Abstract
COVID-19 caused by the SARS-CoV-2 virus, accompanies an unprecedented spike in cytokines levels termed cytokines release syndrome (CRS), in critically ill patients. Clinicians claim that the surge demonstrates a deregulated immune defence in host, as infected cell expression analysis depicts a delay in type-I (interferon-I) and type-III IFNs expression, along with a limited Interferon-Stimulated Gene (ISG) response, which later resume and culminates in elicitation of several cytokines including- IL-6, IL-8, IL-12, TNFα, IL-17, MCP-1, IP-10 and IL-10 etc. Although cytokines are messenger molecules of the immune system, but their increased concentration results in inflammation, infiltration of macrophages, neutrophils and lung injury in patients. This inflammatory response results in the precarious pathogenesis of COVID-19; thus, a complete estimation of the immune response against SARS-CoV-2 is vital in designing a harmless and effective vaccine. In pathogenesis analysis, it emerges that a timely forceful type-I IFN production (18–24 hrs post infection) promotes innate and acquired immune responses, while a delay in IFNs production (3–4 days post infection) actually renders both innate and acquired responses ineffective in fighting infection. Further, underlying conditions including hypertension, obesity, cardio-vascular disease etc may increase the chances of putting people in risk groups, which end up having critical form of infection. This review summarizes the events starting from viral entry, its struggle with the immune system and failure of host immunological parameters to obliterate the infections, which finally culminate into massive release of CRS and inflammation in gravely ill patients.
Keywords: COVID-19, Corona virus, SARS-CoV-2, ACE2, Cytokine Release Storm (CRS), Acute Respiratory Distress Syndrome (ARDS), Deregulated immune response, Immune cell homeostasis
1. Introduction
Corona viruses were discovered in 1960 and one such virus termed as Severe Acute Respiratory Syndrome Corona Virus-2 (SARS-CoV-2) triggers COVID-19 disease. It has resulted in an unusual situation around the globe and has put the Coronavirus family on the forefront of research studies. In the last two decades, we have seen attack of different Corona viruses, notably, SARS or SARS-CoV originated in 2003 in China; MERS-CoV (Middle East Respiratory Syndrome Coronavirus) in 2012 in Saudi Arabia and SARS-CoV-2 emerged in Wuhan (China) in 2019, that resulted into COVID-19 disease and declared a pandemic in March 2020 by WHO [1], [2]. These three viruses belong to the same family of Coronaviridae, have zoonotic origin and share genomic similarities. SARS-CoV-2 reveals 80% sequence distinctiveness with SARS-CoV genome and 50% with MERS causing virus [3]. Although the Coronaviridae family includes four genera: α, β, γ and δ; but only α and β infect humans [4]. SARS-CoV-2 is actually a β-Corona virus and majority of infections might either persist as asymptomatic or result in mild symptoms, including- fever, dry cough, fatigue and shortness of breath like respiratory symptoms. These patients after showing the symptoms generally recover in 10–15 days. The identification of the ailment largely relies on detection of SARS-CoV-2 nucleic acid in nasal dabs of patient by real-time polymerase chain reaction (RT-PCR), or antibody detection in blood, clinical symptoms, epidemiological history, and CT based lung imaging etc [5].
Seldom patients show one or few unusual symptoms including- nausea, vomiting, diarrhoea, rashes, hives, COVID toe, muscle weakness, cardiovascular complications and loss of smell or taste senses etc. Conversely, nearly ~10% of the patients may develop severe complications, coinciding with either limited or all symptoms like acute respiratory distress syndrome (ARDS), pneumonia or pulmonary edema (fluid in the lungs) resulting in lung damage and fibrosis, pulmonary thrombosis, fast heart rate, shortness of breath, chest pain, septic shock or multi-organ failure etc. Clinically speaking, these extremely ill and deceased patients exhibit a hypercytokinemia called as Cytokines Release Syndrome (CRS) or Macrophage Activation Syndrome (MAS), characterized by release of high levels of different cytokines, including- IL-1β, IL-6, IL-7, IL-8, IL-9, TNFα, IL-10, IL-17, MCP-1 (monocyte chemoattractant protein-1), GM-CSF and IFN-γ etc [1], [6]. Moreover, lymphopenia (low lymphocytes levels) and an elevated neutrophil–lymphocyte-ratio (NLR) are also associated with seriousness of the infection in critical cases [7]. Hypertensive patients are prone to develop the severe COVID-19 infection, due to altered rennin-angiotensin system (RAS) and in turn activation of NLRP3 (NOD-like receptor 3) inflammasome [8] . Further, apart from underlying conditions of either hypertension, obesity and cardio-vascular diseases; gender seems to be a risk factor in fatal COVID-19 infections as well. Certain reports suggest male being more prone than females, and could be either due to protective estrogen properties in females or stronger immune response driven elevated cytokines levels in male, but requires additional study-based statistics [9], [10].
Our immune system comprises of both innate and acquired immune response, which corresponds via variable chemo-tactic factors called cytokines. Different cytokines vary in their molecular weights, structure, functions, cells involved in their secretion as well as target etc. The levels of these cytokines swing during different infections, resulting in inflammation and removal of pathogens. Innate immune response represents the first line of defence, which releases the cytokines to activate the acquired (adaptive) immune system, comprising of humoral (B-lymphocytes) and cell-mediated (T-lymphocytes: both CD4+ helper T cells (Th) and CD8+ cytotoxic T cells (Tc)) immunity.
During viral diseases including SARS-CoV-2, innate response directs infected cells to secrete interferons (IFNs) and several pro-inflammatory cytokines. The role of IFNs is to alarm the nearby uninfected cells for developing an antiviral stage, whereas proinflammatory cytokines increases blood flow near infection site and activate macrophage, with other phagocytic cells for clearance of virus, as well as infested cells. The cytokines also activate the CD4+ Th lymphocytes of adaptive responses, which further stimulate B-lymphocytes for secreting specific neutralizing antibodies to counteract the virus and CD8+ Tc cells to initiate programmed cell death of viral laden cells. Thus, innate reaction exposes the virus and directs the acquired response to control infection, but success lies in a well-timed co-ordination between both immune responses. However occasionally, virus after encountering the innate response causes a delay in IFNs production [11]. This delay is unwarranted in controlling virus and results in an unimpeded increase in viral load. Exhaustive viral load renders the adaptive response incompetent in fighting the infection completely. The time loss in IFNs production delay bring along a storm of cytokines release (CRS) including IL-6. IL-8, IL-12, TNFα, IL-17, IFN-γ, IL-1β, MCP-1, IP-10, IL-10, C-reactive proteins (CRP), CXCL10, D-dimer and ferritin etc [6], [12], [13]. This consequently increases the severity of infection including inflammation and its associated complications. Further, a reduction in the counts of natural killer (NK) cells, B and T cells, monocytes, eosinophils and basophils (lymphopenia) is also reported in these critical patients, which results in a delay in viral clearance [7], [13]. These indications in deceased patients suggest that an hyper-inflammatory response finally converts into an immune-suppressive condition and might play a critical role in deterioration of conditions [14]. Looking at these varied consequences, all contemplate a deregulated immune response, and this review is an endeavour to comprehend and elucidate the progression of SARS-CoV-2 infection, which results in severity including CRS and lung injury (ARDS) manifestation.
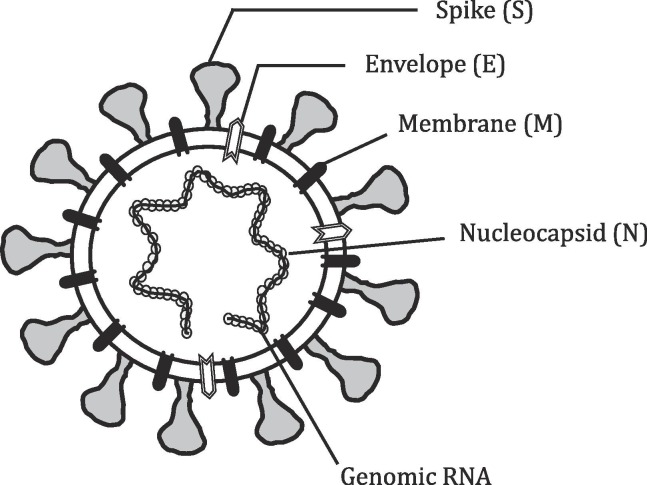
Structure of SARS-CoV-2 and entry: SARS-CoV-2 is an enveloped virus with a positive-sense single-stranded (Ss) RNA strand as genetic material (Fig. 1 ) [4]. Its genome is fully sequenced with 29,903 nucleotides, and have one of the largest RNA genome [15]. Although RNA viruses have an unstable genome, contrastingly SARS-CoV-2 genome is reportedly stable [16]. The virus has 14 open reading frames (ORF) involved in structure, survival and virulence. Architecturally, SARS-CoV-2 structure comprises of four main proteins involving- Spike (S) glycoprotein, envelope (E) glycoprotein, membrane (M) glycoprotein, nucleocapsid (N) protein, and numerous additional proteins (Fig. 1). Spike proteins, S present on membranes, are trans-membranous, form homo-trimers and give it a characteristic crown-like appearance. High sequence similarity between both CoV and CoV-2, led to the proposition that both viruses might utilize the same receptor, ACE2 (angiotensin converting enzyme 2), which is now recognized in two separate studies (Fig. 2 ) [17], [18]. The ACE2 is a membrane bound protein, present on the upper respiratory tract cells involving- oesophagus, alveolar epithelial cells (primary viral target) and other ACE2+ cells present in kidney, heart and gastrointestinal tract etc [3], [11]. Therefore, patients infected with the SARS-CoV-2 also encounter other ailments related to heart, kidneys and intestine etc [19]. MERS however utilizes a different membrane enzyme receptor, DPP4 (dipeptidyl peptidase-4) in the liver, gut, placenta, lung, and kidney cells etc [20].
Fig. 1.
Structure of Coronavirus: Coronavirus are enveloped with Ss RNA genome, which is non-segmented. Virion nucleocapsid along with Ss RNA also has phosphorylated protein (N) in phospholipid bilayer roofed by Spike (S) glycoprotein trimer. The layer protein (M) and envelope (E) protein are also located along with S proteins of the virus envelope.
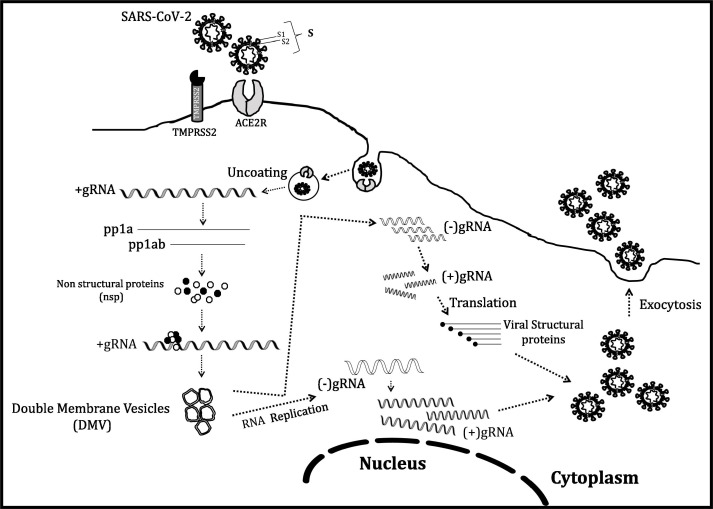
Fig. 2.
Life cycle of virus in an infected cell: After inhalation, virus reaches the respiratory system and internalization of SARS-CoV-2 virus starts with attachment of viral S (spike) protein on ACE2 receptor of humans, followed by TMPRSS2 mediated cleavage of S protein, which lead to internalization of virus. Viral uncoating in cytoplasm releases Ss positive sense genomic RNA (gRNA). The first step is translation of gRNA (+ve strand) into two polypeptides- pp1a and pp1ab, which is cleaved to many non-structural proteins (nsp), including RNA-dependent RNA polymerase, RdRp which is involved in viral genome replication in double membrane vesicles (DMVs). The negative strand, in another round, produces positive (+ve) strand genomic RNA and becomes the genome of the descendant viral cells. The transcribed sub-genomic RNAs is translated into various structural proteins (S, E, M and N) to create the viral progeny. S, E and M proteins enter the endoplasmic reticulum (ER), and the nucleocapsid protein, which joins with the genomic RNA (+ve strand) and combine into complete viral cell in the ER-Golgi compartment, and exocytosed out of the cell.
The virus entrance starts with virus inhalation through respiratory route, followed by binding of viral spike (S) protein on ACE2 receptor of alveolar cells. The attachment of S protein (having S1 and S2 domains) on to ACE2, lead to a change in receptor conformation and subsequently a host enzyme, TMPRSS2 (Transmembrane Serine Protease 2) primes/cleaves the viral S protein, at the S1/S2 site (Fig. 2) triggering viral internalization [21]. This signifies that S1 contains the receptor binding domain (RBD) and is responsible for host determination, while S2 mediates virus fusion with host cell membrane [21]. Viral N protein is heavily phosphorylated and present adjoining RNA, is involved in viral replication cycle and influences host cell response to viral infection (Fig. 1). M protein controls the envelope structure and E protein is involved in the production and maturation of the virus (Fig. 1). Host membranes fusion with virus and internalization happen via receptor mediated endocytosis (Fig. 2) [17]. Hence, both ACE2 and TMPRSS2 are the major contributing factors in the viral entry. Afterwards, SARS-CoV-2 releases its RNA in the cell. The positive nature of genomic RNA allows its direct translation to non-structural proteins (nsp) in very first step. A ribosomal frameshift produces ORF1a and ORF1b, which gets translated into two proteins- pp1a and pp1ab, having overlapping regions (Fig. 2). Subsequently, nsp3 and nsp5 produce two proteases, explicitly- Papain-Like Proteases (PLP) and a Mpro (chymotrypsin-like protease (3CLpro)), which cleave and process pp1a and pp1ab into proteins- 1 to 11 and 12 to 16, respectively [22]. These nsp’s are actually replicase proteins, essentially an assemblage of RNA-dependent RNA polymerase (RdRp)- along with a helicase, vital for polyprotein processing and proficient viral replication [21]. Later, negative-sense strand synthesis occurs as an intermediate product (Fig. 2), tailed by its transcription to mRNAs and utilized for formation of various structural proteins. Replicated genome binds to protein N, which further interacts with structural proteins forming daughter viral particles and leaving the cell through exocytosis (Fig. 2). The daughter viral particles further infect nearby cells and viral dissemination commences. The virus has an apparent symptomless gestation phase amongst 2–14 days, throughout which the viral transmission is still possible [2].
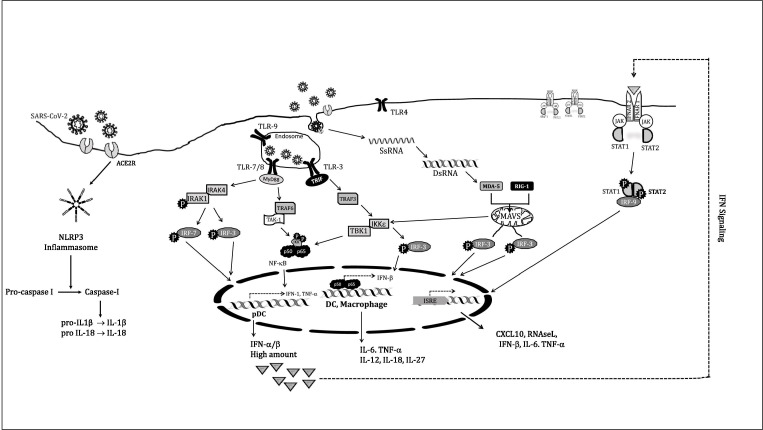
Initial (Innate) Immune Response against Virus: After internalization, the virus comes in contact with respiratory mucosal lining, patrolled by various warriors of innate response including dendritic cells (DCs) and macrophages etc [23]. However, the still evolving perception on SARS-CoV-2 progression suggests that unlike MERS and SARS-CoV-1 illness, where macrophages stand susceptible to virus, SARS-CoV-2 may not directly enter the macrophages. DC seems to be the primary cell of immune system that responds to pathogen and macrophages rather later gets involved in co-production of various cytokines [23]. Infested cells and DC respond to viral infection via IFNs (interferons) secretion, especially type-I IFN (α and β) that help achieving an antiviral state in nearby uninfected cells. It is not difficult to envision that any inadequacy in an intact innate response (IFN production) actually predisposes humans to several fatal viruses’ attack. In CoV-2 disease, IFNs are chiefly secreted by DC’s cells, categorized into- the conventional DC (cDC) and plasmacytoid DC (pDC). The pDC distinctively traverses between blood vessels to the lymphatic system and back and initiates a robust type I IFNs production [24]. cDC although contributes to IFNs production but not vigorous like pDC and rather secrete majority of pro-inflammatory cytokines. In these cells, various PRRs (Pattern Recognition Receptors) provide the patronage for virus detection. These include membrane bound TLRs (Toll like Receptors) and cytoplasm proteins, including MDA-5 (Melanoma Differentiation-Associated protein 5) and RIG-1 (Retinoic Acid-Inducible protein 1) (Fig. 3 ). They identify virus through unique signature elements called PAMP’s (Pathogen-Associated Molecular Patterns) like- dsRNA (an intermediate in certain RNA viruses’ replication) or unique SsRNA (different/ absent 5′-cap than host) etc. Both cDC and pDC have variable TLRs on plasma membrane, as well as inside endosomal membrane, which recognize pathogens (via PAMP) and initiate separate signalling cascades, involving various adaptor proteins and discussed in next section (Fig. 3) [25].
Fig. 3.
Type-I IFNs synthesis happens through multiple cascades (discussed in main text) and is indispensable to induce immunity and its expression operates in various cell types, including DC and macrophages. Chiefly pDC plays a central role in the earliest production of IFN-α/β. In response to viral infection, TLR7 and TLR9 gets stimulated and trigger a robust MyD88-dependent and IRF-7 mediated type-I IFN signalling, involving production of large amounts of IFN-α and IFN-β (IFN-α/β). Stimulation of cytosolic radars RIG-1 and MDA-5, engages the mitochondrial antiviral-signalling MAVS adaptor protein, which results in IRF3-mediated signalling. cDCs and macrophages although not responsible for the IFN-α/β production during the early phase (24 hrs) of infection but generate protective immunity during the effector phase. The IFNs secreted works in an autocrine and paracrine manner via binding to its cognate receptor and initiating JAK-STAT signalling for IFN-γ and cytokines production including IL-6 and TNFα. Virus infected cells show increased inflammation due to activation of the multiprotein inflammasome complex, NLRP3. It activates caspase-I, which triggers the release of pro-inflammatory cytokines IL-1β and IL-18, involved in pyroptosis.
pDCs express TLR-7 and TLR-9 inside endosomes, are more restricted in their antigen uptake, with lower levels of co-stimulatory and MHC (Major Histocompatibility Complex) molecules; but called “professional” DC, as they secrete very large amounts type-I IFNs (α and β both) in a short span (Fig. 3) [24]. Type-I IFNs translation is regulated by three transcription factors namely: c-jun/ATF-2, NF-κB and IFN regulatory factors (IRFs), via distinct mechanisms [7]. Pathogen unmethylated CpG motifs are recognized by TLR-9 and Ss viral RNA binds and activates TLR-7, leading to instigation of a downstream adaptor protein MyD88 (Myeloid differentiation primary response protein 88), which recruit IRAK4 (Interleukin 1R-Associated Kinase) and IRAK1 (through their death domain (DD)), followed by phosphorylation and activation of IRAK1 (Fig. 3). IRAK1 further employs the phosphorylated IRF-3 and IRF-7, which move to the nucleus and produce type-I IFNs. IRF3/7 functions as either homodimers or heterodimers with each other. Though IRF-3 is constitutively expressed in different tissues (IFN-β producers), but IRF-7 is expressed constitutively in pDC, B cells, monocytes; and is an ISG (Interferon Stimulated Gene) required for the expression of most IFN-α subtype, known as a “master regulator” among type-I IFNs [26]. This pathway in pDC is called classical pathway for type-I IFN production (Fig. 3) [24]. All TLRs (except TLR3) of APC’s (antigen presenting cells) can also initiate an IRF- independent pathway for type-I IFNs production, by activating transcription factor NF-κB. Here, MyD88 rather interacts with its downstream effector, a ubiquitin ligase TRAF6 (TNF receptor-associated factor 6), and activates TAK-1 (Transforming Growth Factor-β (TGF-β)-Activating Kinase), thus promoting downstream activation of the IκB Kinases (IKK): IKKα and IKKβ. It is already known that the IKKs phosphorylate inhibitory IκB, sequestering inactive NF-κB in the cytosol. Thus IκB phosphorylation, allow NF-κB translocation to the nucleus, followed by induction of multiple genes encoding pro-inflammatory proteins and co-stimulatory molecules such as type-I IFN, IL-6 and TNFα etc in various cells (Fig. 3).
The IFN-α secretion from pDC, is followed by activation the TLR-3 (endosomal; activated via dsRNA) in cDCs to initiate type-I IFNs production, in a MyD88-independent manner (Fig. 3). TLR-3 employs adaptor protein TRIF (TIR-domain-containing adapter-inducing interferon-β), further binding to TRAF-3 (TNF receptor-associated factors), to induce NF-κB and expression of inflammatory cytokines IL-12, TNFα, IFN-γ, IL-1β, IL-6, IL-18, IL-33 and TGF-β etc (Fig. 3) [27].
The cytoplasmic PRRs: RIG-1 (encode a helicase) and MDA-5, both contribute to type-I IFNs production in a TLR independent mechanism to produce inflammatory cytokines like TNFα, IL-1, IL-6 and IL-12 etc implicated in severity of CoV-2 infections (Fig. 3). Therefore, TLRs, MDA-5 and RIG-1, along with adaptor protein MyD88 are critical for type-I IFNs and chemokines release, which activate macrophages, monocytes, NK cells, neutrophils and pulmonary epithelia etc. Hence, DC along with other cells of immune system, provides both distinct as well as overlapping contributions in host defence against CoV-2. These cells constitute the first line of defence against viral entry and the resultant inflammatory cytokines employ monocytes, various granulocytes and lymphocytes from blood and furthermore activate the adaptive immune system [23], [28], [29]. However, lung natural killers (NK cells) do not exhibit the ACE2 receptor (like macrophages) and improbable to be directly infected by SARS-CoV-2 [23]. Even though antibodies might still stimulate NK cell (via Fc receptor) for cytokines release as well as lyses the infected cells. However, data suggest that NK cells might be more involved in cytokines production than relieving infection [23].
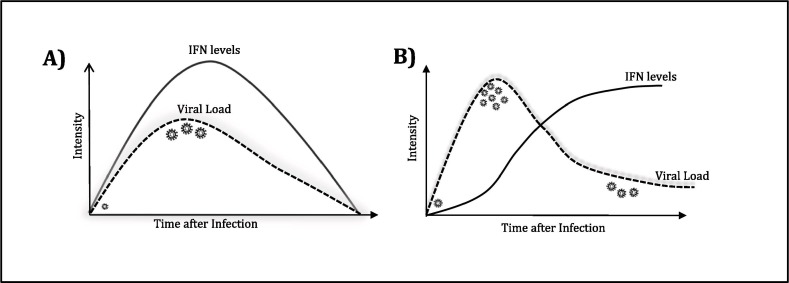
It is reported that in recovered cases, within a few hrs of virus entry, both α and β IFNs (at first day of infection) are rapidly produced and an antiviral state is soon reached [23]. This antiviral stage involves activating enzymes that can cleave both viral RNA (by oligoadenylate synthase (OAS and RNAase L) and prevents translation of structural proteins involved in viral packaging, enabling them to contain viral multiplication. It is seen that the curve of type-I IFNs production and increase in viral titre is largely superimposing each other in milder infections (Fig. 4 A) [30]. Actually, type-I IFN (both α and β) act as signalling molecules, are pleiotropic and produced from a large family of genes. They work as both autocrine and paracrine signalling cytokines, binding to both self and neighbouring nucleated cells (including macrophages, monocytes, T-lymphocytes), through their cognate receptor called IFNAR (interferon α receptor) and initiate the JAK/STAT signalling (Fig. 3). Fundamentally, type-I IFNs binds on their receptor called- IFNAR1 and IFNAR2, resulting in their dimerization, phosphorylation, and activating the Janus Kinase family members- JAK1 and TYK2. These phosphorylate and recruit STAT1/ STAT2, which collectively binds to IRF-9, and together these three (STAT1/STAT2/IRF9) form a trimeric transcription factor called ISGF3, which translocate to the nucleus and binds to ISRE on IFN-stimulated genes (ISGs) including OAS, IRF1, MX1, PKR, MDA-5, RIG-1, ZAP, ISG15, ADAR, viperin and multiple inflammatory genes like TNFα and IL-6 etc (Fig. 3). Thus, a timely type-I IFNs production, followed by JAK-STAT signalling lead to ISGs expression, which effectively contain the virus and instigate the adaptive response. On the contrary, poor outcomes are accompanied by persistent IL-6, TNFα release by various cells, with infiltrating monocytes; which pushes inflammatory reactions and monocyte-derived macrophages to amplify a dysfunctional response, leading to criticalness in patients [31].
Fig. 4.
The defensive function of Type-I IFNs in COVID-19. First panel (A) suggest that when viral load is faint, IFNs generated timely can absolve the virus effectively and patient recovers. Second panel (B) depicts a curve in patients where virus successfully evades the immune response, leading to elevated viral levels and deferred IFNs production could not control viral multiplication, leading to severity of infection.
Independently, increased inflammation is also attained due to internalization of ACE2R in virus infected cells. It reduces ACE2 levels on cell surface and in turn increases Angiotensin II levels, which binds on AT1R and results in an increase in vasoconstriction, ROS levels and NF-κB mediated inflammation [32], [33], [34]. Simultaneously, virus entry activates another class of PRR’s called NOD-like receptors (NLRs) and triggers their homo-oligomerization, which is a multiprotein inflammasome complex called NLRP3 (Nod-like receptor family, pyrin domain-containing 3). It cleaves and activate Caspase-1, which process IL-1β and IL-18 for their secretion (Fig. 3). IL-1β binds to IL-1R on macrophages and in an NF-κB dependent manner bring secretion of IL-6, TNFα and IL-1 from them. IL-18 activates CD8+ T cells to secrete IFN-γ, which mediate macrophage and NK cells differentiation [34]. This severe inflammation can lead to cell death called pyroptosis [35].
Therefore, it is mandatory to examine the different events that lead to both the delay in IFNs production and elevated inflammation, which essentially refers to viral strategies employed to avoid early recognition by the immune system and its subsequent hyper-activation, as discussed in the next section.
Evasion of Immune Response: Like many other positive-strand RNA viruses, SARS-CoV-2 has evolved different mechanisms to dodge the host responses, delay IFNs production and subsequent signalling, which contributes to the exuberant inflammatory response and critical lung immune-pathology (acute respiratory distress syndrome; ARDS) in certain patients [36].
To begin with, the viral encoded nsp3, nsp4 and nsp6 can induce formation of membranous compartments inside cytoplasm, devoted to viral RNA synthesis and let them hide their PAMPs from detection by RIG-1 and MDA5. (Fig. 2). These compartments are double membrane vesicles (DMVs) (~100–300 nm) or viral replication organelles (RO), derived from membranes of the secretory pathway (Fig. 2) [36], [37], [38]. Electron microscopic observations have confirmed that RO like structures exist and its membranes can establish contacts with ER and Golgi membranes for exchange [36], [39].
In another strategy, the virus modifies its mRNA by addition of 5′-cap (m7GpppNm-RNA) catalysed by phosphatases and methyltransferase (MTase) to avoid their detection. In this approach, crystallization studies with SARS-CoV-2, nsp16 and nsp10 have shown that nsp16-nsp10 forms a heterodimer and bring about 2′ O-methylation (MTase) of mRNA 5′-cap [40], [41]. Thus, viral RNA appears akin to the host RNA and averts any PRRs mediated detection [38], [42].
In addition, the virus has other competent strategies to circumvent the immune response and subsequently antagonize the IFNs production. The viral genome contains many accessory proteins (nsp’s), which are not critical for either duplication or discharge of viruses, but rather may have immune-evasion functions. Much of this information has come from studies of earlier spread of SARS-CoV in 2003. Virus can antagonize STAT1 activity by means of three separate mechanisms: first, nsp1, can inhibit phosphorylation of STAT1, without affecting phosphorylation of STAT2, JAK1 or TYK2 [43]. Second, viral ORF6 encodes accessory proteins, that impounds host karyopherins (nuclear import factors) to rough endoplasmic reticulum (RER)/Golgi membrane, required for bringing STAT1 in to nucleus and thus works as an IFN opponent [44]. Third, nsp3 encodes PLP, which process viral poly-peptides into efficient proteins during viral life cycle (mentioned earlier), but also antagonize innate response by deubiquitinating (DUB) and deISGylating host substrates involved in the removal of small regulatory polypeptides, ubiquitin and ISG15, respectively, from many target proteins. These DUB enzymes catalyses the removal of Ub (Ubiquitin) moieties from tagged proteins (including IRF-3 and NF-κB) and affect their stability [22], [45], [46], [47]. Additionally, PLP inhibits the RIG-1 and TLR3 mediated synthesis of the IFN-β. Similarly, deISGylating activity involves removal of ISG-15 (interferon-stimulated gene-15) moieties from marked proteins. ISG15 is a small Ub-like peptide that is covalently attached to some proteins. ISG15 is an Ub-like transformer with pleiotropic outcomes, characteristically produced in host cell immune response [38], [47].
Mitochondria is also capable of responding to viral infection and produces proteins called mitochondrial antiviral signalling (MAVS) proteins that induce cell suicide or apoptosis. This starts with a RIG-1 dependent protein, which binds to MAVS. On one hand, MAVS interacts with ER resident protein STING (Stimulator of IFN Genes), which dimerizes and in turn activate IKKε (Inducible IκB Kinase ε) and NF-κB signalling. Additionally, MAVS also activates IRF-3, making it move to the nucleus and turning on production of type I-IFNs. Conversely, viruses can antagonize the type-I IFNs response mediated by PLP, which inhibits this signalling [46].
Latest details indicate that ORF8 of SARS-CoV-2, can straightforwardly reduce MHC-I molecules surface countenance and interrupts antigen presentation, lowering the detection and eradication of virus-infected cells by Tc cells. Besides this, ORF8 can selectively target lysosomal degeneration by autophagy and in turn CTLs inefficiently eliminate the ORF8-expressing cells [48].
Effective activation of NF-κB leads to a robust production of pro-inflammatory cytokines and chemokines. Therefore, targeting NF-κB for delaying type-I IFNs production is another efficient strategy employed by viruses. NF-κB remains inactive in cytoplasm, due to attached inhibitory molecule IκB. SARS-CoV protein M inhibits phosphorylation and degradation of IκB, thus halting NF-κB relocation to nucleus [38], [49]. Apart from this, M protein antagonizes the MAVS-mediated innate response against virus by inhibiting the phosphorylation of TBK1, IKKα/β, IRF3 and p65, which in turn disrupt nuclear localization of IRF3 thus inhibits type-I IFNs production [50]. ORF3a of CoV-2 is found crucial in apoptosis (like CoV) and opposition of type-I IFNs reactions in infested cells [51]. Besides them, ORF6, ORF8 and nucleocapsid protein also inhibit interferon signalling via different mechanisms [52].
Viral N protein rather interferes with phosphorylation of IRF3, followed by resettlement to nucleus [38], [53]. Additionally, N protein modulates TGF-β signalling to obstruct apoptosis of SARS-CoV diseased host cells, with significant repercussions in tissue fibrosis [54].
SARS-CoV nsp1 can cause host mRNAs translation shutoff by attaching to cellular factors like 40S subunit of ribosomes [42]. Additionally, nsp1 protein causes host mRNA degradation by energising Xrn1 exonuclease and other proteins in cleavage machinery [55].
Besides IFN’s delay, emergent indications also suggest the role of SARS-CoV 3a protein into stimulation of the NLRP3 inflammasome, which in turn trigger caspases mediated IL-1β and IL-18 release, pyroptosis and multi-organ failure, leading to severity of infection [56].
These vital strategies reported in various fatal cases lead to a possible delay in type-I IFNs production, which bargains an early viral constraint, leading to spike in viral load (Fig. 4A & B). This could be the probable reason behind asymptomatic individuals being effective carriers. However, after the initial delay in type-I IFNs production and simultaneous successful viral replications, finally IRF-3, 7 and NF-κB mediate production of type-I IFNs, which activate the nearby cells, including macrophages and in turn helper T (Th) cells, to further secrete IFN-γ and pro-inflammatory cytokines (IL-6 and TNFα) (Fig. 4B) [1], [2]. Developed DCs trigger T cells for their pivotal linkage of onset‐increase, management and continuance of immune reactions, which also stimulates lung immunopathology with reduced survival and weakened antiviral protection [57].
Adaptive (acquired) Immune Response: As discussed, a deferral in IFNs secretion don’t authorise the immune response to obstruct intruder proliferation and viral load reaches at peak on day 10 (second week) in the upper respiratory tract (Fig. 4B) [58], [59]. However, IFNs level finally starts mounting and becomes an immune-modulator, recruiting macrophage, NK and other cells of adaptive response. This delay challenges the adaptive response with the mammoth assignment of clearing the viruses (Fig. 4B). This mismatch in the peaks of IFN secretion and viral load imparts a significant twist in disease progression, with induction of an overactive immune response, which soon gets completely de-regulated with secondary complications and fatal outcomes (Fig. 4A & B) [60]. It is known that acquired immunity involves action of both B-lymphocytes (humoral) and T-lymphocytes (cell mediated). During differentiation, T-lymphocytes express separate markers on their surface, resulting in distinctive sub-populations like CD8+ (Tc), CD4+ (Th), Tregs (regulatory T cells) and Th17 cells etc. Encompassing these cells, a delicately regulated adaptive response monitors differentiation and proliferation of various cells, with production and release of different cytokines. During these developments, the innate immune warriors still remains involved, keep on lending their support. Given this complexity, a clear picture of adaptive response is still not available and our current understanding chiefly based on the previous findings of other coronaviruses, principally SARS-CoV and MERS-CoV. Immune profiling of COVID-19 critical patients display an impaired homeostasis of various immune cells, involving a reduction in total count of T lymphocytes (both Th and Tc cells), B lymphocytes, monocytes and natural killers (NK) [61]. On the contrary, the neutrophils count and CRP levels increase considerably. Thus, it is understandable that immune dysfunction (lymphopenia) would result in immune-suppression. The altered cell homeostasis accompanies an increase in cytokines- IL-6, IL-8, IL-1, IL-17, IL-10 and TGF-β levels etc, which further augments gravity of the illness and discussed next.
The innate response cytokines- IFN-γ, GM-CSF, IL-6, TNFα and IL-2 etc, activate helper T (Th) cells, which particularly play a central role in adaptive immunity. Th0 releases IL-2 to maintain their cell number and the equilibrium between the naive and memory cells. Therefore, it’s not surprising that T-cell reduction is accompanied with reduced IL-2 levels [61]. Cytokines IL-2, IL-6, IL-12 and IL-15 activate the T-lymphocytes to proliferate and differentiate to Th (CD4+) cells and cytotoxic T cells (Tc or CD8+ cells) that can kill viral infected cells. The Th cells further secrete cytokines- IL-4, IL-5, IL-6, IL-13 and IL-21 to initiate the B-lymphocytes mediated humoral response, with production of neutralizing antibody recorded after completion of 10 days. This performs a defensive task by restraining virus in the subsequent stage and thwarts re-infiltration. This is the reason that seroconversion (undetectable stage to production of IgM followed by IgG) in 100% of infected people (with positive virus-specific IgG) is achieved 17–19 days after commencement of indications [7]. It’s daunting to disclose that the antibody’s half-life is found to be limited to a few weeks only and life-long immunity is not achievable [62].
The Th cells secrete IFN-γ and IL-2 to activate CD8+ (Tc) cells, which recognize the viral infected cells and eliminate them by apoptosis. Nevertheless, in acute conditions, lymphopenia sets in, that too specifically biased against CD8+ Tc count (than other lymphocytes), causing a decrease in CD8+ population. It is possible that various mechanisms reported in separate studies either alone or cumulatively lead to lymphopenia. One study reports that the reduction in CD8+ Tc cells is due to reduced IL-2/IL-2R and JAK1-STAT5 signalling [61]. Thus, it makes absolute sense that reduction of CD8+ count comes concurrently with drop in IL-2 and IFN-γ levels in critical patients, than severe and normal patients [61]. Apparently, premature cell death mediated by excessive levels of cytokines like IL-6, IL-10 and TNFα, affect T cells population via affecting DC and neutrophils count [63]. High expression of pro-apoptotic molecules including caspases, FAS and TRAIL also lead to low population of Tc cells [63]. Another report document that, soon after activation, CD8+ cells start expressing repressing receptors like PD1, TIM3, LAG3, CTLA4, NKG2A and CD39, resulting in Tc cell exhaustion [63], [64]. Similar to CD8+ Tc cells, CD4+ Th cells also show fatigue due to activation of markers like CD38 and HLA-DR [63]. Further, immunosuppressive Tregs cells affect immunological homeostasis by inhibiting and suppressing multiple immune cells, such as CD8+, CD4+ T cells, monocytes, NK cells, as well as B cells. Tregs compete with IL‐2 in particular and restrict proliferation of T-lymphocytes to CD8+, CD4+ T cells and also produce IL-10, IL-35 and TGF-β for inhibition of differentiation via unexplored mechanisms [65]. Tregs express inhibitory cytotoxic T lymphocyte antigen-4 (CTLA-4) and lymphocyte-activation gene-3 for deterring innate and acquired immune response. Conversely, IL-2 lead to raised apoptosis of Treg cells in mild cases [66]. Thus, if separate mechanisms contribute to lymphopenia, then its magnitude would definitely be acute and extreme, as T-lymphocytes clears about 80% of virus infected host cells. Reportedly, the memory T cells last for up to 6 years in COVID-19 patients [67].
Besides, lymphocytopenia, the critical COVID-19 patients also show Th cells imbalance, which clearly show favouring a Th2 response. Th cells differentiate and polarise into more Th2 subset than Th1, as serum display increase in levels of Th2 cytokines (IL-4, IL-5 and IL-13) except IL-10 [68]. It is noteworthy as Th2 subset mediated response is pro-inflammatory, while Th1 response is rather crucial in eliminating virus and anti-inflammatory in nature. Allegedly, IL-6 induction of JAK-STAT signalling inhibits Th1 polarization and promotes Th2 formation (2), [63]. Interestingly, elevated levels of cytokines including IL-6 gained too much highlight in serious cases and whether cytokines could become biomarker for assessing criticalness, is worth consideration. The increase in IL-10 correlates positively with IL-6 levels in ICU patients, is actually a Th1 cytokine. Its rise could be substantiated from the fact that being anti-inflammatory, IL-10 could be produced as a self-protective cytokine, attempting inhibition of multiple inflammatory molecules including- IL-4, IL-5, and IL-13 [69].
TNFα is also implicated in the ICU patients, is secreted from NK, macrophages, CD8+ and CD4+ cells. It is a tumor-suppressive cytokine (involved in apoptosis) and acts via two TNF receptors: TNFR1, (Tumor necrosis factor receptor 1) present on all cells; and TNFR2, restricted to immune cells predominantly on Tregs and can directly impact T cell biology [70]. TNFR2 actually lacks a death domain (DD) and once activated, recruits TRAF2 (TNFR-associated factor 2) and cIAP1–cIAP2 (cellular inhibitors of apoptosis proteins). This TRAF2-cIAP1–cIAP2 complex (inhibit caspases and other apoptosis-inducing factors) rather promotes cell proliferation and survival through PI3K/AKT signalling pathway [68]. Thus, promoting survival of viral infested Tc cells as well. This signalling activate NF-κB and STAT5 for secretion of IL-2 and TGF-β/IL-10 respectively [68]. Furthermore, TNFα can also modulate T-cell dynamics by protecting immunosuppressive Tregs cells via similar mechanism.
Another mechanism contributes to TNFα production. When virus enters through ACE2, it reduces the ACE2 levels on the target cells, resulting in increase of the serum AngII levels and deregulation of rennin-angiotensin pathway. AngII is a vasoconstrictor and pro-inflammatory cytokine (through AT1R). Its binding to AT1R, activates the NF-κB and metalloprotease 17 (ADAM17), which generates mature form of TNFα, along with other ligands for EGFR (Epidermal Growth Factor Receptor). ADAM17 process mIL-6R (membrane form) to sIL-6R (soluble form) and with gp-130, it activates STAT3 and NF-κB pathway (discussed later). Now both NF-κB and STAT3 trigger IL-6 amplifier, which ultimately causes hyper-activation of NF-κB and STAT3. This loop secretes IL-6 along with other pro-inflammatory cytokines, and increases COVID-19 mortality [7], [71]. IL-6 implicated as a chief cytokine performer in SARS-CoV-2 infection and CRS, thus requires a deeper understanding.
2. Role of IL-6 in CRS and Th17 response
IL-6 is both pro- and anti-inflammatory, embroiled in several viral infections [72]. In SARS-CoV infection, viral N protein triggers expression of IL-6 by attaching to NF-κB controlling element on the promoter and deletion of C-terminal (in N protein) results in failure of IL-6 activation [14]. The infection of SARS-CoV virus lacking envelope protein (E), reduces IL-6 levels and increases host survival in animal studies [73]. Various cytokines like IL-1β and TNFα also activate IL-6 expression [14], [74], [75].
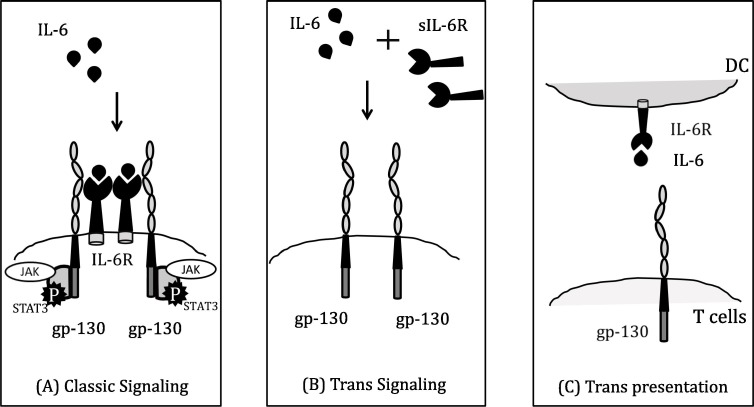
The IL-6 signalling is pleiotropic (multi-effective) and produced from many cells including- B and T lymphocytes, as well non-lymphocytic cells like macrophages, DC, monocytes, fibroblasts, vascular endothelial and mast cells etc [14], [74]. Besides viral infection and immune response, it plays a part in body temperature control, bone protection, and brain operation. It binds to its receptor (IL-6R) and activate STAT3 signalling pathway. IL-6R exists in two forms: one is membrane bound and the other one is soluble form (sIL-6R) [1], [76]. However, mere binding of IL-6 on IL-6R cannot initiate down-stream signalling, and also requires a signal transducer glycoprotein 130 (gp-130) (Fig. 5 (A)) [76]. gp-130 is expressed on most of the cells, whereas IL-6R is limited to hepatocytes, some leukocyte and epithelial cells [72]. IL-6 mediates three pathways of signal transduction- (A) Classical (Cis) signalling pathway: IL-6 interacts with IL-6R, forms a complex, which further binds to gp-130 and initiate signalling. Instigation of cis-signalling results in manifold effects on the acquired immune system (B and T lymphocytes), together with the innate response (including macrophages, neutrophils and NK cells), which contribute towards CRS. (B) Trans signalling pathway: Elevated IL-6 binds to circulating soluble IL-6R (sIL-6R) and composite IL-6/sIL-6R interacts with gp-130 (Fig. 5 (B)) [1], [72]. Thus, IL-6/sIL-6R mediated JAK-STAT3 signalling is stimulated in cells, which do not even express mIL-6R (like endothelial cells) and initiate CRS. This storm brings along release of VEGF (vascular endothelial growth factor), MCP-1, IL-8 and IL-6. VEGF causes increased vascular permeability and leakage, resulting in hypotension and ARDS. Besides this (C) Trans presentation also involves IL-6 uniting with mIL-6R present on immune cells, which forms a multiplex with gp-130 on T helper 17 (Th17) cells, directing towards lowering T-cell signalling and ARDS (Fig. 5 (C)) [74]. In addition, a natural soluble form of gp-130 (sgp-130) is found in the blood at concentrations close to 400 ng/ml [72]. This form of sgp-130 called gp-130-RAPS (translation from alternatively spliced mRNA called Rheumatoid Arthritis antigenic Peptide-bearing Soluble form) only comprises of an Ig-like N-terminal domain and a cytokine binding domain of gp-130. It has shown that like soluble; the shorter gp-130-RAPS might also connect with IL-6/sIL-6R and initiates the JAK/STAT intracellular signalling. In addition, SHP-2 phosphatase is employed to gp-130, becomes phosphorylated by JAK1 and facilitates the stimulation of the RAS-RAF-MAPK signalling route [1]. The JAK/STAT signalling (by both cis and trans signalling) and RAS-RAF, SRC-YAP-NOTCH and AKT-P13K promote the transcription of multiple downstream genes associated with T-cell clonal amplification, B-cell differentiation, acute phase response, cellular signalling activities comprising ligands, receptors, connector proteins and protein kinases etc [1], [19], [74].
Fig. 5.
IL-6 cell signalling can occur three ways: (A) Classic pathway (or cis pathway) involves membrane bound IL-6R and gp-130 whereas; (B) Trans pathway involves IL-6 binding with soluble (sIL-6R) along with gp-130. Both classic and trans pathway mediate JAK/STAT signalling to mediate CRS (C) Trans presentation occurs via dendritic cells (DC) which trans-present IL-6 via their own IL-6R to cognately interact with T cells (termed IL-6 ‘cluster signalling’), resulting in their differentiation to Th17 cells via JAK-STAT signalling. The JAK/STAT signalling (by both cis and trans signalling) and RAS-RAF, SRC-YAP-NOTCH and AKT-P13K promote the transcription of multiple downstream genes associated with T-cell clonal amplification, B-cell differentiation, acute phase response production.
In the initial stage of inflammation, IL-6 is expressed in a restricted region and later it progresses to the liver through the circulatory system, resulting in fast-moving production of a wide variety of acute phase proteins, like C-reactive protein (CRP), serum amyloid A (SAA) and fibrinogen etc. As earlier mentioned, IL-6 promotes differentiation of B-cells and production of antibodies.
IL-6 also plays a major role in Th17 cells production and Treg. IL-6 along with IL-23, TNFα, and a few other cytokines ultimately lead to activation and differentiation of naive Th cells to Th17 [1], [71]. The major cytokine secreted by Th17 cells is IL-17 only, over-produced in COVID-19 patients. IL-17 has broad pro-inflammatory effects; recruits and help in migration of neutrophils and monocytes to the site of infestation and activates other cytokine and chemokines cascades, including IL-1, IL-6, IL-8, IL-21, MCP-1 and TNFα, leading to ARDS [77]. The elevated levels of IL-6 and IL-17, is a common feature in critical COVID-19 patients. It stands clear that IL-6 and IL-17 synergistically promote the expression of pro-survival molecules Bcl-2 and Bcl-xL and prevent cellular apoptosis or obliteration of virus-restricted CD8+ T lymphocytes. The indicators of co-operation seem to be concurrent initiation of STAT3 and NF-κB via separate cytokine- supported pathways. Consequently, IL-6 and IL-17 synergistically promote viral persistence by protecting virus-infected cells.
The IL-6 overproduction and infiltration of Th17 plays a key role in acute lung injury (ALI) causing ARDS. Different cytokines may further leak into circulation causing severe inflammation and multi-organ failure. Thus, while a vigorous immune response should support in fighting the contagion, a surplus lead to accumulation of liquid in the lungs and augments the likelihood of secondary diseases contributing to lethality of infections.
Therapeutics targets and Conclusions: The concerted logical and technical attempts in terms of confirmed 600 clinical trials, are already in progress (SARS-CoV-2 on clinicaltrials.gov). The problem with designing vaccines aganist viral diseases is that animal models cannot reiterate the full clinical manifestations of infection including SARS-CoV-2. It is owing to several disparities with hosts, comprising host restriction factors and the appearance of the receptors required for viral entrance [78]. This impedes the PK/PD (pharmacokinetics/pharmacodynamics) study of vaccine assessment and advancement of a harmless prophylactic, and thus SARS vaccine remains a huge challenge initially. Recently, Sputnik-V vaccine against COVID-19 was developed and launched by Russia, and is administered to medical workers on priority basis. The production, distribution and effectiveness of the vaccine stay behind a contest and mystery to unfold in future. Few others vaccines might become accessible soon in other parts of the world.
The general challenges and safety issues with potential COVID-19 vaccines, includes- cellular immunopathology caused by virus vectors in vaccines. Mere compromised viral challenge during vaccination results in infiltration of lymphocytes, monocytes and eosinophils. It could possibly come from a Th2 and Th17 immune response as; IL-17 implicated in extravasation of eosinophils to lungs and other target organs. Another grave concern is antibody-dependent enhancement (ADE), which involves binding of non-counteracting antibodies to freshly infecting virus, and rather encourage viral entry via FcγR into host cells [77].
Theoretically, different antiviral like Remdesivir and Lopinavir plus Ritonavir can be efficient in fighting the existing outbreak. However, given the complication of CRS, it would be inappropriate to annul the virus single-handedly with the antiviral without targeting CRS in therapy. The unexpected results due to use of immune-suppressants like corticosteroids remains quite contentious as they don’t improve mortality and further delay viral clearance [11]. Lately there have been also apprehensions that steroids may intensify viral shedding and perhaps spearhead towards a greater mortality rate.
It is now clear that early detection and suitable management of this hyper-inflammation is vital in slashing the death of affected people with COVID-19. Redeployment of approved drugs is also popular as these medicines have established safety reports. Given that elevated concentrations of IL-6 are linked with both SARS-CoV plus SARS CoV-2 infections, a compelling possibility could be aiming the release of IL-6 with Tocilizumab (a recombinant humanized anti-human IL-6R monoclonal antibody of the IgG1 subtype) a prescription that targets the IL-6R (both mIL-6R and sIL-6R) and impedes the action of IL-6. It is approved for patients with rheumatoid arthritis (RA) and severe CRS in CART (Chimeric Antigen Receptor) therapy. This medicine lowers irritation and decelerates the sequence of joint damage in RA. Thus, quantity of IL-6 could be a significant biomarker in categorising infection advancement and assessing criticalness amongst COVID-19 patients. Besides this, there are several potential therapies such as inflammatory cytokine blockers (IL-6, IL-1 and IFN), immune cell depletion and stem cell therapy. It may be exploited in the identification, therapy and supervising the disease progression. The IL-6 blockers can dominate only cis- and trans signalling, whereas IL-6R antagonist can suppress trans presentation along with cis and trans signalling [11]. The antibody-based treatment is high-priced and has a limited range of outcomes. Hydroxychloroquine (or chloroquine), Baricitinib (JAK inhibitor) and Anakinra (IL-1R opponent) were suggested as possible remedies for COVID-19 as well [13].
Plasma therapy has also shown positive results in patients. It involves transferring plasma (having antibodies against virus) from recuperated patients of SARS-CoV-2 infection and may shrink death rate. Considering this, many researchers started pursuing the S protein as vaccine targets as well, including Sputnik-V. Although, reports suggest that humoral immunity may not be life lasting as rapid decay of IgG antibodies against S protein RBD was observed with half-life (t1/2) close to 36 days [62].
The other treatment option is around the understanding the delay in IFN production, culminating in release of pro-inflammatory cytokines. In China, besides safety precautions, health workers advised to inhale IFNs and this pre-treatment of cells with IFN counteracts development of SARS-CoV-2. It suggests that exogenic IFN may be adequate to overpower virus-mediated deferral of the IFN reaction. This is an indication that early diagnosis and management could be the best option in management of infection.
Funding
R. Pasrija acknowledges University Grant Commission (F.No. 42–647(2013) SR); Department of Science and Technology (SERB/F/4213/2013–14) and Department of Biotechnology IPLS funding (BT/PR4329/INF’22/1444/2011) to Maharshi Dayanand University.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Prof. Rajbir Singh, honourable Vice Chancellor, Maharshi Dayanand University, Rohtak to allow working from home under lockdown period for which this article could be written.
References
- 1.Zhang C., Wu Z., Li J.-W., Zhao H., Wang G.-Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist Tocilizumab may be the key to reduce mortality. Int. J. Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 3.Hirano T., Murakami M. COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;52:731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 6.Kuppalli K., Rasmussen A.L. A glimpse into the eye of the COVID-19 cytokine storm. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102789. 10.1016/j.ebiom.2020.102789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Sig Transduct Target Ther. 2020;5:84. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen I.-Y., Moriyama M., Chang M.-F., Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin J.-M., Bai P., He W., Wu F., Liu X.-F., Han D.-M., Liu S., Yang J.-K. gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krieger N., Chen J.T., Waterman P.D. Excess mortality in men and women in Massachusetts during the COVID-19 pandemic. Lancet. 2020;395:1829. doi: 10.1016/S0140-6736(20)31234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 12.Zhong J., Tang J., Ye C., Dong L. The immunology of COVID-19: is immune modulation an option for treatment? The Lancet Rheumatol. 2020;2:428–436. doi: 10.1016/S2665-9913(20)30120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soy M., Keser G., Atagündüz P., Tabak F., Atagündüz I., Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumat. 2020;39:2085–2094. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magro G. SARS-CoV-2 and COVID-19: is interleukin-6 (IL-6) the ‘culprit lesion’ of ARDS onset? What is there besides Tocilizumab?SGP130Fc. Cytokine. 2020;2 doi: 10.1016/j.cytox.2020.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new Coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cyranoski D. Profile of a killer: the complex biology powering the Coronavirus pandemic. Nature. 2020;581:22–26. doi: 10.1038/d41586-020-01315-7. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new Coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B., Li M., Zhou Z., Guan X., Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimm. 2020;111 doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mubarak A., Alturaiki W., Hemida M.G. Middle east respiratory syndrome coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J. Immunol. Res. 2019;2019:6491738. doi: 10.1155/2019/6491738. 10.1155/2019/6491738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Astuti I. Ysrafil, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.V. Clemente, P. D’ Arcy, M. Bazzaro, Deubiquitinating Enzymes in Coronaviruses and Possible Therapeutic Opportunities for COVID-19, Int J Mol Sci. 21 (2020) 3492. https://doi.org/doi:10.3390/ijms21103492. [DOI] [PMC free article] [PubMed]
- 23.Vabert N., Britton G.J., Gruber C., Hegde S., Joel K., Kuksin M. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzgerald-Bocarsly P., Feng D. The role of type I interferon production by Dendritic cells in host defense. Biochimie. 2007;89:843–855. doi: 10.1016/j.biochi.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xagorari A., Chlichlia K. Toll-like receptors and viruses: induction of innate antiviral immune responses. Open Microbiol. J. 2008;2:49–59. doi: 10.2174/1874285800802010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ning S., Pagano J.S., Barber G.N. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oeckinghaus A., Ghosh S. The NF-κB family of transcription factors and its regulation. Cold Spring Harbor Perspect. Biol. 2009;1 doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park A., Iwasaki A. Type I and Type III Interferons – Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frieman M., Heise M., Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133:101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vellingiri B., Jayaramayya K., Iyer M., Narayanasamy A., Govindasamy V., Giridharan B., Ganesan S., Venugopal A., Venkatesan D., Ganesan H., Rajagopalan K., Rahman P.K.S.M., Cho S.-G., Kumar N.S., Subramaniam M.D. COVID-19: A promising cure for the global panic. Sci. Total Environ. 2020;725 doi: 10.1016/j.scitotenv.2020.138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bordallo B., Bellas M., Cortez A.F., Vieira M., Pinheiro M. Severe COVID-19: what have we learned with the immunopathogenesis? Adv. Rheumatol. 2020;60:50. doi: 10.1186/s42358-020-00151-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ong J.D.H., Mansell A., Tate M.D. Hero turned villain: NLRP3 inflammasome-induced inflammation during influenza A virus infection. J. Leukoc. Biol. 2017;101:863–874. doi: 10.1189/jlb.4MR0616-288R. [DOI] [PubMed] [Google Scholar]
- 35.Zhao C., Zhao W. NLRP3 inflammasome—a key player in antiviral responses. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.G. Wolff, C.E. Melia, E.J. Snijder, M. Bárcena, Double-Membrane Vesicles as Platforms for Viral Replication, Trends Microbiol. S0966-842X (2020) 30135–9. https://doi.org/10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed]
- 37.Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daphne Y.M., Suthar M.S. Mechanisms of innate immune evasion in re-emerging RNA viruses. Curr Opin Virol. 2015;12:26–37. doi: 10.1016/j.coviro.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus non-structural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. 2013;4:e00524–e613. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Misra A., Park J.-G., Oladunni F., Kovalskyy D., Hromas R.A., Martinez-Sobrido L., Gupta Y.K. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11:3718. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Wiersum G., Kim Y., Jedrzejczak R., Maltseva N.I., Endres M., Jaroszewski L., Godzik A., Joachimiak A., Satchell K.J.F. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. BioRxiv. 2020 doi: 10.1101/2020.04.17.047498. [DOI] [Google Scholar]
- 42.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J. Innate Imun. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the Rough Endoplasmic Reticulum/Golgi Membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K., Baez-Santos Y.M., Wang J., Takayama J., Ghosh A.K., Li K., Mesecar A.D., Baker S.C. Deubiquitinating and interferon antagonism activities of Coronavirus Papain-like proteases. J. Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D.B., Clementz M.A., Banach B.S., Li K., Baker S.C., Chen Z. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S.W., Lai C.C., Ping J.F., Tsai F.-J., Wan L., Lin Y.-J., Kung S.H., Lin C.-W. Severe acute respiratory syndrome Coronavirus papain-like protease suppressed alpha interferon-induced responses through downregulation of extracellular signal-regulated kinase 1-mediated signalling pathways. J. Gen Virol. 2011;92:1127–1140. doi: 10.1099/vir.0.028936-0. [DOI] [PubMed] [Google Scholar]
- 48.Khan M.A.-A.-K., Islam A.B.M.M.K. SARS-CoV-2 proteins exploit host’s genetic and epigenetic mediators for the annexation of key host signaling pathways that confers its immune evasion and disease pathophysiology. BioRxiv. 2020 doi: 10.1101/2020.05.06.050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang X., Gao J., Zheng H., Li B., Kong L., Zhang Y., Wang W., Zeng Y., Ye L. The membrane protein of SARS-CoV suppresses NF-κB activation. J. Med Virol. 2007;79:1431–1439. doi: 10.1002/jmv.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y.-Z., Wang S.-Y., Zheng Z.-Q., Yi Huang W.-W., Li Z.-S., Xu Y.-Y. Wang. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol. Immunol. 2020:1–8. doi: 10.1038/s41423-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.-Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J.Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., Ge X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao X., Nicholls J.M., Chen Y.-G. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-beta signaling. J. Biol. Chem. 2008;283:3272–3280. doi: 10.1074/jbc.M708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakagawa K., Lokugamage K.G., Makino S. Viral and cellular mRNA translation in coronavirus-infected cells. Adv. Virus Res. 2016;96:165–192. doi: 10.1016/bs.aivir.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freeman T.L., Swartz T.H. Targeting the NLRP3 inflammasome in severe COVID-19. Front. Immunol. 2020;11:1518. doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peiris J.S.M., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L.M., Law K.I., Tang B.S.F., Hon T.Y.W., Chan C.S., Chan K.H., Ng J.S.C., Zheng B.J., Ng W.L., Lai R.W.M., Guan Y., Yuen K.Y. Clinical progression and viral load in a community outbreak of Coronavirus-associated SARS pneumonia: a prospective study. The Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheung C.Y., Poon L.L.M., Ng I.H.Y., Luk W., Sia S.-F., Wu M.H.S., Chan K.-H., Yuen K.-Y., Gordon S., Guan Y., Peiris J.S.M. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao V.U.S., Arakeri G., Subash A., Rao J., Jadhav S., Sayeed M.S., Rao G., Brennan P.A. COVID-19: loss of bridging between innate and adaptive immunity? Med. Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.109861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi H., Wang W., Yin J., Ouyang Y., Pang L., Feng Y., Qiao L., Guo X., Shi H., Jin R., Chen D. The inhibition of IL-2/IL-2R gives rise to CD8+ T cell and lymphocyte decrease through JAK1-STAT5 in critical patients with COVID-19 pneumonia. Cell Death Dis. 2020;11:429. doi: 10.1038/s41419-020-2636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild Covid-19. N Engl J Med. 2020;383:1085–1087. doi: 10.1056/NEJMx200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Z., Wherry E.J. T cell responses in patients with COVID-19. Nat Rev Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y., Qi G., Bellanti J.A., Moser R., Ryffel B., Zheng S.G. Regulatory T cells: A potential weapon to combat COVID-19? MedComm. 2020;1:157–164. doi: 10.1002/mco2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stephen-Victor E., Das M., Karnam A., Pitard B., Gautier J.-F., Bayry J. Potential of regulatory T cell-based therapies in the management of severe COVID-19. Eur. Respir. J. 2020;56:2002182. doi: 10.1183/13993003.02182-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. J. Immunol. Res. 2014;59:118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang S., Wang J., Brand D.D., Zheng S.G. Role of TNF–TNF receptor 2 signal in regulatory T cells and its therapeutic implications. Front. Immunol. 2018;9:784. doi: 10.3389/fimmu.2018.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wan S., Yi Q., Fan S., Lv J., Zhang X., Guo L., Lang C., Xiao Q., Xiao K., Yi Z., Qiang M., Xiang J., Zhang B., Chen Y., Gao C. Relationships among lymphocyte subsets, cytokines, and the pulmonary inflammation index in coronavirus (COVID-19) infected patients. Br J Haematol. 2020;189:428–437. doi: 10.1111/bjh.16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mehta A.K., Gracias D.T., Croft M. TNF activity and T cells. Cytokine. 2018;101:14–18. doi: 10.1016/j.cyto.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harbour S.N., DiToro D.F., Witte S.J., Zindl C.L., Gao M., Schoeb T.R., Jones G.W., Jones S.A., Hatton R.D., Weaver C.T. Th17 cells require ongoing classic IL-6 receptor signaling to retain transcriptional and functional identity. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.aaw2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: Importance for the pro-inflammatory activities of IL-6. Int. J. Biol. Sci. 2012;8:1237–1247. doi: 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DeDiego M.L., Nieto-Torres J.L., Regla-Nava J.A., Jimenez-Guardeño J.M., Fernandez-Delgado R., Fett C., Castaño-Rodriguez C., Perlman S., Enjuanes L. Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol. 2014;88:913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Velazquez-Salinas L., Verdugo-Rodriguez A., Rodriguez L.L., Borca M.V. The role of interleukin 6 during viral infections. Front. Microbiol. 2019;10:1057. doi: 10.3389/fmicb.2019.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation. Immun. Disease Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yawata H., Yasukawa K., Natsuka S., Murakami M., Yamasaki K., Hibi M., Taga T., Kishimoto T. Structure-function analysis of human IL-6 receptor: dissociation of amino acid residues required for IL-6-binding and for IL-6 signal transduction through gp130. EMBO J. 1993;12:1705–1712. doi: 10.1002/j.1460-2075.1993.tb05815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hotez P.J., Corry D.B., Bottazzi M.E. COVID-19 vaccine design: the Janus face of immune enhancement. Nat. Rev. Immunol. 2020;20:347–348. doi: 10.1038/s41577-020-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li C.K., Xu X. Molecular Biology of the SARS-Coronavirus. 1st ed., Springer-Verlag; Berlin Heidelberg: 2010. Host Immune Responses to SARS Coronavirus in Humans; pp. 259–278. [Google Scholar]