Abstract
Cardiomyocyte differentiation is a multi‐step process which involves a number of signalling pathways. microRNAs exhibit regulatory functions in various diseases and are involved in the signalling pathways in multiple physiological processes, but the specific functions of particular mRNAs is often not fully understood. of an example of this is that the role of miR‐590‐3p in the differentiation of cardiomyocytes remains unclear. In the current study, RT‐qPCR was used to determine the expression of miR‐590‐3p in cardiomyocytes differentiated from the embryonic carcinoma cell line P19CL6. MTT, EdU, caspase‐3 activity and flow cytometry assays were performed to examine the influence of miR‐590‐3p on cell behaviour. A luciferase assay was used to confirm binding between miR‐590‐3p and PTPN1. Western blotting was used to determine the relationship between the JNK/STAT/NF‐kB pathway and PTPN1. The results inferred that miR‐590‐3p became heavily expressed in differentiated P19CL6. Knockdown miR‐590‐3p suppressed the cell proliferation while at the same time, accelerated apoptosis. Moreover, PTPN1 was identified as the target of miR‐590‐3p. More importantly, PTPN1 overexpression activated the JNK/STAT/NF‐kB pathway and limited the differentiation of P19CL6. Thus the conclusions from this study are that miR‐590‐3p has the potential to regulate the proliferation, apoptosis and differentiation of cardiomyocyte P19CL6 in vitro by targeting PTPN1 via the JNK/STAT/NF‐kB pathway.
Keywords: JNK, MiR‐590‐3p, NF‐kB pathway, P19CL6, PTPN1, STAT
1. INTRODUCTION
During differentiation stem cells mesoderm cells in the beginning. Then it differentiates into cardiomyocyte precursor stem cells and finally functional cardiomyocytes that can spontaneously pulsate. 1 During the differentiation process, gene regulation occurs in which various signalling pathways are involved. 2 Being short non‐coding double‐stranded RNAs, 3 microRNAs (miRNAs) have the capacity to inhibit mRNA expression after transcription. 4 Previous studies concluded that miRNAs exert regulatory functions in different diseases, including myocardial differentiation. 5 , 6 For instance, miR‐98‐5p regulates the myocardial differentiation of mesenchymal stem cells by targeting TBX5. 7 miR‐148a promotes the myocardial differentiation of human bone mesenchymal stromal cells via DNMT1. 8 Selenium deficiency inhibits myocardial development and differentiation when the miR‐215‐5p/CTCF axis gets is targeted. 9 The researchers 10 , 11 noted that miR‐590‐3p is one of the recently updated novel miRNAs. Though its role in cancer has already been investigated, the specific role played by miR‐590‐3p in myocardial differentiation remains unclear.
P19CL6 is a euploid multipotent mouse cell line derived from P19 embryonal carcinoma cells. When treated with dimethyl sulphoxide (DMSO), P19CL6 can differentiate efficiently into cardiac myocytes and exhibits spontaneous heartbeat within 10 days of differentiation. 12 Thus P19CL6 is a helpful in vitro model to investigate the differentiation of cardiomyocytes. 13 The activation of different pathways can also influence the progression of myocardial differentiation. For example, the differentiation of mesenchymal stem cells into cardiomyocytes is regulated by miRNA‐1‐2 via the WNT signalling pathway. 14 Hsp25 and p38 MAPK pathways are involved in the differentiation of cardiomyocytes. 15 JNK, STAT and NF‐kB pathways are involved in the progression of several disease processes but the functions of the JNK/STAT/NF‐kB pathway in myocardial differentiation are still unclear. In the current study, the authors aimed at investigating the influence of miR‐590‐3p on cardiomyocyte P19CL6. The study results demonstrate that miR‐590‐3p has the potential to regulate proliferation, apoptosis and differentiation of cardiomyocyte P19CL6 in vitro by targeting PTPN1 via the JNK/STAT/NF‐kB pathway. Therefore the current study suggests a possible new treatment strategy for illnesses caused due to the lack of myocardial differentiation.
2. MATERIALS AND METHODS
2.1. Cell culture
P19CL6 mouse embryonic carcinoma cells were grown in Minimum Essential Medium, Alpha Medium (a‐MEM; GIBCO) containing 10% Foetal Bovine Serum (GIBCO). The cells were kept under incubation at 37°C in a humidified incubator with 5% CO2 atmosphere. To induce P19CL6 differentiation into cardiomyocytes, P19CL6 cells were seeded at a density of 3.5 × 105 cells/well in a six‐well plate, with the culture medium containing 1% DMSO (Sigma). The conditioned medium was replaced with 2 mL of fresh medium containing 1% DMSO for every 2 days. The days of differentiation were numbered consecutively following the first day of DMSO treatment.
2.2. RT‐qPCR analysis
The authors extracted the total RNA from cells using TRIzol® reagent (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized using Prime Script™ RT Master Mixture (Takara). qRT‐PCR was performed on ABI 7500 Realtime PCR System (ABI) using SYBR Prime Script kit (Takara). The expression levels of mRNA for the tested genes related to GAPDH were analysed using 2−ΔΔCt method.
2.3. Transfection
The authors procured miR‐590‐3p mimics, miR‐590‐3p inhibitor, NC mimics, pcDNA3.1‐PTPN1, NC and pcDNA3.1 from GenePharma company (China). The P19CL6 cells were transfected with the plasmids mentioned above by Lipofectamine2000 (Invitrogen) according to the manufacturer's instructions. The cells were collected for further analysis at 48 hours after transfection.
2.4. MTT assay
At 48 hours after transfection, the cells were cultured in serum‐free DMEM for 0, 24, 48 and 72 hours prior to the addition of MTT (Sigma) to each well. After incubation with MTT for 4 hours at 37°C, the MTT solution was removed and DMSO was added to each well. Then the absorbance was measured at 490 nm using a spectrophotometer.
2.5. EdU assay
The cells were placed into 24‐well plates. EdU kit (Ribobio) was used to evaluate the cell proliferation. EdU and DAPI dyes were utilized to stain the cell nuclei. Finally, fluorescence microscopy (Olympus) was used to take the representative images.
2.6. Caspase‐3 activity
In order to investigate the activation of caspase‐3, the caspase‐3 colorimetric assay kit (BioVision) was used for analysis according to the manufacturer's instructions. Enzyme reactions were performed in a 96‐well microplate, and to each reaction mixture, 5 μL of cell lysate was added. The absorbance was measured at 405 nm using a microplate reader (Tianan).
2.7. Flow cytometry
The cells were harvested, processed and double stained with annexin V‐fluorescein isothiocyanate (FITC) and propidium iodide using Annexin V‐FITC Apoptosis Detection kit (BD Biosciences) following the manufacturer's protocol. Cell apoptosis was analysed with FACScan flow cytometer (BD Biosciences), and the cell apoptotic rates were analysed by bd cellquest pro™ Software (Version 5.1; BD Biosciences).
2.8. Western blot analysis
The cell proteins were extracted using radioimmunoprecipitation assay buffer (Sigma‐Aldrich). The concentration of protein was measured using the BCA kit (Bio‐Rad). Proteins were separated using 10% SDS‐PAGE and transferred to PVDF membranes. The membranes were blocked with 5% skimmed milk for 1 hour at room temperature, followed by incubation with following primary antibodies: Rabbit polyclonal anti‐B‐cell lymphoma 2 (Bcl‐2) antibody (1:1500; cat. no. ab59348; Abcam), rabbit polyclonal anti‐JNK1 (phospho T183; 1:2000; cat. no. ab47337; Abcam), rabbit monoclonal anti‐STAT1 (phospho Y701) antibody (1:1000, cat. no. ab109457; Abcam) and rabbit polyclonal anti‐GAPDH antibody (1:2000, cat. no. ab9485; Abcam) overnight at 4°C. The membranes were subsequently incubated with horseradish peroxidase‐conjugated secondary antibody (1:2000, cat. no. ab205718; Abcam) and visualized with Enhanced Chemiluminescence reagent (Invitrogen) following the manufacturer's protocol.
2.9. Luciferase assay
For luciferase reporter assay, P19CL6 cells were transfected with reporter plasmids using the Lipofectamine 2000 reagent. At 48 hours post‐transfection, firefly and Renilla luciferase activities were measured using Dual Luciferase Assay kit (Promega) as per the manufacturer's instructions.
2.10. Statistical analysis
All the experiments were conducted in triplicates. The data are represented in the next section as mean ± standard deviation (SD). graphpad prism V5.01 Software (GraphPad) was used to determine the group differences by Student's t test or one‐way analysis of variance (ANOVA). P < .05 was considered to indicate statistically significant difference.
3. RESULTS
3.1. High expression of MiR‐590‐3p in the differentiated P19CL6 cells
Figure 1A shows the expression levels of GATA4 and α‐MHC in P19CL6 cells. Interestingly, the expression of GATA4 and α‐MHC can be detected on the fourth day of differentiation. It is critical to note that the expression of GATA4 and α‐MHC increased prominently at 8th and 12th day of differentiation. Meanwhile, the expression of miR‐590‐3p also increased in a time‐based manner among P19CL6 cells (Figure 1A). These results indicate that miR‐590‐3p is involved in the differentiation of P19CL6 cells and may play a regulatory role in this differentiation.
Figure 1.
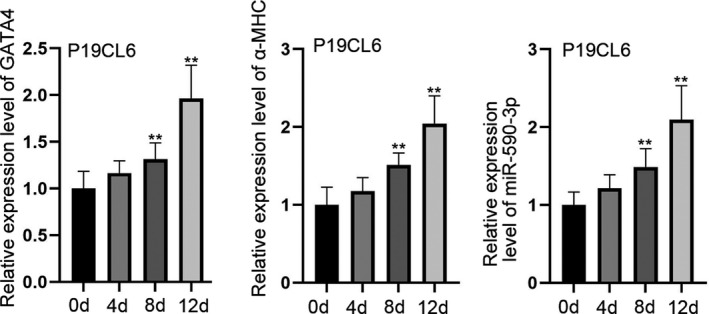
MiR‐590‐3p is highly expressed in differentiated P19CL6 cells. A, RT‐qPCR was utilized to test the expressions of GATA4, α‐MHC and miR‐590‐3p in P19CL6 cells. **P < .01
3.2. Silenced miR‐590‐3p inhibits the proliferation and accelerates the apoptosis of P19CL6 cells
To investigate the effect of miR‐590‐3p on P19CL6 cells, the former was inhibited in P19CL6 cells. RT‐qPCR was used to confirm the interference efficiency of miR‐590‐3p in P19CL6 cells (Figure 2A). Then the MTT and EdU experiments were conducted to determine cell proliferative capacity. The results indicate that the cell viability and the rate of EdU positive cells were decreased. This phenomenon infers that the cell proliferation could be inhibited by miR‐590‐3p knockdown (Figure 2B,C). Then the cell apoptosis was determined by both flow cytometry and caspase‐3 activity assay (Figure 2D,E). The results demonstrate that the silenced miR‐590‐3p promoted apoptosis of P19CL6 cells. Overall, the knockdown of miR‐590‐3p could both restrain proliferation and accelerate apoptosis of P19CL6 cells.
Figure 2.
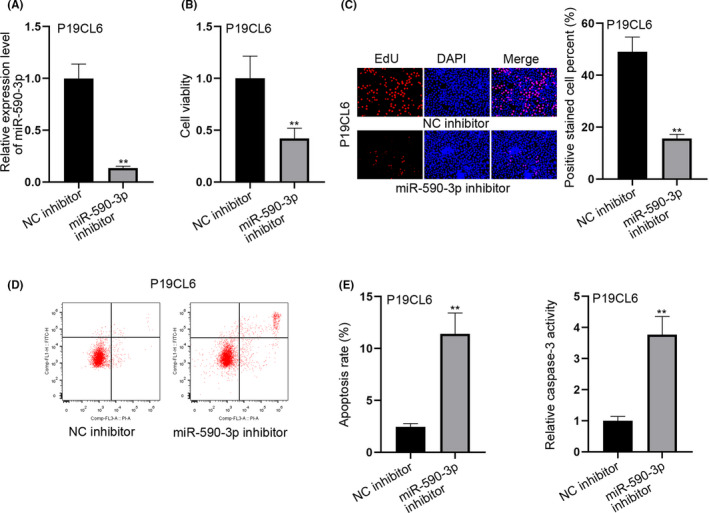
Silenced miR‐590‐3p inhibits proliferation and accelerates apoptosis of P19CL6 cells. A, RT‐qPCR was adopted to test the interference efficiency of miR‐590‐3p in P19CL6 cells. B‐C, MTT and EdU experiments were adopted to test the capability of cell proliferation after silencing miR‐590‐3p. D‐E, The cell apoptotic capability was estimated through flow cytometry and caspase‐3 activity detection assays, when miR‐590‐3p was subjected to inhibition. **P < .01
3.3. PTPN1 acts as the target of miR‐590‐3p in P19CL6 cells
In order to further understand the specific mechanisms of miR‐590‐3p in P19CL6 cells, we analysed the proteins related to miR‐50‐3p through starBase (http://starbase.sysu.edu.cn/). As shown in Figure 3A, the binding sites of miR‐590‐3p and PTPN1 were identified. This indicates that the miR‐590‐3p may react with PTPN1. Further, the luciferase reporter assay was performed to confirm the interaction between miR‐590‐3p and PTPN1. The results indicate that these two molecules could bind to each other at site 1 (Figure 3B). Subsequently, the expression of PTPN1 in P19CL6 was determined. PTPN1 expression decreased in a time‐dependent manner (Figure 3C). The RT‐qPCR results revealed that the expression of PTPN1 increased after miR‐590‐3p knockdown. This indicates that PTPN1 is negatively associated with miR‐590‐3p (Figure 3D). Next, PTPN1 was overexpressed in P19CL6 cells (Figure 3E). Both MTT and EdU assays indicate that cell proliferation was diminished (Figure 3F,G). The results attained in flow cytometry and caspase‐3 activity assays indicate that the cell apoptosis was triggered with the overexpression of PTPN1 (Figure 3H,I). On the whole, it can be established that PTPN1 is the target of miR‐590‐3p and when PTPN1 gets overexpressed, it inhibits cell growth, but induces cell apoptosis.
Figure 3.

PTPN1 acts as the target of miR‐590‐3p in P19CL6 cells. A, The binding sites of miR‐590‐3p and PTPN1. B, Luciferase reporter assays were adopted to test the binding situation of miR‐590‐3p and PTPN1. C, RT‐qPCR was conducted to test the expression of PTPN1 in P19CL6 cells. D, RT‐qPCR was utilized to detect the relationship of miR‐590‐3p and PTPN1. E, RT‐qPCR was carried out to test the overexpression efficiency of PTPN1 in P19CL6 cells. F‐G, MTT and EdU experiments were done to test the cell proliferative capability after PTPN1 got expressed. H‐I, Flow cytometry and caspase‐3 activity detection assays were conducted to estimate cell apoptosis when PTPN1 got expressed. **P < .01
3.4. Upregulation of PTPN1 activates JNK/STAT/NF‐kB pathway and restrains the differentiation of P19CL6 cells
In order to investigate whether the upregulation of PTPN1 can activate the JNK/STAT/NF‐kB pathway, and promote the differentiation of P19CL6 cells, downstream protein expression of JNK and STAT were examined. As shown in Figure 4A, overexpression of PTPN1 reduced the phosphorylation of JNK and STAT. Bcl‐2 expression was also reduced compared to the control group. Since Bcl‐2 is regulated by the NF‐kB pathway, the current study results indicate that the JNK/STAT/NF‐kB pathway could be activated with the overexpression of PTPN1 (Figure 4A). Furthermore, the expression levels of GATA4 and α‐MHC in P19CL6 cells were decreased. This demonstrates that both GATA4 and α‐MHC expressions could be restrained with the upregulation of PTPN1 (Figure 4B,C). Overall, PTPN1 overexpression could activate the JNK/STAT/NF‐kB pathway and restrain the differentiation of P19CL6 cells.
Figure 4.

Upregulation of PTPN1 activates JNK/STAT/NF‐kB pathway and restrains differentiation of P19CL6 cells. A, Western blot experiments were carried out to test the protein levels of phosphorylated JNK, phosphorylated STAT and Bcl‐2. B‐C, RT‐qPCR was utilized to detect the expression of GATA4 and α‐MHC. **P < .01
4. DISCUSSION
Cardiomyocyte differentiation is a multi‐step process that occurs in accordance with a regulated molecular pathway. The different stages of differentiation can be identified based on the expression of specific genes. The expression of GATA4, a specific myocardium transcription factor, is a marker for cell differentiation into cardiomyocytes. It then initiates the expression of structural and functional genes of myocardium such as α‐MHC. With gradual development of the differentiated cardiomyocytes, the cells finally start showing spontaneous rhythmic pulsation. P19CL6 has become a recognized cell model to investigate cardiomyocyte differentiation. 16 Its differentiation potential is mainly manifested in the general direction of mesoderm, and can then induced by DMSO into differentiated cardiomyocytes in an efficient and reproducible way.
miRNAs are short non‐coding double‐stranded RNAs. The main function of miRNAs is to regulate the gene translation by post‐transcriptional binding of the RNA and to prevent ribosomal translation. 17 miRNAs are important in the regulation of many biological processes. MiR‐590‐3p has been shown to be associated with several cardiac diseases. Previous studies 18 already investigated the miR‐590‐3p‐regulated inflammation in experimental autoimmune myocarditis. miR‐590‐3p could regulate proliferation, migration and collagen synthesis of cardiac fibroblasts by targeting ZEB1. 19 The rapid delivery of miR‐590‐3p using targeted exosomes can be used as a potential therapeutic phenomenon to treat acute myocardial infarction. 20 miR‐590‐3p is capable of inducing proliferation of cardiomyocytes and cardiac regeneration. 21 The current study was aimed at investigating the correlations between miR‐590‐3p and cardiomyocyte differentiation. For this an established and recognized cell model for cardiomyocyte differentiatio, was used. The results indicate that miR‐590‐3p was highly expressed in gradually differentiated P19CL16 cells. Knockdown miR‐590‐3p in P19CL6 cells significantly inhibited cell proliferation while at the same time induced apoptosis. Bioinformatic analysis was conducted which identified that PTPN1 was the target of miR‐590‐3p. miRNAs can regulate the expression of mRNA after transcription. In the current study, it was found that PTPN1 has a negative association with miR‐590‐3p. The overexpression of PTPN1 inhibited cell proliferation in P19CL6 cells, but expedited apoptotic ability. The JNK pathway is able to control various physiological processes such as cell differentiation, cell proliferation and protein expressions. 22 The activated JAKs can in turn activate specific STAT family members to form a complex regulatory network. 23 NF‐kB pathway is also involved in different disease progressions. For example, H19 accelerates atherosclerosis by regulating MAPK and NF‐kB signalling pathways. 24 In the current study, the authors identified the relationship between JNK/STAT/NF‐kB pathway and PTPN1. The results indicate that the overexpressed PTPN1 activated JNK/STAT/NF‐kB pathway and therefore inhibited the differentiation of P19CL6 cells.
In summary, the current study demonstrated that miR‐590‐3p has the potential to regulate proliferation, apoptosis and differentiation of cardiomyocyte P19CL6 in vitro by targeting PTPN1 via the JNK/STAT/NF‐kB pathway. The impact of the current study will not be limited to cardiomyocytes, as the results may have a broader significance. Therefore this study may act as a foundation for future studies about the role and mechanism of miRNAs in other diseases.
ETHICAL APPROVAL
Ethical approval was obtained from the institutional review board of Zhongshan Hospital of Fudan University, Shanghai, China.
CONFLICT OF INTEREST
Authors do not have anything to disclose and declare not conflict of interest.
AUTHORS' CONTRIBUTIONS
Fanshun Wang was involved in literature search, data analysis, statistical analysis, manuscript preparation, manuscript review and served as a guarantor. Hongqiang Zhang was involved in concepts, design, definition of intellectual content, literature search, experimental studies, data acquisition and manuscript editing. Chunsheng Wang was involved in data analysis, statistical analysis, manuscript preparation, literature search, experimental studies, data acquisition and manuscript editing.
Wang F, Zhang H, Wang C. MiR‐590‐3p regulates cardiomyocyte P19CL6 proliferation, apoptosis and differentiation in vitro by targeting PTPN1 via JNK/STAT/NF‐kB pathway. Int. J. Exp. Path. 2020;101:196–202. 10.1111/iep.12377
Fanshun Wang and Hongqiang Zhang contribute equal to this article as co‐first author.
DATA AVAILABILITY STATEMENT
All data are provided in this study and raw data can be requested to corresponding author.
REFERENCES
- 1. Paige SL, Plonowska K, Xu A, Wu SM. Molecular regulation of cardiomyocyte differentiation. Circ Res. 2015;116:341‐353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takaya T, Nishi H, Horie T, Ono K, Hasegawa K. Roles of microRNAs and myocardial cell differentiation. Prog Mol Biol Transl Sci. 2012;111:139‐152. [DOI] [PubMed] [Google Scholar]
- 3. Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613‐626. [DOI] [PubMed] [Google Scholar]
- 4. Buglioni S, Vici P, Sergi D, et al. Analysis of the hippo transducers TAZ and YAP in cervical cancer and its microenvironment. Oncoimmunology. 2016;5:e1160187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adamopoulos PG, Tsiakanikas P, Scorilas A. Kallikrein‐related peptidases and associated microRNAs as promising prognostic biomarkers in gastrointestinal malignancies. Biol Chem. 2018;399:821‐836. [DOI] [PubMed] [Google Scholar]
- 6. He T, Chen P, Jin L, et al. miR6605p is associated with cell migration, invasion, proliferation and apoptosis in renal cell carcinoma. Mol Med Rep. 2018;17:2051‐2060. [DOI] [PubMed] [Google Scholar]
- 7. Sun HH, Sun PF, Liu WY. MiR‐98‐5p regulates myocardial differentiation of mesenchymal stem cells by targeting TBX5. Eur Rev Med Pharmacol Sci. 2018;22:7841‐7848. [DOI] [PubMed] [Google Scholar]
- 8. Jiang C, Gong F. MiR‐148a promotes myocardial differentiation of human bone mesenchymal stromal cells via DNA methyltransferase 1 (DNMT1). Cell Biol Int. 2018;42:913‐922. [DOI] [PubMed] [Google Scholar]
- 9. Cai J, Yang J, Liu Q, Gong Y, Zhang Y, Zhang Z. Selenium deficiency inhibits myocardial development and differentiation by targeting the mir‐215‐5p/CTCF axis in chicken. Metallomics. 2019;11:415‐428. [DOI] [PubMed] [Google Scholar]
- 10. Wang WT, Qi Q, Zhao P, Li CY, Yin XY, Yan RB. miR‐590‐3p is a novel microRNA which suppresses osteosarcoma progression by targeting SOX9. Biomed Pharmacother. 2018a;107:1763‐1769. [DOI] [PubMed] [Google Scholar]
- 11. Zu C, Liu S, Cao W, et al. MiR‐590‐3p suppresses epithelial‐mesenchymal transition in intrahepatic cholangiocarcinoma by inhibiting SIP1 expression. Oncotarget. 2017;8:34698‐34708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Young DA, Gavrilov S, Pennington CJ, et al. Expression of metalloproteinases and inhibitors in the differentiation of P19CL6 cells into cardiac myocytes. Biochem Biophys Res Commun. 2004;322:759‐765. [DOI] [PubMed] [Google Scholar]
- 13. Habara‐Ohkubo A. Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct Funct. 1996;21:101‐110. [DOI] [PubMed] [Google Scholar]
- 14. Shen X, Pan B, Zhou H, et al. Differentiation of mesenchymal stem cells into cardiomyocytes is regulated by miRNA‐1‐2 via WNT signaling pathway. J Biomed Sci. 2017;24:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davidson SM, Morange M. Hsp25 and the p38 MAPK pathway are involved in differentiation of cardiomyocytes. Dev Biol. 2000;218:146‐160. [DOI] [PubMed] [Google Scholar]
- 16. Han Y, Fan TB, Peng BT, et al. Effect of miR‐19b on the proliferation and apoptosis of P19CL6 cells during the late‐stage of cardiac differentiation. Zhonghua Yi Xue Za Zhi. 2018;98:617‐621. [DOI] [PubMed] [Google Scholar]
- 17. Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271‐282. [DOI] [PubMed] [Google Scholar]
- 18. Zhao S, Yang G, Liu PN, et al. miR‐590‐3p is a novel MicroRNA in myocarditis by targeting nuclear factor kappa‐B in vivo. Cardiology. 2015;132:182‐188. [DOI] [PubMed] [Google Scholar]
- 19. Yuan X, Pan J, Wen L, et al. MiR‐590‐3p regulates proliferation, migration and collagen synthesis of cardiac fibroblast by targeting ZEB1. J Cell Mol Med. 2020;24(1):227‐237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Ding N, Guan G, et al. Rapid delivery of Hsa‐miR‐590‐3p using targeted exosomes to treat acute myocardial infarction through regulation of the cell cycle. J Biomed Nanotechnol. 2018b;14:968‐977. [DOI] [PubMed] [Google Scholar]
- 21. Eulalio A, Mano M, Dal Ferro M, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376‐381. [DOI] [PubMed] [Google Scholar]
- 22. Kumar A, Singh UK, Kini SG, et al. JNK pathway signaling: a novel and smarter therapeutic targets for various biological diseases. Future Med Chem. 2015;7:2065‐2086. [DOI] [PubMed] [Google Scholar]
- 23. Yang X, Tang Z, Zhang P, Zhang L. Research advances of JAK/STAT signaling pathway in lung cancer. Zhongguo Fei Ai Za Zhi. 2019;22:45‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan JX. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF‐kB signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:322‐328. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are provided in this study and raw data can be requested to corresponding author.


