Abstract
We report that the SARS-CoV-2 nucleocapsid protein (N-protein) undergoes liquid-liquid phase separation (LLPS) with viral RNA. N-protein condenses with specific RNA genomic elements under physiological buffer conditions and condensation is enhanced at human body temperatures (33°C and 37°C) and reduced at room temperature (22°C). RNA sequence and structure in specific genomic regions regulate N-protein condensation while other genomic regions promote condensate dissolution, potentially preventing aggregation of the large genome. At low concentrations, N-protein preferentially crosslinks to specific regions characterized by single-stranded RNA flanked by structured elements and these features specify the location, number, and strength of N-protein binding sites (valency). Liquid-like N-protein condensates form in mammalian cells in a concentration-dependent manner and can be altered by small molecules. Condensation of N-protein is RNA sequence and structure specific, sensitive to human body temperature, and manipulatable with small molecules, and therefore presents a screenable process for identifying antiviral compounds effective against SARS-CoV-2.
Keywords: SARS-CoV-2, Condensation, phase separation, packaging, RNP-MaP, RNA structure, nucleocapsid, coronavirus
Graphical Abstract
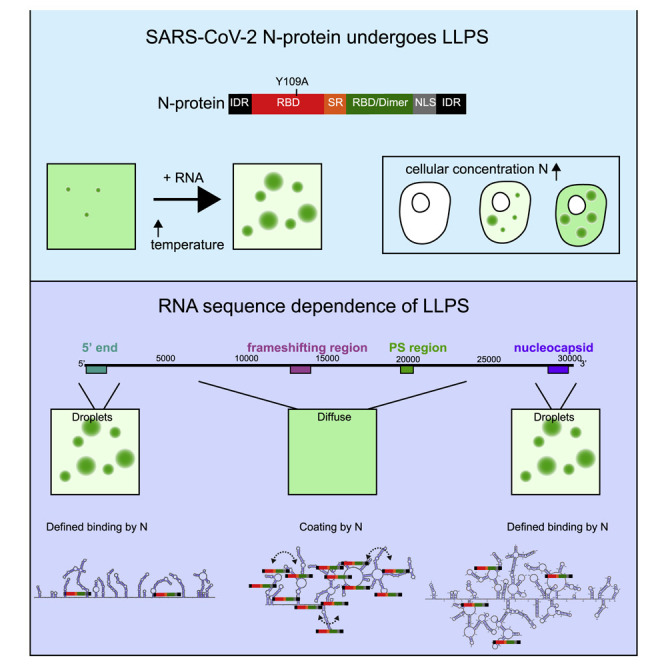
Iserman and Roden et al. demonstrate phase separation (LLPS) of SARS-CoV-2 nucleocapsid (N-protein) with viral RNA. Viral RNA sequences promote or oppose phase separation depending on binding patterns of N-protein with genomic RNA. LLPS-promoting sequences occur at 5′ and 3′ ends of the genome, suggestive of a genome packaging role.
Introduction
Biomolecular condensates are required for multiple cell biological processes and can form through liquid-liquid phase separation (LLPS) of proteins containing intrinsically disordered domains (IDRs) and RNA-binding domains (Brangwynne et al., 2009; Molliex et al., 2015; Nott et al., 2015; Pak et al., 2016; Wang et al., 2018; Zhang et al., 2015). Proteins with IDRs can sample many conformations to engage in weak, multivalent interactions that promote demixing and liquid-like properties of condensates (Holehouse and Pappu, 2018). In many instances, RNA promotes condensate formation (Elbaum-Garfinkle et al., 2015; Maharana et al., 2018; Zhang et al., 2015). Important protein features that determine condensates’ molecular grammar have been discovered based on amino acid sequence composition (Martin et al., 2020; Nott et al., 2015; Patel et al., 2015; Wang et al., 2018). Nucleic acids are integral components of many biomolecular condensates, but very little is understood about their roles. Indeed, only a few examples (Langdon et al., 2018; Ma et al., 2020; Maharana et al., 2018) describe specific RNA sequences and structures that contribute to LLPS.
RNA molecules have a uniform negative charge, leading to the suggestion that RNA acts as an anionic polymer in condensates, driving formation through electrostatic effects (Aumiller et al., 2016; Banerjee et al., 2017; Boeynaems et al., 2019). These conclusions are often based on studies employing RNAs that are far shorter and less chemically complex (i.e., poly(A or U)) than the native condensate RNAs (Aumiller et al., 2016; Banerjee et al., 2017; Boeynaems et al., 2019). Specific native RNA features—including length, sequence, and structure—are not well captured by these experiments. It is likely that native RNA sequences encode important functions such as RNA production timing, storage, transport, and modifications in condensates. As many condensate proteins have low complexity sequences and recognize degenerate RNA-binding motifs, it is unlikely that condensate identity is conferred solely by protein. Major unanswered questions about the role of RNA in LLPS remain. How do RNAs contribute to condensates properties? How are specific RNAs incorporated into specific condensates? Addressing RNA specificity is difficult for cellular condensates as, often, multiple RNAs coexist in the same condensate. This RNA composition complexity makes it challenging to determine which unique RNA elements drive LLPS and specify condensate material properties. However, viral packaging can provide a model to study a single RNA molecule driving condensation. For many RNA viruses, a single long (10–30 kb) RNA genome is packaged, a process which may be driven by LLPS. To examine RNA specificity in LLPS, we explored interaction of SARS-CoV-2 nucleocapsid (N-protein) with its genomic RNA as a model system.
Several viral processes incorporate phase separation. These include viral replication (Heinrich et al., 2018; Nikolic et al., 2017; Rincheval et al., 2017) and packaging, as was recently suggested for measles virus (Guseva et al., 2020). Negative-strand RNA viruses, such as RSV and VSV, replicate in “viral inclusion bodies” which are now thought to have characteristics of phase-separated droplets (Heinrich et al., 2018; Rincheval et al., 2017). Positive-stranded RNA viruses, such as coronaviruses, appear to replicate at small foci associated with cellular membranes (den Boon and Ahlquist, 2010; Knoops et al., 2012; Novoa et al., 2005). In coronaviruses, several studies using both EM and light microscopy have described the initial site of viral capsid and genome assembly as occurring in dynamic, cytoplasmic foci in proximity to membrane structures (Stertz et al., 2007; Verheije et al., 2010) supporting the hypothesis that phase separation plays a role in coronaviruses replication or packaging.
Coronaviruses, including SARS-CoV-2, have large ∼30 kb RNA genomes, and packaging is thought to be highly specific for the complete viral genome (gRNA), excluding host RNA and abundant virus-produced subgenomic RNAs (Masters, 2019). Viral replication and gRNA packaging depends on the N-protein (Grossoehme et al., 2009; McBride et al., 2014). N-protein must find this single gRNA molecule in the midst of many host and viral RNAs and must ensure the large gRNA does not become entangled, as has been observed for long cellular RNAs (Guillén-Boixet et al., 2020; Ma et al., 2020). The N-protein has RNA-binding domains, forms multimers (Cong et al., 2017), and is predicted to contain IDRs (Figure 1 A). N-protein thus has hallmarks of proteins that undergo liquid-liquid phase separation (LLPS), and we hypothesized LLPS, mediated by specific viral RNA sequences, may be important for SARS-CoV-2 processes such as viral genome packaging.
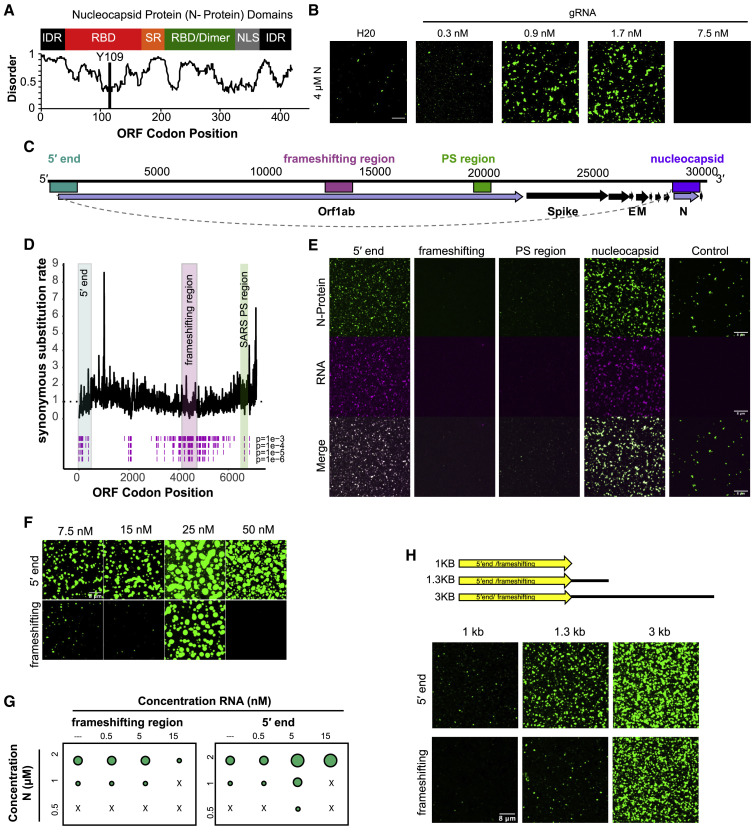
Figure 1.
N-Protein Undergoes LLPS with Specific Viral RNA Sequences
(A) N-protein has characteristics of proteins that undergo LLPS; predicted IDR sequences and multiple RNA-binding domains. Top panel: domain structure of N-protein. Bottom plot: disorder plot (y axis) of N-protein (x axis) (IUPred [Dosztányi, 2018]). IDR, intrinsically disordered regions; RBD, RNA-binding domain; SR, serine, arginine-rich region; dimer, dimerization domain; NLS, nuclear localization signal. Y109 indicates site of mutation used in Figure 3.
(B) N-protein phase separates with RNA containing SARS-CoV-2 genome. 4 μM N-protein undergoes concentration-dependent LLPS with increasing amounts of full-length containing gRNA. Green is N-protein signal.
(C) SARS-CoV-2 genome with regions tested for LLPS color coded: 5′ end (1–1,000; turquoise), frameshifting region (13,401–14,400; magenta), PS region SARS-CoV-2 homolog to published SARS-CoV-1 packaging signal (19,782–20,363; green), nucleocapsid fused to first 75 nt of 5′ end (gray dashed line) (nucleocapsid RNA; 1–75 + 28,273–29,533; purple).
(D) FRESCo (Sealfon et al., 2015) analysis of synonymous substitution restraints in ORF1ab. x axis is position in ORF1ab in codons. y axis is level of synonymous constraint. Significant synonymous constraints at four confidence cutoffs (1e−3, 1e−4, 1e−5, 1e−6) assessed over a ten-codon sliding window are marked by magenta lines. Tested regions correspond to those shown in (C).
(E) LLPS of N-protein is viral RNA sequence dependent. Different RNA regions (magenta signal) from SARS-CoV-2 (at 5 nM) either drive or solubilize N-protein (1 μM) droplets (green signal).
(F) Ability of 5′ end and frameshifting region RNA to drive or solubilize condensation of N-protein (4 μM) over increasing RNA concentrations. Frameshifting region only drives LLPS at 25 nM RNA.
(G) 5′ end promotes LLPS whereas frameshifting region promotes solubilization. Phase diagram of N-protein (green) with either 5′ end or frameshifting region RNA at indicated concentrations. Quantification corresponds to microscopy images in Figure S1D.
(H) Addition of RNA length enhances N-protein LLPS. LLPS was assessed with frameshifting region and 5′ end RNAs extended with non-specific plasmid sequences (at 5 nM RNA, 2 μM N-protein). Scale bar, 8 μm unless otherwise noted.
We find that N-protein phase separates at 37°C with gRNA and that LLPS of N-protein is associated with specific patterns of RNA binding sites within gRNA. Specific RNA elements are correlated with condensate formation or dissolution potentially specifying the number and location of protein binding sites (valency). We further demonstrate that RNA elements encode condensate material properties. Thus, a combination of distinct viral RNA-encoded elements ensures viral condensates of a specific molecular and physical identity. This study of SARS-CoV-2 reveals a new model viral system for uncovering rules for how RNA composition and physical state are specified in condensates and present new assays for screening viral LLPS-disrupting therapeutics.
Results
N-Protein Phase Separates with Viral RNA in a Length-, Sequence-, and Concentration-Dependent Manner
We reconstituted purified N-protein under physiological buffer conditions with RNAs encoding segments of SARS-CoV-2 gRNA and observed that N-protein produced either in mammalian cells (post-translationally modified) or bacteria (unmodified) phase separated with viral RNA (Figures S1A and S1B). Concentrations were chosen in part based on reported N-protein abundance in virions (Bar-On et al., 2020). Unmodified protein yielded larger, more abundant droplets, and the presence of an affinity tag or labeling the protein with dye did not alter behavior (Figure S1B). Since N-protein in SARS-CoV-1 virions is hypophoshorylated (Wu et al., 2009), and packaging (initiated by binding of N-protein to gRNA) first occurs in the cytoplasm (Fehr and Perlman, 2015; Stertz et al., 2007) where N-protein is thought to be in its unphosphorylated state (Fung and Liu, 2018), we used unmodified SARS-CoV-2 N-protein for subsequent experiments.
Pure N-protein demixed into droplets (consistent with results from the Morgan [Carlson et al., 2020] and Fawzi [Myrto Perdikari et al., 2020] labs) and LLPS was enhanced by RNA extracted from the culture medium supernatant of infected cells containing full-length SARS-CoV-2 genome (Figure 1B). To determine whether certain segments of SARS-CoV-2 genome had preferential ability to drive LLPS, we identified regions of the gRNA under synonymous codon constraints. We hypothesized that LLPS occurs specifically with gRNA carrying a viral packaging signal(s), whose exact structure and location in coronaviruses vary and is unknown for SARS-CoV-2. Using the computational algorithm FRESCo (Sealfon et al., 2015), we identified multiple regions with reduced synonymous sequence substitutions, indicative of functional RNA sequences and structures (Figures 1C, 1D, and S1C). In addition to synonymous substitution constraints, we initially focused on regions (1) that also contained predicted conserved structures (RNAz) (Table S1), (2) that are located in ORF1ab RNA (which contains the packaging signals for other Betacoronaviruses [Hsin et al., 2018; Kuo and Masters, 2013; Masters, 2019; Molenkamp and Spaan, 1997; Morales et al., 2013]), (3) that occur in packaged full-length genome sequences, and (4) that are absent from sub-genomic fragments (Kim et al., 2020) (Figures 1C and 1D). We synthesized sequences corresponding to four regions: a region spanning the 5′ end (first 1,000 nts), the frameshifting region (1,000 nts around the frameshifting element), and a PS region sequence corresponding to a proposed SARS-CoV-1 packaging signal (Hsieh et al., 2005). As a control, we also synthesized the highly expressed subgenomic RNA sequence coding for the N-protein (containing the first 75 nucleotides of the 5′ UTR recombined onto the N-protein coding sequence) (Kim et al., 2020) (Figure 1C, referred to as Nucleocapsid RNA).
N-protein LLPS varied as a function of the specific RNA co-component (Figure 1E). The 5′ end and the nucleocapsid RNAs promoted LLPS. In contrast, at the same concentration, the frameshifting region and the PS region RNAs reduced LLPS relative to N-protein alone (Figure 1E). The 5′ end and frameshifting region, which have the same length, displayed consistent near-opposing behaviors across a range of RNA and protein concentrations. The 5′ end generally drives N-protein condensation, whereas the frameshifting region solubilized condensates and promoted LLPS only within a narrow protein and RNA concentration range (Figures 1F, 1G, and S1D). The condensing promoting activity of the 5′ end is sequence specific, as anti-sense RNAs for the 5′ end and frameshifting region behaved similarly to the sense RNA for frameshifting region (Figure S1E). In sum, RNA-mediated LLPS behavior for N-protein shows strong sequence specificity for the 5′ end and sequence at the 3′ end encoding nucleocapsid RNA. Importantly, similar results for viral RNA sequence were observed by the Morgan lab (Carlson et al., 2020).
N-protein binds gRNA in the cytosol in the presence of non-viral RNAs. We therefore assessed how non-viral, lung RNA influences LLPS. Total lung RNA did not alter N-protein-only LLPS; in contrast, when combined with 5′ end RNA, total lung RNA synergized with the 5′ end increasing the condensate size, number, and viral RNA enrichment (Figures S1F–S1H). gRNA is longer than many host RNAs and all subgenomic RNAs and we reasoned that length contributes to an electrostatically driven component of N-protein LLPS given the protein pI (10.07). Addition of 0.3 kb or 2.4 kb of non-viral sequence to the 1 kb 5′ end or frameshifting region RNAs resulted in progressive increase in condensate size/number, with the 5′ end driving enhanced condensates relative to the frameshifting region at all lengths tested (Figure 1H). In sum, N-protein undergoes LLPS under physiological conditions, including in the presence of abundant non-specific RNA, and LLPS is enhanced by viral RNA. Both specific viral RNA sequences (located at the 5′ end) and increased RNA length promotes N-protein to LLPS which raises the possibility that specific packaging of full-length genomic RNA (>30 kb) could occur via LLPS.
Role of Temperature and Material Properties in Dictating N-Protein Condensation with Viral RNA
SARS-CoV-2 replication is most efficient at 33°C (V’kovski et al., 2020) and we therefore assessed the temperature dependence of LLPS. N-protein alone demixed into droplets in a temperature-dependent manner, highly pronounced at fever temperature (40°C) and above (45°C) (Figures 2 A–2D and S2A). Addition of the 5′ end RNA lowered the most efficient condensation temperatures to 37°C and 33°C (which correspond to the exterior lung and upper airway temperatures, respectively) (McFadden et al., 1985). 5′ end RNA droplets included positive changes in their size and abundance in response to temperature, suggesting that temperature may change nucleation, fusion, and/or ripening (Figures 2A and 2B). The decrease in the critical temperature for LLPS was independent of RNA sequence and was also seen in condensates made of N-protein with nucleocapsid RNA (Figure S2A). Further, protein concentration in solution was anti-correlated with surface area occupied by droplets (Figure S2A).
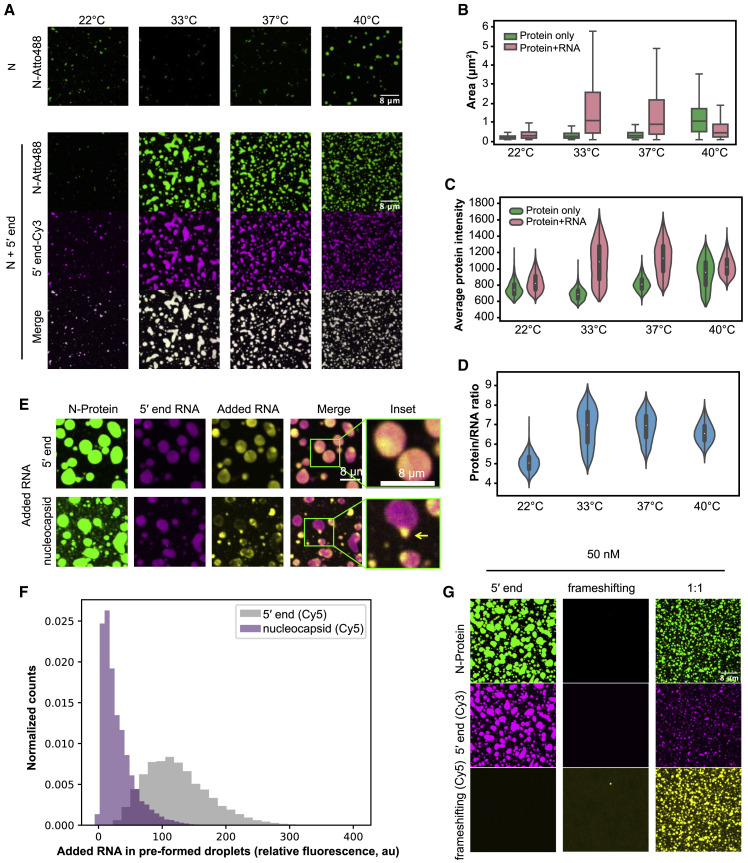
Figure 2.
N-Protein/Viral RNA Condensates Have Temperature- and RNA Sequence-Dependent Material Properties
(A) N-protein (green) alone phase separates in a temperature-dependent manner (4 μM) (upper panel). Temperature dependence is shifted when viral 5′ end RNA (25 nM) (magenta) is present (lower panel).
(B) Quantification of droplet area from (A).
(C) Quantification of average protein signal from (A) based on fluorescence intensity.
(D) Quantification of protein/RNA ratio based on fluorescence intensity from (A).
(E) Sub-genomic nucleocapsid RNA is excluded from preformed 5′ end droplets. 5′ end (yellow, upper panel) is recruited into preformed 5′ end/N-protein droplets (pink and green) but nucleocapsid RNA (yellow, lower panel) is not efficiently recruited and forms separate condensates.
(F) Quantification of (E) showing intensity of second RNA added to preformed droplets. Nucleocapsid RNA (purple) has lower distribution of signal than 5′ end RNA (gray) in regions with high preformed 5′ end RNA signal.
(G) Mixing 5′ end and frameshifting region RNAs makes N-protein condensates with intermediate properties. Left: 5′ end (magenta) and N-protein (green) produced condensates. Middle: frameshifting region (yellow) and N-protein did not produce condensates. Right: Combination of 5′ end and frameshifting region produced smaller condensates than 5′ end alone. Scale bar, 8 μm unless otherwise noted. Violin plots are scaled to have equal widths. Outliers not shown.
In infected cells, subgenomic viral RNAs, like nucleocapsid RNA, are highly abundant species (Kim et al., 2020). We hypothesized that material property differences contribute to specific viral processes such as selective packaging of gRNA and examined N-protein condensates made with RNAs that yielded different material properties. We performed FRAP to examine droplets comprised of 5′ end versus nucleocapsid RNA and observed that N-protein signal recovered faster in 5′ end RNA droplets (t = 1/2, 14 s) than in nucleocapsid RNA droplets (t = 1/2, 28 s) (Figure S2B). The 5′ end RNA promoted larger, more liquid-like condensates; in contrast, the nucleocapsid RNA and a non-viral (luciferase) RNA induced smaller, solid-like, flocculated condensates (Figure S2C). To assess relevance of these material differences to selectivity, we added nucleocapsid RNA to preformed 5′ end droplets. 5′ end RNA readily mixed into preformed N-protein-5′ end condensates; in contrast, subgenomic nucleocapsid RNA was excluded from the preformed 5′ end droplets (nucleocapsid was excluded 10× more than 5′ end) and nucleated separate droplets (Figures 2E and 2F). Thus, material properties of N-protein condensates have clear RNA sequence specificity that excludes other sequences.
Different viral RNAs thus can promote or limit LLPS and yield different material properties (Figures S2A–S2C). We hypothesized some RNA segments might function to maintain liquidity and oppose problematic gelation. Given that the frameshifting region promoted dissolution at most concentrations, we examined whether this RNA could influence the condensation process and solubilize droplets made of other RNAs. Differences in droplet size and abundance reflect changes to the nucleation, coarsening, and/or fusion capacity of condensates while flocculation is evidence of slow relaxation times for droplets that come in contact with one another due to the interplay of viscosity and surface tension (Berry et al., 2018). We mixed frameshifting region RNA with either 5′ end or nucleocapsid RNA. Mixtures containing the 5′ end and frameshifting region produced droplets of intermediate properties, including smaller size and more numerous assemblies. Similarly, frameshifting region RNA made nucleocapsid RNA condensates less flocculated and smaller (Figures 2G, S2D, and S2E). These data suggest that distinct gRNA regions encode different material properties and in combination may yield optimal material properties.
RNA Sequence and Structure Attributes Encode Material Properties
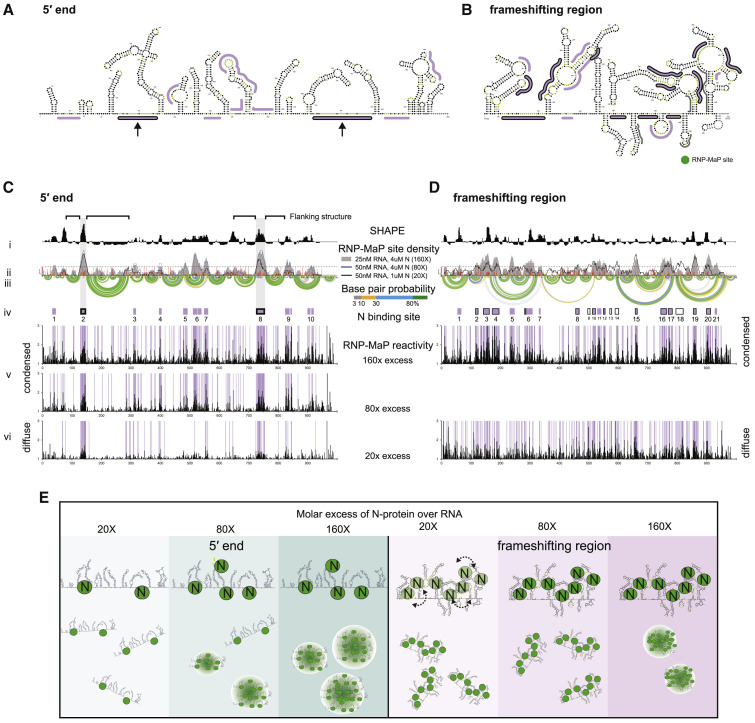
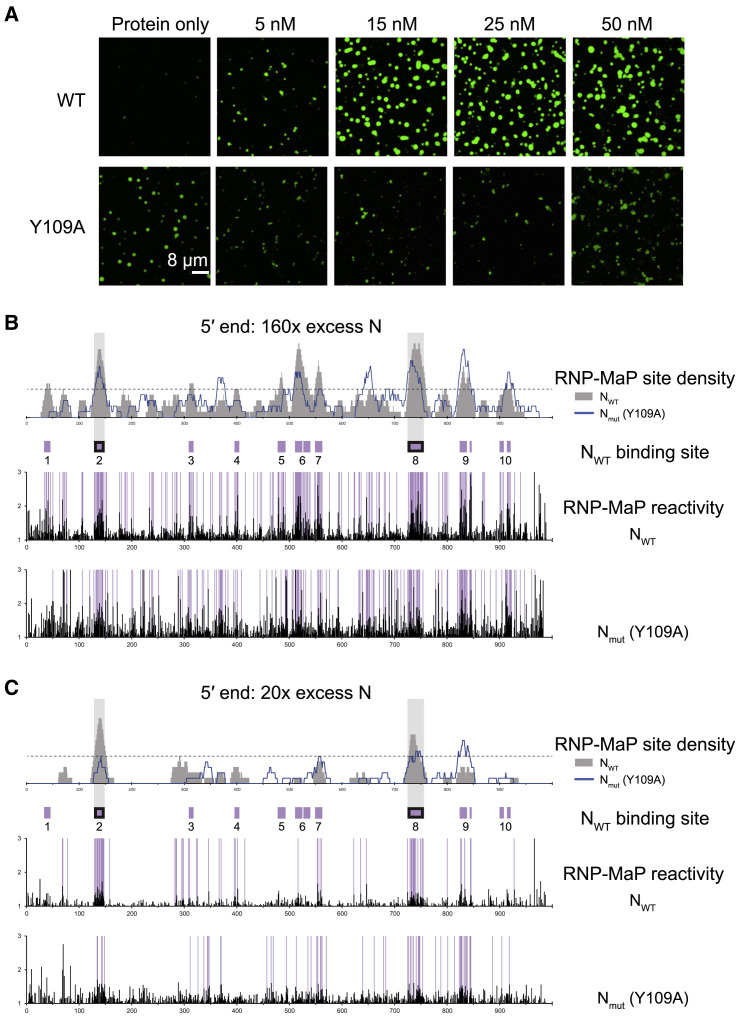
We next examined how the underlying structures of SARS-CoV-2 RNA elements encode distinct LLPS behavior and material properties. We first experimentally assessed and modeled 5′ end and frameshifting region structures using SHAPE-MaP (Figures 3 A–3D, S3A, and S3B; Siegfried et al., 2014). Both RNAs are highly structured. However, the frameshifting region forms a greater number of more complex, multi-helix junction structures and has a higher A/U content (62% versus 52% for 5′ end) (Figures 3A, 3B, S4B, and S4C). We next measured N-protein interactions with viral RNAs using RNP-MaP which selectively crosslinks lysine residues to proximal RNA nucleotides, largely independent of nucleotide identity and local RNA structure (Weidmann et al., 2020). We mapped N-protein interactions at protein:RNA ratios that promote either diffuse or condensed droplets for both the 5′ end and frameshifting region (Figure S4A). For the 5′ end in the diffuse state (20× excess protein), there are two prominent N-protein binding sites, and each occurs in a long A/U-rich unstructured region flanked by strong stem-loop structures (Figures 3A and 3C). In the droplet state (80×/160× excess protein), the two principal sites from the diffuse state remain fully occupied and additional N-protein interaction sites appeared (the valency increased). In contrast, the frameshifting region showed generalized binding across the RNA by N-protein at all ratios (Figures 3B and 3D). Binding was observed in both single-stranded regions and also in A/U-rich structured regions (Figures 3B and 3D). In sum, N-protein interacts specifically with a few preferred sites in the 5′ end in both diffuse and condensed states and interacts more homogeneously across the frameshifting region (Figures 3E and S4D). These highly distinct protein interaction patterns suggest that the different regions have distinct modes of influence on LLPS: (1) specific, multivalent binding at limited sites in the 5′ end that then increase in number during condensation and (2) generalized binding across the frameshifting region at all protein:RNA ratios that is consistent with solubilization (Figure 3E).
Figure 3.
RNA Sequence and Structure Encode Interactions with N-Protein to Specify Condensation or Dissolution
(A and B) SHAPE-Map secondary structure models for the 5′ end (A) or frameshifting region (B). RNP-MaP N-protein binding sites are marked by lines. Two principal binding sites on the 5′ end RNA, both flanked by strong RNA structures, are emphasized with arrows.
(C and D) 5′ end (C) and frameshifting region (D) display condition-specific RNP-MAP reactivity in condensed (80×, 160×) and diffuse (20×) conditions. x axis is the position in nucleotides, y axis is the reactivity (SHAPE or RNP-Map). (i) Windowed (15 nt windows) median SHAPE reactivity (black). (ii) RNP-MaP site density (sites per 15 nt windows); individual nt SHAPE reactivities in colored histograms. (iii) Arcs indicate base pair probabilities (from SHAPE). (iv) N-protein binding sites (boxes: purple, at 160×; with black border, 20×, purple with black border, in 160× and 20×). (v–vi) Raw RNP-MaP reactivity (black) in all conditions. Purple shading highlights RNP-MaP sites.
(E) Model for LLPS. Left panel: 5′ end LLPS coincides with an increase in valency with specific N-protein binding sites. Right panel: frameshifting region RNA has many binding sites (dashed arrows: ensembles of binding sites at lower N concentrations) that enrich N-protein and prevent condensate formation, unless excess N-protein is present to drive LLPS via protein-protein interaction.
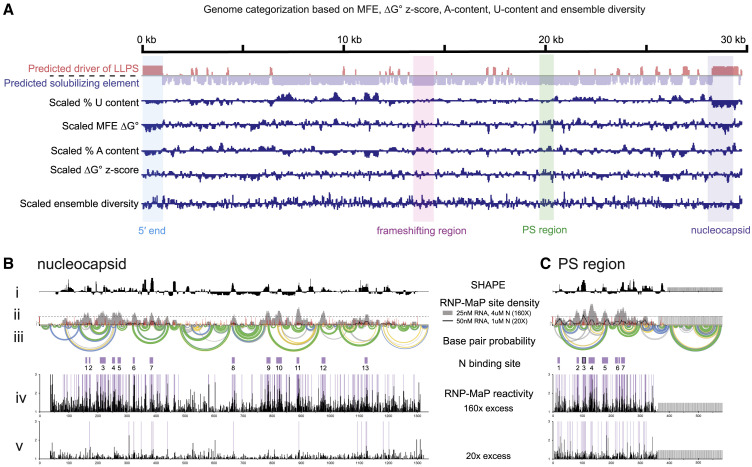
We hypothesized that the gRNA may be a mixture of sequences that promote LLPS (like the 5′ end) and that promote fluidity (frameshifting region). We therefore computationally evaluated sequence and structural properties of the 5′ end and frameshifting region and compared these to the rest of the gRNA. Compared to the 5′ end, most of the RNA genome has higher minimum free energies (MFEs) for predicted structures, a lower ΔG z-score, higher A/U-content, and higher ensemble diversity (more dynamic structures) (Andrews et al., 2020). All of these metrics imply that most of the genome is similar to the frameshifting region (Figures 4 A and S4A–S4C). Interestingly, the two major LLPS-promoting sequences, the 5′ end and nucleocapsid-encoding region at the 3′ end of the gRNA (Figure 1E), are predicted to share multiple features; particularly depletion in U and lower predicted MFE. Nucleocapsid-encoding RNA is predicted to contain a number of highly structured regions although nucleocapsid-encoding RNA is not predicted to be as strongly structured as the 5′ end (scaled ΔG MFE) (Figure 4A). Given that the internal gRNA is more similar to the frameshifting region, internal gRNA sequences may generally act as solubilizing elements (Figures 4A and S5A–S5C). Broadly, different regions of the genome likely make distinct contributions to LLPS of N-protein.
Figure 4.
LLPS Promoting Sequences Are Enriched at the 5′ and 3′ Ends of SARS-CoV-2 Genome
(A) Computational prediction of the similarity of viral genomic sequences (MFE, ΔG z-score, ensemble diversity, A/U content) to 5′ end or frameshifting region. Mean for each feature is computed over all 120 base pair windows with center in the region of interest.
(B and C) Nucleocapsid RNA (predicted 5′ end-like sequence) RNP-MaP reactivities display similar patterns of binding to 5′ end between 20× and 160× conditions. PS region RNA (predicted frameshifting region-like sequence) RNP-map reactivities display similar patterns of binding to frameshifting region between 20× and 160× conditions. (i) Windowed (15 nt windows) median SHAPE reactivity (black). (ii) RNP-MaP site density (sites per 15 nt windows). (iii) Arcs indicate base pair probabilities (from SHAPE). (iv) N-protein binding sites (boxes: purple, at 160×; with black border, 20×, purple with black border, in 160× and 20×). (v–vi) Raw RNP-MaP reactivity (black) in all conditions. Purple shading highlights RNP-MaP sites.
We tested these predictions with RNP-MaP experiments for two additional genome regions: (1) the nucleocapsid RNA which is predicted to share sequence and structural features with the 5′ end and promote LLPS (Figure 1E), and (2) the PS region which shares sequence and structural features with the frameshifting region and similarly limits LLPS. Indeed, these two genome regions showed distinct N-protein binding patterns consistent with their distinct LLPS behaviors (Figures 4B and 4C). Specifically, the nucleocapsid RNA in the diffuse state (20× protein concentration) shows similarities in its N-protein interaction pattern to that of the 5′ end. The nucleocapsid RNA sequence does not support generalized N-protein interaction in the diffuse condition and instead there are few N-protein interactions. These sites of interaction are not as densely occupied as the principle sites in the 5′ end RNA. Thus, the 5′ and 3′ end RNA share notable overall patterns. In contrast, the PS region is homogeneously coated with N-protein in both diffuse and condensed states, similar to the frameshifting region. When combined, these data support a model in which the 5′ and 3′ genome ends interact with N-protein in a localized manner, and specifically drive LLPS, while interior gRNA regions are more uniformly coated in N-protein and have a solubilizing property.
To manipulate N-protein interactions with the RNA, we mutated a conserved residue in the predicted RNA-binding domain of N-protein, Y109, to alanine (Figure 1A). This mutation diminishes N-protein binding to RNA by ∼2,000-fold (Kang et al., 2020) and N-protein with the equivalent amino acid mutation failed to support viral replication in MHV, a related betacoronavirus (Grossoehme et al., 2009). Y109A mutation eliminates 5′ end RNA-driven condensation (Figure 5 A) and interactions between N-protein Y109A and the 5′ end are diminished relative to the WT sequence, in both 20× (40% average binding decrease in N binding intensity) and 160× (50% N binding decrease) conditions. The Y109A mutant also binds at several new sites and overall, binding by the mutant is more diffuse and less punctate than for the wild-type protein (Figures 5B and 5C). N-protein Y109A has a greater propensity to demix in the absence of any added RNA and the frameshifting region RNA retains an ability to dissolve these protein-only condensates (Figure S6). These data indicate that the RNA-binding domain plays a major role in LLPS but that N-protein motifs outside of Y109 contribute to solubilizing interactions.
Figure 5.
Mutation of N-Terminal RNA Binding Domain Blocks RNA-Dependent Phase Separation and Alters RNP-Map Reactivity
(A) 5′ end RNA does not drive LLPS of Y109A mutant N-protein (bottom panel) compared to wild-type N-protein (top panel) at all tested RNA concentrations. Scale bar, 8 μm.
(B and C) At 160× protein (B) and 20× protein (C), Y109A mutant N-protein has altered RNP-map reactivity compared to wild type. 1st panel: RNP-MaP site density (sites per 15 nt windows). 2nd panel: N-protein binding sites (boxes: purple, at 160×; with black border, 20×, purple with black border, in 160× and 20×). 3rd and 4th panels: Raw RNP-MaP reactivity (black). Purple shading highlights RNP-MaP sites.
N-Protein Phase Separates in Mammalian Cells and Can Be Disrupted by Small Molecules in and out of Cells
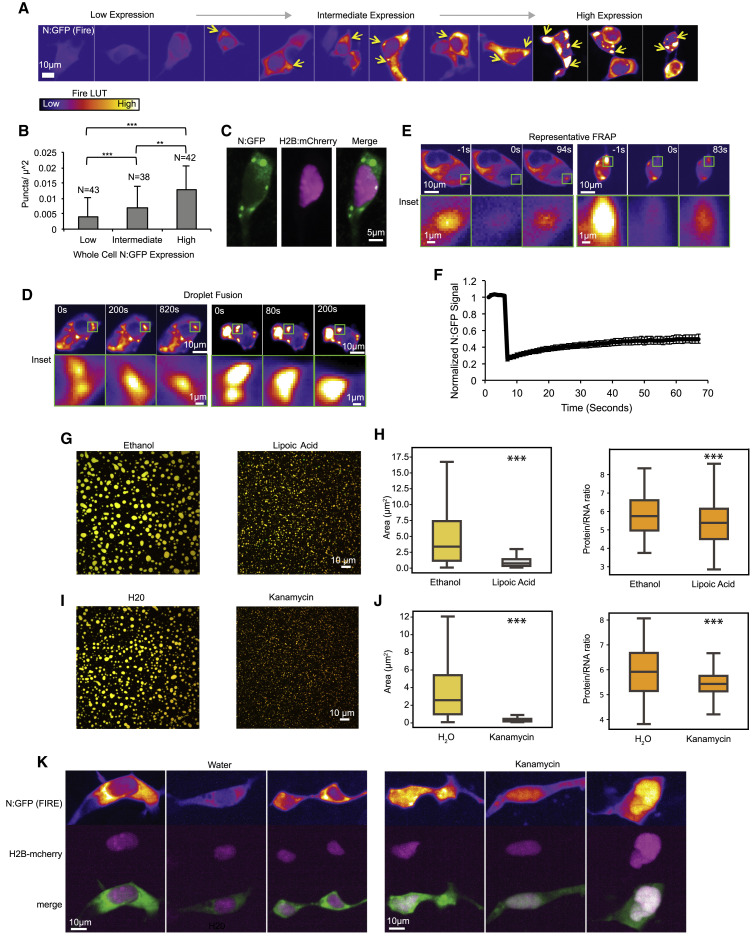
To assess the ability of N-protein to condense in cells, we co-transfected HEK293 cells with N-protein fused to GFPspark and with H2B:mCherry (to mark nuclei in single cells). Cells with higher levels of transfection were more likely to form spherical droplets in the cytoplasm (Figures 6A and 6B), suggesting N-protein condensation is concentration dependent. N-protein signal was generally excluded from the nucleus (Figure 6C). N-protein droplets readily underwent fusion (Figure 6D) and recovered quickly (t = 1/2, 14 s) following FRAP (Figures 6E and 6F) indicating dynamic recruitment of N-protein. These results suggest that N-protein forms cytoplasmic, liquid-like condensates in cells.
Figure 6.
N-Protein Phase Separates in Mammalian Cells and Can Be Disrupted by Small Molecules
(A) N-protein: GFP forms concentration-dependent condensates in HEK293 cells. The fire LUT represents low signal intensity in purple and high signal intensity in yellow. Yellow arrows indicate presence of condensates. Scale bar is 10 μm.
(B) Condensates per μ2 increased significantly with N:GFP expression level. ∗∗p < 0.01 ∗∗∗p < 0.001.
(C) N:GFP shown in green (both diffuse and punctate) was excluded from nuclei (marked with H2B:mCherry shown in magenta) of HEK293 cells. Scale bar is 5 μm.
(D) N:GFP (Fire LUTs) condensates fused in the cytoplasm of HEK293 cells. Top panel: representative cells with 10 μm scale bar. Bottom panel: enlarged of fusion event. Scale bar, 1 μm.
(E) N:GFP condensates recovered partially after FRAP. Top panel shows representative condensate FRAP. Scale bar is 10 μm. Bottom panel: enlargement of N:GFP condensate. Scale bar, 1 μm.
(F) Condensate N-protein exchanges with cytosolic N-protein. Condensates recovered to 24% within 1 min. Error bars show standard error from n = 18 condensates.
(G) 2.38 μg/mL lipoic acid partially prevents N-protein/frameshifting region RNA LLPS relative to ethanol vehicle. Images show merge of protein (green) and RNA (red) signals.
(H) For lipoic acid, size and protein/RNA ratio is reduced relative to vehicle. Left, quantification of condensate area depicted in (G) and right quantification of protein/RNA ratio.
(I) 0.5 mg/mL kanamycin partially prevents N-protein/frameshifting region RNA LLPS relative to water vehicle. Images show merge of protein (green) and RNA (red) signals.
(J) For kanamycin, size and protein/RNA ratio is reduced relative to vehicle. Left, quantification of condensate area depicted in (I) and right quantification of protein to RNA ratio.
(K) 5 mg/mL kanamycin causes relocalization of N:GFP (Fire LUTs) to the cell nucleus (magenta H2B:mCherry signal) in 37% of treated cells (n = 105, 0% in H2O, n = 100). Scale bar, 10 μm unless otherwise noted.
We reasoned that screening for small molecules that increase or decrease N-protein LLPS or altered RNA recruitment could modify N-protein LLPS. We examined 1,6-hexanediol (Kroschwald et al., 2017), lipoic acid (Wheeler et al., 2020), and kanamycin (Blount et al., 2005), each of which potentially alter LLPS by distinctive mechanisms. As a simple positive control which would be useful for future drug screening assays, we examined 1,6-hexanediol which disrupts LLPS (Kroschwald et al., 2017) and, indeed, prevented condensate formation (Figures S7A and S7B). Lipoic acid dissolves cellular stress granules (Wheeler et al., 2020), which, in the absence of N-protein phosphorylation, recruit SARS-CoV-1 N-protein during cellular stress (Peng et al., 2008). Lipoic acid treatment reduced condensate size (Figures 6G and 6H). The aminoglycoside kanamycin binds promiscuously to nucleic acids via electrostatic interactions and was implicated as antiviral in HIV-1 by preventing RNA-protein interactions (Blount et al., 2005). Addition of kanamycin to droplets decreased the size of condensates, decreased the protein/RNA ratio in the reconstitution assay (Figures 6I and 6J), and caused N-protein to relocalize to the nucleus in 37% of treated cells (Figure 6K).
Discussion
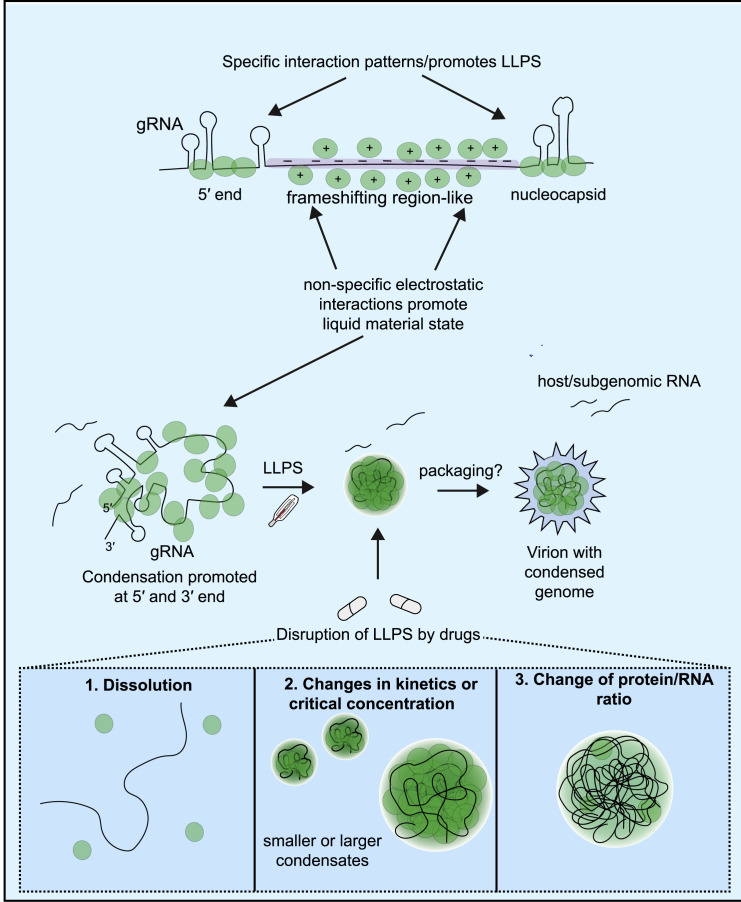
In this work, we show that the SARS-CoV-2 nucleocapsid protein (N-protein) phase separates in an RNA sequence- and structure-dependent manner. We present a potential mechanism for SARS-CoV-2 gRNA packaging through LLPS (Figures 3E and 7 ), where N-protein condensate properties are conferred through specific gRNA sequences, structures, and length. We find that distinct regions of the viral RNA genome either promote LLPS (5′ end region and nucleocapsid-encoding region [nucleocapsid RNA] located at the 3′ end) or act as solubilizing elements (frameshifting region, PS region). Multivalent polymer interactions are a driving force of LLPS and we propose that a punctate N-protein binding pattern enables the 5′ end and the 3′ end regions to promote LLPS. The frameshifting region and PS region, conversely, are more uniformly bound by N-protein in both diffuse and droplet states. These sequences, that are coated by N-protein, have features predicted to be shared across much of the genome (Figure 4A), suggesting that many regions may contribute non-specific electrostatic interactions, likely promoting fluidity and solubilization to limit entanglement of the large gRNA molecule. In this model, the full-length gRNA consists of a mixture of LLPS-promoting and aggregation-dissolving elements to promote regulated, selective LLPS, thereby excluding packaging of host mRNAs (Figure 7).
Figure 7.
LLPS Drives Temperature- and RNA Sequence-Dependent Packaging of the Viral Genome
Packaging of gRNA may be a temperature-dependent LLPS process driven by single-stranded regions flanked by structured regions (5′ end-like) that are stable N-protein binding sites. The majority of the genome resembles the solubilizing frameshifting region, while the region coding for N-protein is similar to the 5′ end. The balance between LLPS-promoting and solubilizing elements may facilitate gRNA packaging. Initial step of packaging (LLPS of N-protein with gRNA) may be targeted by compounds that either (1) induce condensate dissolution (1,6-hexanediol), (2) adjust condensate size through changes in kinetics or critical concentration (example kanamycin, lipoic acid), or (3) adjust protein/RNA ratio (example kanamycin).
LLPS may concentrate components to ensure efficient packaging and may also protect the sensitive gRNA in virions (van Doremalen et al., 2020). The membrane protein (M-protein) is also a known interactor of both gRNA and N-protein and thus N-protein:genome condensates may also specifically interact with the M-protein to facilitate packaging of a single genome per virion (Narayanan et al., 2000). Future experiments are needed to test the proposed model that LLPS governs packaging and to investigate the complex interplay of various interaction partners in N-protein LLPS.
While our model is focused on proposing a role for N-protein LLPS in packaging, N-protein LLPS could also be important for SARS-CoV-2 viral replication. LLPS was previously implicated in replication of other viruses (Alenquer et al., 2019; Heinrich et al., 2018; Nikolic et al., 2017), and RNA sequence, structure, and length could encode both specificity and material N-protein condensate properties that govern functions in viral replication independent of or in addition to membrane encapsulation as might occur in “replication factories.”
The temperature-dependent LLPS of N-protein provides a potential explanation for how SARS-CoV-2 and other coronaviruses spread through the likely reservoir species, Chinese horseshoe bats (Calisher et al., 2006). Bat body temperature lowers during hibernation and goes up during flight. In order to propagate, viral proteins must adapt to bat temperature extremes. It is possible that defects in N-protein LLPS slows viral replication during hibernation. Indeed, it has been observed that coronavirus infection occurs prior to hibernation and persists with mild symptoms during hibernation (Subudhi et al., 2017).
LLPS of N-protein by itself is increased with temperature, which is defined as lower critical solution temperature LLPS (LCST). There are only a handful of biological examples of LCST LLPS (Dao et al., 2018; Iserman et al., 2020; Jiang et al., 2015). LCST LLPS is mainly driven by the presence of aromatic and hydrophobic amino acids (Dao et al., 2018; Jiang et al., 2015; Li et al., 2014). In contrast, LLPS in response to lowered temperature, referred to as upper critical solution temperature (UCST), is mainly driven by polar residues such as arginine (Quiroz and Chilkoti, 2015). Interestingly, the N-protein is rich in certain hydrophobic amino acids (particularly alanine and glycine) and also certain polar amino acids (particularly glutamic acid and arginine), relative to all vertebrate proteins (Table S2), suggesting that a balance of these amino acids, as well as its interactions with RNA, dictates optimal N-protein LLPS. RNA sequence is not likely the major source of LCST, as 5′ end and nucleocapsid RNAs behave similarly, and their structures do not resemble RNA thermometers.
New antivirals are needed for existing, emerging, and drug-resistant viral diseases. We suggest that LLPS could represent a new easily screenable target for antivirals. The two compounds we tested—lipoic acid and kanamycin—were chosen as proof of concept and could serve as positive controls for a screen. Specific RNA sequences and structures which regulate N-protein LLPS may also be targeted directly in the development of antiviral therapies. These straightforward in vitro and in vivo assays comprise a powerful starting point for evaluating compounds to reveal new classes of antiviral strategies that target phase-separation.
Limitations
This study addresses mechanisms of LLPS of components of the SARS-CoV-2 virus. However, because the work involved reconstitution experiments from purified components and expression of viral proteins in mammalian cells rather than in an actual infection, it is still unclear what step(s) in the viral replication cycle may utilize the mechanisms described.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| SARS-CoV2 (USA WA1/2020) | BEI Resources | NR52281 |
| BL21 | NEB | C2527I |
| Chemicals, Peptides, and Recombinant Proteins | ||
| XBA1 | NEB | R0145S |
| NOTI | NEB | R3189S |
| STUI | NEB | R0187S |
| Fugene HD | Promega | E2311 |
| Cy3-UTP | Sigma | PA53026 |
| Cy5-UTP | Sigma | PA55026 |
| DNaseI | NEB | M0303L |
| Millenium RNA ladder | ThermoFisher Scientific | AM7151 |
| TRIzol LS | ThermoFisher Scientific | 10296028 |
| Lysozyme | Fisher Scientific | BP535-10 |
| Roche EDTA-free protease inhibitor cocktail | Millipore Sigma | 11873580001 |
| HisPur™ Cobalt Resin | ThermoFisher Scientific | 89965 |
| RNaseA | QIAGEN | 19101 |
| Atto 488 NHS ester | Millipore Sigma | 41698 |
| 100X/1.49 NA oil | Cargille Lab | 16241 |
| (R)-(+)-α-Lipoic acid | Sigma-Aldrich | 07039 |
| 1,6-Hexanediol | Sigma Aldrich | 240117 |
| Kanamycin | Millipore Sigma | 60615-25G |
| 5-Nitroisatoic Anhydride (5NIA) | AstaTech | 69445 |
| Recombinant SARS-CoV-2 Nucleocapsid Protein with C-terminal His-tag | Ray Biotech | QHD43423 |
| Recombinant 6xHis-TEV-Nucleocapsid Y109A protein | this paper | N/A |
| Recombinant 6xHis-TEV-Nucleocapsid protein | this paper | N/A |
| Recombinant Nucleocapsid protein | this paper | N/A |
| Critical Commercial Assays | ||
| HiScribe™ T7 High Yield RNA Synthesis Kit | NEB | E2040S |
| 1.8 × Mag-Bind TotalPure NGS SPRI beads | Omega Bio-tek | M1378-01 |
| SuperScript II Reverse Transcriptase | Invitrogen | 18064-022 |
| RNeasy MinElute columns | QIAGEN | 74204 |
| Illustra G-50 microspin columns | GE Healthcare | GE27-5330-01 |
| NEBNext second-strand synthesis reaction | NEB | B6117S |
| Nextera XT | Illumina | FC-131-1024 |
| Amicon Ultra-4, PLGC Ultracel-PL Membrane, 10 kDa | Merck | UFC801024 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE162569 |
| Experimental Models: Cell Lines | ||
| Vero E6 | ATCC | CRL-1586 |
| HEK293 | ATCC | CRL-1573 |
| HEK293T | ATCC | CRL-11268 |
| Oligonucleotides | ||
| Sal-N-protein-fw: acgcatcgtcgacATGTCTGATAATGG ACC-CCAAAATCAG |
IDT | N/A |
| Not1-N-protein-rev: tatctatgcggccgc TTAGGCCTGAGTTGAGTCAGC |
IDT | N/A |
| Recombinant DNA | ||
| pJet | ThermoFisher Scientific | K1231 |
| pET30b-6xHis-TEV-Nucleocapsid | This paper | N/A |
| pET30b-6xHis-TEV-Nucleocapsid Y109A | This paper | N/A |
| Nucleocapsid GFP Spark | Sino biological | VG40588-ACGLN |
| Plasmid pUC57-2019-ncov | This paper | N/A |
| pJet- 5′ end | This paper | N/A |
| pJet-frameshifting region | This paper | N/A |
| pJet- PS region | This paper | N/A |
| pJet- Anti-Sense-5′ end | This paper | N/A |
| pJet- Anti-Sense-frameshifting region | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| ShapeMapper2 | Busan and Weeks, 2018 | https://github.com/Weeks-UNC/shapemapper2 |
| RNAStructure | Reuter and Mathews, 2010 | https://rna.urmc.rochester.edu/RNAstructure.html |
| Superfold | Smola et al., 2015 | https://github.com/Weeks-UNC/Superfold |
| VARNA | Darty et al., 2009 | http://varna.lri.fr/index.php?lang=en&page=home&css=varna |
| ImageTank | O’Shaughnessy et al., 2019 | https://www.visualdatatools.com/ImageTank/ |
| Matplotlib | Hunter, 2007 | https://matplotlib.org/ |
| SciPy | Virtanen et al., 2020 | https://www.scipy.org/ |
| FRESCo | Sealfon et al., 2015 | https://www.broadinstitute.org/fresco/running-fresco |
| Muscle 3.8.31 | Edgar, 2004 | https://www.drive5.com/muscle/ |
| RAxML version 8.2.12 with the GTRGAMMA model of nucleotide evolution | Stamatakis, 2014 | https://cme.h-its.org/exelixis/web/software/raxml/ |
| RNAz 2.1 | Gruber et al., 2010 | https://www.tbi.univie.ac.at/software/RNAz/ |
| Scikit-learn | Pedregosa et al., 2011 | https://scikit-learn.org/ |
| ggplot2 3.1.1 | Wickham, 2016 | https://ggplot2.tidyverse.org |
| Genome Viewer | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv/ |
| Other | ||
| SARS-CoV-2 sequence | NC_045512 | https://www.ncbi.nlm.nih.gov/nuccore/1798174254 |
| Severe acute respiratory syndrome coronavirus 2 isolate 2019-nCoV WHU01 | MN988668.1 | https://www.ncbi.nlm.nih.gov/nuccore/MN988668.1 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Amy Gladfelter (amyglad@unc.edu).
Materials Availability
Materials such as plasmid constructs will be available without further restrictions upon request to the Lead Contact (amyglad@unc.edu).
Data and Code Availability
All code is published. Raw and processed sequencing datasets analyzed in this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession number GEO: GSE162569).
Experimental Model and Subject Details
HEK293, HEK293T, and Vero-E6 cells were obtained from ATCC for this study. All cell lines were maintained in DMEM (Corning 10-013-CV) supplemented with 10% Fetal Bovine Serum (Seradigm V500-050) and grown at 37°C. No antibiotics were used.
Method Details
In vitro transcription was carried out according to our established protocols (Langdon et al., 2018). Orf1ab templates were synthesized (IDT) and cloned into pJet (ThermoFisher Scientific K1231) using blunt end cloning. Directionality and sequence were confirmed using Sanger sequencing (GENEWIZ). Plasmid were linearized with XBAI restriction enzyme (NEB R0145S) and gel purified (QIAGEN 28706). Nucleocapsid RNA was produced from pu57 Nucleocapsid, a kind gift from the Sheahan lab, linearized with NOTI (NEB R3189S) and STUI (NEB R0187S). 100 ng of gel purified DNA was used as a template for in vitro transcription (NEB E2040S) carried out according to the manufacturer’s instructions with the addition of 0.1μl of Cy3 (Sigma PA53026) or Cy5 (Sigma PA55026) labeled UTP to each reaction. Following incubation at 37°C for 18 h, in vitro transcription reactions were treated with DNaseI (NEB M0303L) according to the manufacturer’s instructions. Following DNase treatment, reactions were purified with 2.5M LiCL precipitation. Purified RNA amounts were quantified using nanodrop and verified for purity and size using a denaturing agarose gel and Millenium RNA ladder (ThermoFisher Scientific AM7151).
Purification of genomic SARS-CoV-2 RNA (gRNA): Vero E6 cells were cultured to ∼90% confluence in T175 flasks. Immediately prior to infection the culture medium was aspirated and cells were washed with PBS. Flasks were infected at a multiplicity of infection of 3 with SARS-CoV-2 at 37°C for 1 h. After 1 h cells were supplemented with pre-warmed DMEM (GIBCO) with 5% FetalCloneII (HyClone) and 1x Anti-Anti (GIBCO). Cells were then incubated for an additional 24 h at 37°C. After the infection was complete, the cell supernatant was aspirated and concentrated using Millipore Centrifugal Amicon filters to approximately 4 mL total volume. The supernatant was then lysed in TRIzol LS, and viral RNA was extracted from the trizol using chloroform extraction. No size selection was performed.
Recombinant Protein Expression and Purification: For protein purification, full-length N-protein was tagged with an N-terminal 6-Histidine tag (pET30b-6xHis-TEV-Nucleocapsid,) and expressed in BL21 E. coli (New England Biolabs). All steps of the purification after growth of bacteria were performed at 4°C. Cells were lysed in lysis buffer (1.5M NaCl, 20 mM Phosphate buffer pH 7.5, 20 mM Imidazole, 10mg/mL lysozyme, 1 tablet of Roche EDTA-free protease inhibitor cocktail Millipore Sigma 11873580001) and via sonication. The lysate was then clarified via centrifugation (SS34 rotor, 20,000 rpm 30 min) and the supernatant was incubated and passed over a HisPur Cobalt Resin (ThermoFisher Scientific 89965) in gravity columns. The resin was then washed with 4X 10 CV wash buffer (1.5M NaCl, 20 mM Phosphate buffer pH 7.5, 20 mM Imidazole) and protein was eluted with 4 CV Elution buffer (0.25 M NaCl, 20 mM Phosphate buffer pH 7.5, 200 mM Imidazole). The eluate was then dialyzed into fresh storage buffer (0.25 M NaCl, 20 mM Phosphate buffer) and aliquots of protein were flash frozen and stored at −80°C. Protein was checked for purity by running an SDS-PAGE gel followed by Coomassie staining as well as checking the level of RNA contamination via Nanodrop and through running of a native agarose RNA gel. Most experiments were performed with His-tagged N-protein, however LLPS of untagged and His-tagged protein was compared as quality control and results were similar (data not shown). Please note, that while self-purified protein had very low RNA contamination, commercially acquired bacterial expressed N-protein at similar concentrations had a high contamination of RNA that dramatically enhanced LLPS. This enhancement of LLPS was abrogated through addition of RNaseA (QIAGEN 19101).
Dyeing of N-protein: N-protein was dyed by adding (3:1) Atto 488 NHS ester (Millipore Sigma 41698) to purified protein and incubating mix at 30°C for 30 min. Unbound dye was removed by 2 washes with 100X excess of protein storage buffer followed by centrifugation in Amicon® Ultra-4 Centrifugal Filter Units (SIGMA MilliPORE). LLPS of dyed and undyed protein was compared as quality control and results were similar (data not shown).
Mammalian N-protein: was derived from Ray Biotech (Recombinant SARS-CoV-2 Nucleocapsid Protein with C-terminal His-tag, derived from the transfected human HEK293 cells: QHD43423).
Phase separation assays: For in vitro reconstitution LLPS experiments, 15 μl droplet buffer (20 mM Tris pH 7.5, 150 mM NaCl) was mixed with cy3 or cy5 labeled desired RNA and 5 μl protein in storage buffer was added at desired concentration. Similar results were seen with KCl compared to NaCl and we performed assays in NaCl because it is more compatible with potential downstream drug screening assays. The mix was incubated in 384-well plates (Cellvis P384-1.5H-N) for 16 h at 37°C unless indicated otherwise. Droplets already formed after short incubations of 20 min or less, however, they were initially smaller and matured into larger droplets during the overnight incubation step. Imaging of droplets was done on a spinning disc confocal microscope (Nikon CSU-W1) with VC Plan Apo 100X/1.49 NA oil (Cargille Lab 16241) immersion objective and an sCMOS 85% QE 95B camera (Photometrics). Data shown are representative of three or more independent replicates, across several RNA preparations.
Comparison of droplet images to absorbance A280 reading in difuse phase. 15 μl droplet buffer (20 mM Tris pH 7.5, 150 mM NaCl) was mixed with 25nM of cy3 or cy5 labeled desired RNA and 5 μl protein in storage buffer was added to a final concentration of 4uM. The mix was incubated in 384-well plates (Cellvis P384-1.5H-N) for 16 h at 22, 33, 37 or 45°C. Following imaging. 2 μl of difuse phase solution (taken from the top of the well) was nanodroped and absorbance A 280 was recorded. Droplet areas based on 488 (N-protein) signal were quantified using ImageJ (Threshold intensity 230, size > 0.1). Plotted images indicate the average of 2 images for each of 3 techical replicates. For Pearson correlation calculations R was fitted for 12 points (4 temperatures, 3 technical replicates)
Droplet FRAP: Prior to bleaching, 25nM RNA and 4uM protein were incubated at 37°C for 1 h. Droplets were imaged for seven seconds with one second per frame. Following bleaching with 405 nm laser, recovery was monitored for at least one minute with one second frame intervals. Puncta fluorescence recovery was quantified using ImageJ. Fluorescence was relative to both the initial unbleached signal and an unbleached droplet in the same frame. Quantification represents 8 (5′ end) or 9 (nucleocapsid) droplets from 9 movies with error bars depicting standard error. To calculate T1/2 data were fit to a rising single exponential function. (ft(A,k,x) = A∗(1-exp(-x∗k)).
Sequestration experiments: For sequestration experiments, N-protein/5′ end (Cy3) condensates were preformed and after 1.5 h incubation, 5 nM cy5-labeled RNA of interest was added mixed, and incubated for another 14 h before imaging.
Drug treatments of in vitro phase separation assays: For drug treatment of in vitro phase separation assays, droplet buffer was pre-mixed with drugs or vehicles, before RNA and protein were added to the mix. (R)-(+)-α-Lipoic acid (Sigma-Aldrich, cat. number: 07039) was added at 2.38 μg/mL (11 μM) in the presence of excess DTT to reduce its thiole ring and compared to the vehicle ethanol. 1,6-Hexanediol (Sigma Aldrich, cat. number: 240117) was added to a final concentration of 9%. Kanamycin (Millipore Sigma 60615-25G) was added to a final concentration of 0.5 mg/mL (858 μM). 1,6-Hexanediol and Kanamycin were compared to the vehicle H20. We chose this order of component addition, to most closely mimic possible screening conditions, in which drugs would likely be pre-added to multi-well plates. The mixtures were then incubated for 16 h at 37°C before imaging.
RNP-MaP probing of N-Protein-RNA interactions: N-Protein and RNA mixtures were prepared as described in the “Phase Separation Assay” section above and incubated for 1.5 h at 37°C. N-Protein–5′ -End RNA mixtures were prepared in three conditions: (1) 50nM RNA, 1μM protein (diffuse state, 20x excess protein), (2) 50nM RNA, 4μM protein (droplet state, 80x excess protein), and (3) 25nM RNA, 4μM protein (droplet state, 160x excess protein). N-Protein–frameshifting-region RNA, N-Protein–nucleocapsid RNA, N-Protein–PS region RNA, and N-protein Y109A–5′ end RNA mixtures were prepared in two conditions: (1) 50nM RNA, 1μM protein (diffuse state, 20x excess protein) and (2) 25nM RNA, 4μM protein (160x excess protein, droplet state for all but the N-protein Y109A–5′ end RNA mixture, which was diffuse). RNA-only samples were also prepared as a control. After confirmation of phase separation by imaging (Figure S4A) mixtures were immediately subjected to RNP-MaP treatment as described (Weidmann et al., 2020), with modifications described below. Briefly, 200 μl of mixtures were added to 10.5 μl of 200 mM SDA (in DMSO) in wells of a 6-well plate and incubated in the dark for 10 min at 37°C. RNPs were crosslinked with 3 J/cm2 of 365 nm wavelength UV light. To digest unbound and crosslinked N-proteins, reactions were adjusted to 1.5% SDS, 20 mM EDTA, 200mM NaCl, and 40mM Tris-HCl (pH 8.0) and incubated at 37°C for 10 min, heated to 95°C for 5 min, cooled on ice for 2 min, and warmed to 37°C for 2 min. Proteinase K was then added to 0.5 mg/mL and incubated for 1 h at 37°C, followed by 1 h at 55°C. RNA was purified with 1.8 × Mag-Bind TotalPure NGS SPRI beads (Omega Bio-tek), purified again (RNeasy MinElute columns, QIAGEN), and eluted with 14 μl of nuclease-free water.
SHAPE-MaP RNA structure probing: SHAPE-MaP treatment with 5NIA was performed as described (Busan et al., 2019). Briefly, In vitro transcribed 5′ end, frameshifting region, nucleocapsid-encoding, or PS region RNA (1200 ng in 40μL nuclease-free water) was denatured at 95°C for 2 min followed by snap cooling on ice for 2 min. RNA was folded by adding 20 μL of 3.3 × SHAPE folding buffer [333mM HEPES (pH 8.0), 333mM NaCl, 33mM MgCl2] and incubating at 37°C for 20 min. RNA was added to 0.1 volume of 250 mM 5NIA reagent in DMSO (25 mM final concentration after dilution) and incubated at 37°C for 10 min. No-reagent (in neat DMSO) control experiments were performed in parallel. After modification, all RNA samples were purified using RNeasy MiniElute columns and eluted with 14 μl of nuclease-free water.
MaP reverse transcription: After SHAPE and RNP-MaP RNA modification and purification, MaP cDNA synthesis was performed using a revised protocol as described (Mustoe et al., 2019). Briefly, 7 μL of purified modified RNA was mixed with 200 ng of random 9-mer primers and 20 nmol of dNTPs and incubated at 65°C for 10 min followed by 4°C for 2 min. 9 μL 2.22 × MaP buffer [1 × MaP buffer consists of 6 mM MnCl2, 1 M betaine, 50 mM Tris (pH 8.0), 75 mM KCl, 10 mM DTT] was added and the combined solution was incubated at 23°C for 2 min. 1 μL SuperScript II Reverse Transcriptase (200 units, Invitrogen) was added and the reverse transcription (RT) reaction was performed according to the following temperature program: 25°C for 10 min, 42°C for 90 min, 10 × [50°C for 2 min, 42°C for 2 min], 72°C for 10 min. RT cDNA products were then purified (Illustra G-50 microspin columns, GE Healthcare).
Library preparation and Sequencing: Double-stranded DNA (dsDNA) libraries for sequencing were prepared using the randomer Nextera workflow (Smola et al., 2015). Briefly, purified cDNA was added to an NEBNext second-strand synthesis reaction (NEB) at 16°C for 150 min. dsDNA products were purified and size-selected with SPRI beads at a 0.8 × ratio. Nextera XT (Illumina) was used to construct libraries according to the manufacturer’s protocol, followed by purification and size-selection with SPRI beads at a 0.65 × ratio. Library size distributions and purities were verified (2100 Bioanalyzer, Agilent) and sequenced using 2x300 paired-end sequencing on an Illumina MiSeq instrument (v3 chemistry).
Sequence alignment and mutation parsing: FASTQ files from sequencing runs were directly input into ShapeMapper 2 software (Busan and Weeks, 2018) for read alignment, mutation counting, and SHAPE reactivity profile generation. The –random-primer-len 9 option was used to mask RT primer sites with all other values set to defaults. For RNP-MaP library analysis, the protein:RNA mixture samples are passed as the –modified samples and no-protein control RNA samples as –unmodified samples. Median read depths of all SHAPE-MaP and RNP-MaP samples and controls were greater than 50,000 and nucleotides with a read depth of less than 5000 were excluded from analysis.
Secondary structure modeling: The Superfold analysis software (Smola et al., 2015) was used with SHAPE reactivity data to inform RNA structure modeling by RNAStructure (Reuter and Mathews, 2010). Default parameters were used to generate base-pairing probabilities for all nucleotides (with a max pairing distance of 200 nt) and minimum free energy structure models. The local median SHAPE reactivity were calculated over centered sliding 15-nt windows to identify structured RNA regions with median SHAPE reactivities below the global median. Secondary structure projection images were generated using the (VARNA) visualization applet for RNA (Darty et al., 2009).
RNP-MaP reactivity analysis: A custom RNP-MaP analysis script (Weidmann et al., 2020) was used to calculate RNP-MaP “reactivity” profiles from the Shapemapper 2 “profile.txt” output. RNP-MaP “reactivity” is defined as the relative MaP mutation rate increase of the crosslinked protein-RNA sample as compared to the uncrosslinked (no protein control) sample. Nucleotides whose reactivities exceed reactivity thresholds are defined as “RNP-MaP sites.” RNP-MaP site densities were calculated over centered sliding 15-nt windows to identify RNA regions bound by N-protein. An RNP-MaP site density threshold of 5 sites per 15-nt window was used to identify “N-protein binding sites” with boundaries defined by the RNP-MaP site nucleotides.
Mammalian cells methods
Cell Culture: HEK293, HEK293T, and Vero-E6 cells were originally obtained from ATCC. All cell lines were maintained in DMEM (Corning 10-013-CV) supplemented with 10% Fetal Bovine Serum (Seradigm V500-050). No antibiotics were used.
Plasmid Transfection: 24 h prior to transfection, confluent cells were split 1:5. Two h prior to transfection, 2mL of fresh media was added to 10cm dishes. 25ug of plasmid DNA for each Nucleocapsid GFP Spark (Sino biological VG40588-ACGLN) and H2BmCherry (from Jun Lu lab Yale University) was co-transfected using calcium phosphate. Similar results were obtained for Fugene HD (Promega E2311) transfection and for Nucleocapsid fused to mCherry (data not shown). Following transfection, cells were incubated for 72 h prior to imaging or drug treatment.
Drug Treatment: Cells were incubated with drugs for 24 h prior to imaging. Kanamycin (Millipore Sigma 60615-25G) was added to a final concentration of 5 mg/mL with control cells treated with 10% sterile water by volume.
Microscopy and image analysis
Cell Imaging: Cells were imaged using a 40X air objective on a spinning disk confocal microscope (Nikon Ti-Eclipse, Yokogawa CSU-X1 spinning disk). Images were taken with a Photometrics Prime 95B sCMOS camera. Representative cells are taken from at least 6 biological replicates pooled from at least 3 independent rounds of transfection/drug treatment.
Analysis of cell imaging data: Figures 5A, 5E, 5D, and 5M (top) depict maximum intensity projections with Fire LUTs for the low expression cells the depicted range is 0-848. For the high expression cells it is 101-1966. Average fluorescence intensity and area were obtained by thresholding max projections in ImageJ. Number of puncta per cell was manually counted from max projections.
Cell FRAP: Prior to bleaching, cells were imaged for seven seconds with one second per frame. Following bleaching with 488 nm laser, puncta recovery was monitored for at least one minute with one second frame intervals. Puncta fluorescence recovery was quantified using ImageJ. Fluorescence was normalized by subtracting background fluorescence and relative to both the initial unbleached signal and an unbleached puncta in an unbleached cell in the same frame. Quantification represents 18 puncta from 15 movies with error bars depicting standard error.
Quantification of in vitro condensates: For each image, droplets were segmented based on a threshold of 4Xbackground intensity. Any segmented region with an area less than 0.07 μm2 was removed. The average protein and RNA intensity values within each droplet were calculated, and protein/RNA ratios were determined by dividing these averages on a per-droplet basis. For protein intensity and area, a two-sample Kolmogorov-Smirnov (KS) test was applied to compare protein-only with protein+RNA distributions at each temperature. A two-sample KS test was also used to make pairwise comparisons between each of the protein-only distributions, and between each of the protein+RNA distributions. Similarly, a two-sample KS test was performed to compare protein/RNA ratios. Images were processed in ImageTank (O’Shaughnessy et al., 2019) and plotted with Python using Matplotlib and Seaborn. Statistics were performed in Python with SciPy.
Quantification of colocalization: For co-localization analysis, cy5 intensity (signal for 5′ end or nucleocapsid RNA) was plotted for pixels with > 2X background cy3 intensity (signal for 5′ end RNA). Values were represented by a histogram of cy5 intensity.
Computational sequence analysis
FRESCo: Detection of regions of excess synonymous constraint: To detect regions of excess synonymous constraint, we used the FRESCo framework (Sealfon et al., 2015) which detects mutational differences between strains taking into account overlapping features. We scanned for genic translated regions for excess constraint at 10 1, 5, 10, 20, and 50 codon sliding windows. The regions of synonymous constraint were detected based on a set of 44 Sarbecovirus genomes listed in (Jungreis et al., 2020). Genic regions were extracted, translated, and aligned based on the amino acid sequence using Muscle version 3.8.31 (Edgar, 2004). For each gene, sequences with less than 25% identity to the reference SARS-CoV-2 sequence (NC_045512) were removed. A nucleotide-level codon alignment was constructed based on the amino acid alignment, and gene-specific phylogenetic trees were constructed using RAxML version 8.2.12 with the GTRGAMMA model of nucleotide evolution (Stamatakis, 2014). Regions with excess synonymous constraint at a significance level of 1e-5 in ten codon windows were extracted for further analysis. Thirty base pairs of flanking sequence were added on either side of each synonymous constraint element and RNAz 2.1 (Gruber et al., 2010) was used to scan for conserved, stable RNA structures. The rnazWindow.pl script was used to filter alignments and divide into 120 base pair windows. Secondary structure detection was performed for both strands with SVM RNA-class probability set to 0.1.
Computational analysis of RNA composition of 5′ end and frameshifting region versus genome
Percentage AT, mean free energy ΔG, ΔG Z-score, and ensemble diversity was determined in 120bp sliding windows, where mean free energy ΔG, ΔG Z-score, and ensemble diversity are taken from (Andrews et al., 2020). All windows with center ≤1000 were used for the 5′ end region, and all windows with center > = 13401 and < 14401 were used for the frameshifting region.
Genome Analysis: A support vector machine with a linear kernel was trained using the scikit-learn Python library to distinguish between 120-base pair sliding windows in the 5′ end and the frameshifting region of SARS-CoV-2 (NC_045512.2) based on the following features: percent A content, percent U content, mean free energy ΔG, ΔG Z-score, and ensemble diversity, where mean free energy ΔG, ΔG Z-score, and ensemble diversity values were taken from (Andrews et al., 2020). Features were scaled before classification to have mean of 0 and standard deviation of 1. All windows with center ≤1000 were used for the 5′ end, and all windows with center > = 13401 and < 14401 were used for the frameshifting region. The classifier was then applied to all 120 bp windows outside the 5′ end and frameshifting region. The probability estimate for each sliding window of assignment to the 5′ end was plotted, after linearly re-scaling the probabilities for visualization purposes to have maximum of 1 and minimum of −1. Windows in the 5′ end are plotted with their class labels of 1, and windows in the frameshifting region are plotted with their class labels of −1.
Plasmids and sequences used: To create pET30b-6xHis-TEV-Nucleocapsid, the N-protein coding sequence (28273-29533 nt) preceded by the ORFN subgenomic 5′-UTR (1-75 nt) (Plasmid pUC57-2019-ncov, kind gift from Tim Sheahan and Ralph Baric) was cloned into AGB1329 (pET30b-6xHis-TEV) using SALI (NEB R3138S) and NOTI (NEB R3189S) restriction cloning with addition of the restriction sites to pUC57-2019-ncov by PCR using Sal-N-protein-fw (acgcatcgtcgacATGTCTGATAATGGACC-CCAAAATCAG) and Not1-N-protein-rev (tatctatgcggccgcTTAGGCCTGAGTTGAGTCAGC). SARS-CoV-2 genome regions for templates of RNA production (5′ end (1-1000 nt), frameshifting region (13401-14400 nt), SARS-CoV-1 equivalent PS (19782-20363 nt) were derived from MN988668.1 Severe acute respiratory syndrome coronavirus 2 isolate 2019-nCoV WHU01. Sequence of N-protein for purification can be found in Data S1.
Quantification and Statistical Analysis
Images of representative cells are taken from at least 6 biological replicates pooled from at least 3 independent rounds of transfection/drug treatment.
Average fluorescence intensity and area were obtained by thresholding max projections in ImageJ. Number of puncta per cell was manually counted from max projections. For FRAP, fluorescence was normalized by subtracting background fluorescence and relative to both the initial unbleached signal and an unbleached puncta in an unbleached cell in the same frame. Quantification represents 18 puncta from 15 movies with error bars depicting standard error.
For measurements of in vitro condensates, experiments were repeated > 3 times and droplets were segmented based on a threshold of 4∗background intensity. Any segmented region with an area less than 0.07 μm2 was removed. The average protein and RNA intensity values within each droplet were calculated, and protein/RNA ratios were determined by dividing these averages on a per-droplet basis. For protein intensity and area, a two-sample Kolmogorov-Smirnov (KS) test was applied to compare protein-only with protein+RNA distributions at each temperature. A two-sample KS test was also used to make pairwise comparisons between each of the protein-only distributions, and between each of the protein+RNA distributions. Similarly, a two-sample KS test was performed to compare protein/RNA ratios. Images were processed in ImageTank (O’Shaughnessy et al., 2019) and plotted with Python using Matplotlib and Seaborn. Statistics were performed in Python with SciPy.
Acknowledgments
We thank Rick Young, Phil Sharp, Alex Holehouse, Kathleen Hall, Andrea Sorrano, Ahmet Yildez, and their lab members for sharing data and discussions; Timothy Mitchison for discussions and critical reading of the manuscript; David Adalsteinsson for his help with ImageTank software; Ian Seim for analysis consultation and discussions; Benjamin Stormo for critical reading of the manuscript; Alain Laederach for initial discussion on genomic sequence; and James Iserman for essential logistical support. A.S.G., C.I., and C.A.R. were supported by NIH R01GM081506, Fast Grants Award #2139, and an HHMI faculty Scholar Award; C.A.R. was supported by NIH T32 CA 9156-43 and F32GM136164 and L’OREAL USA for Women in Science Fellowship. The work by R.S.G.S., A.S.B., and C.L.T. is supported by NIH grants R01HG005998, U54HL117798, and R01GM071966, HHS grant HHSN272201000054C, and Simons Foundation grant 395506 to O.G.T. K.M.W., M.A.B., and C.A.W. were supported by NIH (R35 GM122532 to K.M.W.). M.A.B. was supported by a Ruth L. Kirschstein Postdoctoral Fellowship (F32 GM128330). T.P.S., E.J.F., and R.S.B. were supported by National Institute of Allergy and Infectious Diseases grants (1U19AI142759; Antiviral Drug Discovery and Development Center). This project was supported in part by the North Carolina Policy Collaboratory at University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly.
Author Contributions
C.A.R. and C.I. contributed equally to this work. Authorship was determined alphabetically. C.A.R., C.I., and A.S.G. conceptualized the project, designed experiments, prepared figures, and drafted and edited the manuscript. C.A.R. and C.I. also performed experiments and analyzed data; M.A.B. designed and performed experiments and computational analyses, analyzed data, prepared figures, and wrote the manuscript. K.M.W. designed experiments, analyzed data, and wrote the manuscript; G.A.M. designed and performed experiments, analyzed data, edited the manuscript, and performed computational analyses; R.S.G.S. designed and performed computational analysis and edited the manuscript; I.J. supported computational analyses; M.K. supported I.J. C.L.T. provided fruitful discussion and edited the manuscript; O.G.T. provided support for R.S.G.S. and C.L.T.;. J.E. generated mutant plasmids; C.A.W. designed experiments and provided fruitful discussion; E.J.F., Y.J.H., T.P.S., and R.S.B. harvested viral RNA.
Declaration of Interests
K.M.W. is an advisor to and holds equity in Ribometrix, to which mutational profiling (MaP) technologies have been licensed. A.S.G. is a scientific advisor of Dewpoint Therapeutics. C.I. is currently employed at Dewpoint Therapeutics. All other authors declare that they have no competing interests.
Published: November 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2020.11.041.
Supplemental Information
References
- Alenquer M., Vale-Costa S., Etibor T.A., Ferreira F., Sousa A.L., Amorim M.J. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat. Commun. 2019;10:1629. doi: 10.1038/s41467-019-09549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews R.J., Peterson J.M., Haniff H.S., Chen J., Williams C., Grefe M., Disney M.D., Moss W.N. An in silico map of the SARS-CoV-2 RNA Structurome. bioRxiv. 2020 doi: 10.1101/2020.04.17.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumiller W.M.J., Pir Cakmak F., Davis B.W., Keating C.D. RNA-Based Coacervates as a Model for Membraneless Organelles: Formation, Properties, and Interfacial Liposome Assembly. Langmuir. 2016;32:10042–10053. doi: 10.1021/acs.langmuir.6b02499. [DOI] [PubMed] [Google Scholar]
- Banerjee P.R., Milin A.N., Moosa M.M., Onuchic P.L., Deniz A.A. Reentrant Phase Transition Drives Dynamic Substructure Formation in Ribonucleoprotein Droplets. Angew. Chem. Int. Ed. Engl. 2017;56:11354–11359. doi: 10.1002/anie.201703191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. eLife. 2020;9:9. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J., Brangwynne C.P., Haataja M. Physical principles of intracellular organization via active and passive phase transitions. Rep. Prog. Phys. 2018;81:046601. doi: 10.1088/1361-6633/aaa61e. [DOI] [PubMed] [Google Scholar]
- Blount K.F., Zhao F., Hermann T., Tor Y. Conformational constraint as a means for understanding RNA-aminoglycoside specificity. J. Am. Chem. Soc. 2005;127:9818–9829. doi: 10.1021/ja050918w. [DOI] [PubMed] [Google Scholar]
- Boeynaems S., Holehouse A.S., Weinhardt V., Kovacs D., Van Lindt J., Larabell C., Van Den Bosch L., Das R., Tompa P.S., Pappu R.V., Gitler A.D. Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. USA. 2019;116:7889–7898. doi: 10.1073/pnas.1821038116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Busan S., Weeks K.M. Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA. 2018;24:143–148. doi: 10.1261/rna.061945.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busan S., Weidmann C.A., Sengupta A., Weeks K.M. Guidelines for SHAPE Reagent Choice and Detection Strategy for RNA Structure Probing Studies. Biochemistry. 2019;58:2655–2664. doi: 10.1021/acs.biochem.8b01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson R.C., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Morgan D.O. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol. Cell. 2020 doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Kriegenburg F., de Haan C.A.M., Reggiori F. Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers. Sci. Rep. 2017;7:5740. doi: 10.1038/s41598-017-06062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T.P., Kolaitis R.M., Kim H.J., O’Donovan K., Martyniak B., Colicino E., Hehnly H., Taylor J.P., Castañeda C.A. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell. 2018;69:965–978.e6. doi: 10.1016/j.molcel.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K., Denise A., Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- Dosztányi Z. Prediction of protein disorder based on IUPred. Protein Sci. 2018;27:331–340. doi: 10.1002/pro.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C.C., Eckmann C.R., Myong S., Brangwynne C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., 3rd, Leibowitz J.L., Giedroc D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A.R., Findeiß S., Washietl S., Hofacker I.L., Stadler P.F. RNAz 2.0: improved noncoding RNA detection. Pac. Symp. Biocomput. 2010:69–79. [PubMed] [Google Scholar]
- Guillén-Boixet J., Kopach A., Holehouse A.S., Wittmann S., Jahnel M., Schlüßler R., Kim K., Trussina I.R.E.A., Wang J., Mateju D. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell. 2020;181:346–361.e17. doi: 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseva S., Milles S., Jensen M.R., Salvi N., Kleman J.P., Maurin D., Ruigrok R.W.H., Blackledge M. Measles virus nucleo- and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci Adv. 2020;6:eaaz7095. doi: 10.1126/sciadv.aaz7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich B.S., Maliga Z., Stein D.A., Hyman A.A., Whelan S.P.J. Phase Transitions Drive the Formation of Vesicular Stomatitis Virus Replication Compartments. MBio. 2018;9:9. doi: 10.1128/mBio.02290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse A.S., Pappu R.V. Functional Implications of Intracellular Phase Transitions. Biochemistry. 2018;57:2415–2423. doi: 10.1021/acs.biochem.7b01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., Huang T.H., Chang M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin W.C., Chang C.H., Chang C.Y., Peng W.H., Chien C.L., Chang M.F., Chang S.C. Nucleocapsid protein-dependent assembly of the RNA packaging signal of Middle East respiratory syndrome coronavirus. J. Biomed. Sci. 2018;25:47. doi: 10.1186/s12929-018-0449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- Iserman C., Desroches Altamirano C., Jegers C., Friedrich U., Zarin T., Fritsch A.W., Mittasch M., Domingues A., Hersemann L., Jahnel M. Condensation of Ded1p Promotes a Translational Switch from Housekeeping to Stress Protein Production. Cell. 2020;181:818–831.e19. doi: 10.1016/j.cell.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Wang S., Huang Y., He X., Cui H., Zhu X., Zheng Y. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 2015;163:108–122. doi: 10.1016/j.cell.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I., Sealfon S., Kellis M. Sarbecovirus comparative genomics elucidates gene content of SARS-CoV-2 and functional impact of COVID-19 pandemic mutations. bioRviv. 2020 doi: 10.1101/2020.06.02.130955. [DOI] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Bárcena M., Limpens R.W., Koster A.J., Mommaas A.M., Snijder E.J. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J. Virol. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., Alberti S. Hexanediol: A Chemical Probe to Investigate the Material Properties of Membrane-Less Compartments. Science Matters. 2017 doi: 10.19185/matters.201702000010. [DOI] [Google Scholar]
- Kuo L., Masters P.S. Functional analysis of the murine coronavirus genomic RNA packaging signal. J. Virol. 2013;87:5182–5192. doi: 10.1128/JVI.00100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon E.M., Qiu Y., Ghanbari Niaki A., McLaughlin G.A., Weidmann C.A., Gerbich T.M., Smith J.A., Crutchley J.M., Termini C.M., Weeks K.M. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 2018;360:922–927. doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N.K., García Quiroz F., Hall C.K., Chilkoti A., Yingling Y.G. Molecular description of the LCST behavior of an elastin-like polypeptide. Biomacromolecules. 2014;15:3522–3530. doi: 10.1021/bm500658w. [DOI] [PubMed] [Google Scholar]
- Ma W., Zhen G., Xie W., Mayr C. Unstructured mRNAs form multivalent RNA-RNA interactions to generate TIS granule networks. bioRxiv. 2020 doi: 10.1101/2020.02.14.949503. [DOI] [Google Scholar]
- Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillén-Boixet J., Franzmann T.M. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E.W., Holehouse A.S., Peran I., Farag M., Incicco J.J., Bremer A., Grace C.R., Soranno A., Pappu R.V., Mittag T. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science. 2020;367:694–699. doi: 10.1126/science.aaw8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden E.R., Jr., Pichurko B.M., Bowman H.F., Ingenito E., Burns S., Dowling N., Solway J. Thermal mapping of the airways in humans. J Appl Physiol (1985) 1985;58:564–570. doi: 10.1152/jappl.1985.58.2.564. [DOI] [PubMed] [Google Scholar]
- Molenkamp R., Spaan W.J. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239:78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales L., Mateos-Gomez P.A., Capiscol C., del Palacio L., Enjuanes L., Sola I. Transmissible gastroenteritis coronavirus genome packaging signal is located at the 5¢ end of the genome and promotes viral RNA incorporation into virions in a replication-independent process. J. Virol. 2013;87:11579–11590. doi: 10.1128/JVI.01836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustoe A.M., Lama N.N., Irving P.S., Olson S.W., Weeks K.M. RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc. Natl. Acad. Sci. USA. 2019;116:24574–24582. doi: 10.1073/pnas.1905491116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrto Perdikari T.M.A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020 doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic J., Le Bars R., Lama Z., Scrima N., Lagaudrière-Gesbert C., Gaudin Y., Blondel D. Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 2017;8:58. doi: 10.1038/s41467-017-00102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D., Baldwin A.J. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa R.R., Calderita G., Arranz R., Fontana J., Granzow H., Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy E.C., Stone O.J., LaFosse P.K., Azoitei M.L., Tsygankov D., Heddleston J.M., Legant W.R., Wittchen E.S., Burridge K., Elston T.C. Software for lattice light-sheet imaging of FRET biosensors, illustrated with a new Rap1 biosensor. J. Cell Biol. 2019;218:3153–3160. doi: 10.1083/jcb.201903019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak C.W., Kosno M., Holehouse A.S., Padrick S.B., Mittal A., Ali R., Yunus A.A., Liu D.R., Pappu R.V., Rosen M.K. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol. Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: Machine Learning in Python. JMLR. 2011;85:2825–2830. [Google Scholar]
- Peng T.Y., Lee K.R., Tarn W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz F.G., Chilkoti A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 2015;14:1164–1171. doi: 10.1038/nmat4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincheval V., Lelek M., Gault E., Bouillier C., Sitterlin D., Blouquit-Laye S., Galloux M., Zimmer C., Eleouet J.F., Rameix-Welti M.A. Functional organization of cytoplasmic inclusion bodies in cells infected by respiratory syncytial virus. Nat. Commun. 2017;8:563. doi: 10.1038/s41467-017-00655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealfon R.S., Lin M.F., Jungreis I., Wolf M.Y., Kellis M., Sabeti P.C. FRESCo: finding regions of excess synonymous constraint in diverse viruses. Genome Biol. 2015;16:38. doi: 10.1186/s13059-015-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried N.A., Busan S., Rice G.M., Nelson J.A., Weeks K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat. Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola M.J., Rice G.M., Busan S., Siegfried N.A., Weeks K.M. Selective 2¢-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 2015;10:1643–1669. doi: 10.1038/nprot.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subudhi S., Rapin N., Bollinger T.K., Hill J.E., Donaldson M.E., Davy C.M., Warnecke L., Turner J.M., Kyle C.J., Willis C.K.R., Misra V. A persistently infecting coronavirus in hibernating Myotis lucifugus, the North American little brown bat. J. Gen. Virol. 2017;98:2297–2309. doi: 10.1099/jgv.0.000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Gultom M., Steiner S., Kelly J., Russeil J., Mangeat B., Cora E. Disparate Temperature-Dependent Virus – Host Dynamics for SARS-CoV-2 and SARS-CoV in the Human Respiratory Epithelium. bioRxiv. 2020 doi: 10.1101/2020.04.27.062315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I. Aerosol and surface stability of HCoV-19 (SARS-CoV-2) compared to SARS-CoV-1. medRxiv. 2020 doi: 10.1101/2020.03.09.20033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Hagemeijer M.C., Ulasli M., Reggiori F., Rottier P.J.M., Masters P.S., de Haan C.A.M. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84:11575–11579. doi: 10.1128/JVI.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., SciPy 1.0 Contributors SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Choi J.M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell. 2018;174:688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann C.A., Mustoe A.M., Jariwala P.B., Calabrese J.M., Weeks K.M. Analysis of RNA-protein networks with RNP-MaP defines functional hubs on RNA. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R.J., Lee H.O., Poser I., Pal A., Doeleman T., Kishigami S., Kour S. Small Molecules for Modulating Protein Driven Liquid-Liquid Phase Separation in Treating Neurodegenerative Disease. bioRxiv. 2020 doi: 10.1101/721001. [DOI] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2016. [Google Scholar]
- Wu C.H., Yeh S.H., Tsay Y.G., Shieh Y.H., Kao C.L., Chen Y.S., Wang S.H., Kuo T.J., Chen D.S., Chen P.J. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 2009;284:5229–5239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Elbaum-Garfinkle S., Langdon E.M., Taylor N., Occhipinti P., Bridges A.A., Brangwynne C.P., Gladfelter A.S. RNA Controls PolyQ Protein Phase Transitions. Mol. Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code is published. Raw and processed sequencing datasets analyzed in this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession number GEO: GSE162569).