Graphical abstract
Keywords: Rhodococcus rhodnii, Shotgun sequencing, Trypanosoma cruzi, Triatominae, Microbiome, Dickeya
Abstract
Trypanosoma cruzi, the causative agent of Chagas disease, colonizes the gut of triatomine insects, including Rhodnius prolixus. It is believed that this colonization upsets the microbiota that are normally present, presumably switching the environment to one more favorable for parasite survival. It was previously thought that one particular bacterium, Rhodococcus rhodnii, was essential for insect survival due to its ability to produce vital B-complex vitamins. However, these bacteria are not always identified in great abundance in studies on R. prolixus microbiota. Here we sequenced the microbiota of the insect anterior midgut using shotgun metagenomic sequencing in order to obtain a high-resolution snapshot of the microbes inside at two different time points and under two conditions; in the presence or absence of parasite and immediately following infection, or three days post-infection. We identify a total of 217 metagenomic bins, and recovered one metagenome-assembled genome, which we placed in the genus Dickeya. We show that, despite Rhodococcus being present, it is not the only microbe capable of synthesizing B-complex vitamins, with the genes required for biosynthesis present in a number of different microbes. This work helps to gain a new insight into the microbial ecology of R. prolixus.
1. Introduction
Trypanosoma cruzi is a protozoan parasite that colonizes the digestive tract of triatomine insect vectors (Reduviidae: Triatominae) [1]. The parasite is responsible for causing Chagas disease, an illness that affects several million people worldwide, predominantly in Latin America. The insect hosts are generally nocturnal feeders, that defecate following feeding. The parasite is excreted in the insect faeces and may enter the bloodstream through the bite wound [1], [2]. Recent evidence also suggests that food contaminated with faeces or urine is increasingly responsible for infection, where ingested parasite is sufficient to cause an acute phase of disease [3], [4].
Colonization of the triatomines by the parasite results in antimicrobial peptide (AMP) production in as little as two hours post-feeding, ultimately leading to a decreased bacterial load. These changes in the microbiota presumably render the gut ecology more favorable for the parasite to replicate [2], [4]. To date, several studies have investigated the community composition of triatomines following parasite colonization. These studies have utilized various methods including denaturing gradient gel electrophoresis [5], [6], RADseq [7] and 16S amplicon sequencing [8], [9], [10], [11], [12], [13], [14], [15]. Results from these studies vary, with some suggesting as few as 20 species make up the community composition [6], while others demonstrate a much higher diversity, despite having clearly dominant members of the microbiota, such as Pectobacterium in R. prolixus and Arsenophonus in the Triatoma brasiliensis complex [8]. No consensus has yet been found regarding microbial members of a “core” community. It is therefore difficult to understand the role of the microbiota with respect to Trypanosoma infection and triatomines in general. One further obstacle is the fact that the blood meal significantly alters the microbiota in triatomines, a detail further complicated by observed differences between male and female insects [9].
Next generation sequencing techniques and their application on triatomines have highlighted the diversity of bacteria associated with different species of triatomine and across different labs. In fact, dominant symbionts can include members of the Proteobacteria, Actinomycetes or Bacilli [5], [6], [7], [8], [9], [10], [11]. What has yet to be determined is the genetic capabilities that are afforded by the individual microbes, which may account for the observed diversity. Early studies indicated, for example, that B-complex vitamins were an essential contribution of the microbiota and, in Rhodnius prolixus, these vitamins are purportedly supplied by the bacterium, Rhodococcus rhodnii [12]. However, it has not yet been closely investigated whether other associated microbes also contribute to essential metabolic processes.
To address the above supposition, we investigated the changes that occur in the microbiome of the R. prolixus anterior midgut three days post-feeding, by using shotgun metagenomic sequencing. This is the first instance of such a study in triatomines with respect to T. cruzi infection and we provide insight into the coding sequence potential of those organisms that are present following parasitic exposure compared to uninfected controls.
2. Methods
2.1. Ethics statement
All experiments were performed in accordance with guidelines on animal experimentation from FIOCRUZ, adhering to all Brazilian legislation regarding animal welfare. Protocols were based upon procedures set out by The Ministry of Science and Technology (CONCEA/MCT) associated with the American Association
for Animal Science (AAAS), the Federation of European Laboratory Animal Science
Associations (FELASA), the International Council for Animal Science (ICLAS) and the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). Ethical approval under license number LW-8/17 was approved by the Committee for Ethics in the Use of Animals, CEUA-FIOCRUZ.
2.2. Insect rearing and dissection
Nymphs used in this study came from a colony maintained at a temperature of 25 ± 1 °C, 60 ± 10% relative humidity and natural illumination by the Vector Behaviour and Pathogen Interaction Group, Belo Horizonte, Brazil. Insects are fed monthly with citrated rabbit blood (10% v/v/) obtained from the Centro de Criação de Animais de Laborató rio (CECAL, Fiocruz, Rio de Janeiro, Brazil) offered in an artificial feeder equipped with a latex membrane and a circulating heated water flow (37 °C), and chicken anesthetized with intraperitoneal injections of a mixture of ketamine (20 mg/kg; Cristália, Brazil) and detomidine (0.3 mg/kg; Syntec, Brazil).
For infection with T. cruzi, a SWR/J mouse was inoculated with 200 µl of triatomine urine containing ~ 5x104 metacyclic trypomastigotes/ml from Dm28c strain. The strain was originally isolated from naturally infected opossum and kept in laboratory culture. The parasites were cultured by two weekly passages in liver-infusion tryptose (LIT) supplemented with 15% fetal bovine serum, 100 mg/ml streptomycin and 100 units/ml penicillin. To prevent loss of activity, parasites were submitted to cycles of triatomine-mice infection every six month. Nine days post-infection, the mouse was anesthetized with ketamine (150 mg/kg; Cristália, Brazil) and xylazine (10 mg/kg; Bayer, Brazil) and the presence of parasite was confirmed by light microscopy. The mice were then offered to R. prolixus fifth instar nymphs in the early morning, which had fasted for approximately 30 days. Nymphs in the same physiological conditions were fed on an anesthetized healthy mouse and used as a control group. Nymphs were fed for the same amount of time and we proceeded on the assumption that small differences in blood taken would not affect results in a major way since samples from each treatment group were subsequently pooled.
Insect gut sections were segregated using a dissecting microscope (Motic, SMZ-168) and sterile instruments immediately after feeding (approx. 5 to 15 min) and three days post-feeding. This time point was chosen because the transformation of the infectious trypomastigote stages of T. cruzi into epimastigote stages, which only occur in the invertebrate hosts, begins as early as a few hours after infection [13]. Three days after the bloodmeal, the colonization should therefore be well established. In total, twelve triatomines were prepared for DNA extraction. Three anterior midguts from exposed nymphs were combined immediately after feeding as well as three anterior midguts from unexposed insects. This section of the intestine was chosen because there is reportedly a higher antimicrobial activity than in the posterior midgut [4]. The same was done three days post-feeding resulting in the four samples ‘T. cruzi infected blood time point 0′ (TcInfT0), ‘T. cruzi infected blood time point 3′ (TcInfT3), ‘T. cruzi control time point 0′ (TcContT0) and ‘T. cruzi control time point 3′ (TcContT3). By combining the anterior midguts of several individuals, enough intestinal DNA material could be obtained for the shotgun metagenomic sequencing.
2.3. DNA extraction and sequencing
Following dissection of R. prolixus, DNA of the four samples, each with three pooled anterior midguts, was extracted using the AllPrep DNA/RNA minikit (Qiagen) following the manufacturer’s recommendations. Purified DNA was shipped to Novogene and four libraries, one for each sample, were prepared by Novogene. Sample preparation was done using the NEB UltraII DNA library preparation kit with four amplification cycles. Paired-end (150 bp) sequencing was performed on an Illumina NovaSeq6000 instrument.
2.4. Sequence analysis
Raw reads of all samples were trimmed using Trimmomatic (v0.33) [14] to remove low quality bases and reads lacking a pair using the following settings: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36. For taxonomic classification, reads were uploaded to the Kaiju[15] web server. Results were downloaded and processed according to scripts available at https://github.com/ntobias-85/metagenome-csbj-2020. Trimmed reads were filtered against the R. prolixus genome (GCA_000181055.3) using bowtie2 (v2.2.5) [16] and the –un-conc option to keep only reads not mapping to the reference. Reads not mapping to R. prolixus were used to generate a co-assembly of all four samples with megahit (v1.1.4) [17] with the --kmin-1pass option activated. Individual samples were then independently mapped to the co-assembly to generate bam files using bowtie2 (v2.2.5) and samtools (v1.2) [18]. In this way, sample-specific information such as the relative abundance of sequences from each bin can be determined.
Anvi’o is a bioinformatic package designed to streamline metagenomic workflows. The co-assembly of the four samples was used to generate a contig database in anvi’o (v5.1) [19], which includes annotation of open reading frames using Prodigal (v2.6.3) [20] as a built-in function. Anvi’o was further used to run hidden Markov models to identify single copy gene collections (anvi-run-hmms) and annotate NCBI COGs (anvi-run-ncbi-cogs). COGs (Clusters of Orthologous Groups) represent families of orthologous protein-coding genes which can be used to study microbial phylogeny and genome annotation. Taxonomy for genes was added by using centrifuge (v1.0.2) [21] and importing the output into the contig database using the script anvi-import-taxonomy-for-genes. Profiles for each single sample were generated, removing all contigs less than 2,500 nucleotides. Finally, the profiles were merged with the anvi-merge command, which includes an automatic binning step with CONCOCT [22]. Amino acid sequences for each gene were exported from the contig database and used as input in the GhostKOALA webserver. The output was parsed back into the contig database using the KEGG-to-anvio script, followed by anvi-import of the resulting output. The automatic binning was manually inspected and refined based on predicted taxonomy, including blastx searches of annotated genes, read coverage and GC-content of contigs. The pangenomic analysis [23] was carried out using anvi’o with settings --minbit = 0.5; --mcl-inflation = 10 and --use-ncbi-blast activated.
For phylogenetic analyses, genomes were downloaded from NCBI and converted to contig databases using the ‘anvi-script-FASTA-to-contigs-db’ script. The program ‘anvi-get-sequences-for-hmm-hits’ was then used to extract ribosomal RNA sequences common among all downloaded genomes and the respective metagenomic bin. Ribosomal proteins were extracted from the database with --concatenate activated. Concatenated protein sequences were then imported into Geneious v6.1.8 and a maximum likelihood tree was constructed with 100 bootstraps using PhyML[24] (version 3.0).
2.5. KEGG analysis
The output from GhostKOALA was used as input for KEGG_decoder (Kyoto Encyclopedia of Genes and Genomes) [25] and subsequently KEGG_encoder, which uses hidden markov models to incorporate some further processes. The outputs were then combined with the python script, Decode_and_Expand.py [25], generating a heatmap of all processes identified for each sample.
3. Results and discussion
3.1. Microbial populations of the R. prolixus anterior midgut
Studies on the microbiome of R. prolixus have so far predominantly profiled bacterial species using the 16S rRNA. Results show that there is little intra-individual diversity associated with the microbiota and insects are often dominated by a single group of organisms [8], [26]. In the case of Rhodnius spp., these are especially Enterobacteria, such as Pectobacteria and Serratia [6], [8], [31]. Furthermore, the Actinobacteria Rhododcoccus, is found not only in Rhodnius, but also in other triatomine genera such as Triatoma [11], [32], [33]. One consequence of only investigating 16S rRNA is that interactions between the insect, fungi, viruses and even other parasites that may be present are totally excluded from analysis. Moreover, bacterial species often possess multiple copies of 16S rRNA genes making a reliable quantification difficult [34]. Another drawback is that sequences can only accurately be assigned to higher level taxonomies, with some difficulty to obtain resolution between species. Nevertheless, 16S amplicon sequencing has several advantages. A large and comprehensive reference database is available facilitating the detection of rare taxa. Additionally, it is significantly cheaper than metagenomic shotgun sequencing and the lower data output is easier to process with regard to bioinformatic pipelines and computational capacity. 16S rRNA amplicon sequencing is therefore highly suitable for taxonomic analysis approaches, such as community composition studies.
Given the advances in sequencing technology and the desire to more fully understand the microbial ecology of triatomine midgut at high-resolution, we proceeded to use metagenomic shotgun sequencing to analyze the anterior midgut of R. prolixus under four different conditions (described in methods). Sequencing resulted in a total 145,812,112 clean reads (TcContT0: 34,750,241 (10.4 GB), TcContT3: 47,062,804 (14.1 GB), TcInfT0: 33,499,209 (10.1 GB) and TcInfT3: 30,499,858 (9.2 GB) for further analysis. To begin with, we used Kaiju [15], which directly BLASTs each read pair against the non-redundant nucleotide database, to profile the composition of microbial communities associated with our samples using the paired end input option for each sample (raw data is available from the European Nucleotide Archive under accession number PRJEB33861). This was done initially to determine the number of raw reads that could be directly attributed to any given organism. Our analysis suggests that, apart from bacterial DNA, a large part of the DNA originates from a mixture of fungi, viruses, parasites and other eukaryotic sequences (Supplementary Fig. 1). We also detected an increase in the proportion of bacterial DNA after the blood meal. This is probably due to the increased supply of nutrients and has been observed several times before [4], [8]. One key difference to keep in mind between this and other studies is the fact that here, we have used a more natural approach to infection, feeding triatomines on mice infected with triatomine urine (with or without parasite), as opposed to other studies that tend to use a significantly higher parasitic load for infection experiments. One final difference is the way in which our bacterial taxonomy was initially determined. We used kaiju from samples originating from the anterior midgut, where the majority of other studies instead use 16S amplicon sequencing from the hindgut (or directly on faeces). Despite these differences, we do see somewhat similar results to previously published work in that there is a moderate proportion of Actinobacteria, Proteobacteria and Firmicutes identified in the samples (Supplementary Fig. 2) [5], [7], [8], [9], [10], [11], [27], [28], [29], [30], [31], [32]. However, the dominant order in our experiment is Chlamydiales, possibly indicative of a dominant microbe in the insect colony used. To more directly compare taxonomic classification of sequencing data with previous works, future experiments should include an additional 16S amplicon sequencing analysis on the same sample used for shotgun sequencing.
3.2. Metagenome assembly and annotation
We were primarily interested in the gene content of the microbiota present in the anterior midgut of R. prolixus. Therefore, initial filtering of our reads was performed against the R. prolixus genome to remove all sequences mapping to the insect DNA. Since the insects originated from the same colony, we predicted that their microbiomes would be relatively similar. We therefore performed a co-assembly with the remaining reads. Co-assembly involves using all available sequence data to get the best assembly possible. The individual samples are then mapped back to this co-assembly to obtain sample specific coverage details. Combining all four samples using megahit, resulted in an assembly containing a total of 480,315 contigs larger than 1,000 bp (max 152,936 bp, average 2,302 bp, N50 2,496 bp). For the annotation, only contigs with 2,500 bp or more were kept (max 152,936 bp, average 6,481 bp, N50 19,857 bp). This cutoff was chosen to ensure a meaningful signal of sequence composition and coverage for subsequent binning steps. The annotation resulted in a total of 357,390 gene calls. Supplementary annotation with the KEGG database allowed us to map the presence or absence of individual pathways to each sample. We then utilized this data to define samples that contained enriched pathways, relative to all other samples. The KEGG decoder and expander script [25] utilizes a hidden Markov database to identify and fill false-negative gaps in pathways that appear incomplete. Based upon this, we screened 162 individual pathways, most of which contained little to no difference between samples. Minor differences could be identified between time point 0 and time point 3 such as in nitrate reduction pathways (Supplementary Fig. 3), but we expect differences would be more evident when including a greater number of samples.
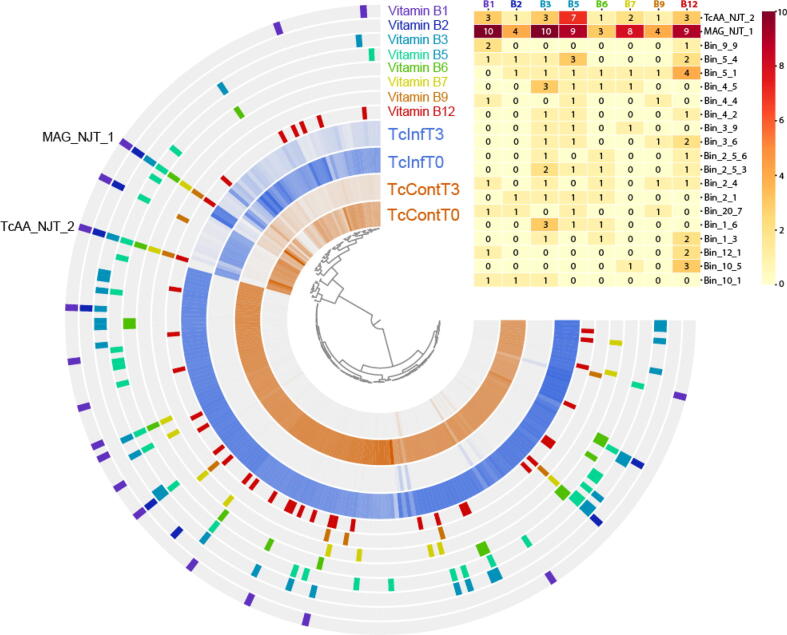
Annotations were useful in refining the automatic binning output from CONCOCT. Although binning algorithms perform well in most cases, manual refinement is still necessary to ensure the accuracy of contig groupings. In total 217 bins were identified that were unrelated to R. prolixus. One draft metagenome-assembled genome (MAG), MAG_NJT_1, was recovered from the genomic bins, which was preliminarily identified as being from the Dickeya genus (more details below). Since Rhodococcus is reportedly an essential symbiont of R. prolixus [33], we also searched for bins containing predominantly Rhodococcus sequence (Fig. 1, annotated with the bin name “TcAA_NJT_2”).
Fig. 1.
Coassembly of metagenomic sequencing samples from anterior midguts of triatomines fed uninfected blood immediately after feeding (TcContT0) or 3 days post-feeding (TcContT3) or infected blood immediately following feeding (TcInfT0) or after 3 days (TcInfT3). In total 217 metagenomic bins were detected (each node on the central tree). The inner four layers represent the relative abundance of sequence in each sample. Layers labeled with vitamin B highlight those metagenomic bins that contain at least one enzyme annotated as being a part of the pathway of the respective vitamin B biosynthesis. Any bins containing 3 or more orthologs are highlighted in the heatmap, with numbers representing the number of orthologs in a given pathway. A full overview can be seen in Supplementary Fig. 7. Also labeled is MAG_NJT_1, a member of the Dickeya genus and TcAA_NJT_2, a Rhodococcus.
Our experiment consisted of DNA samples extracted from either exposed or unexposed insects immediately after feeding and three days post-feeding. We expected that the two samples at time point zero should be similar, while we may see differences between exposed and unexposed samples after three days. For the most part this is true, although the differences associated with three-day time points is limited to just a handful of bins (Fig. 1). We also expected to see changes after three days due to the increase in the expression of various AMPs during this time [2]. However, we saw relatively few differences in the overall relative abundances of microbial profiles between exposed and unexposed (Supplementary Table 1). This may be in part due to the different infection strategy that we utilized (outlined above). The parasite load in this experiment was low, which increases the possibility of a lack of response and may partly explain unexpectedly few differences between the control group and challenged insects. Although further experiments are required to confirm the veracity of our hypothesis, it is possible that microbial changes reported to be associated with parasite entering the insect gut may be, at least in part, due to a higher parasitic load than would naturally occur. Despite this, we observed an increase in sequences derived from MAG_NJT_1, and two other bins (Bin_22_4 and Bin 22_1_1) as well as a decrease in TcAA_NJT_2, which we suspect is Rhodococcus (Supplementary Fig. 4). Eichler and Schaub (2002) also identified such a decrease of Rhodococcus after the blood meal, but the amount of the bacteria recovered quickly and exceeded the starting level after a few days [34]. These differences could be associated with the differential effects of AMPs against different bacterial species [2], yet still raises the question of the importance of Rhodococcus at this time point. Aposymbiotic Rhodnius prolixus, which fail to develop beyond the second larval stage, regenerate when fed with blood containing Rhodococcus [35]. However, since other bacterial symbionts have never been examined in this way, it cannot be determined whether they would have a similar remedying effect.
3.3. Presence of B-complex vitamin biosynthesis pathways
Early research into R. prolixus microbiota provides a narrative that R. rhodnii are essential symbionts in triatomine biology [12] and in the absence of these symbionts, the insects fail to moult [36], [37]. Deliberation of this point includes arguments that the bacteria are used as a food source themselves, with bacterial lysis supplying required nutrients, as well as strong supporting evidence for the role of B-complex vitamins and their production by the Rhodococcus
[37]. Many of these studies were performed several years ago (as early as 1936) and include culture-dependent methodologies. In recent years however, several groups have revisited triatomine microbiomes, with 16S amplicon sequencing emerging as the preferred method for investigations of bacteria.
Amplicon sequencing consistently demonstrates that R. rhodnii are not the only bacterial species present, nor are they regularly the dominant bacteria. Others have demonstrated that dominant symbionts include Actinobacteria (of which Rhodococcus is a member) [11], Serratia [6], Pectobacterium [8] and Arsenophonus[6], [26], [28], which has already been shown to serve as a source of B-vitamins in whiteflies[38]. Furthermore, in other obligate haematophagous insects, symbioses with various B-vitamin producing bacteria have been detected. The Gammaproteobacteria Wigglesworthia spp. is regarded as the main vitamin B provider of the tsetse fly, the aetiological agent of the African trypanosomiasis and Riesia spp. seems to be indispensable for Pediculus humanus humanus, the human body louse. A lack of these symbionts often results in development retardation and a decrease in fecundity [37], [38]. Given this conflicting evidence, we decided to explore the B-vitamin biosynthesis capabilities of the microbiota identified in our study. We extracted all of the gene calls from the co-assembly and uploaded them to GhostKoala to annotate each of our putative coding sequences with KEGG ontology (KO) numbers. Based on the KEGG pathways, we were then able to extract any gene call with a KO number from each of the eight different vitamin B pathways (see Supplementary Table 2 for a list of KO numbers used for each biosynthetic pathway). Unexpectedly, we see that enzymes involved in B-complex vitamin biosynthesis are present in a number of different organisms present within the microbiota (Fig. 1). Therefore, the provision of vitamin B derivatives solely by Rhodococcus seems unlikely. Despite this, only two members contained enzymes present in all eight vitamin biosynthetic pathways. These were TcAA_NJT_2, which is predicted to be Rhodococcus, and have already been shown to possess vitamin B biosynthesis genes including: thiamine (B1), riboflavin (B2), niacin (B3), pantothenate (B5), pyridoxal (B6), biotin (B7), tetrahydrofolate (B9), and cobalamin (B12) [39]. The second bin containing orthologs in each of the vitamin B biosynthetic pathways was MAG_NJT_1.
3.4. Identification of the metagenome-assembled genome
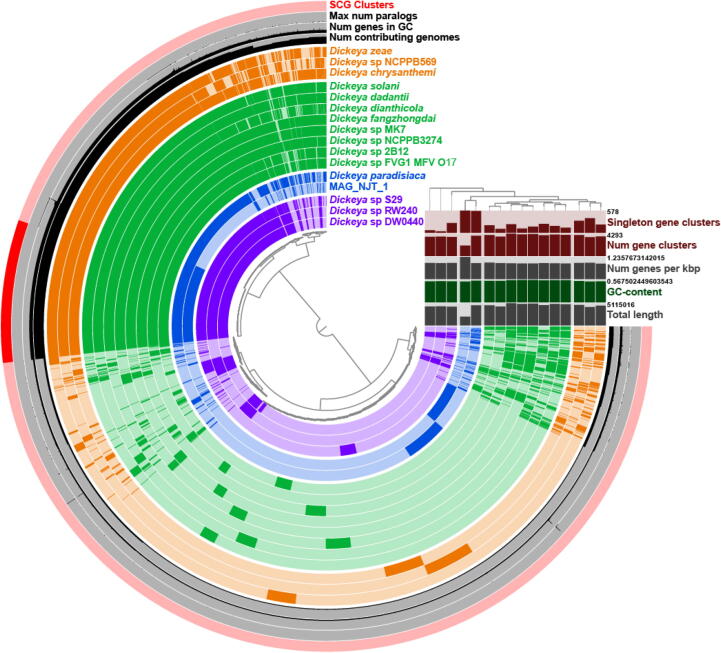
A closer look at MAG_NJT_1 reveals an 89.21% completion and 0% redundancy estimate based on the presence of single copy genes. To identify a likely taxon, we used BLAST to analyze one of the ribosomal subunits, which suggested the bin originated form Pectobacterium. Since some Pectobacterium isolates were reclassified into Brennaria and Dickeya [40], we used concatenated ribosomal proteins from a selection of these three genera, which placed MAG_NJT_1 into the Dickeya genus (Supplementary Fig. 5). Dickeya are a genus of Gammaproteobacteria that typically cause disease in plants. Although these phytopathogens are reported to be widespread in insects [41], it is curious that they should be a dominant taxon in a hematophagous insect. However, this is not the first time that a bacterium predominantly associated with plants has been described as a major taxon associated with R. prolixus, with Methylobacterium identified as the third most abundant microbe in a North American lab colony of R. prolixus [26]. Nevertheless, to check for contamination within this bin, we performed a pangenome analysis using 15 Dickeya isolates and MAG_NJT_1 (Fig. 2). Excluding MAG_NJT_1, other Dickeya isolates appear to have approximately twice the number of gene clusters and are approximately double the length of MAG_NJT_1. Based on this, we estimate that this bin is in fact approximately 50% complete. Of the single core genes (SCGs) present in all 16 isolates, 50 were randomly selected to create a higher resolution phylogeny (Supplementary Fig. 6, Fig. 2). MAG_NJT_1 clustered closely to Dickeya paradisiaca (formerly Brenneria paradisiaca).
Fig. 2.
Pangenomic analysis of 15 Dickeya species together with MAG_NJT_1. The phylogeny on the right represents a maximum likelihood analysis based on the amino acid sequences of 50 randomly selected single-copy core genes (SCG, all SCGs are highlighted in the outermost ring). Comparative statistics on the right represent the respective ring for each genome used in the analysis. Order of genes are based on gene cluster frequencies.
Although it is still likely that B-complex vitamins are essential to Rhodnius development and survival, our data here show that B-vitamins can be contributed to the gut ecology by a number of different microbes. In fact, several of the major bacterial species reported to be present in R. prolixus gut samples such as Arsenophonus and Methylobacterium [8], [26], can also produce vitamin B derivatives [38], [42], [43]. We suggest that the apparent extensive description of Rhodococcus in the early literature may be indicative of an over-reliance on culture-based methods when in fact many microbes are (i) present to fulfill the vitamin B biosynthesis and (ii) are capable of doing so.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the Goethe University Fokus funds and the LOEWE-Centre TBG supported by the Hessen State Ministry of Higher Education, Research and the Arts (HMWK), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, CRA-APQ-00569-15 and CRA-PPM-00162-17), Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular (INCTEM/CNPq, 465678/2014-9), AAG was supported by CNPq productivity grants.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.10.031.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Brener Z. Biology of Trypanosoma Cruzi. Annu Rev Microbiol. 1973;27(1):347–382. doi: 10.1146/annurev.mi.27.100173.002023. [DOI] [PubMed] [Google Scholar]
- 2.Vieira C., Waniek P.J., Mattos D.P., Castro D.P., Mello C.B., Ratcliffe N.A., Garcia E.S., Azambuja P. Humoral responses in Rhodnius prolixus: bacterial feeding induces differential patterns of antibacterial activity and enhances mRNA levels of antimicrobial peptides in the midgut. Parasit Vectors. 2014;7(1):232. doi: 10.1186/1756-3305-7-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shikanai-Yasuda M.A., Carvalho N.B. Oral Transmission of Chagas Disease. Clin Infect Dis. 2012;54(6):845–852. doi: 10.1093/cid/cir956. [DOI] [PubMed] [Google Scholar]
- 4.Castro DP, Moraes CS, Gonzalez MS, Ratcliffe NA, Azambuja P, Garcia ES. Garcia. Trypanosoma cruzi immune response modulation decreases microbiota in Rhodnius prolixus gut and is crucial for parasite survival and development, PLoS ONE; 2012: 7, e36591. [DOI] [PMC free article] [PubMed]
- 5.Gumiel M., da Mota F.F., Rizzo V.d.S., Sarquis O., Castro D.P.d., Lima M.M., Garcia E.d.S., Carels N., Azambuja P. Characterization of the microbiota in the guts of Triatoma brasiliensis and Triatoma pseudomaculata infected by Trypanosoma cruzi in natural conditions using culture independent methods. Parasites Vectors. 2015;8(1) doi: 10.1186/s13071-015-0836-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Mota FF, Marinho LP, Moreira CJ de C, Lima MM, Mello CB, Garcia ES et al. Cultivation-independent methods reveal differences among bacterial gut microbiota in triatomine vectors of Chagas disease. PLoS Negl Trop Dis; 2012, 6:e1631. [DOI] [PMC free article] [PubMed]
- 7.Orantes LC, Monroy C, Dorn PL, Stevens L, Rizzo DM, Morrissey L, et al. Uncovering vector, parasite, blood meal and microbiome patterns from mixed-DNA specimens of the Chagas disease vector Triatoma dimidiata. PLoS Negl Trop Dis; 2018, 12:e0006730. [DOI] [PMC free article] [PubMed]
- 8.Díaz S., Villavicencio B., Correia N., Costa J., Haag K.L. Triatomine bugs, their microbiota and Trypanosoma cruzi: asymmetric responses of bacteria to an infected blood meal. Parasites Vectors. 2016;9(1) doi: 10.1186/s13071-016-1926-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumonteil E., Ramirez-Sierra M.-J., Pérez-Carrillo S., Teh-Poot C., Herrera C., Gourbière S., Waleckx E. Detailed ecological associations of triatomines revealed by metabarcoding and next-generation sequencing: implications for triatomine behavior and Trypanosoma cruzi transmission cycles. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-22455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira JL, Cury JC, Gurgel-Gonçalves R, Bahia AC, Monteiro FA Field-collected Triatoma sordida from central Brazil display high microbiota diversity that varies with regard to developmental stage and intestinal segmentation. PLoS Negl Trop Dis; 2018, 12:e0006709. [DOI] [PMC free article] [PubMed]
- 11.Montoya-Porras L.M., Omar T.-C., Alzate J.F., Moreno-Herrera C.X., Cadavid-Restrepo G.E. 16S rRNA gene amplicon sequencing reveals dominance of Actinobacteria in Rhodnius pallescens compared to Triatoma maculata midgut microbiota in natural populations of vector insects from Colombia. Acta Trop. 2018;178:327–332. doi: 10.1016/j.actatropica.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Brecher G., Wigglesworth V.B. The transmission of Actinomyces rhodnii Erikson in Rhodnius prolixus stål (hemiptera) and its influence on the growth of the host. Parasitology. 1944;35(4):220–224. doi: 10.1017/S0031182000021648. [DOI] [Google Scholar]
- 13.Brenner RR, la Merced Stoka de A. Chagas' Disease Vectors: Anatomic and physiological aspects; 1987.
- 14.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics; 2014, 30:2114–20. [DOI] [PMC free article] [PubMed]
- 15.Menzel P., Ng K.L., Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7(1) doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W, MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics; 2015, 31:1674–6. [DOI] [PubMed]
- 18.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eren A.M., Esen Ö.C., Quince C., Vineis J.H., Morrison H.G., Sogin M.L. Anvi“o: an advanced analysis and visualization platform for ”omics data. PeerJ. 2015;3 doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyatt D., Chen G.-L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11(1) doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D., Song L.i., Breitwieser F.P., Salzberg S.L. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26(12):1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alneberg J., Bjarnason B.S., de Bruijn I., Schirmer M., Quick J., Ijaz U.Z., Lahti L., Loman N.J., Andersson A.F., Quince C. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11(11):1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 23.Delmont TO, Eren AM, Linking pangenomes and metagenomes: the Prochlorococcus metapangenome, PeerJ. 2018; 6:e4320. [DOI] [PMC free article] [PubMed]
- 24.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. Gascuel, New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0, Syst Biol; 2010: 59, 307–21. [DOI] [PubMed]
- 25.Graham E.D., Heidelberg J.F., Tully B.J. Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 2018;12(7):1861–1866. doi: 10.1038/s41396-018-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez-Ruano SM, Škochová V, Rego ROM, Schmidt JO, Roachell W, Hypša V, et al. Microbiomes of North American Triatominae: The Grounds for Chagas Disease Epidemiology, Front Microbiol; 2018, 9:1167. [DOI] [PMC free article] [PubMed]
- 27.Brown J.J., Rodríguez-Ruano S.M., Poosakkannu A., Batani G., Schmidt J.O., Roachell W., Zima J., Jr, Hypša V., Nováková E. Ontogeny, species identity, and environment dominate microbiome dynamics in wild populations of kissing bugs (Triatominae) Microbiome. 2020;8(1) doi: 10.1186/s40168-020-00921-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieran T.J., Arnold K.M.H., Thomas J.C., IV, Varian C.P., Saldaña A., Calzada J.E., Glenn T.C., Gottdenker N.L. Regional biogeography of microbiota composition in the Chagas disease vector Rhodnius pallescens. Parasites Vectors. 2019;12(1) doi: 10.1186/s13071-019-3761-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumonteil E., Pronovost H., Bierman E.F., Sanford A., Majeau A., Moore R., Herrera C. Interactions among Triatoma sanguisuga blood feeding sources, gut microbiota and Trypanosoma cruzi diversity in southern Louisiana. Mol Ecol. 2020;29(19):3747–3761. doi: 10.1111/mec.15582. [DOI] [PubMed] [Google Scholar]
- 30.Šorfová P., Škeříková A., Hypša V. An effect of 16S rRNA intercistronic variability on coevolutionary analysis in symbiotic bacteria: Molecular phylogeny of Arsenophonus triatominarum. Syst Appl Microbiol. 2008;31(2):88–100. doi: 10.1016/j.syapm.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Waltmann A, Willcox AC, Balasubramanian S, Borrini Mayori K, Mendoza Guerrero S, Salazar Sanchez RS, et al. Hindgut microbiota in laboratory-reared and wild Triatoma infestans. PLoS Negl Trop Dis; 2019, 13:e0007383. [DOI] [PMC free article] [PubMed]
- 32.Lima MS, Laport MS, Lorosa ES, Jurberg J, Santos Dos KRN, da Silva Neto MAC, et al. Bacterial community composition in the salivary glands of triatomines (Hemiptera: Reduviidae). PLoS Negl Trop Dis; 2018: 12:e0006739. [DOI] [PMC free article] [PubMed]
- 33.Harington J.S. Studies on Rhodnius prolixus : growth and development of normal and sterile bugs, and the symbiotic relationship. Parasitology. 1960;50(1-2):279–286. doi: 10.1017/S0031182000025373. [DOI] [PubMed] [Google Scholar]
- 34.Eichler S., Schaub G.A. Development of Symbionts in Triatomine Bugs and the Effects of Infections with Trypanosomatids. Exp Parasitol. 2002;100(1):17–27. doi: 10.1006/expr.2001.4653. [DOI] [PubMed] [Google Scholar]
- 35.Durvasula, R.V., Sundaram, R.K., Cordon-Rosales, C., Pennington, P., Beard, C.B, Rhodnius prolixus and its symbiont, Rhodococcus rhodnii: A model paratransgenic control of disease transmission. K. Bourtzis, T.A. Miller. (eds.) Insect Symbiosis. CRC Press, Boca Raton, Florida, USA; 2003.
- 36.Ben-Yakir D. Growth retardation of Rhodnius prolixus symbionts by immunizing host against Nocardia (Rhodococcus) rhodnii. J Insect Physiol. 1987;33(6):379–383. doi: 10.1016/0022-1910(87)90015-1. [DOI] [Google Scholar]
- 37.Biology SBJOE The role of the symbiotic bacteria in the nutrition of Rhodnius prolixus (Hemiptera) Citeseer; 1956.
- 38.Santos-Garcia D, Juravel K, Freilich S, Zchori-Fein E, Latorre A, Moya A, et al. To B or Not to B: Comparative Genomics Suggests Arsenophonus as a Source of B Vitamins in Whiteflies. Front Microbiol; 2018, 9:2254. [DOI] [PMC free article] [PubMed]
- 39.Pachebat J.A., van Keulen G., Whitten M.M.A., Girdwood S., Del Sol R., Dyson P.J., Facey P.D. Draft Genome Sequence of Rhodococcus rhodnii Strain LMG5362, a Symbiont of Rhodnius prolixus (Hemiptera, Reduviidae, Triatominae), the Principle Vector of Trypanosoma cruzi. Genome Announcements. 2013;1(3) doi: 10.1128/genomeA.00329-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samson R, Legendre JB, Christen R, Fischer-Le Saux M, Achouak W, Gardan L, Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int J Syst Evol Microbiol; 2005, 55:1415–27. [DOI] [PubMed]
- 41.Rossmann S, Dees MW, Perminow J, Meadow R, Brurberg MB, Soft Rot Enterobacteriaceae Are Carried by a Large Range of Insect Species in Potato Fields. Appl Environ Microbiol; 2018, 84:1150. [DOI] [PMC free article] [PubMed]
- 42.Ivanova E.G., Fedorov D.N., Doronina N.V., Trotsenko Y.A. Production of vitamin B12 in aerobic methylotrophic bacteria. Microbiology. 2006;75(4):494–496. doi: 10.1134/S0026261706040217. [DOI] [PubMed] [Google Scholar]
- 43.Abanda-Nkpwatt D, Müsch M, Tschiersch J, Boettner M, Schwab W, Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. J Exp Bot; 2006, 57:4025–32. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.