Abstract
Multidrug resistance, especially carbapenem resistance in Acinetobacter bacteria is a global healthcare concern. However, available data on the phenotypic and genotypic characteristics of Acinetobacter isolates from West Africa, including Ghana is scanty. Our aim was to investigate the antibiotic resistance profile and genotypic characteristics of Acinetobacter isolates from Ghana and to characterize carbapenemase producers using whole-genome sequencing (WGS). A total of 36 Acinetobacter isolates collected at three hospitals in Ghana between 2016 and 2017 were analyzed. MICs were determined by commercial antibiotic plates. Acinetobacter baumannii MLST was determined using the Pasteur scheme. WGS of OXA-carbapenemase producers was performed using short- and long-read sequencing strategies. The resistance rate was highest for trimethoprim/sulfamethoxazole (n = 22; 61%). Six (16.7%) and eight (22.2%) isolates were resistant to ceftazidime and colistin, respectively. Two (5.6%) isolates were resistant and one (2.8%) isolate had intermediate sensitivity to three carbapenems. Fifteen STs were identified in 24 A. baumannii isolates including six new STs (ST1467 ∼ ST1472). ST78 was the predominant (n = 6) followed by ST1469 (n = 3). Four carbapenemase-producing A. baumannii isolates also were identified. Isogenic ST103 isolates Ab-B004d-c and Ab-D10a-a harbored blaOXA–23 within Tn2007 on identical plasmids, pAb-B004d-c_3, and pAb-D10a-a_3. ST1472 isolate Ab-C102 and ST107 isolate Ab-C63 carried blaOXA–58 and blaOXA–420, a rare blaOXA–58 variant, respectively, within novel genetic contexts. Our results show that A. baumannii isolates of diverse and unique genotypes, including OXA-carbapenemase producers, are circulating in Ghana highlighting the need for a wider surveillance of antimicrobial resistance.
Keywords: carbapenem-hydrolyzing class D β-lactamases, carbapenem-resistant Acinetobacter, multilocus sequence typing, whole-genome sequencing, Acinetobacter baumannii, OXA-type beta-lactamase
Introduction
Acinetobacter species are important pathogens, responsible for healthcare-related infections. Acinetobacter baumannii is the most clinically prevalent species, but the clinical significance of A. pittii and A. nosocomialis is also increasing (Adams et al., 2008; Chuang et al., 2011; Nemec et al., 2011; Doi et al., 2015). As Acinetobacter spp. have intrinsic resistance to several antibiotic classes and can acquire resistance determinants, carbapenems are an optimal treatment option for infections caused by these bacteria. However, carbapenem resistance is rising worldwide, leaving very limited antimicrobial treatment options (Dijkshoorn et al., 2007; Hamidian and Nigro, 2019). Therefore, carbapenem-resistant Acinetobacter (CRA) is globally recognized as an urgent threat (World Health Organization, 2017; Centers for Disease Control and Prevention, 2019).
Carbapenem resistance in Acinetobacter is mainly caused by the production of carbapenem-hydrolyzing class D β-lactamases (CHDLs) including OXA-23-like, OXA-24-like and OXA-58-like enzymes, or by the production of Ambler class B metallo-β-lactamases (MBLs) such as NDM-, IMP- and VIM-type enzymes (Potron et al., 2015). Until now, only NDM-1-producing A. baumannii has been reported in Accra, the capital of Ghana (Codjoe et al., 2019). In A. baumannii, overexpression of the intrinsic blaOXA–51-like gene mediated by the insertion sequence ISAba1 upstream also confers resistance to carbapenems (Wong et al., 2019). Changes in outer membrane proteins and overexpression of efflux pumps are also associated with carbapenem resistance (Poirel and Nordmann, 2006a; Coyne et al., 2011).
Surveillance of CRA prevalence, associated genotypes, and elucidating the mechanisms underlying resistance are crucial as they enable us to understand how these bacteria disseminate and how best to control their spread. However, due to the poor antimicrobial resistance surveillance in Ghana, data on CRA is limited. Moreover, no data is available yet for CHDL-producing Acinetobacter and their genotypic characteristics.
Our aim was to investigate the antibiotic resistance profile and the genotypic characteristics of clinical isolates of Acinetobacter spp. collected at three regional hospitals in Ghana. Furthermore, we also characterize OXA-carbapenemase producers using both short- and long-read whole-genome sequencing (WGS).
Materials and Methods
Bacterial Isolates and Identification
A total of 36 non-redundant Acinetobacter clinical isolates were recovered from different patients at three hospitals in Ghana (Tamale Teaching Hospital, n = 3; Cape Coast Teaching Hospital, n = 2; and Effia Nkwanta Regional Hospital, n = 31) between 2016 and 2017. Specimen types included urine (n = 12), sputum (n = 8), wound (n = 3), high vaginal swabs (n = 2), blood (n = 1), semen (n = 1) and cerebrospinal fluid (n = 1). For the remaining six samples, specimen types were not found from the medical records. Microbial identification was performed using the MALDI Biotyper (Bruker Daltonics, Karlsruhe, Germany). Acinetobacter baumannii complex (ABC) isolates were further characterized to species level using gyrB multiplex PCR method as previously described (Higgins et al., 2010).
Antibiotic Susceptibility Testing
The MICs of 15 antibiotics including meropenem, colistin, and minocycline were determined by broth microdilution using the commercial microplates DP34 and DP35 (Eiken Chemical Co., Tokyo, Japan). Results were interpreted according to the Clinical and Laboratory Standards Institute guideline document M100-S27 (Clinical and Laboratory Standards Institute, 2017).
Screening for Carbapenemase Genes
DNA was extracted using the Cica GeneusTM DNA Extraction reagent (Kanto Chemical Co., Tokyo, Japan) and subsequently screened for common CHDLs such as blaOXA–23-like, blaOXA–24-like, blaOXA–58-like, and blaOXA–235-like, as well as the A. baumannii native β-lactamase blaOXA–51–like, using multiplex PCR, as previously described (Woodford et al., 2006; Higgins et al., 2013). Screenings for other carbapenemase genes including blaOXA–48, blaNDM, blaKPC, blaIMP, blaVIM, and blaGES were also conducted for all isolates (Dallenne et al., 2010; Wachino et al., 2011).
Multilocus Sequence Typing
Multilocus sequence typing (MLST) was performed for all confirmed A. baumannii isolates using the Pasteur scheme. New allele types and new sequence types (STs) were submitted to PubMLST1 and assigned new numbers. Genetic relatedness of STs was analyzed and visualized using PHYLOVIZ 2.0 goBURST Full MST analysis (Nascimento et al., 2017).
Whole-Genome Sequencing
Short- and long-read WGS was conducted in four CHDL-producing isolates to determine their complete chromosome and plasmid sequences with high accuracy. DNA was extracted with the NucleoSpin tissue kit (Macherey-Nagel, Düren, Germany) for short-read sequencing; in one of the isolates the DNA library was prepared with the Nextera DNA Flex library prep kit and sequenced with Illumina Miseq (Illumina, San Diego, CA, United States); GIEasy FS PCR-Free DNA Library Prep Set was used for the remaining three isolates and sequenced with MGI DNBSEQ (MGI Tech Co., Shenzhen, China). Nanopore MinION (Oxford Nanopore Technologies, Oxford, United Kingdom) was used for long-read sequencing; DNA was extracted with the MagAttract HMW DNA Kit (Qiagen, Hildon, Germany); multiplexed DNA libraries were prepared for all four isolates using the Native Barcoding Kit EXP-NBD104 and the ligation sequence Kit SQK-LSK109; and sequencing was performed on a single R9.4.1 flow cell. Low-quality reads (MinION Q < 10; DNBSEQ Q < 32; and MiSeq Q < 30) and short reads (MinION length <1,000 bp; DNBSEQ and Miseq length <10 bp) were filtered out.
Bioinformatics Analysis
Hybrid de novo genome assembly for WGS data was done with Unicycler v0.4.8 and Flye v2.6 using both short and long reads. Sequence annotation was performed using the online system RAST and BLAST searches. Whole-genome SNP typing was done for four CHDL producers using PathoBacTyper2. Resfinder v3.2 was used to detect acquired resistance genes (Zankari et al., 2012). Insertion sequences were identified using ISfinder3. Plasmid structures and genetic contexts of carbapenemase genes were compared and visualized using Easyfig v2.2.2 (Sullivan et al., 2011).
Phylogenetic Analysis
Forty-five blaOXA–58 plasmids and one blaOXA–420 plasmid in the repository of PLSDB v.2020_03_04 (Galata et al., 2019), as well as two plasmids from this study (pAbC102_1 and pAbC63_1) were used for phylogenetic analysis. Sequence alignment of plasmids was performed using CLC genomic workbench 20 (CLC bio, Aarhus, Denmark). A maximum parsimony tree with 1,000 bootstrap replicates was generated using MEGA X 10.1 (Kumar et al., 2018).
Accession Numbers
Complete chromosomal and plasmid sequences were deposited in GenBank under Bioproject PRJNA473419.
Results
Phenotypic Characterization of Acinetobacter Isolates
Thirty-five out of the 36 isolates belonged to ABC, while the other isolate was identified as A. haemolyticus. Among all ABCs, 24 isolates (68.6%) were characterized as A. baumannii followed by seven A. nosocomialis (20%) and three A. pittii (8.6%). The species for the remaining one ABC isolate (identified using MALDI Biotyper) was not determined due to conflicting results from the gyrB multiplex PCR; this isolate was negative for the blaOXA–51-like.
Resistance to trimethoprim/sulfamethoxazole was the most prevalent (n = 22; 61%), followed by resistance to piperacillin (n = 14; 38.9%), and tobramycin (n = 13; 36.1%) (Table 1). Six (16.7%) isolates were resistant to ceftazidime and eight (22.2%) were resistant to colistin. Two (5.6%) were resistant and one (2.8%) had intermediate susceptibility to three carbapenems. Resistance to amikacin and minocycline was the least frequent (n = 1; 2.8% for each). Resistance to all antibiotics, except for amikacin and colistin, were higher among A. baumannii than non-baumannii Acinetobacter isolates.
TABLE 1.
Antibiotic resistance profile of 36 Acinetobacter isolates.
| Antibiotic |
All isolates (n = 36) |
A. baumannii isolates (n = 24) |
Non-baumannii Acinetobacter isolates (n = 12) |
|||||||||
|
MIC (μg/ml) |
%Ra |
MIC (μg/ml) |
%Ra |
MIC (μg/ml) |
%Ra | |||||||
| Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | ||||
| Piperacillin | 8 to >64 | 32 | >64 | 38.9 | 8 to >64 | >64 | >64 | 50 | 8 to >64 | 32 | >64 | 16.7 |
| Ampicillin-sulbactam | 1/2 to >8/16 | 1/2 | 4/8 | 5.6 | 1/2 to >8/16 | 2/4 | 8/16 | 8.3 | 1/2 to 4/8 | 1/2 | 2/4 | 0 |
| Piperacillin-tazobactam | ≤4/16 to >4/64 | ≤4/16 | >4/64 | 22.2 | ≤4/16 to >4/64 | 4/32 | >4/64 | 29.2 | ≤4/16 to >4/64 | ≤4/16 | 4/32 | 8.3 |
| Ceftazidime | 2 to >16 | 4 | >16 | 16.7 | 4 to >16 | 4 | >16 | 20.8 | 2 to >16 | 4 | 16 | 8.3 |
| Cefepime | 1 to >16 | 4 | >16 | 13.9 | 2 to >16 | 4 | >16 | 16.7 | 1 to >16 | 2 | 16 | 8.3 |
| Imipenem | ≤0.25 to >8 | ≤0.25 | 1 | 5.6 | ≤0.25 to >8 | ≤0.25 | 4 | 8.3 | ≤0.25 to 1 | ≤0.25 | ≤0.25 | 0 |
| Meropenem | ≤0.25 to >8 | 0.5 | 2 | 5.6 | ≤0.25 to >8 | 0.5 | 4 | 8.3 | ≤0.25 to 2 | ≤0.25 | 1 | 0 |
| Doripenem | ≤0.25 to >8 | ≤0.25 | 1 | 5.6 | ≤0.25 to >8 | 0.5 | 4 | 8.3 | ≤0.25 to 1 | ≤0.25 | 1 | 0 |
| Gentamycin | 0.5 to >8 | 0.5 | >8 | 33.3 | 0.5 to >8 | >8 | >8 | 50 | 0.5 to 2 | 0.5 | 2 | 0 |
| Tobramycin | 0.5 to >8 | 1 | >8 | 36.1 | 0.5 to >8 | 8 | >8 | 50 | 0.5 to >8 | 0.5 | 2 | 8.3 |
| Amikacin | ≤1 to >32 | 2 | 4 | 2.8 | 2 to 16 | 2 | 4 | 0 | ≤1 to >32 | 2 | 8 | 1.3 |
| Minocycline | ≤0.25 to >8 | 0.5 | 2 | 2.8 | ≤0.25 to >8 | 0.5 | 4 | 4.2 | ≤0.25 to 4 | ≤0.25 | 1 | 0 |
| Colistin | ≤2 to 8 | ≤2 | 4 | 22.2 | ≤2 to 4 | ≤2 | 4 | 12.5 | ≤2 to >8 | ≤2 | 8 | 41.7 |
| Ciprofloxacin | 0.12 to >2 | 0.5 | >2 | 33.3 | 0.12 to >2 | >2 | >2 | 50 | 0.12 to 1 | 0.25 | 1 | 0 |
| Levofloxacin | ≤1 to >4 | ≤1 | >4 | 16.7 | ≤1 to >4 | 4 | >4 | 25 | ≤1 | ≤1 | ≤1 | 0 |
| Trimethoprim-sulfamethoxazole | ≤19/1 to >38/2 | >38/2 | >38/2 | 61.1 | ≤19/1 to >38/2 | >38/2 | >38/2 | 66.7 | ≤19/1 to >38/2 | >38/2 | >38/2 | 50 |
aResistant.
Detection of Carbapenemase Producers
Two A. baumannii isolates, Ab-D10a-a and Ab-B004d-c, belonging to ST103, and harboring blaOXA–23 were identified. The allelic variant of blaOXA–51 was blaOXA–70 for both isolates. In addition, A. baumannii isolates, Ab-C102A and Ab-C63, belonging to the novel ST1472 and ST107 with blaOXA–51 variants of blaOXA–699 and blaOXA–51 respectively, were also identified and found to harbor CHDLs, blaOXA–58 and blaOXA–420. All four isolates were collected at the Effia Nkwanta Regional Hospital. Other carbapenemase genes, such as MBL genes were not detected by conventional PCR.
Genetic Diversity of Acinetobacter baumannii Isolates
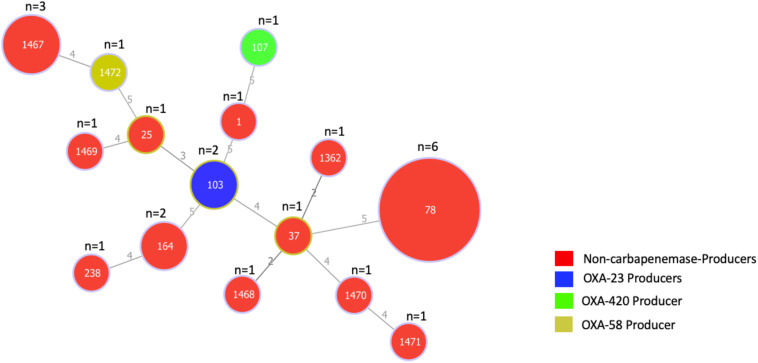
The genetic relatedness of the 24 A. baumannii isolates was assessed using the Pasteur MLST scheme. Isolates were assigned into 15 different STs, six of which were novel STs (ST1467, ST1468, ST1469, ST1470, ST1471, and ST1472). Minimum spanning tree analysis revealed that all STs had no genetic relationship with each other (Figure 1). ST78 (n = 6; 25%) was the most predominant, followed by the novel ST1467 (n = 3; 12.5%).
FIGURE 1.
Minimum spanning tree based on the allelic profiles of 15 STs identified in 24 A. baumannii isolates. The number of isolates for each ST is shown at the top of each node. Node sizes are proportional to the number of isolates for each ST and numbers on connecting lines indicate the number of locus variants determined by pair-wise comparisons. Colors indicate whether isolates are carbapenemase producers and if so, the type of carbapenemase they produce.
Genotypic Characterization of Ab-B004d-c, Ab-D10a-a, and blaOXA–23 Genetic Context
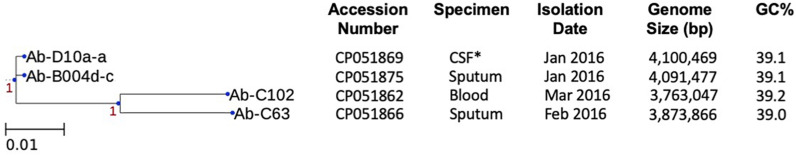
Two OXA-23-producing ST103 isolates, Ab-B004d-c, and Ab-D10a-a, were recovered from sputum and cerebrospinal fluid samples of different patients in January 2016 (Figure 2). Both isolates were resistant to most antibiotics, including carbapenems, but were fully susceptible to colistin (Table 2). Over 4 Gb short-read sequences and about 1.8 Gb long-read sequences were obtained for each isolate after quality filtering (Supplementary Table 1). Using de novo hybrid assembly, two circular chromosomes with 4,091,477 and 4,100,469 bp were generated for Ab-B004d-c and Ab-D10a-a, respectively (Supplementary Table 2). Both isolates were isogenic, differing by only 10 SNPs (Figure 2). Four pairs of identical plasmids ranging in size from 2,697–48,239 bp also were generated for both isolates (Supplementary Table 2). Ab-D10a-a carried an extra 6,619 bp plasmid (Supplementary Table 2). The chromosomes of both isolates carried the aminoglycoside resistance gene aac(3)-IIa, the sulphonamide resistance gene sul1, the intrinsic β-lactamase gene blaOXA–70, the extended-spectrum β-lactamase (ESBL) gene blaCTX–M–15, and the intrinsic AmpC gene blaADC–203. No insertion sequences were detected upstream of blaOXA–70 and blaADC–203. The trimethoprim resistance gene dfrA1, the aminoglycoside resistance gene aac(3)-IId, the tetracycline resistance gene tet(39), and macrolide resistance genes msr(E) and mph(E) were found on two identical plasmids with 48,239 bp (pAb-B004d-c_1 and pAb-D10a-a_1) in both isolates.
FIGURE 2.
Whole-genome SNP analysis of chromosomal sequences of four CHDL-producing isolates and their characteristics. Scale bar indicates nucleotide substitutions per site (1 SNP per 100 sites). Confidence levels are shown close to the branches. *CSF, cerebral spinal fluid.
TABLE 2.
Genotypic and phenotypic antibiotic resistance profiles of four CHDL-producing A. baumannii isolates.
| Isolates | CHDL type | Chromosome/plasmid | Resistance genes |
MIC (mg/L) |
|||||||||||||||
| PIPC | S/A | TAZ/PIPC | CAZ | CFPM | IPM | MEPM | DRPM | GM | TOB | AMK | MINO | CL | CPFX | LVFX | ST | ||||
| Ab-B004d-c | OXA-23 | Chromosome | blaADC-66, blaOXA-70, sul1, ant(3″)-IIa, aac(3)-IIa and blaCTX-M-15 | >64 | >8/16 | >4/64 | >16 | >16 | >8 | >8 | >8 | >8 | >8 | 8 | 0.5 | ≤2 | >2 | >4 | >38/2 |
| Plasmid | dfrA1, mph(E), msr(E),tet(39), aac(3)-IId and blaOXA-23 | ||||||||||||||||||
| Ab-D10a-a | OXA-23 | Chromosome | blaADC-66,blaOXA-70, sul1, ant(3″)-IIa, aac(3)-IIa and blaCTX-M-15 | >64 | >8/16 | >4/64 | >16 | >16 | >8 | >8 | >8 | >8 | >8 | 16 | 0.5 | ≤2 | >2 | >4 | >38/2 |
| Plasmid | dfrA1, mph(E), msr(E), tet(39), aac(3)-IId and blaOXA-23 | ||||||||||||||||||
| Ab-C102 | OXA-58 | Chromosome | ant(3″)-IIa, blaOXA-699 and blaADC-32 | 64 | 1/2 | 4/32 | 8 | 4 | 1 | 1 | 0.5 | 0.5 | 2 | 2 | ≤0.25 | ≤2 | 0.5 | ≤1 | >38/2 |
| Plasmid | aph(6)-Id, aph(3″)-Ib, sul2, aph(3′)-Ia, floR and blaOXA-58 | ||||||||||||||||||
| Ab-C63 | OXA-420 | Chromosome | blaADC-87, blaOXA-51 and ant(3″)-IIa | >64 | 8/16 | >4/64 | 4 | 8 | 4 | 4 | 4 | >8 | >8 | 2 | 0.5 | 4 | >2 | >4 | >38/2 |
| Plasmid | mph(E), msr(E), tet(39), dfrA20, sul2, aph(6)-Id, aph(3″)-Ib, blaOXA-420, ant(2″)-Ia and aph(3’)-Ia | ||||||||||||||||||
Abbreviations: PIPC, piperacillin; S/A, ampicillin-sulbactam; TAZ/PIPC, piperacillin-tazobactam; CAZ, ceftazidime; CFPM, cefepime; IPM, imipenem; MEPM, meropenem; DRPM, doripenem; GM, gentamycin; TOB, tobramycin; AMK, amikacin; MINO, minocycline; CL, colistin, CPFX, ciprofloxacin; LVFX, levofloxacin; ST, trimethoprim-sulfamethoxazole.
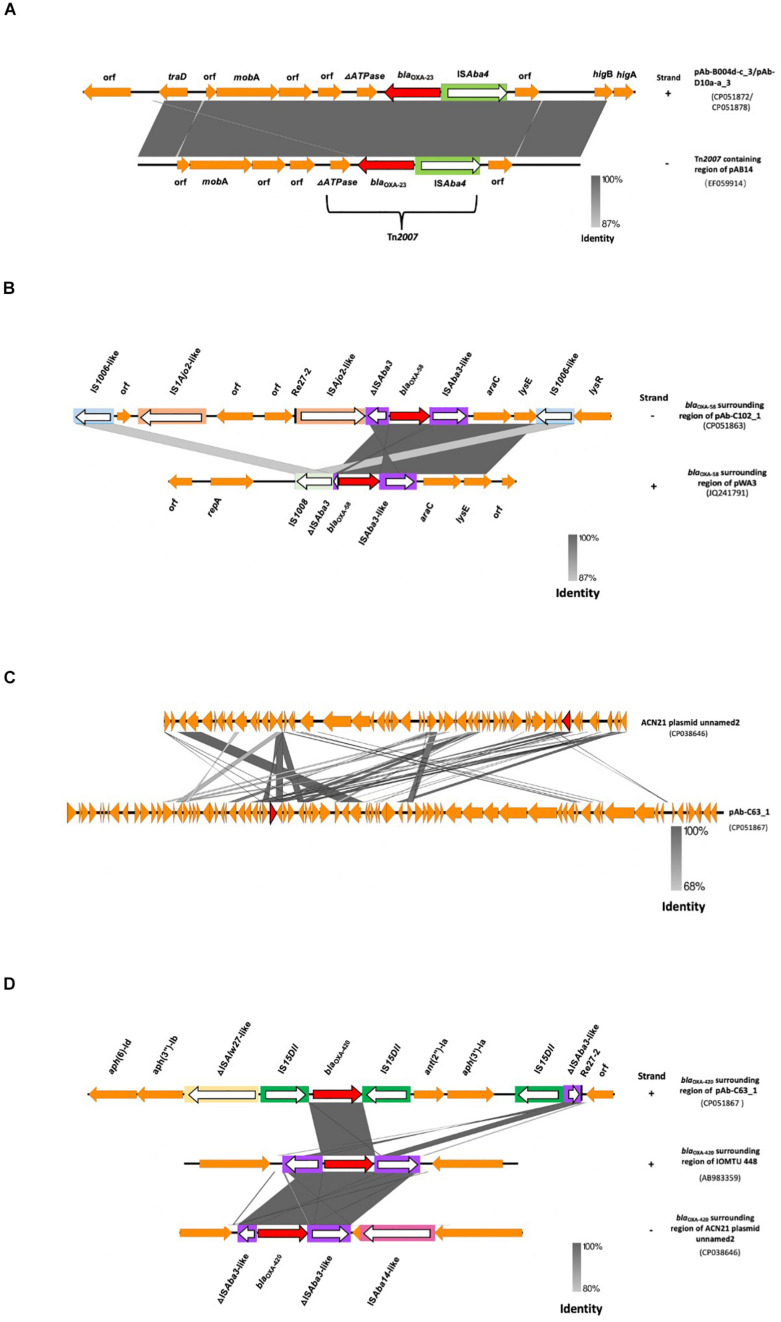
The blaOXA–23 gene was present on two identical 8,215 bp plasmids (pAb-B004d-c_3 and pAb-D10a-a_3) within Tn2007, and flanked upstream by ISAba4 and downstream by a truncated ATPase gene (Figure 3A). No other resistance genes were detected in these plasmids. Annotation results and BLAST searches for ORFs failed to identify any replication origin protein (Rep) gene in these plasmids. The linearized sequences of pAb-B004d-c_3 and pAb-D10a-a_3 were compared with the blaOXA–23-containing region of pAB14 from A. baumannii (accession number: EF059914). Results showed that the genetic structure of Tn2007 was highly identical in pAb-B004d-c_3, pAb-D10a-a_3 and pAB14. In addition, a higBA toxin-antitoxin locus was found downstream of Tn2007.
FIGURE 3.
Genetic contexts of CHDLs in A. baumannii from Ghana. (A) Linearized whole plasmid sequence of blaOXA–23-containing plasmids pAb-B004d-c_3 and pAb-D10a-a_3, compared to the Tn2007-containing region of pAB14. (B) blaOXA–58-surrounding region of pAb-C102_1 compared with that of pWA3. (C) Linearized whole plasmid backbone structure of pAb-C63_1 compared with that of ACN21 plasmid unnamed2. (D) blaOXA–420-surrounding region of pAb-C102_1 compared with that of IOMTU 448 and ACN21 plasmid unnamed2. Carbapenemase genes are represented by red arrows, and other coding sequences are represented in orange. IS elements are inside rectangular boxes. The direction of transposase genes is indicated by white arrows. Same or highly similar IS elements are depicted in the same colors. Re-27 sequences are represented by vertical black bars. Positive and reverse strands are labeled at the right end of each sequence with the symbols “+” and “–”, respectively.
Genotypic Characterization of Ab-C102 and blaOXA–58 Genetic Context
The OXA-58-producing ST1472 isolate Ab-C102 was recovered from the blood of a patient. Ab-C102 was susceptible to almost all antibiotics but displayed a low meropenem MIC (1 mg/L) (Table 2).
Similar to the isolates described before, more than 4 Gb of short-read and near 1 Gb of long-read sequences were obtained by WGS for the isolate Ab-C102, after quality filtering (Supplementary Table 1). A circular chromosome with 3,763,047 bp and three circular plasmids with 19,853, 67,097, and 90,089 bp were generated by hybrid de novo genome assembly (Supplementary Table 2). A novel variant of the intrinsic blaOXA–65 gene with three synonymous nucleotide substitutions, and an intrinsic blaADC–32 gene were identified on the chromosome. No insertion sequences were detected upstream of these genes. The carbapenemase gene blaOXA–58 along with other resistance genes such as aph(6)-Id were identified on a 90,089 bp plasmid (pAb-C102_1; Table 2).
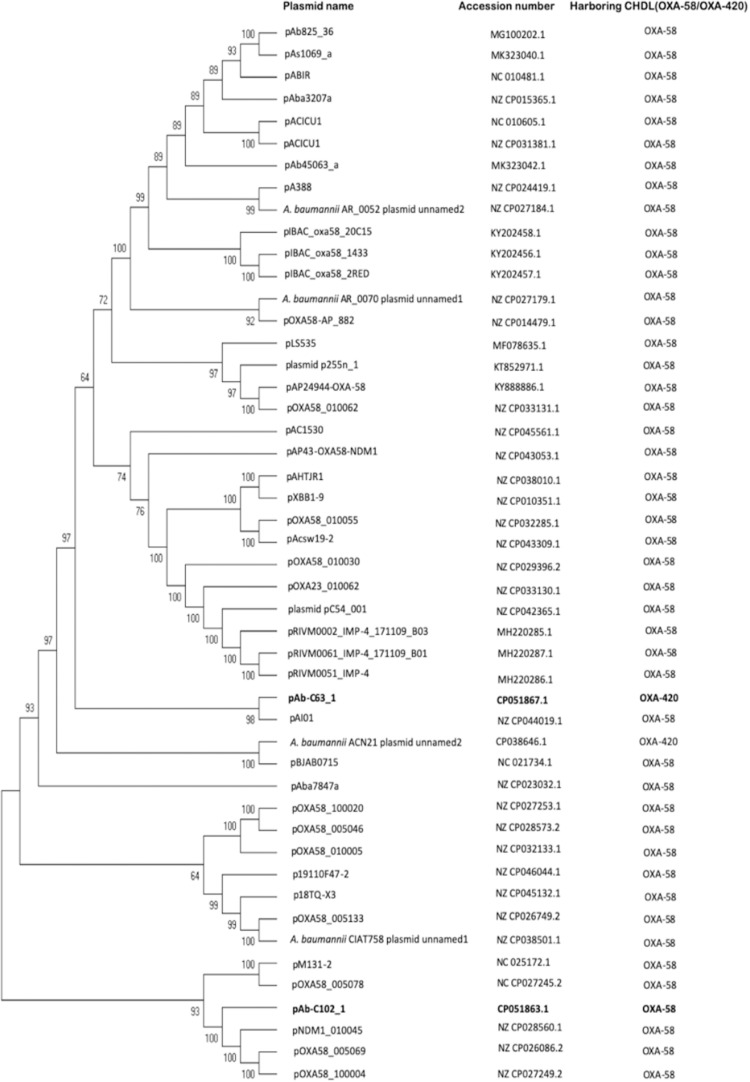
This plasmid encodes a RepB family plasmid Rep that share 100% identity with its counterpart in A. baumannii WB103 (accession number: AZM37906). Phylogenetic analysis showed that the genetically closest plasmid to pAb-C102_1 was pNDM-1_010045, with which it share 46% query cover and 97.25% identity (Figure 4). Interestingly, a BLAST search found that the full plasmid sequence of pAb-C102_1 had the highest homology with the draft WGS data of WB103, sharing 88% of query cover and 99.88% of identity.
FIGURE 4.
Maximum parsimony tree of OXA-58- and OXA-420-containing plasmids. Two plasmids from the current study (highlighted in bold), 45 OXA-58-producing plasmids and one OXA-420-producing plasmid listed in PLSDB database were included. Bootstrap values are shown at each node.
The genetic environment of blaOXA–58 in pAb-C102_1 was comparable to that in pWA3 (accession number: JQ241791), with a similar genetic structure (Figure 3B). In both plasmids, the same ISAba3-like-araC1-lysE structure was found downstream of blaOXA–58 and the same three nucleotide substitutions were present in both ISAba3-like. The ISAba3-like-araC1-lysE was followed by one IS1006-like copy (one nucleotide substitution compared to IS1006) and the lysR gene. Upstream of blaOXA–58, a 427 bp truncated form of ISAba3 was interrupted by a putative novel ISAjo2-like sequence. This contrasted with pWA3 in which a 121 bp truncated form of ISAba3 is interrupted by IS1008. A Re27-2 region, which is a pdif site targeted by the XerC/XerD-like site-specific recombinases, was found 13 bp upstream of the ISAjo2-like sequence. A second copy of ISAjo2-like and IS1006-like (three nucleotide substitutions compared with IS1006) was found upstream of the Re27-2 region. Neither a second copy of Re27-2 nor Re27-1 (single nucleotide variant of Re27-2) were found downstream of blaOXA–58.
Genotypic Characterization of Ab-C63 and blaOXA–420 Genetic Context
Ab-C63 was an isolate recovered from a sputum sample that belonged to ST107 and produced OXA-420, a rare CHDL of the OXA-58 family. This isolate had intermediate susceptibility to carbapenems and was resistant to aminoglycosides, fluoroquinolones and colistin (Table 2).
A 3,873,864 bp circular chromosome and two plasmids with 81,353 and 10,662 bp were generated by hybrid assembly using quality-filtered ∼350 Mb short-read and ∼1.2 Gb long-read sequences (Supplementary Tables 1 and 2). The intrinsic β-lactamase genes blaADC–87 and blaOXA–51, as well as the aminoglycoside resistance gene ant(3″)-IIa were found on the chromosome. The blaOXA–420 gene and other resistance genes such as mph(E) were located on a plasmid with 81,353 bp (pAb-C63_1; Table 2). This plasmid harbored a novel gene encoding a RepB family plasmid Rep which shared the highest similarity (99.7% identity) with proteins from Acinetobacter spp. and A. brisouii (accession numbers: WP_004761033, WP_151711075).
Phylogenetic analysis (Figure 4) showed pAB-C63_1 to be genetically distinct from the blaOXA–420-containing A. baumannii ACN21 plasmid unnamed2 (NZ_CP038646), collected in India, but genetically close to the blaOXA–58-containing pAI01 (NZ_CP044019), with which it shares 42% query cover and 95% identity. Plasmid backbone structure comparisons also revealed that pAb-C63_1 had a novel backbone compared with ACN21 plasmid unnamed2 (Figure 3C). Moreover, a completely distinct genetic context was found surrounding blaOXA–420 in pAbC63_1, in which the gene was flanked by two copies of IS15DII, in contrast with ACN21 plasmid unnamed2 and A. baumannii strain IOMTU in which blaOXA–420 is flanked by ISAba3-like and truncated ISAba3-like elements (Figure 3D). In addition, a third copy of IS15DII was found downstream, interrupting an ISAba3 sequence. The resistance genes ant(2″)-Ia and aph(3′)-Ia were flanked by the second and third copies of IS15DII. The Re27-2 sequence was detected 65 bp downstream of the truncated ISAba3. Moreover, a novel insertion sequence, a ΔISAlw27-like element (coverage = 1268/1282; identities = 1213/1268), was detected upstream of the first copy of IS15DII.
Discussion
Carbapenem resistance in Acinetobacter spp. is a major health threat worldwide. Most of the studies on CRA in Africa were conducted in Northern or Southern regions (Manenzhe et al., 2015), while the current situation in sub-Saharan Africa may be underestimated given the limited laboratory and surveillance capacity. Here, we showed that Acinetobacter species, predominantly A. baumannii, are disseminating in Ghanaian healthcare facilities. A. nosocomialis and A. pittii (previously known as Acinetobacter genomics species 13UT and 3, respectively) were also prevalent. The proportion of these species is parallel to those causing nosocomial bloodstream infections in the United States (Wisplinghoff et al., 2012). Conventional identification methods can not distinguish between ABCs, resulting in misidentification of some species as A. baumannii (Wisplinghoff et al., 2012). Although, gyrB multiplex PCR has been reported as one of the most highly sensitive methods distinguishing species within ABCs (Lee et al., 2014), the conflicting result that we obtained for an ABC isolate using this method has proven the difficulty associated with species identification among ABCs. Multidrug resistance, including to carbapenems, was detected in the Acinetobacter isolates. However, carbapenem resistance was less prevalent (5.6%), in our study (primarily conducted in the Western region of Ghana) compared with a prior study conducted in the country’s capital, Accra, in which seven out of nine A. baumannii isolates from burn wounds were resistant to meropenem (Forson et al., 2017). Higher carbapenem resistance, 51.5 and 10%, have been reported in the West African countries of Nigeria and Sierra Leone, respectively (Nsofor and Chekwube, 2017; Lakoh et al., 2020). Such discrepancy suggests that CRA dissemination varies by region, and may be attributed to the variability in antibiotic choice, infection control, and prevention practices. Moreover, the use of carbapenems in clinical setting in Ghana has been reported as low, and its unavailability as an essential drug on the Ghanaian national health insurance scheme may be associated with low prescription (Labi et al., 2018). We speculate this low carbapenems usage is correlated with low resistance selective pressure which may explain the observed lower carbapenem resistance prevalence in our study. The 22.2% prevalence of colistin resistance among all Acinetobacter isolates as well as 12.5% among A. baumannii isolates in this study are high in comparison with other countries where the resistance prevalence was mostly at single digit levels for A. baumannii (Pormohammad et al., 2020). Such high resistance is alarming especially since colistin has become one of the last-resort antibiotics used for multidrug-resistant gram-negative bacterial infections. The acquired resistance mediated by plasmid originated mcr gene resulted in the global spread of the colistin resistance among Enterobacteriaceae (Wang et al., 2018; Wise et al., 2018). In A. baumannii, colistin resistance is not associated with the mcr gene, rather caused by the overexpressed PetN transferases mediated by two pathways, overexpressed pmrC from mutations in pmrCAB operon and overexpressed pmrC homolog eptA from the insertion of ISAba1 on the upstream (Trebosc et al., 2019). Further investigation remains warranted to elucidate the resistance mechanisms involved in non-baumannii Acinetobacter isolates.
Several global clones (GC) of A. baumannii have emerged in the last decades with GC2, represented by ST2, being the most widespread, followed by GC1, represented by ST1. Multidrug resistance and carbapenemase production are mostly associated with the spread of these specific clones (Hamidian and Nigro, 2019). Here, we showed that A. baumannii isolates recovered in Ghana have high genetic diversity, associated with 15 STs among which six are novel. The predominant GC2 was absent and only a single isolate belonged to GC1. ST78, also known as the “Italian clone” (Carretto et al., 2011), was the predominant sequence type detected in Ghana. However, neither ST1 nor ST78 isolates were associated with carbapenemase production. The CHDL producers found belong to ST103, ST107 and ST1472; Both of ST103, and ST107 were uncommon, while ST1472 is a novel type. OXA-23 production is the leading carbapenem resistance mechanism in A. baumannii, mostly associated with GC2 and GC1 (Hamidian and Nigro, 2019). Although ST103 strains were associated with NDM-2-producing isolates in Egypt, UAE, and Palestine (Kaase et al., 2011; Ghazawi et al., 2012; Sjölander et al., 2014), and an OXA-235-producing strain was reported in Australia (Hamidian et al., 2017), to the best of our knowledge this is the first study to identify ST103 associated with OXA-23 production (isolates Ab-B004d-c and Ab-D10a-a). Based on the occurrence of OXA-58 carbapenemase in several Acinetobacter species and in Proteus mirabilis (Fu et al., 2014; Feng et al., 2016; Lange et al., 2017; Matos et al., 2019), it has been suggested that horizontal gene transfer rather than clonal spread is the main mechanism of blaOXA–58 dissemination. This may also explain the detection of a novel sequence type, ST1472 (isolate Ab-C102). ST107 isolate Ab-C63 carried blaOXA–420, a rare variant of blaOXA–58 that was first described in A. baumannii ST623 and ST32 clinical isolates in Nepal (Shrestha et al., 2015). Ab-C63 was genetically distinct from the Nepalese isolates and other isolates identified in France and Brazil, which had the same ST and produced different CHDLs including OXA-24, OXA-143 and OXA-231 (Jeannot et al., 2014; Rodrigues-Costa et al., 2019).
We identified two isogenic OXA-23 producers, Ab-B004d-c and Ab-D10a-a. These two isolates were recovered from different patients in the same hospital, indicating a possible nosocomial outbreak. The ESBL gene blaCTX–M–15 was also found on the chromosome of both isolates. Third-generation cephalosporin resistance in A. baumannii results mainly from the production of ESBLs of PER, GES and VEB types, or the overexpression of intrinsic AmpC β-lactamases owing to the upstream insertion of specific sequences such as ISAba1 (Hamidian and Hall, 2013; Potron et al., 2015). The ESBL gene blaCTX–M–15 is the main mechanism of resistance to third-generation cephalosporins in Enterobacteriaceae, but not a common one in Acinetobacter. There are only a few report of its occurrence in A. baumannii isolates from India, Haiti, and Brazil (Shakil and Khan, 2010; Potron et al., 2011; Zago et al., 2016). Therefore, by detecting blaCTX–M–15 in isolates from Ghana, our study provides evidence of its wider spread among Acinetobacter species.
The blaOXA–23 gene is the most common carbapenemase gene in A. baumannii and is usually mobilized by Tn2006, Tn2007, Tn2008 (including its variants), Tn2009, Tn6549, and AbaR4 into the chromosome and plasmids. Tn2006 is the most abundant of these mobile elements (Hamidian and Nigro, 2019). Tn2007, which was identified in plasmids pAb-B004d-c_3 and pAb-D10a-a_3 is relatively uncommon with no plasmids listed on PLSDB and only one in GenBank (pAB14) carrying it. However, its detection has been reported in isolates from Algeria, France, Spain, China, and Tanzania (Corvec et al., 2007; Guerrero-Lozano et al., 2015; Wang et al., 2015; Kumburu et al., 2019). The identification of Tn2007 in samples from Ghana, within a plasmid that share a high level of sequence identity with pAB14 (identified in Algeria and Tanzania) strongly supports the importance of plasmids in the spread of carbapenemase genes, especially in Africa.
The blaOXA–58-containing plasmid pAb-C102_1 (isolate Ab-C102) share the same Rep as well as the highest query cover and sequence homology with the draft genome of A. baumannii WB103, an isolate recovered from hospital wastewaters in Singapore and that also carry blaOXA–58 (Chen et al., 2019). It was not possible to compare the genetic structure between the two isolates because WB103 plasmid sequences are not assembled, but the findings suggest that pAb-C102_1 likely derived from a plasmid from WB103.
Diverse genetic environments are described for blaOXA–58. The gene is commonly embedded in structures like ISAba2:ISAba3-blaOXA–58-ISAba3 and IS6 family element-ΔISAba3-blaOXA–58-ISAba3. Usually, these structures are accompanied downstream by araC1 and lysE (Poirel and Nordmann, 2006b; Fu et al., 2014; Cameranesi et al., 2018); the former is involved in blaOXA–58 regulation (Fondi et al., 2010). Mobilization and acquisition of blaOXA–58 is driven by site-specific recombination at XerC/D-like sites (including mediation by Re27) rather than through its surrounding IS elements, leading to diverse structure arrangements (Poirel and Nordmann, 2006b; Cameranesi et al., 2018). We observed a similar pattern in pAb-C102_1, in which an ISAba3 element interrupted by a novel ISAjo2-like sequence was followed by ISAba3-like-araC1-lysE, a structure that commonly surrounds blaOXA–58. In addition, a Re27-2 site was found next to the novel ISAjo2-like element. These results indicate that blaOXA–58 was acquired via a similar XerC/D-like site-specific recombination in pAb-C102_1, and that the second copy of the gene created in the process may have been lost. Moreover, different arrangements in insertion sequences upstream of blaOXA–58 were associated with its overexpression; compared with ISAba3 this resulted in a stronger promoter leading to higher carbapenem resistance levels (Chen et al., 2008, 2010; Fu et al., 2014). It is unclear whether the low carbapenem MIC levels in Ab-C102 is a consequence of ISAjo2-like-ΔISAba3 arrangement and a relatively weak blaOXA–58 promoter. Further investigations are needed to clarify this issue.
The gene blaOXA–420 has three nucleotide substitutions and a single amino acid substitution compared with blaOXA–58, and was first identified in 16 A. baumannii isolates from Nepal (Shrestha et al., 2015). Our BLAST search identified A. baumannii strain ACN21, isolated from India, that also carry blaOXA–420, indicating that this specific variant is mostly spread in South Asia. To the best of our knowledge this study is the first to detect a blaOXA–420-carrying isolate outside of Asia. Isolate Ab-C63 is not genetically related to earlier isolates from Nepal and India. Moreover, the genetic context of blaOXA–420 in pAb-C63_1 is novel while those from India and Nepal are similar. The presence of a Re27-2 sequence, commonly associated with blaOXA–58 acquisition, near blaOXA–420, suggests that this resistance gene also was acquired by XerC/D-mediated site-specific recombination in Ab-C63.
One limitation of our study is the limited sample size. Isolates were collected at three hospitals only, and the number of isolates obtained from two of the hospitals was extremely low. Hence, the results may not reflect the whole situation in Ghana.
In this study, we have demonstrated that the prevalence of carbapenem resistance in Ghana is low and that there is a high level of genetic diversity among clinical isolates of Acinetobacter. Furthermore, we characterized, for the first time in Ghana, OXA- 23-, OXA-58- and OXA-420-producing A. baumannii isolated belonging to ST103, ST1472 and ST107, respectively. By combining short-read and long-read WGS we revealed that blaOXA–23 was within Tn2007 in two plasmids (pAb-B004d-c_3 and pAb-D10a-a_3) and identified novel genetic structures surrounding blaOXA–58 and blaOXA–420 (novel plasmids pAb-C102_1 and pAb-C63_1, respectively). Altogether, our results show that genetically diverse and unique A. baumannii isolates, including OXA-carbapenemase producers, are circulating in Ghana, measures to strengthen and expand surveillance of antimicrobial resistance worldwide remains essential.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of the Faculty of Medicine, Tokyo Medical and Dental University (M2017-208) and the ethics committee of Noguchi Memorial Institute for Medical Research, University of Ghana (FWA 00001824). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
AAy, AAb, and RS conceived and designed the experiments. AAy, AK, MS, WS, SM, IP, MM, and RS performed the experiments. AAy, MS, TH, TS, SI, AAb, and RS analyzed the data. MS, KM, TH, TS, SI, AAb, and RS contributed to reagents, materials, and analysis tools. AAy, MS, AAb, and RS contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the excellent technical support from the Tamale teaching hospital, Cape Coast teaching hospital and Effia Nkwanta regional hospital.
Funding. This work was supported by the Japan Agency for Medical Research and Development (AMED, URL: http://www.amed.go.jp/en/) under Grant Numbers JP19fm0108010 (TH, TS, SI, and RS) and JP20fk0108133 (MS). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.587398/full#supplementary-material
References
- Adams M. D., Goglin K., Molyneaux N., Hujer K. M., Lavender H., Jamison J. J., et al. (2008). Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190 8053–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameranesi M. M., Morán-Barrio J., Limansky A. S., Repizo G. D., Viale A. M. (2018). Site-Specific Recombination at XerC/D Sites Mediates the Formation and Resolution of Plasmid Co-integrates Carrying a blaOXA-58- and TnaphA6-Resistance Module in Acinetobacter baumannii. Front. Microbiol. 9:66. 10.3389/fmicb.2018.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretto E., Barbarini D., Dijkshoorn L., Van Der Reijden T. J., Brisse S., Passet V., et al. (2011). Widespread carbapenem resistant Acinetobacter baumannii clones in Italian hospitals revealed by a multicenter study. Infect. Genet. Evol. 11 1319–1326. 10.1016/j.meegid.2011.04.024 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2019). Antibiotic resistance threats in the United States, 2019. Available online at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf [accessed on April 30, 2020] [Google Scholar]
- Chen H., Gu X., Ng C., Haller L., Rathinam Charles F., Gin K. Y. (2019). Draft genome sequences of a ceftazidime-resistant Acinetobacter baumannii donor and a conjugal Escherichia coli recipient with acquired resistance. Microbiol. Resour. Announc. 8:e00024-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. L., Chang W. C., Kuo S. C., Lee Y. T., Chen C. P., Siu L. K., et al. (2010). Contribution of a plasmid-borne blaOXA-58 gene with its hybrid promoter provided by IS1006 and an ISAba3-like element to beta-lactam resistance in acinetobacter genomic species 13TU. Antimicrob. Agents Chemother. 54 3107–3112. 10.1128/aac.00128-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. L., Wu R. C., Shaio M. F., Fung C. P., Cho W. L. (2008). Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob. Agents Chemother. 52 2573–2580. 10.1128/aac.00393-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y. C., Sheng W. H., Li S. Y., Lin Y. C., Wang J. T., Chen Y. C., et al. (2011). Influence of genospecies of Acinetobacter baumannii complex on clinical outcomes of patients with acinetobacter bacteremia. Clin. Infect. Dis. 52 352–360. 10.1093/cid/ciq154 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2017). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement M100-S27. Wayne, PA: CLSI. [Google Scholar]
- Codjoe F. S., Brown C. A., Smith T. J., Miller K., Donkor E. S. (2019). Genetic relatedness in carbapenem-resistant isolates from clinical specimens in Ghana using ERIC-PCR technique. PLoS One 14:e0222168. 10.1371/journal.pone.0222168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvec S., Poirel L., Naas T., Drugeon H., Nordmann P. (2007). Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51 1530–1533. 10.1128/aac.01132-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne S., Courvalin P., Périchon B. (2011). Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55 947–953. 10.1128/aac.01388-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallenne C., Da Costa A., Decré D., Favier C., Arlet G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrobial. Chemother. 65 490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- Dijkshoorn L., Nemec A., Seifert H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5 939–951. 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- Doi Y., Murray G. L., Peleg A. Y. (2015). Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 36 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Yang P., Wang X., Zong Z. (2016). Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1. OXA-58 and PER-1 by genome sequencing. J. Antimicrob. Chemother. 71 71–75. 10.1093/jac/dkv324 [DOI] [PubMed] [Google Scholar]
- Fondi M., Bacci G., Brilli M., Papaleo M. C., Mengoni A., Vaneechoutte M., et al. (2010). Exploring the evolutionary dynamics of plasmids: the Acinetobacter pan-plasmidome. BMC Evol. Biol. 10:59. 10.1186/1471-2148-10-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forson O. A., Ayanka E., Olu-Taiwo M., Pappoe-Ashong P. J., Ayeh-Kumi P. J. (2017). Bacterial infections in burn wound patients at a tertiary teaching hospital in Accra. Ghana. Ann. Burns Fire Dis. 30 116–120. [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Jiang J., Zhou H., Jiang Y., Yu Y., Zhou J. (2014). Characterization of a novel plasmid type and various genetic contexts of blaOXA-58 in Acinetobacter spp. from multiple cities in China. PLoS One 9:e84680. 10.1371/journal.pone.0084680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galata V., Fehlmann T., Backes C., Keller A. (2019). PLSDB: a resource of complete bacterial plasmids. Nucl. Acids Res. 47 D195–D202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazawi A., Sonnevend A., Bonnin R. A., Poirel L., Nordmann P., Hashmey R., et al. (2012). NDM-2 carbapenemase-producing Acinetobacter baumannii in the United Arab Emirates. Clin. Microbiol. Infect. 18 E34–E36. [DOI] [PubMed] [Google Scholar]
- Guerrero-Lozano I., Fernández-Cuenca F., Galán-Sánchez F., Egea P., Rodríguez-Iglesias M., Pascual Á. (2015). Description of the OXA-23 β-lactamase gene located within Tn2007 in a clinical isolate of Acinetobacter baumannii from Spain. Microb. Drug Resist 21 215–217. 10.1089/mdr.2014.0155 [DOI] [PubMed] [Google Scholar]
- Hamidian M., Hall R. M. (2013). ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J. Antimicrob. Chemother. 68 2682–2683. 10.1093/jac/dkt233 [DOI] [PubMed] [Google Scholar]
- Hamidian M., Nigro S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 5:e000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidian M., Nigro S. J., Hartstein R. M., Hall R. M. (2017). RCH51, a multiply antibiotic-resistant Acinetobacter baumannii ST103IP isolate, carries resistance genes in three plasmids, including a novel potentially conjugative plasmid carrying oxa235 in transposon Tn6252. J. Antimicrob. Chemother. 72 1907–1910. 10.1093/jac/dkx069 [DOI] [PubMed] [Google Scholar]
- Higgins P. G., Lehmann M., Wisplinghoff H., Seifert H. (2010). gyrB multiplex PCR to differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J. Clin. Microbiol. 48 4592–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins P. G., Pérez-Llarena F. J., Zander E., Fernández A., Bou G., Seifert H. (2013). OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 57 2121–2126. 10.1128/aac.02413-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannot K., Diancourt L., Vaux S., Thouverez M., Ribeiro A., Coignard B., et al. (2014). Molecular epidemiology of carbapenem non-susceptible Acinetobacter baumannii in France. PLoS One 9:e115452. 10.1371/journal.pone.0115452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaase M., Nordmann P., Wichelhaus T. A., Gatermann S. G., Bonnin R. A., Poirel L. (2011). NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66 1260–1262. 10.1093/jac/dkr135 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumburu H. H., Sonda T., Van Zwetselaar M., Leekitcharoenphon P., Lukjancenko O., Mmbaga B. T., et al. (2019). Using WGS to identify antibiotic resistance genes and predict antimicrobial resistance phenotypes in MDR Acinetobacter baumannii in Tanzania. J. Antimicrob. Chemother. 74 1484–1493. 10.1093/jac/dkz055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labi A., Obeng-Nkrumah N., Nartey E. T., Bjerrum S., Adu-Aryee N. A., Ofori-Adjei Y. A., et al. (2018). Antibiotic use in a tertiary healthcare facility in Ghana: a point prevalence survey. Antimicrob. Resist. Infect. Control 7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakoh S., Li L., Sevalie S., Guo X., Adekanmbi O., Yang G., et al. (2020). Antibiotic resistance in patients with clinical features of healthcare-associated infections in an urban tertiary hospital in Sierra Leone: a cross-sectional study. Antimicrob. Resist. Infect. Control 9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange F., Pfennigwerth N., Gerigk S., Gohlke F., Oberdorfer K., Purr I., et al. (2017). Dissemination of blaOXA-58 in Proteus mirabilis isolates from Germany. J. Antimicrob. Chemother. 72 1334–1339. [DOI] [PubMed] [Google Scholar]
- Lee M. J., Jang S. J., Li X. M., Park G., Kook J., Kim M. J., et al. (2014). Comparison of rpoB gene sequencing, 16S rRNA gene sequencing, gyrB multiplex PCR, and the VITEK2 system for identification of Acinetobacter clinical isolates. Diagn Microbiol. Infect. Dis. 78 29–34. 10.1016/j.diagmicrobio.2013.07.013 [DOI] [PubMed] [Google Scholar]
- Manenzhe R. I., Zar H. J., Nicol M. P., Kaba M. (2015). The spread of carbapenemase-producing bacteria in Africa: a systematic review. J. Antimicrob. Chemother. 70 23–40. 10.1093/jac/dku356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos A. P., Cayô R., Almeida L. G. P., Streling A. P., Nodari C. S., Martins W. M. B. S., et al. (2019). Genetic Characterization of Plasmid-Borne blaOXA-58 in Distinct Acinetobacter Species. mSphere 4:e00376-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento M., Sousa A., Ramirez M., Francisco A. P., Carriço J. A., Vaz C. (2017). PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33 128–129. 10.1093/bioinformatics/btw582 [DOI] [PubMed] [Google Scholar]
- Nemec A., Krizova L., Maixnerova M., Van Der Reijden T. J., Deschaght P., Passet V., et al. (2011). Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 162 393–404. 10.1016/j.resmic.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Nsofor C. A., Chekwube L. O. (2017). High antibiotics resistance observed in Acinetobacter baumannii isolated from south east Nigeria. EC Microbiol. 8 266–271. [Google Scholar]
- Poirel L., Nordmann P. (2006a). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12 826–836. 10.1111/j.1469-0691.2006.01456.x [DOI] [PubMed] [Google Scholar]
- Poirel L., Nordmann P. (2006b). Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50 1442–1448. 10.1128/aac.50.4.1442-1448.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pormohammad A., Mehdinejadiani K., Gholizadeh P., Nasiri M. J., Mohtavinejad N., Dadashi M., et al. (2020). Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: A systematic review and meta-analysis. Microb. Pathog. 139 103887. 10.1016/j.micpath.2019.103887 [DOI] [PubMed] [Google Scholar]
- Potron A., Munoz-Price L. S., Nordmann P., Cleary T., Poirel L. (2011). Genetic features of CTX-M-15-producing Acinetobacter baumannii from Haiti. Antimicrob. Agents Chemother. 55 5946–5948. 10.1128/aac.05124-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron A., Poirel L., Nordmann P. (2015). Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 45 568–585. 10.1016/j.ijantimicag.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Costa F., Cayô R., Matos A. P., Girardello R., Martins W. M. B. S., Carrara-Marroni F. E., et al. (2019). Temporal evolution of Acinetobacter baumannii ST107 clone: conversion of blaOXA-143 into blaOXA-231 coupled with mobilization of ISAba1 upstream occAB1. Res. Microbiol. 170 53–59. [DOI] [PubMed] [Google Scholar]
- Shakil S., Khan A. U. (2010). Detection of CTX-M-15-producing and carbapenem-resistant Acinetobacter baumannii strains from urine from an Indian hospital. J. Chemother. 22 324–327. 10.1179/joc.2010.22.5.324 [DOI] [PubMed] [Google Scholar]
- Shrestha S., Tada T., Miyoshi-Akiyama T., Ohara H., Shimada K., Satou K., et al. (2015). Molecular epidemiology of multidrug-resistant Acinetobacter baumannii isolates in a university hospital in Nepal reveals the emergence of a novel epidemic clonal lineage. Int. J. Antimicrob. Agents 46 526–531. 10.1016/j.ijantimicag.2015.07.012 [DOI] [PubMed] [Google Scholar]
- Sjölander I., Hansen F., Elmanama A., Khayyat R., Abu-Zant A., Hussein A., et al. (2014). Detection of NDM-2-producing Acinetobacter baumannii and VIM-producing Pseudomonas aeruginosa in Palestine. J. Glob. Antimicrob. Resist. 2 93–97. 10.1016/j.jgar.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebosc V., Gartenmann S., Tötzl M., Lucchini V., Schellhorn B., Pieren M., et al. (2019). Dissecting colistin resistance mechanisms in extensively drug-resistant Acinetobacter baumannii clinical isolates. MBio 10:1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J., Yoshida H., Yamane K., Suzuki S., Matsui M., Yamagishi T., et al. (2011). SMB-1, a novel subclass B3 metallo-beta-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 55 5143–5149. 10.1128/aac.05045-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Yan D., Hou W., Zeng X., Qi Y., Chen J. (2015). Characterization of bla(OxA-23) gene regions in isolates of Acinetobacter baumannii. J. Microbiol. Immunol. Infect. 48 284–290. [DOI] [PubMed] [Google Scholar]
- Wang R., van Dorp L., Shaw L. P., Bradley P., Wang Q., Wang X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise M. G., Estabrook M. A., Sahm D. F., Stone G. G., Kazmierczak K. M. (2018). Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014-2016 as part of the INFORM global surveillance program. PLoS One. 13:e0195281. 10.1371/journal.pone.0195281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H., Paulus T., Lugenheim M., Stefanik D., Higgins P. G., Edmond M. B., et al. (2012). Nosocomial bloodstream infections due to Acinetobacter baumannii. Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J. Infect. 64 282–290. 10.1016/j.jinf.2011.12.008 [DOI] [PubMed] [Google Scholar]
- Wong M. H., Chan B. K., Chan E. W., Chen S. (2019). Over-expression of ISAba1-linked intrinsic and exogenously acquired OXA type carbapenem-hydrolyzing-class D-ß-lactamase-encoding genes is key mechanism underlying carbapenem resistance in Acinetobacter baumannii. Front. Microbiol. 10:2809. 10.3389/fmicb.2019.02809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N., Ellington M. J., Coelho J. M., Turton J. F., Ward M. E., Brown S., et al. (2006). Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27 351–353. 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2017). Global priority list of antibiotic-resistant bacteria to guide research, discover, and development of new antibiotics. Available online at: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf [accessed on April 30, 2020] [Google Scholar]
- Zago M. C., Viana G. F., Ecker A. B., Nishiyama S. A., Zarpellon M. N., Dias J. R., et al. (2016). First report of CTX-M-15-producing Acinetobacter baumannii in Brazil. J. Hosp. Infect. 92 298–299. 10.1016/j.jhin.2015.11.016 [DOI] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.