Summary
We discuss the validation of machine learning models, which is standard practice in determining model efficacy and generalizability. We argue that internal validation approaches, such as cross-validation and bootstrap, cannot guarantee the quality of a machine learning model due to potentially biased training data and the complexity of the validation procedure itself. For better evaluating the generalization ability of a learned model, we suggest leveraging on external data sources from elsewhere as validation datasets, namely external validation. Due to the lack of research attractions on external validation, especially a well-structured and comprehensive study, we discuss the necessity for external validation and propose two extensions of the external validation approach that may help reveal the true domain-relevant model from a candidate set. Moreover, we also suggest a procedure to check whether a set of validation datasets is valid and introduce statistical reference points for detecting external data problems.
Keywords: exploratory data analysis, data science, computational biology, descriptive statistics, scientific method
The Bigger Picture
External validation is critical for establishing machine learning model quality. To improve rigor and introduce structure into external validation processes, we propose two extensions, convergent and divergent validation. Using a case study, we demonstrate how convergent and divergent validations are set up and also discuss technical considerations for gauging performance, including establishment of statistical rigor, how to acquire valid external data, determining the number of times an external validation needs to be performed, and what to do when multiple external validations disagree with each other. Finally, we highlight that external validation remains and will be highly relevant, even to new machine learning paradigms.
External validation is an important step for confirming potential for practical deployments of a machine learning or artificial intelligence algorithm. Unfortunately, this process is not well structured. We discuss how to make external validations more robust and systematic and also introduce two new extensions: convergent and divergent validations.
Introduction
Machine learning is among the most powerful tools for knowledge discovery from data today. If a learned model (trained classifier) correctly captures domain-relevant features (i.e., factors that are explanatory and causal), the model is deemed domain relevant and more likely explainable (i.e., the set of interpretable decision rules is logical and lends itself toward a better domain understanding.) Such models may be more confidently used in a wide variety of practical applications, e.g., predicting credit risk,1 recidivism,2 and the state of charge and health of batteries.3
As there are many different purposes and applications in machine learning, in this paper, we limit ourselves to binary predictive models for about any outcome, with focus on a high-dimensional feature space (i.e., a very large amount of potential predictors). We are also concerned with a statistical feature selection paradigm on the discussions of different issues as well as explanation of approaches. For such settings, model validation is critical for discovering domain-relevant models with better generalization ability, and further implies better interpretability. This is a very specialized context. For general machine learning researches that may incur more modalities (e.g., visions and robotics) where we may encounter many black box models, it is not a trivial case to imply interpretability only from validation performance. There is still a gap to go from those machine learning problems with many black box models. We do not consider these types of problems here.
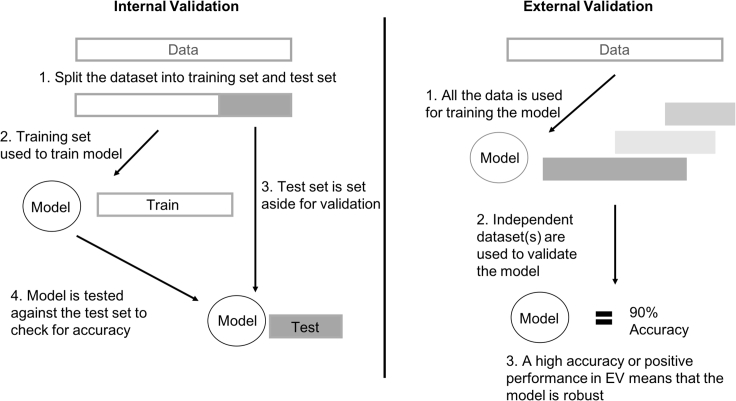
Learned models are evaluated through the process of validation, which is defined as the process of evaluating accuracy—and more robust sensitivity and specificity, although these are less commonly used than accuracy in the machine learning literature—on data that are not part of the training set. Broadly, validation is categorizable into "internal" and "external" (Figure 1). Internal validation (IV) approaches are economical, as they involve splitting one input dataset into parts—with some parts used for training the classifier (training data), and the remainder used for validation (test data). This process is repeated until each part has been used at least once as testing data. This process is also commonly referred to as cross-validation. While considered industry standard and very commonly used, IV procedures are highly heterogeneous and tuneable.4 In particular, it is left to the discretion of the analyst which procedural subtype (e.g., k-fold, bootstrap) to use and what the parameters are (e.g., value of k, number of bootstrap iterations). Another issue is that, if the original input data are, e.g., a biased sample, cross-validation becomes a biased evaluation of a biasedly trained model on a test set that is biased in the same way.
Figure 1.
Validation Methods Broadly Fall into Two Types: Internal and External
Internal validation (IV) involves first splitting the input data into a training set and a test set. The training set is used to train the model while the test set is used to check the accuracy of the model after it has been trained. External validation (EV) uses the entire input data for model training and the validation of the model is done using an independently derived dataset(s).
External validation (EV) involves the use of independently derived datasets (hence, external), to validate the performance of a model that was trained on initial input data. EV is sometimes referred to as independent validation—we find this a misnomer, as the independently derived (by virtue of being sourced from elsewhere) external dataset may not be truly independent of the training data.5 Independent validation is also sometimes used to refer to a validation study by other researchers than the researchers who developed the model.
EV is usually considered important evidence for generalizability. Due to the validation set coming from an independent source, any feature set that was falsely selected due to idiosyncrasies of the input training data (e.g., technical or sampling bias) would likely fail. Hence, a positive performance in EV is regarded as a proof of generalizability.
Good Prediction Accuracy Often Does Not Imply Domain Relevance
Capturing domain-relevant features for model inclusion is important as these lead toward reasonable explanations for the machine learning model. To avoid overfitting or non-generalizability issues, we should strive toward models that use true causal predictors of the outcome. In a health-related example, cancer genes affect many other gene groups, particularly, growth genes.6 A model built on growth genes, while able to differentiate normal tissues from cancer tissues, may also misdiagnose a tissue of rapidly growing cells as cancer.
While domain-relevant features are useful for explaining why a model works well, it is not straightforward to mine these from models. Before we can perform causal interpretation of predictive analytics, the model first needs to give accurate predictions of the outcome. However, it appears that obtaining accurate prediction is fairly easy, even with common validation approaches, such as the bootstrap and cross-validation. However, an accurate model can be naively overfitted: for example, in a medical imaging study, Zech and colleagues7 noticed only after training their neural network that it heavily relied on the word “portable” within an X-ray image, representing the type of X-ray equipment rather than the medical content of the image.
Meaningful and clean data, in the sense that the captured features are informative and that the data are devoid of noise or confounding effects, may yield accurate models. Unfortunately, in practice, any collected data are effectively a sample, and may not be a true representation of the population under study. Before training the model, it is often good to check for sample bias, and where possible check for errors stemming from sample size,8,9 heterogeneity,10 noise,11 and confounding factors.12
Because we cannot rule out that any input data are error-free and unbiased, a good accuracy within a cross-validation is generally insufficient to ascertain whether a model is domain relevant and generalizable. Therefore, it is good practice to repeatedly challenge the learned model with independently sampled data (EV). If the good performance persistently replicates, the more likely the learned model is generalizable.
Why EV Should Be Performed More Often?
Statistical techniques can be used to reason about data domain representativeness and domain relevance, thereby providing a proxy on model interpretability.12,13 But these techniques may not be easy to deploy or may require correct prior knowledge that may not exist.
Since training data may not be truly representative and may not be properly prepared, we should regard any trained model, despite having passed IV (e.g., via cross-validation), as potentially non-generalizable. Before actual deployment into the real world setting, we recommend evaluating a learned model via EV. This is a simple process that involves challenging the model with additional data that are not involved in the original input data or are meaningfully different (for example, in the imaging problem of Zech et al.7 they could have tested for overfitting by using an EV in which the input set was produced using different X-ray equipment).
The logic behind EV is sound: data taken from completely separate sources have less in common, but nonetheless may capture useful domain-relevant aspects. A well-trained model that captures informative features is robust and will continue to exhibit good results even when repeatedly challenged with new data. EV thus provides an assurance that models passing this step are more likely domain-interpretable. There are nonetheless some caveats, as discussed in the remainder of this article.
The Case Study Setup in Support of the EV
As there are many different purposes and applications in machine learning, in this paper, we limit ourselves to binary predictive models for any outcome, with a focus on a high-dimensional feature space (i.e., a very large amount of potential predictors). We are concerned with a statistical feature selection paradigm on the discussions of different issues as well as explanation of approaches. For such settings, model validation is critical for discovering domain-relevant models with better generalization ability, which further implies better interpretability. This is a very specialized context. We illustrate our point using gene signature selection in breast cancer.6,14 A gene signature can be thought of as a set of features where each feature is the expression level of a gene; and the set forms a prediction model, which is a classifier in the context of our discussion here.
We first encountered issues with EV in the Venet et al.15 study of breast cancer prognostic signatures, and explored the implications for machine learning.6,12,14,16 In the Venet et al. study, they evaluated multiple reported gene signatures against a single large dataset, and found that none of these gene signatures could beat randomly generated gene signatures or domain-irrelevant signatures. In other words, each reported breast cancer signature, as a consequence of intensive study, gives rise to a highly accurate model. But an accurate model may not be domain relevant, as the selected features may in themselves be non-causal correlates. This leads to many pretender models (high performance on a dataset, but no relevance).
We propose that a simple way of rooting out pretender models is to evaluate the substitutability of the feature set; i.e., generating randomized feature sets to create null models against which you evaluate your actual model. This may take the following form. Suppose you are developing a model for predicting two classes, A versus B, on a moderately large feature space of 20,000 measured features. Since there are many features, dimensionality reduction is often necessary, and so, from the initial 20,000 feature space, you pick 10 based on class correlation (or any other feature selection approach17), and use these 10 features to train a model X using input training data D1. Model X, when validated on validation data D2, produces an accuracy of 90%, which appears reasonable. This suggests that model X is generalizable, and therefore is domain relevant. However, a randomly selected feature set, also of size 10, is also used to train a model XR on D1, producing an accuracy of 90% on D2. Repeating this experiment many times over shows that the average prediction accuracy of random feature sets is about 92%. The accuracy based on X is scarcely better than the accuracy of XR. In a typical setting, people would not perform this randomization test, and merely report X as good based on the EV prediction accuracy of 90%. While certainly a good result, X has, in fact, little to no information value since random features sets do as well, if not better.
While this technique does not leverage on domain knowledge or carefully constructed explainable models, it can be used straightforwardly for eliminating falsely high-performing models.4 This example also tells us that we cannot trust the accuracy of a single EV as objective evidence of domain relevance.
In a robust model validation strategy, IV and EV should be used together in a tandem configuration where IV is first deployed to provide a quick sense of performance before relying on EV to determine domain-correctness. In particular, IV is unlikely to be able to detect if the training dataset is not domain representative; and non-domain representativeness of training data is not an uncommon situation (cf. https://channels.theinnovationenterprise.com/articles/bad-data-is-ruining-machine-learning-here-s-how-to-fix-it). EV is an essential procedure in counteracting this. Moreover, EV provides robustness to class-prediction accuracy, which in turn acts as a proxy for interpretability. Unfortunately, while it is common to encounter literature covering tutorials and explanations on how to design and execute IV,18, 19, 20 with accompanying studies of its theoretical and statistical properties,21 information on how to conduct EV properly or evaluating the outcome of an EV (when you should or should not believe the results of an EV) is less common. In an advanced search on Nature Publishing Group's website where we searched for publications containing the terms = “cross-validation classifier” and publication date = “2019 to 2020,” we then looked at the first 40 results, after discarding a tutorial and a review article. Of these 40 results, 20 seemed questionable (i.e., there was no evaluation based on clear independent dataset) and 20 seemed appropriate (i.e., had independent datasets or it was not clear that they had an independent dataset). So, one might say that there is some evidence that papers that use cross-validation as part of their study methodology have a tendency (∼50%) to not use an independent validation dataset. The search above returned 1,296 results. After dropping “cross-validation” from the search terms, 2,566 results were returned. So, about half (1,296/2,566) of the papers published by the Nature Group in 2019–2020 that pertain to constructing classifiers (mostly for biomedical applications), rely on cross-validation as part of their methodology. Of these, about half can be considered questionable, as their constructed classifiers were not evaluated on independent datasets. In other words, about 25% (50% ∗ 50%) of the articles published from 2019 to 2020 by Nature Group pertaining to developing classifiers lack sufficient validation.
EV is not as trivial as simply finding some new data to challenge the model. That would assume that any data, as long as sourced elsewhere, would do. Clearly, such looseness lacks rigor. There is also the need for correct interpretive logic to be used alongside evaluation of an EV. For example, when the EV is conducted improperly such that low accuracy is observed, we may mistakenly think the input data were uninformative or that the model was overfitted to the training data. When an EV mistakenly informs that a correctly trained model failed to generalize, it is a waste (or a form of false negative, if you like). Conversely, we should be just as wary when a persistently good EV result is observed, as it may be due to data biases (e.g., the presence of a batch effect),22 false statistical assumptions (the used case requirements of the statistical model are not met),13 or there may be an incompatibility between the input data and the real world (e.g., random sampling biases causing the sample to be non-representative of the population of study).23,24 The phenomenon where irrelevant factors lead to misleading good results is known as the Anna Karenina effect.12 Using such mis-trained models in real world settings can produce undesirable consequences.24
Two Extensions of the EV
IV evaluates performance on data from exactly the same underlying population, whereas EV evaluates performance on data from similar but not identical underlying populations. Different EVs may each contribute data from different underlying populations (e.g., different hospitals, different countries). EV on multiple datasets is important for revealing the heterogeneity in a model's performance, and for fully appreciating the generalizability of a model.25
We propose two procedures that extend the EV. We call these convergent and divergent validations. They have useful properties but require more work to execute26 (see Table 1 and Figure 2 for an overview). For in-depth details on implementations and outcomes of convergent and divergent validations, please refer to, e.g., recent publications of Goh and Wong.6,14
Table 1.
Summary of EV Approaches
| Type of EV | Description | What It Is For | Known Instances of Deployment |
|---|---|---|---|
| Standard | Test 1 classifier on new data | It allows us to study the generalizability of an inferred feature set on one test data | Many. But most prominent are the recent reproducibility initiatives, e.g., Reproducibility Project: Cancer Biology 26 |
| Convergent | Test various independent classifiers on one new data | Evaluating the information value of the various feature sets inferred across various studies on one particular set of class labels | Breast cancer biomarkers 15 |
| Divergent | Test 1 classifier on many new data | It allows us to study the generalizability of an inferred feature set on multiple test data It can also be used to check for issues with validation data | Breast cancer biomarkers14 |
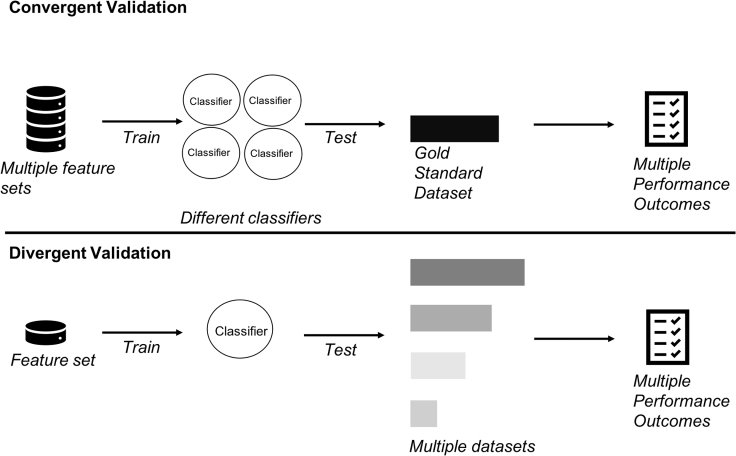
Figure 2.
Two Extensions of the EV: Convergent and Divergent
Convergent validation uses multiple features sets to train multiple models. Each model is, in turn, benchmarked on a gold standard validation dataset. Divergent validation uses a single feature set to train one model followed by repeated challenging with multiple datasets.
There are three key benefits associated with convergent and divergent validations: (1) convergent validation helps us identify better features, leading to better domain-relevant models; (2) divergent validation helps us eliminate domain-irrelevant models because they cannot pass all datasets; and (3) divergent validation helps us identify good EV datasets. These key points are elaborated below.
In convergent validation, you evaluate the information value of multiple feature sets inferred from n datasets, use these to train n models, and then challenge these n models against one validation data. Convergent validation is not a universally adopted procedure (i.e., it is not a mature or regular practice, and not introduced formally to the machine learning field; but it has good potential application.) We propose the idea for convergent validation based on the gene signature profiling work of Venet et al.,15 where they compared the ability of different breast cancer gene signatures (here, these correspond to feature sets) inferred from different studies against one single large high-quality breast cancer dataset, the NKI.27 Venet et al.'s main purpose was to demonstrate that each of the published gene signatures performed poorly in prediction relative to randomly generated signatures. However, the results can also be taken to mean that each of the signatures has a chance to capture an aspect of what is domain relevant. Thus, some signatures have higher information value than others, whereas some had little information value and could not predict the sample classes in the NKI at all. Formally, each time a model is trained on one signature and validated against the NKI counts as one independent validation of that signature. By considering the varied outcomes of each of these independent validations, we may isolate strongly performing signatures from weakly performing ones. In addition, we can identify various levels of commonalities among the most highly performing signatures. This allowed us to isolate a strongly predictive 80-gene breast cancer signature that is essentially inferred from the best performing signatures.6,14 We call this signature the super proliferation set (SPS). Members of this feature set are strongly correlated with breast cancer survival: inclusion of a small number of SPS genes is enough to turn a non-useful signature into a strong predictor and, also, the stronger a published signature the higher its overlap with SPS genes.6,14 These observations suggest that the expression changes of SPS genes are key events in breast cancer survival. In this case study, the feature set garnered from convergent validation has high information value. The procedure is therefore useful for isolating more meaningful explanations in the feature sets, and therefore achieving better interpretability.
In divergent validation, the goal is to understand if a feature set is truly generalizable. The approach involves using a feature set to train a model and then challenging the model repeatedly across multiple validation datasets. Divergent validation is useful for demonstrating that a feature set is universally applicable, and that an initially observed good performance is not due to chance. For example, when SPS was benchmarked across seven new independent datasets, SPS was able to correctly predict the class labels in all seven of these datasets, suggesting that the feature set is domain relevant.6,14 This also suggests that (at least in the context of breast cancer survival), there appears to be "universal" signatures that seem immune to the heterogeneity of test sets. This makes the point that, if one were satisfied that one got a good signature because it worked in one hospital despite failing in others, one would not have identified better and more universal signatures (in the best case) or one would have ended up with a random misleading signature that would likely fail next week (in the worst case).
Technical Considerations for Deploying EV
In this section, we cover four additional technical considerations in deploying EV: (1) generating statistical rigor, (2) considerations for acquiring “valid” external data, (3) how much data are enough, and finally (4) what to do or how to think should the multiple external data exhibit controversial or contradictory results.
Generating Statistical Rigor
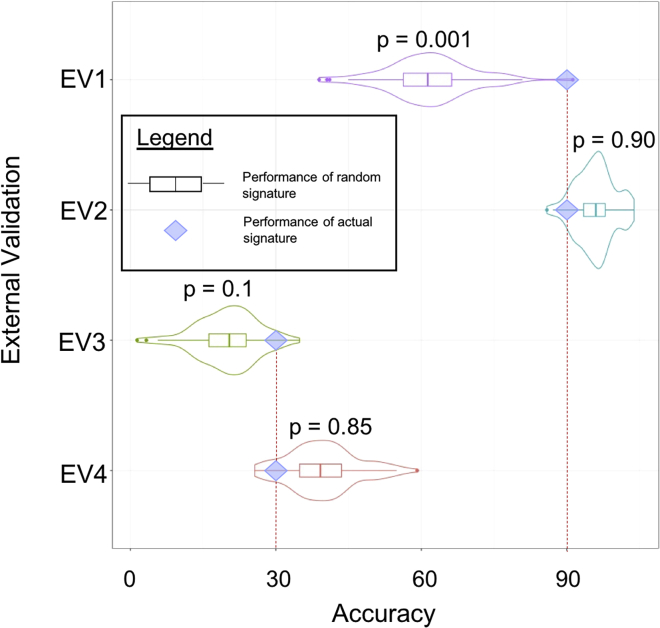
Complementing the above setup, we can make convergent and divergent validations even more meaningful by combining the observed accuracies with empirical and theoretical null distributions as reference points, using the example of Venet et al., in which they compared the predictive performance of each reported signature against randomly generated signatures of the same size.15 The accuracies of these randomly generated signatures constitute an empirical null distribution. This empirical null distribution allows us to understand the information value of a signature, as follows: suppose a validation accuracy of 90% was observed for a model, which is a signature in this example, it is useful to know whether this 90% is meaningful (Figure 3). One way to proxy meaning is to assign a measure of significance to the observed accuracy by checking the number of times a randomly generated signature performs on par or better.28 Suppose only 1 case out of 1,000 random cases equals or exceeds 90%, then the significance value—i.e., the p value—can be represented as a proportion, 1/1,000 or 0.001. This means that the observed accuracy of 90% produced by this feature set is not substitutable by noise. Alternatively, suppose in 900 out of 1,000 cases randomized feature sets perform better. Then, the significance value is 900/1,000, or 0.90. This can mean two things: the feature set on hand is substitutable by noise (and thus has no meaning) or all measured features are essentially class-correlated. Including the significance value allows better evaluation of convergent validation by identifying which feature sets are more likely to be meaningful.
Figure 3.
A Schematic on How to Interpret Observed Validation Accuracies against a Backdrop of Null Accuracies Using p Values
We use four mock external validation (EV1–EV4) scenarios to showcase that high accuracy does not necessarily mean low p values (highly significant outcomes). The violin plots represent null accuracies generated by randomization while the diamond represents the observed accuracy. The p value (p) is the proportion where random accuracies outperforms observed accuracy. Here, EV1 has an observed accuracy of 90%, and this performance is only matched by 1 out of 1,000 random cases. The p value (p) is thus 0.001 (1/1,000) and therefore this feature set is likely meaningful (or high information value). In a second scenario, EV2 has the same observed accuracy as EV1 but this performance is matched by 900 cases out of 1,000 cases. EV2's p is thus 0.90, suggesting that this feature set does not have clear information value. High observed accuracies are not always meaningful: EV3 and EV4 are similar to EV1 and EV2 but with appreciably lower accuracies. The same analysis can also be performed. Low accuracy therefore does not mean low information value.
In divergent validation, a similar tactic can be used as well. Say, a given feature set of size 80 is benchmarked across n validation datasets, then 1,000 random feature sets of the same size are generated and tested across each of these n validation datasets. For each of these 1,000 random feature sets, we count the number of datasets for which it is able to correctly assign the class labels. This forms an empirical null distribution of the number of datasets that a random feature set can perform well on. Then the number of datasets the given feature set performs well on is compared against this empirical null distribution. If a significant fraction, say 5%, of the 1,000 random feature sets perform well on an equal or greater number of datasets than the given feature set, the given feature set is likely substitutable by noise; otherwise, the given feature set is likely not substitutable by noise.
Considerations for Acquiring Valid External Data
It is difficult to establish a priori, based on just the data descriptions, to ascertain if external data are valid. One post-hoc procedure, as discussed later, is to perform divergent validation and check if any of the external data exhibit unusual behaviors.
When training a model, training and EV data should be independently derived. However, just because the training and test data are independently derived does not mean that they make a good training-validation pair. Suppose for some reason the training and validation data are extremely similar, this would guarantee a good validation outcome, yet it informs little on whether a model has been learned intelligently. Such data doppelgängers directly affect a model's performance. For example, Cao and Fullwood5 did a detailed evaluation of existing chromatin interaction prediction systems. The work reveals that the performance of these systems has been overstated because of problems in assessment methodologies when these systems were reported. In particular, these systems were evaluated on validation data that shared a high degree of similarity to training data. While it is good that the problem was discovered, it is surprising that eliminating or minimizing similarity between test and training data is still not a universal practice for model evaluation.
This explanation—that the data involved in the training-validation pair were never checked properly to ensure they were truly dissimilar—also brings in the question of how different do the data need to be and how to check for this.
It is possible to use ordination methods, e.g., principal-component analysis, coupled with scatterplots, to see how the instances are scattered in multi-dimensional space. If the training and validation data points are extremely clustered together, you may expect that the validation results will turn out well. But this does not give us an intuitive or robust way to avoid the doppelgänger effect.
One practice we have observed is to abandon the training-validation distinction altogether and mix the data to derive a larger sample that is hopefully more domain representative, and to test for performance stability instead.29 Alternatively, leveraging on domain-relevant contexts may also work. For example, Cao and Fullwood5 called for more comprehensive and rigorous assessment strategies, based on the particular context of the data being analyzed. In their case, to split training and test data based on individual chromosomes (instead of considering all chromosomes together), and to use different cell types to generate gold standard training-validation pairs, thus establishing better practices/standards in the domain.
It is non-trivial to propose universal measures to guarantee valid EV data. We have to be careful to guard against data doppelgängers. It is also useful to perform multiple rounds of EVs (divergent validation) and to check for consistency. Should any external data behave unusually, it is possible that these data may have issues, and their validity should be called into question.
Determining Sufficient EVs
In our previous simulations with randomly generated feature sets that are then tested against multiple external data, we find that these meaningless feature sets can be used to construct models that somehow work with some but not all external data (an illustration of this is shown in Figure 4). This is expected because we do not expect models constructed from meaningless feature sets to generalize during a divergent validation. However, this assumes that there are sufficient numbers of independent datasets for the divergent validation. In particular, if there are too few independent datasets in the divergent validation, many models constructed from meaningless feature sets may still perform well on all of the independent datasets.
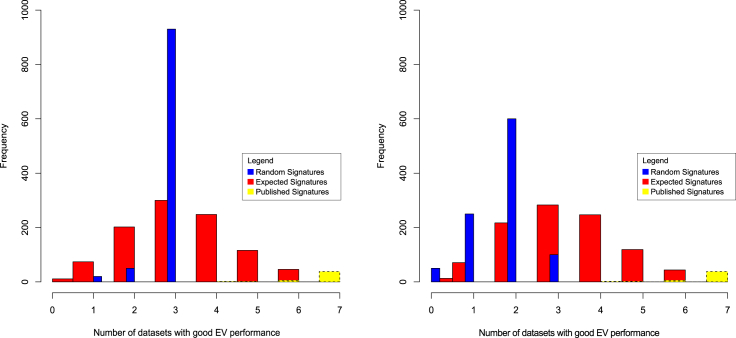
Figure 4.
Divergent Validation Comparing Published Signatures (Yellow), Expected Theoretical Distribution Based on the Binomial Distribution (Red), and Randomized Signatures (Blue) across Seven Datasets
The y axis is a frequency count based on the x axis, which represents the number of validation datasets for which a signature is predictive on. Left illustrates the situation where random signatures do not work well in more than three datasets (although it was later found that it was always the same three datasets that has this issue with random signatures). Right is a repeat experiment using seven new curated datasets with better outcome. The randomized signatures are more evenly distributed although their performance is appreciably lower than purely random (based on the binomial distribution). The fact that it is not always the same datasets that are confounded with random signatures means these curated datasets are more well suited to serve as independent validation data.
Thus, it is important to determine the number of independent datasets needed in a divergent validation to keep models from meaningless feature sets under control. Earlier, we mentioned generating an empirical null distribution using random feature sets on a given test data. This empirical null distribution can be used to infer a bound on the number of independent datasets needed. In particular, the fraction of random feature sets that performs on a par or better than a given feature set on this test data, i.e., the p value, is also the probability of a random feature set performing well on a test data. Therefore, the probability of a random feature set performing well on n independent test data is pn. In other words, if we want no more than say 1% of random feature sets to pass a divergent validation, i.e., pn < 0.01, there should be n > 0.01/log(p) independent datasets. For example, when half the random feature sets can perform well on a test data, i.e., p = 0.5, n = 7 independent test data are needed to keep random feature sets at bay. More generally, binomial distribution is appropriate for constructing the expected behavior for how models trained with random signatures perform in the external divergent validation. Specifically, the probability that a random feature set is able to do well on k independent external datasets follows a binomial distribution prob(k; n, p) = , where n is the total number of external datasets and p is the probability that a random feature set is able to do well in a dataset. Thus the number of random feature sets expected to perform well on k external dataset is given by prob(k; n, p) ∗x, where x is the total number of random feature sets being considered. In our breast cancer example, x = 1,000 random feature sets were generated; about half of the random feature sets was able to perform well on the NKI dataset, thus p = 0.5; this means a random signature has a probability of pn to perform well on n datasets; so n was set to 7 to keep pn < 1%.
As shown in both Figure 4 scenarios, using the binomial distribution as a theoretical model (red bars), it is extremely difficult models trained on random feature sets will work on all seven external data. In the scenario below (Figure 4), we observe that such random models seldom work on more than four EV data. So, four external datasets may be sufficient where external data are in short supply.
What to Do if the EVs Disagree
We may use the information encapsulated in divergent validation (Figure 4) to determine issues in the validation datasets. In the first example (Figure 4, left), across seven validation data, an extremely high blue peak was observed at n = 3, meaning that most randomized feature sets work well on three external data. This deviates from the expected binomial distribution and is suggestive that the seven validation datasets may not be independent of each other and may not be independent of the training data.
Indeed, a closer inspection revealed that it was often the same three datasets contributing to the blue peak in Figure 4 (left). These observations suggest that these validation datasets consistently work with random models. Obviously, such external data should be avoided.
We repeated the divergent validation experiment on a new set of closely curated data (Figure 4, right). This time, the observed performance (blue) is more even, although it still does not match well with the theoretical binomial distribution (red)—suggesting that our estimated value of the probability p is too high. At the same time, it is noteworthy that the models trained on meaningful feature sets worked on all external data (yellow).
These observations lead toward two critical insights: firstly, a meaningful feature set should always work well with any valid external dataset and must exceed the performance of randomized feature sets. Secondly, the application of random feature sets (with zero information value) on divergent validation allows identification of valid external data.
Relevance of EV to New Machine Learning Developments
While convergent and divergent validations have useful properties, they are not panaceas—that is, they will not guarantee that your classifiers are meaningfully generalizable. For example, when most features are correlated with the class labels, EVs will do no better than traditional validation approaches. However, you can detect this issue by adding statistical reference points when conducting the EV (Figure 4).
There are also growing trends in the machine learning area where data from multiple sources are leveraged to learn a model that has the potential to generalize well. For example, federated learning (FL) is a machine learning setup where many devices, acting as nodes in a network, collaboratively train a model under the coordination of a central server.30 This has parallels to convergent validation where each classifier basically has access to only part of the data. However, unlike convergent validation, each node in FL forms part of a large single network-based classifier whose activities are coordinated centrally. Another key difference is control—in convergent validation, unwanted bias is minimized by using a global curated training data, whereas in FL this does not happen (FL is a technological solution focusing on secure data collection). FL and convergent validation also may experience similar issues to local data heterogeneity, such that each node or classifier may have some bias with respect to the true underlying population.
In FL, local models can also be generated instead of a single global model. Here, we speculate an evolution from FL to federated testing, so that data residing in other nodes are used as validation data for divergent validation. This is a tantalizingly applicable scenario for personalized modeling. In this context, there is no global model. Instead, a separate new model is created for all new data (with unknown outcome) using the extended local repository of data until that moment in time. The new model can be dispatched—as a black box with a “good behavior promise”—to federated nodes for divergent validation. In this way, only a summary of the black box's performance on a node is passed back to originating node, and no data from one node is passed to another node.
Other Performance Measures
Although we made our point for EV using the class-prediction accuracy as the primary performance measure, this does not mean that the class-prediction accuracy is a truly useful or objective measure. In fact, it is often argued that accuracy is an uninteresting or limited measure in most applications as it is influenced by class imbalances and other instability issues.4 We do not dispute that. But simply state that it is convenient for use with valid strategies. Moreover, for test sets, class imbalance in itself is not the issue); the issue is class proportion being too different from real life, affecting interpretation of accuracy. If we know what the real life proportion is, we can report a calibrated accuracy that can be more correctly interpreted.4,31
As for whether other measures, such as precision and recall-measure, can objectively prove generalizability where the accuracy cannot, we remain skeptical. There have also been some misleading discussions about Cohen's kappa being resilient against class imbalance issues (https://thedatascientist.com/performance-measures-cohens-kappa-statistic/). Cohen's kappa is a proportion indicating whether a learned model's performance is better than that of a randomly guessing model (which guesses according to the frequency of each class). Cohen's kappa is also sensitive to class imbalance; if class proportion changes, Cohen's kappa can also change dramatically. Say test set 1 has a 20:80 split of positives versus negatives, and sensitivity = specificity = 80%; then kappa = 0.49. Say test set 2 has a 50:50 split and sensitivity = specificity = 80%; then kappa = 0.60. The kappa value does indicate that the performance on test set 2 may be due less to chance than that on test set 1; but it is misleading to think—purely on the basis of kappa—that the performance on test set 2 is higher than on test set 1, as the sensitivity and specificity are actually identical on both test sets.
Sensitivity and specificity together (and thus ROC), since they are not affected by class imbalance, are more robust than kappa. It is in fact possible to compute a confidence interval (CI) for both sensitivity and specificity, to interpret against chance, although these are seldom computed in the machine learning community. For example, the CI of sensitivity and specificity can be computed as a simple asymptotic CI, at least for datasets that are not too small.32 These perspectives can help provide some assurance quality. Although we have not used these ourselves, we think that they may be useful for evaluating EV performance.
Conclusions and Future Directions
Achieving domain-relevant models in machine learning is challenging but necessary for achieving good explainability and generalizability. We may admit better quality models by performing EV better. It is not an error-free process: a good performance in a single independent validation does not guarantee that the model generalizes. EV procedure can be influenced by factors, such as the coverage of the feature set and its correlation with data classes, the presence of technical bias such as batch effects, and whether there is unknown data leakage between the training and validation data.
Designing robust EV technique is highly useful. Because the procedures constitute a core part of machine learning model evaluation, our proposed extensions of the EV are also valid beyond the health and bioscience domains. Given that many datasets and feature sets exist for many modeling problems, it is feasible to perform both convergent and divergent validation methods, as extensions of the traditional independent validation. These two procedural extensions are synergistic, allowing us to take advantage of existing data to develop more robust models: Convergent validation can help to identify domain-relevant feature sets, which provide high explainability, and therefore better interpretable models. But to know which of these models are indeed more likely correct, a single independent validation is insufficient. Divergent validation allows to rigorously test for generalizability. It also allows to detect confounding issues between the training and validation data, as well as between different validation data.
Acknowledgments
W.W.B.G. acknowledges support from the Accelerating Creativity and Excellence (ACE) grant from Nanyang Technological University, Singapore, and an NRF-NSFC grant (grant no. NRF2018NRF-NSFC003SB-006). W.W.B.G. and L.W. acknowledge an MOE Tier 2 (grant no. MOE2019-T2-1-042). L.W. acknowledges support from a Kwan Im Thong Hood Cho Temple Chair Professorship and the National Research Foundation Singapore under its AI Singapore Program (grant no. AISG-GC-2019-002, AISG-100E-2019-027, and AISG-100E-2019-028).
Author Contributions
S.Y.H. and K.P. contributed to the initial drafting of the manuscript and the development of the figures. L.W. and W.W.B.G. conceptualized, supervised, provided critical feedback, and co-wrote the manuscript.
Biographies
About the Authors
Sung Yang Ho is a data scientist in the bio-data science and education laboratory, Nanyang Technological University, focusing on education technology issues.
Kimberly Phua is a student intern in the bio-data science and education laboratory, Nanyang Technological University. Her internship project is concerned with designing better external validation strategies.
Limsoon Wong is Kwan Im Thong Hood Cho Temple chair professor in the School of Computing at the National University of Singapore (NUS). He is also the acting executive director of NUS Graduate School for Integrative Sciences and Engineering. Limsoon is a Fellow of the ACM, named for his contributions to database theory and computational biology.
Wilson Wen Bin Goh is head of the bio-data science and education laboratory, Nanyang Technological University (NTU), program director of NTU’s biomedical data science graduate program, and also assistant chair, School of Biological Sciences, NTU.
Contributor Information
Limsoon Wong, Email: wongls@comp.nus.edu.sg.
Wilson Wen Bin Goh, Email: wilsongoh@ntu.edu.sg.
References
- 1.Hajek P., Michalak K. Feature selection in corporate credit rating prediction. Knowl. Based Syst. 2013;51:72–84. [Google Scholar]
- 2.Tollenaar N., van der Heijden P.G.M. Which method predicts recidivism best? A comparison of statistical, machine learning and data mining predictive models. J. R. Stat. Soc. Ser. A Stat. Soc. 2013;176:565–584. [Google Scholar]
- 3.Ng M.-F., Zhao J., Yan Q., Conduit G.J., Seh Z.W. Predicting the state of charge and health of batteries using data-driven machine learning. Nat. Mach. Intell. 2020;2:161–170. [Google Scholar]
- 4.Ho S.Y., Wong L., Wen Bin Goh W. Avoid oversimplifications in machine learning: going beyond the class-prediction accuracy. Patterns. 2020;1:100025. doi: 10.1016/j.patter.2020.100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao F., Fullwood M.J. Inflated performance measures in enhancer-promoter interaction-prediction methods. Nat. Genet. 2019;51:1196–1198. doi: 10.1038/s41588-019-0434-7. [DOI] [PubMed] [Google Scholar]
- 6.Goh W.W., Wong L. Why breast cancer signatures are no better than random signatures explained. Drug Discov. Today. 2018;23:1818–1823. doi: 10.1016/j.drudis.2018.05.036. [DOI] [PubMed] [Google Scholar]
- 7.Zech J.R., Badgeley M.A., Liu M., Costa A.B., Titano J.J., Oermann E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Button K.S., Ioannidis J.P., Mokrysz C., Nosek B.A., Flint J., Robinson E.S., Munafò M.R. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 9.Krzywinski M., Altman N. Points of significance: power and sample size. Nat. Methods. 2013;10:1139–1140. doi: 10.1038/nmeth.2659. [DOI] [PubMed] [Google Scholar]
- 10.Wang L. Heterogeneous data and big data analytics. Auto. Control Inf. Sci. 2017;3:8–15. [Google Scholar]
- 11.Borodinov N., Neumayer S., Kalinin S.V., Ovchinnikova O.S., Vasudevan R.K., Jesse S. Deep neural networks for understanding noisy data applied to physical property extraction in scanning probe microscopy. NPJ Comput. Mater. 2019;5:25. [Google Scholar]
- 12.Goh W.W., Wong L. Dealing with confounders in omics analysis. Trends Biotechnol. 2018;36:488–498. doi: 10.1016/j.tibtech.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Wong L. Big data and a bewildered lay analyst. Stat. Probab. Lett. 2018;136:73–77. [Google Scholar]
- 14.Goh W.W., Wong L. Turning straw into gold: building robustness into gene signature inference. Drug Discov. Today. 2019;24:31–36. doi: 10.1016/j.drudis.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Venet D., Dumont J.E., Detours V. Most random gene expression signatures are significantly associated with breast cancer outcome. Plos Comput. Biol. 2011;7:e1002240. doi: 10.1371/journal.pcbi.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goh W.W.B., Sng J.C., Yee J.Y., See Y.M., Lee T.S., Wong L., Lee J. Can peripheral blood-derived gene expressions characterize individuals at ultra-high risk for psychosis? Comput. Psychiatry. 2017;1:168–183. doi: 10.1162/CPSY_a_00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blum A.L., Langley P. Selection of relevant features and examples in machine learning. Artif. Intell. 1997;97:245–271. [Google Scholar]
- 18.Little M.A., Varoquaux G., Saeb S., Lonini L., Jayaraman A., Mohr D.C., Kording K.P. Using and understanding cross-validation strategies. Perspectives on Saeb et al. Gigascience. 2017;6:1–6. doi: 10.1093/gigascience/gix020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browne M.W. Cross-validation methods. J. Math. Psychol. 2000;44:108–132. doi: 10.1006/jmps.1999.1279. [DOI] [PubMed] [Google Scholar]
- 20.Tsamardinos I., Greasidou E., Borboudakis G. Bootstrapping the out-of-sample predictions for efficient and accurate cross-validation. Mach Learn. 2018;107:1895–1922. doi: 10.1007/s10994-018-5714-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arlot S., Celisse A. A survey of cross validation procedures for model selection. Stat. Surv. 2009;4:40–79. [Google Scholar]
- 22.Pourhoseingholi M.A., Baghestani A.R., Vahedi M. How to control confounding effects by statistical analysis. Gastroenterol. Hepatol. Bed Bench. 2012;5:79–83. [PMC free article] [PubMed] [Google Scholar]
- 23.Lipton Z.C. The mythos of model interpretability. ACM Queue. 2018;16:31–57. [Google Scholar]
- 24.Kugler L. What happens when big data blunders? Commun. ACM. 2016;59:15–16. [Google Scholar]
- 25.Riley R.D., Ensor J., Snell K.I., Debray T.P., Altman D.G., Moons K.G., Collins G.S. External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ. 2016;353:i3140. doi: 10.1136/bmj.i3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker M., Dolgin E. Cancer reproducibility project releases first results. Nature. 2017;541:269–270. doi: 10.1038/541269a. [DOI] [PubMed] [Google Scholar]
- 27.van 't Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 28.Goh W.W., Wong L. Evaluating feature-selection stability in next-generation proteomics. J. Bioinf Comput. Biol. 2016;14:1650029. doi: 10.1142/S0219720016500293. [DOI] [PubMed] [Google Scholar]
- 29.Tennenholtz G., Zahavy T., Mannor S. Train on validation: squeezing the data lemon. ArXiv. 2018 1802.05846. [Google Scholar]
- 30.Kairouz, P., Brendan McMahan, H., Avent, B., Bellet, A., Bennis, M., Bhagoji, A.N., Bonawitz, K., Charles, Z., Cormode, G., Cummings, R., et al. (2019) Advances and Open Problems in Federated Learning. arXiv:1912.04977. https://www.researchgate.net/publication/337904104_Advances_and_Open_Problems_in_Federated_Learning.
- 31.Riley R.D., Ahmed I., Debray T.P., Willis B.H., Noordzij J.P., Higgins J.P., Deeks J.J. Summarising and validating test accuracy results across multiple studies for use in clinical practice. Stat. Med. 2015;34:2081–2103. doi: 10.1002/sim.6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bland M. An Introduction to Medical Statistics. Oxford University Press; 2015. https://www.researchgate.net/publication/256824854_Martin_Bland_An_Introduction_to_Medical_Statistics_3rd_ed [Google Scholar]