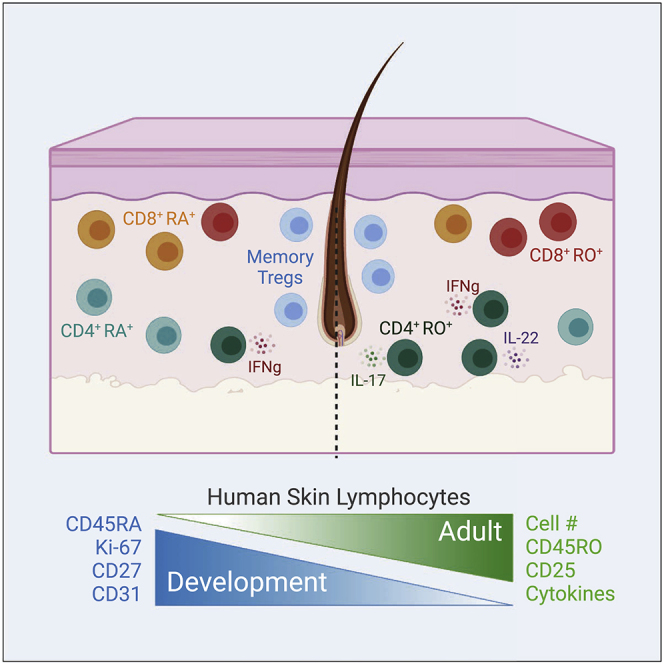
Summary
Lymphocytes in barrier tissues play critical roles in host defense and homeostasis. These cells take up residence in tissues during defined developmental windows, when they may demonstrate distinct phenotypes and functions. Here, we utilized mass and flow cytometry to elucidate early features of human skin immunity. Although most conventional αβ T (Tconv) cells in fetal skin have a naive, proliferative phenotype, a subset of CD4+ Tconv and CD8+ cells demonstrate memory-like features and a propensity for interferon (IFN)γ production. Skin regulatory T cells dynamically accumulate over the second trimester in temporal and regional association with hair follicle development. These fetal skin regulatory T cells (Tregs) demonstrate an effector memory phenotype while differing from their adult counterparts in expression of key effector molecules. Thus, we identify features of prenatal skin lymphocytes that may have key implications for understanding antigen and allergen encounters in utero and in infancy.
Keywords: lymphocytes, human skin, fetal, skin development, hair follicle development, Tregs, memory T cells
Graphical Abstract
Highlights
CyTOF reveals a complex lymphocyte landscape in developing human skin
Developing skin contains CD45RO+ conventional T cells with propensity to produce IFNγ
Regulatory T cells (Tregs) in skin before birth display effector memory properties
Skin Tregs increase in conjunction with initial hair follicle morphogenesis
Dhariwala et al. utilize mass and flow cytometry to elucidate distinguishing features of lymphocytes in developing fetal skin. The early presence of memory-like subsets among conventional and regulatory T cells may have key implications for understanding nascent cutaneous immune function.
Introduction
Each square centimeter of adult human skin contains approximately one million lymphocytes, comprised predominantly of CD4+ and CD8+ alpha beta (αβ) T cells.1 These cells help defend us against cutaneous pathogens and malignancy and also facilitate key homeostatic tissue functions, such as wound healing and hair follicle cycling.2,3 Conversely, they can play a central pathogenic role in common inflammatory and allergic skin diseases. As compared to our relatively nuanced understanding of lymphocytes in adult human skin, little is known about the phenotype or functional capacity of these cells during early life. Deciphering this initial immune landscape has the potential to critically inform our understanding of the role of lymphocytes in normal human skin development as well as in immune-mediated skin diseases that begin early in life.
Human skin begins as a single-cell epithelium during embryogenesis and evolves over the first trimester of fetal life into a stratified epidermis with an overlying periderm. Epidermal differentiation and development of skin appendages occur from 15–20 weeks gestation, followed by acquisition of a functional stratum corneum between 20–24 weeks.4 In utero maturation of skin architecture is accompanied by parallel seeding of the tissue by immune cells. Antigen-presenting cells, found in the skin of 9-week-old embryos, are perhaps the first skin-resident population.5 These are followed by T cells, which exit the fetal thymus around 11–14 weeks,6,7 and are detectable in skin by the second trimester (i.e., around 17–18 weeks).8,9 For many years, studies of human fetal tissues had largely employed immuno-histochemistry or flow cytometry to examine particular immune cell types of interest.8,9 More recently, “human cell atlas” studies have used single-cell RNA sequencing to broadly assess cells in human tissues, including skin, across various ages.10,11 However, we still lack a holistic understanding of the immune landscape in developing fetal skin, especially as relates to the specific identities and phenotypes of fetal skin lymphocytes.
The fetal immune system is not merely a miniature version of that in adults. Rather, it exhibits cellular phenotypes and functions specifically adapted to the needs of the developing fetus and future neonate.12 Perhaps most striking is a propensity toward adaptive immune tolerance, attributable to both lymphocyte-intrinsic and myeloid cell-dependent features,13,14 which help limit excess inflammation to maternal, self, or other in utero antigens. Recent studies of the fetal intestine have identified unique populations of cytokine-producing classical and innate-like effector T cells. These are thought to promote healthy gut development in utero, but may also modulate risk of inflammatory diseases in neonates.15,16
Here, we use a combination of flow and mass cytometry to elucidate the composition and phenotype of lymphocytes in human fetal skin with a focus on the second trimester as a particularly dynamic period. Our findings offer insight into features of fetal skin immunity, including the in utero presence of memory-like conventional T cells and an intimate relationship between accumulation of Tregs and skin morphogenesis. These findings may have important implications for cutaneous immune responses to self and foreign antigens in utero as well as in human infancy.
Results
Mass Cytometry Elucidates Major Immune Cell Subsets in Human Fetal Skin
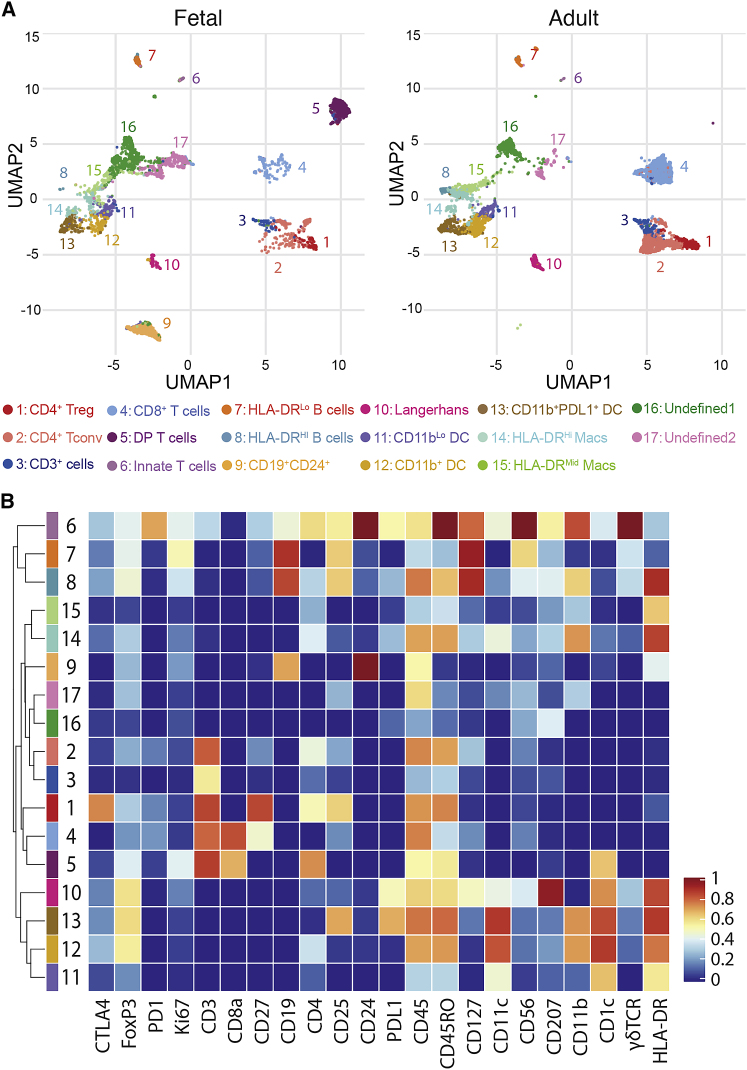
To obtain a broad understanding of the immune cells residing in human fetal skin, we performed mass cytometry (CyTOF) using a 22-antibody panel designed to include major lymphocyte and myeloid markers. 23 weeks gestation was chosen as a mature late second trimester fetal time point, and 5 fetal torso skin samples were processed alongside 5 site-matched adult skin controls. Unbiased clustering of live-CD45+ cells was performed based on relative expression of all markers, and iterative analyses demonstrated that binning into 17 clusters captured the major phenotypic differences present (Figure 1A). Identity assignments for each cluster on uniform manifold approximation and projection (UMAP) plots was performed based on relative expression of key markers as described below (Figures 1B, S1A, and S1B). Although most clusters were present in both fetal and adult skin, a few were found only in fetal tissue (Figures S1A and S1B).
Figure 1.
Mass Cytometry Reveals Immune Cell Heterogeneity between Fetal and Adult Skin
Twenty-three weeks gestational age (g.a.) fetal torso skin along with healthy adult skin torso samples were analyzed in parallel for 22 markers using mass cytometry.
(A) Uniform manifold approximation and projection (UMAP) plots of viable CD45+ cells in fetal versus adult skin, cells colored by cluster identity, and plots annotated with cluster numbers and assigned identities.
(B) Heatmap demonstrating relative expression by cluster of 22 markers in panel, inclusive of both fetal and adult cells.
CD4+ (clusters 1 and 2) and CD8+ (cluster 4) T cells represented the bulk of lymphocytes present in fetal skin and were further expanded at the adult time point. Cluster 3 was CD3+ but negative for CD8 and only marginally positive for CD4. These might represent T cells for which lineage-defining markers were cleaved during tissue digestion, but could also include double-negative mucosal-associated invariant T cells (MAIT).17 Because MAIT markers such as CD161, TRAV1-2, or MR1 tetramer binding were not assessed, a more definitive assignment could not be made. Two of the five fetal skin samples contained a population of CD3+ cells co-expressing both CD4+ and CD8+ (cluster 5). However, flow cytometry performed on an additional 21 fetal skin samples failed to corroborate this as a consistent fetal skin T cell population (Figures S1C and S1D). Cluster 6 expressed CD3, CD56, and γδTCR, likely representing a mixed population containing both γδ T cells and natural killer (NK) T cells, possibly also NK cells or innate lymphoid cells, with the latter being less likely given the positive CD3 expression. These innate type T cells constituted a small fraction of lymphocytes at either age, as has been shown previously for adult skin.18,19
CD19+HLA-DRLo (cluster 7) and CD19+HLA-DRHi (cluster 8) B cells were found at low frequency irrespective of age. In 3 fetal skin samples, however, a third CD19+HLA-DRInt subset was detected that co-expressed CD24+ (cluster 9), a marker typically seen on immature B cells and their precursors.20 Flow cytometry on an additional 4 fetal samples confirmed the presence of this population in the range of 3%–7% of live-CD45 cells (Figures S1E and S1F).
Clusters 10–15, found in both fetal and adult skin demonstrated moderate to high expression of HLA-DR, CD11b, or CD11c, consistent with a myeloid origin. Among these, Langerhans cells (cluster 10) were identifiable based on their expression of CD207. Clusters 11–13 expressed CD11c indicative of classical or monocyte-derived dendritic cells.21,22 Clusters 14 and 15 were low or negative for CD11c but expressed moderate to high HLA-DR and CD11b, suggestive of macrophages.23, 24, 25 Definitive assignment of clusters 16 and 17, which were enriched in fetal as compared to adult skin, was precluded by the absence of subset-defining markers. Staining of separate fetal samples with a myeloid-focused antibody panel (Figure S1G) revealed that, consistent with previous reports,13 macrophages are relatively overrepresented among CD45+ cells in fetal versus adult skin. We did not see significant differences in the prevalence other myeloid populations in the limited number of samples we were able to examine (Figures S1H–S1J).
Fetal Skin Preferentially Contains CD4+ and CD8+ Tconv with a Naive, Proliferative Phenotype
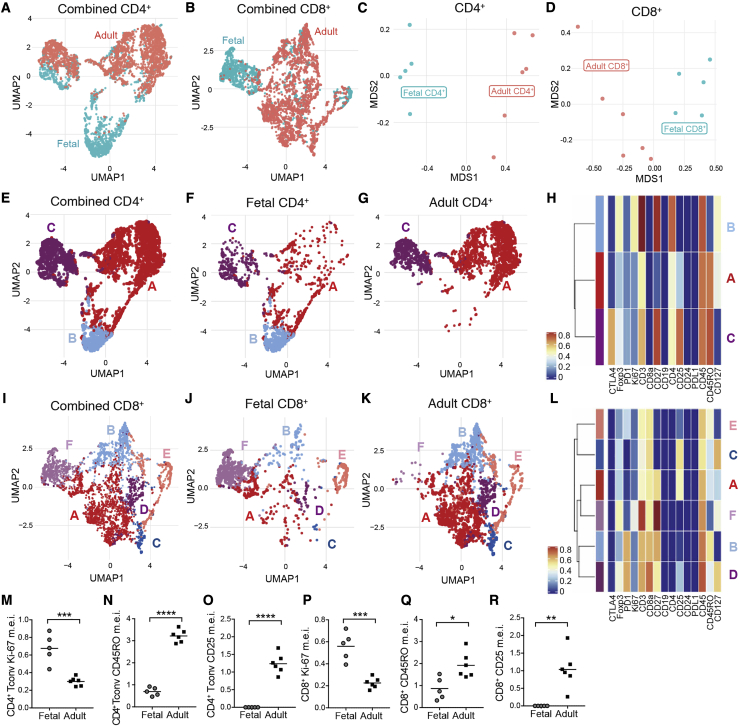
As CD4+ and CD8+ αβ T cells represented the largest lymphocyte population in utero, we next sought a more in-depth understanding of features that might distinguish these populations in fetal versus adult skin. To ensure all CD4+ and CD8+ single-positive T cells were captured in this analysis, we returned to the raw CyTOF data and identified these populations based on a traditional gating strategy with minimum cut-offs for lineage marker expression (Figure S2A). UMAP analysis of CD4+ and CD8+ T cells each demonstrated significant age-dependent clustering (Figures 2A and 2B), which was also reflected in accompanying principal-component analysis (PCA) (Figures 2C and 2D).
Figure 2.
Conventional αβ T Cells in Fetal Skin Largely Demonstrate a Naive, Proliferative Phenotype
Live, CD4+, and CD8+ single-positive cells from 23 weeks fetal skin and adult skin processed for CyTOF were identified by surface markers and analyzed as follows.
(A–D) UMAP plots of combined fetal and adult (A) CD4+ and (B) CD8+ T cells colored by skin age. Principal component analysis (PCA) plots demonstrating the distribution of all (C) CD4+ and (D) CD8+ T cells from each individual sample by age.
(E) UMAP plots of CD4+ T cells from fetal and adult skin, combined. Cells are labeled and colored by cluster, with clusters A and B constituting conventional CD4+ T cells and cluster C representing Tregs.
(F and G) Analogous UMAP plots containing only (F) fetal or (G) adult CD4+ T cells.
(H) Heatmap demonstrating relative expression of key markers by CD4+ clusters A, B, and C.
(I–K) combined and separated UMAP plots of fetal and adult skin CD8+ T cells, colored by cluster.
(L) Heatmap demonstrating relative expression of key markers by each CD8+ cluster.
(M–R) Median expression intensity (m.e.i.) of Ki-67 (M and P), CD45RO (N and Q), and CD25 (O and R) for fetal versus adult CD4+ Tconv and CD8+ T cells as revealed by mass cytometric analyses. Each point in (C), (D), and (M)–(R) represents data from an individual donor. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Sub-clustering of fetal and adult CD4+ T cells revealed three major populations (Figures 2E–2G). Cluster C expressed Foxp3, CD25, and CTLA4 consistent with a regulatory T cell (Treg) phenotype, whereas clusters A and B were identifiable as two subsets of conventional CD4+ T cells (Tconv) (Figures 2H and S2B–S2D). CD4+ Tconv cluster A, which was much more prevalent in adult skin, had greater expression of CD25 and CD45RO. In contrast, CD4+ Tconv cluster B, found exclusively in fetal skin, demonstrated comparatively higher levels of Ki-67, a marker of recent cell cycling, CD3, CD4, Foxp3, and CD27, which is expressed often on naive T cells (Figures 2H, S2E, and S2F).26
For CD8+ T cells, 6 clusters were readily distinguishable (Figures 2I–2K). Clusters A and B, the two largest CD8+ subsets in adult skin, were consistently enriched in adult as compared to fetal samples. Cluster A was denoted by high CD25 and moderate CD45RO expression, and cluster B by high CD45RO and PD1. Cluster C, notable for expression of CD25 and CD127, and cluster D, which expressed high CD127, PD1, and moderate CD25, were each enriched in only one adult skin sample. Cluster E, second only to cluster B in its expression of CD45RO, was a minor population found in equal proportion between fetal and adult skin. Finally, cluster F, the most abundant subset in fetal skin, demonstrated heightened expression of Ki-67, CD3, CD27, and Foxp3 (Figures 2L, S2G, and S2H).
Thus, the largest subsets of both CD4+ Tconv and CD8+ T cells in fetal skin demonstrated a largely naive, proliferative status, whereas those dominating in adult skin were enriched for markers of memory and activation. This general dichotomy was also reflected in statistically significant differences in the mean expression intensity of key markers, especially CD25, CD45RO, and Ki-67, between CD4+ Tconv and CD8+ T cells in fetal and adult skin (Figures 2M–2R and S2).
Subsets of Fetal Skin CD4+ and CD8+ Tconv Are CD45RO+ and Demonstrate Enriched Capacity for IFNγ Production
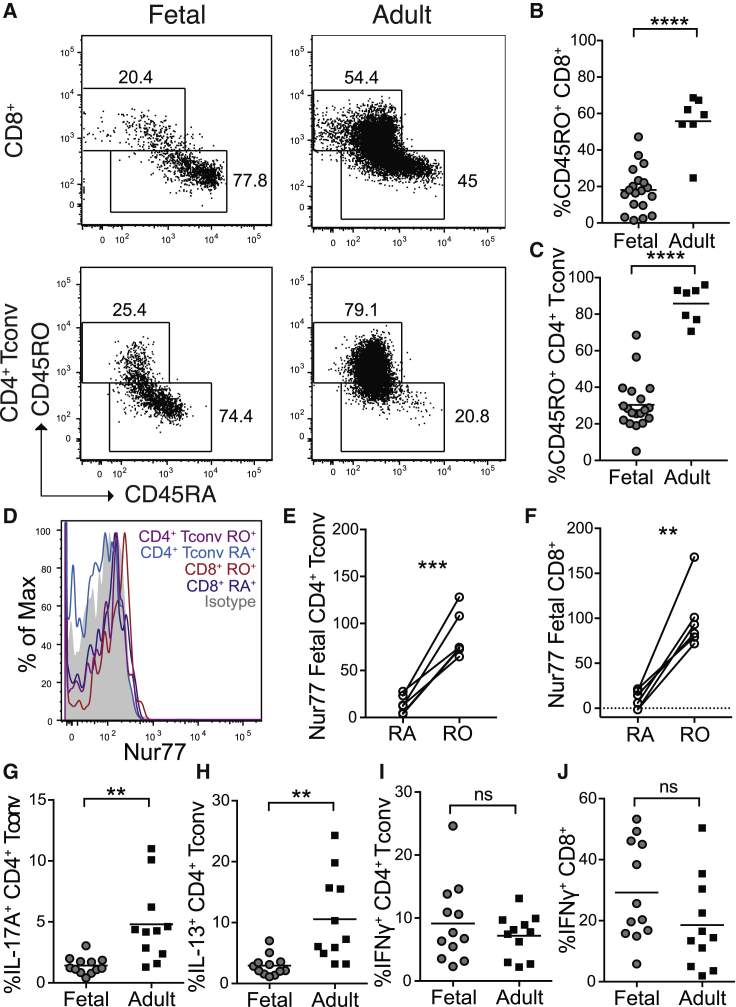
Notably, our CyTOF analysis did reveal cells in fetal skin belonging to subsets of CD4+ Tconv (cluster A) and CD8+ T cells (clusters A, B, and E) with a more activated or memory-like phenotype (Figures 2H, 2L, and S2E–S2H). To corroborate this observation and better quantify the proportion of fetal CD4+ Tconv and CD8+ cells with a naive versus memory phenotype, we performed flow cytometry on a larger number of fetal skin samples including staining for CD45RA and CD45RO, markers respectively found on antigen-naive and antigen-experienced lymphocytes. Strikingly, up to 30% of CD4+ Tconv and slightly fewer CD8+ T cells in fetal skin were CD45RO+ (Figures 3A–3C). Higher levels of Nur77, a specific reporter of T cell receptor (TCR) signaling,27, 28, 29 were detected in fetal CD45RO+ versus CD45RA+ cells, further suggesting antigen stimulation in this putative memory population (Figures 3D–3F).
Figure 3.
Subsets of Fetal Skin CD4+ Tconv and CD8+ T Cells Display a Memory Phenotype and Demonstrate Capacity for IFNγ Production
Cells were isolated from second trimester fetal skin (scalp and/or torso) as well as adult (torso) skin and analyzed by flow cytometry.
(A) Representative plots demonstrating CD45RO expression by fetal CD8+ T cells (gated on live CD3+CD8+CD4neg) and CD4+ Tconv (gated on live CD3+CD4+CD8negFoxp3negCD25lo).
(B and C) Percentage of CD45RO+ (B) CD8+ T cells and (C) CD4+ Tconv in fetal versus adult skin.
(D–F) Representative histogram (D) and quantification (E and F) of Nur77 MFI on fetal skin CD45RO+ versus CD45RA+ CD4+ Tconv and CD8+ cells.
(G–J) Percentage of CD4+ Tconv producing (G) IL-17A, (H) IL-13, and (I) IFNγ after PMA/ionomycin re-stimulation, and (J) percentage of IFNγ-producing CD8+ T cells.
Each point in (B)–(G) represents data from an individual tissue sample; for some fetal samples data from scalp and torso skin from the same fetal donor are included as separate points. ns, not significant (p > 0.05); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
To further characterize the functional potential of fetal skin T cells, we performed ex vivo re-stimulation with PMA and ionomycin followed by intracellular cytokine staining. Memory as compared to naive T cells are known to preferentially produce tumor necrosis factor alpha (TNF-⍺).30 Consistent with their largely naive status, CD4+ Tconv and CD8+ T cells in fetal skin expressed substantially less TNF-⍺ than those in adult skin (Figures S3A and S3B). Production of interleukin (IL)-22, IL-17a, and IL-13 by fetal versus adult CD4+ Tconv was likewise reduced (Figures 3G, 3H, and S3C). In contrast, the percentage of IL-2 producing CD4+ Tconv and IFNγ producing CD4+ Tconv or CD8+ T cells did not differ statistically by age when evaluating these populations as a whole (Figures 3I, 3J, and S3D). To try to account for the different proportions of CD45RO+ versus CD45RA+ cells found in fetal versus adult skin, we normalized production of each cytokine to that of TNF-⍺ on a per-sample basis. This revealed enriched IFNγ production among fetal versus adult skin T cells (Figures S3E and S3F), whereas production of IL-2, IL-13, IL-17, and IL-22 was equivalent or reduced (Figures S3G–S3J). For samples on which such information was available, we also normalized the percentage of IFNγ producing CD4+ and CD8+ T cells to CD45RO+ frequency in these populations and compared IFNγ production by fetal CD45RA+ versus CD45RO+ cells directly (Figures S3K–S3N). This further supported our observation that fetal skin Tconv cells with a memory-like phenotype demonstrate enhanced propensity for IFNγ production.
Fetal Skin Tregs Display Properties of Effector Memory Tregs
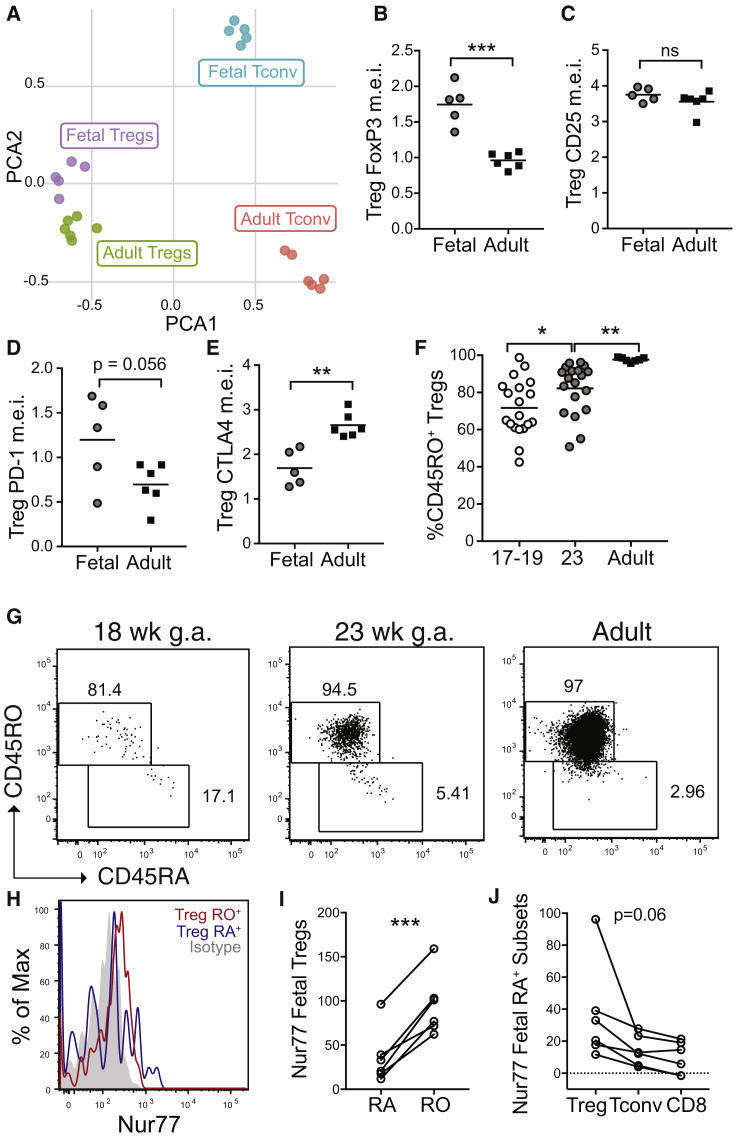
The fetal immune system is poised for immune regulation.31 We therefore returned to our CyTOF analysis to carefully examine the phenotype of Tregs in fetal skin. As noted earlier, Tregs from both fetal and adult skin fell into CD4+ cluster C (Figures 2A–2C and S2A–S2C). Although skin Tregs also segregated by age upon PCA analysis, age-dependent differences were strikingly less than for CD4+ Tconv (Figure 4A). Dissecting this further, we found several markers distinguishing fetal versus adult skin Tregs. Akin to other cell types, Tregs in fetal skin expressed higher levels of Ki-67, CD27, and CD3 (Figures S4A–S4C). CD45RO expression was high among fetal Tregs versus CD4+ T conv cells but lagged slightly behind that of adult Tregs (Figures S4D and S4E). Notably, fetal skin Tregs demonstrated higher levels of the lineage-defining transcription factor, Foxp3 (Figure 4B) and equivalent amounts of the high-affinity IL-2 receptor alpha chain, CD25 (Figure 4C) as compared to adult skin Tregs. With regard to other important Treg activation markers, fetal skin Tregs demonstrated a non-significant trend toward higher PD-1 levels (Figure 4D), but lower amounts of total CTLA4 (Figure 4E). Thus, while differing somewhat in their relative expression of certain markers, Tregs in fetal skin largely demonstrate an effector memory Treg phenotype, akin to Tregs in adult skin.
Figure 4.
Tregs in Human Fetal Skin Demonstrate an Effector Memory Phenotype
Twenty-three weeks g.a. fetal torso skin and healthy adult torso skin samples were analyzed for 22 markers using mass cytometry.
(A) PCA plot of Tregs (cluster C, Figure 2) and CD4+ Tconv (clusters A & B, Figure 2) from 23 week fetal skin versus adult skin based on relative expression of key markers as assessed by mass cytometry.
(B–E) Graphs showing median expression intensity (m.e.i.) of (B) Foxp3, (C) CD25, (D) PD-1, and (E) CTLA4 by mass cytometry on fetal versus adult skin Tregs.
(F and G) Cells were isolated from 17–23 weeks g.a. fetal skin (scalp and/or torso) as well as adult (torso) skin and analyzed by flow cytometry. (F) Percentage of CD45RO+ Tregs in skin by age and (G) accompanying representative flow plots.
(H) Representative histogram and (i) quantification of Nur77 MFI on fetal skin CD45RO+ versus CD45RA+ Tregs.
(J) Nur77 in fetal skin CD45RA+ CD8+, CD4+ Tconv, and Tregs.
Each point in (A)–(E) represents data from an individual donor. Points in (F), (I), and (J) point represent data from an individual tissue sample; for some fetal samples data from scalp and torso skin from the same fetal donor are included as separate points. ns, not significant (p > 0.05); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Tregs leaving the fetal thymus are CD45ROneg.32 Thus, we hypothesized these cells were acquiring their memory phenotype in skin following exposure to self-antigen. To examine this possibility, we performed flow cytometry on additional samples, now including earlier second trimester fetal skin for comparison. In contrast to CD4+ Tconv and CD8+ cells that demonstrated a consistently low percentage of CD45RO+ cells independent of fetal age (Figures S4E and S4F), the proportion of CD45RO+ Tregs progressively increased during the second trimester (Figures 4F–4G). Analogous to conventional T cell subsets, CD45RO+ fetal skin Tregs demonstrated higher Nur77 levels than CD45RA+ Tregs (Figures 4H and 4I). Among all CD45RA+ cells, however, Nur77 expression trended highest among Tregs as compared to CD4+ Tconv or CD8+ counterparts (Figure 4J). Thus, the population of fetal skin Tregs evolves over the second trimester with progressive acquisition of a memory phenotype, perhaps in part driven by detection of cognate antigens.
T Cells Accumulate in Fetal Skin during the Second Trimester via Both Continued Thymic Egress and Local Proliferation
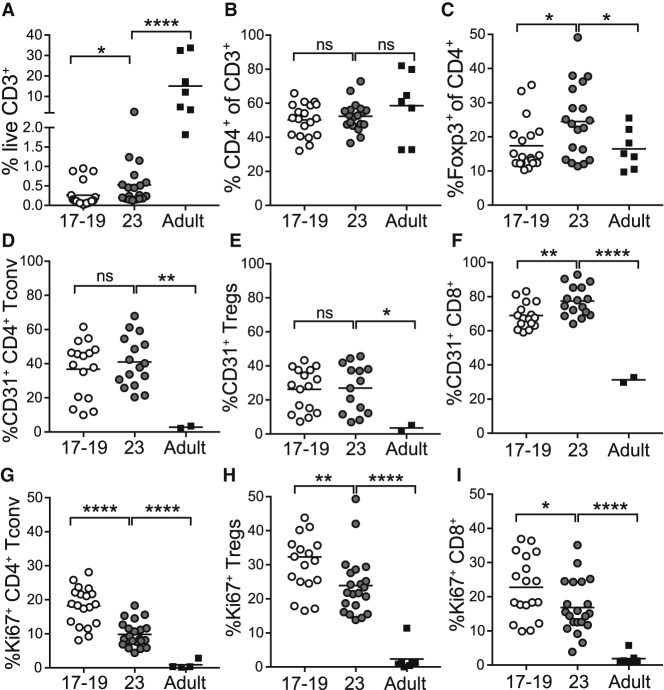
Having delineated features of late second trimester fetal skin lymphocytes, we next sought to better define dynamics of their tissue accumulation. Although CD3+ T cells represented a comparatively small percentage of total events in fetal versus adult skin, they were readily identifiable by flow cytometry in fetal samples as young as 17 weeks and substantially expanded by 23 weeks (Figures 5A and S5A). Although the proportion of CD4+ T cells within the CD3+ T cell compartment was stable across both fetal and adult time points (Figure 5B and S5B), the average percentage of Foxp3+ regulatory T cells (Tregs) among CD4+ cells increased significantly between 17 and 23 weeks gestation and was higher at this later fetal time point than in adult skin (Figures 5C and S5C).
Figure 5.
Lymphocytes Progressively Accumulate in Fetal Skin via a Combination of Thymic Egress and Local Proliferation
Cells were isolated from 17–23 weeks g.a. fetal skin (scalp and/or torso) as well as adult (torso) skin and analyzed by flow cytometry.
(A–C) Percentage of live CD3+ lymphocytes (A), percentage of CD4+ T cells (B), and percentage of Foxp3+ Tregs (C) by age.
(D–F) Percentage of CD31+ cells among CD4+ Tconv cells (D), Tregs (E), and CD8+ T cells (F) by age.
(G–I) Percentage Ki-67+ cells among CD4+ Tconv cells (G), Tregs (H), and CD8+ T (I) cells by age.
Points represent data from an individual tissue sample; for some fetal samples data from scalp and torso skin from the same fetal donor are included as separate points. ns, not significant (p > 0.05); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
To elucidate the contribution of ongoing cellular influx on progressive T cell accumulation in fetal skin, we examined expression of CD31, a cell surface adhesion molecule found in high levels on recent thymic emigrants.33 As compared to adult skin, CD31+ T cells were substantially enriched in fetal skin at both 17–18 and 23 weeks (Figure S5D). Whereas less than 10% of adult CD4+ Tconv cells were CD31+, this marker was seen on up to 60% of fetal skin CD4+ Tconv cells (Figures 5D and 5E). As reported previously, CD8+ cells demonstrated comparative higher CD31 expression at all fetal ages relative to adult (Figure 5F).34 Of note, CD31 expression was significantly higher on CD45ROneg versus CD45RO+ CD4+ Tconv and CD8+ cells in fetal skin, but CD45RO status did not correlate with CD31 expression among fetal skin Tregs (Figures S5F–S5H).
To determine the extent to which in situ proliferation also contributes to rising numbers of fetal skin T cells during the second trimester, we examined Ki-67 expression by flow cytometry (Figure S5E). Corroborating our CyTOF data, Ki-67 was expressed by less than 5% of adult skin T cells. In contrast, it was found on up to 40% of T cells in fetal skin at 17–18 weeks and slightly fewer at 23 weeks (Figures 5G–5I). Whereas CD45RO expression was associated with only slightly higher Ki-67 levels among fetal skin CD4+ Tconv and CD8+ cells, CD45RO+ fetal skin Tregs expressed twice as much Ki-67 compared to their CD45ROneg counterparts (Figures S5I–S5K). The proportion of Ki-67+ cells among skin Tregs was also higher at both fetal time points as compared to CD4+ Tconv or CD8+ T cells (Figure 5H). Thus, although recent thymic emigrants constitute a substantial fraction of T cells in second trimester fetal skin, cell proliferation also contributes to their progressive increase during this period, especially among CD45RO+ cells.
Enrichment in Fetal Skin Tregs Coincides with Initial Hair Follicle Morphogenesis
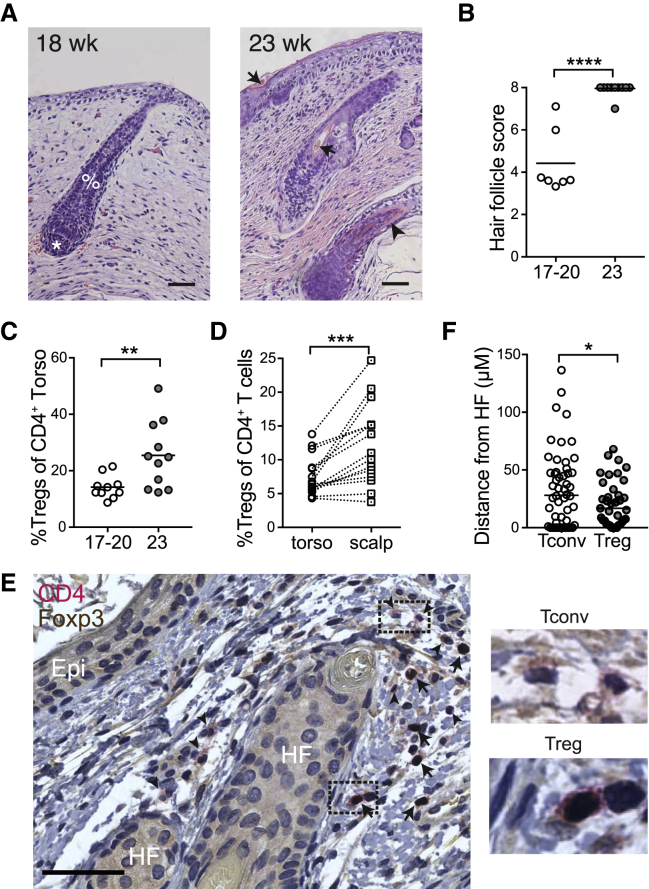
The observation that fetal skin Tregs increase in percentage during the second trimester (Figure 5C) and display unique dynamics of CD31 and Ki-67 expression (Figures 5E and 5H) prompted us to explore the role of tissue-intrinsic factors in their progressive accumulation. Because Tregs in adult human skin localize preferentially around hair follicles and are more abundant in hair-dense skin sites,35,36 we investigated the relationship between the appearance of fetal Tregs and hair follicle development. We first examined histologic sections from our fetal skin torso samples and scored them by stage of hair follicle development.37,38 Consistent with published reports,38 we found that hair follicle development progressed significantly between 20 and 23 weeks gestation (Figures 6A and 6B), which coincided with the increased percentage of Tregs (Figure 6C). Next, we took advantage of the fact that fetal hair follicles mature in a cephalocaudal pattern.39 Using paired skin samples from the scalp and torso of fetuses aged 20 weeks or less, when torso hair follicles are still relatively immature versus those on the scalp, we assessed the density of Tregs irrespective of fetal age. Indeed, for a given fetus, Treg percentages were higher in scalp as compared to torso skin (Figure 6D). Immunohistochemical staining of fetal skin for Foxp3 and CD4 further confirmed the presence of Tregs in close proximity to hair follicles (Figures 6E and 6F). These results demonstrate that skin Tregs increase contemporaneously with fetal hair follicle development in utero.
Figure 6.
Treg Accumulation in Human Fetal Skin Coincides Spatiotemporally with Second Trimester Hair Follicle Development
Fetal skin samples ranging from 17–23 weeks g.a. were processed for histology.
(A) Representative images of tissue sections stained with H&E from 18 and 23 weeks g.a. skin. %, developing fetal hair peg with beginning inner root sheath; ∗, dermal papilla; arrows, hair shafts; arrowhead, outer root sheath. Scale bar, 50 μm.
(B) Hair follicles were scored and graphed with each data point representing cumulative scores from an individual donor.
(C) Percentage Tregs in fetal skin at gestational age 17–20 weeks versus 23 weeks as assessed by flow cytometry.
(D) Percentage Tregs in paired torso versus scalp skin from fetal donors of less than 20 weeks g.a. Each matched set of data points originates from a different fetal skin donor.
(E) Multiplex immunohistochemistry for CD4 (red chromogen, membranous staining) and Foxp3 (brown chromogen, nuclear staining) of human fetal skin. Tregs are indicated by positive brown nuclear and red membranous staining (arrows). Tconv are indicated by negative nuclear staining and positive red membranous staining (arrowheads). Images to the right of the main image represent high-power views of the corresponding dashed boxes. Epi, epidermis; HF, hair follicle. Scale bar, 50 μm.
(F) Graphical results of quantitative image analysis of the distance of Treg or Tconv to the nearest hair follicle epithelial surface. Data represent enumeration of 20 high-power fields (400×) per slide. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Discussion
Here, we provide a holistic view of the cellular immune composition of human fetal skin, with a specific focus on how lymphocyte subsets differ from those present in the adult tissue. Conventional Foxp3negCD4+ and CD8+ T cells in fetal skin are largely distinguished from their adult counterparts by high expression of Ki-67 and CD27 and low expression of CD25 and CD45RO, indicative of a naive phenotype. A subset of CD45RO+ fetal Tconv cells, however, display heightened TCR signaling and IFNγ production consistent with a memory phenotype. Although differing in expression of certain activation molecules, fetal skin Tregs closely resemble their adult counterparts and express markers identifying them as effector memory Tregs. All fetal skin T lymphocytes, but especially CD45ROneg Tconv, are marked by elevated CD31 expression suggesting enrichment for recent thymic emigrants. Local proliferation suggested by Ki-67 is preferentially seen among CD45RO+ cells, in particular Tregs, which also express more Nur77 expression consistent with antigen-dependent TCR signaling. Accumulation of Tregs in fetal skin occurs contemporaneously with hair follicle development, and preferentially at sites of higher hair follicle density within a given fetus, suggesting an early role for hair follicle biology in skin Treg residency. Together, these results illuminate a memory signature among lymphocytes in human fetal skin, which has implications for our understanding of late prenatal and early postnatal immune responses and their role in skin health and disease.
Here, we leveraged CyTOF to simultaneously capture the broad phenotype and prevalence of immune cells within fetal skin. Although differing somewhat in relative abundance, the major leukocyte clusters found in adult skin were also present during early development. We were intrigued to note that certain populations were significantly enriched in or unique to the fetal period. A population of CD24+CD19+ cells, possibly representing B cell precursors as described in human bone marrow,15 were consistently found in fetal specimens. Although B cells have been previously shown to reside in human fetal skin,10 CD24+CD19+ cells were found in even greater abundance. CD4+CD8+ T cells were a minor population in the majority of fetal samples we examined, but constituted a substantial lymphocyte subset in two. Double-positive T cells represent an immature precursor state during lymphocyte thymic development. However, they can be found in blood during lymphocytosis and in skin lesions of atopic dermatitis, systemic sclerosis, and graft versus host disease.40 Analogous to reports of increased double-positive cells T cells in the peripheral lymphoid organs of fetal rodents,41 we speculate that heightened lymphopoiesis during human fetal development may somehow facilitate sporadic escape of these cells to the periphery where they either die or further differentiate into the subsets we see in mature skin. Further work is needed to explore the extent to which these cells are consistently found in other human fetal tissues and at what point during development they “disappear.”
In these studies, we chose to more carefully elucidate the phenotype of lymphocytes, specifically T cells, residing in human fetal skin. Non-conventional T cells represented a small proportion at either age, unfortunately limited cell numbers prevented us from further sub-setting cluster 6 that expressed γδ as well as NK T cell markers. Among CD4+ Tconv and CD8+ T cells, wide segregation by age was primarily driven by differential expression of Ki-67, CD45RO, CD25, and CD27. Preferential Ki-67 expression by fetal T cells indicates a heightened proliferative activity, perhaps influenced by the relatively lymphopenic fetal environment or by recognition of self, maternal, or microbial antigens, as indicated by their Nur77 expression. Reduced proportions of CD45RO+ cells among fetal CD4+ Tconv and CD8+ T cells is consistent with a relative dearth of foreign antigens in utero and corroborates earlier reports of CD45RA expression by most fetal skin CD3+ cells.8,9 Low CD25 and high CD27 expression by fetal cells likely reflects their naive status.26 However, CD25 levels might also indicate limiting IL-2 conditions in fetal tissue.42 Surprisingly, we found that up to 20% of CD4+ Tconv or CD8+ T cells in second trimester fetal skin do express CD45RO. CD45RO+ T cells have also been found in the fetal intestines15 and constitute up to 5% of CD4+ T cells in cord blood from healthy term deliveries.6,43 It is still debated whether the “memory” phenotype of these cells reflects priming to in utero antigens44,45 or acquisition of memory markers secondary to cytokine exposure in the lymphopenic fetal environment.46 Further work is clearly required to define the ontogeny of the CD45RO+ CD4+ Tconv and CD8+ subsets in fetal skin.
To elucidate the distinct functional capacity of fetal versus adult skin Tconv cells, we performed ex vivo re-stimulation and found relatively enriched capacity for IFNγ production as compared to IL-13, IL-17A, and IL-22. Based on higher CD27 levels among fetal T cells and reports that this TNF superfamily co-stimulatory receptor has a cell-intrinsic capacity to promote Th1 at the expense of Th17 responses,47 it is tempting to speculate that CD27 expression might influence the functional capacity of fetal skin T cells. IFNγ- and TNF-α-producing CD45RO+CD4+ Tconv cells have also been identified in the fetal intestine, where they are generally thought to reflect and support healthy gut development.15,48,49 However, they have also been implicated in heightened risk of necrotizing enterocolitis and gastroschisis.15,16 Premature infants also demonstrate different propensities toward diseases in which skin immune cells play a central role, including increased susceptibility to infections by coagulase-negative staphylococci50 and reduced risk of atopic dermatitis (AD).51 Th17 cells are critical players in the normal response to these commensals.52 Our finding of reduced IL-17 production by T cells in second trimester fetal skin may be another key factor, alongside skin barrier immaturity, contributing to prematurity-associated infection risk. Conversely, it is intriguing to consider that heightened propensity toward IFNγ in skin may contribute to the lower incidence of AD in preterm infants. Pediatric AD is characterized by Th2- and Th17-mediated inflammation,53,54 and T cells from the blood of term infants and young children have been shown to display a propensity toward production of type 2 cytokines.55,56 IFNγ levels in cord blood have also been shown to be conversely associated with atopy at 1 year of age.57 If early life exposure to microbial and environmental antigens takes place in an IFNγ-rich environment, this may steer the response away from an atopic diathesis.
Tregs are relatively well-studied in the fetal immune system but have not been carefully examined in skin. We found Tregs in fetal skin to be quite similar to their adult counterparts in demonstrating an effector memory profile.58 Modest age-related differences in key markers, however, may indicate distinct functional capacity of skin Tregs by age. As has been shown for Tregs in pediatric tissues such as the spleen and gut,59 fetal skin Tregs demonstrate higher expression of the master transcriptional regulator Foxp3. PD-1, a co-inhibitory receptor that can mark Tregs with heightened suppressive function as well as stability, was also elevated early.60,61 Conversely, CTLA-4, another key Treg effector molecule, was reduced in fetal Tregs. Although these data provide some early clues, additional work is merited to tease apart the relative suppressive capacity and regulatory mechanisms used by Tregs in young versus mature human tissues.
Treg enrichment in the fetal immune system is thought to be critical for promoting tolerance to self and maternal antigens50 and is supported by multiple mechanisms.13,32,62 This penchant for tolerance extends beyond the fetal period into early childhood, as indicated by enrichment of Tregs in pediatric versus adult tissues59,63 and clinical studies supporting a time-limited early window to promote oral tolerance to food allergens.64 Here, we demonstrate that Tregs progressively increase as a proportion of T cells in fetal skin during the second trimester. More specifically, skin Tregs increase in parallel with terminal hair follicle development, and a relationship between follicle stage and tissue Treg density is preserved at different skin sites within a given fetus. This differs from intestinal Tregs that constitute no more than 10% of CD4+ T cells at all fetal time points studied15 and suggests that tissue-intrinsic factors, not just progressive thymic output, help mediate Treg accumulation. Hair follicle-derived chemokines have been implicated in recruitment of various T cell subsets.65 In neonatal mice, Ccl20 production by hair follicles facilitates postnatal skin Treg accumulation.66 Analogous chemokine-dependent processes in human fetal skin may help recruit recent thymic emigrants. Alternatively, or in addition, an increasing burden of self-antigens contained in terminally differentiated hair follicles may stimulate local Treg induction or proliferation.
In summary, our findings provide insight into cutaneous immune development during the second trimester of fetal life. This marks the start of a critical window extending into early childhood during which specific exposures are thought to influence predisposition to allergic disease. Understanding the developmental trajectory of the skin immune system is of particular relevance given that percutaneous allergen exposure is considered an early catalyst of the atopic march. If barriers to tissue access can be surmounted, an important next step will be to define how skin immune cell populations evolve following the transition out of the womb and how this compares between premature and term neonates. Pairing this with investigations to define the immunologic impact of relevant environmental exposures (i.e., commensal microbes or cutaneous allergens) will inform our understanding of healthy development and hopefully better position us to prevent or treat immune-mediated disease in early life.
Limitations of Study
This study has focused in particular on features of αβ T lymphocytes in fetal versus adult skin. Although a broad survey of other leukocytes was performed, our CyTOF panel did not have sufficient markers to enable deep phenotyping of all cell subsets. For example, measuring expression of CD38 on CD24+CD19+ cells in fetal skin would help in evaluating whether this population reflects a transitional B cell population with regulatory potential.67 Similarly, several subsets of myeloid cells were identified in fetal and adult skin, however, future deeper characterization of dendritic cell, monocyte, and macrophage subsets in developing skin is warranted. Our ability to comment on innate-type T cells (i.e., MAIT, γδ, and NK T cells) in fetal skin was also limited by existing hurdles in combining tetramers with CyTOF and by the total number of fetal skin leukocytes we had to analyze. Based on the lower overall leukocyte abundance in fetal as compared to adult skin, increased numbers of total cells from fetal tissue should be processed to ensure optimal fetal leukocyte numbers for downstream analysis.
Although our data demonstrate robust differences in Treg, CD4+ Tconv, and CD8+ T cell phenotypes between fetal and adult human skin, other functional studies remain to be pursued. Potential differences in the suppressive capacity of fetal versus adult skin Tregs, suggested by their different activation marker profile, could be tested in vitro using traditional suppression assays or in vivo, using humanized mice. Similarly, antigen-specificity of CD45RO+ T cells in human skin might be explicitly tested to look for reactivity to microbial, self, or other antigens. Finally, we were able to demonstrate that Treg accumulation in human skin is synchronous with hair follicle development. However, the functional basis for this temporal association remains to be elucidated, namely is there a specific chemokine signature that drives this enrichment or do other factors contribute. Several of these questions are currently under investigation and have benefitted from the data reported in this manuscript.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Flow Cytometry Antibodies | ||
| Anti-human CD3-BV711 (clone: SK7)-80ug/ml | Biolegend | Cat# 344838; RRID:AB_2565827 |
| Anti-human CD4-PE -Texas Red(clone: MHCD0417) | Life Technologies/ Thermo Fisher Scientific | Cat# MHCD0414; RRID: 10371766 |
| Anti-human CD8-BV650 (clone: SK1) 100ug/ml | Biolegend | Cat# 344730; RRID:AB_2564510 |
| Anti-human CD25-PECy7 (clone: M-A251) | BD Biosciences | Cat# 557741; RRID:AB_396847 |
| Anti-human CD27- PerCP-e710 (clone: 0323) | eBioscience | Cat# 46-0279-42; RRID:AB_1834391 |
| Anti-human CD31- APC-e780 (clone: WM59) | eBioscience | Cat# 47-0319-42; RRID:AB_10730582 |
| Anti-human CD45-APC (clone: HI30) | eBioscience | Cat# 47-0459-41; RRID:AB_1944369 |
| Anti-human CD45RA-BV605(clone: HI100) 25ug/ml | Biolegend | Cat# 304134; RRID:AB_2563814 |
| Anti-human CD45RO-FITC (clone: UCHL1) | eBioscience | Cat# 11045742; RRID:AB_10596999 |
| Anti-human IL2-PE 25ug/ml (MQ1-17H12) | Biolegend | Cat# 500307; RRID:AB_315094 |
| Anti-human CD152/CTLA4-PE (clone: 14D3) | Thermo Fisher Scientific | Cat# 12-1529-42; RRID:AB_10805626 |
| Anti-human CD278/ICOS-APC (clone: ISA-3) | eBiosceince | Cat# 17-9948-42; RRID:AB_10598209 |
| Anti-human FoxP3-e450 (clone: PCH101) | eBioscience | Cat# 48477642; RRID:AB_1834364 |
| Anti-human IFNγ− A700(clone: 4S.B3) | Biolegend | Cat# 502520; RRID:AB_528921 |
| Anti-human IL-13 –FITC (clone: 85BRD) | Thermo Fisher Scientific | Cat# 11-7136-42; RRID: 2572515 |
| Anti-human IL-17A-APC ef-780 (clone: eBio64CAP17) | eBioscience | Cat# 47-7179-41; RRID:AB_11043274 |
| Anti-human CD141-APC (Clone:BDCA-3) | Milteny Biotech | Cat# 130-113-314; RRID:AB_2733313 |
| Anti-human EpCAM-BV711 (9CY) | Biolegend | Cat# 324240; RRID:AB_2734307 |
| Anti-human CD11c-BV605 (B-Ly6) | BD BioSciences | Cat# 563929; RRID:AB_2744276 |
| Anti-human Ki-67- PerCyp Cy5.5 (clone: B56) | BD Biosciences | Cat# 561284; RRID:AB_10611574 |
| Anti-human IL-22-PE (clone: 142928) | R & D Systems | Cat#IC7821P; RRID:AB_495011 |
| Anti-human TNFα−PE-Cy7 (clone: Mab11) | BD BioSciences | Cat#557647; RRID: AB_396764 |
| Anti-human CD14-PE-200ug/ml (MFE2) | Biolegend | Cat# 301806; RRID:AB_314188 |
| Anti-human CD16-BV421 (3G8) | Biolegend | Cat# 302038; RRID:AB_2561578 |
| Anti-human CD1c-PECy7 100ug/ml (clone: L161) | Biolegend | Cat# 331516; RRID:AB_2275574 |
| Anti-human CD19-APC (SJ25C1) | Tonbo Bioscience | Cat# 20-0198-T100; RRID: 2621683 |
| Anti-human CD24-BV711 (ML5) | BD Biosciences | Cat# 563371; RRID:AB_2738164 |
| Anti-human HLA-DR –PercP Cy5.5(clone: Tu36) Anti-human HLA-DR-BV 650 (L243) |
Biolegend Biolegend |
Cat# 361608; RRID:AB_2563198 Cat# 307649; RRID:AB_2562544 |
| Anti-human CD1a-AF700 (H1149) | Biolegend | Cat# 300120; RRID:AB_528764 |
| Anti-mouse Nur77-PE (12.14) | eBioscience | Cat# 12-5965-82; RRID:AB_1257210 |
| Mass Cytometry Antibodies | ||
| Anti-human CD45 (clone: H130, Metal: 89Y) | Fluidigm | Cat# 3089003B; RRID: AB_2661851 |
| Anti-human CD19 (clone: HIB19, Metal: 142Nd) | Fluidigm | Cat# 3142001B; RRID: AB_2651155 |
| Anti-human CD11b (clone: ICRF44, Metal: 144Nd) | Fluidigm | Cat# 3144001B; RRID: AB_2714152 |
| Anti-human CD4 (clone: IRPA-T4, Metal: 145Nd) | Fluidigm | Cat# 3145001B; RRID: AB_2661789 |
| Anti-human CD11c (clone: BU15, Metal: 147Sm) | Fluidigm | Cat# 3147008B; RRID: AB_2687850 |
| Anti-human CD25 (clone: 2A3, Metal: 149Sm) | Fluidigm | Cat# 3149010B; RRID: AB_2756416 |
| Anti-human γδTCR (clone: 11F2, Metal: 152Sm) | Fluidigm | Cat# 3152008B; RRID: AB_2687643 |
| Anti-human CD8α (clone: RPA-T8, Metal: 53Eu) | Biolegend | Cat# 301002; RRID: AB_2661818 |
| Anti-human CD24 (clone: ML5, Metal: 154Sm) | Biolegend | Cat# 311102; RRID:AB_10580644 |
| Anti-human CD56 (clone: B159, Metal: 155Gd) | Fluidigm | Cat# 3155008B; RRID: AB_2861412 |
| Anti-human CD27 (clone: L128, Metal: 158Gd) | Fluidigm | Cat# 3158010B; RRID: AB_2858231 |
| Anti-human PDL1 (clone: 29E.2A3, Metal: 159Tb) | Fluidigm | Cat# 3159029B; RRID:AB_2861413 |
| Anti-human CD152/CTLA4 (clone: 14D3, Metal: 161Dy) | Fluidigm | Cat# 3161004B; RRID: AB_2687649 |
| Anti-human FoxP3 (clone: PCH101, Metal: 162Dy) | Fluidigm | Cat# 3162011A; RRID: AB_2687650 |
| Anti-human CD1c-PECy7 (clone: L161, Metal: 164Dy) | Biolegend | Cat# 331502; RRID: AB_2661820 |
| Anti-human PD1 (clone: EH12.2H7, Metal: 166Er) | Biolegend | Cat# 329902; RRID: AB_940488 |
| Anti-human Ki-67 (clone: Ki67, Metal: 168Er) | Biolegend | Cat# 3168001B; RRID: AB_2810856 |
| Anti-human CD3 (clone: UCHT1, Metal: 170Er) | Fluidigm | Cat# 3170001B; RRID: AB_2811085 |
| Anti-human CD45RO (clone: UCHL1, Metal: 173Yb) | Fluidigm | Cat# 3173016D; RRID: AB_2811052 |
| Anti-human CD207/Langerin (clone: 4C7, Metal: 175Lu) | Fluidigm | Cat# 3175016B; RRID:AB_2861414 |
| Anti-human CD127 (clone: A019D5, Metal: 176Yb) | Fluidigm | Cat# 3176004B; RRID: AB_2687863 |
| Immunohistochemistry Antibodies | ||
| Anti-human CD4 (clone: 41B2) | Agilent | Cat# IS64930-2; RRID:AB_2861415 |
| Anti-human FoxP3 (clone: 236A/E7) | Abcam | Cat# ab20034; RRID: AB_445284 |
| Biological Samples | ||
| De-identified healthy adult and fetal skin tissue | UCSF-affiliated hospitals | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase from Clostridium histolyticum, Type IV | Sigma-Aldrich | Cat# lS00418 |
| DNase | Sigma-Aldrich | Cat# DN25 |
| Fetal bovine serum | Fisher scientific | Cat# SH3054103 |
| Ghost Dye™ Violet 510 Live/Dead Stain | Tonbo Biosciences | Cat#13-0870-T100 |
| FoxP3-staining buffer kit | Invitrogen by Thermofisher | Cat# 00-5523 |
| TONBO cell stimulation cocktail | Tonbo Biosciences | Cat# TNB-4975 |
| Cisplatin | Sigma-Aldrich | Cat# P4394 |
| Intercalator-IR | Fluidigm | Cat# P4394 |
| Cell-ID 20-Plex Pd Barcoding Kit | Fluidigm | Cat# 201060 |
| BOND Polymer Refine Detection Kit | Leica Biosystems | Cat# DS9800 |
| Deposited Data | ||
| Raw data | This paper | Flow repository: ID# FR-FCM-Z3YD (url: https://flowrepository.org/) |
| Software and Algorithms | ||
| GraphPad Prism | GraphPad Software, Inc | https://www.graphpad.com:443/scientific-software/prism/ |
| FlowJo v10.5.0 | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo |
| R Statistical Computing Software | The R Foundation; https://www.r-project.org/ | https://www.r-project.org/ |
| CyTOF Workflow | Nowicka et al., 2017 | https://www.bioconductor.org/packages/release/workflows/html/cytofWorkflow.html |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tiffany Scharschmidt (Tiffany.Scharschmidt@ucsf.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The mass cytometry dataset generated in this study is deposited and publicly available at Flow repository: ID# FR-FCM-Z3YD (url: https://flowrepository.org/).
Method Details
Human skin specimens
Human fetal tissues (17-23 weeks gestational age) were obtained from Zuckerberg San Francisco General Hospital from terminations of pregnancy after maternal written informed consent with approval from the UCSF Research Protection program (Data S1). Samples were excluded in the case of (1) known maternal infection, (2) intrauterine fetal demise, and/or (3) known or suspected chromosomal abnormality. Normal adult human skin was obtained from patients at UCSF undergoing surgeries in which healthy skin was discarded as a routine procedure (Data S2). Skin from both genders was included for fetal and adult tissue. In all instances, samples were obtained in de-identified manner and constituted non-human subjects research. The study was conducted in accordance with the Declaration of Helsinki principles.
Skin tissue processing
Skin samples were stored at 4°C in a sterile container with PBS and gauze until the time of digestion. Subcutaneous fat was removed, and skin was minced finely with dissection scissors and mixed in a 6-well plate with 3 mL of digestion buffer consisting of 0.8 mg/ml Collagenase Type 4 (4188; Worthington), 0.02 mg/ml DNase (DN25-1G; Sigma-Aldrich), 10% FBS, 1% HEPES, and 1% penicillin/streptavidin in RPMI medium. Samples were incubated overnight in 5% CO2 and harvested with wash buffer (2% FBS, 1% penicillin/streptavidin in RPMI medium), then filtered twice through a 100-μm filter, centrifuged, and counted.
Flow cytometry
Single cell suspensions were stained for surface antigens and a live/dead marker (Ghost Dye Violet 510, Tonbo Biosciences) in FACS buffer (PBS with 2% fetal bovine serum) for 30 min at 4°C. They were then fixed and permeabilized using the FOXP3-staining buffer kit (eBioscience) before staining for intracellular markers. To measure intracellular cytokine production, cells suspensions were stimulated for 4 hours ex vivo using a commercial cell stimulation cocktail (Tonbo Biosciences, catalog no. TNB-4975) prior to staining for flow cytometry as above. Antibodies were purchased from Biolegend, BD Biosciences, eBiosciences and R&D as listed in the Key Resources Table. For longitudinal experiments comparing mean fluorescent intensities, voltages were standardized using SPHERO Rainbow calibration particles (BD Biosciences). Samples were run on a Fortessa (BD Biosciences) in the UCSF Flow Cytometry Core. FlowJo software (FlowJo LLC) was used to analyze flow cytometry data.
Mass Cytometry (CyTOF)
Single cell suspensions were incubated for 1 minute with 25 μM Cisplatin (Sigma-Aldrich, P4394) to allow subsequent cell viability measurement, then fixed in 1.5% paraformaldehyde (Electron Microscopy Sciences) and frozen down in the presence of 10% di-methyl sulfoxide (DMSO) and 10% bovine serum albumin (BSA) at stored at −80°C for subsequent staining. Individual vials were thawed and 2x106 cells were barcoded using the Cell-ID 20-Plex Pd Barcoding Kit (Fluidigm, product number 201060) for 15 minutes at room temperature. Palladium based barcoding was used as this has been reported to not only enable simultaneous staining of up to 20 different samples but also enable doublet discrimination.68 All the barcoded samples were then combined into a single tube and cells were stained with metal conjugated antibodies, purchased either from BD Biosciences or Fluidigm as listed in the Key Resources Table. Surface antigens were stained in cell staining media (0.5% BSA, 0.02% Sodium azide in PBS) for 30 minutes at room temperature with gentle agitation. For intracellular staining, cells were permeabilized using the FOXP3-staining buffer kit (eBioscience) and then incubated for another 30 minutes at room temperature with intracellular antibodies in the presence of the FOXP3-staining kit permeabilization buffer. Cells were stored overnight at 4°C in a buffer containing iridium and then run on a Helios CyTOF system (Fluidigm) in the UCSF, Parnassus flow cytometry core facility. FCS files were first processed in FlowJo (FlowJO LLC) to gate on populations of interest, e.g., CD45+ (singlet live-CD45+), CD4+ T cells (CD3+CD4+CD8neg CD56neg), CD8+ (CD3+CD8+CD4negCD56neg). Further analysis was performed in R using the HDCyto and CATALYST packages found on Bioconductor. Generation of graphs was drawn from the CytofWorkflow pipeline similarly found on Bioconductor.69 Multidimensional Scaling analogous to PCA analysis based on median expression intensity of antibody markers was used as a first pass at dimensionality reduction and visualization of relatedness of samples. Uniform Manifold Approximation and Projection (UMAP), an improved method of dimensionality reduction based on greater mathematical rigor than t-SNE, was used to visualize relationships among the single cells on a 2-D plane.70 Unlike t-SNE, distances between groups on UMAP plots reflect their degree of relatedness capturing the global relations. Clustering into subpopulations of T cells was performed71 taking into account all markers in the panel except CD207, CD56, CD1c, CD11c, γδTCR, CD11b, and HLA-DR. Clustering was performed using FlowSOM72 and CosnensusClusteringPlus73 for metaclustering. Cluster C in the CD4+ T cells (Figure 2), identified as Tregs based on their expression profile, was separated from the Tconv groups using R scripts to filter data points by cluster.
Tissue Processing for Histopathology
For histologic analysis of fetal skin, the tissue was fixed in 10% formalin for 24 hours and submitted as research specimens to the UCSF Dermatopathology service, where they were embedded in paraffin, cut into 4 μM sections, and stained with stained with hematoxylin and eosin to reveal endogenous cellular structures. Staging of hair follicle morphogenesis was performed by the investigators in consultation with a dermatopathologist according to previously published criteria,37 which grade developing hair follicles on a 0 to 8 scale based on histopathologic features of the developing hair germ, hair peg and eventual hair follicle.
Immunohistochemistry
Formalin-fixed paraffin-embedded sections of 4 μm thickness underwent multiplex staining with antibodies specific for Foxp3 (1:800, Abcam), and CD4 (predilute, Dako) on an automated immunostainer (Bond, Leica Biosystems).
Whole Slide Scanning and Digital Image Analysis
Slides were scanned at 400x resolution with an Aperio AT2 scanner (Leica Biosystems) using a 20x/0.75NA Plan Apo objective with a 2x optical mag changer to generate digital images. Image resolution was 400x: 0.25 μM/pixel. Quantitative analysis was performed on images generated from sections stained with the dual antibody combination of CD4 (red chromogen) and Foxp3 (brown chromogen). Image files were viewed with Aperio ImageScope software (Leica Biosystems). The “ruler” tool was used to manually quantify the distance from the leading edge of either Treg (brown nuclear positive/red membrane positive) or Tconv (brown nuclear negative/red membrane positive) to the nearest hair follicle epithelial surface. Twenty random high-power fields (400x) were analyzed per slide.
Quantification and Statistical analysis
Significance was determined using either two-tailed unpaired Student’s t test (for measuring differences between separate groups), or one-way analysis of variance (ANOVA) (for multiple comparisons) in GraphPad Prism Software. Data points in all graphs represent individual skin samples. P values correlate with symbols as follows: ns, not significant (p > 0.05); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Acknowledgments
We appreciate support from lab members of Trevor Burt, M.D., Melissa Ng, and Ventura Mendoza, who assisted in acquisition of fetal skin samples, and UCSF surgeons, Isaac Neuhaus, Scott Hansen, Hobart Harris, and Ester Kim who provided surgically discarded adult skin samples. Flow cytometry and CyTOF data were generated in the UCSF Parnassus Flow Cytometry Core, which is supported by the Diabetes Research Center (DRC) (NIHP30 DK063720). We thank Matt Spitzer for input and expertise in development of CyTOF methods for human skin. We appreciate Tim McCalmont’s consultation on histologic examination of fetal hair follicles. The graphical abstract was created using BioRender. This work was funded by Doris Duke Clinician-Scientist Development Award 2017070. T.C.S. receives salary support from NIH (5K08AR68409), although fetal work was not funded through this mechanism.
Author Contributions
M.O.D. and T.C.S. drafted the manuscript and figures. M.O.D., A.W., K.S.V., E.G.L., J.N.C., S.C., K.T., M.M.L., and S.F. performed experiments and analyzed data. M.L.P. contributed materials. M.O.D., M.M.L., M.P., J.F.A., M.D.R., and T.C.S. oversaw experimental design. D.K. and M.O.D. performed bioinformatic analysis.
Declaration of Interests
M.D.R. is a founder and consultant for TRex Bio and Sitryx Bio and receives research funding from Abbvie, TRex, and LEO Pharma. K.T. is currently an employee of Amgen. E.G.L. is currently an employee of SentiBio. All other authors declare no competing interests.
Published: November 3, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100132.
Supplemental Information
References
- 1.Clark R.A., Chong B., Mirchandani N., Brinster N.K., Yamanaka K.-I., Dowgiert R.K., Kupper T.S. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- 2.Ali N., Zirak B., Rodriguez R.S., Pauli M.L., Truong H.-A., Lai K., Ahn R., Corbin K., Lowe M.M., Scharschmidt T.C. Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell. 2017;169:1119–1129. doi: 10.1016/j.cell.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison O.J., Linehan J.L., Shih H.-Y., Bouladoux N., Han S.-J., Smelkinson M., Sen S.K., Byrd A.L., Enamorado M., Yao C. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 2019;363:eaat6280. doi: 10.1126/science.aat6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoath S.B., Maibach H.I. CRC Press; 2003. Neonatal Skin: Struture and Function. [Google Scholar]
- 5.Schuster C., Vaculik C., Fiala C., Meindl S., Brandt O., Imhof M., Stingl G., Eppel W., Elbe-Bürger A. HLA-DR+ leukocytes acquire CD1 antigens in embryonic and fetal human skin and contain functional antigen-presenting cells. J. Exp. Med. 2009;206:169–181. doi: 10.1084/jem.20081747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhivaki D., Lo-Man R. In utero development of memory T cells. Semin. Immunopathol. 2017;39:585–592. doi: 10.1007/s00281-017-0650-0. [DOI] [PubMed] [Google Scholar]
- 7.Spencer J., Dillon S.B., Isaacson P.G., MacDonald T.T. T cell subclasses in fetal human ileum. Clin. Exp. Immunol. 1986;65:553–558. [PMC free article] [PubMed] [Google Scholar]
- 8.Di Nuzzo S., Pavanello P., Masotti A., Giordano G., De Panfilis G. Densities, distribution and phenotypic expression of T cells in human fetal skin. Arch. Dermatol. Res. 2009;301:753–755. doi: 10.1007/s00403-009-0943-9. [DOI] [PubMed] [Google Scholar]
- 9.Schuster C., Vaculik C., Prior M., Fiala C., Mildner M., Eppel W., Stingl G., Elbe-Bürger A. Phenotypic characterization of leukocytes in prenatal human dermis. J. Invest. Dermatol. 2012;132:2581–2592. doi: 10.1038/jid.2012.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popescu D.-M., Botting R.A., Stephenson E., Green K., Webb S., Jardine L., Calderbank E.F., Polanski K., Goh I., Efremova M. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W. Construction of a human cell landscape at single-cell level. Nature. 2020;581:303–309. doi: 10.1038/s41586-020-2157-4. [DOI] [PubMed] [Google Scholar]
- 12.Adkins B., Leclerc C., Marshall-Clarke S. Neonatal adaptive immunity comes of age. Nat. Rev. Immunol. 2004;4:553–564. doi: 10.1038/nri1394. [DOI] [PubMed] [Google Scholar]
- 13.McGovern N., Shin A., Low G., Low D., Duan K., Yao L.J., Msallam R., Low I., Shadan N.B., Sumatoh H.R. Human fetal dendritic cells promote prenatal T-cell immune suppression through arginase-2. Nature. 2017;546:662–666. doi: 10.1038/nature22795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mold J.E., McCune J.M. Immunological tolerance during fetal development: from mouse to man. Adv. Immunol. 2012;115:73–111. doi: 10.1016/B978-0-12-394299-9.00003-5. [DOI] [PubMed] [Google Scholar]
- 15.Schreurs R.R.C.E., Baumdick M.E., Sagebiel A.F., Kaufmann M., Mokry M., Klarenbeek P.L., Schaltenberg N., Steinert F.L., van Rijn J.M., Drewniak A. Human Fetal TNF-α-Cytokine-Producing CD4+ Effector Memory T Cells Promote Intestinal Development and Mediate Inflammation Early in Life. Immunity. 2019;50:462–476. doi: 10.1016/j.immuni.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Halkias J., Rackaityte E., Hillman S.L., Aran D., Mendoza V.F., Marshall L.R., MacKenzie T.C., Burt T.D. CD161 contributes to prenatal immune suppression of IFNγ-producing PLZF+ T cells. J. Clin. Invest. 2019;129:3562–3577. doi: 10.1172/JCI125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Constantinides M.G., Link V.M., Tamoutounour S., Wong A.C., Perez-Chaparro P.J., Han S.-J., Chen Y.E., Li K., Farhat S., Weckel A. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019;366:eaax6624. doi: 10.1126/science.aax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bos J.D., Teunissen M.B., Cairo I., Krieg S.R., Kapsenberg M.L., Das P.K., Borst J. T-cell receptor gamma delta bearing cells in normal human skin. J. Invest. Dermatol. 1990;94:37–42. doi: 10.1111/1523-1747.ep12873333. [DOI] [PubMed] [Google Scholar]
- 19.Gober M.D., Fishelevich R., Zhao Y., Unutmaz D., Gaspari A.A. Human natural killer T cells infiltrate into the skin at elicitation sites of allergic contact dermatitis. J. Invest. Dermatol. 2008;128:1460–1469. doi: 10.1038/sj.jid.5701199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duperray C., Boiron J.M., Boucheix C., Cantaloube J.F., Lavabre-Bertrand T., Attal M., Brochier J., Maraninchi D., Bataille R., Klein B. The CD24 antigen discriminates between pre-B and B cells in human bone marrow. J. Immunol. 1990;145:3678–3683. [PubMed] [Google Scholar]
- 21.Collin M., Bigley V. Human dendritic cell subsets: an update. Immunology. 2018;154:3–20. doi: 10.1111/imm.12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Keeffe M., Mok W.H., Radford K.J. Human dendritic cell subsets and function in health and disease. Cell. Mol. Life Sci. 2015;72:4309–4325. doi: 10.1007/s00018-015-2005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGovern N., Schlitzer A., Gunawan M., Jardine L., Shin A., Poyner E., Green K., Dickinson R., Wang X.N., Low D. Human dermal CD14+ cells are a transient population of monocyte-derived macrophages. Immunity. 2014;41:465–477. doi: 10.1016/j.immuni.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bujko A., Atlasy N., Landsverk O.J.B., Richter L., Yaqub S., Horneland R., Øyen O., Aandahl E.M., Aabakken L., Stunnenberg H.G. Transcriptional and functional profiling defines human small intestinal macrophage subsets. J. Exp. Med. 2018;215:441–458. doi: 10.1084/jem.20170057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilling D., Fan T., Huang D., Kaul B., Gomer R.H. Identification of markers that distinguish monocyte-derived fibrocytes from monocytes, macrophages, and fibroblasts. PLoS ONE. 2009;4:e7475. doi: 10.1371/journal.pone.0007475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larbi A., Fulop T. From “truly naïve” to “exhausted senescent” T cells: when markers predict functionality. Cytometry A. 2014;85:25–35. doi: 10.1002/cyto.a.22351. [DOI] [PubMed] [Google Scholar]
- 27.Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., Hogquist K.A. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashouri J.F., Weiss A. Endogenous Nur77 Is a Specific Indicator of Antigen Receptor Signaling in Human T and B Cells. J. Immunol. 2017;198:657–668. doi: 10.4049/jimmunol.1601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashouri J.F., Hsu L.-Y., Yu S., Rychkov D., Chen Y., Cheng D.A., Sirota M., Hansen E., Lattanza L., Zikherman J., Weiss A. Reporters of TCR signaling identify arthritogenic T cells in murine and human autoimmune arthritis. Proc. Natl. Acad. Sci. USA. 2019;116:18517–18527. doi: 10.1073/pnas.1904271116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Fliedner V., Miescher S., Gérain J., Gallati H., Barras C., Heumann D., Cerottini J.C. Production of tumor necrosis factor-alpha by naive or memory T lymphocytes activated via CD28. Cell. Immunol. 1992;139:198–207. doi: 10.1016/0008-8749(92)90112-3. [DOI] [PubMed] [Google Scholar]
- 31.Burt T.D. Fetal regulatory T cells and peripheral immune tolerance in utero: implications for development and disease. Am. J. Reprod. Immunol. 2013;69:346–358. doi: 10.1111/aji.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cupedo T., Nagasawa M., Weijer K., Blom B., Spits H. Development and activation of regulatory T cells in the human fetus. Eur. J. Immunol. 2005;35:383–390. doi: 10.1002/eji.200425763. [DOI] [PubMed] [Google Scholar]
- 33.Kohler S., Thiel A. Life after the thymus: CD31+ and CD31- human naive CD4+ T-cell subsets. Blood. 2009;113:769–774. doi: 10.1182/blood-2008-02-139154. [DOI] [PubMed] [Google Scholar]
- 34.Douaisi M., Resop R.S., Nagasawa M., Craft J., Jamieson B.D., Blom B., Uittenbogaart C.H. CD31, a Valuable Marker to Identify Early and Late Stages of T Cell Differentiation in the Human Thymus. J. Immunol. 2017;198:2310–2319. doi: 10.4049/jimmunol.1500350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez Rodriguez R., Pauli M.L., Neuhaus I.M., Yu S.S., Arron S.T., Harris H.W., Yang S.H.-Y., Anthony B.A., Sverdrup F.M., Krow-Lucal E. Memory regulatory T cells reside in human skin. J. Clin. Invest. 2014;124:1027–1036. doi: 10.1172/JCI72932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulman J.M., Pauli M.L., Neuhaus I.M., Sanchez Rodriguez R., Taravati K., Shin U.S., McCalmont T.H., Rosenblum M.D. The distribution of cutaneous metastases correlates with local immunologic milieu. J. Am. Acad. Dermatol. 2016;74:470–476. doi: 10.1016/j.jaad.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paus R., Müller-Röver S., Van Der Veen C., Maurer M., Eichmüller S., Ling G., Hofmann U., Foitzik K., Mecklenburg L., Handjiski B. A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J. Invest. Dermatol. 1999;113:523–532. doi: 10.1046/j.1523-1747.1999.00740.x. [DOI] [PubMed] [Google Scholar]
- 38.Holbrook K.A., Minami S.I. Hair follicle embryogenesis in the human. Characterization of events in vivo and in vitro. Ann. N Y Acad. Sci. 1991;642:167–196. [PubMed] [Google Scholar]
- 39.Holbrook K.A., Odland G.F. Structure of the human fetal hair canal and initial hair eruption. J. Invest. Dermatol. 1978;71:385–390. doi: 10.1111/1523-1747.ep12556818. [DOI] [PubMed] [Google Scholar]
- 40.Overgaard N.H., Jung J.W., Steptoe R.J., Wells J.W. CD4+/CD8+ double-positive T cells: more than just a developmental stage? J. Leukoc. Biol. 2015;97:31–38. doi: 10.1189/jlb.1RU0814-382. [DOI] [PubMed] [Google Scholar]
- 41.Jiménez E., Sacedón R., Vicente A., Hernández-López C., Zapata A.G., Varas A. Rat peripheral CD4+CD8+ T lymphocytes are partially immunocompetent thymus-derived cells that undergo post-thymic maturation to become functionally mature CD4+ T lymphocytes. J. Immunol. 2002;168:5005–5013. doi: 10.4049/jimmunol.168.10.5005. [DOI] [PubMed] [Google Scholar]
- 42.Kim H.P., Imbert J., Leonard W.J. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006;17:349–366. doi: 10.1016/j.cytogfr.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X., Mozeleski B., Lemoine S., Dériaud E., Lim A., Zhivaki D., Azria E., Ray C.L., Roguet G., Launay O. CD4 T cells with effector memory phenotype and function develop in the sterile environment of the fetus. Sci. Transl. Med. 2014;6:238ra72. doi: 10.1126/scitranslmed.3008748. [DOI] [PubMed] [Google Scholar]
- 44.Frascoli M., Coniglio L., Witt R., Jeanty C., Fleck-Derderian S., Myers D.E., Lee T.-H., Keating S., Busch M.P., Norris P.J. Alloreactive fetal T cells promote uterine contractility in preterm labor via IFN-γ and TNF-α. Sci. Transl. Med. 2018;10:eaan2263. doi: 10.1126/scitranslmed.aan2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rackaityte E., Halkias J., Fukui E.M., Mendoza V.F., Hayzelden C., Crawford E.D., Fujimura K.E., Burt T.D., Lynch S.V. Viable bacterial colonization is highly limited in the human intestine in utero. Nat. Med. 2020;26:599–607. doi: 10.1038/s41591-020-0761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jameson S.C., Lee Y.J., Hogquist K.A. Innate memory T cells. Adv. Immunol. 2015;126:173–213. doi: 10.1016/bs.ai.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coquet J.M., Middendorp S., van der Horst G., Kind J., Veraar E.A.M., Xiao Y., Jacobs H., Borst J. The CD27 and CD70 costimulatory pathway inhibits effector function of T helper 17 cells and attenuates associated autoimmunity. Immunity. 2013;38:53–65. doi: 10.1016/j.immuni.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Li N., van Unen V., Abdelaal T., Guo N., Kasatskaya S.A., Ladell K., McLaren J.E., Egorov E.S., Izraelson M., Chuva de Sousa Lopes S.M. Memory CD4+ T cells are generated in the human fetal intestine. Nat. Immunol. 2019;20:301–312. doi: 10.1038/s41590-018-0294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stras S.F., Werner L., Toothaker J.M., Olaloye O.O., Oldham A.L., McCourt C.C., Lee Y.N., Rechavi E., Shouval D.S., Konnikova L. Maturation of the Human Intestinal Immune System Occurs Early in Fetal Development. Dev. Cell. 2019;51:357–373. doi: 10.1016/j.devcel.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Dong Y., Speer C.P. The role of Staphylococcus epidermidis in neonatal sepsis: guarding angel or pathogenic devil? Int. J. Med. Microbiol. 2014;304:513–520. doi: 10.1016/j.ijmm.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 51.Barbarot S., Gras-Leguen C., Colas H., Garrot E., Darmaun D., Larroque B., Roze J.C., Ancel P.Y. Lower risk of atopic dermatitis among infants born extremely preterm compared with higher gestational age. Br. J. Dermatol. 2013;169:1257–1264. doi: 10.1111/bjd.12581. [DOI] [PubMed] [Google Scholar]
- 52.Belkaid Y., Tamoutounour S. The influence of skin microorganisms on cutaneous immunity. Nat. Rev. Immunol. 2016;16:353–366. doi: 10.1038/nri.2016.48. [DOI] [PubMed] [Google Scholar]
- 53.Brunner P.M., Israel A., Zhang N., Leonard A., Wen H.-C., Huynh T., Tran G., Lyon S., Rodriguez G., Immaneni S. Early-onset pediatric atopic dermatitis is characterized by TH2/TH17/TH22-centered inflammation and lipid alterations. J. Allergy Clin. Immunol. 2018;141:2094–2106. doi: 10.1016/j.jaci.2018.02.040. [DOI] [PubMed] [Google Scholar]
- 54.Esaki H., Brunner P.M., Renert-Yuval Y., Czarnowicki T., Huynh T., Tran G., Lyon S., Rodriguez G., Immaneni S., Johnson D.B. Early-onset pediatric atopic dermatitis is TH2 but also TH17 polarized in skin. J. Allergy Clin. Immunol. 2016;138:1639–1651. doi: 10.1016/j.jaci.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 55.Hebel K., Weinert S., Kuropka B., Knolle J., Kosak B., Jorch G., Arens C., Krause E., Braun-Dullaeus R.C., Brunner-Weinzierl M.C. CD4+ T cells from human neonates and infants are poised spontaneously to run a nonclassical IL-4 program. J. Immunol. 2014;192:5160–5170. doi: 10.4049/jimmunol.1302539. [DOI] [PubMed] [Google Scholar]
- 56.Prescott S.L., Macaubas C., Holt B.J., Smallacombe T.B., Loh R., Sly P.D., Holt P.G. Transplacental priming of the human immune system to environmental allergens: universal skewing of initial T cell responses toward the Th2 cytokine profile. J. Immunol. 1998;160:4730–4737. [PubMed] [Google Scholar]
- 57.Tang M.L., Kemp A.S., Thorburn J., Hill D.J. Reduced interferon-gamma secretion in neonates and subsequent atopy. Lancet. 1994;344:983–985. doi: 10.1016/s0140-6736(94)91641-1. [DOI] [PubMed] [Google Scholar]
- 58.Rosenblum M.D., Way S.S., Abbas A.K. Regulatory T cell memory. Nat. Rev. Immunol. 2016;16:90–101. doi: 10.1038/nri.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thome J.J.C., Bickham K.L., Ohmura Y., Kubota M., Matsuoka N., Gordon C., Granot T., Griesemer A., Lerner H., Kato T., Farber D.L. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat. Med. 2016;22:72–77. doi: 10.1038/nm.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao H., Liao X., Kang Y. Tregs: Where We Are and What Comes Next? Front. Immunol. 2017;8:1578. doi: 10.3389/fimmu.2017.01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asano T., Meguri Y., Yoshioka T., Kishi Y., Iwamoto M., Nakamura M., Sando Y., Yagita H., Koreth J., Kim H.T. PD-1 modulates regulatory T-cell homeostasis during low-dose interleukin-2 therapy. Blood. 2017;129:2186–2197. doi: 10.1182/blood-2016-09-741629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mold J.E., Michaelsson J., Burt T.D., Muench M.O., Beckerman K.P., Busch M.P., Lee T.H., Nixon D.F., McCune J.M. Maternal Alloantigens Promote the Development of Tolerogenic Fetal Regulatory T Cells in Utero. Science. 2008;322:1562–1565. doi: 10.1126/science.1164511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cordoro K.M., Hitraya-Low M., Taravati K., Sandoval P.M., Kim E., Sugarman J., Pauli M.L., Liao W., Rosenblum M.D. Skin-infiltrating, interleukin-22-producing T cells differentiate pediatric psoriasis from adult psoriasis. J. Am. Acad. Dermatol. 2017;77:417–424. doi: 10.1016/j.jaad.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du Toit G., Roberts G., Sayre P.H., Bahnson H.T., Radulovic S., Santos A.F., Brough H.A., Phippard D., Basting M., Feeney M., LEAP Study Team Randomized trial of peanut consumption in infants at risk for peanut allergy. N. Engl. J. Med. 2015;372:803–813. doi: 10.1056/NEJMoa1414850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adachi T., Kobayashi T., Sugihara E., Yamada T., Ikuta K., Pittaluga S., Saya H., Amagai M., Nagao K. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat. Med. 2015;21:1272–1279. doi: 10.1038/nm.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scharschmidt T.C., Vasquez K.S., Pauli M.L., Leitner E.G., Chu K., Truong H.-A., Lowe M.M., Sanchez Rodriguez R., Ali N., Laszik Z.G. Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host Microbe. 2017;21:467–477. doi: 10.1016/j.chom.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Masson A., Le Buanec H., Bouaziz J.-D. Purification and immunophenotypic characterization of human B cells with regulatory functions. Methods Mol. Biol. 2014;1190:45–52. doi: 10.1007/978-1-4939-1161-5_4. [DOI] [PubMed] [Google Scholar]
- 68.Zunder E.R., Finck R., Behbehani G.K., Amir A.D., Krishnaswamy S., Gonzalez V.D., Lorang C.G., Bjornson Z., Spitzer M.H., Bodenmiller B. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nowicka M., Krieg C., Crowell H.L., Weber L.M., Hartmann F.J., Guglietta S., Becher B., Levesque M.P., Robinson M.D. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 2017;6:748. doi: 10.12688/f1000research.11622.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 71.Crowell H.L., Zanotelli V., Chevrier S., Robinson M.D., Bodenmiller B. 2020. CATALYST: Cytometry dATa anALYSis Tools. R package.https://github.com/HelenaLC/CATALYST [Google Scholar]
- 72.Van Gassen S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., Saeys Y. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 73.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass cytometry dataset generated in this study is deposited and publicly available at Flow repository: ID# FR-FCM-Z3YD (url: https://flowrepository.org/).