Abstract
Synovitis refers to the inflammation of the synovial membrane and is commonly detected in patients with osteoarthritis (OA). Recent reports have suggested that microRNAs (miRNAs) could be a promising target for diagnosis and prognosis in OA. This study examines the effect of microRNA-10a (miR-10a) in fibroblast-like synoviocyte (FLS)-mediated synovitis obtained from patients with OA. Expression of miR-10a is negatively associated with the severity of synovitis. miR-10a inhibited proliferation, migration, and secretion of pro-inflammatory cytokines of OA-FLS that were obtained from OA patients in vitro. By using a patient-derived xenograft (PDX) model, miR-10a repressed proliferation of OA-FLSs and production of OA synovium-derived pro-inflammatory cytokines in vivo. Twist Family BHLH Transcription Factor 1 (TWIST1) and mitogen-activated protein kinase kinase kinase 7 (MAP3K7) were identified as an upstream regulator and direct target of miR-10a in OA-FLSs, respectively. Nuclear factor κB (NF-κB) signaling pathway, a downstream pathway of MAP3K7, was also repressed by miR-10a in OA-FLSs. To summarize, the TWIST1-miR-10a-MAP3K7-NF-κB pathway mediates the development of synovitis in OA. miR-10a functions as an anti-inflammatory mediator in OA-FLS.
Keywords: miR-10a, OA, FLS, synovitis, TWIST1, MAP3K7, NF-κB
Graphical Abstract
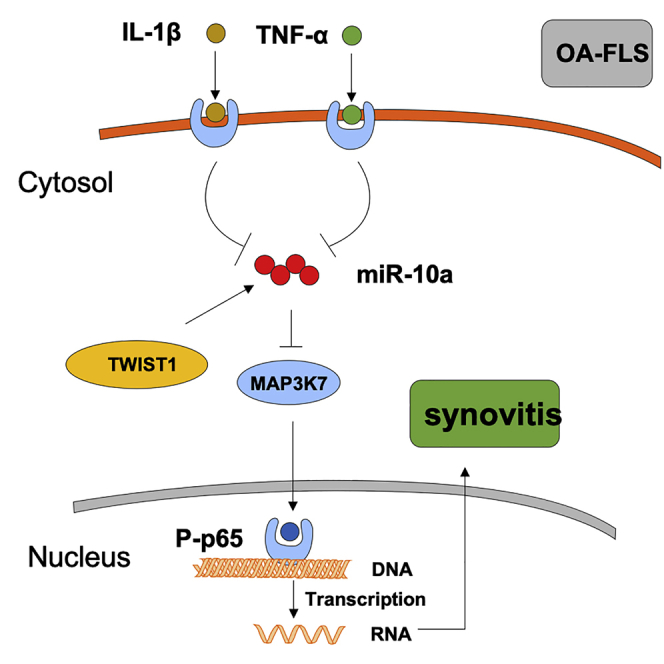
Synovitis refers to the inflammation of the synovial membrane and is commonly detected in patients with osteoarthritis (OA). Dr. Zhu and colleagues demonstrate the essential role of TWIST1-miR-10a-MAP3K7-nuclear factor κB (NF-κB) axis in fibroblast-like synoviocyte (FLS)-mediated synovitis of OA patients, which provides novel understanding for treating patients with OA synovitis.
Introduction
Osteoarthritis (OA) is a chronic progressive disease characterized by cartilage degradation, bone erosion, and synovitis.1 The research field of OA mainly focuses on the pathological mechanisms of cartilage degradation and bone erosion, whereas the role of synovitis in OA pathogenesis has been long overlooked. Recently, researchers took note of the essential role of synovitis in OA and investigated the effects of synovitis on joint degeneration in the disease.2
MicroRNAs (miRNAs) are a class of small non-coding RNAs that repress their target genes at the post-transcriptional level. miRNAs have been shown to play versatile roles in many physiological and pathological processes. Recently, miRNAs were shown to be involved in inflammation of the joint synovium in patients suffering from rheumatoid arthritis (RA).3 However, the potential role of miRNAs in OA synovitis has not yet been elucidated. MicroRNA-10a (miR-10a), a member of the miR-10 family, plays an essential role in various cancers.4 miR-10a-5p is the main nature form of miR-10a precursor.5 Recently, a few key studies have identified the anti-inflammatory role of the miR-10 family in RA6,7 and patients with ankylosing spondylitis.8 However, there have been no studies undertaken to investigate the specific role of miR-10a-5p in OA synovitis until now. Due to the long-term underrated role of synovitis in OA and the anti-inflammatory effects of miR-10a-5p in other autoimmune diseases, we try to illustrate the role of miR-10a-5p in synovitis in OA.
Fibroblast-like synoviocytes (FLSs) are one of the main cellular components involved in OA synovitis.9 The pro-inflammatory role of FLSs has been extensively demonstrated in the joint synovium of RA patients. Activated FLSs secrete pro-inflammatory chemokines to recruit immunocytes and produce cytokines to promote angiogenesis, as well as extracellular matrix degradation.10 Although FLS-mediated inflammation in OA is not as severe as in RA, OA-FLSs have demonstrated obvious pro-inflammatory properties compared with those of healthy synovium.2,11 Because both miR-10a and FLSs are essential components in joint synovitis, the present study aimed to investigate the role of miR-10a in FLS-mediated OA synovitis.
Results
The Expression of miR-10a in Blood, Synovium, and FLSs of OA
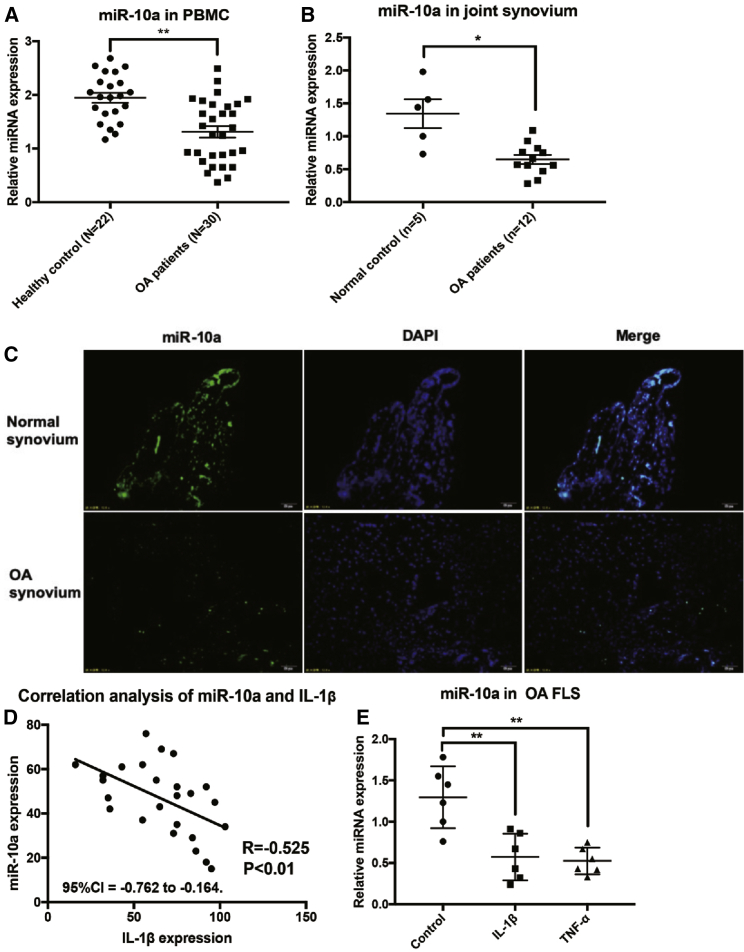
First, the expression of mature miR-10a was quantified in peripheral blood mononuclear cells (PBMCs) from OA patients and healthy controls by qRT-PCR. The relative expression of miR-10a was significantly repressed in the PBMCs from OA patients compared with healthy controls (Figure 1A). In contrast, we demonstrated that in synovial tissue, miR-10a expression was significantly decreased in OA patients compared with control patients (Figure 1B). Fluorescence in situ hybridization (FISH) results further validated the relatively lower expression of miR-10a in OA synovium compared with the normal control (Figure 1C). Hematoxylin and eosin (H&E) staining was used to show the tissue architecture of normal and OA synovia (Figure S1). To assess whether synovial expression of miR-10a reflects disease activity, we examined the relationship of synovial tissue expressions between miR-10a and IL-1β, a key pro-inflammatory marker of synovitis. Synovial tissue expression of miR-10a was inversely correlated to production of IL-1β (Figure 1D). To further illustrate the regulatory role of miR-10a in synovitis in OA-FLS, we quantified the expression of miR-10a in OA-FLSs following stimulation with two pro-inflammatory cytokines, interleukin-1β (IL-1β) and tumor necrosis factor alpha (TNF-α). Both IL-1β and TNF-α significantly repressed endogenous expression of miR-10a in OA-FLS (Figure 1E). Together, these results suggested that miR-10a might play an anti-inflammatory role in synovitis of OA.
Figure 1.
The Expression of miR-10a in Blood, Synovium, and FLSs of OA
(A) The expression of miR-10a in blood from OA patients (n = 30) and healthy controls (n = 22). (B) The expression of miR-10a in synovium from OA patients (n = 12) and controls (n = 3). (C) The localization of miR-10a in the synovium from OA patients (n = 3) and controls (n = 3). (D) The correlation (Pearson’s correlation coefficient) of miR-10a expression and OA severity (n = 25). (E) The response of miR-10a to IL-1β (10 ng/mL) and TNF-α (20 ng/mL) treatment in OA-FLSs for 48 h (n = 6). Data represent means ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, as determined by Student’s t test.
The Effects of miR-10a on Human OA-FLSs In Vitro
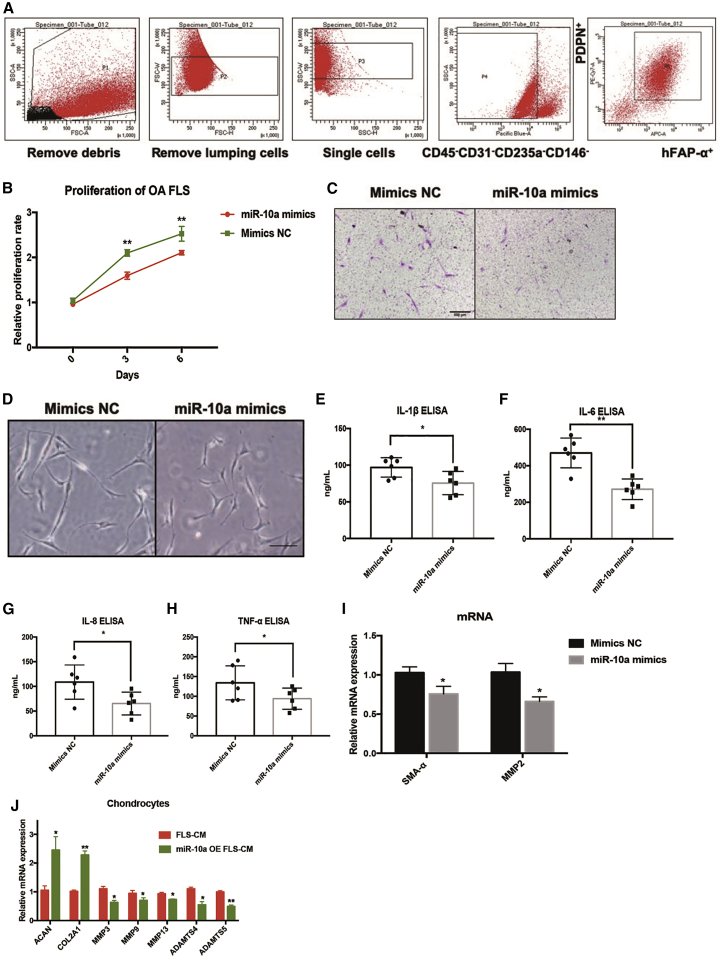
To investigate the contribution of miR-10a to OA-FLS pro-inflammatory mechanisms, we sorted OA-FLSs (CD45−CD31−CD235a−CD146−PDPN+FAPα+; myeloid cells were excluded with CD45, endothelial cells were excluded with CD31, red blood cells were excluded with CD235a, and pericytes were excluded with CD146; then two established FLS markers, Podoplanin [PDPN] and FARα,12 were used to sort FLSs) and cultured them in vitro (Figure 2A). OA-FLSs were transfected with miR-10a mimics or negative control of mimics (mimics NC). A significant upregulation of miR-10a expression was detected by qRT-PCR in OA-FLSs (Figure S2). Next, we assessed the proliferation of OA-FLSs following transfection with miR-10a mimics or mimics NC. miR-10a overexpression significantly decreased OA-FLS proliferation (Figure 2B). A transwell assay was further used to investigate the effect of miR-10a on migration of OA-FLSs. miR-10a mimics repressed the migratory ability of OA-FLSs (Figure 2C). In addition, the morphological elongation of the fibroblasts indicated better migratory ability. The miR-10a mimics inhibited the elongated shape of OA-FLSs (Figure 2D), suggesting that miR-10a could repress migration of OA-FLSs. To elucidate the role of miR-10a in regulating the pro-inflammatory phenotype of OA-FLSs, we quantified the secretion of key pro-inflammatory mediators by enzyme-linked immunosorbent assays (ELISAs). In OA-FLSs transfected with miR-10a mimics, levels of IL-1β, IL-6, IL-8, and TNF-α were downregulated compared with OA-FLSs transfected with mimics NC (Figure 2E–2H). In addition, two fibrotic markers, α-smooth muscle actin (α-SMA) and matrix metallopeptidase 2 (MMP2), were also repressed by miR-10a in OA-FLSs (Figure 2I). The interaction between OA-FLS and chondrocyte is essential for OA cartilage pathogenesis.13 To evaluate the effects of miR-10a-overexpressing (OE) OA-FLSs on OA-chondrocyte, we collected the conditional medium (CM) from miR-10a-OE OA-FLSs to culture OA-chondrocyte in vitro. miR-10a-OE OA-FLS CM induced chondrogenic genes ACAN and COL2A1 and repressed OA pathogenesis genes MMP3, MMP9, MMP13, ADAMTS4, and ADAMTS5 in primary chondrocytes (Figure 2J). Taken together, these results suggested that miR-10a could repress the aggressive properties of OA-FLS.
Figure 2.
The Effects of miR-10a on Human OA-FLSs In Vitro
(A) The sorting strategy of OA-FLSs from replaced OA synovial tissues (n = 3). (B) miR-10a represses proliferation of human OA-FLSs in vitro (n = 3). (C) miR-10a inhibits migration of human OA-FLSs in vitro (n = 3). (D) miR-10a represses elongation of OA-FLSs in vitro (n = 3). (E–H) miR-10a reduces the production of pro-inflammatory cytokines in human OA-FLSs in vitro (n = 6). (I) miR-10a represses mRNA expressions of fibrotic markers in OA-FLSs (n = 3). (J) miR-10a-OE OA-FLS CM induced chondrogenic genes and repressed OA pathogenesis genes in primary chondrocytes. Data represent means ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, as determined by the Student’s t test. (E–I) *p < 0.05, **p < 0.01, compared to mimic NC group; (J) *p < 0.05, **p < 0.01, compared to FLS-CM.
The Effects of miR-10a on OA Synovitis in a PDX Model In Vivo
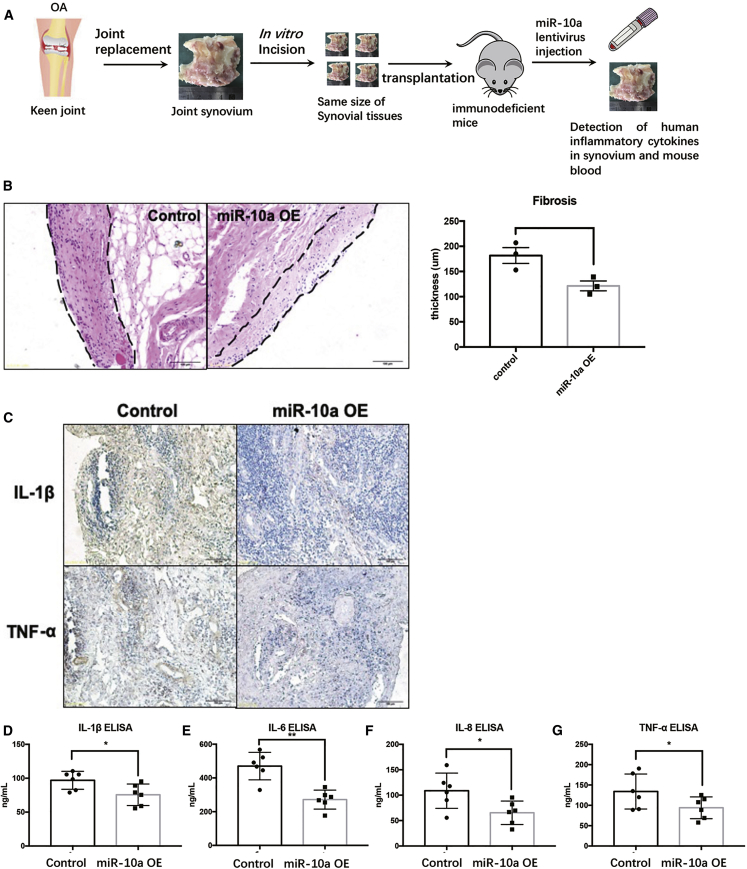
To further evaluate the ameliorative effects of miR-10a on OA synovitis in vivo, we established a patient-derived xenograft (PDX) model (Figure 3A). Severe combined immunodeficiency (SCID) mice were engrafted with human OA synovial tissue and treated with miR-10a-OE lentiviruses. Three days after transplantation of OA synovial tissue into SCID mice, miR-10a-OE lentiviruses and control lentiviruses were injected into the transplanted synovium (Figure S3). Two weeks later, engraft synovial tissues were collected for analysis. Then H&E staining was carried out to identify the severity of synovitis in the transplanted synovium. The thickness of the FLS layer was less in miR-10a-OE OA synovium (Figure 3B). In addition, immunohistochemistry (IHC) and ELISA were used to detect the expression of human pro-inflammatory cytokines in engrafted synovial tissues and mice serum. Results demonstrated that human IL-1β, IL-6, IL-8, and TNF-α were clearly present in the serum of the engrafted mice and downregulated after miR-10a overexpression (Figure 3C–3G). Overall, these data showed that the severity of OA synovitis decreased after miR-10a overexpression in vivo.
Figure 3.
The Effects of miR-10a on OA Synovitis in an In Vivo PDX Model
(A) The workflow of the PDX model in vivo. (B) H&E staining in miR-10a-OE OA synovium and control OA synovium. (C) IHC staining shows the expression of human pro-inflammatory cytokines in engrafted synovial tissues. (D–G) ELISA was used to detect human pro-inflammatory cytokines in mice sera. Data represent means ± SEM (each group has six mice); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, as determined by the Student’s t test. (D–G) *p < 0.05, **p < 0.01, compared to control group.
Validation of MAP3K7 as a Target of miR-10a in OA-FLSs
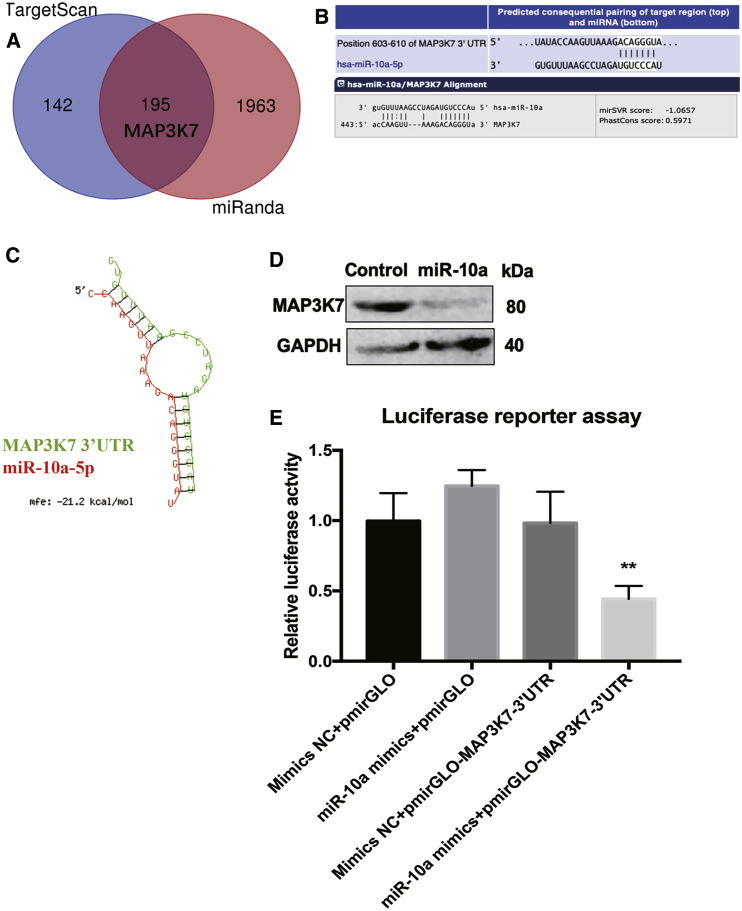
To identify functional candidate targets of miR-10a that may be involved in mediating the pro-inflammatory effect of OA-FLSs, we used miRNA target prediction algorithms TargetScan (http://www.targetscan.org/vert_71/) and miRanda (http://www.microrna.org/microrna/home.do) to predict potential targets of miR-10a (Figure 4A, total of 195 overlapped targets). Among these, mitogen-activated protein kinase kinase kinase 7 (MAP3K7) was of particular interest given its pro-inflammatory role in arthritis (Figure 4B).14 MAP3K7 knockdown (KD) could significantly inhibit IL-1β-induced production of pro-inflammatory cytokines, including IL-6 and IL-8 in FLSs from RA patients, suggesting that MAP3K7 is a potential pro-inflammatory target. Therefore, we validated the effect of miR-10a on MAP3K7 in OA-FLSs at the first place. RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) results showed that the binding possibility between miR-10a and the 3′ untranslated region (UTR) of MAP3K7 was quite high (Figure 4C). Transfection of OA-FLSs with miR-10a mimics strongly reduced MAP3K7 protein expression (Figure 4D). In addition, a luciferase reporter gene assay was performed to validate the direct binding between miR-10a and 3′ UTR of MAP3K7. The 3′ UTR of MAP3K7 was cloned into a dual-luciferase reporter vector pmirGLO. Co-transfection of miR-10a mimics with the MAP3K7 3′ UTR luciferase reporter vector into HEK293T cells reduced the luciferase activity (Figure 4E), indicating that miR-10a directly binds to the 3′ UTR of MAP3K7.
Figure 4.
Validation of MAP3K7 as a Target of miR-10a in OA-FLSs
(A) Two miRNA target prediction algorithms, TargetScan and miRanda, were used to predict potential targets of miR-10a. (B) The binding site between MAP3K7 3'UTR and miR-10a. (C) RNAhybrid results showed the binding possibility between miR-10a and the 3′ UTR of MAP3K7. (D) miR-10a mimics strongly reduced endogenous protein expression of MAP3K7 in OA-FLSs (n = 3). (E) Luciferase reporter assays showed that miR-10a directly binds to the 3′ UTR of MAP3K7 (n = 3). Data represent means ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, as determined by the Student’s t test. (E) **p < 0.01, compared to miR-10a mimics+pmirGLO group.
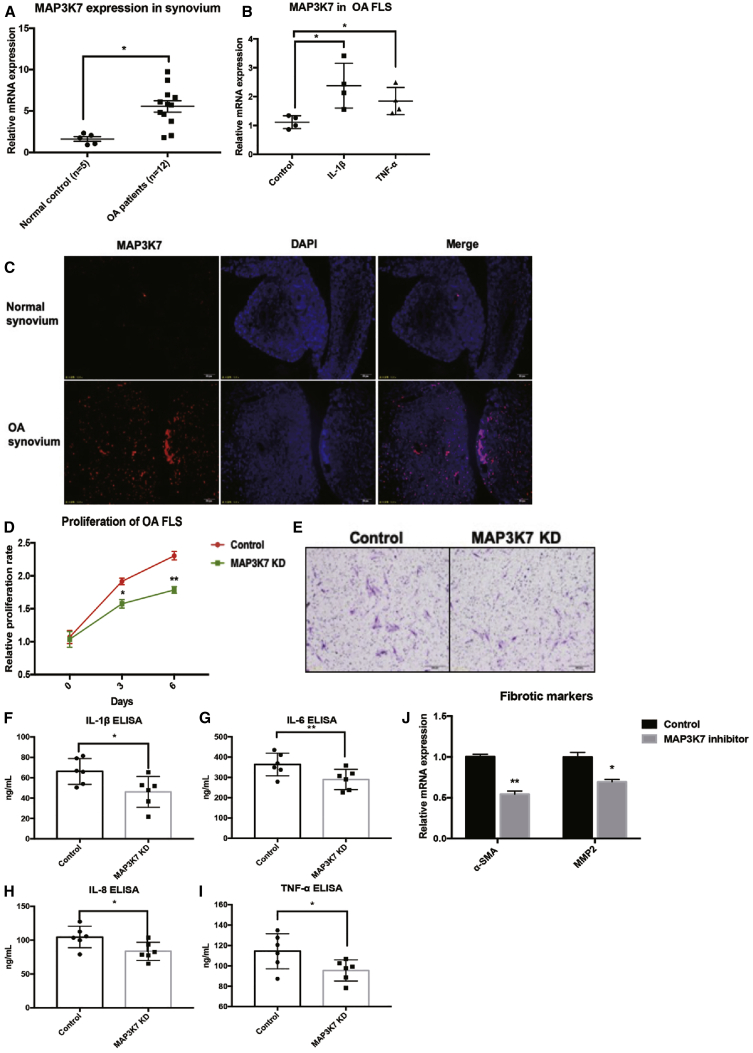
Next, we investigated the role of MAP3K7 in OA-FLSs. The expression of MAP3K7 was elevated in synovial tissues from OA patients compared with controls, as observed by quantitative PCR (qPCR) and immunofluorescence (Figures 5A and 5C). In contrast with miR-10a, ectopic expression of IL-1β and TNF-α could induce expression of MAP3K7 in OA-FLSs (Figure 5B). To understand whether MAP3K7 is directly responsible for inflammation in OA-FLSs, we used small hairpin RNA (shRNA) to knock down the expression of MAP3K7 (Figure S4). Overall, silencing of MAP3K7 led to the similar phenotype of miR-10a-OE OA-FLSs, including proliferation, migration, and expression of pro-inflammatory cytokines and fibrotic markers (Figures 5D–5J), implying that miR-10a restrains synovitis in OA-FLSs directly by targeting MAP3K7.
Figure 5.
The Role of MAP3K7 in OA-FLSs
(A) The mRNA expression of MAP3K7 was elevated in the synovial tissues of OA patients (n = 12) compared with controls (n = 3) by qPCR. (B) IL-1β and TNF-α treatment could induce expression of MAP3K7 in OA-FLSs (n = 3). (C) The protein expression of MAP3K7 was elevated in the synovial tissues of OA patients by immunostaining (n = 3). (D) The effects of MAP3K7 knockdown on proliferation of OA-FLSs (n = 3). (E) The effects of MAP3K7 knockdown on migration of OA-FLSs (n = 3). (F–I) The effects of MAP3K7 knockdown on the secretion of pro-inflammatory cytokines in OA-FLSs (n = 6). (J) MAP3K7 knockdown represses mRNA expressions of fibrotic markers in OA-FLSs (n = 3). Data represent means ± SEM; ∗p < 0.05, ∗∗p < 0.01, as determined by the Student’s t test. (F–J) *p < 0.05, **p < 0.01, compared to control group.
RNA Sequencing (RNA-Seq) Identified an Associated Pathway of miR-10a in OA-FLSs
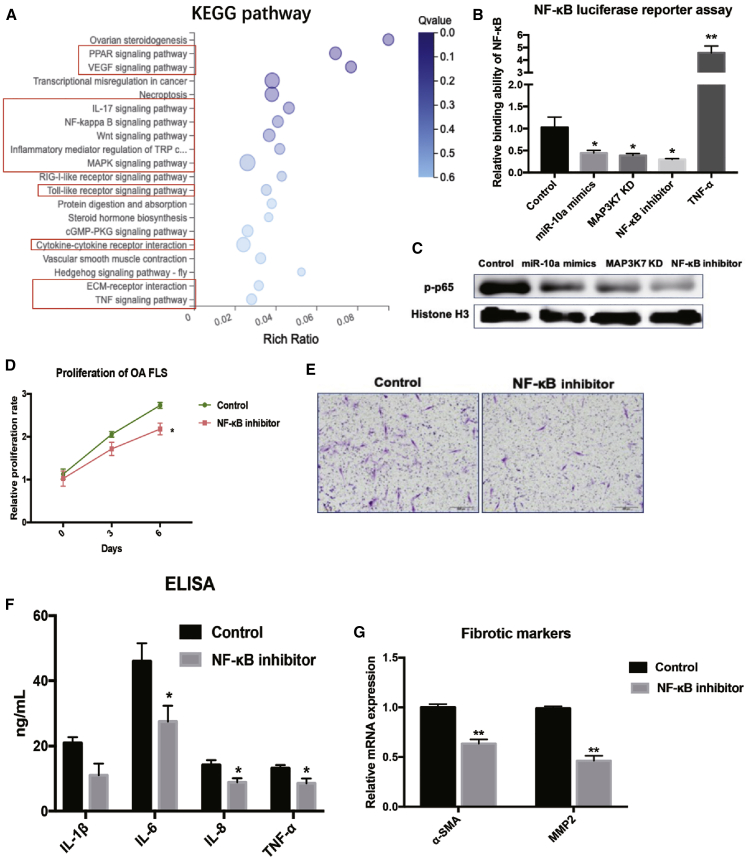
To identify the associated pathway of miR-10a in OA-FLSs, we analyzed the whole mRNA of miR-10a-OE OA-FLSs and control OA-FLSs by RNA-seq (Figure S5). Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway enrichment analysis revealed the association of miR-10a with several arthritis-related pathways (Figure 6A). Signaling pathways such as Peroxisome proliferator-activated receptors (PPAR), vascular endothelial growth factor (VEGF), nuclear factor κB (NF-κB), Wnt, mitogen-activated protein kinase (MAPK), and TNF are critical in arthritis (red rectangles). Among these potential associated pathways, the NF-κB signaling pathway was highlighted because MAP3K7 is one of its central regulators.15 To confirm the relevance of this pathway to miR-10a, we performed a pathway reporter assay and found that miR-10a mimics, MAP3K7 KD, and a NF-κB inhibitor (Bay11-7082) could repress activation of the NF-κB signaling pathway in OA-FLSs (Figure 6B). Moreover, western blot results further confirmed that p-p65, the key component in the NF-κB pathway, was repressed by miR-10a overexpression, MAP3K7 KD, and NF-κB blockage in OA-FLSs (Figure 6C). In addition, some other associated pathways, including c-Jun N-terminal kinase (JNK), extracellular-signal-regulated kinase (ERK), and transforming growth factor β (TGF-β), were also measured in miR-10a OE OA-FLSs. The results of reporter assays showed that miR-10a did not affect JNK, TGF-β, or ERK pathways in OA-FLSs (Figure S6). To further prove the role of the NF-κB signaling pathway in OA-FLSs, we also used the pan-inhibitor of the NF-κB signaling pathway (Bay11-7082) and specific MAP3K7 (Takinib) to block the NF-κB signaling pathway in OA-FLSs. As expected, results showed that proliferation, migration, and production of pro-inflammatory cytokines were repressed in NF-κB signaling pathway-blocked OA-FLSs (Figures 6D–6G; Figure S7). To summarize, these data suggested that the miR-10a-MAP3K7-NF-κB regulatory axis exerts anti-inflammatory effects in OA-FLSs.
Figure 6.
RNA-Seq-Identified NF-κB Pathway Is Associated with miR-10a in OA-FLSs
(A) KEGG signaling pathway enrichment analysis identified the most relevant pathways of miR-10a in OA-FLSs. (B) miR-10a repressed NF-κB reporter activity in OA-FLSs. The stars indicated the significant differences from control (n = 3). (C) miR-10a repressed nucleus (NE) p65 in OA-FLSs (n = 3). (D) The effects of blockage of the NF-κB pathway on proliferation of OA-FLSs (n = 3). (E) The effects of blockage of the NF-κB pathway on migration of OA-FLSs (n = 3). (F) The effects of inhibition of the NF-κB pathway on the secretion of pro-inflammatory cytokines in OA-FLS (n = 3). (G) The effects of blockage of NF-κB pathway mRNA expressions of fibrotic markers in OA-FLSs (n = 3). Data represent means ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, as determined by the Student’s t test.
Twist Family BHLH Transcription Factor 1 (TWIST1) Is an Upstream Regulator of miR-10a in OA-FLSs
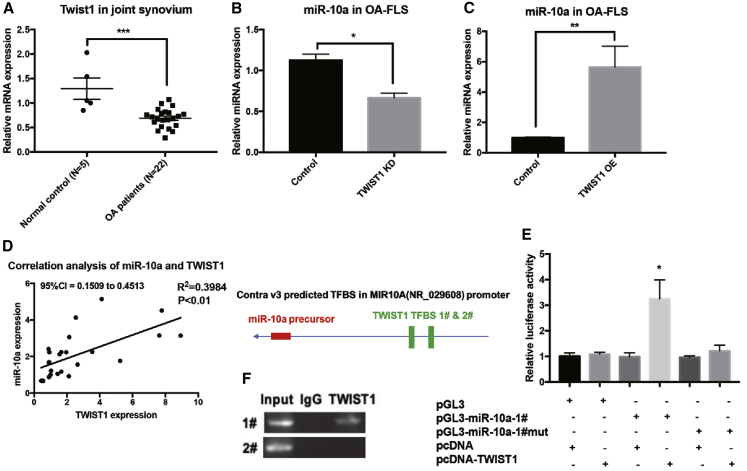
The MAP3K7-NF-κB pathway was identified as a downstream target and associated pathway of miR-10a in OA-FLSs. The interaction of TWIST1 and NF-κB pathway has been reported in inflammation.16 As a transcriptional factor, TWIST1 played an essential role in various diseases, including inflammation and cancer.17 As expected, TWIST1 was also downregulated in synovial tissues from OA patients when compared with normal synovium (Figure 7A). To test the regulatory effects of TWIST1 on miR-10a, we used a shRNA and OE lentivirus of TWIST1 (Figure S8). miR-10a was repressed and induced in TWIST1 KD and OE OA-FLSs, respectively (Figures 7B and 7C). In addition, synovial tissue expression of miR-10a was positively correlated to that of TWIST1 (Figure 7D). Interestingly, two TWSIT1 binding sites (E box) were predicted in the promoter region of miR-10a gene. To validate this direct regulation, we transfected OA-FLSs with a reporter construct driven by the miR-10a promoter. TWIST1 overexpression repressed promoter activity of miR-10a (Figure 7E). When a reporter construct harboring mutation in one out of the two E box motifs in the miR-10a promoter was used, suppression of miR-10a promoter activity by TWIST1 was attenuated accordingly (Figure 7E). In addition, we determined whether TWIST1 directly binds to the endogenous miR-10a promoter in OA-FLSs. Chromatin immunoprecipitation (ChIP) assays demonstrated the binding of TWIST1 to miR-10a promoter that contains an E box (Figure 7F).
Figure 7.
TWIST1 Is an Upstream Regulator of miR-10a in OA-FLSs
(A) The expression of TWIST1 in synovium from OA patients (n = 12) and controls (n = 5). (B and C) The expression of miR-10a in TWIST1 KD (B) or OE (C) OA-FLSs (n = 3). (D) The correlation (Pearson’s correlation coefficient) of TWIST1 and miR-10a in OA synovium (n = 25). (E) Luciferase reporter assays showed that TWIST1 directly binds to the E box in the promoter region of miR-10a, a Renilla luciferase-encoding internal control plasmid for normalization (n = 3). (F) ChIP assays demonstrated the binding of TWIST1 to miR-10a promoter that contains an E box. Data represent means ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, as determined by the Student’s t test.
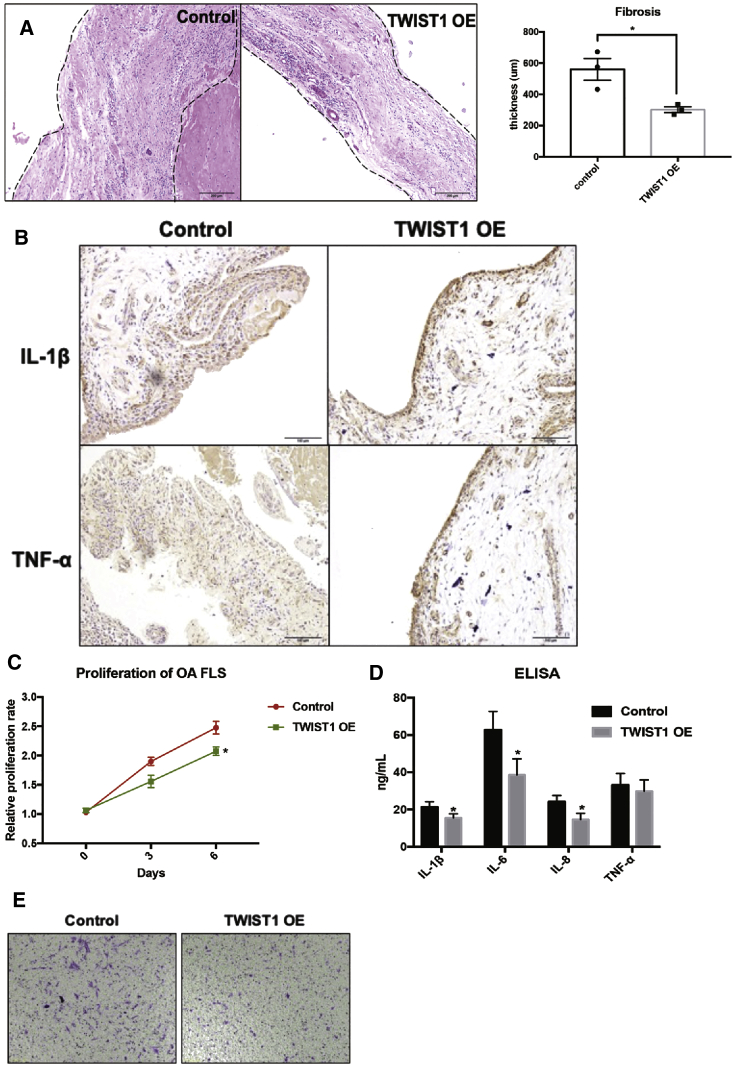
In addition, a PDX model was used to investigate the role of TWIST1 on synovitis in vivo. Two weeks later after transplantation of OA synovium in SCID mice, the injection of TWIST1 OE lentiviruses ameliorated the thickness of the FLS layer in engrafted synovium (Figure 8A). Results of IHC and ELISA showed that TWIST1 OE repressed the production of human pro-inflammatory cytokines (IL-1β and TNF-α) in engrafted synovium (Figure 8B). As expected, TWIST1 also inhibited the aggressive cellular functions, such as proliferation, migration, and production of inflammatory cytokines, of OA-FLSs in vitro (Figures 8C–8E). In addition, blockage of MAP3K7 or NF-κB could reverse the TWIST1 KD-induced pro-inflammatory effects in OA-FLSs (Figure S9). Collectively, TWIST1 was proved as a directly upstream regulator of miR-10a in OA-FLSs and reduced the severity of OA-FLS-mediated synovitis both in vivo and in vitro.
Figure 8.
The Effects of TWIST1 on OA Synovitis In Vivo and In Vitro
(A) H&E staining in TWIST1-OE OA synovium and control OA synovium (n = 6). (B) IHC staining shows the expression of human pro-inflammatory cytokines in engrafted synovial tissues (n = 6). (C–E) The effects of OE of TWIST1 on proliferation, migration, and production of inflammatory cytokines of OA-FLSs (n = 3). Data represent means ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, as determined by the Student’s t test. (C and D) *p < 0.05, **p < 0.01, compared to control group.
Discussion
In the past, research on OA has been focused on the pathogenesis of degenerative damage of cartilage and bone.18 The joint synovial tissue of healthy people is very difficult to obtain, so researchers typically use joint synovium from OA patients as so-called normal controls for RA patients with more severe synovitis. However, this is not consistent with the clinical situation: clinical OA patients are often accompanied by a certain degree of synovitis, and the synovitis symptoms of some OA patients are also very serious.19,20 Furthermore, there are a few studies that have suggested that synovitis is an important factor causing OA cartilage damage.9,21 Therefore, it is necessary to investigate the pathogenesis of OA synovitis, instead of simply using it as a “control group” for RA.
Previously, research reports on miR-10 families, including miR-10a and miR-10b, have focused on the field of oncology.4 Recently, some reports have shown that miR-10a plays an important anti-inflammatory role in RA,6,7 and miR-10b is an important inflammatory regulation factor in ankylosing spondylitis.8 In the present study, we found that miR-10a was significantly decreased in OA patients’ blood samples and synovial tissues compared with the normal control group.
However, because of the difficulty of obtaining normal controls, this study collected only three cases of normal joint synovium in 18 months. Despite being compared with a large number of synovial tissues from OA patients, the three normal synovial tissues consistently showed higher expression of miR-10a. The limited number of normal controls is indeed a limitation of this study, and we will continue to work with surgeons from departments of general surgery and orthopedics to maximize the number of normal controls in our future study.
Although the joint synovium of normal people is difficult to obtain, the peripheral blood of normal people is a substitute, especially for the detection of miRNAs (small size and with diagnostic potential). In this study, 22 cases of peripheral blood from healthy donors were included as normal controls of peripheral blood from OA patients. The results showed a significant reduction of miR-10a expression in PBMCs, isolated from peripheral blood, in OA patients compared with normal controls (consistent with results from synovial tissues), which indicated the following: (1) miR-10a is a potential diagnostic biomarker of OA, and (2) PBMCs consist of lymphocytes (such T lymphocytes and B lymphocytes) and other mononuclear cells (such as monocytes). Therefore, miR-10a may also play a role in these immune cells that are infiltrated in the joint synovium of OA patients. Does miR-10a exert its anti-inflammatory function both in immune cells and in OA-FLSs within the joint synovium? Is it possible that immune cells and OA-FLSs interact with each other via miR-10a? This is another important direction for our future study.
The expression of miR-10a was positively associated with the severity of synovitis in OA patients, suggesting that miR-10a may also play an anti-inflammatory role in OA synovitis. We further sorted primary OA-FLSs (CD45−CD31−CD235a−CD146−PDPN+FAPα+) and cultured them in vitro. Two essential pro-inflammatory cytokines, IL-1β and TNF-α, repressed endogenous expression of miR-10a in OA-FLSs, implying that miR-10a is likely to be involved in OA-FLS-mediated synovitis.
Further, we confirmed that miR-10a could inhibit the inflammatory phenotypes of OA-FLSs at in vitro and in vivo levels. In particular, we used a PDX model to evaluate the anti-inflammatory effects of miR-10a on OA synovitis in vivo. The PDX model has proved to be a very useful model for studying pathogenesis and drug screening, and has been widely used to investigate many inflammatory immune diseases.22,23 Although this in vivo model requires more synovial tissues than in vitro research, the advantages of this model are obvious when compared with in vitro OA-FLS culture: (1) it mimics an in vivo environment for synovial tissues; (2) this model provides dynamic production of new blood vessels in the transplanted synovial tissue; (3) it could be used to study the dynamic changes in the human-derived inflammatory factors in the circulation of mice; and (4) this model is an in vivo drug screening platform for OA.
Recent studies have found that FLSs have different functions in different areas of the joint synovium in RA patients, but little is known about the molecular mechanisms of FLSs involved in the development of synovitis in OA. Using ChIP and luciferase reporter assays, we confirmed TWIST1 and MAP3K7 are a direct upstream activator and downstream target for miR-10a and could initiate the repression of the downstream NF-κB pathway. These data demonstrated that the TWIST1-miR-10a-MAP3K7 cascade can block the NF-κB signaling pathway and ultimately achieve the effect of inhibition of OA-FLS-mediated inflammation. Therefore, targeting miR-10a is a potential anti-inflammatory therapy for OA synovitis that is definitely worth further investigation. In summary, we found that the TWIST1-miR-10a-MAPK7-NF-κB signaling axis can inhibit OA-FLS-mediated inflammation by combining in vitro models, in vivo models, and clinical data. This paper showed that miR-10a can be used as a promising target for treating patients with OA synovitis.
Materials and Methods
Patients, Ethics, and Inclusion/Exclusion Criteria
Synovial tissues were obtained from OA patients (all Chinese) upon joint replacement or synovectomy at the First Affiliated Hospital of USTC, Hefei, China. This study was approved by the Ethics Committee of the First Affiliated Hospital of USTC. The diagnosis of primary knee OA (28 women and 2 men; average age, 56.12 years; age range, 48–73 years) conformed to the criteria stipulated by the American College of Rheumatology (ACR),24 and radiographic changes were grade III or IV according to the Kellgren and Lawrence classification method.25 Patients with OA induced by trauma, inflammation, tuberculosis, or sepsis were excluded. Blood samples from 22 healthy volunteers (10 women and 12 men; average age, 43.50 years; range, 32–57 years) and synovial samples from five patients (five men; average age, 43.2 years; all normal synovia were from knee operations) with non-inflammatory knee joint diseases were used as controls. For the human blood samples-related experiments, the sample size (n = 22) was predicted based on a preliminary experiment that included six healthy donors and six OA patients.
Isolation and Culture of Primary OA-FLSs and OA-Chondrocyte
The method used for the isolation of FLSs from synovial tissue was modified from a method previously described.26 In brief, the synovium was removed from joints of patients with OA. Isolated hyperplastic synovial tissues were washed and minced in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies), supplemented with 10% fetal bovine serum (GIBCO) and penicillin and streptomycin (GIBCO). Dissected synovial tissue was digested in culture medium containing 1 mg/mL collagenase type 4 (Sigma) and 0.1 mg/mL deoxyribonuclease I (Sigma) and incubated for 1.5 h at 37°C. Tissues were then vortexed and resuspended in fresh medium. The medium containing the digested tissue was centrifuged at 3,000 rpm for 5 min, and the cell pellet was resuspended in fresh medium for cell sorting. The sorted OA-FLSs were cultured at 37°C in 5% CO2. Culture medium was replenished every 2 days, and cells were subcultured when they reached 90% confluence.
For the primary culture of OA-chondrocyte, the cartilage from OA patients was minced into small pieces prior to collagenase II (Sigma) digestion under continuous agitation (200 rpm) for 8 h at 37°C. Cell suspension was passed through a cell strainer (40-μm pore size; BD), washed twice with PBS, centrifuged for 10 min (1,000 rpm), and resuspended in 10 mL culture medium (DMEM [GIBCO] supplemented with 10% FBS [GIBCO] and penicillin/streptomycin). Primary OA-chondrocyte was sub-cultured every 3 days.
Flow Cytometry
OA-FLSs were incubated with the following fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, or allophycocyanin (APC)-conjugated antibodies at 4°C for 1 h: CD45-FITC, CD31-FITC, CD146-FITC, CD235a-FITC, PDPN-PE, or human fibroblast activation protein (hFAP)-APC. All flow cytometry antibodies were from Miltenyi. After washing thrice with pre-cooled PBS, the cells were analyzed using a flow cytometer (BD FACSAria II) and BD FACSDiva software. Unlabeled cells were used in other fluorescence-activated cell sorting (FACS) assays as NCs.
Treatment and Transfection of OA-FLSs
OA-FLSs were cultured in Roswell Park Memorial Institute 1640 medium and transfected with 100 nM miR-10a mimics (Two seqeuences: 5′-UACCCUGUAGAUCCGAAUUUGUG-3′ and 5′-CAAAUUCGGAUCUACAGGGUAuu-3′) or corresponding scramble control (GenePharma) using Lipofectamine RNAiMAX Reagent (Invitrogen) in accordance with the manufacturer’s instructions. Following 24–72 h of transfection, cells were directly lysed for further experiments. MAP3K7 and TWIST1 small hairpin RNAs (shRNAs) were designed using the online design program from MIT (http://sirna.wi.mit.edu/home.php): MAP3K7 5′-GCCCTTCAATGGAGGAAAT-3′ and TWIST1 5′-GAGACCTAGATGTCATTGT-3′. The hairpin-type shRNAs, with a nine-nucleotide loop, were cloned into pLVTHM (Addgene). The stem-loop sequence of miR-10a was cloned into pLVTHM for miR-10a overexpression. The lentivirus was packaged in HEK293T cells. The overexpression lentivirus of TWIST1 was generated and packaged by Genechem. For all experiments, OA-FLSs were used between passages 2 and 6. For the stimulation experiments, cells were stimulated with NF-κB pathway blocker Bay11-7082 (10 μM/mL) or TNF-α (10 ng/mL) and IL-1β (10 ng/mL).
Mice
Adult female SCID mice (8–12 weeks, 20–22 g) were acquired from the Model Animal Research Center of Nanjing University (China). All mice were housed in individually ventilated cages and maintained in a pathogen-free room (specific-pathogen-free [SPF] level, bedding material: Corn Cob; the husbandry conditions: light/dark cycle 12/12 h; animal room temperature: 22°C ± 2°C; relative humidity: 45% ± 5%; access to mouse maintenance food and water ad libitum). To allow for acclimatization to the experimental conditions, we randomly selected animals and kept them in individual cages for at least 4 days before the experiment. During housing, animals were monitored once daily for health status and possible adverse events. No adverse events were observed during the experimental process. All experimental mice were handled in compliance with the guidelines of the institutional ethics committee. All care was taken to cause no pain to the mice. Before all surgical procedures, ketamine-xylazine was used for anesthesia and analgesia (ketamine: 75–150 mg/kg + xylazine: 16–20 mg/kg intraperitoneally [i.p.]). Overdose of chemical anesthetics (three to four times anesthetic dose) was used for euthanasia, and cervical dislocation was used for confirmation of euthanasia. Biopsy samples (synovial tissues from OA patients) were transplanted into the anesthetized SCID mice. Specifically, after swabbing the skin in the neck region with 70% ethanol, a small incision was made in the dorsal skin behind the ear of each mouse, the tissue (5 × 5 × 5 mm) was inserted subcutaneously, and the wound was closed with suture clips. Then, 12 two-tissues-bearing mice were divided into two groups: control group (six mice were injected with control lentivirus) and miR-10a- or TWIST1-OE group (six mice were injected with miR-10a or TWIST1-OE lentivirus). A single dose of 108 particles of lentiviruses was directly injected into the transplanted synovium subcutaneously. After 2 weeks, all mice were sacrificed, and the synovial tissues were extracted for subsequent experiments. All sections of this report adhere to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines for reporting animal research.
ELISA
To assess the effect of miR-10a expression on OA-FLS cytokine production, we collected supernatants following 48 h of transfection, and TNF-α, IL-1β, IL-6, and IL-8 were measured using targeted ELISAs according to the manufacturer’s conditions (R&D Systems).
Migration Assay
Transwell chambers (CORING) were used to assess OA-FLS migration in response to TWIST1-OE, miR-10a-OE, MAP3K7 KD, and NF-κB pathway inhibition. TWIST1-OE, miR-10a-OE, MAP3K7 KD, and NF-κB pathway-blocked OA-FLSs were seeded at a density of 20,000 cells/well in the migration chamber on 8-μm membranes. Migrating cells attached to the lower membrane were fixed with 1% glutaraldehyde and stained with 0.1% crystal violet. Cells from six random high-power fields for each well were counted to assess the average number of migrating cells.
RNA Extraction and Real-Time PCR Assays
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the standard protocol. The concentration and quality of all RNA samples were evaluated using NanoDrop 2000 (Thermo). Reverse transcription of mRNA was performed using a MasterMix kit (Takara), and miRNA reverse transcription was performed using a TaqMan reverse transcription kit (Life Technologies) following the manufacturer’s instructions. qPCR of genes was performed using Universal SYBR Green Master mix (Applied Biosystems), and miRNA qPCR was performed using TaqMan-specific microRNA probe (Life Technologies) on a 7500 real-time PCR system (Applied Biosystems). Gene expression was normalized to GAPDH, and miRNA expression was normalized to U6 small nucleolar RNA (snoRNA) unless otherwise stated. Primer sequences are listed in the Supplemental Information.
Western Blot
For protein detection, total proteins were electrophoresed and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were individually incubated with antibodies for MAP3K7 (Abcam), p-p65 (Abcam), or histone H3 (Santa Cruz). The blots were developed using a horseradish peroxidase-conjugated secondary antibody, and the immunoreactive proteins were visualized with an enhanced chemiluminescence system (GE System).
RNA-Seq
Total RNA was quantified using a NanoDrop2000 spectrophotometer (NanoDrop Technologies). OA-FLSs were collected 48 h after transfection of miR-10a mimics and mimics NC. Two passing samples were included; the RNA Integrity Number (RIN) values were 8.0 and 7.8 and quality control (QC) (q30) were 88.5 and 88.6. PE150 paired-end sequencing and the MGI-2000 platform were used. The clean reads obtained by sequencing will be uploaded to NCBI later. For detection of whole mRNA expression, RNA-seq was performed by BGI (China) according to standard procedure. In brief, the sequencing data were filtered with SOAPnuke (v.1.5.2)27 by: (1) removing reads containing sequencing adaptor; (2) removing reads whose low-quality base ratio (base quality less than or equal to 5) is more than 20%; and (3) removing reads whose unknown base (‘N’ base) ratio is more than 5%, and afterward clean reads were obtained and stored in FASTQ format. The clean reads were mapped to the reference genome using HISAT2 (v.2.0.4).28 Bowtie2 (v.2.2.5)29 was applied to align the clean reads to the reference coding gene set; then the expression level of gene was calculated by RNA-Seq by Expectation-Maximization (RSEM) (v.1.2.12).30 The heatmap was drawn by pheatmap (v.1.0.8)31 according to the gene expression in different samples. Essentially, differential expression analysis was performed using the PossionDis32 with false discovery rate (FDR) ≤0.001 and |log2ratio| ≥1. To take insight to the change of phenotype, Gene Ontology (GO) (http://geneontology.org/) and KEGG (https://www.kegg.jp/) enrichment analysis of annotated different expressed genes was performed by Phyper (https://en.wikipedia.org/wiki/Hypergeometric_distribution) based on hypergeometric test; the significant levels of terms and pathways were corrected by Q value with a rigorous threshold (Q ≤ 0.05) by Bonferroni.
IHC Staining
Paraffin-embedded OA synovia were sliced and then blocked in 5% goat serum for 30 min. The tissue was incubated with primary IL-1β and TNF-α antibodies (Abcam) overnight at 4°C. The sections were then washed with PBS and stained using a streptavidin-peroxidase kit (Abcam) according to the manufacturer’s protocol. Then the sections were visualized through reactions with 3,3′-Diaminobenzidine (DAB). The IHC staining was performed as reported previously.5
FISH
FISH for miR-10a was performed on 4-mm-thick OCT-embedded tissue sections. The miRNA FISH detection probe for miR-10a was purchased from GenePharma. FISH was performed according to a standard protocol for miRNA FISH from GenePharma.
Luciferase Reporter Assay
The pmirGLO miRNA expression reporter vector system (Promega) was used to construct the reporter plasmid containing 400 bp of the 3′ UTR of human MAP3K7 mRNA encompassing the sequence complementary to the miR-10a seed sequence. The 3′ UTR of MAP3K7 was connected to the end of firefly luciferase, and Renilla luciferase was used as an internal control. HEK293T cells were transfected with 1 mg of reporter plasmid and 20 nM miRNA mimics or mimics NC in 12-well plates using Lipofectamine 2000 according to the manufacturer’s protocol. The luciferase activity assay was performed 48 h after transfection, using the Dual-Luciferase Reporter Assay System (Promega) and GloMax 20/20 LUMINOMETER (Promega).
Luciferase reporter plasmid pGL3-Basic vector (Promega) containing a 2.5-kb fragment of the miR-10a predictive promoter (−500 to +2,000 bp relative to transcription initiation site) was used for TWIST1 binding test. A reporter construct mutated in one out of the two binding sites present in this promoter was generated using the Phusion Site-Directed Mutagenesis Kit (Thermo). TWIST1-OE OA-FLSs and control OA-FLSs were co-transfected with luciferase reporter constructs or an empty control vector and a Renilla luciferase-encoding internal control plasmid. Forty-eight hours after transfection, luciferase activity was measured by using a GlutaMAX machine.
OA-FLSs were transfected with a NF-κB, JNK, ERK, or TGF-β luciferase reporter using Lipofectamine 2000 (Invitrogen) and NF-κB reporter kit (NF-κB pathway), SRE reporter kit (ERK pathway), SBE reporter kit (TGF-β pathway), and AP1 reporter kit (JNK pathway) (BPS Bioscience). The luciferase assay was performed using the dual-luciferase reporter assay system (Promega), according to the manufacturer’s instructions.
ChIP
ChIP assay was performed using a ChIP kit (Abcam) using an anti-Twist1 antibody (Abcam) according to the manufacturer’s instructions.33 OA-FLSs were cultured to collect chromatin, which was immunoprecipitated with either Twist1 or control antibodies. Extracted DNA was amplified by PCR using primer pairs specific for the E box in the promoter region of miR-10a.
Statistical Analysis
Statistical analyses were performed using SPSS software. The normality was assessed by SPSS. All samples follow normal distribution. Therefore, t test was used in the following analysis. For the analysis with more than two groups, an ANOVA was performed and p < 0.05. Therefore, a following Dunnett’s t test was used for the following comparison between two groups, with each experimental group compared back to the control group. For the correlation analysis, we first assessed normality and then used Pearson’s analysis. The p values were considered not significant (n.s.) or significant at ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Ethics Statement
This study was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of USTC. This usage of clinical samples was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of USTC with written informed consents obtained from all of the patients.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81871788 and 31900616); the project for Science and Technology Leader of Anhui Province (grant 2018H177); the Scientific Research Fund of Anhui Education (grant 2017jyxm1097); and the Anhui Provincial Postdoctoral Science Foundation (grant 2019B302).
Author Contributions
J.T., Z.Y., and C.Z. designed the experiments. W.H., W.Z., and J.M. conducted the experiments. W.H. wrote the paper.
Declaration of Interest
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.020.
Contributor Information
Zhiying Yu, Email: lizheyzy@163.com.
Chen Zhu, Email: zhuchena@ustc.edu.cn.
Supplemental Information
References
- 1.Xia B., Di Chen, Zhang J., Hu S., Jin H., Tong P. Osteoarthritis pathogenesis: a review of molecular mechanisms. Calcif. Tissue Int. 2014;95:495–505. doi: 10.1007/s00223-014-9917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathiessen A., Conaghan P.G. Synovitis in osteoarthritis: current understanding with therapeutic implications. Arthritis Res. Ther. 2017;19:18. doi: 10.1186/s13075-017-1229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong W., Zhang P., Wang X., Tu J., Wei W. The Effects of MicroRNAs on Key Signalling Pathways and Epigenetic Modification in Fibroblast-Like Synoviocytes of Rheumatoid Arthritis. Mediators Inflamm. 2018;2018:9013124. doi: 10.1155/2018/9013124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lund A.H. miR-10 in development and cancer. Cell Death Differ. 2010;17:209–214. doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- 5.Tu J., Cheung H.-H., Lu G., Chen Z., Chan W.-Y. MicroRNA-10a promotes granulosa cells tumor development via PTEN-AKT/Wnt regulatory axis. Cell Death Dis. 2018;9:1076. doi: 10.1038/s41419-018-1117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussain N., Zhu W., Jiang C., Xu J., Geng M., Wu X., Hussain S., Wang B., Rajoka M.S.R., Li Y. Down-regulation of miR-10a-5p promotes proliferation and restricts apoptosis via targeting T-box transcription factor 5 in inflamed synoviocytes. Biosci. Rep. 2018;38 doi: 10.1042/BSR20180003. BSR20180003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mu N., Gu J., Huang T., Zhang C., Shu Z., Li M., Hao Q., Li W., Zhang W., Zhao J. A novel NF-κB/YY1/microRNA-10a regulatory circuit in fibroblast-like synoviocytes regulates inflammation in rheumatoid arthritis. Sci. Rep. 2016;6:20059. doi: 10.1038/srep20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L., Al-Mossawi M.H., Ridley A., Sekine T., Hammitzsch A., de Wit J., Simone D., Shi H., Penkava F., Kurowska-Stolarska M. miR-10b-5p is a novel Th17 regulator present in Th17 cells from ankylosing spondylitis. Ann. Rheum. Dis. 2017;76:620–625. doi: 10.1136/annrheumdis-2016-210175. [DOI] [PubMed] [Google Scholar]
- 9.Sokolove J., Lepus C.M. Role of inflammation in the pathogenesis of osteoarthritis: latest findings and interpretations. Ther. Adv. Musculoskelet. Dis. 2013;5:77–94. doi: 10.1177/1759720X12467868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeser R.F., Collins J.A., Diekman B.O. Ageing and the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016;12:412–420. doi: 10.1038/nrrheum.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wenham C.Y.J., Conaghan P.G. The role of synovitis in osteoarthritis. Ther. Adv. Musculoskelet. Dis. 2010;2:349–359. doi: 10.1177/1759720X10378373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizoguchi F., Slowikowski K., Wei K., Marshall J.L., Rao D.A., Chang S.K., Nguyen H.N., Noss E.H., Turner J.D., Earp B.E. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun. 2018;9:789. doi: 10.1038/s41467-018-02892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guilak F., Nims R.J., Dicks A., Wu C.L., Meulenbelt I. Osteoarthritis as a disease of the cartilage pericellular matrix. Matrix Biol. 2018;71–72:40–50. doi: 10.1016/j.matbio.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh A.K., Umar S., Riegsecker S., Chourasia M., Ahmed S. Regulation of Transforming Growth Factor β-Activated Kinase Activation by Epigallocatechin-3-Gallate in Rheumatoid Arthritis Synovial Fibroblasts: Suppression of K(63)-Linked Autoubiquitination of Tumor Necrosis Factor Receptor-Associated Factor 6. Arthritis Rheumatol. 2016;68:347–358. doi: 10.1002/art.39447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adhikari A., Xu M., Chen Z.J. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene. 2007;26:3214–3226. doi: 10.1038/sj.onc.1210413. [DOI] [PubMed] [Google Scholar]
- 16.Kasinathan N.K., Subramaniya B., Sivasithamparam N.D. NF-κB/twist mediated regulation of colonic inflammation by lupeol in abating dextran sodium sulfate induced colitis in mice. J. Funct. Foods. 2018;41:240–249. [Google Scholar]
- 17.Qin Q., Xu Y., He T., Qin C., Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma A.R., Jagga S., Lee S.S., Nam J.S. Interplay between cartilage and subchondral bone contributing to pathogenesis of osteoarthritis. Int. J. Mol. Sci. 2013;14:19805–19830. doi: 10.3390/ijms141019805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Hunter D.J., Jin X., Ding C. The importance of synovial inflammation in osteoarthritis: current evidence from imaging assessments and clinical trials. Osteoarthritis Cartilage. 2018;26:165–174. doi: 10.1016/j.joca.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Scanzello C.R., Goldring S.R. The role of synovitis in osteoarthritis pathogenesis. Bone. 2012;51:249–257. doi: 10.1016/j.bone.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monemdjou R., Fahmi H., Kapoor M. Synovium in the pathophysiology of osteoarthritis. Therapy. 2010;7:661–668. [Google Scholar]
- 22.Sanmamed M.F., Chester C., Melero I., Kohrt H. Defining the optimal murine models to investigate immune checkpoint blockers and their combination with other immunotherapies. Ann. Oncol. 2016;27:1190–1198. doi: 10.1093/annonc/mdw041. [DOI] [PubMed] [Google Scholar]
- 23.Yu X., Petersen F. A methodological review of induced animal models of autoimmune diseases. Autoimmun. Rev. 2018;17:473–479. doi: 10.1016/j.autrev.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Altman R., Asch E., Bloch D., Bole G., Borenstein D., Brandt K., Christy W., Cooke T.D., Greenwald R., Hochberg M., Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Arthritis Rheum. 1986;29:1039–1049. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 25.Kellgren J.H., Lawrence J.S. Radiological assessment of osteo-arthrosis. Ann. Rheum. Dis. 1957;16:494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardy R.S., Hülso C., Liu Y., Gasparini S.J., Fong-Yee C., Tu J., Stoner S., Stewart P.M., Raza K., Cooper M.S. Characterisation of fibroblast-like synoviocytes from a murine model of joint inflammation. Arthritis Res. Ther. 2013;15:R24. doi: 10.1186/ar4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R., Li Y., Kristiansen K., Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 28.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolde R. 2019. Pheatmap: pretty heatmaps. R package version 1.0.12. pp. 1–8. [Google Scholar]
- 32.Audic S., Claverie J.M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 33.Sharif M.N., Šošić D., Rothlin C.V., Kelly E., Lemke G., Olson E.N., Ivashkiv L.B. Twist mediates suppression of inflammation by type I IFNs and Axl. J. Exp. Med. 2006;203:1891–1901. doi: 10.1084/jem.20051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.