Abstract
Background
Component resolved diagnosis, recently redefined as precision allergy medicine diagnosis — PAMD@, may help understanding allergic cross-reactivity patterns among polysensitized patients and their clinical implication.
Objective
We aimed to investigate similarities among allergens by empirically determining the occurrence of co-sensitization patterns and to relate them to clinical features, in particular to asthma.
Methods
A retrospective cohort study in 1057 participants suspected to have allergic sensitization was performed in Vienna. To define cross-reactivity patterns, cluster analysis for 671 patients who showed reaction to at least one of the allergens in ISAC112 was performed and followed by multivariate logistic regression analysis to relate clusters and clinical symptoms, in particular current asthma.
Results
We determined 18 cross-reactivity clusters, comprising of 6 food, 10 respiratory, and 2 other clusters of allergens. Overall, 14% of the cohort patients were positive for 1 cross-reactivity cluster and 23% to 2 or more clusters. Multisensitized patients who were sensitized to PR-10 allergen proteins in addition to Bermuda timothy grass pollen clusters showed the highest association with asthma (odds ratio, 4.22 and 95% CI: 2.32–7.68) and an increase of 10 years of the duration of allergy increased the odds for a combined sensitization to PR-10 cluster and Bermuda-timothy cluster by 1.27 (95% CI: 1.06–1.53).
Conclusion
Similarities among IgE positivity patterns determined by ISAC112 revealed 18 cross-reactivity clusters. This PAMD@ approach allowed prediction of clinical features and revealed that certain cross-reactivity patterns are related to duration of allergic symptoms.
Keywords: IgE, Molecular diagnosis, Sensitization, Cluster analysis, Asthma, Duration of allergy
Abbreviations: BMI, Body mass index; CCDs, Cross-reactive carbohydrate determinants; HDM, House dust mites; IgE, Immunoglobulin E; ISAC, Immuno-solid phase allergen chip; PAMDA@, Precision allergy molecular diagnostic applications; PR-10, Pathogenesis related protein family 10; sIgE, specific IgE; SPT, Skin prick test
Introduction
According to the World Health Organization (WHO),1 globally approximately 300 million individuals suffer from asthma and 400 million from allergic rhinitis. In addition, over 250 million individuals are expected to have a food allergy. It is estimated that by 2025 half of the European population may have one or another form of an allergy.2 Therefore, diagnosis and treatment of allergy are a major public health issue.
In 1989, molecular allergy diagnosis evolved by cloning allergen-encoding complementary DNAs, which led to improvement of the IgE-mediated allergy diagnosis.3 Comprehensive examination of reaction pattern to diverse recombinant allergens could assist physicians in decisions regarding the most suitable treatment modality or allergen specific immunotherapy.4 Interactions between host factors, including genetics and epigenetics, skin barrier, microbiome, infectious disease history, and environmental factors contribute to the individuals’ specific IgE profile.5 Molecular diagnosis provides a new way to recognize cross reactivity phenomena and co-sensitization in a “sensitization profile” of the IgE mediated responses of patients. As a method of precision allergy medicine diagnosis (PAMD@), it has increasingly entered daily clinical routine.6
A comprehensive assessment of the IgE reaction patterns could not only help to establish the IgE cross-reactivity on a molecular basis but also to determine similarities of the various allergens. It has been documented that PAMD@ may provide a more accurate and detailed test in IgE mediated allergic patients in comparison to skin prick tests.7 To the best of our knowledge only 1 study has applied the method of cluster analysis on the larger sets of reactions to allergen molecules.8 We conducted cluster analyses within 3 groups of the allergen molecules of ISAC112 stratified as food, respiratory, and other allergens.
Clusters of reactivity could be used in 2 ways: direct analysis of molecular similarities between allergens and detection of multisensitized and polysensitized patients in association with the clinical appearance.
Because of the significant number of allergens, it is not feasible to check all molecular similarities between them, eg, for the 112 allergens of ISAC the number of pairs to check is 6216; therefore, guiding comparisons by empirically determining the occurrence of co-sensitization patterns would be of great help. On the other hand, relationships between clusters of reactivity to food and respiratory allergens could elucidate the process of developing multisensitization and polysensitization.
In this report, we aimed to not only describe the association between polysensitization and multisensitization but also to link them with the duration of clinical allergy and asthma. Hence, we bridge specific IgE determination with a precise clinical evaluation.6 The main research questions we attempted to answer were:
-
1.
Is sensitization to food and respiratory allergen clusters related to occurrence of asthma?
-
2.
How does duration of allergy affect the sensitization pattern and development of cross-reactivity and multisensitization?
Methods
Study design and population
This retrospective cohort and single-center study was conducted during 2012–2015 in an allergy clinic in Vienna. Of the 1412 suspected allergic patients, the whole set of data as well as ISAC112 results were available for 1057 patients. The study protocol was approved by the Ethics Committee of the Medical University Vienna (EK 2002/2012).
Allergic sensitization
ImmunoCAP ISAC112
Specific IgE levels of the patients' sera samples were measured against 112 allergen molecules according to the manufacturer's protocol (Thermo Fisher Scientific Inc, Phadia AB, Uppsala, Sweden). Semi-quantitative results of the sIgE levels were defined as negative for values less than 0.3 ISU and otherwise as positive. The 112 ISAC allergens were categorized into food allergens including: nuts, wheat, soy, apple, peach, kiwi, egg, milk, chicken, cod fish, shrimp, and airborne allergens including: birch pollen, timothy and Bermuda grass pollen, olive, mugwort, plantain, ragweed, pellitory, plane tree, cat, dog, horse, cow, mouse, molds, house dust mites, and other allergens including: latex, honey bee, and wasp.
Skin prick test
Skin prick test was carried out for 10% of the patients. In the skin provocation test, whole extracts of the respiratory and food allergen panels have been used (ALK Abello, Hoersholm, Denmark).
Based on clinical history and results of the ISAC112, alder, birch, hazel, ash, grasses, mugwort, ragweed (ambrosia), buckhorn plantain, house dust mites, cat, dog, and Alternaria among airborne allergens and hazelnut, peanut, wheat flour, egg, cow milk, soy, and cod fish from nutritional allergens were tested in skin prick test as reported previously.9
Statistical methods
Cluster analysis
Analyses were performed using the anonymized data file applying SPSS 25.0 (IBM Corp. USA) and Stata 13.1 (StataCorp, USA). Specific IgE concentrations were dichotomized at a cut-off of ≥0.3 ISU. Allergens were grouped into respiratory, food, and other allergens. Within these groups of allergens dissimilarity indices for pairs of allergens were computed using the Lance & Williams metric. Bottom-up agglomerative clustering with complete linkage as amalgamation rule was applied. A distance cut-off of 0.2 was arbitrarily chosen (similarity of 80%). This procedure led to the extraction of 6 food reactivity clusters, 10 respiratory, and 2 other cross reactivity and co-sensitization clusters. Assignment of a patient to a cluster was positive if the patient was positive to at least 80% of the allergens within a cluster. In addition, we performed supervised cluster analysis restricting allergens to those belonging to the tropomyosin, PR-10, or profilin type similar to the analysis performed by Scala E et al (2011) and Scala E et al (2010)8,10 and added those of the cupin-, prolamin-, or lipocalin-superfamilies. Furthermore, since patients could not be clustered in distinct groups aligned to these types of allergens, we performed cluster analyses within types following the same methodology as described above.
Statistical evaluation of clinical features
Clusters, to which more than 5% of the patients reacted, were selected for further analysis. Frequency of these cluster assignments were determined and presented as percent positivity.
The relationship between a history of asthma or other clinical features as the outcome variable and cluster assignments was analyzed by multiple logistic regression adjusted for sex, age, family history of atopy, smoking, and BMI. Cluster positivity in relation to duration of the allergy was also analyzed by multiple logistic regression adjusted for age at onset of allergy. In the subset of patients for whom skin prick test results were available, descriptive analyses of positivity to allergen clusters from ISAC and SPT results are presented. Results of logistic regression are presented as odds ratios (OR) and 95% confidence intervals. Results with associated p-values below 5% were considered significant.
Results
Characteristics of the cohort
Overall, between 2012 and 2015 n = 1057 patients (544 male and 513 female) presenting with allergy symptoms and screened for specific IgE using the ISAC112 microarray, were included. Subjects' age ranged from newborn to elderly (0–100 years) with a median age of 38 years (interquartile range IQR; 21–50). Duration of allergy as by patients’ reports varied between less than 1 year to more than 70 years. Clinical history of asthma has been recorded for 12.6% of the patients. Overall, 63% were positive to at least 1 allergen, 14% reacted to 1 cross-reactivity cluster and were considered polysensitized. Multisensitization accounted for 22.9% who reacted to at least 2 clusters. (Table 1).
Table 1.
Characteristics of the cohort (n = 1057)
| Median (IQR) or n (%) | |
|---|---|
| Age at first visit (years) | 38; (21–50) |
| Sex (Male) (%) | 544 (51.5) |
| Body mass index-mean (BMI kg/m2) | 23.29 (20.06–26.29) |
| Family history of the allergy | |
| Father | 198 (18.7) |
| Mother | 240 (22.7) |
| Mother and father | 72 (6.8) |
| Smoking status | |
| Never smoker | 72.1 |
| Former smoker | 14.1 |
| Smoker | 13.2 |
| Duration of the allergy (years) | 104, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23 |
| Age at onset of the allergy (years) | 209, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 |
| Asthma | 133 (12.6) |
| Comorbidities | |
| Diabetes | 27 (2.6) |
| Gastroesophageal reflux | 184 (17.4) |
| Gastritis | 74 (7.0) |
| Migraine | 177 (16.7) |
| Hypertension | 125 (11.8) |
| Positive to any ISAC112 allergen | 671 (63.5) |
| Multisensitized | 242 (22.9) |
| Polysensitized | 148 (14.0) |
Polysensitization and/multisensitization to food and respiratory clusters (cross-reactivity or co-sensitization).
The cluster analysis of 671 allergic patients, who showed reaction to at least 1 of the allergen molecules in ISAC112, revealed 18 sensitization clusters: 6 clusters of food allergens, to which 152 of all patients reacted, in addition, 379 subjects reacted to at least 1 cluster of respiratory allergens. Furthermore, 11 patients showed a positive reaction to 2 clusters of neither respiratory nor food allergens that were grouped into “other” allergens. We introduced clusters with the name of the allergen sources. Details of the clusters including structure of the allergen molecules, cross-reactivity, and co-sensitization and molecular spreading patterns are shown in Table 2 and Fig. 1.
Table 2.
Results of cluster analysis
| no | Clusters name | Allergen molecules | Sensitization pattern | Biochemical name and function |
|---|---|---|---|---|
| Food clusters | ||||
| 132 | Soybean-nuts-fruits | Ara h 8, Cor a 1.0101, Cor a 1.0401, Gly m 4, Mal d 1, Pru p 1 | Cross reactivity | PR-10 proteins |
| 2 | Cod-walnut-sesame | Gad c 1, Jug r 1, Ses i 1 | Co- sensitization | Beta parva albumin- 2S albumin |
| 2 | Peanut-soybean | Ara h 3, Gly m 5, Gly m 6 | Cross-reactivity | Cupin- Beta conglycinin- Glycinin |
| 7 | Peanut | Ara h 2, Ara h 6 | Molecular spreading | Conglutin (2S albumin) |
| 7 | Hazelnut-peach | Cor an 8, Pru p 3 | Cross reactivity | nsLTP |
| 2 | Kiwi-chicken | Act d 1, Gal d 3 | Co- sensitization | Cystein protease, Ovotransferrin |
| Respiratory clusters | ||||
| 76 | House dust mite | Der f 1, Der f 2, Der p 1, Der p 2 | Cross reactivity and Molecular spreading | Cysteine protease, Neimann-Pick-type C2 protein |
| 150 | Bermuda-timothy | Cyn d 1, Phl p 1, Phl p 4 | Cross-reactivity and Molecular spreading | Beta expansins, Glycoprotein (Berberine bridge enzyme) |
| 53 | Birch-timothy-mercury | Bet v 2, Phl p 12, Mer a 1 | Cross reactivity | Profilin |
| 20 | Horse-mouse | Equ c1, Mus m 1 | Cross reactivity | Lipocalin |
| 3 | Dog-cat | Can f 3, Fel d 2 | Cross reactivity | Serum albumin |
| 223 | Alder-birch | Aln g 1, Bet v 1 | Cross reactivity | PR-10 proteins |
| 1 | Cattel-horse | Bos d 6, Equ c 3 | Cross reactivity | Serum albumin |
| 121 | Timothygrasspollen | Phl p 5, Phl p 6 | Molecular spreading | |
| 22 | Birch-timothy | Bet v 4, Phl p 7 | Cross reactivity | Polcalcin |
| 38 | Cypress-planetree | Cup a 1, Pla a 1 | Co- sensitization | Pectate lyase, Putative invertase inhibitor |
| Others | ||||
| 10 | Herring worm-cockroach | Ani s 3, Bla g 7 | Co- sensitization | Tropomyosin, Nitrile specifier |
| 1 | Latex | Hev b 5, Hev b 6.01 | Co- sensitization | Acidic protein, Hevein precursor |
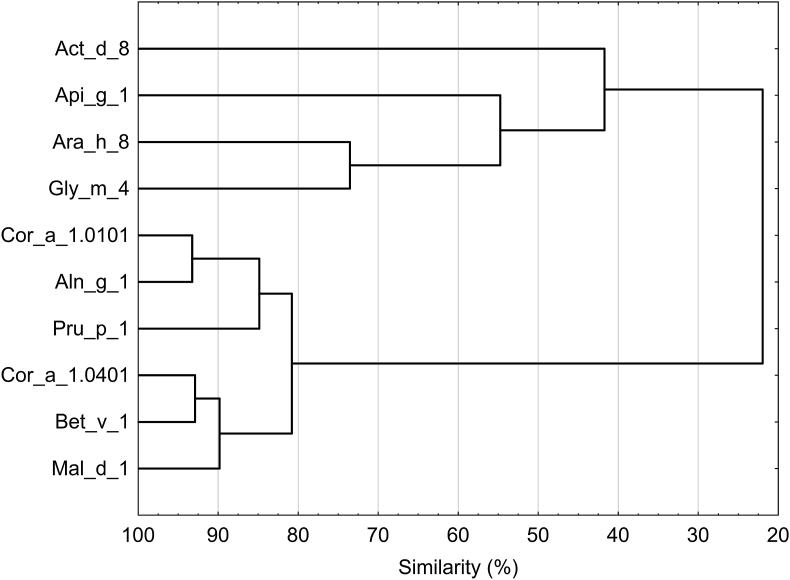
Fig. 1.
Dendrogram of the 18 cross-reactivity and co-sensitization clusters. The strongest correlation was found between PR-10 food pattern and PR-10 respiratory cluster
Polysensitization to food, cross reactivity, and co-sensitization
The most common food cross-reactivity pattern comprised peanut-, hazelnut-, soybean-, and apple-peach allergens assigned the name “soybean-nuts-fruits” (Ara h 8, Cor a 1.0101, Cor a 1.0401, Gly m 4, Mal d 1, Pru p 1) to which 132 subjects exhibited a positive reaction. The 5 other polysensitization patterns comprising of 20 patients included: “cod-walnut-sesame” group (Gad c 1, Jug r 1, Ses i 1), “peanut-soybean” (Ara h 3, Gly m 5, Gly m 6), “peanut” (Ara h 2, Ara h 6), “hazelnut-peach” (Cor an 8, Pru p 3), “kiwi-chicken” (Act d 1, Gal d 3).
Polysensitization to respiratory allergens, cross reactivity, and co-sensitization
Overall, sensitization to respiratory clusters was frequently seen; the highest prevalence belonged to “alder-birch” (Aln g 1, Bet v 1) with 223 patients. The second and third common cross reactivity clusters were “Bermuda-timothy” (Cyn d 1, Phl p 1, Phl p 4) with 150 patients and “timothy grass pollen” (Phl p 5, Phl p 6) with 121 patients. Overall, 76 individuals reacted to the house dust mite group and 53 to the “birch-timothy-mercury” pattern (Bet v 2, Phl p 12, Mer a 1). To “cypress-plane tree” (cup a 1, Pla a 2) 38 were found positive and to other clusters 11 patients (Table 2).
Relationship between clusters
Correlation between food and respiratory allergen clusters revealed a remarkable relationship between “alder-birch” and “soybeen-nuts-fruits” clusters (Fig. 1). In other words, 130 patients reacted to the “soybean-nuts-fruits” cluster and to “alder-birch” simultaneously. Furthermore, 43% of the subjects who reacted to “soybean-nuts-fruits” also showed sensitization to the Bermuda-timothy cluster, 29% to timothy grass pollen, and 23% to birch-timothy-mercury. All patients positive for any PR-10 allergens were also positive for Bet v 1.
The number of patients who were sensitized to both Bermuda-timothy and timothy grass pollen clusters was 83. But 44.7% of the patients who reacted to Bermuda-timothy did not react to timothy grass pollen and 31% of those sensitized to the Phl p 5- Phl p 6 group (timothy grass pollen) were not co-sensitized to Bermuda-timothy (Cyn d 1- Phl p 1- Phl p 4). Further analysis identified that multisensitization was common (84%) among patients, who reacted to “Bet v 2, Phl p 12, Mer a 1”. (See also Fig. 1).
Supervised cluster analysis for PR-10 molecules
In addition to unsupervised cluster analysis, we performed a supervised hierarchical clustering on PR-10 molecules including respiratory and food PR-10 allergens. The findings suggested reactions to these molecules in 2 distinct clusters: 1 cluster comprising the kiwi, peanut, celery, and soybean allergens and the other cluster hazelnut, tree pollen, birch pollen, peach, and apple allergens. Similarity between these 2 clusters was less than 30% (Fig. 3).
Fig. 3.
Dendrogram of the PR-10 allergens. There are two distinct clusters linked at about 22% similarity only
Relationship between clusters and clinical symptoms (asthma, duration of allergic symptoms).
For the analysis of a relationship to clinical features only clusters were considered to which at least 5 percent of the subjects were sensitized.
Except for the “Birch-timothy-mercury” pattern that showed a statistically not significantly increased risk for asthma (odds ratio 1.87; 95% CI: 0.91–3.91), all other clusters were significantly related. (Table 3). A particularly strong relationship was found for PR-10 food and respiratory proteins in combination with Bermuda-timothy (odds ratio 4.22; 95% CI:2.32–7.68). (Table 3). Smoking did not confound these relationships.
Table 3.
Results of logistic regression analysis of cluster positivity and asthma risk
| Sensitization/co-sensitization to clusters | Crude OR (95% CI) | Adjusteda OR (95% CI) |
|---|---|---|
| Soybean-nuts-fruits | 2.75 (1.76–4.29) | 2.67 (1.68–4.25) |
| House dust mite | 2.15 (1.21–3.81) | 2.43 (1.32–4.48) |
| Bermuda- timothy | 2.05 (1.31–3.21) | 2.02 (1.27–3.21) |
| Birch- timothy- mercury | 1.66 (0.81–3.40) | 1.87 (0.91–3.91) |
| Alder- birch | 2.05 (1.38–3.05) | 1.98 (1.31–2.98) |
| Timothy grass pollen | 1.65 (0.99–2.73) | 1.76 (1.05–2.96) |
| Soybean-nuts- fruits & Bermuda- timothy | 4.24 (2.38–7.56) | 4.22 (2.32–7.68) |
| Soybean-nuts- fruits & Alder- birch | 2.81 (1.80–4.39) | 2.74 (1.73–4.36) |
| Bermuda- timothy & Alder- birch | 2.60 (1.53–4.44) | 2.60 (1.51–4.50) |
| Alder- birch & Timothy grass pollen | 2.60 (1.37–4.92) | 2.77 (1.44–5.35) |
| Bermuda- timothy & Timothy grass pollen | 2.06 (1.18–3.61) | 2.27 (1.28–4.04) |
CI: Confidence Interval, OR: Odds Ratio
adjusted for sex, age at first visit, family history of allergy, smoking and BMI
Fig. 2 shows frequency of the reactions to ISAC112 allergens in patients with asthma.
Fig. 2.
Percent positivity of reactions to ISAC112 allergens in patients with asthma (n = 133)
Additional multivariate analyses showed that sensitization to “Bermuda-Timothy”, “alder-birch” and reaction to “soybean-nuts-fruits” and “Bermuda-Timothy” was significantly associated with increasing duration of the allergy. (Table 4). In addition, there was a highly significant (p < 0.001) relationship between duration of allergy and the number of sensitizations with about one additional sensitization by 10 years increase of the duration of allergy.
Table 4.
Results of logistic regression analysis of various sensitization patterns on duration of allergy. Odds ratios for an increase of the risk of having the respective sensitization pattern by an increase of 10 years of the duration of allergy
| Sensitization/co-sensitization to clusters | Crude OR (95% CI) | Adjusteda OR (95% CI) |
|---|---|---|
| Soybean-nuts- fruits | 1.20 (1.05–1.38) | 1.14 (0.99–1.32) |
| House dust mite | 0.98 (0.80–1.19) | 0.88 (0.72–1.08) |
| Bermuda- timothy | 1.28 (1.12–1.46) | 1.16 (1.01–1.33) |
| Birch- timothy- mercury | 0.92 (0.74–1.16) | 0.82 (0.65–1.02) |
| Alder- birch | 1.23 (1.09–1.38) | 1.21 (1.07–1.38) |
| Timothy grass pollen | 1.02 (0.87–1.18) | 0.91 (0.78–1.07) |
| Soybean-nuts- fruits & Bermuda- timothy | 1.38 (1.16–1.65) | 1.27 (1.06–1.53) |
| Soybean-nuts- fruits & Alder- birch | 1.21 (1.05–1.39) | 1.14 (0.99–1.32) |
| Bermuda- timothy & Alder- birch | 1.36 (1.16–1.59) | 1.25 (1.07–1.48) |
| Alder- birch & Timothy grass pollen | 1.14 (0.93–1.40) | 1.04 (0.85–1.28) |
| Bermuda- timothy & Timothy grass pollen | 1.08 (0.91–1.28) | 0.97 (0.81–1.16) |
CI: Confidence Interval, OR: Odds Ratio
adjusted for age at onset of allergy
Discussion
Findings of our study demonstrate reactivity clusters to various allergen molecules and correlation between food and respiratory cross-reactivity patterns. Moreover, we defined several polysensitization and multisensitization clusters.
Grass pollen (Phl p1 and Cyn d1) and Bet v1 were the most prevalent airborne allergens among our large cohort of suspected allergic patients, which is consistent with a previous study among adolescents in Salzburg, Austria.11 We found sensitization to Fel d 1 and Ole e 1 as the second common major airborne allergens. Mal d 1, Pru p 1 and Ara h 8, in accordance with a study in southern Sweden, accounted for the most common food allergen sensitization. Our data also support the observation that sensitization to major food allergens is not as common as those to respiratory allergens.11,12
The association between sensitizations to allergens we obtained in our cohort, could often be confirmed by their biological structures. Most clusters could be established as due to cross-reactivity between allergens, co-sensitization, or molecular spreading. Nevertheless, in some instances no cross-reactivity between allergen molecules within a cluster could be found, for instance, Fel d 1 and Ole e 1. Major cross-reactivity clusters between PR-10 food allergens consist of Ara h 8, Cor a 1.0101, Cor a 1.0401, Gly m 4, Mal d 1 and Pru p 1. In this group, patients sensitized to 7 food allergen molecules in addition to cluster 12, which included 2 pollen allergens from the PR-10 protein family, namely Bet v 1, the most prevalent PR- 10 allergen, and Aln g 1. Associations between PR-10 molecules have been explained previously,13, 14, 15 however, in our study cross-reaction to Gly m 4 was very common in combination with apple and hazelnut sensitization. The second food-allergen cluster comprising major fish allergen (Gad c 1), walnut (Jug r 1), and sesame (Ses i 1), could be due structural similarities between the nut and fish allergens 2S albumin and parvalbumin beta homologues. Cross reactivity between walnut and sesame has been described.13 Since co-sensitization of the cod fish allergens and nuts has not been reported yet, we evaluated its similarity according to the UniProt knowledgebase: We detected 22 identical positions and 40 similar positions between Gad c1 and Jug r 1.
A multicenter prospective study in Europe described about 60% of children being allergic to nuts showing co-sensitization to peanut, tree nut, and sesame.16 Nonetheless, in order to find an explanation for the empirical association between the allergens belonging to albumin proteins in cluster 2, more studies along with in vitro investigations are required. We present cross-reactivity between Ara h 3 major peanut allergen and soybean allergens Gly m 5 and Gly m 6 in cluster 3. This concurs with previous findings that demonstrate sequence similarities of the legumin like proteins17 and are also consistent with a previous study among 66 patients with peanut allergy in Austria.18
Molecular spreading phenomena have already been explained for grass pollen sensitization and house dust mites. Peanut comprises of 16 types of allergen molecules from 8 different protein families. We found Ara h 2 and Ara h 6 in one cluster, which are known as 2S albumin proteins. Sensitization to 2 molecules from the same family could suggest “molecular spreading”. Ara h 2 has been classified up to now as the most potent allergen in this family.19, 20, 21 In a previous study in Austrian peanut-allergic patients more than 70% were sensitized to Ara h 2 and Ara h 6 molecules.18
Nonspecific lipid transfer protein cross-reactivity between Cor a 8 and Pru p 3 could be assumed to be the basis for our fifth cluster. This is in good agreement with Spanish and Italian studies suggesting Pru p 3 as a strong sensitizer in combination with peanut and hazelnut sensitization.22,23
Interestingly, the last cluster among food allergens that was identified consists of kiwi and egg white proteins. Allergy to egg and chicken among children have been frequently reported but there are only few studies about the cross-reactivity or co-sensitization for egg white allergens such as Gal d 3. Act d 1 is one of the main allergens from 13 types of green kiwi fruits allergen molecules that have already been recognized. It is a molecule with 30 kDa belonging to the cysteine proteases.24 Kiwi fruit allergens not only could act as a trigger for different symptoms but they could also increase the risk of sensitization to other allergens. So far cross-reactivity between kiwi fruits and a variety of fruits, vegetables, nuts and seeds has been reported.25 We found no previous report about an association between Act d 1 and Gal d 3. However, about 119 similar and 65 identical positions between these two proteins were found according to UniProt database. Hence, further studies are needed to assess the sequence similarity between these molecules and their three-dimensional structures.
Consistent with previous studies, cluster 7 illustrates both phenomena, molecular spreading and cross reactivity, among sensitized patients. Dermatophagoides pteronyssinus has been reported as an important allergen in asthma.26, 27, 28 Because ISAC112 chip comprises only Der p 1, Der p 2, Der f 1 and Der f 2, we could not determine the association between all different potential HDM allergens.
Cluster 8 shows co-sensitizations among oligo molecular sensitization to timothy grass pollens Phl p1, Phl p 4, and Bermuda pollen allergens. Previous studies proposed cross-reactivity between Cyn d 1 and Phl p 1, besides Ph l p 1 is an initiator allergen in at least 75% of cases.27,29, 30, 31 We confirmed these findings but also found Phl p 5 and Phl p 6 in a separate cluster, possibly due to molecular spreading and with lower tendency to cross reactivity with Bermuda pollen.
Within profilin cross-reactivity we observed IgE sensitization to Bet v 2, Phl p 2, and Mer a 1. This is in close agreement with previous studies.31,32 In a Manchester cohort study, Hev b 8 together with Mer a 1 was a dominant allergen in the profilin group in accordance with our findings.
Cluster 10 displays reactivity to lipocalins. Common lipocalin reactivity allergens like Can f 1, Can f 2, Equ c 1, Fel d 4 and Mus m 1 were reported.33 In a study in West Sweden the most prevalent cross-reactivity lipocalin allergens were Fel d 4 and Equ c 1.34 Among lipocalin allergens, we demonstrate cross-reactivity between Equ c 1 and Mus m 1. Sensitizations to furry animals are prevalent and the second most common group are serum albumin allergens of dander and fluid. There are minor allergens belonging to mammalian animals and include Equ c 3 (horse), Bos d 6 (bovine), Can f 3 (dog), and Fel d 2 (cat) and Sus s 1 (pig).35 Our study provides additional support for cross-reactivity among mammalian albumin allergens and confirms reactivity between Can f 3 and Fel d 2 as a typical association between minor allergens in cluster 11.36
Cluster 13 represents a new finding about the relation between Equ c 3 (equine) and Bos d 6 (bovine). In addition to the cat-pork syndrome, clinical co-sensitization between cow meat and milk allergy and association between horse allergy and cat or dog sensitization have been proposed.37 In one study, more than half of the patients who reacted to Equ c 3 were also sensitized to cat or dog.38
Our cluster of “Aln g 1 and Bet v1” is in complete agreement with previous findings.39 Cluster 15 shows cross-reactivity between birch pollen (Bet v 4) and grass pollen (Phl p 7), calcium binding allergens, which are restricted to pollen allergens. Phl p 7 as a minor allergen in the timothy group is highly cross reactive with other polcalcin allergens (like Bet v 4) and hence our results support earlier studies.40,41 We also observed co-sensitization between cypress (Cup a 1) and plane tree (Pla a 1) in cluster 16. Major allergen Cup a 1, belonging to pectate lyase along with nonglycosylated major allergen Pla a1 could suggest co-sensitization by tree pollen allergens.42 Tropomyosin cross-reactivity between Ani s 3 (worm allergen) and Bla g 7 (German cockroach) cluster supports other findings about similarities between tropomyosin.43 The latex cluster probably illustrates co-sensitization between Hev b 5 as a major allergen, heat stable and acidotic protein with unknown biologic function and Hev b 6.01 the major allergen belonging to chitinases.44
The relationship between clusters, especially between food and respiratory clusters, suggests a powerful correlation between PR-10 food and respiratory allergens. We found that all patients sensitized to the “soybean-nuts-fruits” cluster reacted to “alder-birch”. Only less than 40% of patients reacting to Bet v 1 and Aln g 1 did not show sensitization to the PR-10 food cluster.
The results of previous reports indicated different patterns of sensitization, which are related to rhinitis and asthma. In a cohort study among children at school age it has been demonstrated that polysensitization to respiratory allergens is associated with asthma.45 A Korean study found an association between polysensitization to airborne allergens and severity of asthma.46 As proposed in another cohort from Melbourne, co-sensitization to common food and airborne allergens is significantly related to respiratory allergic diseases.47 House dust mites cross-reactivity increased the risk of rhinitis and asthma as reported from the Manchester asthma and allergy birth cohort study.28 A recent epidemiologic study in Stockholm in children 16 years old showed an increasing risk for persistent rhinitis after sensitization to Bet v 1 pollen allergen besides increasing the number of PR-10 food allergens.13
Several risk factors are recognized for asthma; however, a comprehensive risk assessment associated with the sensitization patterns in allergic patients has rarely been done. We assessed sensitization to all clusters prevalent in more than 5% of the cohort and in asthmatic and symptomatic allergic patients. Approximately 12% of the patients had a clinical history of asthma. The highest odd's ratio for asthma was found for multisensitized patients with PR-10 allergens and Bermuda-timothy cluster positivity. Besides sensitization to house dust mite cluster (odd's ratio, 2.43), PR-10 allergens (odd's ratio, 2.67), Bermuda-timothy cluster (odd's ratio, 2.02) and alder-birch cluster (odd's ratio, 1.98) have been confirmed as significant risk factors for asthma.
We addressed duration of allergy and sensitization patterns among allergic patients probably for the first time. We found a remarkable correlation between polysensitization to PR-10 clusters and Bermuda-timothy cross-reactivity cluster with increasing duration of the allergy. These findings suggest that with increasing disease duration the pattern of sensitization is broadening. On the other hand, these results imply that expansion of sensitization to multiple allergens could be responsible for the maintenance of asthma symptomatology. If this is true, desensitization therapy should commence, if feasible, in asthmatic patients as early as possible.
As lifestyle factors may play a role in sensitization to allergens, we included besides demographic characteristics such as age and sex and family history of asthma also BMI and active smoking in multivariate analysis to adjust for these possible confounders. As shown in Table 3. These factors had little effect on the association between sensitization clusters and asthma. There were, however, some statistically significant differences (results not shown) between smokers and overweight/obese individuals and sensitization patterns. Smokers were significantly less often sensitized to birch-timothy-mercury cluster allergens and overweight/obese individuals were significantly less often sensitized to alder-birch cluster allergens.
Our study has some limitations. Since we relied on results from ISAC112, our analysis is restricted to the allergens covered by this procedure. We are, however, aware that the “Bermuda-timothy” cluster could be affected by CCD recognition, as natural (n) Cyn d 1, nPhl p 4, like nApi g 5, nCup a 1, and MUXF3, express cross-reactive carbohydrate determinants (CCDs), which could lead in principal to nonspecific IgE binding.48,49 Our results are from 1 large allergy clinic but may have a different composition of patients than seen in other institutions, therefore, further progress can be expected from multicenter studies. Another limitation is the fact that we included cases of asthma diagnosed by a physician but without having access to records of their clinical assessment.
This study has highlighted that increasing the duration of the allergy is related with development of cross reactivity and polysensitization. Furthermore, polysensitization to food and/or respiratory allergens are related to manifestation of asthma.
In conclusion, similarities among IgE positivity patterns determined by ISAC112 revealed 18 cross-reactivity clusters. In line with the PAMD@ concept,6 this novel procedure allowed prediction of clinical features and showed cross-reactivity patterns and expansion of the reactivity to different allergens is related to duration of symptoms.
Funding
The study was supported by the Austrian Science Fund FWF, grants SFB F4606–B28 and in part by the Swiss Messerli Foundation to EJJ; EU was supported by FWF grant KLI 284-B00.
Authors' contributions
SGD analyzed all data and compiled the manuscript. ANJ elaborated data on clinical symptoms of patients and skin prick test results. NML took responsibility for the ethics' approval and together with EU diagnosed and treated the patients. MK and EJJ designed the study and supported data analysis and data presentation. All authors contributed to the manuscript writing and editing, and approved the final version.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the Medical University Vienna (EK 2002/2012).
Consent for publication
All authors have seen and approved the last version and agreed to publication of the work.
Availability of data and material
The data sets used and analyzed during this study are available from the corresponding author on reasonable request.
Potential competing interests
The authors report no competing interests.
Acknowledgements
We would like to especially thank Mrs. Bellinda Neuherz for excellent clinical assistance, and Mag. Daniela Hallmann for kind patient administration.
Footnotes
Full list of author information is available at the end of the article
References
- 1.Bousquet J., Mantzouranis E., Cruz A.A. Uniform definition of asthma severity, control, and exacerbations: document presented for the World Health Organization Consultation on Severe Asthma. J Allergy Clin Immunol. 2010;126(5):926–938. doi: 10.1016/j.jaci.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 2.Manifesto A. 2015. Tackling the allergy crisis in europe-concerted policy action needed. Available at: EAACI website: https://www eaaci.org/outreach/public-declarations/3243-advocacy-manifesto-tackling-the-allergy-crisis-in-europe-2015 html. Accessed. [Google Scholar]
- 3.Curin M., Garib V., Valenta R. Single recombinant and purified major allergens and peptides: how they are made and how they change allergy diagnosis and treatment. Ann Allergy Asthma Immunol. 2017;119(3):201–209. doi: 10.1016/j.anai.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canonica G., Bachert C., Hellings P. Allergen immunotherapy (AIT): a prototype of precision medicine. World Allergy Organization Journal. 2015;8(1):1–10. doi: 10.1186/s40413-015-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valenta R., Karaulov A., Niederberger V. vol. 138. Elsevier; 2018. pp. 195–256. (Molecular Aspects of Allergens and Allergy. Advances in Immunology). [DOI] [PubMed] [Google Scholar]
- 6.Ansotegui I.J., Melioli G., Canonica G.W. A WAO—ARIA—GA2LEN consensus document on molecular-based allergy diagnosis (PAMD@): update 2020. World Allergy Organization Journal. 2020;13(2):100091. doi: 10.1016/j.waojou.2019.100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canonica G.W., Ansotegui I.J., Pawankar R. A WAO-ARIA-GA2LEN consensus document on molecular-based allergy diagnostics. World Allergy Organization Journal. 2013;6(1):1–17. doi: 10.1186/1939-4551-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scala E., Alessandri C., Palazzo P. IgE recognition patterns of profilin, PR-10, and tropomyosin panallergens tested in 3,113 allergic patients by allergen microarray-based technology. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mothes-Luksch N., Jordakieva G., Hinterhölzl L., Jensen A., Hallmann P., Kundi M. Allergy diagnosis from symptoms to molecules, or from molecules to symptoms: a comparative clinical study. World Allergy Organization Journal. 2018;11(1):22. doi: 10.1186/s40413-018-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scala E., Alessandri C., Bernardi M. Cross-sectional survey on immunoglobulin E reactivity in 23 077 subjects using an allergenic molecule-based microarray detection system. Clin Exp Allergy. 2010;40(6):911–921. doi: 10.1111/j.1365-2222.2010.03470.x. [DOI] [PubMed] [Google Scholar]
- 11.Stemeseder T., Klinglmayr E., Moser S. Cross-sectional study on allergic sensitization of Austrian adolescents using molecule-based IgE profiling. Allergy. 2017;72(5):754–763. doi: 10.1111/all.13071. [DOI] [PubMed] [Google Scholar]
- 12.Sterner T., Uldahl A., Svensson Å. IgE sensitization in a cohort of adolescents in southern Sweden and its relation to allergic symptoms. Clin Mol Allergy. 2019;17(1):6. doi: 10.1186/s12948-019-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westman M., Lupinek C., Bousquet J. Early childhood IgE reactivity to pathogenesis-related class 10 proteins predicts allergic rhinitis in adolescence. J Allergy Clin Immunol. 2015;135(5):1199–1206. doi: 10.1016/j.jaci.2014.10.042. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersen M.-B.S., Hall S., Dragsted L.O. Identification of European allergy patterns to the allergen families PR-10, LTP, and profilin from Rosaceae fruits. Clin Rev Allergy Immunol. 2011;41(1):4–19. doi: 10.1007/s12016-009-8177-3. [DOI] [PubMed] [Google Scholar]
- 15.Ebner C., Hirschwehr R., Bauer L. Identification of allergens in fruits and vegetables: IgE cross-reactivities with the important birch pollen allergens Bet v 1 and Bet v 2 (birch profilin) J Allergy Clin Immunol. 1995;95(5):962–969. doi: 10.1016/s0091-6749(95)70096-x. [DOI] [PubMed] [Google Scholar]
- 16.Brough H.A., Caubet J.-C., Mazon A. Defining challenge-proven coexistent nut and sesame seed allergy: a prospective multicenter European study. J Allergy Clin Immunol. 2020;145(4):1231–1239. doi: 10.1016/j.jaci.2019.09.036. [DOI] [PubMed] [Google Scholar]
- 17.Chruszcz M., Maleki S.J., Majorek K.A. Structural and immunologic characterization of Ara h 1, a major peanut allergen. J Biol Chem. 2011;286(45):39318–39327. doi: 10.1074/jbc.M111.270132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ackerbauer D., Bublin M., Radauer C. Component-resolved IgE profiles in Austrian patients with a convincing history of peanut allergy. Int Arch Allergy Immunol. 2015;166(1):13–24. doi: 10.1159/000371422. [DOI] [PubMed] [Google Scholar]
- 19.Otsu K., Guo R., Dreskin S. Epitope analysis of Ara h 2 and Ara h 6: characteristic patterns of IgE-binding fingerprints among individuals with similar clinical histories. Clin Exp Allergy. 2015;45(2):471–484. doi: 10.1111/cea.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z., Zhou N., Xiong F. Allergen composition analysis and allergenicity assessment of Chinese peanut cultivars. Food Chem. 2016;196:459–465. doi: 10.1016/j.foodchem.2015.09.070. [DOI] [PubMed] [Google Scholar]
- 21.Valcour A., Jones J.E., Lidholm J., Borres M.P., Hamilton R.G. Sensitization profiles to peanut allergens across the United States. Ann Allergy Asthma Immunol. 2017;119(3):262–266. e1. doi: 10.1016/j.anai.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Javaloyes G., Goikoetxea M.J., Nuñez I.G. Pru p 3 acts as a strong sensitizer for peanut allergy in Spain. J Allergy Clin Immunol. 2012;130(6):1432–1434. doi: 10.1016/j.jaci.2012.08.038. e3. [DOI] [PubMed] [Google Scholar]
- 23.Schulten V., Nagl B., Scala E. Pru p 3, the nonspecific lipid transfer protein from peach, dominates the immune response to its homolog in hazelnut. Allergy. 2011;66(8):1005–1013. doi: 10.1111/j.1398-9995.2011.02567.x. [DOI] [PubMed] [Google Scholar]
- 24.Bublin M., Pfister M., Radauer C. Component-resolved diagnosis of kiwifruit allergy with purified natural and recombinant kiwifruit allergens. J Allergy Clin Immunol. 2010;125(3):687–694. doi: 10.1016/j.jaci.2009.10.017. e1. [DOI] [PubMed] [Google Scholar]
- 25.Wang J., Vanga S.K., McCusker C., Raghavan V. A comprehensive review on kiwifruit allergy: pathogenesis, diagnosis, management, and potential modification of allergens through processing. Compr Rev Food Sci Food Saf. 2019;18(2):500–513. doi: 10.1111/1541-4337.12426. [DOI] [PubMed] [Google Scholar]
- 26.Posa D., Hofmaier S., Arasi S., Matricardi P.M. Natural evolution of IgE responses to mite allergens and relationship to progression of allergic disease: a review. Curr Allergy Asthma Rep. 2017;17(5):28. doi: 10.1007/s11882-017-0697-y. [DOI] [PubMed] [Google Scholar]
- 27.Custovic A., Sonntag H.-J., Buchan I.E., Belgrave D., Simpson A., Prosperi M.C. Evolution pathways of IgE responses to grass and mite allergens throughout childhood. J Allergy Clin Immunol. 2015;136(6):1645–1652. doi: 10.1016/j.jaci.2015.03.041. e8. [DOI] [PubMed] [Google Scholar]
- 28.Weghofer M., Thomas W., Kronqvist M. Variability of IgE reactivity profiles among European mite allergic patients. Eur J Clin Invest. 2008;38(12):959–965. doi: 10.1111/j.1365-2362.2008.02048.x. [DOI] [PubMed] [Google Scholar]
- 29.Hatzler L., Panetta V., Lau S. Molecular spreading and predictive value of preclinical IgE response to Phleum pratense in children with hay fever. J Allergy Clin Immunol. 2012;130(4):894–901. doi: 10.1016/j.jaci.2012.05.053. e5. [DOI] [PubMed] [Google Scholar]
- 30.Matricardi P.M., Dramburg S. Molecular diagnosis of allergy: the pediatric perspective. Frontiers in Pediatrics. 2019;7:369. doi: 10.3389/fped.2019.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson A., Lazic N., Belgrave D.C. Patterns of IgE responses to multiple allergen components and clinical symptoms at age 11 years. J Allergy Clin Immunol. 2015;136(5):1224–1231. doi: 10.1016/j.jaci.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wölbing F., Kunz J., Kempf W., Grimmel C., Fischer J., Biedermann T. The clinical relevance of birch pollen profilin cross-reactivity in sensitized patients. Allergy. 2017;72(4):562–569. doi: 10.1111/all.13040. [DOI] [PubMed] [Google Scholar]
- 33.Virtanen T., Kinnunen T., Rytkönen-Nissinen M. Mammalian lipocalin allergens–insights into their enigmatic allergenicity. Clin Exp Allergy. 2012;42(4):494–504. doi: 10.1111/j.1365-2222.2011.03903.x. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki S., Nwaru B.I., Ekerljung L. Characterization of sensitization to furry animal allergen components in an adult population. Clin Exp Allergy. 2019;49(4):495–505. doi: 10.1111/cea.13355. [DOI] [PubMed] [Google Scholar]
- 35.Hilger C., van Hage M., Kuehn A. Diagnosis of allergy to mammals and fish: cross-reactive vs. specific markers. Curr Allergy Asthma Rep. 2017;17(9):64. doi: 10.1007/s11882-017-0732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konradsen J.R., Fujisawa T., Van Hage M. Allergy to furry animals: new insights, diagnostic approaches, and challenges. J Allergy Clin Immunol. 2015;135(3):616–625. doi: 10.1016/j.jaci.2014.08.026. [DOI] [PubMed] [Google Scholar]
- 37.Wilson J.M., Platts-Mills T.A. Meat allergy and allergens. Mol Immunol. 2018;100:107–112. doi: 10.1016/j.molimm.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uriarte S., Sastre J. Clinical relevance of molecular diagnosis in pet allergy. Allergy. 2016;71(7):1066–1068. doi: 10.1111/all.12917. [DOI] [PubMed] [Google Scholar]
- 39.Valenta R., Breiteneder H., Pettenburger K. Homology of the major birch-pollen allergen, Bet v I, with the major pollen allergens of alder, hazel, and hornbeam at the nucleic acid level as determined by cross-hybridization. J Allergy Clin Immunol. 1991;87(3):677–682. doi: 10.1016/0091-6749(91)90388-5. [DOI] [PubMed] [Google Scholar]
- 40.Muehlmeier G., Maier H. Polysensitization to pollen due to profilin and calcium-binding protein: distribution of IgE antibodies to marker allergens in grass and birch pollen allergic rhinitis patients in southern Germany. Eur Arch Oto-Rhino-Laryngol. 2014;271(4):719–725. doi: 10.1007/s00405-013-2609-7. [DOI] [PubMed] [Google Scholar]
- 41.San Nicoló M., Braun T., Eder K., Berghaus A., Gröger M. Clinical relevance of IgE to profilin and/or polcalcin in pollen-sensitized patients. Int Arch Allergy Immunol. 2016;169(2):101–107. doi: 10.1159/000444279. [DOI] [PubMed] [Google Scholar]
- 42.Asam C., Hofer H., Wolf M., Aglas L., Wallner M. Tree pollen allergens—an update from a molecular perspective. Allergy. 2015;70(10):1201–1211. doi: 10.1111/all.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reese G., Ayuso R., Lehrer S.B. Tropomyosin: an invertebrate pan–allergen. Int Arch Allergy Immunol. 1999;119(4):247–258. doi: 10.1159/000024201. [DOI] [PubMed] [Google Scholar]
- 44.Yeang H., Arif S.A.M., Yusof F., Sunderasan E. Allergenic proteins of natural rubber latex. Methods. 2002;27(1):32–45. doi: 10.1016/S1046-2023(02)00049-X. [DOI] [PubMed] [Google Scholar]
- 45.Skypala I., Bull S., Deegan K. The prevalence of PFS and prevalence and characteristics of reported food allergy; a survey of UK adults aged 18–75 incorporating a validated PFS diagnostic questionnaire. Clin Exp Allergy. 2013;43(8):928–940. doi: 10.1111/cea.12104. [DOI] [PubMed] [Google Scholar]
- 46.Ha E.K., Baek J.H., Lee S.-Y. Association of polysensitization, allergic multimorbidity, and allergy severity: a cross-sectional study of school children. Int Arch Allergy Immunol. 2016;171(3-4):251–260. doi: 10.1159/000453034. [DOI] [PubMed] [Google Scholar]
- 47.Alduraywish S.A., Standl M., Lodge C.J. Is there a march from early food sensitization to later childhood allergic airway disease? Results from two prospective birth cohort studies. Pediatr Allergy Immunol. 2017;28(1):30–37. doi: 10.1111/pai.12651. [DOI] [PubMed] [Google Scholar]
- 48.Einhorn L., Hofstetter G., Brandt S. Molecular allergen profiling in horses by microarray reveals Fag e 2 from buckwheat as a frequent sensitizer. Allergy. 2018;73(7):1436–1446. doi: 10.1111/all.13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Homann A., Schramm G., Jappe U. Glycans and glycan-specific IgE in clinical and molecular allergology: sensitization, diagnostics, and clinical symptoms. J Allergy Clin Immunol. 2017;140(2):356–368. doi: 10.1016/j.jaci.2017.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and analyzed during this study are available from the corresponding author on reasonable request.