Summary
T cells use highly diverse receptors (TCRs) to identify tumor cells presenting neoantigens arising from genetic mutations and establish anti-tumor activity. Immunotherapy harnessing neoantigen-specific T cells to target tumors has emerged as a promising clinical approach. To assess whether a comprehensive peripheral mononuclear blood cell analysis predicts responses to a personalized neoantigen cancer vaccine combined with anti-PD-1 therapy, we characterize the TCR repertoires and T and B cell frequencies in 21 patients with metastatic melanoma who received this regimen. TCR-α/β-chain sequencing reveals that prolonged progression-free survival (PFS) is strongly associated with increased clonal baseline TCR repertoires and longitudinal repertoire stability. Furthermore, the frequencies of antigen-experienced T and B cells in the peripheral blood correlate with repertoire characteristics. Analysis of these baseline immune features enables prediction of PFS following treatment. This method offers a pragmatic clinical approach to assess patients’ immune state and to direct therapeutic decision making.
Keywords: TCR repertoire, flow cytometry, T cell clonality, immunotherapy, cancer vaccines, predictive biomarkers, melanoma, NEO-PV-01, anti-PD-1
Graphical Abstract
Highlights
Pre-treatment blood-based factors predict response to immunotherapy
TCR repertoire clonality and stability associate with improved clinical outcomes
Baseline T and B cell memory phenotypes associate with improved clinical outcomes
Combined baseline TCR repertoire and PBMC phenotypes predict immunotherapy response
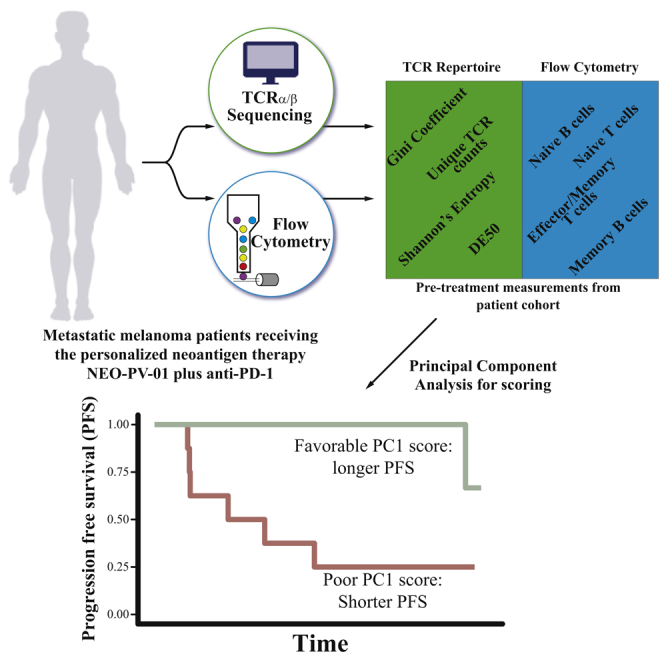
Poran et al. study peripheral blood cells from metastatic melanoma patients before, during, and after treatment with personalized neoantigen therapy plus anti-PD-1. The combination of T cell receptor repertoire profiling and immunophenotyping of blood cells collected pre-treatment presents a strong predictor for response to treatment in a minimally invasive manner.
Introduction
Evidence has been accumulating that the immune system can recognize, prevent, and control tumor growth.1, 2, 3 It is now widely accepted that somatic mutations in tumor cells can lead to neoantigen presentation, recognition by cytotoxic T cells through their T cell receptor (TCR), and tumor cell killing.4, 5, 6 We recently reported the first clinical trial combining a personalized neoantigen vaccine (NEO-PV-01) with PD-1 inhibition in multiple solid tumors and showed robust de novo induction of neoantigen-specific T cells.7 TCR repertoires have previously been used as potential biomarkers for response to immune checkpoint inhibitors (ICI), but often with low predictive confidence.8, 9, 10, 11, 12, 13 A recent study demonstrated how the peripheral blood TCR repertoire response to one cycle of anti-PD-1 is informative of later treatment effectiveness.14 In addition, phenotyping of immune populations from peripheral blood mononuclear cells (PBMCs)15, 16, 17, 18, 19, 20 has been previously used as ICI response predictors. However, associating PBMC TCR repertoires with immune cell phenotypes could prove to be a strong response predictor for ICI therapy. We developed a highly predictive model based on the combined analysis of peripheral blood TCR repertoire features and frequencies of T and B cell subpopulations at the pre-treatment time point. This model indicates an underlying susceptible immune state that is distinct for metastatic melanoma patients who had a favorable response to personalized neoantigen vaccine plus anti-PD-1 treatment.
Results
Patients with Longer PFS Exhibit Increased Peripheral TCR Clonality prior to and throughout Treatment
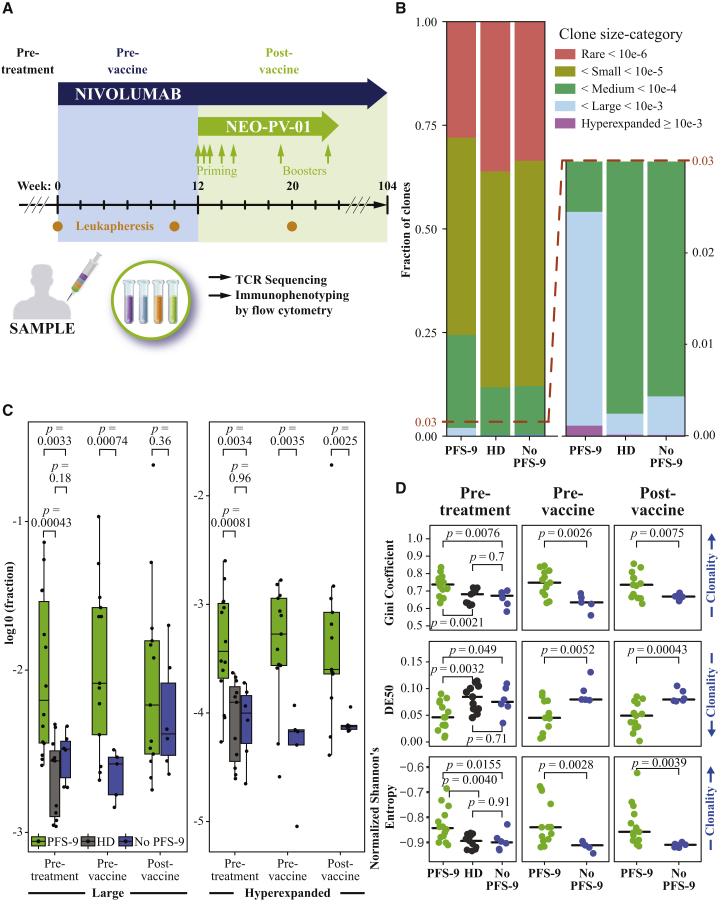
Metastatic melanoma patients enrolled in the neoantigen vaccine trial NT-001 (NCT02897765) received nivolumab combined with the personalized neoantigen vaccine NEO-PV-01.7 Three leukapheresis samples were taken at week 0 (pre-treatment, pre-T), week 10 (pre-vaccination, pre-V), and week 20 (post-vaccination, post-V) (Figure 1A). First, RNA was extracted from CD3+ pan-T cells from these leukapheresis samples and subjected to T cell receptor α-chain and β-chain (TCR-α and TCR-β) sequencing.21 We analyzed a total of 57 samples from 21 melanoma patients in the trial for whom we had samples available from at least one time point. Patients were classified into two groups: those who achieved progression-free survival (PFS) 9 months post-initiation of nivolumab therapy and those who did not (PFS-9 and no PFS-9, respectively). Of the 21 patients analyzed in this study, 14 had PFS-9, and seven did not7 (Table S1).
Figure 1.
Patients with Lack of Progression at 9 Months Have a Higher Peripheral TCR Repertoire Clonality prior to and throughout Study Treatment
(A) Treatment schedule outline of the single-arm clinical trial of nivolumab plus personalized neoantigen vaccine (NEO-PV-01). Timings are indicated for nivolumab (blue arrow), personalized vaccine (NEO-PV-01, green arrow), and leukaphereses (orange dots). Leukaphereses from three time points (pre-treatment, pre-vaccine, and post-vaccine) are used for CD3+ T cell isolation for TCR-α/β sequencing and PBMCs for immunophenotyping by flow cytometry.
(B) The proportion of clones belonging to each clone size category, averaged across patients (or HDs), and time points. An inset focusing on larger bins is provided (right). The legend defines frequency-based categories.
(C) The (log–) fraction of clones belonging to the large (left) or hyperexpanded (right), for patients with and without PFS-9 at each time point or HDs. Boxplots indicate 25%, 50%, and 75% percentiles, and whiskers extend to the smallest/largest value within 1.5 times the interquartile range. p values are derived from a two-tailed Student’s t test.
(D) The skewedness of the TCR-β repertoire frequency distribution measured by the Gini coefficient, DE50, and normalized Shannon’s entropy of each HD and patient across time points. The black line indicates median. p values are derived from a two-tailed Student’s t test.
We assigned each TCR-β amino acid (AA) complementary determining region 3 (CDR3) clone to a category based on its observed frequency: rare, small, medium, large, or hyperexpanded22 (STAR Methods). The relative fractions of each category in each sample were averaged across the aggregate samples of patients with and without PFS-9 (Figure 1B). Notable differences between the two groups were observed, particularly in the large and hyperexpanded categories (Figure 1B, right). Similarly, we analyzed samples from 11 healthy donors (HDs) to compare with the TCR-β repertoires of melanoma patients. The clonal composition of the HDs resembled those of patients without PFS-9. We compared the fractions of clones in each category of patients with and without PFS-9 at each time point (Figure 1C; Figure S1A). Interestingly, at pre-T and pre-V, the large and hyperexpanded categories were increased in patients with PFS-9 compared with patients without PFS-9, at the expense of the categories of less abundant clones, particularly the rare clones. At post-V, only the hyperexpanded category showed a significant difference, suggesting a possible vaccine-specific effect (Figure 1C, right panel). These results demonstrate that samples from patients with PFS-9 have a significantly increased proportion of high frequency clones, especially hyperexpanded clones. This resembles the increased clonality of peripheral TCR repertoires of patients who have a chronic viral infection.23 These increases in high-frequency categories were also detected when comparing patients with PFS-9 with HDs.
To further characterize the distribution of clonal frequencies in the repertoires, we quantified the skewedness/evenness of the frequency distributions using clonality and diversity parameters at each time point. We calculated the Gini coefficient,10,24 Diversity-Evenness (DE50),9 normalized Shannon’s entropy,25,26 sum of squares, Lorentz curves,27 and the number of unique nucleotide and AA TCR-β CDR3 sequences10 and tested their association with PFS-9 status (Figure 1D; Figures S1B–S1D). The difference in Gini coefficient and DE50 between patients with and without PFS-9 at the pre-T time point were reported in Ott et al., 2020.7 These parameters indicated an increased clonality of the T cell repertoire in PFS-9 patients at all three time points. While repertoire clonality increases with age,22 there were no significant age differences between patients with and without PFS-9. Similarly, the two groups had identical proportions of males and females (Table S1). Therefore, patient age and sex are unlikely to confound the clonality differences. We detected no difference in clonality between patients without PFS-9 and HDs, although the HD population is younger on average than the patient population. We found no differences between either the average CDR3 length or the total count of aligned TCR molecules of patients with and without PFS-9 (Figure S1C). In addition, we determined the reactivity to the common viral antigens CMV, EBV, or influenza (CEF) by an interferon (IFN)-γ ELISpot assay to assess the impact of exposure to these viruses on the measured TCR repertoire characteristics. We detected no difference in CEF reactivity between patients with and without PFS-9 (Figure S1E). Taken together, these data demonstrate that peripheral TCR repertoire clonality was increased in melanoma patients with PFS-9 even prior to initiation of treatment. These findings support that TCR clonality from PBMCs may represent a minimally invasive biomarker predicting response to the combination therapy.
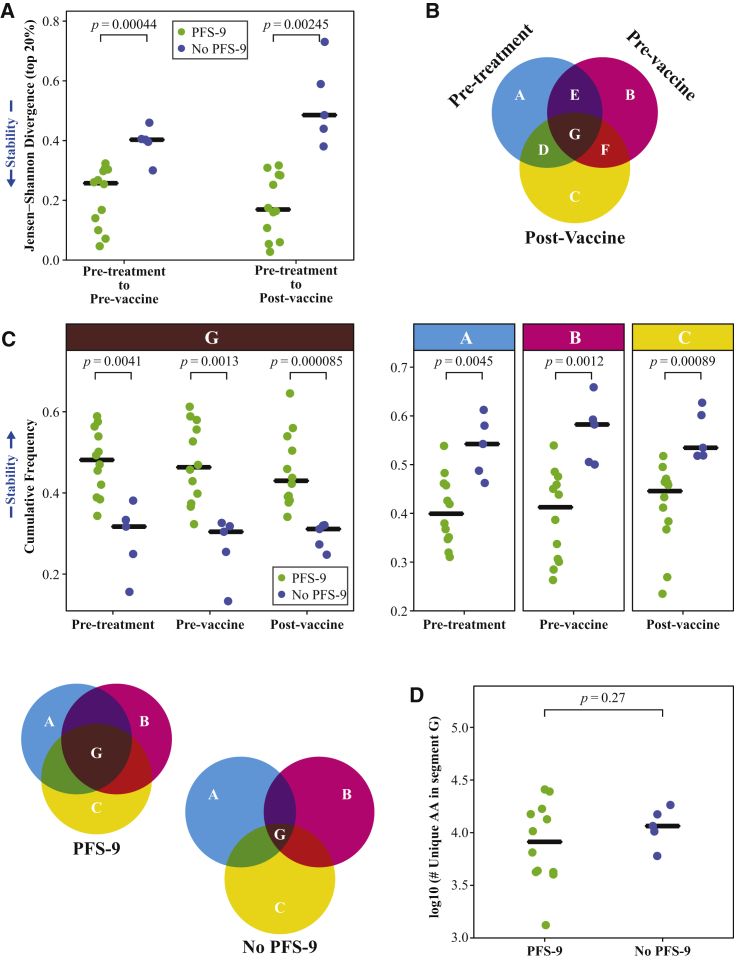
Peripheral TCR Repertoires in Patients with Prolonged PFS Are More Stable over Time
Notably, the overall clonality measurements did not change significantly in response to treatment. We therefore sought to understand the extent to which the identities of the TCR-β repertoires changed over time. We first tested whether turnover rates, as measured by the Jensen-Shannon Divergence (JSD),28,29 correlated with PFS-9 status. Analyzing the most frequent clones covering the top 20% of each repertoire, we compared the JSD of pre-T to either the pre-V or post-V time points. Both comparisons demonstrated lower JSD values, indicating lower TCR turnover, in patients with PFS-9 (Figure 2A). The same was true when comparing the post-V to the pre-V time point (Figure S2A), indicating that the differences in turnover do not depend on the choice of baseline. The difference in turnover between patients with and without PFS-9 remains significant regardless of the fraction of the repertoire used for the calculation (Figure S2B). Of note, comparing pre-V with post-V turnover (relative to pre-T) in patients with PFS-9 shows no difference, whereas the turnover of TCR clones in the repertoires of patients without PFS-9 continues to increase from pre-V to post-V (Figure S2B).
Figure 2.
Peripheral TCR Repertoires Are More Stable over Time in Patients Who Do Not Progress at 9 Months
(A) Jensen-Shannon Divergence (JSD) of TCR-β CDR3 sequences accounting for the top 20% of the repertoire frequency between pre-V (left) or post-V (right) and baseline (pre-T) of patients with and without PFS-9. Low JSD values represent repertoire stability as indicated by the arrow (left). Black line indicates median. p values are derived from a two-tailed Student’s t test.
(B) Schematic of a 3-way repertoire comparison. Segments in the Venn diagram represent the cumulative frequencies of the TCR-β CDR3 sequences detected in the intersecting time points in that segment.
(C) Comparison of the cumulative frequencies of TCR-β CDR3 sequences between patients with and without PFS-9, at each time point. High cumulative frequency of G represents repertoire stability is as indicated by the arrow (left). Black line indicates median. p values are derived from a two-tailed Student’s t test. Venn diagram on the bottom left illustrate repertoire stability of patients with and without PFS-9.
(D) The number of unique TCR-β CDR3 AA sequences detected at all three time points in patients with and without PFS-9. The black line indicates median. Black line indicates median. p value derived from a two-tailed Student’s t test.
See also Figure S2.
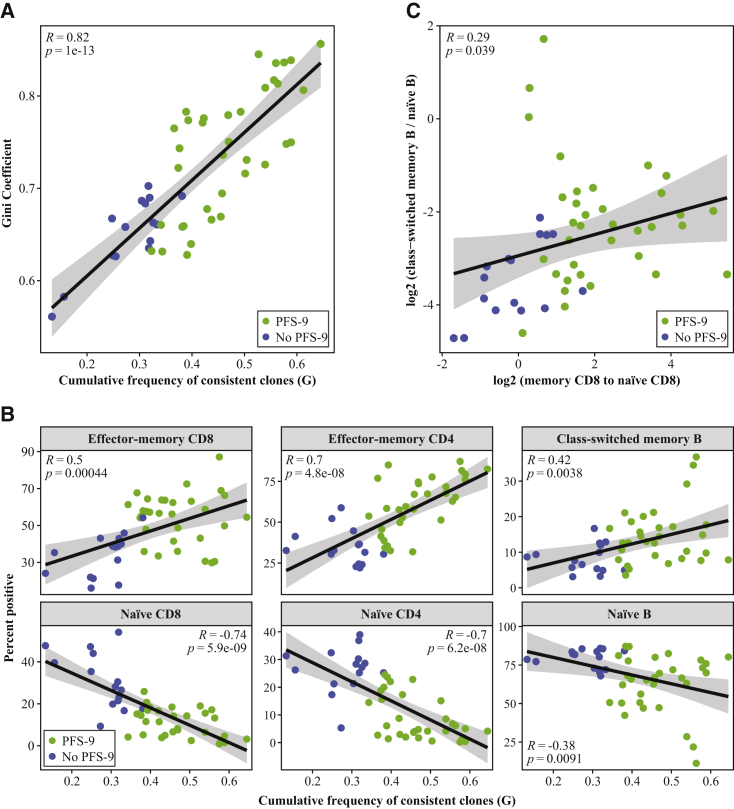
To further characterize repertoire stability, we tested TCR-β overlap across all three time points (Figure 2B). We calculated the cumulative frequencies of clones detected at a single time point (Venn diagram segment A, B, or C), two time points (segment D, E, or F), and persistent clones, which were found in all three samples over a 5-month treatment period (segment G). The cumulative frequency of persistent T cell clones (in segment G) is significantly increased in patients with PFS-9 compared with patients without PFS-9 (Figure 2C, left). This increase comes at the expense of the frequencies of clones detected at a single time point (segments A, B, and C, Figure 2C, right) but not of clones found at two time points (Figure S2C). No difference was detected in the absolute number of unique clones in segment G in patients with or without PFS-9 (Figure 2D). This indicates that the increased cumulative frequency of this segment is driven by the clone sizes, rather than the number of individual clones. These findings demonstrate an increased longitudinal stability of the TCR-β repertoire in patients with PFS-9. To test whether repertoire clonality and the long-term stability are correlated, we analyzed the association between the Gini coefficient (Figure 1D) and the cumulative frequency of the persistent clones (G segment). We found a strong correlation between these values (Figure 3A), indicating that repertoire clonality and stability are linked, and the expanded clones are persistent across the three time points. Conversely, we observed a negative correlation comparing TCR-β clonality with the cumulative frequency of clones that were only detected at a single time point (segments A, B, and C).
Figure 3.
Peripheral TCR Repertoires Stability and Diversity Are Correlated and Positively Correlate with Effector/Memory Phenotypes
(A) The Gini coefficient of each patient versus the cumulative frequency of the G segment from each time points. Pearson's correlation coefficient (R) and the associated p value are indicated.
(B) The percentage of positive effector-memory CD8+, memory CD4+ T cell, class-switched memory B cell, naive CD8+, naive CD4+ T cell, and naive B cell populations versus the cumulative frequency of the G segment (the persistent TCR-β clones). Pearson's correlation coefficient (R) and the associated p value are indicated.
(C) The ratio of class-switched memory B cells to naive B cells versus the ratio of CD8+ effector-memory T cells to naive CD8+ T cells. Pearson's correlation coefficient (R) and the associated p value are indicated.
TCR-α-Chain Sequencing Is Indicative of Highly Consistent Repertoire Clonality and Stability
TCR specificity is determined by the TCR-α and TCR-β-chain pairing. While the pairing of these chains is not preserved in bulk RNA sequencing, we evaluated whether the CDR3 sequences of the TCR-α chain (CDR3α) support our findings regarding the clonality and stability of the TCR-β repertoires. To that end, we sequenced the CDR3α of all 21 patients and 11 HDs for which materials were available (Table S1). TCR-α sequencing showed increased large and hyperexpanded clones in PFS-9 versus no PFS-9 patient samples, matching the TCR-β sequencing results. Reproducible results were also observed for the various clonality/diversity measurements, as well as repertoire stability (Figures S2D–S2F). This concordance between TCR-α and TCR-β repertoire sequencing supports that either can represent the clonal structure in a sample.30
Peripheral TCR Repertoire Stability and Diversity Correlate with Effector/Memory Phenotypes
We next hypothesized that the increased TCR repertoire clonality and stability in patients with PFS-9 is related to the phenotype of the T cells of these patients. We compared the cumulative frequency of the persistent TCR-β clones (G segment) with the frequencies of T cell subpopulations determined by flow cytometry (Figures S3A and S3B). We observed a strong positive correlation between the cumulative frequency of persistent clones and the frequency of effector-memory/memory CD8+ and CD4+ T cells across patients. We also observed a strong negative correlation with naive T cells, and to a lower extent with CTLA-4-expressing T cells (Figure 3B; Figure S3C). A comparable enrichment of effector-memory/memory T cells has been reported in patients with chronic viral infections,23 again suggesting similarities in immune phenotypes of patients with PFS-9 and individuals with a chronic viral infection. Interestingly, we also detected a positive correlation between the persistent TCR-β clones and the memory B cell compartment, and a negative correlation with the naive B cell compartment (Figure 3B). Immunophenotyping of PBMCs showed different subpopulation frequencies between patients with and without PFS-9, consistent with reports from independent cohorts.15,16 Together, these findings suggest that TCR repertoire features can inform the B cell compartment and perhaps other immune cell phenotypes. This is further supported by the correlation between the ratio of effector-memory to naive CD8 T cells and the ratio of class-switched to naive B cells (Figure 3C). These results suggest that a broad view of a patient’s adaptive immune system is relevant in predicting the clinical efficacy of immunotherapy.
To directly test for a relationship between the frequency of a clone, its phenotype and antigen specificity, we sorted and sequenced the TCR-αβ of single neoantigen-specific T cells for a few patients with sample availability (STAR Methods). We validated via transduction eight TCR-αβ sequences (STAR Methods), which target six epitopes from across five patients (Table S2). Three TCRs, originating from patients M1 and M13, were previously reported in Ott et al., 2020.7 Of the validated TCRs, four were not detectable at the pre-T time point (M1−TCR3,7 M4−TCR1, M13-TCR4, and M13-TCR67) and significantly expanded only following vaccination, reaching medium (n = 2) or large (n = 2) frequency categories post-vaccine, suggesting vaccine-induced expansion. Two validated TCRs (M1-TCR3 and M4-TCR1) were specifically selected for validation based on their expansion following vaccine treatment. Excluding these two, three of the six remaining neoantigen-specific clones (M6−TCR1.1, M14-TCR1, M14-TCR2) were found in the large frequency category of the pre-T peripheral TCR-β repertoire, indicating that at least some large clones at baseline are neoantigen specific (Figure S4). The frequency of these validated TCR-α and TCR-β in the periphery correlated to the number of IFN-γ spot forming cells as reported in Ott et al., 2020.7 Immunophenotyping showed that, in all cases, the population from which the neoantigen-specific T cells were obtained was of memory/effector-memory phenotype7 (Figure S4).
Together, the results from this small set of validated TCRs allow us to integrate the TCR sequence, clone size, functionality, and phenotype. These data show that there are cases in which larger clones at the pre-T time point are neoantigen specific and are putatively IFN-γ producing with a memory phenotype, while in other cases we detect clones that expanded following vaccine to have these same features.
Combined Baseline Peripheral TCR-α/β Repertoire Features and Immunophenotyping Predict PFS
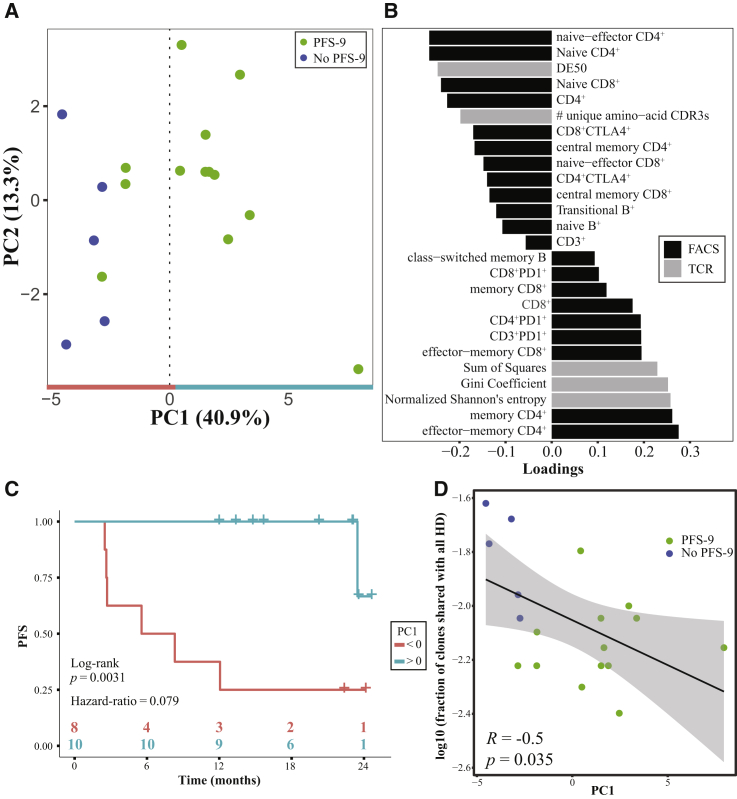
We sought to determine the strength of the combined pre-treatment measurements from PBMCs in predicting PFS, and which individual measurements are key to the prediction. For this analysis, we aggregated over 25 PFS-9-informative baseline features per patient, which allowed us to build a predictive model (Table S3). To identify dominant biomarkers, we used principal-component analysis (PCA), an unsupervised algorithm that represents the data along their axes with the largest variance. Importantly, PCA was performed without providing the algorithm with the explicit labels of the patients’ PFS-9 status. The algorithm separated patients with and without PFS-9 along PC1, the axis representing the highest variance in the data (Figure 4A). In order to better understand the key features driving this separation, we examined the impact of the features (loadings) on PC1 (Figure 4B). Among the top features correlating with PC1 are a clonal TCR repertoire, Gini coefficient, and a memory T cell phenotype, including PD-1+ T cells. Conversely, TCR repertoire diversity (as indicated by DE50) and naive T cell phenotypes negatively correlate with PC1. A class-switched memory B cell phenotype also correlated with PC1, which might be related to recent findings of the involvement of these cells in the tumor microenvironment in patients responding to ICI.31 The relative contribution of each of these features to PC1 demonstrated by the loadings (Figure 4B) points to the measurements that are most important in the makeup of the PC1 score.
Figure 4.
PCA of Baseline Peripheral TCR-β Repertoire Features and Immunophenotyping Separates Patients by PFS Status at 9 Months
(A) First 2 components from a PCA of the aggregate peripheral measurements from the TCR-β repertoire and immunophenotyping.
(B) The contributions (loadings) of the measured features to PC1. Color indicates source of data.
(C) Kaplan-Meier curves for PFS of patients with PC1 >0 (teal) versus patients with PC1 <0 (red).
(D) The fraction of public TCR-β CDR3 sequences (shared with all 11 HDs) versus the PC1 score of each patient. Pearson's correlation coefficient (R) and the associated p value are indicated.
When categorizing patients as having either a positive or a negative PC1 value, we saw a significant difference between the median PFS of the two groups (6.93 months for PC1 <0 while at 21.6 months the median has not yet been reached for PC1 >0, hazard ratio = 0.079, p = 0.0031) (Figure 4C), and overall a significant correlation between PC1 and PFS (Figure S5A). When repeating the analysis with the same features but replacing the TCR-β repertoire values by those derived from the TCR-α sequencing, both the PCA plot and the median PFS analysis reach a similar patient stratification (Figures S5C–S5E). These findings demonstrate a marked difference in peripheral blood samples of patients with and without PFS-9. A clonal T cell repertoire and an antigen-experienced PBMC phenotype were the features that correlated most with longer PFS following combined anti-PD-1 and neoantigen therapy.
To assess repertoire overlap between the samples from melanoma patients and HDs, we screened TCR-β CDR3 sequences from each patient for their presence in the HD samples. We quantified the proportion of TCR-β sequences (out of the 1,000 most frequent clones at baseline) that were shared with the 11 HD repertoires, and the number of HDs with which they were shared. The proportion of highly public clones (shared with all 11 healthy donor repertoires) was decreased in patients with PFS-9 compared to patients without PFS-9 (Figure S5B). Furthermore, we observed a negative correlation between the overall proportion of the public clones and the PC1 score of each patient (Figure 4D). These data indicate that the increased PC1 scores reflect a decreased publicness of the TCR repertoire, suggesting that the expanded circulating T cell clones in patients with PFS-9 are not commonly found in HDs, and are potentially private, tumor-related clones. This additional characteristic might reflect the patient immune state and could function as a clinically meaningful predictor of response to immunotherapy.
Discussion
Here, we show that data integration from a comprehensive analysis of PBMC samples enables an accurate response prediction. These analyses indicate that PFS in metastatic melanoma patients can be predicted with pre-treatment peripheral blood samples, strongly suggesting a pre-existing immune state that is amenable to immune modulation through ICI and/or neoantigen therapy. Characteristics of this immune state include high effector-memory T and B cell frequencies, as well as a clonal TCR repertoire with low turnover and decreased levels of highly public TCRs. A diverse, non-clonal TCR baseline repertoire detected in both the no PFS-9 patient samples and the young HD samples suggests a reduced tumor antigen-induced immune modulation in patients without PFS-9. These findings support the hypothesis that, in some patients, metastatic cancer leads to changes in the peripheral blood immune repertoire, rendering these patients more likely to respond to immunotherapy. It should be noted that varying levels of differences between a patient’s tumor TCR repertoire and peripheral blood repertoire could be expected, due to the unique conditions within the tumor microenvironment, T cell trafficking, and antigen exposure.32,33
We propose a minimally invasive approach that could facilitate patient selection and improve decision making regarding the appropriate course of treatment.
Limitations of Study
This study presents a cohort of patients treated with both NEO-PV-01 and nivolumab, with a limited set of patients and available samples. Due to the unique treatment settings, the study currently lacks a validation set and additional studies would need to be performed to validate these results and potentially extend them to other cancer indications and immunotherapies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD107a-BV786 | BD Biosciences | Cat # 563869; RRID:AB_2738458 |
| CD137-BV650 | BD Biosciences | Cat # 564092; RRID:AB_2738586 |
| CD138-PE | BD Biosciences | Cat # 552026; RRID:AB_394323 |
| CD14-FITC | BioLegend | Cat # 301804; RRID:AB_400086 |
| CD152 (CTLA-4)-PE Cy5 | BD Biosciences | Cat # 555854; RRID:AB_396177 |
| CD19-APC-Cy7 | BD Biosciences | Cat # 557791; RRID:AB_396873 |
| CD19-BUV496 | BD Biosciences | Cat # 564655; RRID:AB_2744311 |
| CD1c-BV421 | BD Biosciences | Cat # 565050; RRID:AB_2744319 |
| CD20-BUV805 | BD Biosciences | Cat # 612905; RRID:AB_2870192 |
| CD22-BV510 | BioLegend | Cat # 302526; RRID:AB_2721532 |
| CD24-BV650 | BD Biosciences | Cat # 563720; RRID:AB_2632388 |
| CD25-BV605 | BD Biosciences | Cat # 562660; RRID:AB_2744343 |
| CD25-PerCPCy5.5 | BD Biosciences | Cat # 560503; RRID:AB_1727453 |
| CD267-APC | BioLegend | Cat # 311912; RRID:AB_2565423 |
| CD269-PE-CF594 | BioLegend | Cat # 357512; RRID:AB_2566531 |
| CD26-APC | BioLegend | Cat # 302710; RRID:AB_10916120 |
| CD26-PECF594 | BD Biosciences | Cat # 565158; RRID:AB_2739085 |
| CD27-BV711 | BD Biosciences | Cat # 356430; RRID:AB_2650751 |
| CD27-BV786 | BD Biosciences | Cat # 563327; RRID:AB_2744353 |
| CD3-BV421 | BD Biosciences | Cat # 563797; RRID:AB_2744383 |
| CD38-BV711 | BioLegend | Cat # 303528; RRID:AB_2563811 |
| CD3-FITC | BioLegend | Cat # 300306; RRID:AB_314042 |
| CD45RA-Alexa Fluor 700 | BD Biosciences | Cat # 560673; RRID:AB_1727496 |
| CD45RO-BV605 | BD Biosciences | Cat # 562791; RRID:AB_2744411 |
| CD4-BUV496 | BD Biosciences | Cat # 564652; RRID:AB_2744422 |
| CD56-FITC | BioLegend | Cat # 362546; RRID:AB_2565964 |
| CD62L-FITC | BD Biosciences | Cat # 555543; RRID:AB_395927 |
| CD69-BV786 | BD Biosciences | Cat # 563834; RRID:AB_2738441 |
| CD69-PE Cy7 | BD Biosciences | Cat # 557745; RRID:AB_396851 |
| CD79a-PE-Cy7 | BioLegend | Cat # 333510; RRID:AB_2564595 |
| CD8-BUV805 | BD Biosciences | Cat # 564912; RRID:AB_2744465 |
| Foxp3-PE Cy7 | Life Technologies | Cat # 25-4776-42; RRID:AB_10804638 |
| Gamma-9-APC | BioLegend | Cat # 331310; RRID:AB_2057504 |
| HLA-DR-PE-Cy5 | BioLegend | Cat # 307608; RRID:AB_314686 |
| ICOS-BUV395 | BD Biosciences | Cat # 564777; RRID:AB_2738946 |
| Ig kappa light chain-Alexa Fluor 700 | BD Biosciences | Cat # 561319; RRID:AB_10611867 |
| Ig lamda light chain-PerCPCy5.5 | BioLegend | Cat # 316618; RRID:AB_2561512 |
| IgD-BV605 | BD Biosciences | Cat # 563313; RRID:AB_2738134 |
| IgM-BUV396 | BD Biosciences | Cat # 563903; RRID:AB_2721269 |
| LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit | ThermoFisher | Cat # L10119 |
| PD-1-BV510 | BD Biosciences | Cat # 563076; RRID:AB_2737990 |
| PD-1-PE-CF594 | Invitrogen | Cat # 61-2799-42; RRID:AB_2574598 |
| TCF7-PE | BD Biosciences | Cat # 564217; RRID:AB_2687845 |
| Biological Samples | ||
| Patient PBMCs from NT-001 trial | Ott et al., 2020 | N/A |
| Healthy Donor PBMCs | Precision for Medicine | Cat # 93000-10M |
| Healthy Donor PBMCs | StemExpress | Cat # LE010F |
| Healthy Donor PBMCs | AllCells | https://www.allcells.com/tissue-products/leukopak/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CEF viral peptide pool | JPT | Cat # PM-CEF-E |
| Benzonase | Millipore Sigma | Cat # 70746 |
| Assay peptides | Ott et al., 2020 | N/A |
| MHC class I tetramers | Ott et al., 2020 | N/A |
| Critical Commercial Assays | ||
| Pan T cell Isolation Kit, human | Miltenyi Biotec | Cat # 130-096-535 |
| RNeasy Plus Micro Kit | QIAGEN | Cat # 74034 |
| RNeasy Plus Mini Kit | QIAGEN | Cat # 74134 |
| Qubit RNA HS Assay Kit | ThermoFisher | Cat # Q32855 |
| KAPA Library Quantification Kit | Roche | Cat # KK4824 |
| MiSeq Reagent Kit v2 | Illumina | Cat # MS-102-2003 |
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat # 554714 |
| Foxp3/Transcription Factor Staining Buffer Set | eBioscience | Cat # 00-5523-00 |
| AEC substrate-chromogen | BD Biosciences | Cat # 551951 |
| IFN gamma Human ELISPOT Kit | Invitrogen | Cat # 88-7386-88 |
| CD3 MicroBeads, human | Miltenyi Biotec | Cat # 130-050-101 |
| Experimental Models: Cell Lines | ||
| Jurkat, Clone E6-1 | ATCC | Cat # ATCC-TIB-152 |
| A375 | ATCC | Cat # ATCC-CRL-1619 |
| 293FT Cell Line | Thermo Fisher | Cat # R70007 |
| Software and Algorithms | ||
| MiXCR (version 3.0.12) | Bolotin et al., 2015 | N/A |
| FlowJo (version 10) | FlowJo Software (for Windows) Version 10. Becton, Dickinson and Company; 2019. | https://www.flowjo.com/ |
| GraphPad Prism (version 7.01) | GraphPad Software, La Jolla California, USA | https://www.graphpad.com |
| R (version 3.5.1) | CRAN | https://www.r-project.org/ |
| DescTools (R package) | Signorell et al., 2020 | N/A |
| Philentropy (R package) | Drost et al., 2018 | N/A |
| gplots (R package) | Warnes et al., 2015 | N/A |
| Stats (R package) | R Core Team 2018 | N/A |
| Survival (R package) | Therneau et al., 2000 | N/A |
| Survminer (R package) | Kassambara et al., 2019 | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed and will be fulfilled by the Lead Contact, Asaf Poran (asaf.poran@biontech.us).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
Patient TCR sequencing information will be provided for patients consistent with their institutional informed consent. The code supporting the findings of this study is available upon request from the Lead Contact.
Experimental Model and Subject Details
Human Subjects and Study Design
Detailed protocols of the clinical study are reported in Ott et al., 20207. Patients who were 18 years of age or older with histologically or cytologically confirmed unresectable or metastatic melanoma were eligible for enrollment in this multicenter phase I clinical trial (NCT02897765). The gender and age of all patients evaluated in this manuscript can be found in Table S1. No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. This study was conducted in accordance with the Declaration of Helsinki and with approval by Institutional Review Board at each participating site and written informed consent was obtained for all patients. Key eligibility criteria included an Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1 (on a 5-point scale, with higher scores indicating greater disability) and life expectancy of > 6 months. Excluded from the study were patients with untreated central nervous system (CNS) metastases, active or history of autoimmune disease, and who had received previous therapy with PD-1, or PD-L1 inhibitors other than CTLA-4 (cytotoxic T lymphocyte-associated antigen 4). NEO-PV-01 is a personalized vaccine composed of up to 20 synthesized peptides that are approximately 14 to 35 amino acids in length. The peptides are divided into up to four individual pools, each pool consisting of 1 to 5 peptides. On the day of vaccination, each pool of NEO-PV-01 was mixed with polyinosinic polycytidylic acid polylysine carboxymethylcellulose (poly ICLC; Hiltonol®) for a concentration of 300 μg/mL of each peptide. Each patient was scheduled to receive 5 doses of vaccination during the priming phase, on days 1 and 4 of week 12 and then weekly for 3 weeks (weeks 13, 14, and 15) followed by 2 booster vaccinations on weeks 19 and 23. Each pool was administered by subcutaneous injection to one of the four limbs or left or right midriff as an alternative anatomical location. The anatomical site of injection for each pool was constant through the vaccination regimen. All patients, regardless of disease status, were allowed to start vaccination with NEO-PV-01 + adjuvant poly-ICLC. Nivolumab was administered on study day 1 at dose 240 mg by IV over 30 min every 2 weeks.
All healthy donor (HD) material used for this study was obtained with written informed consent through external vendors identified in the Key Resources Table.
Clinical assessments
Adverse events, laboratory values, ECG, and vital signs were assessed regularly and graded according to the National Cancer Institute Common Terminology Criteria for Adverse Events, version 4.0. Response was assessed at weeks 8, 12, 24, 36, 52, 64, 76, 88, and 100 according to Response Evaluation Criteria in Solid Tumors (RECIST), version 1.1, by the investigator.
Peripheral Samples
Leukaphereses were obtained from patients at pre-treatment, pre-vaccine (week 10-12) and post-vaccine (week 20). Healthy donor leukaphereses were obtained from various external vendors identified in the Key Resources Table. PBMCs were isolated within 4h after collection using Ficoll-paque (GE Healthcare) density-gradient centrifugation and cryopreserved in Recovery Cell Culture Freezing Medium (Invitrogen) or cell freezing media (90% FBS and 10% DMSO). PBMCs were stored in vapor-phase liquid nitrogen until time of analysis.
Method Details
T Cell and RNA Isolation for TCR Sequencing
PBMCs were thawed and subjected to negative selection using the Pan T Cell Isolation Kit (Miltenyi) according to manufacturer’s protocol. T cells were counted, centrifuged for 15 minutes at 300xg then flash frozen and stored as dry pellets at −80°C.
RNA isolation was performed using the RNeasy Plus Micro Kit (QIAGEN) on the QIAcube (QIAGEN; Protocol: Purification of total RNA using gDNA Eliminator and RNeasy MinElute spin columns) according to manufacturer’s protocol. RNA concentration was measured using the Qubit™ RNA HS Assay Kit (ThermoFischer) according to manufacturer’s protocol. Eluted RNA was stored at −80°C.
TCR Library Preparation and Sequencing
TCRα and TCRβ libraries were prepared from isolated RNA using the Long Read iR-Profile Reagent System (iRepertoire) either in-house or at the iRepertoire headquarters according to manufacturer’s protocol. For in-house sequencing, libraries were pooled, and concentrations were calculated using the KAPA Library Quantification Kit (Roche) on the Lightcycler® 480 II (Roche). Pooled libraries were sequenced using the MiSeq Reagent Kit v2 500-cycles (Illumina), either in-house or at the iRepertoire headquarters according to manufacturer’s protocol. In-house sequencing runs included a 10% Phi-X spike-in. Throughout the study, the number of samples per pool was designed to maintain equal sequencing depths across samples.
Phenotypic analysis of peripheral samples
Patient and healthy donor PBMCs were thawed into FBS, followed by a wash with X-Vivo media (Lonza). Cells were then treated with benzonase (Millipore) for 30 minutes at a 1:1000 dilution in media at 37°C. Cells were washed with media and counted using the Guava easyCyte Flow cytometer. A total of 2 x10ˆ6 cells per sample were plated for antibody staining and washed once with FACS buffer (PBS + 0.5% BSA). Cells were then incubated with cell surface antibody for 30 minutes on ice, followed by a wash with FACS buffer. Cells were fixed and permeabilized for intracellular staining using either the BD cytofix/cytoperm kit (BD Biosciences; used for B cell panel) or Foxp3/Transcription Factor Staining Buffer Set (eBioscience; used for T cell panel) for 20 minutes on ice according the manufacturer’s instructions. Cells were washed with the corresponding permeabilization wash buffer according to the manufacturer’s instructions. Cells were then incubated with intracellular antibodies in the corresponding permeabilization wash buffer for 30 minutes on ice, washed with the appropriate permeabilization wash buffer, followed by a final wash with FACS buffer. Cells were stored in FACS buffer at 4°C until acquisition on a BD LSR Fortessa instrument.
TCR Repertoire Generation
TCR repertoires were generated by running a licensed copy of MiXCR34 on the paired-end raw sequencing fastq files. The parameters included the species specifications (Human, hsa), starting material (RNA), 5′ and 3′ primers (v and c primers, respectively) with no adapters, and searching for either TCRα (tra) or TCRβ chains (trb).
TCRα or TCRβ CDR3 clonotypes were filtered by removal of non-functional sequences (out-of-frame sequences or those containing stop codons). Clonal frequency was calculated based on the count for each clone out of the total count.
TCR Repertoire Diversity/Clonality Analysis
Clone size category (Figures 1B and 1C; Figure S1) was based on clonal frequency, Fi as follows: rare (Fi < 1e-6), small (1e-6 ≤ Fi < 1e-5), medium (1e-5 ≤ Fi < 1e-4), large (1e-4 ≤ Fi < 1e-3), and hyperexpanded (1e-3 ≤ Fi).
The unique number of nucleotide /amino acid TCRα or TCRβ CDR3s was calculated per sample. Global diversity/clonality coefficients have been calculated as follows:
DE50 – AA CDR3 clonotypes were sorted based on their frequency in descending order. The cumulative frequencies of this sorted frequency vector were calculated. The rank of the first value that was equal or larger than 0.50 was divided by the total number of unique aa CDR3 clonotypes to obtain the DE50 value. For example, if the 40 most frequent clones (but not 39) of a repertoire covers 50% of the total counts of the clones in that repertoire, consisting of 1000 clones, the DE50 value would be 0.04.
Gini Coefficient – ranges between 0 (all clones are equally frequent – repertoire diversity) and 1 (frequency dominated by one clone, repertoire clonality). Calculated using the “Gini” function from the “DescTools” R package35.
Normalized Shannon’s Entropy – higher values represent higher inequality of the frequencies25,26.
Lorentz curves – similarly to the DE50 estimate, but a continuous curve between DE0 and DE100. Calculated using the “Lc” function from the “DescTools” R package35.
Sum of squares – the sum of squares measurement is calculated as the sum of the frequencies of the aa CDR3 clonotypes, each squared.
TCR Repertoire Stability
Assessment of the 2-way repertoire turnover was performed using the Jensen-Shannon Divergence28,29. This measurement ranges between 0 (completely overlapping repertoires – no turn-over) and 1 (completely disjoint repertoires – high turn-over). This calculation was performed using the “JSD” function from the R package “philentropy”36, either using the pre-treatment time-point as baseline, and comparing pre-vaccine and post-vaccine samples to that baseline (Figure 2A), or by comparing consecutive time-points to each other (Figure S2A). Calculation was based on the most frequent aa CDR3 clonotypes covering 20% of the total counts in each repertoire.
This analysis was performed for varying levels of repertoire coverage, from 20% to 80% (Figure S2B). Significance was calculated from a repeated-measures analysis for modeling, with AR(1) covariance structure.
Assessment of the 3-way repertoire stability was performed using a Venn diagram approach (Figure 2B). The aa CDR3 sequences from the 3 time-points in each patient were intersected using the “venn” function from the R package “gplots”37; the size of each of the 7 segments (A-G) was defined as the sum of the frequencies of the clones in that segment. For segments shared by 2 (D-F) or 3 (G) segments, each clone has 2 or 3 frequency values, corresponding to each time-point it was detected in. Therefore, segments D-G have 2 or 3 size values. In addition, we also calculated the intersection of the unique aa CDR3 sequences across the 3 time-points, regardless of their frequencies (Figure 2D).
Principal Component Analysis
Select measurements taken at baseline from either the TCR repertoire analysis or the immunophenotyping of the PBMCs was aggregated in one matrix. The matrix was centered and scaled, and PCA was calculated using the R function “prcomp” from the “stats” R package38. The loadings, or contributions of the different measurements to PC1, were retrieved from the rotation matrix (Figure 4B).
Kaplan-Meier analysis was performed based on categorizing patients as belonging to PC1 < 0 or PC1 > 0. Calculation was performed using the “survfit” function from the “survival” R package39 and plotted using the “ggsurvplot” function from the “survminer” R package40. P value was calculated using the log-ratio test and hazard-ration calculated using a univariate Cox proportional hazards regression model.
Public TCR Sequence Analysis
The 11 HDs were used to represent healthy population peripheral TCR repertoires. For each patient, each of the top 1000 most frequent TCRβ aa CDR3 sequence found in the pre-treatment time-point was scored by the number of HD repertoires in which it was detected (ranging from 0- no one, to 11 - all). For each score, the fraction of clones with that score in each patient was calculated (Figure S5B).
scTCRseq of neoantigen-specific TCRs
Single-cell TCR sequencing (scTCRseq) was performed as previously described7. Briefly, patient PBMCs were sorted into 96-well plates either based on tetramer staining or staining with T cell activation markers (i.e., CD69, CD107a, CD25, PD-1) either immediately after overnight rest (ex vivo) or after neoantigen peptide exposure for 5-6 days and recall for 6-24h (stim). Cells were stained with T cell activation markers as described in the peripheral immunophenotyping section, with anti-CD107a antibody added 6h prior to the end of co-culture during peptide recall. Final sort gates for neoantigen-reactive subpopulations were determined based on control tetramer staining or control co-cultures with DMSO. Plates were submitted to iRepertoire for scTCRseq (iPair-TCR). Paired sequencing information was used to determine clonal composition of sorted cell populations and candidate alpha/beta chain pairs were tested for neoantigen-reactivity after lentivirus-based T cell transductions41.
For the transductions, second generation lentivirus vectors were generated on 293FT producer cell lines using shuttle plasmids (GenScript) containing TCR beta and alpha chains under the control of a SFFV promoter and separated by a furin cleavage site and a P2A ribosomal skip sequence. Human TCR variable regions were fused to cysteine-modified mouse constant regions for both chains resulting in recombinant mTCR42,43. Neoantigen-reactivity of mTCR Jurkat cells was determined by IL-2 secretion measured by electrochemiluminescence (MSD), according to the manufacturer’s instructions, in 24h co-culture assays of mTCR transduced Jurkat cells with neoantigen peptide and allele matched antigen presenting cell lines or CD3+ T cell depleted autologous patient PBMCs using CD3 microbeads (Miltenyi).
IFN-γ ELISpot assay for CEF Reactivity
IFN-γ ELISpot assays to assess reactivity to CMV, EBV, or influenza (CEF) were performed using 96-well MultiScreen Filter Plates (Millipore) and the Ready-Set-Go! Human IFN-γ ELISpot Kit (Invitrogen) according to manufacturer’s instructions and as previously described7. Plates were coated overnight at 4°C with anti-IFNγ Capture Antibody diluted in 1X Coating Buffer, washed with 1X Coating Buffer and blocked with X-Vivo media (Lonza) containing 1% penicillin/streptomycin (GIBCO) for 1h. PBMCs plated in triplicate with 1 × 106 cells per well after they were rested overnight in X-Vivo media (Lonza) supplemented with 1% penicillin/streptomycin (GIBCO) at 37°C. CEF viral peptide pools were added to ELISpot wells at a 5 μM final concentration. Each plate included a healthy donor positive and negative control with the CEF viral peptide pool (JPT) and Cell Stimulation Cocktail (Life Technologies) to confirm reagent performance. Plates were incubated overnight at 37°C. Plates were washed 3 times using PBS with 0.05% Tween-20 and detection antibody diluted in 1X ELISpot diluent was added to wells for 2h. After washing 3 times with PBS with 0.05% Tween-20, Avidin-HRP was diluted in 1X ELISpot diluent and added to wells for 45 min. Plates were washed with both PBS with Tween-20 and PBS 3 times. AEC substrate-chromogen (BD Biosciences) was then added for 20 min. Plates were rinsed with deionized water 3 times and allowed to dry at room temperature overnight. Spots were imaged and enumerated using an Immunospot analyzer (Cellular Technology Limited).
Quantification and Statistical Analysis
Statistical Analysis
All analyses were performed in the R language and environment for statistical computing (version 3.5.1). Likewise, all figures were created using an R script relying on the “ggplot2” package44. Unless otherwise specified, P values are derived from a two-tailed Student’s t test.
Additional Resources
All patient data presented in this manuscript is from the multicenter phase I clinical trial NCT02897765. https://clinicaltrials.gov/
Acknowledgments
We thank all the patients and their families who participated in the study. We thank Drs. M. Malloy, S. Chhangawala, and M. DeMario for their comments, suggestions, and assistance. We thank A. Lamb, J. Tepper, and M. Moles for clinical operations support, and T. Kohler for assistance in the revision process. We thank all BioNTech US (formerly Neon Therapeutics, Inc.) employees for their dedication that enabled this study. We thank John Welle of Acumen Medical Communications for graphic design support. We dedicate this publication to our beloved friend, colleague, and a brilliant scientist Y. Sonia Ting (1984–2019), who brightened our days with her smile and was a beacon of innovation. This study was funded by Neon Therapeutics/BioNTech US.
Author Contributions
Conceptualization, Formal Analysis, Investigation, Visualization, and Writing – Original Draft, A.P.; Conceptualization, Methodology, Investigation, Visualization, and Writing – Original Draft, J.S.; Investigation, Methodology, Visualization and Writing – Review and Editing, M.E.B.; Investigation and Methodology, R.B.; Investigation, Methodology, and Visualization, K.N.B.; Data Curation, A.W.; Investigation, R.G.W. and J.P.; Clinical Investigators on the Study, P.A.O. and S.H.-L.; Biostatistical Analysis and Data Curation, Z.S.K.; Supervision, Funding Acquisition, and Writing – Review and Editing, R.B.G.; Supervision, Conceptualization, and Writing – Review and Editing, M.S.R.; and Supervision, Conceptualization, and Writing – Review and Editing, L.S.
Declaration of Interests
A.P., J.S., M.E.B., R.B., K.N.B., A.W., R.G.W., J.P., Z.S.K., M.S.R., and L.S. are/were all employees and/or equity holders at BioNTech US (formerly Neon Therapeutics, Inc.). P.A.O.: research funding paid to the institution: BMS, Merck, AstraZeneca, Celldex, CytomX, Glaxo Smith Kline, ARMO Biosciences; Neon Therapeutics, Consultant: Array, BMS, Merck, Genentech, Pfizer, Novartis, Neon Therapeutics, CytomX, Celldex. S.H.-L.: consultant to Amgen, BMS, Genmab, Xencor; Research support from BMS, Merck and Vaccinex. R.B.G.: Board of Directors, Alkermes plc and Infinity Pharmaceuticals and Scientific Advisory Board, Leap Therapeutics; stockholder and employee of BioNTech US (formerly Neon Therapeutics, Inc.).
Published: November 17, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100141.
Contributor Information
Asaf Poran, Email: asaf.poran@biontech.us.
Lakshmi Srinivasan, Email: lakshmi.srinivasan@biontech.us.
Supplemental Information
References
- 1.Coley W.B. II. Contribution to the Knowledge of Sarcoma. Ann. Surg. 1891;14:199–220. doi: 10.1097/00000658-189112000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrlich P. Über den jetzigen Stand der Chemotherapie. Ber. Dtsch. Chem. Ges. 1909;42:17–47. [Google Scholar]
- 3.Finn O.J. Immuno-oncology: understanding the function and dysfunction of the immune system in cancer. Ann. Oncol. 2012;23:viii6–viii9. doi: 10.1093/annonc/mds256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsushita H., Vesely M.D., Koboldt D.C., Rickert C.G., Uppaluri R., Magrini V.J., Arthur C.D., White J.M., Chen Y.-S., Shea L.K. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schumacher T.N., Schreiber R.D. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 6.Gubin M.M., Artyomov M.N., Mardis E.R., Schreiber R.D. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J. Clin. Invest. 2015;125:3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott P.A., Hu-Lieskovan S., Chmielowski B., Govindan R., Naing A., Bhardwaj N., Margolin K., Awad M.M., Hellmann M.D., Lin J.J. A Phase Ib Trial of Personalized Neoantigen Therapy Plus Anti-PD-1 in Patients with Advanced Melanoma, Non-small Cell Lung Cancer, or Bladder Cancer. Cell. 2020;183:347–362. doi: 10.1016/j.cell.2020.08.053. [DOI] [PubMed] [Google Scholar]
- 8.Postow M.A., Manuel M., Wong P., Yuan J., Dong Z., Liu C., Perez S., Tanneau I., Noel M., Courtier A. Peripheral T cell receptor diversity is associated with clinical outcomes following ipilimumab treatment in metastatic melanoma. J. Immunother. Cancer. 2015;3:23. doi: 10.1186/s40425-015-0070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosoi A., Takeda K., Nagaoka K., Iino T., Matsushita H., Ueha S., Aoki S., Matsushima K., Kubo M., Morikawa T. Increased diversity with reduced “diversity evenness” of tumor infiltrating T-cells for the successful cancer immunotherapy. Sci. Rep. 2018;8:1058. doi: 10.1038/s41598-018-19548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirsch I., Vignali M., Robins H. T-cell receptor profiling in cancer. Mol. Oncol. 2015;9:2063–2070. doi: 10.1016/j.molonc.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairfax B.P., Taylor C.A., Watson R.A., Nassiri I., Danielli S., Fang H., Mahé E.A., Cooper R., Woodcock V., Traill Z. Peripheral CD8+ T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat. Med. 2020;26:193–199. doi: 10.1038/s41591-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogan S.A., Courtier A., Cheng P.F., Jaberg-Bentele N.F., Goldinger S.M., Manuel M., Perez S., Plantier N., Mouret J.-F., Nguyen-Kim T.D.L. Peripheral Blood TCR Repertoire Profiling May Facilitate Patient Stratification for Immunotherapy against Melanoma. Cancer Immunol. Res. 2019;7:77–85. doi: 10.1158/2326-6066.CIR-18-0136. [DOI] [PubMed] [Google Scholar]
- 13.Hopkins A.C., Yarchoan M., Durham J.N., Yusko E.C., Rytlewski J.A., Robins H.S., Laheru D.A., Le D.T., Lutz E.R., Jaffee E.M. T cell receptor repertoire features associated with survival in immunotherapy-treated pancreatic ductal adenocarcinoma. JCI Insight. 2018;3:e122092. doi: 10.1172/jci.insight.122092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valpione S., Galvani E., Tweedy J., Mundra P.A., Banyard A., Middlehurst P., Barry J., Mills S., Salih Z., Weightman J. Immune-awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat. Can. 2020;1:210–221. doi: 10.1038/s43018-019-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan S.A., Levesque M.P., Cheng P.F. Melanoma Immunotherapy: Next-Generation Biomarkers. Front. Oncol. 2018;8:178. doi: 10.3389/fonc.2018.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nixon A.B., Schalper K.A., Jacobs I., Potluri S., Wang I.-M., Fleener C. Peripheral immune-based biomarkers in cancer immunotherapy: can we realize their predictive potential? J. Immunother. Cancer. 2019;7:325. doi: 10.1186/s40425-019-0799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krieg C., Nowicka M., Guglietta S., Schindler S., Hartmann F.J., Weber L.M., Dummer R., Robinson M.D., Levesque M.P., Becher B. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 18.Lalani A.A., Xie W., Martini D.J., Steinharter J.A., Norton C.K., Krajewski K.M., Duquette A., Bossé D., Bellmunt J., Van Allen E.M. Change in Neutrophil-to-lymphocyte ratio (NLR) in response to immune checkpoint blockade for metastatic renal cell carcinoma. J. Immunother. Cancer. 2018;6:5. doi: 10.1186/s40425-018-0315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacdalan D.B., Lucero J.A., Sacdalan D.L. Prognostic utility of baseline neutrophil-to-lymphocyte ratio in patients receiving immune checkpoint inhibitors: a review and meta-analysis. OncoTargets Ther. 2018;11:955–965. doi: 10.2147/OTT.S153290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber R., Fleming V., Hu X., Nagibin V., Groth C., Altevogt P., Utikal J., Umansky V. Myeloid-Derived Suppressor Cells Hinder the Anti-Cancer Activity of Immune Checkpoint Inhibitors. Front. Immunol. 2018;9:1310. doi: 10.3389/fimmu.2018.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C., Sanders C.M., Yang Q., Schroeder H.W., Jr., Wang E., Babrzadeh F., Gharizadeh B., Myers R.M., Hudson J.R., Jr., Davis R.W., Han J. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc. Natl. Acad. Sci. USA. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Britanova O.V., Putintseva E.V., Shugay M., Merzlyak E.M., Turchaninova M.A., Staroverov D.B., Bolotin D.A., Lukyanov S., Bogdanova E.A., Mamedov I.Z. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 2014;192:2689–2698. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 23.Lindau P., Mukherjee R., Gutschow M.V., Vignali M., Warren E.H., Riddell S.R., Makar K.W., Turtle C.J., Robins H.S. Cytomegalovirus Exposure in the Elderly Does Not Reduce CD8 T Cell Repertoire Diversity. J. Immunol. 2019;202:476–483. doi: 10.4049/jimmunol.1800217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas P.G., Handel A., Doherty P.C., La Gruta N.L. Ecological analysis of antigen-specific CTL repertoires defines the relationship between naive and immune T-cell populations. Proc. Natl. Acad. Sci. USA. 2013;110:1839–1844. doi: 10.1073/pnas.1222149110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson A.L., Nel H.J., Bradbury L., Phipps J., Thomas R., Lê Cao K.A., Kenna T.J., Brown M.A. Altered Repertoire Diversity and Disease-Associated Clonal Expansions Revealed by T Cell Receptor Immunosequencing in Ankylosing Spondylitis Patients. Arthritis Rheumatol. 2020;72:1289–1302. doi: 10.1002/art.41252. [DOI] [PubMed] [Google Scholar]
- 26.Jia Q., Zhou J., Chen G., Shi Y., Yu H., Guan P., Lin R., Jiang N., Yu P., Li Q.-J., Wan Y. Diversity index of mucosal resident T lymphocyte repertoire predicts clinical prognosis in gastric cancer. OncoImmunology. 2015;4:e1001230. doi: 10.1080/2162402X.2014.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thapa D.R., Tonikian R., Sun C., Liu M., Dearth A., Petri M., Pepin F., Emerson R.O., Ranger A. Longitudinal analysis of peripheral blood T cell receptor diversity in patients with systemic lupus erythematosus by next-generation sequencing. Arthritis Res. Ther. 2015;17:132. doi: 10.1186/s13075-015-0655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris H., DeWolf S., Robins H., Sprangers B., LoCascio S.A., Shonts B.A., Kawai T., Wong W., Yang S., Zuber J. Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci. Trans. Med. 2015;7:272ra10. doi: 10.1126/scitranslmed.3010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putintseva E.V., Britanova O.V., Staroverov D.B., Merzlyak E.M., Turchaninova M.A., Shugay M., Bolotin D.A., Pogorelyy M.V., Mamedov I.Z., Bobrynina V. Mother and child T cell receptor repertoires: deep profiling study. Front. Immunol. 2013;4:463. doi: 10.3389/fimmu.2013.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oakes T., Heather J.M., Best K., Byng-Maddick R., Husovsky C., Ismail M., Joshi K., Maxwell G., Noursadeghi M., Riddell N. Quantitative Characterization of the T Cell Receptor Repertoire of Naïve and Memory Subsets Using an Integrated Experimental and Computational Pipeline Which Is Robust, Economical, and Versatile. Front. Immunol. 2017;8:1267. doi: 10.3389/fimmu.2017.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helmink B.A., Reddy S.M., Gao J., Zhang S., Basar R., Thakur R., Yizhak K., Sade-Feldman M., Blando J., Han G. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochsenreither S., Fusi A., Wojtke S., Busse A., Nüssler N.C., Thiel E., Keilholz U., Nagorsen D. Comparison of T-cell receptor repertoire restriction in blood and tumor tissue of colorectal cancer patients. J. Transl. Med. 2010;8:35. doi: 10.1186/1479-5876-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reuben A., Zhang J., Chiou S.-H., Gittelman R.M., Li J., Lee W.-C., Fujimoto J., Behrens C., Liu X., Wang F. Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat. Commun. 2020;11:603. doi: 10.1038/s41467-019-14273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 35.Signorell A. 2020. DescTools: Tools for Descriptive Statistics.R package version 0.99.32. [Google Scholar]
- 36.Drost H.-G. Philentropy: Information Theory and Distance Quantification with R. J. Open Source Softw. 2018;3:765. [Google Scholar]
- 37.Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Liaw W.H.A., Lumley T., Maechler M., Magnusson A., Moeller S., Schwartz M. 2015. gplots: Various R Programming Tools for Plotting Data. [Google Scholar]
- 38.R Core Team . R Foundation for Statistical Computing; 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 39.Therneau T.M., Grambsch P.M. Springer; 2000. Modeling Survival Data: Extending the Cox Model. [Google Scholar]
- 40.Kassambara A., Kosinski M., Biecek P. 2019. survminer: Drawing Survival Curves using “ggplot2”. R package version 0.4.6. [Google Scholar]
- 41.Banu N., Chia A., Ho Z.Z., Garcia A.T., Paravasivam K., Grotenbreg G.M., Bertoletti A., Gehring A.J. Building and optimizing a virus-specific T cell receptor library for targeted immunotherapy in viral infections. Sci. Rep. 2014;4:4166. doi: 10.1038/srep04166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen C.J., Li Y.F., El-Gamil M., Robbins P.F., Rosenberg S.A., Morgan R.A. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res. 2007;67:3898–3903. doi: 10.1158/0008-5472.CAN-06-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuball J., Dossett M.L., Wolfl M., Ho W.Y., Voss R.-H., Fowler C., Greenberg P.D. Facilitating matched pairing and expression of TCR chains introduced into human T cells. Blood. 2007;109:2331–2338. doi: 10.1182/blood-2006-05-023069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickham H. Springer; 2009. Ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Patient TCR sequencing information will be provided for patients consistent with their institutional informed consent. The code supporting the findings of this study is available upon request from the Lead Contact.