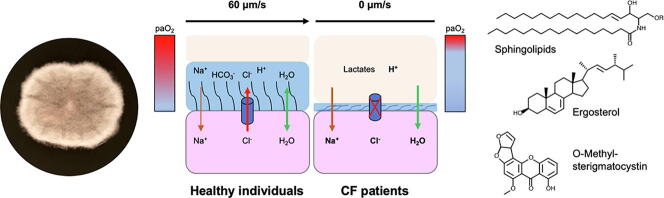
Graphical abstract
Keywords: Scedosporium apiospermum, Cystic fibrosis, Adaptation, Plasma membrane, Secondary metabolites, Degradation of aromatic compounds
Abstract
Scedosporium species rank second among the filamentous fungi colonizing the lungs of patients with cystic fibrosis (CF). Apart from the context of immunodeficiency (lung transplantation), the colonization of the CF airways by these fungi usually remains asymptomatic. Why the colonization of the lower airways by Scedosporium species is fairly tolerated by CF patients while these fungi are able to induce a marked inflammatory reaction in other clinical contexts remains questionable. In this regards, we were interested here in exploring the transcriptional reprogramming that accompanies the adaptation of these fungi to the particular microenvironment encountered in the airways of CF patients. Cultivation of Scedosporium apiospermum in conditions mimicking the microenvironment in the CF lungs was shown to induce marked transcriptional changes. This includes notably the down-regulation of enzymes involved in the synthesis of some major components of the plasma membrane which may reflect the ability of the fungus to evade the host immune response by lowering the biosynthesis of some major antigenic determinants or inhibiting their targeting to the cell surface through alterations of the membrane fluidity. In addition, this analysis revealed that some genes encoding enzymes involved in the biosynthesis of some mycotoxins were down-regulated suggesting that, during the colonization process, S. apiospermum reduces the production of some toxic secondary metabolites to prevent exacerbation of the immune system response. Finally, a strong up-regulation of many genes encoding enzymes involved in the degradation of aromatic compounds was observed, suggesting that these catabolic properties would predispose the fungus to particular patterns of human pathogenicity. Together these data provide new insights into the adaptative mechanisms developed by S. apiospermum in the CF lungs, which should be considered for identification of potential targets for drug development, but also for the experimental conditions to be used in in vitro susceptibility testing of clinical isolates to current antifungals.
1. Introduction
Scedosporium species are worldwide distributed filamentous fungi usually living as saprophytes in polluted soils and water [1]. Nevertheless, these fungi may also cause in Human a large variety of infections, ranging from localized infections such as subcutaneous mycetoma and bone or joint infections resulting from traumatic inoculation of some fungal elements, to disseminated infections in immunocompromised hosts, particularly in solid organ transplant recipients [2]. Moreover, these fungi have gained attention since the past two decades, mainly because of their worldwide recognition as significant pathogens in patients with cystic fibrosis [3].
Cystic fibrosis (CF) which is the most common genetic inherited disease in Caucasian populations, is caused by mutations in the gene CFTR (for cystic fibrosis transmembrane conductance regulator). The encoded protein is located at the apical membrane of numerous epithelial cell types where it is involved in the efflux of chloride and bicarbonate anions. Nevertheless, prognosis in CF essentially depends on the lesions of the lungs, which are the main target organs of the disease. Indeed, the respiratory tract of patients with CF is often colonized by microorganisms, mainly bacteria, but also yeasts and filamentous fungi, sometimes causing respiratory infections which are the major cause of morbidity and mortality in these patients. With a frequency ranging between 4.5 and 15.9%, species of the Scedosporium genus rank second among the filamentous fungi colonizing the CF airways, after Aspergillus fumigatus, and among Scedosporium species, the most common is Scedosporium apiospermum or Scedosporium boydii, depending on the country [4], [5], [6], [7], [8], [9], [10], [11], [12].
In the CF context, colonization of the respiratory tract by Scedosporium species is usually asymptomatic, although some cases of bronchitis or allergic broncho-pulmonary mycoses have been reported [13], [14]. Nevertheless, as described for Aspergillus fumigatus [15], all epidemiological studies that have been conducted so far demonstrated the usual chronicity of the colonization of the airways by Scedosporium species, each patient being colonized by a single genotype conserved over time despite the antifungal treatment [9], [16], [17]. Therefore, considering the propensity of these fungi to cause severe and often fatal disseminated infections in case of immunodeficiency [2], [3], all efforts should be made to detect this fungal colonization as early as possible, since lung or heart–lung transplantation still remains the ultimate treatment of patients with CF. Likewise, studies should be conducted to elucidate why these fungi are so difficult to eradicate.
Failure to eradicate bacteria in the CF lungs has been attributed to impairment of the local immune response, more precisely to a reduced synthesis of the major isoform of nitric oxide synthase (NOS-2) in the CF airway epithelium [18] because of the marked upregulation of the protein inhibitor of activated STAT-1 which is required for NOS-2 transcription [19]. In addition, persistence of bacteria in the CF mucus progressively leads to an acute inflammation with an influx of neutrophils and the release of elastase in the airways, leading to structural damage of the lungs which contribute to maintaining bacterial persistence. Finally, the sharp oxygen concentration gradient between the airway lumen and the depth of the CF bronchial mucus may provide advantage to the microorganisms. Bacteria respond to this hypoxic environment by an increased production of extracellular matrix components which contribute to bacterial persistence by limiting access of antibiotics and of the host antimicrobial components to bacterial cells [20].
However, beside hypoxia, the defect in efflux of chloride and bicarbonate anions resulting from mutations in the gene CFTR lead to many other changes in the physico-chemical properties of the bronchial mucus: decreased osmotic pressure, acid pH, increased carbon dioxide pressure, and increased concentration of lactates resulting from the fermentative activity of the cells. All these environmental conditions are known to affect physiology and morphogenesis in various fungal species as well as their virulence, for example production of capsule polysaccharides in Cryptococcus neoformans [21], [22], [23], or synthesis of structural polysaccharides and width of the cell wall, adhesin synthesis, and biofilm formation in the yeast Candida albicans [24], [25], [26] (for reviews see references [27], [28], [29], [30], [31]). Taking advantage of the recent availability of S. apiospermum genome [32], this study therefore was designed to investigate the transcriptional changes in response to the particular environmental conditions encountered by the fungus in the CF sticky mucus.
2. Materials and methods
2.1. Microorganism and culture conditions
This study was conducted using the reference strain S. apiospermum IHEM (Institute of Hygiene and Epidemiology-Mycology section) 14462, originally isolated from sputum sample from a French CF patient, which is publicly available at Sciensano (Brussels, Belgium) and was previously used for genome sequencing [32]. The strain, preserved as freeze-dried, was first grown on yeast extract-peptone-dextrose-agar plates containing chloramphenicol 0.5 g/L.
Then the fungus was cultivated into the synthetic medium Yeast Nitrogen Base (YNB) with 2% glucose, buffered at pH 7 with phosphate, with incubation under normal atmospheric conditions (80% N2 – 20% O2 – 0.039% CO2) as the reference culture condition (A condition). Six other culture conditions were investigated in parallel: (i) to mimic a decreased osmolarity, sucrose was changed to glucose as the carbon source (B condition); (ii) increased lactate concentration was investigated by mixing an equal amount of glucose and lactate, 1% each (C condition); (iii) lowering the pH was done by buffering YNB-glucose broth at 6.4 with phosphate (D condition); (iv and v) YNB-glucose pH 7 was also incubated under 95% N2 – 5% O2 – 0.039% CO2 or under 75% N2 – 20% O2 – 5% CO2 to reproduce hypoxia (E condition) and hypercapnia (F condition); and (vi) the fungus also was cultivated in CF Synthetic Medium (G condition) described by Palmer et al. [33] which mimics the nutritional conditions found in the CF mucus, and which takes into account several parameters studied independently in the above-mentioned conditions, particularly low osmolarity, presence of lactate, and decreased pH.
Triplicate cultures were incubated for 5 days at 37 °C with constant shaking. Then the mycelia were harvested and ground in liquid nitrogen.
2.2. Sample preparation and sequencing
Total RNAs were extracted from the homogenates with a RNA Plant kit (Macherey-Nagel). Total RNA in these extracts were quantified by fluorometry and their integrity was evaluated by micro-electrophoresis. Libraries were prepared in the paired-end strand specific mode with the HiSeq SBS Kit v4. 125-bp long reads were sequenced by Eurofins using the Illumina HiSeq 2500 platform.
2.3. RNA-seq analysis
Transcript assembly and quantification was performed using the Hisat2/StringTie/Ballgown pipeline as described by Pertea et al. [34]. Reads were first trimmed with Trimmomatic using the default parameters for paired-end reads [35] and subsequently mapped onto S. apiospermum genome sequence with Hisat2 in the strand specific mode RF [36]. Maximum intron length was set at 4000 and both discordant and mixed alignments were discarded. Each sample was first analyzed separately. Aligned reads were grouped into transcribed fragments (transfrags) with StringTie [37] to generate 21 individual assemblies. The published reference annotation of S. apiospermum was used as a guide to assemble reads into larger sequences. Individual assemblies were next merged with StringTie to generate a global assembly with the following parameters: minimum input transcript of 25 transcripts per million (TPM) (-T), minimum isoform fraction at 0.2 (-f), minimum input transcript length of 200 nt (-m) and a minimum input transcript coverage of 10 (-c).
Transcript annotation was performed by blasting sequences against Uniprot database with Blast+ [38] and by scanning domains from the PfamA database with Hmmerscan [39]. Searches were performed in parallel using the perl scripts made by Brian Haas (http://hpcgridrunner.github.io/).
Merged transcribed fragments were finally quantified in each sample using original alignments. Quantification files were processed with Ballgown [40] in R [41]. We also quantify transcript abundance by pseudo-aligning reads on the resulting transcriptome in the SMEM-based lightweight alignment mode of Salmon v0.7.2 [42]. We used the R package DESeq2 [43] to detect significantly and differentially expressed genes in all pairwise comparison with the A culture condition using negative binomial based linear models and raw counts. Transcripts with an absolute log2 fold change above 2 and a FDR-adjusted p-value <0.001 were retained as differentially expressed in the given comparison. Significantly enriched functions were detected with an hypergeometric distribution test in R.
2.4. Quantitative PCR
Quantitative PCR (qPCR) experiments were carried out as described by Le Govic et al. [44]. Primers used in this study are presented in Table 1.
Table 1.
Primers used in this study.
| Primer name | Sequence (5′ to 3′) |
|---|---|
| 4161-1F | CCAAGACCTTGACCACCGATC |
| 4161-1R | GAGTGAGGGAGCGGGTGACAA |
| 4161-2F | CCAAACGCAACGATTCAGGCGG |
| 4161-2R | GTCGGCGTCTTCGTGTGTAATC |
| 4161-3F | CAGGCCTTTCGAGGTCACATGC |
| 4161-3R | CCATGTCACTGTGGTTGGCAG |
| 4161-4F | GTCCGTTCTAGGTGCAGGAC |
| 4161-4R | CCTAGCATTCTCTGGAGCCCT |
| 4161-5F | GTTGATTGGTGGTGCGTTGGTT |
| 4161-5R | CATCACCACCACCAAAGTCATC |
| 6569-1F | CTGGGACAGTATGGCCACTC |
| 6569-1R | TTCTCCTCGAGCGTGAGCTC |
| 6569-2F | GTTTGGCACCCTCGCAATTG |
| 6569-2R | CAGCGATGAGGGTATTGCTC |
| 6569-3F | GTCCGAGCAGTCTGATCTCC |
| 6569-3R | CGCGAAGTTGCCGTATCGGTG |
| 6569-4F | CCATTTCGCGTATCGCAGGGC |
| 6569-4R | CTAGGCGACCTTCGAAAGCTC |
| 6569-5F | GGACGTATCGCTACCGTTCG |
| 6569-5R | GCCAGGCCAACAGCATCAGT |
2.5. Co-expression analysis
A global co-expression network was inferred from the Transcript Per Million expression matrix obtained with Salmon (see above) containing the abundance of the 14,327 predicted transfrags. Distance between all pairwise gene combinations was calculated with Pearson Correlation Coefficient together with the Highest Reciprocal Ranking (HRR) procedure as described in Liesecke et al. [45]. The global co-expression network contained 14,086 genes and 61,829 connections with an HRR value below 20. This large graph was further cut into smaller subgraphs with a fast greedy algorithm to detect communities of more densely connected genes [46]. Communities with more than 10 transcripts were further analyzed.
3. Results
3.1. Transcriptome assembly from RNA-seq reads
A total of 21 samples representing 7 experimental conditions (Table 2) were sequenced from paired-end strand specific libraries using Illumina short reads. Each RNA-seq run was used to assemble transcripts with StringTie after mapping them on the S. apiospermum genome sequence with Hisat2. Reads mapped on the genome sequence at a very good rate (> 95%) (Table 3). Individual assemblies were merged into a global one using the draft genome annotation as a support for transcript reconstruction. According to previous annotation, S. apiospermum genome comprises 10,920 genes, 8375 being validated or predicted protein-coding genes and the remaining being considered as pseudogenes [32]. Using the reference annotation, we found new isoforms, i.e. new transcripts differing in structure from the predicted ones. A total of 14,327 transcribed sequences (=transfrags) (Supplemental Table S1) were assembled using this procedure which corresponded to 10,873 different genomic loci. Among the 14,327 transfrags, 8375 predicted transcripts were found (representing 8063 loci with complete match), as well as 2674 potentially new isoforms and 1998 new transcripts (Table 3).
Table 2.
RNA-seq run description.
| Condition | RNA-seq accession | Number of reads | % overall mapping |
|---|---|---|---|
| A (control) | 1 | 11,454,114 | 96.54 |
| A (control) | 2 | 8,960,682 | 96.78 |
| A (control) | 3 | 3,860,262 | 96.85 |
| B (low osmolarity) | 4 | 18,365,999 | 95.96 |
| B (low osmolarity) | 5 | 29,215,410 | 93.92 |
| B (low osmolarity) | 6 | 13,621,825 | 95.49 |
| C (elevated lactate) | 16 | 13,240,120 | 96.02 |
| C (elevated lactate) | 17 | 6,153,612 | 96.14 |
| C (elevated lactate) | 18 | 29,138,378 | 96.87 |
| D (low pH) | 7 | 19,233,452 | 96.38 |
| D (low pH) | 8 | 4,701,204 | 97.28 |
| D (low pH) | 9 | 2,120,142 | 96.81 |
| E (hypoxia) | 19 | 16,933,269 | 96.15 |
| E (hypoxia) | 20 | 10,095,065 | 96.35 |
| E (hypoxia) | 21 | 14,768,183 | 95.84 |
| F (hypercapnia) | 10 | 10,759,382 | 97.04 |
| F (hypercapnia) | 11 | 6,941,592 | 95.75 |
| F (hypercapnia) | 12 | 8,569,169 | 96.74 |
| G (CSFM) | 13 | 1,092,941 | 96.25 |
| G (CSFM) | 14 | 16,313,276 | 96.49 |
| G (CSFM) | 15 | 3,640,226 | 95.17 |
Table 3.
Comparison between predicted and RNA-seq inferred transfrag predictions.
| Transfrag type | Number of transfrags |
|---|---|
| Complete match of intron chain | 8375 |
| A transfrag overlapping a reference exon (pre-mRNA) | 137 |
| A transfrag falling entirely within a reference intron | 5 |
| Potentially novel isoform: at least one splice junction is shared with a reference transcript | 2674 |
| Generic exonic overlap with a reference transcript (transfrags in exons) | 110 |
| Possible polymerase run-on fragment | 360 |
| An intron of the transfrag matches a reference intron on the opposite strand | 146 |
| Unknown, intergenic transcript | 1998 |
| Exonic overlap with reference on the opposite strand | 522 |
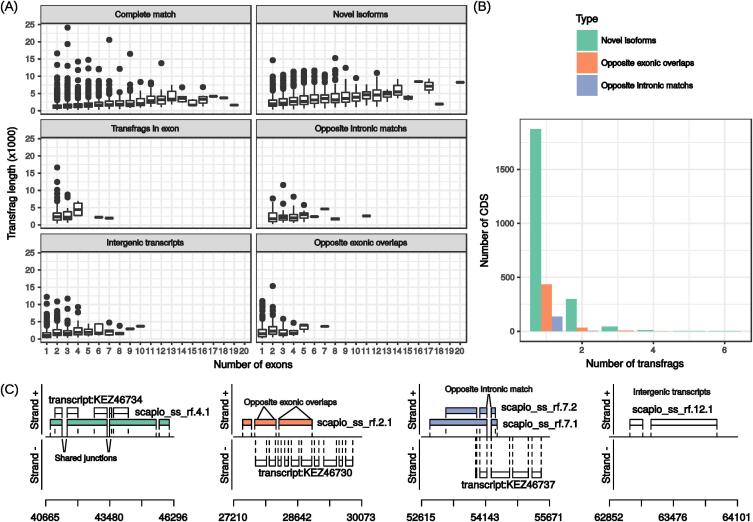
In silico gene prediction may sometimes fail to correctly predict transcription start sites as well as exon–intron junctions. We therefore looked at the length and exonic structure of newly predicted transfrags. On average, new isoforms were longer than previously predicted transcripts (3143 vs. 1641 nt) while new intergenic transcripts were similar in size (1590 nt) (Fig. 1A). They also had more exons (4.5 in average vs. 3.6 for reference transcripts) suggesting that many genes display alternatively spliced transcripts. Exon number was only slightly but significantly correlated with transfrag length (Spearman’s rho > 0.28, p-value < 2e-16) revealing that longer transcripts do not contain longer 3′ UTR regions only.
Fig. 1.
RNA-seq guided gene prediction. Transcribed fragments (transfrags) assembled with the Hisat2/Stringtie pipeline were compared to predicted genes in the reference annotation of S. apiospermum. In (A), transfrag length is plotted againt exon number for the different transfrag classes. Transfrags were detected within already identified coding sequences (CDS) or in intergenic sequences. In (B), transfrag number per CDS is represented according to transfrag nature (potential isoform containing at least one matching intron, opposite intronic match or opposite exonic overlap). Specific examples of 4 transfrags (isoform, opposite exonic overlap, opposite intronic match and new intergenic) are depicted on scaffold SEQ_SAPIO_0011 in (C). Each box represents an exon. X-axis indicates genomic locations on the positive strand. Reference transcripts are indicated with “transcript:” and RNA-seq assembled transcripts with “scapio_ss_rf”.
We found that the new 2674 isoforms concerned 2233 coding sequences (CDS) out of 8375 indicating that some CDS corresponded to at least two isoforms (Fig. 1B). Most of these CDS (1875) had only one isoform in addition to the reference transcript and the highest number of isoforms was 6. A potential isoform sharing two spliced junctions is depicted on the left part of Fig. 1C. In this case, the isoform scapio_ss_rf.4.1 was longer than the reference transcript KEZ46734 (SAPIO_CDS0010) on scaffold SEQ_SAPIO_0011. Antisense transcripts were also detected by transfrags overlapping exons or matching introns on the opposite strand (Table 3, Fig. 1). The most frequent antisense transfrags corresponded to overlaps with exons on the opposite strand, although both types (exon overlap or intron matching) were longer in average than the reference transcripts (2501 nt and 2226 nt, respectively).
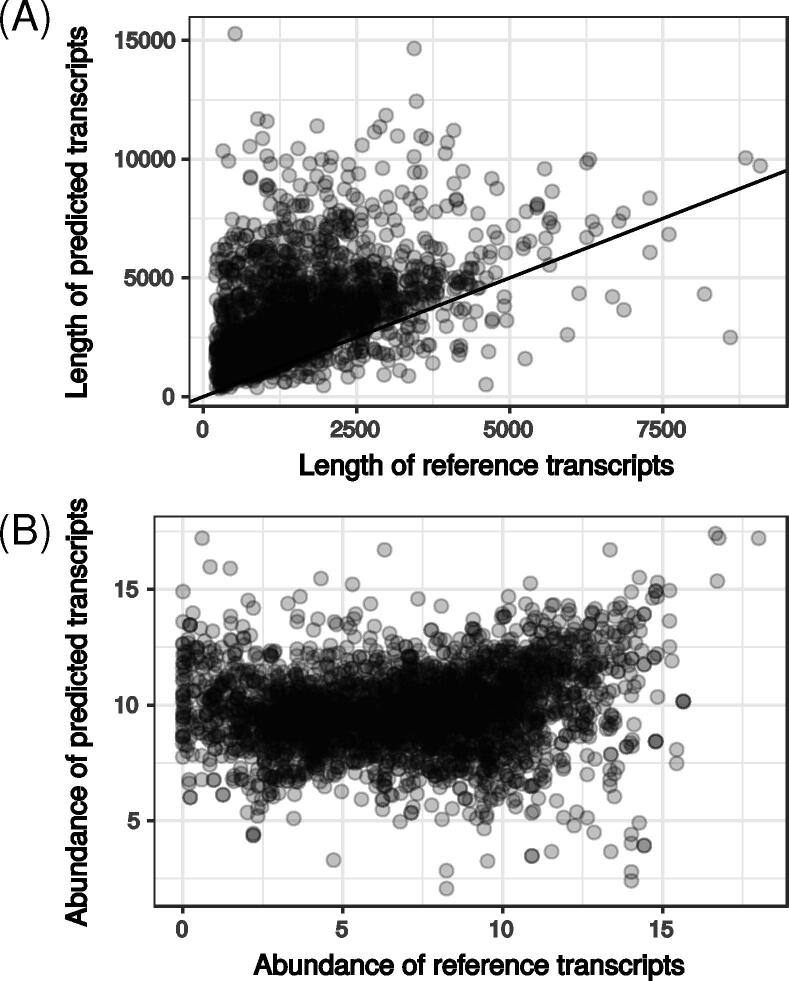
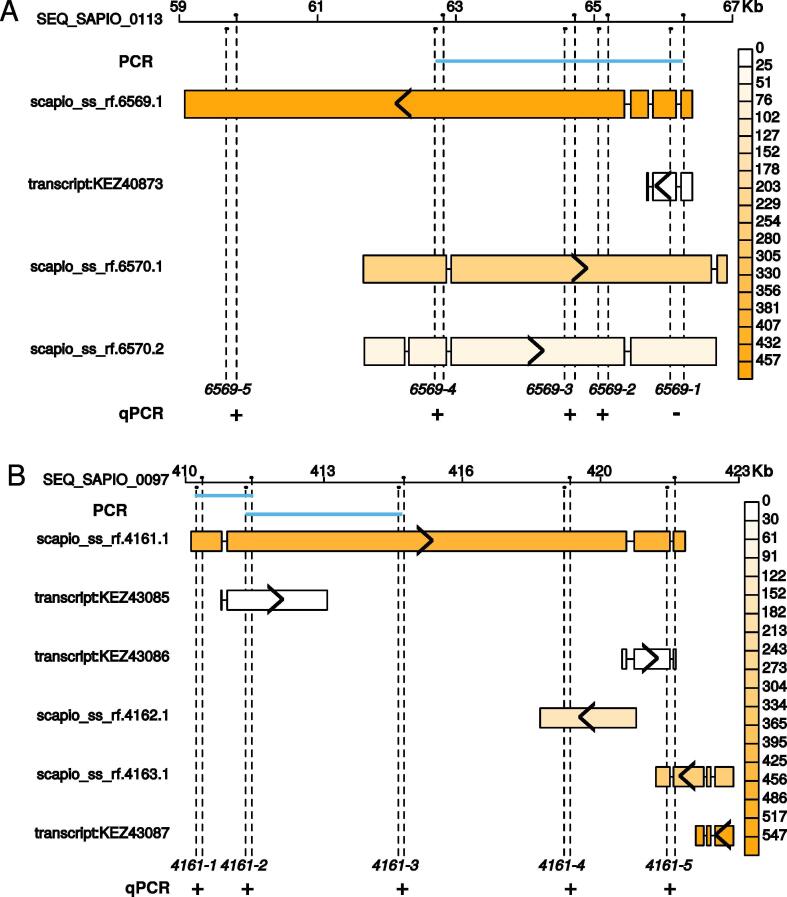
Among the 10,873 loci inferred from the RNA-seq analysis, 2541 had several transcripts including reference and newly predicted transcripts. As indicated above, predicted transcripts from our assembly were generally longer than reference transcripts (Fig. 2A). We summed TPM over the 21 samples to estimate a global abundance for each transcript revealing a small but significant positive relationship between abundances of predicted and of reference transcripts (Spearman’s rho = 0.18, p-value < 2e-16) (Fig. 2B). This could reveal that isoforms and reference transcripts had different expression patterns. Abundance of 79 reference transcripts was very low (TPM < 1.5) while their isoforms had higher abundance (TPM > 32). This suggests that many reference transcripts are not necessarily the main gene expression products. This was typically illustrated on the scapio_ss_rf.6569 locus (Fig. 3A). This locus corresponded to several transcripts with both sense and antisense orientations. The predicted global expression of KEZ40873 was very low, while that of scapio_ss_rf.6569.1 which was much longer appeared to be higher. The existence of such a long transcript was confirmed by qPCR experiments on cDNA using the primer pairs 6569-2, 6569-3, 6569-4 and 6569-5, and with the primer pair 6569-1F and 6569-4R. Although we were unable to amplify a long sequence with primers 6569-4F and 6569-5R, the amplicon obtained with the primer pair 6569-5 confirmed the existence of this transcribed fragment.
Fig. 2.
Comparison of reference and predicted transcripts. For each locus on the genome containing at least one reference and one RNA-seq predicted transcript, we compared either their length (A) or their abundance (B) expressed as the log2 of the Fragment Per Kilobase per Million of reads (FPKM) sum calculated over all the 21 samples. In (A), the line indicates length predicted = length of the reference.
Fig. 3.
Detailed examples of predicted transcript structures. Two examples are shown with loci scapio_ss_rf.6569 (A) and scapio_ss_rf.4161 (B). The genomic loci and coordinates are represented in the top of each panel. Each transcript is represented with its exons (rectangles) separated by introns (lines) and is colored according to its global expression level measured as the sum of transcripts per million (TPM) on the 21 samples. > and < symbols indicate sense or antisense orientation, respectively. Primers used for validation purposes are indicated on the genomic loci and their names are indicated in the bottom of the vertical dashed lines showing primer positions. Forward (F) primers are above the genomic locus line and reverse (R) primers are below this line. These primers were used for qPCR (a+ showing a positive amplification, a− showing no amplification) and PCR (indicated by blue lines when amplification was made with primers belonging to different couples) amplification. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A detailed inspection of the 2541 multi-transcript loci revealed that 291 corresponded to two and sometimes three reference transcripts. As an example, locus scapio_ss_rf.4161 corresponded to two reference transcripts KEZ43085 and KEZ43086 (Fig. 3B). In this locus, a transfrag (scapio_ss_rf.4161.1) simultaneously contained these two transcripts suggesting the existence of a polycistron. Successful amplifications on cDNAs with the 5 primer pairs 4161-1, 4161-2, 4161-3, 4161-4 and 4161-5 confirmed the existence of this long transcript, particularly the amplification with primers 4161-3 and the cross-amplification using 4161-3F and 4161-2R. Although we were unable to amplify a fragment using the 4161-5F/4161-4R primer pair, the positive qPCR amplification with primers 4161-5 indicated a log2FCqPCR (condition G vs. A) = 1.97 (in agreement with the use of primers 4161-3 showing a log2FCqPCR = 3.74). The amplicon generated with primers 4161-5 was unlikely to result only from the amplification of scapio_ss_rf.4161.3 because this transcript was shown to be down-regulated by RNA-seq (log2FCRNA-seq(G vs. A) = −0.5). As a consequence, such polycistronic transcripts are likely to occur in S. apiospermum, although their biological meaning still remains to be determined.
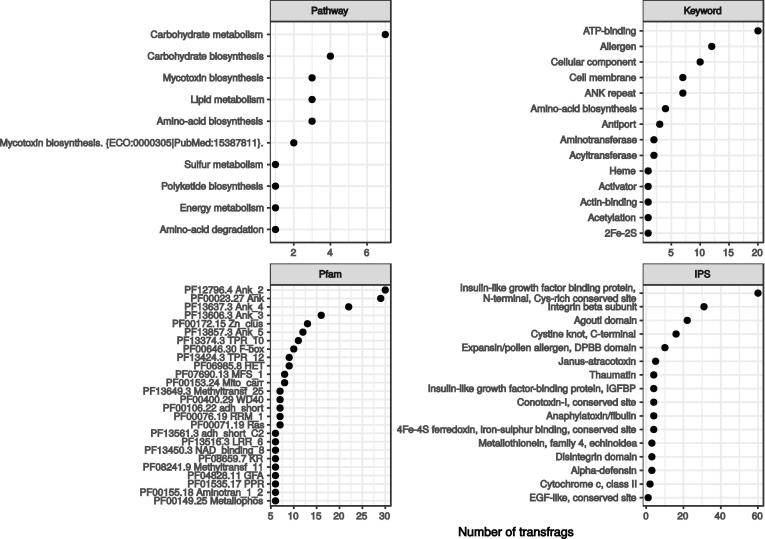
Transfrags annotated as intergenic transcripts could be potentially considered as new genes. Among the 1998 predicted intergenic, 310 had homologies with one Uniprot accession by Blastx (Fig. 4). These transcripts encoded proteins related to primary metabolism (carbohydrates and lipids), but also to mycotoxin biosynthesis. Many of these proteins were also associated with ATP-binding and allergen motifs. To a larger extent, 547 out of the 1998 transcripts had protein sequence matching with at least one Pfam domain in PfamA as found by Hmmscan. Many (109) of these domains were related to ankyrin repeat domains but enzymatic activities such as alcohol dehydrogenases and methyltransferases were also detected. For the remaining 1073 sequences with neither homolog in Uniprot nor evident Pfam domains, specific signatures were searched with Interproscan. Only 144 out of these 1073 intergenic transcripts had such signatures with a marked presence of cysteine-rich domains. As an example, we were able to amplify scapio_ss_rf.7481.1 from cDNA. We verified by both RNA-seq and qPCR that this transcript was significantly more expressed in culture condition E. As observed by Blastx against NCBI nr database, the resulting protein is predicted to contain an adenosylmethionine-8-amino-7-oxononanoate aminotransferase domain suggesting an amino-acid transferase activity.
Fig. 4.
Functional annotation of predicted intergenic sequences. Pathway and keywords were retrieved from Uniprot annotations for transcripts having homologs in this database. Pfam domains were detected with Hmmscan against PfamA. For proteins with neither homolog in Uniprot nor Pfam domain, Interproscan (IPS) signatures were searched.
We next used transfrag expression levels to perform a differential expression analysis and to highlight transcripts that characterized the best each experimental condition. Finally, we determined whether or not newly assembled transfrags improved current reference transcript annotation.
3.2. Cellular processes affected by the different culture conditions
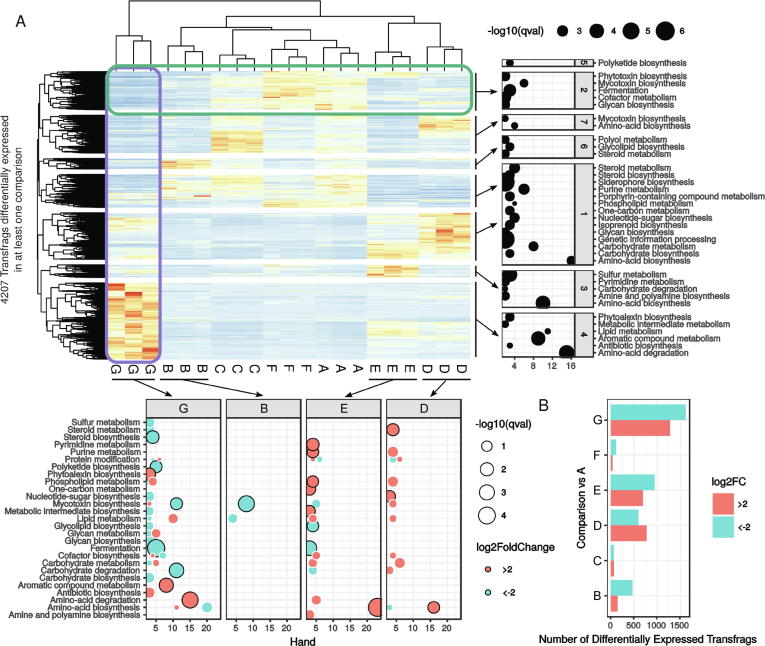
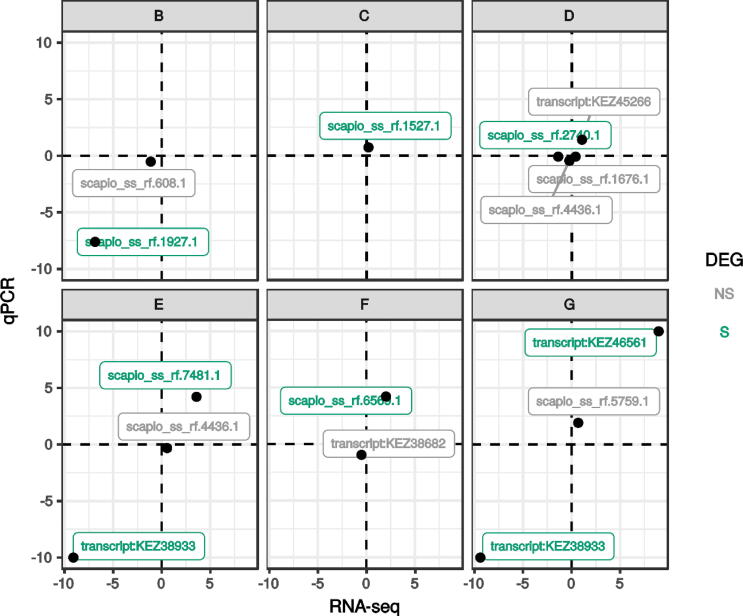
The full RNA-seq assembled transcriptome was first filtered to remove transfrags with very low variance (< 1) and summed raw counts (< 10) across the 21 samples. A total of 13,810 transcripts (96%) were retained for further analysis. Differentially expressed transcripts were detected in 6 conditions compared to the reference culture condition A using a general linear model following a negative binomial to estimate true transcript abundance. This analysis revealed 4207 transfrags that were differentially expressed in at least one of the 6 comparisons (Fig. 5A). Expression of several genes was monitored by qPCR to confirm the validity of the differential approach analysis (Fig. 6). A very good correlation between the two technologies was observed, confirming the reliability of transcript abundance estimated from RNA-seq data. Transcripts detected as weakly expressed in RNA-seq data also displayed very weak expression levels as measured by qPCR for scapio_ss_rf.608.1 and scapio_ss_rf.4436.1 for example. Down-regulated transcripts such as scapio_ss_rf.1927.1 and up-regulated transcripts such as KEZ46561 also had very comparable induction folds measured by RNA-seq and qPCR.
Fig. 5.
Differential transfrag expression analysis. Abundance was used to estimate differentially expressed transfrags in 6 comparisons (condition A being the reference). (A) The heatmap contains the 4207 transfrags (rows) that are differentially expressed in at least one condition (columns, adjusted p-value < 0.001 and absolute log2 fold change > 2). Conditions were clustered according to the 4207 transfrag centered and scaled expression. Transfrags were annotated using Uniprot pathways (bottom panels: one panel indicates functions represented by significantly differentially expressed transfrags in one condition, e.g. G culture condition as represented by the purple box). In bottom panels, significantly enriched Uniprot pathways are surrounded by a black circle. Transfrags were also clustered according to their expression profiles, allowing the identification of 7 clusters that were also characterized according to the Uniprot pathways (right panels: one panel indicates functions represented by a cluster of genes, as represented for example by the green box). In bottom and right panels, only pathways significantly overrepresented (adjusted p-value <0.05) are depicted. (B) Barplot indicating the total number of differentially expressed transfrags in each comparison. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Concordance between RNA-seq and qPCR measurement of gene expression levels. Several differentially (green, S) and not differentially (grey, NS) expressed genes were analyzed by qPCR and compared to in silico quantification by RNA-seq. Data are normalized log2 fold change over the A condition. B: low osmolarity; C: elevated lactate; D: low pH; E: hypoxia; F: hypercapnia; and G: CF synthetic medium (CFSM). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Surprisingly, our analysis first revealed that cultivation of S. apiospermum under hypercapnia or in the presence of an elevated lactate concentration did not induce as much changes as expected. Indeed, only 41 and 117 transcripts were up- and down-regulated under hypercapnic conditions while 71 and 69 transcripts were up- and down-regulated when the fungus was cultivated in the presence of 1% lactate (Fig. 5B, supplemental Table S2).
Likewise, low osmolarity triggered a moderate transcriptional reprogramming since we detected 146 and 467 transfrags up- and down-regulated, respectively, under this culture condition (Fig. 5B, supplemental Table S2). Among the down-regulated transcripts, several encoded proteins were related to lipid metabolism or mycotoxin biosynthesis (Fig. 5A, Fig. 5B, supplemental Table S3), suggesting that, as reported in other fungal models, low osmolarity induces change in the plasma membrane composition, but also regulates some secondary metabolic pathways in S. apiospermum [47].
By contrast, a large series of genes was predicted to be involved in the adaptation of the fungus to growth at acid pH. Indeed at pH 6.4, 774 transcripts were found to be up-regulated and 596 were down-regulated (Fig. 5B, supplemental Table S2). As illustrated in Fig. 5A, the main pathways affected in this culture condition were associated to the biogenesis of the cell membrane (metabolism of lipids, phospholipids, and steroids) and to both primary (purines, nucleotide-sugars, amino-acids, proteins, carbohydrates) and secondary (mycotoxins) metabolisms (Fig. 5A, Fig. 5B, supplemental Table S3). This indicated that the sole lowering the pH of a single unit is sufficient to induce strong transcriptional changes in S. apiospermum.
Our data also revealed marked changes in gene expression when the mold was cultivated in hypoxic conditions. We also identified 695 and 946 S. apiospermum transcripts up- and down-regulated, respectively, under hypoxia (Fig. 5B, supplemental Table S2). These transfrags corresponded to genes mainly involved in general processes and metabolisms (fermentation, protein modification, carbohydrate metabolism and degradation, amine and polyamine biosynthesis, nitrogenous base metabolisms), membrane composition (lipid, phospholipid, and glycolipid metabolism), and mycotoxin biosynthesis (Fig. 5A, Fig. 5B, supplemental Table S3). Likewise, hypoxic conditions triggered a coordinate over-expression of a series of more than 20 genes involved in amino-acid biosynthesis (Fig. 5A, Fig. 5B, supplemental Table S3). Although a positive effect of hypoxia on global amino-acid biosynthesis was recently observed in Saccharomyces cerevisiae [48], the significance of this regulation is still unclear since molecular oxygen does not appear to be required for this process.
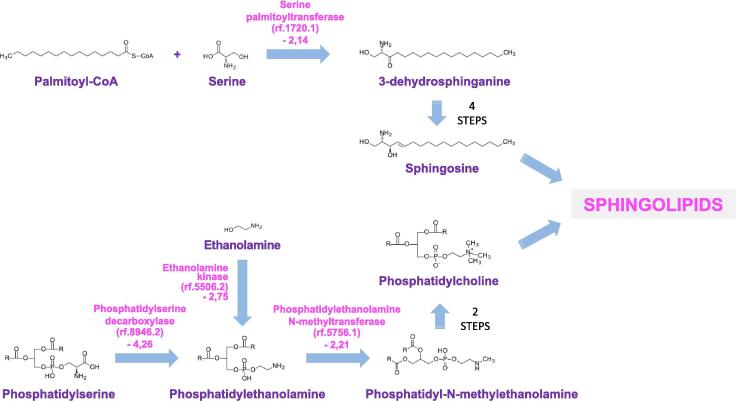
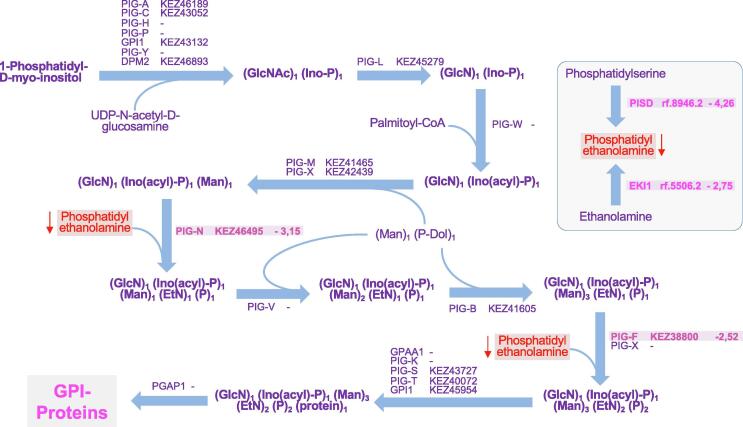
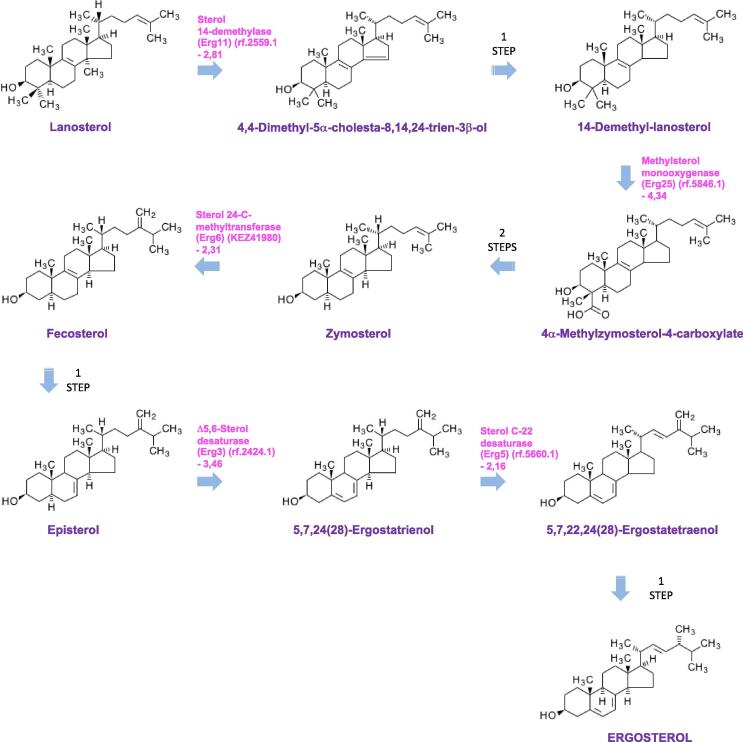
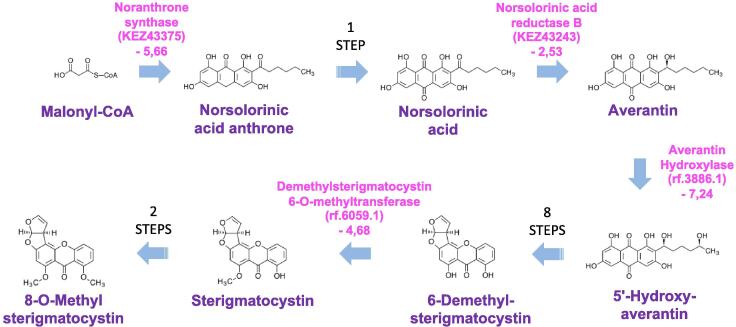
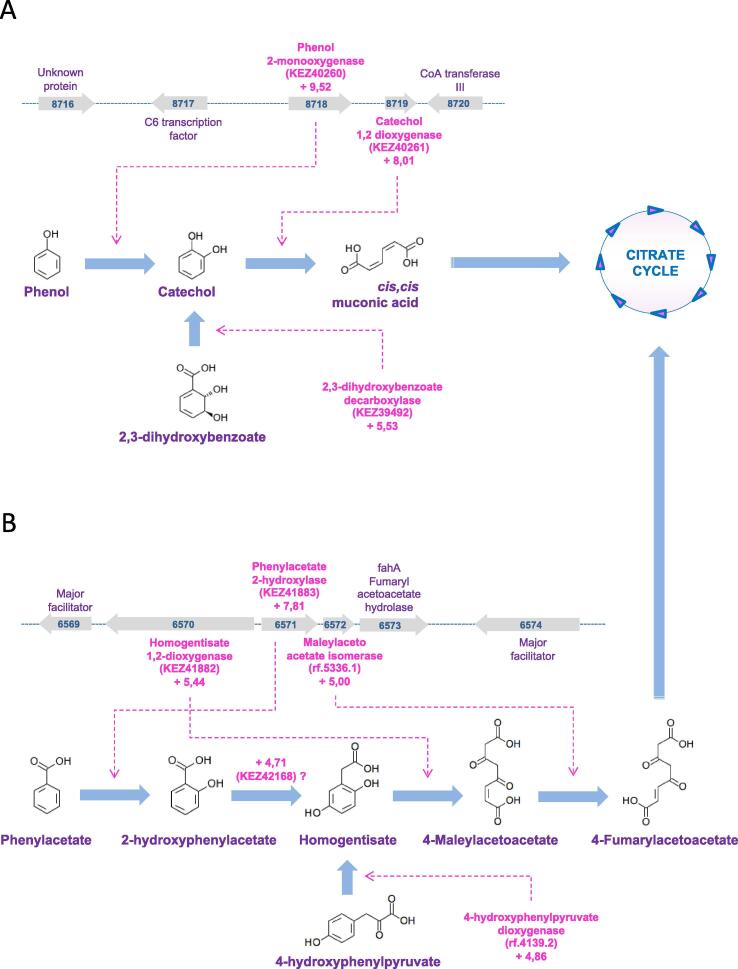
Above all, the strongest transcriptional reprogramming in S. apiospermum was obtained by comparing the control culture condition with the CFSM culture condition. Indeed, 1277 transcripts were enriched in CFSM compared to the control condition, and 1616 were down-regulated (Fig. 5B, supplemental Table S2). Beside the numerous general processes influenced by acid pH, elevated lactate concentration or hypoxia (Fig. 5A, Fig. 5B, supplemental Table S3), several additional pathways were found to be specifically regulated when S. apiospermum was cultivated in this medium which mimics the CF bronchial mucus. First, the amino-acid metabolism appeared strongly impacted in this culture condition since 20 transcripts encoding enzymes involved in amino-acid biosynthesis were down-regulated and 15 transcripts encoding enzymes involved in amino-acid degradation were up-regulated (Fig. 5A, Fig. 5B, supplemental Table S3). This could indicate that the fungus uses amino-acids from the synthetic medium as primary source of carbon, nitrogen, and sulfur for some biosynthesis. We also observed a dramatic shift in the composition of the cell envelop. This is attested by the down-regulation of a series of 4 transcripts encoding enzymes involved in early steps of the sphingolipid biosynthetic pathway [the serine palmitoyltransferase (rf.1720.1), the phosphatidylserine decarboxylase (rf.8946.2), the ethanolamine kinase (rf.5506.2), and the phosphatidylethanolamine N-methyltransferase (rf.5756.1) (Fig. 7)], of two transcripts encoding key enzymes of the glycosylphosphatidylinositol (GPI)-anchor protein biosynthetic pathway [the GPI ethanolamine phosphate transferase 1 (KEZ46495) and the GPI-anchor biosynthesis protein Pig-f (KEZ38800)] (Fig. 8), and of transcripts corresponding to 5 of the ERG gene series involved in the conversion of squalene to ergosterol (the C-14-alpha sterol demethylase Erg11p (rf.2559.1), C-4 methyl sterol oxidase Erg25p (rf.5846.1), sterol C24-methyltransferase Erg6p (KEZ41980), delta 5,6-sterol desaturase Erg3p (rf.2424.1), cytochrome P450 sterol C-22 desaturase Erg5p (rf.5660.1)] (Fig. 9). Our data also revealed the down-regulation of some transcripts encoding enzymes involved in the biosynthesis of aflatoxin-like mycotoxins including notably the polyketide synthase noranthrone synthase (KEZ43375), the norsolorinic acid reductase B (KEZ43243), the averantin hydroxylase (rf.3886.1), and the demethylsterigmatocystin 6-O-methyltransferase (rf.6059.1) (Fig. 10). Also interesting, this analysis revealed the up-regulation of a list of transcripts encoding enzymes involved in the degradation pathways of aromatic compounds. This includes in particular three enzymes required for the conversion of both phenol and 2,3-dihydroxybenzoate to cis,cis muconic acid [the phenol 2-monooxygenase (KEZ40260), the 2,3-dihydroxybenzoate decarboxylase (KEZ39492), and the catechol 1,2 dioxygenase (KEZ40261)] (Fig. 11A), but also 4 enzymes implicated in the conversion of phenylacetate and 4-hydroxyphenylpyruvate to 4-fumarylacetoacetate [the phenylacetate 2-hydroxylase (KEZ41883), the homogentisate 1,2-dioxygenase (KEZ41882), the maleylacetoacetate isomerase (rf.5336.1), and the 4-hydroxyphenylpyruvate dioxygenase (rf.4139.2) (Fig. 11B). This could indicate an active recycling of the aromatic compounds towards the citrate cycle when the fungus is cultivated in CFSM.
Fig. 7.
Some transcripts encoding enzymes involved in sphingolipid biosynthesis are down-regulated in the CF specific medium (CFSM) culture condition (G condition). Only key metabolic intermediates are shown. Differentially regulated transcripts are indicated in pink as follows: name of the predicted protein (reference number of the transcript) and Log of fold-change of the transfrag versus condition A. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
Some transcripts encoding enzymes involved in the glycosylphosphatidylinositol (GPI)-anchor protein biosynthetic pathway are down-regulated in the CF specific medium (CFSM) culture condition (G condition). Only key metabolic intermediates are shown. Differentially regulated transcripts are indicated in pink as follows: name of the predicted protein (reference number of the transcript) and Log of fold-change of the transfrag versus condition A. When identified in the genome of S. apiospermum, reference numbers of transcripts homologs to the PIG series are indicated. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
Some transcripts encoding enzymes involved in ergosterol biosynthesis are down-regulated in the CF specific medium (CFSM) culture condition (G condition). Only key metabolic intermediates are shown. Differentially regulated transcripts are indicated in pink as follows: Name of the predicted protein (reference number of the transcript) and Log of fold-change of the transfrag versus condition A. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 10.
Some transcripts encoding enzymes involved in aflatoxin-like mycotoxins are down-regulated in the CF specific medium (CFSM) culture condition (G condition). Only key metabolic intermediates are shown. Differentially regulated transcripts are indicated in pink as follows: name of the predicted protein (reference number of the transcript) and Log of fold-change of the transfrag versus condition A. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 11.
Some transcripts encoding enzymes involved in the degradation of aromatic compounds are up-regulated in the CF specific medium (CFSM) culture condition (G condition). A. The phenol catabolism cluster identified at contig 122 [1]. B. The tyrosine catabolism cluster identified in this study at contig 105. Differentially regulated transcripts are indicated in pink as follows: name of the predicted protein (reference number of the transcript) and Log of fold-change of the transfrag versus condition A. For each cluster, grey arrows represent predicted CDS. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Global gene co-expression network
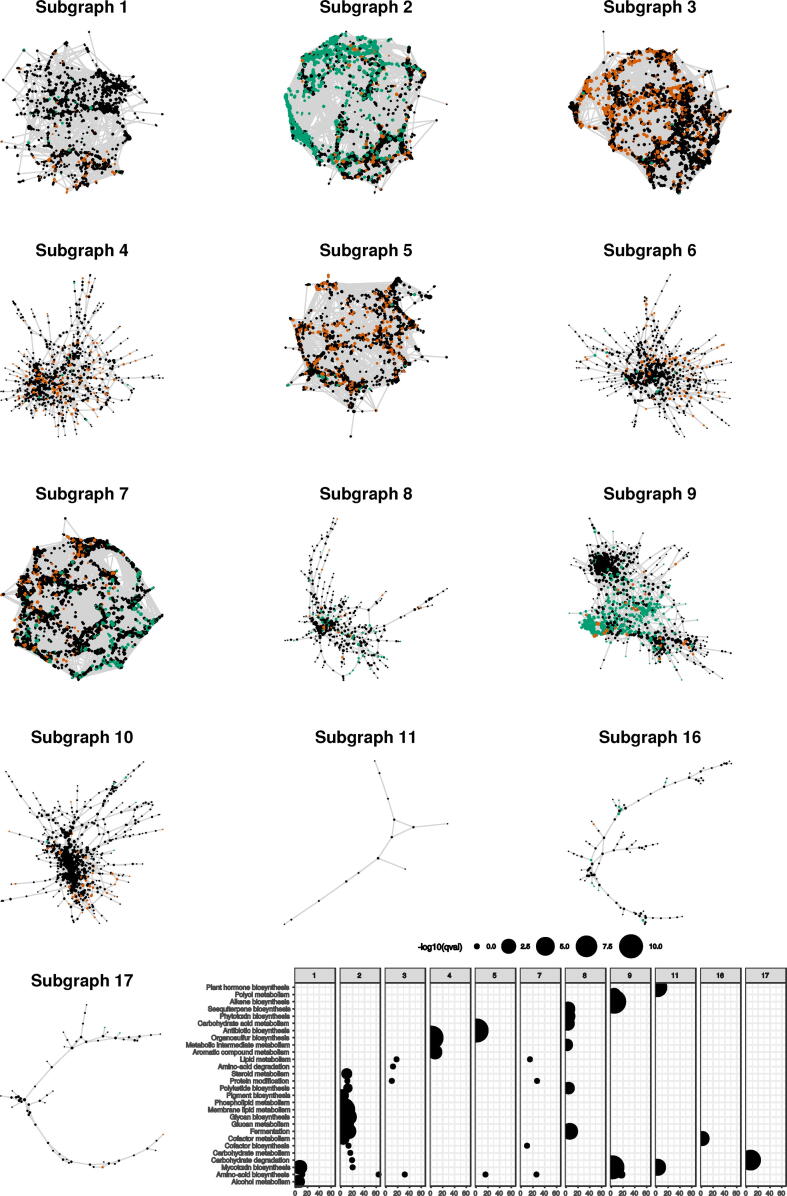
To further investigate how metabolic activities of S. apiospermum are coordinated to face specific culture conditions, we constructed a gene co-expression network from our dataset. Expression profiles were compared for all pairwise combinations of the 14,327 transcripts. Relationships displaying an HRR below 10 were considered as significant. The global network was split into specific communities, of which the 13 largest are represented in Fig. 12. Each subgraph contained both non-differentially and differentially expressed genes. Firstly, two specific topologies were observed. Subgraphs 1, 2, 3, 5 and 7 had very dense structures of highly connected genes and were therefore more likely to contain genes encoding enzymes from the primary metabolism or for basal cell activities. Many steps related to amino-acid biosynthesis and lipid metabolism indeed were found in these subgraphs as revealed by the significant enrichment of this process (Fig. 12). This observation highlights how strongly different culture conditions are accompanied with large transcriptional changes. In other subgraphs, many genes had a node degree below < 3 showing their low connectivity. Specific functions dedicated to aromatic compound metabolism or mycotoxin biosynthesis were significantly enriched in these subgraphs, which may reveal fine-tuned adaptations to each growth conditions sensed by S. apiospermum as environmental cues. Secondly, differentially expressed genes mainly clustered into subgraphs 2, 3, 5, 7 and 9. This suggests that transcriptional responses observed in the present work involve only few transcriptional modules containing thousands of genes.
Fig. 12.
Co-expression networks of Scedosporium apiospermum transcripts. Transcripts in black correspond to non-differentially expressed genes. In red are shown transcripts significantly more expressed in at least one condition versus condition A, and in green, transcripts significantly less expressed in at least one condition versus condition A. The bottom panel describes general functions represented in each subgraph together with their enrichment (hypergeometric test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
All 13 subgraphs combined, a total of 243 transcripts displaying a PFAM “Fungal specific transcription factor” domain (PF04082.15 and PF11951.5) were identified in the networks among which 98 were differentially expressed in at least one comparison. Unfortunately, many of these transcription factors had no or weak similarities to Uniprot Fungi accessions. However, they are likely to be good candidates for further characterization of metabolic regulation in S. apiospermum. Some of them could potentially be master coordinators of metabolic pathways, playing essential roles in adaptation to environmental growth conditions.
4. Discussion
Contaminating spores (conidia) of S. apiospermum are rarely found in the air compared to other environmental molds. However, this species ranks second among the filamentous fungi that are able to colonize the respiratory tract of patients with CF. Nevertheless, beside the context of lung transplantation, the colonization of the airways by S. apiospermum remains rather well tolerated by the patients. Although some cases of respiratory infections or allergic bronchopulmonary mycosis due to S. apiospermum were previously reported, clinical expression inherent to the colonization of the CF airways is usually from low to absent. This raises the question of why the colonization process of the respiratory tract by S. apiospermum is fairly tolerated by CF patients while it is known that in other clinical contexts, the fungus is able to cause marked symptoms. In this context, we investigated the transcriptional reprogramming that accompanies the exposure of the fungus to the particular microenvironment encountered in the CF bronchial mucus. In this aim, the transcriptome of the S. apiospermum reference strain (IHEM 14462) was analyzed after cultivation of the fungus in a synthetic medium corresponding to the nutritional composition of CF sputum (CFSM) [33], but also in different growth conditions that independently mimic the various physico-chemical constraints encountered in the bronchial mucus of CF patients including hypercapnia, hypoxia, acid pH, low osmolarity, and increased lactate concentration.
Although our analysis first revealed that growth under hypercapnia or in the presence of an elevated lactate concentration did not induce marked transcriptional changes, low osmolarity, hypoxic or acid pH conditions of culture were shown to trigger a moderate to marked transcriptional reprogramming in the fungus. In these last conditions, most of the transfrags regulated were associated to changes in general cell processes (fermentation, protein modification), membrane composition modification (lipid, phospholipid, and glycolipid metabolism), and regulation of primary (nitrogenous bases, nucleotide-sugars, amino-acids, proteins, carbohydrates, amines, and polyamines) and secondary (mycotoxins) metabolisms.
Above all, the most important transcriptional reprogramming was induced by cultivation of the fungus in CFSM. We first observed a shift in the global cell envelop composition (down-regulation of sphingolipid, GPI-anchored protein, and ergosterol biosynthesis) when the fungus was grown in this medium. This adaptative process may reflect the fungal capacity of evading the host immune system by lowering the biosynthesis of antigenic determinants (sphingolipids and GPI-anchored proteins) or limiting their targeting at the cell surface through down-regulation of the ergosterol biosynthetic pathway and an altered membrane fluidity. It is also important to highlight that the apparent reduction in the ergosterol content in condition mimicking the CF sputum could also underlie the discrepancy between in vitro susceptibility of the fungus towards antifungals and in vivo inefficiency of the therapeutic molecules. In this regards, it should be considered in the future the use of CFSM instead of the classical RPMI culture medium to determine the in vitro susceptibility to antifungals for Scedosporium isolates from sputum samples from patients with CF.
In addition, this analysis revealed that some genes encoding enzymes involved in the biosynthesis of aflatoxin-like mycotoxins are down-regulated when the fungus was cultured in CFSM and, to a lesser extent, under low osmolarity and hypoxic conditions. Although true aflatoxins have never been described in the genus Scedosporium, some precursor metabolites occurring in these secondary metabolic pathways such as 8-O-methylsterigmatocystin were previously isolated in the closely related species S. boydii [49]. Given the fact that it has been reported that sterigmatocystin induces apoptosis in human pulmonary cells in vitro and displays pro-inflammatory properties on mouse alveolar macrophages [50], [51], it is thus possible to hypothesize that during the lung colonization process, S. apiospermum reduces the production of toxic secondary metabolites related to sterigmatocystin to prevent exacerbation of the host immune response, notably the macrophage-mediated oxidative burst.
Finally, we observed a strong up-regulation of many genes encoding enzymes involved in the degradation of aromatic compounds, especially when S. apiospermum was grown in CFSM. These include notably genes that are partially clustered in the genome of the fungus for phenol or tyrosine catabolism. The capacity of S. apiospermum to degrade aromatic hydrocarbons is already well documented [1], [52], [53], [54], [55], [56], [57] and it has been hypothesized that this metabolic trait would predispose this fungus to particular patterns of human pathogenicity [58], [59]. We thus here provide first experimental data that support this assumption.
In conclusion, this study allowed us to define for the first time the whole transcriptome of S. apiospermum. These data which represent a breakthrough for elucidation of the pathogenic mechanisms of the fungus, largely confirmed the draft genome annotation since transcripts were found for almost all the protein-encoding CDS. They also revealed that most of the CDS considered as pseudogenes actually were misidentified, because of the lack of introns, and demonstrated the possibility of isoforms and polycistronic mRNA. More importantly, comparison of the transcriptional response of the fungus to the particular abiotic environment found in the CF bronchial mucus revealed the adaptative mechanisms developed by the fungus to dissimulate itself towards the host immune defenses, particularly a reduced production of some toxic secondary metabolites and a dramatic shift in the global composition of the cell envelop. Biochemical studies should be conducted to confirm these data which may have important clinical significance, since these may explain the lack of efficiency of antifungal treatment despite in vitro data suggestive of efficacy.
Conflict of interest
Declarations of interest: none.
Authors’ contribution
PV and JPB: Conceptualization of the study and funding acquisition; PV: Performed cultural studies, RNA extractions and PCR verifications; PV and TDG: Data curation and formal analysis. PV and NP: project administration and supervision; PV, TDG and NP: Writing the original draft of the manuscript. SLG, GN and JPB: Contributed to the discussion and review and editing of the manuscript.
Acknowledgments
This work was funded by the French patient organizations against cystic fibrosis, Association Grégory Lemarchal and Vaincre la Mucoviscidose (RF20150501375/1/1/110) which are gratefully acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.10.034.
Contributor Information
Nicolas Papon, Email: nicolas.papon@univ-angers.fr.
Jean-Philippe Bouchara, Email: jean-philippe.bouchara@univ-angers.fr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Rougeron A., Giraud S., Alastruey-Izquierdo A., Cano J., Rainer J. Ecology of Scedosporium species: present knowledge and future research. Mycopathologia. 2018;183:185–200. doi: 10.1007/s11046-017-0200-2. [DOI] [PubMed] [Google Scholar]
- 2.Ramirez-Garcia A., Pellon A., Rementeria A., Buldain I., Barreto-Berguer E. Scedosporium complex and Lomentospora prolificans: an updated overview of underrated opportunist. Med Mycol. 2018;56(Suppl. 1):102–127. doi: 10.1093/mmy/myx113. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz C., Vandeputte P., Rougeron A., Giraud S., Dugé de Bernonville T. Developing collaborative works for faster progress on fungal respiratory infections in cystic fibrosis. Med Mycol. 2018;56(Suppl. 1):46–59. doi: 10.1093/mmy/myx106. [DOI] [PubMed] [Google Scholar]
- 4.Cimon B., Carrère J., Chazalette J.P., Giniès J.L., Chabasse D. Mycoses bronchopulmonaires au cours de la mucoviscidose. Résultats d'une étude épidémiologique longitudinale sur une période de 5 ans. J Mycol Méd. 2000;10:128–135. [Google Scholar]
- 5.Horré R., Marklein G., Siekmeier R., Nidermajer S., Reiffert S.M. Selective isolation of Pseudallescheria and Scedosporium species from respiratory tract specimens of cystic fibrosis patients. Respiration. 2009;77:320–324. doi: 10.1159/000167419. [DOI] [PubMed] [Google Scholar]
- 6.Blyth C.C., Harun A., Middleton P.G., Sleiman S., Lee O. Detection of occult Scedosporium species in respiratory tract specimens from patients with cystic fibrosis by use of selective media. J Clin Microbiol. 2010;48:314–316. doi: 10.1128/JCM.01470-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blyth C.C., Middleton P.G., Harun A., Sorrell T.C., Meyer W. Clinical associations and prevalence of Scedosporium spp. in Australian cystic fibrosis patients: identification of novel risk factors? Med Mycol. 2010;48(Suppl 1):S37–S44. doi: 10.3109/13693786.2010.500627. [DOI] [PubMed] [Google Scholar]
- 8.Harun A., Blyth C.C., Gilgado F., Middleton P., Chen S.C. Development and validation of a multiplex PCR for detection of Scedosporium spp. in respiratory tract specimens from patients with cystic fibrosis. J Clin Microbiol. 2011;49(4):1508–1512. doi: 10.1128/JCM.01810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zouhair R., Rougeron A., Razafimandimby B., Kobi A., Bouchara J.P. Distribution of the different species of the Pseudallescheria boydii/Scedosporium apiospermum complex in French patients with cystic fibrosis. Med Mycol. 2013;51:603–613. doi: 10.3109/13693786.2013.770606. [DOI] [PubMed] [Google Scholar]
- 10.Masoud-Landgraf L., Badura A., Eber E., Feierl G., Marth E. Modified culture method detects a high diversity of fungal species in cystic fibrosis patients. Med Mycol. 2014;52:179–186. doi: 10.3109/13693786.2013.792438. [DOI] [PubMed] [Google Scholar]
- 11.Sedlacek L., Graf B., Schwarz C., Albert F., Peter S. Prevalence of Scedosporium species and Lomentospora prolificans in patients with cystic fibrosis in a multicenter trial by use of a selective medium. J Cyst Fibros. 2015;14(2):237–241. doi: 10.1016/j.jcf.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Chen M., Kondori N., Deng S., Gerrits van den Ende A.H.G., Lackner M. Direct detection of Exophiala and Scedosporium species in sputa of patients with cystic fibrosis. Med Mycol. 2018;56(6):695–702. doi: 10.1093/mmy/myx108. [DOI] [PubMed] [Google Scholar]
- 13.Cimon B., Carrère J., Vinatier J.F., Chazalette J.P., Chabasse D. Clinical significance of Scedosporium apiospermum in patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis. 2000;19:53–56. doi: 10.1007/s100960050011. [DOI] [PubMed] [Google Scholar]
- 14.Noni M., Katelari A., Kapi A., Stathi A., Dimopoulos G. Scedosporium apiospermum complex in cystic fibrosis; should we treat? Mycoses. 2017;60:594–599. doi: 10.1111/myc.12634. [DOI] [PubMed] [Google Scholar]
- 15.Cimon B., Symoens F., Zouhair R., Chabasse D., Nolard N. Molecular epidemiology of airway colonization by Aspergillus fumigatus in cystic fibrosis patients. J Med Microbiol. 2001;50:367–374. doi: 10.1099/0022-1317-50-4-367. [DOI] [PubMed] [Google Scholar]
- 16.Defontaine A., Zouhair R., Cimon B., Carrère J., Bailly E. Genotyping study of Scedosporium apiospermum isolates from patients with cystic fibrosis. J Clin Microbiol. 2002;40:2108–2114. doi: 10.1128/jcm.40.6.2108-2114.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matray O., Mouhajir A., Giraud S., Godon C., Gargala G. Semi-automated repetitive sequence-based PCR amplification for species of the Scedosporium apiospermum complex. Med Mycol. 2016;54:409–419. doi: 10.1093/mmy/myv080. [DOI] [PubMed] [Google Scholar]
- 18.Kelley T.J., Drumm M.L. Inducible nitric oxide synthase expression is reduced in cystic fibrosis murine and human airway epithelial cells. J Clin Invest. 1998;102:1200–1207. doi: 10.1172/JCI2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley T.J., Elmer H.L. In vivo alterations of IFN regulatory factor-1 and PIAS1 protein levels in cystic fibrosis epithelium. J Clin Invest. 2000;106:403–410. doi: 10.1172/JCI9560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chmiel J.F., Davis P.B. State of the Art: Why do the lungs of patients with cystic fibrosis become infected and why can't they clear the infection? Respir Res. 2003;4:8. doi: 10.1186/1465-9921-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granger D.L., Perfect J.R., Durack D.T. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Invest. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaragoza O., Fries B.C., Casadevall A. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2. Infect Immun. 2003;71:6155–6164. doi: 10.1128/iai.71.11.6155-6164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahn Y.S., Cox G.M., Perfect J.R., Heitman J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr Biol. 2005;15:2013–2020. doi: 10.1016/j.cub.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 24.Ene I.V., Adya A.K., Wehmeier S., Brand A.C., MacCallum D.M. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol. 2012;14:1319–1335. doi: 10.1111/j.1462-5822.2012.01813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ene I.V., Heilmann C.J., Sorgo A.G., Walker L.A., de Koster C.G. Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans. Proteomics. 2012;12:3164–3179. doi: 10.1002/pmic.201200228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ene I.V., Cheng S.C., Netea M.G., Brown A.J. Growth of Candida albicans cells on the physiologically relevant carbon source lactate affects their recognition and phagocytosis by immune cells. Infect Immun. 2013;81:238–248. doi: 10.1128/IAI.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/mmbr.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell A.P. Fungal CO2 sensing: a breath of fresh air. Curr Biol. 2005;15:R934–R936. doi: 10.1016/j.cub.2005.10.064. [DOI] [PubMed] [Google Scholar]
- 29.Han T.L., Cannon R.D., Villas-Bôas S.G. The metabolic basis of Candida albicans morphogenesis and quorum sensing. Fungal Genet Biol. 2011;48:747–763. doi: 10.1016/j.fgb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Grahl N., Shepardson K.M., Chung D., Cramer R.A. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot Cell. 2012;11:560–570. doi: 10.1128/EC.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler G. Hypoxia and gene expression in eukaryotic microbes. Annu Rev Microbiol. 2013;67:291–312. doi: 10.1146/annurev-micro-092412-155658. [DOI] [PubMed] [Google Scholar]
- 32.Vandeputte P, Ghamrawi S, Rechenmann M, Iltis A, Giraud S, et al. (2014) Draft genome sequence of the pathogenic fungus Scedosporium apiospermum. Genome Announc 2(5). pii: e00988-14. doi: 10.1128/genomeA.00988-14. [DOI] [PMC free article] [PubMed]
- 33.Palmer K.L., Aye L.M., Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eddy S.R. Accelerated profile HMM searches. PLOS Comput Biol. 2011;7(10) doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frazee A.C., Pertea G., Jaffe A.E., Langmead B., Salzberg S.L. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33:243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A language and environment for statistical computing. http://www.R-project.org/ [Google Scholar]
- 42.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Govic Y., Papon N., Le Gal S., Lelièvre B., Bouchara J.P. Genomic organization and expression of iron metabolism genes in the emerging pathogenic mold Scedosporium apiospermum. Front Microbiol. 2018;9:827. doi: 10.3389/fmicb.2018.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liesecke F., Daudu D., Dugé de Bernonville R., Besseau S., Clastre M. Ranking genome-wide correlation measurements improves microarray and RNA-seq based global and targeted co-expression networks. Sci Rep. 2018;8:10885. doi: 10.1038/s41598-018-29077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clauset A., Newman M.E., Moore C. Finding community structure in very large networks. Phys Rev E. 2004;70 doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 47.Duran R., Cary J.W., Calvo A.M. Role of the osmotic stress regulatory pathway in morphogenesis and secondary metabolism in filamentous fungi. Toxins (Basel) 2010;2:367–381. doi: 10.3390/toxins2040367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bendjilali N, MacLeon S, Kalra G, Willis SD, Hossian AK, et al. (2017) Time-course analysis of gene expression during the Saccharomyces cerevisiae hypoxic response. G3 (Bethesda) 7(1):221-231. doi: 10.1534/g3.116.034991. [DOI] [PMC free article] [PubMed]
- 49.Lan W.J., Wang K.T., Xu M.Y., Zhang J.J., Lam C.K. Secondary metabolites with chemical diversity from the marine-derived fungus Pseudallescheria boydii F19–1 and their cytotoxic activity. RSC Adv. 2016;6:76206. doi: 10.1039/C6RA06661E. [DOI] [Google Scholar]
- 50.Rand T.G., Dipenta J., Robbins C., Miller J.D. Effects of low molecular weight fungal compounds on inflammatory gene transcription and expression in mouse alveolar macrophages. Chem Biol Interact. 2011;190:139–147. doi: 10.1016/j.cbi.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 51.Cui J., Wang J., Huang S., Jiang X., Li Y. Sterigmatocystin induced apoptosis in human pulmonary cells in vitro. Exp Toxicol Pathol. 2017;69:695–699. doi: 10.1016/j.etp.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 52.April T.M., Abbott S.P., Foght J.M., Currah R.S. Degradation of hydrocarbons in crude oil by the ascomycete Pseudallescheria boydii (Microascaceae) Can J Microbiol. 1998;44:270–278. doi: 10.1139/w97-152. [DOI] [PubMed] [Google Scholar]
- 53.Martín-Gil J., Navas-Gracia L.M., Gómez-Sobrino E., Correa-Guimaraes A., Hernández-Navarro S. Composting and vermicomposting experiences in the treatment and bioconversion of asphaltens from the Prestige oil spill. Bioresour Technol. 2008;99:1821–1829. doi: 10.1016/j.biortech.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 54.Reyes-César A., Absalón Á.E., Fernández F.J., González J.M., Cortés-Espinosa D.V. Biodegradation of a mixture of PAHs by non-ligninolytic fungal strains isolated from crude oil-contaminated soil. World J Microbiol Biotechnol. 2014;30:999–1009. doi: 10.1007/s11274-013-1518-7. [DOI] [PubMed] [Google Scholar]
- 55.Lladó S., Covino S., Solanas A.M., Petruccioli M., D'annibale A. Pyrosequencing reveals the effect of mobilizing agents and lignocellulosic substrate amendment on microbial community composition in a real industrial PAH-polluted soil. J Hazard Mater. 2015;283:35–43. doi: 10.1016/j.jhazmat.2014.08.065. [DOI] [PubMed] [Google Scholar]
- 56.Morales L.T., González-García L.N., Orozco M.C., Restrepo S., Vives M.J. The genomic study of an environmental isolate of Scedosporium apiospermum shows its metabolic potential to degrade hydrocarbons. Stand Genomic Sci. 2017;12:71. doi: 10.1186/s40793-017-0287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan X., Zhang X., Chen X., Kong D., Liu X. Synergistic degradation of crude oil by indigenous bacterial consortium and exogenous fungus Scedosporium boydii. Bioresour Technol. 2018;264:190–197. doi: 10.1016/j.biortech.2018.05.072. [DOI] [PubMed] [Google Scholar]
- 58.Prenafeta-Boldú F.X., Guivernau M., Gallastegui G., Viñas M., de Hoog G.S. Fungal/bacterial interactions during the biodegradation of TEX hydrocarbons (toluene, ethylbenzene and p-xylene) in gas biofilters operated under xerophilic conditions. FEMS Microbiol Ecol. 2012;80:722–734. doi: 10.1111/j.1574-6941.2012.01344.x. [DOI] [PubMed] [Google Scholar]
- 59.Blasi B., Poyntner C., Rudavsky T., Prenafeta-Boldú F.X., Hoog S. Pathogenic yet environmentally friendly? Black fungal candidates for bioremediation of pollutants. Geomicrobiol J. 2016;33:308–317. doi: 10.1080/01490451.2015.1052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.