SLP76 is a key molecule involved in the T cell signaling pathway; therefore, SLP76 deficiency can lead to a severe type of immunodeficiency. By reporting on a patient with germline mutation in SLP76, deficiency in this gene is linked, for the first time, to a human disease.
Abstract
The T cell receptor (TCR) signaling pathway is an ensemble of numerous proteins that are crucial for an adequate immune response. Disruption of any protein involved in this pathway leads to severe immunodeficiency and unfavorable clinical outcomes. Here, we describe an infant with severe immunodeficiency who was found to have novel biallelic mutations in SLP76. SLP76 is a key protein involved in TCR signaling and in other hematopoietic pathways. Previous studies of this protein were performed using Jurkat-derived human leukemic T cell lines and SLP76-deficient mice. Our current study links this gene, for the first time, to a human immunodeficiency characterized by early-onset life-threatening infections, combined T and B cell immunodeficiency, severe neutrophil defects, and impaired platelet aggregation. Hereby, we characterized aspects of the patient's immune phenotype, modeled them with an SLP76-deficient Jurkat-derived T cell line, and rescued some consequences using ectopic expression of wild-type SLP76. Understanding human diseases due to SLP76 deficiency is helpful in explaining the mixed T cell and neutrophil defects, providing a guide for exploring human SLP76 biology.
Introduction
Studies of human immunodeficiencies have greatly increased our understanding of the fundamental functions of key proteins involved in the immune response (Fischer, 2007). Central aspects of leukocyte receptor signaling are disrupted in a wide array of disorders, leading to focal phenotypes both clinically and experimentally. In the case of SCIDs, specific disruptions of TCR signaling were found in a variety of settings that can be particularly informative (Cirillo et al., 2015; Notarangelo, 2013). Following the engagement of the TCR by the peptide/MHC, lymphocyte-specific protein tyrosine kinase (LCK) is activated and phosphorylates CD3 immunoreceptor tyrosine-based activation motifs (ITAMs). Phosphorylated ITAMs recruit and activate ζ-associated protein of 70 kD (ZAP-70). In turn, ZAP-70 phosphorylates two key adaptor molecules, linker of activated T cells (LAT; Balagopalan et al., 2015) and SLP76 (Jordan and Koretzky, 2010). SLP76 is recruited to membrane-bound phospho-LAT by Gads (Yablonski, 2019) and facilitates the activation of IL-2–inducible tyrosine kinase (ITK; Bogin et al., 2007). ITK-dependent phosphorylation of phospholipase C-γ1 (PLC-γ1) at the membrane leads to hydrolysis of phosphatidylinositol (4,5) diphosphate to inositol (1,4,5) trisphosphate and diacylglycerol as well as release of Ca2+ from endoplasmic reticulum stores. This results in massive Ca2+ influx, activation of Erk, and reorganization of the actin cytoskeleton, leading to cellular activation (Notarangelo, 2014). The aim of this process is to develop a fully activated T cell in a context-dependent manner that will lead to diverse outcomes, such as promotion of thymic selection, T cell differentiation, migration, and release of effector cytokines during acute responses.
Genetic defects causing severe immunodeficiencies have been reported in many of the proteins involved in the initiation of this cascade, including the CD3 subunits (CD3δ, CD3ε, and CD3ζ), CD45, ZAP-70, LCK, LAT, PLC-γ2, and ITK (Bacchelli et al., 2017; Dadi et al., 2003; Fischer et al., 2005; Hauck et al., 2012; Huck et al., 2009; Keller et al., 2016; Ombrello et al., 2012; Serwas et al., 2014). Deficiency of SLP76, a 76-kD protein (Jackman et al., 1995), has not yet been directly associated with a human disease. SLP76 is an adaptor protein that lacks intrinsic enzymatic activity but contains multiple protein-binding domains. It has three distinct domains: an NH2-terminal domain (amino acids 1–155), a central proline-rich domain (amino acids 156–421) that includes a Gads binding site (amino acids 224–244), and a C-terminal SH2 domain (amino acids 422–533; Kumar et al., 2002). Although SLP76 is expressed throughout hematopoietic compartments (Clements et al., 1998a), its function as a signal transducer downstream of the TCR was elucidated over the years, mainly using the SLP76-deficient Jurkat-derived human leukemic cell line, J14 (Yablonski et al., 1998). SLP76 is encoded in mice by the lymphocyte cytosolic protein 2 (Lcp2) gene. SLP76/Lcp2-deficient mice have provided considerable insights into the role of SLP76 in several hematopoietic lineages (Clements et al., 1998b; Pivniouk et al., 1998). While 60% of these mice die prenatally, the remaining have various T cell, neutrophil, mast cell, and platelet defects, as well as vascular development abnormalities (Clements et al., 1999; Newbrough et al., 2003; Pivniouk et al., 1998, 1999).
Here, we describe for the first time a severe human immunodeficiency due to biallelic mutations in SLP76. The disease is characterized by early onset of life-threatening infections, combined T and B cell immunodeficiency, impaired platelet aggregation, and a severe neutrophil defect.
Results and discussion
Clinical description
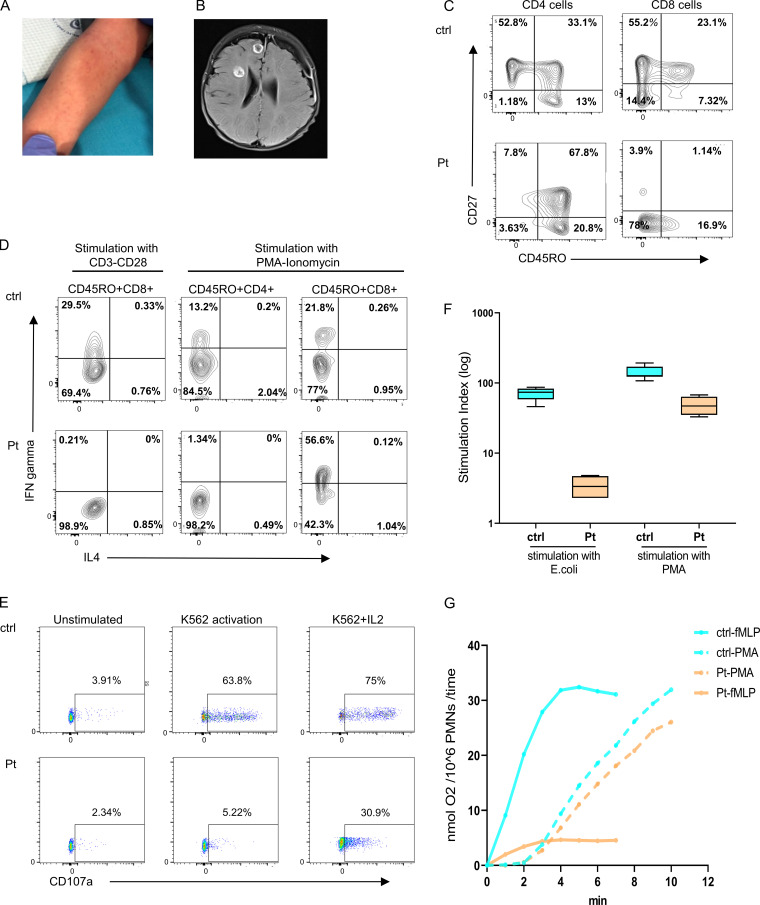
The index case is the first and only child born to first-cousin parents of Palestinian origin without any family history of immunodeficiency. Birth was at term, weight was 3.5 kg, and neonatal course was uneventful. He presented at the age of 2 mo with recurrent skin abscesses and skin rash. Initial workup at his hometown hospital revealed Coombs-positive hemolytic anemia that was treated successfully with systemic corticosteroids and blood transfusions. Over the next month, he failed to gain weight and developed respiratory distress, even when the initial infectious workup was negative. He was transferred to our care for further evaluation at 3 mo of age. On physical examination, the patient was cachectic and hypotonic, with enlarged spleen and liver but no other dysmorphic features. A petechial rash was seen on his skin (Fig. 1 A), the bacille Calmette-Guerin vaccine site was red and edematous, and a local lymph node was swollen. The patient developed a generalized seizure at 4 mo of age, and brain imaging revealed multiple abscesses (Fig. 1 B). These lesions were aspirated and were found to be positive for Aspergillus fumigatus. In addition, positive Aspergillus galactomannan antigen (1.73 units, negative < 0.01 units) and a high titer of CMV (631,255 cp/ml immediate early gene, negative < 1,000 cp/ml immediate early gene) were detected in his peripheral blood. During his admission, he was treated with broad-spectrum antibiotics, anti–bacille Calmette-Guerin medications (rifampin, ethambutol, and isoniazid), i.v. ganciclovir, trimethoprim/sulfamethoxazole, and voriconazole, in addition to receiving monthly intravenous immunoglobulin (i.v. Ig). At age 10 mo, the patient underwent haplo-identical hematopoietic stem cell transplantation using peripheral blood from his mother, following a conditioning regimen containing ATG, treosulfan, fludarabine, thiotepa, and MabThera. Unfortunately, the patient died of transplant-related complications 30 d after the procedure.
Figure 1.
Clinical and immunologic presentation of patient with SLP76 mutation. (A) Nonblanching, petechia-like, small lesions in the patient's forearm. (B) Brain magnetic resonance imaging revealed several round lesions with peripheral enhancement in temporal, frontal, and insular lobes with subdural collection in the peripheral part of the left frontal lobe. (C) Immunophenotyping of patient’s (Pt) CD4 and CD8 cells measured by flow cytometry. Left: The CD4 cells have a central memory phenotype (CD27+CD45RO+). Right: The CD8 cells have a TEMRA phenotype (CD27−CD45RO−) compared with age-matched healthy control (ctrl). The experiment was performed once. (D) PBMCs isolated from the patient and travel control were stimulated with either CD3 and CD28 overnight or with PMA and ionomycin for 5 h and stained for IFN-γ and IL-4. Shown are gated on CD45RO+CD4+ and CD45RO+CD8+ cells as measured via flow cytometry. This experiment was performed once. (E) NK cell degranulation in PBMCs from patients and age-matched healthy control was measured by the surface expression of CD107a without stimulation or after stimulation with K562 cells alone and with IL-2. One representative experiment out of two is shown. (F) Neutrophils oxidative burst, determined by flow cytometry analysis of dihydrorhodamine assay, in peripheral blood cells obtained from the patient and age-matched healthy control was measured after stimulation with E. coli bacteria or with PMA. Average data from four different experiments are shown. Statistical analysis was performed using unpaired one-tailed t tests. (G) Superoxide production in neutrophils from the patient and age-matched healthy control was measured as superoxide dismutase–inhibitable reduction of ferricytochrome c. (nmol O2/106PMNs/time) in response to stimulation with PMA or with fMLP. Dashed line indicates stimulation with PMA, and full line indicates stimulation with fMLP. One experiment with seven repeats.
Immune phenotype of the reported patient
Initial hematologic investigations revealed a normal complete blood cell count with white blood cells of 12.5 k/μl, neutrophil count of 5.67 k/μl, and lymphocyte count of 8.2 k/μl (Table 1). Platelet counts and coagulation studies were normal. Nevertheless, platelet aggregation was reduced, specifically in response to collagen (Table S1). IgG levels were not measured before first i.v. Ig infusion, subsequent IgM measurements were elevated, and lymphocyte immune phenotyping showed a high proportion of CD8 T cells and a very low proportion of CD4 T cells (Table 1). Transplacental-acquired maternal T cells were excluded using fluorescence in situ hybridization analysis of the X and Y chromosome probes. The patient’s residual CD4 T cells had abnormally diminished, naive phenotypes, skewing toward a predominantly central memory phenotype (CD45RO+CD27+CCR7+). In contrast, the patient’s CD8 T cell phenotype was most consistent with terminally differentiated T-effector memory RA cells (TEMRA) phenotype (CD45RO−CD27−CCR7−; Fig. 1 C and Fig. S1 A). Thymic activity, as determined by TCR excision circles quantification, was low (Table 1). Whereas there was an increase in the percentage of CD25+T cells, possibly due to enhanced peripheral activation, CD25+ FOXP3+ expression (regulatory T cells) appeared relatively intact (Table 1). Peripheral lymphocyte proliferation, as determined by either thymidine incorporation or CFSE, was absent in response to PHA or anti-CD3, with or without anti-CD28 (Table S2 and Fig. S1 B). Interestingly, exogenous IL-2 partially restored anti-CD3 and anti-CD28 responses, resulting in increased cell proliferation (Table S2), suggesting a possible defect in TCR/CD28-induced IL-2 production. Cytokine production upon ex vivo stimulation with anti-CD3/CD28 was absent (Fig. 1 D). In contrast, PMA/ionomycin stimulation showed reduced, but not absent, IFN-γ production and no substantial Th2 cytokine production within CD4 T cells. In addition, elevated IFN-γ production was demonstrated among CD8 T cells (Fig. 1 D), as expected, given the preponderance of TEMRA cells (Hamann et al., 1997). Taken together, this suggests a defect downstream of the TCR but upstream of PLC-γ1 (Yablonski et al., 1998). In expanded T cell lymphoblasts, PMA/ionomycin-induced TNFα and IL-2 production within CD4 T cells appeared to be similar to that of the control, but IFN-γ production was reduced (Fig. S1 C). Total B cell counts were within normal ranges. Nevertheless, B cell immune phenotype revealed reduced class switched memory B cells (defined as IgM−IgD−CD19+CD27+) and transitional B cells (IgM2+CD19+CD382+), along with elevated naive B cells (defined as IgD+CD27−CD19+) and immature B cells (defined as IgD+CD38+CD19+; Fig. S1 D). Total natural killer (NK) cell numbers were also within normal range, but K562-induced degranulation was diminished, as reflected in degranulation-associated surface expression of CD107a, and partially rescued in response to IL-2 (Fig. 1 E).
Table 1. Immunologic workup of the SLP76-deficient patient.
| Patient | Normal rangea | |
|---|---|---|
| CBC (cells/ml × 103) | ||
| WBC | 12.5 | 5.2–11 |
| Neutrophils | 5.6 | 1–8.5 |
| Monocytes | 0.2 | 0.2–1 |
| Lymphocytes | 8.2 | 3.4–7.6 |
| Eosinophils | 0.01 | 0–0.7 |
| Hemoglobin (g/dl) | 9.6 | 11–14 |
| Platelets | 243 | 150–400 |
| Lymphocyte subsets (cells/ml × 103) | ||
| T cells (CD3+) | 7.6 | 2.5–5.5 |
| T helper cell (CD4+) | 0.65 | 1.6–4 |
| Cytotoxic T cell (CD8+) | 6.65 | 0.6–1.7 |
| Double negative (CD4−CD8−αβ+; % of WBC) | 0.4% | <1% |
| αβ TCR | 57% | 26–100% |
| γδ TCR | 2% | 1–38% |
| Regulatory T cell (CD4+CD25+FOXP3+) | 4% | >5% |
| B cells (CD45+CD19+) | 0.36 (6%) | 0.05–0.3 (5–25%) |
| NK (CD16+CD56+) | 36% | 6–30% |
| TREC (copies per 0.5 mg DNA) | 36 | >400 |
| Serum Ig (mg/dl) | ||
| IgA | 36 | 38-222 |
| IgM | 434 | 56-208 |
| IgG | 1,220 | 590-1,430 |
CBC, complete blood count; TREC, TCR excision circles; WBC, white blood cell.
aHealthy donors, age 1–2 yr, with percentages/counts presented as median (10th and 90th percentiles).
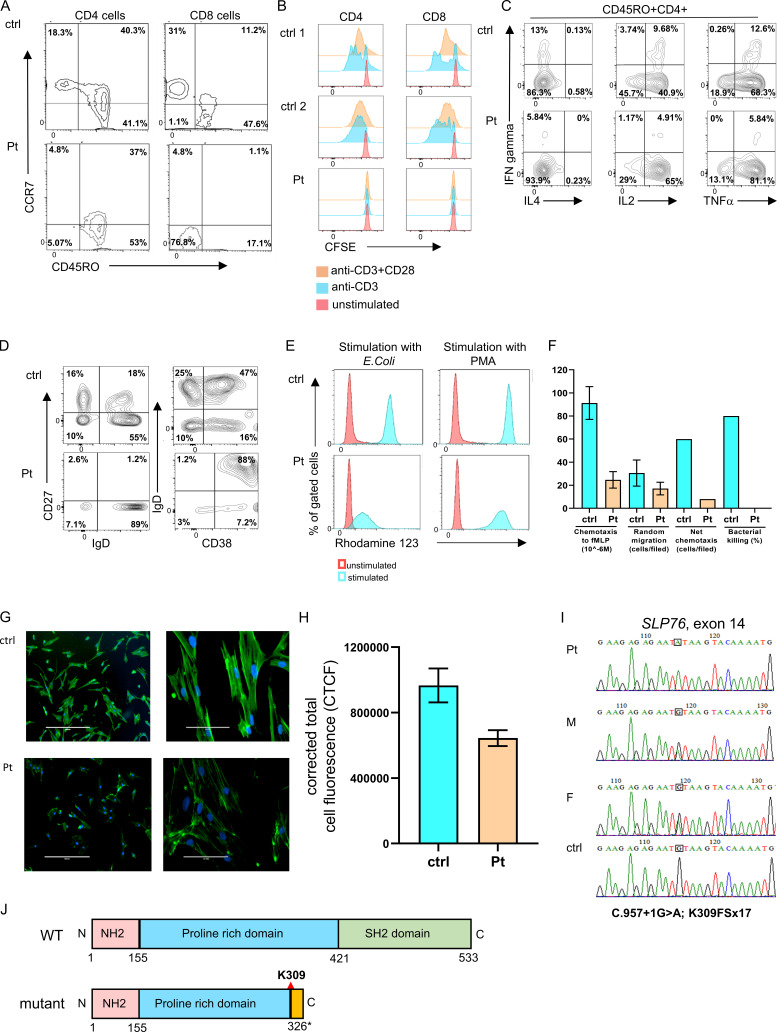
Figure S1.
Immune phenotype and genetic characterization of the SLP76-deficient patient. (A) Immunophenotyping of patient’s CD4 and CD8 cells measured by flow cytometry. The CD4 cells have a central memory phenotype (CCR7+CD45RO+; left), and the CD8 cells have a TEMRA phenotype (CCR7−CD45RO−; right) compared with age-matched healthy control (ctrl). The experiment was performed once. (B) T cell proliferation: T cell proliferation using CFSE fluorescence cell incorporation assay 4 d after stimulation with anti-CD3 or anti-CD3/CD28. These data show reduced proliferation in the patient CD4 and CD8 cells compared with the two healthy controls. The experiment was performed once. (C) Flow cytometric analysis of cytokine response: Expanded T cells lymphoblasts were stimulated with plate-bound OKT3 (10 μg/ml) and soluble CD28 (2 μg/ml) for 12 h., then 20 ng/ml PMA and 1 μmol/liter ionomycin in the presence of brefeldin A for 5 h. IFN-γ, IL-4, IL-2, and TNFα production among CD45RO+CD4+ cells was assessed via flow cytometry. One representative experiment out of two is shown. (D) Peripheral B cell immunophenotyping:B cell immunophenotyping measured by flow cytometry revealed decreased frequencies of naive (CD27−IgD+) and class-switched (CD27+IgD−) B cells, along with elevated immature B cells (CD38+IgD+) in the patient pheripheral blood lymphocytes compared with control. One representative experiment out of two is shown. (E) Neutrophils oxidative burst: Dihydrorhodamine assay was performed in peripheral blood from the patient and age-matched healthy controls after stimulation with E. coli bacteria or PMA using flow cytometry. One representative experiment out of four is presented. (F) Neutrophils function: Neutrophil chemotaxis, random migration, net chemotaxis, and bacterial killing were markedly reduced in the patient’s neutrophils compared with age-matched healthy controls. For the chemotaxis and random migration experiments, each bar represents the average of two experiments, and for the net chemotaxis and bacterial killing assays, the bars represent data from one experiment. Statistical analyses were performed using unpaired one-tailed t test. (G) Fluorescence studies of actin polymerization: The actin polymerization in fibroblasts from the patient and control was determined using phalloidin staining. Fluorescent microscopy images of FITC phalloidin staining demonstrate clear fluorescence attenuation in the patient’s cells compared with control. Phalloidin stains green and the nuclei (DAPI) stain blue. Images of ×10 magnification (left) and ×40 magnification (right) are shown. One representative experiment out of three is shown. (H) Quantification of the corrected total cell fluorescence (CTCF) in the patient and control fibroblasts using ImageJ software. Results are the average of three replicates, with error bars indicating the SD. (I) Genetic analysis of SLP76 mutation: Sanger sequencing confirmed the presence of a donor splice site mutation in the patient, which fully segregated with the parents (M, mother; F, father). The mutated nucleotide is boxed. A normal sequence of a healthy control is also presented. (J) SLP76 domains: Schematic representation of WT and mutant SLP76 protein domains. The splicing mutation causes a putative frame shift following lysine 309 (K309) in the central proline-rich domain (blue) and a premature stop codon 17 amino acids downstream of the mutation (yellow). This results in a deletion of the C-terminal SH2 domain (green).
The fungal brain abscess led to a concern for neutrophil dysfunction. Interestingly, dihydrorhodamine analysis showed a markedly reduced response to Escherichia coli stimulation, whereas neutrophil function was only slightly reduced in response to PMA (Fig. 1 F and Fig. S1 E). In addition, neutrophil superoxide production was absent in response to N-formylmethionyl-leucyl-phenylalanine (fMLP), while PMA-mediated induction was near normal (Fig. 1 G). Moreover, neutrophil chemotaxis, random migration, net chemotaxis, and bacterial killing were markedly reduced in cells obtained from the patient compared with a healthy control (Fig. S1 F). These results suggest a substantial defect in proximal neutrophil signaling, which may be partially bypassed by PMA stimulation.
Finally, we used phalloidin, a bicyclic peptide that selectively binds to and stabilizes actin filaments, to stain polymerized actin in fibroblasts obtained from the patient and a healthy control. Reduced fluorescence intensity was observed in the patient cells compared with the control, suggesting inhibition of cytoskeletal assembly (Fig. S1, G and H).
Taken together, the patient displayed a clinical and immunologic phenotype suggestive of combined immunodeficiency, impaired very proximal T cell and neutrophil signaling, abnormal platelet function, and reduced actin polymerization.
Genetic evaluation
Given parental consanguinity, autosomal recessive disease was suspected. Whole-exome sequencing revealed 7,489 homozygous variants. This list of variants was subsequently reduced to 37 rare variants by filtering out variants present in ≥0.01 of our in-house exomes (n = 3,797) and present with a minor allele frequency of ≥0.01 in gnomAD (https://gnomad.broadinstitute.org/), the 1000 Genomes Project (https://www.internationalgenome.org/home) or dbSNP 135 database, or the National Heart, Lung, and Blood Institute Exome Sequencing Project (http://evs.gs.washington.edu/EVS/). The only variant among these candidates that was predicted to be pathogenic and could significantly affect the immune system was a novel homozygous mutation C.957+1G>A; K309FSx17 in the SLP76 gene that affects a donor splice site at the beginning of intron 14 (Fig. 2 A). The combined annotation-dependent depletion (CADD) score of the mutation was 33, suggesting pathogenicity. Whole-exome sequencing did not reveal any other mutations known to be involved in immunodeficiency or immune dysregulation. The mutation was confirmed in the patient using direct Sanger sequencing, and family segregation has found that both parents are heterozygote carriers (Fig. S1 I). Furthermore, cDNA was prepared from the patient, his parents, and controls cells, followed by PCR using primers from exons 13 and 16. The resulting smaller fragment in the patient's cDNA, compared with control, suggested an exon skipping due to the homozygote splice mutation (Fig. 2 B). Indeed, Sanger sequencing of both fragments revealed a skipping of exon 14, resulting in a putative frame shift following lysine 309 and a premature stop codon 17 amino acids downstream. The mutation is located in the central proline-rich domain and results in the deletion of the C-terminal SH2 domain (Fig. S1 J).
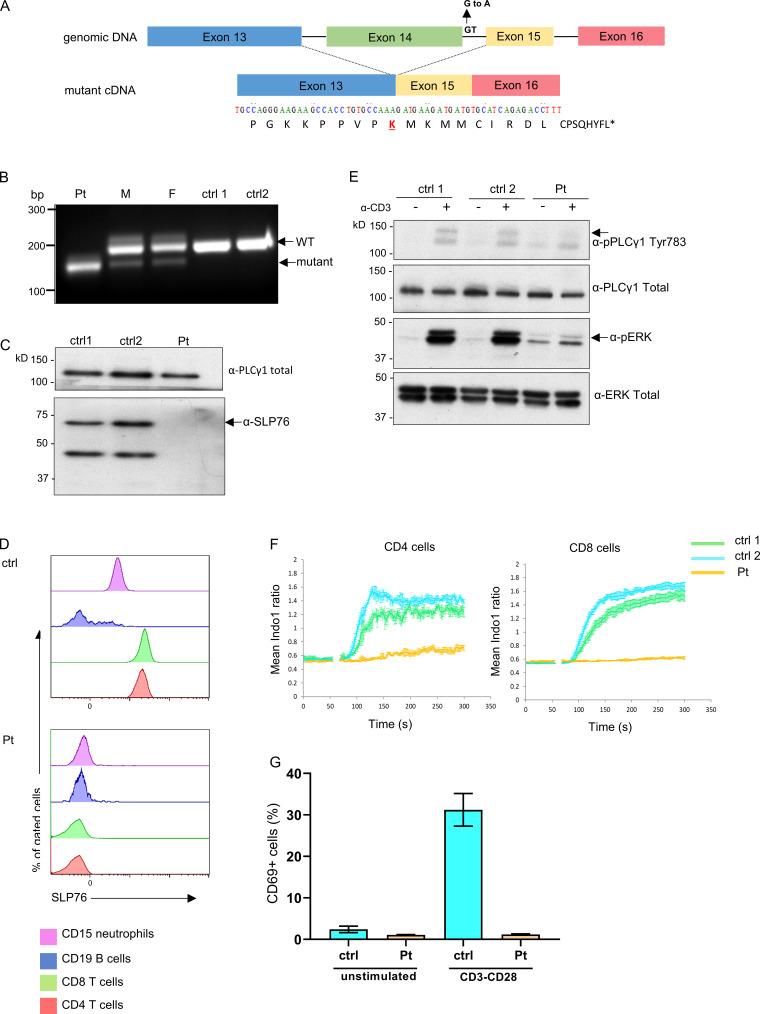
Figure 2.
Genetic, cellular, and functional characterization of SLP76 mutation. (A) Top: Schematic presentation of genomic DNA of SLP76 where the splice site mutation at the beginning of intron 14 is shown. Bottom: The mutated form of the cDNA is presented, where the skipping of exon 14 results in a putative frame shift after lysine (K) 309 of SLP76 (marked in red). The putative 17 amino acids and premature termination codon (asterisk) of the shifted sequence are shown. (B) PCR was performed on cDNA obtained from the patient, his parents, and two healthy controls (ctrl) using primers from exons 13 and 16 of SLP76. A single smaller fragment (190 bp), which suggests an exon skipping due to the splice mutation, is seen in the patient’s cells compared with a single higher band (221 bp) in the anticipated length in the healthy controls. The mother (M) and father (F) show both bands, as can be expected with the two being heterozygote carriers to the mutation. One representative gel out of three is shown. (C) Western blot analysis of SLP76 expression in expanded T cell lymphoblasts from the patient and two age-matched healthy controls. Total PLC-γ1 expression serves as a control for protein loading. One representative blot out of three is shown. (D) Flow-cytometric analysis determines the expression of SLP76 in T cells (CD4 and CD8), B cells (CD20), and neutrophils (CD15) from healthy control (top) and the patient (bottom). One representative experiment out of three is shown. (E) Western blot analysis of phosphorylated PLC-γ1 and phosphorylated ERK1/2 with (+) or without (−) stimulation with anti-CD3 in expanded T cell lymphoblasts from the patient and two age-matched healthy controls. Total PLC-γ1 and total ERK1/2 were used as a loading control for the patient and two age-matched healthy controls, respectively. One representative blot out of two is shown. (F) Expanded T cell lymphoblasts from the patient and two age-matched healthy controls were barcoded, mixed together, loaded with indo-1-AM, and stained for CD4 and CD8. Intracellular Ca2+ concentration was measured using flow cytometry at 37°C, with cross-linked anti-CD3 added at the 60-s time point. Mean ratiometric Ca2+ measurements are presented separately for the CD4+ (left) and CD8+ (right) populations. Results are the average of three replicates, with error bars indicating the SD. One representative experiment out of two is shown. (G) PBMCs from the patient and healthy age-matched controls were either stimulated with anti-CD3/CD28 or left unstimulated. CD69 expression was measured using flow cytometry and the results of three different analyses are summarized in the graph. Statistical analysis was performed using unpaired one-tailed t tests.
The biologic effect and signaling capacity of the naturally occurring novel SLP76 mutation
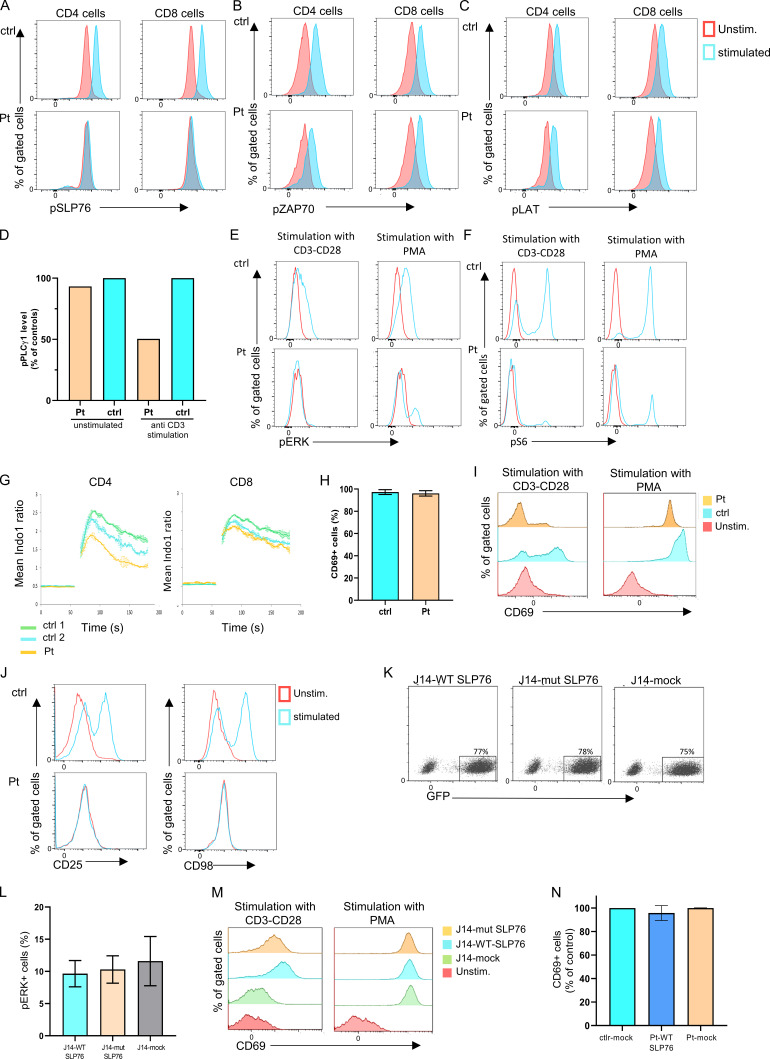
Complete loss of SLP76 protein expression was detected by both Western blot (Fig. 2 C) and flow cytometry analyses (Fig. 2 D) in peripheral blood mononuclear cells (PBMCs). Next, we addressed the effect of the mutated SLP76 on TCR signaling in primary T cells of the patient after stimulation with anti-CD3. As expected, phosphorylated SLP76 was not detected in either CD4 or CD8 patient’s cells following stimulation (Fig. S2 A), confirming loss of function of the SLP76 protein. The phosphorylation of upstream molecules, including ZAP-70 and LAT, was not affected in the patient’s cells (Fig. S2, B and C). In contrast, downstream events, including the phosphorylation of ERK, PLC-γ1, and phospho-S6, all of which are dependent on the assembly of the LAT and SLP76 complex, were significantly reduced or absent (Fig. 2 E and Fig. S2, D–F). Stimulation with PMA, which bypasses LAT and SLP76 signaling, led to modestly improved activation of Erk and phospho-S6 (Fig. S2, E and F). The basal pERK noted is likely an artifact of non–TCR-mediated cellular activation by mitogens in the presence of clonal proliferation and/or activation of other cell types that do not express or require SLP76. In addition, Ca2+ mobilization in CD4+ or CD8+ T lymphoblast cells from the patient was completely absent following stimulation with anti-CD3, while ionomycin stimulation led to normal response (Fig. 2 F and Fig. S2 G). Similarly, anti–CD3/CD28-mediated activation led to poor upregulation of surface activation markers of CD69, CD25, and CD98 in the patient’s T cells, with relatively normal response to PMA (Fig. 2 G and Fig. S2, H–J).
Figure S2.
The biologic effect on the TCR signaling due to the SLP76 mutation. (A-C) TCR-induced signaling in SLP76 mutated cells: Flow cytometry analyses of intracellular levels of phosphorylated SLP76 (A), phosphorylated ZAP-70 (B), and phosphorylated LAT (C) in unstimulated and stimulated peripheral CD4 and CD8 T cells from the patient and control are shown. No phosphorylation of SLP76 was detected in the patient's cells, and the phosphorylation of ZAP-70 and LAT was normal compared with control. One representative experiment out of two is shown. (D) Quantification of the level of phospho-PLC-γ1. The level of phospho-PLC-γ1 in the patient and control cells as presented in the Western blot (Fig. 2 E) was quantified by densitometry analysis using ImageJ software. The level of phospho-PLC-γ1 in the patient cells with or without anti-CD3 stimulation was calculated as the percentage from control. The bars for the control are the average of two controls (ctrl 1 and ctrl 2), and it is set to 100%. Reduction in the patient’s phospho-PLC-γ1 level was detected. (E and F) TCR signaling in SLP76-mutated T cells: Phosphorylation of pERK1/2 (E) and pS6 (F) in patient and control T cells, either without stimulation or after stimulation with anti-CD3/CD28 or with PMA, was measured using flow cytometry. Stimulation with anti-CD3/CD28 did not lead to phosphorylation in pERK1/2 and pS6 in the patient’s cells, while PMA stimulation led to a modestly improved response comparable to control. One representative experiment out of two is shown. (G) Ca2+ mobilization: Expanded T cell lymphoblasts from the patient and age-matched healthy controls were loaded with indo-1-AM. Ca2+ concentration was measured following stimulation with ionomycin. Ca2+ mobilization was determined in CD4 and CD8 T cells. Results are the average of two replicates, with error bars indicating the SD. One representative experiment out of two is shown. (H-J) Expression of surface activation markers. (H) PBMCs from the patient and age-matched healthy controls were stimulated with PMA. CD69 expression was measured using flow cytometry and the results of three different analyses are summarized in the graph (P value = NS). Statistical analysis was performed using unpaired one-tailed t tests. (I) The actual flow cytometry results of CD69 expression in unstimulated and stimulated PBMCs from the patient and control are presented. One representative experiment out of four is presented. (J) Flow cytometric expression of CD25 and CD98 in unstimulated and stimulated T cells from the patient and age-matched control reveals lack of upregulation of these markers in the patient cells compared with control. One representative experiment out of two is shown. (K) Flow cytometric analysis showing equal amounts of GFP+ cells in J14-transduced experiments. J14 cells were transduced with an IRES-GFP-tagged retroviral vector encoding FLAG-tagged WT or mutant SLP76, or with the mock vector. All showed similar levels of transduction rate determined by GFP+ cells. (L and M) Correction of the SLP76 mutant phenotype in J14. (L) SLP76-deficient Jurkat-derived human leukemic T cell line (J14) retrovirally transduced with the WT or mutant form of SLP76, or with the empty mock vector. Phosphorylation of ERK1/2 was measured using flow cytometry after PMA stimulation. The stimulation with PMA, which bypasses SLP76 signaling, causes a normal response in all cells (P value = NS). The results of three different analyses are summarized in the graph. Statistical analysis was performed using unpaired one-tailed t tests. (M) Flow cytometry measurements of CD69 expression in unstimulated and stimulated J14 cells retrovirally transduced with empty vector, WT, or the mutant form of SLP76. One representative experiment out of four is presented. (N) Correction of the SLP76 mutant phenotype in expanded T cell lymphoblasts. Expanded T cell lymphoblasts from the patient were retrovirally transduced with either WT SLP76 or empty mock vector tagged with GFP. Expanded T cells from age-matched healthy controls were retrovirally transduced only with empty mock vector tagged with GFP. These cells were stimulated with PMA, and the CD69 expression was measured on GFP+ cells using flow cytometry. For the control, the bar represents the average of two experiments, and for the patient, the bars represent data from two experiments, which include four independent samples. Stimulation with PMA revealed normal CD69 expression in all the expanded cells.
Study of mutation in SLP76-deficient Jurkat cells (J14)
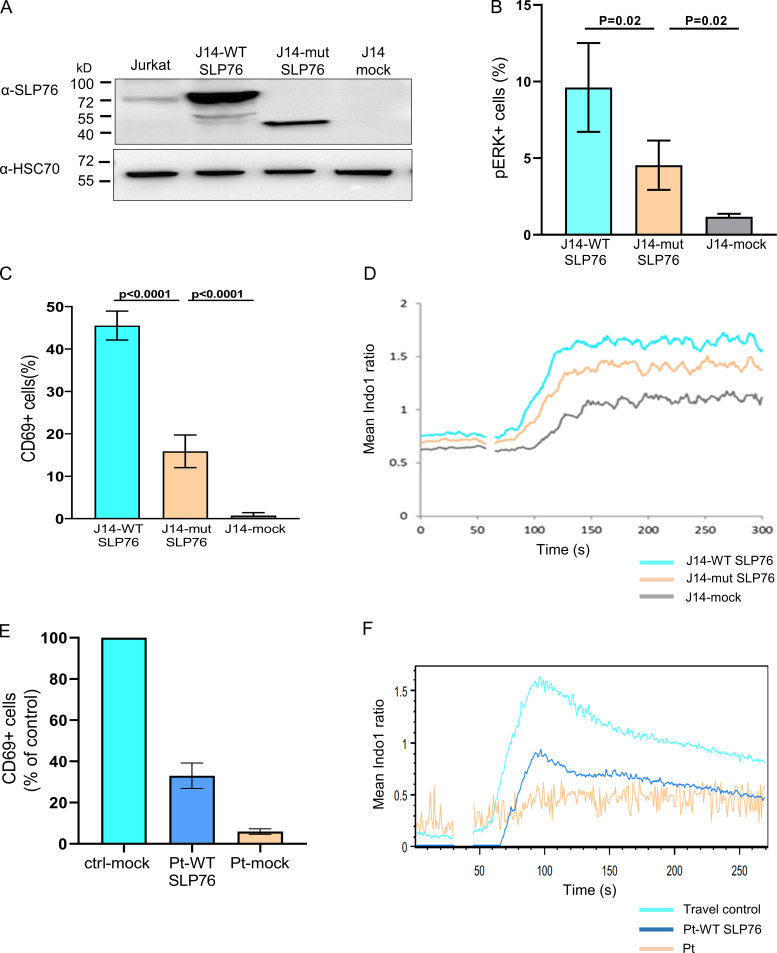
To examine the downstream effects of the specific SLP76 mutation found in our patient, we decided to model the mutation and examine some of its effects in a SLP76-deficient Jurkat-derived cell line (J14). J14 cells were transduced with an internal ribosome entry site (IRES)-GFP–tagged retroviral vector encoding FLAG-tagged WT or mutant SLP76, or with the mock vector, which all showed similar levels of transduction rate determined by GFP+ cells (75%; Fig. S2 K). Consistent with the frameshift mutation following K309, a truncated form of SLP76 was detected by Western blot analysis in the mutant reconstituted cells, whereas WT SLP76 reconstituted cells expressed full-length SLP76 (Fig. 3 A). Moreover, lower expression levels of the truncated SLP76 protein were detected, suggesting an unstable protein. The absence of mutant protein in primary cells, despite the presence in transduced Jurkat cells, likely reflects a combination of effects. First, the use of fixed promoter-driven cDNA expression in the transduction system. Second, the patient’s peripheral blood was overwhelmingly composed of clonally expanded, terminally differentiated T cells, a population that has diminished levels of SLP76 (Hussain et al., 2002).
Figure 3.
Modeling and correction of the SLP76 mutant phenotype. SLP76-deficient Jurkat-derived human leukemic T cell line (J14) retrovirally transduced with the WT or mutant form of SLP76 or with the empty mock vector. (A) Western blot analysis to determine SLP76 expression. The truncated SLP76 is expected to be 37 kD. The expression of SLP76 in the common Jurkat cell line serves as a reference control, and HSC70 was used as a loading control. One representative blot out of three is shown. (B) Phosphorylated ERK1/2 was measured using flow cytometry in either anti-TCR (C305) stimulated cells or unstimulated cells. The analysis was done only on transduced cells by gating on GFP+ cells. The results of three different analyses are summarized in the graph. Statistical analysis was performed using unpaired one-tailed t tests. (C) CD69 expression was measured using flow cytometry in either anti-TCR (C305) stimulated cells or unstimulated cells. The analysis was done only on transduced cells by gating on GFP+ cells. The results of four different analyses are summarized in the graph. Statistical analysis was performed using unpaired one-tailed t tests. (D) Reconstituted J14 cells were barcoded, mixed together, and loaded with indo-1-AM. Shown is the mean ratiometric Ca2+ measurement, with anti-TCR (C305) added at the 60-s time point. Results are the average of three replicates. One representative experiment out of two is shown. (E) Expanded T cell lymphoblasts from the patient were retrovirally transduced with either WT SLP76 or empty mock vector tagged with GFP. Expanded T cell lymphoblasts from two-age matched healthy controls were retrovirally transduced with empty mock vector tagged with GFP. Both cells from the patient and controls were either stimulated with anti-CD3/CD28 or left unstimulated, and CD69 expression was measured on GFP+ cells using flow cytometry. The result of CD69 expression in the patient’s cells that were reconstituted with WT SLP76 is shown as a percentage from healthy control (ctrl-mock), where the healthy control is set for 100%. For the control, each bar represents the average of three experiments, and for the patient, the bars represent data from two experiments, which include four independent samples. Statistical analyses were performed using unpaired one-tailed t test. (F) WT SLP76–transfected patient T cells, patient T cells, and travel control T cells were loaded with indo-1-AM. Ca2+ influx in response to anti-CD3 stimulation was determined by the ratio of the fluorescent signals at 405 nm (Ca2+-bound dye) to 485 nm (Ca2+-free dye) over time via flow cytometry. The experiment was performed once.
We also analyzed the downstream effects of the SLP76 mutation. These analyses were performed while gating on GFP+ cells, which are the cells expressing WT or mutant SLP76. ERK phosphorylation (Fig. 3 B and Fig. S2 L), CD69 expression (Fig. 3 C and Fig. S2 M), and Ca2+ mobilization (Fig. 3 D) were moderately reduced in the mutant SLP76-expressing Jurkat cells, and this reduction was statistically significant.
Reconstitution of patient’s cells with WT SLP76
To further link the specific SLP76 mutation to the immune phenotype and the abnormal signaling capacity, the patient's primary T lymphoblasts were retrovirally transduced with WT SLP76 or with empty vector. Following stimulation with anti-CD3/CD28, the patient’s cells that were reconstituted with WT SLP76 and gated on GFP+ cell showed fivefold increased CD69 expression compared with vector-reconstituted patient cells (mock). Furthermore, compared with healthy control cells (ctrl-mock), there was a 33% rescue (percentage of healthy control cells; Fig. 3 E). In addition, WT SLP76 partially rescued the Ca2+ influx in the patient’s T cells upon stimulation with anti-CD3/CD28 (Fig. 3 F). As previously shown with the patient’s PBMCs, stimulation with PMA revealed normal CD69 expression in the patient’s T lymphoblasts, even without the introduction of WT SLP76 (Fig. S2 N).
Effect of SLP76 deficiency on T and B cell receptor repertoire
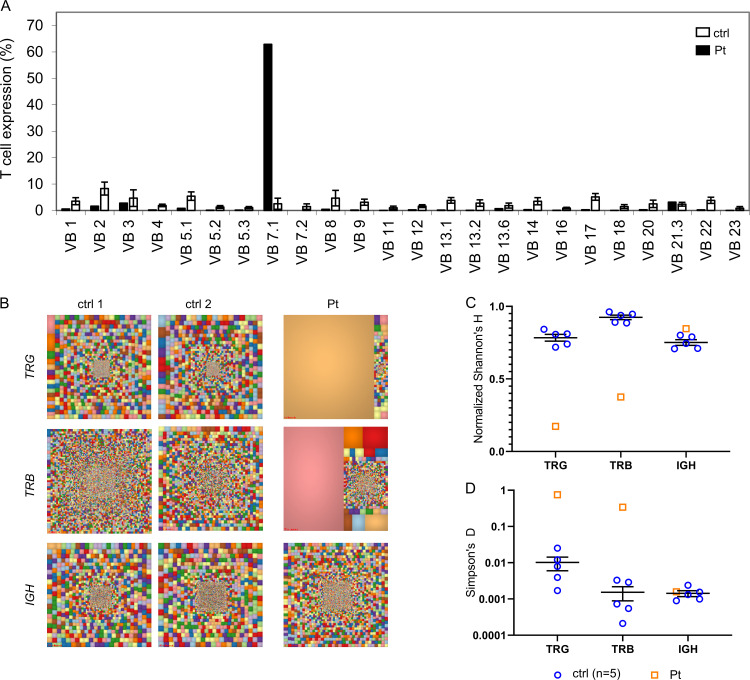
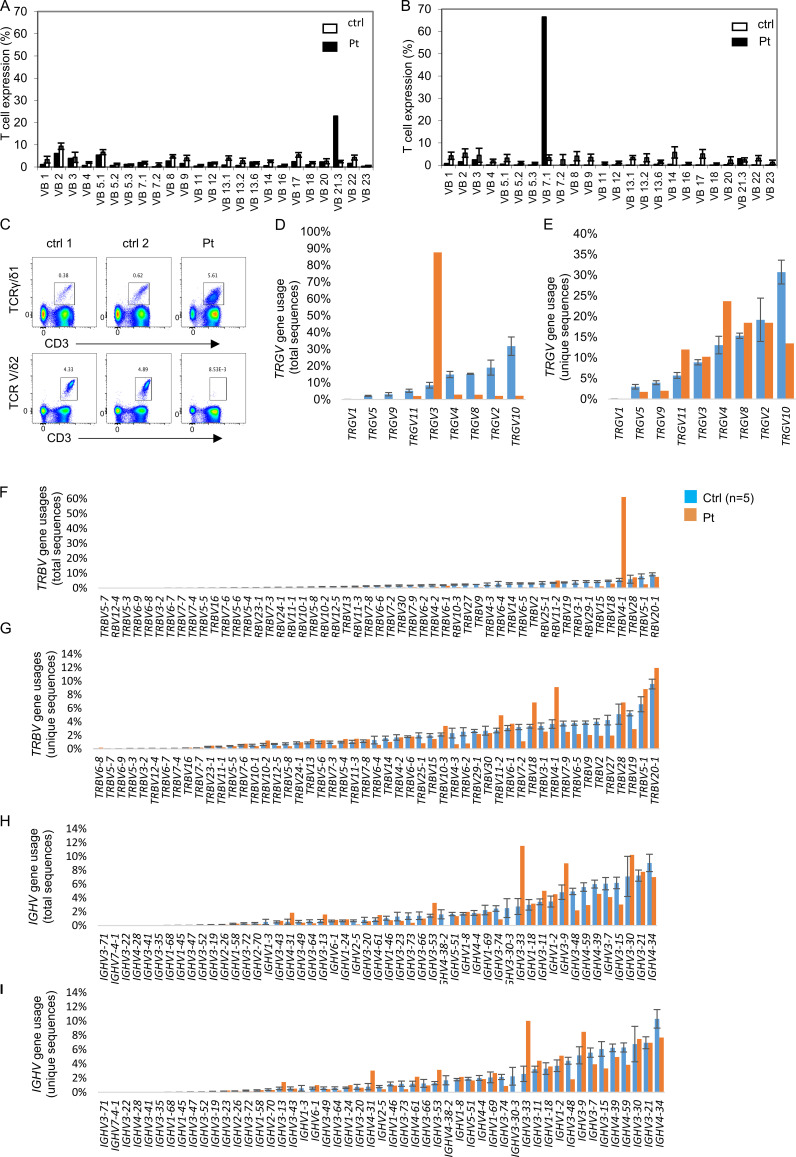
To assess the impact of SLP76 deficiency on TCR diversity, we first analyzed the patient’s cells for the repertoire of Vβ usage using flow cytometry (Fig. 4 A). The patient’s TCR repertoire showed clonal expansion, with a skewing toward Vβ7.1, particularly in CD8 cells (Fig. S3, A and B). Analysis of γδ T cells showed a marked skewing toward Vδ-1 and away from Vδ-2 (Fig. S3 C). In addition, we analyzed TCR γ (TRG) and TCR β (TRB) in T cells and Ig heavy (IGH) chain on B cells using next-generation sequencing technology. Graphical presentation of the sampled immune repertoire treemaps showed decreased diversity and prominent clonal expansions in TRG and TRB, but not in IGH (Fig. 4 B). Calculation of normalized Shannon’s H, which measures the overall diversity of a given repertoire, clearly showed that the patient’s TRG and TRB repertoires are decreased compared with control, whereas the normalized Shannon’s H for the patient’s IGH repertoire was comparable to control (Fig. 4 C). In addition, we calculated and measured the degree of clonal expansions in TRG, TRB, and IGH repertoires using Simpson’s D. The Simpson’s D indices for the patient’s TRG and TRB repertoires were higher compared with control, whereas the patient’s Simpson’s index for the IGH repertoire was comparable to control (Fig. 4 D). Thus, the TRG and TRB repertoires of the patient demonstrated restricted repertoires with dramatic clonal expansions, whereas the diversity of the patient’s IGH repertoire was comparable to control repertoires.
Figure 4.
Immune repertoire determined by next-generation sequencing for the SLP76-mutated patient. (A) Flow cytometry analysis of surface membrane expression of 24 different TRB variable gene families in the patient’s CD3 cells (black bars) compared with healthy controls (ctrl; white bars, n = 85, provided by the kit). One representative result out of two is shown. (B) Treemap representation of TRG, TRB, and B cell receptor (IGH) repertoire in PBMCs from the patient and two age-matched healthy controls. Each square represents a unique V to J joining, and the size of the square represents relative frequency within that sample. Two representative controls out of four are shown. (C and D) Quantification of the diversity and unevenness of the TRG, TRB, and IGH repertoire using the Shannon’s H index of diversity (C) and the Simpson’s D index of unevenness (D) in five healthy controls and in the SLP76-mutated patient.
Figure S3.
T and B cell receptor repertoire in the SLP76-deficient patient. (A and B) Flow cytometry analysis of TCRVβ repertoire: Flow cytometry analysis of surface membrane expression of 24 different TRB variable gene families in the patient’s CD4 cells (A) or CD8 cells (B; black bars) compared with healthy control (white bars, n = 85, provided by the kit). The nomenclature of the TCR V genes is based on Wei et al. (1994). (C) Analysis of γδ T cells: Flow cytometry analysis of CD3+TCRγδ1+ and CD3+TCRVδ2+ cells. Analysis of the γδ T cells showed a marked skewing toward VD-1 and away from VD-2. Results are expressed as the percentage of positive cells. (D–I) Differential V gene uses in TRG, TRB, and IGH repertoires. Percentage of different V gene usages for the TRG (D and E), TRB (F and G), and IGH (H and I) repertoires is demonstrated in unique and total sequences. For the patient, each bar represents the percentage of specific gene use based on either total (D, F, and H) or unique (E, G, and I) sequences. For the control, the bar represents the average of gene uses (n = 5), and error bars represent the SE. The nomenclature of the V genes is based on IMGT (Lefranc et al., 1999).
Next, we analyzed for preferential V gene uses in TRG, TRB, and IGH repertoires on both total and unique sequence datasets. The preferential utilization of TRGV3 and TRBV4-1 gene segments in the patient was observed only in the total sequence dataset due to clonal expansions (Fig. S3, D and E). TRBV4-1 (IMGT nomenclature, Lefranc et al., 1999) corresponds to Vβ7.1 (Wei et al. nomenclature, Wei et al., 1994), which showed prominent clonal expansion by the TCRVβ repertoire (Fig. S3 B). Although the diversity of the IGH repertoire of the patient was comparable to control, surprisingly, IGHV gene uses in the patient were not comparable to control (Fig. S3, F–I), with robust preferential utilization of IGHV3-9 and IGHV3-33, both in unique and total sequence datasets. Taken together, the overall diversity of the TRG and TRB repertoires is clonally expanded, yet without major gene preference during the primary process of VDJ recombination and selection, but with preferential TRGV and TRBV gene uses in the periphery. The diversity of the IGH repertoire was not affected with major IGHV gene preferences in the peripheral blood of the patient with SLP76 deficiency.
Defective pre-TCR/TCR signaling underlies some of the most severe types of T cell immunodeficiencies (Notarangelo, 2014). Here, we provide evidence for the consequences of a potentially hypomorphic loss of SLP76 function in humans, which leads to T cell immune deficiency, with neutrophil and platelet defects as well.
In the SLP76-deficient mouse model, thymocyte development was blocked at the double-negative 3 stage, resulting in the absence of peripheral T cells. In contrast, our patient had peripheral T cells, albeit skewed toward expanded memory and CD8 TEMRA cells with an abnormal TCR repertoire. We assume that our patient did not completely lack the entire SLP76 protein and thus may have expressed either an unstable truncated protein lacking the C terminus as a result of the frameshift mutation at residue 309, or a low level of the full-length WT protein due to a small degree of leakiness of the splicing mutation. Both alternatives may explain the presence of peripheral T cells, though the ability to measure the mutant SLP76 is hampered by the lack of typical naive or memory T cells in the patient’s periphery (Hussain et al., 2002). Consistent with the first option, TCR signaling could be partially reconstituted in the SLP76-deficient J14 cell line upon introduction of a C-terminally deleted SLP76 construct, lacking all residues after 314 (Yablonski et al., 2001). Furthermore, homozygous splice mutants of Lcp2 in mice that reduced the quantity of WT SLP76 protein by 90% displayed only a partial block in thymocyte development and enabled the presence of peripheral, dysregulated T cells, which, similar to those of our patient, had a skewed expansion of CD8 memory cells and increased IFN-γ production (Siggs et al., 2015). SLP76 has a role in pre–B cell receptor signaling at the pre–B cell stage as well (Su and Jumaa, 2003), thus explaining our observation regarding the elevation in immature B cells seen in the periphery of the patient. SLP76-deficient neutrophils have shown decreased Fcγ receptor-dependent and adhesion-dependent production of reactive oxygen intermediates and decreased integrin-dependent spreading (Newbrough et al., 2003). In addition, SLP76 deficiency was shown to affect actin dynamics, as determined by reduced phalloidin staining (Wood et al., 2006). These neutrophil and actin phenotypes were similarly observed in our patient. Our patient developed fungal brain abscesses, likely due to a specific defect in neutrophil function, as well as viremia typically associated with T cell defects. The bleeding tendency that was observed in our patient is probably due to decreased collagen-induced platelet aggregation and granule release, which was previously suggested to be a feature of SLP76 deficiency in mice (Clements et al., 1999). It is known that SLP76 is required for signaling downstream of glycoprotein VI, which is an ITAM-bearing collagen receptor expressed by platelets (Gross et al., 1999).
LAT and ITK are close binding and signaling partners of SLP76 in T cells. Completely LAT-deficient patients present with SCID, with low T cells and low Ig (Bacchelli et al., 2017; Keller et al., 2016), all of which contrast with SLP76 deficiency. This difference may reflect the possibility that the patient retained some level of function, exhibited either by truncated SLP76 or by residual full-length SLP76 protein due to a small degree of leakiness of the mutation. The predicted C-terminally truncated protein contains the Gads binding site, suggesting that it could be recruited to LAT by Gads. It also contains the N-terminal tyrosine phosphorylation sites, which can mediate signaling; however, recent studies suggest a role of the C-terminal SH2 of SLP76 in mediating the formation of LAT microclusters (Lewis et al., 2018; Liu et al., 2013). Therefore, we speculate that the immune deficiency may relate specifically to reduced LAT microclustering in the presence of truncated SLP76, which was expressed at very low levels. Similar to LAT deficiency, SLP76 deficiency affected downstream events, including phosphorylation of PLC-γ1 and ERK and Ca2+ mobilization, suggesting that this event depends on the normal assembly of LAT and SLP76. As expected, both LAT and SLP76 deficiencies do not affect phosphorylation of the upstream molecule ZAP-70. Yet, in contrast to LAT deficiency, which leads to absent tyrosine phosphorylation of SLP76, SLP76 deficiency had no impact on LAT phosphorylation. In addition, ITK-deficient patient cells have impaired but present Ca2+ flux and clinically have an EBV-triggered lymphoproliferative syndrome with progressive loss of CD4 T cells and Ig without clear evidence of immune deficiency outside of EBV-mediated/triggered pathology (Serwas et al., 2014). Therefore, SLP76 serves a nonredundant role in human TCR-mediated calcium signaling and cytokine production and proliferation, but unlike LAT and ITK deficiency, it is not required to generate normal numbers of T cells and Ig. It is also important to note that the patient’s clinical and lymphocyte phenotypes might have been similar to ITK deficiency had the patient contracted EBV. As such, in other scenarios the clinical presentation of SLP76 deficiency, whil unique in that it combines T cell and neutrophil host defense defects, may not be limited to what was seen in this patient.
Interestingly, our patient displayed early-onset autoimmune hemolytic anemia, which is similar to previously reported patients with genetic alterations in TCR signaling (e.g., LAT, ZAP-70, ITK, or LCK), suggesting that reduced, but not absent, TCR-induced signaling can lead to defects in immune tolerance as well.
This study describes the consequences of SLP76 deficiency in humans. Knowing these consequences can be helpful in explaining T+B+NK+ SCID and mixed neutrophil and T cell defects, but also provides a guide for a better understanding of human SLP76 biology. In addition, studying immune defects can identify targets for immune suppression and expected phenotypes of immune suppression. The observations made in this patient could guide the type of suppression that would result from SLP76 inhibition, which could be useful for inhibiting mast cell activity (Wu et al., 2004) in addition to T cell and neutrophil-mediated pathology.
Materials and methods
Patient
The patient was evaluated at the Edmond and Lily Safra Children’s Hospital, Sheba Medical Center, Israel. The patient’s parents gave written, informed consent for this study, which was approved by the local institutional review boards at Sheba Medical Center and the Ministry of Health.
Flow cytometry and antibodies
For analysis of cell surface markers, PBMCs were used as starting material or 100 µl of whole blood in EDTA was lysed using RBC Lysis Buffer (BD Pharm Lyse; BD Biosciences). Cells were washed twice in FACS buffer (PBS with 5% FBS) and resuspended in 100 µl of FACS buffer with antibodies for 30 min. Cells were washed in FACS buffer and resuspended in 500 µl of FACS buffer. For the intracellular staining of SLP76, the PerFix-nc kit (Beckman Coulter) was used. Measurement and analysis were performed using flow cytometry (NAVIOS; Beckman Coulter) and Kaluza software (Beckman Coulter), or using LSRFortessa with Diva 6.1.3 software (BD Biosciences) and analyzed with FlowJo 10 software (TreeStar).
The following antibodies were used: PacB anti-CD3 (SK7; Biolegend), BUV395 anti-CD3 (SP34-2; BD Biosciences), Alexa Fluor 700 anti-CD3 (UCHT1; BD Biosciences), BV711 anti-CD4 (SK3; BD Biosciences), FITC anti-CD4 (RM4-5; Invitrogen), BUV395 anti-CD4 (RPA-T4; BD Biosciences), APC anti-CD4 (13B8.2; Beckman Coulter), APC-H7 anti-CD8 (SK1; BD Biosciences), BV510 anti-CD8 (SK; BD Biosciences), Pacific blue anti-CD8 (B9.11; Beckman Coulter), APC anti-CD45RA (BUV737; BD Biosciences), ECD anti-CD45RO (IM2712U; Beckman Coulter), Krome orange anti-CD45 (J33; Beckman Coulter), PE anti-CD25 (M-A251; BD Biosciences), PE anti-CD56 (N901; Beckman Coulter), PerCP/Cy5.5 anti-CD19 (HIB19; Biolegend), APC Alexa Flour 750 anti-CD19 (J3-119; Beckman Coulter), BV421 anti-IgD (IA6-2; BD Biosciences), PE anti-CD27 (L128; BD Biosciences), PE-CD197 (CCR7; G043H7; Beckman Coulter), APC anti-CD38 (HB7; BD Biosciences), V450 mouse anti-human CD27 (clone M-T271), PE mouse anti-human TCRγδ-1 (clone 11F2; all BD Biosciences), PerCP anti-human TCR Vδ2 (Biolegend), CD45RO-TRPE (Beckman Coulter), and PE anti-SLP76 (REA427; Miltenyi Biotec).
Immunologic evaluation
Lymphocyte proliferative response to mitogens and the amount of TCR excision circles were determined as previously described (Amariglio et al., 2010). Proliferative response was measured by labeling PBMCs with 2.5 mM CFSE (Thermo Fisher Scientific), 7-aminoactinomycin D (BD Biosciences), PacB anti-CD3 (SK7; Biolegend), PE anti-CD25 (M-A251; BD Biosciences), APC-H7 anti-CD8 (SK1; BD Biosciences), and BV711 anti-CD4 (SK3; BD Biosciences) 4 d after stimulation. Activation response was determined by CD25 levels in stimulated PBMCs. The analysis of TCR Vβ expression was determined according to manufacturer’s manual (Beta Mark TCR Vβ Repertoire Kit; Beckman Coulter). NK degranulation was assessed by CD107a surface staining without stimulation (medium-cultured cells) and 3 h after stimulation with K562 cells at a ratio of 1:1 as previously described (Bryceson et al., 2012). The erythroleukemic cell line K562 (CCL-243; ATCC) was used as a target cell line. NK cells were cultured in medium containing 600 U/ml IL-2 (Novartis) for 48 h to assess the degranulation of activated NK cells. For surface staining, the following antibodies were used: PacB anti-CD3 (SK7; Biolegend), APC-H7 anti-CD8 (SK1; BD Biosciences), FITC anti-CD56 (NCAM16.2; BD Biosciences), and PE anti-CD107a (H4A3; BD Biosciences).
Flow cytometric analysis of cytokine response
Patient and age-matched healthy control PBMCs were resuspended at 106 cells/ml in RPMI 1640 supplemented with 10% FBS, penicillin, streptomycin, and L-glutamine (Gibco); aliquoted (1 ml/well) into a 48-well plate; and were stimulated with either plate-bound CD3 (5 µg/ml OKT; Invitrogen) and soluble CD28 (1 µg/ml L293; BD Biosciences) for 30 h, with brefeldin A (10 µg/ml) added 24 h after stimulation; or 20 ng/ml PMA (Sigma-Aldrich) and 1 μmol/liter ionomycin (Sigma-Aldrich) for 5 h, with brefeldin A (10 µg/ml) added after 2.5 h. Cells were then fixed and permeabilized with BD Cytofix/Cytoperm reagents (BD Biosciences) according to the manufacturer’s protocol and stained with Alexa Fluor 700 mouse anti-human CD3 (clone UCHT1), BUV395 mouse anti-human CD4 (clone RPA-T4), BV510 mouse anti-human CD8 (clone SK; BD Biosciences), CD45RO-TRPE (Beckman Coulter), PerCP-Cy5.5 rat anti-human IL-2 (clone MQ1-17H12), FITC rat anti-human IL-4 (clone MP4-25D2), PE rat anti-human IL-5 (clone TRFK5), BV711 rat anti-human IL-13 (clone JES10-5A2), V450 mouse anti-human IFN-γ (clone B27), and APC mouse anti-human TNF (clone MAb11; BD Biosciences). Cells were gated on CD45RO+CD4+ or CD45RO+CD8+ cells.
Neutrophil studies
Neutrophils were isolated (98% purity and viability) from heparinized blood using a negative-selection isolation kit (EasySep; StemCell Technologies). The oxidative burst of neutrophils (dihydrorhodamine assay) after stimulation with E. coli bacteria or PMA was measured by using the FagoFlowEx kit (Exbio Diagnostics). Superoxide production was measured as superoxide dismutase–inhibitable reduction of ferricytochrome c. The neutrophils were stimulated by PMA (Sigma-Aldrich) or chemoattractant fMLP (Sigma-Aldrich). Chemotaxis was assessed in a 48-well chemotaxis chamber through a 3-µm pore size filter and induced by 1 µM fMLP. Random migration was conducted in the presence of M199 medium (Biological Industries). Net chemotaxis was calculated by subtracting the random from the chemotactic migration (fMLP-stimulated neutrophils). All procedures were performed in quadruplicate.
Platelet aggregation
Citrated whole blood samples (9:1) were centrifuged (110 g for 10 min) to obtain platelet-rich plasma, followed by further centrifugation (1,900 g for 10 min) to obtain platelet-poor plasma. Light transmission aggregometry (AggRAM; Hellena) was performed on platelet-rich plasma samples upon addition of various platelet agonists (final concentration: 10 μM adenosine diphosphate, 10 μg/ml collagen, 50 μM epinephrine, and 1.5 mg/ml ristocetin). Changes in the light transmission of PRO over baseline (platelet-poor plasma) were recorded for 5 min and calculated as maximal aggregation.
Cell lines
The SLP76-deficient cell line J14 is derived from the Jurkat T cell line and has been described previously (Yablonski et al., 1998). The J14 cell line was cultured in RPMI 1640 medium supplemented with 10% FBS, penicillin, streptomycin, and glutamine. Fibroblasts were obtained from forearm skin biopsy specimens from the patient and from foreskins of healthy humans (Vilboux et al., 2013). Patient fibroblasts were cultured in DMEM supplemented with 10% FBS, glutamine, and antibiotics.
Whole-exome and Sanger sequencing
High-throughput sequencing for whole-exome sequencing was performed on genomic DNA samples from the patient. Coding regions were enriched with a SureSelect Human All Exon V5 Kit (Agilent Technologies) and then sequenced as 100-bp paired-end runs on an Illumina HiSeq 2500 (Illumina). We used the BWA mem algorithm (version 0.7.15) for alignment of the sequence reads to the human reference genome (hg19). The HaplotypeCaller algorithm of Genomic Analysis ToolKit (GATK version 3.4) was applied for variant calling, as recommended in the best practice pipeline (DePristo et al., 2011). KGG-seq (version 1.0) was used for annotation of identified variants (Li et al., 2017), and in-house scripts were applied for filtering based on family pedigree and local datasets of variants detected in previous sequencing projects. The SLP76 mutation was validated by using dideoxy Sanger sequencing in the carriers and healthy control. Data were evaluated using Sequencer software (version 5.0; Gene Codes Corporation).
RNA and cDNA preparation
Total RNA was isolated from 2–4 × 106 cells from peripheral blood from the patient, his parents, and healthy control using the TRIzol reagent (Sigma-Aldrich) according to the manufacturer’s instructions. Full-length cDNA was prepared from 1 µg total RNA using the high-capacity cDNA kit (Applied Biosystems). PCR was performed with primers that flanked the mutation (exons 13 and 16).
Western blot
Whole-cell lysates were prepared from age-matched healthy controls and patient-derived T cell lymphoblasts. Cell lysates from one million cells per lane or 80 µg of protein were loaded on 10% polyacrylamide gel and separated by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membrane using the Trans-Blot Turbo System (Bio-Rad). Blots were probed overnight with specific primary antibodies—for instance, polyclonal anti-human SLP-76 previously described (Gonen et al., 2005), anti-human SLP76 (ab17029; Abcam), anti–PLC-γ1 (sc-81; Santa Cruz), anti–phospho-PLC-γ Tyr783 (AT-7142; MBL), anti-human phosphor-MAPK (T202\Y204-Erk1/2; #9101; Cell Signaling Technology), and anti-human p44/42 MAPK-Erk1/2 (#9102; Cell Signaling Technology). Stimulation was induced with anti-CD3 (10 µg/ml OKT; Invitrogen). Anti-HSC70 (sc-7298; Santa Cruz Biotechnology) was used as a loading control. For imaging, appropriate HRP-labeled secondary antibodies (1:10,000) were used. Bands were revealed using enhanced chemiluminescence (Biological industries). Densitometry was performed on scanned immunoblot images using ImageJ software.
Assessment of intracellular Ca2+ flux
2.5 million cells from each donor were barcoded with cell trace far red as described previously (Sukenik et al., 2017) and mixed together. Barcoded cells were then resuspended in RPMI with 10% FCS and 20 mM Hepes, pH 7.4, loaded with the fluorescent Ca2+ indicator dye, Indo-1-AM, at 37°C and then washed two times in calcium buffer consisting of 25 mM Hepes (pH 7.4), 1 mM CaCl2, 125 mM NaCl, 5 mM KCl, 1 mM Na2HPO4, 0.5 mM MgCl2, 1 g/liter glucose, and 1 mg/ml bovine serum albumin. Cells were then stained in calcium buffer for 15 min on ice with PE anti-CD4 (13B8.2; Beckman Coulter) and Alexa Flour 700 anti-CD8 (B9.11; Beckman Coulter), and finally washed once more and resuspended in calcium buffer. An LSR Fortessa (BD Biosciences) was used for ratiometric, kinetic measurement of intracellular Ca2+, with the temperature maintained at 37°C, and TCR stimulation was induced by pre-cross-linked anti-CD3 (5 µg/ml OKT3; Invitrogen) and goat anti-mouse (2.5 µg/ml), which was added at the 60-s time point. Excitation was at 355 nm and emission was measured using the bandpass filters 405/20 (Ca-bound indo1) and 485/22 (free indo1), with data presented as the ratio between these two values. Data analysis was performed on each subpopulation by gating on CD4 or CD8 cells within each barcoded population.
Analysis of surface activation markers
PBMCs from the patient and age-matched healthy controls (0.5 × 106) were either left untreated or stimulated with CD3/CD28 (ImmunoCult human T cell activator; StemCell Technologies), with plate-bound CD3 (5 μg/ml OKT; Invitrogen) and soluble CD28 (1 μg/ml CD28.2; Invitrogen), or with PMA (50 ng/ml; Sigma-Aldrich) in RPMI medium overnight at 37°C. J14 cells (0.5 × 106) were either left untreated or stimulated with the monoclonal antibody C305 (specific for the Jurkat Ti β chain; Weiss and Stobo, 1984) or with PMA (50 ng/ml; Sigma-Aldrich) in RPMI medium overnight at 37°C. Cells were then washed with PBS and stained for surface activation markers with the following antibodies: PE anti-CD69 (TP1.55.3; Beckman Coulter), Alexa Fluor 700 anti-CD69 (FN50), FITC anti-CD98 antibody (UM7F8), and PE-Cy7 anti-CD25 (M-A251; BD Biosciences).
Ex vivo T cell expansion
Blood from the patient and age-matched healthy controls was subjected to a Ficoll density gradient (Ficoll-Paque PLUS; GE Healthcare) centrifugation, after which PBMCs were collected. T cells were stimulated with 5 ng/ml PMA and 1 µM ionomycin (Sigma-Aldrich) and 100 U/ml recombinant human IL-2 (PeproTech) for 48 h at 106 cells/ml in a 24-well plate. After 48 h, cells were counted and seeded at 0.5 × 106 cells/ml in medium with IL-2 (100 U/ml) continued cell expansion for another 2 wk.
T cell signaling
Expanded T cell lymphoblasts (0.5 × 106) were stimulated by cross-linking of 4 µg/ml anti-CD3 (OKT; Invitogen) and 5 µg/ml anti-CD28 (CD28.2; Invitrogen) with 10 µg/ml goat anti-mouse IgG (Polyclonal; BD Biosciences) for 5–30 min in 37°C or left untreated. Cells were fixed in 1.6% paraformaldehyde or with fix and Perm Medium A (Molecular Probes) for 10 min and permeabilized with cold methanol for 30 min. J14 cells (0.5 × 106) were stimulated with the monoclonal antibody C305 (specific for the Jurkat Ti β chain; Weiss and Stobo, 1984) or with PMA (50 ng/ml) for 2 min at 37°C or left untreated. Cells were fixed in intracellular fixation buffer (Invitrogen) for 30 min and permeabilized with cold methanol for 30 min. Permeabilized cells were stained with the following intracellular antibodies detecting: SLP-76 phosphorylated at S376 (anti-pSLP76; #92711; Cell Signaling Technology), ERK1/2 phosphorylated at T202 and Y204 (anti-pERK1/2; #4370; Cell Signaling Technology), phospho-ERK1/2 (20A; BD Biosciences), phospho-ERK1/2 (MILANAR8R, Invitrogen), ZAP-70 phosphorylated at T319 (anti-pZAP70; #2717; Cell Signaling Technology), LAT phosphorylated at T191 (anti-pLAT; #3584; Cell Signaling Technology), and phospho-S6 phosphorylated at pS235 and pS236 (N7–548; BD Biosciences). Measurement and analysis were performed using flow cytometry (NAVIOS; Beckman Coulter) and Kaluza software (Beckman Coulter).
Fluorescence microscopy analysis of phalloidin staining
Fibroblasts were grown on coverslips in a 6-well plate untilthey reached 70–80% confluence. Cells were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.1% Triton X-100 for 3 min, washed with PBS, and stained with 1 µg/ml FITC-phalloidin (Sigma-Aldrich) for 60 min at room temperature. The slides were mounted using immunofluore-containing DAPI (CytoCell) for nuclear staining and analyzed by fluorescent microscopy (Olympus). For the quantification of the corrected total cell fluorescence (CTCF = integrated density − [area of selected cell × mean fluorescence of background readings]), 80 cells from patient fibroblasts and 80 cells from control fibroblasts were counted. The analysis was done using ImageJ software. The results summarize three different experiments.
TCR and B cell receptor repertoire analysis
At least 150 ng of genomic DNA from the patient’s peripheral blood was used to generate the TCR and B cell receptor libraries. Specifically, primers for conserved regions of V and J genes in the TRG, TRB, and IGH chain loci were used according to the manufacturer’s protocol (LymphoTrack; Invivoscribe Technologies). The libraries were quantified, pooled, and sequenced using Mi-Seq Illumina technology (Illumina). Raw data of the sequences were subjected to bioinformatic analyses. First, the sequences were converted to FASTA format using the analysis package of LymphoTrack and Geneious software (Biomatters). Second, the FASTA sequences were submitted to the IMGT HighV-QUEST webserver (http://www.imgt.org). Third, the statistics file from IMGT HighV-QUEST were used to generate hierarchical treemaps using Treemap software (Macrofocus GmbH). In addition, Shannon’s H and Simpson’s D diversity indices were calculated using the PAST program (free web from http://priede.bf.lu.lv/ftp/pub/TIS/datu_analiize/PAST/2.17c/download.html) based on the following equations:
where R is total sequences, i is unique sequences; and pi is the proportion of total sequences belonging to the “i”th unique sequence.
The frequencies of the different gene uses were summarized from the IMGT HighV-QUEST statistics files using Excel.
Generation of retroviral plasmid
N-terminally FLAG-tagged human SLP76 cDNA was expressed from the pMIGR retroviral vector, as previously described (pMIGR1-FLAG-SLP76; Sela et al., 2011). This plasmid was modified to generate pMIGR1-FLAG-SLP76-mut by deleting the portion of the SLP76 cDNA corresponding to exon 14, using the site-directed mutagenesis kit (Q5 Site-Directed Mutagenesis Kit; New England Biolabs).
The pMIGR-FLAG-SLP76-mut plasmid was subjected to Sanger sequencing to confirm the deletion of exon 14.
Retroviral infections
The retroviruses were generated using the Amphotrophic Phoenix retrovirus packaging cell line as described previously (Pear et al., 1993). Specifically, the retroviral plasmid was transfected to the Amphotrophic Phoenix cell line using calcium phosphate–mediated transfection (CAPHOS; Sigma-Aldrich), and the culture medium containing the retrovirus was collected 48–72 h after transfection. Primary T cells from the patient were cultured with pMIGR1-FLAG-SLP76 retrovirus, and the J14 Jurkat cell line was cultured with pMIGR1-FLAG-SLP76-mut retrovirus. In addition, both T cell and J14 Jurkat cell lines were cultured with pMIGR1-FLAG retrovirus as a negative control. Since the pMIGR1 backbone plasmid allows for the coexpression of GFP protein, all the retroviruses used contained GFP sequences, allowing for the selection of the transduced cells for further analysis.
Transfection of PBMCs
PBMCs from the patient and age-matched healthy controls were transfected with pcDNA3.1+/C-(K)-DYK plasmid DNA expressing human WT SLP76 (GenScript) using the Amaxa human T cell nucleofector kit (VPA-1002; Lonza), according to the manufacturer’s protocol for unstimulated human T cells with Amaxa nucleofection program U-014. Cells were cultured 12 h after transfection before assays were performed.
Online supplemental material
Fig. S1 shows extended immunologic and genetic analyses, including immuno-phenotyping of patient’s CD4 and CD8 cells measured by flow cytometry, T cell proliferation, flow cytometric analysis of cytokine response, peripheral B cell immuno-phenotyping, neutrophil function, fluorescence studies of actin polymerization, and genetic analysis of SLP76 mutation and SLP76 domains. Fig. S2 shows extended workup of the consequences of SLP76 mutation, including TCR-induced signaling in SLP76 mutated cells, quantification of the level of phosphor-PLC-γ1, TCR signaling in SLP76-mutated T cells, Ca2+ mobilization, and expression of surface activation markers. Also shown is flow cytometric analysis showing equal amounts of GFP+ cells in J14 transduced experiments and correction of the SLP76 mutant phenotype in J14 or in expanded T cell lymphoblasts. Fig. S3 shows extended workup of TCR and B cell receptor repertoires, including flow cytometric analysis of TCRVβ repertoire, analysis of γδ T cells, and differential V gene uses in TRG, TRB, and IGH repertoires. Table S1 contains data on platelet aggregation. Table S2 contains data on lymphocyte proliferation.
Supplementary Material
contains data on platelet aggregation.
contains data on lymphocyte proliferation.
Acknowledgments
We thank the family for participating in this study. The study was performed in partial fulfillment of the requirements for the PhD degree of A. Lev at the Mina and Everard Goodman Faculty of Life Sciences, Bar-Ilan University.
This work was supported by the Jeffrey Modell Foundation; Ministry of Health, State of Israel; and Israel Science Foundation Israel Precision Medicine Program grant 3115/19 to R. Somech, A. Hendel, and Y.N. Lee. This work was supported in part, by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Author contributions:D. Yablonski, J.D. Milner, and R. Somech conceptualized the study, designed the experiments, supervised results, and wrote the manuscript. A. Lev., Y.N. Lee, A.J. Simon, and A. Hendel designed, performed, analyzed, and interpreted experiments, and contributed to manuscript preparation. Y.N. Lee and A. Lev performed and analyzed experiments related to next-generation sequencing. G. Sun, E. Hallumi, K.S. Zrihen, S. Levy, T. Beit Halevi, M. Papazian, N. Shwartz, I. Somekh, H. Vernitsky, E. Javasky, C.A. Ma, and Y. Zhang performed experiments and collected and assembled data. S. Levy-Mendelovich performed experiments related to platelet function. B. Wolach and R. Gavrieli performed experiments related to neutrophil function. O. Barel, N. Amariglio, and G. Rechavi performed and analyzed experiments related to whole-exome sequencing. T. Stauber and R. Somech treated the patient.
References
- Amariglio, N., Lev A., Simon A., Rosenthal E., Spirer Z., Efrati O., Broides A., Rechavi G., and Somech R.. 2010. Molecular assessment of thymus capabilities in the evaluation of T-cell immunodeficiency. Pediatr. Res. 67:211–216. 10.1203/PDR.0b013e3181c6e554 [DOI] [PubMed] [Google Scholar]
- Bacchelli, C., Moretti F.A., Carmo M., Adams S., Stanescu H.C., Pearce K., Madkaikar M., Gilmour K.C., Nicholas A.K., Woods C.G., et al. 2017. Mutations in linker for activation of T cells (LAT) lead to a novel form of severe combined immunodeficiency. J. Allergy Clin. Immunol. 139:634–642.e5. 10.1016/j.jaci.2016.05.036 [DOI] [PubMed] [Google Scholar]
- Balagopalan, L., Kortum R.L., Coussens N.P., Barr V.A., and Samelson L.E.. 2015. The linker for activation of T cells (LAT) signaling hub: from signaling complexes to microclusters. J. Biol. Chem. 290:26422–26429. 10.1074/jbc.R115.665869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogin, Y., Ainey C., Beach D., and Yablonski D.. 2007. SLP-76 mediates and maintains activation of the Tec family kinase ITK via the T cell antigen receptor-induced association between SLP-76 and ITK. Proc. Natl. Acad. Sci. USA. 104:6638–6643. 10.1073/pnas.0609771104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson, Y.T., Pende D., Maul-Pavicic A., Gilmour K.C., Ufheil H., Vraetz T., Chiang S.C., Marcenaro S., Meazza R., Bondzio I., et al. 2012. A prospective evaluation of degranulation assays in the rapid diagnosis of familial hemophagocytic syndromes. Blood. 119:2754–2763. 10.1182/blood-2011-08-374199 [DOI] [PubMed] [Google Scholar]
- Cirillo, E., Giardino G., Gallo V., D’Assante R., Grasso F., Romano R., Di Lillo C., Galasso G., and Pignata C.. 2015. Severe combined immunodeficiency--an update. Ann. N. Y. Acad. Sci. 1356:90–106. 10.1111/nyas.12849 [DOI] [PubMed] [Google Scholar]
- Clements, J.L., Ross-Barta S.E., Tygrett L.T., Waldschmidt T.J., and Koretzky G.A.. 1998a. SLP-76 expression is restricted to hemopoietic cells of monocyte, granulocyte, and T lymphocyte lineage and is regulated during T cell maturation and activation. J. Immunol. 161:3880–3889. [PubMed] [Google Scholar]
- Clements, J.L., Yang B., Ross-Barta S.E., Eliason S.L., Hrstka R.F., Williamson R.A., and Koretzky G.A.. 1998b. Requirement for the leukocyte-specific adapter protein SLP-76 for normal T cell development. Science. 281:416–419. 10.1126/science.281.5375.416 [DOI] [PubMed] [Google Scholar]
- Clements, J.L., Lee J.R., Gross B., Yang B., Olson J.D., Sandra A., Watson S.P., Lentz S.R., and Koretzky G.A.. 1999. Fetal hemorrhage and platelet dysfunction in SLP-76-deficient mice. J. Clin. Invest. 103:19–25. 10.1172/JCI5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadi, H.K., Simon A.J., and Roifman C.M.. 2003. Effect of CD3delta deficiency on maturation of alpha/beta and gamma/delta T-cell lineages in severe combined immunodeficiency. N. Engl. J. Med. 349:1821–1828. 10.1056/NEJMoa031178 [DOI] [PubMed] [Google Scholar]
- DePristo, M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43:491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, A. 2007. Human primary immunodeficiency diseases. Immunity. 27:835–845. 10.1016/j.immuni.2007.11.012 [DOI] [PubMed] [Google Scholar]
- Fischer, A., de Saint Basile G., and Le Deist F.. 2005. CD3 deficiencies. Curr. Opin. Allergy Clin. Immunol. 5:491–495. 10.1097/01.all.0000191886.12645.79 [DOI] [PubMed] [Google Scholar]
- Gonen, R., Beach D., Ainey C., and Yablonski D.. 2005. T cell receptor-induced activation of phospholipase C-gamma1 depends on a sequence-independent function of the P-I region of SLP-76. J. Biol. Chem. 280:8364–8370. 10.1074/jbc.M409437200 [DOI] [PubMed] [Google Scholar]
- Gross, B.S., Lee J.R., Clements J.L., Turner M., Tybulewicz V.L., Findell P.R., Koretzky G.A., and Watson S.P.. 1999. Tyrosine phosphorylation of SLP-76 is downstream of Syk following stimulation of the collagen receptor in platelets. J. Biol. Chem. 274:5963–5971. 10.1074/jbc.274.9.5963 [DOI] [PubMed] [Google Scholar]
- Hamann, D., Baars P.A., Rep M.H., Hooibrink B., Kerkhof-Garde S.R., Klein M.R., and van Lier R.A.. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 186:1407–1418. 10.1084/jem.186.9.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, F., Randriamampita C., Martin E., Gerart S., Lambert N., Lim A., Soulier J., Maciorowski Z., Touzot F., Moshous D., et al. 2012. Primary T-cell immunodeficiency with immunodysregulation caused by autosomal recessive LCK deficiency. J. Allergy Clin. Immunol. 130:1144–1152.e11. 10.1016/j.jaci.2012.07.029 [DOI] [PubMed] [Google Scholar]
- Huck, K., Feyen O., Niehues T., Rüschendorf F., Hübner N., Laws H.J., Telieps T., Knapp S., Wacker H.H., Meindl A., et al. 2009. Girls homozygous for an IL-2-inducible T cell kinase mutation that leads to protein deficiency develop fatal EBV-associated lymphoproliferation. J. Clin. Invest. 119:1350–1358. 10.1172/JCI37901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, S.F., Anderson C.F., and Farber D.L.. 2002. Differential SLP-76 expression and TCR-mediated signaling in effector and memory CD4 T cells. J. Immunol. 168:1557–1565. 10.4049/jimmunol.168.4.1557 [DOI] [PubMed] [Google Scholar]
- Jackman, J.K., Motto D.G., Sun Q., Tanemoto M., Turck C.W., Peltz G.A., Koretzky G.A., and Findell P.R.. 1995. Molecular cloning of SLP-76, a 76-kDa tyrosine phosphoprotein associated with Grb2 in T cells. J. Biol. Chem. 270:7029–7032. 10.1074/jbc.270.13.7029 [DOI] [PubMed] [Google Scholar]
- Jordan, M.S., and Koretzky G.A.. 2010. Coordination of receptor signaling in multiple hematopoietic cell lineages by the adaptor protein SLP-76. Cold Spring Harb. Perspect. Biol. 2:a002501. 10.1101/cshperspect.a002501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, B., Zaidman I., Yousefi O.S., Hershkovitz D., Stein J., Unger S., Schachtrup K., Sigvardsson M., Kuperman A.A., Shaag A., et al. 2016. Early onset combined immunodeficiency and autoimmunity in patients with loss-of-function mutation in LAT. J. Exp. Med. 213:1185–1199. 10.1084/jem.20151110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, L., Pivniouk V., de la Fuente M.A., Laouini D., and Geha R.S.. 2002. Differential role of SLP-76 domains in T cell development and function. Proc. Natl. Acad. Sci. USA. 99:884–889. 10.1073/pnas.022619199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc, M.P., Giudicelli V., Ginestoux C., Bodmer J., Müller W., Bontrop R., Lemaitre M., Malik A., Barbié V., and Chaume D.. 1999. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 27:209–212. 10.1093/nar/27.1.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.B., Scangarello F.A., Murphy J.M., Eidell K.P., Sodipo M.O., Ophir M.J., Sargeant R., Seminario M.C., and Bunnell S.C.. 2018. ADAP is an upstream regulator that precedes SLP-76 at sites of TCR engagement and stabilizes signaling microclusters. J. Cell Sci. 131:jcs215517. 10.1242/jcs.215517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Li J., Li M.J., Pan Z., Hsu J.S., Liu D.J., Zhan X., Wang J., Song Y., and Sham P.C.. 2017. Robust and rapid algorithms facilitate large-scale whole genome sequencing downstream analysis in an integrative framework. Nucleic Acids Res. 45:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Thaker Y.R., Stagg L., Schneider H., Ladbury J.E., and Rudd C.E.. 2013. SLP-76 sterile α motif (SAM) and individual H5 α helix mediate oligomer formation for microclusters and T-cell activation. J. Biol. Chem. 288:29539–29549. 10.1074/jbc.M112.424846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbrough, S.A., Mocsai A., Clemens R.A., Wu J.N., Silverman M.A., Singer A.L., Lowell C.A., and Koretzky G.A.. 2003. SLP-76 regulates Fcgamma receptor and integrin signaling in neutrophils. Immunity. 19:761–769. 10.1016/S1074-7613(03)00305-4 [DOI] [PubMed] [Google Scholar]
- Notarangelo, L.D. 2013. Partial defects of T-cell development associated with poor T-cell function. J. Allergy Clin. Immunol. 131:1297–1305. 10.1016/j.jaci.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarangelo, L.D. 2014. Immunodeficiency and immune dysregulation associated with proximal defects of T cell receptor signaling. Curr. Opin. Immunol. 31:97–101. 10.1016/j.coi.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ombrello, M.J., Remmers E.F., Sun G., Freeman A.F., Datta S., Torabi-Parizi P., Subramanian N., Bunney T.D., Baxendale R.W., Martins M.S., et al. 2012. Cold urticaria, immunodeficiency, and autoimmunity related to PLCG2 deletions. N. Engl. J. Med. 366:330–338. 10.1056/NEJMoa1102140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear, W.S., Nolan G.P., Scott M.L., and Baltimore D.. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 90:8392–8396. 10.1073/pnas.90.18.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivniouk, V., Tsitsikov E., Swinton P., Rathbun G., Alt F.W., and Geha R.S.. 1998. Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell. 94:229–238. 10.1016/S0092-8674(00)81422-1 [DOI] [PubMed] [Google Scholar]
- Pivniouk, V.I., Martin T.R., Lu-Kuo J.M., Katz H.R., Oettgen H.C., and Geha R.S.. 1999. SLP-76 deficiency impairs signaling via the high-affinity IgE receptor in mast cells. J. Clin. Invest. 103:1737–1743. [PMC free article] [PubMed] [Google Scholar]
- Sela, M., Bogin Y., Beach D., Oellerich T., Lehne J., Smith-Garvin J.E., Okumura M., Starosvetsky E., Kosoff R., Libman E., et al. 2011. Sequential phosphorylation of SLP-76 at tyrosine 173 is required for activation of T and mast cells. EMBO J. 30:3160–3172. 10.1038/emboj.2011.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwas, N.K., Cagdas D., Ban S.A., Bienemann K., Salzer E., Tezcan I., Borkhardt A., Sanal O., and Boztug K.. 2014. Identification of ITK deficiency as a novel genetic cause of idiopathic CD4+ T-cell lymphopenia. Blood. 124:655–657. 10.1182/blood-2014-03-564930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggs, O.M., Miosge L.A., Daley S.R., Asquith K., Foster P.S., Liston A., and Goodnow C.C.. 2015. Quantitative reduction of the TCR adapter protein SLP-76 unbalances immunity and immune regulation. J. Immunol. 194:2587–2595. 10.4049/jimmunol.1400326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y.W., and Jumaa H.. 2003. LAT links the pre-BCR to calcium signaling. Immunity. 19:295–305. 10.1016/S1074-7613(03)00202-4 [DOI] [PubMed] [Google Scholar]
- Sukenik, S., Frushicheva M.P., Waknin-Lellouche C., Hallumi E., Ifrach T., Shalah R., Beach D., Avidan R., Oz I., Libman E., et al. 2017. Dimerization of the adaptor Gads facilitates antigen receptor signaling by promoting the cooperative binding of Gads to the adaptor LAT. Sci. Signal. 10:eaal1482. 10.1126/scisignal.aal1482 [DOI] [PubMed] [Google Scholar]
- Vilboux, T., Lev A., Malicdan M.C., Simon A.J., Järvinen P., Racek T., Puchalka J., Sood R., Carrington B., Bishop K., et al. 2013. A congenital neutrophil defect syndrome associated with mutations in VPS45. N. Engl. J. Med. 369:54–65. 10.1056/NEJMoa1301296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S., Charmley P., Robinson M.A., and Concannon P.. 1994. The extent of the human germline T-cell receptor V beta gene segment repertoire. Immunogenetics. 40:27–36. 10.1007/BF00163961 [DOI] [PubMed] [Google Scholar]
- Weiss, A., and Stobo J.D.. 1984. Requirement for the coexpression of T3 and the T cell antigen receptor on a malignant human T cell line. J. Exp. Med. 160:1284–1299. 10.1084/jem.160.5.1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, J.E., Schneider H., and Rudd C.E.. 2006. TcR and TcR-CD28 engagement of protein kinase B (PKB/AKT) and glycogen synthase kinase-3 (GSK-3) operates independently of guanine nucleotide exchange factor VAV-1. J. Biol. Chem. 281:32385–32394. 10.1074/jbc.M604878200 [DOI] [PubMed] [Google Scholar]
- Wu, J.N., Jordan M.S., Silverman M.A., Peterson E.J., and Koretzky G.A.. 2004. Differential requirement for adapter proteins Src homology 2 domain-containing leukocyte phosphoprotein of 76 kDa and adhesion- and degranulation-promoting adapter protein in FcepsilonRI signaling and mast cell function. J. Immunol. 172:6768–6774. 10.4049/jimmunol.172.11.6768 [DOI] [PubMed] [Google Scholar]
- Yablonski, D. 2019. Bridging the Gap: Modulatory Roles of the Grb2-Family Adaptor, Gads, in Cellular and Allergic Immune Responses. Front. Immunol. 10:1704. 10.3389/fimmu.2019.01704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonski, D., Kuhne M.R., Kadlecek T., and Weiss A.. 1998. Uncoupling of nonreceptor tyrosine kinases from PLC-gamma1 in an SLP-76-deficient T cell. Science. 281:413–416. 10.1126/science.281.5375.413 [DOI] [PubMed] [Google Scholar]
- Yablonski, D., Kadlecek T., and Weiss A.. 2001. Identification of a phospholipase C-gamma1 (PLC-gamma1) SH3 domain-binding site in SLP-76 required for T-cell receptor-mediated activation of PLC-gamma1 and NFAT. Mol. Cell. Biol. 21:4208–4218. 10.1128/MCB.21.13.4208-4218.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains data on platelet aggregation.
contains data on lymphocyte proliferation.