Abstract
Orobanchaceae root parasitic weeds cause serious agricultural damage worldwide. Although numerous studies have been conducted to establish an effective control strategy for the growth and spread of root parasitic weeds, no practical method has been developed so far. Previously, metabolomic analyses were conducted on germinating seeds of a broomrape, Orobanche minor, to find novel targets for its selective control. Interestingly, planteose metabolism was identified as a possible target, and nojirimycin (NJ) selectively inhibited the germination of O. minor by intercepting planteose metabolism, although its precise mode of action was unclear. Here, transcriptome analysis by RNA-Seq was conducted to obtain molecular insight into the effects of NJ on germinating O. minor seeds. Differential gene expression analysis results suggest that NJ alters sugar metabolism and/or signaling, which is required to promote seed germination. This finding will contribute to understanding the effect of NJ and establishing a novel strategy for parasitic weed control.
Keywords: broomrape, nojirimycin, Orobanche minor, parasitic weed, sugar signaling, transcriptome
Introduction
Witchweeds and broomrapes in the parasitic Orobanchaceae family cause serious agricultural damage worldwide.1) Since the first infestation of a witchweed, Striga asiatica, in agricultural fields in the United States, numerous studies have been conducted to establish effective control strategies for the growth and spread of root parasitic weeds.2,3) Biochemical studies of the germination process of S. asiatica revealed that ethylene triggers its germination.4) This finding enabled the eradication of S. asiatica in the United States through ethylene treatment of fields to force the germination of root parasitic weeds without host crops.2) Germination without host crops is fatal, because root parasitic weeds are obligate parasites that completely depend on the host. This strategy is called “suicidal germination”, and the development of germination stimulants is a major research focus in parasitic weed management.5) However, the application of ethylene has only been successful in the United States because the areas infected with Striga in other countries are too large for this method to be efficient. Further, broomrapes do not generally respond to ethylene.
Strigolactones (SLs) are key chemicals that induce suicidal germination in both witchweeds and broomrapes.5,6) Since SLs have been revealed to be key chemicals in the symbiotic relationship with arbuscular mycorrhiza and in the regulation of plant morphology, many SL-related physiology and biochemistry studies have been conducted in the last decade.6) A deep understanding of SL biochemistry has enabled the development of promising SL mimics for practical use as inducers of suicidal germination. Sasaki et al. developed a carbamate-type SL mimic, T-010, which can be synthesized through a simple, low-cost method.7) T-010 not only reduced Striga hermonthica emergence, but also increased the yield of the host crop, sorghum (Sorghum bicolour), in field experiments.8) Recently, sphynolactone-7 (SPL-7), which induces the germination of S. hermonthica in the femtomolar range, was developed based on the biochemistry of the agonists binding to a high-affinity SL receptor, Striga HYPOSENSITIVE TO LIGHT/KARRIKIN INSENSITIVE2 7 (ShHTL/KAI2 7). It was shown that 100 pM SPL7 reduced the emergence of S. hermonthica artificially infested with maize in pot experiments.9)
Since SL-dependent germination is a unique feature of root parasitic weeds, the investigation of the physiological and biochemical processes in the germination process will contribute to the identification of novel molecular targets for the selective control of root parasitic weeds. In line with this concept, metabolomic analyses were conducted on the germinating broomrape, Orobanche minor, and planteose metabolism was identified as a possible target for its control.10) Planteose, a trisaccharide composed of galactose, glucose, and fructose, is present in a limited number of plant species, including the seeds of several mints,11) tobacco,12) sesame (Sesamum indicum),13) and chia (Salvia hispanica).14) Neither the physiological role of planteose nor its metabolic pathway in plants has been well studied. In the seeds of O. minor, planteose is accumulated as a storage sugar and, after the detection of SLs, is metabolized rapidly via sucrose to the monosaccharides, glucose and fructose. Our previous study revealed that nojirimycin (NJ) selectively inhibited the germination of O. minor by intercepting planteose metabolism.10) When NJ was applied to synthetic SL (GR24)-treated seeds, the second step of the planteose metabolic pathway, the hydrolysis of sucrose by invertase, was significantly inhibited. Because NJ, an iminosugar, was originally isolated as a potent inhibitor of glucosidase,15) it was postulated that NJ directly inhibited invertases in O. minor seeds. However, in vitro assays revealed that NJ was not a potent inhibitor of invertases, but the activity of invertase in NJ-treated O. minor seeds was significantly decreased as compared to a control.10) Thus, we hypothesized that NJ could inhibit the post-translational modifications of invertases required for their activation through an unknown mode of action (MoA).
Invertase, one of the key enzymes involved in sugar metabolism, irreversibly hydrolyzes sucrose into glucose and fructose; thus, it has a pivotal role in plant development.16–19) Invertases are classified into three classes according to their subcellular localizations and optimal pHs; neutral/alkaline cytoplasmic invertases (CINs), acid vacuolar invertases (VINs), and acid cell wall invertases (CWINs).19) Because the hydrolysis of sucrose occurs in many important physiological processes, such as osmotic regulation and phloem unloading during sugar translocation, the regulation of invertase activity is crucial for plant seed germination. In Arabidopsis thaliana, the expression of genes encoding VINs and CWINs is induced by gibberellin synthesized after the detection of red light in the germination process.20) The expression and activation of three classes of invertases during germination have also been confirmed in several broomrape species, indicating their important roles in sugar metabolism in the germination process.10,21,22)
Because the acid invertases, VINs and CWINs, are transported to acidic compartments through vesicular trafficking, post-translational modifications, such as glycosylation and processing, are important for regulating their activity.18) When N-glycosylation was inhibited by tunicamycin in bright yellow-2 (BY-2) tobacco cells, CWI was degraded prior to sorting in the Golgi network.23) In A. thaliana, the precursor protease vesicle-localized vacuolar processing enzyme-γ (VPEγ) is involved in the regulation of VIN activity through its turnover.17,24) Additionally, inhibitor proteins are also involved in the regulation of invertase activity.17,19,25) Invertase inhibitors are localized to the cell wall or vacuole and directly inhibit CWIN or VIN by forming stable complexes in a pH-dependent manner.19,26) Multiple post-translational regulatory mechanisms suggest that the activation/deactivation of invertase has a significant effect on plant development and growth.
Moreover, the involvement of invertases in sugar signaling has recently been highlighted. When a vacuolar invertase gene, GhVIN1, was suppressed in cotton, a fibreless seed phenotype was observed. Since the seed weight of GhVIN1-RNAi was increased, the fibreless phenotype was not caused by a change in nutritional status. The application of hexokinase (HXK) inhibitors resulted in the same fibreless phenotype, indicating that GhVIN1 is involved in sugar signaling mediated by HXK.27) Recently, CWINs in A. thaliana were revealed as positive regulators of ovule initiation through sugar signaling, possibly mediated by extracellular receptor-like-kinases (RLKs) and hexose transporters.28)
Here, transcriptome analysis was conducted to obtain molecular insights into the effect of NJ on germinating seeds of O. minor. The transcriptome in germinating O. minor seeds was assembled de novo by RNA-Seq and was found to reflect the loss of photosynthesis. Interestingly, differentially expressed genes (DEGs) in NJ-treated seeds involved homologues of invertase inhibitors, sugar transporters, phosphatases, and kinases, suggesting that NJ treatment affects sugar signaling during germination in O. minor.
Materials and Methods
1. Plant material and germination treatment
O. minor seeds were collected from mature plants grown in colonies in Yokohama, Japan, in June 2013 and stored at 4°C. Seed germination was induced as reported previously.10) The seed surface was sterilized with a solution containing 1% sodium hypochlorite and 0.1% (w/v) Tween 20 for 2 min, rinsed with distilled water, and dried under a vacuum. Then, the seeds were conditioned on two layers of glass filters (Whatman GF/D; GE Healthcare, Chicago, IL, USA) fully moistened with distilled water in a Petri dish in the dark at 23°C for one week. After conditioning, the upper layer of the glass filter with the seeds was transferred to a new Petri dish containing a fresh glass filter (Whatman GF/D). Germination was induced by the application of SL solution (1 mg/L rac-GR24) with or without NJ (10 µmol/L). Seeds were collected just after the conditioning, and 0.5, 3, 24, and 48 hr after treatment (HAT) with GR24. Seeds were frozen in liquid nitrogen and stored at −80°C until use.
2. RNA isolation and sequencing
Total RNA was isolated using the PureLink® RNA Mini Kit (Thermo Fischer Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions. Preparation of RNA-Seq libraries and sequencing using the HiSeq 2000 (Illumina, San Diego, CA, USA) were performed at BGI Japan (Kobe, Japan). Raw paired-end reads (90 bp each, ca. 50 million reads in each library) were submitted to the DNA Data Bank of Japan (DDBJ) (Accession number: DRA10691).
3. RNA-Seq data processing
Transcriptome assembly was performed using DDBJ Read Annotation Pipeline.29) The trinity software package (version r2013-02-25) was used to construct the transcriptome.30,31) The quality of the assembled transcriptome was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO) version 3.1.032) with the lineage dataset embryophyta-odb9. The DAVID bioinformatics tool33,34) was used to evaluate the results of the BUSCO analysis. Open reading frame (ORF) prediction in the assembled transcriptome was conducted using TransDecoder (https://github.com/TransDecoder/TransDecoder/wiki). Read mapping to the reference transcriptome and differential expression analysis were conducted with OmicsBox software (BioBam Bioinformatics, Valencia, Spain). The Pairwise Differential Expression Analysis (Without Replicates) module, based on the software package NOISeq, was used with the default parameters.35,36) Genes with a probability higher than 0.9 were considered to be differentially expressed between NJ-treated and non-treated seeds.
Results
1. De novo assembly of O. minor seed RNA-Seq data
The total RNAs were extracted from O. minor seeds after conditioning and at 0.5, 3, 24, and 48 HAT with or without NJ and purified. After quality control of the RNA samples, RNA-Seq libraries were constructed and sequenced using HiSeq 2000. De novo assembly of the transcriptome was performed using reads obtained from RNA samples after conditioning and at 48 HAT without NJ. A total of 119,181 contigs (length >200 bp) with an average size of 951 bp and N50 of 1.5 kbp were obtained. The assembled transcriptome contained 68.9% of complete and 6.0% of fragmented BUSCOs, while 25.1% (361 of 1440 orthologs total) were missing. The high number of missing BUSCOs could represent the loss of photosynthesis in the holoparasitic O. minor. To evaluate this hypothesis, gene ontology (cellular compartment) enrichment analysis was conducted on representative A. thaliana orthologs corresponding to the missing BUSCOs in the O. minor transcriptome (Supplemental Table S1). As expected, more than half of the missing BUSCOs were genes encoding chloroplast-related proteins (Table 1), indicating that the high number of missing BUSCOs represents the loss of photosynthesis. Based on this result, we judged that the quality of the assembled transcriptome was acceptable for further analysis.
Table 1. Gene Ontology term (cellular component) enrichment analysis of representative A. thaliana orthologs corresponding to the missing BUSCOs in O. minor transcriptome.
| GO term | Counta) | %b) | P value | Fold enrichment |
|---|---|---|---|---|
| Chloroplast | 171 | 51.0 | 1.6E−55 | 3.4 |
| Chloroplast thylakoid membrane | 48 | 14.3 | 7.9E−31 | 9.2 |
| Chloroplast thylakoid | 27 | 8.1 | 2.0E−18 | 10.2 |
| Chloroplast thylakoid lumen | 18 | 5.4 | 2.2E−16 | 17.5 |
| Mitochondrion | 100 | 29.9 | 9.6E−16 | 2.3 |
| Chloroplast stroma | 38 | 11.3 | 4.5E−14 | 4.5 |
| Thylakoid lumen | 13 | 3.9 | 1.7E−12 | 19.8 |
| Thylakoid | 20 | 6.0 | 2.5E−11 | 7.5 |
| Chloroplast envelope | 28 | 8.4 | 2.5E−09 | 4.0 |
| Plastid chromosome | 6 | 1.8 | 5.1E−06 | 25.9 |
| Nucleoid | 7 | 2.1 | 2.5E−06 | 15.5 |
| Photosystem II oxygen evolving complex | 6 | 1.8 | 1.2E−05 | 19.4 |
| Chloroplast membrane | 11 | 3.3 | 1.2E−05 | 6.2 |
| Extrinsic component of membrane | 6 | 1.8 | 4.1E−04 | 9.5 |
| Plastid-encoded plastid RNA polymerase complex | 3 | 0.9 | 1.6E−03 | 46.6 |
| Chloroplast inner membrane | 6 | 1.8 | 3.0E−03 | 6.1 |
| NAD(P)H dehydrogenase complex (plastoquinone) | 3 | 0.9 | 1.2E−02 | 17.9 |
| Plastoglobule | 4 | 1.2 | 7.2E−02 | 4.1 |
a) Number of the orthologs related to the GO term. b) Percentage of the orthologs in the total representative A. thaliana orthologs (Supplemental Table S1).
2. Effect of nojirimycin on the transcriptome of O. minor seeds
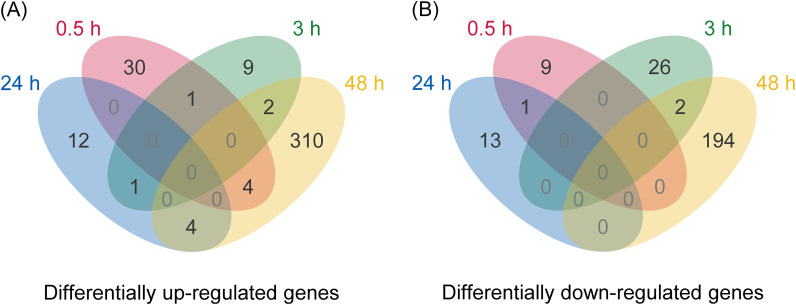
Using TransDecoder, 86,857 ORFs were predicted in the assembled transcriptome. Functions of the proteins encoded by the predicted ORFs were annotated using OmicsBox with Blast and InterProScan using default parameters. After removing duplicated sequences, 16,672 contigs with annotations were used as a reference transcriptome for differential expression analysis. Cleaned reads obtained from each sample were mapped against the reference transcriptome, and a count table was created with OmicsBox using default parameters. Differential expression analysis was performed for each pair of transcripts (NJ-treated vs. non-treated) in the seeds at the same sampling point. As a result, NJ was revealed to alter the expression of a small number of genes at all time points until 24 HAT (Table 2, Supplemental Tables S2–S4). On the other hand, a total of 516 genes were differentially expressed in the NJ-treated seeds at 48 HAT, which might reflect the secondary effects of NJ (Table 2, Supplemental Table S5). A few genes were differentially expressed at multiple time points (Fig. 1, Supplemental Table S6).
Table 2. Number of genes differentially expressed in NJ-treated seeds of O. minor compared to non-treated seeds.
| Time after GR24 treatment (hr) | Number of up-regulated genes | Number of down-regulated genes |
|---|---|---|
| 0.5 | 35 | 10 |
| 3 | 13 | 28 |
| 24 | 17 | 14 |
| 48 | 320 | 196 |
Fig. 1. Venn diagram illustrating the number of differentially (A) up-regulated and (B) down-regulated genes in NJ-treated seeds of O. minor as compared to non-treated seeds. The genes differentially expressed at multiple time points are listed in Supplemental Table S6.
Among the DEGs, candidate genes for the MoA of NJ are listed in Tables 3 and 4. Some genes were differentially expressed at multiple time points, such as comp33755_c0_seq1, whose expression was up-regulated at 0.5 and down-regulated at 3 and 48 HAT (Table 3, Supplemental Table S6). Interestingly, this gene encodes a plant invertase/pectin methylesterase inhibitor superfamily protein. Another gene (comp68818_c0_seq1) encoding a sugar transporter that might be involved in sugar utilization was also up-regulated at 0.5 and down-regulated at 3 HAT (Table 3). A gene encoding protein phosphatase 2C family protein (comp65219_c0_seq4) was up-regulated at 0.5 and 48 HAT (Table 3, Supplemental Table S6), and genes encoding protein kinase superfamily protein (comp232524_c0_seq1) and receptor like protein 54 homolog (comp68094_c0_seq11) were also up-regulated at 0.5 HAT (Table 3), indicating that the expression of these signaling-related genes could be directly induced by NJ treatment. Genes encoding proteins involved in cell wall assembly or modification, such as COBRA-LIKE PROTEIN-7-PRECURSOR homolog (comp34798_c0_seq1) and GARACTURONOSYL TRANSFERASE-LIKE 9 homolog (comp59432_c0_seq2), also showed increased expression at 0.5 HAT.
Table 3. Representative up-regulated genes in the NJ-treated seed of O. minora).
| HATb) | Contig | Fold change c) | Descriptiond) |
|---|---|---|---|
| 0.5 | comp34798_c0_seq1 (down-regulated at 3 HAT) | 8.7 | SEC61 BETA1, COBRA-LIKE PROTEIN-7 PRECURSOR, putative membrane-anchored cell wall protein |
| comp65219_c0_seq4 (up-regulated at 48 HAT) | 3.0 | PP2C CLADE D 5, protein phosphatase 2C family protein | |
| comp33755_c0_seq1 (down-regulated at 3 and 48 HAT) | 9.2 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| comp59432_c0_seq2 | 5.7 | GARACTURONOSYL TRANSFERASE-LIKE 9, encodes a protein with putative galacturonosyltransferase activiy | |
| comp68818_c0_seq1 (down-regulated at 3 HAT) | 6.2 | SUGAR TRANSPORTER 1, encodes a H+/hexose cotransporter | |
| comp232524_c0_seq1 (down-regulated at 3 HTA) | 4.9 | Protein kinase superfamily protein | |
| comp68094_c0_seq11 | 5.4 | RECEPTOR LIKE PROTEIN 54 | |
| 3 | comp92283_c0_seq1 (up-regulated at 48 HAT) | 6.7 | MYB55, encodes a putative transcription factor |
| 48 | comp34269_c0_seq1 | 3.2 | DROUGHT-INDUCED 8, RESPONSIVE TO ABA 18, belongs to the dehydrin protein family, ABA- and drought-induced glycine-rice dehydrin protein |
| comp68110_c0_seq1 | 5.3 | LATE EMBRYOGENESIS ABUNDANT 1, encodes an ABA-induced protein that accumulates during seed maturation | |
| comp64405_c0_seq5 | 2.2 | RNA-BINDING PROTEIN 25, an alternative splicing factor involved in mediation of abiotic stress and ABA responses | |
| comp33799_c0_seq1 | 2.4 | Leucin-rich repeat (LRR) family protein | |
| comp47352_c0_seq1 | 2.2 | KEEP ON GOING, encodes a RING E3 ligase involved in ABA signaling | |
| comp67868_c0_seq5 | 2.6 | Leucin-rich repeat (LRR) family protein |
a) Full list of the differentially expressed genes is available as supplemental materials. b) Hours after GR24 treatment. c) Ratio of normalized count of contig in the NJ-treated O. minor seed to that in the non-treated seed. d) Description for homolog in A. thaliana in TAIR database.
Table 4. Representative down-regulated genes in the NJ-treated seed of O. minora).
| HATb) | Contig | Fold change c) | Descriptiond) |
|---|---|---|---|
| 3 | comp51024_c0_seq1 | 0.19 | Protein kinase superfamily protein |
| comp167704_c0_seq1 | 0.10 | Protein kinase superfamily protein | |
| comp34407_c0_seq1 | 0.33 | ALG6, ALG8 glycosyltransferase family | |
| comp57003_c1_seq1 | 0.25 | CELLULOSE SYNTAHSE INTERACTIVE 3, encodes a plasma membrane, microtubule associated protein with sequence similarity to CSI1 that is involved in cellulose biosynthesis and cell elongation | |
| comp69973_c0_seq1 | 0.38 | 1,3-β-glucan synthase component (DUF1218) | |
| 24 | comp65908_c0_seq1 | 0.29 | AKT1 INTERACTING PROTEIN PHOSPHATASE 1, HIGHLY ABA-INDUCED PP2C GENE 2, encodes a member of the group A PP2C family that is responsible for negatively regulating seed dormancy |
| 48 | comp82286_c0_seq1 | 0.02 | Plant invertase/pectin methylesterase inhibitor superfamily protein |
| comp35073_c0_seq1 | 0.11 | EXPANSIN 11, a member of α-expansin gene family | |
| comp55148_c0_seq1 | 0.34 | PECTIN METHYLESTERASE 12, plant invertase/pectin methylesterase inhibitor superfamily | |
| comp31267_c0_seq1 | 0.29 | Leucin-rich repeat (LRR) family protein | |
| comp35657_c0_seq1 | 0.23 | Leucin-rich receptor-like protein kinase family protein | |
| comp58051_c0_seq1 | 0.37 | CYP707A1, encodes a protein with ABA 8′-hydroxylase activity, involved in ABA catabolism | |
| comp61702_c0_seq1 | 0.32 | PGLR, polygalacturonase | |
| comp60161_c0_seq1 | 0.32 | Encodes a DUF579 containing protein essential for normal xylan synthesis and deposition in the secondary cell wall | |
| comp46319_c1_seq1 | 0.13 | Galactosyltransferase family protein | |
| comp68899_c0_seq1 | 0.26 | Protein kinase superfamily protein | |
| comp71322_c0_seq1 | 0.42 | FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 8, possibly involved in embryogenesis and seed development | |
| comp69696_c0_seq1 | 0.30 | EXPANSIN B2, a member of β-expansins | |
| comp34691_c0_seq1 | 0.40 | PYL4, encodes a member of the PYR/PYL/RCAR family proteins function as ABA sensors | |
| comp69112_c0_seq1 | 0.41 | Protein-tyrosine phosphatase | |
| comp68531_c0_seq1 | 0.28 | Pectinesterase | |
| comp70281_c0_seq1 | 0.45 | XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 9, encodes a member of xyloclucan endotransglucosylase/hydrolases that catalyze the cleavage and molecular grafting of xyloglucan chains function in loosing and rearrangement of the cell wall | |
| comp69670_c0_seq1 | 0.45 | EXPANSIN A15, a member of α-expansin gene family | |
| comp69069_c0_seq1 | 0.28 | FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 13, possibly involved in embryogenesis and seed development | |
| comp34126_c0_seq1 | 0.43 | UGT85A2, UDP-glucosyl transferase 85A2 |
a) Full list of the differentially expressed genes is available as supplemental materials. b) Hours after GR24 treatment. c) Ratio of normalized count of contig in the NJ-treated O. minor seed to that in the non-treated seed. d) Description for homolog in A. thaliana in TAIR database.
In later stages, the effect of NJ treatment was remarkable on abscisic acid (ABA) signaling. Genes encoding ABA-related proteins, such as RESPONSIVE TO ABA 18 homolog (comp34269_c0_seq1), a homolog of RNA-BINDING PROTEIN 25, an alternative splicing factor involved in mediation of abiotic stress and ABA responses (comp64405_c0_seq5), and a homolog of KEEP ON GOING, encoding a RING E3 ligase involved in ABA signaling (comp47352_c0_seq1), had increased expression at 48 HAT. In contrast, a homolog of HIGHLY ABA-INDUCED PP2C GENE 2, a negative regulator of dormancy, (comp65908_c0_seq1) showed decreased expression at 24 HAT; and a homolog of CYP707A1, ABA 8′-hydroxylase, which is involved in ABA catabolism (comp58051_c0_seq1), and a homolog of PYL4, an ABA receptor (comp34691_c0_seq1), were down-regulated at 48 HAT (Table 4). Additionally, many signaling-related genes encoding kinases and leucine-rich repeat (LRR) family proteins, as well as cell wall genes encoding expansins, glycosyl transferases, and glycosylases, were down-regulated in the NJ-treated seeds from 3 to 48 HAT (Table 4).
Discussion
In this study, a total of 119,181 contigs were obtained by de novo assembly of RNA-Seq data. BUSCO analysis revealed that one quarter of plant BUSCOs (361 of 1440 orthologs total) were missing in the assembled transcriptome (Supplemental Table S1), and one half of the missing BUSCOs were chloroplast genes (Table 1). Since O. minor is a holoparasite, photosynthetic machinery was lost during its evolution.37,38) In Orobanche cernua and Orobanche (Phelipanche) ramosa, the genes encoding the Rubisco large subunit (rbcL) had become pseudogenes.39) This could have been caused by mutations under relaxed selective pressure for photosynthesis, as indicated by reconfigured plastomes and altered chromosomal architectures in the holoparasitic broomrape family.40) The missing chloroplast-related BUSCOs in the assembled transcriptome in this study might also reflect global alterations in chromosomes caused by the loss of photosynthesis in O. minor.
Differential gene expression analysis revealed that NJ alters the expression of a restricted number (<50) of genes until 24 HAT, indicating that the MoA of NJ is not non-specific toxicity (Table 2, Fig. 1). Previously, the activity of acid invertases, VINs and CWINs, was shown to be lowered in NJ-treated O. minor seeds.10) Our transcriptomic data showed that the expression of comp33755_c0_seq1, encoding plant invertase/pectin methylesterase inhibitor superfamily protein, was decreased by GR24 in germinating seeds but increased by NJ (Table 3, Supplemental Fig. S1A). This result strongly suggests that NJ alters sugar signaling in O. minor seeds. Since invertase inhibitors are key regulators of invertase activities,17,19,25) their increase might be one reason for low invertase activity in NJ-treated O. minor seeds.10) A similar expression pattern was observed for comp68818_c0_seq1 (SUGAR TRANSPORTER 1, STP1, homolog) which also supports this hypothesis (Table 3, Supplemental Fig. S1B). Recently, ovule-specific CWIN2 and CWIN4 in A. thaliana were silenced by microRNAs, and RNA-Seq analysis was conducted on flower buds from the silenced plant. Gene expression of STP2, STP6, and STP9, together with other signaling molecules like protein kinases, was decreased in the silenced plant, suggesting that the status of sugars at apoplasts and/or CWINs influences the transcription of these genes.28) Genes comp65219_c0_seq4 (protein phosphatase 2C family protein), comp232524_c0_seq1 (protein kinase superfamily protein), comp68094_c0_seq1 (receptor like protein), and comp68094_c0_seq11 (MYB55 homolog), which also had altered expression, may also be involved in sugar signaling in germinating O. minor seeds. Although NJ is an iminosugar and is recognized as a β-glucosidase inhibitor, it is possible that NJ acts as a glucose mimic in the sugar signaling pathway through its close structural similarity. Taken together, we hypothesized that NJ inhibited the germination of O. minor by disrupting the sugar signaling pathway, a process essential for promoting germination.
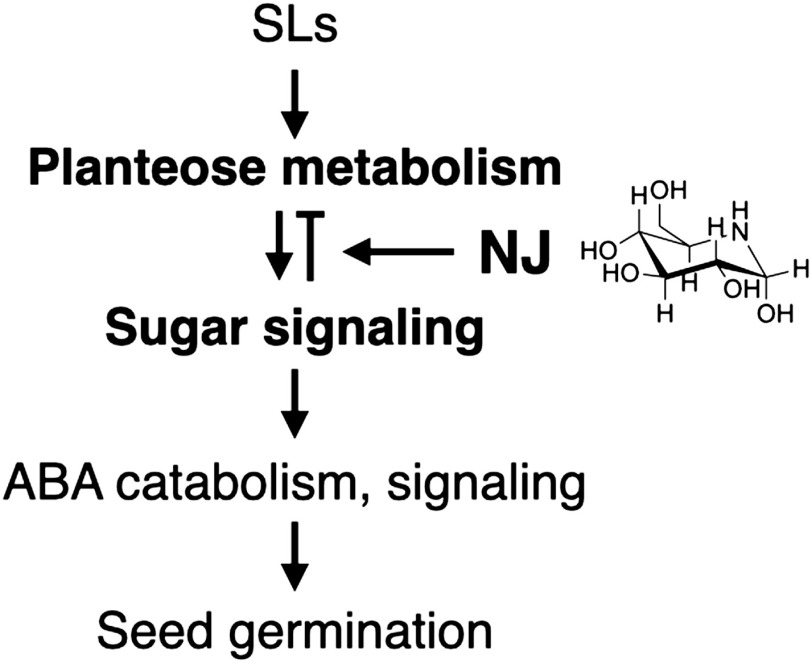
As compared to the restricted effect of NJ on gene expression until 24 HAT, the expression of a wider range of genes was affected at 48 HAT (Table 2, Fig. 1). Since the germination process was suppressed in NJ-treated seeds, DEGs at this time point might be involved in promoting germination. Changing the balance of gibberellin and ABA doses during germination ends dormancy and promotes germination.41,42) The ABA catabolic enzyme PrCYP707A1 is a key component in Phelipanche ramosa germination.43) During conditioning, the DNA methylation status of the PrCYP707A1promoter was modulated, enabling it to respond to SL.44) These observations indicate that ABA catabolism is a key process in broomrape germination. Our DEG analysis revealed that the gene expression of CYP707A1 homolog (comp58051_c0_seq1) and ABA receptor PYL4 homolog (comp34691_c0_seq1) was decreased in the NJ-treated seeds at 48 HAT (Table 4). This result indicates that sugar metabolism and/or signaling promotes ABA catabolism in germinating O. minor seeds. There is a close link between sugar status and ABA response in plants.45,46) Exogenous glucose suppresses the expression of CYP707A2, another ABA catabolic enzyme, in A. thaliana.47) As shown previously,10) since glucose is depleted in NJ-treated seeds, NJ, as a glucose mimic, might suppress CYP707A1 expression in O. minor seeds. Genes with increased expression were involved in ABA signaling (comp34269_c0_seq1, comp68110_c0_seq1, comp64405_c0_seq5, and comp47352_c0_seq1 in Table 4), which also suggestes ABA catabolism is not induced in NJ-treated seeds. Taken together, a hypothetical germination process scheme of O. minor and MoA of NJ is illustrated in Fig. 2.
Fig. 2. Hypothetical scheme of the germination process of O. minor and the MoA of NJ. SLs induce planteose metabolism to provide glucose required to promote the gemination process. There might be a feedback regulation of planteose metabolism by products through sugar signaling. NJ, as a glucose mimic, disrupts sugar signaling and ongoing planteose metabolism. Accordingly, ABA catabolism and signaling, key processes in germination, are suppressed.
Additionally, genes related to cell walls were differentially expressed in NJ-treated O. minor seeds. During plant germination, cell wall degradation and synthesis are coordinately regulated for radicle emergence and cell expansion.48) Since polysaccharides are major components in the cell wall, the sugar status of seeds may affect cell wall metabolism. Because of the complex structure of the cell wall and the large number of components involved in its metabolism, the regulatory mechanisms of cell wall metabolism through sugar signaling have not been studied intensively in root parasitic weeds or in model plants. Our transcriptomic data indicates that sugar status affects the expression of diverse cell wall-related genes in O. minor.
Our transcriptome analysis offers new insights into the inhibitory effect of NJ on the germination of O. minor. The changes in the gene expression profile of NJ-treated seeds suggest that NJ acts as a glucose mimic, disrupting sugar signaling. Furthermore, ABA catabolism, a key process in germination, was suppressed in NJ-treated seeds (Fig. 2). Precise quantification of some key genes, together with metabolic profiling involving ABA and its catabolite, will increase the understanding of the effect of NJ on germination and help to establish a novel strategy for parasitic weed control.
Acknowledgements
This research is supported in part by JST/JICA SATREPS (JPMJSA1607 to A.O., T.W., and Y.S.) and JSPS KAKENHI Grant-in-Aid for Scientific Research (B) (20H02924 to A.O. and D.O.).
Electronic supplementary materials
The online version of this article contains supplementary materials (Supplemental Tables S1–S6, Fig. S1), which are available at https://www.jstage.jst.go.jp/browse/jpestics/
References
- 1) C. Parker: “Parasitic Orobanchaceae,” eds. by D. M. Joel, J. Gressel and L. J. Musselman, Springer-Verlag, Berlin, Heidelberg, Germany, pp. 313–344, 2013.
- 2) R. E. Eplee and R. S. Norris: “Parasitic Weeds in Agriculture,” ed. by L. J. Mussleman, CRC Press, Boca Raton, FL, USA, pp. 173–182, 1987.
- 3) A. Okazawa and T. Wakabayashi: “Discovery and Synthesis of Crop Protection Products,” eds. by P. Maienfisch and T. M. Stevenson, American Chemical Society, Washington DC, USA, pp. 317–330, 2015.
- 4) G. H. Egley and J. E. Dale: Weed Sci. 18, 586–589 (1970). [Google Scholar]
- 5) B. Zwanenburg and D. Blaco-Ania: J. Exp. Bot. 69, 2205–2218 (2018). [DOI] [PubMed] [Google Scholar]
- 6) K. Yoneyama: J. Pestic. Sci. 45, 45–53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7) M. Sasaki, Y. Sugimoto, H. Takikawa, H. Miyake and N. Matsuo: (Kobe University and Sumitomo Chemical Co., Ltd.): US Pat. US8822383B2 (2011).
- 8) H. Samejima, A. G. Babiker, H. Takikawa, M. Sasaki and Y. Sugimoto: Pest Manag. Sci. 72, 2035–2042 (2016). [DOI] [PubMed] [Google Scholar]
- 9) D. Uraguchi, K. Kuwata, Y. Hijikata, R. Yamaguchi, H. Imaizumi, A. M. Sathiyanarayanan, C. Rakers, N. Mori, K. Akiyama, S. Irle, P. McCourt, T. Kinoshita, T. Ooi and Y. Tsuchiya: Science 362, 1301–1305 (2018). [DOI] [PubMed] [Google Scholar]
- 10) T. Wakabayashi, B. Joseph, S. Yasumoto, T. Akashi, T. Aoki, K. Harada, S. Muranaka, T. Bamba, E. Fukusaki, Y. Takeuchi, K. Yoneyama, T. Muranaka, Y. Sugimoto and A. Okazawa: J. Exp. Bot. 66, 3085–3097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11) D. French, R. W. Youngquist and A. Lee: Arch. Biochem. Biophys. 85, 471–473 (1959). [DOI] [PubMed] [Google Scholar]
- 12) D. French: J. Am. Chem. Soc. 77, 1024–1025 (1955). [Google Scholar]
- 13) P. M. Dey: FEBS Lett. 114, 153–156 (1980). [Google Scholar]
- 14) X. Xing, Y. S. Y. Hsieh, K. Yap, M. E. Ang, J. Lahnstein, M. R. Tucker, R. A. Burton and V. Bulone: Carbohydr. Polym. 175, 231–240 (2017). [DOI] [PubMed] [Google Scholar]
- 15) T. Niwa, S. Inouye, T. Tsuruoka, Y. Koaze and T. Niida: Agric. Biol. Chem. 34, 966–968 (1970). [Google Scholar]
- 16) A. Sturm and G.-Q. Tang: Trends Plant Sci. 4, 401–407 (1999). [DOI] [PubMed] [Google Scholar]
- 17) K. Koch: Curr. Opin. Plant Biol. 7, 235–246 (2004). [DOI] [PubMed] [Google Scholar]
- 18) T. Roitsch and M.-C. González: Trends Plant Sci. 9, 606–407 (2004). [DOI] [PubMed] [Google Scholar]
- 19) Y.-L. Ruan, Y. Jin, Y.-J. Yang, G.-J. Li and J. S. Boyer: Mol. Plant 3, 942–955 (2010). [DOI] [PubMed] [Google Scholar]
- 20) W. Mitsuhashi, S. Sasaki, A. Kanazawa, Y.-Y. Yang, Y. Kamiya and T. Toyomasu: Biosci. Biotechnol. Biochem. 68, 602–608 (2004). [DOI] [PubMed] [Google Scholar]
- 21) R. Draie, T. Péron, J.-B. Pouvreau, C. Véronési, S. Jégou, P. Delavault, S. Thoiron and P. Simier: Mol. Plant Pathol. 12, 638–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22) Z. Farrokhi, H. Alizadeh and H. Alizadeh: Plant Physiol. Biochem. 142, 8–14 (2019). [DOI] [PubMed] [Google Scholar]
- 23) S. Pagny, L.-A. Denmat-Ouisse, V. Gomord and L. Faye: Plant Cell Physiol. 44, 173–182 (2003). [DOI] [PubMed] [Google Scholar]
- 24) E. Rojo, J. Zouhar, C. Carter, V. Kovaleva and N. V. Raikhel: Proc. Natl. Acad. Sci. U.S.A. 100, 7389–7394 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25) T. Rausch and S. Greiner: Biochim. Biophys. Acta 1696, 253–261 (2004). [DOI] [PubMed] [Google Scholar]
- 26) M. Hothorn, W. Van den Ende, W. Lammens, V. Rybin and K. Scheffzek: Proc. Natl. Acad. Sci. U.S.A. 107, 17427–17432 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27) L. Wang, A. Cook, J. W. Patrick, X.-Y. Chen and Y.-L. Ruan: Plant J. 78, 686–696 (2014). [DOI] [PubMed] [Google Scholar]
- 28) S. Liao, L. Wang, J. Li and Y.-L. Ruan: Plant Physiol. 183, 1126–1144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29) H. Nagasaki, T. Mochizuki, Y. Kodama, S. Saruhashi, S. Morizaki, H. Sugawara, H. Ohyanagi, N. Kurata, K. Okubo, T. Takagi, E. Kaminuma and Y. Nakamura: DNA Res. 20, 383–390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30) M. G. Grabherr, B. J. Haas, M. Yassour, J. J. Z. Levin, D. A. Thompson, I. Amit, X. Adiconis, L. Fan, R. Raychowdhury, Q. Zeng, Z. Chen, E. Mauceli, N. Hacohen, A. Gnirke, N. Rhind, F. di Palma, B. W. Birren, C. Nusbaum, K. Lindblad-Toh, N. Friedman and A. Regev: Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31) B. J. Haas, A. Papanicolaou, M. Yassour, M. Grabherr, P. D. Blood, J. Bowden, M. B. Couger, D. Eccles, B. Li, M. Lieber, M. D. MacManes, M. Ott, J. Orvis, N. Pochet, F. Strozzi, N. Weeks, R. Westerman, T. William, C. N. Dewey, R. Henschel, R. D. LeDuc, N. Friedman and A. Regev: Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32) M. Seppey, M. Manni and E. M. Zdobnov: “Gene Prediction. Methods in Molecular Biology, vol. 1962,” eds. by M. Kollman, Humana, New York, NY, USA, pp. 227–245, 2019. [DOI] [PubMed]
- 33) D. W. Huang, B. T. Sherman and R. A. Lempicki: Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 34) D. W. Huang, B. T. Sherman and R. A. Lempicki: Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35) S. Tarazona, F. García-Alcalde, J. Dopazo, A. Ferrer and A. Conesa: Genome Res. 21, 2213–2223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36) S. Tarazona, P. Furió-Tarí, D. Turrà, A. Di Pietro, M. J. Nueda, A. Ferrer and A. Conesa: Nucleic Acids Res. 43, e140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37) J. H. Westwood, J. I. Yoder, M. P. Timko and C. W. dePamphilis: Trends Plant Sci. 15, 227–235 (2010). [DOI] [PubMed] [Google Scholar]
- 38) H. S. Heide-Jørgensen: “Parasitic Orobanchaceae,” eds. by D. M. Joel, J. Gressel and L. J. Musselman, Springer-Verlag, Berlin, Heidelberg, Germany, pp. 1–18, 2013.
- 39) A. D. Wolfe and C. W. dePamphilis: Plant Cell Rep. 33, 965–977 (1997). [DOI] [PubMed] [Google Scholar]
- 40) S. Wicke, K. F. Müller, C. W. dePamphilis, D. Quandt, N. J. Wickett, Y. Zhang, S. S. Renner and G. M. Schneeweiss: Plant Cell 25, 3711–3725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41) K. Shu, W. Zhou, F. Chen, X. Luo and W. Yang: Front. Plant Sci. 27, 416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42) B. Vishal and P. P. Kumar: Front. Plant. Sci. 9, 838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43) M.-M. Lechat, J.-B. Pouvreau, T. Péron, M. Gauthier, G. Montiel, C. Véronési, Y. Todoroki, B. L. Bizec, F. Monteau, D. Macherel, P. Simier, S. Thoiron and P. Delavault: J. Exp. Bot. 63, 5311–5322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44) M.-M. Lechat, G. Brun, G. Montiel, C. Véronési, P. Simier, S. Thoiron, J.-B. Pouvreau and P. Delavault: J. Exp. Bot. 66, 3129–3140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45) R. R. Finkelstain and S. I. Gibson: Curr. Opin. Plant Biol. 5, 26–32 (2001). [DOI] [PubMed] [Google Scholar]
- 46) S. Gazzarrini and P. McCourt: Curr. Opin. Plant Biol. 4, 387–391 (2001). [DOI] [PubMed] [Google Scholar]
- 47) G. Zhu, Y. Liu, N. Ye, R. Liu and J. Zhang: Physiol. Plant. 143, 375–384 (2011). [DOI] [PubMed] [Google Scholar]
- 48) W. J. Barnes and C. T. Anderson: Mol. Plant 11, 31–46 (2018). [DOI] [PubMed] [Google Scholar]