, 1.7 µm particle size, 170 Å pore size, Waters, Manchester, UK) as analytical column. RPC was used with a linear 60 min acetonitrile gradient from 2–30% for peptide separation. (Solvent A: 0.1% FA in water; Solvent B: 0.1% FA in acetonitrile; Flow rate of 250 nL/min).
MS/MS data acquisition was performed in data dependent mode for the 15 most abundant precursor ions. Precursor ions with charge stages between 2+ and 5+ were selected for fragmentation if they exceeded an intensity threshold of 100,000. For MS/MS spectra acquisition the set AGC-target was 100,000 with a maximal ion injection time of 50 ms. Precursor ions were fragmented at a normalized collision energy (NCE) of 25 and the fragment ions were measured with a resolution of 17,500 at 200 m/z. To avoid redundant precursor sampling a dynamic exclusion was applied for 20 s.
5.4. Data Analysis
For protein identification, the generated raw data were processed using the Proteome Discoverer™ Software 2.0.0.802. Database search was performed with the SEQUEST algorithm against an Ophiophagus hannah (txid:8665, King cobra) protein database (UniProt), since it represents the closest sequence database to the analyzed samples. Carbamidomethylation of cysteine was used as fixed modification. Furthermore, oxidation of methionine, conversion of glutamine to pyro-glutamic acid at peptide N-termini, loss of N-terminal methionine and the acetylation of protein N-termini were considered as variable modifications. Precursor and fragment ion tolerance were set at 10 ppm and 0.02 Da, respectively. Peptide-spectra matches with a maximum delta Cn of 0.05 were used by Percolator for FDR estimation.
Unidentified spectra were exported to a new mgf file and de novo sequencing was performed with Novor [174] via DenovoGUI 1.16.2 [175]. Modifications and allowed mass tolerances were identical to the database search approach. Hits with a Novor score above 80 were considered for a protein BLAST approach. Protein BLAST for the sequenced peptides was conducted with the NCBI BLAST p algorithm (2.9.0+) with default settings against non-redundant protein sequences (nr) narrowed down to serpents (taxid: 8570). Alignments were chosen according to the max Score, the query coverage and if the homologous proteins were related to venom activity. With this information, a venom specific peptide database was created to support database searching for further analyses. Similarly, the data search was also performed against Serpents protein data base from UniProt.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [176] with the dataset identifier PXD018726 and 10.6019/PXD018726.
Venom components were classified according to protein families and their relative abundances calculated as, reported previously [51]. Briefly, the proteins analyzed were sorted into different groups of protein families. The relative abundance of each family was calculated as percent of total number of venom proteins detected by the mass spectrometer. The mathematical relationship below was used to calculate the relative abundance of each protein group. Pie chart (Figure 3) and Table 1, presents the percentage relative abundance of proteins.
| (1) |
Acknowledgments
The authors acknowledge financial support by the Cluster of Excellence “Advanced Imaging of Matter” of the Deutsche Forschungsgemeinschaft (DFG)-EXC 2056-project ID 390715994 and BMBF via project 05K16GUA. AM would like to thank Patrick Spencer, (Centro de Biotecnologia, Instituto de Pesquisas Energéticas e Nucleares, Avenue Lineu Prestes 2242, São Paulo 05508-000, Brazil) for critically reviewing the manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6651/12/11/669/s1 Table S1: De novo peptide sequencing Naja naja venom. Table S2: De novo peptide sequencing Naja oxiana venom. Table S3: Proteomic data of Naja naja venom searched against Serpents Uniprot protein data base. Table S4: Proteomic data of Naja naja venom searched against King cobra Uniprot protein data base. Table S5: Proteomic data of Naja oxiana venom searched against Serpents Uniprot protein data base. Table S6: Proteomic data of Naja oxiana venom searched against Serpents Uniprot protein data base.
Author Contributions
Conceptualization, A.M. and C.B.; methodology, H.S., B.D., and A.B.; Venom milking, Z.M.; software, H.S., A.M., B.D., and A.B.; validation, H.S., A.M., C.B., S.A.A., and B.D.; formal analysis, A.M., S.A.A., A.U., and A.A.; investigation, A.M., S.A.A., B.D., A.B., A.A., and A.U.; resources, C.B. and H.S.; data curation, A.M. and B.D.; writing—original draft preparation, A.M.; writing—review and editing, All co-authors; supervision, C.B.; project administration, A.M. and C.B.; All authors have read and agreed to the published version of the manuscript.
Funding
This work is in part supported by the Cluster of Excellence ‘The Hamburg Centre for Ultrafast Imaging’ of the Deutsche Forschungsgemeinschaft (DFG)-EXC 1074-project ID 194651731. A part of the project was supported by higher education commission (HEC), Pakistan, (NRPU/R&D/HEC-No: 20-3891) and (HEC-No: 7709/Federal/ NRPU/R&D/HEC/ 2017).
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
The present study describes a comprehensive overview of the venom proteome of Naja naja and Naja oxiana. A few protein fragments were found to be N-terminal acetylated. The identification of Ras-like proteins in the venom of Naja naja indicates the presence of extracellular vesicles in the venom.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ali W., Javaid A., Hussain A., Bukhari M.S. Diversity and habitat preferences of amphibians and reptiles in pakistan: A review. J. Asia Pac. Biodivers. 2018;11:173–187. doi: 10.1016/j.japb.2018.01.009. [DOI] [Google Scholar]
- 2.Khan M.S. Frankfurt Contributions to Natural History. Volume 16 Chimaira; Frankfurt, Germany: 2002. A guide to the snakes of pakistan. [Google Scholar]
- 3.Wallach V., Kenneth L.W., Boundy J. Snakes of the World. A Catalogue of Living and Extinct Species. CRC Press; Boca Raton, FL, USA: 2014. [Google Scholar]
- 4.Panagides N., Jackson T.N., Ikonomopoulou M.P., Arbuckle K., Pretzler R., Yang D.C., Ali S.A., Koludarov I., Dobson J., Sanker B., et al. How the cobra got its flesh-eating venom: Cytotoxicity as a defensive innovation and its co-evolution with hooding, aposematic marking, and spitting. Toxins. 2017;9:103. doi: 10.3390/toxins9030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wuster W. The cobras of the genus naja in india. Hamadryad. 1998;23:15–32. [Google Scholar]
- 6.Wuster W., Thorpe R.S. Asiatic cobras: Population systematics of the naja naja species complex (serpentes: Elapidae) in india and central asia. Herpetologica. 1992;48:69–85. [Google Scholar]
- 7.Gutierrez J.M., Williams D., Fan H.W., Warrell D.A. Snakebite envenoming from a global perspective: Towards an integrated approach. Toxicon. 2010;56:1223–1235. doi: 10.1016/j.toxicon.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Wong K.Y., Tan C.H., Tan N.H. Venom and purified toxins of the spectacled cobra (naja naja) from pakistan: Insights into toxicity and antivenom neutralization. Am. J. Trop Med. Hyg. 2016;94:1392–1399. doi: 10.4269/ajtmh.15-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization . Rabies and Envenomings-A Neglected Public Health Issue. World Health Organization; Geneva, Switzerland: 2007. [Google Scholar]
- 10.Gutierrez J.M., Burnouf T., Harrison R.A., Calvete J.J., Kuch U., Warrell D.A., Williams D.J., Global Snakebite I. A multicomponent strategy to improve the availability of antivenom for treating snakebite envenoming. Bull. World Health Organ. 2014;92:526–532. doi: 10.2471/BLT.13.132431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheske L., Ruitenberg J., Bissumbhar B. Needs and availability of snake antivenoms: Relevance and application of international guidelines. Int. J. Health Policy Manag. 2015;4:447–457. doi: 10.15171/ijhpm.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan M.S. The snakebite problem in pakistan. Bull. Chic. Herp. Soc. 2014;49:165–167. [Google Scholar]
- 13.Khan R. Treating snakebites, one snake at a time. [(accessed on 11 March 2019)];Express Trib. 2019 Available online: https://tribune.com.pk/story/1926889/treating-snakebites-one-snake-time. [Google Scholar]
- 14.Ralph R., Sharma S.K., Faiz M.A., Ribeiro I., Rijal S., Chappuis F., Kuch U. The timing is right to end snakebite deaths in south asia. BMJ. 2019;364:k5317. doi: 10.1136/bmj.k5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faisal T., Tan K.Y., Sim S.M., Quraishi N., Tan N.H., Tan C.H. Proteomics, functional characterization and antivenom neutralization of the venom of pakistani russell’s viper (daboia russelii) from the wild. J. Proteom. 2018;183:1–13. doi: 10.1016/j.jprot.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Hashmi S.U., Alvi A., Munir I., Perveen M., Fazal A., Jackson T.N.W., Ali S.A. Functional venomics of the big-4 snakes of pakistan. Toxicon. 2020;179:60–71. doi: 10.1016/j.toxicon.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Mackessy S.P. The field of reptile toxinology: Snakes, lizards, and their venoms. In: Mackessy S.P., Taylor and Francis Group, editor. Handbook of Venoms and Toxins of Reptiles. CRC Press; Boca Raton, FL, USA: 2009. pp. 3–23. [Google Scholar]
- 18.Daltry J.C., Ponnudurai G., Shin C.K., Tan N.H., Thorpe R.S., Wuster W. Electrophoretic profiles and biological activities: Intraspecific variation in the venom of the malayan pit viper (calloselasma rhodostoma) Toxicon. 1996;34:67–79. doi: 10.1016/0041-0101(95)00122-0. [DOI] [PubMed] [Google Scholar]
- 19.Alape-Giron A., Sanz L., Escolano J., Flores-Diaz M., Madrigal M., Sasa M., Calvete J.J. Snake venomics of the lancehead pitviper bothrops asper: Geographic, individual, and ontogenetic variations. J. Proteome Res. 2008;7:3556–3571. doi: 10.1021/pr800332p. [DOI] [PubMed] [Google Scholar]
- 20.Tan K.Y., Tan C.H., Fung S.Y., Tan N.H. Venomics, lethality and neutralization of naja kaouthia (monocled cobra) venoms from three different geographical regions of southeast Asia. J. Proteom. 2015;120:105–125. doi: 10.1016/j.jprot.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Amorim F.G., Costa T.R., Baiwir D., De Pauw E., Quinton L., Sampaio S.V. Proteopeptidomic, functional and immunoreactivity characterization of bothrops moojeni snake venom: Influence of snake gender on venom composition. Toxins. 2018;10:177. doi: 10.3390/toxins10050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Augusto-de-Oliveira C., Stuginski D.R., Kitano E.S., Andrade-Silva D., Liberato T., Fukushima I., Serrano S.M., Zelanis A. Dynamic rearrangement in snake venom gland proteome: Insights into bothrops jararaca intraspecific venom variation. J. Proteome Res. 2016;15:3752–3762. doi: 10.1021/acs.jproteome.6b00561. [DOI] [PubMed] [Google Scholar]
- 23.Rex C.J., Mackessy S.P. Venom composition of adult western diamondback rattlesnakes (crotalus atrox) maintained under controlled diet and environmental conditions shows only minor changes. Toxicon. 2019;164:51–60. doi: 10.1016/j.toxicon.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee A.K., Maity C.R. The composition of naja naja venom samples from three districts of west bengal, india. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 1998;119:621–627. doi: 10.1016/S1095-6433(97)00475-3. [DOI] [PubMed] [Google Scholar]
- 25.Quraishi N.A., Qureshi H.I., Simpson I.D. A contextual approach to managing snake bite in pakistan: Snake bite treatment with particular reference to neurotoxieity and the ideal hospital snake bite kit. J. Pak. Med. Assoc. 2008;58:325–331. [PubMed] [Google Scholar]
- 26.Asad M.H., Murtaza G., Ubaid M., Durre S., Sajjad A., Mehmood R., Mahmood Q., Ansari M.M., Karim S., Mehmood Z., et al. Naja naja karachiensis envenomation: Biochemical parameters for cardiac, liver, and renal damage along with their neutralization by medicinal plants. BioMed Res. Int. 2014;2014:970540. doi: 10.1155/2014/970540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomonte B., Calvete J.J. Strategies in ‘snake venomics’ aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim Toxins Incl. Trop. Dis. 2017;23:26. doi: 10.1186/s40409-017-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vonk F.J., Casewell N.R., Henkel C.V., Heimberg A.M., Jansen H.J., McCleary R.J., Kerkkamp H.M., Vos R.A., Guerreiro I., Calvete J.J., et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Von Reumont B.M. Studying smaller and neglected organisms in modern evolutionary venomics implementing rnaseq (transcriptomics)-a critical guide. Toxins. 2018;10:292. doi: 10.3390/toxins10070292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fry B.G. From genome to “venome”: Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15:403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerkkamp H.M., Kini R.M., Pospelov A.S., Vonk F.J., Henkel C.V., Richardson M.K. Snake genome sequencing: Results and future prospects. Toxins. 2016;8:360. doi: 10.3390/toxins8120360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiezel G.A., Shibao P.Y.T., Cologna C.T., Morandi Filho R., Ueira-Vieira C., De Pauw E., Quinton L., Arantes E.C. In-depth venome of the brazilian rattlesnake crotalus durissus terrificus: An integrative approach combining its venom gland transcriptome and venom proteome. J. Proteom. Res. 2018;17:3941–3958. doi: 10.1021/acs.jproteome.8b00610. [DOI] [PubMed] [Google Scholar]
- 33.Rokyta D.R., Margres M.J., Calvin K. Post-transcriptional mechanisms contribute little to phenotypic variation in snake venoms. G3 Bethesda. 2015;5:2375–2382. doi: 10.1534/g3.115.020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melani R.D., Skinner O.S., Fornelli L., Domont G.B., Compton P.D., Kelleher N.L. Mapping proteoforms and protein complexes from king cobra venom using both denaturing and native top-down proteomics. Mol. Cell Proteom. 2016;15:2423–2434. doi: 10.1074/mcp.M115.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munawar A., Ali S.A., Akrem A., Betzel C. Snake venom peptides: Tools of biodiscovery. Toxins. 2018;10:474. doi: 10.3390/toxins10110474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simoes-Silva R., Alfonso J., Gomez A., Holanda R.J., Sobrinho J.C., Zaqueo K.D., Moreira-Dill L.S., Kayano A.M., Grabner F.P., da Silva S.L., et al. Snake venom, a natural library of new potential therapeutic molecules: Challenges and current perspectives. Curr. Pharm. Biotechnol. 2018;19:308–335. doi: 10.2174/1389201019666180620111025. [DOI] [PubMed] [Google Scholar]
- 37.King G.F. Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert Opin. Biol. 2011;11:1469–1484. doi: 10.1517/14712598.2011.621940. [DOI] [PubMed] [Google Scholar]
- 38.Koh D.C., Armugam A., Jeyaseelan K. Snake venom components and their applications in biomedicine. Cell Mol. Life Sci. 2006;63:3030–3041. doi: 10.1007/s00018-006-6315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciolek J., Reinfrank H., Quinton L., Viengchareun S., Stura E.A., Vera L., Sigismeau S., Mouillac B., Orcel H., Peigneur S., et al. Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease. Proc. Natl. Acad. Sci. USA. 2017;114:7154–7159. doi: 10.1073/pnas.1620454114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdel-Ghani L.M., Rahmy T.R., Tawfik M.M., Kaziri I., Al-Obaidi A., Rowan E.G., Plevin R., Abdel-Rahman M.A. Cytotoxicity of nubein6.8 peptide isolated from the snake venom of naja nubiae on melanoma and ovarian carcinoma cell lines. Toxicon. 2019;168:22–31. doi: 10.1016/j.toxicon.2019.06.220. [DOI] [PubMed] [Google Scholar]
- 41.Wuster W. Taxonomic changes and toxinology: Systematic revisions of the asiatic cobras (naja naja species complex) Toxicon. 1996;34:399–406. doi: 10.1016/0041-0101(95)00139-5. [DOI] [PubMed] [Google Scholar]
- 42.Asad M.H.H.B., McCleary R.J.R., Salafutdinov I., Alam F., Shah H.S., Bibi S., Ali A., Khalid S., Hasan S.M.F., Sabatier J.M., et al. Proteomics study of southern punjab pakistani cobra (naja naja: Formerly naja naja karachiensis) venom. Toxicol. Environ. Chem. 2019 doi: 10.1080/02772248.2019.1619743. Ahead of Print. [DOI] [Google Scholar]
- 43.Ali S.A., Yang D.C., Jackson T.N.W., Undheim E.A.B., Koludarov I., Wood K., Jones A., Hodgson W.C., McCarthy S., Ruder T., et al. Venom proteomic characterization and relative antivenom neutralization of two medically important pakistani elapid snakes (bungarus sindanus and naja naja) J. Proteom. 2013;89:15–23. doi: 10.1016/j.jprot.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 44.Wong K.Y., Tan C.H., Tan K.Y., Quraishi N.H., Tan N.H. Elucidating the biogeographical variation of the venom of naja naja (spectacled cobra) from pakistan through a venom-decomplexing proteomic study. J. Proteom. 2018;175:156–173. doi: 10.1016/j.jprot.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 45.Chanda A., Patra A., Kalita B., Mukherjee A.K. Proteomics analysis to compare the venom composition between naja naja and naja kaouthia from the same geographical location of eastern india: Correlation with pathophysiology of envenomation and immunological cross-reactivity towards commercial polyantivenom. Expert Rev. Proteom. 2018;15:949–961. doi: 10.1080/14789450.2018.1538799. [DOI] [PubMed] [Google Scholar]
- 46.Chanda A., Kalita B., Patra A., Senevirathne W., Mukherjee A.K. Proteomic analysis and antivenomics study of western india naja naja venom: Correlation between venom composition and clinical manifestations of cobra bite in this region. Expert Rev. Proteom. 2019;16:171–184. doi: 10.1080/14789450.2019.1559735. [DOI] [PubMed] [Google Scholar]
- 47.Chanda A., Mukherjee A.K. Quantitative proteomics to reveal the composition of southern india spectacled cobra (naja naja) venom and its immunological cross-reactivity towards commercial antivenom. Int. J. Biol. Macromol. 2020;160:224–232. doi: 10.1016/j.ijbiomac.2020.05.106. [DOI] [PubMed] [Google Scholar]
- 48.Tan C.H., Tan K.Y., Fung S.Y., Tan N.H. Venom-gland transcriptome and venom proteome of the malaysian king cobra (ophiophagus hannah) Bmc Genom. 2015;16:687. doi: 10.1186/s12864-015-1828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W., Xu Y., Li Z., Fan J., Yang Y. Genome-wide mining of microsatellites in king cobra (ophiophagus hannah) and cross-species development of tetranucleotide ssr markers in chinese cobra (naja atra) Mol. Biol. Rep. 2019;46:6087–6098. doi: 10.1007/s11033-019-05044-7. [DOI] [PubMed] [Google Scholar]
- 50.Suryamohan K., Krishnankutty S.P., Guillory J., Jevit M., Schroder M.S., Wu M., Kuriakose B., Mathew O.K., Perumal R.C., Koludarov I., et al. The indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 2020;52:106–117. doi: 10.1038/s41588-019-0559-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kunalan S., Othman I., Syed Hassan S., Hodgson W.C. Proteomic characterization of two medically important malaysian snake venoms, calloselasma rhodostoma (malayan pit viper) and ophiophagus hannah (king cobra) Toxins. 2018;10:434. doi: 10.3390/toxins10110434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munawar A., Zahid A., Negm A., Akrem A., Spencer P., Betzel C. Isolation and characterization of bradykinin potentiating peptides from agkistrodon bilineatus venom. Proteome Sci. 2016;14:1. doi: 10.1186/s12953-016-0090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munawar A., Trusch M., Georgieva D., Hildebrand D., Kwiatkowski M., Behnken H., Harder S., Arni R., Spencer P., Schluter H., et al. Elapid snake venom analyses show the specificity of the peptide composition at the level of genera naja and notechis. Toxins. 2014;6:850–868. doi: 10.3390/toxins6030850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munawar A., Trusch M., Georgieva D., Spencer P., Frochaux V., Harder S., Arni R.K., Duhalov D., Genov N., Schluter H., et al. Venom peptide analysis of vipera ammodytes meridionalis (viperinae) and bothrops jararacussu (crotalinae) demonstrates subfamily-specificity of the peptidome in the family viperidae. Mol. Biosyst. 2011;7:3298–3307. doi: 10.1039/c1mb05309d. [DOI] [PubMed] [Google Scholar]
- 55.Moura-da-Silva A.M., Almeida M.T., Portes-Junior J.A., Nicolau C.A., Gomes-Neto F., Valente R.H. Processing of snake venom metalloproteinases: Generation of toxin diversity and enzyme inactivation. Toxins. 2016;8:183. doi: 10.3390/toxins8060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maiga A., Mourier G., Quinton L., Rouget C., Gales C., Denis C., Lluel P., Senard J.M., Palea S., Servent D., et al. G protein-coupled receptors, an unexploited animal toxin targets: Exploration of green mamba venom for novel drug candidates active against adrenoceptors. Toxicon. 2012;59:487–496. doi: 10.1016/j.toxicon.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 57.Blanchet G., Collet G., Mourier G., Gilles N., Fruchart-Gaillard C., Marcon E., Servent D. Polypharmacology profiles and phylogenetic analysis of three-finger toxins from mamba venom: Case of aminergic toxins. Biochimie. 2014;103:109–117. doi: 10.1016/j.biochi.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Tasoulis T., Isbister G.K. A review and database of snake venom proteomes. Toxins. 2017;9:290. doi: 10.3390/toxins9090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsetlin V.I., Karlsson E., Utkin Yu N., Pluzhnikov K.A., Arseniev A.S., Surin A.M., Kondakov V.V., Bystrov V.F., Ivanov V.T., Ovchinnikov Yu A. Interaction surfaces of neurotoxins and acetylcholine receptor. Toxicon. 1982;20:83–93. doi: 10.1016/0041-0101(82)90171-4. [DOI] [PubMed] [Google Scholar]
- 60.Kreienkamp H.J., Weise C., Raba R., Aaviksaar A., Hucho F. Anionic subsites of the catalytic center of acetylcholinesterase from torpedo and from cobra venom. Proc. Natl. Acad Sci. USA. 1991;88:6117–6121. doi: 10.1073/pnas.88.14.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hucho F., Weise C., Kreienkamp H.J., Tsetlin V., Utkin Y., Machold J. Mapping the functional topography of a receptor. Bioorg. Khim. 1992;18:1319–1329. [PubMed] [Google Scholar]
- 62.Nickitenko A.V., Michailov A.M., Betzel C., Wilson K.S. Three-dimensional structure of neurotoxin-1 from naja naja oxiana venom at 1.9 a resolution. Febs Lett. 1993;320:111–117. doi: 10.1016/0014-5793(93)80073-4. [DOI] [PubMed] [Google Scholar]
- 63.Kini R.M., Koh C.Y. Snake venom three-finger toxins and their potential in drug development targeting cardiovascular diseases. Biochem. Pharm. 2020:114105. doi: 10.1016/j.bcp.2020.114105. [DOI] [PubMed] [Google Scholar]
- 64.Chang C.C., Lee C.Y. Isolation of neurotoxins from the venom of bungarus multicinctus and their modes of neuromuscular blocking action. Arch. Int. Pharm. 1963;144:241–257. [PubMed] [Google Scholar]
- 65.Changeux J.P., Kasai M., Lee C.Y. Use of a snake venom toxin to characterize the cholinergic receptor protein. Proc. Natl. Acad. Sci. USA. 1970;67:1241–1247. doi: 10.1073/pnas.67.3.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jerusalinsky D., Kornisiuk E., Alfaro P., Quillfeldt J., Ferreira A., Rial V.E., Duran R., Cervenansky C. Muscarinic toxins: Novel pharmacological tools for the muscarinic cholinergic system. Toxicon. 2000;38:747–761. doi: 10.1016/S0041-0101(99)00196-8. [DOI] [PubMed] [Google Scholar]
- 67.Chu N.S. Contribution of a snake venom toxin to myasthenia gravis: The discovery of alpha-bungarotoxin in taiwan. J. Hist. Neurosci. 2005;14:138–148. doi: 10.1080/096470490881770. [DOI] [PubMed] [Google Scholar]
- 68.Xu Y.L., Kou J.Q., Wang S.Z., Chen C.X., Qin Z.H. Neurotoxin from naja naja atra venom inhibits skin allograft rejection in rats. Int. Immunopharmacol. 2015;28:188–198. doi: 10.1016/j.intimp.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 69.Kryukova E.V., Shelukhina I.V., Kolacheva A.A., Alieva A.K., Shadrina M.I., Slominsky P.A., Kasheverov I.E., Utkin Y.N., Ugrumov M.V., Tsetlin V.I. Possible involvement of neuronal nicotinic acetylcholine receptors in compensatory brain mechanisms at early stages of parkinson’s disease. Biomed. Khim. 2017;63:241–247. doi: 10.18097/PBMC20176303241. [DOI] [PubMed] [Google Scholar]
- 70.Tsai P.C., Fu Y.S., Chang L.S., Lin S.R. Cardiotoxin iii inhibits hepatocyte growth factor-induced epithelial-mesenchymal transition and suppresses invasion of mda-mb-231 cells. J. Biochem. Mol. Toxicol. 2016;30:12–21. doi: 10.1002/jbt.21735. [DOI] [PubMed] [Google Scholar]
- 71.Ebrahim K., Vatanpour H., Zare A., Shirazi F.H., Nakhjavani M. Anticancer activity a of caspian cobra (naja naja oxiana) snake venom in human cancer cell lines via induction of apoptosis. Iran J. Pharm Res. 2016;15:101–112. [PMC free article] [PubMed] [Google Scholar]
- 72.Blanchet G., Alili D., Protte A., Upert G., Gilles N., Tepshi L., Stura E.A., Mourier G., Servent D. Ancestral protein resurrection and engineering opportunities of the mamba aminergic toxins. Sci. Rep. 2017;7:2701. doi: 10.1038/s41598-017-02953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Modahl C.M., Roointan A., Rogers J., Currier K., Mackessy S.P. Interspecific and intraspecific venom enzymatic variation among cobras (naja sp. And ophiophagus hannah) Comp. Biochem Physiol. C Toxicol. Pharm. 2020;232:108743. doi: 10.1016/j.cbpc.2020.108743. [DOI] [PubMed] [Google Scholar]
- 74.Deka A., Gogoi A., Das D., Purkayastha J., Doley R. Proteomics of naja kaouthia venom from north east india and assessment of indian polyvalent antivenom by third generation antivenomics. J. Proteom. 2019;207:103463. doi: 10.1016/j.jprot.2019.103463. [DOI] [PubMed] [Google Scholar]
- 75.Tan K.Y., Wong K.Y., Tan N.H., Tan C.H. Quantitative proteomics of naja annulifera (sub-saharan snouted cobra) venom and neutralization activities of two antivenoms in africa. Int. J. Biol. Macromol. 2020;158:605–616. doi: 10.1016/j.ijbiomac.2020.04.173. [DOI] [PubMed] [Google Scholar]
- 76.Tan C.H., Wong K.Y., Chong H.P., Tan N.H., Tan K.Y. Proteomic insights into short neurotoxin-driven, highly neurotoxic venom of philippine cobra (naja philippinensis) and toxicity correlation of cobra envenomation in asia. J. Proteom. 2019;206:103418. doi: 10.1016/j.jprot.2019.103418. [DOI] [PubMed] [Google Scholar]
- 77.Tan C.H., Wong K.Y., Tan N.H., Ng T.S., Tan K.Y. Distinctive distribution of secretory phospholipases a(2) in the venoms of afro-asian cobras (subgenus: Naja, afronaja, boulengerina and uraeus) Toxins. 2019;11:116. doi: 10.3390/toxins11020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hodgson W.C., Dal Belo C.A., Rowan E.G. The neuromuscular activity of paradoxin: A presynaptic neurotoxin from the venom of the inland taipan (oxyuranus microlepidotus) Neuropharmacology. 2007;52:1229–1236. doi: 10.1016/j.neuropharm.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 79.Armugam A., Gong N., Li X., Siew P.Y., Chai S.C., Nair R., Jeyaseelan K. Group ib phospholipase a2 from pseudonaja textilis. Arch. Biochem. Biophys. 2004;421:10–20. doi: 10.1016/j.abb.2003.09.045. [DOI] [PubMed] [Google Scholar]
- 80.Martin J.K., Luthra M.G., Wells M.A., Watts R.P., Hanahan D.J. Phospholipase a2 as a probe of phospholipid distribution in erythrocyte membranes. Factors influencing the apparent specificity of the reaction. Biochemistry. 1975;14:5400–5408. doi: 10.1021/bi00696a003. [DOI] [PubMed] [Google Scholar]
- 81.Vargas L.J., Londono M., Quintana J.C., Rua C., Segura C., Lomonte B., Nunez V. An acidic phospholipase a(2) with antibacterial activity from porthidium nasutum snake venom. Comp. Biochem Physiol. B Biochem. Mol. Biol. 2012;161:341–347. doi: 10.1016/j.cbpb.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 82.Marques P.P., Esteves A., Lancellotti M., Ponce-Soto L.A., Marangoni S. Novel acidic phospholipase a2 from porthidium hyoprora causes inflammation with mast cell rich infiltrate. Biochem. Biophys. Rep. 2015;1:78–84. doi: 10.1016/j.bbrep.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fernandez J., Gutierrez J.M., Angulo Y., Sanz L., Juarez P., Calvete J.J., Lomonte B. Isolation of an acidic phospholipase a2 from the venom of the snake bothrops asper of costa rica: Biochemical and toxicological characterization. Biochimie. 2010;92:273–283. doi: 10.1016/j.biochi.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 84.Mora-Obando D., Fernandez J., Montecucco C., Gutierrez J.M., Lomonte B. Synergism between basic asp49 and lys49 phospholipase a2 myotoxins of viperid snake venom in vitro and in vivo. PLoS ONE. 2014;9:e109846. doi: 10.1371/journal.pone.0109846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Laustsen A.H. Toxin synergism in snake venoms. J. Toxin Rev. 2016;35:165–170. doi: 10.1080/15569543.2016.1220397. [DOI] [Google Scholar]
- 86.Jimenez-Charris E., Montealegre-Sanchez L., Solano-Redondo L., Castro-Herrera F., Fierro-Perez L., Lomonte B. Divergent functional profiles of acidic and basic phospholipases a2 in the venom of the snake porthidium lansbergii lansbergii. Toxicon. 2016;119:289–298. doi: 10.1016/j.toxicon.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 87.Kini R.M. Excitement ahead: Structure, function and mechanism of snake venom phospholipase a2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 88.Nicolas J.P., Lin Y., Lambeau G., Ghomashchi F., Lazdunski M., Gelb M.H. Localization of structural elements of bee venom phospholipase a2 involved in n-type receptor binding and neurotoxicity. J. Biol. Chem. 1997;272:7173–7181. doi: 10.1074/jbc.272.11.7173. [DOI] [PubMed] [Google Scholar]
- 89.Arita H., Hanasaki K., Nakano T., Oka S., Teraoka H., Matsumoto K. Novel proliferative effect of phospholipase a2 in swiss 3t3 cells via specific binding site. J. Biol. Chem. 1991;266:19139–19141. [PubMed] [Google Scholar]
- 90.Fry M.R., Ghosh S.S., East J.M., Franson R.C. Role of human sperm phospholipase a2 in fertilization: Effects of a novel inhibitor of phospholipase a2 activity on membrane perturbations and oocyte penetration. Biol. Reprod. 1992;47:751–759. doi: 10.1095/biolreprod47.5.751. [DOI] [PubMed] [Google Scholar]
- 91.Nakajima M., Hanasaki K., Ueda M., Arita H. Effect of pancreatic type phospholipase a2 on isolated porcine cerebral arteries via its specific binding sites. Febs Lett. 1992;309:261–264. doi: 10.1016/0014-5793(92)80785-F. [DOI] [PubMed] [Google Scholar]
- 92.Malih I., Ahmad R.M.R., Tee T.Y., Saile R., Ghalim N., Othman I. Proteomic analysis of moroccan cobra naja haje legionis venom using tandem mass spectrometry. J. Proteom. 2014;96:240–252. doi: 10.1016/j.jprot.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 93.Yap M.K., Fung S.Y., Tan K.Y., Tan N.H. Proteomic characterization of venom of the medically important southeast asian naja sumatrana (equatorial spitting cobra) Acta Trop. 2014;133:15–25. doi: 10.1016/j.actatropica.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 94.Huang H.W., Liu B.S., Chien K.Y., Chiang L.C., Huang S.Y., Sung W.C., Wu W.G. Cobra venom proteome and glycome determined from individual snakes of naja atra reveal medically important dynamic range and systematic geographic variation. J. Proteom. 2015;128:92–104. doi: 10.1016/j.jprot.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 95.Shan L.L., Gao J.F., Zhang Y.X., Shen S.S., He Y., Wang J., Ma X.M., Ji X. Proteomic characterization and comparison of venoms from two elapid snakes (bungarus multicinctus and naja atra) from china. J. Proteom. 2016;138:83–94. doi: 10.1016/j.jprot.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 96.Sintiprungrat K., Watcharatanyatip K., Senevirathne W.D., Chaisuriya P., Chokchaichamnankit D., Srisomsap C., Ratanabanangkoon K. A comparative study of venomics of naja naja from india and sri lanka, clinical manifestations and antivenomics of an indian polyspecific antivenom. J. Proteom. 2016;132:131–143. doi: 10.1016/j.jprot.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 97.Choudhury M., McCleary R.J.R., Kesherwani M., Kini R.M., Velmurugan D. Comparison of proteomic profiles of the venoms of two of the ‘big four’ snakes of india, the indian cobra (naja naja) and the common krait (bungarus caeruleus), and analyses of their toxins. Toxicon. 2017;135:33–42. doi: 10.1016/j.toxicon.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 98.Lauridsen L.P., Laustsen A.H., Lomonte B., Gutierrez J.M. Exploring the venom of the forest cobra snake: Toxicovenomics and antivenom profiling of naja melanoleuca. J. Proteom. 2017;150:98–108. doi: 10.1016/j.jprot.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 99.Hus K.K., Buczkowicz J., Petrilla V., Petrillova M., Lyskowski A., Legath J., Bocian A. First look at the venom of naja ashei. Molecules. 2018;23:609. doi: 10.3390/molecules23030609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Silva-de-Franca F., Villas-Boas I.M., Serrano S.M.T., Cogliati B., Chudzinski S.A.A., Lopes P.H., Kitano E.S., Okamoto C.K., Tambourgi D.V. Naja annulifera snake: New insights into the venom components and pathogenesis of envenomation. PLoS Negl. Trop. Dis. 2019;13:e0007017. doi: 10.1371/journal.pntd.0007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gutierrez J.M., Rucavado A. Snake venom metalloproteinases: Their role in the pathogenesis of local tissue damage. Biochimie. 2000;82:841–850. doi: 10.1016/S0300-9084(00)01163-9. [DOI] [PubMed] [Google Scholar]
- 102.Takeda S., Takeya H., Iwanaga S. Snake venom metalloproteinases: Structure, function and relevance to the mammalian adam/adamts family proteins. Biochim. Biophys. Acta. 2012;1824:164–176. doi: 10.1016/j.bbapap.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 103.Markland F.S., Jr., Swenson S. Snake venom metalloproteinases. Toxicon. 2013;62:3–18. doi: 10.1016/j.toxicon.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 104.Gutierrez J.M., Escalante T., Rucavado A., Herrera C. Hemorrhage caused by snake venom metalloproteinases: A journey of discovery and understanding. Toxins. 2016;8:93. doi: 10.3390/toxins8040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bernardoni J.L., Sousa L.F., Wermelinger L.S., Lopes A.S., Prezoto B.C., Serrano S.M., Zingali R.B., Moura-da-Silva A.M. Functional variability of snake venom metalloproteinases: Adaptive advantages in targeting different prey and implications for human envenomation. PLoS ONE. 2014;9:e109651. doi: 10.1371/journal.pone.0109651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gutierrez J.M., Escalante T., Rucavado A. Experimental pathophysiology of systemic alterations induced by bothrops asper snake venom. Toxicon. 2009;54:976–987. doi: 10.1016/j.toxicon.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 107.Razi M.T., Asad M.H., Khan T., Chaudhary M.Z., Ansari M.T., Arshad M.A., Saqib Q.N. Antihaemorrhagic potentials of fagonia cretica against naja naja karachiensis (black pakistan cobra) venom. Nat. Prod. Res. 2011;25:1902–1907. doi: 10.1080/14786419.2010.490785. [DOI] [PubMed] [Google Scholar]
- 108.Costal-Oliveira F., Stransky S., Guerra-Duarte C., Naves de Souza D.L., Vivas-Ruiz D.E., Yarleque A., Sanchez E.F., Chavez-Olortegui C., Braga V.M.M. L-amino acid oxidase from bothrops atrox snake venom triggers autophagy, apoptosis and necrosis in normal human keratinocytes. Sci. Rep. 2019;9:781. doi: 10.1038/s41598-018-37435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fox J.W. A brief review of the scientific history of several lesser-known snake venom proteins: L-amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon. 2013;62:75–82. doi: 10.1016/j.toxicon.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 110.Izidoro L.F., Sobrinho J.C., Mendes M.M., Costa T.R., Grabner A.N., Rodrigues V.M., da Silva S.L., Zanchi F.B., Zuliani J.P., Fernandes C.F., et al. Snake venom l-amino acid oxidases: Trends in pharmacology and biochemistry. BioMed Res. Int. 2014;2014:196754. doi: 10.1155/2014/196754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Costa T.R., Burin S.M., Menaldo D.L., de Castro F.A., Sampaio S.V. Snake venom l-amino acid oxidases: An overview on their antitumor effects. J. Venom. Anim. Toxins Incl. Trop. Dis. 2014;20:23. doi: 10.1186/1678-9199-20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suhr S.M., Kim D.S. Identification of the snake venom substance that induces apoptosis. Biochem. Biophys. Res. Commun. 1996;224:134–139. doi: 10.1006/bbrc.1996.0996. [DOI] [PubMed] [Google Scholar]
- 113.Ande S.R., Kommoju P.R., Draxl S., Murkovic M., Macheroux P., Ghisla S., Ferrando-May E. Mechanisms of cell death induction by l-amino acid oxidase, a major component of ophidian venom. Apoptosis. 2006;11:1439–1451. doi: 10.1007/s10495-006-7959-9. [DOI] [PubMed] [Google Scholar]
- 114.Vogel C.W., Muller-Eberhard H.J. Cobra venom factor: Improved method for purification and biochemical characterization. J. Immunol. Methods. 1984;73:203–220. doi: 10.1016/0022-1759(84)90045-0. [DOI] [PubMed] [Google Scholar]
- 115.Laursen N.S., Andersen K.R., Braren I., Spillner E., Sottrup-Jensen L., Andersen G.R. Substrate recognition by complement convertases revealed in the c5-cobra venom factor complex. Embo J. 2011;30:606–616. doi: 10.1038/emboj.2010.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vogel C.W., Fritzinger D.C. Cobra venom factor: Structure, function, and humanization for therapeutic complement depletion. Toxicon. 2010;56:1198–1222. doi: 10.1016/j.toxicon.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 117.Morgan B.P., Harris C.L. Complement therapeutics; history and current progress. Mol. Immunol. 2003;40:159–170. doi: 10.1016/S0161-5890(03)00111-1. [DOI] [PubMed] [Google Scholar]
- 118.Wang B., Xu H., Li J., Gao H.M., Xing Y.H., Lin Z., Li H.J., Wang Y.Q., Cao S.H. Complement depletion with cobra venom factor alleviates acute hepatic injury induced by ischemiareperfusion. Mol. Med. Rep. 2018;18:4523–4529. doi: 10.3892/mmr.2018.9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Y.L., Kuo J.H., Lee S.C., Liu J.S., Hsieh Y.C., Shih Y.T., Chen C.J., Chiu J.J., Wu W.G. Cobra crisp functions as an inflammatory modulator via a novel zn2+ -and heparan sulfate-dependent transcriptional regulation of endothelial cell adhesion molecules. J. Biol. Chem. 2010;285:37872–37883. doi: 10.1074/jbc.M110.146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chang L.S., Liou J.C., Lin S.R., Cheng Y.C. Purification and characterization of taiwan cobra venom proteins with weak toxicity. Toxicon. 2005;45:21–25. doi: 10.1016/j.toxicon.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 121.Wang J., Shen B., Guo M., Lou X., Duan Y., Cheng X.P., Teng M., Niu L., Liu Q., Huang Q., et al. Blocking effect and crystal structure of natrin toxin, a cysteine-rich secretory protein from naja atra venom that targets the bkca channel. Biochemistry. 2005;44:10145–10152. doi: 10.1021/bi050614m. [DOI] [PubMed] [Google Scholar]
- 122.Wang F., Li H., Liu M.N., Song H., Han H.M., Wang Q.L., Yin C.C., Zhou Y.C., Qi Z., Shu Y.Y., et al. Structural and functional analysis of natrin, a venom protein that targets various ion channels. Biochem. Biophys. Res. Commun. 2006;351:443–448. doi: 10.1016/j.bbrc.2006.10.067. [DOI] [PubMed] [Google Scholar]
- 123.Zhou Q., Wang Q.L., Meng X., Shu Y., Jiang T., Wagenknecht T., Yin C.C., Sui S.F., Liu Z. Structural and functional characterization of ryanodine receptor-natrin toxin interaction. Biophys. J. 2008;95:4289–4299. doi: 10.1529/biophysj.108.137224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Osipov A.V., Levashov M.Y., Tsetlin V.I., Utkin Y.N. Cobra venom contains a pool of cysteine-rich secretory proteins. Biochem. Biophys. Res. Commun. 2005;328:177–182. doi: 10.1016/j.bbrc.2004.12.154. [DOI] [PubMed] [Google Scholar]
- 125.Serrano S.M. The long road of research on snake venom serine proteinases. Toxicon. 2013;62:19–26. doi: 10.1016/j.toxicon.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 126.Kang T.S., Georgieva D., Genov N., Murakami M.T., Sinha M., Kumar R.P., Kaur P., Kumar S., Dey S., Sharma S., et al. Enzymatic toxins from snake venom: Structural characterization and mechanism of catalysis. Febs J. 2011;278:4544–4576. doi: 10.1111/j.1742-4658.2011.08115.x. [DOI] [PubMed] [Google Scholar]
- 127.Katzir I., Shani J., Goshen G., Sela J., Ninary E., Dogonovski A.M., Shabashov D., Inoue S., Ikeda K., Hayashi K., et al. Characterization of nerve growth factors (ngfs) from snake venoms by use of a novel, quantitative bioassay utilizing pheochromocytoma (pc12) cells overexpressing human trka receptors. Toxicon. 2003;42:481–490. doi: 10.1016/S0041-0101(03)00225-3. [DOI] [PubMed] [Google Scholar]
- 128.Kostiza T., Meier J. Nerve growth factors from snake venoms: Chemical properties, mode of action and biological significance. Toxicon. 1996;34:787–806. doi: 10.1016/0041-0101(96)00023-2. [DOI] [PubMed] [Google Scholar]
- 129.McCleary R.J., Kini R.M. Non-enzymatic proteins from snake venoms: A gold mine of pharmacological tools and drug leads. Toxicon. 2013;62:56–74. doi: 10.1016/j.toxicon.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 130.Uzair B., Khan B.A., Sharif N., Shabbir F., Menaa F. Phosphodiesterases (pdes) from snake venoms: Therapeutic applications. Protein Pept. Lett. 2018;25:612–618. doi: 10.2174/0929866525666180628160616. [DOI] [PubMed] [Google Scholar]
- 131.Thirawatananond P., McPherson R.L., Malhi J., Nathan S., Lambrecht M.J., Brichacek M., Hergenrother P.J., Leung A.K.L., Gabelli S.B. Structural analyses of nudt16-adp-ribose complexes direct rational design of mutants with improved processing of poly(adp-ribosyl)ated proteins. Sci. Rep. 2019;9:5940. doi: 10.1038/s41598-019-39491-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yin J., Chen S., Zhang N., Wang H. Multienzyme cascade bioreactor for a 10 min digestion of genomic DNA into single nucleosides and quantitative detection of structural DNA modifications in cellular genomic DNA. Acs. Appl. Mater. Interfaces. 2018;10:21883–21890. doi: 10.1021/acsami.8b05399. [DOI] [PubMed] [Google Scholar]
- 133.Lai W., Lyu C., Wang H. Vertical ultrafiltration-facilitated DNA digestion for rapid and sensitive uhplc-ms/ms detection of DNA modifications. Anal. Chem. 2018;90:6859–6866. doi: 10.1021/acs.analchem.8b01041. [DOI] [PubMed] [Google Scholar]
- 134.Willmann L., Erbes T., Krieger S., Trafkowski J., Rodamer M., Kammerer B. Metabolome analysis via comprehensive two-dimensional liquid chromatography: Identification of modified nucleosides from rna metabolism. Anal. Bioanal. Chem. 2015;407:3555–3566. doi: 10.1007/s00216-015-8516-6. [DOI] [PubMed] [Google Scholar]
- 135.Carregari V.C., Rosa-Fernandes L., Baldasso P., Bydlowski S.P., Marangoni S., Larsen M.R., Palmisano G. Snake venom extracellular vesicles (svevs) reveal wide molecular and functional proteome diversity. Sci. Rep. 2018;8:12067. doi: 10.1038/s41598-018-30578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ogawa Y., Kanai-Azuma M., Akimoto Y., Kawakami H., Yanoshita R. Exosome-like vesicles in gloydius blomhoffii blomhoffii venom. Toxicon. 2008;51:984–993. doi: 10.1016/j.toxicon.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 137.Xu R., Greening D.W., Zhu H.J., Takahashi N., Simpson R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016;126:1152–1162. doi: 10.1172/JCI81129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yanez-Mo M., Siljander P.R., Andreu Z., Zavec A.B., Borras F.E., Buzas E.I., Buzas K., Casal E., Cappello F., Carvalho J., et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Raposo G., Stahl P.D. Extracellular vesicles: A new communication paradigm? Nat. Rev. Mol. Cell Biol. 2019;20:509–510. doi: 10.1038/s41580-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 140.Maas S.L.N., Breakefield X.O., Weaver A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Osier N., Motamedi V., Edwards K., Puccio A., Diaz-Arrastia R., Kenney K., Gill J. Exosomes in acquired neurological disorders: New insights into pathophysiology and treatment. Mol. Neurobiol. 2018;55:9280–9293. doi: 10.1007/s12035-018-1054-4. [DOI] [PubMed] [Google Scholar]
- 142.Chong S.Y., Lee C.K., Huang C., Ou Y.H., Charles C.J., Richards A.M., Neupane Y.R., Pavon M.V., Zharkova O., Pastorin G., et al. Extracellular vesicles in cardiovascular diseases: Alternative biomarker sources, therapeutic agents, and drug delivery carriers. Int. J. Mol. Sci. 2019;20:3272. doi: 10.3390/ijms20133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Klein-Scory S., Tehrani M.M., Eilert-Micus C., Adamczyk K.A., Wojtalewicz N., Schnolzer M., Hahn S.A., Schmiegel W., Schwarte-Waldhoff I. New insights in the composition of extracellular vesicles from pancreatic cancer cells: Implications for biomarkers and functions. Proteome Sci. 2014;12:50. doi: 10.1186/s12953-014-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Luhtala N., Aslanian A., Yates J.R., 3rd, Hunter T. Secreted glioblastoma nanovesicles contain intracellular signaling proteins and active ras incorporated in a farnesylation-dependent manner. J. Biol. Chem. 2017;292:611–628. doi: 10.1074/jbc.M116.747618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cufaro M.C., Pieragostino D., Lanuti P., Rossi C., Cicalini I., Federici L., De Laurenzi V., Del Boccio P. Extracellular vesicles and their potential use in monitoring cancer progression and therapy: The contribution of proteomics. J. Oncol. 2019;2019:1639854. doi: 10.1155/2019/1639854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kriebel P.W., Majumdar R., Jenkins L.M., Senoo H., Wang W., Ammu S., Chen S., Narayan K., Iijima M., Parent C.A. Extracellular vesicles direct migration by synthesizing and releasing chemotactic signals. J. Cell Biol. 2018;217:2891–2910. doi: 10.1083/jcb.201710170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr. Biol. 2018;28:R435–R444. doi: 10.1016/j.cub.2018.01.059. [DOI] [PubMed] [Google Scholar]
- 148.Willms E., Cabanas C., Mager I., Wood M.J.A., Vader P. Extracellular vesicle heterogeneity: Subpopulations, isolation techniques, and diverse functions in cancer progression. Front. Immunol. 2018;9:738. doi: 10.3389/fimmu.2018.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Olamendi-Portugal T., Batista C.V.F., Pedraza-Escalona M., Restano-Cassulini R., Zamudio F.Z., Benard-Valle M., de Roodt A.R., Possani L.D. New insights into the proteomic characterization of the coral snake micrurus pyrrhocryptus venom. Toxicon. 2018;153:23–31. doi: 10.1016/j.toxicon.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 150.Wolfenden R.N. On the nature and action of the venom of poisonous snakes: Ii. A note upon the venom of the indian viper (daboia russellii) J. Physiol. 1886;7:357–364. doi: 10.1113/jphysiol.1886.sp000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shao J., Shen H., Havsteen B. Purification, characterization and binding interactions of the chinese-cobra (naja naja atra) serum antitoxic protein csap. Biochem J. 1993;293:559–566. doi: 10.1042/bj2930559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang X., Buck F., Havsteen B. Elucidation of a new biological function of an old protein: Unique structure of the cobra serum albumin controls its specific toxin binding activity. Int. J. Biochem. Cell Biol. 1998;30:225–233. doi: 10.1016/S1357-2725(97)00113-1. [DOI] [PubMed] [Google Scholar]
- 153.Ohkura N., Inoue S., Ikeda K., Hayashi K. The two subunits of a phospholipase a2 inhibitor from the plasma of thailand cobra having structural similarity to urokinase-type plasminogen activator receptor and ly-6 related proteins. Biochem. Biophys Res. Commun. 1994;204:1212–1218. doi: 10.1006/bbrc.1994.2592. [DOI] [PubMed] [Google Scholar]
- 154.Okumura K., Masui K., Inoue S., Ikeda K., Hayashi K. Purification, characterization and cdna cloning of a phospholipase a2 inhibitor from the serum of the non-venomous snake elaphe quadrivirgata. Biochem. J. 1999;341:165–171. doi: 10.1042/bj3410165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ullah A., Masood R. The sequence and three-dimensional structure characterization of snake venom phospholipases b. Front. Mol. Biosci. 2020;7:175. doi: 10.3389/fmolb.2020.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Georgieva D., Seifert J., Ohler M., von Bergen M., Spencer P., Arni R.K., Genov N., Betzel C. Pseudechis australis venomics: Adaptation for a defense against microbial pathogens and recruitment of body transferrin. J. Proteome Res. 2011;10:2440–2464. doi: 10.1021/pr101248e. [DOI] [PubMed] [Google Scholar]
- 157.Takahashi H., Hattori S., Iwamatsu A., Takizawa H., Shibuya M. A novel snake venom vascular endothelial growth factor (vegf) predominantly induces vascular permeability through preferential signaling via vegf receptor-1. J. Biol. Chem. 2004;279:46304–46314. doi: 10.1074/jbc.M403687200. [DOI] [PubMed] [Google Scholar]
- 158.Yamazaki Y., Matsunaga Y., Tokunaga Y., Obayashi S., Saito M., Morita T. Snake venom vascular endothelial growth factors (vegf-fs) exclusively vary their structures and functions among species. J. Biol. Chem. 2009;284:9885–9891. doi: 10.1074/jbc.M809071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Toivanen P.I., Nieminen T., Laakkonen J.P., Heikura T., Kaikkonen M.U., Yla-Herttuala S. Snake venom vegf vammin induces a highly efficient angiogenic response in skeletal muscle via vegfr-2/nrp specific signaling. Sci. Rep. 2017;7:5525. doi: 10.1038/s41598-017-05876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Norton R.S., Chandy K.G. Venom-derived peptide inhibitors of voltage-gated potassium channels. Neuropharmacology. 2017;127:124–138. doi: 10.1016/j.neuropharm.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 161.Yang W., Feng J., Wang B., Cao Z., Li W., Wu Y., Chen Z. Bf9, the first functionally characterized snake toxin peptide with kunitz-type protease and potassium channel inhibiting properties. J. Biochem. Mol. Toxicol. 2014;28:76–83. doi: 10.1002/jbt.21538. [DOI] [PubMed] [Google Scholar]
- 162.Pung Y.F., Wong P.T., Kumar P.P., Hodgson W.C., Kini R.M. Ohanin, a novel protein from king cobra venom, induces hypolocomotion and hyperalgesia in mice. J. Biol. Chem. 2005;280:13137–13147. doi: 10.1074/jbc.M414137200. [DOI] [PubMed] [Google Scholar]
- 163.Aird S.D. Ophidian envenomation strategies and the role of purines. Toxicon. 2002;40:335–393. doi: 10.1016/S0041-0101(01)00232-X. [DOI] [PubMed] [Google Scholar]
- 164.Dhananjaya B.L., D’Souza C.J. The pharmacological role of nucleotidases in snake venoms. Cell Biochem. Funct. 2010;28:171–177. doi: 10.1002/cbf.1637. [DOI] [PubMed] [Google Scholar]
- 165.Vaiyapuri S., Wagstaff S.C., Watson K.A., Harrison R.A., Gibbins J.M., Hutchinson E.G. Purification and functional characterisation of rhiminopeptidase a, a novel aminopeptidase from the venom of bitis gabonica rhinoceros. PLoS Negl. Trop. Dis. 2010;4:e796. doi: 10.1371/journal.pntd.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brillard-Bourdet M., Nguyen V., Ferrer-di Martino M., Gauthier F., Moreau T. Purification and characterization of a new cystatin inhibitor from taiwan cobra (naja naja atra) venom. Biochem. J. 1998;331 Pt 1:239–244. doi: 10.1042/bj3310239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fry B.G., Vidal N., Norman J.A., Vonk F.J., Scheib H., Ramjan S.F., Kuruppu S., Fung K., Hedges S.B., Richardson M.K., et al. Early evolution of the venom system in lizards and snakes. Nature. 2006;439:584–588. doi: 10.1038/nature04328. [DOI] [PubMed] [Google Scholar]
- 168.Gutierrez J.M., Lomonte B., Leon G., Rucavado A., Chaves F., Angulo Y. Trends in snakebite envenomation therapy: Scientific, technological and public health considerations. Curr. Pharm. Des. 2007;13:2935–2950. doi: 10.2174/138161207782023784. [DOI] [PubMed] [Google Scholar]
- 169.Zhao H., Gan T.X., Liu X.D., Jin Y., Lee W.H., Shen J.H., Zhang Y. Identification and characterization of novel reptile cathelicidins from elapid snakes. Peptides. 2008;29:1685–1691. doi: 10.1016/j.peptides.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 170.Oh J.H., Hyun J.Y., Varshavsky A. Control of hsp90 chaperone and its clients by n-terminal acetylation and the n-end rule pathway. Proc. Natl. Acad. Sci. USA. 2017;114:E4370–E4379. doi: 10.1073/pnas.1705898114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shemorry A., Hwang C.S., Varshavsky A. Control of protein quality and stoichiometries by n-terminal acetylation and the n-end rule pathway. Mol. Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Drazic A., Myklebust L.M., Ree R., Arnesen T. The world of protein acetylation. Biochim Biophys. Acta. 2016;1864:1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 173.Ree R., Varland S., Arnesen T. Spotlight on protein n-terminal acetylation. Exp. Mol. Med. 2018;50:90. doi: 10.1038/s12276-018-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ma B. Novor: Real-time peptide de novo sequencing software. J. Am. Soc. Mass Spectrom. 2015;26:1885–1894. doi: 10.1007/s13361-015-1204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Muth T., Weilnbock L., Rapp E., Huber C.G., Martens L., Vaudel M., Barsnes H. Denovogui: An open source graphical user interface for de novo sequencing of tandem mass spectra. J. Proteome Res. 2014;13:1143–1146. doi: 10.1021/pr4008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The pride database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.