Abstract
The genetic alteration underlying the great majority of primary angioedema with normal C1 inhibitor (nl-C1-INH-HAE) cases remains unknown. To search for variants associated with nl-C1-INH-HAE, we genotyped 133 unrelated nl-C1-INH-HAE patients using a custom next-generation sequencing platform targeting 55 genes possibly involved in angioedema pathogenesis. Patients already diagnosed with F12 alterations as well as those with histaminergic acquired angioedema were excluded. A variant pathogenicity curation strategy was followed, including a comparison of the results with those of genotyping 169 patients with hereditary angioedema due to C1-inhibitor deficiency (C1-INH-HAE), and only filtered-in variants were studied further. Among the examined nl-C1-INH-HAE patients, carriers of neither the ANGPT1 p.Ala119Ser nor the KNG1 p.Met379Lys variant were found, whereas the PLG p.Lys330Glu was detected in four (3%) unrelated probands (one homozygote). In total, 182 different variants were curated, 21 of which represented novel mutations. Although the frequency of variants per gene was comparable between nl-C1-INH-HAE and C1-INH-HAE, variants of the KNG1 and XPNPEP1 genes were detected only in nl-C1-INH-HAE patients (six and three, respectively). Twenty-seven filtered variants in 23 different genes were detected in nl-C1-INH-HAE more than once, whereas 69/133 nl-C1-INH-HAE patients had compound heterozygotes of filtered variants located in the same or different genes. Pedigree analysis was performed where feasible. Our results indicate the role that alterations in some genes, like KNG1, may play in disease pathogenesis, the complex trait that is possibly underlying in some cases, and the existence of hitherto unrecognized disease endotypes.
Keywords: next-generation sequencing, pedigree analysis, primary angioedema, primary angioedema with normal C1 inhibitor
1. Introduction
Primary angioedema is defined as localized and self-limiting edema of the subcutaneous and submucosal tissue occurring in the absence of wheals and of a causative factor. According to the criteria of the Hereditary Angioedema International Working Group [1], all hereditary forms as well as the two idiopathic forms of acquired angioedema (histaminergic and non-histaminergic) can be considered as primary angioedema. Hereditary angioedema due to C1-INH deficiency (C1-INH-HAE), the prototype of primary angioedema, is an autosomal dominant disease caused by deleterious mutations in the SERPING1 gene, leading to quantitative and/or functional C1 inhibitor (C1-INH) deficiency [2]. Normal C1-INH levels and function characterize all other forms of primary angioedema, which clinically present with individual attacks indistinguishable from C1-INH-HAE attacks, despite differing from C1-INH-HAE in many aspects [3]. Until recently, the only genetic defects known to be associated with the hereditary forms of primary angioedema with normal C1-INH levels were mutations in the F12 gene [4,5]. All other familial cases of angioedema with normal C1-INH levels were characterized as unknown angioedema. In 2019, next-generation sequencing technologies provided new insights into the genetics of primary angioedema with normal C1 inhibitor (nl-C1-INH-HAE). Two new missense mutations in ANGPT1 (c.807G>T, p.Ala119Ser) and PLG (c.988A>G, p.Lys330Glu) genes were detected in association with the disease, whereas family segregation and meticulous functional studies have proved their pathogenicity [6,7,8]. Recently, Bork et al. [9] reported a hitherto unknown variant in exon 10 of the KNG1 gene (c.1136T>A, p.Met379Lys) co-segregated with clinical symptoms of hereditary angioedema (HAE) with normal C1-INH levels in three generations of a large German family.
Interestingly, the recently discovered pathogenic variants expanded our concept of nl-C1-INH-HAE pathophysiology beyond the contact system indicating new disease endotypes [6,10,11]. Moreover, a series of patients misdiagnosed as idiopathic non-histaminergic acquired angioedema (InH-AAE) have been reported in the literature, who, after genotyping, were proved to be suffering nl-C1-INH-HAE associated with F12, PLG, or ANGPT1 mutations [12]. Thus, further uncovering the genetic basis of nl-C1-INH-HAE is expected not only to facilitate a better understanding of disease pathophysiology that could drive the discovery of new therapeutic targets but also to provide useful indicators for the clinical management of the disease. To this aim, here, we applied a custom next-generation sequencing (NGS) platform targeting a series of genes entangled in the metabolism and function of bradykinin to detect candidate genes involved in the pathogenesis of nl-C1-INH-HAE.
2. Experimental Section
2.1. Patients
Patients fulfilling the diagnostic criteria of primary angioedema according to the Hereditary Angioedema International Working Group [1] and presenting with normal C1-INH plasma levels were included in the study. Beyond those diagnosed with hereditary angioedema with normal C1-INH, patients with idiopathic non-histaminergic acquired angioedema were included in this group, since they represent a temporary exclusion diagnosis that does not rule out either the appearance of angioedema in the next generation or the presence of a yet unidentified genetic background [12]. Patients already diagnosed with hereditary angioedema with normal C1-INH and factor XII mutation (FXII-HAE), as well as those with histaminergic acquired angioedema, were excluded.
In total, 133 unrelated patients (53 Hungarian, 32 Italian, 27 Spanish, 12 Greek, and 9 Polish) (35 male; age 40.8 ± 17.4 years) were enrolled in the study. Their mean (±SD) age at disease onset was 27.0 ± 16.4 years (median: 24 years). One in 133 patients had suffered only one angioedema attack during their life; the other 132/133 had a mean frequency of angioedema attacks of 7.8 per year (median: 5 per year). Of the 133 patients, 104 presented with a family history of angioedema, whereas 31/133 patients were on long-term prophylaxis with tranexamic acid. A further 169 patients with C1-INH-HAE were genotyped as controls to search for the presence of variants common in the two forms of angioedema that possibly affect the clinical expression of the disease.
The study was carried out according to the principles of good clinical practice and adhered to the ethical standards of the Declaration of Helsinki with written informed consent from all subjects. The Ethics Committee of the University of Thessaly approved the protocol of the study.
2.2. Genotyping
A custom NGS panel was designed using the Ion AmpliSeq Thermo Fisher Scientific Designer (Thermo Scientific, Waltham, Massachusetts, US) to analyze 55 genes (all coding regions and exon–intron splice junctions) (Supporting Information, Table S1) possibly involved in angioedema pathogenesis and/or the clinical phenotype. The gene list was compiled from literature data on angioedema and genetic predisposition, protein–protein interaction networks, and pathway analysis. In total, 825 amplicons in two primer pools provide 99.61% coverage of all targeted regions.
To construct DNA libraries for each sample using the Ion AmpliSeq Library Kit 2.0 (Thermo Scientific, Waltham, MA, USA), 10 ng of gDNA per primer pool was used. The produced libraries were indexed with a unique adapter using the Ion Xpress barcode adapter kit (Thermo Scientific, Waltham, MA, USA). Barcoded libraries were purified using the Agencourt AMPure XP Beads (Beckman Coulter, Brea, CA, USA), quantified with a Qubit 2.0 fluorometer (Thermo Scientific, Waltham, MA, USA), diluted to 100 pM and pooled in equimolar proportion. Template preparation, enrichment, and chip loading were carried out on the Ion Chef system (Thermo Scientific, Waltham, MA, USA). Sequencing was performed on S5XL on 520 and 530 chips, using the Ion 510, Ion 520, and Ion 530 Kit - Chef (Thermo Scientific, Waltham, MA, USA). All procedures were performed according to the manufacturer’s instructions.
Base calling, demultiplexing, and alignment to the hg19 reference genome (GRCh37) of the raw sequencing data were performed in Torrent Suite 5.10 software (Thermo Scientific, Waltham, MA, USA) using the default parameters. Variant calling was performed by the VariantCaller v.5.8.0.19 plug-in and coverage analysis by the CoverageAnalysis v.5.8.0.8 plug-in in Torrent Suite 5.10.
Confirmatory Sanger sequencing was appropriately performed where necessary. Since the causative ANGPT1 variant (c.807G>T) had not been described at the time of the design of our NGS panel, this gene was not included among those analyzed by this method. Thus, ANGPT1 genotyping was performed by Sanger sequencing as previously described [6].
2.3. Variant Pathogenicity Curation
All variants detected after alignment to the hg19 genome using the VariantCaller plug-in were annotated in Ion Reporter software v.5.6 (Thermo Scientific, Waltham, MA, USA) with the gene name and for their possible presence in the Single Nucleotide Polymorphism Database (v135) [13], the Exome Aggregation Consortium (ExAC) [14], the 1000 Genomes project [15], and the ClinVar [16] according to the recommendations of the Human Genome Variation Society (HGVS) [17]. SIFT [18] and PolyPhen version 2 [19] bioinformatics tools were used for in silico pathogenicity prediction of the variants. Alignments and all obtained sequences were visually inspected using the Integrative Genomics Viewer (IGV) v.2.2 (Broad Institute, Cambridge, MA, USA).
Variants with a worldwide frequency of >1% (1000 Genomes Global Minor Allele Frequency, ExAC) and polymorphisms for which no disease associations are reported in the ClinVar database, as well as synonymous and intronic single-nucleotide variants (SNVs), were excluded from further analysis.
3. Results and Discussion
Among the nl-C1-INH-HAE patients, no carriers of the ANGPT1 p.Ala119Ser variant were found, indicating that at least this mutation of the ANGPT1 gene represents very rare causative genetic damage. However, the PLG p.Lys330Glu variant was detected in four (3%) unrelated probands (one homozygote), which have already been described in detail elsewhere [20]. Pedigree analysis of these cases confirmed the incomplete penetrance of this alteration. Including our cases, more than 100 patients with nl-C1-INH-HAE due to this mutation have been reported in the literature since its first description [21].
Among the variants identified in the 55 analyzed genes, 182 different variants were filtered in and included in further analysis. Twelve alterations occurred in the 5′ untranslated region (UTR) (6.6%) and 18 in the 3′-UTR (10%). Missense mutations corresponded to 76.6% of the total, followed by small insertions/deletions leading to frameshift (1%), non-sense (3.3%), splice site (0.5%), stop-loss (0.5%), and non-frameshift insertions/deletions (1.5%). A list containing all variants is found in Table S2 (Supporting Information). Of the 182 mutations (indicated in black in Table S2), 21 were not previously reported in population databases (novel mutations). The frequency of variants per gene was not significantly different between nl-C1-INH-HAE and C1-INH-HAE patients, with the exception of KNG1 and XPNPEP1 genes, where six and three variants were detected, respectively, in the nl-C1-INH-HAE group but none in the C1-INH-HAE group.
A series of 27 filtered variants in 23 different genes was detected in our material more than once. As shown in Table 1, in a proportion of these variants, their allele frequencies among nl-C1-INH-HAE patients were significantly different from those in the European population or even in our C1-INH-HAE cohort. According to the guidelines of the American College of Medical Genetics and Genomics [22], this is a criterion in favor of the pathogenicity of the variants. The exact contribution of each one of these variants in the pathogenesis or in the clinical phenotype of the disease is difficult to envisage. However, a finding that merits particular attention is the frequency of filtered androgen receptor gene (AR) variants, which, among nl-C1-INH-HAE patients, is significantly higher than that in both the European population and in the cohort of C1-INH-HAE controls. Further studies are worth undertaking to investigate the possible correlation of these variants with the estrogen-dependence of the disease’s clinical phenotype.
Table 1.
Variants with a worldwide allelic frequency of <1% that were detected in our material more than once. EMAF: European minor allele frequency; ExAC ENFAF: ExAC European non-Finnish allele frequency; nl-C1-INH-HAE AF: allele frequency among nl-C1-INH-HAE patients; C1-INH-HAE AF: allele frequency among C1-INH-HAE patients, P1: EMAF vs. nl-C1-INH-HAE AF, P2: EMAF vs. C1-INH-HAE AF, P3: ni-C1-INH-HAE AF vs. C1-INH-HAE AF.
| Genes | Coding | Amino Acid Change | dbSNP | SIFT | PolyPhen | EMAF | ExAC ENFAF | nl-C1-INH-HAE AF | C1-INH-HAE AF | P1 | P2 | P3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BDKRB1 | c.721G>A | p.Gly241Arg | rs45528332 | tolerated | probably damaging | 0.0037 | 0.0052 | 0.0113 | 0.0148 | 0.1500 | 0.0350 | 0.7000 |
| MME | c.674G>C | p.Gly225Ala | rs147564881 | tolerated | probably damaging | 0.0023 | 0.0033 | 0.0113 | 0.0030 | 0.0310 | 0.7400 | 0.2100 |
| PLAUR | c.802A>G | p.Met268Val | rs138492321 | tolerated | possibly damaging | 0.0062 | 0.0045 | 0.0188 | 0.0000 | 0.0440 | 0.1500 | 0.0110 |
| C1S | c.943G>A | p.Asp315Asn | rs117907409 | deleterious | probably damaging | 0.0053 | 0.0052 | 0.0113 | 0.0059 | 0.2400 | 0.8300 | 0.4700 |
| F13B | c.1025T>C | p.Ile342Thr | rs17514281 | deleterious | possibly damaging | 0.0097 | 0.0098 | 0.0263 | 0.0059 | 0.0380 | 0.4900 | 0.0390 |
| F2 | c.*97G>A | rs1799963 | 0.0080 | 0.0263 | 0.0148 | 0.0130 | 0.2600 | 0.3100 | ||||
| TLR4 | c.842G>A | p.Cys281Tyr | rs137853920 | deleterious | probably damaging | 0.0044 | 0.0027 | 0.0150 | 0.0030 | 0.0420 | 0.7900 | 0.1000 |
| KRT1 | c.1669A>G | p.Ser557Gly | rs77846840 | tolerated | benign | 0.0019 | 0.0263 | 0.0296 | 0.8000 | |||
| SERPINE1 | c.*180C>T | rs41334349 | 0.0110 | 0.0226 | 0.0266 | 0.1400 | 0.0400 | 0.7500 | ||||
| AR | c.-207C>A | rs189146053 | 0.0000 | 0.0188 | 0.0030 | <0.0001 | 0.0844 | 0.0500 | ||||
| AR | c.1174C>T | p.Pro392Ser | rs201934623 | tolerated | benign | 0.0000 | 0.0041 | 0.0113 | 0.0000 | 0.0007 | 0.0500 | |
| TPSAB1 | c.407A>G | p.His136Arg | rs201820654 | tolerated | benign | 0.0034 | 0.0113 | 0.0089 | 0.7600 | |||
| TPSG1 | c.508G>A | p.Gly170Arg | rs117769620 | tolerated | benign | 0.0065 | 0.0073 | 0.0188 | 0.0118 | 0.0757 | 0.3893 | 0.4832 |
| ELANE | c.770C>T | p.Pro257Leu | rs17216663 | tolerated | benign | 0.0108 | 0.0080 | 0.0188 | 0.0030 | 0.3062 | 0.1775 | 0.0530 |
| F12 | c.418C>G | p.Leu140Val | rs35515200 | tolerated | possibly damaging | 0.0042 | 0.0033 | 0.0075 | 0.0030 | 0.4533 | 0.7904 | 0.4287 |
| F12 | c.530C>T | p.Ala177Val | rs144821595 | tolerated | benign | 0.0002 | 0.0001 | 0.0075 | 0.0000 | <0.0001 | 0.7948 | 0.1103 |
| ACE | c.1453C>G | p.Pro485Ala | rs202178737 | deleterious | benign | 0.0000 | 0.0001 | 0.0075 | 0.0000 | 0.0059 | 0.1103 | |
| BDKRB1 | c.844C>T | p.Arg282Ter | rs145322761 | 0.0035 | 0.0038 | 0.0075 | 0.0030 | 0.4533 | 0.7904 | 0.4287 | ||
| PLG | c.266G>A | p.Arg89Lys | rs143079629 | tolerated | benign | 0.0100 | 0.0108 | 0.0075 | 0.0030 | 0.7164 | 0.2177 | 0.4287 |
| KLK3 | c.629C>G | p.Ser210Trp | rs61729813 | deleterious | probably damaging | 0.0110 | 0.0109 | 0.0075 | 0.0178 | 0.6223 | 0.3319 | 0.2748 |
| DPP4 | c.796G>A | p.Val266Ile | rs56179129 | tolerated | benign | 0.0060 | 0.0045 | 0.0075 | 0.0000 | 0.7755 | 0.1547 | 0.1103 |
| PLAU | c.1048T>C | p.Tyr350His | rs72816325 | deleterious | probably damaging | 0.0058 | 0.0059 | 0.0075 | 0.0000 | 0.7755 | 0.1547 | 0.1103 |
| PLAUR | c.-87C>T | rs147665588 | 0.0060 | 0.0075 | 0.0089 | 0.7755 | 0.5702 | 0.8550 | ||||
| F13A1 | c.1730C>T | p.Thr577Met | rs143711562 | tolerated | benign | 0.0029 | 0.0020 | 0.0075 | 0.0000 | 0.2930 | 0.3149 | 0.1103 |
| TNF | c.251C>T | p.Pro84Leu | rs4645843 | tolerated | benign | 0.0030 | 0.0028 | 0.0075 | 0.0030 | 0.2930 | 0.9945 | 0.4287 |
| GPER1 | c.14C>T | p.Ser5Phe | rs117290655 | tolerated | benign | 0.0048 | 0.0045 | 0.0075 | 0.0089 | 0.6173 | 0.4193 | 0.8550 |
| MPO | c.2031-2A>C | rs35897051 | 0.0072 | 0.0071 | 0.0075 | 0.0059 | 0.9227 | 0.8391 | 0.8096 |
Sixty-nine of the examined nl-C1-INH-HAE patients were heterozygous for more than one and up to nine filtered variants located in the same or different genes (compound heterozygotes). No correlation was found between the number of heterozygous variants carried by patients and their age at disease onset or the frequency of attacks.
Family segregation studies were performed when feasible and provided useful information. Firstly, the variants p.Leu140Val and p.Ala177Val of the F12 gene proved to be non-pathogenic. However, the novel PLG p.Val728Glu (c.2183T>A) variant was found to co-segregate with angioedema symptoms in a Greek patient (male, 15 years old) and their suffering father (52 years old) but not in his unaffected mother. His 10-year-old sister also carries the variant but she has not demonstrated disease symptoms as yet. The p.Val728Glu variant is located inside the plasmin serine protease domain (residues 562–791), which is an active serine protease with a wide substrate specificity [23]. Thus, the p.Val728Glu substitution could eventually affect functional interrelationships between the plasminogen/plasmin system and the kinin pathway, leading to an alteration in vasopermeability.
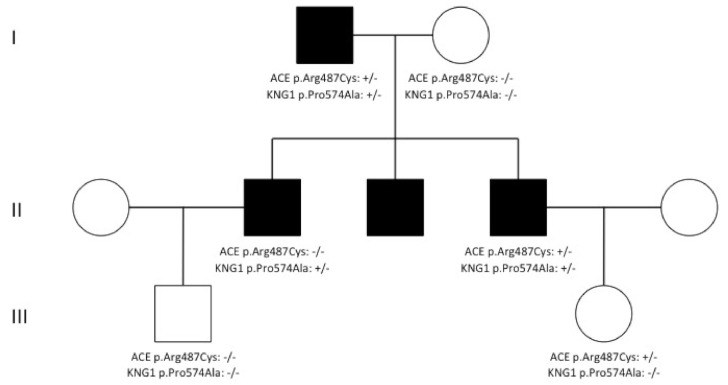
The recently reported KNG1 p.Met379Lys variant [9] was not detected in any of our nl-C1-INH-HAE patients. However, the KNG1 p.Pro574Ala (c.1720C>G) variant was detected in three affected members (two brothers and their father) of an Italian family but not in three asymptomatic relatives. Two of the patients suffered typical disease with repeated angioedema attacks, whereas the third had only experienced one attack during his life following a viral infection. Interestingly, the two patients who suffered repeated attacks were also carriers of the ACE p.Arg487Cys (c.1459C>T) variant. The same variant, despite it being predicted as deleterious by bioinformatics tools, was also detected in one of the three analyzed asymptomatic relatives (Figure 1). It seems that the KNG1 p.Pro574Ala variant presents with incomplete penetrance or that its possible pathogenicity depends upon its compound heterozygosity with the ACE p.Arg487Cys variant. In conjunction with the abovementioned high frequency of filtered KNG1 variants observed among nl-C1-INH-HAE patients, these findings indicate that variations in the KNG1 gene could contribute to the pathogenesis of the disease; thus, they deserve further consideration.
Figure 1.
Pedigree demonstrating co-segregation of the missense mutation of the KNG1 p.Pro574Ala (c.1720C>G) and the ACE p.Arg487Cys (c.1459C>T) variants with nl-C1-INH-HAE.
The genes encoding for tryptases (TPSAB1, TPSD1, and TPSG1) were included in the panel of analyzed genes because raised serum tryptase has been occasionally observed in cases of acquired angioedema [24,25]. The variant p.Arg158Gln (c.473G>A) of the TPSG1 gene was detected in all three affected women in three generations and in one of the three examined asymptomatic first-degree relatives of an Italian family. Should this finding be confirmed by further studies, it would implicate new pathways or cells (e.g., mastocytes) in the pathogenesis of nl-C1-INH-HAE.
A final remarkable finding was that the two suffering members (a mother and her daughter) of a Hungarian family were carriers of the same series of novel or rare variants in different genes: BDKRB1 p.Arg282Ter, CPN1 p.Glu407Lys, SERPING1 c.*57C>G (3′UTR), PLAUR p.Met268Val, MASP1 p.Val680Ala, TLR4 p.Cys281Tyr, and MPO p.Arg524His. This observation suggests that, at least in certain cases, nl-C1-INH-HAE could be the result of the cumulative effect of multiple gene variations.
Taken together, the above observations clearly demonstrate that, genetically, nl-C1-INH-HAE is an extremely complex disorder. The relatively small size of the examined cohort of patients and the non-availability of families for pedigree analysis represent the main limitations of the study, which is true for all rare disease studies. Thus, strong evidence of the causative effect of certain variants has not been provided. Nevertheless, the results of the study helpfully highlight the role that alterations in some genes, like KNG1, may play in the pathogenesis of the disease, the complex trait that is possibly underlying some cases, and the existence of hitherto unrecognized disease endotypes. Finally, it must be underlined that every day, contemporary genomic approaches discover new genes associated with the disease, indicating the involvement of new pathways in its pathogenesis (e.g., the ANGPT1 [6] and the MYOF [26] genes). Therefore, beyond the genes examined in this study, there are many other candidate disease genes remaining to be examined, like many endothelium-associated ones.
Acknowledgments
We would like to acknowledge the following colleagues for their collaboration in collecting patients’ samples and clinical information: Tiziana De Pasquale, Department of Allergy, Hospital of Civitanova Marche, Italy; Alessandra Zoli, Department of Clinical Immunology, Ospedali Riuniti, Ancona, Italy; Anna Radice, Unit of Immunoallergology, Azienda Ospealiera Universitarta Careggi, Florence, Italy; Stefano Pizzimenti, Department of Allergy, University of Torino, Italy; Emmanouil Manoussakis, Allergy Unit, 2nd Pediatric Clinic, University of Athens, P & A Kyriakou Children’s Hospital, Athens, Greece.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/9/11/3402/s1: Table S1: The genes analyzed by our custom NGS panel and their coverages; Table S2: The filtered-in variants detected among nl-C1-INH-HAE patients. Variants not previously reported in population databases (novel mutations) are indicated in bold.
Author Contributions
Conceptualization, A.E.G., G.L., M.Z., C.S., and H.F.; clinical evaluation and investigation, M.B., T.G.-Q., F.P. (Fotis Psarros), G.P., M.S., D.F., S.d.G., M.M., and A.Z.; genetic investigation, G.L., F.P. (Faidra Parsopoulou), M.Z., D.C., and S.V.; writing—original draft preparation, A.E.G., G.L., and F.P. (Faidra Parsopoulou); writing—review and editing, M.S. and H.F.; supervision, A.E.G.; project administration, A.E.G.; funding acquisition, A.E.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an Investigator Initiated Research grant (IIR-GRC-000905) from Shire International GmbH, a member of the Takeda group of companies.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cicardi M., Aberer W., Banerji A., Bas M., Bernstein J.A., Bork K., Caballero T., Farkas H., Grumach A., Kaplan A.P., et al. Classification, diagnosis, and approach to treatment for angioedema: Consensus report from the Hereditary Angioedema International Working Group. Allergy. 2014;69:602–616. doi: 10.1111/all.12380. [DOI] [PubMed] [Google Scholar]
- 2.Bork K., Davis-Lorton M. Overview of hereditary angioedema caused by C1-inhibitor deficiency: Assessment and clinical management. Eur. Ann. Allergy Clin. Immunol. 2013;45:7–16. [PubMed] [Google Scholar]
- 3.Bork K. Hereditary angioedema with normal C1 inhibitor. Immunol. Allergy Clin. N. Am. 2013;33:457–470. doi: 10.1016/j.iac.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Bork K., Wulff K., Witzke G., Hardt J. Hereditary angioedema with normal C1-INH with versus without specific F12 gene mutations. Allergy. 2015;70:1004–1012. doi: 10.1111/all.12648. [DOI] [PubMed] [Google Scholar]
- 5.Magerl M., Germenis A.E., Maas C., Maurer M. Hereditary angioedema with normal C1 inhibitor: Update on evaluation and treatment. Immunol. Allergy Clin. N. Am. 2017;37:571–584. doi: 10.1016/j.iac.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Bafunno V., Firinu D., D’Apolito M., Cordisco G., Loffredo S., Leccese A., Bova M., Barca M.P., Santacroce R., Cicardi M., et al. Mutation of the angiopoietin-1 gene (ANGPT1) associates with a new type of hereditary angioedema. J. Allergy Clin. Immunol. 2018;141:1009–1017. doi: 10.1016/j.jaci.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Bork K., Wulff K., Steinmüller-Magin L., Braenne I., Staubach-Renz P., Witzke G., Hardt J. Hereditary angioedema with a mutation in the plasminogen gene. Allergy. 2018;73:442–450. doi: 10.1111/all.13270. [DOI] [PubMed] [Google Scholar]
- 8.Dewald G. A missense mutation in the plasminogen gene, within the plasminogen kringle 3 domain, in hereditary angioedema with normal C1 inhibitor. Biochem. Biophys. Res. Commun. 2018;498:193–198. doi: 10.1016/j.bbrc.2017.12.060. [DOI] [PubMed] [Google Scholar]
- 9.Bork K., Wulff K., Rossmann H., Steinmüller-Magin L., Braenne I., Witzke G., Hardt J. Hereditary angioedema cosegregating with a novel kininogen 1 gene mutation changing the N-terminal cleavage site of bradykinin. Allergy. 2019;74:2479–2481. doi: 10.1111/all.13869. [DOI] [PubMed] [Google Scholar]
- 10.Zuraw B.L. Hereditary angioedema with normal C1 inhibitor: Four types and counting. J. Allergy Clin. Immunol. 2018;141:884–885. doi: 10.1016/j.jaci.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 11.D’Apolito M., Santacroce R., Colia A.L., Cordisco G., Maffione A.B., Margaglione M. Angiopoietin-1 haploinsufficiency affects the endothelial barrier and causes hereditary angioedema. Clin. Exp. Allergy. 2019;49:626–635. doi: 10.1111/cea.13349. [DOI] [PubMed] [Google Scholar]
- 12.Firinu D., Loffredo S., Bova M., Cicardi M., Margaglione M., Del Giacco S. The role of genetics in the current diagnostic workup of idiopathic non-histaminergic angioedema. Allergy. 2019;74:810–812. doi: 10.1111/all.13667. [DOI] [PubMed] [Google Scholar]
- 13.dbSNP. [(accessed on 20 August 2020)]; Available online: http://www.ncbi.nlm.nih.gov/projects/SNP/
- 14.ExAC. [(accessed on 1 October 2019)]; Available online: http://exac.broadinstitute.org.
- 15.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ClinVar. [(accessed on 20 August 2020)]; Available online: http://www.ncbi.nlm.nih.gov/clinvar/
- 17.HGVS. [(accessed on 20 August 2020)]; Available online: http://www.hgvs.org/mutnomen/
- 18.SIFT. [(accessed on 20 August 2020)]; Available online: http://sift.jcvi.org/
- 19.PolyPhen-2. [(accessed on 20 August 2020)]; Available online: http://genetics.bwh.harvard.edu/pph2/
- 20.Germenis A.E., Loules G., Zamanakou M., Psarros F., González-Quevedo T., Speletas M., Bork K., Wulff K., Steinmüller-Magin L., Braenne I., et al. On the pathogenicity of the plasminogen K330E mutation for hereditary angioedema. Allergy. 2018;73:1751–1753. doi: 10.1111/all.13324. [DOI] [PubMed] [Google Scholar]
- 21.Recke A., Massalme E.G., Jappe U., Steinmüller-Magin L., Schmidt J., Hellenbroich Y., Hüning I., Gillessen-Kaesbach G., Zillikens D., Hartmann K. Identification of the recently described plasminogen gene mutation p.Lys330Glu in a family from Northern Germany with hereditary angioedema. Clin. Transl. Allergy. 2019;9:1–4. doi: 10.1186/s13601-019-0247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hedge M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponting C.P., Marshall J.M., Cederholm-Williams S.A. Plasminogen: A structural review. Blood Coagul. Fibrinolysis. 1992;3:605–614. doi: 10.1097/00001721-199210000-00012. [DOI] [PubMed] [Google Scholar]
- 24.Regner K.R., Riegert-Johnson D.L., Volcheck G.W. Serial measurement of serum tryptase in angiotensin-converting enzyme inhibitor-associated angioedema. Mayo Clin. Proc. 2003;78:655–656. doi: 10.4065/78.5.655-b. [DOI] [PubMed] [Google Scholar]
- 25.Fok J.S., Hissaria P., Giri P., Heddle R., Smith W. Acquired angioedema with raised serum tryptase. Ann. Allergy Asthma Immunol. 2013;110:59–60. doi: 10.1016/j.anai.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Ariano A., D’Apolito M., Bova M., Bellanti F., Loffredo S., D’Andrea G., Intrieri M., Petraroli A., Maffione A.B., Spadaro G., et al. A myoferlin gain-of-function variant associates with a new type of hereditary angioedema. Allergy. 2020 doi: 10.1111/all.14454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.