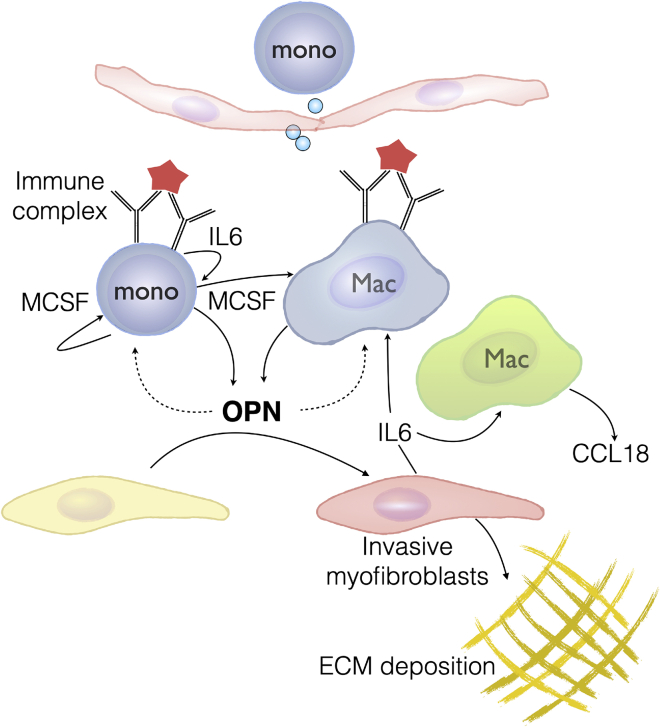
Summary
Progressive lung fibrosis is a major cause of mortality in systemic sclerosis (SSc) patients, but the underlying mechanisms remain unclear. We demonstrate that immune complexes (ICs) activate human monocytes to promote lung fibroblast migration partly via osteopontin (OPN) secretion, which is amplified by autocrine monocyte colony stimulating factor (MCSF) and interleukin-6 (IL-6) activity. Bulk and single-cell RNA sequencing demonstrate that elevated OPN expression in SSc lung tissue is enriched in macrophages, partially overlapping with CCL18 expression. Serum OPN is elevated in SSc patients with interstitial lung disease (ILD) and prognosticates future lung function deterioration in SSc cohorts. Serum OPN levels decrease following tocilizumab (monoclonal anti-IL-6 receptor) treatment, confirming the connection between IL-6 and OPN in SSc patients. Collectively, these data suggest a plausible link between autoantibodies and lung fibrosis progression, where circulating OPN serves as a systemic proxy for IC-driven profibrotic macrophage activity, highlighting its potential as a promising biomarker in SSc ILD.
Keywords: systemic sclerosis, SSc, fibrosis, osteopontin, SPP1, IL-6, macrophages, immune complex, ILD, biomarker
Graphical Abstract
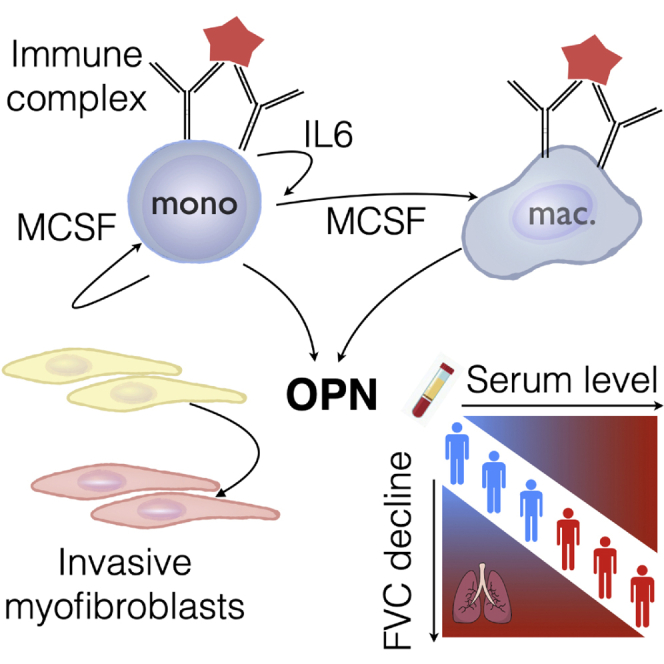
Highlights
Immune complexes induce osteopontin (OPN) secretion from macrophages via MCSF and IL-6
OPN is expressed predominantly by macrophages in fibrotic interstitial lung disease
Circulating OPN levels are elevated and predict disease progression in systemic sclerosis
IL-6 receptor blockade reduces levels of circulating OPN in systemic sclerosis patients
Gao et al. show that immune complexes induce expression of osteopontin in macrophages/monocytes via IL-6 and MCSF. Osteopontin is expressed by macrophages in fibrotic lung tissue. Serum osteopontin levels predict fibrosis progression in systemic sclerosis patients and is decreased following IL-6 receptor blockade, highlighting its potential as a biomarker.
Introduction
Systemic sclerosis (SSc) is an autoimmune disease characterized by a distinct pathogenic triad of microvascular damage, dysregulation of innate and adaptive immunity, and fibrosis involving skin and many internal organs, such as the heart, kidney, and lungs.1 Although mortality because of renal involvement has significantly decreased since the adoption of angiotensin pathway blockade, the proportion of deaths because of interstitial lung disease has continued to increase, making it the leading cause of death of SSc patients.2 With limited therapeutic options to stabilize or reverse SSc-associated lung fibrosis, biomarkers that help identify patients at a greater risk of progressive disease are considered vital for successful clinical development of novel treatment modalities.3
Several distinct autoantibodies have been described in SSc patients, many correlating with specific clinical presentations.4 For instance, the presence of anti-topoisomerase I (Scl-70) antibodies has been associated with diffuse skin involvement, significant pulmonary fibrosis, and overall worse survival;5 anti-centromere antibodies, on the other hand, are associated with limited skin disease and pulmonary hypertension without fibrosis. Given that the targets for these antibodies are intracellular nuclear antigens, there is limited evidence of their direct pathogenicity.6,7 Although “functional” autoantibodies have been reported in sera of SSc patients,8 distinguished by their binding to accessible, pathobiologically relevant targets such as platelet derived growth factor receptor (PDGFR)9, their activity has been called into question10,11. It is conceivable that autoantibodies may complex with their cognate antigens, made accessible in tissue by cell death or other environmental stimuli, and perpetuate chronic, inflammatory sequelae that underlie their association with distinct clinical presentations. Indeed, numerous studies have documented the presence of immune complexes (ICs) in sera, lungs, and bronchoalveolar lavage (BAL) fluid of SSc patients, potentially implicating them in the pathogenesis.12, 13, 14, 15
Monocyte/macrophage activation has been observed in fibrotic SSc skin and lung tissue.16, 17, 18 Although the upstream activators remain unknown, it is plausible that ICs are among the key triggers of activating myeloid cells and sustaining the chronic inflammation and fibrosis in SSc tissue. Here we sought to understand the profibrotic potential of IC-activated monocytes using in vitro cell culture systems and establish their in vivo relevance using sera and tissue from SSc patients. We demonstrate that osteopontin (OPN) is a disease-relevant surrogate of IC-activated profibrotic monocytes/macrophages and highlight its potential as a systemic biomarker to predict future lung function decline using three independent cohorts of SSc patients.
Results
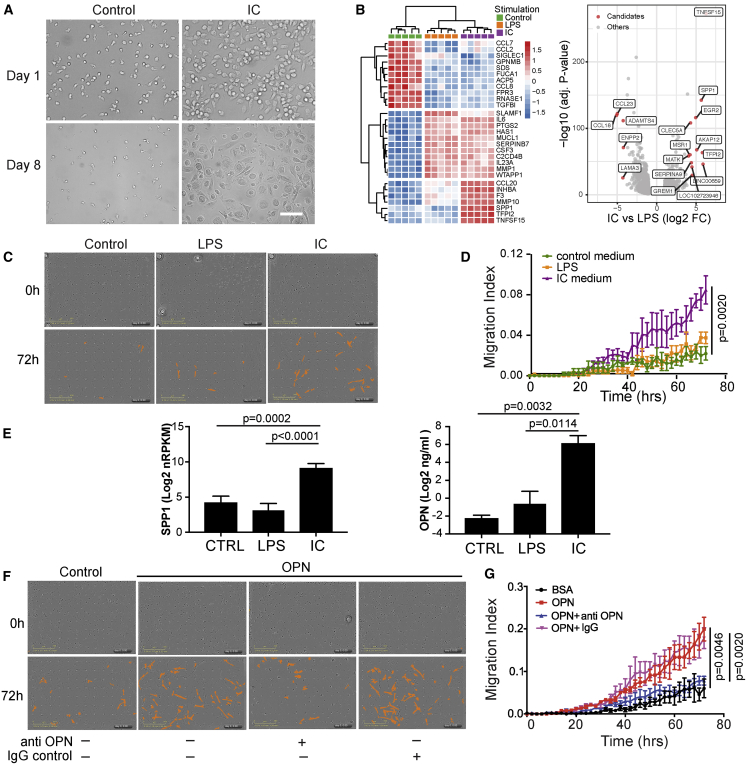
We initially sought to characterize the effects of IC activation of monocytes using a simple in vitro model that emulates antigen:antibody complexes immobilized in tissue. In this model, ICs are formed by addition of antibodies against an antigen immobilized on a tissue culture plate well to which cells of interest are exposed.19 Human monocytes adopted an activated, macrophage-like morphology following stimulation with the plate-bound IC (Figure 1A). Using this protocol, blood monocytes isolated from five human donors were subjected to IC activation, and genes that were differentially modulated were queried by RNA sequencing (RNA-seq). IC-activated monocytes revealed a consistent and striking profile that partially overlapped with that imparted by lipopolysaccharide (LPS) stimulation, used as a canonical positive control. IC and LPS activation resulted in upregulation of several pro-inflammatory genes, including IL6, CSF3, and PTGS2. However, IC activation also upregulated a suite of genes that were distinct from those induced by LPS, suggesting their relative selectivity for IC activation (e.g., SPP1, MMP10, and TNFSF15; Figure 1B; Table S1). In parallel experiments, we observed that cell culture medium conditioned by IC-stimulated monocytes were capable of augmenting migration of primary human lung fibroblasts (Figures 1C and 1D), but media from LPS-stimulated wells failed to do so. Given that SPP1 was robustly induced by IC stimulation and reports implicating OPN) (encoded by SPP1) in various fibrotic diseases,20, 21, 22, 23, 24 we queried whether OPN could directly mediate the migratory effect induced by IC-conditioned medium. Indeed, OPN protein levels were highly elevated in IC-induced monocyte supernatants (Figure 1E). Additionally, recombinant human OPN directly promoted fibroblast migration through the Transwell, and this effect was neutralized by a monoclonal antibody specific to OPN (Figures 1F and 1G). However, OPN did not significantly affect proliferation or collagen secretion by lung fibroblasts in our experimental setting (Figures S1A and 1B), suggesting that OPN’s principal profibrotic effect on fibroblasts may be via facilitation of migration to areas adjacent to the fibrotic niche, thereby aiding fibrosis progression.
Figure 1.
IC Activation Elicits OPN Production in Monocytes
(A) Morphology of primary human monocytes after 24-h immune complex (lC) stimulation on day 1 and day 8. Scale bar, 50 μm.
(B) Heatmap of the top genes expressed variably between monocytes from 5 donors stimulated with IC or LPS compared with the unstimulated control. A volcano plot depicts all differentially expressed genes under IC versus LPS conditions (|log2 fold change| > 2, false discovery rate [FDR] p < 0.05).
(C and D) Representative images of transmigrated primary human lung fibroblasts in response to conditioned medium from monocytes stimulated with IC or LPS. Cells migrated to the bottom surface were identified (colored orange) and (D) quantified (N = 3) by periodic imaging over 72 h of culture and automated analysis by IncuCyte software and expressed as migration index (described in STAR Methods).
(E) OPN mRNA expression (N = 5) and protein secretion from human monocytes stimulated with IC or LPS (N = 3). Values are mean ± SEM, log transformed and analyzed by two-tailed paired t test.
(F and G) Representative images and quantification of transmigrated primary human lung fibroblasts (N = 3) in response to rhOPN (10 μg/mL) and with or without neutralizing anti-OPN antibody (Ab).
Statistical significance in (D) and (G) was assessed using a Kolmogorov-Smirnov test.
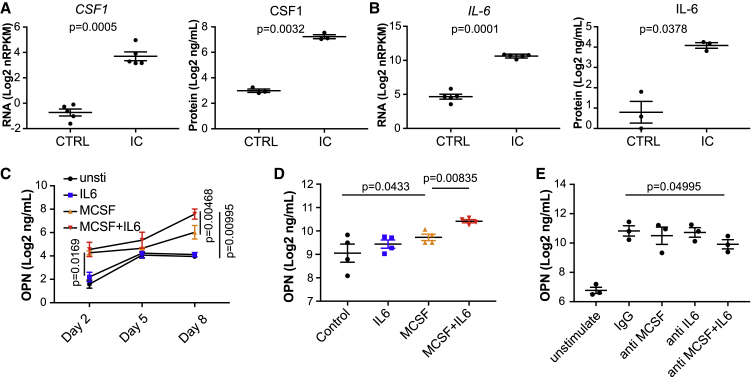
Given the rapid phenotypic transition of IC-activated cells into a macrophage-like morphology, we hypothesized a potential role for an autocrine macrophage differentiation factor, such as MCSF (CSF1) in this process. Indeed, IC-stimulated monocytes rapidly turned on transcription of CSF1, and the growth factor was abundant in IC-stimulated supernatants (Figure 2A). In addition, we noticed that interleukin-6 (IL-6), an inflammatory cytokine that has been shown previously to induce OPN in a cell line,25 was also induced at the transcript and protein levels (Figure 2B). Therefore, we tested whether MCSF and/or IL-6 are necessary and sufficient to induce OPN in our experimental context. Purified recombinant MCSF was sufficient to stimulate OPN production in human monocytes (Figure 2C). Although IL-6 was not sufficient to do so in isolation, it significantly amplified OPN production by MCSF, a trend that increased over time. This effect was also evident in monocytes differentiated into macrophages by treatment with MCSF for 10 days; although these macrophages secreted high levels of OPN at baseline, the levels increased further in the presence of IL-6 and MCSF (Figure 2D). Next we tested whether IL-6 and MCSF were required for IC-mediated OPN production. To temporally decouple the stimuli, we transferred monocytes from IC-coated plates to new plates after 24 h and monitored OPN in the supernatants over time in the presence or absence of neutralizing antibodies to IL-6R and/or MCSFR. Strikingly, despite removal of the IC stimulus, the monocytes differentiated into macrophage-like cells that secreted high levels of OPN by day 9. Although this induction seemed unaffected by blocking IL-6R or MCSFR, neutralizing both receptors partially diminished OPN levels in the supernatants (Figure 2E). These data suggest that, although secondary factors, such as IL-6 and MCSF, may participate in the amplification, OPN induction by IC stimulation may not be entirely dependent on them.
Figure 2.
Autocrine MCSF and IL-6 Amplify OPN Production in Monocytes/Macrophages
(A and B) CSF1 (MCSF) and (B) IL6 expression in primary human monocytes (N = 5) and their protein levels (N = 3) in supernatant after stimulation with IC. Values are mean ± SEM, log transformed and analyzed by two-tailed paired t test.
(C) Monocyte production of OPN in response to MCSF and/or IL-6 on days 2, 5, and 8 (N = 3). OPN in supernatants were measured by ELISA.
(D) OPN production by monocyte-derived macrophages stimulated with MCSF and/or IL-6 at 24 h (N = 4).
(E) 24-h IC-stimulated monocytes were tested for OPN production in the presence or absence of neutralizing Abs to CSF1R and/or IL-6R on day 9 after stimulation. Values are mean ± SEM, log transformed and analyzed by one-tailed paired t test.
All experiments were repeated from 3–5 different donors as specified.
Although OPN was sufficient to mediate the pro-migratory effects of IC-conditioned medium, OPN was not required for this effect because a monoclonal antibody that was effective in blocking OPN activity failed to abrogate the effect in IC-conditioned medium, suggesting redundant mechanisms underlying this activity (Figure S2). Although these observations support OPN’s profibrotic activity because of its inherent effect on fibroblasts, they also suggest a potential role as a biomarker, should this pathway of macrophage activation occur in vivo, in human fibrotic tissue.
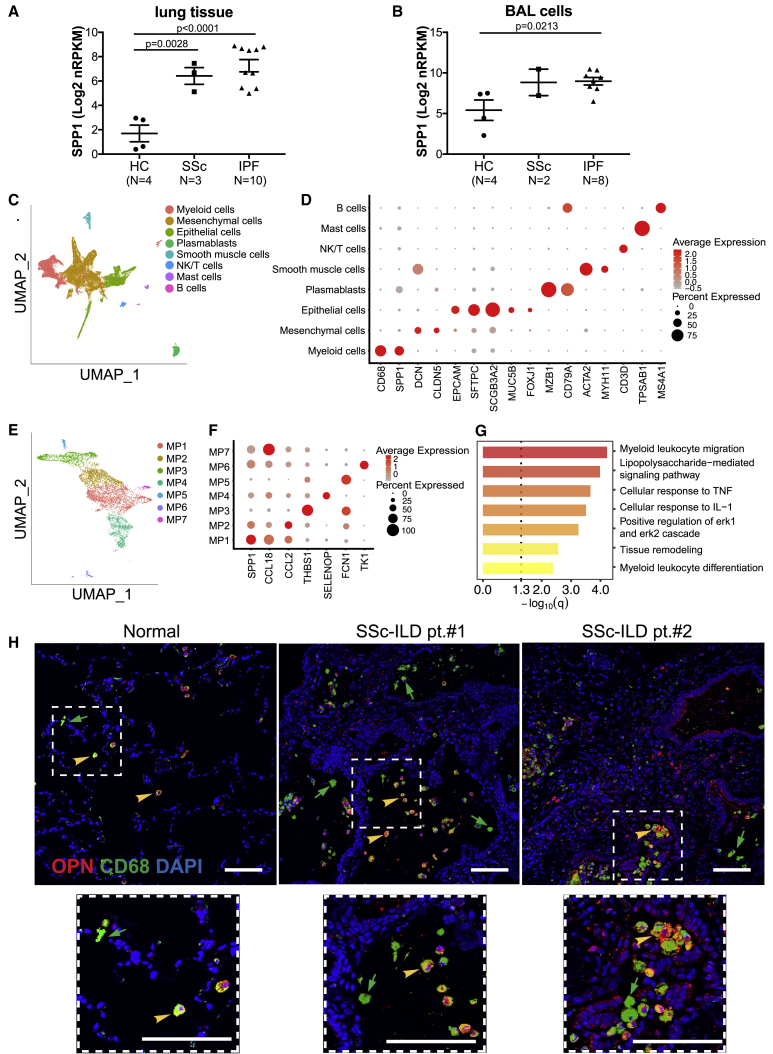
Next we measured OPN expression in explanted lung tissues from patients with SSc interstitial lung disease (ILD) and compared them with non-diseased control and idiopathic pulmonary fibrosis (IPF) subjects, in whom OPN is known to be upregulated.24. Bulk RNA-seq showed that OPN encoding SPP1 transcripts was significantly elevated in SSc ILD tissue, similar to that in IPF lungs (Figure 3A). In addition, BAL cells, which are typically enriched for alveolar macrophages, also recapitulated some of this expression difference between diseased and non-diseased samples, suggesting that OPN can be upregulated in interstitial and alveolar compartments (Figure 3B).
Figure 3.
Expression of OPN in Human Fibrotic Lungs
(A and B) OPN RNA expression in (A) lung tissue biopsy (N = 17) and (B) BAL cells (N = 14) from healthy control (HC) subjects and individuals with SSc or IPF, measured by RNA-seq. Values are mean ± SEM and were analyzed by one-way ANOVA.
(C) Cell clusters identified using canonical markers in single-cell RNA-seq data from cell suspensions of digested lung explants from individuals with SSc ILD (N = 3), visualized using a UMAP plot.
(D) Expression pattern for SPP1 and other selected canonical cell type marker genes among the clusters.
(E) Subclustering of the macrophages into seven discrete subtypes (MP1–MP7).
(F) Expression profile of SPP1 and CCL18 among various macrophage subsets typified by a top marker gene.
(G) Gene Ontology biological process enrichment among genes that are differentially expressed in SPP1-enriched and CCL18-enriched clusters (MP1 versus MP7).
(H) Representative immunofluorescent staining of OPN (red), CD68 (green), and DAPI (blue) in a normal lung and two lungs from individuals with SSc ILD.
Although various cell types have been shown to produce OPN,26 in the context of idiopathic lung fibrosis, prior reports have considered epithelial cells to be a predominant source.24 In addition, certain myeloid cell populations, such as eosinophils27 and CD1A-expressing antigen-presenting cells,28 can be important sources of OPN in the context of chronic lung disease. To further define the cellular context of OPN production in SSc lung tissue, cell suspensions derived from explanted lung tissue were subjected to single-cell RNA-seq. Over 29,000 cells in total, isolated from three SSc ILD subject lungs, were sequenced, batch corrected, aligned, clustered, and annotated with common cell types using canonical marker genes. Although the SPP1 signal was detectable in many cell types, the dominant signal was attributable to a cluster that was identified as macrophages because of their expression of a combination of canonical markers such as CD68, MARCO, and MSR1, etc. (Figure 3D; Table S2).
Given that various developmental lineages and upstream activation programs can impart distinct expression profiles in macrophages29 and the earlier observation of differential OPN induction in vitro, we postulated that SPP1 expression may be enriched in certain subset(s) of macrophages (Figure 3E). Indeed, bioinformatically dissecting the broadly defined macrophage population into smaller subclusters revealed that OPN expression was distributed unevenly. SPP1 transcripts were relatively enriched in the subcluster MP1, whereas expression at a lower prevalence was also noted in MP2, MP6, and MP7 (Figures 3E and 3F). The MP1 macrophage population also displayed relative upregulation of genes associated with lipid metabolism and lipoprotein recognition (APOC1, APOE, and TREM2) as well as genes associated with degradation of lipids and extracellular matrix (CYP27A1, LIPA, and MMP9). Secretion of matrix metalloproteinases (e.g., MMP9) by macrophages is a key component of their extracellular matrix turnover function,30 implying that the phenotype of the MP1 population may be biased toward tissue remodeling. The average log fold changes and percent detected values for the top 10 marker genes defining subcluster MP1 are shown in Figure S3, and full information is provided in Table S3. In particular, CCL18, a chemokine known to be expressed by lung macrophages and shown to be a prognostic biomarker in individuals with IPF and SSc ILD,31, 32, 33 was expressed predominantly by the MP7 cluster and less so by the MP1 cluster. It is plausible that CCL18 and SPP1 expression mark functionally related macrophages (shared profibrotic connotation) that are spatiotemporally distinct, reflecting divergent upstream activation pathways. This is suggested by canonical pathway analysis, contrasting SPP1-hi MP1 and CCL18-hi MP7 subclusters, which showed significant enrichment for Gene Ontology terms such as myeloid cell migration and differentiation as well as responses to various types of proinflammatory signaling (Figure 3G; Table S4). Immunofluorescent staining independently confirmed the scRNA-seq data, showing prominent OPN expression in CD68+ myeloid cells in SSc lung explants (Figure 3H). Interestingly, macrophage subsets expressing high levels of SPP1 were also observed in IPF lungs, based on the recently published evidence from several independent labs,34, 35, 36, 37, 38 suggesting the possibility that macrophage expression of OPN may indeed be a common profibrotic denominator across fibrotic ILDs involving distinct etiopathogenic mechanisms.
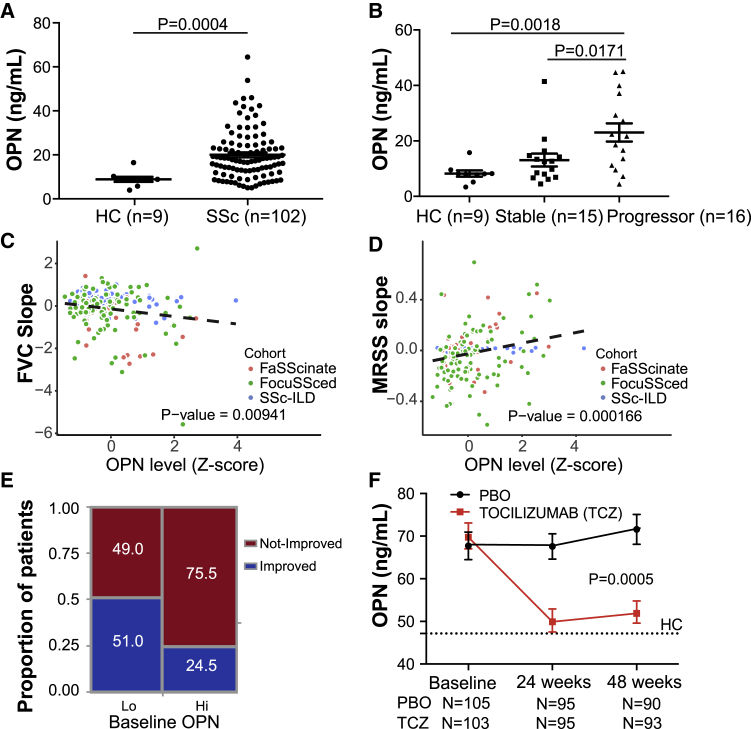
Given that OPN protein levels are easily quantifiable in the periphery and have been shown to be prognostic in a cohort of individuals with IPF,24 we sought to investigate whether serum OPN levels are clinically informative in individuals with SSc. We initially explored serum OPN protein levels in an observational SSc ILD cohort recruited at the University of Michigan that included individuals with SSc and high-resolution computed tomography (HRCT)-confirmed lung involvement (Table S5). Serum OPN levels were substantially higher in individuals with SSc relative to non-diseased control subjects; in particular, the levels were higher in individuals who were categorized as progressors compared with those with relatively stable lung function (Figures 4A and 4B). We extended our analyses to two additional well-characterized SSc clinical trial cohorts: faSScinate39 and focuSSced40 cohorts that predominantly included individuals with early diffuse cutaneous SSc at risk of lung disease. Across these three cohorts, higher OPN levels at baseline were correlated significantly with greater lung function decline, even after adjusting for baseline age, sex, and forced vital capacity (FVC) parameters (Figure 4C). Moreover, given that skin fibrosis is a common feature in SSc, we tested whether serum OPN levels were informative regarding skin disease progression, using modified Rodnan skin score (MRSS) measurements over time. Indeed, higher OPN levels at baseline correlated with MRSS worsening (Figure 4D), except in the Michigan cohort (a combination of individuals with limited and diffuse cutaneous SSc and longer disease duration; Table S6). These data suggest that circulating OPN may be useful to predict the course of fibrotic activity across multiple organs. In fact, a significantly higher proportion of individuals with higher baseline OPN met the categorical outcome of “not improving” (Figure 4E), based on the novel composite response index in SSc (ACR-CRISS), which incorporates clinical metrics (FVC and MRSS changes) and clinicians’ assessments and outcomes reported by the subjects.41
Figure 4.
OPN Is a Prognostic Biomarker in SSc
(A) Serum OPN protein levels in an observational cohort of individuals with SSc ILD (N = 102, Michigan cohort) and healthy donors (N = 9).
(B) OPN levels in the Michigan cohort, clinically stratified by disease severity and progression status. Clinical demographic information and categorization definitions are provided in Table S5. Values are mean ± SEM and were analyzed by Mann-Whitney test.
(C) Prognostic effect of serum OPN levels for future FVC change in three SSc cohorts. Shown is a linear regression model of percent predicted FVC (ppFVC) slope adjusted for cohorts, age, sex, and baseline ppFVC with available measurements during 1 (faSScinate, focuSSced) to 2 years (Michigan) of follow-up. Refer to Table S6 for individual cohort analyses.
(D) Prognostic effect of baseline serum OPN levels for skin thickening (MRSS) change in the three SSc cohorts over time. Shown is a linear regression model of ppMRSS slope adjusted for cohorts, age, sex, and baseline MRSS with available measurements. Refer to Table S6 for individual cohort analyses.
(E) Categorical CRISS response proportions of subjects from the focuSSced cohort (placebo arm), stratified by baseline serum OPN, split at median. Fisher’s exact test, 2-tailed, p = 0.0081.
(F) Pharmacodynamic effect on OPN in SSc patients treated with TCZ or placebo (PBO) in the focuSSced cohort. A dotted line represents median OPN levels in age- and sex-matched HC subjects. Values are mean ± SEM. The p value was calculated based on a linear regression model, with the OPN level as response variable and time, treatment, and interaction of time and treatment as covariates.
Given our observation that IL-6 amplified macrophage OPN production in vitro, we tested whether a correlation existed in vivo between OPN and IL-6 levels in individuals with SSc. We observed that baseline serum IL-6 did indeed correlate with serum OPN (ρ = 0.41, p = 7.92e–10), adding plausibility to this relationship in individuals with SSc (Figure S4). We next tested whether circulating OPN levels were modulated pharmacodynamically by neutralization of IL-6 activity in vivo by longitudinally assessing OPN levels in the focuSSced cohort. Indeed, in individuals treated with tocilizumab (TCZ), an anti-IL-6R monoclonal antibody, we observed a robust reduction in circulating OPN levels, whereas they remained elevated in the placebo arm (Figure 4F).
Discussion
Although autoantibodies, ICs, myeloid activation, OPN production, and fibroblast activity have been implicated independently in SSc in the past, our observations in this study postulate a scenario where these individual factors cooperate in a coherent manner to promote fibrosis. It is plausible that inflammatory monocytes are exposed in injured tissue to locally deposited ICs that could trigger OPN production from these cells. This IC activation may drive further monocyte migration and differentiation into tissue macrophages through autocrine and paracrine effects of factors such as IL-6 and MCSF, which, in turn, can further augment OPN production throughout this process (Figure 5). Although autoantibodies are implicated in rheumatic diseases such as SSc, many fibrotic lung diseases, such as IPF, lack clear autoimmune drivers.42,43 However, many ILDs histopathologically feature tertiary lymphoid follicles that are known to harbor plasma cells,44 which may produce antibodies to locally encountered antigens. These antibodies against persistent antigens may provide chronic IC triggers, although the antigens may not necessarily be autoantigens.
Figure 5.
Proposed Schematic of OPN Induction and Profibrotic Activity in SSc
Tissue macrophages activated by IC trigger inflammation and OPN secretion. Infiltrating monocytes are activated by IC and differentiate into macrophages, aided by autocrine and paracrine MCSF, amplified by IL-6, and further contribute to OPN production. OPN facilitates tissue remodeling, in part by sensitizing and mobilizing fibroblasts toward other fibrogenic growth factors, and thereby contribute to fibrosis progression.
The finding that myeloid differentiation factors can trigger OPN production directly from monocytes suggests that macrophage-derived OPN may be relevant in non-IC scenarios where these growth factors are produced in response to other injury/inflammatory stimuli. Although we show that OPN can be profibrotic by its direct effect on fibroblasts, this does not preclude other ways in which it could contribute to tissue remodeling, which may include feedback activity on other immune cells,26 influencing local epithelial cell differentiation, etc.45
Macrophages at chronic inflammatory sites likely experience disparate upstream activation triggers, and distinct subsets may be protective or pathogenic at any given time. Data from our scRNA-seq results and from others point to this important functional heterogeneity, which is still in the process of elucidation. Precisely targeting macrophage subsets to treat fibrosis will require a greater understanding of their functional diversity and temporal plasticity46 with the aid of clinically translatable biomarkers. Although our intent has been focused on characterizing OPN-expressing macrophages in fibrotic lungs, we showed that these cells are, in part, distinct from macrophages predominantly expressing CCL18, a chemokine that also displayed a systemic pharmacodynamic effect upon blocking the IL-6 pathway.39 The pharmacodynamic traits illustrated here in the context of SSc and IL-6 pathway inhibition position OPN as a biomarker for therapeutic strategies designed to modulate macrophage activity, and with its prognostic property, it has potential to provide insights not only regarding the clinical activity of drugs being tested but of the overall fibrotic disease process itself. Considering that OPN is elevated in progressors and reduced by treatment that also significantly slows lung function decline in randomized placebo-controlled clinical trials, it seems plausible that macrophage-derived OPN may have a causal role in SSc disease progression. Further exploration is warranted to unravel OPN’s molecular mechanism of action and potential as a therapeutic target in fibrotic disease.
Limitations of Study
Some components of the hypothesized model, such as the role of ICs in activation of myeloid cells for the excess OPN detected in individuals with SSc, are primarily based on our in vitro observations and cross-sectional reports from prior literature and therefore need to be tested formally in vivo. Furthermore, the prognostic effects we demonstrate across our SSc cohorts are from post hoc exploratory analyses using research-grade assays; therefore, additional validation using clinical-grade assays and prespecified analysis plans, ideally in a prospective setting, will be necessary to validate OPN as a useful prognostic biomarker in fibrotic disease.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-CD68 Monoclonal Antibody | Abcam | Cat# ab955, RRID:AB_307338 |
| Rabbit anti-Osteopontin antibody | Abcam | Cat# ab8448, RRID:AB_306566 |
| Mouse anti M-CSF R antibody | R & D Systems | Cat# MAB3291, RRID:AB_2292260 |
| IL6R antibody (Tocilizumab) | Roche | N/A |
| Rabbit anti-Albumin Antibody | Sigma-Aldrich | Cat# A0433, RRID:AB_257887 |
| Goat anti Osteopontin antibody | R & D Systems | Cat# AF1433, RRID:AB_354791 |
| Biological Samples | ||
| Human Serum samples | University of Michigan | N/A |
| Human Serum samples | Genentech clinic trial for Tociilizmab | FaSScinate(NCT01532869)/FcuSSced (NCT02453256) cohorts |
| Human blood samples | Genentech | N/A |
| Human healthy lung samples | Northern California Transplant Donor Network | N/A |
| Human SSc lung samples | UCSF | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant human OPN | Sigma-Aldrich | SRP3131 |
| Human serum albumin | Sigma-Aldrich | A9080 |
| Recombinant human IL6 | R & D Systems | 206-IL |
| Recombinant human MCSF | R & D Systems | 216-MC |
| Critical Commercial Assays | ||
| Human Osteopontin Quantikine ELISA Kit | R & D Systems | DOST00 |
| Simple Plex Cartridge Kit for OPN | Bio Techne | SPCKB-PS-000232 |
| Bio-Plex Pro Human Cytokine 48-plex assay | Bio Rad | 12007283 |
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | G7570 |
| Chromium Single Cell 3′ Library and Gel bead kit v2 | 10x Genomics | PN-120267 |
| Pan monocytes isolation kit, human | Miltenyi Biotec | 130-096-537 |
| Deposited Data | ||
| RNA seq for IC stimulated monocytes | GSE155687 | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155687 |
| scRNA seq for fibrotic lung | GSE159354 | |
| Experimental Models: Cell Lines | ||
| Primary human lung fibroblast cells | ATCC | PCS201013 |
| Software and Algorithms | ||
| JMP | SAS Institute | SCR_014242 |
| Prism 7 | GraphPad | SCR_005375 |
| Seurat R package version 3.1.1 | https://satijalab.org/seurat | |
Resource Avaliability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Thirumalai R. Ramalingam (Ramalingam.Thirumalai@gene.com)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The accession number for the RNA seq data from monocytes stimulated by IC generated reported in this paper is GSE155687.
The accession numnber for the scRNA seq data supporting the current study is made available at GEO, under the accession number GSE159354.
Experimental Model and Subject Details
Primary Cell Culture
Primary human lung fibroblast cells were purchased from ATCC (PCS201013) and cultured in serum free fibroblast culture medium (PCS201040).
The PBMCs were isolated from 50 mL of heparinized blood of four healthy donors by Ficoll-paque. Cd14+ Primary human monocytes were purified from PBMC by Miltenyi Pan Monocyte Isolation Kit according to the manufacturer’s instructions and cultured in RPMI with 10% ultra-low IgG FCS, 10 mM HEPES and L-glutamine.
Human lung for scRNA seq
Explanted lung tissues for scRNA seq were obtained from patients with scleroderma associated interstitial lung disease (SSc-ILD) who met American College of Rheumatology criteria for scleroderma. Written informed consent was obtained from all subjects and the study was approved by the UCSF institutional review board. Human lungs not used by the Northern California Transplant Donor Network were used as controls; studies indicate that these lungs are physiologically and pathologically normal.
Clinical cohorts
An observational SSc-ILD cohort included 102 SSc patients with HRCT confirmed ILD (Michigan). The study protocol was approved by the University of Michigan Medical School Institutional Review Board. Written consent was given by all patients prior to participation.
The FaSScinate cohort included 36 patients assigned to the placebo arm of a phase 2 clinical trial (NCT01532869) where baseline sera were available for research purposes.
Focusced cohort (NCT02453256) included patients with serum available for research purposes.
Demographic and clinic characteristics of patients are presented in Table S5.
Method Details
Immune complex stimulation
The RIA/EIA 96 well plates were coated with 100 ul 20ug/mL of Human serum albumin (HAS, Sigma A9080) in 100 mM sodium carbonate at 4°C overnight. The coated plates were washed once with 200 ul of wash buffer (PBS+ 10% Ultra-low IgG FBS) and block with wash buffer at 37°C for 1 hr. After blocking, the plates were incubated with 100 ul per well of 10 ug/mL rabbit anti-HSA (Sigma A0433) in washing buffer at 37°C for 2-4 hr. The Immune complex (IC) coated plates were washed once with 200 ul of wash buffer and ready to use for stimulation.
Primary human monocytes were seeded in the IC coated plate at density of 200k cells/well and cultured for the mentioned duration. The RNA of the cells was extracted and sequenced. Primary monocytes were also stimulated with IC for 24 hours and moved to another plate without IC. Human CSF1 R blocking antibody (R&D MAB3291) and human IL6R antibody (Roche, Tocilizumab) and IgG control were added in the medium for 9 days to neutralize activities of M-CSF and IL6 respectively after IC stimulation. Media was replenished every two days. The supernatants were collected and OPN protein was measured with Human Osteopontin (OPN) Quantikine ELISA Kit (DOST00). CSF1 and IL6 protein levels were measured using a Luminex kit (Bio-Rad).
CSF1 and IL-6 stimulation
For monocytes stimulation, primary monocytes were stimulated by adding 10 ng/ml CSF1 and 20ng/ml IL6 for 10 days. Supernatants at days 2, 5 and 8 were collected.
For macrophage stimulation, primary monocytes were differentiated into macrophage by stimulating with 50ng/ml CSF1 for 10 days. Differentiated cells were washed, counted and reseeded and stimulated with 10 ng/ml CSF1 and/or 20ng/ml IL6 for 24 hr.
Fibroblast migration assay
Primary human fibroblasts cells were assayed using commercially available 96-well cell migration plate (IncucyteTM ClearView, Essen BioScience). The cells were plated in the top chamber of the IncuCyte® ClearView plate at a density of 1000 cells/well. For IC stimulation, 200ul conditional medium collected from unstimulated, LPS and IC stimulated primary monocytes were added in the bottom of the insert. For OPN, the plate inserts were coated with 10ug/ml OPN and BSA control O/N at 4°C. 200ul of DMEM medium with 0.5% FBS and PDGF was added to the bottom chamber as a chemoattractant. The cells were incubated at 37°C in a humidified incubator with 5% CO2 and 95% air and monitored and by Incucyte for 72 hr. with repeat scanning (10x) for every 2 hours. Whole-well images of cell on both bottom and top of the ClearView plate membrane are captured at user-defined intervals. Images are processed and analyzed by the IncuCyte® ZOOM software using automated algorithms to quantify cell area on each side of the membrane.
Fibroblast growth rate assay and collagen secretion
Lung fibroblasts seeded in 96-well culture plates at a cell density of 10K cells/well and incubated in serum free fibroblast culture medium at 37°C O/N. After 12 h, the medium was replaced by DMEM medium with 0.1% FBS alone as control or 0.1% FBS plus increasing concentrations of Osteopontin (2 and 5ug/ml) or 10% FBS as positive control. The cells were maintained in culture for another 48 h. Cell viability was determined using the CellTiter-Glo® Luminescent Cell Viability Assay. All assays were performed in triplicate. For collagen secretion, 10ug/ml OPN were coated on 24 well plate and fibroblast were cultured in the wells for 48hrs after staved in serum free medium O/N. The total RNA were extracted from cells using RNeasy mini kit (QIAGEN) and cDNA were synthesized by high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific). Taqman real-time quantitative assays (Thermo Fisher Scientific) were used to detect the expression of collagens and fibrogenetic genes. Expression data were normalized to the geometric mean of housekeeping gene GAPDH to control the variability in expression levels and were analyzed using the 2-ΔΔCT method.
Human serum OPN and linear regression analysis
Osteopontin levels of all the serum samples in patients from SSc-ILD and FaSScinate cohorts and in age/sex matched healthy donors were quantified with Human Osteopontin (OPN) Quantikine ELISA Kit (DOST00). Sera were diluted 40-fold with calibrator diluent RD5-24 in the kit and measured following the manufacturer’s protocol. Osteopontin levels in patients from Focusced cohort were measured using the Simple Plex platform (Protein Simple).
For Michigan SSc-ILD cohort, baseline FVC (BLFVC) was calculated based on the mean of FVC measurements between ± 90 days from baseline blood sampling. Slope of FVC/MRSS change was calculated for each subject using linear regression model with FVC/MRSS as response variable and time points in duration up to 730 days. For FaSScinate and Focusced cohorts, the slope of FVC change was calculated for each subject using the FVC measured between baseline and 48 weeks. The slope of MRSS change was calculated for each subject using their MRSS measured between baseline and 48 weeks. To alleviate platform/cohort specific batch effects in pooled analyses of the three cohorts, OPN levels were transformed to a standardized score (z-score) prior to analyses. Prognostic effect on slope of FVC or MRSS change was analyzed using linear regression model adjusted for age, sex, cohort, and baseline FVC/MRSS with FVC/MRSS slope as response variable.
Single cell RNA seq
After bronchoalveolar lavage, fresh lung explant tissue was stored in complete media on wet ice overnight. The tissue was washed in HBSS and then thoroughly minced in digestion buffer (HBSS, 2.5mg/mL Collagenase D, 100ug/mL DNase). Minced tissue was rocked 45 minutes at 37°C. Residual tissue material was transferred into fresh digestion buffer and rocked another 45 minutes at 37°C. Single cells from both rounds of digest were combined and utilized for downstream analyses. Single cell suspensions were sorted by FACS with markers for CD45, EPCAM and CD31. Sample processing for single-cell RNA-seq was done using Chromium Single Cell 3′ Library and Gel bead kit v2 following manufacturer’s guide (10x Genomics). The cell density and viability of single-cell suspension were determined by Vi-CELL XR cell counter (Beckman Coulter). All of the processed samples had a very high percentage of viable cells. The cell density was used to infer the volume of single cell suspension needed in the reverse transcription (RT) master mix, aiming to achieve ∼6,000 cells per sample. cDNAs and libraries were prepared following the manufacturer’s user guide (10x Genomics). Libraries were profiled by Bioanalyzer High Sensitivity DNA kit (Agilent Technologies) and quantified using Kapa Library Quantification Kit (Kapa Biosystems). Each library was sequenced in one lane of HiSeq4000 (Illumina) following the manufacturer’s sequencing specification (10x Genomics).
Sequencing data were aligned to human genome GRCh38 using Cell Ranger version 2.0.3 (10x Genomics). The data were processed using the Seurat R package version 3.1.1. For each sample, counts were normalized using NormalizeData function of Seurat (method = “LogNormalize”) and variable genes for dimensionality reduction were selected using FindVariableFeatures function of Seurat (selection.method = “vst,” nfeatures = 4000). After removal of IG (immunoglobulin) and TR (T cell receptor) genes, anchors among the samples were identified using FindIntegrationAnchors function, and these anchors were used to integrate all sample datasets together with IntegrateData function of Seurat. Then we performed graph-based unsupervised clustering of this integrated data using FindNeighbors (reduction = “pca,” dims = 1:30) and FindClusters (resolution = 0.2) function of Seurat. Visualization of the clusters on a 2D map was performed with uniform manifold approximation and projection (UMAP) using RunUMAP function of Seurat based on networks of the shared-nearest neighbor (SNN) graph. Differentially expressed genes of each cluster were identified using FindAllMarkers function of Seurat. Cell types for each cluster were annotated via canonical cell type markers.
For myeloid cell analysis, cells from CD45+ sort and annotated as myeloid cells were selected for reclustering, followed by data analysis using Seurat as described above. The graph-based clustering was performed using FindNeighbors (reduction = “pca,” dims = 1:20) and FindClusters (resolution = 0.4) function. Visualization of the clusters was performed using RunUMAP function, and differentially expressed genes of each cluster were identified using FindAllMarkers. Clusters which expressed markers of dendritic cells or NK/T cells were removed, and the remaining cells were annotated as macrophage subsets. Differentially expressed genes between two macrophage subsets of interest were performed, followed by GO analysis (http://geneontology.org/) on biological process terms.
Immunofluorescence (IF) staining
Normal human and SSc lungs, which were obtained under Institutional Review Board–approved protocols from donors were fixed in 4% (wt./vol.) PFA at 4C for 3 h. Samples were embedded in paraffin, sectioned at 7 μm. Antigen retrieval on paraffin sections used 10-mM sodium citrate butter, pH 6, in a pressure cooker–like device. Sections were incubated with primary antibodies, diluted in blocking solution (10% donkey serum, 3% BSA, 0.1% Triton X-100 in PBS), overnight at 4°C. After washing, sections were incubated with secondary antibodies diluted in 5% donkey serum, 0.1% Triton X-100 in PBS for 2 hours at room temperature and then washed and counterstained with DAPI. Antibodies were mouse CD68 (1:200; ab955; Abcam) and rabbit Osteopontin (1:1000; ab8448; Abcam).
Quantification and Statistical Analysis
Statistical analyses were performed using JMP Pro 14 SAS or Prism 7. For the small numbers of samples of in vitro experiments in Figures 1 and 2, the raw data were log transformed to normalize the distribution. Normality was validated with the Shapiro Wilk test. Exact test used to determine statistical significance depended upon factors including, but not limited to, the hypothesis being tested, sample size, data distribution, and is specified in the figure legends. All the values are mean ± SEM. Significance level for all statistics was set to p < 0.05.
Additional Resources
The registration numbers for FaSScinate and FocuSSced clinic trials are NCT01532869 and NCT02453256 respectively.
The link for FaSScinate trial is https://clinicaltrials.gov/ct2/show/NCT01532869
The link for FocuSSced trial is https://clinicaltrials.gov/ct2/show/NCT02453256
Acknowledgments
This work was supported by Genentech Inc. D.K. was funded by NIH/NIAMS R01 AR-07047.
Author Contributions
T.R.R. was responsible for the overall study concept and design. X.G., A.G., D.J.D., K.B.M., and N.R. acquired data. X.G., G.J., K.-H.S., and J.A.V.H. performed data analysis. Z.M., D.K., A.J., J.R.A., and P.J.W. contributed resources. X.G. and T.R.R. wrote the first draft of the manuscript. All authors read and approved the manuscript.
Declaration of Interests
All authors except P.J.W. and D.K. are current or past employees of Genentech, a member of the Roche group, and may hold Roche stock or stock options. D.K. is a consultant to Actelion, Acceleron, Arena, Bayer, Boehringer Ingelheim, Bristol-Myer Squibb, CSL Behring, Corbus, Cytori, GSK, Genentech/ Roche, Galapagos, Merck, Mitsubishi Tanabi, and UCB. He has received grants as part of investigator-initiated trials (to the University of Michigan) from Bayer, Bristol-Myer Squibb, and Pfizer and has stock options in Eicos Sciences, Inc. A.J. is a former employee of Genentech and is now employed by Gilead Sciences (Foster City, CA, USA).
Published: November 17, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100140.
Contributor Information
Dinesh Khanna, Email: khannad@med.umich.edu.
Thirumalai R. Ramalingam, Email: ramalingam.thirumalai@gene.com.
Supplemental Information
References
- 1.Denton C.P., Khanna D. Systemic sclerosis. Lancet. 2017;390:1685–1699. doi: 10.1016/S0140-6736(17)30933-9. [DOI] [PubMed] [Google Scholar]
- 2.Steen V.D., Medsger T.A. Changes in causes of death in systemic sclerosis, 1972-2002. Ann. Rheum. Dis. 2007;66:940–944. doi: 10.1136/ard.2006.066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furst D., Pope J., Seibold J., Bombardieri S., Denton C., Distler O., Kahaleh B., Kennedy A., Khanna D., Lafyatis R. Progress and priorities in systemic sclerosis: the next 10 years – report from the World Scleroderma Foundation. JSRD. 2016;1:7–9. [Google Scholar]
- 4.Nihtyanova S.I., Denton C.P. Autoantibodies as predictive tools in systemic sclerosis. Nat. Rev. Rheumatol. 2010;6:112–116. doi: 10.1038/nrrheum.2009.238. [DOI] [PubMed] [Google Scholar]
- 5.Walker U.A., Tyndall A., Czirják L., Denton C., Farge-Bancel D., Kowal-Bielecka O., Müller-Ladner U., Bocelli-Tyndall C., Matucci-Cerinic M. Clinical risk assessment of organ manifestations in systemic sclerosis: a report from the EULAR Scleroderma Trials And Research group database. Ann. Rheum. Dis. 2007;66:754–763. doi: 10.1136/ard.2006.062901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta H., Goulet P.-O., Nguyen V., Pérez G., Koenig M., Senécal J.-L., Sarfati M. Topoisomerase I peptide-loaded dendritic cells induce autoantibody response as well as skin and lung fibrosis. Autoimmunity. 2016;49:503–513. doi: 10.1080/08916934.2016.1230848. [DOI] [PubMed] [Google Scholar]
- 7.Kayser C., Fritzler M.J. Autoantibodies in systemic sclerosis: unanswered questions. Front. Immunol. 2015;6:167. doi: 10.3389/fimmu.2015.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Günther J., Rademacher J., van Laar J.M., Siegert E., Riemekasten G. Functional autoantibodies in systemic sclerosis. Semin. Immunopathol. 2015;37:529–542. doi: 10.1007/s00281-015-0513-5. [DOI] [PubMed] [Google Scholar]
- 9.Baroni S.S., Santillo M., Bevilacqua F., Luchetti M., Spadoni T., Mancini M., Fraticelli P., Sambo P., Funaro A., Kazlauskas A. Stimulatory autoantibodies to the PDGF receptor in systemic sclerosis. N. Engl. J. Med. 2006;354:2667–2676. doi: 10.1056/NEJMoa052955. [DOI] [PubMed] [Google Scholar]
- 10.Classen J.-F., Henrohn D., Rorsman F., Lennartsson J., Lauwerys B.R., Wikström G., Rorsman C., Lenglez S., Franck-Larsson K., Tomasi J.-P. Lack of evidence of stimulatory autoantibodies to platelet-derived growth factor receptor in patients with systemic sclerosis. Arthritis Rheum. 2009;60:1137–1144. doi: 10.1002/art.24381. [DOI] [PubMed] [Google Scholar]
- 11.Loizos N., Lariccia L., Weiner J., Griffith H., Boin F., Hummers L., Wigley F., Kussie P. Lack of detection of agonist activity by antibodies to platelet-derived growth factor receptor alpha in a subset of normal and systemic sclerosis patient sera. Arthritis Rheum. 2009;60:1145–1151. doi: 10.1002/art.24365. [DOI] [PubMed] [Google Scholar]
- 12.Sato S., Fujimoto M., Hasegawa M., Takehara K., Tedder T.F. Altered B lymphocyte function induces systemic autoimmunity in systemic sclerosis. Mol. Immunol. 2004;41:1123–1133. doi: 10.1016/j.molimm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Silver R.M., Metcalf J.F., LeRoy E.C. Interstitial lung disease in scleroderma. Immune complexes in sera and bronchoalveolar lavage fluid. Arthritis Rheum. 1986;29:525–531. doi: 10.1002/art.1780290410. [DOI] [PubMed] [Google Scholar]
- 14.Seibold J.R., Medsger T.A., Jr., Winkelstein A., Kelly R.H., Rodnan G.P. Immune complexes in progressive systemic sclerosis (scleroderma) Arthritis Rheum. 1982;25:1167–1173. doi: 10.1002/art.1780251004. [DOI] [PubMed] [Google Scholar]
- 15.Siminovitch K., Klein M., Pruzanski W., Wilkinson S., Lee P., Yoon S.J., Keystone E. Circulating immune complexes in patients with progressive systemic sclerosis. Arthritis Rheum. 1982;25:1174–1179. doi: 10.1002/art.1780251005. [DOI] [PubMed] [Google Scholar]
- 16.Stifano G., Christmann R.B. Macrophage Involvement in Systemic Sclerosis: Do We Need More Evidence? Curr. Rheumatol. Rep. 2016;18:2. doi: 10.1007/s11926-015-0554-8. [DOI] [PubMed] [Google Scholar]
- 17.Higashi-Kuwata N., Jinnin M., Makino T., Fukushima S., Inoue Y., Muchemwa F.C., Yonemura Y., Komohara Y., Takeya M., Mitsuya H., Ihn H. Characterization of monocyte/macrophage subsets in the skin and peripheral blood derived from patients with systemic sclerosis. Arthritis Res. Ther. 2010;12:R128. doi: 10.1186/ar3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christmann R.B., Sampaio-Barros P., Stifano G., Borges C.L., de Carvalho C.R., Kairalla R., Parra E.R., Spira A., Simms R., Capellozzi V.L., Lafyatis R. Association of Interferon- and transforming growth factor β-regulated genes and macrophage activation with systemic sclerosis-related progressive lung fibrosis. Arthritis Rheumatol. 2014;66:714–725. doi: 10.1002/art.38288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behnen M., Leschczyk C., Möller S., Batel T., Klinger M., Solbach W., Laskay T. Immobilized immune complexes induce neutrophil extracellular trap release by human neutrophil granulocytes via FcγRIIIB and Mac-1. J. Immunol. 2014;193:1954–1965. doi: 10.4049/jimmunol.1400478. [DOI] [PubMed] [Google Scholar]
- 20.Matsue Y., Tsutsumi M., Hayashi N., Saito T., Tsuchishima M., Toshikuni N., Arisawa T., George J. Serum osteopontin predicts degree of hepatic fibrosis and serves as a biomarker in patients with hepatitis C virus infection. PLoS ONE. 2015;10:e0118744. doi: 10.1371/journal.pone.0118744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binder M., Christoph S., Sehnert B., Uhl M., Peter H.-H., Voll R.E., Warnatz K., Kollert F. Elevated serum osteopontin levels in idiopathic retroperitoneal fibrosis. Clin. Exp. Rheumatol. 2012;30:772–775. [PubMed] [Google Scholar]
- 22.Wu M., Schneider D.J., Mayes M.D., Assassi S., Arnett F.C., Tan F.K., Blackburn M.R., Agarwal S.K. Osteopontin in systemic sclerosis and its role in dermal fibrosis. J. Invest. Dermatol. 2012;132:1605–1614. doi: 10.1038/jid.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzen J.M., Krämer R., Meier M., Werfel T., Wichmann K., Hoeper M.M., Riemekasten G., Becker M.O., Haller H., Witte T. Osteopontin in the development of systemic sclerosis--relation to disease activity and organ manifestation. Rheumatology (Oxford) 2010;49:1989–1991. doi: 10.1093/rheumatology/keq223. [DOI] [PubMed] [Google Scholar]
- 24.Pardo A., Gibson K., Cisneros J., Richards T.J., Yang Y., Becerril C., Yousem S., Herrera I., Ruiz V., Selman M., Kaminski N. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med. 2005;2:e251. doi: 10.1371/journal.pmed.0020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchibori T., Matsuda K., Shimodaira T., Sugano M., Uehara T., Honda T. IL-6 trans-signaling is another pathway to upregulate Osteopontin. Cytokine. 2017;90:88–95. doi: 10.1016/j.cyto.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Icer M.A., Gezmen-Karadag M. The multiple functions and mechanisms of osteopontin. Clin. Biochem. 2018;59:17–24. doi: 10.1016/j.clinbiochem.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Morimoto Y., Hirahara K., Kiuchi M., Wada T., Ichikawa T., Kanno T., Okano M., Kokubo K., Onodera A., Sakurai D. Amphiregulin-Producing Pathogenic Memory T Helper 2 Cells Instruct Eosinophils to Secrete Osteopontin and Facilitate Airway Fibrosis. Immunity. 2018;49:134–150.e6. doi: 10.1016/j.immuni.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 28.Shan M., Yuan X., Song L.Z., Roberts L., Zarinkamar N., Seryshev A., Zhang Y., Hilsenbeck S., Chang S.H., Dong C. Cigarette smoke induction of osteopontin (SPP1) mediates T(H)17 inflammation in human and experimental emphysema. Sci. Transl. Med. 2012;4:117ra9. doi: 10.1126/scitranslmed.3003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wynn T.A., Vannella K.M. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray P.J., Wynn T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neighbors M., Cabanski C.R., Ramalingam T.R., Sheng X.R., Tew G.W., Gu C., Jia G., Peng K., Ray J.M., Ley B. Prognostic and predictive biomarkers for patients with idiopathic pulmonary fibrosis treated with pirfenidone: post-hoc assessment of the CAPACITY and ASCEND trials. Lancet Respir. Med. 2018;6:615–626. doi: 10.1016/S2213-2600(18)30185-1. [DOI] [PubMed] [Google Scholar]
- 32.Prasse A., Probst C., Bargagli E., Zissel G., Toews G.B., Flaherty K.R., Olschewski M., Rottoli P., Müller-Quernheim J. Serum CC-chemokine ligand 18 concentration predicts outcome in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2009;179:717–723. doi: 10.1164/rccm.200808-1201OC. [DOI] [PubMed] [Google Scholar]
- 33.Prasse A., Pechkovsky D.V., Toews G.B., Schäfer M., Eggeling S., Ludwig C., Germann M., Kollert F., Zissel G., Müller-Quernheim J. CCL18 as an indicator of pulmonary fibrotic activity in idiopathic interstitial pneumonias and systemic sclerosis. Arthritis Rheum. 2007;56:1685–1693. doi: 10.1002/art.22559. [DOI] [PubMed] [Google Scholar]
- 34.Reyfman P.A., Walter J.M., Joshi N., Anekalla K.R., McQuattie-Pimentel A.C., Chiu S., Fernandez R., Akbarpour M., Chen C.-I., Ren Z. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valenzi E., Bulik M., Tabib T., Morse C., Sembrat J., Trejo Bittar H., Rojas M., Lafyatis R. Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann. Rheum. Dis. 2019;78:1379–1387. doi: 10.1136/annrheumdis-2018-214865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habermann A.C., Gutierrez A.J., Bui L.T., Yahn S.L., Winters N.I., Calvi C.L., Peter L., Chung M.-I., Taylor C.J., Jetter C. Single-cell RNA-sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 2020;6:eaba1972. doi: 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams T.S., Schupp J.C., Poli S., Ayaub E.A., Neumark N., Ahangari F., Chu S.G., Raby B.A., DeIuliis G., Januszyk M. Single Cell RNA-seq reveals ectopic and aberrant lung resident cell populations in Idiopathic Pulmonary Fibrosis. Sci. Adv. 2020;6:eaba1983. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morse C., Tabib T., Sembrat J., Buschur K.L., Bittar H.T., Valenzi E., Jiang Y., Kass D.J., Gibson K., Chen W. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 2019;54:1802441. doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khanna D., Denton C.P., Jahreis A., van Laar J.M., Frech T.M., Anderson M.E., Baron M., Chung L., Fierlbeck G., Lakshminarayanan S. Safety and efficacy of subcutaneous tocilizumab in adults with systemic sclerosis (faSScinate): a phase 2, randomised, controlled trial. Lancet. 2016;387:2630–2640. doi: 10.1016/S0140-6736(16)00232-4. [DOI] [PubMed] [Google Scholar]
- 40.Khanna D., Lin C.J.F., Furst D.E., Goldin J., Kim G., Kuwana M., Allanore Y., Matucci-Cerinic M., Distler O., Shima Y., focuSSced investigators Tocilizumab in systemic sclerosis: a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Respir. Med. 2020;8:963–974. doi: 10.1016/S2213-2600(20)30318-0. [DOI] [PubMed] [Google Scholar]
- 41.Khanna D., Berrocal V.J., Giannini E.H., Seibold J.R., Merkel P.A., Mayes M.D., Baron M., Clements P.J., Steen V., Assassi S. The American College of Rheumatology Provisional Composite Response Index for Clinical Trials in Early Diffuse Cutaneous Systemic Sclerosis. Arthritis Rheumatol. 2016;68:299–311. doi: 10.1002/art.39501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamiya H., Panlaqui O.M. Systematic review and meta-analysis of clinical significance of autoantibodies for idiopathic pulmonary fibrosis. BMJ Open. 2019;9:e027849. doi: 10.1136/bmjopen-2018-027849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J.S., Kim E.J., Lynch K.L., Elicker B., Ryerson C.J., Katsumoto T.R., Shum A.K., Wolters P.J., Cerri S., Richeldi L. Prevalence and clinical significance of circulating autoantibodies in idiopathic pulmonary fibrosis. Respir. Med. 2013;107:249–255. doi: 10.1016/j.rmed.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiller H.B., Mayr C.H., Leuschner G., Strunz M., Staab-Weijnitz C., Preisendörfer S., Eckes B., Moinzadeh P., Krieg T., Schwartz D.A. Deep Proteome Profiling Reveals Common Prevalence of MZB1-Positive Plasma B Cells in Human Lung and Skin Fibrosis. Am. J. Respir. Crit. Care Med. 2017;196:1298–1310. doi: 10.1164/rccm.201611-2263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Lopategi A., Ge X., Lu Y., Kitamura N., Urtasun R., Leung T.-M., Fiel M.I., Nieto N. Osteopontin induces ductular reaction contributing to liver fibrosis. Gut. 2014;63:1805–1818. doi: 10.1136/gutjnl-2013-306373. [DOI] [PubMed] [Google Scholar]
- 46.Duffield J.S., Forbes S.J., Constandinou C.M., Clay S., Partolina M., Vuthoori S., Wu S., Lang R., Iredale J.P. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J. Clin. Invest. 2005;115:56–65. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA seq data from monocytes stimulated by IC generated reported in this paper is GSE155687.
The accession numnber for the scRNA seq data supporting the current study is made available at GEO, under the accession number GSE159354.