Abstract
Coagulopathy and syncytial formation are relevant effects of the SARS-CoV-2 infection, but the underlying molecular mechanisms triggering these processes are not fully elucidated. Here, we identified a potential consensus pattern in the Spike S glycoprotein present within the cytoplasmic domain; this consensus pattern was detected in only 79 out of 561,000 proteins (UniProt bank). Interestingly, the pattern was present in both human and bat the coronaviruses S proteins, in many proteins involved in coagulation process, cell–cell interaction, protein aggregation and regulation of cell fate, such as von Willebrand factor, coagulation factor X, fibronectin and Notch, characterized by the presence of the cysteine-rich EGF-like domain. This finding may suggest functional similarities between the matched proteins and the CoV-2 S protein, implying a new possible involvement of the S protein in the molecular mechanism that leads to the coagulopathy and cell fusion in COVID-19 disease.
Subject terms: Viral proteins, Infectious diseases
The severe acute respiratory syndrome coronavirus (SARS-1) of 2002, the Middle East respiratory syndrome coronavirus (MERS-CoV) of 2012, and now the SARS-2 of 2019 (causing the COVID-19 disease) are all due to coronaviruses of the beta subgroup1. This positive-sense single-stranded RNA virus family possesses the structural proteins spike (S), membrane (M) and envelope (E) proteins, along with the nucleocapsid (N) protein2.
SARS spike glycoprotein is a trimeric protein that shows a large mass of 500 kDa for trimer and, striking, this appears like a three-bladed propeller with a radius of 90 Å3 (see Fig. 1). The SARS spike protein is characterized by the presence of four structural domains (Fig. 1). While the two large ecto-domains S1 and S2 are responsible for receptor binding and membrane fusion respectively, the cytoplasmic domain (CD) has an important function in the assembly of several enveloped viruses, as described for other viral membrane proteins. In the case of alphaviruses, for instance, the CD of the E2 glycoprotein plays a pivotal role in the interaction with the capsid protein during particle formation4,5. A critical role for the cytoplasmic tails in this process has been reported for several members including Simian virus 5 (SV5)6,7, Sendai virus8,9, and measles virus10. In Sendai virus the matrix protein was found to interact independently of the cytoplasmic tails of the HN and F glycoproteins11,12. In orthomyxovirus influenza A, the cytoplasmic tails of the two glycoproteins, HA and NA, influence budding efficiency as well as particle morphology. Their separate removal caused only limited effects while the lack of both tails resulted in severely impaired formation of deformed particles13–16. Interestingly, many viral ectodomain fragments of fusion protein without transmembrane (TM) and ENDO domains fold into a post-fusion states17,18, suggesting that membrane-anchoring parts help maintain functional metastable high energy conformations. In the case of HIV-1-gp41, for instance, Lu et al. report that antibodies (IgG) against LLP1–2 and LLP2 (lentivirus lytic peptide α helix 1 and 2) regions inhibited HIV-1 Envelope-mediated cell fusion and bound to the interface between effector and target cells suggesting that LLP1–2, especially the LLP2 region located inside the viral membrane is transiently exposed on the membrane surface during the fusion process19.
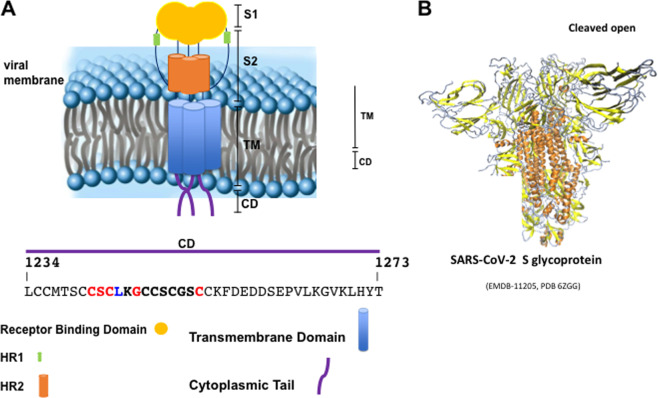
Fig. 1. SARS-CoV-2 S protein and cytoplasmic-domain.
Schematic representation of the SARS-CoV-2 Spike glycoprotein in the viral membrane (a). Cartoon backbone representation of the three-dimensional structure 6ZGG pdb of the S1 and S2 regions in pre-fusion native state in the furin-cleaved open conformation (b)49. The cytoplasmic tail sequence is reported and the analyzed sequence is highlighted. In red, the consensus pattern is shown. S (subunit), TM (Transmembrane Domain), CD (Cytoplasmic Domain), and HR (heptad repeat region).
For coronaviruses is not entirely clear how the intra-virion parts of the fusion protein influence reactions that are carried out by the much larger exterior portion of the protein.
In the carboxy-terminal domain of the Coronaviral S protein there are two areas of conservation: one is at the transition of TM and ectodomain, i.e. where the S protein exits the viral membrane and it is characterized by a conspicuous, highly conserved 8-residues sequence (KWPWY/WVWL), probably important for membrane fusion but not for S protein incorporation into particles. The other area is located in the membrane-proximal part of the CD and it shows a conserved abundance of cysteines (Fig. 1). The carboxy-terminal truncations reveal that it is this specific domain that mediates particle assembly of the coronaviral spikes.
The importance of the cysteine-rich region for membrane fusion has already been established20,21. A spike mutant, with part of the cysteine-rich region deleted, was able to promote hemi-fusion, but was blocked in fusion pore formation. Whether this effect was due by preventing acylation is not completely clear, but it is possible that the membrane-inserted hydrophobic acyl chains are implicated in fusion pore formation. A positive role of cysteine palmitoylation in cell fusion has been reported for influenza virus HA protein22,23, while a negative role was observed for Vesicular Stomatitis virus (VSV)24, influenza virus25,26 and the Murine leukemia virus fusion protein27. In CoV S protein, the cysteines, and/or their palmitate adducts of the endo-domain, can change the rate-limiting step of the membrane fusion reaction28. Therefore, in this contest the role of the CD should be better investigated.
We analyzed the CD sequence of CoV-2 S protein in order to identify potential consensus sequence patterns (Fig. 1). A bioinformatic analysis using PattInProt v5.4up (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_pattinprot.html)29, setting 100% of similarity, was performed identifying, in 79 proteins out of 561,000 proteins of the UniProt bank, a new potential amino acid pattern. The consensus pattern was C-[TS]-C-h-X-G-X(4,6)-C, herein called CAF-motif (Cysteine Aggregation Fusion), where h is a hydrophobic residue and X any other residue. The consensus pattern and the sequence alignments between the proteins and the pattern are shown in Fig. 2 and the PattInProt analysis is shown in Fig. 1S.
Fig. 2. Selected Protein Sequences with their UniProt code that show the CAF-motif.
The analysis was performed with PattInProt program29. In blue square brackets are indicated proteins present in Extracellular matrix, in red ones those implicated in coagulation, in gray ones regulators of cell fate and in green ones those involved in other biological processes.
Interestingly, the only viral proteins that showed the CAF-motif were S proteins of human coronaviruses SARS and SARS -2 and the S proteins of bat coronavirus. Moreover, other proteins involved in coagulation, extracellular recognition and cell fate presented the same pattern. Intriguingly, the CAF-motif occurs in proteins such as coagulation factor X, von Willebrand factor, platelet endothelial aggregation receptor 1 and some pro-thrombin activators venom toxins that are involved in the coagulation process. The identification of a common pattern could suggest a new function of the S protein in the pathological effects of the SARS -2 infection.
Biopsy and autopsy studies on COVID-19 patients showed pulmonary pathology alveolar damage with diffuse thickening of the alveolar wall, formation of hyaline membranes and macrophages and mononuclear cells infiltration30–36. Moreover, according to recent reports, the most severely ill patients show relevant coagulopathy37,38. Clinical studies have revealed that 71.4% of non-survivors of COVID-19 matched the grade of overt disseminated intravascular coagulation (≥5 points according to the International Society on Thrombosis and Haemostasis criteria) and showed abnormal coagulation results during later stages of the disease such as particularly increased levels of D-dimer and other fibrin degradation products that were significantly associated with poor prognosis39,40. The molecular mechanisms at the base of coagulopathy in COVID-19 disease are not yet identified so that the identification of this common pattern could suggest a similar molecular mechanism in the coagulation induction.
Other matched proteins are found in the extracellular matrix (ECM) involved in cellular adhesion to the matrix, cell–cell interaction, and cell signaling such as: fibropellin, fibronectin, versican, cadherins, and proteins responsible for cell fate regulation such as spondin, nidogen, and the cell-surface receptor Notch, which is also involved in fusion cell fate41 and its ligand Dll4 (Delta-like 4).
Among the protein list are metallothioneins and Keratin-associated protein 5-5 (KRTAP 5-5); the first ones are a family of small, highly conserved, cysteine-rich metal-binding proteins important for zinc and copper homeostasis, buffering against toxic heavy metals and protection from oxidative stress, while KRTAP 5-5 belongs to a large protein family involved in crosslinking keratin intermediate filaments during hair formation process.
Intriguingly, this consensus pattern is located in the epidermal growth factor (EGF)-like domain present in the great part of the found proteins and characterized by six cysteines which form disulfide bonds within the domain (C1–C3, C2–C4, and C5–C6).
The EGF-like domain is involved in receptor–ligand interactions, extracellular matrix formation, cell adhesion and signal transduction, and chemotaxis42 and in many proteins such as coagulation factors the EGF-like domains are known to bind calcium ions with D and E residues allowing Ca2+ mediated protein–protein interactions43,44. In Notch, for example, the EGF motifs show multiple functions, such as the prevention of constitutive activation, reciprocal interaction with the ligands, and lateral interaction for homodimerization, playing a crucial role in Notch signaling system45. Moreover, Notch signaling, a major regulator of cardiovascular function and inflammation, is also implicated in several biological processes mediating viral infections such as SARS-CoV-2, playing an important role in developing of myocarditis, heart failure, and lung inflammation in COVID-19 patients46. In macrophages Dll1,4/Notch signaling promotes the inflammatory cytokines storm, interleukin-6 (IL-6) among those, which in turn increases the expression of notch ligands (Dll1,4), thus amplifying the signal establishing a feedback loop46.
In view of the above, we propose a hypothetic active role of the Coronavirus S protein cytoplasmic domain in protein–protein aggregation for clots formation and cell–cell fusion SARS-2-S protein-driven47. Therefore, our findings suggest a new potential molecular mechanism linked to the infection in which after virus-cell fusion, the infected cells expose on their surface the CAF-motif, leading to clots formation and cell–cell fusion by protein–protein aggregation processes. Moreover, it should not to be rolled out the ability of CD’s S protein to coordinate Ca2+ ions, similarly to EGF-like domain, for mediating cell–cell fusion, which is responsible for instance of syncytia formation48.
The identification of this new consensus pattern provides a first evidence of a functional similarity between the CoV S protein and proteins involved in coagulation, in regulation of cell fate and cell–cell interaction and fusion that are at the base of coagulopathy and syncytia formation in COVID-19 disease.
Supplementary information
Figure 1S. The original PattInProt analysis
Acknowledgements
We thank Edoardo Trotta and Gabriella Santoro for the critical discussion of the results.
Author contributions
S.M.: conceptualization, investigation, and manuscript writing; S.B.: investigation and manuscript writing.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by R. A. Knight
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41420-020-00372-1) contains supplementary material, which is available to authorized users.
References
- 1.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beniac DR, Andonov A, Grudeski E, Booth TF. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suomalainen M, Liljestrom P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 1992;66:4737–4747. doi: 10.1128/JVI.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H, Lindqvist B, Garoff H, von Bonsdorff CH, Liljestrom P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitt AP, He B, Lamb RA. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J. Virol. 1999;73:8703–8712. doi: 10.1128/JVI.73.10.8703-8712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waning DL, Schmitt AP, Leser GP, Lamb RA. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J. Virol. 2002;76:9284–9297. doi: 10.1128/JVI.76.18.9284-9297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouillot-Coriou N, Roux L. Structure-function analysis of the Sendai virus F and HN cytoplasmic domain: different role for the two proteins in the production of virus particle. Virology. 2000;270:464–475. doi: 10.1006/viro.2000.0291. [DOI] [PubMed] [Google Scholar]
- 9.Takimoto T, Bousse T, Coronel EC, Scroggs RA, Portner A. Cytoplasmic domain of Sendai virus HN protein contains a specific sequence required for its incorporation into virions. J. Virol. 1998;72:9747–9754. doi: 10.1128/JVI.72.12.9747-9754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cathomen T, Naim HY, Cattaneo R. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 1998;72:1224–1234. doi: 10.1128/JVI.72.2.1224-1234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali A, Avalos RT, Ponimaskin E, Nayak DP. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 2000;74:8709–8719. doi: 10.1128/JVI.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanderson CM, Wu HH, Nayak DP. Sendai virus M protein binds independently to either the F or the HN glycoprotein in vivo. J. Virol. 1994;68:69–76. doi: 10.1128/JVI.68.1.69-76.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin H, Leser GP, Lamb RA. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 1994;13:5504–5515. doi: 10.1002/j.1460-2075.1994.tb06885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin H, Leser GP, Zhang J, Lamb RA. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitnaul LJ, Castrucci MR, Murti KG, Kawaoka Y. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J. Virol. 1996;70:873–879. doi: 10.1128/JVI.70.2.873-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Pekosz A, Lamb RA. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 2000;74:4634–4644. doi: 10.1128/JVI.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl Acad. Sci. USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markosyan RM, Cohen FS, Melikyan GB. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell. 2003;14:926–938. doi: 10.1091/mbc.e02-09-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu L, et al. Surface exposure of the HIV-1 env cytoplasmic tail LLP2 domain during the membrane fusion process: interaction with gp41 fusion core. J. Biol. Chem. 2008;283:16723–16731. doi: 10.1074/jbc.M801083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bos EC, Heijnen L, Luytjes W, Spaan WJ. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology. 1995;214:453–463. doi: 10.1006/viro.1995.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang KW, Sheng Y, Gombold JL. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology. 2000;269:212–224. doi: 10.1006/viro.2000.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naeve CW, Williams D. Fatty acids on the A/Japan/305/57 influenza virus hemagglutinin have a role in membrane fusion. EMBO J. 1990;9:3857–3866. doi: 10.1002/j.1460-2075.1990.tb07604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakai T, Ohuchi R, Ohuchi M. Fatty acids on the A/USSR/77 influenza virus hemagglutinin facilitate the transition from hemifusion to fusion pore formation. J. Virol. 2002;76:4603–4611. doi: 10.1128/JVI.76.9.4603-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitt MA, Rose JK. Fatty acid acylation is not required for membrane fusion activity or glycoprotein assembly into VSV virions. Virology. 1991;185:875–878. doi: 10.1016/0042-6822(91)90563-Q. [DOI] [PubMed] [Google Scholar]
- 25.Naim HY, Amarneh B, Ktistakis NT, Roth MG. Effects of altering palmitylation sites on biosynthesis and function of the influenza virus hemagglutinin. J. Virol. 1992;66:7585–7588. doi: 10.1128/JVI.66.12.7585-7588.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinhauer DA, Wharton SA, Skehel JJ, Wiley DC, Hay AJ. Amantadine selection of a mutant influenza virus containing an acid-stable hemagglutinin glycoprotein: evidence for virus-specific regulation of the pH of glycoprotein transport vesicles. Proc. Natl Acad. Sci. USA. 1991;88:11525–11529. doi: 10.1073/pnas.88.24.11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang C, Compans RW. Analysis of the cell fusion activities of chimeric simian immunodeficiency virus-murine leukemia virus envelope proteins: inhibitory effects of the R peptide. J. Virol. 1996;70:248–254. doi: 10.1128/JVI.70.1.248-254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shulla A, Gallagher T. Role of spike protein endodomains in regulating coronavirus entry. J. Biol. Chem. 2009;284:32725–32734. doi: 10.1074/jbc.M109.043547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Combet C, Blanchet C, Geourjon C, Del‚age G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, et al. COVID-19 infection: the China and Italy perspectives. Cell Death Dis. 2020;11:438. doi: 10.1038/s41419-020-2603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatakrishnan AJ, et al. Benchmarking evolutionary tinkering underlying human-viral molecular mimicry shows multiple host pulmonary-arterial peptides mimicked by SARS-CoV-2. Cell Death Discov. 2020;6:96. doi: 10.1038/s41420-020-00321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gebicki J, Wieczorkowska M. COVID-19 infection: mitohormetic concept of immune response. Cell Death Discov. 2020;6:60. doi: 10.1038/s41420-020-00297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J, et al. Digestive symptoms of COVID-19 and expression of ACE2 in digestive tract organs. Cell Death Discov. 2020;6:76. doi: 10.1038/s41420-020-00307-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Y, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuyama T, Kubli SP, Yoshinaga SK, Pfeffer K, Mak TW. An aberrant STAT pathway is central to COVID-19. Cell Death Differ. 2020;oct 9:1–17. doi: 10.1038/s41418-020-00633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi CS, Nabar NR, Huang NN, Kehrl JH. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.China National Health Commision. Diagnosis and treatment of novel coronavirus pneumonia in China (trial version 7). https://www.who.int/docs/default-source/wpro--documents/countries/china/covid-19-briefing-nhc/1-clinical-protocols-forthediagnosis-and-treatment-of-covid-19v7.pdf?sfvrsn=c6cbfba4_2. Accessed 14 April 2020.
- 38.Yao XH, et al. A pathological report of three COVID-19 cases by minimal invasive autopsies. Zhonghua Bing Li Xue Za Zhi. 2020;49:411–417. doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]
- 39.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iba T, Levy JH, Levi M, Connors JM, Thachil J. Coagulopathy of coronavirus disease 2019. Crit. Care Med. 2020;48:1358–1364. doi: 10.1097/CCM.0000000000004616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Artero RD, Castanon I, Baylies MK. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development. 2001;128:4251–4264. doi: 10.1242/dev.128.21.4251. [DOI] [PubMed] [Google Scholar]
- 42.Ma J, et al. Role of a novel EGF-like domain-containing gene NGX6 in cell adhesion modulation in nasopharyngeal carcinoma cells. Carcinogenesis. 2005;26:281–291. doi: 10.1093/carcin/bgh312. [DOI] [PubMed] [Google Scholar]
- 43.Rao Z, et al. Crystallization of a calcium-binding EGF-like domain. Acta Crystallogr. D Biol. Crystallogr. 1995;51:402–403. doi: 10.1107/S0907444994009881. [DOI] [PubMed] [Google Scholar]
- 44.Elíes, J. et al. Calcium Signaling. Advances in experimental medicine and biology (ed. Islam M.) Vol. 1131. 183–213 (Springer, Cham., 2020). [DOI] [PubMed]
- 45.Sakamoto K, Chao WS, Katsube K, Yamaguchi A. Distinct roles of EGF repeats for the Notch signaling system. Exp. Cell Res. 2005;302:281–291. doi: 10.1016/j.yexcr.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 46.Rizzo P, et al. COVID-19 in the heart and the lungs: could we “Notch” the inflammatory storm? Basic Res. Cardiol. 2020;115:31. doi: 10.1007/s00395-020-0791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffmann M, Kleine-Weber H, Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen EH, Grote E, Mohler W, Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 49.Wrobel AG, et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1S. The original PattInProt analysis