Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is currently creating a global health emergency. This crisis is driving a worldwide effort to develop effective vaccines, prophylactics, and therapeutics. Nucleic acid (NA)-based treatments hold great potential to combat outbreaks of coronaviruses (CoVs) due to their rapid development, high target specificity, and the capacity to increase druggability. Here, we review key anti-CoV NA-based technologies, including antisense oligonucleotides (ASOs), siRNAs, RNA-targeting clustered regularly interspaced short palindromic repeats-CRISPR-associated protein (CRISPR-Cas), and mRNA vaccines, and discuss improved delivery methods and combination therapies with other antiviral drugs.
Keywords: coronavirus (CoV), antisense oligonucleotide (ASO), lipid-ASO nanomicelle, siRNA, RNA-targeting CRISPR-Cas, mRNA vaccines
Coronaviruses and Nucleic Acid-Based Technologies
Coronaviruses (CoVs) are positive-sense, single-stranded RNA (ssRNA) viruses from the Coronaviridae family in the order Nidovirales [1]. CoVs are organized into four main genera: Alpha-, Beta-, Gamma-, and Delta-coronaviruses. Many zoonotic (see Glossary) pathogenic CoVs belong to the Betacoronavirus genus, including the severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 that is responsible for the current coronavirus disease 2019 (COVID-19) pandemic [2]. Other major zoonotic CoVs, such as the human coronaviruses (HCoVs), including HCoV-229E and HCoV-NL63 (in the Alphacoronavirus genus), and HCoV-OC43 and HCoV-HKU1 (in the Betacoronavirus genus), cause common colds in humans [3,4]. The polycistronic CoV RNA genome is approximately 30 kb long and encodes 14 open reading frames (ORFs). The 5′ proximal end of the genome contains two large ORFs that comprise two-thirds of the viral genome. These ORFs are translated into two large polyproteins that encode 16 nonstructural proteins (nsps) that are mainly involved in viral replication and transcription [5]. The remaining one-third of the viral genome encodes four major structural proteins: spike surface glycoprotein (S), membrane (M), nucleocapsid (N), envelope (E), and the accessory proteins, which are required for viral entry into the host cells and viral budding [6].
Nucleic acid (NA)-based therapies involve the use of synthetic NA molecules as tools or therapeutic agents for disease intervention mainly by modulating the target gene expression through sequence-specific recognition. Therapeutic nucleic acids (TNAs) are considered to be one of the most important contemporary drug discovery platforms in addition to small molecules and antibody therapeutics. Current TNAs include antisense oligonucleotides (ASOs), clustered regularly interspaced short palindromic repeats-CRISPR-associated protein (CRISPR-Cas) and other gene-editing tools, siRNAs, mRNAs, ribozymes, aptamers, and more [7]. These molecules act with different mechanisms of action, including modulation of gene expression by RNA interference (RNAi) or catalytic cleavage of the transcripts, and inhibition of protein–protein interactions by binding to specific peptide domains [8]. In addition, NA-based technology can be employed as vaccines by encoding viral proteins that act as antigens. TNA therapies have been developed for treatments such as cancers, central nervous system disorders, and virus infections with more than 2000 NA therapies currently undergoing clinical trials with, to date, ten NA drugs having gained approval by the FDA [7,9].
Due to lack of approved effective therapeutics and vaccines, numerous licensed or experimental drugs are currently being investigated for SARS-CoV-2 infection [10]. However, most of these drugs have thus far demonstrated only modest outcomes, with only remdesivir authorized by the FDA for emergency use during the COVID-19 pandemic [11]. On account of the current urgent situation, developing anti-CoV NA-based agents could represent a promising solution, since they are highly specific (see also druggability), easy to design, and less time consuming to develop compared with conventional therapies. Crucially, by targeting highly conserved sequences, an NA-based drug could be used to combat multiple viral strains and overcome problems of drug resistance. In this review, we evaluate several key anti-CoV NA-based technologies, including ASOs, siRNAs, RNA-targeting CRISPR-Cas systems, and mRNA vaccines (Figure 1 ) and discuss improved delivery methods and combination therapies with other antiviral drugs.
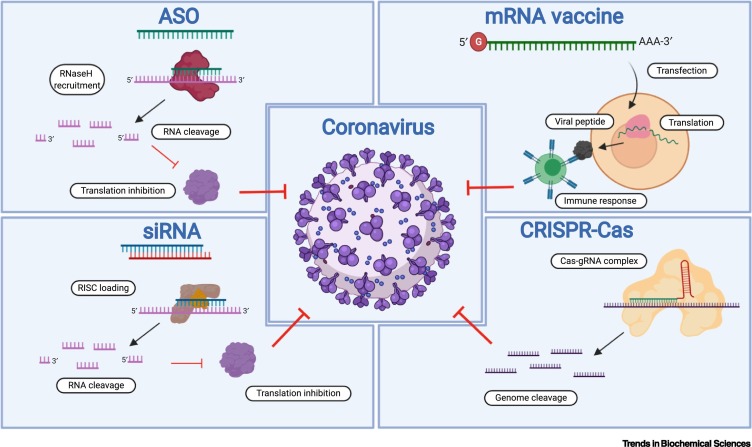
Figure 1.
Key Nucleic Acid (NA)-Based Technologies Targeting Coronavirus (CoV).
Several key NA-based strategies against CoV, including antisense oligonucleotides (ASOs), siRNA, RNA-targeting clustered regularly interspaced short palindromic repeats-CRISPR-associated protein (CRISPR-Cas) systems, and mRNA vaccines were discussed in this review. In particular, ASOs, siRNAs, and CRISPR-Cas technologies can be used as anti-CoV therapeutics through sequence-specific targeting of the RNA viral genome or host cellular genes involved in crucial viral activities (also see Figure 2, Figure 3, Figure 4). Once mRNA vaccines are delivered into the host cells, the mRNAs can be translated into viral proteins that subsequently trigger immune responses (also see Figure 5). Abbreviation: RISC, RNA-induced silencing complex.
Strategies against Ongoing and Future CoV Outbreaks
Therapeutic Targets
In principle, any region in the CoV genome can be considered as a potential therapeutic target for antiviral NA-based drugs. However, TNAs should be designed to target highly conserved sequences amongst CoV genomes to prevent potential selective pressures that generate drug-resistant virus mutants. Another benefit of this strategy is to broaden the application of these NA-based drugs to target multiple CoV strains using the same compounds. Analysis of SARS-CoV-2 genomes has revealed highly conserved viral regions, including sequences encoding RNA-dependent RNA polymerase (RdRp) and N protein, making them promising targets for developing anti-CoV TNAs [12]. Indeed, a bioinformatics analysis demonstrated that a set of six CRISPR-associated RNAs (crRNAs) designed within these conserved sites are predicted to target 91% of currently sequenced CoVs [12]. In addition, targeting host factors using different TNA strategies is worth exploring, as this may result in a cellular environment that is unfavorable for viral replication [13,14]. Moreover, host-targeted TNAs for CoV inhibition can minimize the chance of generating drug-resistant mutants because of the genetic stability of host factors [15]. Previous studies reported success in blocking viral entry using TNAs to suppress expression of host proteins that are critical for viral infection and progression, such as membrane receptor transmembrane protease serine 2 (TMPRSS2) and angiotensin-converting enzyme 2 (ACE2) [16,17].
An optimal therapeutic cocktail of multiple NA-based drugs designed to target either a single sequence or multiple sequences could be considered to enhance antiviral effects [18]. Targeting multiple viral sequences and several different viruses simultaneously has recently been successfully demonstrated for treatment of swine enteric CoVs (SECoVs) [19]. A cocktail of TNAs designed to target both viral and host genes could be used in combination. Moreover, TNAs can be used in combinational therapies with other antivirals such as nucleoside analogs (e.g., remdesivir, EIDD-2801) [20,21], protease inhibitors (e.g., ritonavir/lopinavir) [22], and cysteine protease inhibitors (e.g., MDL-28170, ONO 5334) [23]. This strategy may improve the effectiveness at a lower dose through drug synergy, thereby limiting side effects and reducing the likelihood of drug resistance. Recently, a nucleoside-modified mRNA vaccine expressing different conserved influenza virus antigens has been developed as a universal influenza virus vaccine candidate. This multiple-targeting mRNA vaccine can trigger strong immune responses in a mouse model, protecting against a panel of group 1 influenza A viruses [24]. A similar mRNA vaccination strategy holds promise for CoVs as well.
Strategies for Anti-CoV TNA Delivery
Most of TNAs to date, including approved therapeutics, are locally administered (eyes, spinal cord); a few of them are systemically delivered or targeted to the liver [9]. As the lungs are a primary site of organ damage during SARS-CoV infection, they have been one of the main targets for therapeutic compounds to impede virus progression [25]. An optimal strategy to increase the efficacy of anti-CoV TNAs is to deliver them directly to the lungs via aerosol [26]. For example, an anti-transforming growth factor (TGF)-β1 siRNA delivered via inhalation significantly inhibited bleomycin-induced pulmonary fibrosis in both acute and chronic mouse models in a dose-dependent manner [27]. The current aerosol/inhalation administered ASOs in preclinical development and in clinical trials are listed in Table 1 [28., 29., 30., 31., 32.]. Together, these results show the great potential of delivering NA drugs via aerosol administration directly to the lungs.
Table 1.
Studies Describing ASOs Administered via Aerosol
| Indication | Target | Dose/route | Clinical trial status | Experimental model | Chemistry | Observations | Refs |
|---|---|---|---|---|---|---|---|
| Cystic fibrosis | CFTRa protein | Intranasal | Phase Ib | Human | PTOb and 2′OMec | Phase Ib passed | [28] |
| Asthma | IL-4Rαd Protein | Intranasal | Preclinical/in vivo | Mice/monkeys | PTO | Attenuation of airway hyperactivity in mouse model | [29] |
| Cystic fibrosis | ENaCe proteins constituent | Intranasal | Preclinical/in vivo | Mice | Constrained ethyl (cEt) | ASO drugs can be delivered to the lungs at low doses and low volumes in simple water-based solutions | [30] |
| Pulmonary fibrosis | STAT1f protein | Intranasal | Preclinical/in vivo | Rats | PTO - liposome | Reduction of extension of alveolitis and fibrosis, reduction of the release of inflammatory cytokines | [31] |
| Asthma | CCR3g protein | Intratracheal | Preclinical/in vivo | Rats | PTO | Topical delivery of two ASOh synergizes for potent efficacy at low doses | [32] |
Cystic fibrosis transmembrane conductance regulator.
Phosphorothioate.
2′O-Methyl.
Interleukin 4 receptor α.
Epithelial sodium channel.
Signal transducer and activator of transcription 1.
C–C chemokine receptor 3.
Antisense oligonucleotide.
As many antiviral compounds are hydrophobic or only slightly soluble [33], they require encapsulation within a nanoparticle to enable their delivery. Nanomicelle technology holds great promise for combinational therapy against CoVs, where ASOs and small molecule antivirals would be conjugated together within the same vector for delivery. Lipid-modified ASOs (LASOs) are able to self-assemble into nanomicelles that significantly improved cellular entry without aid of transfection agents [34., 35., 36.]. In particular, hydrophobic chemotherapy drugs can be conjugated with these nanomicelles, facilitating their delivery [37]. Holding this advantageous property, a nanomicelle can be encapsulated with anti-CoV small molecules and applied as a combinational therapy that can be administered by aerosol inhalation. ASOs forming nanomicelles can be designed to act on the viral genomes, resulting in synergistic effects of both ASOs and encapsulated drugs in CoV inhibition (Figure 2 ). Moreover, this strategy could possibly enhance the potency of antiviral nucleoside analogs by combining with ASOs that downregulate ExoN proofreading activity of CoVs through targeting the nsp14 protein [20]. In short, the combination of LASO and hydrophobic drugs can represent a new class of therapeutic cocktail to inhibit CoVs, including SARS-CoV-2.
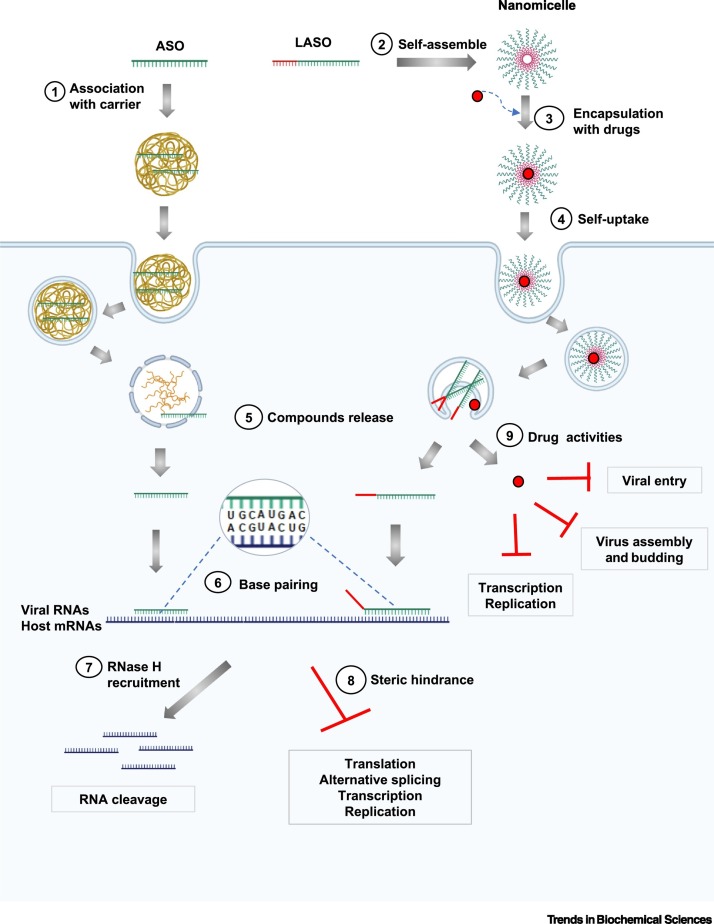
Figure 2.
Antisense Oligonucleotide (ASO) Mechanism of Action and Strategies to Combat Coronaviruses.
Association of ASOs with carriers enhances their cellular delivery (1). Alternatively, lipid-modified ASOs (LASO) can self-assemble into nanomicelles (2), which have the ability to encapsulate hydrophobic drugs such as antiviral small molecules (3) and enter the cells unaided (4). Once delivered into the cells, ASOs, LASOs, and drugs can be released (5). ASOs/LASOs base-pair with the target RNA sequences, which could be host cellular mRNA or a viral RNA genome (6). The formed DNA:RNA hybrids can induce cleavage of the RNA in the heteroduplexes (through RNase H recruitment and activity), leading to degradation of the targeted sequences (7). Binding of ASOs/LASOs to the target RNA sequences also can form a steric hindrance blocking translation or modulating alternative splicing, thereby shutting down the gene expression of host genes or disrupting RNA-based viral functions such as translation, replication, and transcription (8). In addition, the conjugated drugs can exert their activities via inhibition of viral processes such as transcription/replication, host entry, and virus assembly and budding (9). The figure was modified from Robson et al. [20], with permission, and created with BioRender.
Regarding the delivery of mRNA vaccines, several strategies already being tested in clinical trials appear promising. CureVac is currently evaluating a mRNA vaccine platform based on protamine-formulated RNA complexes for the delivery of lung and prostate cancer vaccines [38,39]. Recently, Moderna has put forward a Phase I clinical trial for an mRNA vaccine against Zika virus delivered by lipid nanoparticle (LNP) (NCT03323398). These mRNA vaccine formulations provide multiple advantages, including high potency, ease and cost-effectiveness of production, and safe administration, and could potentially be applied to the delivery of CoV mRNA vaccines into the body.
ASO Technology against CoVs
ASOs are defined as short, synthetic, single-stranded oligodeoxynucleotides (usually 15–25 nucleotides in length) that can target transcripts through Watson–Crick hybridization [40]. The resulting DNA:RNA hybrids can lead to cellular RNase H recruitment and cleavage of the RNA in the heteroduplexes by RNase H [41,42]. Alternatively, ASOs can function in an RNase H-independent manner by disrupting translation of the targeted transcripts via steric hindrance or by modulating pre-mRNA alternative splicing through blocking splicing cis-elements and/or affecting RNA structure (known as splice switching oligonucleotides) [43,44] (Figure 2). Currently, numerous ASOs are undergoing clinical trials for treatment of a variety of infectious, metabolic, and neurological human diseases, and eight ASO drugs have been approved [40].
Several early studies reported success using ASO technology to target SARS-CoV. Phosphorodiamidate morpholino-oligomers (PMO), a class of ASOs chemically modified to be uncharged, water soluble, and nuclease resistant, knocked down the target sequences by disruption of either translation or splicing of the pre-mRNA [45,46]. PMO conjugated with HIV Tat peptide, that had previously demonstrated significantly enhanced cellular delivery [47], was found to target the 5′ end of a murine CoV replicase polyprotein containing the AUG translation start site region [48]. These peptide-conjugated PMOs (P-PMO) were shown to effectively enter host cells, block translation of the ORF1ab replicase polyprotein, and inhibit CoV growth and proliferation [48]. Further study using P-PMOs specifically targeting conserved RNA elements critical for viral replication or translation, such as the transcription regulatory sequences (TRS) located in the 5′ untranslated region (UTR) of the SARS-CoV genome, demonstrated effective antiviral potency [49]. Another study reported strong antiviral effects of a P-PMO that was designed to base-pair with the 5′ terminus of the CoV RNA genome (called 5TERM PMO) [50]. Accordingly, this ASO reduced viral titers in both cell culture and in vivo mouse models [50].
siRNA against CoVs
siRNA technology takes advantage of the cellular RNAi pathway that uses RNA molecules to interact with complementary mRNAs and subsequently inhibit the expression of a particular gene [51]. Accordingly, synthetic 21–25 nucleotide double-stranded (ds) RNA molecules designed to target the gene of interest are recognized by the host RNA-induced silencing complexes (RISC) which then separate the two strands of RNA. The RISC-associated antisense strand guides the complex to the specific target sequences, leading to their cleavage and degradation [51] (Figure 3 ). siRNAs have been widely explored to combat various viruses [18] and several antiviral siRNAs have undergone clinical trials [52,53]. There have been two FDA-approved siRNAs and many clinical trials are underway for a multitude of pathologies, and translation times from bench to bedside will hopefully become shorter as the technology matures [54,55].
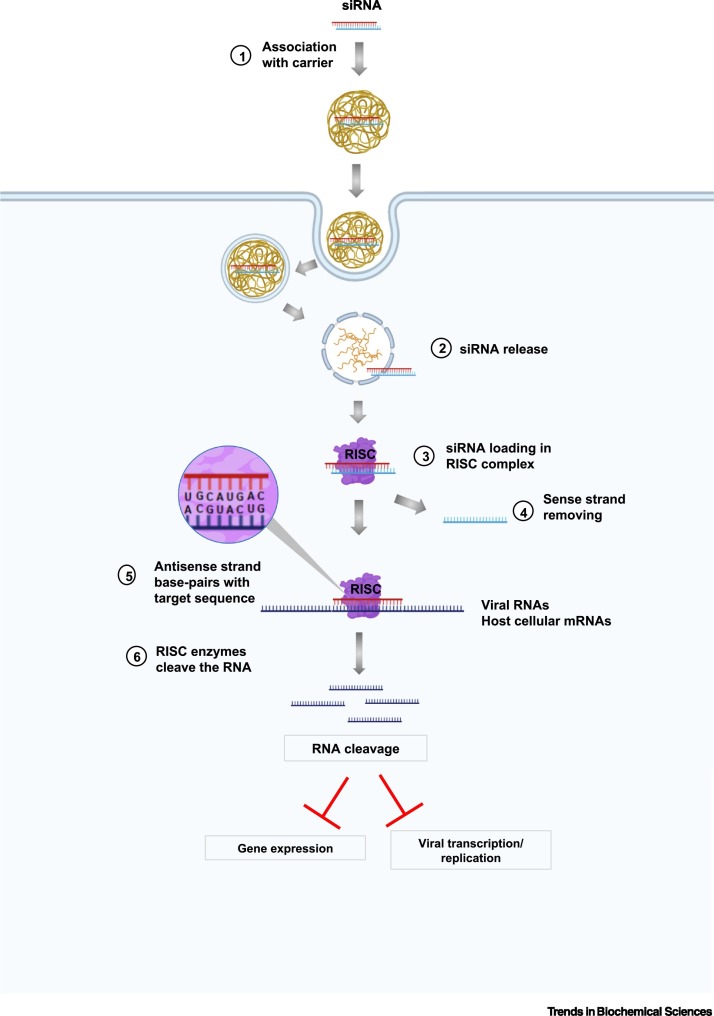
Figure 3.
Mechanism of siRNA Technology and Strategies to Downregulate Coronaviruses.
siRNA molecules are associated with carriers that facilitate their delivery into the cells (1). The siRNAs are recognized and loaded into the RNA-induced silencing complexes (RISC), which subsequently separate two strands of associated siRNAs and release the sense strands (3, 4). The RISC-associated antisense strand directs the complex to the target matching RNA sequences, which could be viral RNAs or host cellular transcripts (5), leading to RNA degradation catalyzed by RISC enzyme (6). As a consequence, these siRNAs can downregulate the expression of target host/viral genes crucial for viral activities and interrupt viral replication/transcription. Created with BioRender.
The CoV structural proteins and RdRp have been targeted by siRNAs in a number of studies due to their importance in viral replication and disease progression. Akerström et al. employed RNAi technology to lower the expression of viral proteins through targeting subgenomic RNAs (sgRNAs) [56]. They developed three siRNAs for targeting accessory proteins 7a/7b, 3a/3b, and the structural protein S. In SARS-CoV infected Vero E6 cells, a significant decrease of virus yield was observed when expressing each siRNA separately, suggesting that the accessory proteins, in addition to the well-characterized S protein, may play an important role during SARS-CoV replication [56]. Moreover, this study reported that the siRNAs can specifically inhibit target sgRNAs without affecting related sequences such as viral genomic RNA or other sgRNAs that contain only two or more mismatches (17/19 bp) with the designed siRNA, demonstrating the high sequence-specificity of the technique [56]. Another strategy aimed to target several structural proteins (S, M, E, and N) synchronously using a set of siRNAs. He et al. reported that a combination of siRNAs at very low concentration, inhibiting two structural proteins simultaneously in SARS-CoV infected cells, displayed a synergistic effect on reducing viral accumulation, which was not observed with a single siRNA at high concentration [57]. Another study explored the antiviral effect of targeting two genes synchronously by using two different small hairpin RNAs (shRNAs) targeting the M gene of porcine epidemic diarrhea virus (PEDV) and swine acute diarrhea syndrome coronavirus (SADS-CoV) and the N gene of porcine deltacoronavirus (PDCoV), respectively. Simultaneous expression of these two shRNAs displayed significant downregulation of M and N protein expression and prevented the replication of PEDV, SADS-CoV, and PDCoV simultaneously [19].
RNA-Targeting CRISPR-Cas Systems against CoVs
CRISPR-Cas9, the type II CRISPR Cas system from Streptococcus pyogenes, is one of the most frequently used RNA-guided DNA endonuclease systems that determines target sites for dsDNA cleavage via RNA–DNA hybridization between guide RNA (gRNA) and specific DNA target sequences [58., 59., 60.]. CRISPR-Cas9 as an antiviral agent can exert its effects through either targeting directly the viral genome or disrupting the expression of host proteins involved in crucial viral activities [61]. The canonical CRISPR-Cas9 system can target dsDNA viruses such as herpesviruses, human papilloma virus (HPV) and hepatitis B virus (HBV), or RNA viruses that have dsDNA intermediates during their life cycle, such as HIV [62].
The native CRISPR-Cas9 form targets DNA exclusively because its binding and catalysis relies on recognition of a protospacer adjacent motif (PAM), a short DNA sequence next to and on the strand opposite the 20-nucleotide target site in dsDNA [59,61]. However, studies have engineered CRISPR-Cas9 systems to cleave ssRNA by simultaneously delivering PAM-presenting oligonucleotides (PAMmers) that base-pair to specific sites of target ssRNA, mimicking the double-stranded target sequence [59,61]. In addition, the RNA-targeting CRISPR-Cas13d system, based on the RNA-guided RNA endonuclease derived from Ruminococcus flavefaciens XPD3002, has opened up potential therapeutic opportunities to combat RNA viruses [63,64]. The gRNA or crRNAs containing a customizable 22-nucleotide spacer sequence leads the associated Cas13d to the RNA target for RNA cleavage and gene silencing [12]. Nguyen et al. have recently presented a strategy to combat SARS-CoV-2 using the CRISPR-Cas13d system with gRNAs consisting of spacer sequences complementary to the target sequences [65]. They have also designed 10 333 potential gRNAs targeting ten coding regions of SARS-CoV-2, which specifically recognize the viral RNA genome without disrupting the human transcriptome. As a safe and effective viral vector for gene therapy in clinical experiments, adeno-associated virus (AAV) was considered as a vehicle to deliver the Cas13d effector to the patients [65]. Due to the small size, the Cas13d effector can be packed in the same AAV particle along with up to three different gRNAs, resulting in high efficacy, specificity, and a high barrier to drug resistance (Figure 4 ).
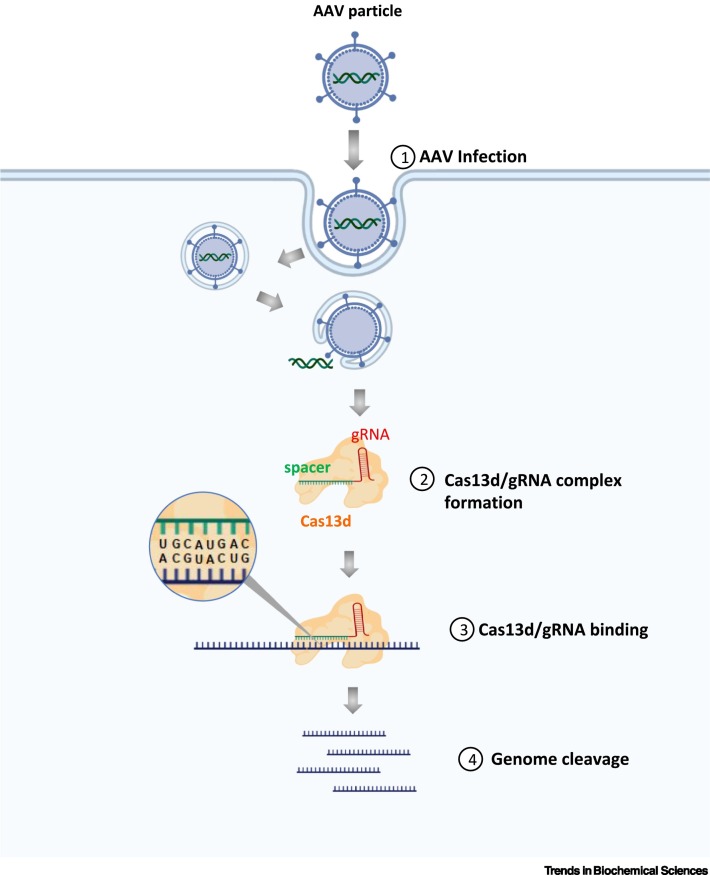
Figure 4.
RNA-Targeting Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Systems for Coronavirus Inhibition.
The adeno-associated virus (AAV) serves as a carrier to deliver vector constructs consisting of both Cas13d effector and guide RNAs (gRNAs) (1). Once inside the cell, Cas13d protein and gRNA are expressed and Cas13d protein forms a complex with gRNA (2). The spacer sequence acts as a guide to the Cas13d effector by matching to the complementary sequences in the viral RNA genome (3), thus allowing the associated Cas13d effector to cleave the viral RNA (4), therefore disrupting viral functions. Created with BioRender.
Recently, a prophylactic antiviral CRISPR-based system (called PAC-MAN) has been explored as a genetic tool to inhibit SARS-CoV-2 and influenza A virus (IAV) [12]. Using a bioinformatics pipeline, the authors identified highly conserved viral sequences across multiple SARS-CoV-2 genomes and designed potential crRNAs for targeting these regions by CRISPR-Cas13d. The Cas13d PAC-MAN effectively recognized and degraded synthesized RNA fragments of SARS-CoV-2. Due to the lack of available live SARS-CoV-2, the authors tested the potential of Cas13d PAC-MAN on an H1N1 strain of IAV. For this, a pool of IAV-targeting crRNAs was designed to act on various IAV strains and demonstrated effective inhibition of H1N1 IAV infection in human lung epithelial cells [12]. Further studies on these antiviral CRISPR-Cas systems are warranted and will accelerate the identification of safe and potent CRISPR-based antiviral strategies for treating CoV infections.
mRNA-Based Vaccines
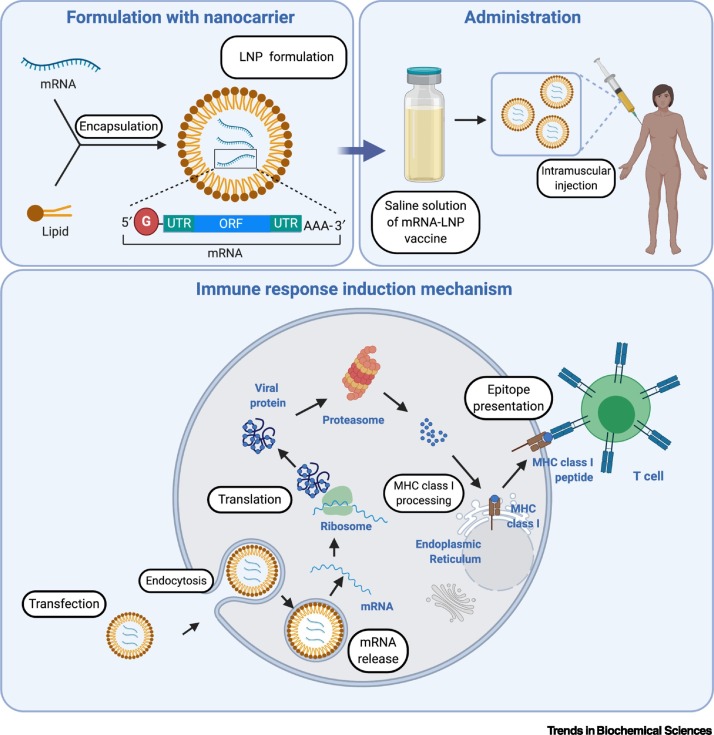
NA vaccines, consisting of either DNA or RNA encoding for antigenic proteins, employ the host’s transcriptional and translational machinery to produce a disease-specific antigen [66]. Once DNA vaccine-encoding plasmids are delivered into the host cells, the associated genes are transcribed in the nucleus under the control of eukaryotic promoters and translated in the cytoplasm [67]. By contrast, mRNA vaccines are transcribed from antigen-coding template DNA by in vitro transcription [67]. Once delivered into the cells, the mRNAs are translated in the cytoplasm to synthesize antigens of interest. The resulting proteins are subsequently processed, presented, and recognized by the immune system, thereby activating a strong humoral and T cell immune response [68] (Figure 5 ).
Figure 5.
Basic Mechanism of Action of mRNA Vaccines.
The naked mRNAs are encapsulated with a carrier such as a lipid nanoparticle (LNP), which enhances cellular delivery and stability of mRNAs. The LNP-mRNA vaccines are subsequently delivered into the body by intramuscular injection. Once LNP-mRNAs are present in the host cells, the mRNAs are released and translated by the host protein synthesis machinery. The proteasomal degradation of the generated proteins produces peptides that are subsequently associated with MHC class I molecules and presented on the surface of host antigen-presenting cells. The peptide-MHC I complexes are recognized by the CD8+ T cells, stimulating cellular immune responses. Created with BioRender. Abbreviations: MHC, major histocompatibility complex; ORF, open reading frame; UTR, untranslated region.
Numerous studies have demonstrated the safety, tolerance, and high potency of NA vaccines [67,69,70]. Moreover, the NA vaccines have a number of advantages over traditional vaccines, including their ability to deliver multiple antigens with one immunization and to stimulate both humoral and cellular immune responses, which makes, for example, tumor escape less probable [67]. mRNA vaccines are potential prophylactic and therapeutic strategies against various infectious diseases [69,70] and are attractive candidates for responding to the COVID-19 pandemic as they can be rapidly developed and their production can be scaled up within a short period of time [69,70]. RNA vaccines have several advantages over DNA vaccines, making them a more viable alternative [67,71]. HPLC-purified mRNAs containing modified nucleosides could suppress RNA-mediated immune activations and significantly enhanced translation rates compared with unpurified mRNAs [72,73]. Moreover, self-amplifying RNA (saRNA) vaccines that encode both the antigen of interest and proteins enabling RNA vaccine replication, induce immune responses with considerably lower mRNA inputs [70].
A number of mRNA vaccines against SARS-CoV-2 are currently under development and have demonstrated encouraging results, with six candidates in clinical trials (Table 2 ) [74,75]. Accordingly, an LNP mRNA-1273 vaccine developed by Moderna encodes a SARS-CoV-2 full length S protein and is currently in Phase III study after demonstrating the induction of anti-SARS-CoV-2 immune responses and acceptable safety in Phase I clinical trials [76,77]. BioNTech and Pfizer have recently reported the promising results of Phase I/II clinical trials of two LNP-formulated mRNA vaccines against SARS-CoV-2; BNT162b1 (encoding for S receptor-binding domain) and BNT162b2 (encoding for SARS-CoV-2 full-length S, stabilized in the prefusion conformation), which showed the ability to stimulate both humoral and cell-mediated antiviral mechanisms [78,79]. Due to greater safety compared with BNT162b1, BNT162b2 was finally selected for further international trial evaluations [79] and has newly achieved success in the first interim analysis from the Phase III study, with greater than 90% effectiveness in prevention of infection and no serious safety concerns thus far. Recently, repRNA-CoV2S, a lipid inorganic nanoparticles-formulated, self-amplifying RNA vaccine encoding the SARS-CoV-2 S protein was reported to stimulate SARS-CoV-2 neutralizing antibody and T cell responses in mice and non-human primates [80]. Finally, a different strategy was employed by Lu et al., their vaccine RQ3013-VLP produces SARS-CoV-2 virus-like particles in vivo by using a cocktail of mRNAs encoding three structural proteins: S, M, and E. This mRNA vaccine was incorporated with modified nucleosides and packed in LNP and demonstrated the ability to stimulate robust humoral and T cell immune response in mice [81].
Table 2.
SARS-CoV-2 mRNA Vaccine Candidates Currently in Clinical Trials
| mRNA vaccines | Sponsors/collaborators | Target | Characteristics | Current stage/study identifier |
|---|---|---|---|---|
| mRNA-1273 | ModernaTX, Inc. | Full-length, prefusion stabilized Sa protein | LNPc-encapsulated mRNA | Phase III (NCT04470427) |
| BNT162b2 | BioNTech SE + Pfizer | Full-length, prefusion stabilized S protein | LNP-encapsulated nucleoside-modified mRNA | Phase II/III (NCT04368728) |
| BNT162b1 | BioNTech SE | RBDb of S protein | LNP-encapsulated nucleoside-modified mRNA | Phase I/II (NCT04368728) |
| CVnCoV | CureVac AG | Full length S protein | LNP-encapsulated mRNA | Phase II (NCT04515147) |
| LNP-nCoVsaRNA | Imperial College London | Prefusion stabilized S protein | LNP-encapsulated self-amplifying mRNA | Phase I (ISRCTN17072692) |
| ARCoV | People’s Liberation Army (PLA) Academy of Military Sciences, Suzhou Abogen Biosciences + Walvax Biotechnology | RBD of S protein | LNP-encapsulated mRNA | Phase I (ChiCTR2000034112) |
| ARCT-021 | Arcturus Therapeutics, Inc. | Prefusion S protein | LNP-encapsulated self-replicating mRNA | Phase I/II (NCT04480957) |
Spike.
Receptor-binding domain.
Lipid nanoparticle.
Challenges and Concerns
The main challenges of TNAs in clinical applications include their safe and efficient in vivo delivery, as most NA-molecules exhibit nuclease susceptibility, poor cellular uptake, rapid clearance from circulation, and immunogenicity, as indicated in Table 3 (summarizing the pros and cons of each TNA strategy) [82., 83., 84.]. Extensive chemical modifications are required to enhance NA drug stability and delivery, thereby improving their drug-like properties [9]. For example, 2′-O-methyl (2′-OMe) modifications of the ribose sugar can significantly enhance nuclease resistance of TNAs and reduce innate immune responses [9,84,85]. Of note, up to eight of ten FDA-approved TNAs are administered without any conjugates or delivery vehicles and only rely on chemical modifications to enhance their delivery efficiency, demonstrating the great potency and importance of chemical modifications [9]. Furthermore, many current TNA investigations focus on associating TNAs with conjugations such as monoclonal antibodies (mAb), aptamers, or with nanocarriers such as LNP to facilitate their tissue distribution (Table 3).
Table 3.
Pros and Cons of Each TNA Strategy
| Therapeutic strategies | Advantages | Disadvantages | Refs |
|---|---|---|---|
| ASOs | - Easy to synthesize, lower cost - Different modes of action: transcript cleavage, steric blocking - Sufficient intracellular uptake through endocytosis without an additional delivery vehicle (all of 7 FDA- approved ASOs were not designed to associate with a delivery agent) - Low immunoreactivity - Excellent target specificity, capacity toward ‘small molecule or protein-undruggable targets’ - Advances in chemical modifications of the backbone and nucleotides increase stability from nuclease degradation and reduce toxicity - Rapid development, timely production, especially for patient-customized ASOs - Have a long history of clinical development. There have been 8 approved ASOs since 1998, demonstrating safety of ASO drugs |
- Unmodified ASOs are less stable than siRNAs - Main concerns for effective clinical applications include stability to nuclease degradation, delivery, off-target events. - In many cases, approved ASOs are extremely expensive - Accumulate in the highest concentration in liver and kidney - Host gene targeting-ASOs may cause adverse effects through affecting endogenous biological functions involved target genes |
[9,83,85., 86., 87., 88., 89., 90.] |
| siRNAs | - Natural duplex siRNAs are more stable than ASOs - High target specificity, capacity toward small molecule or protein-undruggable targets - Chemical modifications have been developed to enhance the stability and activity and reduce immune stimulation and toxicity as well - Conjugation with a ligand and/or encapsulation with a nanocarrier such as LNP enhances delivery - Two approved siRNAs including patisiran (LNPa formulation) and givosiran (GalNAc conjugate) |
- More expensive production than ASOs - Poor intracellular uptake without delivery aids - Main problems for their effective clinical application involve delivery, stability, and off-target effects - Inducing the innate immune responses by siRNA itself and its associated delivery vehicles - Accumulate in the highest concentration in liver and kidney |
[9,83,85,91., 92.] |
| CRISPR/Cas | - Powerful tool for gene editing and precision medicine - Achieve both DNA and RNA targeting - The use of synthetic gRNAs enables incorporation of chemical modifications, avoiding nuclease degradation and cellular toxicity - Nonviral delivery methods (such as LNP) can facilitate transient Cas expression, reduce off-target effects, lower immune activation, and achieve tissue-specific targeting |
- More expensive production than ASOs - Inducing immune stimulation (both innate and adaptive immune responses), poor intracellular uptake, off-target events - Adaptive immune activation and even pre-existing immune responses in human against Cas9 - Delivery of both gRNAs and Cas (4-kb gene) are crucially required - Viral delivery methods have limited packaging capacity, high immunogenic potential (especially for repeated doses), high frequency of off-target events (due to longevity of Cas expression) |
[93., 94., 95., 96.] |
| mRNA vaccines | - Safety, tolerance, and high potency - Rapid development, ease of production, and scalability - Formulating mRNA into nanocarriers such as LNP enhances efficient in vivo delivery - Safer profile than DNA vaccine. Low risk of oncogenic potential due to genomic integration as for DNA vaccines - Availability of numerous technologies can be applied to enhance stability and translation rate and reduce immunostimulatory effects of mRNA vaccines, such as: + The use of modified nucleosides reduces innate immune activations + Optimization of poly(A) tail, regulatory elements sequence, and/or codon optimization to enhance stability and translation rate + Self-amplifying RNA (saRNA) vaccines induce immune responses with considerably lower mRNA inputs |
- Naked mRNA vaccine is sensitive to nuclease degradation and exhibits poor intracellular uptake - Immunogenicity - Deep-frozen storage is required - New technology (there has not been any approved mRNA vaccine so far). Need to establish safety regulations and legislation |
[67,69., 70., 71.,97., 98., 99.] |
Lipid nanoparticle.
The safety of TNAs in clinical applications is a high priority, as potential side effects can originate from multiple factors, including chemical modifications, treatment duration, dose, or sequence. In particular, host targeted-TNAs may cause unintended toxicities as inhibition of ACE2, for example, could be proinflammatory and potentially increase mortality in COVID-19 [100]. Undesired immunosuppression events were reported for NA molecules containing specific 2′OMe motifs [84,85] and severe thrombocytopenia has been observed for two ASOs containing 2′-O-methoxy-ethyl phosphorothioate (2′-MOE PS) modifications, inotersen and volanesorsen [83]. Therefore, additional studies on optimizing chemical modifications and delivery methods that could enhance the potency and safety profiles of TNAs are highly warranted.
Concluding Remarks
Several outbreaks of serious zoonotic CoV diseases have occurred in the 21st century, such as SARS-CoV (2002–2003), MERS-CoV (ongoing since 2012), and SARS-CoV-2 (started in 2019). There are currently no effective therapeutics combating SARS-CoV-2, the infectious agent that has caused the current global pandemic, COVID-19. Scientists worldwide are engaged in a search for treatments and prophylactics against SARS-CoV-2 and related viruses. NA-based therapies that can specifically target CoVs are rationally designed based on available knowledge of the viral genome.
It is worth designing TNAs that target highly conserved sequences among CoVs, so that they can be used against a variety of viral strains and to avoid drug resistance due to selection pressure. Conserved sequences compromise functions that are central to virus life cycles; therefore, mutations occurring in these regions are unlikely to be viable. TNAs can also be designed to target host cellular factors crucial for viral progression. In addition, targeting multiple factors crucial for viral pathogenesis simultaneously by using optimal therapeutic cocktails of TNAs designed to several sequences, or using TNAs along with other antiviral drugs, are approaches worth considering. This strategy may enhance antiviral effects, allow for lower drug doses, reduce the likelihood of drug resistance occurring, and stimulate robust immune response.
However, some limitations of TNAs, such as safety profiles and delivery methods, have hindered their clinical applications. Although diverse chemical modifications and delivery vehicles have enhanced the pharmacological properties of TNAs, they can also potentially cause adverse side effects. Therefore, more studies are warranted to provide safer TNAs and more effective distribution (see Outstanding Questions). Antiviral TNAs can be administered directly to lungs by aerosol, making them promising therapeutics against respiratory CoVs. Encapsulation of antiviral drugs with nanomicelles of LASO can provide a distinct combinational treatment combating CoV infections, delivered directly to the lungs via aerosol. mRNA-based vaccines, with the encouraging results of several clinical trials, are remarkably promising bioproducts to combat the ongoing and future CoV outbreaks.
Outstanding Questions.
To what extent do host-targeted TNAs cause side effects to host cellular biological processes?
There are several CoV replication steps that are potentially targetable. Which steps are the most viable targets for TNAs to achieve therapeutically relevant viral suppression?
Despite the possibility to administer NA-based drugs via aerosol, the main limitation of TNAs is their poor distribution to other tissues or organs except the lungs. How can the targeted distribution of TNAs be improved?
TNAs generally poorly reach their target on their own. In order to enhance cellular penetration, administrations of TNAs have focused on formulating with a nanocarrier (such as liposome or exosome), or conjugation with cell-penetrating peptides, aptamers, or antibodies. What are the optimal strategies to improve cellular uptake of anti-CoV TNAs into the infected cells?
Alt-text: Outstanding Questions
Acknowledgments
This review was created by a consortium of volunteer scientists from around the world who signed up to the Crowdfight COVID-19 initiative (https://crowdfightcovid19.org) to help in the global effort against the COVID-19 pandemic. We thank our coordinator Alfonso Pérez Escudero and all at Crowdfight COVID-19 for making this review possible. W.L.N. acknowledges funding support from CUHK (the ‘Improvement on competitiveness in hiring new faculties funding scheme’, a seed fund from the Faculty of Medicine, and a PIEF grant) and the Croucher Foundation (start-up fund). P.R. acknowledges funding support from Amidex Fondation ‘Emergence & Innovation’ project.
Glossary
- Aptamers
structured, short single-stranded oligonucleotides (typically ~20–100 nucleotides) that fold into defined architectures and act as ligands binding to targets such as proteins.
- Druggability
in drug discovery, druggability refers to a biological target that is known to or is predicted to bind with high affinity to a drug-like molecule.
- Lipid nanoparticle (LNP)
lipid-based nanoparticles are widely used in nanomedicine due to their biocompatible properties; these kinds of particles are easy to prepare and allow an encapsulation of various moieties, including drugs or nucleic acids. Lipid core nanocarriers feature high-capacity reservoirs for lipophilic drug entrapment. The three main types of lipid nanoparticles are liposomes (defined as phospholipid vesicles consisting of one or more concentric lipid bilayers with an aqueous internal cavity), solid lipid nanoparticles (defined as spherical solid particles dispersed in aqueous solution), and micelles (hydrophobic solid core coated with a phospholipid monolayer).
- Nanomicelle
colloidal dispersions belonging to a group of association or amphiphilic colloids, which form spontaneously under certain concentration and temperature phases.
- Nucleoside-modified mRNA
mRNA molecules containing modified nucleosides [such as pseudouridine, 5-methylcytidine, N6-methyladenosine], which allows reduction of innate immune responses induced by mRNAs.
- Ribozymes
RNA enzymes; RNA molecules with catalytic activity or RNA–protein complexes in which solely the RNA performs the enzymatic function.
- RNA interference (RNAi)
a naturally occurring mechanism for gene silencing mediated by short fragments of double-strand RNA, which hybridize to homogenous sequences in the target mRNAs and stimulate their catalytic cleavage.
- Zoonotic pathogen
an infectious disease (zoonosis)-causing pathogen that has jumped from non-human animals to humans; zoonotic pathogens can be bacterial, viral, parasitic, or prion.
References
- 1.Sahul Hameed A.S., et al. ICTV virus taxonomy profile: Nodaviridae. J. Gen. Virol. 2019;100:3–4. doi: 10.1099/jgv.0.001170. [DOI] [PubMed] [Google Scholar]
- 2.Chan J.F.W., et al. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su S., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J., et al. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42:3–11. [PubMed] [Google Scholar]
- 7.Weng Y., et al. Improved nucleic acid therapy with advanced nanoscale biotechnology. Mol. Ther. Nucleic Acids. 2020;19:581–601. doi: 10.1016/j.omtn.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asha K., et al. Advancements in nucleic acid based therapeutics against respiratory viral infections. J. Clin. Med. Res. 2018;8 doi: 10.3390/jcm8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts T.C., et al. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh S., et al. siRNA could be a potential therapy for COVID-19. EXCLI J. 2020;19:528–531. doi: 10.17179/excli2020-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beigel J.H., et al. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbott T.R., et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–876. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S., et al. CRISPR-Cas targeting of host genes as an antiviral strategy. Viruses. 2018;10:40. doi: 10.3390/v10010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koujah L., et al. CRISPR-Cas based targeting of host and viral genes as an antiviral strategy. Semin. Cell Dev. Biol. 2019;96:53–64. doi: 10.1016/j.semcdb.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar N., et al. Host-directed antiviral therapy. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Böttcher-Friebertshäuser E., et al. Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide-conjugated morpholino oligomer targeting the hemagglutinin-activating protease TMPRSS2. J. Virol. 2011;85:1554–1562. doi: 10.1128/JVI.01294-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu C.-Y., et al. siRNA silencing of angiotensin-converting enzyme 2 reduced severe acute respiratory syndrome-associated coronavirus replications in Vero E6 cells. Eur. J. Clin. Microbiol. Infect. Dis. 2008;27:709–715. doi: 10.1007/s10096-008-0495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qureshi A., et al. A review on current status of antiviral siRNA. Rev. Med. Virol. 2018;28 doi: 10.1002/rmv.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li K., et al. Significant inhibition of re-emerged and emerging swine enteric coronavirus in vitro using the multiple shRNA expression vector. Antivir. Res. 2019;166:11–18. doi: 10.1016/j.antiviral.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robson F., et al. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell. 2020;79:710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo H.S., et al. Simeprevir potently suppresses SARS-CoV-2 replication and synergizes with remdesivir. bioRxiv. 2020 doi: 10.1101/2020.05.26.116020. Published online September 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao B., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe COVID-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riva L., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freyn A.W., et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., et al. New understanding of the damage of SARS-CoV-2 infection outside the respiratory system. Biomed. Pharmacother. 2020;127:110195. doi: 10.1016/j.biopha.2020.110195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu C.-X., et al. Poly(ester amine)-mediated, aerosol-delivered Akt1 small interfering RNA suppresses lung tumorigenesis. Am. J. Respir. Crit. Care Med. 2008;178:60–73. doi: 10.1164/rccm.200707-1022OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Alessandro-Gabazza C.N., et al. Development and preclinical efficacy of novel transforming growth factor-β1 short interfering RNAs for pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2012;46:397–406. doi: 10.1165/rcmb.2011-0158OC. [DOI] [PubMed] [Google Scholar]
- 28.Beumer W., et al. Evaluation of eluforsen, a novel RNA oligonucleotide for restoration of CFTR function in in vitro and murine models of p.Phe508del cystic fibrosis. PLoS One. 2019;14 doi: 10.1371/journal.pone.0219182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fey R.A., et al. Local and systemic tolerability of a 2’O-methoxyethyl antisense oligonucleotide targeting interleukin-4 receptor-α delivery by inhalation in mouse and monkey. Inhal. Toxicol. 2014;26:452–463. doi: 10.3109/08958378.2014.907587. [DOI] [PubMed] [Google Scholar]
- 30.Crosby J.R., et al. Inhaled ENaC antisense oligonucleotide ameliorates cystic fibrosis-like lung disease in mice. J. Cyst. Fibros. 2017;16:671–680. doi: 10.1016/j.jcf.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Wang W.J., et al. The effects of aerosolized STAT1 antisense oligodeoxynucleotides on rat pulmonary fibrosis. Cell. Mol. Immunol. 2009;6:51–59. doi: 10.1038/cmi.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allakhverdi Z., et al. Multitargeted approach using antisense oligonucleotides for the treatment of asthma. Ann. N. Y. Acad. Sci. 2006;1082:62–73. doi: 10.1196/annals.1348.047. [DOI] [PubMed] [Google Scholar]
- 33.Chauhan G., et al. Nanotechnology for COVID-19: therapeutics and vaccine research. ACS Nano. 2020;14:7760–7782. doi: 10.1021/acsnano.0c04006. [DOI] [PubMed] [Google Scholar]
- 34.Karaki S., et al. Lipid-oligonucleotide conjugates improve cellular uptake and efficiency of TCTP-antisense in castration-resistant prostate cancer. J. Control. Release. 2017;258:1–9. doi: 10.1016/j.jconrel.2017.04.042. [DOI] [PubMed] [Google Scholar]
- 35.Barthélémy, P. et al. Institut National de la Sante et de la Recherche Medicale (INSERM). Hydrophobically modified antisense oligonucleotides comprising a triple alkyl chain, 001516
- 36.Barthélémy, P. et al. Institut National de la Sante et de la Recherche Medicale (INSERM). Hydrophobically modified antisense oligonucleotides comprising a ketal group, 001517
- 37.Barthélémy, P. et al. Institut National de la Sante et de la Recherche Medicale (INSERM). Evaluation de l’autonanovectorisation de l'oligonucléotide anti-TCTP comme stratégie innovante pour le traitement des cancers de prostate résistants à la castration, 16349
- 38.Sebastian M., et al. Phase Ib study evaluating a self-adjuvanted mRNA cancer vaccine (RNActive®) combined with local radiation as consolidation and maintenance treatment for patients with stage IV non-small cell lung cancer. BMC Cancer. 2014;14:748. doi: 10.1186/1471-2407-14-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rausch S., et al. mRNA vaccine CV9103 and CV9104 for the treatment of prostate cancer. Hum. Vaccin. Immunother. 2014;10:3146–3152. doi: 10.4161/hv.29553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhuri K., et al. Antisense oligonucleotides: an emerging area in drug discovery and development. J. Clin. Med. Res. 2020;9:2004. doi: 10.3390/jcm9062004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scoles D.R., et al. Antisense oligonucleotides: a primer. Neurol. Genet. 2019;5 doi: 10.1212/NXG.0000000000000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karaki S., et al. In: Antisense Therapy. Sharad S., Kapur S., editors. IntechOpen; 2019. Antisense oligonucleotides, a novel developing targeting therapy. [Google Scholar]
- 43.Ottesen E.W. ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl. Neurosci. 2017;8:1–6. doi: 10.1515/tnsci-2017-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh N.N., et al. Pre-mRNA splicing modulation by antisense oligonucleotides. Methods Mol. Biol. 2018;1828:415–437. doi: 10.1007/978-1-4939-8651-4_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hudziak R.M., et al. Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev. 1996;6:267–272. doi: 10.1089/oli.1.1996.6.267. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh C., et al. Evaluation of antisense mechanisms of action. Methods Enzymol. 2000;313:135–143. doi: 10.1016/s0076-6879(00)13008-3. [DOI] [PubMed] [Google Scholar]
- 47.Moulton H.M., et al. HIV Tat peptide enhances cellular delivery of antisense morpholino oligomers. Antisense Nucleic Acid Drug Dev. 2003;13:31–43. doi: 10.1089/108729003764097322. [DOI] [PubMed] [Google Scholar]
- 48.Neuman B.W., et al. Antisense morpholino-oligomers directed against the 5’ end of the genome inhibit coronavirus proliferation and growth. J. Virol. 2004;78:5891–5899. doi: 10.1128/JVI.78.11.5891-5899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neuman B.W., et al. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 2005;79:9665–9676. doi: 10.1128/JVI.79.15.9665-9676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burrer R., et al. Antiviral effects of antisense morpholino oligomers in murine coronavirus infection models. J. Virol. 2007;81:5637–5648. doi: 10.1128/JVI.02360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dana H., et al. Molecular mechanisms and biological functions of siRNA. Int. J. Biomed. Sci. 2017;13:48–57. [PMC free article] [PubMed] [Google Scholar]
- 52.Choi J.H., Croyle M.A. Emerging targets and novel approaches to Ebola virus prophylaxis and treatment. BioDrugs. 2013;27:565–583. doi: 10.1007/s40259-013-0046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeVincenzo J., et al. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim Y.-K. RNA therapy: current status and future potential. Chonnam Med. J. 2020;56:87–93. doi: 10.4068/cmj.2020.56.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saw P.E., Song E.-W. siRNA therapeutics: a clinical reality. Sci. China Life Sci. 2020;63:485–500. doi: 10.1007/s11427-018-9438-y. [DOI] [PubMed] [Google Scholar]
- 56.Akerström S., et al. Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S. Antivir. Res. 2007;73:219–227. doi: 10.1016/j.antiviral.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He M.-L., et al. Kinetics and synergistic effects of siRNAs targeting structural and replicase genes of SARS-associated coronavirus. FEBS Lett. 2006;580:2414–2420. doi: 10.1016/j.febslet.2006.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garneau J.E., et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 59.O’Connell M.R., et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiedenheft B., et al. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 61.Soppe J.A., Lebbink R.J. Antiviral goes viral: harnessing CRISPR/Cas9 to combat viruses in humans. Trends Microbiol. 2017;25:833–850. doi: 10.1016/j.tim.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 62.Bayat H., et al. The impact of CRISPR-Cas system on antiviral therapy. Adv. Pharm. Bull. 2018;8:591–597. doi: 10.15171/apb.2018.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Konermann S., et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan W.X., et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell. 2018;70:327–339. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen T.M., et al. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020;30:189–190. doi: 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Restifo N.P., et al. The promise of nucleic acid vaccines. Gene Ther. 2000;7:89–92. doi: 10.1038/sj.gt.3301117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McNamara M.A., et al. RNA-based vaccines in cancer immunotherapy. J Immunol Res. 2015;2015:794528. doi: 10.1155/2015/794528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yi C., et al. mRNA vaccines: possible tools to combat SARS-CoV-2. Virol. Sin. 2020;35:259–262. doi: 10.1007/s12250-020-00243-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang C., et al. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogel A.B., et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verbeke R., et al. Three decades of messenger RNA vaccine development. Nano Today. 2019;28:100766. [Google Scholar]
- 72.Pardi N., Weissman D. Nucleoside modified mRNA vaccines for infectious diseases. Methods Mol. Biol. 2017;1499:109–121. doi: 10.1007/978-1-4939-6481-9_6. [DOI] [PubMed] [Google Scholar]
- 73.Karikó K., et al. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Locht C. Vaccines against COVID-19. Anaesth. Crit. Care Pain Med. 2020;39:703–705. doi: 10.1016/j.accpm.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le T.T., et al. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- 76.Jackson L.A., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Corbett K.S., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sahin U., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 79.Walsh E.E., et al. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Erasmus J.H., et al. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu J., et al. A COVID-19 mRNA vaccine encoding SARS-CoV-2 virus-like particles induces a strong antiviral-like immune response in mice. Cell Res. 2020;30:936–939. doi: 10.1038/s41422-020-00392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dammes N., Peer D. Paving the road for RNA therapeutics. Trends Pharmacol. Sci. 2020;41:755–775. doi: 10.1016/j.tips.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin W., Rogge M. Targeting RNA: a transformative therapeutic strategy. Clin. Transl. Sci. 2019;12:98–112. doi: 10.1111/cts.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alharbi A.S., et al. Rational design of antisense oligonucleotides modulating the activity of TLR7/8 agonists. Nucleic Acids Res. 2020;48:7052–7065. doi: 10.1093/nar/gkaa523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Khvorova A., Watts J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stebbins C.C., et al. Immunogenicity for antisense oligonucleotides: a risk-based assessment. Bioanalysis. 2019;11:1913–1916. doi: 10.4155/bio-2019-0133. [DOI] [PubMed] [Google Scholar]
- 87.Kim J., et al. Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 2019;381:1644–1652. doi: 10.1056/NEJMoa1813279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chi X., et al. Safety of antisense oligonucleotide and siRNA-based therapeutics. Drug Discov. Today. 2017;22:823–833. doi: 10.1016/j.drudis.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 89.Prasad V. Nusinersen for spinal muscular atrophy: are we paying too much for too little? JAMA Pediatr. 2018;172:123–125. doi: 10.1001/jamapediatrics.2017.4360. [DOI] [PubMed] [Google Scholar]
- 90.Geary R.S., et al. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 91.Gaglione M., Messere A. Recent progress in chemically modified siRNAs. Mini Rev. Med. Chem. 2010;10:578–595. doi: 10.2174/138955710791384036. [DOI] [PubMed] [Google Scholar]
- 92.Meng Z., Lu M. RNA interference-induced innate immunity, off-target effect, or immune adjuvant? Front. Immunol. 2017;8:331. doi: 10.3389/fimmu.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Basila M., et al. Minimal 2′-O-methyl phosphorothioate linkage modification pattern of synthetic guide RNAs for increased stability and efficient CRISPR-Cas9 gene editing avoiding cellular toxicity. PLoS One. 2017;12 doi: 10.1371/journal.pone.0188593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilbie D., et al. Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc. Chem. Res. 2019;52:1555–1564. doi: 10.1021/acs.accounts.9b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Charlesworth C.T., et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xing H., Meng L.-H. CRISPR-Cas9: a powerful tool towards precision medicine in cancer treatment. Acta Pharmacol. Sin. 2020;41:583–587. doi: 10.1038/s41401-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pardi N., et al. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boo S.H., Kim Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020;52:400–408. doi: 10.1038/s12276-020-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 100.McLachlan C.S. The angiotensin-converting enzyme 2 (ACE2) receptor in the prevention and treatment of COVID-19 are distinctly different paradigms. Clin. Hypertens. 2020;26:14. doi: 10.1186/s40885-020-00147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]