Summary
Increasing evidence suggests Alzheimer's disease (AD) pathophysiology is influenced by primary and secondary bile acids, the end product of cholesterol metabolism. We analyze 2,114 post-mortem brain transcriptomes and identify genes in the alternative bile acid synthesis pathway to be expressed in the brain. A targeted metabolomic analysis of primary and secondary bile acids measured from post-mortem brain samples of 111 individuals supports these results. Our metabolic network analysis suggests that taurine transport, bile acid synthesis, and cholesterol metabolism differ in AD and cognitively normal individuals. We also identify putative transcription factors regulating metabolic genes and influencing altered metabolism in AD. Intriguingly, some bile acids measured in brain tissue cannot be explained by the presence of enzymes responsible for their synthesis, suggesting that they may originate from the gut microbiome and are transported to the brain. These findings motivate further research into bile acid metabolism in AD to elucidate their possible connection to cognitive decline.
Keywords: Alzheimer's disease, bile acids, cholesterol metabolism, transcriptomics, metabolomics, genome-scale metabolic models, transcriptional regulatory networks
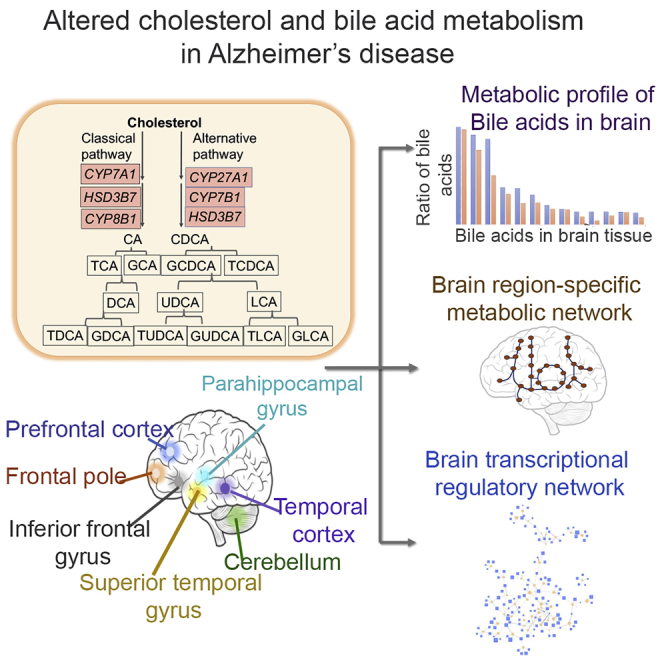
Graphical Abstract
Highlights
Altered cholesterol and bile acid metabolism linked to Alzheimer disease (AD)
Evidence for alternative bile acid pathway genes expression in post-mortem brains
Metabolomics analysis shows secondary bile acids associated with cognitive decline
Genome-scale metabolic networks of brain regions identify reactions linked to AD
Baloni et al. use a systems biology approach to identify alterations in cholesterol and bile acid metabolism in Alzheimer disease (AD). Expression of alternative bile acid and neural cholesterol clearance pathway along with transporters of taurine and bile acids suggest the role of the gut-brain axis in AD.
Introduction
Alzheimer's disease (AD), the leading cause of dementia, is a progressive, multifactorial disease1,2 in which the onset and progression of symptoms varies significantly among individuals. Recent studies have shown that metabolic dysfunction is one of the factors associated with neurodegenerative disorders.3,4 Various physiological processes such as lipid metabolism, immune function, amyloid precursor protein metabolism, oxidative stress, neurotransmitter function, and mitochondrial functions are altered in AD, which can affect metabolism.5, 6, 7 Interest in the transport of biochemical compounds between the brain and the gut and their possible role in regulating metabolic changes centrally and peripherally has increased recently across several neurodegenerative diseases.8,9 There is increasing evidence to suggest a role in AD for primary and secondary bile acids (BAs).7,10,11 BAs are amphipathic molecules and primary BAs are derived from cholesterol, mostly in the liver, whereas secondary BAs are typically produced by bacteria in the gut.12 Increased levels of secondary BAs and ratios to their primary BA educts have been linked to AD and cognitive decline.7
Cholesterol metabolism and transport have been studied extensively and are clearly linked with AD.1,13,14 Cholesterol clearance leads to the production of BAs that carry out lipid absorption and cholesterol homeostasis and also function as signaling molecules.15 Primary BAs such as cholic acid (CA) and chenodeoxycholic acid (CDCA) are synthesized as a result of cholesterol efflux and then conjugated with glycine or taurine for secretion into bile and later metabolized by gut bacteria.12 There are two major BA biosynthetic pathways: the classical pathway (neutral pathway) and the alternative pathway (acidic pathway). The classical pathway in mammalian liver is initiated by cholesterol 7α-hydroxylase (CYP7A1) and subsequently requires 12α-hydroxylase (CYP8B1), among numerous other enzymes, for the synthesis of CA, whereas CDCA is produced in the absence of CYP8B1.16 Sterol 27-hydroxylase (CYP27A1) is required for the initiation of an alternative BA pathway.17 In the brain, sterol 24-hydroxylase (CYP46A1) converts cholesterol to 24S-hydroxycholesterol (systematic name cholest-5-en-3β,24S-diol), and subsequently, 7α-hydroxylation is carried out by 24-hydroxycholesterol 7α-hydroxylase (CYP39A1)18 (Figure 1). Studies in human and mouse brain samples, as well as cell lines, have shown that BAs can cross the blood-brain barrier (BBB) and bind to nuclear receptors, causing physiological changes.19,20 There is limited information on the role of BAs in the human brain and their association with cognitive decline in AD pathophysiology. Systematic analysis of omics data derived from blood and post-mortem brain samples of AD and cognitively normal (CN) or control individuals has the potential to identify differences in cholesterol and BA metabolism and how they contribute to AD pathogenesis.
Figure 1.
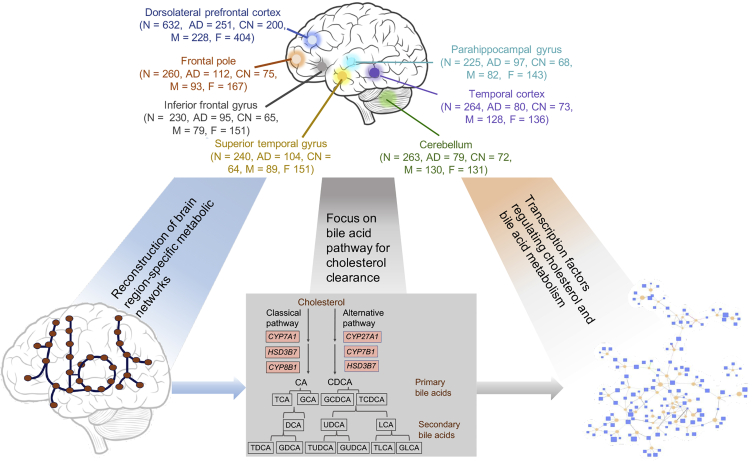
Graphical Overview of Analyses Described Herein to Study Altered Cholesterol and Bile Acid (BA) Metabolism in AD
Numbers of samples from each brain region are indicated along with AD and control samples and male/female breakdown in parentheses. We used the post-mortem brain sample transcriptome data to generate region-specific metabolic networks. We used these networks to study BA and cholesterol metabolism. Using the brain transcriptional regulatory network, we identified transcription factors that regulate genes in cholesterol and BA metabolism. See also Table S1.
In this study, we analyzed a large number of transcriptome data from the Religious Orders Study and Memory and Aging Project (ROSMAP), the Mayo Clinic, and the Mount Sinai Brain Bank that had a total of 2,114 post-mortem brain samples from 7 different brain regions. We reconstructed metabolic networks using the data from these 3 cohorts and studied the role of circulating BAs that may contribute to AD and altered cholesterol metabolism in these individuals. We also generated targeted metabolomics data of primary and secondary BAs from the post-mortem brain samples of 111 AD patients and controls.
Various genomic studies have reported transcriptional regulatory changes in neurodegenerative diseases.21,22 The biological significance of these transcription factors (TFs) regulating metabolic changes is not completely understood. The brain-specific metabolic and transcriptional regulatory networks proved useful in identifying candidate metabolites and genes involved in the disease manifestation. A schematic representation of the study is represented in Figure 1. Our study used the following approaches to investigate the role of BAs in AD:
-
(1)
Transcriptional profiling of genes that are involved in cholesterol and BA metabolism using publicly available data from post-mortem brain samples
-
(2)
Reconstruction and analysis of genome-scale metabolic networks of various brain regions to identify genes and reactions that are significant in AD versus CN
-
(3)
Transcriptional regulatory network analysis of brain samples to predict candidate TFs regulating metabolically important genes
In summary, our study addresses an important need to better understand the potential roles of BAs in AD pathophysiology.
Results
In recent studies, cytotoxic and neuroprotective BAs were identified in AD and their probable link to cognitive decline in the individuals was reported.7,23 To further investigate the role of primary and secondary BAs in AD and CN individuals, we analyzed 2,114 post-mortem brain samples from 3 independent cohorts for 7 brain regions (Table S1) and selected genes involved in cholesterol and BA metabolism.
Here, we studied the role of BAs in AD pathology in the context of genome-scale metabolic and transcriptional regulatory networks (Figure 1).
Transcriptomic Analysis of Genes Encoding Enzymes Associated with BA Metabolism
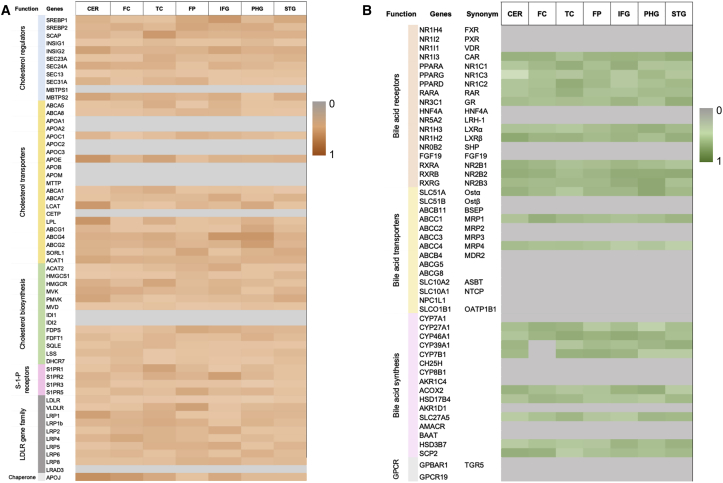
BAs are products of cholesterol metabolism. To identify cholesterol and BA genes that are expressed in the brain, we curated a list of regulators, transporters, and biosynthesis genes in these three independent cohorts. Cholesterol biosynthesis regulators SREBF1 and SREBF2 were expressed in post-mortem brain samples, and recent studies have identified variants of SREBP2, the protein encoded by SREBF2, and their probable link with AD.14,24,25 The expression of genes involved in cholesterol transport—ABCA1, ABCA5, ABCA7, APOE, LPL, and LCAT and members of the low-density lipoprotein receptor (LDLR) gene family (LDLR, VLDLR, LRP1, LRP2, LRP4, LRP5, LRP6, LRP8, LRAD3) in the brain samples suggests the active transport of cholesterol and cholesterol homeostasis in the brain (Figure 2). ABCA7, a cholesterol transporter belonging to the class of ATP-binding cassette transporters that has been identified as a risk factor for late-onset AD,17 is not found in the existing Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and was manually curated into our models. ABCA7 was expressed in the post-mortem brain samples. We also probed genes encoding for receptors linked with the classical and alternative BA pathway and found expression of PPARA, PPARG, LXRα/β, RAR, and RXRs (RXRA, RXRB, RXRG) in the samples but no evidence of expression of the farnesoid X receptor/BA receptor (FXR).
Figure 2.
Transcriptomic Analysis of Genes Associated with Cholesterol and BA Metabolism
Heatmap of cholesterol (A) and BA metabolism (B) genes. The color gradient is based on ubiquity scores calculated for the genes and the gray color represents genes having no expression data in the brain regions from the 3 cohorts. Brain regions represented in the plot are cerebellum (CER), prefrontal cortex (FC), temporal cortex (TC), frontal pole (FP), inferior frontal gyrus (IFG), parahippocampal gyrus (PHG), and superior temporal gyrus (STG). The function of genes is indicated on the left side of each heatmap.
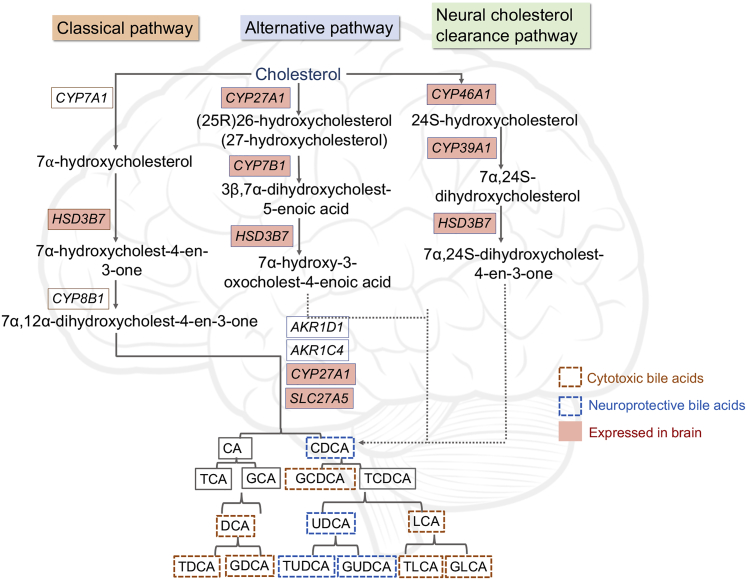
We observed the consistent expression of CYP27A1 and CYP7B1, which are involved in the initial steps of the alternative BA pathway depicted in Figure 3, from the analysis of transcriptomic data of post-mortem brain samples from three independent cohorts (Table S1). In Figure 3, the BAs are marked as cytotoxic and neuroprotective,7,23 but all BAs become toxic at elevated concentrations because of their ability to solubilize membranes.23 We did not observe expressions of CYP7A1 and CYP8B1, suggesting that the classical BA biosynthesis pathway is not prevalent in the brain samples. The classical pathway is known to be most active in the liver.26 It has been reported that neural cholesterol clearance through BA synthesis is mediated by CYP46A1, and subsequently by CYP39A1 in the liver, leading to the synthesis of CDCA.11,27 In addition to genes involved in the alternative BA pathway, we also observed the expression of brain-specific CYP46A1 and CYP39A1 genes in all of the cohorts. This analysis suggested that the brain uses an alternative and neural cholesterol clearance pathway of BA synthesis11,27 and not the classical pathway.23
Figure 3.
Schematic Representation of BA Synthesis Pathway in Humans
Genes expressed in brain samples from our analysis are highlighted in pink. The order of enzymatic reactions can vary. Based on the results from MahmoudianDehkordi et al.,7 BAs have been marked as neuroprotective or cytotoxic.
Metabolomics Analysis of Post-mortem Brain Samples to Identify Levels of Primary and Secondary BAs
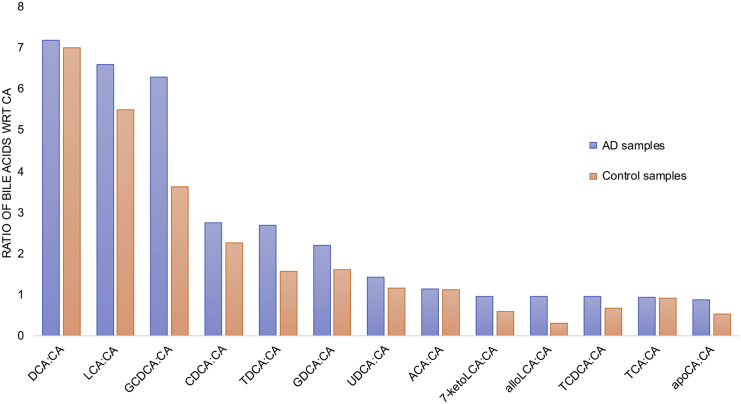
BAs were quantified from 111 post-mortem brain samples from the dorsolateral prefrontal cortex of individuals with AD, mild cognitive impairment (MCI), and CN in the ROSMAP study (https://www.synapse.org/#!Synapse:syn10235594) (Table S2). Although the genes involved in the production of CA were not expressed in the brain samples, the detection of CA from the metabolomics analysis suggested that CA may enter the brain from the periphery, as previously shown in other studies.20,28,29 We compared the levels of primary and secondary BAs in individuals with a Consortium to Establish a Registry for Alzheimer's Disease (CERAD) score of 1–4, in which 1, 2, 3, and 4 indicate definitive AD, probable AD, possible AD, and no evidence of AD, respectively. The ratio of primary conjugated and secondary BAs with respect to CA showed that deoxycholic acid (DCA), lithocholic acid (LCA), glycochenodeoxycholate (GCDCA), CDCA, taurodeoxycholic acid (TDCA), glycodeoxycholic acid (GDCA), ursodeoxycholic acid (UDCA), allolithocholate (alloLCA) and taurocholic acid (TCA) were higher in individuals with AD (CERAD score 1–3) compared to controls (Figure 4). Similar results were reported in the serum metabolomics samples of AD and CN individuals.7,10 Allo-cholic acid (ACA) is a steroid BA that has been studied in the context of signaling mechanisms related to differentiation, proliferation, or apoptosis of hepatocytes.30 The CDCA:CA ratio was calculated, and it showed a higher value for AD compared to CN individuals in the study (Table S3). This finding suggests that the alternative BA pathway is more active in AD versus CN individuals. Also, the higher ratio of primary BAs, like TCA, and secondary BAs, such as DCA, LCA, TDCA, and GDCA, in AD individuals indicated that these BAs may be associated with cognitive function.
Figure 4.
Metabolomics Analysis of Post-mortem Brain Samples to Identify Levels of Primary and Secondary BAs
Bar plots representing the ratio of BAs with respect to cholic acid (CA) (primary BA) are shown here. The primary and secondary BAs measured from 111 brain samples from the ROSMAP study are represented here. The blue bars represent AD samples and the light orange bars represent control samples. See also Table S3.
The primary BAs are conjugated with glycine or taurine for secretion into bile.17 In addition to the primary and secondary BAs, we also measured levels of taurine in serum samples in AD and CN individuals. In the serum, we observed that AD patients had higher levels of serum taurine compared to controls. Taurine is required for the conjugation of primary and secondary BAs. This is an interesting observation, and we need to explore the transport and physiological levels of taurine in the brains of individuals with AD.
Metabolic Reconstruction of Brain Regions and Pathway-Level Analysis
We reconstructed metabolic networks for brain region-specific samples in the three independent cohorts. The seven brain regions in this study included cerebellum (CER), prefrontal cortex (FC), temporal cortex (TC), frontal pole (FP), inferior frontal gyrus (IFG), parahippocampal gyrus (PHG), and superior temporal gyrus (STG). We used transcriptome data from post-mortem brain samples for reconstructing metabolic networks (see STAR Methods for more details). The brain region-specific metabolic networks consisted of ∼5,600–6,300 reactions, 2,800–4,000 metabolites, and each model had genes varying from 1,500 to 1,757 in these networks. Figure S1A provides information about the numbers of reactions, metabolites, and genes present in each of the brain region-specific networks, and Figure S1B compares the gene content overlaps across each of these networks. We have made the detailed content of all of these models available to the scientific community (Table S4. Details of Reactions and Metabolites in the Cerebellum Metabolic Network, Related to STAR Methods and Figure S1, Table S5. Details of Reactions and Metabolites in the Frontal Cortex Metabolic Network, Related to STAR Methods and Figure S1, Table S6. Details of Reactions and Metabolites in the Temporal Cortex Metabolic Network, Related to STAR Methods and Figure S1, Table S7. Details of Reactions and Metabolites in the Frontal Pole Metabolic Network, Related to STAR Methods and Figure S1, Table S8. Details of Reactions and Metabolites in the Inferior Frontal Gyrus Metabolic Network, Related to STAR Methods and Figure S1, Table S9. Details of Reactions and Metabolites in the Parahippocampal Gyrus Metabolic Network, Related to STAR Methods and Figure S1, Table S10. Details of Reactions and Metabolites in the Superior Temporal Gyrus Metabolic Network, Related to STAR Methods and Figure S1).
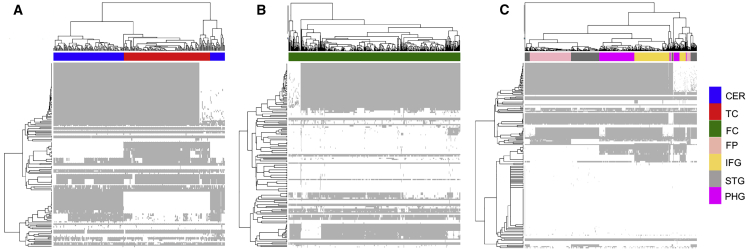
We tested each model using 16 brain-specific in silico tests meant to mimic the experimental evidence of metabolic functions in the brain (“metabolic tasks”) (Table S11) that were obtained from a recently published work on human reconstruction.31 These metabolic tasks represent a set of reactions that are brain specific, and the metabolic networks generated passed 65%–85% of the tasks (see STAR Methods). The metabolic tasks are listed in Table S11 and the models are provided in SBML in https://github.com/PriceLab/Bile_acid_AD. In addition to generating brain region-specific metabolic networks, we also used the transcriptome data of 2,114 post-mortem brain samples and obtained personalized networks for each sample in the study. Of the 2,114 brain samples, 818 samples corresponded to individuals with AD, 138 to possible AD, 137 to probable AD, and 617 controls. The dataset also consisted of 12 samples from other dementias, 163 samples with progressive supranuclear palsy, 58 samples with pathologic aging, and 2 samples that were uncharacterized. From our metabolic networks, we identified 518 reactions that were involved in cholesterol metabolism, BA synthesis, and transport of BAs between different compartments in the metabolic networks. The personalized metabolic networks had distinct sets of BA reactions active in the brain regions (details in STAR Methods). Since the post-mortem brain samples for the brain regions were collected by three independent cohorts having different sequencing protocols and depth, the flux results were analyzed separately for these cohorts. The data suggest that the CER and TC have similar sets of BA reactions that can be active in the personalized metabolic networks (Figure 5).
Figure 5.
Metabolic Reconstruction of Brain Regions and Analysis of Reactions Involved in BA Metabolism
Clustergram for 518 reactions involved in BA metabolism.
(A) Mayo clinic cohort (CER and TC).
(B) ROSMAP cohort (FC).
(C) Mount Sinai Brain Bank (FP, IFG, STG, and PHG).
The rows correspond to BA reactions in the network and the columns are colored based on the brain regions.
We analyzed the reaction fluxes and found a similar set of BA reactions carrying fluxes in metabolic networks of these independent cohorts. We used this information to carry out statistical analyses and identify reactions that are significantly different (p < 0.05) in brain regions of AD versus the CN individuals and to identify reactions that were significant in males versus females with AD. We found that reactions carrying out the transport of taurine and cholesterol were significant in the dorsolateral prefrontal cortex, TC and PHG. Taurine is an abundant amino acid present at roughly 1.2 mM in the brain.32 SLC6A6 (TAUT) and SLC36A1 (PAT1) function as taurine transporters, and increased transport of taurine across the BBB has been reported for oxidative stress conditions.33 We found expressions of both of these genes in the brain transcriptome dataset, suggesting that these genes are expressed in the brain and involved in the transport of taurine. Table 1 provides details for significant BA reactions in brain regions identified from our analysis.
Table 1.
List of Bile Acid (BA) Reactions from Our Metabolic Analysis of Brain Regions
| Reaction (VMH ID) | Genes Associated | Subsystem | p Value |
|---|---|---|---|
| Frontal Cortex | |||
| AKR1C41 | AKR1C4 | BA synthesis | 0.033 |
| r2505 | ABCC1 | transport, endoplasmic reticular | 0.030 |
| r2146 | SLCO1A2 | transport, extracellular | 0.023 |
| TAUBETAtc | SLC6A6 | transport, extracellular | 0.009 |
| Temporal Cortex | |||
| HMR_1685 | CYP27A1 | BA synthesis | 0.0067 |
| CHSTEROLt | ABCA1, ABCG5, ABCG8 | transport, extracellular | 0.0073 |
| TAUPAT1c | SLC36A1 | transport, extracellular | 0.0165 |
| TCHOLABCtc | ABCA8 | transport, extracellular | 0.0324 |
| 3DHCDCHOLt2 | SLC10A1, SLC10A2 | transport, extracellular | 0.0185 |
| EBP1r | EBP | cholesterol metabolism | 0.0433 |
| HMGLx | HMGCL | cholesterol metabolism | 0.0293 |
| DHCR241r | DHCR24 | cholesterol metabolism | 0.0413 |
| EBP2r | EBP | cholesterol metabolism | 0.0277 |
| PHG | |||
| HSD3B7P | HSD3B7 | BA synthesis | 0.003 |
| r1051 | transport, endoplasmic reticular | 0.047 | |
| r1052 | transport, lysosomal | 0.047 | |
| r2146 | SLCO1A2 | transport, extracellular | 0.009 |
| RE1796R | HSD3B1, HSD3B2 | BA synthesis | 0.003 |
| TAUPAT1c | SLC36A1 | transport, extracellular | 0.015 |
The reactions are represented in their VMH IDs, and information related to the genes and subsystems are also shown in the table. p values calculated by Fisher’s exact test are indicated in the table, and only those reactions with p < 0.05 are represented here.
From our analysis, we identified reactions with CYP27A1, required by the neural cholesterol clearance pathway, the classical pathway, and the alternative BA pathway, as being significant in AD versus CN brains. Other than BA synthesis, reactions involving metabolites such as 7α-hydroxycholesterol (Virtual Metabolic Human [VMH], www.vmh.life, VMH: xol7a), 7α-hydroxy-5β-cholestan-3-one (VMH: xol7ah), 3α,7α-dihydroxy-5β-cholestane (VMH: xol7ah2), and 7α-hydroxy-cholestene-3-one (VMH: xol7aone) were also identified as being significantly different between AD and CN (p values for these reactions reported in Table 1). The transport of BAs such as taurolithocholic acid 3-sulfate (VMH: HC02198), UDCA (VMH: HC02194), TCA (VMH: tchola), and 3-dehydroxychenodeoxycholic acid (VMH: 3dhchchol) can also be probed further to understand the role of these BAs in AD. Thus, in silico analysis of brain region-specific metabolic networks provides insights into reactions that may be involved in metabolic changes in AD that can be validated from experimental data.
Identifying Transcriptional Regulators Responsible for Altered Metabolism in AD
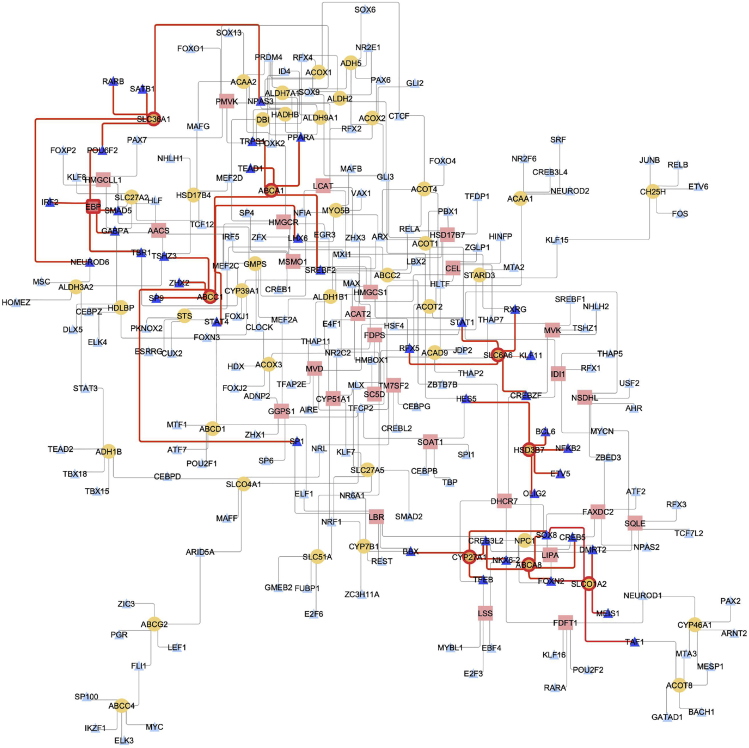
TFs are one important aspect of metabolic regulation that operate by adjusting the expression of genes encoding enzymes. Using a transcriptional regulatory network informed from the same Mayo TC bulk RNA-seq samples used for the metabolic reconstruction, we identified candidate TFs that interact with metabolic genes in cholesterol and BA metabolism. We selectively studied those genes that belonged to reactions that were significantly differentially expressed in AD versus controls to study their role in AD. For example, one gene that came up from our metabolic analysis of AD and controls was emopamil-binding protein (EBP) (Figure 6). EBP is involved in cholesterol metabolism, as it is responsible for one of the final steps in the production of cholesterol. Our brain transcriptional regulatory network (TRN) analysis identified POU6F2, IRF2, SMAD5, GABPA, and TBR1 as the top candidate TF regulators for EBP. Regulation by these TFs can help in understanding their role in altered cholesterol metabolism in AD, particularly in evaluating the summation of coordinated changes since these TFs control other genes as well. CYP27A1, as mentioned earlier, is part of the alternative BA synthesis pathway, and CREB3L2 and SOX8 are putative TFs that regulate the expression of this gene. CREB3L2 (cyclic AMP [cAMP]-responsive element binding protein 3-like 2) is induced as a result of endoplasmic reticulum (ER) stress and may function in unfolded protein response signaling in neurons.34 Other than the metabolism-related genes, we also evaluated the interactions of BA transporters such as SLC6A6, SLCO1A2, ABCC1, ABCA1, SLC36A1, and ABCA8 and their transcriptional regulation. As seen in Figure 6, SREBF2 was found to interact with ABCA1, and recently there were reports of variants of SREBP2 that have been linked with AD.25 Increased SREBF2 expression leads to higher cholesterol levels and presumably oxysterol and cholestenoic acid levels, which are ligands of the liver X receptor (LXR). The peroxisome proliferator-activated receptors (PPARs) regulate various physiological processes and are expressed in the central nervous system. PPARA regulates genes involved in fatty acid metabolism and has been reported to regulate neuronal ADAM10 expression, in turn affecting the proteolysis of the amyloid precursor protein.35 PPARA was identified as a putative regulator of ABCA1 in our brain transcriptional regulatory network. ABCA1 plays a role in cholesterol metabolism and transport and is a candidate risk gene for late-onset AD (LOAD).36 SLC6A6, involved in the transport of taurine, was found to be putatively regulated by STAT1, a TF reported to play an important role in spatial learning and memory formation,37 and RXRG, which forms heterodimers with retinoic acid (RA), LXRs, and vitamin D receptors (VDRs).38 Neuronal differentiation 6 (NEUROD6) functions in neuronal development, differentiation, and survival in AD.39 The regulation of SLC36A1 by NEUROD6 indicated that this TF plays a role in controlling the transport of taurine in the brain. These interactions can be probed further to understand their role in AD pathophysiology.
Figure 6.
Transcriptional Regulators Responsible for Altered Cholesterol and BA Metabolism in AD
Transcriptional regulatory network of brain highlighting transcription factors (TFs) and metabolic genes involved in cholesterol and BA metabolism. TFs are represented as blue triangles, BA metabolism genes as yellow circles, and cholesterol metabolism genes as pink rectangles. The significant genes are highlighted with a red border and TFs in a darker shade of blue. The red edges represent interactions between genes and TFs.
In summary, the brain transcriptional regulatory network analysis led to the identification of candidate TFs that regulate genes in cholesterol and BA metabolism, providing clues toward possible roles in BA dysregulation in AD.
Discussion
We carried out a systematic study to identify alterations in cholesterol and BA metabolism in AD versus CN controls using patient-derived post-mortem transcriptomics and metabolomics data. The primary findings of our study are (1) alternative and neural cholesterol clearance pathway of BA synthesis pathway genes were expressed in the brain samples included in this study, indicating that these pathways are prevalent in the brain as compared to the classical BA synthesis pathway; (2) targeted metabolomics analysis of post-mortem brain samples identified primary and secondary BAs and higher ratios of GCDCA:CA, and secondary BAs such as DCA, LCA, TDCA, CDCA, and GDCA in AD versus controls suggest that these BAs may be associated with cognitive decline in AD; (3) the presence of secondary BAs in metabolomics data suggests a possible role of the gut microbiome in AD and highlights the need to study the gut-brain axis to understand changes in AD; (4) transporters associated with taurine and cholesterol metabolism showed different usage based on our genome-scale metabolic network analysis of three independent cohorts; and (5) transcriptional regulatory network analysis identified TFs, including PPARA, RXRG, and SREBF2, regulating BA and cholesterol genes in the brain.
Role of BAs in AD Pathophysiology and Use of Genome-Scale Metabolic Models
BAs are derived from cholesterol, and their synthesis is regulated by complex feedback mechanisms.12,18 Recent studies have identified BAs in brain samples and linked them with cognitive decline in AD.7,10,19 To understand the physiological role of BAs in the brain of AD and CN individuals, we analyzed transcriptome data from post-mortem brain samples obtained from three independent cohorts and identified that genes involved in the alternative BA pathway were expressed compared to the classical pathway in the brain. The alternative BA pathway is initiated by CYP27A1, which catalyzes the steroid side-chain oxidation and in the subsequent step forms C24-BAs. It is also known that cholesterol is converted to 24-hydroxycholesterol by cholesterol 24-hydroxylase (CYP46A1) in the brain, and the gene was found to be expressed in the brain samples. The primary BAs conjugate with glycine and taurine to form conjugated BAs. Taurine plays a neuroprotective role in the brain, and BAs conjugated with taurine are found to be present in the brain. Metabolomics data of serum samples showed that AD patients had higher levels of serum taurine compared to controls, indicating that taurine transport across the BBB may be affected in AD. The presence of secondary BAs in the post-mortem brain samples suggests that these BAs are either endogenously present in the brain or they are transported through the BBB. BAs such as UDCA, TCA, taurolithocholic acid 3-sulfate, and 3-dehydrochenodeoxycholic acid were also identified from our analysis, and the role of these BAs can be probed further. Based on an association study, taurolithocholic acid was predicted to be a cytotoxic BA, whereas chenodeoxycholic acid and UDCA were predicted to be neuroprotective BAs.7 Our analysis of the transcriptome data of 2,114 samples mapped into the metabolic networks of brain regions implicated reactions involved in the production of metabolites such as 7α-hydroxycholesterol, 7α-hydroxy-5β-cholestan-3-one, 7α-hydroxycholestene-3-one, and other derivatives that are formed through CYP7A1 being significantly different (p values for these reactions reported in Table 1) between AD and CN. Although CYP7A1 was not expressed in the post-mortem brain samples, the difference in abundance of these metabolites in AD versus CN suggests that we should explore the possibility of these metabolites entering the brain through the periphery. Our brain-tissue metabolic models can be used by the community to capture in silico changes and possibly identify metabolic biomarkers ahead of disease manifestation, making them useful in understanding interactions and mechanisms between different classes of metabolites and AD pathophysiology.
Transcriptional Regulation of BA and Cholesterol Genes
Metabolism is influenced by the regulation of TFs and metabolic genes. In this study, we used a transcriptional regulatory network of brain (and selected brain regions) to identify candidate TFs that may interact with genes in cholesterol and BA metabolism. We identified SREBF2, PPARA, RXRG, and other TFs, some of which have been studied and implicated in AD. SREBF2 expression enhances cholesterol levels40 and presumably oxysterol and cholestenoic acid levels, which are ligands of LXR.41 LXRs and the genes regulated by LXRs such as ABCA1, ABCG1, and APOE, modulate intracellular cholesterol content and cholesterol efflux and have been associated with AD pathogenesis.42 Our analysis also identified PPARA as a putative regulator of ABCA1, and recent studies have demonstrated that PPAR pathway activation increases ABCA1 levels, which in turn led to APOE lipidation and amyloid β clearance.43 We also identified the transport of taurine as an important factor from the metabolic analysis. SLC6A6 (neurotransmitter transporter, taurine) and SLC36A1 (neutral amino acid/proton symporter) play a role in taurine transport. Although there was a 1.02- to 1.3-fold change in the expression of these transporters in AD compared with control samples of the three cohorts across four tested brain regions, this difference was only found to be statistically significant in CER (Table S1). The integration of expression data with the metabolic networks of brain regions identified reactions involving taurine transporters that were statistically significant in AD versus controls, further supporting their potential role. We had also identified STAT1 as a putative TF of SLC6A6 from our analysis of brain regulatory networks. Studies have suggested that the increased STAT1 may be involved in inflammation in the AD brain.37,44 NEUROD6 regulates the activity of SLC36A1, a proton-coupled amino acid transporter. NEUROD6 is a basic helix-loop-helix TF, and SNPs in NEUROD6 have been associated with AD, especially in APOE4+ women.45 Our analysis has been able to capture metabolic genes and putative TFs that regulate them. These findings can be further strengthened by the generation of higher-quality footprint data from brain samples.
Studying the Gut-Brain Axis to Understand Physiological Changes in AD
Increasing evidence from experimental and clinical data suggests the influence of the gut-brain axis and gut microbiota in neurodegenerative diseases.46,47 From our metabolic analysis, we identified taurolithocholic, 3-dehydrochenodeoxycholic, and UDCA, secondary BAs, that are significant in AD compared to CN,48 suggesting a possible connection to the gut microbiome. Recently, the BA deconjugation and biotransformation pathways have been reconstructed in a resource of genome-scale reconstructions of >800 human gut microbes.49,50 Of these, only 23 species could synthesize 3-dehydrochenodeoxycholic acid, only 4 could synthesize LCA, and only 3 could synthesize UDCA.50 For instance, the species Ruminococcus (Blautia) gnavus and Collinsella aerofaciens synthesize 3-dehydroxychenodeoxycholic and UDCA, Eggerthella lenta synthesizes 3-dehydrochenodeoxycholic, and several Clostridiales representatives synthesize LCA,50 indicating that these species may play a role in AD. Interestingly, increased LCA in plasma has recently been proposed as a potential biomarker for AD.51 Recent reports have shown the influence of BAs in the host metabolism via alterations of the bacterial community structure.52 The personalized brain models developed in this study could be joined with the personalized microbial community models established previously.50,53 In future efforts, such combined host-microbe metabolic modeling will yield more insight into mechanisms underlying altered BA metabolism in AD.
Limitations of Study
Despite using three independent cohorts, transcriptomics data are insufficient to fully parametrize the metabolic models. If denser longitudinal omics data become available, then they will help improve the predictions from these in silico models. Although our analysis identifies reactions that are significant in these conditions, the directionality of the reactions can only be solidly determined if we have additional time-series metabolomics data (and ideally, isotopic labeling experiments). Methods are being developed to obtain cell-type-specific data54,55 from which we can gain additional information regarding distinct cell-type specific metabolic changes in AD. These studies have shown that microglia and neuronal cells had different transcriptional responses in AD versus control. Integrating such cell-specific data will help in refining the models and making more accurate predictions.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Extracts of brain tissue | Rush Alzheimer’s Disease Center (RADC) Research Resource Sharing Hub | https://www.radc.rush.edu/ |
| Extracts of brain tissue | NIH Brain Tissue Repository, Ichan School of Medicine at Mount Sinai | https://icahn.mssm.edu/research/nih-brain-tissue-repository |
| Extracts of brain tissue | Mayo Clinic Brain Bank | https://www.brainsupportnetwork.org/brain-donation/mayo-clinic/ |
| Deposited Data | ||
| Post-mortem brain transcriptome data | Synapse | https://www.synapse.org/#!Synapse:syn2580853/ |
| Metabolomics data | Synapse | https://www.synapse.org/#!Synapse:syn10235594/ |
| Software and Algorithms | ||
| MATLAB R2018a | MathWorks | https://www.mathworks.com/products/matlab.html |
| COBRA toolbox v3.0 | Heirendt et al.77 | https://opencobra.github.io/cobratoolbox/stable/ |
| Gurobi optimizer v7.5 | Gurobi | https://www.gurobi.com/academia/ |
| IBM CPLEX v12.7.1 | IBM | https://www.ibm.com/analytics/cplex-optimizer |
| mCADRE algorithm | Wang et al.63 | http://bmcsystbiol.biomedcentral.com/articles/10.1186/1752-0509-6-153 |
| iMAT algorithm | Zur et al.74 | https://doi.org/10.1093/bioinformatics/btq602 |
| TReNA package | Bioconductor | https://rdrr.io/bioc/TReNA/ |
| Cytoscape 3.7.1 | Cytoscape | https://cytoscape.org/ |
| Other | ||
| Codes for model generation and simulation | GitHub | https://github.com/PriceLab/Bile_acid_AD |
Resource Availability
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Nathan Price (nathan.price@systemsbiology.org)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The post-mortem brain transcriptome data used in this study can be downloaded from https://www.synapse.org/#!Synapse:syn2580853. The targeted metabolomics data of bile acids used in this study can be downloaded from https://www.synapse.org/#!Synapse:syn10235594. The code used for reconstruction and model generation and simulation are provided in GitHub https://github.com/PriceLab/Bile_acid_AD
Experimental Model and Subject Details
Details of sample collection, description of post-mortem brain samples, RNA extraction, library preparation and sequencing are provided in previously published work.56, 57, 58 The combined samples from three independent cohorts consisted of 2114 post-mortem samples (Table S1). There were 265 samples of temporal cortex (TC) and cerebellum (CER), 632 samples of frontal cortex (FC), 303 samples of frontal pole (FP), superior temporal gyrus (STG), inferior frontal gyrus (IFG) and parahippocampal gyrus (PHG) with pathologies such as AD, MCI, Parkinson’s and control. Summary of sample sizes of AD cases and controls are shown in Table S1 and represented in Figure 1. Details of sample collection, preparation, and metabolite quantification are provided in previously published work.59 Sample details and quantified bile acids are provided in Table S2.
Method Details
Transcriptome analysis of post-mortem brain samples
Transcriptome data was obtained from post-mortem brain samples of AD patients and cognitively normal individuals from Religious Orders Study and Memory and Aging Project (ROSMAP), Mayo Clinic, University of Florida, Institute for Systems Biology and Mount Sinai Brain Bank (MSBB). 265 samples of temporal cortex (TC) and cerebellum (CER), 632 samples of frontal cortex (FC), 303 samples of frontal pole (FP), superior temporal gyrus (STG), inferior frontal gyrus (IFG) and parahippocampal gyrus (PHG) with pathologies such as AD, MCI, Parkinson’s and control were analyzed and used for construction of brain region-specific metabolic models. ROSMAP data can be requested via the Rush Alzheimer’s Disease Center website (https://www.radc.rush.edu/). RNA-seq libraries were prepared by different methods such as poly-A enriched, strand-specific and ribo-zero. The RNaseq data from different centers were uniformly processed using a consensus set of tools as described in Logsdon et al.,60 with only library type-specific parameters varying between pipelines. The authors performed library normalization using fixed/mixed effects modeling. To summarize the method in Logsdon et al.60 for normalization, the genes were filtered and only those genes that had more than 1 CPM in 50% of samples were retained for analysis. Conditional quantile normalization was done to account for variations in gene length and GC content and principal component analysis was used to detect sample outliers. Weighted linear models were used to estimate the confidence of sampling abundance by using the voom-limma package in Bioconductor. This workflow was used to account for differences between the samples, experimental batch effects, and technical variations due to RNA-Seq. We have used these uniformly-processed data for analysis in our study. Table S1 has information on the number of patients with various pathologies and controls and methods used for RNA-sequencing. The data used in the preparation of this article were downloaded from Synapse (https://www.synapse.org/#!Synapse:syn2580853/). We performed two-tailed t test with Benjamini-Hochberg correction to identify differentially expressed genes with corresponding p values. The differential expression analysis for transcriptome data from three independent cohorts is presented in Table S1.
Bile acid sample preparation and analysis
Participants of the Religious Orders Study (ROS) are comprised of Catholic brothers, nuns, and priests who were cognitively normal at study entry and agreed to annual clinical examinations and brain donation at time of death. The Rush Memory and Aging Project (MAP) is a companion study that includes community-dwelling older adults that all agreed to evaluations similar to ROS. Quantification of bile acid concentration was performed at the University of Hawaii cancer center. The bile acid-free matrix (BAFM) was used to prepare bile acid calibrators. Extracts of brain tissue along with bile acid reference standards were subjected to instrumental analysis.61,62 All of the 57 bile acid standards were obtained from Steraloids Inc. (Newport, RI) and TRC Chemicals (Toronto, ON, Canada) and 9 stable isotope-labeled standards were obtained from C/D/N Isotopes Inc. (Quebec, Canada) and Steraloids Inc. (Newport, RI). A Waters ACQUITY ultra performance LC system coupled with a Waters XEVO TQ-S mass spectrometer was used for all analyses. Chromatographic separations were performed with an ACQUITY BEH C18 column. UPLC-MS raw data obtained with negative mode were analyzed using TargetLynx applications manager to obtain calibration equations and the quantitative concentration (μM) of each bile acid. Bile acids were measured from the dorsolateral prefrontal cortex of 111 individuals with brain pathology (28, 33, 10, 22 and 18 with CERAD score of 1, 2, 3, 4 and 9 (missing), respectively). Metabolomics data can be accessed with permission at https://www.synapse.org/#!Synapse:syn10235594. We calculated the ratio of primary and secondary bile acids measured in metabolomics study and performed two-tailed t test to calculate p value for each bile acid.
Brain region-specific metabolic reconstruction
We used transcriptome data (https://www.synapse.org/#!Synapse:syn2580853/) derived from post-mortem brain samples of three independent cohorts: Mayo clinic, ROSMAP and Mount Sinai Brain Bank. These cohorts contained information of different brain regions (CER, FC, TC, FP, STG, IFG and PHG) and the data was used to generate brain region-specific metabolic networks. We converted the transcriptome data to binary by considering transcripts with expression values less than 25th percentile in the matrix as 0 otherwise 1. We calculated ubiquity scores for genes in each brain region separately and used those for implementing mCADRE workflow.63 The ubiquity score of a gene is equal to the sum of samples in which the gene is expressed divided by the total number of samples. The Recon 3D model31 of human metabolism was used as template to reconstruct brain region-specific metabolic networks as this model had information of reactions related to the primary and conjugated primary acids additionally added to refine the model. The mCADRE workflow required two inputs to build region-specific metabolic models: (1) a global metabolic reconstruction, which in this case was the Recon 3D model, and (2) region-specific gene expression data from many individuals. Using the mCADRE workflow, we generated the draft reconstructions for each brain region. We used functions in COBRA toolbox such as detectDeadEnds to identify dead end metabolites and identifyBlockedRxns to compute all blocked reactions in the draft reconstructions. We used reactions from the Recon 3D model for filling gaps in the metabolic network. We carried out this step for each metabolic network reconstructed for brain regions. We also removed the reactions belonging to drug metabolism from the network as they were not related to functions in the brain. Only the partial urea cycle is reported to be active in the brain, and so we identified enzymes in the urea cycle that are present in the brain64 and included the reactions related to these genes in the metabolic networks. We also included exchange reactions for metabolites identified in the cerebrospinal fluid (CSF) (by metabolomics data as well as the whole-body metabolism reconstruction53 and metabolites that can be taken up across the blood brain barrier (BBB) from blood into the CSF.65, 66, 67, 68, 69, 70 The list of metabolites known to pass the BBB is provided in Table S12. Using the removeUnusedGenes function in the COBRA toolbox we removed genes that were not used in any reaction in the reconstructions. Then we carried out manual curation for genes present in the metabolic network using information from Human Protein Atlas71 for genes expressed in the brain. This effort helped in providing further evidence for genes being present in the metabolic networks for brain regions. The information for reactions, metabolites and genes in metabolic networks of brain regions is provided in Table S4. Details of Reactions and Metabolites in the Cerebellum Metabolic Network, Related to STAR Methods and Figure S1, Table S5. Details of Reactions and Metabolites in the Frontal Cortex Metabolic Network, Related to STAR Methods and Figure S1, Table S6. Details of Reactions and Metabolites in the Temporal Cortex Metabolic Network, Related to STAR Methods and Figure S1, Table S7. Details of Reactions and Metabolites in the Frontal Pole Metabolic Network, Related to STAR Methods and Figure S1, Table S8. Details of Reactions and Metabolites in the Inferior Frontal Gyrus Metabolic Network, Related to STAR Methods and Figure S1, Table S9. Details of Reactions and Metabolites in the Parahippocampal Gyrus Metabolic Network, Related to STAR Methods and Figure S1, Table S10. Details of Reactions and Metabolites in the Superior Temporal Gyrus Metabolic Network, Related to STAR Methods and Figure S1. We tested our models for 16 metabolic tasks (Table S11) that are brain specific and the models passed 65%–85% of those tests. As astrocytes are predominantly involved in maintaining brain physiology,72 we used objective function of astrocytes for our brain metabolic networks. We constrained bounds of exchange reactions using information from a published work on metabolic interactions between cell types in the brain.73 Details of metabolites involved in objective function and bounds for constrained reactions are given in Table S4. Details of Reactions and Metabolites in the Cerebellum Metabolic Network, Related to STAR Methods and Figure S1, Table S5. Details of Reactions and Metabolites in the Frontal Cortex Metabolic Network, Related to STAR Methods and Figure S1, Table S6. Details of Reactions and Metabolites in the Temporal Cortex Metabolic Network, Related to STAR Methods and Figure S1, Table S7. Details of Reactions and Metabolites in the Frontal Pole Metabolic Network, Related to STAR Methods and Figure S1, Table S8. Details of Reactions and Metabolites in the Inferior Frontal Gyrus Metabolic Network, Related to STAR Methods and Figure S1, Table S9. Details of Reactions and Metabolites in the Parahippocampal Gyrus Metabolic Network, Related to STAR Methods and Figure S1, Table S10. Details of Reactions and Metabolites in the Superior Temporal Gyrus Metabolic Network, Related to STAR Methods and Figure S1. We integrated expression data with the brain region-specific metabolic networks and generated context-specific personalized metabolic networks for each sample in the study using iMAT algorithm.74 Flux variability analysis75 was carried out to evaluate minimum and maximum flux for each reaction in the metabolic networks. The context-specific metabolic networks were sampled using Markov Chain Monte Carlo (MCMC) sampling method as described in Bordbar et al.76 The sampling distribution for each reaction in the network was considered as significantly different between AD and CN if the two distributions overlapped by less than 5%. Table S13 has the results of sampling analysis. The codes used for reconstruction and model generation and simulation are provided in GitHub (https://github.com/PriceLab/Bile_acid_AD). We used COBRA toolbox v3.077 for metabolic analysis that was implemented in MATLAB R2018a and academic licenses of Gurobi optimizer v7.5 and IBM CPLEX v12.7.1 were used to solve LP and MILP problems.
Reaction and pathway-level analysis
We carried out flux variability analysis75 for each context-specific personalized metabolic network and used the values for predicting metabolic changes in AD versus CN individuals and sex of the individuals. FVA results were used to generate a matrix in which reactions for which both minimum (vmin) and maximum (vmax) FVA flux are 0, were considered to be non-active and assigned a state of 0, while the remaining reactions were assigned a state of 1. We carried out this analysis for all context-specific metabolic networks. Thus, our matrix contained binary values for all reactions in 2114 context-specific personalized metabolic networks for seven brain regions. We used this scheme to classify the reactions and obtain information not only on the basis of flux measurements but also their activity in each network. From all the reactions in the metabolic networks, we selected only those that belonged to cholesterol metabolism, bile acid synthesis and transport reactions associated with the bile acid metabolites. We applied Fisher’s exact test on the binarized values of reactions to identify those reactions with p value < 0.05 in AD versus CN. These reactions were identified as significant reactions in these groups.
Metabolic regulatory network
The transcriptional regulatory network analysis (TReNA) package (https://rdrr.io/bioc/TReNA/) was used for identifying transcription factors (TFs) that are part of the co-expression modules of interest. Brain-specific transcriptional regulatory network was constructed78 using information from ENCODE. We downloaded the DNase Hypersensitivity (DHS) fastq files from ENCODE for all available brain samples and aligned the sequences using the SNAP method.79 We performed two alignments using seed size of 16 and 20bp. The length of sequence data was > 50 bp. The regions of open chromatin were identified using peak calling algorithm, F-Seq.80 Footprints were generated using default parameters for Wellington81 and HINT.82 Our method generated individual gene models and those footprints that are within the proximal promoter region (+/−5 kb of the transcription start site) are considered as priors in assessing the relationship between the expression of the TF and target genes. We prioritized putative TF regulators for each gene in the model using LASSO regression techniques, Pearson and Spearman correlation and random forest methods and projected the scores from these approaches into PCA space. The principal components were summed together to obtain a single composite score called pcaMax. This process is part of the trena package in Bioconductor (https://rdrr.io/bioc/TReNA/) and we applied the method to the post-mortem samples from the temporal cortex from Mayo Clinic. We used metabolic genes identified from reaction-level analysis involved in bile acid and cholesterol metabolism and mapped top five transcription factors that interact with these metabolic genes and created an interaction network. These interaction networks gave information for transcription factors that regulate metabolic genes and are involved in significant reactions in AD versus cognitively normal individuals. Cytoscape 3.7.183 was used for visualizing the brain transcriptional regulatory network.
Quantification and Statistical Analysis
For DEG analysis, two-tailed t test with Benjamini-Hochberg correction was used for comparison between AD and CN samples. For targeted metabolomics data, we used the ratio of primary and secondary bile acids and performed two-tailed t test to calculate p values. Fisher’s exact test was used on the binarized values of reactions and p value was calculated. A p value of < 0.05 is considered statistically significant. We used LASSO regression, Pearson and Spearman correlation and random forest for calculating scores for putative TF regulators for each gene in the model,
Acknowledgments
The authors would like to thank the AMP-AD Consortium for funding the project and the AMP-AD Knowledge Portal for sharing the data. The authors would also like to thank members of the Hood-Price group at ISB for their support and help. Funding for the ADMC (Alzheimer’s Disease Metabolomics Consortium, led by R.K.-D. at Duke University) was provided by National Institute on Aging (NIA) grant no. R01AG046171, a component of the Accelerated Medicines Partnership for AD (AMP-AD) Target Discovery and Preclinical Validation Project (https://www.nia.nih.gov/research/dn/amp-ad-target-discovery-and-preclinical-validation-project) and NIA grant no. RF1 AG0151550, a component of the M2OVE-AD Consortium (Molecular Mechanisms of the Vascular Etiology of AD–Consortium (https://www.nia.nih.gov/news/decoding-molecular-ties-between-vascular-disease-and-alzheimers). The Religious Orders and the Rush Memory and Aging studies were supported by NIA grant nos. P30AG10161, R01AG15819, R01AG17917, andU01AG46152. Additionally, M.A., R.K.-D., and G.K. are supported by NIA grant nos. RF1 AG058942 and R01 AG057452. M.A. and G.K. are also supported by funding from the Qatar National Research Fund NPRP8-061-3-011. K.N. is supported by NIA grant nos. NLM R01 LM012535 and NIA R03 AG054936. A.J.S. is supported by NIH grants, including P30 AG010133, R01 AG019771, and R01 CA129769. W.J.G. is supported by funding from the UK Biotechnology and Biological Sciences Research Council (grant nos. BB/I001735/1 and BB/N015932/1) and the Engineering and Physical Sciences Research Council via an Impact Acceleration Account to Swansea University. J.T.Y. is supported by the Institute for Systems Biology’s Translational Research Fellows Program.
Author Contributions
N.D.P. and R.K.-D. conceived and supervised the study. P.B. reconstructed the brain region-specific metabolic networks; P.B., C.C.F., and J.Y. analyzed the transcriptomics data for post-mortem brain samples downloaded from the AMP-AD knowledge portal; A.K.-P. analyzed the metabolomics data of the brain; I.T. provided the list of metabolites that can cross the BBB; W.J.G. and A.K.-P. provided valuable comments about the BA analysis; W.J. measured the BAs from the post-mortem brain samples; the AMP-AD Consortium and the Alzheimer Disease Metabolomics Consortium collected the transcriptomics and metabolomics data; J.T.Y. and P.B. did the sampling of the metabolic networks; A.H. and I.T. provided the information about the gut microbiome; P.B., C.C.F., J.T.Y., J.Y., A.K.-P., K.N., A.H., S.M., G.L., A.J.S., M.A., G.K., W.J.G., I.T., R.K.-D., and N.D.P. contributed to the writing of this paper. All of the authors reviewed and edited the paper.
Declaration of Interests
The authors declare no competing interests.
Published: November 17, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100138.
Contributor Information
Rima Kaddurah-Daouk, Email: rima.kaddurahdaouk@duke.edu.
Nathan D. Price, Email: nathan.price@systemsbiology.org.
The Alzheimer’s Disease Metabolomics Consortium:
Rima Kaddurah-Daouk, Alexandra Kueider-Paisley, Gregory Louie, P. Murali Doraiswamy, Colette Blach, Arthur Moseley, J. Will Thompson, Siamak Mahmoudiandehkhordi, Kathleen Welsh-Balmer, Brenda Plassman, Andrew Saykin, Kwangsik Nho, Gabi Kastenmüller, Matthias Arnold, Sudeepa Bhattacharyya, Xianlin Han, Rebecca Baillie, Oliver Fiehn, Dinesh Barupal, Peter Meikle, Sarkis Mazmanian, Mitchel Kling, Leslie Shaw, John Trojanowski, Jon Toledo, Cornelia van Duijin, Thomas Hankemier, Ines Thiele, Almut Heinken, Nathan Price, Cory Funk, Priyanka Baloni, Wei Jia, David Wishart, Roberta Brinton, Rui Chang, Lindsay Farrer, Rhoda Au, Wendy Qiu, Peter Würtz, Lara Mangravite, Jan Krumsiek, John Newman, Bin Zhang, and Herman Moreno
Supplemental Information
The total number of post-mortem brain samples for different regions, method of preparation of RNA-sequencing library preparation, number of samples for different pathologies, gender and APOE status of the individuals is described in the table. Related to STAR Methods and Figure 1.
References
- 1.Di Paolo G., Kim T.-W. Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat. Rev. Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel J.W., Vachon-Presseau E., Pichet Binette A., Tam A., Orban P., La Joie R., Savard M., Picard C., Poirier J., Bellec P., Alzheimer’s Disease Neuroimaging Initiative and the PREVENT-AD Research Group Brain properties predict proximity to symptom onset in sporadic Alzheimer’s disease. Brain. 2018;141:1871–1883. doi: 10.1093/brain/awy093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai H., Cong W.N., Ji S., Rothman S., Maudsley S., Martin B. Metabolic dysfunction in Alzheimer’s disease and related neurodegenerative disorders. Curr. Alzheimer Res. 2012;9:5–17. doi: 10.2174/156720512799015064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattson M.P., Arumugam T.V. Hallmarks of Brain Aging: Adaptive and Pathological Modification by Metabolic States. Cell Metab. 2018;27:1176–1199. doi: 10.1016/j.cmet.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaddurah-Daouk R., Zhu H., Sharma S., Bogdanov M., Rozen S.G., Matson W., Oki N.O., Motsinger-Reif A.A., Churchill E., Lei Z., Pharmacometabolomics Research Network Alterations in metabolic pathways and networks in Alzheimer’s disease. Transl. Psychiatry. 2013;3:e244. doi: 10.1038/tp.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Genomics of Alzheimer’s Disease Consortium (IGAP) Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimers Dement. 2015;11:658–671. doi: 10.1016/j.jalz.2014.05.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MahmoudianDehkordi S., Arnold M., Nho K., Ahmad S., Jia W., Xie G., Louie G., Kueider-Paisley A., Moseley M.A., Thompson J.W., Alzheimer’s Disease Neuroimaging Initiative and the Alzheimer Disease Metabolomics Consortium Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease-An emerging role for gut microbiome. Alzheimers Dement. 2019;15:76–92. doi: 10.1016/j.jalz.2018.07.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghaisas S., Maher J., Kanthasamy A. Gut microbiome in health and disease: linking the microbiome-gut-brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol. Ther. 2016;158:52–62. doi: 10.1016/j.pharmthera.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tremlett H., Bauer K.C., Appel-Cresswell S., Finlay B.B., Waubant E. The gut microbiome in human neurological disease: a review. Ann. Neurol. 2017;81:369–382. doi: 10.1002/ana.24901. [DOI] [PubMed] [Google Scholar]
- 10.Nho K., Kueider-Paisley A., MahmoudianDehkordi S., Arnold M., Risacher S.L., Louie G., Blach C., Baillie R., Han X., Kastenmüller G., Alzheimer’s Disease Neuroimaging Initiative and the Alzheimer Disease Metabolomics Consortium Altered bile acid profile in mild cognitive impairment and Alzheimer’s disease: Relationship to neuroimaging and CSF biomarkers. Alzheimers Dement. 2019;15:232–244. doi: 10.1016/j.jalz.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMillin M., DeMorrow S. Effects of bile acids on neurological function and disease. FASEB J. 2016;30:3658–3668. doi: 10.1096/fj.201600275R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths W.J., Sjövall J. Bile acids: analysis in biological fluids and tissues. J. Lipid Res. 2010;51:23–41. doi: 10.1194/jlr.R001941-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martins I.J., Berger T., Sharman M.J., Verdile G., Fuller S.J., Martins R.N. Cholesterol metabolism and transport in the pathogenesis of Alzheimer’s disease. J. Neurochem. 2009;111:1275–1308. doi: 10.1111/j.1471-4159.2009.06408.x. [DOI] [PubMed] [Google Scholar]
- 14.Fonseca A.C.R.G., Resende R., Oliveira C.R., Pereira C.M.F. Cholesterol and statins in Alzheimer’s disease: current controversies. Exp. Neurol. 2010;223:282–293. doi: 10.1016/j.expneurol.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Houten S.M., Watanabe M., Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25:1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 17.Chiang J.Y.L. Bile acid metabolism and signaling. Compr. Physiol. 2013;3:1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang J.Y. Recent advances in understanding bile acid homeostasis. F1000Res. 2017;6:2029. doi: 10.12688/f1000research.12449.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan X., Elliott C.T., McGuinness B., Passmore P., Kehoe P.G., Hölscher C., McClean P.L., Graham S.F., Green B.D. Metabolomic Profiling of Bile Acids in Clinical and Experimental Samples of Alzheimer’s Disease. Metabolites. 2017;7:E28. doi: 10.3390/metabo7020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinn M., McMillin M., Galindo C., Frampton G., Pae H.Y., DeMorrow S. Bile acids permeabilize the blood brain barrier after bile duct ligation in rats via Rac1-dependent mechanisms. Dig. Liver Dis. 2014;46:527–534. doi: 10.1016/j.dld.2014.01.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ament S.A., Pearl J.R., Cantle J.P., Bragg R.M., Skene P.J., Coffey S.R., Bergey D.E., Wheeler V.C., MacDonald M.E., Baliga N.S. Transcriptional regulatory networks underlying gene expression changes in Huntington’s disease. Mol. Syst. Biol. 2018;14:e7435. doi: 10.15252/msb.20167435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearl J.R., Colantuoni C., Bergey D.E., Funk C.C., Shannon P., Basu B., Casella A.M., Oshone R.T., Hood L., Price N.D., Ament S.A. Genome-Scale Transcriptional Regulatory Network Models of Psychiatric and Neurodegenerative Disorders. Cell Syst. 2019;8:122–135.e7. doi: 10.1016/j.cels.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths W.J., Abdel-Khalik J., Yutuc E., Roman G., Warner M., Gustafsson J.-Å., Wang Y. Concentrations of bile acid precursors in cerebrospinal fluid of Alzheimer’s disease patients. Free Radic. Biol. Med. 2019;134:42–52. doi: 10.1016/j.freeradbiomed.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah S.A., Yoon G.H., Chung S.S., Abid M.N., Kim T.H., Lee H.Y., Kim M.O. Novel osmotin inhibits SREBP2 via the AdipoR1/AMPK/SIRT1 pathway to improve Alzheimer’s disease neuropathological deficits. Mol. Psychiatry. 2017;22:407–416. doi: 10.1038/mp.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horton J.D., Goldstein J.L., Brown M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Björkhem I., Araya Z., Rudling M., Angelin B., Einarsson C., Wikvall K. Differences in the regulation of the classical and the alternative pathway for bile acid synthesis in human liver. No coordinate regulation of CYP7A1 and CYP27A1. J. Biol. Chem. 2002;277:26804–26807. doi: 10.1074/jbc.M202343200. [DOI] [PubMed] [Google Scholar]
- 27.Mertens K.L., Kalsbeek A., Soeters M.R., Eggink H.M. Bile Acid Signaling Pathways from the Enterohepatic Circulation to the Central Nervous System. Front. Neurosci. 2017;11:617. doi: 10.3389/fnins.2017.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeMorrow S. Bile Acids in Hepatic Encephalopathy. J. Clin. Exp. Hepatol. 2019;9:117–124. doi: 10.1016/j.jceh.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie G., Wang X., Jiang R., Zhao A., Yan J., Zheng X., Huang F., Liu X., Panee J., Rajani C. Dysregulated bile acid signaling contributes to the neurological impairment in murine models of acute and chronic liver failure. EBioMedicine. 2018;37:294–306. doi: 10.1016/j.ebiom.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendoza M.E., Monte M.J., Serrano M.A., Pastor-Anglada M., Stieger B., Meier P.J., Medarde M., Marin J.J. Physiological characteristics of allo-cholic acid. J. Lipid Res. 2003;44:84–92. doi: 10.1194/jlr.m200220-jlr200. [DOI] [PubMed] [Google Scholar]
- 31.Brunk E., Sahoo S., Zielinski D.C., Altunkaya A., Dräger A., Mih N., Gatto F., Nilsson A., Preciat Gonzalez G.A., Aurich M.K. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 2018;36:272–281. doi: 10.1038/nbt.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffin J.W.D., Bradshaw P.C. Amino Acid Catabolism in Alzheimer’s Disease Brain: Friend or Foe? Oxid. Med. Cell. Longev. 2017;2017:5472792. doi: 10.1155/2017/5472792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang Y.-S., Ohtsuki S., Takanaga H., Tomi M., Hosoya K., Terasaki T. Regulation of taurine transport at the blood-brain barrier by tumor necrosis factor-α, taurine and hypertonicity. J. Neurochem. 2002;83:1188–1195. doi: 10.1046/j.1471-4159.2002.01223.x. [DOI] [PubMed] [Google Scholar]
- 34.Kondo S., Saito A., Hino S., Murakami T., Ogata M., Kanemoto S., Nara S., Yamashita A., Yoshinaga K., Hara H., Imaizumi K. BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol. Cell. Biol. 2007;27:1716–1729. doi: 10.1128/MCB.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corbett G.T., Gonzalez F.J., Pahan K. Activation of peroxisome proliferator-activated receptor α stimulates ADAM10-mediated proteolysis of APP. Proc. Natl. Acad. Sci. USA. 2015;112:8445–8450. doi: 10.1073/pnas.1504890112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupton M.K., Proitsi P., Lin K., Hamilton G., Daniilidou M., Tsolaki M., Powell J.F. The role of ABCA1 gene sequence variants on risk of Alzheimer’s disease. J. Alzheimers Dis. 2014;38:897–906. doi: 10.3233/JAD-131121. [DOI] [PubMed] [Google Scholar]
- 37.Hsu W.-L., Ma Y.-L., Hsieh D.-Y., Liu Y.-C., Lee E.H.Y. STAT1 negatively regulates spatial memory formation and mediates the memory-impairing effect of Aβ. Neuropsychopharmacology. 2014;39:746–758. doi: 10.1038/npp.2013.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang P., Chandra V., Rastinejad F. Retinoic acid actions through mammalian nuclear receptors. Chem. Rev. 2014;114:233–254. doi: 10.1021/cr400161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satoh J., Yamamoto Y., Asahina N., Kitano S., Kino Y. RNA-Seq data mining: downregulation of NeuroD6 serves as a possible biomarker for Alzheimer’s disease brains. Dis. Markers. 2014;2014:123165. doi: 10.1155/2014/123165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferris H.A., Perry R.J., Moreira G.V., Shulman G.I., Horton J.D., Kahn C.R. Loss of astrocyte cholesterol synthesis disrupts neuronal function and alters whole-body metabolism. Proc. Natl. Acad. Sci. USA. 2017;114:1189–1194. doi: 10.1073/pnas.1620506114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths W.J., Abdel-Khalik J., Yutuc E., Morgan A.H., Gilmore I., Hearn T., Wang Y. Cholesterolomics: an update. Anal. Biochem. 2017;524:56–67. doi: 10.1016/j.ab.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koldamova R., Lefterov I. Role of LXR and ABCA1 in the pathogenesis of Alzheimer’s disease - implications for a new therapeutic approach. Curr. Alzheimer Res. 2007;4:171–178. doi: 10.2174/156720507780362227. [DOI] [PubMed] [Google Scholar]
- 43.Liao F., Yoon H., Kim J. Apolipoprotein E metabolism and functions in brain and its role in Alzheimer’s disease. Curr. Opin. Lipidol. 2017;28:60–67. doi: 10.1097/MOL.0000000000000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitamura Y., Shimohama S., Ota T., Matsuoka Y., Nomura Y., Taniguchi T. Alteration of transcription factors NF-kappaB and STAT1 in Alzheimer’s disease brains. Neurosci. Lett. 1997;237:17–20. doi: 10.1016/s0304-3940(97)00797-0. [DOI] [PubMed] [Google Scholar]
- 45.Fowler K.D., Funt J.M., Artyomov M.N., Zeskind B., Kolitz S.E., Towfic F. Leveraging existing data sets to generate new insights into Alzheimer’s disease biology in specific patient subsets. Sci. Rep. 2015;5:14324. doi: 10.1038/srep14324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kowalski K., Mulak A. Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J. Neurogastroenterol. Motil. 2019;25:48–60. doi: 10.5056/jnm18087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang C., Li G., Huang P., Liu Z., Zhao B. The Gut Microbiota and Alzheimer’s Disease. J. Alzheimers Dis. 2017;58:1–15. doi: 10.3233/JAD-161141. [DOI] [PubMed] [Google Scholar]
- 48.Ridlon J.M., Harris S.C., Bhowmik S., Kang D.-J., Hylemon P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7:22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magnúsdóttir S., Heinken A., Kutt L., Ravcheev D.A., Bauer E., Noronha A., Greenhalgh K., Jäger C., Baginska J., Wilmes P. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 2017;35:81–89. doi: 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- 50.Heinken A., Ravcheev D.A., Baldini F., Heirendt L., Fleming R.M.T., Thiele I. Personalized modeling of the human gut microbiome reveals distinct bile acid deconjugation and biotransformation potential in healthy and IBD individuals. bioRxiv. 2017 doi: 10.1101/229138. [DOI] [Google Scholar]
- 51.Marksteiner J., Blasko I., Kemmler G., Koal T., Humpel C. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer’s disease. Metabolomics. 2018;14:1. doi: 10.1007/s11306-017-1297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian Y., Gui W., Koo I., Smith P.B., Allman E.L., Nichols R.G., Rimal B., Cai J., Liu Q., Patterson A.D. The microbiome modulating activity of bile acids. Gut Microbes. 2020;11:979–996. doi: 10.1080/19490976.2020.1732268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thiele I., Sahoo S., Heinken A., Heirendt L., Aurich M.K., Noronha A. When metabolism meets physiology: Harvey and Harvetta. bioRxiv. 2018 doi: 10.1101/255885. [DOI] [Google Scholar]
- 54.Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J.Z., Menon M., He L., Abdurrob F., Jiang X. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohammadi S., Davila-Velderrain J., Kellis M. Reconstruction of Cell-type-Specific Interactomes at Single-Cell Resolution. Cell Syst. 2019;9:559–568.e4. doi: 10.1016/j.cels.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allen M., Wang X., Burgess J.D., Watzlawik J., Serie D.J., Younkin C.S., Nguyen T., Malphrus K.G., Lincoln S., Carrasquillo M.M. Conserved brain myelination networks are altered in Alzheimer’s and other neurodegenerative diseases. Alzheimers Dement. 2018;14:352–366. doi: 10.1016/j.jalz.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Jager P.L., Ma Y., McCabe C., Xu J., Vardarajan B.N., Felsky D., Klein H.U., White C.C., Peters M.A., Lodgson B. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data. 2018;5:180142. doi: 10.1038/sdata.2018.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang M., Beckmann N.D., Roussos P., Wang E., Zhou X., Wang Q., Ming C., Neff R., Ma W., Fullard J.F. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci. Data. 2018;5:180185. doi: 10.1038/sdata.2018.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J., Wei R., Xie G., Arnold M., Kueider-Paisley A., Louie G., Mahmoudian Dehkordi S., Blach C., Baillie R., Han X. Peripheral serum metabolomic profiles inform central cognitive impairment. Sci. Rep. 2020;10:14059. doi: 10.1038/s41598-020-70703-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Logsdon, B.A., Perumal, T.M., Swarup, V., Wang, M., Funk, C., Gaiteri, C., et al. Meta-analysis of the human brain transcriptome identifies heterogeneity across human AD coexpression modules robust to sample collection and methodological approach. bioRxiv 10.1101/510420.
- 61.Xie G., Wang Y., Wang X., Zhao A., Chen T., Ni Y., Wong L., Zhang H., Zhang J., Liu C. Profiling of serum bile acids in a healthy Chinese population using UPLC-MS/MS. J. Proteome Res. 2015;14:850–859. doi: 10.1021/pr500920q. [DOI] [PubMed] [Google Scholar]
- 62.Xie G., Zhong W., Li H., Li Q., Qiu Y., Zheng X., Chen H., Zhao X., Zhang S., Zhou Z. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. 2013;27:3583–3593. doi: 10.1096/fj.13-231860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y., Eddy J.A., Price N.D. Reconstruction of genome-scale metabolic models for 126 human tissues using mCADRE. BMC Syst. Biol. 2012;6:153. doi: 10.1186/1752-0509-6-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Braissant O., McLin V.A., Cudalbu C. Ammonia toxicity to the brain. J. Inherit. Metab. Dis. 2013;36:595–612. doi: 10.1007/s10545-012-9546-2. [DOI] [PubMed] [Google Scholar]
- 65.Redzic Z. Molecular biology of the blood-brain and the blood-cerebrospinal fluid barriers: similarities and differences. Fluids Barriers CNS. 2011;8:3. doi: 10.1186/2045-8118-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pardridge W.M. Transport of nutrients and hormones through the blood-brain barrier. Diabetologia. 1981;20(Suppl):246–254. [PubMed] [Google Scholar]
- 67.Smith Q.R. Transport of glutamate and other amino acids at the blood-brain barrier. J. Nutr. 2000;130(4S Suppl):1016S–1022S. doi: 10.1093/jn/130.4.1016S. [DOI] [PubMed] [Google Scholar]
- 68.Pardridge W.M. The blood-brain barrier: bottleneck in brain drug development. NeuroRx. 2005;2:3–14. doi: 10.1602/neurorx.2.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pardridge W.M., Mietus L.J. Palmitate and cholesterol transport through the blood-brain barrier. J. Neurochem. 1980;34:463–466. doi: 10.1111/j.1471-4159.1980.tb06621.x. [DOI] [PubMed] [Google Scholar]
- 70.Spector R. Fatty acid transport through the blood-brain barrier. J. Neurochem. 1988;50:639–643. doi: 10.1111/j.1471-4159.1988.tb02958.x. [DOI] [PubMed] [Google Scholar]
- 71.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 72.Martín-Jiménez C.A., Salazar-Barreto D., Barreto G.E., González J. Genome-Scale Reconstruction of the Human Astrocyte Metabolic Network. Front. Aging Neurosci. 2017;9:23. doi: 10.3389/fnagi.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lewis N.E., Schramm G., Bordbar A., Schellenberger J., Andersen M.P., Cheng J.K., Patel N., Yee A., Lewis R.A., Eils R. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat. Biotechnol. 2010;28:1279–1285. doi: 10.1038/nbt.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zur H., Ruppin E., Shlomi T. iMAT: an integrative metabolic analysis tool. Bioinformatics. 2010;26:3140–3142. doi: 10.1093/bioinformatics/btq602. [DOI] [PubMed] [Google Scholar]
- 75.Gudmundsson S., Thiele I. Computationally efficient flux variability analysis. BMC Bioinformatics. 2010;11:489. doi: 10.1186/1471-2105-11-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bordbar A., Yurkovich J.T., Paglia G., Rolfsson O., Sigurjónsson Ó.E., Palsson B.O. Elucidating dynamic metabolic physiology through network integration of quantitative time-course metabolomics. Sci. Rep. 2017;7:46249. doi: 10.1038/srep46249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heirendt L., Arreckx S., Pfau T., Mendoza S.N., Richelle A., Heinken A., Haraldsdóttir H.S., Wachowiak J., Keating S.M., Vlasov V. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 2019;14:639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Funk C.C., Jung S., Richards M.A., Rodriguez A., Shannon P., Donovan-Maiye R., Heavner B., Chard K., Xiao Y., Glusman G. Atlas of Transcription Factor Binding Sites from ENCODE DNase Hypersensitivity Data Across 27 Tissue Types. Cell Rep. 2020;32:108029. doi: 10.1016/j.celrep.2020.108029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zaharia M., Bolosky W.J., Curtis K., Fox A., Patterson D., Shenker S., Stoica I., Karp R.M., Sittler T. Faster and More Accurate Sequence Alignment with SNAP. arXiv. 2011 http://arxiv.org/abs/1111.5572 1111.5572v1. [Google Scholar]
- 80.Boyle A.P., Guinney J., Crawford G.E., Furey T.S. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Piper J., Elze M.C., Cauchy P., Cockerill P.N., Bonifer C., Ott S. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 2013;41:e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gusmao E.G., Dieterich C., Zenke M., Costa I.G. Detection of active transcription factor binding sites with the combination of DNase hypersensitivity and histone modifications. Bioinformatics. 2014;30:3143–3151. doi: 10.1093/bioinformatics/btu519. [DOI] [PubMed] [Google Scholar]
- 83.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The total number of post-mortem brain samples for different regions, method of preparation of RNA-sequencing library preparation, number of samples for different pathologies, gender and APOE status of the individuals is described in the table. Related to STAR Methods and Figure 1.
Data Availability Statement
The post-mortem brain transcriptome data used in this study can be downloaded from https://www.synapse.org/#!Synapse:syn2580853. The targeted metabolomics data of bile acids used in this study can be downloaded from https://www.synapse.org/#!Synapse:syn10235594. The code used for reconstruction and model generation and simulation are provided in GitHub https://github.com/PriceLab/Bile_acid_AD