Abstract
B cells in human food allergy have been studied predominantly in the blood. Little is known about IgE+ B cells or plasma cells in tissues exposed to dietary antigens. We characterized IgE+ clones in blood, stomach, duodenum and esophagus of 19 peanut allergic patients, using high-throughput DNA sequencing. IgE+ cells in allergic patients are enriched in stomach and duodenum, and have a plasma cell phenotype. Clonally-related IgE+ and non-IgE-expressing cell frequencies in tissues suggest local isotype switching, including transitions between IgA and IgE isotypes. Highly similar antibody sequences specific for peanut allergen Ara h 2 are shared between patients, indicating that common immunoglobulin genetic rearrangements may contribute to pathogenesis. These data define the gastrointestinal tract as a reservoir of IgE+ B-lineage cells in food allergy.
One Sentence Summary:
Peanut allergic patient GI tissues implicate local IgE class-switching and common IgH genetic rearrangements in pathogenesis.
Introduction
Food allergy currently affects 3–6% of the US population (1) and is increasing in prevalence (2). Allergy to peanuts is marked by severe reactions and persistence into adulthood (3). The type I hypersensitivity responses of classic allergic reactions are mediated by allergen crosslinking of IgE bound to FcεRI receptors on the surface of tissue mast cells and blood basophils. IgE-expressing B cells are rare in the blood, and serum IgE levels are tightly regulated in non-atopic individuals but elevated in most allergic patients. Key knowledge gaps in human allergy include the triggers leading to dysregulation of IgE production; the anatomic sites where IgE-expressing B-lineage cells develop; and how IgE antibody repertoires differ between allergic and healthy individuals. Few studies have examined allergen-specific or IgE+ B cells in non-blood tissues in human food allergic patients, due to the difficulty of sampling these sites.
IgE+ B cells in mice are reported to be short-lived antibody-secreting cells derived from IgG1+ precursors (for high-affinity IgE) or IgM+ precursors (for low-affinity IgE) (4, 5). In humans, class-switch recombination (CSR) DNA excision circles and somatic hypermutation (SHM) patterns within B cell clones indicate that IgE+ B cells can originate from CSR of IgG+ or, less often, IgM+ B cells (6–8). Isotype switching studies have focused on secondary lymphoid tissues such as lymph nodes and tonsils, but there is some evidence that IgE isotype switching can occur in the nasal mucosa of aeroallergy patients (6, 9, 10). IgE antibodies have been detected in intestinal secretions and stool from food-allergic subjects (11, 12), suggesting that IgE-producing plasma cells may reside in gastrointestinal (GI) tissues. We tested this possibility directly in GI biopsies of peanut allergic patients through DNA sequencing of expressed immunoglobulin transcripts, and immunofluorescence microscopy, and find abundant IgE+ plasma cells. We find a striking co-occurrence of IgE-expressing and non-IgE-expressing clonally-related B-lineage cells in the same GI biopsy samples, suggesting local class switching in these tissues. We further identify common convergent heavy chain sequences of peanut allergen-specific B cells shared between allergic donors, indicating that some portion of the pathogenic IgE repertoire in peanut allergy may serve as a predictable biomarker of disease.
Results
Detection of IgE+ B-lineage cells in the allergic GI tract
We used high-throughput DNA sequencing (HTS) to analyze immunoglobulin heavy chain (IgH) transcripts in mucosal GI biopsies from proximal esophagus (PE), medial esophagus (ME), distal esophagus (DE), stomach (S) and duodenum (D), as well as peripheral blood from 19 peanut-allergic (PA) adults with diagnoses verified by placebo-controlled double-blinded food challenge (13) (Fig. 1A and Table S1). All allergic patients were actively avoiding peanut prior to food challenge. A subset of the peanut-allergic individuals also had allergies to other foods (Table S1). Control stomach biopsies (n = 11), duodenal biopsies (n = 5) and peripheral blood samples (n = 13) were obtained from nonallergic (NA) individuals (Fig. 1A and Table S1). To further evaluate for features characteristic of peanut allergy, we obtained stomach (n = 4) and duodenal (n = 3) biopsies from individuals with other non-food allergies (OA) including aeroallergies (Table S1). IgH libraries amplified from cDNA were sequenced to a median depth of 467,547 reads per sample (Table S2). IgM, IgD, IgG, IgA and IgE isotypes were amplified separately to prevent the formation of inter-isotype PCR chimeric sequence artifacts. IGHV and IGHJ gene usage, complementarity-determining region 3 (CDR-H3) length, clone size and average IGHV SHM frequencies (9) were similar for all tissues in PA, OA and NA subjects (Fig. S1).
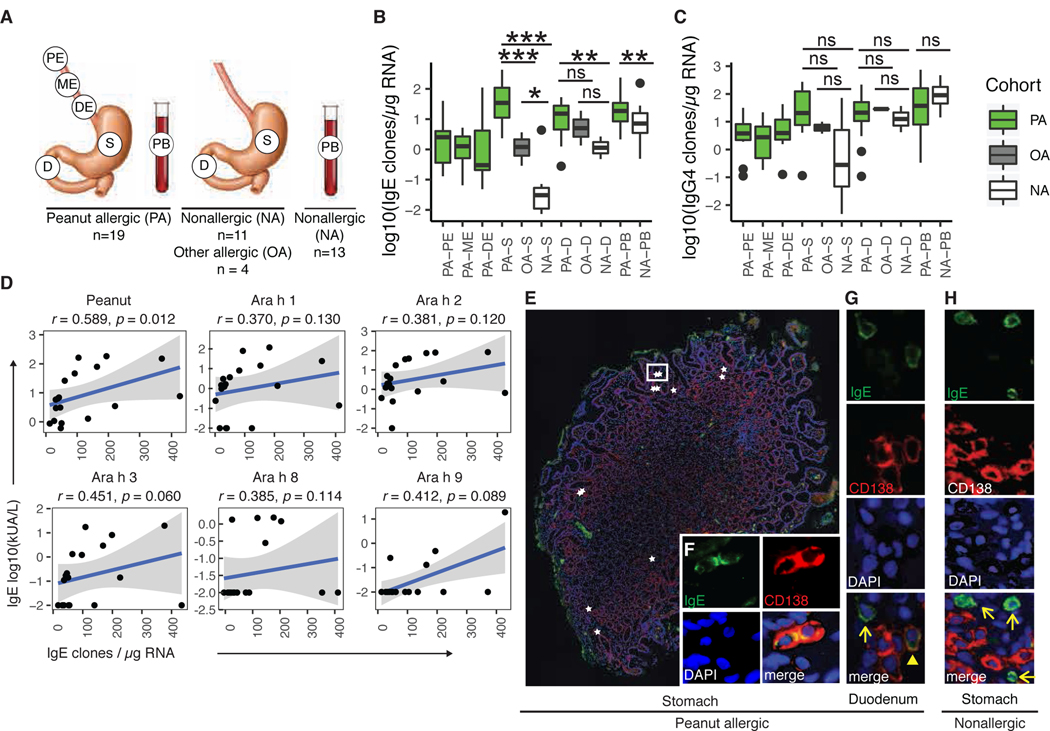
Figure 1. Stomach and duodenum are IgE+ B-lineage cell reservoirs.
(A) Gut mucosal and peripheral blood samples were obtained from donors with peanut allergy (PA), other non-food allergies (OA) or no allergies (NA). (B-C) Normalized frequencies of B cell clones containing IgE (B) or IgG4 (C) in biopsies from PA, OA and NA donors (Wilcoxon-Mann-Whitney (WMW) test: *p < 0.05; **p < 0.01; ***p < 0.001; ns = not significant). (D) Spearman’s correlation of peanut allergen-specific serum IgE (y-axis) to normalized IgE clone counts in stomach and duodenum (x-axis) (shaded area = 95% confidence intervals). (E-H) Immunofluorescence of stomach (E-F) and duodenal (G) biopsies from PA donors or from nonallergic stomach (H) (IgE (green), plasma cell marker CD138 (red) and nuclei (DAPI; blue)). (E) IgE+CD138+ plasma cells (stars) localized singly and in clusters between gastric glands; a white rectangle outlines two IgE+CD138+ plasma cells, for which single-channel staining is shown in (F). (G) IgE+CD138+ plasma cell (arrowhead) and IgE+CD138- putative mast cell (arrow). (H) A representative image from a NA donor is shown; IgE+CD138- putative mast cells were observed (arrows), but IgE+CD138+ plasma cells were absent.
CDR-H3 is the most diverse region of the antibody heavy chain and a major determinant of antigen specificity (14). We clustered IgH sequences into inferred clonal B cell lineages based on IGHV and IGHJ gene segments, CDR-H3 length and nucleotide sequence as previously described (15, 16). The number of unique IgE-expressing clones, normalized to the total amount of RNA isolated from each sample, was markedly increased in stomach, duodenum and peripheral blood in PA subjects compared to NA controls (Fig. 1B). Interestingly, OA stomach and duodenum exhibited intermediate numbers of IgE+ clones, between those of PA and NA donors. The induction of allergen-specific IgG4 responses has been associated with allergen tolerance and densensitization (17, 18). In our PA cohort, IgG4 clone counts were equivalent to those found in NA or OA control tissues (Fig. 1C), suggesting that the allergic phenotype of these PA patients is not due to low IgG4 clone frequencies in the GI tract. More total B cell or plasma cell clones of all isotypes combined were found in PA stomach biopsies compared to OA and NA controls (Fig. S2A). Frequencies of IgE+ or IgG4+ clones in tissues from participants with more than one food allergy did not differ from those with peanut allergy alone (Fig. S2B-S2C). IgE+ clone counts from stomach and duodenum, but not peripheral blood, correlated well with serum IgE titers for total peanut protein (r = 0.589, p = 0.012), but did not reach significance for individual peanut allergen components (Figs.1D, S2D and Table S3) (19). Real-time quantitative PCR (RT-qPCR) showed higher frequencies of rearranged IgK light chain transcripts, compared to reference genes GAPDH and ß-actin, in stomach and duodenal biopsies compared to the esophagus, and also in stomach and duodenal biopsies of PA compared to NA and OA donors (Fig. S2E), indicating that peanut allergy is associated with elevated concentrations of B cell or plasma-cell derived Ig transcripts.
We next evaluated the cellular phenotype of IgE-transcript-expressing cells in the gut by performing immunofluorescence (IF) staining on formalin-fixed, paraffin-embedded gut biopsies from four PA subjects (Fig. 1E–1H and Fig. S3A-S3B). IgE+ plasma cells (PC), identified by double staining for IgE and CD138, were detected in scattered small foci in the lamina propria of stomach (Fig. 1E and 1F) and duodenum (Fig. 1G). IgE+CD138- cells with bright IgE cytoplasmic membrane staining were also seen and morphologically identified as mast cells (Fig. 1G). B cells can be distinguished from PC based on their smaller diameter and nucleus-to-cytoplasmic ratios. We did not detect IgE+ cells with the morphological characteristics of small B lymphocytes, but it is possible that IgE expression was below the limit of detection on these cells (19). No IgE+CD138+ cells were found in esophageal biopsies of PA subjects (Fig. S3A and S3B) or in stomach and duodenal biopsies from NA control tissue (Fig. S3A and S3C). Staining with secondary antibody alone was performed as a control and showed low background (Fig. S3D and S3E).
IgE+ cells form clonally-expanded lineages in stomach, duodenum and blood
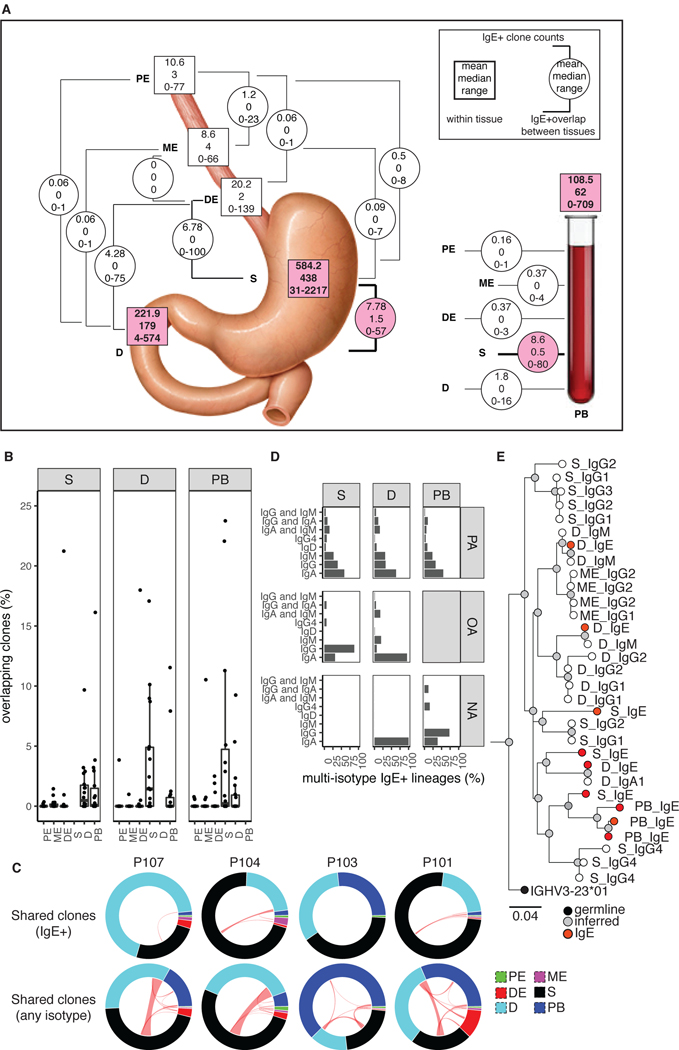
Members of B cell clones expressing non-IgE isotypes in the GI tract of NA subjects can be detected at low frequencies in peripheral blood (20), bone marrow (21), mammary glands (22), spleen (23), lung (23) and other sites in the gut (23–25). Limited numbers of clonally-related IgE+ B cells in peripheral blood and bone marrow (26) or nasal mucosa (9, 27) have been described in aeroallergy patients. We observed rare clones containing IgE+ members in more than one GI or blood tissue site (Fig. 2A, 2B and 2C, upper row) in PA subjects, most commonly between the stomach and duodenum or peripheral blood. These sites also shared increased numbers of clonally-related B cells expressing other isotypes (Fig. 2C, bottom row). We note that the esophageal sites had few IgE+ clones and that detection of clonal overlap between these sites and other tissues is limited by lack of data. Up to 23% of IgE+ clone populations in stomach, duodenum or peripheral blood had detectable IgE+ members in other tissues in PA subjects (Fig. 2B). IgA was the most common other isotype detected in multi-isotype IgE+ clones in all tissues from PA individuals (Fig. 2D), while IgG was most common in the stomach of individuals with non-food allergy. No multi-isotype IgE+ clones were detected in the stomach biopsies of NA donors. IgA was also the most frequent other isotype in IgE+ clones in PA peripheral blood, whereas IgG was more frequent in NA peripheral blood. IgM was also frequently found in IgE+ clones from PA stomach, duodenum and peripheral blood. In the duodenum, which contains high frequencies of IgA+ B cells, IgA was the most common isotype in IgE+ clones, regardless of allergy status. Large, somatically mutated clonal lineages that had expanded to multiple tissue sites and contained IgE together with several other non-IgE isotypes were also identified in PA subjects (Fig. 2E).
Figure 2. Expanded IgE+ clonal lineages in multiple tissue sites.
(A) IgE+ clone counts in tissue (squares) and overlapping between tissues (circles). (B) Percent of clones with IgE members in both indicated tissues, with denominator tissue indicated in the panel upper label. Dots represent PA patients. One outlier value was removed for plotting (42.1% overlap of P107 PB and stomach). (C) Clonal sharing between tissues (arc segments). Red lines denote clones with IgE+ members (top row) or any isotype (bottom row). Four representative PA patients are shown. (D) Isotype composition of multi-isotype IgE+ clones. (E) A representative clonal lineage from P115. Tissue and isotype are indicated for member nodes. Scale bar indicates IGHV SHM frequency.
Local IgE class-switching in the human GI tract
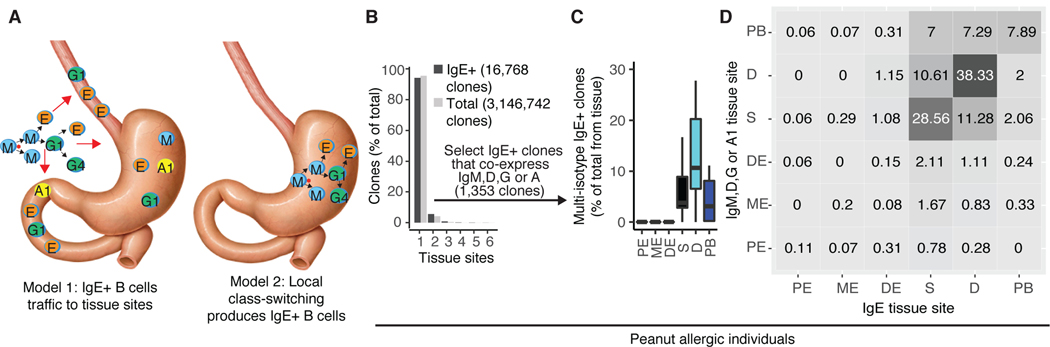
IgE+ B-lineage cells in the GI tract could arise either from trafficking of IgE-switched cells from other tissue sites such as lymph nodes, or by local class-switching to IgE+ in situ (Fig. 3A). CSR to IgE can occur in human B cells expressing the upstream isotypes IgM, IgD, IgG and IgA1, but not IgA2, which is downstream of IgE. IgE+ B cells could, however, undergo CSR to express IgA2. We reasoned that if IgE+ B cells were generated locally, the potential precursors (IgM, IgD, IgG or IgA1-expressing B cells) from the same clonal lineages would be more abundant in the same tissue biopsy than in samples from other spatially-separated tissues. Conversely, if IgE CSR occurs elsewhere and the cells later transit to the GI tract, clone members expressing IgE, IgM, IgD, IgG or IgA1 would be more evenly distributed between tissue sites (Fig. 3A). More than 90% of total or IgE-expressing clones in our dataset were detected in only one tissue (Fig. 3B). Of the IgE+ clones that we identified, 1,353 clones contained at least one other isotype. These multi-isotype clones were detected primarily in the duodenum, stomach and peripheral blood, where IgE+ clone counts and B cell frequencies were highest (Fig. 3C). Paired stomach and duodenal biopsies were obtained from OA (n = 3) and NA (n = 5) donors; however, no IgE+ clones were detected in both stomach and duodenal tissue from these donors (Fig. S4A), despite having detectable clonal expansion as evidenced by the presence of other isotypes in these IgE+ clones (Fig. S4B).
Figure 3. Evidence for local IgE CSR in GI tissues.
(A) Two models for the generation of GI-resident IgE+ B cells. Arrows denote class-switching events (black) or transit of cells to the GI tract (red). (B) Percentage of IgE+ or total clones found in single or multiple tissue sites. (C) Percentage of IgE+ clones that contain members expressing other isotypes in the same tissue. (D) Analysis of tissue distribution of IgE sequences (x-axis) and IgM, IgD, IgG or IgA1-expressing members in the same clone (y-axis). Numbers in the boxes represent the mean number of clones identified per PA individual (n=18 for stomach and duodenum; n=19 for esophagus and blood). Shading of each box indicates the number of clones, with darker shading for increasing abundance.
For the multi-isotype IgE+ clones found in PA individuals, we quantified the co-occurrence of IgE and IgM, IgD, IgG or IgA1 expressing clone members in pairwise tissue site combinations. IgE and non-IgE members co-occurred most often in the same tissue site (Fig. 3D). These results were highly significant for stomach, duodenum, peripheral blood and proximal esophagus, as assessed by label permutation testing (p = 0.001) (Fig. S4C and S4D). Stomach and duodenum were the second most common non-IgE member tissue sites for IgE+ clones from duodenum and stomach, respectively (Fig. 3D). Normalizing clonal overlaps to account for sequence read depth in each tissue (Fig. S5A) or analyzing non-IgE isotypes individually (Fig. S5B) produced similar results. Taken together, the simplest interpretation of these data is that IgE+ B-lineage cells in the stomach and duodenum are generated through local CSR in those tissues. The IgE+ B cell clones found in the circulation may be emigrants from GI tissues, or could be derived from CSR of B cells in secondary lymphoid tissue sites not sampled in this study. In support of the latter possibility, a recent study of six deceased organ donors reported that B cell clones (without isotype information) of the GI tract and the blood-related tissues (blood, spleen, lung and bone marrow) comprise two largely separate “networks” of shared clones (23).
Shared somatic mutation patterns between IgE+ and IgA+ clone members in GI tissues
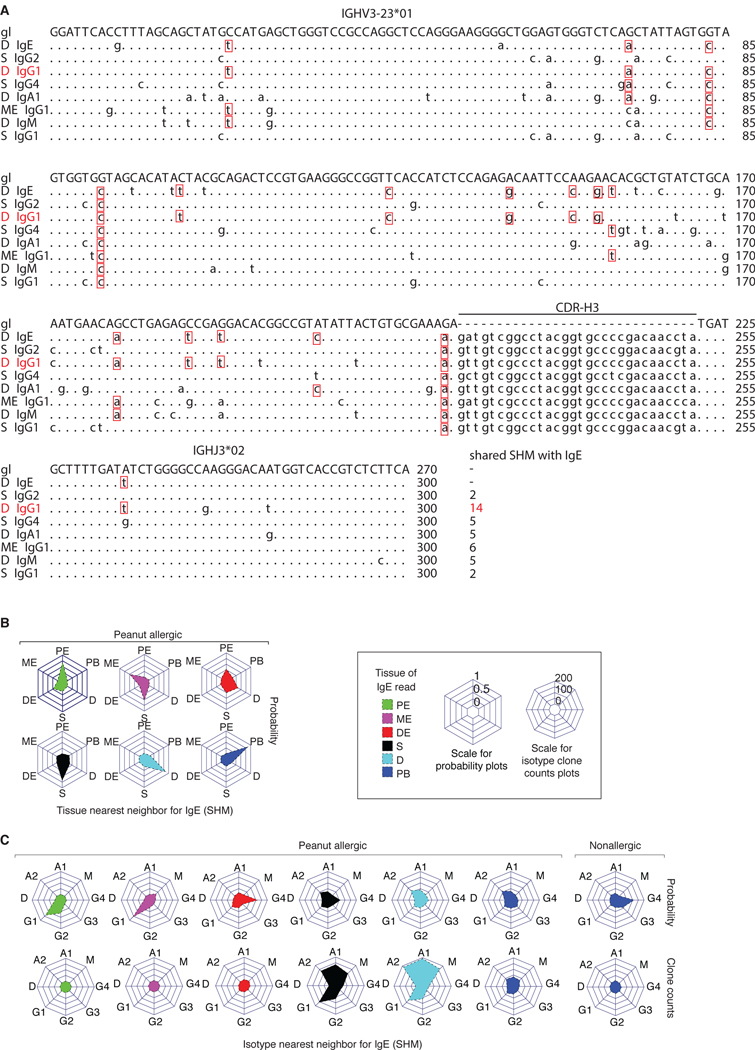
Most of the IgE+ clones that we observed in PA GI tissues showed SHM (Fig. S1 and Fig. S6A), allowing us to evaluate potential relationships of mutation inheritance between B cells expressing IgE, IgA and other isotypes within clones (8) (Fig. 4A). We identified the isotype and tissue of the sequence that had the most shared SHM changes with an IgE+ clone member (the “nearest neighbor”) and normalized the counts of these relationships to adjust for differences in sequencing depth in each tissue and isotype (see Methods). The resulting value was expressed as a nearest neighbor probability and scaled to 1. For IgE sequences found in the stomach, duodenum, and peripheral blood, the nearest neighbor was overwhelmingly found in the same tissue for PA (Fig. 4B) as well as OA and NA donors (Fig. S6B), providing further support for local class-switching to IgE in the stomach and duodenum. The nearest neighbor isotype to IgE, normalized for isotype read frequencies, was G4>A2>G1>A1 in the stomach, A2>A1>G1>G4 in the duodenum and A2>G3>G1>A1 in the peripheral blood (Fig. 4C, upper row). IgE nearest neighbors in OA stomach and duodenum and NA duodenum were also most frequently A2, with an increased probability of G1 in the OA stomach (Fig. S6C). In contrast to the IgE+ clones found in PA tissues, the nearest neighbor isotype to IgE in NA peripheral blood was G4>G3>G2>G1. We additionally compared the frequency, in counts of clones, of each nearest neighbor isotype event without normalization to read depth, and found that IgA1 and IgA2 were frequent nearest neighbors for IgE in all PA tissues, with the highest frequency in the duodenum (Fig. 4C, lower row). IgM and IgG1 nearest neighbors for IgE were also observed in the stomach and duodenum of PA subjects at lower frequencies. IgE and clonally related nearest SHM neighbors, including IgM sequences, had similar overall IGHV SHM values (Fig. S6D). Consistent with the notion that IgE+ B cells could be derived from CSR of IgA1 clone members, all PA subjects contained low but detectable levels of serum IgA specific for total peanut protein; nonallergic controls who lack peanut-specific IgE also showed low but detectable peanut-specific serum IgA (Table S3).
Figure 4. SHM patterns in GI tissues are consistent with local IgE CSR.
(A) Nucleotide sequence alignment of members of an IgE-containing clonal lineage. Red boxes show shared SHM with the illustrated IgE clone member. The sequence with the most shared SHM, 14 in this case, is the nearest neighbor. (B) Collision probabilities that the most closely related clone member to IgE is found in a given tissue (polar axes) indicated by the height of the filled region on the central axis. The panels indicate the nearest neighbor for IgE sequences found in each of the six tissues (indicated by color). Plots show the sum of clones for PA individuals. (C) Nearest isotype neighbors for IgE. Nearest neighbor isotype collision probabilities (top row) or unnormalized clone counts (bottom row) are shown.
Common IgH rearrangements in IgE-expressing Ara h 2-binding clones
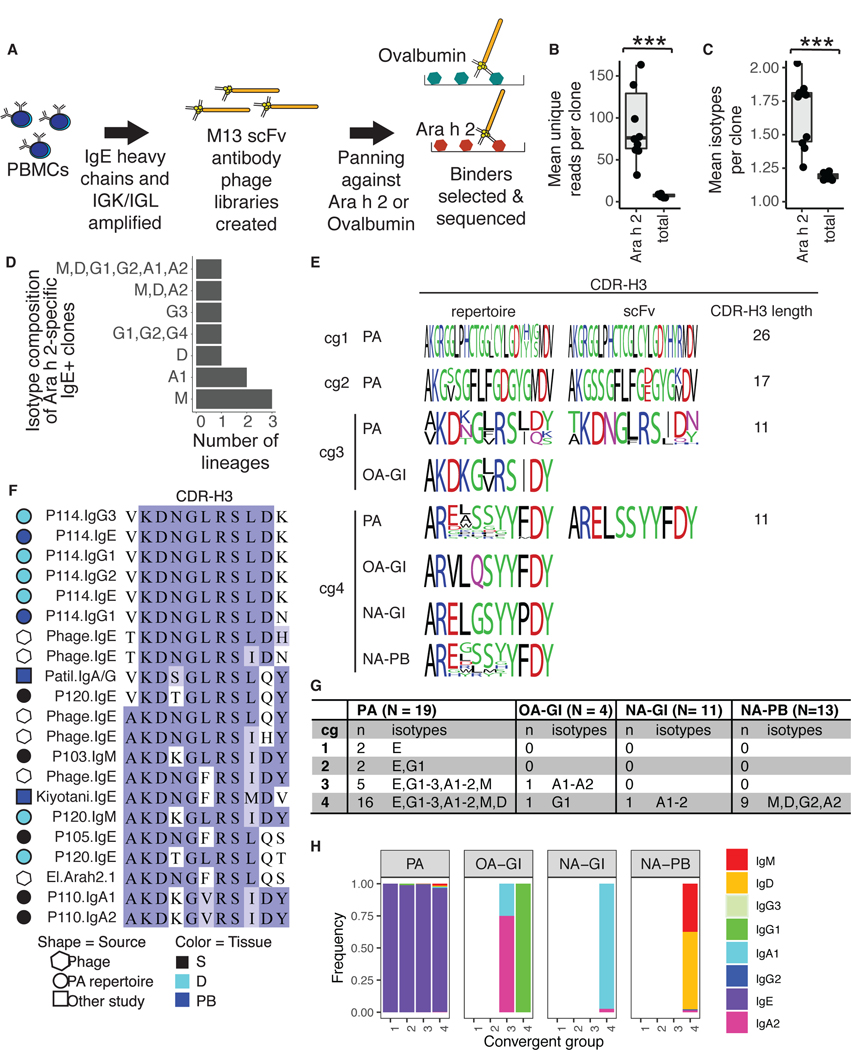
To assess the antigen specificity of GI tract IgE+ sequences, we generated single-chain variable region (scFv) phage display libraries (28, 29, 30) from IgE heavy chains and light chains isolated from stomach and duodenal biopsies from 10 PA individuals. As monosensitization to Ara h 2 is highly predictive of peanut allergy (31) and all but one (P102) of the 10 PA donors had serum IgE against Ara h 2 (Table S3), we panned GI scFv libraries against purified Ara h 2 protein to identify allergen-specific IgE. Libraries were also panned against ovalbumin as a negative control (Fig. 5A). A caveat of this approach, which has been used to identify aeroallergen-specific IgE in prior studies (9, 32), is that scFv libraries are generated with combinatorial pairing of heavy and light chains. HTS was used to identify scFv library inserts before and after the first and second rounds of panning. Ara h 2-binding scFv sequences were enriched for several heavy and light chain V gene segments including IGHV1-18, IGLV1-51 and IGLV2-14 (Fig. S7A and S7B). Clustering with HTS IgH repertoire sequences identified 463 Ara h 2-specific IgE clones that showed five-fold or greater enrichment in read counts after Ara h 2 panning and no enrichment after panning against ovalbumin. ELISA experiments (28, 33) with randomly-selected scFv clones isolated after Ara h 2 panning verified Ara h 2-specificity for seven distinct CDR-H3 amino acid sequences and six clonally-related groups (Fig. S7C and S7D). Ara h 2-binding B cell clones from the blood and GI tract were more clonally expanded than total clones from the same individuals, both by the number of unique reads (Fig. 5B) and the number of isotypes per clone (Fig. 5C). 10 of the Ara h 2-specific clones contained at least one other isotype in addition to IgE (Fig. 5D and Table S4).
Figure 5. Identification of convergent Ara h 2-specific IgE CDR-H3 sequences.
(A) Panning of scFv phage libraries generated from IgE and IgK or IgL transcripts was used to identify Ara h 2-specific clones. (B-C) The mean number of (B) unique reads or (C) isotypes found in Ara h 2-specific or total clones per donor is shown (WMW test: ***p < 0.001). For plotting, one outlier (P105) with 604 unique reads per Ara h 2-specific clone was excluded. (D) Isotypes expressed in Ara h 2-specific clones containing IgE+ members. (E) Comparison of convergent group CDR-H3 sequences from PA, OA and NA HTS repertoires to scFv phage sequences. Sequence logos were generated from the most abundant CDR-H3 from each participant. The relative heights of the letters within a stack are proportional to their frequencies and are colored according to chemical properties: polar (green); basic (blue); acidic (red); neutral (purple); or hydrophobic (black). (F) cg3 CDR-H3 sequences, colored by BLOSUM62 scores. The experimental source (indicated by symbol shape) and tissue origin (symbol color) of sequences are illustrated. (G) Isotypes expressed by Ara h 2-binding convergent clone group members. (H) Isotype frequencies for cg1 to cg4, collapsed by unique reads.
Although human antibody repertoires are highly diverse, antigen-driven clonal proliferation can cause strong selection and reveal highly similar “convergent” antibodies in multiple individuals, as reported for viral infections (34) or vaccinations (35, 36). Potential convergent antibody responses have begun to be observed in PA blood samples (19, 37), but with limited evidence for convergent IgE. We hypothesized that convergent IgE sequences specific for peanut allergens would be detectable in the abundant IgE+ clone datasets obtained from the GI tract samples. We searched isotype data from the 19 PA patients for IgH with 80% CDR-H3 amino acid identity and identical CDR-H3 length as Ara h 2-binding IgE scFv clones. Four convergent groups (‘cg1’ to ‘cg4’) contained IgE+ members in at least two allergic individuals (Fig. 5E). Notably, the cg3 group encompassed examples reported in independent prior studies from Ara h 2-specific B cells (37) or bulk PBMCs (38) isolated from peanut allergic individuals (Fig. 5F). Most cg sequences in PA individuals expressed IgE (Fig. 5G and 5H). cg3 and cg4 sequences were also detected in OA and NA individuals, but only as non-IgE isotypes (Fig. 5G and 5H). Taken together, these results suggest that convergent Ara h 2-specific B cell responses are made by both allergic and nonallergic individuals, but are only expressed as IgE in subjects with allergy to peanut. The detection of cg3 sequences in PA patient data from other independent studies further highlights these clonotypes as a common kind of antibody gene rearrangement repeatedly associated with human peanut allergy.
Discussion
The GI tract is the primary route of exposure for food allergen proteins, and a site of interaction with a major reservoir of lymphocytes and plasma cells in the body. We find that PA patient stomach and duodenum harbor large numbers of somatically mutated, clonally-expanded, allergen-specific IgE+ B-lineage cells, including cells with a plasma cell phenotype, whereas non-allergic controls have few to no IgE+ B-lineage clones in GI sites, and patients with other non-food allergies have intermediate numbers of IgE+ clones. Stomach and duodenal IgE+ clone counts in PA individuals correlate well with peanut allergen-specific serum IgE concentrations. These findings indicate that GI tissues are an important reservoir for allergen-specific IgE+ plasma cells in PA subjects, and could contribute significantly to allergen-specific serum IgE in the tissues and perhaps systemically. We did not detect IgE+ cells with morphological features of memory B cells, but IgE expression levels in such B cells may be below the limit of detection for IF in formalin-fixed tissues, potentially due to membrane IgE receptor downregulation (19). Further studies will be required to evaluate whether memory IgE+ cells are present in human GI tissues. One implication of our findings is that local IgE+ plasma cells in the GI tissues of PA patients could contribute disproportionately to the FcεRI-bound IgE of nearby mast cells in the same tissues. Regional differences of mast cell IgE loading by different IgE+ plasma cell clones in GI tissues could contribute to the clinical heterogeneity of patient symptoms and sensitivity to peanut allergen exposure.
Our analysis of over 1300 multi-isotype IgE+ clonal lineages from PA patients shows that IgE+ clone members are most often found in the same tissue biopsy as non-IgE+ members of the same clones. In addition, the greatest sharing of SHM changes is seen between clone members in the same tissue, consistent with IgE+ B cell development via local CSR in these sites. We detected a high frequency of clones containing IgE and IgA1 or IgA2 members in the GI tract, which opens the possibility that IgA1 CSR to IgE and IgE CSR to IgA2 are common events in PA individuals. This may be an important difference between IgE+ B cell development in humans and mice, as mice cannot switch from IgA to IgE due to the ordering of isotypes in their IgH locus. Prior data from peripheral blood B cells in limited numbers of individuals with aeroallergies indicated that IgE clone members were usually most closely related to IgG1 members (8). These data therefore suggest that B cell differentiation pathways in patients who develop food allergy differ from those in patients with aeroallergies, and potentially that food allergy sensitization or allergen-specific B cell clonal expansion may occur in oral or gastrointestinal mucosa. Our results do not address whether IgE+ B cells participate in GI-tract germinal center reactions, or whether class-switching to IgE might occur after affinity maturation has already taken place in GI-resident IgA, IgG or IgM cells. Similarly, our data do not address whether T cell help is required for GI-tract IgE CSR or which microanatomical locations or niches could be involved. CSR events could potentially occur in duodenal Peyer’s patch germinal centers or the small aggregates of lymphoid cells that have been observed in human gastric biopsies (39). Alternatively, CSR to IgE could occur in mesenteric lymph nodes followed by preferential trafficking of clonally-related cells to distinct GI-tract anatomical sections. In addition, our data do not address whether the IgE+ plasma cells we detect in patient GI tissues are long-lived or short-lived, requiring continuous production to maintain their numbers. Further evaluation of these topics, as well as analysis of changes that may occur during food allergy oral immunotherapy in allergen-specific IgE+ and other isotype-expressing B-lineage cells in the GI tract, should be high priorities for future research.
Comparing total IgH repertoire data to Ara h 2-specific clones identified with phage display and PA patient Ara h 2-specific sequences reported in prior literature, we identified four convergent IgE Ara h 2-binding CDR-H3 antibody sequence groups shared between multiple PA individuals. The cg4 motif was found in a majority of allergic donors as well as OA and NA controls, but only appeared as IgE in PA patients, suggesting that class-switching of peanut-specific B cells from IgA/G/M isotypes to IgE, rather than the generation of these V(D)J gene rearrangements, may distinguish PA subjects from those without peanut allergy. A second convergent antibody group, cg3, was observed almost entirely as IgE in PA patients, including patients from two previous independent studies, but was only observed as non-IgE isotypes in non-food allergic control subjects, including in OA controls who had numerous IgE clones in their sampled tissues. Together, these findings indicate that recognizable and common IgE antibody gene rearrangements are a hallmark of peanut allergy. Such sequence patterns, if replicated and extended beyond our relatively small clinical study, could begin to define a diagnostic set of IgE antibody types that may be specific for particular allergies, and could be of value in tracking patient responses to treatment.
Materials and Methods
Study Design
The objective of this study was to characterize the IgE+ B cell repertoires in peripheral blood and gut mucosa biopsies from adult individuals with clinically-defined peanut allergy, other non-food allergies and nonallergic controls. Most of the experiments consist of analysis of targeted IgH Illumina Miseq 600-cycle sequencing data from cDNA libraries prepared from bulk RNA extracted from PBMC samples or GI biopsies. To identify peanut allergen-specific IgE+ B cell clones, scFv libraries were constructed from the IgE+ B cell repertoires from 10 PA study individuals, and specific binders were identified by panning against the major peanut allergen, Ara h 2. Control PBMC samples were isolated from adult and juvenile patients, and no PBMC donor had any known non-drug allergy. Control stomach and duodenal tissue samples were isolated either from adult patients with no known non-drug allergy (NA cohort) or with known non-food allergies (OA cohort). No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to the disease status of the samples. No outliers were removed for statistical analyses.
Study Approval
The protocol for this study was reviewed and approved by the Institutional Review Board of Stanford University. Written informed consent was obtained for all participants before entering the study. The POISED study is registered at www.clinicaltrials.gov (NCT02103270). Only baseline pre-treatment specimens from participants enrolled in the POISED study were used.
Study Participants
19 adult patients with peanut allergy, assessed by double-blind placebo-controlled food challenge (DBPCFC) as described (13), consented to sampling of peripheral blood and endoscopic biopsy of five sites in the GI tract: proximal esophagus, medial esophagus, distal esophagus, stomach and duodenum. At the time of sampling, patients were not receiving immunotherapy for allergy and were avoiding peanut-containing foods. In addition to allergic individuals, we obtained control peripheral blood samples from 13 nonallergic participants with no history of allergic reaction, control stomach biopsies from 11 participants with no known food allergy or aeroallergy, and control duodenal biopsies from five participants with no known food allergy or aeroallergy (courtesy of Dr. Nielsen Fernandez-Becker and Stanford Tissue Bank). A small additional cohort of four individuals with aeroallergy or drug allergy was also recruited and provided informed consent for collection of stomach and duodenal samples (courtesy of Dr. Nielsen Fernandez-Becker and Stanford Tissue Bank). Paired stomach and duodenal samples were obtained for three OA subjects, and five NA subjects. The median age and sex breakdown of these patient groups was 28 years and 26% female (peanut allergic), 36 years and 50% female (non-food allergic) and 46 years and 83% female (nonallergic). Other demographic data for the study cohort are shown in Table S1.
Collection of samples and processing
Upper endoscopy was performed by Stanford University gastroenterologists using the Pentax or Olympus adult upper endoscope. Biopsies were taken using a Boston Scientific Radial Jaw 4 Large Capacity Needle. Specimens were placed immediately in empty tubes or in tubes containing RNAlater (Thermo Fisher Scientific) and stored at −80°C until processing. PBMCs were isolated from whole blood samples from allergic and nonallergic donors by density gradient centrifugation on Histopaque-1077 (Sigma-Aldrich). For allergic donors and a subset of nonallergic donors, the plasma was also collected and stored at −80°C. PBMCs were cryopreserved and stored in liquid nitrogen until processing.
RNA extraction and generation of cDNA libraries
RNA was extracted from PBMCs using RNeasy mini spin columns (Qiagen) according to the manufacturer’s protocols. To extract RNA from GI biopsies, samples were removed from RNAlater and homogenized in TRIzol (Invitrogen) using a BeadBug (Benchmark Scientific) with stainless steel, 2.8 mm acid-washed beads. After visual confirmation of homogenization, 100 μL of chloroform was added, the sample was vortexed, incubated for two minutes at room temperature, and spun down at 12000 x g for 15 minutes at 4°C. The transparent upper phase was transferred to a new tube and an equal volume of 70% ethanol was added. This mixture was then transferred to an RNeasy mini spin column for RNA extraction according to the manufacturer’s protocols. Stomach and duodenum libraries were accidentally mixed for PA participant P106 during processing. This participant was excluded from analyses where stomach and duodenum were considered separately.
cDNA was synthesized from 100 ng of RNA, when available, using SuperScript III (Thermo Fisher Scientific) and primed with random hexamers (Promega) according to manufacturer’s protocols. IgH genes were amplified by multiplexed RT-PCR using Biomed2 IGHV primers in the FR1 framework region (40) and isotype-specific primers for IgM, IgD, IgG, IgA and IgE located in the CH1 constant region. Each isotype was amplified separately to prevent the formation of cross-isotype chimeric PCR products. Light chains were not amplified or analyzed. The primer sequences have been described previously (16). Primers contained eight nucleotide barcodes used to identify samples as well as half of the Illumina adapter sequence needed for cluster generation and sequencing on the MiSeq instrument. RT-PCR was performed with gene-specific primers for 35 cycles, and then an additional 12 cycles were completed with primers to complete the Illumina adapters. For Illumina cluster recognition, four randomized nucleotides were included in the primers immediately after the Illumina adapter sequence in the constant region primers. The PCR programs were as follows: First PCR: 94°C for 7 min.; 35 cycles of (94°C 30 s, 58°C 45 s, 72°C 120 s); and final extension at 72°C for 10 min.; Second PCR: 0.4 μL of first PCR product was used as template for this 20 μL reaction; Conditions: 94°C for 15 min.; 12 cycles of (94°C 30 s, 60°C 45 s, 72°C 90 s); and final extension at 72°C for 10 min. The products of each second PCR reaction were analyzed by agarose gel electrophoresis and pooled in equal amounts based on gel band intensity. The pooled library was size selected by agarose gel electrophoresis to remove primer dimers and small products and purified with QIAquick Gel Extraction Kits (Qiagen). IgH libraries were sequenced on an Illumina MiSeq using 600-cycle sequencing kits.
Primer design for RT-qPCR
To estimate the frequency of B cells in donor samples, we designed primers to amplify mRNA transcripts from rearranged IgK light chains. The sequences of human Ig kappa J-segment genes (J00242, J00242, Z70260, U95246, Z46620, J00242, J00242, AF103571 and J00242) and constant region genes (J00241, M11736, M11737, AF017732, AF113887) were obtained from IMGT/GENE-DB (41). Forward primers specific for IGKJ-segment genes IGKJ1–5 were designed by reverse complementing the BIOMED-2 IGKJ gene-specific primers Igk-J-A-1, Igk-J-B-1 and Igk-J-C-1 (40). A compatible reverse primer in the IGKC constant region genes was then designed using Primer3 software (v.0.4.0) (42). Two reference genes, ß-actin and GAPDH, were included for normalization. Primers for ACTB (NM_001101) and GAPDH (NM_002046) reference genes were purchased from Sigma (H_ACTB_1 and H_GAPDH_2, respectively). Primer specificity for all three genes was validated using PCR followed by gel electrophoresis and by melt curve analysis. All primers pairs were selected to span introns to reduce artifacts originating from contaminating genomic DNA.
RT-qPCR
RT-qPCR for relative quantification of IGKC expression by comparative threshold cycle (CT) was performed in 10 μL reaction volumes in 96-well plates using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, A25780) according to manufacturer’s instructions. Cycling was performed on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) using the following conditions: 50°C for 2 min for UDG activation, 95°C for 2 min for polymerase activation, followed by 40 cycles of 95°C for 15 s for denaturation, 60°C for 15 s for annealing and 72°C for 1 min for elongation. Each sample’s three gene amplifications were completed in the same run. No-template controls and inter-plate control reactions containing the same template for all reaction plates were performed in each run. All reactions were run in duplicate, including no-template and inter-plate controls.
Analysis of RT-qPCR data
Data from multiple plates were analyzed as a single data set. Baseline and threshold (Cq) values were assigned for all plates using the StepOne Software v2.3 (Thermo Fisher Scientific) and were manually reviewed before further analysis. Fold expression of IgK transcripts over reference genes was calculated as 2(-ΔCqmean), where ΔCqmean = CqTmean – (CqR1mean+CqR2mean)/2, and CqT, CqR1, and CqR2 are the Cq mean values of the target gene or the reference genes.
IgH sequence analysis
Paired-end reads were merged by FLASH and sequences with perfect or one-off matches to full-length VH and CH primers were demultiplexed according to the primer-encoded barcodes indicating the sample and participant from which each read-pair was derived. For isotype calling, we required perfect or one-off matches to constant region gene sequences upstream of the CH primers, to remove artefactual sequences generated by cross-priming in the CH1 region and sequences with low-certainty subisotype calls (IgG1-IgG4, IgA1-IgA2). Out-of-frame rearrangements, rearrangements containing stop codons, sequence reads with no isotype call, and potential PCR contaminant low read-count IgE sequences with identical nucleotide sequences found in more than one subject were removed from analysis. Reads were aligned using a local installation of IgBLAST to identify features including germline IGHV, IGHD and IGHJ genes, framework and complementarity determining regions, and positions containing mutations. Sequences were subsequently clustered into inferred clonal lineages via single linkage clustering as described (16), with restrictions that the sequences must be from the same individual, use the same IGHV gene and the same IGHJ gene (disregarding allele call), have the same CDR-H3 length, and have at least 90% identity in the nucleotides encoding the CDR-H3 region. Unless otherwise stated, identical reads sequenced from the same cell aliquot were collapsed to avoid overcounting B cells represented by multiple RNA transcripts or PCR-generated copies in the sequencing pool.
Clone counts
Productive IgH sequence reads passing quality filters were collapsed into inferred clonal lineages, as described above. IgM, IgD, IgG, IgE and IgA-containing clones were defined as those that contained at least one member of the indicated isotype. The number of distinct clones containing any isotype was designated ‘total clones.’ Normalized clone counts were obtained by dividing the number of observed clones by the amount of RNA isolated from tissue specimens. Linear regression models and 95% confidence intervals for correlations with peanut allergen-specific serum measurements were computed using the geom_smooth package in ggplot (method = “lm”) in the R statistical programming language.
Antibody measurements
Total IgE and allergen-specific IgE and IgA were measured by ImmunoCAP 250 assay (Thermo Fisher Scientific). IgE and IgA antibodies against total peanut protein as well as IgE antibodies against peanut allergen component proteins, Ara h 1, Ara h 2, Ara h 3, Ara h 8 and Ara h 9 were measured. Values <0.01 kUA/L for IgE measurements, or <0.01 mgA/L for IgA measurements, were considered below the limit of detection of the assay.
Immunofluorescence staining and imaging
Allergic biopsies from proximal, medial and distal esophagus, and stomach and duodenum were obtained fresh from the operation room, fixed in 10% formalin for four hours and then embedded in paraffin. 4 μm sections were cut and baked at 60°C one hour prior to staining. Slides were stained with prediluted rabbit anti-human IgE (Abcam, ab75673) for two hours, followed by mouse anti-human CD138 at 1:2000 (Thermo Fisher Scientific, MA1–10091) for one hour. Secondary staining was performed for 30 minutes with Alexa Fluor 555 goat anti-rabbit IgG H+L (Invitrogen A-21429) and Alexa Fluor 647 donkey anti-mouse IgG H+L (Invitrogen A-31571). Nuclei were stained with ProLong Gold Antifade with DAPI (Invitrogen P36941). Secondary-only control staining was performed as above, except slides were incubated with 1x PBS dilution buffer instead of primary antibody. Imaging and analysis were performed using Ariol v4.0 software (Leica Biosystems). Slides were scanned at 20x magnification with 3 z-stacks for Alexa Fluor 555, Alexa Fluor 647 and DAPI. Aperio ImageScope software (Leica Biosystems) was used to measure the largest width and height found in each biopsy. These dimensions were then multiplied to obtain an estimate of the imaged area. Scoring of IgE+CD138+ cells was performed by a board-certified pathologist blinded to sample identity and disease category.
Clonal B cell lineage trees
The three clonally-related sequences from the same tissue and expressing the same isotype that had the highest read counts were selected as representative sequences for lineage tree plotting. Lineage trees were inferred from the IGHV gene sequences using maximum parsimony via PHYLIP (version 3.695) and visualized using FigTree software (version 1.4.3).
Permutation testing
The significance of the number of IgE and IgM/D/G or A1 events that co-localized in the same tissue was tested using permutation tests. The tissue localization of clones was randomized using the shuffle() method of the python module, random. Data were permuted 1000 times to generate a permutation distribution. The p-value for the observed number of co-localizing same-tissue clones was obtained by counting the number of permutations that had a number of co-localizing same tissue clones as large or larger than the value observed in the original data.
IgE nearest neighbor analysis
Multi-isotype clonal lineages containing at least one IgE member and at least one member that was not IgE (IgM, IgD, IgG or IgA) were identified. The distance between the most abundant IgE-expressing clone member from a given sample and all IgM/D/G/A members of the same clone was calculated based on the number of shared mutations from germline in the IGHV and IGHJ gene segments. The calculation of the probability of the nearest neighbor falling in a specific tissue or a specific isotype compartment (the collision probability) is discussed below.
Calculation of the collision probability
To compare the degree of similarity between clonal populations in different tissues or isotypes in a manner independent of sequencing depth, we used a metric that has been applied previously to estimate clone size using deep-sequenced replicate DNA libraries (16, 34, 43). The collision probability is the probability that arbitrarily chosen sequence pairs from different cellular aliquots of a single sample are clonally related (44). The collision probability was calculated as previously reported (8) with the modification that identical sequences from the same cellular aliquot were counted just once to prevent overestimating cell abundance because of multiple mRNA copies from the same cell. The collision probability was calculated as:
where Nij and Nik are the copy numbers of uniquely observed members of clone i observed in B cell populations, j and k; and Tj and Tk are total unique read numbers in the corresponding cDNA libraries. In Figure 4B and S6B, j and k are the B cell subsets in cDNA libraries generated from aliquots of template RNA from independently sampled tissue biopsies. In Figure 4C and S6C, j and k are the B cell subsets expressing different isotypes in a cDNA library generated from the same cell aliquot. In Figure S5A, j and k are the B cell subsets expressing a non-IgE read or IgE, from cDNA libraries generated from RNA from independently sampled tissue biopsies.
Sequence alignments
Nucleotide sequences for rearranged IgH VDJ genes were aligned and visualized using the msa package in R. Amino acid CDR-H3 sequences were aligned using ClustalW and visualized using Jalview v10.2.5.
Seqlogo generation
Seqlogos were generated using the WebLogo tool (https://weblogo.berkeley.edu).
Statistics
Statistics were calculated in the R statistical language using indicated packages. For boxplots, plots show the median value as the central line, boxes show the 25–75% range, and whiskers encompass values to 1.5 times the interquartile range. No outliers were removed for statistical analyses.
Construction of M13 scFv phage display libraries
Separate M13 scFv phage display libraries were generated from stomach and duodenal tissue from 10 peanut-allergic subjects (donors 101 to 110) at the start of their enrollment in the POISED study. scFvs were expressed in a 5’-IGH-linker-IGK/IGL-3’ orientation as an N-terminal pIII fusion protein (28, 30). Sequences encoding IgE heavy chain variable regions were selectively amplified from tissue-derived cDNAs in separate reactions using modified forward primers (29) and a modified “IGHE CH1 ver. 3” reverse primer (45) (Table S5). IgE variable regions were then amplified in a second PCR reaction and IgK and IgL variable regions were separately amplified from tissue-derived cDNA using modified primers (29) (Table S5). Overlap extension PCR then was used to fuse an individual’s IgE variable regions and IgK or IgL variable regions via the connecting linker sequence. The scFv overlap products from a single person were combined in a ratio of 1:2 lambda to kappa overlap products to reflect human light chain frequencies (46). The overlap products from the 10 participants were next combined together in equal amounts based on band intensity on an agarose gel to form either a stomach or duodenum pool and each pool was purified using a QIAquick PCR Purification Kit (Qiagen). Detailed PCR protocols are provided in Table S5.
The stomach and duodenum pools, along with pSEX81 phagemid vector (Progen), were digested with excess NcoI-HF and NotI-HF (New England Biolabs) and the cut pSEX81 vector was treated with phosphatase. After purification with a QIAquick Gel Extraction Kit (Qiagen), cut pSEX81 was ligated separately to the stomach or duodenum scFv insert pools and ligation products were electroporated into electrocompetent SS320 E. coli cells (Lucigen). The next day, colonies on dilution plates were counted, the transformants on the undiluted plates were scraped into 2xYT broth with 15% glycerol and aliquots were stored at −80°C. Quantitation of the dilution plates gave estimated library diversities of 4 × 107 (stomach) or 5.2 × 107 (duodenum) transformants. Quantitation of control reactions indicated that 6% of these transformants may be uncut or re-ligated pSEX81.
scFv panning to isolate Ara h 2-binding antibodies
Stomach and duodenum scFv libraries were panned against natural purified Ara h 2 protein (Indoor Biotechnologies) or negative control ovalbumin (OVA) (Fisher Scientific) using a protocol modified from published work (28, 30, 47). Two panning rounds were performed. 100 ng/μL Ara h 2 or OVA in PBS was used in the first round (pan1), while 10 ng/μL was used in the second round (pan2). Pan1 began with 2 × 1012 phage (1012 phage from each scFv library), while pan2 started with 1012 phage. Wells were washed five times using PBS with 0.1% (v/v) Tween 20 in pan1 and were washed 10 times in pan2. Enriched phage were eluted with trypsin and recovered after infection of SS320 E. coli. 105 to 106 colonies were recovered after pan1, while 106 to 3 × 106 colonies were recovered after pan2. Long-term panning stocks were made by scraping colonies into 2xYT broth with 15% glycerol and aliquots were stored at −80°C.
Identification of Ara h 2-specific scFv by HTS analysis
Phagemids from the pre-panned libraries, as well as after the two panning steps, were purified from E. coli using a QIAprep Spin Miniprep Kit (Qiagen). Targeted amplicons from scFv were PCR-amplified using primers in the IGHV FR2 framework region (40) and a reverse primer in the pSEX81vector downstream of the light chain (Table S5). A second PCR was performed to complete the Illumina MiSeq adapters as previously published (48), but used 15 cycles with a 1.5-minute extension time. The pooled library was sequenced using an Illumina MiSeq instrument with a 600-cycle kit. Because these scFv amplicons were longer than 600 nucleotides, we first isolated the sequences that did not merge with FLASH. Eight-nucleotide barcodes were used to assign the sequences to their originating sample. Sequences in which IgBLAST identified an IgK or IgL sequence in read 2 and an IgH sequence in read 1 were analyzed further. After binning sequences by IGHV gene identity and CDR-H3 length, cd-hit was used with a 90% CDR-H3 nucleotide or amino acid identity cut-off value to cluster the scFv IgH sequences with total IgH repertoire data from the tissue sequencing of the 10 individuals represented in the phage libraries. We identified and further analyzed sequences that were at least five-fold enriched after binding Ara h 2 in panning round 1 or 2 relative to their frequency in the pre-panned libraries, and which showed no enrichment when panned with OVA in the first or second panning round.
Confirmation of Ara h 2-specific binding by phage ELISA
scFv binding to Ara h 2 was confirmed by phage ELISA (28, 30, 33). Full-length colony PCR products of monoclonal scFvs derived from the first or second panning frozen stocks were sequenced by Sanger sequencing. Individual scFv species with unique CDR3-H3s were then expressed and tested individually for binding to 10 ng/μL Ara h 2 or OVA in replicate. scFvs with seven distinct CDR-H3s were tested and confirmed as Ara h 2 binders in this manner (Fig. S7C and S7D).
Supplementary Material
Table S2. Per-person read and clone counts.
Table S1. Patient demographics.
Fig. S1. Overall IGH repertoire properties are similar in allergic and nonallergic individuals.
Fig. S2. Supplementary material for B cell clone counts in tissues.
Figure S3. Immunofluorescence microscopy analysis of IgE+CD138+ cells in tissues.
Figure S4. Permutation testing for the frequency of co-occurrence of IgE and clonal precursors in tissue sites.
Fig. S5. Analysis of multi-isotype IgE+ clones in multiple tissue sites.
Fig. S6. Supplementary material for nearest neighbor analysis.
Fig. S7. Analysis of Ara h 2-specific scFv heavy chain sequences.
Table S3. ImmunoCAP serum antibody measurements.
Table S4. Nearest neighbors for allergen-specific IgE clones.
Table S5. Phage primers and PCR protocols.
Acknowledgments:
The authors thank the participants in this study. We also thank Dr. Katherine J.L. Jackson for sharing analysis scripts and for critical reading of the manuscript.
Funding: NIH/NIAID grants U19 AI104209 (SJG, KCN, SDB, RSC, NAF-B), R01 AI125567 (SDB, SJG), R01 AI140134 (KCN), R01 HL118612 (KCN), U01 AI140498 (KCN), UM1 AI130839 (KCN), a Big Data for Human Health Seed Grant from the Li Ka Shing Foundation (SDB and SAJ), an endowment from the Crown Family Foundation (SDB), the Sean N. Parker Center for Allergy and Asthma Research at Stanford University Medical Center (KCN), Food Allergy Research and Education Center of Excellence (KCN), Myra Reinhard Foundation (KCN), End Allergies Together (KCN), the Hartman Family Foundation (KCN), the Naddisy Foundation (KCN), and the Levin Family Foundation (KCN).
Footnotes
Competing interests: KCN has received funding from, is currently funded by, or cofounded the following: the NIH, Food Allergy Research & Education, End Allergies Together, Before Brands, Alladapt Immunotherapeutics, Adare Pharmaceuticals, AstraZeneca, Novartis, Genentech, Astellas, DBV Technologies, ForTra, Aimmune Therapeutics, Regeneron, Nestle, Sanofi, ImmuneWorks, Cour, Allergenis, Ukko, the Environmental Protection Agency, Myra Reinhard Foundation, Hartman Family Foundation, Naddisy Foundation, and Levin Family Foundation; RSC has received funding from, is currently funded by, or consults for the following: the NIAID, CoFAR, End Allergies Together, Alladapt Immunotherapeutics, AstraZeneca, Novartis, Astellas, AnaptysBio, DBV Technologies, Aimmune Therapeutics, AllerGenis, Regeneron, Guidepoint, GI Innovation, and Stanford Maternal and Child Health Research Institute. SJG receives funding from the NIH (NIAID grants U19AI104209, R01AI125567); SDB has received funding from or is currently funded by the NIH (NIAID grants R01AI125567, U19AI104209, R01AI127877, R01AI130398), the Bill and Melinda Gates Foundation, the Ellison Medical Foundation, DARPA, the Stanford Maternal and Child Health Research Institute, and an endowment from the Crown Family Foundation. SDB has consulted for Regeneron and Sanofi as an expert witness, and owns stock in CareDx, and AbCellera.
Data and materials availability: All DNA sequence data used in the study are available in the SRA archive with Bioproject ID PRJNA602630.
References and Notes
- 1.Sicherer SH, Epidemiology of food allergy. J Allergy Clin Immunol 127, 594–602 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Liu AH et al. , National prevalence and risk factors for food allergy and relationship to asthma: results from the National Health and Nutrition Examination Survey 2005–2006. J Allergy Clin Immunol 126, 798–806 e713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savage JH, Limb SL, Brereton NH, Wood RA, The natural history of peanut allergy: Extending our knowledge beyond childhood. J Allergy Clin Immunol 120, 717–719 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Gould HJ, Wu YB, IgE repertoire and immunological memory: compartmental regulation and antibody function. Int Immunol 30, 403–412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akdis M, Akdis CA, IgE class switching and cellular memory. Nat Immunol 13, 312–314 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Takhar P. et al. , Allergen drives class switching to IgE in the nasal mucosa in allergic rhinitis. J Immunol 174, 5024–5032 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Berkowska MA et al. , Human IgE(+) B cells are derived from T cell-dependent and T cell-independent pathways. J Allergy Clin Immunol 134, 688–697 e686 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Looney TJ et al. , Human B-cell isotype switching origins of IgE. J Allergy Clin Immunol 137, 579–586 e577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin M. et al. , Persistence and evolution of allergen-specific IgE repertoires during subcutaneous specific immunotherapy. J Allergy Clin Immunol 137, 1535–1544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coker HA, Durham SR, Gould HJ, Local somatic hypermutation and class switch recombination in the nasal mucosa of allergic rhinitis patients. J Immunol 171, 5602–5610 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Li H. et al. , Transcytosis of IgE-antigen complexes by CD23a in human intestinal epithelial cells and its role in food allergy. Gastroenterology 131, 47–58 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Berin C, Wood S, Lopez-Exposito I, Sampson H, Nowak-Wegrzyn A, Detection Of Milk-specific IgE And IgA In Stool Samples From Children With Food Allergy. Journal of Allergy and Clinical Immunology 127, AB181 (2011). [Google Scholar]
- 13.Chinthrajah RS et al. , Development of a tool predicting severity of allergic reaction during peanut challenge. Ann Allergy Asthma Immunol 121, 69–76 e62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu JL, Davis MM, Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13, 37–45 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Glanville J. et al. , Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci U S A 108, 20066–20071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roskin KM et al. , IgH sequences in common variable immune deficiency reveal altered B cell development and selection. Sci Transl Med 7, 302ra135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Veen W. et al. , IgG4 production is confined to human IL-10-producing regulatory B cells that suppress antigen-specific immune responses. J Allergy Clin Immunol 131, 1204–1212 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Nouri-Aria KT et al. , Grass pollen immunotherapy induces mucosal and peripheral IL-10 responses and blocking IgG activity. J Immunol 172, 3252–3259 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Croote D, Darmanis S, Nadeau KC, Quake SR, Human IgE producing B cells have a unique transcriptional program and generate high affinity, allergen-specific antibodies. bioRxiv, (2018). [Google Scholar]
- 20.Nair N. et al. , High-dimensional immune profiling of total and rotavirus VP6-specific intestinal and circulating B cells by mass cytometry. Mucosal Immunol 9, 68–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bemark M. et al. , Limited clonal relatedness between gut IgA plasma cells and memory B cells after oral immunization. Nature Communications 7, 12698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindner C. et al. , Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat Immunol 16, 880–888 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Meng W. et al. , An atlas of B-cell clonal distribution in the human body. Nat Biotechnol 35, 879–884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn-Walters DK, Boursier L, Spencer J, Hypermutation, diversity and dissemination of human intestinal lamina propria plasma cells. Eur J Immunol 27, 2959–2964 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Di Niro R. et al. , Responsive population dynamics and wide seeding into the duodenal lamina propria of transglutaminase-2-specific plasma cells in celiac disease. Mucosal Immunol 9, 254–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin M, Levander F, Palmason R, Greiff L, Ohlin M, Antibody-encoding repertoires of bone marrow and peripheral blood-a focus on IgE. J Allergy Clin Immunol 139, 1026–1030 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Wu YC et al. , Influence of seasonal exposure to grass pollen on local and peripheral blood IgE repertoires in patients with allergic rhinitis. J Allergy Clin Immunol 134, 604–612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rondot S, Koch J, Breitling F, Dubel S, A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol 19, 75–78 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Marks JD et al. , By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol 222, 581–597 (1991). [DOI] [PubMed] [Google Scholar]
- 30.Nielsen SCA et al. , Shaping of infant B cell receptor repertoires by environmental factors and infectious disease. Sci Transl Med 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kukkonen AK, Pelkonen AS, Makinen-Kiljunen S, Voutilainen H, Makela MJ, Ara h 2 and Ara 6 are the best predictors of severe peanut allergy: a double-blind placebo-controlled study. Allergy 70, 1239–1245 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Steinberger P, Kraft D, Valenta R, Construction of a combinatorial IgE library from an allergic patient. Isolation and characterization of human IgE Fabs with specificity for the major timothy grass pollen allergen, Phl p 5. J Biol Chem 271, 10967–10972 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Bradbury ARM, Marks JD, in Phage Display T. Clackson HB Lowman Eds. (Oxford University Press, 2009), pp. 277–279. [Google Scholar]
- 34.Parameswaran P. et al. , Convergent antibody signatures in human dengue. Cell Host Microbe 13, 691–700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson KJ et al. , Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe 16, 105–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joyce MG et al. , Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 166, 609–623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patil SU et al. , Peanut oral immunotherapy transiently expands circulating Ara h 2-specific B cells with a homologous repertoire in unrelated subjects. J Allergy Clin Immunol 136, 125–134 e112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiyotani K. et al. , Characterization of the B-cell receptor repertoires in peanut allergic subjects undergoing oral immunotherapy. J Hum Genet 63, 239–248 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Carney JA, Gastric mucosal lymphoid follicles: histology, distribution, frequency, and etiologic features. Am J Surg Pathol 34, 1019–1024 (2010). [DOI] [PubMed] [Google Scholar]
- 40.van Dongen JJ et al. , Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia 17, 2257–2317 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Giudicelli V, Chaume D, Lefranc MP, IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res 33, D256–261 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Untergasser A. et al. , Primer3--new capabilities and interfaces. Nucleic Acids Res 40, e115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyd SD et al. , Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med 1, 12ra23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnabel ZE, The Estimation of Total Fish Population of a Lake. The American Mathematical Monthly 45, 348–352 (1938). [Google Scholar]
- 45.Andreasson U. et al. , The human IgE-encoding transcriptome to assess antibody repertoires and repertoire evolution. J Mol Biol 362, 212–227 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Mole CM, Bene MC, Montagne PM, Seilles E, Faure GC, Light chains of immunoglobulins in human secretions. Clin Chim Acta 224, 191–197 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Rader C, Steinberger P, Barbas CF 3rd, in Phage Display: A Laboratory Manual, Barbas CF 3rd, Burton DR, Scott JK, Silverman GJ, Eds. (Cold Spring Harbor Laboratory Press, 2001), pp. 10.12–10.15. [Google Scholar]
- 48.Hoh RA et al. , Single B-cell deconvolution of peanut-specific antibody responses in allergic patients. J Allergy Clin Immunol 137, 157–167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Per-person read and clone counts.
Table S1. Patient demographics.
Fig. S1. Overall IGH repertoire properties are similar in allergic and nonallergic individuals.
Fig. S2. Supplementary material for B cell clone counts in tissues.
Figure S3. Immunofluorescence microscopy analysis of IgE+CD138+ cells in tissues.
Figure S4. Permutation testing for the frequency of co-occurrence of IgE and clonal precursors in tissue sites.
Fig. S5. Analysis of multi-isotype IgE+ clones in multiple tissue sites.
Fig. S6. Supplementary material for nearest neighbor analysis.
Fig. S7. Analysis of Ara h 2-specific scFv heavy chain sequences.
Table S3. ImmunoCAP serum antibody measurements.
Table S4. Nearest neighbors for allergen-specific IgE clones.
Table S5. Phage primers and PCR protocols.